Abstract
MYOD is a master regulator of the skeletal myogenic program. But what regulates expression of Myod? More than 20 years ago, studies established that Myod expression is largely controlled by just two enhancer regions located within a region 24 kb upstream of the transcription start site in mammals, which regulate Myod expression in the embryo, fetus and adult. Despite this apparently simple arrangement, Myod regulation is complex, with different combinations of transcription factors acting on these enhancers in different muscle progenitor cells and phases of differentiation. A range of epigenetic modifications in the Myod upstream region also play a part in activating and repressing Myod expression during development and regeneration. Here the evidence for this binding at Myod control regions is summarized, giving an overview of our current understanding of Myod expression regulation in mammals.
Keywords: Myod, Myogenesis, Mammalian embryo, Transcription factor, Epigenetic regulation
Introduction
Myogenic determination gene number 1 (Myod1, usually simply referred to as Myod) was isolated on account of to its ability to induce skeletal muscle differentiation in fibroblasts, and a variety of other non-skeletal muscle cells (Choi et al. 1990; Davis et al. 1987; Weintraub et al. 1989). Due to this ability to reprogramme other cells to muscle and because it is expressed in cells of the early muscle lineage, Myod is often referred to as a master regulator (Chan and Kyba 2013; Weintraub et al. 1989), although it should be noted that Myod expression is not sufficient to reprogramme all non-muscle cells (e.g. Albini et al. 2013).
Myod encodes a basic helix-loop-helix transcription factor belonging to a larger family of myogenic regulatory factors (MRFs) that together control determination and differentiation of all skeletal muscle cells. Myogenic Factor 5 (Myf5), Myogenic Regulatory Factor 4 (Mrf4, also known as Myf6) and Myod act to direct cells into the skeletal myogenic lineage (Kassar-Duchossoy et al. 2004; Rudnicki et al. 1993). In the absence of all three factors myoblasts, and consequently skeletal muscles, do not form in the embryo (Kassar-Duchossoy et al. 2004; Rudnicki et al. 1993). While Myod and Myf5 can functionally substitute for each other to maintain myogenesis during development, in the absence of Myf5 and Myod, Mrf4 cannot drive fetal skeletal muscle differentiation (Kassar-Duchossoy et al. 2004). Expression of a fourth MRF, Myogenin (Myog), which is required for terminal differentiation of skeletal muscle cells in the embryo, is induced by the other three MRFs (Hasty et al. 1993; Nabeshima et al. 1993). Likewise, the MRFs regulate myogenesis in satellite cells, a stem cell population that mediates muscle homeostasis, growth and repair in adult muscle (reviewed in Zammit 2017).
Since MRFs act to drive myogenesis, their expression must be under strict spatiotemporal control during development and regeneration. In the case of Myod, studies dating back over two decades have shown that its expression in the embryo and adult is controlled by two cis-regulatory elements located upstream of the transcription start site (TSS) together with a minimal promoter region (Asakura et al. 1995; Chen et al. 2001, 2002; Faerman et al. 1995; Goldhamer et al. 1992, 1995; L’Honore et al. 2003; Tapscott et al. 1992). These elements bind both activators and repressors of transcription in different combinations depending on the cell type. The locus is also subject to epigenetic regulation, with modification of histones, DNA methylation and interaction with noncoding RNA all contributing to the correct regulation of expression. This review summarises the factors that have been shown to bind upstream of Myod and the evidence that they are involved in regulation of Myod expression in mammals.
Skeletal muscle development and regeneration
The muscles of the limbs and trunk are derived from somites, epithelial structures that bud off in a rostral-caudal sequence from the paraxial mesoderm (reviewed in Tajbakhsh and Buckingham 2000). In mouse, the first somite pair forms at around embryonic day 8 (E8) and there after a new pair of somites forms caudally every 2 h or so until approximately 65 somites are formed (Forsberg et al. 1998; Tam 1981). The dorsal part of the somite forms the dermomyotome (DM), from which the dermis and skeletal muscle of the trunk and limbs will develop. Soon after the somite forms, progenitor cells at the edges (lips) of the DM migrate under the DM and form the myotome (Venters et al. 1999). Subsequently, the DM begins to lose its epithelial character and cells from the central portion of the DM also move into the myotome, contributing further to its growth (Relaix et al. 2005). Cells of the dorsomedial myotome will form the epaxial (back) muscles while those in the ventrolateral region will form hypaxial muscles—the diaphragm, limb and ventral body wall muscles. At the level of the limb buds, cells in ventrolateral lip of the DM migrate into the limbs, where they then proliferate and differentiate (Houzelstein et al. 1999). Muscles of the head are primarily derived from the cranial mesoderm, which comprises cells originating from the anterior lateral plate, unsegmented paraxial mesoderm (rostral to the somite forming paraxial mesoderm) and prechordal mesoderm, although some neck, jaw and tongue muscle progenitors migrate to the head from the most rostral (occipital) somites (Michailovici et al. 2015).
This wave of primary myogenesis is complete by approximately E14.5 in mouse and is closely followed by a second, fetal, wave of myogenesis, where myoblasts proliferate and fuse with pre-existing primary myofibres or form de novo myofibres through fusion with each other. During this phase, which lasts until birth, some cells remain undifferentiated, forming a pool of progenitors, or satellite cells, which become located under the basal lamina surrounding each myofibre. These satellite cells are quiescent in adult muscle, but can be induced to proliferate and differentiate in response to various stimuli, including exercise and injury (Relaix and Zammit 2012).
The first myogenic genes to be transcribed are Myf5 and Mrf4, in the DM and then the myotome, shortly followed by branchial arch expression. Myf5 expression is also activated in the limb muscle precursors following their migration to the limb (reviewed in Comai and Tajbakhsh 2014; Vicente-García and Carvajal 2018). Expression of Myod follows Myf5 and continues in muscle cells until birth when expression is downregulated, although a low level of Myod transcription endures in certain adult skeletal muscles. Expression of Myod is also reactivated in activated satellite cells during muscle regeneration.
Other models of myogenesis
In addition to in vivo investigation, many studies of myogenic development make use of cell lines that are easily differentiated to myotubes or which express high levels of MRFs. In particular, C2 cell line clones, such as C2C12 and C2.7, undergo proliferation in high serum growth conditions, but differentiate and form contractile myofibres when serum is limited. These cells most likely represent a model of satellite cell differentiation, since they are derived from injured adult mouse muscle (Yaffe and Saxel 1977). In addition, Rhabdomyosarcoma cells are characterized by expression of MRFs, including Myod (Clark et al. 1991), and are a model for understanding regulation of MRFs (Taberlay et al. 2011).
Cis-regulatory elements controlling Myod expression
Gene expression is regulated, in part, by enhancers, genomic regulatory elements that bind transcription factors, interact with target promoters, recruit RNA Polymerase II (RNAPII) and promote transcription. Since enhancers are functional even outside their endogenous context, reporter assays are often used to test their ability to activate gene expression in cell culture or transgenic animals.
A series of experiments using reporter assays in vitro and in vivo showed that at a region extending 24 kb upstream of the human MYOD gene can essentially recapitulate the endogenous Myod expression pattern in the mouse embryo, fetus, and adult muscle, including regenerating satellite cells. Remarkably, a large amount of this expression is controlled by just two enhancers, which have come to be known as the Core Enhancer (CE) and the Distal Regulatory Region (DRR), together with a proximal regulatory region (PRR) immediately upstream of the transcription start site (TSS; Fig. 1a; Asakura et al. 1995; Chen et al. 2001, 2002; Faerman et al. 1995; Goldhamer et al. 1992, 1995; L’Honore et al. 2003; Tapscott et al. 1992).
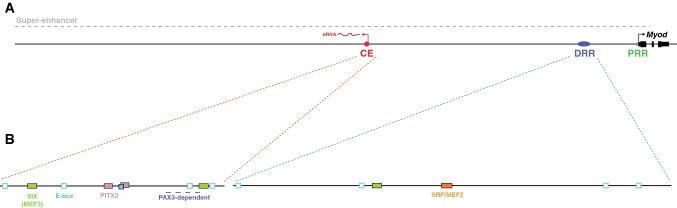
Fig. 1.
Sequences that regulate Myod expression. a Schematic of a region 50 kb upstream of the Myod gene, including the core enhancer (CE, in red), the distal regulatory region (DRR, in blue) and the proximal regulatory region (PRR, in green) immediately upstream of the Myod transcription start site (TSS, black arrow). This region lies within a super enhancer, associated with high levels of transcription factors occupancy, H3K27ac marks and multiple enhancer RNAs (eRNAs). One of these is an eRNA is transcribed from the CE that plays a role in regulating Myod expression through promoting chromatin accessibility at the Myod promoter. b Transcription factor binding sites for bHLH factors (E-box) including MYOD, MYF5, CLOCK, BMAL2, SIM2, MSC and TCF21 are shown in blue, for SIX and EYA factors in green (MEF3 site), for PITX2 in pink (Paired binding site) and for SRF/MEF2 in orange (CaRG box). Closed boxes show sites that have been shown to bind these factors in electromobility shift or other assays are shown as closed boxes, while open boxes show predicted sites. The E-box (closed) that binds CLOCK/BMAL2 overlaps a PIXT2 binding site. A region that is PAX3 dependent in somites and limbs, and is required for expression in myotomally-derived muscles, is indicated by the dashed purple line. (Color figure online)
The CE, which was first identified as a 258 bp region located 20 kb upstream of the TSS of the human MYOD gene, shows 89% sequence identity with a similar region 23 kb upstream of the mouse Myod gene (Fig. 1a; Goldhamer et al. 1995). The CE drives initiation of Myod expression in muscle precursor cells and nascent myocytes of the DM and myotome, branchial arches and limb buds in a pattern that mirrors the endogenous gene (Chen et al. 2001; Goldhamer et al. 1995). However, the CE is not sufficient for continued maintenance of reporter gene expression in differentiated skeletal muscle, since the transgene activity decreases during fetal and postnatal development until it becomes largely undetectable in adult muscle (Faerman et al. 1995). Linker-scan mutagenesis of the CE reveals elements within it that are essential for expression in all skeletal muscle progenitors, including the head, and separate elements that are required only for expression in the epaxial muscles and the hypaxial muscles of the body wall (Kucharczuk et al. 1999).
Although sufficient to drive early expression, deletion of the CE within the context of the 24 kb MYOD reporter construct results merely in reduced expression of the reporter in the hypaxial myotome together with a delay in expression in branchial arches and limb buds (Chen et al. 2001), indicating other regions in the 24 kb construct can compensate in part for the loss of the CE. Indeed, deletion of the CE within the mouse genome shows the CE is not absolutely required to initiate endogenous Myod expression in somites, since this does not change timing or pattern of expression, although it does lead to weaker expression of Myod. However, initiation of Myod expression in branchial arch and limb buds is delayed, confirming the CE controls the timely initiation of Myod expression in these muscles (Chen and Goldhamer 2004).
The DRR is a 714 bp region located approximately 5 kb upstream of mouse Myod, and also shares high sequence conservation with its human counterpart (Fig. 1a; Chen et al. 2001; Tapscott et al. 1992). In transgenic mouse reporter studies, the DRR drives expression at a later stage than the CE, in differentiated skeletal muscle cells of the myotome, head and limb (Asakura et al. 1995; Chen et al. 2001; Kablar et al. 1997). In addition, it continues to drive reporter expression in adult skeletal muscle cells in a pattern that resembles endogenous Myod expression (Hughes et al. 1993), and upregulation in activated satellite cells in regenerating muscle (L’Honore et al. 2003). Deletion of the DRR in the mouse genome causes a reduction of early Myod expression before cells differentiate, while later expression of Myod in the embryo and fetus is unaffected, results that are unexpected in light of the transgenic studies showing it drives expression in differentiated cells, and suggesting redundancy with other sequences in the Myod locus at later stages. However, the DRR is required to maintain levels of Myod expression in adult skeletal muscle, consistent with transgenic assays (Chen et al. 2002). Taken together these studies indicate that in general the CE is required for initiation of Myod expression and the DRR with maintenance of expression and adult skeletal muscle expression, but that redundancy with each other and other regions exists.
The PRR, which extends 275 bp upstream of the Myod TTS, is unable to direct specific expression of Myod, but acts as a minimal promoter in combination with the DRR or CE (Fig. 1a).
Transcription factor binding to Myod cis-regulatory elements: activators
Despite a role for Myod in all skeletal muscle groups, genetic studies show that the factors acting upstream to regulate expression are varied and differ depending on the origin of the cells and the phase of muscle formation (reviewed in Buckingham 2017). Summarised below, for those factors that are implicated in direct regulation of Myod expression, is evidence for binding of these factors to the CE and DRR, and other regions within in the proximal upstream region of Myod.
MRFs and PAX3/PAX7
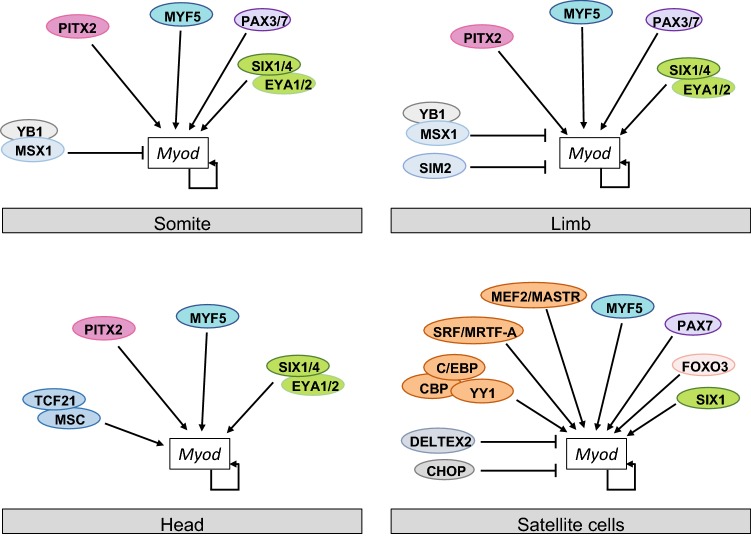
In the somites and limbs, Pax3 and Myf5 act in parallel pathways to regulate Myod expression during the first wave of myogenesis (Tajbakhsh et al. 1997). Pax3 expression precedes Myod in the DM and cells that migrate to the limb. Pax7 becomes expressed a little later in the central DM and plays a redundant role in embryonic myogenesis with Pax3. Indeed, although the early myotome forms in Pax3; Pax7 mutant mice, no further muscle forms after this (Relaix et al. 2005). Consistent with this, Myod expression is almost completely absent in Pax3/Pax7 double mutant embryos at later embryonic stages (E13.5; Relaix et al. 2005).
As mentioned above, Myf5 and Mrf4 expression also precedes Myod expression. While loss of Myf5 alone does not impact Myod expression, loss of Myf5 and Mrf4, in a mutant where targeted disruption of the Myf5 locus also results in lack of Mrf4 expression due to the genes being closely adjacent to each other (henceforth referred to as Myf5(Mrf4) mutants), causes an approximately 2-day delay in expression of Myod in the somites (Kablar et al. 1997; Kassar-Duchossoy et al. 2004; Tajbakhsh et al. 1997). Similarly, loss of Pax3 alone does not affect Myod expression, whereas Pax3;Myf5(Mrf4) mutants do not express Myod in somites and limb, indicating these factors compensate for each other in regulating Myod in the trunk of the embryo (Kassar-Duchossoy et al. 2004; Fig. 2). Craniofacial Myod expression on the other hand is seen in these mutants, since Pax3 is not expressed in cranial mesoderm, although Pax7 is (Tajbakhsh et al. 1997; Fig. 2). Instead, PITX2 and other transcription factors play a role in regulating Myod expression in the head mesoderm (see sections below).
Fig. 2.
Transcription factor binding and regulation of Myod in different cell types. Factors that regulate Myod expression and have been shown to bind the CE, PRR, DRR or adjacent regions are shown for different cells types. See text and Table 1 for details
A subset of Pax3/Pax7 expressing cells from the embryonic somites will become adult satellite cells and continue to express Pax7, which is required for satellite cell function in muscle homeostasis and regeneration (Kuang et al. 2006; Oustanina et al. 2004; Relaix et al. 2006; von Maltzahn et al. 2013). Pax7 is able to activate the expression of Myod, and after satellite cell activation both Myf5 and Myod are required for normal muscle regeneration, since when Myf5 and Myod are together knocked out in adult muscle, satellite cells fail to reliably enter the myogenic pathway and do not differentiate (Gayraud-Morel et al. 2007; Megeney et al. 1996; Ustanina et al. 2007; Yamamoto et al. 2018; Zammit et al. 2006).
Linker-scan mutagenesis of the CE suggests a region near the 3′ end (LS14 and 15, Kucharczuk et al. 1999) is dependent on PAX3 activity in the somites and limbs (Fig. 1b), however, direct binding of PAX3 (or PAX7) had not been shown at the CE, or indeed the DRR, which may indicate these regions are indirectly regulated by PAX3/PAX7. However, several cell-based studies have shown binding of PAX/PAX7 to Myod upstream regions other than the CE or DRR. In myoblasts transfected with tagged PAX3 or PAX7, PAX7 was shown by chromatin immunoprecipitation (ChIP) to bind approximately 40 kb upstream of the TSS (Fig. 2; Table 1; Soleimani et al. 2012a). Other experiments using an antibody which recognises both PAX3 and PAX7 showed that in partnership with FOXO3, PAX3/PAX7 binds a region between − 1.5 and − 1.6 kb upstream of the Myod TSS to synergistically regulate Myod expression (Table 1; Fig. 2; Hu et al. 2008). This binding is associated with increased recruitment of RNAPII to the Myod promoter (Hu et al. 2008). FOXO3 is further implicated as a regulator of endogenous Myod expression, since in mice mutant for Foxo3, Myod is down-regulated in regenerating muscle (Fig. 2; Hu et al. 2008).
Table 1.
Protein binding identified at the Myod proximal regulatory region (PRR), distal regulatory region (DRR), core enhancer (CE) and adjacent regions
| Region | Interacting protein | Binding shown by | Binding sitea | References |
|---|---|---|---|---|
| PRR | MSC | ChIP-qPCR in mouse embryo branchial arch tissue | Not characterised | Moncaut et al. (2012) |
| TCF21 | ChIP-qPCR in mouse embryo branchial arch tissue | Not characterised. | Moncaut et al. (2012) | |
| RAD21 | ChIP-qPCR in Rhabdomyosarcoma cells | Not characterised | Taberlay et al. (2011) | |
| p300 | ChIP-qPCR in C2C12 cells | Not characterised | Hamed et al. (2013) | |
| MYOD | ChIP-seq in C2C12 cells and primary myoblasts | Not characterised | Mousavi et al. (2013) and Umansky et al. (2015) | |
| MYF5 | ChIP-seq in mouse embryonic fibroblasts transduced with MYF5 | Not characterised | Conerly et al. (2016) | |
| PITX2 | ChIP-PCR in C2C12 cells and extraocular eye muscle cell line | Not characterised | Zacharias et al. (2011) | |
| DELTEX2 | ChIP-qPCR in C2C12 cells | Not characterised | Luo et al. (2017) | |
| Inter PRR–DRR | PITX2 | ChIP-qPCR in hindlimb buds and myotome. EMSA with in vitro translated PITX2 | PITX2 binding site at − 615 bp | L’Honore et al. (2010) |
| MYOD | EMSA with nuclear extracts from hindlimb muscle, G8 myoblasts and C3H/lOT1/2 fibroblasts | Two E boxes at − 1750 and − 865 bp | Zingg et al. (1994) | |
| GLI2 | ChIP-pPCR in P19 cells | GLI consensus binding sites at − 1.6 kb and − 0.6 kb | Voronova et al. (2013) | |
| FOXO3 | ChIP-PCR antibody in C2C12 cells. EMSA with recombinant FOXO3 | Two Forkhead Regulatory Elements at − 940 bp and − 1598 bp | Hu et al. (2008) | |
| PAX3/7 | ChIP-PCR in C2C12 cells. EMSA with recombinant PAX3 and PAX7 | Paired binding site at − 1502 bp | Hu et al. (2008) | |
| CHOP | ChIP-PCR with C2C12 cells | Not characterised. Primers bind at − 3 kb and − 4 kb | Alter and Bengal (2011) | |
| DRR | SIX1/4 | ChIP-seq and -qPCR in C2C12 cells and satellite cell derived myoblasts. EMSA with in vitro translated SIX1 and SIX4 | MEF3 site in DRR | Le Grand et al. (2012), Liu et al. (2010) and Relaix et al. (2013) |
| EYA1/2 | ChIP-qPCR in mouse embryo trunk muscle precursor cells | Not characterised, likely to be via SIX binding at MEF3 site | Relaix et al. (2013) | |
| PITX2 | ChIP-qPCR in mouse embryo myotome | Not characterised | L’Honore et al. (2010) | |
| MSC | ChIP-qPCR in mouse embryo branchial arch tissue | Not characterised | Moncaut et al. (2012) | |
| TCF21 | ChIP-qPCR in mouse embryo branchial arch tissue | Not characterised | Moncaut et al. (2012) | |
| SRF | ChIP-qPCR in C2C12 cells. EMSA with nuclear extracts from C2.7 cells | Divergent CaRG box (a hybrid SRF/MEF2 binding site) | L’Honore et al. (2003) and L’Honore et al. (2007) | |
| MEF2 | ChIP-qPCR in C2C12 cells.. EMSA with nuclear extracts from C2C12 cells | Divergent CaRG box (a hybrid SRF/MEF2 binding site), and a canonical MEF2 binding site | L’Honore et al. (2007) and Mokalled et al. (2012) | |
| YY1 | EMSA with nuclear extracts from C2.7 cells | Divergent CaRG box (a hybrid SRF/MEF2 binding site) | L’Honore et al. (2003) | |
| MRTF-A | ChIP-qPCR in C2C12 cells | Divergent CaRG box (a hybrid SRF/MEF2 binding site) | Mokalled et al. (2012) | |
| MASTR | ChIP-qPCR in C2C12 cells | Divergent CaRG box (a hybrid SRF/MEF2 binding site), and a canonical MEF2 binding site | Mokalled et al. (2012) | |
| C/EBP | EMSA with nuclear extracts from C2.7 cells | Divergent CaRG box (a hybrid SRF/MEF2 binding site) | L’Honore et al. (2003) | |
| p300 | EMSA with nuclear extracts from C2.7 cells. ChIP-qPCR in C2C12 cells | Divergent CaRG box (a hybrid SRF/MEF2 binding site) | L’Honore et al. (2003) and Hamed et al. (2013) | |
| MYOD | ChIP-seq in C2C12 cells and primary myoblasts. | Not characterised | Mousavi et al. (2013) and Umansky et al. (2015) | |
| MYF5 | ChIP-seq in mouse embryonic fibroblasts transduced with MYF5 | Not characterised | Conerly et al. (2016) | |
| DELTEX2 | ChIP-qPCR in C2C12 cells. | Not characterised | Luo et al. (2017) | |
| Inter DRR–CE | MYOD | ChIP-seq in C2C12 cells and primary myoblasts | Not characterised. Chip-seq peaks map to region around − 10.5 kb and − 12 kb | Conerly et al. (2016), Cui et al. (2017) and Umansky et al. (2015) |
| MYF5 | ChIP-seq in mouse embryonic fibroblasts transduced with MYF5 | Not characterised. Chip-seq peaks map to region around − 10.5 kb | Conerly et al. (2016) | |
| CE | SIX1/4 | ChIP -seq and -qPCR in C2C12 cells. EMSA with in vitro translated SIX1 and SIX4 | Two MEF3 sites | Liu et al. (2010) and Relaix et al. (2013) |
| EYA1/2 | ChIP-qPCR in mouse embryo trunk muscle precursor cells | Not characterised, likely to be via SIX binding at MEF3 site | Relaix et al. (2013) | |
| PITX2 | ChIP-qPCR in hindlimb buds and myotome. EMSA with in vitro translated PITX2 | Two PITX2 binding sites | L’Honore et al. (2010) | |
| RAD21 | ChIP-qPCR in Rhabdomyosarcoma cells | Not characterised | Taberlay et al. (2011) | |
| LSD1 | ChIP-qPCR antibody in C2C12 cells | Not characterised | Scionti et al. (2017) | |
| CLOCK | ChIP-PCR in muscles from mice kept in a 12 h light–dark cycle. EMSA with extract from C2C12 cells overexpressing CLOCK and BMAL | Divergent E-box (CAGCTT) | Andrews et al. (2010) and Zhang et al. (2012) | |
| BMAL2 | ChIP-PCR in muscles from mice kept in a 12 h light–dark cycle. EMSA with extract from C2C12 cells overexpressing CLOCK and BMAL | Divergent E-box (CAGCTT) | Andrews et al. (2010) and Zhang et al. (2012) | |
| MYOD | ChIP-seq in mouse embryonic fibroblasts transduced with Myod, in C2C12 cells, and primary myoblasts | Not characterised | Conerly et al. (2016), Cui et al. (2017), Mousavi et al. (2013) and Umansky et al. (2015) | |
| MYF5 | ChIP-seq with in mouse embryonic fibroblasts transduced with MYF5 | Not characterised | Conerly et al. (2016) | |
| YB1 | ChIP-PCR and -qPCR in C2C12 cells expressing | Not characterised | Lee et al. (2004) and Song and Lee (2010) | |
| MSX1 | ChIP-PCR in C2C12 cells and mouse embryo limbs | Not characterised | Lee et al. (2004), Song and Lee (2010) and Wang et al. (2011) | |
| SIM2 | ChIP-PCR in mouse embryo limb buds | Not characterised | Havis et al. (2012) | |
| Upstream of CE | GLI2 | ChIP-pPCR in P19 cells overexpressing GLI2 | GLI consensus binding sites at − 32 kb and − 36 kb | Voronova et al. (2013) |
| PAX7 | Chromatin Tandem Affinity Purification (ChTAP) in satellite cell-derived myoblasts | Not characterised. Chip-seq peaks map to region around − 40 kb | Soleimani et al. (2012b) |
aCharacterized binding sites are those sites where physical binding and/or function had been validated via EMSA and/or luciferase assays, including mutation of the binding site. For some ChIP-seq studies binding was determined through analysis of files downloaded from the Gene Expression Omnibus (Conerly et al. 2016; GEO accession: GSE75370; Cui et al. 2017; GEO accession: GSE63716; Umansky et al. 2015, GEO accession: GSE56131)
MYF5, MRF4, and MYOD itself, regulate the expression of Myod through the DRR, since the DRR cannot drive reporter gene expression in Myod;Myf5(Mrf4) mutant embryos (Kablar et al. 1999). In these embryos the CE drives weak expression in the somites and delayed expression in the limb (Kablar et al. 1999). MYOD can also regulate its own expression in vitro via binding E-boxes, binding sites for bHLH factors, in the promoter region of Myod (Fig. 2; Table 1; Zingg et al. 1994). Although the DRR and CE contain E-boxes, direct binding of MRFs has not been shown as yet in embryos and whether these factors differentially bind to the CE and DRR is not known. However, several genome-wide studies have shown MYOD and MYF5 bind the CE and/or the DRR and adjacent sites in myoblasts in culture, although occupancy at these sites differs depending on the cell type and specific assay, so how this binding translates to functional regulation is still to be investigated (Table 1; Conerly et al. 2016; Cui et al. 2017; Mousavi et al. 2013; Soleimani et al. 2012b; Umansky et al. 2015).
SIX and EYA
Six homeodomain (SIX) transcription factors and their co-factors, members of the Eyes absent (EYA) family, are also upstream regulators of Myod in both trunk and head skeletal muscle. Mice mutant for Six1/4 or Eya1/2 show a reduction in Myod expression in muscle precursor cells of the somite and limb (Grifone et al. 2007; Laclef et al. 2003). Both the CE and DRR bind SIX1/SIX4 and SIX2 in vitro and EYA proteins in vivo (Table 1; Fig. 2; Liu et al. 2010; Relaix et al. 2013), and consistent with a direct role in regulating the CE, when SIX binding sites in the CE are mutated, the CE is unable to drive robust reporter expression in the trunk (Relaix et al. 2013). Despite expression of Myod in the head in Six1/4 and Eya1/2 mutants, mutation of the SIX binding sites in the CE results in loss of reporter expression in the head. This suggests another SIX factor, likely to be SIX2, activates expression of Myod in the head in the absence of Six1 and Six4 (Fig. 2; Relaix et al. 2013). SIX1 also appears to regulate Myod expression in regenerating adult muscles cells, since SIX1 is bound to the DRR in differentiating satellite cells derived from adult mouse, and when SIX1 activity is knocked down in these cells Myod expression is reduced (Table 1; Fig. 2; Le Grand et al. 2012).
PITX2
PITX2, a paired-like homeodomain protein, is another factor that regulates Myod expression in both the trunk and head (Diehl et al. 2006; Dong et al. 2006; L’Honore et al. 2010). In the limb and myotome PITX2 cooperates with MYF5 and MYF4 to regulate Myod expression, while in the extraocular eye muscles of the head it acts upstream of Myf5 and Mrf4 (reviewed in Hernandez-Torres et al. 2017). PITX2 binds the CE (but not the DRR) in hindlimb buds and is able to activate expression of a reporter gene in cell based assays, while mutation of two paired binding sites within the CE abolishes the reporter activity (Table 1; L’Honore et al. 2010), indicating PITX2 binding to the CE is required for limb expression. In contrast, myotomal expression of Myod does not require Pitx2, but in Pitx2;Myf5(Mrf4) mutants almost all myotomal Myod expression is absent, suggesting redundancy between these factors (L’Honore et al. 2010). In support of a direct role in regulating MyoD expression in somites, chromatin immunoprecipitation experiments show PITX2 binding to the CE, DRR and just upstream of the PRR in mouse embryonic myotome (Table 1; Fig. 2; L’Honore et al. 2010). In the head conditional inactivation of Pitx2 in the extraocular eye muscle precursors results in downregulation of Myod expression. ChIP of PITX2 in C2C12 cells and in an extraocular eye muscle derived cell line indicates that PITX2 binds the PRR but not the CE (the DRR was not tested; Table 1; Fig. 2; Zacharias et al. 2011). These data indicate that Myod expression is regulated through PITX2 binding a different combination of regulatory regions in the limbs (CE) compared to the myotome (CE, DRR, upstream of the PRR) and extraocular eye muscles (PRR), although how PITX2 discriminates is currently unknown.
MUSCULIN and TCF21
The bHLH transcription factors, MUSCULIN (MSC; also known as MYOR) and TCF21 (CAPSULIN) are together required for formation of some first branchial arch-derived skeletal muscles, where they are transiently expressed (Lu et al. 2002). Myod expression is absent in these cells in Msc/Tcf21 mutants and both factors bind the PRR and DRR (but not the CE) in E10.5 branchial arch cells, suggesting they directly regulate Myod expression (Table 1; Fig. 2; Moncaut et al. 2012).
GLI2
Sonic hedgehog (Shh) signalling, which is mediated by the activating transcription factors GLI1 and GLI2, plays a role in regulating Myod expression both in the embryo and in regenerating muscle. Shh is required for expression of Myod in the epaxial myotome and limb and for initiation of Myod expression in the limb (Borycki et al. 1999; Hu et al. 2012). It is also required for the upregulation of Myod in activated satellite cells (Straface et al. 2009). In a cell culture model in which P19 cells are differentiated to myotubes, GLI2 activity was found to regulate Myod expression, possibly through GLI2 binding to sites ~ 35 kb upstream of the CE and ~ 1 kb upstream of the MyoD TSS (Table 1; Voronova et al. 2013), raising the possibility that GLI2 directly regulates Myod expression in vivo.
SRF, MEF2 and cofactors
Serum response factor (SRF), a transcription factor of the MADS box family, plays a role in skeletal, cardiac and smooth muscle formation (reviewed in Coletti et al. 2016). In skeletal muscle it is not required for embryonic Myod expression and myogenesis, but plays a role in adult muscle homeostasis. Members of the Myocardin family of transcription factors, including MRTF and MASTR, act as co-factors for SRF and MEF2 factors, also MADS box family members, respectively, and consequently also play important roles in muscle formation (Creemers et al. 2006; Olson and Nordheim 2010). MASTR and MRTF-A are up-regulated in satellite cells in response to injury, and are required for differentiation of satellite cells during growth and regeneration in the adult (Mokalled et al. 2012). Consistent with the DRR driving expression of Myod in adult muscle, all these factors bind the DRR (Table 1; Fig. 2). SRF binds to a divergent CArG box in the DRR, in complexes that include YY1, a regulator of enhancer–promoter interactions, CBP, a transcriptional coactivator, C/EBP family factors and MRTF-A (Table 1; Fig. 2; L’Honore et al. 2003, 2007; Mokalled et al. 2012; Weintraub et al. 2017). This CArG box is required for expression of a DRR-reporter construct in differentiating myoblasts in culture and in activated satellite cells after muscle lesion in vivo (L’Honore et al. 2003). The divergent CArG box is a combined MEF/SRF binding site and is also bound by a complex of MASTR and MEF2 family factors in myoblasts (Table 1; Fig. 2; L’Honore et al. 2007). It appears that a switch occurs as cells differentiate, such that while SRF binding initiates Myod expression, as cells differentiate MEF2 binding replaces SRF and maintains expression (L’Honore et al. 2007).
CLOCK and BMAL1
In adult skeletal muscle, Myod mRNA exhibits circadian rhythm which is abolished in mice mutant for Clock or Bmal1, two bHLH-PAS domain transcription factors that heterodimerize to positively regulate the circadian clock through binding E-boxes in target gene loci. The CE is required to regulate periodic Myod expression in vivo, and both CLOCK and BMAL1 bind the CE in muscle (Andrews et al. 2010; Zhang et al. 2012). This binding appears to be via a conserved non-canonical E-box in the CE (Fig. 1b; Table 1; Zhang et al. 2012), and consistent with the CE regulating periodic activation of Myod transcription, CLOCK and BMAL1 binding to the CE in cultured cells is most enriched when Myod transcripts are at their highest (Andrews et al. 2010).
Transcription factor binding to Myod cis-regulatory elements: repressors
Myod expression must also be repressed in a controlled temporal and spatial fashion. Reporter assays show that the DRR drives reporter gene expression in non-myogenic cells, suggesting that repressor sequences may be found outside the DRR region (Asakura et al. 1995). Some linker-scan mutations in the CE also result in ectopic expression, which suggests the CE contains regions that bind repressors (Kucharczuk et al. 1999). The evidence for repressor binding in the Myod upstream region is summarised below.
SIM2
The bHLH-PAS domain transcription factor, Sim2 is expressed in muscle progenitors shortly after they have migrated into the limb and before Myod expression is upregulated (Coumailleau and Duprez 2009; Havis et al. 2012). In Sim2 mutants Myod expression in the limb is upregulated compared to wild type levels (Havis et al. 2012). Sim2 appears to prevent entry into the myogenic program through repression of Myod expression, since it binds to the CE in embryonic mouse limb buds and represses Myod transcription when overexpressed in primary limb bud myoblasts (Table 1; Fig. 2; Havis et al. 2012).
DELTEX2
Deltex2, which encodes an E3 ubiquitin ligase, is expressed during regeneration in adult muscle in myogenic progenitor cells and inhibits myogenic differentiation (Luo et al. 2017). In cell differentiation assays, exogenously expressed DELTEX2 is able to bind the DRR and PRR, but not the CE (Table 1; Fig. 2), and leads to an enrichment of dimethyl lysine 9 of histone H3 (H3K9me2), a repressive chromatin mark, at the Myod TSS. This is likely to be through inhibiting the activity of JMJD1C, a lysine demethylase (Luo et al. 2017).
C/EBP homology protein
C/EBP homology protein (CHOP, also known as DDIT3) is expressed in quiescent satellite cells in vivo and is transiently induced during myoblast differentiation in vitro (Alter and Bengal 2011; Fukada et al. 2007). Knockdown of CHOP in C2C12 cells results in premature expression of myogenic genes, and conversely overexpression of CHOP results in inhibition of the myogenic program and reduced nuclei numbers in myotubes that do form (Alter and Bengal 2011). CHOP seems to act by binding at approximately 3 kb upstream of the Myod TSS and repressing expression of Myod (Table 1; Fig. 2). Interestingly, this binding is associated with a decrease in Histone H4 acetylation (an active histone mark) near the DRR and may indicate CHOP acts through regulating the activity of a histone deacetylase (Table 1). Further evidence for this comes from experiments in 293 T cells where CHOP was shown to interact physically with Histone Deacetylase 1 (HDAC1; Alter and Bengal 2011).
Epigenetic regulation of Myod regulatory regions
Whilst transcription factors direct the activation of gene expression, DNA must be accessible for them to bind. The accessibility of enhancer and promoter sequences to transcription factor binding is controlled through multiple epigenetic mechanisms including DNA methylation, histone modification and interactions with non-coding RNAs, all of which are at play in Myod regulation.
Methylation
DNA methylation at cytosine (C) residues within CpG dinucleotides is generally associated with repression of gene expression. It has long been known that treating a mouse embryonic fibroblast cell line (CHC3H 10T1/2) with 5-azacytidine, which inhibits DNA methylation, can convert these cells to skeletal muscle, suggesting that DNA methylation is important in maintaining repression of myogenic genes in non-muscle cells (Constantinides et al. 1977). Indeed, DNA hypomethylation is associated with active enhancers (e.g. Blattler et al. 2014; Hon et al. 2013). This is seen at the CE, where studies in adult tissues and cell culture show that myoblasts and myotubes have low levels of methylation at the CE, while non-muscle tissues have higher levels (Brunk et al. 1996; Carrio et al. 2016; Ehrlich et al. 2016). Consistent with this, in the mouse embryo the presomitic mesoderm shows higher levels of methylation at the CE compared to the somites, with demethylation of CE preceding Myod expression (Brunk et al. 1996). However methylation of the CE may not be sufficient for repression of Myod expression, since a demethylated CE transgene is not activated precociously or ectopically, suggesting other mechanisms may also be responsible for active repression of Myod expression (Brunk et al. 1996).
Nucelosomes and histone modification
In eukaryotes, DNA is wrapped around a core complex of histone proteins, the nucleosome, helping to package long DNA molecules into the nucleus but reducing the accessibility of transcriptional machinery to the DNA. Modification, such as acetylation and methylation, of amino acids in the N-terminal tails of histones alter how tightly the DNA interacts with the nucleosome and thereby regulates accessibility of transcription factor binding. As such nucleosome position and histone modification at enhancers and promoters are important regulators of gene expression. For instance, genome-wide studies using chromatin immunoprecipitation of histones have shown active promoters and enhancers often contain nucleosome-depleted regions, while nucleosomes downstream of promoters are associated with high levels of Histone H3 lysine 4 tri methylation (H3K4me3), and nucleosomes at enhancer regions are preferentially marked by high levels of H3K4me1 (Bernstein et al. 2006; Heintzman et al. 2007, 2009; Rada-Iglesias et al. 2011). The presence of H3K27ac marks promoters and enhancers as transcriptionally active, while H3K27me3, H3K9me2/3 and H4K20me3 are associated with repressed genes (Creyghton et al. 2010; Heintzman et al. 2007, 2009; Rada-Iglesias et al. 2011; Wang et al. 2008; Young et al. 2011). Acetylation of lysine residues in the Histone H4 tail are also associated with transcriptional activation (Wang et al. 2008).
These associations are reflected in studies of histone modification in muscle and non-muscle cells. Cell culture experiments show that myoblasts and myotubes both have active histone marks at the Myod promoter, DRR and CE, as well as other regions upstream of these, as does skeletal muscle, while non-muscle tissues have repressive marks in the same region upstream of the Myod gene (Ehrlich et al. 2016). For instance one study in a Rhabdomyosarcoma cell line, has shown that both the CE and the Myod TSS region are nucleosome free, and are associated with high levels of H3K4me1 and H3K4me3 respectively, while in colorectal cancer cells, where Myod is not expressed, both the CE and promoter contain nucleosomes (Table 2; Taberlay et al. 2011).
Table 2.
Histone marks identified at the Myod proximal regulatory region (PRR), distal regulatory region (DRR) and core enhancer
| Region | Histone | Type of mark | Binding shown by | Comments | References |
|---|---|---|---|---|---|
| PRR | H3K4me3 | Active | ChIP-qPCR in Rhabdomyosarcoma cells | Taberlay et al. (2011) | |
| H3K27ac | Active | ChIP-qPCR in C2C12 cells | Levels increase as cells differentiate | Hamed et al. (2013) | |
| H4Kac | Active | ChIP-PCR in mouse embryo limbs | Lower levels than at CE | Havis et al. (2012) | |
| H3K9ac | Active | ChIP-qPCR in C2C12 cells | Levels increase as cells differentiate | Yang et al. (2011) and Hamed et al. (2013) | |
| H3.3 | Active | ChIP-qPCR in C2C12 cells | Lower levels than CE. Levels increase as cells differentiate | Yang et al. (2011) | |
| DRR | H3K4me3 | Active | ChIP-qPCR in C2C12 cells | Levels increase as cells differentiate | Yang et al. (2011) |
| H3K27ac | Active | ChIP-qPCR in C2C12 cells | Levels increase as cells differentiate | Hamed et al. (2013) | |
| H3K9ac | Active | ChIP-qPCR in C2C12 cells | Levels increase as cells differentiate | Yang et al. (2011) and Hamed et al. (2013) | |
| H3.3 | Active | ChIP-qPCR in C2C12 cells. | Lower levels than CE. Levels increase as cells differentiate | Yang et al. (2011) | |
| H3K20me2 | Repressive | ChIP-qPCR in satellite cells isolated from mouse muscle | Levels decrease in Suv4-20h1 knockout cells | Boonsanay et al. (2016) | |
| H3K9me2 | Repressive | ChIP-qPCR in C2C12 cells | Luo et al. (2017) | ||
| CE | H3K4me3 | Active | ChIP-qPCR in C2C12 cells | Levels increase as cells differentiate | Scionti et al. (2017) and Yang et al. (2011) |
| H3K27ac | Active | ChIP-qPCR in and differentiated C2C12 cells | Levels increase greatly as cells differentiate | Hamed et al. (2013) | |
| H4Kac | Active | ChIP-PCR in mouse embryo limbs | Higher levels than at PRR | Havis et al. (2012) | |
| H3K4me1 | Enhancer | ChIP-qPCR in Rhabdomyosarcoma cells | Taberlay et al. (2011) | ||
| H3K9ac | Active | ChIP-qPCR in C2C12 cells | Levels increase as cells differentiate. Levels decrease when MSX1 expressed | Yang et al. (2011), Hamed et al. (2013) and Lee et al. (2004) | |
| H3.3 | Active | ChIP-qPCR in C2C12 | Higher levels than DRR and PRR. Levels increase as cells differentiate | Yang et al. (2011) | |
| H3K9me2 | Repressive | ChIP-qPCR in C2C12 | Levels increase when MSX1 expressed. | Lee et al. (2004) | |
| HB1 | Repressive | ChIP-qPCR in C2C12 | Levels increase when MSX1 expressed | Lee et al. (2004) |
Enhancers are also often associated with P300, a transcriptional coactivator that contains a histone acetyltransferease (HAT) domain (Visel et al. 2009). P300 binding at the CE, DRR and PRR is found to increase as myoblast cells differentiate towards myotubes in C2 cell culture, with a corresponding increase in H3K27ac and H3K9ac (transcription elongation) levels (Table 2; Gates et al. 2017; Hamed et al. 2013). Whilst mouse mutants carrying an allele with reduced P300 HAT activity exhibit reduced Myod expression (Roth et al. 2003), suggesting that acetylation of histones at Myod regulatory regions is required for robust expression.
In addition to modification of core histones, variant histone isoforms, which have particular regulatory functions, are differentially incorporated into nucleosomes. For instance the Histone H3 variant, H3.3, is associated with active genes and enhancers (Ahmad and Henikoff 2002; Jin et al. 2009). At the Myod locus a study in C2 cells has shown that levels of H3.3 increase at the CE, DRR and PRR as myoblasts differentiate, and when levels of H3.3 are reduced at these regulatory regions through inhibiting the activity of the H3/H4 histone chaperone, Histone regulator A (HIRA), Myod transcription is reduced (Fig. 2; Yang et al. 2011), suggesting that transcription of Myod is facilitated by integration of Histone H3.3.
Few studies have looked at histone binding at the Myod locus in vivo, however Havis et al. (2012) showed that acetylated Histone H4 is present at the CE and PRR but not at the DRR in mouse embryos (Table 2). Interestingly, this reflects the situation seen in transgenic mice where the CE drives limb bud expression but the DRR does not (Asakura et al. 1995; Chen et al. 2001; Goldhamer et al. 1995).
DNA looping
In active chromatin, loops of DNA are formed that bring the promoter and enhancer close together for activation of gene expression. The cohesion complex, which includes the RAD21 subunit, mediates this looping and thus many enhancers and promoters are associated with RAD21 binding. This is true of the Myod CE and promoter in Rhabdomyosarcoma cells, in which Myod is highly expressed, which are both associated with RAD21 binding (Fig. 2; Taberlay et al. 2011).
Heterochromatin
In quiescent satellite cells levels of heterochromatin, which represses gene activation, are high and decrease as cells are activated and enter the myogenic pathway (Hawke and Garry 2001). In proliferating C2 cells, in which Myod expression is low, the CE is associated with high levels of H3K9me2/3 (Scionti et al. 2017), while in satellite cells isolated from adult mouse muscle the DRR is associated with high levels of H3K20me2, a substrate for further methylation to H3K20me3, which decrease as cells differentiate (Table 2; Boonsanay et al. 2016). This repression of Myod via deposition of H3K20me2 appears to be mediated by SUV4-20H1, a dimethyltransferase, since in satellite cells mutant for Suv4-20h1, Myod transcription increases while H3K20me2 is reduced and H3K4me3 is increased at the DRR (Table 2; Boonsanay et al. 2016).
Unlike the other histones, H1 sits atop the nucleosome, keeping the DNA wrapped around the core histone complex, and thus also plays an important role in regulating accessibility to the DNA (Izzo and Schneider 2016). During myogenesis, integration of the Histone H1b variant at the CE, mediated by binding of MSH Homeobox 1 (MSX1) and Y-box binding protein 1 (YB1) at the CE, is also associated with increased H3K9 methylation (Tables 1, 2; Lee et al. 2004; Song and Lee 2010). This is consistent with the role of MSX1 in repression of muscle differentiation in the hypaxial DM and migrating limb muscle progenitor cells in the mouse embryo (Fig. 2; Houzelstein et al. 1999).
Enhancer RNAs
Another method of regulating accessibility of chromatin at the Myod locus is via an enhancer RNA that is transcribed from the CE (CEeRNA) (Mousavi et al. 2013; Scionti et al. 2017). Enhancer RNAs are a relatively recently discovered class of non-coding RNAs identified from whole genome studies. They are transcribed from active enhancers and appear to have important roles in regulating transcription (reviewed in Liu 2017). The mechanism by which the CEeRNA regulates Myod expression is still being investigated, but it seems to promote accessibility of the Myod promoter, since if it is depleted via siRNA then chromatin accessibility, as measured by DNAse1 sensitivity, is reduced, together with reduced Myod transcription levels (Mousavi et al. 2013). Expression of the CEeRNA during differentiation in C2 cells is promoted by binding of Lysine Specific Demethylase 1 (LSD1) to the CE (Fig. 1), whilst in mouse embryos a conditional knockout of Lsd1 in muscle progenitor cells results in a strong reduction of CEeRNA expression in the limb (Scionti et al. 2017), indicating Lsd1 is involved in CEeRNA expression in the embryo. Interestingly, this Lsd1 conditional knockout also results in a strong reduction of initial Myod expression in the limbs and a mild reduction in somites, which recovers later, reminiscent of the Myod expression seen when CE is deleted in vivo (Chen and Goldhamer 2004). This suggests transcription of the CEeRNA is also required for CE activity in driving the initiation of Myod expression.
Super-enhancers
Super enhancers are large regions of the genome that are characterised by high levels transcription factor occupancy, H3K27ac marks, mediator complex, and transcription of multiple enhancer RNAs (Khan et al. 2018). Interestingly, a region extending approximately 40–50 kb upstream of Myod, in both mouse and human locus, has been described that has characteristics of a super-enhancer (Ehrlich et al. 2016; Mousavi et al. 2013), which further implicates a larger region beyond 24 kb upstream of the Myod TSS in expression regulation (Fig. 1). Interestingly PAX3 has been shown to promote chromatin accessibility, and may act to do so in a region from the Myod TSS up to approximately 40 kb upstream, since in mouse embryoid bodies induced to express Pax3, both binding of PAX3 and accessibility of chromatin increases in this region during skeletal muscle differentiation (Magli et al. 2019).
Conclusion
At first glance the Myod upstream region, with two main enhancer elements may seem uncomplicated. However, this belies a complex regulatory landscape where tissue and temporal specificity are merged, in the most part, into two these elements. This is in contrast to Myf5 regulation, which has multiple enhancer regions located in a region 140 kb upstream of its TSS that drive distinct expression patterns in the somite, limb and head, and Myog which has just one 133 bp regulatory region located immediately upstream of its TSS (reviewed in Carvajal and Rigby 2010). While we have knowledge of many (but, undoubtedly, nowhere near all) of the transcription factors that bind Myod regulatory elements and the histone marks that decorate the region, we are far from understanding the details of how they interact with each other to bring about the precise spatial and temporal expression of Myod. For instance, while mutant studies can show that different transcription factors compensate for each other, what is their relative contribution in the wildtype situation? Similarly, what is the relative contribution of the CE and DRR and (how) do these regions communicate with each other to drive precise Myod expression? What is the relationship of transcription factor binding with epigenetic marks? Many studies have shown a correlation between transcription factor binding and epigenetic marks, but which comes first and interactions between all of them is yet to be fully explored. Non-coding RNAs, including the recent discovery of enhancer RNAs, have added another level of regulation that must be integrated into our picture of how all these different factors interact with each other to regulate Myod transcription.
Acknowledgements
FCW would like to thank Dr Robert Knight, Prof Peter Zammit and anonymous reviewers for their very helpful comments on the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Albini S, Coutinho P, Malecova B, Giordani L, Savchenko A, Forcales SV, Puri PL. Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres. Cell Rep. 2013;3:661–670. doi: 10.1016/j.celrep.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter J, Bengal E. Stress-induced C/EBP homology protein (CHOP) represses MyoD transcription to delay myoblast differentiation. PLoS ONE. 2011;6:e29498. doi: 10.1371/journal.pone.0029498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews JL, et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci USA. 2010;107:19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura A, Lyons GE, Tapscott SJ. The regulation of MyoD gene expression: conserved elements mediate expression in embryonic axial muscle. Dev Biol. 1995;171:386–398. doi: 10.1006/dbio.1995.1290. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blattler A, Yao L, Witt H, Guo Y, Nicolet CM, Berman BP, Farnham PJ. Global loss of DNA methylation uncovers intronic enhancers in genes showing expression changes. Genome Biol. 2014;15:469. doi: 10.1186/s13059-014-0469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonsanay V, et al. Regulation of skeletal muscle stem cell quiescence by Suv4-20h1-dependent facultative heterochromatin formation. Cell Stem Cell. 2016;18:229–242. doi: 10.1016/j.stem.2015.11.002. [DOI] [PubMed] [Google Scholar]
- Borycki AG, Brunk B, Tajbakhsh S, Buckingham M, Chiang C, Emerson CP., Jr Sonic hedgehog controls epaxial muscle determination through Myf5 activation. Development. 1999;126:4053–4063. doi: 10.1242/dev.126.18.4053. [DOI] [PubMed] [Google Scholar]
- Brunk BP, Goldhamer DJ, Emerson CP., Jr Regulated demethylation of the myoD distal enhancer during skeletal myogenesis. Dev Biol. 1996;177:490–503. doi: 10.1006/dbio.1996.0180. [DOI] [PubMed] [Google Scholar]
- Buckingham M. Gene regulatory networks and cell lineages that underlie the formation of skeletal muscle. Proc Natl Acad Sci USA. 2017;114:5830–5837. doi: 10.1073/pnas.1610605114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrio E, Magli A, Munoz M, Peinado MA, Perlingeiro R, Suelves M. Muscle cell identity requires Pax7-mediated lineage-specific DNA demethylation. BMC Biology. 2016;14:30. doi: 10.1186/s12915-016-0250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal JJ, Rigby PWJ. Regulation of gene expression in vertebrate skeletal muscle. Exp Cell Res. 2010;316:3014–3018. doi: 10.1016/j.yexcr.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Chan SS, Kyba M. What is a master regulator? J Stem Cell Res Ther. 2013 doi: 10.4172/2157-7633.1000e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Goldhamer DJ. The core enhancer is essential for proper timing of MyoD activation in limb buds and branchial arches. Dev Biol. 2004;265:502–512. doi: 10.1016/j.ydbio.2003.09.018. [DOI] [PubMed] [Google Scholar]
- Chen JC, Love CM, Goldhamer DJ. Two upstream enhancers collaborate to regulate the spatial patterning and timing of MyoD transcription during mouse development. Dev Dyn. 2001;221:274–288. doi: 10.1002/dvdy.1138. [DOI] [PubMed] [Google Scholar]
- Chen JC, Ramachandran R, Goldhamer DJ. Essential and redundant functions of the MyoD distal regulatory region revealed by targeted mutagenesis. Dev Biol. 2002;245:213–223. doi: 10.1006/dbio.2002.0638. [DOI] [PubMed] [Google Scholar]
- Choi J, Costa ML, Mermelstein CS, Chagas C, Holtzer S, Holtzer H. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc Natl Acad Sci USA. 1990;87:7988–7992. doi: 10.1073/pnas.87.20.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J, et al. Expression of members of the myf gene family in human rhabdomyosarcomas. Br J Cancer. 1991;64:1039–1042. doi: 10.1038/bjc.1991.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coletti D, Daou N, Hassani M, Li Z, Parlakian A. Serum response factor in muscle tissues: from development to ageing. Eur J Transl Myol. 2016;26:6008. doi: 10.4081/ejtm.2016.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai G, Tajbakhsh S. Molecular and cellular regulation of skeletal myogenesis. Curr Top Dev Biol. 2014;110:1–73. doi: 10.1016/B978-0-12-405943-6.00001-4. [DOI] [PubMed] [Google Scholar]
- Conerly ML, Yao Z, Zhong JW, Groudine M, Tapscott SJ. Distinct activities of Myf5 and MyoD indicate separate roles in skeletal muscle lineage specification and differentiation. Dev Cell. 2016;36:375–385. doi: 10.1016/j.devcel.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides PG, Jones PA, Gevers W. Functional striated-muscle cells from non-myoblast precursors following 5-azacytidine treatment. Nature. 1977;267:364–366. doi: 10.1038/267364a0. [DOI] [PubMed] [Google Scholar]
- Coumailleau P, Duprez D. Sim1 and Sim2 expression during chick and mouse limb development. Int J Dev Biol. 2009;53:149–157. doi: 10.1387/ijdb.082659pc. [DOI] [PubMed] [Google Scholar]
- Creemers EE, Sutherland LB, Oh J, Barbosa AC, Olson EN. Coactivation of MEF2 by the SAP domain proteins myocardin and MASTR. Mol Cell. 2006;23:83–96. doi: 10.1016/j.molcel.2006.05.026. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, et al. Muscle-relevant genes marked by stable H3K4me2/3 profiles and enriched MyoD binding during myogenic differentiation. PLoS ONE. 2017;12:e0179464. doi: 10.1371/journal.pone.0179464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Diehl AG, Zareparsi S, Qian M, Khanna R, Angeles R, Gage PJ. Extraocular muscle morphogenesis and gene expression are regulated by Pitx2 gene dose. Invest Ophthalmol Vis Sci. 2006;47:1785–1793. doi: 10.1167/iovs.05-1424. [DOI] [PubMed] [Google Scholar]
- Dong F, et al. Pitx2 promotes development of splanchnic mesoderm-derived branchiomeric muscle. Development. 2006;133:4891–4899. doi: 10.1242/dev.02693. [DOI] [PubMed] [Google Scholar]
- Ehrlich KC, Paterson HL, Lacey M, Ehrlich M. DNA hypomethylation in intragenic and intergenic enhancer chromatin of muscle-specific genes usually correlates with their expression yale. J Biol Med. 2016;89:441–455. [PMC free article] [PubMed] [Google Scholar]
- Faerman A, Goldhamer DJ, Puzis R, Emerson CP, Jr, Shani M. The distal human myoD enhancer sequences direct unique muscle-specific patterns of lacZ expression during mouse development. Dev Biol. 1995;171:27–38. doi: 10.1006/dbio.1995.1257. [DOI] [PubMed] [Google Scholar]
- Forsberg H, Crozet F, Brown NA. Waves of mouse Lunatic fringe expression, in four-hour cycles at two-hour intervals, precede somite boundary formation. Curr Biol. 1998;8:1027–1030. doi: 10.1016/s0960-9822(07)00424-1. [DOI] [PubMed] [Google Scholar]
- Fukada SI, et al. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- Gates LA, et al. Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J Biol Chem. 2017;292:14456–14472. doi: 10.1074/jbc.M117.802074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayraud-Morel B, Chretien F, Flamant P, Gomes D, Zammit PS, Tajbakhsh S. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev Biol. 2007;312:13–28. doi: 10.1016/j.ydbio.2007.08.059. [DOI] [PubMed] [Google Scholar]
- Goldhamer DJ, Faerman A, Shani M, Emerson CP., Jr Regulatory elements that control the lineage-specific expression of myoD. Science. 1992;256:538–542. doi: 10.1126/science.1315077. [DOI] [PubMed] [Google Scholar]
- Goldhamer DJ, Brunk BP, Faerman A, King A, Shani M, Emerson CP., Jr Embryonic activation of the myoD gene is regulated by a highly conserved distal control element. Development. 1995;121:637–649. doi: 10.1242/dev.121.3.637. [DOI] [PubMed] [Google Scholar]
- Grifone R, et al. Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev Biol. 2007;302:602–616. doi: 10.1016/j.ydbio.2006.08.059. [DOI] [PubMed] [Google Scholar]
- Hamed M, Khilji S, Chen J, Li Q. Stepwise acetyltransferase association and histone acetylation at the Myod1 locus during myogenic differentiation. Sci Rep. 2013;3:2390. doi: 10.1038/srep02390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- Havis E, et al. Sim2 prevents entry into the myogenic program by repressing MyoD transcription during limb embryonic myogenesis. Development. 2012;139:1910–1920. doi: 10.1242/dev.072561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawke TJ, Garry DJ. Myogenic satellite cells: physiology to molecular biology. J Appl Physiol (2001) 2001;91:534–551. doi: 10.1152/jappl.2001.91.2.534. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Torres F, Rodriguez-Outeirino L, Franco D, Aranega AE. Pitx2 in embryonic and adult myogenesis. Front Cell Dev Biol. 2017;5:46. doi: 10.3389/fcell.2017.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzelstein D, et al. The homeobox gene Msx1 is expressed in a subset of somites, and in muscle progenitor cells migrating into the forelimb. Development. 1999;126:2689–2701. doi: 10.1242/dev.126.12.2689. [DOI] [PubMed] [Google Scholar]
- Hu P, Geles KG, Paik JH, DePinho RA, Tjian R. Codependent activators direct myoblast-specific MyoD transcription. Dev Cell. 2008;15:534–546. doi: 10.1016/j.devcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JK, McGlinn E, Harfe BD, Kardon G, Tabin CJ. Autonomous and nonautonomous roles of Hedgehog signaling in regulating limb muscle formation. Genes Dev. 2012;26:2088–2102. doi: 10.1101/gad.187385.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SM, Taylor JM, Tapscott SJ, Gurley CM, Carter WJ, Peterson CA. Selective accumulation of MyoD and myogenin mRNAs in fast and slow adult skeletal muscle is controlled by innervation and hormones. Development. 1993;118:1137–1147. doi: 10.1242/dev.118.4.1137. [DOI] [PubMed] [Google Scholar]
- Izzo A, Schneider R. The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics. Biochim Biophys Acta. 2016;1859:486–495. doi: 10.1016/j.bbagrm.2015.09.003. [DOI] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Ying C, Asakura A, Tapscott SJ, Rudnicki MA. MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development. 1997;124:4729–4738. doi: 10.1242/dev.124.23.4729. [DOI] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Ying C, Tapscott SJ, Goldhamer DJ, Rudnicki MA. Myogenic determination occurs independently in somites and limb buds. Dev Biol. 1999;206:219–231. doi: 10.1006/dbio.1998.9126. [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in Myf5: Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- Khan A, Mathelier A, Zhang XG. Super-enhancers are transcriptionally more active and cell type-specific than stretch enhancers. Epigenetics. 2018;13:910–922. doi: 10.1080/15592294.2018.1514231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, Charge SB, Seale P, Huh M, Rudnicki MA. Distinct roles for Pax7 and Pax3 in adult regenerative myogenesis. J Cell Biol. 2006;172:103–113. doi: 10.1083/jcb.200508001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharczuk KL, Love CM, Dougherty NM, Goldhamer DJ. Fine-scale transgenic mapping of the MyoD core enhancer: MyoD is regulated by distinct but overlapping mechanisms in myotomal and non-myotomal muscle lineages. Development. 1999;126:1957–1965. doi: 10.1242/dev.126.9.1957. [DOI] [PubMed] [Google Scholar]
- Laclef C, Souil E, Demignon J, Maire P. Thymus, kidney and craniofacial abnormalities in Six 1 deficient mice. Mech Dev. 2003;120:669–679. doi: 10.1016/s0925-4773(03)00065-0. [DOI] [PubMed] [Google Scholar]
- Le Grand F, et al. Six1 regulates stem cell repair potential and self-renewal during skeletal muscle regeneration. J Cell Biol. 2012;198:815–832. doi: 10.1083/jcb.201201050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Habas R, Abate-Shen C. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science. 2004;304:1675–1678. doi: 10.1126/science.1098096. [DOI] [PubMed] [Google Scholar]
- L’Honore A, Lamb NJ, Vandromme M, Turowski P, Carnac G, Fernandez A. MyoD distal regulatory region contains an SRF binding CArG element required for MyoD expression in skeletal myoblasts and during muscle regeneration. Mol Biol Cell. 2003;14:2151–2162. doi: 10.1091/mbc.E02-07-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Honore A, Rana V, Arsic N, Franckhauser C, Lamb NJ, Fernandez A. Identification of a new hybrid serum response factor and myocyte enhancer factor 2-binding element in MyoD enhancer required for MyoD expression during myogenesis. Mol Biol Cell. 2007;18:1992–2001. doi: 10.1091/mbc.E06-09-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Honore A, Ouimette JF, Lavertu-Jolin M, Drouin J. Pitx2 defines alternate pathways acting through MyoD during limb and somitic myogenesis. Development. 2010;137:3847–3856. doi: 10.1242/dev.053421. [DOI] [PubMed] [Google Scholar]
- Liu F. Enhancer-derived RNA: a primer. Genomics Proteom Bioinform. 2017;15:196–200. doi: 10.1016/j.gpb.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Chu A, Chakroun I, Islam U, Blais A. Cooperation between myogenic regulatory factors and SIX family transcription factors is important for myoblast differentiation. Nucleic Acids Res. 2010;38:6857–6871. doi: 10.1093/nar/gkq585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JR, et al. Control of facial muscle development by MyoR and capsulin. Science. 2002;298:2378–2381. doi: 10.1126/science.1078273. [DOI] [PubMed] [Google Scholar]
- Luo D, de Morree A, Boutet S, Quach N, Natu V, Rustagi A, Rando TA. Deltex2 represses MyoD expression and inhibits myogenic differentiation by acting as a negative regulator of Jmjd1c. Proc Natl Acad Sci USA. 2017;114:E3071–E3080. doi: 10.1073/pnas.1613592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magli A, et al. Time-dependent Pax3-mediated chromatin remodeling and cooperation with Six4 and Tead2 specify the skeletal myogenic lineage in developing mesoderm. PLoS Biol. 2019;17:e3000153. doi: 10.1371/journal.pbio.3000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- Michailovici I, Eigler T, Tzahor E. Craniofacial muscle development. Cranio Dev. 2015;115:3–30. doi: 10.1016/bs.ctdb.2015.07.022. [DOI] [PubMed] [Google Scholar]
- Mokalled MH, Johnson AN, Creemers EE, Olson EN. MASTR directs MyoD-dependent satellite cell differentiation during skeletal muscle regeneration. Genes Dev. 2012;26:190–202. doi: 10.1101/gad.179663.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncaut N, Cross JW, Siligan C, Keith A, Taylor K, Rigby PW, Carvajal JJ. Musculin and TCF21 coordinate the maintenance of myogenic regulatory factor expression levels during mouse craniofacial development. Development. 2012;139:958–967. doi: 10.1242/dev.068015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi K, et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I, Nabeshima Y. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oustanina S, Hause G, Braun T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004;23:3430–3439. doi: 10.1038/sj.emboj.7600346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, Zammit PS. Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development. 2012;139:2845–2856. doi: 10.1242/dev.069088. [DOI] [PubMed] [Google Scholar]
- Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- Relaix F, et al. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, et al. Six homeoproteins directly activate Myod expression in the gene regulatory networks that control early myogenesis. PLoS Genet. 2013;9:e1003425. doi: 10.1371/journal.pgen.1003425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth JF, et al. Differential role of p300 and CBP acetyltransferase during myogenesis: p300 acts upstream of MyoD and Myf5. EMBO J. 2003;22:5186–5196. doi: 10.1093/emboj/cdg473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PNJ, Stead RH, Braun T, Arnold HH, Jaenisch R. Myod or Myf-5 is required for the formation of skeletal-muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Scionti I, et al. LSD1 Controls timely MyoD expression via MyoD core enhancer transcription. Cell Rep. 2017;18:1996–2006. doi: 10.1016/j.celrep.2017.01.078. [DOI] [PubMed] [Google Scholar]
- Soleimani VD, et al. Transcriptional dominance of Pax7 in adult myogenesis is due to high-affinity recognition of homeodomain motifs. Dev Cell. 2012;22:1208–1220. doi: 10.1016/j.devcel.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani VD, et al. Snail regulates MyoD binding-site occupancy to direct enhancer switching and differentiation-specific transcription in myogenesis. Mol Cell. 2012;47:457–468. doi: 10.1016/j.molcel.2012.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YJ, Lee H. YB1/p32, a nuclear Y-box binding protein 1, is a novel regulator of myoblast differentiation that interacts with Msx1 homeoprotein. Exp Cell Res. 2010;316:517–529. doi: 10.1016/j.yexcr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Straface G, et al. Sonic hedgehog regulates angiogenesis and myogenesis during post-natal skeletal muscle regeneration. J Cell Mol Med. 2009;13:2424–2435. doi: 10.1111/j.1582-4934.2008.00440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlay PC, et al. Polycomb-repressed genes have permissive enhancers that initiate reprogramming. Cell. 2011;147:1283–1294. doi: 10.1016/j.cell.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajbakhsh S, Buckingham M. The birth of muscle progenitor cells in the mouse: spatiotemporal considerations. Curr Top Dev Biol. 2000;48:225–268. doi: 10.1016/s0070-2153(08)60758-9. [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–138. doi: 10.1016/s0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- Tam PPL. The control of somitogenesis in mouse embryos. Development. 1981;65:103–128. [PubMed] [Google Scholar]
- Tapscott SJ, Lassar AB, Weintraub H. A novel myoblast enhancer element mediates MyoD transcription. Mol Cell Biol. 1992;12:4994–5003. doi: 10.1128/mcb.12.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umansky KB, Gruenbaum-Cohen Y, Tsoory M, Feldmesser E, Goldenberg D, Brenner O, Groner Y. Runx1 transcription factor is required for myoblasts proliferation during muscle regeneration. PLoS Genet. 2015;11:e1005457. doi: 10.1371/journal.pgen.1005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustanina S, Carvajal J, Rigby P, Braun T. The myogenic factor Myf5 supports efficient skeletal muscle regeneration by enabling transient myoblast amplification. Stem Cells. 2007;25:2006–2016. doi: 10.1634/stemcells.2006-0736. [DOI] [PubMed] [Google Scholar]
- Venters SJ, Thorsteinsdottir S, Duxson MJ. Early development of the myotome in the mouse. Dev Dyn. 1999;216:219–232. doi: 10.1002/(SICI)1097-0177(199911)216:3<219::AID-DVDY1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Vicente-García C, Carvajal JJ. Transcription factors in mammalian myogenesis. In: Riviere JE, editor. Reference module in biomedical sciences. Amsterdam: Elsevier; 2018. [Google Scholar]
- Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Maltzahn J, Jones AE, Parks RJ, Rudnicki MA. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc Natl Acad Sci USA. 2013;110:16474–16479. doi: 10.1073/pnas.1307680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronova A, et al. Hedgehog signaling regulates MyoD expression and activity. J Biol Chem. 2013;288:4389–4404. doi: 10.1074/jbc.M112.400184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. The Msx1 homeoprotein recruits polycomb to the nuclear periphery during development. Dev Cell. 2011;21:575–588. doi: 10.1016/j.devcel.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci USA. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub AS, et al. YY1 is a structural regulator of enhancer-promoter loops. Cell. 2017;171(1573–1588):e1528. doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe D, Saxel O. Serial passing and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, et al. Loss of MyoD and Myf5 in skeletal muscle stem cells results in altered myogenic programming and failed regeneration. Stem Cell Rep. 2018;10:956–969. doi: 10.1016/j.stemcr.2018.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, et al. Myogenic transcriptional activation of MyoD mediated by replication-independent histone deposition. Proc Natl Acad Sci USA. 2011;108:85–90. doi: 10.1073/pnas.1009830108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, et al. ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res. 2011;39:7415–7427. doi: 10.1093/nar/gkr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias AL, Lewandoski M, Rudnicki MA, Gage PJ. Pitx2 is an upstream activator of extraocular myogenesis and survival. Dev Biol. 2011;349:395–405. doi: 10.1016/j.ydbio.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS. Function of the myogenic regulatory factors Myf5, MyoD, myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin Cell Dev Biol. 2017;72:19–32. doi: 10.1016/j.semcdb.2017.11.011. [DOI] [PubMed] [Google Scholar]
- Zammit PS, Relaix F, Nagata Y, Ruiz AP, Collins CA, Partridge TA, Beauchamp JR. Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci. 2006;119:1824–1832. doi: 10.1242/jcs.02908. [DOI] [PubMed] [Google Scholar]
- Zhang X, Patel SP, McCarthy JJ, Rabchevsky AG, Goldhamer DJ, Esser KA. A non-canonical E-box within the MyoD core enhancer is necessary for circadian expression in skeletal muscle. Nucleic Acids Res. 2012;40:3419–3430. doi: 10.1093/nar/gkr1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg JM, Pedraza-Alva G, Jost JP. MyoD1 promoter autoregulation is mediated by two proximal E-boxes. Nucleic Acids Res. 1994;22:2234–2241. doi: 10.1093/nar/22.12.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]