Abstract
A Galactomyces geotrichum strain with lipolytic activity was isolated and identified by the analysis of internal transcribed spacer (ITS) sequence of 18 s rDNA. Full-length lipase gene of this stain is composed of 1692 base pairs (bp) without intron, which encodes a 563-amino-acid protein. A catalytic triad (Ser217–Glu354–His463) was found by constructing the three-dimensional structure of the lipase. In shake flasks, the lipase (LIP) catalytic activity in the supernatant of the recombinant Pichia pastoris increased 48.7% by codon optimization. LIP purified by anion exchange column showed a single protein band on 12% SDS-PAGE. The molecular weight (MW) of LIP was approximately 62 kDa. The specific activity of purified LIP reached 1257.9 U/mg. The optimum temperature and pH of LIP catalysis were 45 °C and pH 8.2, respectively. LIP was stable over the pH range of 4.2–11.2. LIP maintained its activity constantly at 40 °C and 50 °C for 120 min. Zn2+ inhibited LIP activity; Ba2+, Mn2+, Ca2+ and EDTA increased the enzyme activity. Referring the amount of hydrolyzed olive oil by LIP as 100%, various oils including lard, peanut oil, rapeseed oil, sunflower oil, soybean oil and linseed oil were efficiently hydrolyzed by 17.24 ± 1.34%, 40.34 ± 2.56%, 105.86 ± 2.78%, 115.51 ± 2.32%, 116.21 ± 2.15%, 120.69 ± 1.98%, respectively. The characteristics allow LIP as a potential biocatalyst in various fields of industry.
Keywords: Galactomyces geotrichum, Lipase, Pichia pastoris, Codon optimization, Characterization
Introduction
Lipases (triacylglycerol acylhydrolase, EC.3.1.1.3) have a catalytic effect on the hydrolysis, esterification, interesterification, acidolysis and alcoholysis of esters (Mohamed et al. 2011). Lipases are widely applied in the industries of textile, detergent, paper, food processing, tea processing and the production of biodiesel (Hasan et al. 2006). Lipases can be isolated from animals, plants and microorganisms. Lipases produced by bacteria and fungi are extensively available (Mazhar et al. 2017). Fungal lipase is an important enzyme for its industrial application (Singh and Mukhopadhyay 2012).
The lipase from Geotrichum candidum has shown the substrate specificity for unsaturated long-chain fatty acids containing double bond(s) at the position of cis-9 or cis-9, 12 (Baillargeon et al. 1989). Galactomyces geotrichum is the teleomorph of G. candidum (Yan et al. 2007). And G. geotrichum lipases are also specified for esters of unsaturated long-chain fatty acids (Phillips et al. 1995). Galactomyces geotrichum lipases have great potential in hydrolysis of various oils (Maldonado et al. 2016). However, naturally produced G. geotrichum lipases in the original strains usually show low lipase productivity and poor tolerance to normal factors like temperature, pH and ions for industries (Shu et al. 2010).
Heterologous expression is a strategy to improve the yield of lipase production. Commonly used system for heterogenous expression is the eukaryotic expression system, such as Pichia pastoris and Saccharomyces cerevisiae (Zheng et al. 2019; Ueda et al. 2002). Literaturally, a lipase gene from G. candidum Y162 has been cloned and successfully expressed in P. pastoris. The lipase activity reached 55 U/mL (Yan et al. 2008). In general, expression level of the exogenous gene is low due to less frequently used codons in P. psatoris. Codon usage bias of P. pastoris has been reported based on the genome sequence data (De Schutter et al. 2009). The common method to improve expression level of the exogenous gene is to replace less frequently used codons with preferable codons. The production of heterologous proteins encoded by exogenous genes has been successfully improved by codon optimization (Wang et al. 2018; Lu et al. 2016; Sun et al. 2016) and secretion helper factors (Wang et al. 2019). There is much elevation potential for improvement of the productivity of G. geotrichum lipases for an industrial biocatalyst.
In this study, a lipase-producing strain was selected and identified as G. geotrichum. The lipase gene of this strain was expressed in P. pastoris by codon optimization. Then, LIP was purified by anion exchange column. And biochemical characteristics of LIP were analyzed. Different types of oils were selected to explore the hydrolysis ability of LIP.
Materials and methods
Strains, vectors, and chemicals
Escherichia coli (E. coli) Top10 competent cell (Beijing, China), pSURE-T vector (Beijing, China), yeast expression vector pPICZαA (preserved by our laboratory) and P. pastoris X-33 (preserved by our laboratory) were used in this experiment. Yeast extract and peptone were purchased from Oxoid (Hampshire, England). LA Taq DNA polymerase, dNTPs, T4 DNA ligase, Xba I, EcoRI, Sac I, BspHI were bought from TaKaRa (Otsu, Japan). All other chemicals used in this study which were not specifically mentioned were analytical grade and commercially available.
Isolation of the lipase-producing strain
Soil contaminated with oil was collected from the canteen drain in Ministry Agriculture of Feed Industry Centre (MAFIC) of Chinese Agricultural University. The sample was incubated in the enrichment culture (0.50% olive oil, 0.05% MgSO4, 0.05% NaCl, 0.20% yeast extract, 0.15% KH2PO4; pH 8.0) at 28 °C, 150 rpm for 48 h. Then, the diluted liquid was plated on the Potato Dextrose Agar (PDA) medium. Single colonies were picked up and cultured on new PDA plates containing 0.01% of Rhodamine B and 0.50% of olive oil for 48 h. Strains showed transparent rings were fermented in enzyme producing medium (6.00% soy flour, 4.00% soybean oil, 0.20% (NH4)2SO4, 0.10% KH2PO4, 0.05% MgSO4, 1.00% glucose; pH 8.0) at 28 °C and 150 rpm for 72 h.
Lipase activity assay
Alkali titration method was used to determine the lipase activity (Yu et al. 2007). Olive oil was used as the substrate. Olive oil was emulsified by polyving akohol for 8–10 min as pretreatment. The reaction was carried out at pH 8.2 and 45 °C for 15 min. The amount of fatty acids was titrated with 0.05 M NaOH. One unit of lipase activity was defined as the amount of enzyme that released 1 μmol free acid per minute at the measuring condition.
Identification of the lipase-producing strain
Genomic DNA was extracted from the lipase-producing strain. Internal transcribed spacer 1 and 2 (ITS1 and ITS2) of rDNA were amplified by using primers ITS1-F (5′-TCCGTAGGTGAACCTGCGG-3′)/ITS1-R (5′-GCTGCGTTCTTCATCGATGC-3′) and ITS2-F (5′-GTATCGATGAAGAACGCAGC-3′)/ITS2-R (5′-TCCTCCGCTTATTGATATGC-3′), with the procedure of reaction: 94 °C for 3 min, 30 cycles (94 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min), finally 72 °C for 5 min. The PCR products were recovered from 0.8% of agrose gel and then ligated into pSURE-T vector. The ligation products were transformed into E. coli Top10. The positive colonies were selected and cultured on Luria–Bertani (LB) plates (1.00% tryptone, 0.50% NaCl, 0.50% yeast extract, and 1.50% agar) containing 100 μg/mL of Ampicillin. The recombinant plasmids were analyzed with universal M13 primers.
Cloning of lipase gene from G. geotrichum Mafic-0601
Total DNA of G. geotrichum Mafic-0601 was used as the template for the amplification of the lipase gene. The primers, Lip-F (5′-ATGGAWTCCAAAAGC-3′) and Lip-R (5′-TTAACCGTAGAGATT AA-3′), were designed according to reported G. geotrichum lipase gene sequences (GenBank accession no. DQ313172 and JX074060). The PCR reaction procedure was set as a denaturation step at 95 °C for 3 min, 35 cycles of 94 °C 30 s, 58 °C 30 s, 72 °C 1 min, followed by an extension step at 72 °C for 5 min. Then, the purified PCR products were ligated into pSURE-T vector. The ligation products were transformed into E. coli Top10. Recombinant plasmid, pSURE-T-lip, was analyzed by using M13 primers.
Construction of lipase engineering strains
According to the full-length gene sequence of G. geotrichum Mafic-0601, Lip-mat-F (5′-CGGAATTCCAGGCCCCCACGGCCGTT-3′)/Lip-mat-R (5′-GCTCTAGATTAACCGTAGAGATTA AGC-3′) were synthesized to clone the mature lipase gene (lip). The pSURE-T-lip was used as the template for PCR amplification. The PCR reaction was performed for denaturation at 95 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 58 °C for 30 s and 72 °C for 1 min and extension at 72 °C for 5 min. The mature lip that digested with EcoRI and Xba I was ligated into pPICZαA vector that pre-treated with the same enzymes. The recombinant plasmid, pPIC-lip-wt, was identified by enzymes digestion and sequence analysis. The mature lip gene sequence was optimized according to the codon usage bias of P. pastoris by DNA2.0 (Comso Bio Ltd) and synthesized by Tsingke corp (Beijing). The amino acid sequence of mature lip was not changed after code optimization. The optimized gene was ligated into pPICZαA vector to construct recombinant plasmid, pPIC-lip-opt.
To determine the three-dimensional (3-D) structure of the lipase, deduced amino acid sequence was input and the structure was predicted by using SWISS-MODEL (https://www.swissmodel.expasy.org/). The Protein Data Bank (PDB; http://www.rcsb.org/pdb/) was used as the template sources.
Transformation and fermentation in shake flasks
pPIC-lip-wt linearized with BspHI and pPIC-lip-opt linearized with Sac I were transformed into P. pastoris X-33 competent cell by electroporation (2000 V, 5 ms), respectively. Transformants were cultured on YPDS plates (1% yeast extract, 2% peptone, 2% dextrose, 1 M sorbitol, and 2% agar) with 100 μg/mL of zeocin. Recombinant strains, X-33/lip-wt and X-33/lip-opt were picked up and kept on new YPD plates.
Single colonies of X-33/lip-wt and X-33/lip-opt were incubated in 20 mL of buffered glycerol-complex (BMGY) medium (100 mM potassium phosphate, 0.34% YNB, 4 × 10−5% biotin, 1.00% glycerol, 1.00% yeast extract, and 2.00% peptone; pH 6.0) at 28 °C and 250 rpm for 24 h. The cells were harvested by centrifugation at 7500 × g for 5 min at room temperature. The cell pellet was re-suspended in buffered methanol-complex (BMMY) medium (100 mM potassium phosphate, 0.34% YNB, 4 × 10−5% biotin, 0.50% methanol, 1.00% yeast extract, and 2.00% peptone; pH 6.0). The cultivation was maintained at 28 °C, 250 rpm for 72 h. Methanol (0.5%, v/v) was added to the culture every 24 h to induce protein expression. After collecting the supernatant of the culture, the expression of the protein was confirmed by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE).
Protein purification and identification
The culture supernatant was harvested by centrifugation at 7500×g for 5 min, then precipitated with 70% saturated ammonium sulfate on ice. The precipitate was collected by centrifugation at 15,000×g for 30 min and then resuspended in 5 mL of buffer A (20 mM Tris–HCl, pH 8.0). The solution dialyzed overnight at 4 °C was centrifuged at 3000 × g for 30 min and then purified using UNOspHere Q Strong Anion Exchange Support (1 cm × 20 cm, Bio-Rad). The column was pre-equilibrated by adding five volumes of column buffer A. 2 mL of dialysis liquid was loaded in the column. The non-absorbed protein was washed by buffer A. The linear gradient of buffer B (0–1 mol/L NaCl, 20 mmol/L Tris–HCl, pH 8.0) was applied at a flow rate of 1.5 mL/min. The protein concentration was measured by using Micro-BCA Protein Assay Reagent (Pierce USA).
Protein digestion was performed using FASP method with modifications (Wisniewski et al. 2009). Nanospray ESI–MS was performed on a Thermo Q-Exactive high-resolution mass spectrometer (Thermo Scientific, Waltham, MA, USA) with 70,000 MS scan resolution, 17,500 MS/MS scan resolution and top-10 MS/MS selection. Raw data from the mass spectrometer were preprocessed with Mascot Distiller 2.4 for peak picking. The resulted peak lists were searched in the NCBI database using a Mascot 2.5 search engine.
Enzyme properties
The effect of pH on LIP activity was investigated at 45 °C with pH ranging from 2.2 to 8.2 (Na2HPO4-citric acid) and 9.2 to 11.2 (glycine-sodium hydroxide buffer). The pH stability was determined by incubating LIP in buffers of different pH (from pH 2.2 to 11.2) for 12 h and 24 h at room temperature. Then, the residual activity was measured at 45 °C and pH 8.2.
To determine the optimum temperature of LIP, the relative activity of LIP was measured at pH 8.2 from 30 to 60 °C. To determine the thermal stability, LIP was incubated at 40 °C and 50 °C for 30, 60, 90 and 120 min, 55 °C for 10, 20 and 30 min, 60 °C for 5, 10, 15 and 20 min. The samples were collected at different time intervals for the measurement of residual activity as mentioned above.
To determine the effects of chemicals on LIP activity, the enzyme was incubated in 10 mM/L solutions of CuSO4, KCl, ZnSO4, MnSO4, MgCl2, FeSO4, MgCl2, CaCl2, BaCl2 and EDTA for 2 h, respectively. The residual activity was determined under the optimal condition (45 °C and pH 8.2).
Comparison of hydrolysis rate on various substrates
Various oils including lard, rapeseed oil, sunflower oil, soybean oil, peanut oil and linseed oil were purchased from Chaoshifa supermarket of Beijing. These food grade lipids were used as substrates to determine the hydrolysis ability of LIP. All lipids were adjusted to the emulsions of the same concentration. One milliliter of emulsified oil was mixed with an equal volume of buffer (pH 8.2) and 4 U of the enzyme was added. The mixture was kept in a bath for 10 min at 45 °C. The amount of free fatty acids was measured by using the titration method (0.05 M NaOH). Each experiment was performed in triplicate.
Nucleotide sequence accession number
The nucleotide sequence of lipase gene from G. geotrichum Mafic-0601 was assigned to GenBank accession number KJ607958.
Results and discussion
Isolation and identification of the lipase-producing strain
Strain Mafic-0601 was isolated according to its ability to hydrolyze olive oil. The strain was identified as G. geotrichum by ITS sequence analysis. The sequence of this strain is 100% homologous to G. geotichum partial ITS sequences (GenBank accession numbers KF768312, KF713521 and HG532089; data not shown). Mafic-0601 was incubated in enzyme producing medium for 72 h. The lipase activity of this strain reached 22.77 ± 0.8 U/mL.
Many fungi including Penicillium aurantiogriseum, Rhizopus, Aspergillus carneu, Candida cylindracea can all produce lipases (Lima et al. 2003; Cordova et al. 1998; Kaushik et al. 2006; Kim and Hou 2006). The lipase activities of the natural strains generally range from 10 to 40 U/mL. For example, Cihangir and Sarikaya (2004) isolated a lipase-producing strain from the soil of Turkey. The lipase activity of this strain reached 17 U/mL. Colen et al. (2006) isolated a lipase-producing strain from the soil of Brazilian savanna. The lipase activity of the strain reached 27.7 U/mL. The lipase activity of Mafic-0601 was 22.77 ± 0.8 U/mL, which was in the range of lipase activities of natural strains.
Lipase gene cloning of Mafic-0601
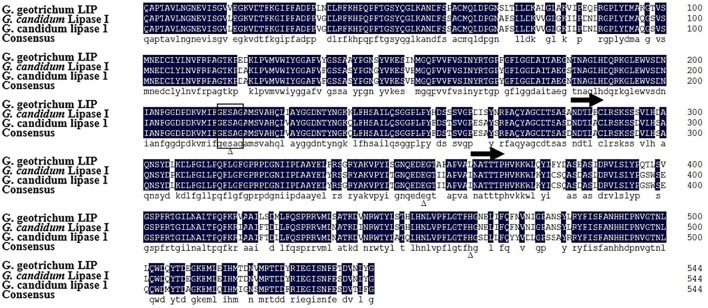
The full-length sequence of lip gene was amplified by PCR. The lip gene with no intron is composed of 1692 base pairs (bp). The protein deduced from DNA sequence consisted of 563 amino acid residues with a predicted molecular weight of 61.5 kDa. A signal peptide (19 amino acids) presented at the N-terminus of the protein was predicted (http://www.cbs.dtu.dk/services/SignalP/). Two N-glycosylation sites (N-X-S/T) have been identified by amino acid sequence analysis (one-way arrows represented in Fig. 1). The protein sequence of LIP was submitted to GenBank (accession no. AJF11715). LIP showed 89% identical to the lipase of G. candidum FZ-4 (GenBank accession no. ALJ02547), and 85% identical to the lipase of G. candidum ch-3 (GenBank accession no. ABN64097). The amino acid sequence analysis revealed a conserved substrate binding region (Gly-Glu-Ser-Ala-Gly), the common sequence feature of the serine hydrolase (Fig. 1).
Fig. 1.
Alignment of amino acid sequence of mature mafic-0601 lipase with other lipases. Identical amino acids were shown with lowercase letters at the bottom. The sequence in box was the conservative motif G–x–S–x–G. Catalytic sites that containing Ser217, Glu354and His463 were marked by triangles. Two glycosylation sites (N-X-S/T) have been identified by amino acid sequence analysis (one-way arrows represented)
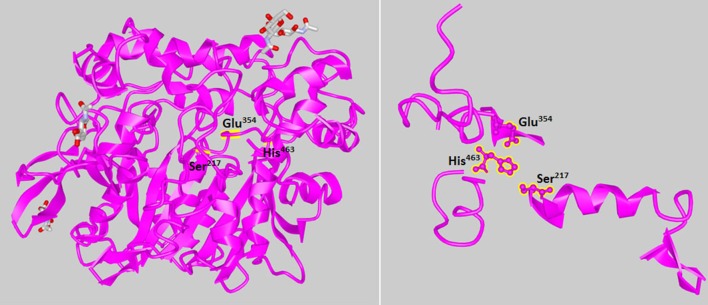
By searching the SWISS-MODEL, a crystal structure of the G. candidum lipase (Accession Number of 1THG) was a preferred template for homology modeling, since their amino acid sequence shared 99% identity. The 3D structure of LIP was shown in Fig. 2. The lipases from different sources share limited amino acid homology but similar structures, contain eight sheets, six helixes and a catalytic triad (Ser-Asp/Glu-His) (Fischer and Pleiss 2003). LIP also contained this catalytic triad (Ser217–Glu354–His463).
Fig. 2.
3D model of mafic-0601 lipase generated SWISS-MODEL
LIP over-expression by codon optimization in P. pastoris
To reach high expression level of LIP, mature lip gene was optimized according to the codon bias of P. pastoris. 348 bp of the 1635 bp were replaced (Fig. 3). The synthetic optimized sequence (lip-opt) shared 78.7% identity to the wild-type lipase gene (lip-wt). The enzyme activities of the strains harboring lip-wt and lip-opt gene were compared in shake flasks. After 72 h of methanol induction, the lipase activity of X-33/lip-opt (93.7 ± 1.5 U/mL) was higher than that of X-33/lip-wt (63.0 ± 0.9 U/mL). However, there was no significant difference in biomass between X-33/lip-wt and X-33/lip-ot (data not shown). And the expression of proteins in X-33/lip-wt and X-33/lip-opt was analyzed by SDS-PAGE (Fig. 4). Methylotrophic yeast P. pastoris has been demonstrated as an ideal heterologous expression system due to its high expression level, efficient secretion ability and post-transcription modification (appropriate folding and glycosylation). Codon optimization has been successfully utilized to express Aspergillus niger lipase (lip2) gene in P. pastoris (Yang and Liu 2010). In this study, the lipase activity of X-33/lip-opt also effectively increased by codon optimization.
Fig. 3.
Alignment of wild type mafic-0601 lipase gene (lip-wt) and optimized gene (lip-opt). Identical bases were highlighted in black
Fig. 4.
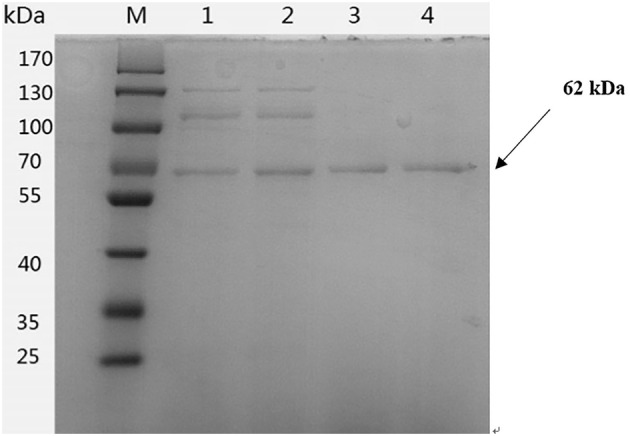
SDS-PAGE analysis of different expressed recombinant X-33/lip-opt and X-33/lip-wt proteins. Lane M: protein marker; Lane 1–2: recombinant X-33/lip-opt protein in the culture supernatant, Lane 3–4: recombinant X-33/lip-wt protein in the culture supernatant. wt wild-type, opt codon-optimized
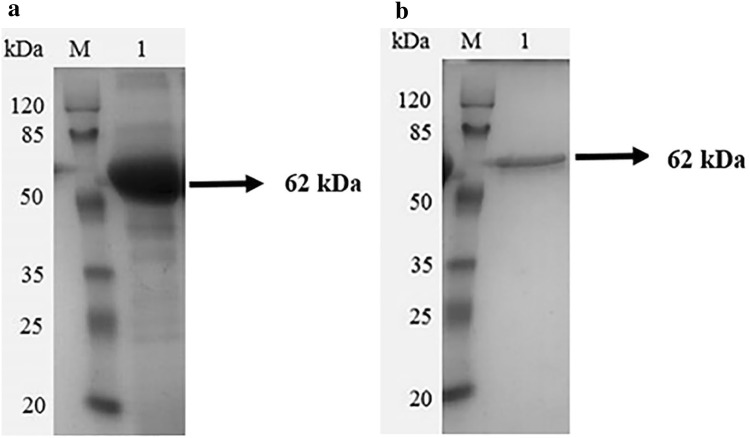
The recombinant lipase (LIP) was precipitated from culture by ammonium sulfate and then purified by anion exchange column. The single protein band was observed on a 12% SDS-PAGE, showed its purity and homogeneity (Fig. 5). SDS-PAGE analysis showed that the molecular weight (MW) of LIP was approximately 62 kDa. The MW of LIP was consistent with reported G. geotrichum lipases (50–70 kDa) (Bertolini et al. 1995). Since P. pastoris secrets low level of native proteins and expression plasmid with α-factor (coding signal peptide) was used in this study. Exogenous LIP was secreted into the supernatant of the culture medium of P. pastoris. Thus, LIP was approximately free from contamination of endogenous protein. The specific activity of LIP purified by anion exchange column reached 1258.21 U/mg (Table 1). The protein band excised from SDS-PAGE was confirmed by LC/mass/mass. Peptide mass fingerprints analysis were identical to the predicted amino acid of LIP (data not shown). The specific activity of LIP was higher than reported G. geotrichum lipases (Fernandez et al. 2006; Yan et al. 2007; Qiao et al. 2017a, b) expressed in P. pastoris (165.90–813.20 U/mg). The high specific activity of LIP is meaningful for its potential application in various fields such as food, feed manufacture, cosmetic and detergent.
Fig. 5.
SDS-PAGE analysis of the purified mafic-0601 lipase that expressed by recombinant P. pastoris. a Lane M: protein marker; Lane 1: crude recombinant lipase of X-33/lip-opt; b Lane M: protein marker; Lane 1, purified recombinant lipase of X-33/lip-opt. opt: codon-optimized
Table 1.
Enzyme activities and protein concentration of LIP during the course of purification of the enzyme protein
| Purification step | Total protein (mg) | Total activity (U) | Specific activity (U/mg) | Fold purification | Yield (%) |
|---|---|---|---|---|---|
| Culture supernatant | 144.00 ± 2.00 | 32,217 ± 128 | 223.73 ± 2.22 | 1.00 | 100.00 |
| (NH4)2SO4 precipitation | 91.90 ± 2.30 | 17,905 ± 153 | 194.83 ± 3.21 | 0.87 | 55.58 |
| Q-SeqHacel | 0.67 ± 0.03 | 843 ± 58 | 1258.21 ± 30.30 | 5.38 | 2.62 |
Values were expressed as mean ± standard deviation
Characterization of the recombinant lipase
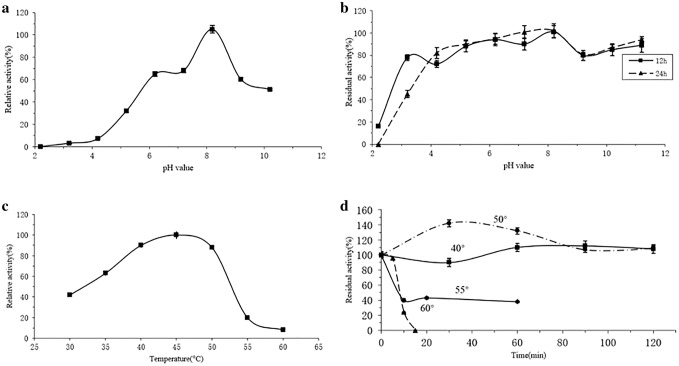
The optimal pH of LIP was pH 8.2 at 45 °C (Fig. 6a). In the pH range of 6.0–10.0, LIP showed above 60% catalytic activity. When pH decreased to 4.0, relatively low activity of LIP was detected. The pH stability was also determined by incubating LIP in buffers for 12 h and 24 h at various pH. The residual activity of LIP remained above 70% in the range of 4.2–11.2 (Fig. 6b). Generally, the optimum pH of Geotrichum lipases were from 6.0 to 7.0 (Maldonado et al. 2016). Compared with the properties of reported Geotrichum lipases, LIP has shown higher optimum pH (8.2) and broader pH stability (4.2–11.2). LIP exhibited a stronger preference for high pH than most G. geotrichum lipases (Yan et al. 2007; Qiao et al. 2017a). And alkaline lipases were considered popular in the industries because of its tolerance towards alkalinity (Bora et al. 2013). Besides its alkaline resistance, LIP in this study also showed higher acid resistance than reported Geotrichum lipases (Maldonado et al. 2016). Acidic lipases now are rarely reported. Although acidic lipases are highly useful in fields of industries, such as the assistance of fat digestion in animal feed (Zhang et al. 2019).
Fig. 6.
a Effect of pH on the enzyme activity of LIP. The LIP activity was investigated at 45 °C by varying pH from 2.2 to 11.2. b pH stability of LIP. The enzyme was incubated at pH ranging from 2.2 to 11.2 for 12 h and 24 h at room temperature (about 25 °C). Then the residual activity was measured at 45 °C and pH 8.2. c Effect of temperature on the enzyme activity of LIP. The relative activity of the enzyme was measured at pH 8.2 from 30 to 60 °C. d Temperature stability of LIP. The enzyme was incubated at 40 °C and 50 °C for 120 min, 55 °C for 60 min, and 60 °C for 15 min, respectively. Then the residual activity was measured at 45 °C and pH 8.2. Values were expressed as mean ± standard deviation
The optimal temperature of LIP was 45 °C (Fig. 6c). From 30 to 55 °C, above 40% relative activity of LIP was detected. For investigating the temperature stability, LIP was incubated at different temperatures (40–60 °C) for different time duration (Fig. 6d). At 40 and 50 °C, LIP maintained its activity consistency within 120 min. When kept at 55 °C, the enzyme activity remained 40% after 10 min. The activity of LIP was lost after incubation for 15 min at 60 °C. Literaturally, Geotrichum lipases showed optimum temperatures between 20 and 40 °C (Maldonado et al. 2016). Deactivation of reported G. geotrichum lipases expressed in P. pastoris occurred rapidly when the temperature was higher than 60 °C (Yan et al. 2007; Qiao et al. 2017a). In this study, the activity of LIP maintained for 15 min at 60 °C. The selection of the lipase for application is due to its stability under different circumstances. This significant characteristic allows LIP superior as a thermostable lipase for potentially applied in industries, with low-cost input and high productive output.
Effects of various metal ions and EDTA (10 mM) on enzyme activity were detected (Table 2). Zn2+ inhibited LIP activity by 23.4 ± 1.9%. Ba2+, Mn2+, Ca2+ and EDTA increased the enzyme activity by 17.3 ± 2.2%, 10.5 ± 2.1%, 16.6 ± 1.8%, 18.5 ± 2.3%, respectively. The reason why the enzyme activity was activated or inhibited by ions and EDTA has not been elucidated. Qiao et al. (2017a) reported a possible reason for Ca2+ was formation of fatty acid calcium salts, which might facilitate the hydrolysis of esters. And it also has been reported that the effect of EDTA on the lipase activity may be stimulatory or null (Carvalho et al. 2013).
Table 2.
Effects of metal ions and EDTA (final concentration: 10 mmol/L) on catalytic activity of LIP
| Chemical agents | Relative activity (%) |
|---|---|
| No addition | 100.0 ± 1.2 |
| CuSO4 | 101.9 ± 2.3 |
| KCl | 109.2 ± 2.5 |
| ZnSO4 | 76.6 ± 1.9 |
| MnSO4 | 110.5 ± 2.1 |
| MgCl2 | 105.7 ± 2.3 |
| BaCl2 | 117.3 ± 2.2 |
| CaCl2 | 116.6 ± 1.8 |
| FeSO4 | 103.8 ± 1.8 |
| EDTA | 118.5 ± 2.3 |
Values were expressed as mean ± standard deviation
Comparison of hydrolysis rate on various substrates
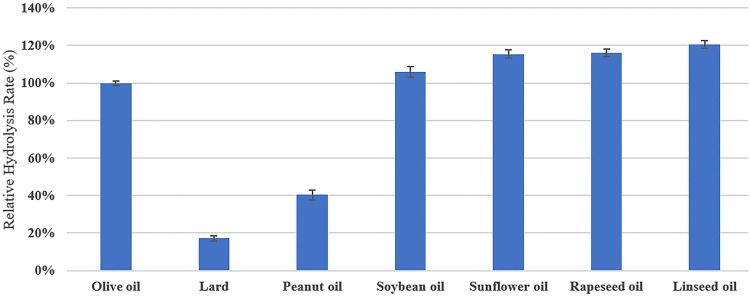
In this study, lard, peanut oil, rapeseed oil, sunflower oil, soybean oil and linseed oil were hydrolyzed by LIP. The relative hydrolysis rates of these oils were compared with that of olive oil regarded as 100%. From the result (Fig. 7), the relative hydrolysis rates of lard, peanut oil, rapeseed oil, sunflower oil, soybean oil and linseed oil were 17.24 ± 1.34%, 40.34 ± 2.56%, 105.86 ± 2.78%, 115.51 ± 2.32%, 116.21 ± 2.15%, 120.69 ± 1.98%, respectively.
Fig. 7.
Comparison of hydrolysis capability by LIP on various oils. Hydrolysis rate of olive oil was regarded as 100%. Values were expressed as mean ± standard deviation
The oils from animals or vegetables are composed of different fatty acids. It has been demonstrated that hydrolysis rates of oils were influenced by the oil type and lipase source (Santos et al. 2013). Gupta et al. (2012) found a maximum hydrolyzing ability for olive oil of 62.37 U/mg, which was more than that of castor oil (24.26 U/mg). The hydrolytic activity of the lipase isolated from C. gloesporioides was low for lard and high for tributyrin (Colen et al. 2006). The lipase from Penicillium camembertii Thom PG-3 showed the high hydrolysis degree for cotton seed oil and low for beef tallow (Tan et al. 2004). Generally, the hydrolysis rate of the lipase toward vegetable oil is higher than animal oil. It may be associated with the length of carbon chains (Pogori et al. 2008). And it also has been reported that G. geotrichum lipases might have great potential in utilizing various vegetable oils because of its high specificity for unsaturated long chain fatty acids and esters (Maldonado et al. 2016). Our results in this study are consistent with the reported results. Isolated lipase-producing fungus is effective for hydrolyzing a broad spectrum of substrates, which agrees with Yan et al. (2007). LIP was considered to have potential application prospects in the biocatalysis industry.
Conclusion
In this study, G. geotrichum Mafic-0601 that produces lipase was isolated and identified by ITS sequence. A lipase gene (GenBank accession number KJ607958) was cloned from this strain and expressed in P. pastoris by codon optimization. The lipase yield of LIP (93.7 ± 1.5 U/mL) was 4.1 times higher than the original strain (22.77 ± 0.8 U/mL). LIP exhibited optimum activity at pH 8.2 and 45 °C. Compared with other G. geotrichum lipases, LIP has shown higher temperature stability (40–60 °C) and broader pH stability (4.2–11.2). Ba2+, Mn2+, Ca2+ and EDTA enhanced LIP activity. LIP could also be effective for hydrolyzing a broad spectrum of substrates. Thus, LIP showed potential application in industries processing. The potential for G. geotrichum lipases application could also effectively increase.
Acknowledgements
The authors sincerely thank Dr. Zhen Li for providing help on protein identification. This work was supported by National Science and Technology Support Program (2013BAD10B01) and the National High Technology Research and Development Program (2012AA022208).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Baillargeon MW, Bistline RG, Jr, Sonnet PE. Evaluation of strains of Geotrichum candidum for lipase production and fatty acid specificity. Appl Microbiol Biot. 1989;30:92–96. doi: 10.1007/BF00256003. [DOI] [Google Scholar]
- Bertolini MC, Schrag JD, Cygler M, Ziomek E, Thomas DY, Vernet T. Expression and characterization of Geotrichum candidum lipase I gene. Comparison of specificity profile with lipase II. Eur J Biochem. 1995;228:863–869. doi: 10.1111/j.1432-1033.1995.0863m.x. [DOI] [PubMed] [Google Scholar]
- Bora L, Gohain D, Das R. Recent advances in production and biotechnological applications of thermostable and alkaline bacterial lipases. J Chem Technol Biot. 2013;88:1959–1970. [Google Scholar]
- Carvalho NB, Barbosa JMP, Oliveira MVS, Fricks AT, Lima AS, Soares CMF. Biochemical properties of Bacillus sp. ITP-001 lipase immobilized with a sol gel process. Quim Nova. 2013;36:52–58. doi: 10.1590/S0100-40422013000100010. [DOI] [Google Scholar]
- Cihangir N, Sarikaya E. Investigation of lipase production by a new isolate of Aspergillus sp. World J Microb Biot. 2004;20:193–197. doi: 10.1023/B:WIBI.0000021781.61031.3a. [DOI] [Google Scholar]
- Colen G, Junqueira RG, Moraes-Santos T. Isolation and screening of alkaline lipase-producing fungi from Brazilian savanna soil. World J Microb Biot. 2006;22:881–885. doi: 10.1007/s11274-005-9118-9. [DOI] [Google Scholar]
- Cordova J, Nemmaoui M, Ismaili-Alaoui M, Morin A, Roussos S, Raimbault M, Benjilali B. Lipase production by solid state fermentation of olive cake and sugar cane bagasse. J Mol Catal B-Enzym. 1998;5:75–78. doi: 10.1016/S1381-1177(98)00067-8. [DOI] [Google Scholar]
- De Schutter K, Lin YC, Tiels P, Van Hecke A, Glinka S, Weber-Lehmann J, Rouze P, Van de Peer Y, Callewaert N. Genome sequence of the recombinant protein production host Pichia pastoris. Nat Biotechnol. 2009;27:561–566. doi: 10.1038/nbt.1544. [DOI] [PubMed] [Google Scholar]
- Fernandez L, Perez-Victoria I, Zafra A, Benitez PL, Morales JC, Velasco J, Adrio JL. High-level expression and characterization of Galactomyces geotrichum (BT107) lipase I in Pichia pastoris. Protein Expr Purif. 2006;49:256–264. doi: 10.1016/j.pep.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Fischer M, Pleiss J. The lipase engineering database: a navigation and analysis tool for protein families. Nucleic Acids Res. 2003;31:319–321. doi: 10.1093/nar/gkg015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Ingole PG, Singh K, Bhattacharya A. Comparative study of the hydrolysis of different oils by lipase-immobilized membranes. J Appl Polym Sci. 2012;124:E17–E26. doi: 10.1002/app.35400. [DOI] [Google Scholar]
- Hasan F, Shah AA, Hameed A. Industrial applications of microbial lipases. Enzyme Microb Technol. 2006;39:235–251. doi: 10.1016/j.enzmictec.2005.10.016. [DOI] [Google Scholar]
- Kaushik R, Saran S, Isar J, Saxena RK. Statistical optimization of medium components and growth conditions by response surface methodology to enhance lipase production by Aspergillus carneus. J Mol Catal B-Enzym. 2006;40:121–126. doi: 10.1016/j.molcatb.2006.02.019. [DOI] [Google Scholar]
- Kim BS, Hou CT. Production of lipase by high cell density fed-batch culture of Candida cylindracea. Bioprocess Biosyst Eng. 2006;29:59–64. doi: 10.1007/s00449-006-0058-z. [DOI] [PubMed] [Google Scholar]
- Lima VMG, Krieger N, Sarquis MIM, Mitchell DA, Ramos LP, Fontana JD. Effect of nitrogen and carbon sources on lipase production by Penicillium aurantiogriseum. Food Technol Biotech. 2003;41:105–110. [Google Scholar]
- Lu Y, Fang C, Wang Q, Zhou Y, Zhang G, Ma Y. High-level expression of improved thermo-stable alkaline xylanase variant in Pichia pastoris through codon optimization, multiple gene insertion and high-density fermentation. Sci Rep. 2016;6:37869. doi: 10.1038/srep37869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado RR, Lopes D, Oliveira EA, Kamimura ES, Macedo GA. A review on Geotrichum lipases: production, purification, immobilization, and applications. Chem Biochem Eng Q. 2016;30:439–454. doi: 10.15255/CABEQ.2016.907. [DOI] [Google Scholar]
- Mazhar H, Abbas N, Ali S, Sohail A, Hussain Z, Ali SS. Optimized production of lipase from Bacillus subtilis PCSIRNL-39. Afr J Biotechnol. 2017;16:1106–1115. doi: 10.5897/AJB2017.15924. [DOI] [Google Scholar]
- Mohamed SA, Abdel-Mageed HM, Tayel SA, EI-Nabrawi MA, Fahmy AS. Characterization of Mucor racemosus lipase with potential application for the treatment of cellulite. Process Biochem. 2011;46:642–648. doi: 10.1016/j.procbio.2010.11.002. [DOI] [Google Scholar]
- Phillips A, Pretorius GH, van Rensburg HG. Molecular characterization of a Galactomyces geotrichum lipase, another member of the cholinesterase/lipase family. Biochim Biophys Acta. 1995;1252:305–311. doi: 10.1016/0167-4838(95)00126-F. [DOI] [PubMed] [Google Scholar]
- Pogori N, Cheikhyoussef A, Xu Y, Wang D. Production and biochemical characterization of an extracellular lipase from Rhizopus chinensis CCTCC M201021. Bio/Technology. 2008;7:710–717. [Google Scholar]
- Qiao H, Zhang F, Guan W, Zuo J, Feng D. Optimisation of combi-lipases from Aspergillus niger for the synergistic and efficient hydrolysis of soybean oil. Anim Sci J. 2017;88:772–780. doi: 10.1111/asj.12718. [DOI] [PubMed] [Google Scholar]
- Qiao H, Zhang W, Guan W, Chen F, Zhang S, Deng Z. Enhanced expression of lipase I from Galactomyces geotrichum by codon optimisation in Pichia pastoris. Protein Expr Purif. 2017;138:34–45. doi: 10.1016/j.pep.2017.05.005. [DOI] [PubMed] [Google Scholar]
- Santos KC, Cassimiro DMJ, Avelar MHM, Hirata DB, de Castro HF, Fernandez-Lafuente R, Mendes AA. Characterization of the catalytic properties of lipases from plant seeds for the production of concentrated fatty acids from different vegetable oils. Ind Crop Prod. 2013;49:462–470. doi: 10.1016/j.indcrop.2013.05.035. [DOI] [Google Scholar]
- Shu Z, Jiang H, Lin R, Jiang Y, Lin L, Huang J. Technical methods to improve yield, activity and stability in the development of microbial lipases. J Mol Catal B-Enzym. 2010;62:1–8. doi: 10.1016/j.molcatb.2009.09.003. [DOI] [Google Scholar]
- Singh AK, Mukhopadhyay M. Overview of fungal lipase: a review. Appl Biochem Biotechnol. 2012;166:486–520. doi: 10.1007/s12010-011-9444-3. [DOI] [PubMed] [Google Scholar]
- Sun F, Bai R, Yang H, Wang F, He J, Wang C, Tu M. Heterologous expression of codon optimized Trichoderma reesei Cel6A in Pichia pastoris. Enzyme Microb Technol. 2016;92:107–116. doi: 10.1016/j.enzmictec.2016.07.004. [DOI] [PubMed] [Google Scholar]
- Tan T, Zhang M, Xu J, Zhang J. Optimization of culture conditions and properties of lipase from Penicillium camembertii Thom PG-3. Process Biochem. 2004;39:1495–1502. doi: 10.1016/S0032-9592(03)00296-6. [DOI] [Google Scholar]
- Ueda M, Takahashi S, Washida M, Shiraga S, Tanaka A. Expression of Rhizopus oryzae lipase gene in Saccharomyces cerevisiae. J Mol Catal B-Enzym. 2002;17:113–124. doi: 10.1016/S1381-1177(02)00018-8. [DOI] [Google Scholar]
- Wang Y, Luo D, Zhao Y, Tian S, Deng W, Li C, Ma L. High-level expression and characterization of solvent-tolerant lipase. J Biosci Bioeng. 2018;125:23–29. doi: 10.1016/j.jbiosc.2017.06.012. [DOI] [PubMed] [Google Scholar]
- Wang J, Wu Z, Zhang T, Wang Y, Yang B. High-level expression of Thermomyces dupontii thermophilic lipase in Pichia pastoris via combined strategies. 3 Biotech. 2019;9:62. doi: 10.1007/s13205-019-1597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Yan J, Yang J, Xu L, Yan Y. Gene cloning, overexpression and characterization of a novel organic solvent tolerant and thermostable lipase from Galactomyces geotrichum Y05. J Mol Catal B-Enzym. 2007;49:28–35. doi: 10.1016/j.molcatb.2007.07.006. [DOI] [Google Scholar]
- Yan J, Yang J, Xu L, Yan Y. Cloning and overexpression of lipase gene from Geotrichum candidum Y162. Acta Microbiol Sinica. 2008;2:184–190. [PubMed] [Google Scholar]
- Yang J, Liu L. Codon optimization through a two-step gene synthesis leads to a high-level expression of Aspergillus niger lip2 gene in Pichia pastoris. J Mol Catal B-Enzym. 2010;63:164–169. doi: 10.1016/j.molcatb.2010.01.011. [DOI] [Google Scholar]
- Yu M, Qin S, Tan T. Purification and characterization of the extracellular lipase lip2 from Yarrowia lipolytica. Process Biochem. 2007;42:384–391. doi: 10.1016/j.procbio.2006.09.019. [DOI] [Google Scholar]
- Zhang X, Ai Y, Xu Y, Yu X. High-level expression of Aspergillus niger lipase in Pichia pastoris: characterization and gastric digestion in vitro. Food Chem. 2019;274:305–313. doi: 10.1016/j.foodchem.2018.09.020. [DOI] [PubMed] [Google Scholar]
- Zheng J, Lan X, Li X, Huang L, Zhang Y, Wang Z. High-level expression and characterization of a stereoselective lipase from Aspergillus oryzae in Pichia pastoris. Protein Expr Purif. 2019;155:1–7. doi: 10.1016/j.pep.2018.10.012. [DOI] [PubMed] [Google Scholar]