Abstract
Unconventional T lymphocyte populations are emerging as important regulators of tumor immunity. Despite this, the role of TCRαβ+CD4–CD8–NK1.1– innate αβ T-cells (iαβTs) in pancreatic ductal adenocarcinoma (PDA) has not been explored. We found that iαβTs represent ~10% of T-lymphocytes infiltrating PDA in mice and humans. Intra-tumoral iαβTs express a distinct TCR-repertoire and profoundly immunogenic phenotype compared to their peripheral counterparts and conventional lymphocytes. iαβTs comprised ~75% of the total intra-tumoral IL-17+ cells. Moreover, iαβT cell adoptive transfer is protective in both murine models of PDA and human organotypic systems. We show iαβT cells induce a CCR5-dependent immunogenic macrophage reprogramming, thereby enabling marked CD4+ and CD8+ T cell expansion/activation and tumor protection. Collectively, iαβTs govern fundamental intra-tumoral crosstalk between innate and adaptive immune populations and are attractive therapeutic targets.
Keywords: Pancreatic cancer, Kras, T cells, Anti-tumor immunity, Immunotherapy
Introduction
The phenotypic composition of inflammatory cells infiltrating pancreatic ductal adenocarcinoma (PDA) influences disease outcome (1). Depending on the signals released, innate immune cells entrain the adaptive immune compartment towards an immunogenic or tolerogenic response, affecting PDA growth and metastasis. Classically activated M1-like macrophages secrete pro-inflammatory mediators, such as TNFα, IFNγ, and IL-12, which in turn polarize CD4+ T cells towards a Th1 phenotype and activate cytotoxic CD8+ T cells. More commonly, alternatively activated M2-like macrophages promote tolerogenic Th2 and T regulatory cell (Treg) differentiation (2). However, this relationship is bidirectional as adaptive immune cells can in turn influence innate immune programming. For example, we previously showed that CD4+ T cells can augment tolerogenic macrophage polarization via IL-4 (3). Similarly, FoxP3+ Tregs can influence dendritic cell capacity for cross-presentation of tumor antigen (4). These data suggest bidirectional crosstalk between innate and adaptive immune subsets in the PDA tumor microenvironment (TME).
There is growing evidence illustrating the contribution of unconventional T lymphocytes in shaping the immune milieu in PDA. γδT cells, characterized by expression of TCRγδ, expand early in pancreatic oncogenesis. We showed that γδT cells exert direct immune-suppressive influences on conventional T cells in PDA via the PD-L1-PD1 axis (5). By contrast, NKT cells have recently been shown to protect against PDA by modulating the phenotype of tumor-associated macrophages (TAMs) through mPGES-1 and 5-LOX (6).
We observed that CD3+TCRαβ+CD4–CD8–NK1.1– cells represented ~10% of T cells in murine and human PDA. Given the emerging role for unconventional T cells in PDA, we were spurred to investigate the function of these iαβTs in this disease. iαβTs exhibited a unique cellular program that was influenced by diverse microbial and sterile stimuli within the TME. We further hypothesized that iαβTs may have important immune-modulatory functions in PDA. We found that iαβTs exhibited marked tumor-protective properties in both murine and human PDA by driving a CCR5-dependent immunogenic macrophage polarization, resulting in conventional T cell activation in situ. Collectively, our work uncovers fundamental intra-tumoral crosstalk between iαβTs and the innate and adaptive immune compartments, and suggests that strategies targeting iαβTs may be effective for reprogramming of the TME in human cancer.
Results
iαβTs are a prominent T cell population in murine and human PDA
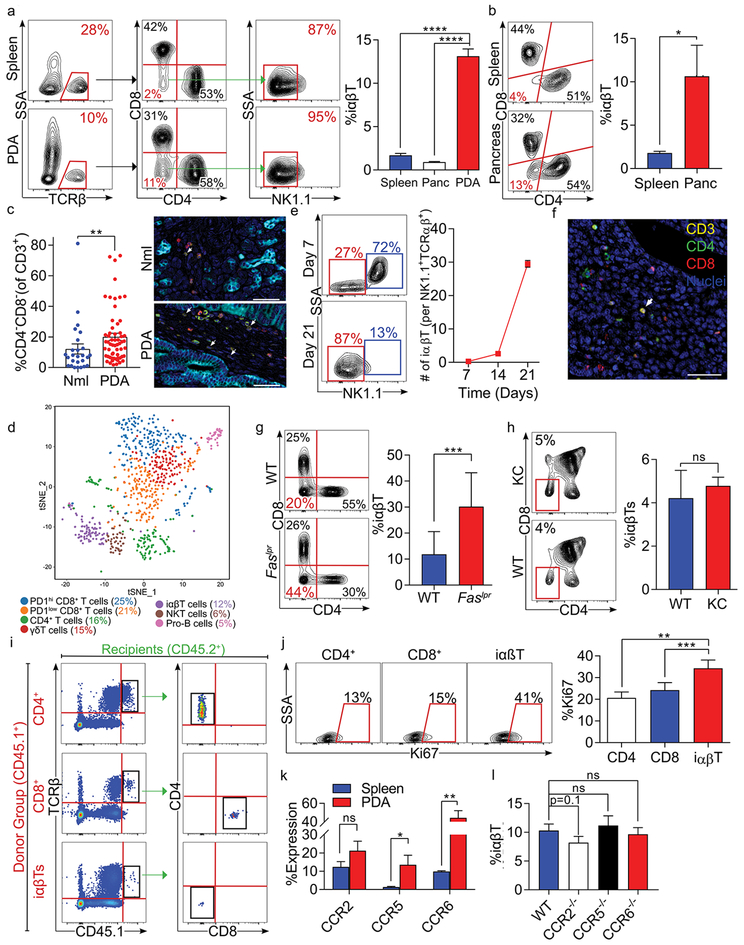
To assess the significance of iαβTs in PDA, we determined their prevalence in an invasive orthotopic model of PDA using tumor cells derived from Pdx1Cre;KrasG12D;Tp53R172H (KPC) mice that express mutant Kras and p53 in their pancreatic progenitor cells, a slowly progressive pre-invasive autochthonous model of PDA using p48Cre;KrasG12D (KC) mice whose pancreata express oncogenic Kras alone, and human disease. We discovered that CD4–CD8–NK1.1– iαβTs constitute ~10% of TCRαβ+ T cells in orthotopic KPC tumors (Figures 1a, S1a) and KC pancreata (Figure 1b). Human PDA similarly possessed an increased CD4–CD8– T cell infiltrate compared to normal adjacent pancreas or PBMC (Figures 1c, S1b). By contrast, iαβTs were scarce in murine spleen and human PBMCs. PDA-infiltrating iαβTs did not express characteristic functional markers of NKT cells and ~10% bound an MR1 specific tetramer, compared to ~40% in the gut (Figure S1c–f). Single cell RNAseq of PDA-infiltrating CD3+ cells further confirmed that this population is transcriptomically distinct from CD4+, CD8+, γδT, and NKT cell populations (Figure 1d). iαβTs upregulated Ccr7 expression, and exhibited reduced Il2rb in comparison to other lymphocyte populations (Figure S1g, h). These observations were confirmed by flow cytometry (Figure S1i, j). In depth analysis comparing iαβT to NKT cells revealed that iαβT cells downregulated the NKT markers Klrk1, Klra7, Klrd1, NKG7, Klrc2, and Ly6c2 but upregulated the transcription factors Socs3 and Junb (Figure S1k). Interestingly, iαβTs increased as a fraction of TCRαβ+CD4–CD8– T cells as tumors progressed (Figure 1e). γδT cells remained stable over the course of oncogenesis, as previously reported1. Multiplex immunohistochemistry suggested that iαβTs were interspersed among CD4+ and CD8+ T cells within the TME (Figure 1f). Of note, PDA-infiltrating iαβTs constituted ~30% of TCRαβ+ cells in Faslpr mice (Figure 1g), known to accumulate iαβTs in secondary lymphoid organs (7). To determine whether thymic production of iαβTs is accelerated during pancreatic oncogenesis, we interrogated T cell populations in the thymus of 6-month-old KC mice. Thymic iαβTs were not increased in prevalence in KC mice compared to WT (Figure 1h). Adoptive transfer tracking experiments suggested that neither CD4+ nor CD8+ T cells converted to the iαβT phenotype in PDA. Likewise, iαβTs did not gain CD4 or CD8 expression (Figure 1i). Interestingly, cellular proliferation was higher in PDA-infiltrating iαβTs than in either CD4+ or CD8+ T cells (Figure 1j). We recently reported that unconventional T cells, particularly γδT cells, are recruited to the PDA tumor microenvironment via diverse chemokine signaling networks (5). We observe that PDA-infiltrating iαβTs express CCR2, CCR5, and CCR6 (Figure 1k) and CCR2 deletion trended to mitigate iαβT recruitment in PDA (Figure 1l).
Figure 1. iαβTs expand in PDA.
(a) CD45+ leukocytes infiltrating day 21 orthotopic KPC tumors, normal pancreas, and spleens in WT mice were gated and tested for the frequency of TCRβ+CD4–CD8–NK1.1– iαβTs. Representative contour plots and quantitative data are shown (n=10). (b) CD45+TCRβ+ NK1.1– leukocytes from pancreata and spleens of 6 month-old KC mice were gated and tested for co-expression CD4 and CD8. Representative contour plots are shown (n=5). (c) Multiplex IHC of human PDA and adjacent normal pancreas were stained for CK19, CD3, CD4, and CD8. The frequency of CD3+CD4–CD8– cells were quantified and representative images are shown. (d) Orthotopic KPC tumors were harvested from WT mice on day 21. CD45+CD3+ leukocytes were purified by FACS and analyzed by single cell RNAseq. The distribution of cellular clusters was determined using the t-Distributed Stochastic Neighbor Embedding (t-SNE) algorithm. Each cluster is identified by a distinct color. Percent cellular abundance in each cluster is indicated. (e) Orthotopic KPC tumors were harvested from WT mice on days 7, 14, or 21 after tumor cell implantation and tumor-infiltrating CD45+TCRβ+CD4–CD8– leukocytes were gated and tested for expression of NK1.1. Representative contour plots from days 7 and 21 and quantitative data comparing frequency of tumor-infiltrating iαβT per NKT cells at all time points are shown (n=5/time point). (f) Paraffin-embedded sections made from tumors of mice serially treated with anti-TCRγ/δ and NK1.1 depleting antibodies were tested for co-expression of Hematoxylin, CD3, CD4, and CD8 in the PDA TME. (g) CD45+TCRβ+NK1.1– leukocytes infiltrating orthotopic KPC tumors in WT and Faslpr mice were gated and tested for expression of CD4 and CD8. Representative contour plots and quantitative data are shown (n=5/group). (h) The thymus from 6-month old WT and KC mice were harvested and CD45+TCRβ+ thymocytes were gated and tested for expression of CD4 and CD8. The frequency of iαβTs in the thymus was calculated (n=5/group). (i) CD4+ T cells, CD8+ T cells, or iαβTs were harvested from CD45.1 mice and transferred i.v. to orthotopic PDA-bearing CD45.2 mice. PDA tumors were harvested at 96 hours and CD45.1+ cells were gated and tested for CD4 and CD8 expression. Representative contour plots are shown (n=5/group). (j) WT mice were orthotopically administered KPC tumor cells and sacrificed on day 21. PDA-infiltrating CD4+ T cells, CD8+ T cells, and iαβTs were assayed for Ki67 proliferative index. Representative contour plots and quantitative data are shown (n=5). (k) Splenic and orthotopic PDA-infiltrating iαβTs were tested on day 21 for expression of CCR2, CCR5, and CCR6 (n=5). (l) The frequency of PDA-infiltrating iαβTs was tested on day 21 in WT, CCR2–/–, CCR5–/–, and CCR6–/– hosts (n=5/group). All experiments were repeated at least 3 times (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
iαβTs exhibit a distinctive phenotype in PDA and are targets of immunotherapy
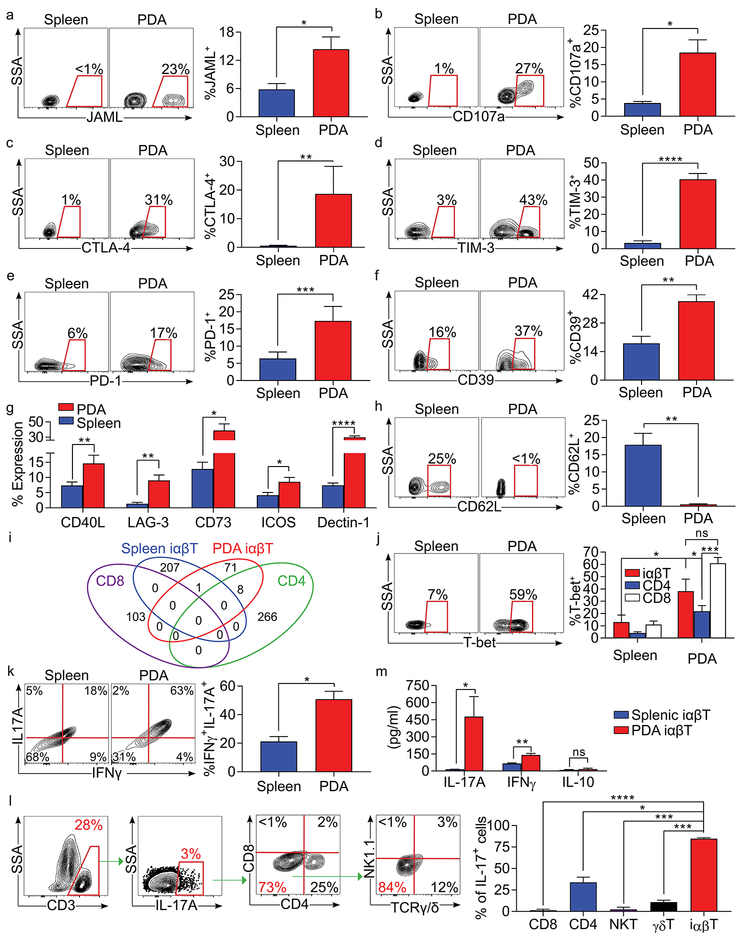
PDA-infiltrating iαβTs acquire a distinctive phenotype relative to the periphery. Tumor-penetrating iαβT expressed the adhesion ligand JAML and the cytotoxic marker CD107a (Figure 2a, b). Additionally, iαβTs in the TME upregulated the checkpoint receptors CTLA-4, TIM-3, PD-1, and LAG-3, ectonucleotidases CD39 and CD73, costimulatory receptors CD40L and ICOS, and the C-type lectin receptor Dectin-1 (Figures 2c–g). PDA-associated iαβTs were also highly activated compared to their splenic counterparts, downregulating CD62L (Figure 2h). Moreover, the TCR nucleotide sequences of tumor-infiltrating iαβTs exhibited minimal overlap with those of splenic iαβTs or conventional T cells infiltrating PDA (Figure 2i and Table S1). iαβTs in the TME were further distinct from both intra-tumoral CD4+ and CD8+ T cells and splenic iαβT in their high expression of the Th1-family transcription factor T-bet (Figure 2j). STAT1 signaling, associated with Th1-differentiation, was similarly upregulated in iαβTs in PDA (Figure S2a). Expression of Th17-family transcription factor RORγt was also increased (Figure S2b). Accordingly, PDA-infiltrating iαβTs expressed high IFNγ and TNFα compared to their splenic counterparts and to both intra-tumoral CD4+ and CD8+ T cells (Figure S2c, d). IFNγ and IL-17 were distinctively co-expressed in the iαβT cell population (Figure 2k). Notably, iαβTs in tumor accounted for ~75% of the total IL-17+ immune cells in PDA. (Figure 2l). By contrast, FoxP3 and IL-10 were minimally expressed in iαβTs in PDA, whereas TGFβ was upregulated (Figure S2e–g). Analysis of iαβT cell conditioned media confirmed high IL-17 and IFNγ, but low IL-10, secretion (Figure 2m).
Figure 2. iαβTs infiltrating PDA are phenotypically distinct.
WT mice bearing orthotopic KPC tumors were sacrificed on day 21. (a-g) Splenic and PDA-infiltrating iαβTs were tested for expression of (a) JAML, (b) CD107a, (c) CTLA-4, (d) TIM-3, (e) PD-1, (f) CD39, (g) CD40L, LAG-3, CD73, ICOS, and Dectin-1. (h) Splenic and PDA-infiltrating iαβTs were tested for expression of CD62L. (i) TCR sequencing of splenic iαβTs and PDA-infiltrating iαβTs, CD4+ and CD8+ T cells was performed in triplicate and assessed for overlapping clones between populations. (j) Splenic and PDA-infiltrating iαβTs, CD4+ T cells, and CD8+ T cells were tested for expression of T-bet. (k) Splenic and orthotopic PDA-infiltrating iαβTs were tested for co-expression of IFNγ and IL-17A. (l) PDA-infiltrating CD3+IL-17+ cells were gated and tested for the frequency of CD4+ T cells, CD8+ T cells, NKT cells, γδT cells, and iαβTs. (m) Splenic and PDA-infiltrating iαβTs were cultured in vitro for 24h and cell culture supernatant was harvested and assayed for IL-17, IFNγ, and IL-10 (n=5/group). Flow cytometry experiments were repeated more than 4 times with similar results (n=5 mice for each replicate experiment; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
Immunotherapy has emerged as an efficacious treatment option in select subsets of PDA patients (8). Since iαβTs express high levels of costimulatory molecules and checkpoint receptors in PDA, we postulated that immune-based therapies targeting these entities would activate iαβTs in situ. Accordingly, we found that ICOS ligation upregulated IFNγ and IL-17 expression in splenic iαβTs in vitro (Figure S3a, b). Further, treatment of PDA-bearing mice with an ICOS agonist was tumor-protective and was associated with marked iαβT cellular expansion and activation (Figure S3c–e). Given our observation that PDA-infiltrating iαβTs upregulate PD-1, we also tested the effect of PD-L1 blockade on iαβT phenotype (Figure 2e). αPD-L1 mAb treatment promoted iαβT activation in the TME (Figure S3f). These data suggest that iαβTs are distinctive targets of costimulatory and checkpoint receptor based therapy in PDA whose impact must be considered in immune-based therapeutics. Moreover, the combination of αPD-L1 treatment and iαβT cell transfer offered additional protection compared to either monotherapy (Figure S3g).
To investigate whether PDA-infiltrating iαβTs are MHC I or MHC II restricted, we administered orthotopic PDA tumor to β2-microglobulin−/− and MHC II−/− mice, respectively. PDA tumors in MHC II−/− hosts did not alter iαβT cell recruitment (Figure S3h), whereas β2-microglobulin deficient mice exhibited reduced iαβT cell recruitment to the PDA tumor microenvironment and lower cellular expression of T-bet and TNFα (Figure S3i–k). In contrast to CD8+ T cells, however, iαβT infiltrating Ova-expressing PDA tumors lacked Ova-pentamer staining and were thus not MHC I antigen-restricted, lending support to their innate immune properties (Figure S3l, m). Accordingly, intra-tumoral iαβTs were less clonal than both CD4+ or CD8+ T cells (Figure S3n).
iαβT cell phenotype in PDA is influenced by diverse inflammatory and microbial signals
Since CCR2 recruits innate T cells to the PDA TME (Figure 1k) (5), we postulated that CCR2 signaling also modulates iαβT cellular phenotype in situ. Accordingly, intra-tumoral iαβT cell expression of T-bet and CD44 was reduced in CCR2–/– hosts, expression of ICOS, CTLA-4, and TIM3 was upregulated, whereas cytokine expression was not altered (Figure S4a). Since we previously reported that Dectin-1 can regulate cytokine secretion in unconventional T cells (9), we postulated that, unlike CCR2, Dectin-1 signaling may govern iαβT cell cytokine profile. PDA-infiltrating iαβTs in Dectin-1–/– mice expressed reduced IL-17 and IFNγ, while TNFα expression was unchanged. (Figure S4b). Accordingly, ligating Dectin-1 upregulated IFNγ and IL-17 in naïve splenic iαβTs (Figure S4c). Collectively, these data suggest that CCR2 and Dectin-1 signals influence distinct aspects of iαβT cell programming in PDA.
We recently reported that the host microbiome in PDA exerts suppressive influences on intra-tumoral CD4+ and CD8+ T cell differentiation (10). We therefore postulated that the PDA microbiome has similar tolerogenic effects on iαβTs. To test this, we ablated the microbiome in PDA-bearing mice using an established oral antibiotic regimen (Figure S4d) (11). Consistent with our hypothesis, microbial ablation activated iαβTs, increasing their expression of T-bet, CD44, LFA-1, ICOS, IL-17, IFNγ, and TNFα (Figure S4e). Mechanistically, we reported that the microbiome directs immune suppression in PDA via TLR4 signaling (10,12). Interestingly, we found that iαβTs upregulate TLR4 expression in PDA (Figure S4f). Consistent with our analysis of PDA-infiltrating CD4+ and CD8+ T cells (10), TLR4 ligation mitigated iαβT cell activation (Figure S4g).
iαβTs protect against PDA and induce immunogenic CD4+ and CD8+ T cell activation in mouse and human models
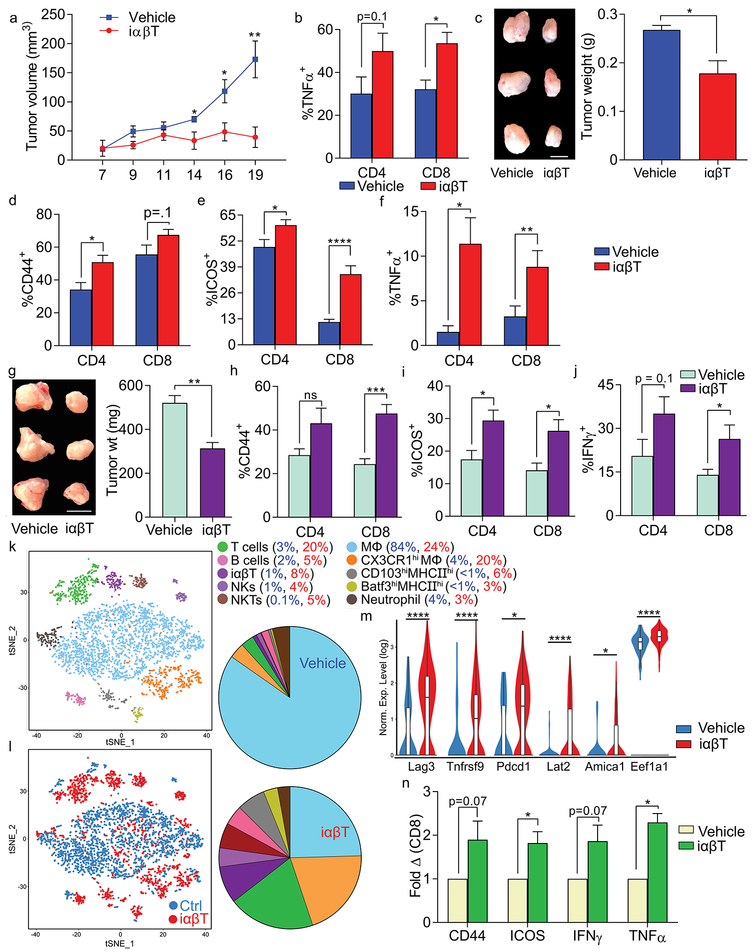
To determine the influence of iαβTs on pancreatic oncogenesis, we adoptively transferred iαβTs to mice coincident with SQ tumor challenge. iαβT cell administration mitigated pancreatic tumor growth (Figure 3a). iαβT cell transfer resulted in intra-tumoral CD8+ T cell activation (Figure 3b). Similarly, intra-pancreatic iαβT cellular transfer with orthotopically implanted KPC tumor cells protected against tumor growth (Figure 3c) and again activated CD4+ and CD8+ T cells in the TME, inducing higher expression of CD44, ICOS, and TNFα (Figure 3d–f). To determine whether iαβTs are suitable agents for cellular therapy, we serially administered iαβTs i.v. to mice bearing established orthotopic PDA tumors. iαβTs administration protected against tumor growth and immunogenically reprogrammed the adaptive TME (Figure 3g–j). Furthermore, analysis of tumors treated with iαβTs by single cell RNAseq suggested a marked CD4+ and CD8+ T cell expansion compared to controls. Specifically, conventional T cell populations increased from 3% to 20% of leukocytes with iαβT cell treatment (Figure 3k, l). Other lymphocyte subsets were also expanded. Consistent with our flow cytometry data, conventional T cells in tumors of iαβT cell treated mice expressed higher activation markers (lag3, tnfrsf9, pdcd1, lat2), indicators of cytotoxicity (Amica1), and Th1-related transcription factors (Eef1a1) (Figures 3m, S5a).
Figure 3. iαβTs protect against PDA and enhance intra-tumoral T cell immunity.
(a) WT mice were administered KPC tumor cells subcutaneously, either alone or admixed with iαβTs. Tumor growth was serially measured (n=5/group). (b) WT mice were administered KPC tumor cells subcutaneously, either alone or admixed with PDA-infiltrating iαβTs. On day 21, tumor-infiltrating CD8+ T cells were analyzed for expression of TNFα (n=5/group). (c-f) WT mice were orthotopically administered KPC tumor cells, either alone or admixed with iαβTs. Tumors were harvested on day 21. (c) Representative pictures of tumors and quantitative analysis of tumor weight are shown. (d) PDA-infiltrating CD4+ and CD8+ T cells were analyzed for expression of CD44, (e) ICOS, and (f) TNFα (n=5/group). Each mouse experiment was repeated more than 3 times. (g-j) Mice with established orthotopic KPC tumor were serially transferred i.v. twice weekly with iαβTs or vehicle beginning on day 5 (n=5). Mice were sacrificed at day 21 after tumor implantation. (g) Representative pictures of tumors and quantitative analysis of tumor weight are shown. (h) PDA-infiltrating CD4+ and CD8+ T cells were analyzed for expression of CD44, (i) ICOS, and (j) IFNγ. Subcutaneous and orthotopic tumor experiments were each repeated at least 4 times. (k-m) Mice with established orthotopic KPC tumor were serially transferred i.v. twice weekly with iαβTs or vehicle beginning on day 5. Mice were sacrificed at day 21 and single cell RNAseq performed on FACS-purified CD45+ tumor-infiltrating leukocytes. (k) The distribution of cellular clusters was determined using the t-SNE algorithm. Each cluster is identified by a distinct color. (l) A t-SNE plot overlay of tumor-infiltrating leukocytes in tumors of mice treated with iαβTs (red) vs vehicle (blue) is shown. Percent cellular abundance in each cluster is depicted in pie charts and specified in the accompanying legend. (m) Violin plots comparing normalized log expression of select genes in the T cell cluster for both treatment groups are shown. (n) PDOTS derived from resected human tumors were treated with autologous iαβTs or vehicle. At 72h, CD8+ T cells were analyzed for expression of CD44, ICOS, IFNγ, and TNFα. Data is indicated as fold change in the iαβT cell-treated group compared to vehicle-treatment (n=5 patients; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
To determine whether iαβTs have the capacity to immunogenically reprogram adaptive immunity in human disease, we generated patient-derived organotypic tumor spheroids (PDOTS) from freshly harvested human tumors using a microfluidic-based system that we recently described (13). We selectively added FACS-sorted autologous iαβTs or vehicle to the PDOTS and analyzed the system at 3 days. Consistent with our murine data, iαβTs induced marked conventional T cell activation in the human PDOTS system (Figures 3n and S5b).
PDA-infiltrating iαβTs induce effector T cell-dependent tumor-protection, but paradoxically suppress conventional T cells in vitro
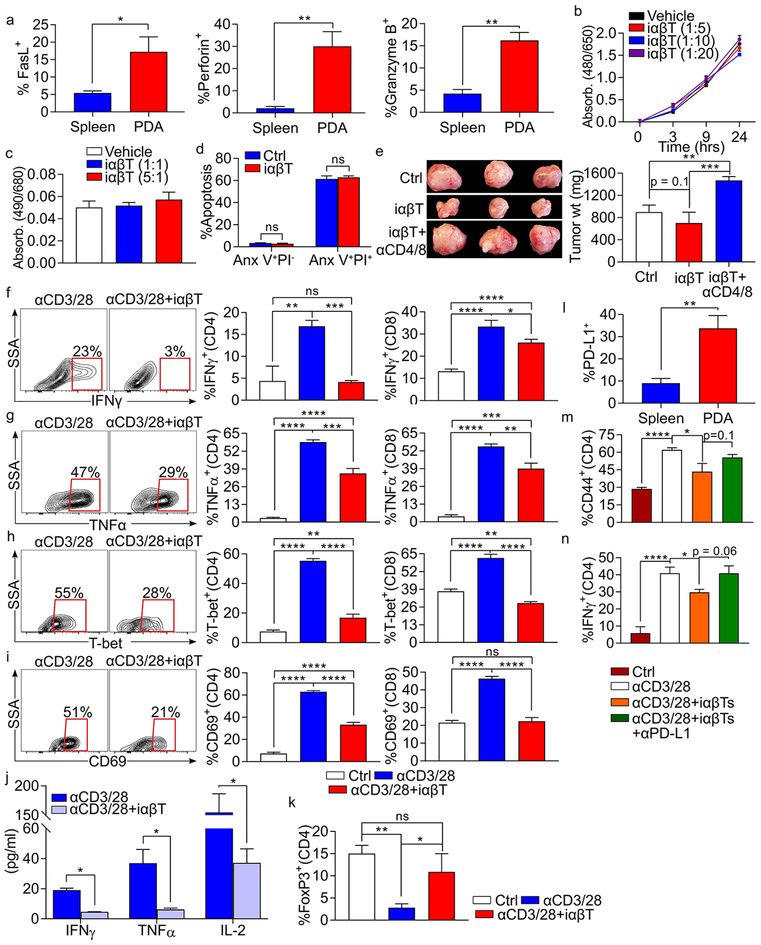
In addition to expressing high levels of the cytotoxicity marker CD107a (Figure 2b), PDA-associated iαβTs also upregulate FasL, Perforin, and Granzyme B (Figure 4a). We therefore postulated that iαβTs may be directly cytotoxic to PDA tumor cells. However, iαβTs did not mitigate tumor cell proliferation, induce tumor cell lysis, or promote apoptosis (Figure 4b–d). Since iαβT cellular transfer enhanced T cell immunity in situ, we speculated that iαβTs mediate tumor-protection by directly activating CD4+ and CD8+ T cells in the PDA microenvironment. Accordingly, whereas CD4+/CD8+ T cell depletion did not accelerate tumor progression in control mice as we previously reported (5,10), CD4+/CD8+ T cell depletion abrogated the tumor-protection associated with iαβT transfer (Figure 4e). We therefore used in vitro modeling to determine whether iαβTs directly enhance αβT cell immunogenicity. Belying our in vivo data, we found that PDA-infiltrating iαβTs were highly inhibitory to CD4+ and CD8+ T cell activation in vitro and prevented their upregulation of IFNγ, TNFα, T-bet, CD69, and CD44 in response to CD3/CD28 co-ligation (Figures 4f–i and S5c). iαβTs similarly mitigated polyclonal T cell secretion of IFNγ, TNFα, and IL-2 in cell culture supernatant (Figure 4j). Further, iαβTs promoted upregulation of FoxP3 expression in CD4+ T cells (Figure 4k). The inhibitory effects of iαβTs on conventional T cells were independent of TGFβ secretion (Figure S5d–f). We previously reported that γδT cells can inhibit CD4+ and CD8+ T cell activation in cancer via the PD-L1-PD-1 axis (5). Therefore, we postulated that iαβTs, which upregulate PD-L1 in PDA (Figure 4l), suppress effector T cells through PD-L1-PD1 interactions. PD-L1 blockade partially reversed the inhibitory effects of PDA-infiltrating iαβTs on effector T cell activation (Figure 4m, n).
Figure 4. iαβTs induce T cell dependent tumor immunity but are directly suppressive to conventional T cells.
(a) WT mice bearing orthotopic KPC tumors were sacrificed on day 21. Splenic and PDA-infiltrating iαβTs were tested for expression of FasL, Perforin, and Granzyme B (n=5 mice). (b, c) iαβTs were harvested by FACS from orthotopic PDA tumors and cultured in various ratio with KPC tumor cells. (b) Proliferation of KPC tumors cells was tested using the XTT assay. (c) Cytotoxicity against KPC tumor cells was determined in an LDH release assay. (d) iαβTs were harvested by FACS from orthotopic PDA tumors and cultured in 1:1 ratio with KPC tumor cells. KPC tumor cell apoptosis was determined by co-staining for Annexin V and PI. (e) WT mice were orthotopically administered KPC tumor cells admixed with iαβTs. Cohorts were either serially depleted of both CD4+ and CD8+ T cells or administered isotype control before sacrifice on day 21. Representative images and quantitative analysis of tumor weights are shown (n=5/group). This experiment was repeated twice. (f-i) Polyclonal splenic CD4+ or CD8+ T cells were cultured without stimulation, stimulated by CD3/CD28 co-ligation, or stimulated by CD3/CD28 co-ligation in co-culture with iαβTs. CD4+ and CD8+ T cell activation were determined at 72h by their expression of (f) IFNγ, (g) TNFα, (h) T-bet, and (i) CD69. Representative contour plots and quantitative data are shown. (j) Polyclonal splenic CD3+ T cells were stimulated by CD3/CD28 co-ligation, either alone or in co-culture with iαβTs. Cell culture supernatant was tested for expression of IFNγ, TNFα, and IL-2 at 72h. (k) Polyclonal splenic CD4+ T cells were cultured without stimulation, stimulated by CD3/CD28 co-ligation, or stimulated by CD3/CD28 co-ligation in co-culture with iαβTs. CD4+ T cells were tested for expression of FoxP3 at 72h. (l) Spleen and PDA-infiltrating iαβTs were tested for expression of PD-L1 by flow cytometry. (m, n) Polyclonal splenic CD4+ T cells from PD-L1–/– mice were cultured without stimulation, stimulated by CD3/CD28 co-ligation, or stimulated by CD3/CD28 co-ligation in co-culture with WT iαβTs, either alone or with an αPD-L1 neutralizing mAb. CD4+ T cells were tested for expression of (m) CD44 and (n) IFNγ at 72h. Experiments were performed in replicates of 5 and repeated at least 4 times (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
PDA-associated iαβTs promote immunogenic macrophage polarization via CCR5 activation
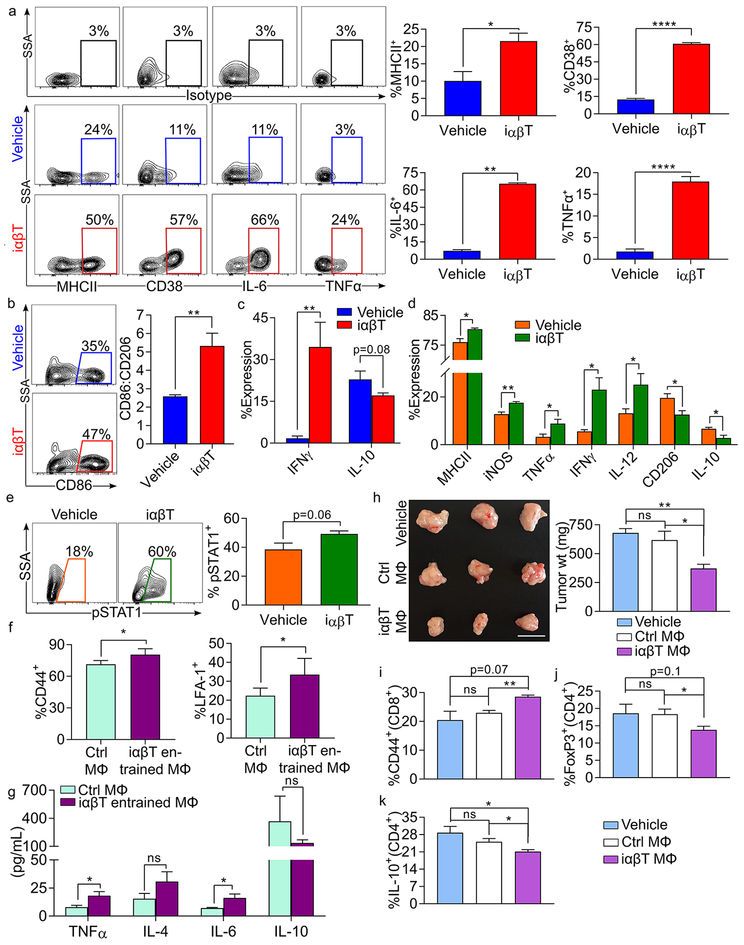
We previously showed that macrophage programming governs the balance between T cell tolerance and immunogenicity in PDA (5,14). Therefore, we postulated that iαβTs may indirectly augment intra-tumoral adaptive immunity by inducing immunogenic macrophage polarization, which in turn reprograms effector T cell populations. Accordingly, iαβTs directly upregulated macrophage expression of MHCII, CD38, IL-6, and TNFα in co-culture experiments (Figure 5a). iαβTs also promoted increased CD86 and IFNγ expression in macrophages and downregulated CD206 and IL-10 (Figure 5b, c). In vivo iαβT cell transfer in PDA also induced higher MHCII, iNOS, TNFα, IFNγ, and IL-12 expression in tumor-associated macrophages (TAMs), while CD206, and IL-10 expression were reduced (Figure 5d). iαβT cell transfer also significantly increased STAT1 signaling in TAMs, which is linked to M1-like macrophage polarization (Figure 5e). Further, our single cell RNAseq data indicated a marked contraction of F480hi macrophages in tumor after serial iαβT cell transfer. Specifically, macrophages represented 84% of leukocytes in control tumors compared to only 24% in iαβT cell treated mice (Figure 3l, m). Moreover, consistent with our flow cytometry data, iαβT cell transfer resulted in upregulation of genes related to antigen-presentation (e.g. H2-family genes, cd74, B2m), T cell chemoattraction (ccl5, cxcl9, cxcl10), IFN signaling (stat1, ifnar1, ifngr1, irf5, irf7, irf8), and M1 polarization (tnf, ccl9, il6ra, ccr5) on macrophages, whereas M2-associated transcription factors were downregulated (stat6, socs3, il1b) (Figure S6a–e). Ingenuity pathway analysis of upstream regulators indicated that STAT1, IFNγ, and TNFα signaling were significantly activated in TAMs infiltrating tumors of iαβT cell treated mice relative to controls (Figure S6f–h). Further, Gene Set Enrichment Analysis (GSEA) indicated significant enrichment of “KEGG_Antigen processing and presentation” and “KEGG_Chemokine signaling pathway” in macrophages of iαβT cell-treated tumors (Figure S6i, j). Finally, we noted the expansion of a distinct CX3CR1hiF480hi macrophage population in the TME of iαβT cell treated mice (Figure 3l, m). This population upregulated ccl5, but did not exhibit upregulation of the other genes and pathways related to antigen-presentation and pro-inflammatory signaling observed in the broader macrophage population (Figure S6k). Collectively, these data show that iαβTs induce profound immunogenic macrophage programming in PDA.
Figure 5. iαβTs induce immunogenic reprogramming of macrophages.
(a-c) Splenic macrophages were cultured alone or co-cultured with iαβTs for 24h. Macrophages were then harvested and tested for expression of (a) MHC II, CD38, IL-6, TNFα, (b) CD86, CD206, (c) IFNγ, and IL-10. Select contour plots and quantitative data are shown. This experiment was repeated 5 times. (d, e) KPC tumor-bearing mice were adoptively transferred with iαβTs. Tumors were harvested on day 21 and TAMs were analyzed for expression of (d) MHCII, iNOS, TNFα, IFNγ, IL-12, CD206, IL-10, and (e) pSTAT1. This experiment was repeated twice (n=5/group). (f, g) iαβT cell-entrained splenic macrophages and control splenic macrophages were pulsed with Ova323–339 peptide and used to stimulated Ova-restricted CD4+ T cells. T cell activation was determined at 96h by expression of (f) CD44 and LFA-1. (g) Cell culture supernatant was harvested and tested for expression of TNFα, IL-4, IL-6, and IL-10. In vitro experiments were performed in replicates of 5 and repeated 3 times. (h-k) Cohorts of WT mice were administered orthotopic KPC tumor either alone, admixed with control macrophages or macrophages that had been co-cultured with iαβTs. Pancreatic tumors were harvested on day 21 (n=5/group). (h) Tumor weights were recorded. (i) Tumor infiltrating CD8+ T cells were analyzed for expression of CD44. (j) Tumor infiltrating CD4+ T cells were analyzed for expression of FoxP3 and (k) IL-10. This experiment was repeated twice (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
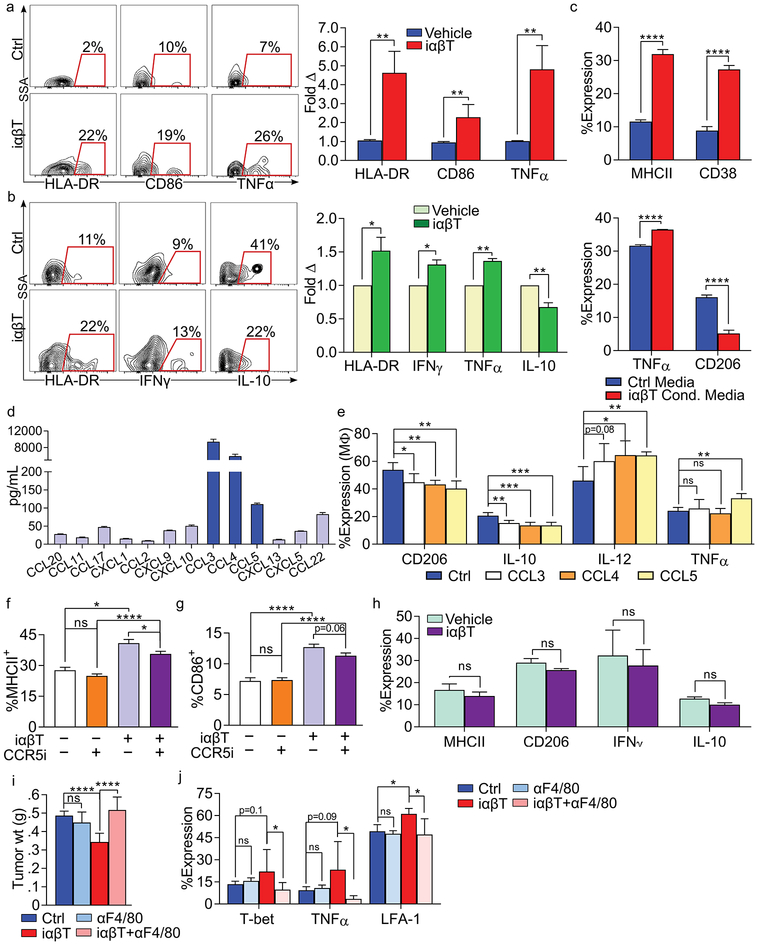
We postulated that iαβTs secondarily augment adaptive T cell immunity in situ via their immunogenic influence on macrophage differentiation. To test this, antigen-pulsed macrophages entrained by co-culture with iαβTs were used to stimulate antigen-restricted CD4+ T cells. Consistent with our hypothesis, iαβT-entrained macrophages were more efficient than controls at antigen-presentation based on T cell expression of activation markers and secreted cytokines (Figure 5f, g). Moreover, in vivo adoptive transfer of macrophages entrained by iαβT co-culture was markedly protective against PDA, whereas transfer of control macrophages was not. (Figure 5h). Adoptive transfer of iαβT-entrained macrophages also activated intratumoral conventional T cells and reduced Treg differentiation in situ (Figure 5i–k). Collectively, these data suggest that iαβTs can induce macrophage-mediated adaptive anti-tumor immunity in PDA. To determine whether iαβTs also activate macrophages in human systems, we co-cultured FACS-sorted iαβTs with human PBMC-derived macrophages. Consistent with our murine data, iαβTs activated human macrophages (Figure 6a). Furthermore, autologous treatment of organotypic 3D models of human PDA with iαβTs similarly resulted in marked activation of TAMs (Figure 6b).
Figure 6. iαβTs induce immunogenic macrophage programming via CCR5 activation.
(a) iαβTs were FACS-sorted from 3 healthy volunteers and co-cultured with autologous naïve PBMC-derived macrophages. At 24h, macrophage expression of HLA-DR, CD86, and TNF-α were determined by flow cytometry. Experiments for each individual were performed in triplicate. Representative contour plots are shown and quantitative data is presented as fold-change in expression compared with macrophages cultured alone. (b) Human PDOTS were treated with autologous iαβTs or vehicle. Tumor-associated macrophages were tested for expression of HLA-DR, IFNγ, TNFα, and IL-10. Representative contour plots are shown and quantitative data is indicated as fold-change in expression compared to vehicle treatment (n=5 patients). (c) Naive macrophages were cultured with iαβT cell conditioned media or control media. After 24h, macrophage expression of MHCII, CD38, TNF-α, and CD206 were determined by flow cytometry. This experiment was repeated 3 times. (d) iαβTs were FACS-sorted from orthotopic PDA tumors and cultured for 24h. Cell culture supernatant was analyzed in a chemokine array. This experiment was repeated twice (n=5). (e) WT bone marrow-derived macrophages were treated with rCCL3, rCCL4, rCCL5, or vehicle and assessed at 24h. (f, g) WT derived macrophages were cultured alone or co-cultured with PDA-infiltrating iαβTs for 24h. A CCR5 small molecule inhibitor (CCR5i) or vehicle was added to select wells. Macrophage expression of (f) MHC II and (g) CD86. This experiment was repeated 3 times (n=5/group). (h) CCR5–/– macrophages were cultured with PDA-infiltrating iαβTs for 24h. Macrophage expression of MHC II, CD206, IFNγ, and IL-10 were determined. This experiment was repeated 4 times in replicates of 5. (i, j) Orthotopic PDA tumors were harvested from control WT mice or WT mice treated with αF4/80, iαβT cell transfer, or αF4/80 plus iαβT cell transfer. (i) tumor weights were measured and (j) CD8+ T cell activation was determined by expression of T-bet, TNFα, and LFA-1 (n=7/group; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
Mechanistically, we postulated that iαβTs secrete specific inflammatory mediators that may drive immunogenic macrophage programming. To test this, we cultured naive splenic macrophages with iαβT cell conditioned-media or control media. iαβT conditioned-media upregulated MHC II, CD38, and TNFα in macrophages and downregulated CD206, suggesting that iαβT-derived secreted factors are sufficient to induce an activated macrophage phenotype (Figure 6c). To determine the mechanism through which iαβTs regulate macrophage polarization, we performed unbiased analysis of inflammatory mediators secreted by iαβTs. We found that iαβTs produced markedly high levels of CCL3, CCL4, and CCL5, each of which ligate CCR5 (Figure 6d) (15). Consistent with our hypothesis, treatment of macrophages with rCCL3, rCCL4, and rCCL5 induced M1-like differentiation (Figure 6e). Further, CCR5 inhibition mitigated immunogenic macrophage polarization induced by iαβTs (Figure 6f, g). CCR5–/– macrophages similarly failed to become activated in co-culture with iαβTs (Figure 6h). Collectively, these data suggest that iαβTs program macrophages by secretion of factors that engage the CCR5 axis. Corroboratively, in vivo depletion of macrophages abrogated the tumor-protection and effector T cell activation observed with iαβT cell adoptive transfer (Figure 6i, j).
Discussion
We show that TCRαβ+CD4–CD8–NK1.1– represent ~10% of T cells in the mouse and human PDA TME. These cells differ from innate lymphoid cells (ILC) by expression of the CD3-TCR complex and are distinguished from NKT cells by absence of obligate marker expression on transcriptomic and flow cytometric analyses. Interestingly, our iαβTs share some phenotypic features with mucosal associated invariant T cells (MAIT cells), a subset of CD3+ T cells that can express CD4 and CD8, but are classically CD4–CD8–. MAIT cells predominate in the gut, where they defend against microbial infection, but have been described in the blood, liver, and lungs (16). Given that PDA-infiltrating iαβTs and MAIT cells both exhibit high expression of cytokines, including IL-17 and IFNγ, as well as production of cytolytic molecules such as Perforin and Granzyme B, we probed the antigen specificity of PDA-infiltrating iαβT and gut-derived MAIT cells (17). The MHC class I-like protein, MR1, is responsible for presenting bacterially-produced vitamin B metabolites to MAIT cells. In turn, recognition of antigen results in MAIT cell stimulation and secretion of pro-inflammatory cytokines (18). However, we found only ~10% of iαβT in PDA recognize a murine MR1 specific tetramer, as compared to ~40% of CD3+CD4−CD8− cells in the gut. CD4+ and CD8+ T cells in PDA expressed similar levels of the MR1 specific tetramer compared to iαβT cells. Thus, although the cytokine profiles remain similar, we are averse to define our population as MAIT cells and prefer their more general description as iαβTs. We maintain the possibility that iαβTs in PDA encompass a diversity of phenotypes, as do most broadly described T cell populations, but the precise cellular ontogeny is beyond the scope of our work. Rather, our investigations focus on the impact of these cells in interfacing with other innate and adaptive immune subsets, as novel targets of immunotherapy, and their role in combating oncogenic progression.
The PDA microenvironment is regulated by diverse cellular and biochemical inflammatory signals that modulate tumor growth. However, the cross-talk between cellular subsets in the inflammatory TME remains incompletely understood. The importance of unconventional T cells subsets in this immune interplay is becoming increasingly recognized in both hematologic and solid organ malignancies. For example, in PDA we showed that γδT cells cripple conventional CD4+ and CD8+ T cell immunogenicity via upregulation of PD-L1 and Galectin-9 (5). By contrast, NKT cells exhibit protective effects in pre-invasive models of PDA via monocytic cross-talk (6). Consistent with the role of NKT cells as first responders in inflammation (19,20), we found that NKT cell levels steadily declined in PDA as tumors progressed, whereas iαβTs sharply increased in the TME over the course of disease progression. This latter observation is consistent with high levels of CCL2 in mature PDA tumors (21), as we discovered that iαβTs are recruited to the TME via CCR2 signaling.
We found that iαβTs are distinctly pro-inflammatory in PDA, promiscuously expressing high costimulatory (ICOS, CD40L) and checkpoint (CTLA-4, PD-1, TIM-3, LAG-3) receptors and co-expressing both Th1 and Th17 families of fate-determining transcription factors (T-bet, RORγt) and cytokines (IFNγ, IL-17). However, in contrast to the tolerogenic role of iαβTs in the context of organ transplantation (19), PDA-infiltrating iαβTs minimally express Foxp3 and IL-10, consistent with their more immunogenic function. Interestingly, the tumor-protective effects of iαβTs belies the fact that these cells are the highest producer of IL-17 in PDA, a cytokine that has been shown to have directly proliferative effects on tumor cells (20). Though we observe that iαβTs possess a cytotoxic phenotype in PDA (expression of perforins, granzymes, and FasL), they were incapable of directly inhibiting tumor proliferation or inducing tumor cell death in vitro. This contrasts with findings in melanoma and lymphoma where iαβTs exhibit potent tumor-specific cytotoxicity (22,23). This observation could in theory be due to the paucity of tumor-associated antigens (TAA) and neoantigens in PDA relative to other cancers and the consequent limitation of MHCI-dependent killing. However, we observed that iαβTs in PDA do not recognize tumor antigen; therefore their lack of directly tumoricidal properties is unlikely solely due to the scarcity of TAA.
We found that iαβTs delimit PDA progression by shaping the innate and adaptive immunological landscape. Specifically, iαβTs promote macrophage reprogramming to a distinctly immunogenic phenotype, which in turn enhances effector CD4+ and CD8+ T cell function. Several studies have shown that TAMs drive tumor progression in PDA by possessing potent immunosuppressive properties (24–26). Interestingly, we show that treatment with iαβTs not only results in global contraction of TAMs in the PDA TME, but also confers significant pro-inflammatory phenotypic changes to the remaining macrophages, leading to expansion of effector T cell populations. We also found that deletion of TAMs abrogated the tumor-protection and enhancement in adaptive immunity associated with iαβT cell transfer. Programs upregulated in TAMs by treatment with iαβTs included antigen-presentation, T cell chemoattraction, and IFN, TNF, and STAT1 signaling. In this way, iαβTs may shift the balance toward anti-tumor immunity by dually abrogating immune-suppression and promoting increased antigen presentation capacity to effector T cells and their activation (Figure S7). We found that the iαβT cell – macrophage crosstalk was linked to the high levels of CCR5 ligands secreted by iαβTs. Moreover, considering the central role of TAMs in regulating the adaptive immune program and the protective tumor-immunity associated with iαβT cellular transfer, our data suggest that iαβTs may be attractive vehicles for cell therapy in PDA.
This work also highlights paradoxical effects regarding the influence of iαβTs on CD4+ and CD8+ T cells. We show that iαβTs directly inhibit CD4+ and CD8+ T cells in vitro in co-culture experiments via the PD-L1-PD1 axis. This parallels the suppressive effects of γδT cells in PDA (5). Conversely, the broader in vivo effect of iαβTs is a downstream immunogenic activation of effector T cells via macrophage activation. These findings are consistent with previous work suggesting that TAM programming is the dominant driver of the adaptive immune landscape in PDA (14,24). This functional hierarchy is even more plausible considering that the iαβT-mediated suppressive effect on effector T cells requires direct cell contact via PD-L1–PD1 interaction, whereas macrophage activation can occur remotely via secretion of CCL3, CCL4, and CCL5 (CCR5 ligands). The contact dependent inhibitory effects of iαβTs on effector T cells, activating effects (including PD1 upregulation) observed after iαβT cell transfer on effector T cells, and the observed activation of iαβTs in response to αPD-L1 therapy in vivo, suggests that immunotherapy targeting the PD-L1–PD-1 axis could synergistically accentuate the tumor-protective and immunological benefits of iαβT cell directed therapies.
The signals that drive the distinctive phenotype of iαβTs in PDA are diverse and modifiable and collectively account for their complex features. Signaling via Dectin-1, a pattern recognition receptor capable of binding Galectin-9 and fungal wall β-glucans, promoted IFNγ and IL-17 production in iαβTs, but did not appreciably modulate iαβT surface phenotype. By contrast, CCR2 signaling activated iαβT cell surface phenotype and upregulated expression of costimulatory and checkpoint receptors, but did not alter cytokine production. The PDA-associated microbiome and LPS have broadly suppressive effects on iαβTs, reducing both cytokine and surface activation marker expression. Therefore, tailored approaches can be used to modulate iαβT cell phenotype in situ or ex vivo to optimize phenotypic outcomes. Finally, the use of iαβTs as an adoptive cell therapy into human organotypic PDOT systems of pancreatic cancer is the first application of its kind. The ability for iαβTs to induce significant immunogenic activation in these 3D human systems illustrates their potential as a novel cell therapy. In aggregate, our work suggests that iαβT cell cross-talk serves to connect innate and adaptive immunity in PDA by reprogramming of macrophages that leads to conventional CD4+ and CD8+ T cell activation. This work thus describes a critical new cellular entity in the landscape of the TME and suggests that iαβTs can be utilized for adoptive cell therapy or as targets of activation for immune-based therapies in situ in PDA.
Materials and Methods
Animals and In Vivo Procedures
C57BL/6 (H-2Kb), B6.MRL-Faslpr/J, Dectin-1–/–, CCR2–/–, CCR5–/–, CCR6–/–, β2m–/–, MHCII–/–, OT-II, and CD45.1 mice were purchased from Jackson Labs (Bar Harbor, ME) and bred in-house. KC mice, which express KrasG12D in the progenitor cells of the pancreas, were a gift of Dafna Bar-Sagi (New York University) (1). Both male and female mice were used, but animals were gender-matched within each experiment. For orthotopic pancreatic tumor challenge, 8–10 week old mice were administered intra-pancreatic injections of FC1242 tumor cells derived from KPC mice, as we previously described(5). In select experiments we utilized KPC tumor cells, which we engineered to express Ovalbumin using the pCIneo-OVA vector (Addgene, Cambridge, MA). PDA cells (1x105) were suspended in PBS with 50% Matrigel (BD Biosciences, Franklin Lakes, NJ) and were injected into the body of the pancreas via laparotomy. Mice were sacrificed 3 weeks later for analysis. Alternatively, PDA cells (3x105) were implanted subcutaneously and tumor growth was serially measured. In some experiments, iαβTs or macrophages were mixed with tumor cells in a 1:3 ratio before orthotopic or subcutaneous injection. In other experiments, iαβTs were harvested by FACS and administered i.v. (5x106) twice weekly to orthotopic PDA bearing mice beginning on day 5 after tumor implantation. For tracking experiments, iαβTs from CD45.1 hosts were co-injected with PDA cells into pancreata of CD45.2 mice and harvested 96 hours later. In select experiments, animals were treated with neutralizing mAbs directed against CD4 (GK1.5), CD8 (Lyt 2.1), F4/80 (CI:A3–1), TCRγ/δ (UC3–10A6), NK1.1 (PK136), and PD-L1 (10F.9G2; all BioXCell, West Lebanon, NH) using regimens we have previously described(5,10). Alternatively, mice were treated with an agonizing ICOS mAb (7E.17G9, 100μg, Days 4, 7 and 10 post-tumor challenge; BioXcell) or TLR4 ligand (LPS, 5μg, i.p., 3x/week; Invivogen, San Diego, CA). Anti-microbial ablation was performed as previously described (10). Briefly, mice were administered an antibiotic cocktail by oral gavage daily for five consecutive days. Controls were gavaged with PBS. The oral gavage cocktail contained Vancomycin (50mg/ml; Sigma, St. Louis, MO), Neomycin (10mg/ml; Sigma), Metronidazole (100mg/ml; Santa Cruz Biotech, Dallas, TX) and Amphotericin (1mg/ml; MP Biomedicals, Santa Ana, CA). Additionally, for the duration of the experiments, mouse drinking water was mixed with Ampicillin (1mg/ml; Santa Cruz Biotech), Vancomycin (0.5mg/ml; Sigma), Neomycin (0.5mg/ml; Sigma), Metronidazole (1mg/ml; Santa Cruz Biotech) and Amphotericin (0.5μg/ml; MP Biomedicals). All studies were approved by the New York University School of Medicine Institutional Animal Care and Use Committee.
Sequential Immunohistochemistry, Image Acquisition and Processing for Human Tissues
Human tissues used for immunohistochemical analysis were obtained with written informed consent in accordance with the Declaration of Helsinki and were acquired through the Oregon Pancreas Tissue Registry. All studies were approved by an Institutional Review Board under Oregon Health & Science University IRB protocol #3609. Human PDA and murine orthotopic PDA tissues were fixed with 10% buffered formalin, dehydrated in ethanol, and embedded with paraffin. Sequential IHC was performed on 5 μm FFPE sections using an adapted protocol based on methodology we previously described (27). Briefly, slides were deparaffinized and stained with hematoxylin (S3301, Dako, Santa Clara, CA), followed by whole-slide scanning at 20X magnification on an Aperio AT2 (Leica Biosystems, Wetzlar, Germany). Tissues then underwent 15 minutes heat-mediated antigen retrieval in pH 6.0 Citra solution (BioGenex, Fremont, CA), followed by 20 minutes endogenous peroxidase blocking in 0.6% H2O2 (mouse) or 10 minutes blocking in Dako Dual Endogenous Enzyme Block (S2003, Dako, Santa Clara, CA) (human), then 10 minutes protein blocking with 5% normal goat serum and 2.5% BSA in TBST. For staining of mouse tissue, primary antibody incubations were carried out overnight at 4°C with CD4 (D7D2Z, 1:50, Cell Signaling Technology, Danvers, MA), CD3 (SP7, 1:300, Thermo Fisher), and CD8 (4SM15, 1:100, eBioscience). For staining of human tissues, primary antibody incubations were carried out for 30 minutes at room temperature using CD4 (SP35, 1:4, Ventana), CD68 (PG-M1, 1:50, Abcam), CD3 (SP7, 1:150, Thermo Fisher), and Pan Cytokeratin (AE1/AE3, 1:2000, Abcam), and 60 minutes at room temperature using CD45 (HI30, 1:100, Thermo Fisher). After washing off primary antibody in TBST, either anti-rat, anti-mouse, or anti-rabbit Histofine Simple Stain MAX PO horseradish peroxidase (HRP)- conjugated polymer (Nichirei Biosciences, Tokyo, Japan) was applied for 30 minutes at room temperature, followed by AEC chromogen (Vector Laboratories, Burlingame, CA). Slides were digitally scanned following each chromogen development, and the staining process was repeated starting at the Citra step for all subsequent staining cycles.
Scanned images were registered in MATLAB version R2018b using the SURF algorithm in the Computer Vision Toolbox (The MathWorks, Inc., Natick, MA). Image processing and cell quantification were performed using FIJI (FIJI Is Just ImageJ) (28), CellProfiler Version 3.5.1 (29), and FCS Express 6 Image Cytometry RUO (De Novo Software, Glendale, CA). AEC signal was extracted for quantification and visualization in FIJI using a custom macro for color deconvolution. Briefly, the FIJI plugin Color_Deconvolution [H AEC] was used to separate hematoxylin, followed by postprocessing steps for signal cleaning and background elimination. AEC signal was extracted in FIJI using with the NIH plugin RGB_to_CMYK. For visualization, signal-extracted images were overlaid in pseudo-color in FIJI. Color deconvoluted images were processed in CellProfiler to quantify single cell mean intensity signal measurements for every stained marker, and human T cells were identified and quantified by image cytometry in FCS Express based on expression of known markers as follows: Pan Cytokeratin- CD45+ CD68- CD3+, CD4+/-, CD8+/-.
Murine and Human Cellular Isolation, Flow Cytometry, and FACS
Single cell suspensions of mouse PDA tumors were prepared for flow cytometry as described previously with slight modifications (5). Briefly, pancreata were placed in cold 2% FACS (PBS with 2% FBS) with Collagenase IV (1 mg/mL; Worthington Biochemical, Lakewood, NJ), Trypsin inhibitor (1mg/mL; EMD Millipore, Billerica, MA) and DNase I (2 U/mL; Promega, Madison, WI), and minced with scissors to sub-millimeter pieces. Tissues were then incubated at 37°C for 20 minutes with gentle shaking every 5 minutes. Specimens were passed through a 70μm mesh and centrifuged at 350g for 5 minutes. Splenocytes and thymocytes were prepared by manual disruption, as we described (5). Cell pellets were re-suspended and cell labeling was performed after blocking FcγRIII/II with an anti-CD16/CD32 mAb (eBiosciences, San Diego, CA) by incubating 1x106 cells with 1 μg of fluorescently conjugated mAbs directed against mouse CD45 (30-F11), CD3 (17A2), CD4 (RM4–5), CD8 (53–6.7), Tcrβ (H57–597), CD62L (MEL-14), FasL (MFL3), NK1.1 (PK136), CD39 (Duha59), CCR2 (K036C2), CCR5 (HM-CCR5), CCR6 (292L17), CD44 (IM7), CD206 (C068C2), CD107a (1D4B), JAML (4E10), CD86 (GL-1), Gr1 (RB6–8C5), MHC II (M5/114.15.2), PD-1 (29F.1A12), ICOS (15F9), TNFα (MP6-XT22), IL-17A (TC11–18H10.1), TGFβ (TW7–16B4), LFA-1 (H155–78), IL-10 (JES5–16E3), IFN-γ (XMG1.2), Granzyme B (12-8898-80), PD-L1 (10F.9G2), B7–2 (PO3), CTLA-4 (UC10–4B9), Tim-3 (RMT3–23), CD40L (24–31), LAG-3 (C9B7W), TLR4 (UT41), CD73 (TY/11.8), TGFβ (TW7–16B4), Dectin-1 (RH1), Ki67 (16A8), IL-6 (MP5–20F3), TCR Vβ7 (TR310), TCR Vβ2 (B20.6), α-GalCer:CD1d complex (L363), pSTAT1 (A15158B; all Biolegend, San Diego, CA), CD45.1 (A20), RORγt (AFKJS-9), Perforin (eBioOMAK-D), T-bet (eBio4B10), and Foxp3 (FJK-16s; all eBioscience, San Diego, CA). Tetramer staining was performed as previously described, using a murine MR1 specific tetramer courtesy of the NIH Core Tetramer Facility (Atlanta, GA) (30). Intracellular staining was performed using the Foxp3 Fixation/Permeabilization Solution Kit (eBiosciences). Ova-specific CD8+ T cells were identified using a SIINFEKL Pentamer (Proimmune, Oxford, UK). Mouse bone marrow-derived macrophages were generated as we described (31). Human pancreatic tumor and PBMC were collected under an IRB approved protocol. Human pancreatic leukocytes were prepared in a similar manner to that of mice. PBMC were isolated by overlaying whole blood diluted 1:1 in PBS over an equal amount of Ficoll (GE Healthcare, Princeton, NJ). Cells were then spun at 2200 RPM and the buffy coat was harvested, as we have described (32). Human PBMC Macrophages were cultured plated in complete RPMI containing human FBS supplemented with human GM-CSF (2μg/ml) and left in culture for 3 days until they used in experiments. Analysis of human cells was performed using fluorescently conjugated antibodies directed against CD45 (2DI), TCRαβ cells (IP26), CD4 (A161A1), CD8 (HIT1A), TCR Vα7.2 (3C10), and CD56 (HCD56; all Biolegend). Dead cells were excluded from analysis using zombie yellow (Biolegend). Flow cytometry was performed on the LSR-II (BD Biosciences) or Attune NxT Flow Cytometer (Thermo Fisher, Waltham, MA). FACS-sorting was performed on the SY3200 (Sony, Tokyo, Japan). Data were analyzed using FlowJo (Treestar, Ashland, OR). Chemokine and cytokine levels in cell culture supernatant were analyzed using Legendplex arrays, as per the manufacturer’s protocol (Biolegend).
Co-culture and In-Vitro Experiments
For mouse or human macrophage polarization assays, iαβTs were co-cultured with naïve splenic or PMBC-derived macrophages, respectively, for 36 hours in a 1:4 ratio. In select experiments, 36-hour conditioned media from iαβTs was added to macrophage cultures. In some experiments, a CCR5 small molecule inhibitor or vehicle was added to the co-culture well (Maraviroc, 20μM; Tocris Biosciences, Bristol, UK). Alternatively, iαβTs were cultured alone and treated with either Dectin-1 ligand (depleted zymosan, 10μg/ml; Invivogen) or ICOSL Fc (10μg/ml; Novoprotein, Summit, NJ). iαβT–KPC cell co-culture experiments were performed in various ratios for 24 hours. Tumor cell proliferation was measured using the XTT assay kit according to the manufacturer’s protocol (Sigma). Tumor cell lysis was measured using an LDH Cytotoxicity Assay Kit (Pierce, Rockford, IL) and apoptosis was determined using Annexin V and Propium Iodide staining (both Biolegend). For antibody-based T cell proliferation assays, T cells were activated using CD3/CD28 co-ligation in 96 well plates, as we previously described(5). Alternatively, cells were activated using αGalCer (0.1ug/ml) in 96 well plates, as we described (33). For antigen-restricted T cell stimulation assays, CD4+ OT-II T cells were cultured with macrophages pulsed with Ova323–339 peptide in a 5:1 ratio. T cell activation was determined at 72h by flow cytometry. In selected wells, iαβTs were added to T cell activation assays in a 1:4 ratio. TGFβ (AF-101-NA) and PD-L1 (10F.9G2, both 10μg/ml; R&D, Minneapolis, MN) were selectively neutralized in these assays.
Fluorescence In Situ Hybridization (FISH)
The EUB338 16S rRNA gene probe labeled with the fluorophore Cy3 (extinction wavelength, 555 nm; emission wavelength, 570 nm; Molecular Probes, Eugene, OR) was used to detect the bacterial colonization within mouse pancreatic tissues by FISH. Fluorescence microscopic analysis was conducted with Nikon Eclipse 90i confocal microscope (Nikon, Melville, NY) using a Cy3 labeled-probe at 50 pmol/ml as described (34–36).
PCR Analysis
Genomic DNA was extracted using a DNAeasy mini kit (Qiagen, Valencia, CA). PCR was performed in duplicate for each sample using the BioRad Real-Time PCR System (BioRad, Hercules, CA). The primer sequences for Ova was F-GTGTTTAGCTCTTCAGCCAATCT, R-CTGCATGGACAGCTTGAGATA. Amplified PCR products were evaluated by agarose gel electrophoresis and subsequent ethidium bromide staining.
Sequential Immunohistochemistry and Image Acquisition
For histological analysis, murine orthotopic PDA tissues were fixed with 10% buffered formalin, dehydrated in ethanol, and embedded with paraffin. Sequential IHC was performed on 5 μm FFPE murine PDA using a staining methodology we previously described (27). Briefly, slides were deparaffinized and stained with hematoxylin (S3301, Dako, Santa Clara, CA), followed by whole-tissue scanning at 20X magnification on an Aperio AT2 (Leica Biosystems, Wetzlar, Germany). Slides then underwent 20 minutes endogenous peroxidase blocking in 0.6% H2O2 followed by 15 minutes heat-mediated antigen retrieval in pH 6.0 Citra solution (BioGenex, Fremont, CA), followed by 10 minutes protein blocking with 5% normal goat serum and 2.5% BSA in TBST. Primary antibody incubations were carried out overnight at 4°C using rabbit anti-CD4 (D7D2Z, 1:50, Cell Signaling Technology, Danvers, MA), rabbit anti-CD3 (SP7, 1:300, Thermo Fisher), and rat anti-CD8 (4SM15, 1:100, eBioscience). After washing off primary antibody in TBST, either anti-rat or anti-rabbit Histofine Simple Stain MAX PO horseradish peroxidase (HRP)-conjugated polymer (Nichirei Biosciences, Tokyo, Japan) was applied for 30 minutes at room temperature, followed by AEC chromagen (Vector Laboratories, Burlingame, CA). Slides were digitally scanned following chromagen development, and the staining process was repeated starting at Citra step for all subsequent staining cycles. Images were co-registered using MATLAB software, followed by AEC color deconvolution and pseudocoloring in Fiji software.
Single Cell RNAseq Data Pre-Processing
Sequencing results were demultiplexed and converted to FASTQ format using Illumina bcl2fastq software. The Cell Ranger Single-Cell Software Suite (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger) was used to perform sample demultiplexing, barcode processing, and single-cell 3’ gene counting. The cDNA insert was aligned to the mm10/GRCm38 reference genome. Only confidently mapped non-PCR duplicates with valid barcodes and UMIs were used to generate the gene-barcode matrix. Further analysis including the identification of highly variable genes, dimensionality reduction, standard unsupervised clustering algorithms, and the discovery of differentially expressed genes was performed using the Seurat R package (37). To exclude low quality cells, cells that were extreme outliers in terms of library complexity, or cells that may possibly be multiple cells or doublets, we calculated the distribution of genes detected per cell and removed any cells in the top and bottom 2% quantiles. We additionally removed cells with more than 10% of the transcripts coming from mitochondrial genes.
Integrated Analysis of Single Cell Datasets
To account for technical batch differences between the three libraries, we utilized the Seurat alignment method for data integration, which specifically does not expect that confounding variables have uniform effects on all cells in a dataset and allows for global transcriptional shifts between datasets. Seurat uses a variant of canonical correlation analysis (CCA) to find linear combinations of features and identifies shared correlation structures across datasets. For each dataset, we identified variable genes, while controlling for the strong relationship between variability and average expression. We took the union of the top 2,000 genes with the highest dispersion from both datasets and ran a CCA to determine the common sources of variation between datasets. We then aligned the subspaces based on the first 15 canonical correlation vectors, generating a new dimensionality reduction that was then used for further analysis. For single cell analysis of lymphocytes in PDA, from an unbiased pool of CD45+ tumor-infiltrating leukocytes, we refined our analysis to include CD3d/CD3de expressing cells that expressed >1000 detected genes. We then normalized the data by the total expression, multiplied this by a scale factor of 10,000, and log-transformed the result. The final dataset included 807 cells with a median of 1,530 detected genes. For analysis of the total leukocyte population in tumors treated with iαβTs, we normalized the data by the total expression, multiplied this by a scale factor of 10,000, and log-transformed the result. The final dataset included 3,839 cells with a median of 1,148 detected genes.
Visualization and Clustering of Single Cell RNAseq Data
To visualize the data, we further reduced the dimensionality of the dataset to project the cells in two-dimensional space using PCA followed by t-distributed Stochastic Neighbor Embedding (tSNE) based on the aligned CCA. Aligned CCA was also used as a basis for partitioning the dataset into clusters using a smart local moving (SLM) community detection algorithm (https://arxiv.org/ftp/arxiv/papers/1308/1308.6604.pdf). To find markers that define individual clusters, we performed differential expression analysis using Wilcoxon rank sum test for each cluster compared to all other cells for genes detected in at least 20% of the cluster cells. The initial analysis of PDA-infiltrating lymphocytes yielded 7 clusters. We assigned cell type identities based on the expression of known population markers as follows: PD1hiCD8+ T cells – Pdcd1hiCd4loCd8ahiCcl1hi; PD1loCD8+ T Cells – Pdcd1loCd4loCd8ahiCcl1loCcl5hi; CD4+ T Cells – Cd4hiCd8aloCd74hiPdcd4hi; iαβTs – Cd4loCd8aloCcr7hi Il2rblo Nkg7lo Klrd1lo Klrc2lo; NKT Cells – Cd4loCd8aloNkg7hi Klrc2hi; γδ T Cells – Tcrg-c2hiCd4loCd8aloCd8bmed GzmahiCcl5hi; Pro-B Cells – Ccnb2hiHmgb2hiStmn1hiCd3dlo. The initial analysis for the tumor-infiltrating leukocytes consisted of 14 clusters. We then assigned them cell type identities based on the expression of known population markers, identifying 10 distinct populations in the following manner: T cells – Cd3dhiCd3ghi Cd3ehiCd4hiCd8ahi; B Cells – Cd19hi Ebf1 hi; iαβTs – Cd3dhiCd3ghi Cd3ehiCd4loCd8aloLy6c1hi Klrk1low Klra7low; NKT Cells – Cd3dhiCd3ghi Cd3ehi Klrk1 hi Klra7hi; NK Cells – Cd3dlowCd3glow Cd3elowKlrk1 hi Klra7hi; Macrophages – ItgamhiAdgre1hiCd177lo; CX3CR1+ Macrophages – ItgamhiAdgre1hiCx3cr1hi; CD103hi MHCIIhi – ItgaehiH2-Ab1hiCD74hiItgamlow; Batf3hi MHCIIhi – Batf3hi CD74hiH2-Ab1hi ItgaxloItgamlow; Neutrophils – Cd177hiItgamhi. GSEA (Gene Set Enrichment Analysis) was performed on differentially expressed genes in the Macrophage cluster. Upregulated and downregulated sets of genes were ranked based on their average and normalized log2 fold change between treatment and control group and each gene set was assessed for enrichment in the KEGG_2016 geneset library [http://amp.pharm.mssm.edu/Enrichr/#stats] using python package gseapy [https://pypi.org/project/gseapy/] for analyses.
PDOTS preparation, treatment, and analysis
PDOTS were prepared as we previously described with slight modifications (38). Briefly, human surgically resected tumor specimens were received fresh in DMEM media on ice and minced in 10cm dishes. Minced tumors were resuspended in DMEM + 10% FBS with 100 U/mL collagenase type IV to obtain spheroids. Partially digested samples were pelleted and then re-suspended in fresh DMEM +10% FBS and strained over both 100 mm and 40 mm filters to generate S1 (>100 mm), S2 (40–100 mm), and S3 (<40 mm) spheroid fractions, which were subsequently maintained in ultra-low-attachment tissue culture plates. An aliquot of the S2 fraction was pelleted and re-suspended in type I rat tail collagen and mixed with PBMC-derived allogeneic iαβTs that had been FACS sorted and activated overnight. This spheroid-iαβT mixture was then injected into the center gel region of the DAX-1 3D microfluidic cell culture chip. Control spheroids were also plated (Aim Biotech, Singapore, Malaysia). After 30 minutes at 37° C, collagen hydrogels containing PDOTS were hydrated with media in the side channels. After 72 hours, spheroids were digested with collagenase and single cell suspensions were generated for flow cytometry. Fluorescently labelled antibodies for CD45 (2D1), TCRαβ (IP26), CD4 (A161A1), CD8 (HIT1a), HLA-DR (L243), TNFα (Mab11), CD11B (ICR F44), IFN-γ (4S.B3), IL-10 (JES3–9D7), ICOS (C398.4A), CD3 (OKT3), CD44 (IM7; all Biolegend) and CD86 (2331FUN-1; BD Biosciences) we used to stain for analysis. Human biological samples were sourced ethically, and their research use was in accord with the terms of the informed consents under an IRB approved protocol.
T Cell Receptor Sequencing
CD4+, CD8+, and iαβTs were isolated by FACS and genomic DNA was extracted using a DNAeasy mini kit (Qiagen). Mouse TCR sequencing was performed using the immunoSEQ Assay (Adaptive Biotechnologies, Seattle, WA). V, D, and J Segments of the TCR were identified by multiplex PCR using forward primers in each V segment and reverse primers in each J segment. Detected template reads were normalized to total DNA content. Assessment of T cell clonality and sequence overlap analyses were performed on immunoSEQ ANALYZER 3.0 software (Adaptive Biotechnologies).
Statistical Analysis
Data is presented as mean +/- standard error. Statistical significance was determined by the Student’s t test and the log-rank test using GraphPad Prism 7 (GraphPad Software, La Jolla, CA). P-values ≤0.05 were considered significant. Significance for GSEA analysis and differential gene expression based on single cell RNAseq was determined using the Wilcoxon rank sum test with Bonferroni multiple-comparison correction.
Supplementary Material
Significance.
We found that innate αβ T cells (iαβTs) are a profoundly activated T cell subset in PDA that slow tumor growth in murine and human models of disease. iαβTs induce a CCR5-dependent immunogenic TAM program, T cell activation and expansion, and should be considered as novel targets for immunotherapy.
Acknowledgments
Grant Support: This work was supported by NIH grants CA168611 (GM), CA203105 (GM), CA215471 (GM), CA19311 (GM), and DK106025 (GM), the Knight Cancer Institute NCI P30 CA069533 (LMC, RCS), the Brenden-Colson Center for Pancreatic Care (LMC, SML, KRL, JL, RCS, SS, JG), Stand Up To Cancer – Lustgarten Foundation, Pancreatic Cancer Convergence Dream Team Translational Research Grant (SU2-AACR-DT14–14) (LMC, SML), Stand Up to Cancer is a Division of the Entertainment Industry Foundation administered by the American Association for Cancer Research, the Scientific Partner of SU2C; and resident research fellowship from the American Society for Colorectal Surgery (MH).
Footnotes
The authors declare no competing interests.
References
- 1.Guerra C, Schuhmacher AJ, Canamero M, Grippo PJ, Verdaguer L, Perez-Gallego L, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 2007;11(3):291–302 doi 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 2.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol 2010;11(10):889–96 doi 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 3.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 2009;16(2):91–102 doi 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jang JE, Hajdu CH, Liot C, Miller G, Dustin ML, Bar-Sagi D. Crosstalk between Regulatory T Cells and Tumor-Associated Dendritic Cells Negates Anti-tumor Immunity in Pancreatic Cancer. Cell Rep 2017;20(3):558–71 doi 10.1016/j.celrep.2017.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daley D, Zambirinis CP, Seifert L, Akkad N, Mohan N, Werba G, et al. gammadelta T Cells Support Pancreatic Oncogenesis by Restraining alphabeta T Cell Activation. Cell 2016;166(6):1485–99 e15 doi 10.1016/j.cell.2016.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janakiram NB, Mohammed A, Bryant T, Ritchie R, Stratton N, Jackson L, et al. Loss of natural killer T cells promotes pancreatic cancer in LSL-Kras(G12D/+) mice. Immunology 2017;152(1):36–51 doi 10.1111/imm.12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balomenos D, Shokri R, Daszkiewicz L, Vazquez-Mateo C, Martinez AC. On How Fas Apoptosis-Independent Pathways Drive T Cell Hyperproliferation and Lymphadenopathy in lpr Mice. Front Immunol 2017;8:237 doi 10.3389/fimmu.2017.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang X, Seo YD, Chang JH, Coveler A, Nigjeh EN, Pan S, et al. Long-lived pancreatic ductal adenocarcinoma slice cultures enable precise study of the immune microenvironment. Oncoimmunology 2017;6(7):e1333210 doi 10.1080/2162402X.2017.1333210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao R, Graffeo CS, Gulati R, Jamal M, Narayan S, Zambirinis CP, et al. Interleukin 17-producing gammadeltaT cells promote hepatic regeneration in mice. Gastroenterology 2014;147(2):473–84 e2 doi 10.1053/j.gastro.2014.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov 2018. doi 10.1158/2159-8290.CD-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reikvam DH, Erofeev A, Sandvik A, Grcic V, Jahnsen FL, Gaustad P, et al. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PloS one 2011;6(3):e17996 doi 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ochi A, Nguyen AH, Bedrosian AS, Mushlin HM, Zarbakhsh S, Barilla R, et al. MyD88 inhibition amplifies dendritic cell capacity to promote pancreatic carcinogenesis via Th2 cells. J Exp Med 2012;209(9):1671–87 doi 10.1084/jem.20111706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, Zhou CW, et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discov 2018;8(2):196–215 doi 10.1158/2159-8290.CD-17-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seifert L, Werba G, Tiwari S, Giao Ly NN, Alothman S, Alqunaibit D, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature 2016;532(7598):245–9 doi 10.1038/nature17403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu L, LaRosa G, Kassam N, Gordon CJ, Heath H, Ruffing N, et al. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J Exp Med 1997;186(8):1373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franciszkiewicz K, Salou M, Legoux F, Zhou Q, Cui Y, Bessoles S, et al. MHC class I-related molecule, MR1, and mucosal-associated invariant T cells. Immunological Reviews 2016;272(1):120–38 doi doi: 10.1111/imr.12423. [DOI] [PubMed] [Google Scholar]
- 17.Le Bourhis L, Martin E, Péguillet I, Guihot A, Froux N, Coré M, et al. Antimicrobial activity of mucosal-associated invariant T cells. Nature Immunology 2010;11:701 doi 10.1038/ni.1890 [DOI] [PubMed] [Google Scholar]
- 18.van Wilgenburg B, Scherwitzl I, Hutchinson EC, Leng T, Kurioka A, Kulicke C, et al. MAIT cells are activated during human viral infections. Nature Communications 2016;7:11653 doi 10.1038/ncomms11653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juvet SC, Zhang L. Double negative regulatory T cells in transplantation and autoimmunity: recent progress and future directions. J Mol Cell Biol 2012;4(1):48–58 doi 10.1093/jmcb/mjr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McAllister F, Bailey JM, Alsina J, Nirschl CJ, Sharma R, Fan H, et al. Oncogenic Kras activates a hematopoietic-to-epithelial IL-17 signaling axis in preinvasive pancreatic neoplasia. Cancer Cell 2014;25(5):621–37 doi 10.1016/j.ccr.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nywening TM, Wang-Gillam A, Sanford DE, Belt BA, Panni RZ, Cusworth BM, et al. Targeting tumour-associated macrophages with CCR2 inhibition in combination with FOLFIRINOX in patients with borderline resectable and locally advanced pancreatic cancer: a single-centre, open-label, dose-finding, non-randomised, phase 1b trial. Lancet Oncol 2016;17(5):651–62 doi 10.1016/S1470-2045(16)00078-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voelkl S, Moore TV, Rehli M, Nishimura MI, Mackensen A, Fischer K. Characterization of MHC class-I restricted TCRalphabeta+ CD4- CD8- double negative T cells recognizing the gp100 antigen from a melanoma patient after gp100 vaccination. Cancer Immunol Immunother 2009;58(5):709–18 doi 10.1007/s00262-008-0593-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young KJ, Kay LS, Phillips MJ, Zhang L. Antitumor activity mediated by double-negative T cells. Cancer research 2003;63(22):8014–21. [PubMed] [Google Scholar]
- 24.Daley D, Mani VR, Mohan N, Akkad N, Ochi A, Heindel DW, et al. Dectin 1 activation on macrophages by galectin 9 promotes pancreatic carcinoma and peritumoral immune tolerance. Nat Med 2017;23(5):556–67 doi 10.1038/nm.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daley D, Mani VR, Mohan N, Akkad N, Pandian G, Savadkar S, et al. NLRP3 signaling drives macrophage-induced adaptive immune suppression in pancreatic carcinoma. J Exp Med 2017;214(6):1711–24 doi 10.1084/jem.20161707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerriero JL. Macrophages: The Road Less Traveled, Changing Anticancer Therapy. Trends Mol Med 2018. doi 10.1016/j.molmed.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsujikawa T, Kumar S, Borkar RN, Azimi V, Thibault G, Chang YH, et al. Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell Rep 2017;19(1):203–17 doi 10.1016/j.celrep.2017.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 2012;9(7):676–82 doi 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 2006;7(10):R100 doi 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corbett AJ, Eckle SBG, Birkinshaw RW, Liu L, Patel O, Mahony J, et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 2014;509:361 doi 10.1038/nature13160 [DOI] [PubMed] [Google Scholar]
- 31.Wang W, Marinis JM, Beal AM, Savadkar S, Wu Y, Khan M, et al. RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer. Cancer Cell 2018;34(5):757–74 e7 doi 10.1016/j.ccell.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rehman A, Hemmert KC, Ochi A, Jamal M, Henning JR, Barilla R, et al. Role of fatty-acid synthesis in dendritic cell generation and function. J Immunol 2013;190(9):4640–9 doi 10.4049/jimmunol.1202312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ibrahim J, Nguyen AH, Rehman A, Ochi A, Jamal M, Graffeo CS, et al. Dendritic cell populations with different concentrations of lipid regulate tolerance and immunity in mouse and human liver. Gastroenterology 2012;143(4):1061–72 doi 10.1053/j.gastro.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sunde PT, Olsen I, Gobel UB, Theegarten D, Winter S, Debelian GJ, et al. Fluorescence in situ hybridization (FISH) for direct visualization of bacteria in periapical lesions of asymptomatic root-filled teeth. Microbiology 2003;149(Pt 5):1095–102 doi 10.1099/mic.0.26077-0. [DOI] [PubMed] [Google Scholar]
- 35.Thimm T, Tebbe CC. Protocol for rapid fluorescence in situ hybridization of bacteria in cryosections of microarthropods. Applied and environmental microbiology 2003;69(5):2875–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi YS, Kim YC, Baek KJ, Choi Y. In Situ Detection of Bacteria within Paraffin-embedded Tissues Using a Digoxin-labeled DNA Probe Targeting 16S rRNA. Journal of visualized experiments : JoVE 2015(99):e52836 doi 10.3791/52836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36(5):411–20 doi 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenkins RW, Aref AR, Lizotte PH, Ivanova E, Stinson S, Zhou CW, et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discov 2017. doi 10.1158/2159-8290.CD-17-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.