Abstract
DNA markers for pancreatic ductal adenocarcinoma (PDAC) are urgently needed for detection of minimally invasive disease. The epigenetic relevance of the cysteine dioxygenase 1 gene (CDO1) has been never investigated in PDAC. Three studies, including cellular experiments, tissue validation, and pilot testing for pancreatic cytology, were carried out. Promoter DNA methylation value (MV) of CDO1 was quantified by quantitative methylation‐specific PCR. CDO1 expression was consistent with its promoter DNA methylation in 7 PDAC cell lines. In 160 retrospectively collected primary PDAC tumor tissues, MV was significantly higher compared to the corresponding noncancerous pancreas (area under the receiver operating characteristic curve [AUC] = 0.97, P < .0001), and CDO1 hypermethylation was highly specific to PDAC tumor tissues. CDO1 hypermethylation group (MV over 19) was significantly associated with diverse prognostic factors in PDAC. Surprisingly, it was significantly higher in prospectively collected PDAC cytology samples (n = 37), including both pancreatic juice (n = 12) and endoscopic ultrasound‐fine needle aspiration (EUS‐FNA) cytology (n = 25) compared to pancreatic benign diseases (AUC = 0.96, P < .0001). Detection of PDAC was confirmed by DNA testing in 35 of 37 patients (95% sensitivity); thus, it was more sensitive than cytology (33%) or EUS‐FNA cytology (88%). Promoter DNA methylation of CDO1 is extremely specific for PDAC tumors, and accumulates with PDAC tumor progression. It could be a definitive diagnostic marker of PDAC in pancreatic juice or EUS‐FNA cytology.
Keywords: CDO1, diagnosis, methylation, pancreatic cancer, prognosis
1. INTRODUCTION
Pancreatic cancer ranks 15th among malignant cancers in terms of incidence (337 872 cases in 2012) and is the seventh leading cause of cancer‐related death (330 391 deaths in 2012) worldwide.1 Pancreatic ductal adenocarcinoma (PDAC), a dominant histological type of pancreatic cancer, represents one of the most fatal malignancies, is the fourth leading cause of cancer‐related deaths in the United States in 2014, and without any substantive improvement in curative therapies, is anticipated to be the second leading cause of cancer‐related deaths by 2030.2 One explanation behind its poor medical advancement is its nonspecific symptoms, resulting in delayed diagnosis and dismal prognosis. At most, only 20% of patients with PDAC present with resectable tumors.3 Surgical resection is the only alternative for cure or long‐term survival of patients with PDAC.4 Nevertheless, the 5‐year survival rate remains at approximately 10%, even after curative surgery with the best adjuvant chemotherapy.5 To improve the prognosis of pancreatic cancer, early diagnosis is believed to be mandatory, and a simple and less invasive surveillance system has been in high demand for early diagnosis.
Pancreatic juice obtained from the endoscopic retrograde pancreatography (ERP) test is considered to be a body fluid in which PDAC cells are the most densely concentrated, and therefore, is a promising tool to diagnose PDAC. However, cytology testing using this fluid (pancreatic juice) is disappointing, because the detection rates are much lower than those for endoscopic ultrasound‐fine needle aspiration (EUS‐FNA) cytology or histology, which is the most popular test to confirm PDAC at present. The diagnostic accuracy of EUS‐FNA histology is also estimated to be high (sensitivity and specificity of 84% and 97%, respectively).6 However, EUS‐FNA is an invasive clinical tool associated with peritoneal dissemination, and noninvasive cancer biomarkers for the early detection of PDAC have been highly anticipated.
Cancer‐specific genomic or epigenetic alterations are promising for such noninvasive approaches. K‐ras mutation is frequently seen in PDAC, and a meta‐analysis of the diagnostic value of detecting K‐ras mutation in the pancreatic juice of patients with PDAC revealed that the pooled sensitivity and specificity were 59% and 87%, respectively, which was inferior to EUS‐FNA.7 Furthermore, novel DNA methylation markers for PDAC were explored in the context of discovery, tissue validation, and pilot testing in pancreatic juice, and cluster of differentiation 1d molecule (CD1D) methylation was detected in 75% of PDAC with 95% specificity (area under the receiver operating characteristic curve [AUC] = 0.92) in pancreatic juice.8 These findings suggested that promoter DNA methylation is more promising than K‐ras mutation in pancreatic juice diagnosis.
The cysteine dioxygenase 1 gene (CDO1) is a tumor suppressor gene in human cancer that was identified by a pharmacological unmasking microarray.9, 10, 11 CDO1 encodes a nonheme iron enzyme that converts cysteine to cysteine sulfinic acid that affects mitochondria function, while it suppresses the production of glutathione from cysteine and induces reactive oxygen species generation, subsequently promoting apoptosis.12 CDO1 plays a role as a tumor suppressor gene. As it is a methylation‐specific gene in human cancer, CDO1 methylation has been recently reported in a variety of cancers, such as esophageal,11, 13, 14 lung,15 gastric,11, 16 breast,11, 12, 17 biliary tract,18 colorectal,11, 19 kidney,20 prostate,21 bladder,11 penile,22 and uterine cancers.23 However, there has been no report on the involvement of CDO1 in PDAC. To the best of our knowledge, this study represents the first investigation of the clinical relevance of methylation of the CDO1 promoter DNA in PDAC tissues and its diagnostic potential in pancreatic juice.
2. MATERIALS AND METHODS
2.1. Collection of samples (primary tumor tissues, noncancerous pancreatic tissues, and pancreatic cytology solutions) from patients with PDAC
We initially analyzed 160 patients with primary PDAC who underwent surgical resection of primary tumors with no prior chemotherapy at the Kitasato University Hospital (Sagamihara, Japan) between 1986 and 2013. Clinicopathological characteristics of these patients are shown in Table 1. The TNM classification was used according to the 7th edition of the UICC staging system. This study was approved by the Institutional Review Board for Observation and Epidemiological Studies, Kitasato University Medical Ethics Organization (approval no. B18‐017). All patients gave written informed consent for any pathological investigation.
Table 1.
Univariate and multivariate prognostic analysis in 160 patients with pancreatic ductal adenocarcinoma who underwent pancreatectomy
| Variable | Number | Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|---|---|
| RR | 95% CI | P valuea | RR | 95% CI | P valueb | |||
| Age, years | ||||||||
| <65/over 65 | 80/80 | 0.8 | 0.6‐1.2 | .4538 | ND | |||
| Gender | ||||||||
| Male/female | 84/76 | 1.2 | 0.8‐1.8 | .2467 | ND | |||
| Lymphatic invasion | ||||||||
| Absence/presence | 23/137 | 1.9 | 1.1‐3.6 | .0245 | 1.5 | 0.8‐2.9 | NS | |
| Venous invasion | ||||||||
| Absence/presence | 15/145 | 3.1 | 1.3‐10 | .0169 | 1.3 | 0.5‐4.5 | NS | |
| Intrapancreatic nerve invasion | ||||||||
| Absence/presence | 16/144 | 3.6 | 1.6‐10.2 | .0024 | 2.1 | 0.9‐6.3 | NS | |
| Retropancreatic tissue invasion | ||||||||
| Absence/presence | 61/99 | 1.7 | 1.2‐2.7 | .0062 | 1.3 | 0.7‐2.2 | NS | |
| Portal venous system invasion | ||||||||
| Absence/presence | 130/30 | 1.8 | 1.1‐2.8 | .0142 | 2.1 | 1.2‐3.6 | .0073 | |
| Extrapancreatic nerve plexus invasion | ||||||||
| Absence/presence | 126/34 | 2.2 | 1.4‐3.3 | .0002 | 1.3 | 0.8‐2.2 | NS | |
| Arterial system invasion | ||||||||
| Absence/presence | 155/5 | 3.8 | 1.3‐8.6 | .0020 | 3.5 | 1.2‐8.6 | .0300 | |
| Preoperative value of serum CA19‐9 | ||||||||
| <37/over 37 | 39/121 | 3.5 | 2.1‐6.4 | <.0001 | 2.9 | 1.6‐5.5 | .0003 | |
| CDO1 methylation value | ||||||||
| <19/over 19 | 81/79 | 1.5 | 1.1‐2.2 | .0242 | 1.2 | 0.8‐1.9 | NS (0.3) | |
| Dissected pancreatic tissue margin | ||||||||
| Negative/positive | 100/60 | 2.9 | 2.0‐4.3 | <.0001 | 2.4 | 1.5‐3.7 | .0002 | |
| Stage (7th UICC) | ||||||||
| 0 | 1 | Ref. | <.0001 | Ref. | .0027 | |||
| I | 8 | 2.6 × 106 | 0.019 | 15 × 107 | ||||
| II | 121 | 8.6 | 1.9‐152 | 0.2 | 0.01‐1.2 | |||
| III | 7 | 5.9 | 2.4‐12 | 4.2 | 1.4‐11 | |||
| IV | 23 | 0.5 | 0.2‐1.3 | 0.6 | 0.2‐1.7 | |||
Bold values indicate significance.
Log‐rank test.
Cox proportional hazards model.
CA, carbohydrate antigen; CI, confidence interval; ND, not done; NS, not significant; Ref., reference; RR, relative risk.
To diagnose PDAC, pancreatic juice cytology was carried out along with ERP or EUS‐FNA. We prospectively registered 43 patients from whom pancreatic cytology samples of PDAC with no prior chemotherapy were collected (n = 37) and those suffering from pancreatic benign disease (n = 6), chronic pancreatitis (n = 4), and autoimmune pancreatitis (n = 2). Among the 37 patients with PDAC, 28 suffered from distant metastatic disease and 9 did not have any distant metastasis and underwent pancreatectomy. Pancreatic cytology samples from patients with PDAC (n = 37) were obtained by ERP (n = 12) or EUS‐FNA cytology (n = 25) solutions at the Kitasato University Hospital between 2017 and 2018. The EUS‐FNA cytology samples were obtained from washing solutions after EUS‐FNA tissues were removed from the inner needle, and differed from EUS‐FNA histology. The eligibility criteria of the study were as follows: (a) patients suspected with PDAC; (b) patients who had a high‐risk factor of PDAC including chronic pancreatitis; (c) patients who underwent ERP for a definitive diagnosis of PDAC; or (d) patients who underwent endoscopic EUS‐FNA for a definitive diagnosis of PDAC. This study was also approved by the Institutional Review Board for Observation and Epidemiological Studies, Kitasato University Medical Ethics Organization (approval no. B16‐105), and informed consent was obtained from all participants separately for tissue samples.
Both studies (approval numbers B16‐105 and B18‐017) were carried out in accordance with the Declaration of Helsinki.
2.2. Cell lines
Two PDAC cell lines, PK‐8 and KLM‐1, and 1 colorectal cancer cell line, DLD1, were kindly provided from the Cell Resource Centre for Biomedical Research Institute of Development, Aging and Cancer, Tohoku University (Sendai, Japan). Five other PDAC cell lines, PK‐59, PK‐45 H, PK‐45P, MIA Paca2, and PANC‐1, and the hepatocellular carcinoma cell line, HepG2, were purchased from RIKEN BioResource Centre. All cell lines except MIA Paca2 were maintained in RPMI‐1640 medium (Gibco) and MIA Paca2 was maintained in DMEM (Gibco) containing 10% FBS.
2.3. DNA extraction and bisulfite treatment
Tissue sections from primary tumors and corresponding noncancerous pancreas tissues were stained with H&E and dissected under a microscope. Genomic DNA was extracted from formalin‐fixed paraffin‐embedded (FFPE) PDAC tissues or cell lines using a QIAamp DNA FFPE Tissue Kit or a QIAamp DNA Mini Kit (Qiagen), respectively.
The pancreatic cytology solutions were aliquoted in a 200 μL volume. Samples were immediately stored at −80°C. DNA was extracted from samples using a QIAamp DNA Micro Kit (Qiagen) into 40 μL distilled water. Bisulfite treatment was done by using an EZ DNA Methylation‐Gold Kit (Zymo Research) and the bisulfited DNA was subsequently amplified by quantitative methylation‐specific PCR (Q‐MSP). Primer sequences were designed to recognize the DNA alterations as previously described.19
2.4. Quantitative methylation‐specific PCR
Quantitative methylation‐specific PCR was carried out using iQ Supermix (Bio‐Rad) in triplicate on a C1000 Touch Thermal Cycler CFX96 Real Time System (Bio‐Rad). Serial dilutions of bisulfite modified DNA from DLD1 was used to construct the calibration curve on each plate as a methylation positive control, and HepG2 served as a methylation negative control as reported.19 The methylation value (MV) was defined by a ratio of amplified signal value of methylated CDO1 normalized to β‐actin, then multiplied by 100 (Q‐MSP value).
2.5. RNA extraction and RT‐PCR
Total RNA from cell lines was extracted using RNeasy Mini Kit (Qiagen), and was reverse‐transcribed with a Super Script III reverse transcriptase kit (Invitrogen). Primers sequences for CDO1 and β‐actin were described previously.18 Reverse transcription‐PCR was undertaken by 30 cycles of 95°C for 1 minute, 58°C for 1 minute, and 72°C for 1 minute. The PCR products were separated on 1.5% agarose gel, and then visualized by ethidium bromide staining. β‐Actin was used as an internal control.
2.6. Cell treatment with 5‐aza‐2′‐deoxycytidine and trichostatin A
Cells (1 × 106 cells/T‐75 flask) were treated with 1 or 5 μmol/L of the demethylating agent, 5‐aza‐2′‐deoxycytidine (5‐Aza‐dC) (Sigma‐Aldrich), dissolved in 50% acetic acid or mock‐treated with PBS dissolved in the same amount of acetic acid every 24 hours for 4 days. When combining with the histone deacetylase inhibitor trichostatin A (TSA) (Sigma‐Aldrich), 300 nmol/L TSA was added to the medium for the final 24 hours.
2.7. Immunohistochemistry
Immunostaining was carried out on FFPE sections (4 μm thick). Sections were incubated using the anti‐CDO1 rabbit polyclonal Ab (dilution of 1:100) (Atlas Antibodies). Immune complexes were detected with a Histofine Simple Stain MAX PO (MULTI) (Nichirei), following the manufacturer's protocol, and visualized using the 3,3′‐diaminobenzidine substrate. Sections were counter‐stained with hematoxylin solution.
2.8. Statistical analysis
Student's t test was used for analysis of continuous variables, and the χ2 test was used for analysis of categorical variables. Clinicopathological characteristics and follow‐up data were analyzed in terms of the disease‐specific survival (DSS), which was measured from the date of operation to the date of cancer‐specific death or last follow‐up. The DSS was calculated by the Kaplan‐Meier method, and survival differences were assessed using the log‐rank test. Variables suggested to be prognostic factors in univariate analysis (P < .05) were subjected to multivariate analysis using a Cox proportional hazards model. A P value < .05 indicated statistical significance. All statistical analyses were undertaken using the SAS software package JMP Pro14 (SAS Institute).
3. RESULTS
3.1. Methylation and expression profiles of CDO1 in PDAC cell lines
We initially examined 7 PDAC cell lines to examine the expression status of CDO1. CDO1 expression was not detected at the mRNA level in any of the PDAC cell lines, unlike HepG2 cells (Figure 1A). We then examined the promoter DNA methylation status of CDO1 in all the PDAC cell lines by bisulfite treatment followed by Q‐MSP analysis (Figure 1B). Promoter DNA of CDO1 was found to be hypermethylated in all 7 PDAC cell lines; however, it was not hypermethylated in HepG2 cells. These findings indicated that the DNA methylation status of CDO1 is tightly correlated with CDO1 expression in PDAC cell lines.
Figure 1.
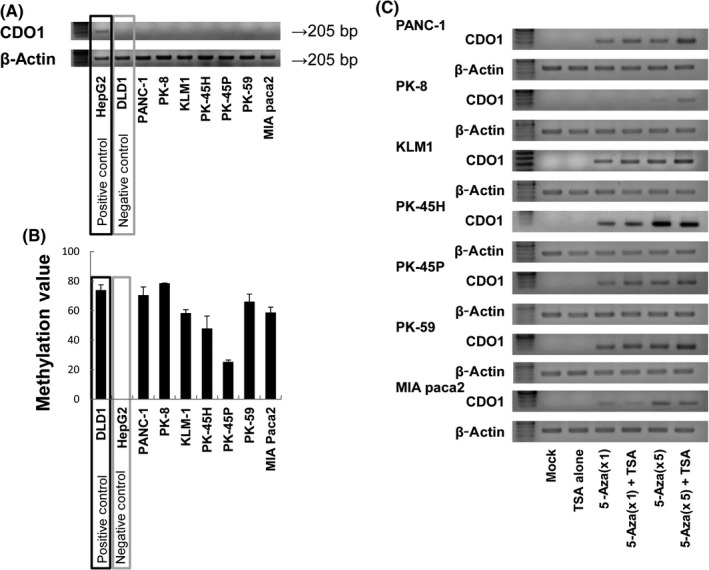
Methylation and expression profiles of CDO1 in pancreatic ductal adenocarcinoma (PDAC) cell lines. A, CDO1 mRNA expression was assessed by RT‐PCR. The positive control for RT‐PCR was the HepG2 cell line, and the negative control was the DLD1 cell line. CDO1 mRNA expression was not detected in any of the PDAC cell lines. B, Promoter DNA methylation of CDO1 was quantified by quantitative methylation‐specific PCR (Q‐MSP) (mean ± SD). The positive control for Q‐MSP was the DLD1 cell line, and the negative control was the HepG2 cell line. In all PDAC cell lines, CDO1 promoter DNA was hypermethylated. C, CDO1 mRNA expression after treatment with the demethylating agents, 5‐aza‐2′‐deoxycytidine (5‐Aza‐dC, 1 or 5 μmol/L) in the presence or absence of trichostatin A (TSA), a histone deacetylase inhibitor, by RT‐PCR. It was confirmed that CDO1 expression was regulated by epigenetics. AUC, area under the receiver operating characteristic curve
Furthermore, to confirm that CDO1 was regulated by methylation, demethylation treatments using 5‐Aza‐dC and TSA were undertaken for the PDAC cell lines (Figure 1C). Demethylation treatments reactivated CDO1 expression in all PDAC cells, indicating that silenced expression of CDO1 in PDAC cells is regulated in an epigenetic manner.
3.2. Methylation of CDO1 promoter DNA in 160 patients with PDAC and its relationship with prognosis
Next, to clarify the clinical significance of MV of the CDO1 promoter DNA, Q‐MSP assessment of PDAC tumor tissues and corresponding noncancerous pancreas tissue (CN) was also carried out on 160 patients with PDAC. The mean MV in primary PDAC tumor tissues was 23.9 (range, 0.0–87.5), and the mean MV in CN was 0.08 (range, 0.0–3.4) (Figure 2A). The PDAC tissues showed significantly higher MV of CDO1 promoter DNA compared to the CN (Figure 2B, P < .0001), and the difference was robust to discriminate tumor from CN (AUC = 0.97, P < .0001) (Figure 2C). We confirmed the CDO1 protein expression by immunostaining (Figure 2D). In normal pancreas tissue, CDO1 protein was expressed in islet and acinar cells in addition to pancreatic tubules. In contrast, in PDAC tissue, CDO1 protein was not expressed in tumor cells, if the tumor showed hypermethylation of CDO1.
Figure 2.
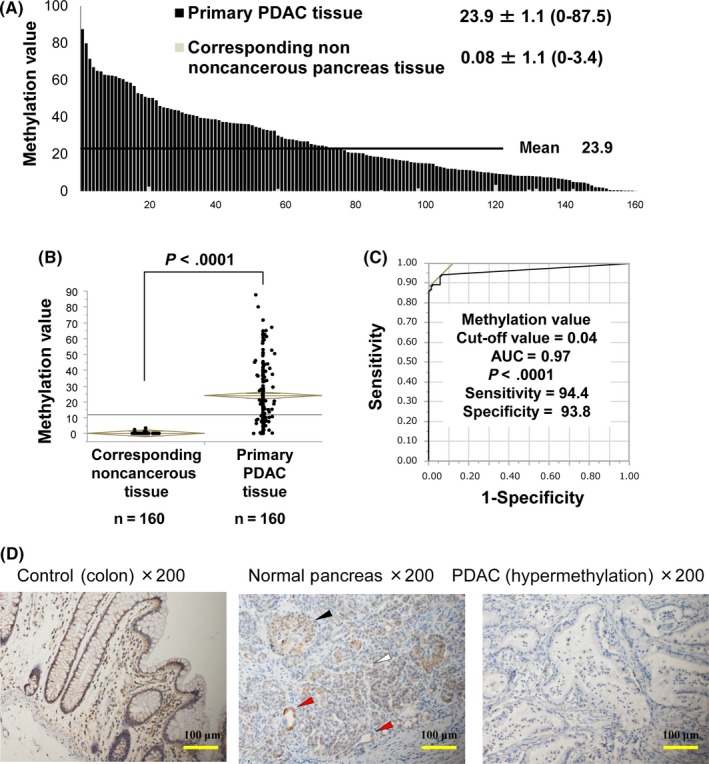
Methylation value (MV) of CDO1 promoter DNA in 160 primary pancreatic ductal adenocarcinoma (PDAC) tissues. A, Quantitative methylation‐specific PCR assessment was used to examine the MV in 160 primary PDAC tissues and 160 corresponding noncancerous pancreatic tissues (CN). B, MV of CDO1 was significantly different between primary PDAC tissue and CN (P < .0001). C, Receiver operating characteristic curve of CDO1 methylation to differentiate primary PDAC tumors from CN. The area under the curve (AUC) represents the accuracy in discriminating normal from tumor tissue in terms of sensitivity and specificity (AUC = 0.97, P < .0001). D, Representative images of immunostaining with an anti‐CDO1 Ab are shown. Colon tissue is shown as a positive control. In NC, CDO1 protein was expressed in epithelium (red arrows), islet (black arrow), and acinar cells (white arrow). In the PDAC tissue, CDO1 protein was not expressed in epithelium
We further investigated whether CDO1 MV could predict prognostic outcomes of PDAC. A Kaplan‐Meier curve of DSS was constructed for the 160 patients according to the CDO1 MV, and P value and relative risk (RR) were plotted to analyze survival differences between CDO1 MV above and below the best optimized cut‐off values, determined by using the log‐rank test (Figure 3A). We determined the best optimal cut‐off value for prognostic stratification using the log‐rank plot analysis.24, 25 We thereby defined the optimal cut‐off value of the CDO1 MV as 19.0, which indicated the highest RR with statistical significance (P < .05). Using this cut‐off value to divide the patients into hyper‐ and hypomethylation groups, the CDO1 hypermethylation group showed a 5‐year survival rate of 16.3% (n = 79), whereas the CDO1 hypomethylation group showed a 5‐year survival rate of 24.5% (n = 81). The prognostic difference between these 2 groups was highly significant (P = .024) (Figure 3B).
Figure 3.
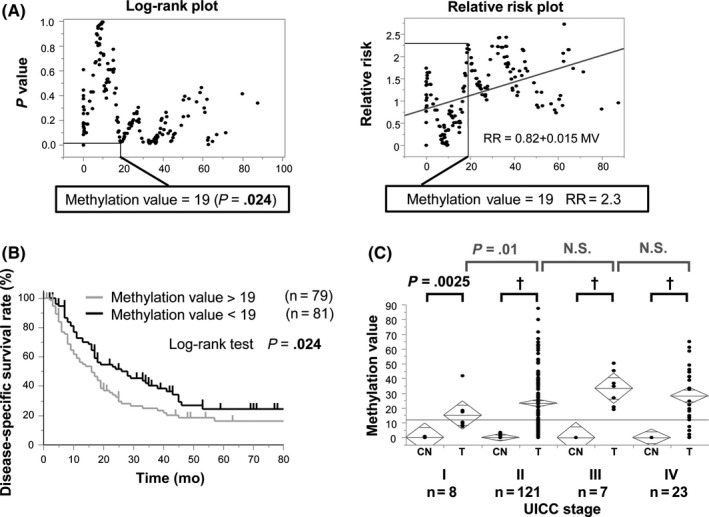
Prognostic analysis of the methylation of CDO1 promoter DNA in 160 patients with pancreatic ductal adenocarcinoma (PDAC). A, Identification of an optimal cut‐off value for the prognosis using log‐rank plot analysis and relative risk (RR) plot. Note that RR became high as the methylation value of CDO1 increased. B, Kaplan‐Meier curve for CDO1 methylation status with value above or below 19.0 in primary PDAC. The prognostic difference between these 2 groups showed high statistical significance (P = .024). C, Correlation of CDO1 methylation level to UICC stage. Methylation levels of CDO1 showed significant differences between Stage I and Stage II. There was no significant difference between other stages of PDAC. †P < .0001. CN, corresponding noncancerous pancreatic tissue; N.S., not significant; T, PDAC tumor tissue
3.3. Univariate and multivariate prognostic analyses including CDO1 methylation status in patients with PDAC
The characteristics of 160 patients with PDAC and the univariate prognostic factors are summarized in Table 1. Univariate prognostic factors involved lymphatic permeation factor (P = .025), vascular permeation factor (P = .017), intrapancreatic nerve invasion factor (P = .00024), retropancreatic tissue invasion factor (P = .0062), portal venous system invasion factor (PV; P = 0.014), extrapancreatic nerve plexus invasion factor (P = .0002), arterial system invasion factor (P = 0.002), preoperative serum CA19‐9 level (P < .0001), CDO1 MV (P = .024), dissected peripancreatic tissue margin factor (DPM; P < .0001), and UICC stage (P < .0001). The clinicopathologic factors related to prognosis were then examined in the multivariate analysis. We found that PV (P = .0073, RR = 2.1), arterial system invasion factor (P = .03, RR = 3.5), preoperative serum CA19‐9 level (P = .0003, RR = 2.9), and DPM positive (P = .0002, RR = 2.4) were independent of UICC stage (P = .0027) in 160 patients with PDAC. Moreover, high CDO1 MV was not an independent prognostic factor (P = 0.3). However, CDO1 methylation status was associated with UICC stage. The CDO1 MV at each UICC stage is shown in Figure 3C. Stage 0 is not described because the MV of cases of stage 0 was 0. The mean MV of the other stages was: Stage I, 15.2; II, 23.3; III, 33.4; and IV, 28.3. The CDO1 MV showed significant differences between Stage I and Stage II. However, there were no significant differences among other stages. Furthermore, the MV of each stage was compared with CN. In Stage I, PDAC tissues had significantly higher MV compared to the CN (P = .0025). This result suggested that the methylation of CDO1 promoter DNA was correlated with UICC stage determination factors.
3.4. Correlation of CDO1 methylation level to prognostic factors in patients with PDAC
The correlation of promoter DNA methylation of CDO1 to clinicopathologic factors of PDAC by the χ2 test is shown in Table 2. High CDO1 MV groups were significantly related to pathological factors that are involved in UICC stage determination, such as T factor and N factor. In addition, high CDO1 MV groups were related to pathological factors, such as intrapancreatic nerve invasion, retropancreatic tissue invasion factor, PV, arterial system invasion, extrapancreatic nerve plexus invasion, and DPM, which were negative prognostic factors in PDAC as revealed by univariate analysis. This might explain elimination of CDO1 hypermethylation as an independent prognostic factor in PDAC.
Table 2.
Correlation of clinicopathologic characteristics of patients with pancreatic ductal adenocarcinoma who underwent pancreatectomy and CDO1 methylation
| Variable | >19 | <19 | P valuea | Variable | >19> | <19 | P valuea | ||
|---|---|---|---|---|---|---|---|---|---|
| UICC stage | 0 | 0 | 1 | .0070 | v | Negative | 4 | 11 | .060 |
| I | 1 | 7 | Positive | 75 | 70 | ||||
| II | 57 | 64 | ne | Negative | 4 | 12 | .036 | ||
| III | 7 | 0 | Positive | 75 | 69 | ||||
| IV | 14 | 9 | CH | Negative | 41 | 36 | .300 | ||
| T | 1 | 1 | 4 | .0018 | Positive | 38 | 45 | ||
| 2 | 2 | 7 | DU | Negative | 45 | 45 | .500 | ||
| 3 | 39 | 53 | Positive | 34 | 36 | ||||
| 4 | 37 | 16 | RP | Negative | 21 | 40 | .003 | ||
| N | Negative | 14 | 30 | .0060 | Positive | 58 | 41 | ||
| Positive | 65 | 51 | PV | Negative | 58 | 72 | .010 | ||
| Infiltration | INFa | 1 | 0 | .2000 | Positive | 21 | 9 | ||
| INFb | 49 | 59 | A | Negative | 56 | 70 | .016 | ||
| INFg | 29 | 22 | Positive | 23 | 11 | ||||
| Location | Ph | 54 | 60 | .4000 | PL | Negative | 56 | 70 | .016 |
| Pb | 25 | 21 | Positive | 23 | 11 | ||||
| Histology | mod | 32 | 30 | .9000 | CA19‐9 | <37 | 19 | 20 | .900 |
| pap | 3 | 3 | Over 37 | 60 | 61 | ||||
| por | 13 | 11 | DPM | Negative | 43 | 57 | .037 | ||
| well | 31 | 36 | Positive | 36 | 24 | ||||
| ly | Negative | 8 | 15 | .1000 | |||||
| Positive | 71 | 66 | |||||||
Bold values indicate significance.
χ2 test.
A, arterial system invasion; CA, carbohydrate antigen; CH, distal bile duct invasion; DPM, dissected pancreatic tissue margin; DU, duodenal invasion; ly, lymphatic invasion; mod, moderate; ne, intrapancreatic nerve invasion; pap, papillary; Pb, body of pancreas; Ph, head of pancreas; PL, extrapancreatic nerve plexus invasion; por, poor; PV, portal venous system invasion; RP, retropancreatic tissue invasion; v, venous invasion.
3.5. Potential utility of CDO1 methylation as a tumor diagnostic marker
Methylation of CDO1 promoter DNA was significantly higher in pancreatic cytology sample solutions obtained from patients with PDAC compared to those from patients with pancreatic benign diseases (P = .018) (Figure 4A), where PDAC with MV of 5.0 or higher was found to be positive in 35 (94.6%) of 37 patients with 100% specificity (0% in benign pancreatic disease) during detection of CDO1 hypermethylation (Figure 4B, AUC = 0.96, P < .0001) in contrast to conventional ERP cytology (4/12, 33%), EUS‐FNA cytology (22/25, 88%), and EUS‐FNA histology (24/25, 96%). β‐Actin was used for normalization. Thirty‐seven pancreatic cytology sample solutions from patients with PDAC consisted of 12 ERP cytology solutions and 25 EUS‐FNA cytology solutions, and their detection rates were 11/12 (91.7%) and 24/25 (96%), respectively. In 1 patient whose cancer could not be determined by EUS‐FNA histology, methylation of CDO1 promoter DNA could not detect cancer, either. The MV of patients with PDAC in the cytology solutions is shown in Figure 4C. The mean MV in patients with PDAC was 43.9 (range, 0–172.8), which was rather higher than the primary tumors. The MV for each UICC stage in the cytology solutions is shown in Figure 4D. Stage I included just 1 case, however, MV showed a high value of 18. Intriguingly, mean CDO1 MVs in pancreatic cytology samples were comparable with those in primary PDAC tissues, suggesting that the abundance of cancer cells in pancreatic cytology samples are as high as primary PDAC tissues.
Figure 4.
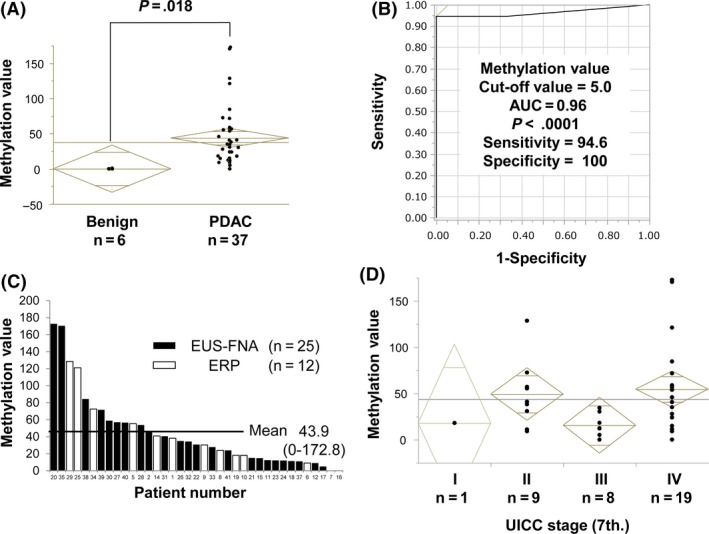
Methylation value (MV) of CDO1 promoter DNA in pancreatic cytology sample solutions. A, In the pancreatic cytology sample solutions, quantitative methylation‐specific PCR (Q‐MSP) assessment was carried out. There was a significant difference in CDO1 MV between pancreatic cytology sample solutions from patients with pancreatic ductal adenocarcinoma (PDAC) and those from pancreatic benign diseases (P = .018). B, Receiver operating characteristic curve of MV for the detection of PDAC. The area under the curve (AUC) represents the accuracy in discriminating PDAC from benign pancreatic diseases with high sensitivity and high specificity (AUC = 0.96, P < .0001). C, CDO1 MVs in all pancreatic cytology solutions from patients with PDAC. Mean of CDO1 MVs was 43.9 (range, 0.0‐172.8). Of those, 12 samples were collected from endoscopic retrograde pancreatography (ERP) cytology (white bars) and 25 samples were collected from endoscopic ultrasound‐fine needle aspiration (EUS‐FNA) cytology (black bars). Two cases were detected to be negative by Q‐MSP. One of them belonged to ERP cytology and the other belonged to EUS‐FNA cytology. D, Correlation of CDO1 methylation level to UICC stage (I‐IV) in pancreatic cytology sample solutions
4. DISCUSSION
As K‐ras mutation occurs in the precancerous lesion of PDAC,26 epigenetic alterations might be more promising than genetic alterations for the development of simple and less invasive surveillance systems for PDAC.7 Moreover, recent rigorous exploration of DNA methylation markers identified extraordinarily cancer‐specific and prevalent aberrations of DNA methylation in human cancer,9, 10, 11, 27 and revealed that the length range of promoter DNA methylation was larger than genetic alterations. DNA methylation is predominantly recognized around the wide range of the 5′‐position of cytosine residues (5mC or 5‐methyl cytosine) followed by guanine dinucleotide sequences across the CpG islands region in the genome. These CpG islands are usually unmethylated in normal cells, and allow active transcription of the gene involved. However, in cancer cells, they are frequently targeted for hypermethylation, an alteration that causes transcriptional repression of the associated gene, including tumor suppressors. Thus, tumor suppressor genes in primary tumor tissues are targets of cancer‐specific methylation, and such landmarks coincide with functional aspects.28
In this study, for the first time, we showed that CDO1 methylation in PDAC was cancer‐specific and extremely distinctive, because CDO1 methylation is hardly seen in the CN. This finding suggested CDO1 methylation occurs in PDAC cancer tissues in a very specific manner, and detection of CDO1 methylation is derived from a pinpoint tumor clone. The development of an endoscopic technique for collection of pancreatic fluid, termed endoscopic pancreatic function testing, has led to improved understanding of these alterations and is particularly helpful in characterizing pancreatic cancer. Moreover, its frequencies in PDAC tumor tissues are so high that it presents extraordinary potential to detect cancer cells in pancreatic juice in this current study. The AUC was beyond 0.9, and sensitivity of cancer detection was at least 95% in the pancreatic juice. This is remarkable for the diagnosis of PDAC because, for the same samples, cytology testing diagnosed PDAC in only 33% of samples using conventional biopsy and in 88% of samples using EUS‐FNA cytology, both of which are inferior to our current result.
In hepatopancreaticobiliary cancer, pancreaticoduodenectomy is needed for cure but it has a high mortality rate,29 and surgeons prefer to definitively confirm cancer preoperatively. However, biopsy samples of the hepatopancreaticobiliary tumors cannot be obtained directly without invasive procedures, because tumors of this type are located off the luminal mucosa. Pancreatic secretions thus play an important role in obtaining information with regard to a variety of pathophysiological mechanisms in the context of exocrine pancreatic disease. Cytological testing of the ERP wash solution is currently the main possible way to diagnose the tumor preoperatively. Investigators have found endoscopically collected pancreatic fluid to be a valuable biofluid for the purposes of translational science for PDAC.
Techniques such as proteomics, cytokines, genetic mutation, DNA methylation, and microRNA analyses can be utilized to gain a better understanding of the molecular characteristics of pancreatic diseases.30 In pancreatic cancer, recently, 4 sequential case‐control studies (discovery, technical validation, biological validation, and clinical piloting) were carried out to determine the diagnostic utility of the methylation of highly relevant gene CD1D (encoding a member of the CD1 family of transmembrane glycoproteins) methylation.8 Results of these studies showed that CD1D methylation in the pancreatic juice yielded an AUC value of 0.92 for patients with pancreatic cancer compared to patients with normal pancreas and chronic pancreatitis. CD1D methylation in the pancreatic juice detected pancreatic cancer with 75% sensitivity and 95% specificity. In our current study, CDO1 methylation in the pancreatic juice is likely to be superior to CD1D methylation, because its sensitivity of cancer detection is as frequent as 95% in the pancreatic juice. Endoscopic collection of pancreatic fluid is safe and relatively straightforward, permitting opportunities for longitudinal analysis of these translational markers throughout the course of disease.
4.1. Limitations
As an early diagnostic marker, it is necessary to examine lesions that are difficult to distinguish from malignant pancreatic tumors, such as intraductal papillary mucinous neoplasm and mucinous cystic neoplasm. We should clarify them in future experiments. Similarly, as a useful diagnostic marker for providing an early therapeutic opportunity and better outcomes, it might be better if MV could be used to diagnose Stage I PDAC. Unfortunately, in this study, there was only 1 Stage I patient, but the result was promising. The next studies are expected to solve these issues.
In conclusion, although our data are still at a pilot study stage, and a large‐scale validation study is needed, methylation of CDO1 promoter DNA is extremely specific for PDAC, and accumulates with PDAC tumor progression. It could therefore be a definitive diagnostic marker of PDAC in ERP solutions and could facilitate early detection of PDAC, which is one of the most dismal among human cancers.
DISCLOSURE
The authors have no conflict of interest.
ACKNOWLEDGMENTS
We thank Miss Tomomi Miyake for her technical assistance. This research was supported by the Japan Society for the Promotion of Science KAKENHI (Grant No. JP18K16373 and JP18K08656). This study was also supported by the Integrative Research Program grant of the Graduate School of Medical Science, Kitasato University, and a Parents’ Association Grant of Kitasato University School of Medicine. We would like to thank Editage for English language editing.
Nishizawa N, Harada H, Kumamoto Y, et al. Diagnostic potential of hypermethylation of the cysteine dioxygenase 1 gene (CDO1) promoter DNA in pancreatic cancer. Cancer Sci. 2019;110:2846–2855. 10.1111/cas.14134
Nishizawa and Harada equally contributed to this work.
Clinical Trial Registration: UMIN‐CTR Clinical Trial number: UMIN000036413.
Funding information The Integrative Research Program grant of the Graduate School of Medical Science, Kitasato University; Japan Society for the Promotion of Science, Grant/Award Number: JP18K16373; Parents’ Association Grant of Kitasato University School of Medicine
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87‐108. [DOI] [PubMed] [Google Scholar]
- 2. Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913‐2921. [DOI] [PubMed] [Google Scholar]
- 3. Lopez NE, Prendergast C, Lowy AM. Borderline resectable pancreatic cancer: definitions and management. World J Gastroenterol. 2014;20:10740‐10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605‐1617. [DOI] [PubMed] [Google Scholar]
- 5. Oettle H, Neuhaus P, Hochhaus A, et al. Adjuvant chemotherapy with gemcitabine and long‐term outcomes among patients with resected pancreatic cancer: the CONKO‐001 randomized trial. JAMA. 2013;310:1473‐1481. [DOI] [PubMed] [Google Scholar]
- 6. Eloubeidi MA, Chen VK, Eltoum IA, et al. Endoscopic ultrasound‐guided fine needle aspiration biopsy of patients with suspected pancreatic cancer: diagnostic accuracy and acute and 30‐day complications. Am J Gastroenterol. 2003;98:2663‐2668. [DOI] [PubMed] [Google Scholar]
- 7. Yang J, Li S, Li J, et al. A meta‐analysis of the diagnostic value of detecting K‐ras mutation in pancreatic juice as a molecular marker for pancreatic cancer. Pancreatology. 2016;16:605‐614. [DOI] [PubMed] [Google Scholar]
- 8. Kisiel JB, Raimondo M, Taylor WR, et al. New DNA methylation markers for pancreatic cancer: discovery, tissue validation, and pilot testing in pancreatic juice. Clin Cancer Res. 2015;21:4473‐4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamashita K, Upadhyay S, Osada M, et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2:485‐495. [DOI] [PubMed] [Google Scholar]
- 10. Kim MS, Chang X, Yamashita K, et al. Aberrant promoter methylation and tumor suppressive activity of the DFNA5 gene in colorectal carcinoma. Oncogene. 2008;27:3624‐3634. [DOI] [PubMed] [Google Scholar]
- 11. Brait M, Ling S, Nagpal JK, et al. Cysteine dioxygenase 1 is a tumor suppressor gene silenced by promoter methylation in multiple human cancers. PLoS ONE. 2012;7:e44951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jeschke J, O'Hagan HM, Zhang W, et al. Frequent inactivation of cysteine dioxygenase type 1 contributes to survival of breast cancer cells and resistance to anthracyclines. Clin Cancer Res. 2013;19:3201‐3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ushiku H, Yamashita K, Katoh H, et al. Promoter DNA methylation of CDO1 gene and its clinical significance in esophageal squamous cell carcinoma. Dis Esophagus. 2017;30:1‐9. [DOI] [PubMed] [Google Scholar]
- 14. Kojima K, Yamashita K, Ushiku H, et al. The clinical significance of cysteine dioxygenase type 1 methylation in Barrett esophagus adenocarcinoma. Dis Esophagus. 2017;30:1‐9. [DOI] [PubMed] [Google Scholar]
- 15. Ooki A, Maleki Z, Tsay JJ, et al. A panel of novel detection and prognostic methylated DNA markers in primary non‐small cell lung cancer and serum DNA. Clin Cancer Res. 2017;23:7141‐7152. [DOI] [PubMed] [Google Scholar]
- 16. Ushiku H, Yamashita K, Ema A, et al. DNA diagnosis of peritoneal fluid cytology test by CDO1 promoter DNA hypermethylation in gastric cancer. Gastric Cancer. 2017;20:784‐792. [DOI] [PubMed] [Google Scholar]
- 17. Minatani N, Waraya M, Yamashita K, et al. Prognostic significance of promoter DNA hypermethylation of cysteine dioxygenase 1 (CDO1) gene in primary breast cancer. PLoS ONE. 2016;11:e0144862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Igarashi K, Yamashita K, Katoh H, et al. Prognostic significance of promoter DNA hypermethylation of the cysteine dioxygenase 1 (CDO1) gene in primary gallbladder cancer and gallbladder disease. PLoS ONE. 2017;12:e0188178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kojima K, Nakamura T, Ohbu M, et al. Cysteine dioxygenase type 1 (CDO1) gene promoter methylation during the adenoma‐carcinoma sequence in colorectal cancer. PLoS ONE. 2018;13:e0194785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deckers IA, Schouten LJ, Van Neste L, et al. Promoter methylation of CDO1 identifies clear‐cell renal cell cancer patients with poor survival outcome. Clin Cancer Res. 2015;21:3492‐3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meller S, Zipfel L, Gevensleben H, et al. CDO1 promoter methylation is associated with gene silencing and is a prognostic biomarker for biochemical recurrence‐free survival in prostate cancer patients. Epigenetics. 2016;11:871‐880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feber A, Arya M, de Winter P, et al. Epigenetics markers of metastasis and HPV‐induced tumorigenesis in penile cancer. Clin Cancer Res. 2015;21:1196‐1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang RL, Su PH, Liao YP, et al. Integrated epigenomics analysis reveals a DNA methylation panel for endometrial cancer detection using cervical scrapings. Clin Cancer Res. 2017;23:263‐272. [DOI] [PubMed] [Google Scholar]
- 24. Mandelker DL, Yamashita K, Tokumaru Y, et al. PGP9.5 promoter methylation is an independent prognostic factor for esophageal squamous cell carcinoma. Cancer Res. 2005;65:4963‐4968. [DOI] [PubMed] [Google Scholar]
- 25. Ooki A, Yamashita K, Kikuchi S, et al. Potential utility of HOP homeobox gene promoter methylation as a marker of tumor aggressiveness in gastric cancer. Oncogene. 2010;29:3263‐3275. [DOI] [PubMed] [Google Scholar]
- 26. Laghi L, Orbetegli O, Bianchi P, et al. Common occurrence of multiple K‐RAS mutations in pancreatic cancers with associated precursor lesions and in biliary cancers. Oncogene. 2002;21:4301‐4306. [DOI] [PubMed] [Google Scholar]
- 27. Yamashita K, Sakuramoto S, Watanabe M. Genomic and epigenetic profiles of gastric cancer: potential diagnostic and therapeutic applications. Surg Today. 2011;41:24‐38. [DOI] [PubMed] [Google Scholar]
- 28. Koch A, Joosten SC, Feng Z, et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol. 2018;15:459‐466. [DOI] [PubMed] [Google Scholar]
- 29. Kimura W, Miyata H, Gotoh M, et al. A pancreaticoduodenectomy risk model derived from 8575 cases from a national single‐race population (Japanese) using a web‐based data entry system: the 30‐day and in‐hospital mortality rates for pancreaticoduodenectomy. Ann Surg. 2014;259:773‐780. [DOI] [PubMed] [Google Scholar]
- 30. Hart PA, Topazian M, Raimondo M, et al. Endoscopic pancreas fluid collection: methods and relevance for clinical care and translational science. Am J Gastroenterol. 2016;111:1258‐1266. [DOI] [PMC free article] [PubMed] [Google Scholar]


