Abstract
Many in silico predictors of genetic variant pathogenicity have been previously developed, but there is currently no standard application of these algorithms for variant assessment. Using 4,094 ClinVar-curated missense variants in clinically actionable genes, we evaluated the accuracy and yield of benign and deleterious evidence in 5 in silico meta-predictors, as well as agreement of SIFT and PolyPhen2, and report the derived thresholds for the best performing predictor(s). REVEL and BayesDel outperformed all other meta-predictors (CADD, MetaSVM, Eigen), with higher positive predictive value, comparable negative predictive value, higher yield, and greater overall prediction performance. Agreement of SIFT and PolyPhen2 resulted in slightly higher yield but lower overall prediction performance than REVEL or BayesDel. Our results support the use of gene-level rather than generalized thresholds, when gene-level thresholds can be estimated. Our results also support the use of 2-sided thresholds, which allow for uncertainty, rather than a single, binary cut-point for assigning benign and deleterious evidence. The gene-level 2-sided thresholds we derived for REVEL or BayesDel can be used to assess in silico evidence for missense variants in accordance with current classification guidelines.
Subject terms: Statistical methods, Genetic testing
Introduction
In silico prediction of variant pathogenicity is one of the eight evidence categories recommended by the American College of Medical Genetics and American College of Pathologist (ACMG/AMP) guidelines1. Many in silico algorithms have been developed to predict the degree of sequence conservation and functional impact of missense variants, each of which generates a score based on tolerance to variation. Assessment of in silico evidence based on the observed concordance of multiple scores yields a high rate of discordant predictions, even among well-classified variants, suggesting that such an approach is not ideal2,3. Recently, several in silico meta-predictors have been developed from analyses of multiple individual scores, and have demonstrated superior performance to that of individual predictors, although robust, systematic comparison of these approaches is lacking4–6.
Furthermore, in silico meta-predictors have primarily been trained on large numbers of variants selected from genome-wide data. Yet, variants from same gene share common features and gene-level transcription facilitates the events that drive the development and progression of disease, as well as response to therapy7. Global applications of in silico scores may result in a large number of false predictions as a one-size-fits-all approach8,9, but gene-level thresholds for assessing benign and deleterious evidence are unavailable, in practice. In addition, while many algorithms recommend binary thresholds for assessing in silico evidence, clinical recommendations based on thresholds for “likely benign” and “likely pathogenic” are 2-sided10.
Given the wide variety of in silico algorithms and approaches available, there is currently no standard application of these tools for variant assessment; many laboratories use different tools and thresholds for evidence of pathogenic or benign classification11,12. As such, the 2015 ACMG/AMP guidelines assign only a supporting level of evidence to in silico predictions1. In this study, we aimed to assess in silico evidence for pathogenicity of missense variants using gene-level and generalized (all genes combined) 2-sided thresholds, compare prediction performance among 5 commonly used meta-predictors, and identify the thresholds corresponding to the meta-predictor(s) with the best overall prediction performance that can be used to assess in silico evidence in accordance with the ACMG/AMP guidelines.
Methods
Data
To evaluate the prediction performance of various in silico scores, we compiled a dataset of 4,094 classified missense variants in 66 clinically relevant genes from ClinVar. Variants were included if reviewed by expert panel or classified by any of 6 submitters that consistently provide assertion criteria: Ambry Genetics, Emory Genetics Laboratory, GeneDx, InSiGHT, InVitae and Sharing Clinical Reports Project. For each variant, we defined its consensus class as the most supported category among benign/likely benign (B/LB), pathogenic/likely pathogenic (P/LP) and variants of uncertain significance (VUS), i.e. pathogenic/likely pathogenic if NP/LP ≥ max(NVUS, 1) and NB/LB = 0 or benign/likely benign if NB/LB ≥ max(NVUS, 1) and NP/LP = 0. Variants with conflicting classes (i.e., NP/LP ≥ 1 and NB/LB ≥ 1) or classified as VUS by the majority of submitters (i.e., NVUS > max(NB/LB, NP/LP)) were excluded from analysis.
In silico scores were obtained for 5 meta-predictors (CADD4, MetaSVM5, Eigen13, REVEL14 and BayesDel6) and 2 individual predictors (SIFT15 and PolyPhen216) from the dbNSFP v3.5c database (August 6, 2017)17 and related websites (Supplementary Table S1). Datasets included classified variants in 66 genes (Supplementary Table S2) and a subset of variants in 20 genes, each containing at least 10 B/LB and 10 P/LP variants. Approximately 5.1%, 0.6%, 0.1%, and 2.1% of SIFT, PolyPhen2, MetaSVM, and Eigen values, respectively, were missing across the 4,094 variants. CADD, REVEL, and BayesDel had no missing values (Supplementary Table S3).
Gene-level and generalized thresholds
To derive gene-level thresholds for the 20 genes with ≥10 B/LB and ≥10 P/LP variants, we fit models using Firth logistic regression18 for variants within each gene. Firth logistic regression was used to ensure model robustness under the scenarios of separation or sparse benign/likely benign and pathogenic/likely pathogenic outcomes19,20. Gene-level 2-sided thresholds for in silico evidence were derived at the predicted probabilities 0.2 and 0.8, respectively. These predicted probabilities were projected from the regression model, depend in part on the proportion of pathogenic variants in the dataset constructed, and were not based on estimation in any patient population; the probabilities were chosen to achieve ≥90% overall predictive accuracy for most meta-predictors. Thus, in silico data was assessed for benign evidence, deleterious evidence or no evidence using gene-level thresholds for the 20 genes. For comparison of gene-level thresholds to those estimated with a broader approach aggregating variation across all genes, we further estimated generalized thresholds for the 20 genes using Firth logistic regression and predicted probabilities, as outlined above. Moreover, there were 46 genes for which gene-level thresholds were not estimable, primarily due to lack of sufficient numbers of B/LB or P/LP classified variants for analysis. As an exploratory analysis, we further derived generalized thresholds for the combined set of 66 genes, using the methods described above. For all analyses, variants with missing scores were assigned no evidence for the corresponding in silico predictor. To assess the robustness of the gene-level thresholds we derived for benign and deleterious evidence, we estimated 90% confidence intervals (CI) from 10,000 bootstrapping replicates stratified by classification status.
Cross-validation and sensitivity analysis
To account for potential overfitting, the predictive performance of each meta-predictor was evaluated using leave-one-out cross-validation, in which the assigned evidence was compared to the B/LB and P/LP status from ClinVar consensus classification. Performance statistics were evaluated using positive predictive value (PPV), negative predictive value (NPV) and yield rate (YR). YR was defined as the proportion of variants received benign or deleterious evidence among all evaluated variants. To assess the joint performance of accuracy and yield, we computed overall prediction performance (OPP), defined as the root mean square of PPV, NPV and YR, i.e., OPP = . Differences in PPV, NPV, or YR between predictors were each tested by Fisher’s exact test. Differences in OPP were tested by a Monte Carlo permutation test with 10,000 permutations that each randomly exchanged all assigned evidence among comparator predictors. Performance statistics for gene-level and generalized thresholds using 20 genes are shown in Table 1. For the meta-predictor demonstrating the highest OPP statistic, we report the optimized gene-level thresholds in Table 2. As an exploratory analysis, performance statistics for generalized thresholds using all 66 genes were also compared to those obtained using a combination of gene-level and generalized thresholds, and are provided in Supplementary Table S5.
Table 1.
Prediction performance of in silico evidence assignment (2,153 variants in 20 genes).
| Method | Performance statistic (Rank)a | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| TP | TN | FP | FN | NE | PPV | NPV | YR | OPP | |
| SIFT/PolyPhen2 agreement | 888 | 649 | 177 | 54 | 385 | 0.834 (11) | 0.923 (9) | 0.821 (1) | 0.861 (10) |
| Gene-level thresholds | |||||||||
| CADD | 746 | 707 | 60 | 38 | 602 | 0.926 (7) | 0.949 (7) | 0.720 (9) | 0.871 (8) |
| MetaSVM | 848 | 746 | 57 | 39 | 463 | 0.937 (5) | 0.950 (6) | 0.785 (4) | 0.894 (4) |
| Eigen | 850 | 761 | 51 | 27 | 464 | 0.943 (3) | 0.966 (2) | 0.784 (5) | 0.901 (3) |
| REVEL | 858 | 784 | 40 | 33 | 438 | 0.955 (2) | 0.960 (3) | 0.797 (3) | 0.907 (2) |
| BayesDel | 859 | 798 | 39 | 40 | 417 | 0.957 (1) | 0.952 (5) | 0.806 (2) | 0.908 (1) |
| Generalized thresholds | |||||||||
| CADD | 563 | 525 | 75 | 18 | 972 | 0.882 (10) | 0.967 (1) | 0.549 (11) | 0.819 (11) |
| MetaSVM | 848 | 697 | 75 | 64 | 469 | 0.919 (8) | 0.916 (11) | 0.782 (6) | 0.875 (6) |
| Eigen | 747 | 684 | 80 | 31 | 611 | 0.903 (9) | 0.957 (4) | 0.716 (10) | 0.865 (9) |
| REVEL | 846 | 673 | 52 | 60 | 522 | 0.942 (4) | 0.918 (10) | 0.758 (7) | 0.876 (5) |
| BayesDel | 825 | 672 | 58 | 52 | 546 | 0.934 (6) | 0.928 (8) | 0.746 (8) | 0.874 (7) |
aAll performance statistics, except those for SIFT/PolyPhen2 agreement, were evaluated by leave-one-out cross-validation. Ranks in parentheses were the descending orders of performance statistics among comparison methods. The p-values of Monte Carlo permutation tests for differences of OPP statistics between evidence of gene-level versus generalized thresholds were 0.0005, 0.09, 0.002, 0.006 and 0.003 for CADD (OPP: 0.871 vs. 0.819), MetaSVM (OPP: 0.894 vs. 0.875), Eigen (OPP: 0.901 vs. 0.865), REVEL (OPP: 0.907 vs. 0.876) and BayesDel (OPP: 0.908 vs. 0.874), respectively. TN, true negative; FN, false negative; TP, true positive; FP, false positive; NE, no evidence; PPV, positive predictive value; NPV, negative predictive value; YR, yield rate; OPP, overall prediction performance.
Table 2.
Gene-level thresholds for assigning benign and deleterious in silico evidence in missense variantsa.
| Gene | Thresholds of REVEL scores | Thresholds of BayesDel scores | ||
|---|---|---|---|---|
| TBE | TDE | TBE | TDE | |
| ATM | 0.359 | 0.689 | −0.180 | 0.216 |
| ATP7B | 0.514 | 0.731 | −0.076 | 0.248 |
| BRCA1 | 0.628 | 0.824 | 0.147 | 0.425 |
| BRCA2 | 0.581 | 0.974 | 0.080 | 0.500 |
| CFTR | 0.438 | 0.727 | −0.032 | 0.277 |
| COL3A1 | 0.515 | 0.762 | 0.026 | 0.329 |
| FBN1 | 0.326 | 0.597 | −0.328 | 0.047 |
| KCNH2 | 0.417 | 0.649 | −0.176 | 0.127 |
| MLH1 | 0.109 | 0.815 | 0.107 | 0.423 |
| MSH2 | 0.562 | 0.862 | 0.085 | 0.426 |
| MSH6 | 0.556 | 0.881 | 0.095 | 0.419 |
| MUTYH | 0.214 | 0.661 | −0.078 | 0.263 |
| MYBPC3 | 0.013 | 0.511 | −0.531 | 0.012 |
| NF1 | 0.261 | 0.605 | −0.191 | 0.077 |
| NSD1 | 0.400 | 0.705 | −0.082 | 0.268 |
| RET | 0.481 | 0.732 | −0.122 | 0.300 |
| RYR2 | 0.349 | 0.597 | −0.233 | 0.038 |
| SCN5A | 0.425 | 0.704 | −0.108 | 0.180 |
| TP53 | 0.536 | 0.667 | −0.003 | 0.132 |
| TSC2 | 0.703 | 0.970 | 0.244 | 0.561 |
aThe 2-sided thresholds, denoted as TBE and TDE, are the lower and upper limits of REVEL or BayesDel scores for assigning BE and DE, respectively. Gene-level thresholds for BE and DE were estimated at probabilities of pathogenicity 0.2 and 0.8, respectively. BE, benign evidence; DE, deleterious evidence.
In an effort to minimize the overlap of variants included in this analysis and those used to train any of the 5 meta-predictor models, we also performed a sensitivity analysis in 20 genes to assess meta-predictor performance among only those variants evaluated by all submitters in ClinVar during the period from September 2015 to August 2017.
Alternative methods
For comparison, we evaluated the performance of in silico prediction using generalized thresholds from a combination of 20 genes and that of SIFT/PolyPhen2 agreement (Table 1). Evidence for SIFT/PolyPhen2 agreement was assessed as deleterious if SIFT < 0.05 and PolyPhen2 = “possibly/probably damaging”, or benign if SIFT ≥ 0.05 and PolyPhen2 = “benign”. All analyses were conducted with R for Statistical Computing version 3.3.3.
Results
Evidence assignment using gene-level and generalized thresholds
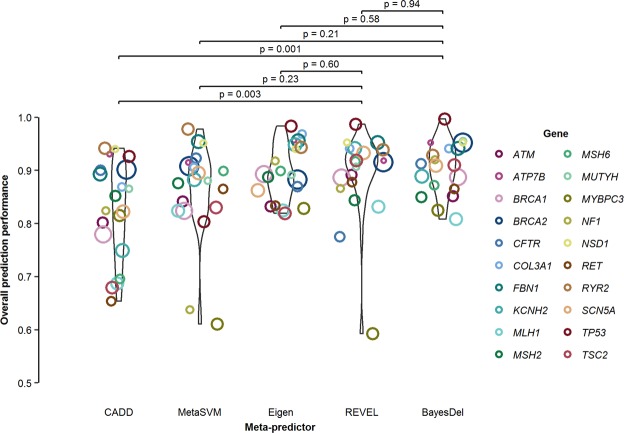
Using gene-level thresholds, REVEL and BayesDel achieved the highest OPP, 0.907 and 0.908, respectively (Table 1). Compared to CADD, MetaSVM, and Eigen, predictions using REVEL had equivalent NPV (−0.6% to 1.1% relative change, all p > 0.33) and improved PPV (1.3% to 3.2% higher, all p < 0.28), YR (1.5% to 10.6% higher, all p < 0.37) and OPP (0.6% to 4.1% higher, all p < 0.60) (Table 1; Fig. 1). Like REVEL, BayesDel outperformed CADD, MetaSVM, and Eigen. REVEL and BayesDel had nearly equivalent PPV, NPV, YR and OPP (all p > 0.44 for differences between the two). For each of 5 meta-predictors in 20 genes, evidence assignment using gene-level thresholds consistently outperformed that of generalized thresholds (2.2% to 5.9% higher OPP, all p < 0.09) (Table 1). While REVEL and BayesDel were concordant for 86.4% of variants and similarly outperformed the other meta-predictors, they did not agree on assigned evidence for 13.6% of variants; 6.6% were better predicted with REVEL and 7.0% with BayesDel (Supplementary Table S4).
Figure 1.
Assessment of in silico evidence in missense variants. The OPP statistics were reported in each of the 20 genes using gene-level thresholds. The OPP in 20 genes combined were 0.871, 0.894, 0.901, 0.907 and 0.908 for CADD, MetaSVM, Eigen, REVEL and BayesDel, respectively. P-values for pairwise comparisons were each estimated from Monte Carlo permutation test with 10,000 permutations. OPP, overall prediction performance.
Similar trends in prediction performance were observed for thresholds estimated across 66 genes compared to the 20-gene analysis. Using a combination of gene-level and generalized thresholds, REVEL and BayesDel achieved higher OPP than those of CADD, MetaSVM and Eigen (0.8% to 9.7% higher, all p < 0.32) (Supplementary Table S5). REVEL and BayesDel were discordant on assigned evidence for 14.5% of variants, roughly half of which were better predicted with REVEL and half with BayesDel (Supplementary Table S6).
Evidence assignment by alternative methods
Our 20-gene analysis also demonstrated that evidence assigned using REVEL or BayesDel gene-level and generalized thresholds each had consistently better prediction performance than those derived using SIFT/PolyPhen2 agreement (Table 1). Evidence assigned using REVEL vs. SIFT/PolyPhen2 agreement was concordant for 1,463 (68.0%) variants (Supplementary Table S7). Among the 690 variants for which evidence assignment was discordant, use of thresholds based on SIFT/PolyPhen2 agreement resulted in 27.5% false positives/negatives, in contrast to REVEL’s 4.6% false positives/negatives. Proportions of true and false predictions similarly favored BayesDel over SIFT/PolyPhen2 agreement (Supplementary Table S8). Using REVEL or BayesDel thresholds achieved 4.9% or 5.6% more correct predictions than SIFT/PolyPhen2, respectively (Supplementary Tables S7 and S8). Comparisons of REVEL or BayesDel with SIFT/PolyPhen2 agreement for all 66 genes yielded similar results (Supplemental Tables S9 and S10). Using REVEL or BayesDel thresholds yielded 12.4% or 11.7% more correct predictions than SIFT/PolyPhen2, respectively.
Sensitivity analysis
To assess the impact of classified variants that may overlap between meta-predictor training datasets and our evaluation dataset, we validated the prediction performance of 5 meta-predictors in a subset of 870 missense variants in 20 genes that were unlikely to have been previously used in the training datasets of meta-predictors. Similar to the results observed in the full dataset, REVEL and BayesDel had nearly identical performance statistics, and both had higher OPP than CADD, MetaSVM and Eigen (0.8% to 5.6% higher, all p < 0.68) (Supplementary Table S11).
Thresholds for assigning in silico evidence
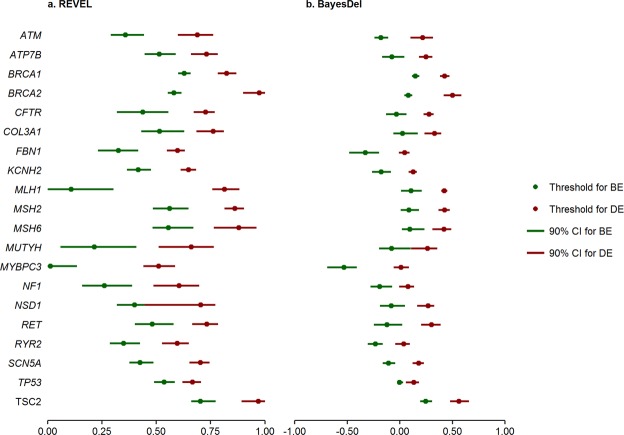
Gene-level 2-sided thresholds of REVEL and BayesDel scores for assessment of benign and deleterious in silico evidence in missense variants are provided in Table 2. A single, binary cut-point of 0.5 and 0 is often used for REVEL and BayesDel, respectively, to distinguish pathogenic evidence from benign2. However, our derived thresholds for benign evidence were above these cut-points for several genes (Fig. 2). For both REVEL and BayesDel, there was substantial variation among thresholds across genes, highlighting the importance of using gene-level thresholds whenever possible. In addition, the 90% CI for gene-level benign and deleterious thresholds were non-overlapping in all except one gene (NSD1, REVEL-based thresholds only), supporting the robustness of gene-level thresholds (Supplementary Table S12; Fig. 2).
Figure 2.
Variation in thresholds for assigning benign and deleterious in silico evidence across 20 genes. (a) Gene-level 2-sided thresholds and their 90% confidence intervals (CI) for REVEL. (b) Gene-level 2-sided thresholds and their 90% confidence intervals (CI) for BayesDel. Thresholds for BE and DE were represented by green and red dots, respectively. BE, benign evidence; DE, deleterious evidence.
While these findings support the use of gene-level thresholds for in silico evidence, many clinically actionable genes currently lack a sufficient number of classified variants for gene-level analysis. Given the NPV, PPV, and OPP for generalized thresholds are high (all >90%; Supplementary Table S5), and their prediction performance is superior to that of SIFT/PolyPhen2 agreement, generalized thresholds derived from the full set of 66 genes and their 90% CI are also presented (Supplementary Table S12). For REVEL, the 90% CI estimated for generalized thresholds for benign and deleterious evidence overlap with those estimated at the gene-level for 7/20 and 8/20 genes, respectively. For BayesDel, overlapping 90% CI for generalized and gene-level thresholds indicating benign or deleterious evidence were observed for 9/20 and 7/20 genes, respectively. We note that for REVEL, the generalized threshold’s 90% CI estimated for benign evidence overlaps with that estimated for gene-level pathogenic evidence for 2 genes (MYBPC3 and NSD1); likewise, the 90% CI around the generalized threshold estimated for pathogenic evidence overlapped with that estimated for gene-level benign evidence for 1 gene (TSC2). No overlap between the generalized threshold’s 90% CI for benign evidence and CIs estimated for gene-level pathogenic evidence was observed for any genes using BayesDel, however, the 90% CI around the generalized threshold for pathogenic evidence overlapped with that for gene-level benign evidence for TSC2.
Discussion
In this study, we evaluated and compared the performance of 5 well known meta-predictors often used for in silico assessment in variant classification. Our results were consistent with studies that found meta-predictors better assessed variant pathogenicity than concordance of individual predictors2,4–6. Our findings suggest that REVEL and BayesDel outperform the other 3 meta-predictors (Fig. 1). We also observed superior performance of gene-level thresholds compared to generalized thresholds. When evaluated on a gene-by-gene basis, REVEL and BayesDel achieved the highest OPP. In addition, sensitivity analysis in a subset of missense variants unlikely to have been included in the training datasets of the meta-predictors yielded similar results.
The strengths of our approach include generation of gene-level thresholds for variant assessment in 20 clinically actionable genes, and comparison to generalized thresholds estimated from a broader set of 66 such genes. Both gene-level and generalized thresholds yielded NPV, PPV and overall performance statistics ≥90%, and whether estimated from REVEL or BayesDel, both outperformed SIFT/PolyPhen2 agreement. However, our results also demonstrated improved prediction performance when assigning in silico evidence using gene-level thresholds compared to generalized thresholds, which highlights the importance of gene-specific assessment of variant pathogenicity21. We further observed wide variation in gene-level thresholds, and that the 90% CI around the generalized thresholds overlapped with those of the gene-level thresholds for <50% of the 20 genes examined in this study. Taken together, our findings support the use of gene-level thresholds for clinical variant assessment, when available.
An additional strength of our approach is the use of 2-sided thresholds for evidence assignment, as opposed to the current binary thresholds recommended for meta-predictors such as REVEL and BayesDel2. The use of 2-sided thresholds for evidence assignment is a data-driven solution to the problem of uncertainty; a single threshold necessarily assigns evidence to all variants but at the cost of high false positive or false negative rates. However, we acknowledge the trade-off between accuracy and yield, as thresholds quantified from Firth logistic regression models may be overly conservative in protecting against false predictions at the expense of the proportion of variants for which benign or pathogenic evidence can be assigned. Future methods development may include non-linear models or a weighted mechanism for adaptive balance between accuracy and yield designed to improve the overall performance of evidence assignment.
Our findings are based on the use of ClinVar-curated variants, a rapidly growing database with confidently annotated disease-causing alterations, as a training set to calibrate in silico scores on a gene-by-gene basis. In view of potential circularity issues arising from the testing of variants available in ClinVar that may have been previously used to train the meta-predictors we analyzed in the present study, we performed a sensitivity analysis to assess the impact of these variants. However, we were precluded from fully accounting for all such variants due to the fact that some of the variants included in our sensitivity analysis may have been deposited in ClinVar prior to 2015 and/or included in HGMD or ExAC at the time the models were developed. Nonetheless, our sensitivity analysis results were consistent with those of the larger analysis based on the full set of variants.
Conclusions
Assigning in silico evidence using gene-level 2-sided thresholds from REVEL or BayesDel scores achieved higher predictive accuracy and yield compared to the use of other in silico predictors. The REVEL and BayesDel thresholds we report can serve as a viable resource for assigning in silico evidence to missense variants in the genes examined herein, to be used in conjunction with other lines of evidence in variant assessment as recommended in ACMG/AMP guidelines.
Supplementary information
Acknowledgements
This study makes use of data from ClinVar and dbNSFP databases. A list of contributing groups of ClinVar and dbNSFP data can be found at https://www.ncbi.nlm.nih.gov/clinvar/ and https://sites.google.com/site/jpopgen/dbNSFP, respectively.
Author Contributions
D.Q. and T.P. designed the study. Y.T., S.L. and A.C. collected data. Y.T. and D.Q. evaluated data. Y.T., D.Q. and M.H.B. wrote the manuscript. All authors discussed the results and contributed to the final manuscript.
Data Availability
All the data necessary to produce the results of this article is included in Supplementary data.
Competing Interests
All of the authors are employed by and receive a salary from Ambry Genetics.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-49224-8.
References
- 1.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghosh R, Oak N, Plon SE. Evaluation of in silico algorithms for use with ACMG/AMP clinical variant interpretation guidelines. Genome Biol. 2017;18:225. doi: 10.1186/s13059-017-1353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mesbah-Uddin M, Elango R, Banaganapalli B, Shaik NA, Al-Abbasi FA. In-silico analysis of inflammatory bowel disease (IBD) GWAS loci to novel connections. PLoS One. 2015;10:e0119420. doi: 10.1371/journal.pone.0119420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong C, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng BJ. PERCH: A unified framework for disease gene prioritization. Hum. Mutat. 2017;38:243–251. doi: 10.1002/humu.23158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray D, Doran P, MacMathuna P, Moss AC. In silico gene expression analysis–an overview. Mol. Cancer. 2007;6:50. doi: 10.1186/1476-4598-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crockett DK, et al. Utility of gene-specific algorithms for predicting pathogenicity of uncertain gene variants. J. Am. Med. Inform. Assoc. 2012;19:207–211. doi: 10.1136/amiajnl-2011-000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, et al. Gene-specific function prediction for non-synonymous mutations in monogenic diabetes genes. PLoS One. 2014;9:e104452. doi: 10.1371/journal.pone.0104452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson BA, et al. Calibration of multiple in silico tools for predicting pathogenicity of mismatch repair gene missense substitutions. Hum. Mutat. 2013;34:255–265. doi: 10.1002/humu.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bean LJH, Hegde MR. Clinical implications and considerations for evaluation of in silico algorithms for use with ACMG/AMP clinical variant interpretation guidelines. Genome Med. 2017;9:111. doi: 10.1186/s13073-017-0508-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amendola LM, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am. J. Hum. Genet. 2016;98:1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ionita-Laza I, McCallum K, Xu B, Buxbaum JD. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 2016;48:214–220. doi: 10.1038/ng.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ioannidis NM, et al. REVEL: An ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 16.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: A one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum. Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80:27–38. doi: 10.1093/biomet/80.1.27. [DOI] [Google Scholar]
- 19.Heinze G, Schemper M. A solution to the problem of separation in logistic regression. Stat. Med. 2002;21:2409–2419. doi: 10.1002/sim.1047. [DOI] [PubMed] [Google Scholar]
- 20.Wang, X. Firth logistic regression for rare variant association tests. Frontiers in Genetics5 (2014). [DOI] [PMC free article] [PubMed]
- 21.Wang M, Wei L. iFish: predicting the pathogenicity of human nonsynonymous variants using gene-specific/family-specific attributes and classifiers. Sci Rep. 2016;6:313–321. doi: 10.1038/srep31321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data necessary to produce the results of this article is included in Supplementary data.