Abstract
Kinesin family member C1 (KIFC1) is implicated in the clustering of multiple centrosomes to maintain tumor survival and is thought to be an oncogene in several kinds of cancers. In our experiments, we first performed bioinformatics analysis to investigate the expression levels of KIFC1 in bladder cancer (BC) specimens and normal bladder epitheliums and then, using our samples, verified findings by quantitative real‐time PCR and western blotting assays. All data showed that KIFC1 was significantly upregulated in BC specimens at both the mRNA and protein levels. Immunohistochemical studies in a cohort of 152 paraffin‐embedded BC tissues displayed that upregulated expression of KIFC1 clearly correlated with pT status (P = .014) and recurrent status (P = .002). Kaplan‐Meier survival analysis and log‐rank test indicated that patients with BC with high KIFC1 expression had both shorter cancer‐specific survival (P < .001) and recurrence‐free survival time (P < .001) than those with low KIFC1 expression. Furthermore, ectopic downregulation of KIFC1 weakened BC cell proliferation and migration both in vitro and in vivo, whereas upregulation of KIFC1 enhanced this in vitro. Overexpression of KIFC1 phosphorylated GSK3β and promoted Snail through activating AKT (protein kinase B0) to induce proliferation and epithelial–mesenchymal transition (EMT) and, therefore, substantially promoted BC migration and metastasis. Our study revealed an oncogenic role for KIFC1 to promote BC cell proliferation and EMT via Akt/GSK3β signaling; KIFC1 might be a promising prognostic biomarker as well as a therapeutic target for BC.
Keywords: AKT/GSK3β, bladder cancer, epithelial–mesenchymal transition, KIFC1, proliferation
1. INTRODUCTION
With its high rate of morbidity and mortality, bladder cancer (BC) caused c. 549 000 new cases and 200 000 reported deaths worldwide in 2018.1 Moreover, BC was the fourth most common malignancy in American males and the sixth most common in Chinese males.2, 3 Urothelial carcinoma of the bladder, which is the most common histopathologic type of BC, can be categorized into two types. While nonmuscle‐invasive BC (NMIBC) has a high tendency to relapse and progress into muscle‐invasive BC (MIBC), 25%‐80% of MIBC will eventually develop distant metastases, resulting in a 5‐y survival rate of <50%, despite surgery and adjuvant therapies including radiotherapy and chemotherapy.4, 5, 6, 7 Therefore, there is an urgent need to determine the underlying molecular mechanisms in the tumorigenesis and/or metastasis of BC and to develop some new targeting drugs.
Kinesin family member C1 (KIFC1), also known as HSET, is a minus end‐directed motor protein that is related to microtubule transport, centrosome clustering, and spindle formation during mitosis.8, 9, 10, 11 KIFC1 is located on chromosome 6p21.32 and belongs to the kinesin‐14 family of motor proteins that also includes KIFC2 and KIFC3.12 KIFC1 has been previously reported to participate in several biological functions including vesicular and organelle trafficking,8 oocyte development,13 spermiogenesis,14 and double‐stranded DNA transportation.15 Rath et al16, 17, 18 reported that, although KIFC1 was nonessential in normal somatic cells, it was indispensable for tumor cells to prevent aneuploidy and cell death via its centrosome clustering mechanism. Therefore, KIFC1 was considered to be a promising chemotherapy target that led to the development of several inhibitors such as AZ82 and CW069.12 To date, KIFC1 has been reported to be overexpressed in various cancers. Xiao et al19 suggested that KIFC1 facilitated the proper cell division of a human seminoma. Other groups have reported that KIFC1 promoted hepatocellular carcinoma epithelial–mesenchymal transition (EMT) and metastasis through gankyrin/AKT signaling.20 Similarly, KIFC1 was also correlated with metastasis in breast cancer, ovarian adenocarcinoma, and brain metastasis of non‐small‐cell lung cancer.21, 22, 23 Moreover, KIFC1 was able to induce drug resistance against taxane in prostate cancer,24 tamoxifen, and docetaxel in breast cancer.25, 26 Furthermore, upregulation of KIFC1 has been identified in several solid human cancers including esophageal squamous cell carcinoma, gastric cancer, and renal cell carcinoma.27, 28, 29 These data, together, suggested that KIFC1 plays a crucial oncogenic role in tumorigenesis.
However, to date, the biological roles and underling molecular mechanisms of KIFC1 in the pathogenesis of BC remain largely unknown. In the present study, we first investigated the expression pattern of KIFC1 in BC tissues and then demonstrated the relationship between KIFC1 expression and the clinicopathological features of patients with BC. Next, we upregulated or downregulated the expression of KIFC1 to determine its roles in proliferation, migration, and metastasis of BC cells. Furthermore, we explored the possible roles of KIFC1 in EMT, and also uncovered potential molecular mechanisms.
2. MATERIALS AND METHODS
2.1. Bioinformatics analysis of human publicly available datasets
mRNA‐seq data of BC patients from The Cancer Genome Atlas (TCGA)30 were downloaded from http://firebrowse.org/ (a Broad Institute TCGA genome data analysis center) at the 2016_01_28 run, and data from GSE13507 were obtained from Gene Expression Omnibus (GEO)31 datasets, which are found in the National Center for Biotechnology Information Search database (NCBI) (https://www.ncbi.nlm.nih.gov/).
2.2. Cell lines and cell culture
Human normal bladder cell SV‐HUC‐1 and BC cell lines (T24, 5637, TCCSUP, UMUC3, J82 and RT4) were acquired from the American Type Culture Collection (ATCC). BIU87 was obtained from the China Center for Type Culture Collection. All cells were cultured in Dulbecco's modified Eagle's medium (DMEM) or RPMI 1640 medium containing 10% fetal bovine serum (Gibco), 100 U/mL penicillin and 100 U/mL streptomycin and then were incubated in an humidified atmosphere with 5% CO2 in air at 37°C.
2.3. Patients and specimens
This study was approved by the ethics committee of the Sun Yat‐sen University Cancer Center (SYSUCC) and conformed to provisions of the Declaration of Helsinki. Written informed consent was acquired from each patient before the study. Paired fresh BC tumors and adjacent normal tissues, obtained from the SYSUCC Bio‐bank, were used for quantitative real‐time polymerase chain reaction (qPCR) and western blotting. Paraffin‐embedded BC tissues and normal bladder specimens were acquired from 152 BC patients who had undergone radical cystectomy between 2000 and 2013. Patients without a definite pathological diagnosis, without availability of resection sample, and without follow‐up data were excluded from the study. The median follow‐up time of these BC patients was 52.5 mo (range from 0.2 to 197.0 mo). Detailed clinicopathological characteristics of these BC patients are recorded in Table 1. Clinical stage of BC was defined based on the tumor‐nodes‐metastasis (TNM) system (AJCC/UICC 2007), while the histopathological grade of BC was assessed according to World Health Organization classification (2004 edition).
Table 1.
Relationship between KIFC1 expression and clinicopathological parameters in 152 BC cases
| Clinical parameters | KIFC1 expression | P‐valuea | ||
|---|---|---|---|---|
| All | Low | High | ||
| Total n (%) | 152 | 79 (52.0) | 73 (48.0) | |
| Age (y) | ||||
| ≤60 | 75 | 40 (53.3) | 35 (46.7) | .741 |
| >60 | 77 | 39 (50.6) | 38 (49.4) | |
| Gender | ||||
| Male | 132 | 68 (51.5) | 64 (48.5) | .771 |
| Female | 20 | 11 (55.0) | 9 (45.0) | |
| Tumor multiplicity | ||||
| Unifocal | 73 | 41 (56.2) | 32 (43.8) | .320 |
| Multifocal | 79 | 38 (48.1) | 41 (51.9) | |
| Tumor size (cm) | ||||
| ≤3 | 64 | 29 (45.3) | 35 (54.7) | .161 |
| >3 | 88 | 50 (56.8) | 38 (43.2) | |
| pT status | ||||
| T1‐T2 | 72 | 45 (62.5) | 27 (37.5) | .014 |
| T3‐T4 | 80 | 34 (42.5) | 46 (57.5) | |
| pN status | ||||
| pN− | 114 | 62 (54.4) | 52 (45.6) | .303 |
| pN+ | 38 | 17 (44.7) | 21 (55.3) | |
| Histological grade | ||||
| Low | 17 | 11 (64.7) | 6 (35.3) | .265 |
| High | 135 | 68 (50.4) | 67 (49.6) | |
| Recurrence | ||||
| No | 96 | 59 (61.5) | 37 (38.5) | .002 |
| Yes | 56 | 20 (35.7) | 36 (64.3) | |
Chi‐squared test.
Bold values represent P < .05.
2.4. RNA extraction, reverse transcription, and qPCR
Total RNA was extracted using TRIzol reagent (Invitrogen) in accordance with the manufacturer's instructions and then reverse‐transcribed into cDNA using a PrimeScript™ Master Mix (RR036A, TaKaRa). cDNA was then run on standard qPCR instruments. qPCR primer sequences used in our experiments are listed as follows: KIFC1 forward primer (F): 5′‐ACTACAGTGCCACAGACA‐3′; KIFC1 reverse primer (R): 5′‐CCTGATGTGCCAGACTTC‐3′; GAPDH F: 5′‐GGAGCGAGATCCCTCCAAAAT‐3′; GAPDH R; 5′‐GGCTGTTGTCATACTTCTCATGG‐3′; E‐cadherin F: 5′‐CGAGAGCTACACGTTCACGG‐3′; E‐cadherin R: 5′‐GGGTGTCGAGGGAAAAATAGG‐3′; α‐Catenin F: 5′‐GGGGATAAAATTGCGAAGGAGA‐3′; α‐Catenin R: 5′‐GTTGCCTCGCTTCACAGAAGA‐3′; β‐Catenin F: 5′‐AAAGCGGCTGTTAGTCACTGG‐3′; β‐Catenin R: 5′‐CGAGTCATTGCATACTGTCCAT‐3′; N‐cadherin F: 5′‐TCAGGCGTCTGTAGAGGCTT‐3′; N‐cadherin R: 5′‐ATGCACATCCTTCGATAAGACTG‐3′; Vimentin F: 5′‐GACGCCATCAACACCGAGTT‐3′; Vimentin R: 5′‐CTTTGTCGTTGGTTAGCTGGT‐3′; Fibronectin F: 5′‐CGGTGGCTGTCAGTCAAAG‐3′; Fibronectin R: 5′‐AAACCTCGGCTTCCTCCATAA‐3′; Snail F: 5′‐TCGGAAGCCTAACTACAGCGA‐3′; Snail R: 5′‐AGATGAGCATTGGCAGCGAG‐3′; Slug F: 5′‐CGAACTGGACACACATACAGTG‐3′; Slug R: 5′‐CTGAGGATCTCTGGTTGTGGT‐3′. Expression data were showed as 2−[(Ct of gene) − (Ct of GAPDH)] and then analyzed.
2.5. Immunohistochemistry (IHC) analysis
Immunohistochemistry studies were carried out to determine different KIFC1 protein expression patterns in our cohorts of 152 human BC specimens using a method similar to that reported previously.32 First, all paraffin‐embedded BC specimens were cut into 4‐μm sections and baked at 65°C for 1 h. Sections were deparaffinized with xylene (once for 15 min, three times), these specimens were then rehydrated in concentration gradient ethanol solutions (100%, 95%, 70% and 50%, each for 3 min). Then, EDTA antigenic retrieval buffer (pH 8.0) and high pressure were used to retrieve antigen. BC samples were then processed with 3% hydrogen peroxide for 10 min to extinguish any endogenous peroxidase. Next, 5% bovine serum albumin was used to block nonspecific sites. Subsequently, rabbit monoclonal anti‐KIFC1 antibody (ab172620, 1:250 dilution, Abcam) was prepared and tissue slides were incubated overnight at 4°C. PBS was used as the negative control. Slides were washed and processed with prediluted anti‐rabbit secondary antibody (Dako), then slides were immunostained using diaminobenzidine (Dako) and counterstained with 10% Mayer's hematoxylin.
Immunohistochemistry staining for KIFC1 was assessed using a semi‐quantitative scoring system by recording staining intensity as well as the proportion of positively stained cells. Specifically, the intensity of staining was scored as 0 (no staining), 1 (weak staining, light yellow), 2 (moderate staining, yellow brown) or 3 (strong staining, brown). The proportion of positively stained cells was graded as follows: 0 (0%), 1 (1%‐9%), 2 (10%‐29%), 3 (30%‐49%) and 4 (≥50%). Immunoreactivity score (IS) was separately evaluated by two independent pathologists who were blinded to the clinical parameters; IS was then recorded as the product of staining intensity and the proportion of positively stained cells. As the median score of these 152 BC tissues was 3, scores ≤ 3 were defined as low KIFC1 expression, while scores > 3 were indicated as high KIFC1 expression.
2.6. Protein extraction and western blotting analysis
Total protein from frozen tissues and cultured cells was harvested using RIPA buffer (Beyotime Technology) and adding proteinase inhibitor cocktail (Roche). The Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific) was used to quantify protein concentration; 30 μg protein was used for loading. The following primary antibodies were used: anti‐KIFC1 (ab172620, Abcam); anti‐GAPDH (ProteinTech Group Inc.); anti‐E‐cadherin, anti‐β‐catenin) and anti‐vimentin (Becton Dickinson Transduction Laboratories); anti‐Snail, anti‐AKT, anti‐phosphorylated Akt (pAkt), anti‐GSK3β and anti‐phosphorylated GSK3β (pGSK3β) (Cell Signaling Technology).
2.7. Construction of the recombinant lentiviral vector
Commercialized lentiviral vectors expressing KIFC1 short hairpin RNAs (shRNAs) were purchased from GeneCopoeia. Target sequences were as follows: sh1: 5′‐GCAACATCCGTGTATTCTGCC‐3′; sh2: 5′‐CCAGGGCTATCAAATAAAGAA‐3′. A KIFC1‐overexpressed (KIFC1‐OE) plasmid was constructed by our group using transcripts NM_002263.3 and NP_002254.2. Lipofectamine 2000 reagent (Thermo Fisher Scientific) was used to transfect the relevant BC cells in accordance with the manufacturer's instructions. Stable BC cell lines were picked following incubation with 0.5‐1.0 μg/mL puromycin for 7 d.
2.8. Cell counting kit‐8 (CCK‐8) assay
CCK‐8 reagent (Beyotime Technology) was adopted to measure cell viability according to the manufacturer's protocols. A density of 2000 cells per well was seeded onto 96‐well plates, the viabilities of which were recorded every 24 h up to day 5.
2.9. Colony formation assay
In total, 500 cells per well were inoculated onto 6‐well plates and then cultured for c. 2 wk. The generated colonies were then fixed with methanol, and stained with 0.1% crystal violet to calculate the number and size of colonies.
2.10. Cell wound‐healing assay
Bladder cancer cells were first seeded onto 6‐well plates and grown until confluent. Each well was then scratched with a sterile 100‐mL pipette tip to create three to five artificial homogeneous wounds, remaining cells were then cultured in a serum‐free medium. Cell migration was captured using an inverted microscope at 0 h and 24 h.
2.11. Transwell migration assay
Here, 5 × 104 BC cells were seeded in serum‐free medium in the upper chamber while 10% FBS medium was added to the lower chamber . Cells were incubated at 37°C for 24 h. Invading cells were fixed, stained and then counted under a microscope (Nikon Eclipse 80i).
2.12. Immunofluorescent staining assay
Bladder cancer cells were fixed, membranes were permeabilized with 0.5% Triton‐X‐100, and then cells incubated with mouse anti‐E‐cadherin and anti‐vimentin antibodies (1:200 dilution, Becton Dickinson Transduction Laboratories) overnight at 4°C. Cells were washed thoroughly and then incubated with Key Fluor goat anti‐mouse IgG (1:200 dilution, KeyGEN BioTECH) for 1 h and mounted with DAPI stain (Vector Laboratories).
2.13. Experimental metastasis model
Sixteen 4‐wk‐old BALB/c nude mice were divided equally into two groups, in which each mouse was injected intravenously through tail vein with 1 × 106 BC cells. After 8 wk, all mice were sacrificed to count the number of tumor nodules formed on the lung surfaces. In addition, lungs of mice were excised and embedded in paraffin for further examination. All procedures had been approved by the animal ethics committee of SYSUCC.
2.14. Statistical analysis
All functional experiments were carried out three times, results are represented as mean ± standard deviation (X ± SD). Student's two‐tailed t test and Wilcox test were, respectively, adopted to assess differences in continuous and discrete data. Correlations between KIFC1 expression and clinicopathological features were carried out using the chi‐squared test. Survival curves were built using the Kaplan‐Meier method and then analyzed using log‐rank test, while the Cox proportional hazards regression model was used for both univariate and multivariate survival analysis. SPSS 21.0 software (IBM Corp.) and GraphPad Prism 7.0 (GraphPad Software) were used for statistical assessments and plotting. A P‐value < .05 was considered as statistically significant.
3. RESULTS
3.1. KIFC1 was frequently upregulated in BC
To investigate the status of KIFC1 expression in BC, we first performed bioinformatics analysis on human BC datasets publicly available from TCGA and GEO. In both datasets, KIFC1 mRNA expression levels were markedly upregulated in BC tissues compared with normal bladder epitheliums (P < .001, Figure 1A,B). Analogously, in a cohort of 41 paired fresh BC tissues and normal adjacent epitheliums from our center, the KIFC1 mRNA expression level measured by qPCR was dramatically upregulated compared with the adjacent normal tissues (P < .001, Figure 1C). Furthermore, western blotting assay demonstrated that KIFC1 protein expression was significantly upregulated in 15 out of 17 human BC tissues compared with the paired normal tissues (P = .002, Figure 1D). These preliminary data demonstrated that KIFC1 was usually upregulated in BC.
Figure 1.
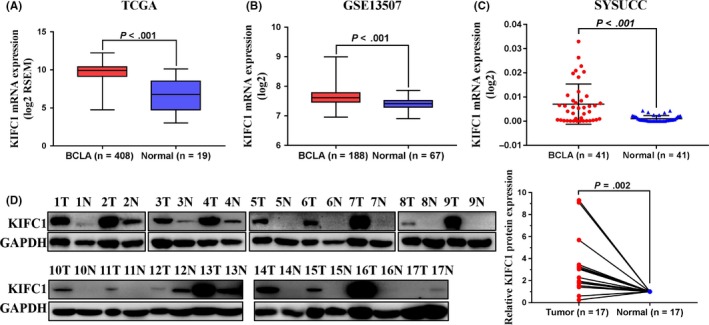
KIFC1 was upregulated in BC samples. A and B, KIFC1 mRNA expression levels of BC tissues in TCGA and GEO datasets (GSE13507). C, mRNA expression levels of KIFC1 were detected by qPCR in 41 pairs of BC tissues and the adjacent normal specimens from our center. D, Western blotting analyses suggested that KIFC1 was increased in 15 of 17 paired BC tissues. Expression of GAPDH was used as a loading control. BCLA, bladder cancer; Normal, normal bladder epithelium; SYSUCC, Sun Yat‐sen University Cancer Center
3.2. KIFC1 was closely associated with poorer prognosis in BC
To evaluate the potential clinical and prognostic values of KIFC1 expression in BC patients, we examined the KIFC1 protein expression level using IHC in a large cohort of 152 BC patients. Results showed that 73/152 (48.0%) of BC cases examined had overexpression of KIFC1 (Table 1), whereas KIFC1 expression in the non‐neoplastic bladder tissues was absent or at low levels. KIFC1 was mostly positioned in the nucleus of the BC tissues. KIFC1 staining of representative samples of BC and normal bladder tissues, and the distribution of KIFC1 staining intensity in 152 patients with BC are shown in Figure 2A,B. In correlation analysis between KIFC1 expression and clinicopathological parameters of BC patients, we found that high KIFC1 expression was apparently associated with tumor pT status (P = .014) and recurrence status (P = .002) (Table 1). However, no significant association was observed between KIFC1 expression and other clinical features such as age, gender, tumor multiplicity, tumor size, pN status, and histological grade.
Figure 2.
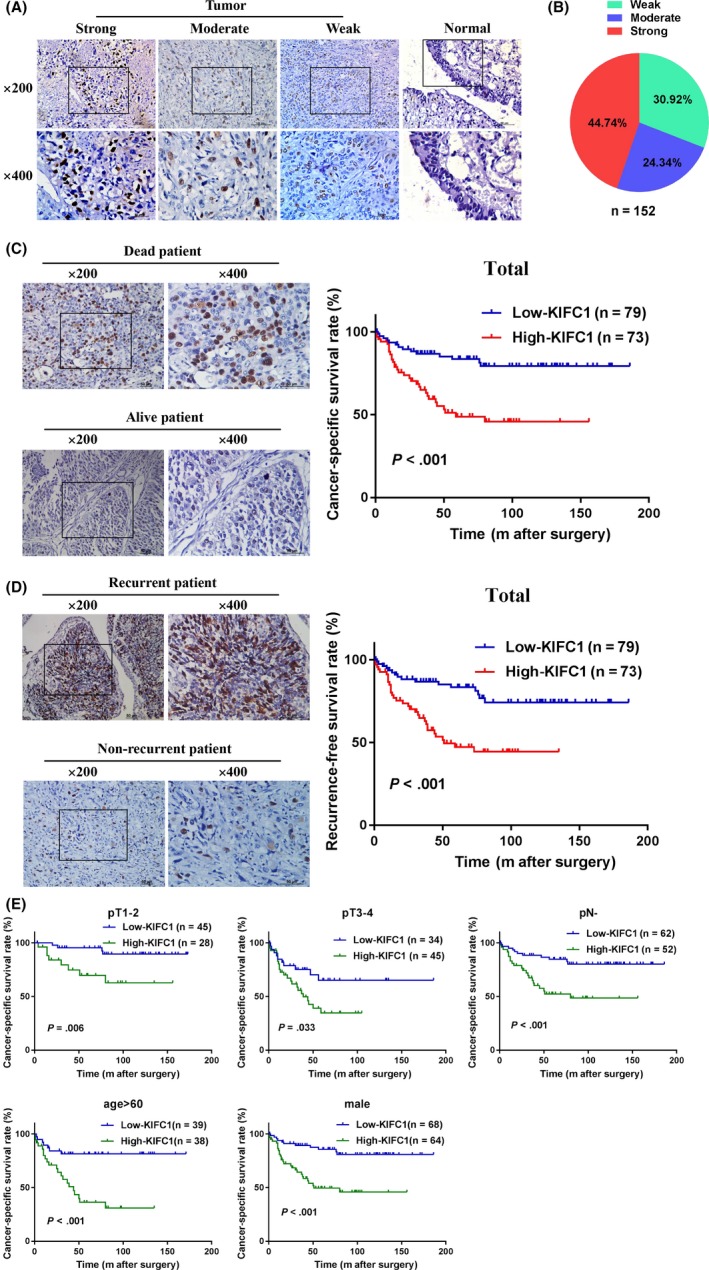
Upregulation of KIFC1 was closely related to poor prognosis of BC. A, Representative images of KIFC1 staining in paraffin‐embedded BC and normal tissues. Staining intensity in the tumor group was scored as strong, moderate, and weak. KIFC1 was mostly positioned in the nucleus of the bladder specimens. B, Distribution of KIFC1 staining intensity in 152 BC samples. C and D, Kaplan‐Meier cancer‐specific survival (CSS) and recurrence‐free survival (RFS) analysis in the entire cohort of BC patients. Representative immunohistochemical images were also shown. E, Kaplan‐Meier subgroups survival analysis revealed that BC patients with high KIFC1 expression had a lower CSS in different T stages, pN−, age > 60 and male
Moreover, Kaplan‐Meier survival analysis and log‐rank test determined that patients with BC and with high KIFC1 expression had shorter cancer‐specific survival times (CSS, P < .001) and less recurrence‐free survival (RFS, P < .001) than patients who had low KIFC1 expression (Figure 2C,D). Furthermore, subgroups of different T stages, pN−, age > 60 and male patient with BC revealed that patients with BC and with high KIFC1 expression had a lower CSS than patients with low KIFC1 expression (Figure 2E). Additionally, univariate analyses indicated that age (P = .047), pT status (P < .001) and KIFC1 expression (P < .001) were significantly associated with the CSS of patients with BC (Table 2). Further multivariate Cox regression analysis confirmed tumor pT status (hazards ratio: 2.96; confidence interval: 1.46‐5.99; P = .003) and KIFC1 expression (hazards ratio: 2.50; confidence interval: 1.33‐4.72; P = .005) as independent prognostic factors in BC patients (Table 2). These above results suggested that high KIFC1 expression was significantly correlated with poor prognosis in patients with BC.
Table 2.
Univariate and multivariate analysis of prognostic parameters in 152 BC patients
| Prognostic parameters | Median overall survival (mo) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | P‐value | HR (95% CI) | P‐value | ||
| Age (y) (>60 vs ≤60) | 40.0 vs 71.0 | 1.80 (1.01, 3.22) | .047 | 1.71 (0.92, 3.19) | .089 |
| Gender (male vs female) | 56.0 vs 34.5 | 0.95 (0.40, 2.23) | .903 | 0.93 (0.39, 2.24) | .876 |
| Tumor multiplicity (multifocal vs unifocal) | 56.0 vs 43.0 | 1.09 (0.62, 1.93) | .759 | 1.20 (0.66, 2.18) | .545 |
| Tumor size (cm) (>3 vs ≤3) | 57.5 vs 43.5 | 1.17 (0.65, 2.09) | .604 | 1.32 (0.72, 2.39) | .366 |
| pT status (T3‐T4 vs T1‐T2) | 33.0 vs 80.0 | 3.73 (1.96, 7.10) | <.001 | 2.96 (1.46, 5.99) | .003 |
| pN status (N+ vs N−) | 30.0 vs 68.5 | 1.47 (0.76, 2.85) | .256 | 1.03 (0.50, 2.15) | .929 |
| Histological grade (high vs low) | 51.0 vs 59.0 | 1.51 (0.54, 4.20) | .431 | 1.21 (0.42, 3.46) | .720 |
| KIFC1 expression (high vs low) | 33.0 vs 76.0 | 3.12 (1.71, 5.71) | <.001 | 2.50 (1.33, 4.72) | .005 |
Bold values represent P < .05.
3.3. KIFC1 promoted the proliferation and migration of BC cells in vitro
To explore the functional role of KIFC1 in BC cells, we first detected KIFC1 expression in normal bladder cell line SV‐HUC‐1 and seven BC cell lines using western blotting. KIFC1 was overexpressed in TCCSUP and BIU87 cells, and expressed less in SV‐HUC‐1 and T24 cells (Figure 3A). Therefore, SV‐HUC‐1 and T24 cells were selected to construct KIFC1 knockdown cells and T24 cells were selected to construct KIFC1‐OE knockdown cells. The efficiency of ectopic expression was evaluated through western blotting (Figure 3B). Next, cell proliferation assays, including CCK‐8 and colony formation assays, showed that upregulation of KIFC1 increased the viability of BC cells and distinctly generated more colonies compared with the control cells, whereas downregulation of KIFC1 had the opposite effect (Figure 3C,D). In addition, wound‐healing and transwell migration assays demonstrated that the migration rate was increased in KIFC1‐OE T24 cells and reduced in the KIFC1‐silenced cells TCCSUP and BIU87 (Figure 4A,B). These above data indicated that KIFC1 was required for cell proliferation and migration of BC cells.
Figure 3.
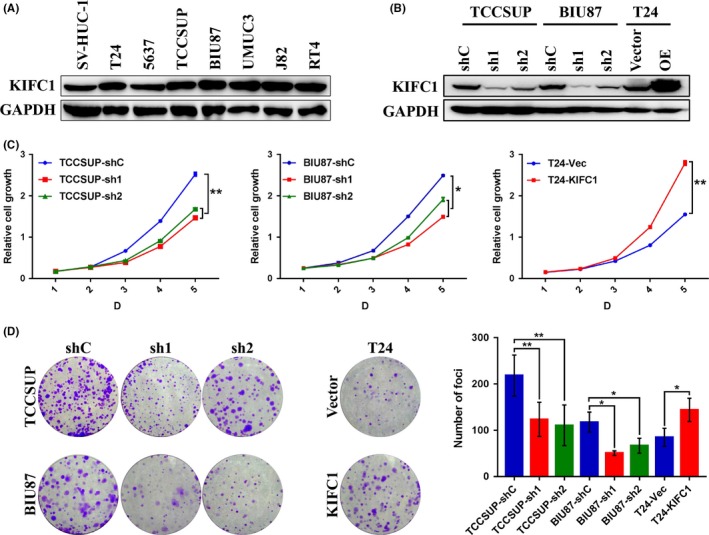
Overexpression of KIFC1 promoted the proliferation of BC cells in vitro. A, Expression levels of KIFC1 protein in normal bladder cell SV‐HUC‐1 and seven BC cell lines by western blotting. Glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) was adopted as the loading control. B, Western blotting revealed that KIFC1 was stably knocked down or overexpressed in corresponding BC cells. C and D, The proliferation ability of stably transfected BC cells were determined by CCK‐8 and colony forming assays. sh, short hairpin RNA; *P < .05; **P < .01
Figure 4.
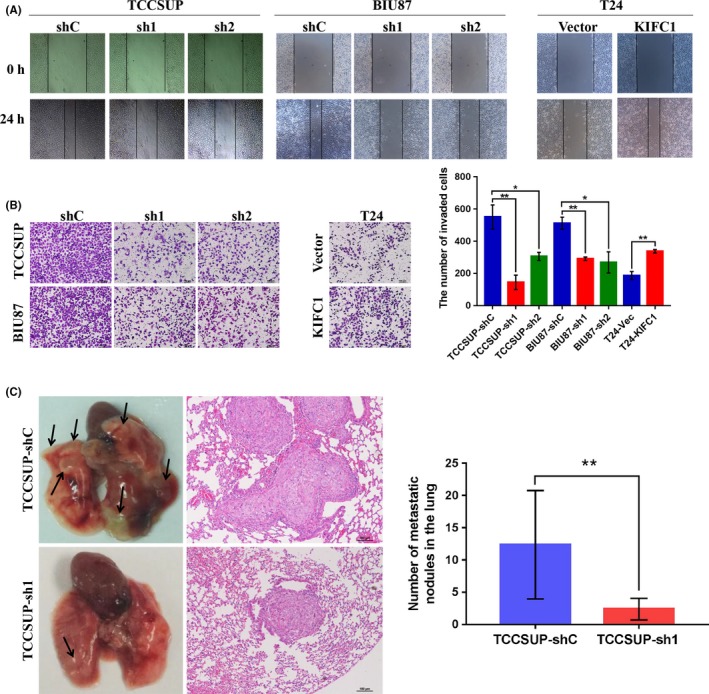
KIFC1 affected the aggressive capacity of BC cells both in vitro and vivo. A and B, Cell wound‐healing assay and transwell migration assay were carried out to measure the migration ability of BC cells with ectopic KIFC1 expression. C, Representative images of lungs derived from nude mice showing metastatic nodules (indicated by arrows) and hematoxylin and eosin (H&E) staining of lung metastatic nodules are shown. sh, short hairpin RNA; *P < .05; **P < .01
3.4. Downregulation of KIFC1 inhibits metastatic potential of BC cells in vivo
To determine if KIFC1 plays a role in BC metastasis in vivo, two groups of 4‐wk‐old BALB/c nude mice were injected intravenously with either TCCSUP‐shC cells or TCCSUP‐sh1 cells. At 8 wk after cell injection, all mice were sacrificed and metastatic lung nodules were examined. Results showed that the numbers of metastatic lung nodules in mice injected with TCCSUP‐sh1 cells were less than those in the control cells; this result was confirmed by subsequent hematoxylin and eosin (H&E) staining of metastatic lesions in the lungs (Figure 4C).
3.5. KIFC1 could induce EMT in BC
EMT is one of the key steps during invasion and metastasis of tumors. In our study, as KIFC1 was closely related to BC recurrence and promoted the migration of BC cells, we then wanted to see using EMT if KIFC1 affected the invasive phenotype of BC cells. qPCR demonstrated upregulated expression of three mesenchymal markers (N‐cadherin, fibronectin and vimentin) and two transcription factors (Snail and Slug) were expressed in KIFC1‐OE cells compared with vector cells. Expression levels of three epithelial markers (E‐cadherin, α‐catenin, and β‐catenin) were downregulated, contrasting with opposite findings in KIFC1‐silenced cells (Figure 5A).
Figure 5.
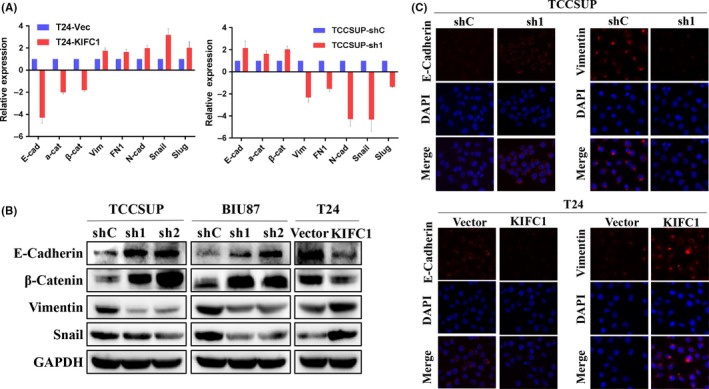
KIFC1 could induce the EMT course of BC. A, Relative mRNA expressions of epithelial markers (E‐cadherin, α‐catenin and β‐catenin), mesenchymal markers (vimentin, fibronectin and N‐cadherin) and EMT‐related transcription factors (Snail and Slug) were compared by qPCR between KIFC1‐OE, KIFC1‐silenced, and their corresponding control BC cells. B, Western blotting compared KIFC1 ectopic expression BC cells with their respective control cells in relative expression of EMT protein. C, Immunofluorescence staining showed an upregulated expression of epithelial marker E‐cadherin and a downregulated expression of mesenchymal marker vimentin in TCCSUP‐shKIFC1 cell. Opposite results were seen in T24‐KIFC1‐OE cells. Nuclei were counterstained with DAPI. (magnification: ×600)
As determined by western blotting assay, KIFC1 could upregulate the expression of mesenchymal‐related markers (vimentin and Snail) and inhibit epithelial markers (E‐cadherin and β‐catenin) in KIFC1‐OE T24 cells, whereas downregulation of mesenchymal‐related markers and upregulation of epithelial markers were observed in KIFC1‐silenced TCCSUP and BIU87 cells (Figure 5B). Immunofluorescence staining confirmed that overexpression of KIFC1 impaired E‐cadherin expression and increased vimentin expression, while downregulated expression of KIFC1 increased E‐cadherin expression and decreased vimentin expression (Figure 5C).
3.6. KIFC1 promoted AKT and GSK3β phosphorylation to induce EMT and proliferation
Activation of AKT has an important role in inducing EMT by phosphorylating GSK3β, thereby leading to Snail's stabilization, as well as nuclear localization and finally triggering cell migration and EMT.33 Liu and colleagues reported that Maelstrom promoted hepatocellular carcinoma metastasis by inducing EMT via Akt/GSK3β/Snail signaling.34 In our study, results from western blotting showed that, in the KIFC1‐silenced BC cells, expression of pAkt, pGSK3β, and Snail were consistently downregulated, whereas these genes were upregulated in KIFC1‐OE cells (Figure 6A), which indicated that KIFC1 might also induce EMT through the same signaling.
Figure 6.
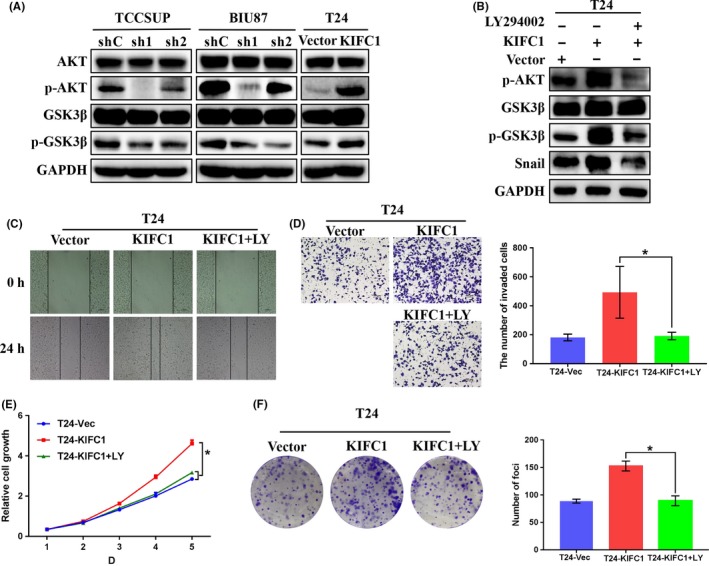
KIFC1 activated Akt/GSK3β signaling to induce proliferation and EMT of BC. A, The expression of total Akt, pAkt, total GSK3β and pGSK3β were detected by western blotting. GAPDH was used as the loading control. B, Western blotting analysis indicated that PI3K inhibitor LY294002 (LY) could effectively decrease expression levels of p‐AKT, p‐GSK3β and Snail induced by KIFC1. C‐F, Wound‐healing and transwell migration assay showed that LY could inhibit the migration ability of KIFC1‐OE cells, and CCK‐8 and colony formation assays revealed the suppressive effect in proliferation of BC cells of LY. *P < .05
To confirm our hypothesis, a PI3K inhibitor LY294002 was used to inhibit AKT activity in a rescue experiment. KIFC1‐OE cells were first treated with LY294002 (final concentration 20 μmol/L), and then cultured for 24 h for further study. Western blotting assay demonstrated that LY294002 could distinctly decrease the expression levels of pAkt, pGSK3β and Snail, which were previously upregulated by KIFC1 (Figure 6B). Transwell migration and wound‐healing assays, which were associated with the malignant cancer phenotype, showed that LY294002 could effectively decrease the migration ability of KIFC1‐OE cells (P < .05, Figure 6C,D). Furthermore, both CCK‐8 and colony formation assays demonstrated that, to some extent, LY294002 could also impair the proliferation of BC cells through inhibiting phosphorylation of AKT and GSK3β (P < .05, Figure 6E,F). In summary, KIFC1 promoted the migration, metastasis, and proliferation abilities of BC via Akt/GSK3β signaling.
4. DISCUSSION
Due to frequent tumor recurrence and distant metastasis, BC is considered a high economic‐burden malignancy with unsatisfactory prognosis. To date, many basic research studies have been conducted to find biomarkers for BC, only a tiny minority of which have been widely applied to clinical practice. Hence, there is still a need to find effective biomarkers for BC diagnosis and to predict its prognosis.
KIFC1, which plays an important role in the bipolar mitotic division of cancer cells, is greatly significant in maintaining cancer cell survival, but is redundant in normal cells.12 KIFC1 had been reported to drive tumor proliferation, metastasis, and drug resistance in several cancers,20, 21, 22, 23, 24, 25, 26, 27, 28, 29 and is related to shorter overall survival rate and poor prognosis, suggesting that KIFC1 might be a promising therapeutic target. However, the relationship between KIFC1 expression and prognosis of BC remained unclear.
In our current study, we first performed bioinformatics analysis on human publicly available BC datasets as the learning cohort to investigate mRNA expression in KIFC1. Findings were validated using BC fresh tissues and paraffin‐embedded specimens from our center. Results showed that KIFC1 was upregulated at both the mRNA and protein levels in BC tissues, consistent with the expression pattern of KIFC1 in other reported tumors.19, 20, 21, 22, 27, 29 We investigated the relationship between KIFC1 expression and the clinicopathological features of patients with BC who had undergone radical cystectomy by performing IHC on 152 BC specimens. Chi‐squared test revealed that KIFC1 expression was positively correlated with pT status and recurrence of BC. Kaplan‐Meier survival analysis and log‐rank test demonstrated that high expression of KIFC1 predicted poor CSS and RFS. High KIFC1 expression in patients with BC and with characteristics of pT status, pN−, age > 60 and male sex had a poorer CSS than those with low KIFC1 expression. In addition, univariate and multivariate analyses confirmed that pT status and KIFC1 expression were independent prognostic factors for BC.
Next, we wondered if KIFC1 played a crucial role in the malignant potential of BC. KIFC1 strengthened the proliferation and migration abilities of the BC cells in vitro. Furthermore, an experimental metastasis model demonstrated that knockdown of KIFC1 clearly inhibited metastasis of BC cells in vivo. Therefore, we focused our further studies on the migration and/or invasion abilities of BC cells and investigated roles for KIFC1 in EMT. KIFC1 was closely associated with recurrence in patients with BC, leading to a poor prognosis. EMT is an important mechanism responsible for invasion of malignant tumors. To be specific, EMT confers tumor cells with various malignancy characteristics including increased mobility, aggressiveness, a stem‐like phenotype, as well as evasion from apoptosis.35 Assays including qPCR, western blotting and immunofluorescence staining showed that EMT‐related epithelial markers were decreased and mesenchymal markers as well as transcription factors were increased in KIFC1‐OE BC cells; findings were opposite in KIFC1‐silenced BC cells. KIFC1 therefore plays an important role in the EMT of BC, and could dramatically accelerate this process.
Epithelial−mesenchymal transition had been a popular academic research direction for many years. More recently, Han et al20 concentrated on searching upstream of KIFC1, and reported that KIFC1 was regulated by miR‐532‐3p and then promoted EMT and metastasis of hepatocellular carcinomas via gankyrin/AKT signaling. Our center previously demonstrated that Maelstrom promoted hepatocellular carcinoma metastasis by inducing EMT through Akt/GSK3β/Snail signaling34 downstream of KIFC1, prompting further studies. We measured the expression levels of related proteins involved in Akt/GSK3β/Snail signaling, showing that KIFC1 played a crucial part. Subsequently, we performed rescue experiments to confirm our hypothesis using a PI3K inhibitor LY294002, which inhibited the AKT activation. Western blotting showed that, after treatment with LY294002, the levels of related proteins involved in this signaling pathway, and upregulated by KIFC1, then distinctly decreased. Cellular experiments demonstrated that LY294002 could revert the migration and proliferation abilities of BC cells; this finding further confirmed our hypothesis.
In conclusion, our present study demonstrated that high KIFC1 expression, frequently seen in BC tissues, was significantly associated with poor prognosis of patients with BC and might be a potential prognostic biomarker and therapeutic target for BC. KIFC1 plays an important oncogenic role by promoting the aggressiveness of BC.
Bladder cancer cells proliferate, migrate, and metastasize both in vitro and vivo, due to KIFC1 induction of EMT via Akt/GSK3β signaling and enhanced proliferation, migration, and metastasis of BC.
DISCLOSURE
The authors declare that they have no competing interests.
ACKNOWLEDGMENTS
We are very grateful to Ting Zhou for her great help in statistical calculations. This work was supported by grants from the National Key R&D Program of China (No. 2017YFC1309001), National Natural Science Foundation of China (Nos. 81672530 and 81730072) and Natural Science Foundation of Guangdong Province of China (No. 2017A030313686). We thank all the patients who provided precious pathological specimens and our investigators who did a great job in this experiments.
Xiao K‐H, Teng K, Ye Y‐L, et al. Kinesin family member C1 accelerates bladder cancer cell proliferation and induces epithelial–mesenchymal transition via Akt/GSK3β signaling. Cancer Sci. 2019;110:2822‐2833. 10.1111/cas.14126
Kang‐hua Xiao, Kai Teng, and Yun‐lin Ye contributed equally to this work.
Contributor Information
Zi‐ke Qin, Email: qinzk@sysucc.org.cn.
Dan Xie, Email: xiedan@sysucc.org.cn.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115‐132. [DOI] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7‐30. [DOI] [PubMed] [Google Scholar]
- 4. Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer. 2015;15(1):25‐41. [DOI] [PubMed] [Google Scholar]
- 5. Choi W, Czerniak B, Ochoa A, et al. Intrinsic basal and luminal subtypes of muscle‐invasive bladder cancer. Nat Rev Urol. 2014;11(7):400‐410. [DOI] [PubMed] [Google Scholar]
- 6. Alfred Witjes J, Lebret T, Compérat EM, et al. Updated 2016 EAU guidelines on muscle‐invasive and metastatic bladder cancer. Eur Urol. 2017;71(3):462‐475. [DOI] [PubMed] [Google Scholar]
- 7. Prasad SM, Decastro GJ, Steinberg GD. Urothelial carcinoma of the bladder: definition, treatment and future efforts. Nat Rev Urol. 2011;8(11):631‐642. [DOI] [PubMed] [Google Scholar]
- 8. Nath S, Bananis E, Sarkar S, et al. Kif5B and Kifc1 interact and are required for motility and fission of early endocytic vesicles in mouse liver. Mol Biol Cell. 2007;18(5):1839‐1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ando A, Kikuti YY, Kawata H, et al. Cloning of a new kinesin‐related gene located at the centromeric end of the human MHC region. Immunogenetics. 1994;39(3):194‐200. [DOI] [PubMed] [Google Scholar]
- 10. DeLuca JG, Newton CN, Himes RH, Jordan MA, Wilson L. Purification and characterization of native conventional kinesin, HSET, and CENP‐E from mitotic HeLa cells. J Biol Chem. 2001;276(30):28014‐28021. [DOI] [PubMed] [Google Scholar]
- 11. Yu K, Hou L, Zhu J, Ying X, Yang W. KIFC1 participates in acrosomal biogenesis, with discussion of its importance for the perforatorium in the Chinese mitten crab Eriocheir sinensis . Cell Tissue Res. 2009;337(1):113‐123. [DOI] [PubMed] [Google Scholar]
- 12. Xiao Y, Yang W. KIFC1: a promising chemotherapy target for cancer treatment? Oncotarget. 2016;7(30):48656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hall VJ, Compton D, Stojkovic P, et al. Developmental competence of human in vitro aged oocytes as host cells for nuclear transfer. Hum Reprod. 2007;22(1):52‐62. [DOI] [PubMed] [Google Scholar]
- 14. Yang W, Sperry AO. C‐terminal kinesin motor KIFC1 participates in acrosome biogenesis and vesicle transport1. Biol Reprod. 2003;69(5):1719‐1729. [DOI] [PubMed] [Google Scholar]
- 15. Farina F, Pierobon P, Delevoye C, et al. Kinesin KIFC1 actively transports bare double‐stranded DNA. Nucleic Acids Res. 2013;41(9):4926‐4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Basto R, Brunk K, Vinadogrova T, et al. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133(6):1032‐1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kwon M, Godinho SA, Chandhok NS, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22(16):2189‐2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rath O, Kozielski F. Kinesins and cancer. Nat Rev Cancer. 2012;12(8):527‐539. [DOI] [PubMed] [Google Scholar]
- 19. Xiao Y, Shen H, She Z, et al. C‐terminal kinesin motor KIFC1 participates in facilitating proper cell division of human seminoma. Oncotarget. 2017;8(37):61373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Han J, Wang F, Lan Y, et al. KIFC1 regulated by miR‐532‐3p promotes epithelial‐to‐mesenchymal transition and metastasis of hepatocellular carcinoma via gankyrin/AKT signaling. Oncogene. 2019;38:406‐420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pawar S, Donthamsetty S, Pannu V, et al. KIFCI, a novel putative prognostic biomarker for ovarian adenocarcinomas: delineating protein interaction networks and signaling circuitries. J Ovarian Res. 2014;7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pannu V, Rida PCG, Ogden A, et al. HSET overexpression fuels tumor progression via centrosome clustering‐independent mechanisms in breast cancer patients. Oncotarget. 2015;6(8):6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grinberg‐Rashi H, Ofek E, Perelman M, et al. The expression of three genes in primary non‐small cell lung cancer is associated with metastatic spread to the brain. Clin Cancer Res. 2009;15(5):1755‐1761. [DOI] [PubMed] [Google Scholar]
- 24. Martin SK, Kyprianou N. Exploitation of the androgen receptor to overcome taxane resistance in advanced prostate cancer. Adv Cancer Res. 2015;127:123‐158. [DOI] [PubMed] [Google Scholar]
- 25. Zou JX, Duan Z, Wang J, et al. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Mol Cancer Res. 2014;12(4):539‐549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De S, Cipriano R, Jackson MW, Stark GR. Overexpression of kinesins mediates docetaxel resistance in breast cancer cells. Cancer Res. 2009;69(20):8035‐8042. [DOI] [PubMed] [Google Scholar]
- 27. Imai T, Oue N, Yamamoto Y, et al. Overexpression of KIFC1 and its association with spheroid formation in esophageal squamous cell carcinoma. Pathol Res Pract. 2017;213(11):1388‐1393. [DOI] [PubMed] [Google Scholar]
- 28. Oue N, Mukai S, Imai T, et al. Induction of KIFC1 expression in gastric cancer spheroids. Oncol Rep. 2016;36(1):349‐355. [DOI] [PubMed] [Google Scholar]
- 29. Li G, Chong T, Yang J, Li H, Chen H. Kinesin motor protein KIFC1 is a target protein of miR‐338‐3p and associated with poor prognosis and progression of renal cell carcinoma. Oncol Res. 2018;27:125‐137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weinstein JN, Collisson EA, Mills GB, et al. The cancer genome atlas pan‐cancer analysis project. Nat Genet. 2013;45(10):1113‐1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clough E, Barrett T. The Gene Expression Omnibus database. Methods Mol Biol. 2016;1418:93‐110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen M, Wu R, Li G, et al. Motor neuron and pancreas homeobox 1/HLXB9 promotes sustained proliferation in bladder cancer by upregulating CCNE1/2. J Exp Clin Cancer Res. 2018;37(1):154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou BP, Deng J, Xia W, et al. Dual regulation of Snail by GSK‐3β‐mediated phosphorylation in control of epithelial–mesenchymal transition. Nat Cell Biol. 2004;6(10):931‐940. [DOI] [PubMed] [Google Scholar]
- 34. Liu L, Dai Y, Chen J, et al. Maelstrom promotes hepatocellular carcinoma metastasis by inducing epithelial–mesenchymal transition by way of Akt/GSK‐3beta/Snail signaling. Hepatology. 2014;59(2):531‐543. [DOI] [PubMed] [Google Scholar]
- 35. Sato M, Shames DS, Hasegawa Y. Emerging evidence of epithelial‐to‐mesenchymal transition in lung carcinogenesis. Respirology. 2012;17(7):1048‐1059. [DOI] [PubMed] [Google Scholar]


