Summary
Enterovirus A71 (EV-A71) infection causes hand-foot-and-mouth disease (HFMD) and fatal neurological diseases, and there are no effective treatments. Host factors play key roles in establishing viral infection and determining the disease progression and outcome of antiviral therapies. In this study, we found that the expression of Pim1 was significantly upregulated in EV-A71 infection. Ectopic expression or silencing of Pim1 promoted or inhibited EV-A71 replication through two distinct mechanisms. Pim1 enhanced viral IRES activity by increasing viral 2A protease-mediated eIF4G cleavage and blocked AUF1, a suppressor of IRES, translocation from the nucleus to cytosol. More importantly, we discovered that Pim1 inhibitors (SGI-1776, AZD-1208, and CX-6258) reduced EV-A71 reproduction. Particularly, CX-6258 remarkably reduced EV-A71 reproduction more than 1,000 times, providing a potential therapeutic agent for EV-A71 treatment.
Subject Areas: Biological Sciences, Biochemistry, Microbiology, Virology, Viral Microbiology, Molecular Microbiology, Cell Biology
Graphical Abstract
Highlights
-
•
Pim1 is a key positive regulator of EV-A71 infection
-
•
Pim1 enhances viral IRES activity by increasing 2A protease activity
-
•
Pim1 blocks AUF1 translocation from the nucleus to cytosol
-
•
Pim1 inhibitors potently inhibit EV-A71 reproduction more than 1,000 times
Biological Sciences; Biochemistry; Microbiology; Virology; Viral Microbiology; Molecular Microbiology; Cell Biology
Introduction
Enterovirus A71 (EV-A71), a typical positive single-stranded RNA virus, was first identified in 1969 in patients with central nervous system disease (Manki et al., 1997, Schmidt et al., 1974). EV-A71 belongs to the Picornaviridae family of viruses, with a genome of nearly 7.4 kb encoding four structural proteins (VP1, VP2, VP3, and VP4) and seven non-structural proteins (2A, 2B, 2C, 3A, 3B, 3C, and 3D) (Lin et al., 2009a, Lu et al., 2012). EV-A71 has also been recognized as an agent for hand-foot-and-mouth disease (HFMD). HFMD is usually a mild and self-limiting disease; however, sometimes it causes severe neurological disorders or even death due to the limited effective treatment for EV-A71 infection (Hamaguchi et al., 2008, Yi et al., 2011). Furthermore, EV-A71 has shown repeated outbreaks worldwide in the last decade and infects millions of people annually (Yi et al., 2011).
It is well known that the initiation of EV-A71 protein translation the first step for initiating viral replication after entry into host cells is driven by the internal ribosome entry site (IRES) in the 5′ UTR. This occurs in a cap-independent manner and is regulated by many positive and negative regulators (Lee et al., 2017). EV-A71 contains a type I IRES with five stem loops (domains II–VI), which binds to the cleaved eukaryotic initiation factor eIF4G (Thompson and Sarnow, 2003). The viral translation process is highly promoted by viral protein 2A protease (2Apro). The protein 2Apro cleaves eIF4G, and then the cleaved eIF4G binds and enhances viral IRES activity (Glaser and Skern, 2000). Although a number of IRES trans-acting factors (ITAFs), such as AU-rich element-binding factor 1 (AUF1) (Lin et al., 2014), upstream element-binding protein (FBP) 1–3 (Huang et al., 2011), hnRNP K (Lin et al., 2008), and hnRNP A1 (Lin et al., 2009b) have been identified, current knowledge on their regulation is very limited.
Previous studies have uncovered several host factors required for EV-A71 replication. For example, the virus first attaches to the cell membrane factors heparan sulfate glycosaminoglycans (Tan et al., 2013) and PSGL1 (Nishimura et al., 2009). It then releases the viral genome for the production of viral components helped by host factors such as AUF1 (Lin et al., 2014), FBP1 (Huang et al., 2011), and hnRNP A1 (Lin et al., 2009b) and is subsequently assembled for the release of the virus from cells. It has been shown that some pan protein kinases, such as phosphatidylinositol 3-kinase and MERK/ERK kinases, play crucial roles in EV-A71 replication (Duan et al., 2017, Hung et al., 2014, Wang et al., 2017, Wong et al., 2005). The inhibitors of these kinases potently inhibited viral replication (Sun et al., 2016, Zhang et al., 2018). However, results from anticancer clinical studies of these inhibitors showed strong side effects. It would be a great idea to target a specific kinase to treat cancers and viral infections.
Pim1, a member of the calmodulin-dependent protein kinase family together with another two kinases, is a serine-threonine kinase identified as the preferential site of integration for the Moloney murine leukemia virus (Selten et al., 1985). Pim1 is elevated and plays crucial roles in several types of human cancers such as prostate adenocarcinoma, mantle cell lymphomas, and hematopoietic malignancies (Nawijn et al., 2011). Recently, several groups reported that Pim1 affects viral transcription activation and modulates the latency and productive infections (De Vries et al., 2015, Park et al., 2015, Vries et al., 2016). However, to date there have been no reports that show any response or effect of Pim1 in EV-A71 replication. Therefore, in this study, we analyzed the expression profile of Pim1 after EV-A71 infection. We showed that Pim1 was upregulated in response to EV-A71 infection. More importantly, we not only demonstrated the importance of Pim1 in EV-A71 infection but also revealed the potent antiviral effects of Pim1 inhibitors, potential agents for antiviral treatment as well.
Results
Stimulation of Pim1 mRNA Level by EV-A71 Infection
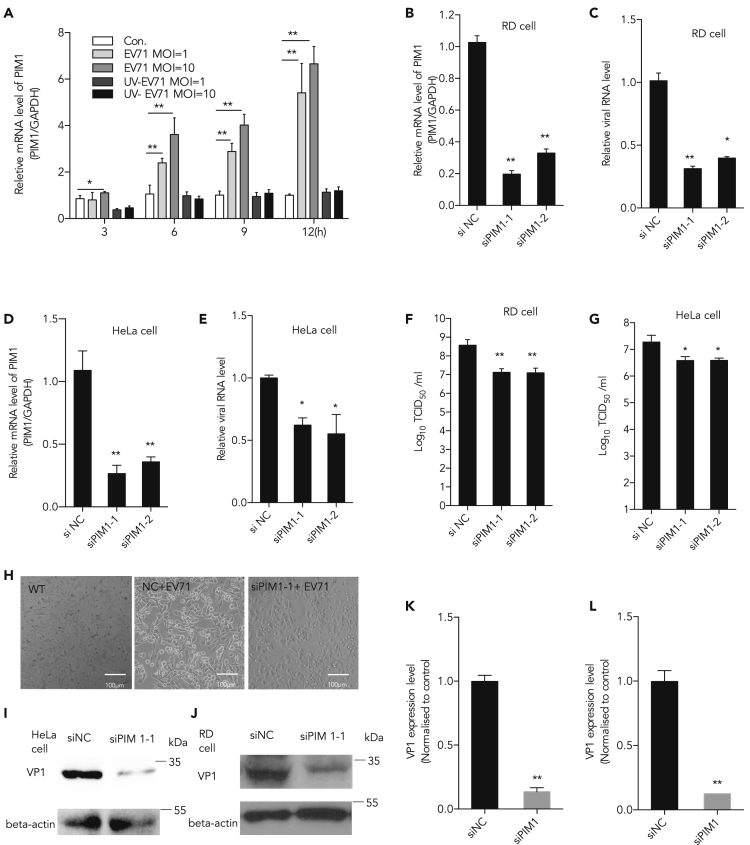
To reveal the regulation pattern of Pim1 in EV-A71 infection, the mRNA level of Pim1 was analyzed during the process of viral infection. RD cells were infected with EV-A71 or inactive EV-A71 (UV light-inactive virus) at MOI of 1 and 10, and cellular mRNA was extracted at different time points (post infection, p.i.). The results showed that the mRNA level of Pim1 notably increased as early as 3 h p.i. at a high MOI of 10, and significant upregulation was observed at 6 and 9 h p.i. both at MOIs 1 and 10, when compared with the control group (Figure 1A). Notably, although virus treated with UV would not affect virus entry (Dong et al., 2018, Dan et al., 2019), it did not stimulate Pim1 response.
Figure 1.
Inhibiting EV-A71 Replication by Knockdown of Pim1
(A) RD cells were infected with EV-A71 or inactive EV-A71 (UV light-inactive virus, negative control) at MOI of 10, and cellular mRNA was extracted at different time points p.i. Pim1 mRNA level was determined by qRT-PCR.
(B and D) Pim1 was silenced by two individual siRNA duplexes si-Pim1-1 and si-Pim1-2. The relative cellular Pim1 mRNA level was quantified by qRT-PCR at 48 h after transfection in both (B) RD and (D) HeLa cells. GAPDH was used as the internal control.
(C and E) (C) RD and (E) HeLa cells treated with Pim1-specific siRNA and infected with EV-A71 at MOI 0.01. The extracellular viral RNA level was calculated at 48 h.
(F and G) Viral titer was further determined in both (F) RD and (G) HeLa cells.
(H) Cell survival image was taken in RD cells.
(I and J) Pim1 was silenced in EV-A71-infected (J) RD and (I) HeLa cells.
(K) The quantification results of (I).
(L) The quantification results of (J). Protein levels of viral VP1 protein were detected and quantitated. The density of VP1 to β-actin was set as 1, and the relative fold change is indicated.
Data are represented as mean ± SD (n = 3). Student's t test, *p < 0.05, compared with mock group; **p < 0.01, compared with mock group.
Inhibition of EV-A71 Replication by Knockdown of Pim1
To determine whether Pim1 was required during EV-A71 infection, an RNAi approach was employed to knockdown the level of Pim1; 48 h after small interfering RNA (siRNA) transfection, the level of Pim1 mRNA was reduced in RD and HeLa cells by 80% and 70% by siPim1-1- and siPim1-2-specific siRNAs, respectively, when compared with non-targeting sequences, as quantified by qRT-PCR (p < 0.01) (Figures 1B and 1D). MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays were performed to determine cell viability upon Pim1 silencing. There was no significant difference in cell number between siPim1-transfected cells and non-target RNA-transfected cells at 24 and 48 h post transfection (data not shown).
RD or HeLa cells were transfected with siPim1-1- or siPim1-2-specific siRNA at 40 nM or non-target siRNA as control. After 24-h transfection, cells were infected with EV-A71 at an MOI of 0.01. As shown in Figures 1C and 1E, extracellular virion RNA levels were strongly decreased in Pim1-specific siRNA-treated cells when compared with cells treated with non-targeting sequences (p < 0.01). The viral titer was also determined after silencing Pim1. The results showed that the viral titer was significantly decreased in both RD and HeLa cells after knockdown of Pim1 (Figures 1F and 1G). Knockdown of Pim1 protected the cytopathic effects induced from EV-A71 infection in RD cells (Figure 1H). The viral protein (VP1) expression level was determined in both RD and HeLa cells. Results showed that VP1 was significantly decreased in both RD and HeLa cells after silencing Pim1 (Figures 1I–1L).
Promoting EV-A71 Replication by Ectopic Expression of Pim1
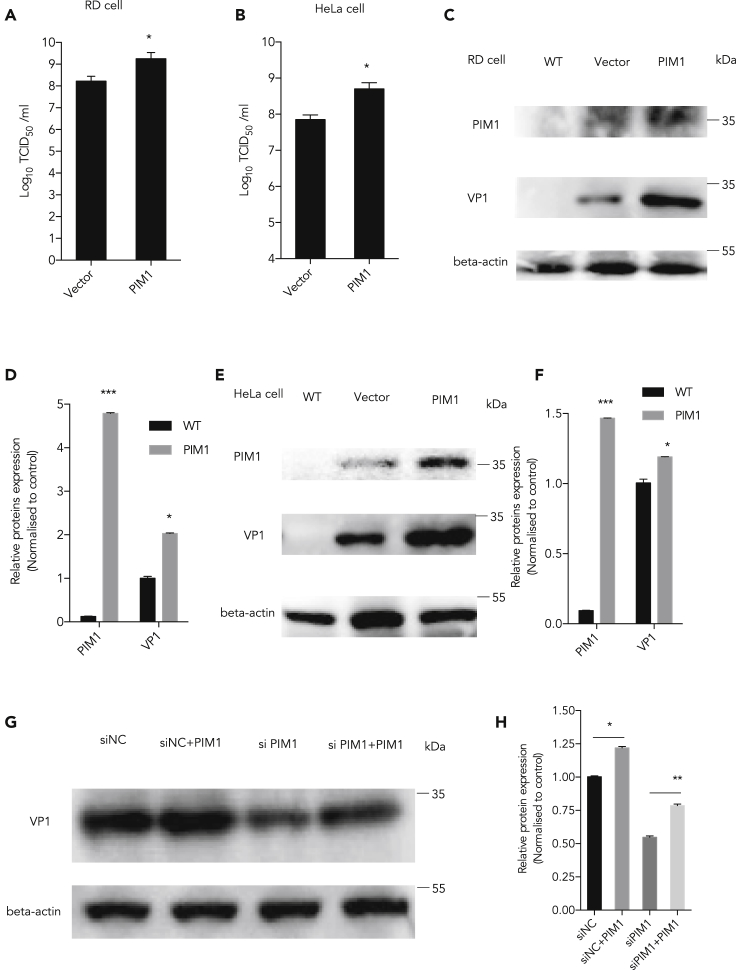
To further validate the function of Pim1 on virus replication, we conducted gain-of-function studies by ectopic expression of Pim1 by transfection of a Pim1-expressing plasmid into both RD and HeLa cells. We showed that ectopic expression of Pim1 increased viral titers both in RD and HeLa cells (Figures 2A and 2B). The viral VP1 protein level also significantly increased in the cells with ectopic expression of Pim1 (Figures 2C–2F). Furthermore, we performed a rescue experiment to show the function of Pim1 in EV-A71 infection. We first knocked down the endogenous Pim1 expression by an siRNA that specifically targets the 3′ UTR in RD cells, ectopically expressed Pim1 (resistance to siRNA) with a plasmid for 48 h, and then infected with EV-A71 at MOI = 1 for 24 h. Our results showed that the reduced VP1 level by Pim1 depletion was restored by the ectopic Pim1 expression, excluding the possibility that the VP1 decrease by siRNA is due to off-target effects (Figures 2G and 2H).
Figure 2.
Promoting EV-A71 Replication by Ectopic Expression of Pim1
(A and B) After ectopic expression of Pim1, viral titer was determined following the below-mentioned methods in (A) RD and (B) HeLa cells.
(C and E) Pim1 was ectopically expressed in EV-A71-infected cells at MOI 1 for 24 h in (C) RD and (E) HeLa cells. Protein levels of Pim1 and viral protein VP1 were detected. β-Actin was used as the internal control.
(D) The quantification results of (C).
(F) The quantification results of (E).
(G) siRNA targeting Pim1 3′ UTR at 40 nM was co-transfected with the Pim1 expression plasmid for 48 h in RD cells and then infected with EV71 at MOI 1 for 24 h.
(H) The quantification results of (G). The cell lysates were collected for western blot assay. Protein levels were detected and quantitated. The density of individual protein to β-actin in the control was set as 1, and the relative fold change is indicated.
Data are represented as mean ± SD (n = 3). *p < 0.05, compared with mock group; **p < 0.01, compared with mock group.
Enhancing Viral IRES Activity by Pim1
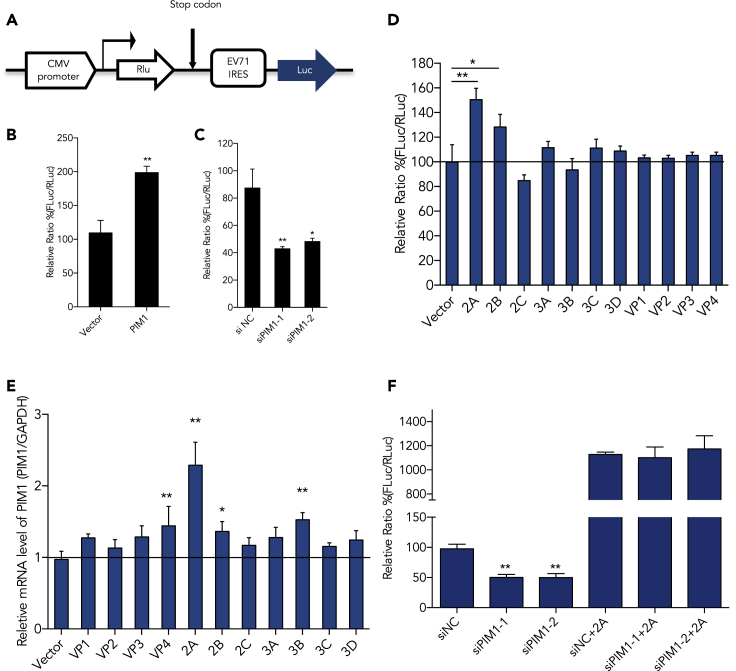
It is well known that enterovirus protein translation is initiated through viral IRES elements in a cap-independent manner. A dicistronic reporter plasmid was used to evaluate EV-A71 IRES activity when Pim1 was modulated (Figure 3A). The relative IRES activity is represented by calculating the ratio of FLuc expression to RLuc expression. As indicated in the Methods section, Pim1 was over-expressed or silenced by transfecting with a Pim1-expressing plasmid or siRNA for 24 h and then transfected with pIRES plasmid. The IRES activity was analyzed 48 h after first round transfection. As shown in Figures 3B and 3C, over-expression of Pim1 increased the EV-A71 IRES activity to 200% of the control, whereas knockdown of endogenous Pim1 expression decreases EV-A71 IRES activity to 40% of the control. We later co-transfected pIRES and a viral protein-expressing plasmid individually. Our results showed that viral 2Apro was the most effective protein that increased the IRES activity (Figure 3D). After we transfected all viral protein expression plasmids into 293T cells, we found that viral 2Apro as well as VP4, 2B, and 3B stimulated Pim1 expression, whereas only 2Apro significantly increased the mRNA level of Pim1 by more than two times (Figure 3E). To further address our findings, we then performed the rescue experiments. We silenced Pim1 and then ectopically expressed 2Apro. We demonstrated that 2Apro rescued the reduction of IRES activity after knockdown of Pim1 (Figure 3F).
Figure 3.
Enhancing Viral IRES Activity by Pim1
(A) A diagram of the dicistronic reporter plasmid, which was used to evaluate EV-A71 IRES activity when Pim1 was modulated.
(B and C) (B) The relative IRES activity after ectopic expression of Pim1 or (C) knockdown of Pim1 in 293T cells.
(D) After co-transfection of the reporter plasmid and individual viral protein expression plasmid into 293T cells for 48 h, the RLuc and FLuc activities were analyzed.
(E) The relative mRNA level of Pim1 was determined by qRT-PCR assay after transfection of the individual viral protein expression plasmid into 293T cells for 48 h.
(F) IRES activities after ectopic expression of 2Apro, with or without silencing of Pim1. Results are represented as mean ± SD from three independent experiments. *p < 0.05, **p < 0.01.
Suppression of EV-A71 Infection by Pim1 Inhibitors
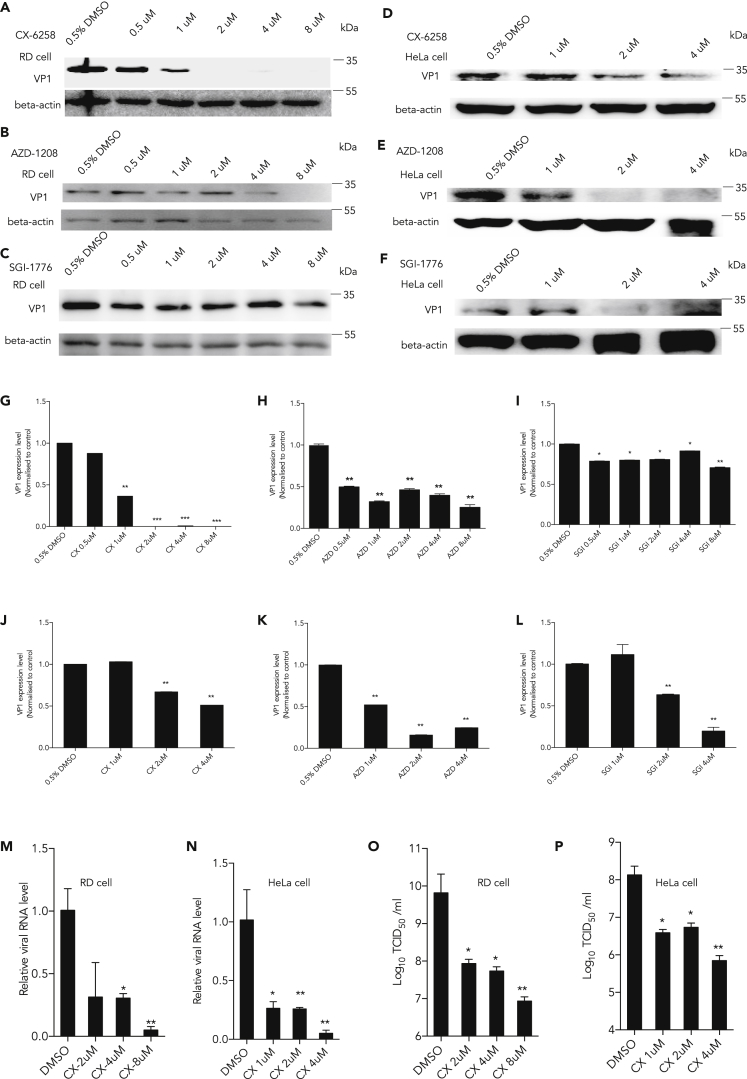
Highly selective Pim1 inhibitors have been developed for anti-tumor activity in the past decade. CX-6258 ((3E)-5-chloro-3-[[5-[3-[(hexahydro-4-methyl-1H-1,4-diazepin-1-yl)carbonyl]phenyl]-2-furanyl]methylene]-1,3-dihydro-2H-indol-2-one), SGI-1776 (N-[(1-methyl-4-piperidinyl)methyl]-3-[3-(trifluoromethoxy)phenyl]-imidazo[1,2-b]pyridazin-6-amine), and AZD-1208 ((5Z)-[[2-[(3R)-3-amino-1-piperidinyl][1,1′-biphenyl]-3-yl]methylene]-2,4-thiazolidinedione) are novel, orally bioavailable inhibitors for treating patients with cancer in clinical trials (Haddach et al., 2012, Chen et al., 2009, Harada et al., 2015). To further test whether Pim1 could be a potential drug target against EV-A71 infection, we examined the effects of these inhibitors on EV-A71 infection in both RD and HeLa cells. We first tested all the inhibitors on cell viability in 293T, RD, and HeLa cells by MTT assay. Our results showed that SGI-1776 and CX-6258 inhibited cell growth at a higher concentration, whereas AZD-1208 did not affect cell growth (Figure S1). We pretreated RD or HeLa cells for 2 h with indicated inhibitors and concentrations and then infected with EV-A71 for 24 h at MOI 1. As shown in Figures 4A–4L, the VP1 expression level was remarkably repressed by Pim1 inhibitors in a dose-dependent manner. CX-6258 showed the best inhibitory effects in RD cells (Figures 4A and 4G). We further used CX-6258 as a Pim1 inhibitor to determine the extracellular viral mRNA levels, as mentioned above. The results showed that after inhibiting Pim1, viral mRNA levels were significantly decreased both in RD and HeLa cells (Figures 4M and 4N). In addition, the viral titer was also decreased by more than 103 after inhibiting Pim1 activity with CX-6258 in RD and HeLa cells (Figures 4O and 4P).
Figure 4.
Suppressing EV-A71 Infection by Pim1 Inhibitors
(A–F) Different Pim1 inhibitors CX-6258 (A and D), AZD-1208 (B and E), and SGI-1776 (C and F) were added in the culture media at the indicated concentration for 2 h with 10% fetal bovine serum and then cells were infected with EV-A71 at MOI 1 for 24 h; 0.5% DMSO was loaded in the control group. The VP1 protein level was determined, and beta-actin was loaded as internal control.
(G and J) The quantification results of (A) and (D), respectively.
(H and K) The quantification results of (B) and (E), respectively.
(I and L) The quantification results of (C) and (F), respectively.
(M and N) RD and HeLa Cells were firstly treated with CX-6258 at the indicated concentrations for 2 h, then infected with EV-A71 at MOI 0.01 for 72 h. The extracellular viral RNA levels in the supernatant were determined by qRT-PCR.
(O and P) RD and HeLa Cells were firstly treated with CX-6258 at the indicated concentration for 2 h, then infected EV-A71 at MOI 0.01 for 72 h. The viral titer in the supernatant was measured by TCID50 assay. Protein levels were detected and quantitated by image density. The density of individual protein to β-actin in the control was set as 1, and the relative fold change is indicated.
Results are represented as mean ± SD from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S1.
Inhibition of 2Apro-Mediated eIF4G Cleavage by Inhibition of Pim1
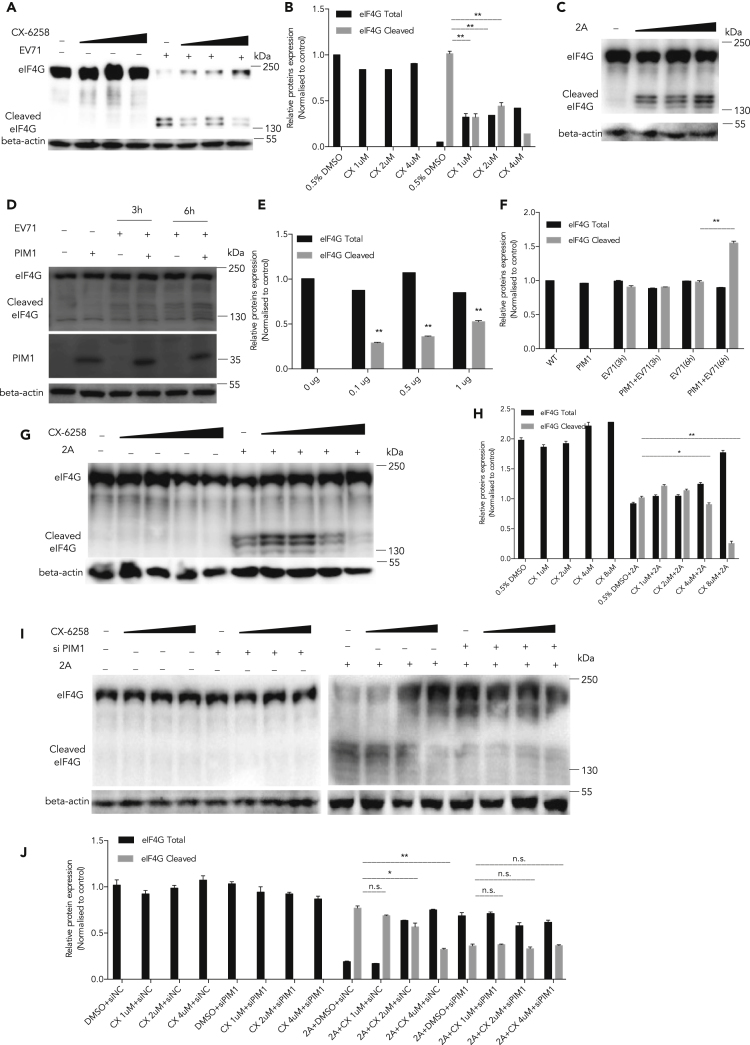
To address how Pim1 affects the IRES activity regulated by 2Apro, we checked the effect of Pim1 on 2Apro cleavage activity. We and others previously reported that the IRES-driven translation was activated after the cleavage of eIF4G by 2Apro (Lu et al., 2011, Dong et al., 2018, Dan et al., 2019). We first tested whether Pim1 inhibitors could affect the cleavage of eIF4G after virus infection. As shown in Figures 5A and 5B, RD cells were treated with CX-6258 at different concentrations and then infected with EV-A71 at MOI 10. After 9 h p.i., the cleavage of eIF4G was significantly inhibited. We confirmed the expression plasmid 2Apro cleavage activity in 293T cells (Figures 5C and 5E). To further demonstrate the underlying mechanism, HEK293T cells were first treated with CX-6258 for 2 h and then co-transfected with a 2Apro-expressing plasmid for 36 h. We found that the cleavage of eIF4G was also inhibited with Pim1 inhibitor in a dose-dependent manner (Figures 5G and 5H). Similarly, when 293T cells were transfected with the Pim1 expression plasmid for 48 h, and then infected with EV-A71 at MOI 10 for the indicated time points, the cleavage of eIF4G was upregulated in the Pim1 ectopically expressed cells (Figures 5D and 5F). To test whether the Pim1 inhibitor would directly affect 2Apro activity, we further examined the status of 2Apro -mediated eIF4G cleavage in Pim1 knockdown cells treated with CX-6258 at different concentrations (1, 2, and 4 μM). Our results showed that CX-6258 did not further affect 2Apro activity in the Pim1-depleted cells (Figures 5I and 5J), demonstrating that the inhibitory effects of 2Apro-mediated eIF4G cleavage were mediated through the suppressed Pim1 kinase activity.
Figure 5.
Inhibiting 2Apro-mediated eIF4G Cleavage by CX-6258
(A) RD cells were treated with different concentrations of CX-6258 and then infected with EV-A71 at MOI 10. After culture for 9 h, cell lysates were collected and the cleavage of eIF4G was determined.
(B) The quantification results of (A).
(C) HEK293T cells were transfected with p2Apro (0.1, 0.5, and 1 μg) for 36 h, and the cleavage of eIF4G was determined.
(D) HEK293T cells were first treated with different concentrations of CX-6258 for 2 h and transfected with 1 μg 2Apro-expressing plasmid for 36 h. The cleavage of eIF4G was determined.
(E and F) The quantification results of (C) and (D), respectively.
(G) 293T cells were transfected with a Pim1-expressing plasmid for 48 h and then infected with EV-A71 at MOI 10 for the indicated time. The cleavage of eIF4G was determined.
(H) The quantification results of (G).
(I) 293T cells were first transfected with Pim1-specific siRNA for 48 h and then treated with CX-6258 at the indicated concentrations for 2 h, followed by transfection of a 2Apro-expressing plasmid at 48 h. Cell lysates were collected, and the status of eIF4G cleavage was examined.
(J) The quantification results of (I). Protein levels were detected and quantitated by image density. The density of individual protein to β-actin or GAPDH in the control was set as 1, and the relative fold change is indicated.
Results are represented as mean ± SD from three independent experiments. *p < 0.05, **p < 0.01.
Induction of the Cytoplasmic Accumulation of AUF1 by Inhibition of Pim1
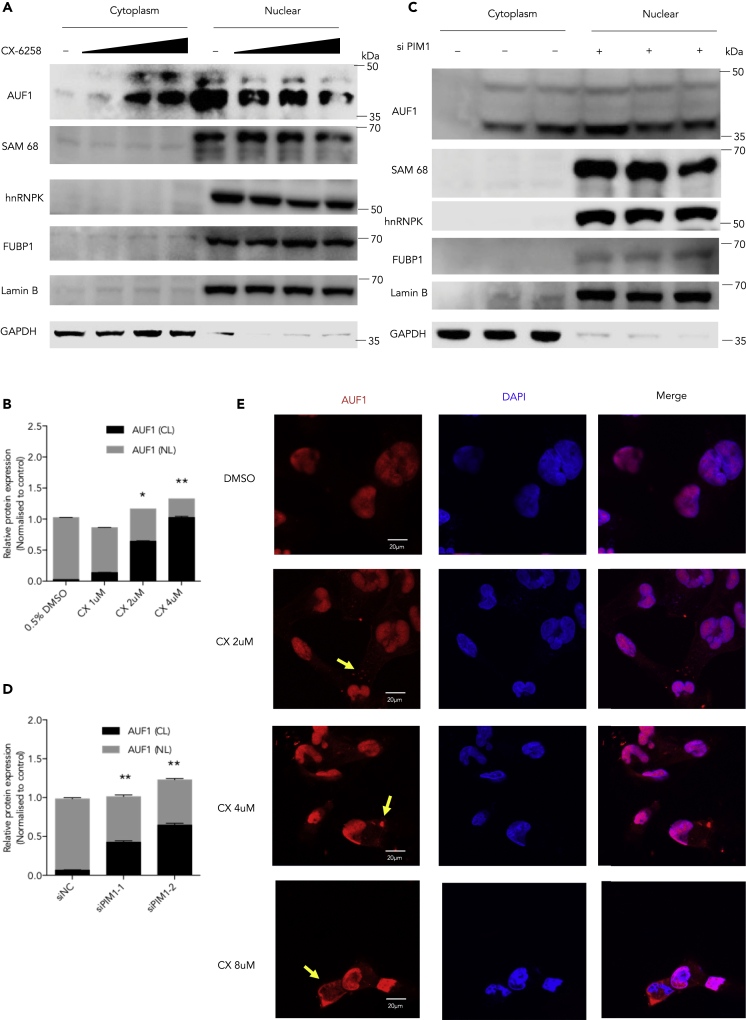
We further explored whether other mechanism(s) of Pim1 are involved in the regulation of the viral IRES activity. After treating cells with the Pim1 inhibitor CX-6258 for 2 h without viral infection or 2A expression, we isolated the cytoplasm and nuclear proteins and applied western blot assays. After examining the translocation of ITAF factors, including AUF1, hnRNP K, FBP1, and Sam68, we found that only AUF1, a suppressor of the IRES, was affected by the Pim1 inhibitor. The accumulation of AUF1 in the cytoplasm was significantly increased (Figures 6A and 6B). Consistently, only AUF1, but not hnRNP K, FBP1, or Sam68, translocated from the nucleus to the cytosol after knockdown of Pim1 by target-specific siRNA (Figures 6C and 6D). We also used confocal images to examine the cellular localization of AUF1. AUF1 accumulation was shown in the cytoplasm in the cells treated with the Pim1 inhibitor (Figure 6E). We also noticed that AUF1 appeared to form puncta in the cytoplasm at 2 and 4 μM concentrations of CX-6258, which correlated with the stress response of Pim1 and AUF1 (Braso-Maristany et al., 2016, Roggli et al., 2012).
Figure 6.
Inducing AUF1 Cytoplasmic Accumulation by CX-6258
(A) RD cells were treated with CX-6258 at the indicated concentrations for 2 h, and then the cytoplasmic and nuclear proteins were extracted. The cytoplasm and nuclear distribution of AUF1, Sam68, hnRNP K and FUBP1 were determined by western blot assay.
(B) The quantification results of (A).
(C) Pim1-specific siRNAs were transfected into RD cells for 48 h. The cytoplasmic and nuclear proteins were extracted. The protein levels of AUF1, Sam68, hnRNP K, and FUBP1 were examined by western blot assays.
(D) The quantification results of (C).
(E) The cytoplasmic and nuclear distribution of AUF1 was detected in RD cells after treating with CX-6258 at indicated concentrations for 2 h under a fluorescence microscope. The yellow arrows indicated the representative cells of AUF1 translocated in the cytosole.
Results are represented as mean ± SD from three independent experiments. *p < 0.05, **p < 0.01.
Discussion
In this study, we have provided evidence that Pim1 is a positive regulator for EV-A71 replication. Pim1 elevated EV-A71 replication by upregulating IRES activity by increasing 2Apro-mediated eIF4G cleavage and blocking AUF1 translocation from the nucleus to the cytosol. More importantly, we discovered that Pim1 inhibitors potently repressed EV-A71 infection. Particularly, CX-6258 reduced the viral titer over 1,000 times, a strong indication as a potential antiviral drug against enterovirus infections.
Previously, different expression patterns were observed, and many novel host genes regulated by EV-A71 infection have been identified, including those involved in the immune response, endoplasmic reticulum stress, and vesicular trafficking. In 2004, Shih et al. observed the upregulation of apoptotic genes in response to EV-A71 infection by analyzing 10,692 gene profiles in infected human SF268 cells (Shih et al., 2004). In addition, Leong et al. found that there is a trend to inhibit cell apoptosis and cell growth arrest by analyzing 7,600 genes in EV-A71 (strain MS/7423/87, B2 sub-genotype)-infected RD cells (Leong and Chow, 2006). In 2011, Kan et al. used RNAi screening and identified 256 host factors involved in EV-A71 replication in RD cells (Wu et al., 2016). However, the function and importance of these host factors in EV-A71 replication are largely unclear. In our previous studies, we demonstrated that several heat shock proteins, including ERp57, Hsc70, and Hsp27, play a crucial role in EV-A71 infection by regulating viral IRES activities. In these studies, we showed that heat shock protein inhibitors markedly suppressed virus replication by decreasing IRES activities (Wang et al., 2016, Dong et al., 2018, Dan et al., 2019).
It was particularly interesting to find that Pim1, an important kinase involved in human cancers and never reported to be involved in the process of EV-A71 infection, was upregulated following EV-A71 infection (Figure 1A). We postulated that Pim1 must have played important role in EV-A71 replication because the process of virus replication often shares similar signaling cascades to those involved in cancer cell growth, migration, or cell death. By loss- and gain-of-function studies, we showed that Pim1 played crucial role in facilitating EV-A71 replication (Figures 1 and 2). Our data demonstrated the importance of Pim1 in EV-A71 infection.
IRES plays a key role in the synthesis of viral proteins during EV-A71 infection (Gao et al., 2014). In this study, we further demonstrated that Pim1 promoted IRES activity upon EV-A71 infection (Figure 3). EV-A71 infection increased Pim1 expression level (Figures 1 and 3), and the increased Pim1 expression promoted EV-A71 replication (Figure 2). We then tested if Pim1 inhibitors restricted EV-A71 replication. All three Pim1-specific inhibitors (SGI-1776, CX-6258, AZD-1208) showed a high potency to suppress viral infectivity (Figure 4). We chose the best antiviral inhibitor CX-6258 and tested its effects on both intracellular and extracellular viral RNA levels, which reflected viral replication and reproduction, respectively. We showed that both viral RNA levels and viral titer dramatically decreased in a dose-dependent manner (Figure 4).
EV-A71 recruits many host factors to facilitate viral protein synthesis via IRES-mediated translation. We previously showed that Hsc70 binds the full length of viral RNA and promotes IRES activity. Hsc70 regulates IRES activity and serves as an antiviral target of EV-A71 infection (Dong et al., 2018). It is interesting to test whether Pim1 kinase could enhance IRES activity in other ways. We first found that 2Apro rescued the reduced IRES activity in Pim1-silenced cells. 2Apro is one of the well-known proteases that cleaves eIF4G to promote viral replication (Glaser and Skern, 2000). The cleaved eIF4G separates into two parts, and the C-terminal part directly binds IRES to initiate the viral translation process (Ohlmann et al., 1996). However, it was not known if Pim1 could affect IRES activity by regulating 2Apro activity. Surprisingly, we did observe that inhibition of Pim1 by either both siRNAs or the inhibitor (CX-6258) inhibited the cleavage of eIF4G cleavage upon EV-A71 infection (Figure 5). The Pim1 inhibitor suppressed 2Apro-mediated eIF4G cleavage in a dose-dependent manner (Figure 5). More importantly, the Pim1 inhibitor showed no effect on 2Apro-mediated eIF4G cleavage after knockdown of Pim1, demonstrating the high specificity of the inhibitor to Pim1 kinase, instead of a possible effect on 2A protease. In addition, we revealed that Pim1 (possibly the dominant p40 isoform) played a crucial role in the control of AUF1 cytosol translocation (Figure 6).
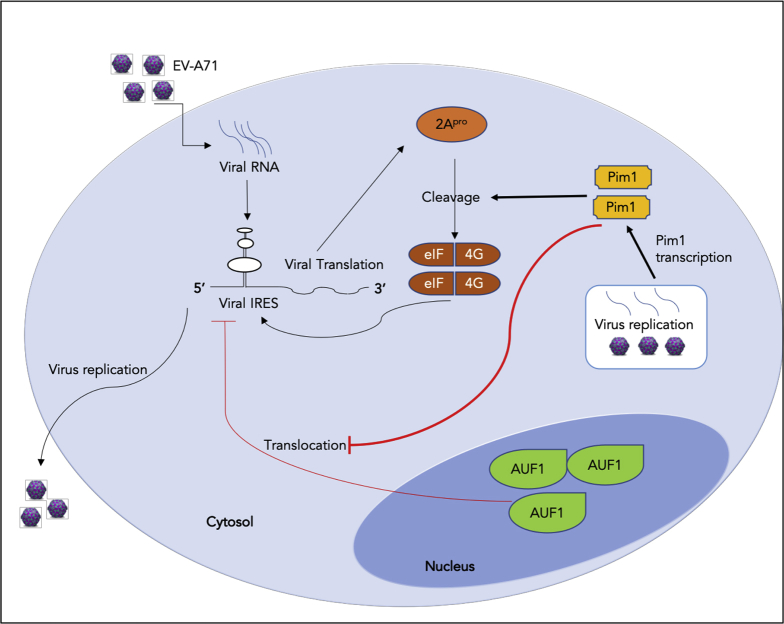
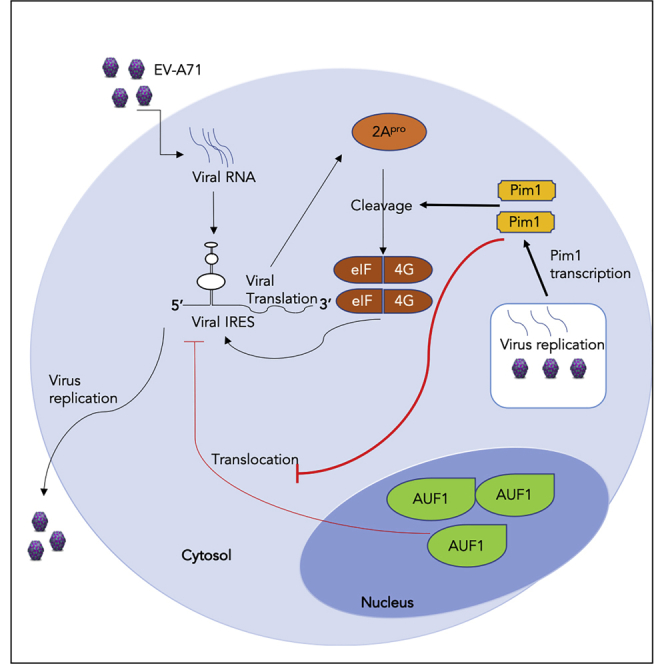
In conclusion, we have demonstrated that Pim1 contributes to EV-A71 infection through two distinct mechanisms. Pim1 exhibits a positive role in EV-A71 infection by enhancing IRES activity by stimulating 2Apro-mediated eIF4G cleavage and blocking AUF1 cytosol translocation (Figure 7). Most importantly, we provide evidence that Pim1 inhibitors, particularly CX-6258, decrease viral reproduction by over 1,000 times, providing a potent antiviral agent for potential clinical settings in the future.
Figure 7.
Diagram of Pim1 Function in EV-A71 Infection
Limitations of the Study
Our study has demonstrated that Pim1 exhibits a positive role in EV-A71 replication by enhancing IRES activity by two distinct mechanisms, i.e., enhancing 2Apro-mediated eIF4G cleavage and blocking AUF1 cytosol translocation. We have performed several experiments such as western blot, qRT-PCR, and mass spectrum analysis to see if there are interaction sites between these proteins. However, we failed to determine whether Pim1 directly binds eIF4G or affects eIF4G phosphorylation. The interplay between VP4, 2B, 3B, and Pim1 cannot be excluded, and further studies are required in the future. Nevertheless, as anticancer agents, Pim1 inhibitors show great antiviral activities that are very attractive for the treatment of patients with HFMD in the future.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The work was partially supported by grants from National Science Foundation of China [81671995], grants from The Science Technology and Innovation Committee of Shenzhen Municipality [JCYJ20170818100531426, JCYC20180507181627057], the General Research grant of Hong Kong [11100215], and Strategic funds from City University of Hong Kong.
Author Contributions
F.Z., J.L., Q.W., and Y.C. performed the experiments; M.-L.H. designed and supervised the research; and F.Z. and M.-L.H. analyzed data and wrote the manuscript.
Declaration of Interests
The authors declare no competing financial interests.
Published: September 27, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.08.008.
Supplemental Information
References
- Braso-Maristany F., Filosto S., Catchpole S., Marlow R., Quist J., Francesch-Domenech E., Plumb D.A., Zakka L., Gazinska P., Liccardi G. PIM1 kinase regulates cell death, tumor growth and chemotherapy response in triple-negative breast cancer. Nat. Med. 2016;22:1303–1313. doi: 10.1038/nm.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.S., Redkar S., Bearss D., Wierda W.G., Gandhi V. Pim kinase inhibitor, SGI-1776, induces apoptosis in chronic lymphocytic leukemia cells. Blood. 2009;114:4150–4157. doi: 10.1182/blood-2009-03-212852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan X., Wan Q., Yi L., Lu J., Jiao Y., Li H., Song D., Chen Y., Xu H., He M.L. Hsp27 responds to and facilitates enterovirus A71 replication by enhancing viral internal ribosome entry site-mediated translation. J. Virol. 2019;93 doi: 10.1128/JVI.02322-18. e02322–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries M., Smithers N.P., Howarth P.H., Nawijn M.C., Davies D.E. Inhibition of Pim1 kinase reduces viral replication in primary bronchial epithelial cells. Eur. Respir. J. 2015;45:1745–1748. doi: 10.1183/09031936.00206514. [DOI] [PubMed] [Google Scholar]
- Dong Q., Men R., Dan X., Chen Y., Li H., Chen G., Zee B., Wang M.H.T., He M.L. Hsc70 regulates the IRES activity and serves as an antiviral target of enterovirus A71 infection. Antiviral. Res. 2018;150:39–46. doi: 10.1016/j.antiviral.2017.11.020. [DOI] [PubMed] [Google Scholar]
- Duan H., Zhu M., Xiong Q., Wang Y., Xu C., Sun J., Wang C., Zhang H., Xu P., Peng Y. Regulation of enterovirus 2A protease-associated viral IRES activities by the cell's ERK signaling cascade: implicating ERK as an efficiently antiviral target. Antiviral. Res. 2017;143:13–21. doi: 10.1016/j.antiviral.2017.03.018. [DOI] [PubMed] [Google Scholar]
- Gao M., Duan H., Liu J., Zhang H., Wang X., Zhu M., Guo J., Zhao Z., Meng L., Peng Y. The multi-targeted kinase inhibitor sorafenib inhibits enterovirus 71 replication by regulating IRES-dependent translation of viral proteins. Antiviral. Res. 2014;106:80–85. doi: 10.1016/j.antiviral.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Glaser W., Skern T. Extremely efficient cleavage of eIF4G by picornaviral proteinases L and 2A in vitro. FEBS Lett. 2000;480:151–155. doi: 10.1016/s0014-5793(00)01928-1. [DOI] [PubMed] [Google Scholar]
- Haddach M., Michaux J., Schwaebe M.K., Pierre F., O'brien S.E., Borsan C., Tran J., Raffaele N., Ravula S., Drygin D. Discovery of CX-6258. A potent, selective, and orally efficacious pan-pim kinases inhibitor. ACS Med. Chem. Lett. 2012;3:135–139. doi: 10.1021/ml200259q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaguchi T., Fujisawa H., Sakai K., Okino S., Kurosaki N., Nishimura Y., Shimizu H., Yamada M. Acute encephalitis caused by intrafamilial transmission of enterovirus 71 in adult. Emerg. Infect. Dis. 2008;14:828–830. doi: 10.3201/eid1405.071121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M., Benito J., Yamamoto S., Kaur S., Arslan D., Ramirez S., Jacamo R., Platanias L., Matsushita H., Fujimura T. The novel combination of dual mTOR inhibitor AZD2014 and pan-PIM inhibitor AZD1208 inhibits growth in acute myeloid leukemia via HSF pathway suppression. Oncotarget. 2015;6:37930–37947. doi: 10.18632/oncotarget.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.N., Lin J.Y., Locker N., Kung Y.A., Hung C.T., Lin J.Y., Huang H.I., Li M.L., Shih S.R. Far upstream element binding protein 1 binds the internal ribosomal entry site of enterovirus 71 and enhances viral translation and viral growth. Nucleic Acids Res. 2011;39:9633–9648. doi: 10.1093/nar/gkr682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H.C., Shih S.R., Chang T.Y., Fang M.Y., Hsu J.T. The combination effects of licl and the active leflunomide metabolite, A771726, on viral-induced interleukin 6 production and EV-A71 replication. PLoS One. 2014;9:e111331. doi: 10.1371/journal.pone.0111331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.M., Chen C.J., Shih S.R. Regulation mechanisms of viral IRES-driven translation. Trends Microbiol. 2017;25:546–561. doi: 10.1016/j.tim.2017.01.010. [DOI] [PubMed] [Google Scholar]
- Leong W.F., Chow V.T. Transcriptomic and proteomic analyses of rhabdomyosarcoma cells reveal differential cellular gene expression in response to enterovirus 71 infection. Cell Microbiol. 2006;8:565–580. doi: 10.1111/j.1462-5822.2005.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.Y., Chen T.C., Weng K.F., Chang S.C., Chen L.L., Shih S.R. Viral and host proteins involved in picornavirus life cycle. J. Biomed. Sci. 2009;16:103. doi: 10.1186/1423-0127-16-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.Y., Li M.L., Brewer G. mRNA decay factor AUF1 binds the internal ribosomal entry site of enterovirus 71 and inhibits virus replication. PLoS One. 2014;9:e103827. doi: 10.1371/journal.pone.0103827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.Y., Li M.L., Huang P.N., Chien K.Y., Horng J.T., Shih S.R. Heterogeneous nuclear ribonuclear protein K interacts with the enterovirus 71 5' untranslated region and participates in virus replication. J. Gen. Virol. 2008;89:2540–2549. doi: 10.1099/vir.0.2008/003673-0. [DOI] [PubMed] [Google Scholar]
- Lin J.Y., Shih S.R., Pan M., Li C., Lue C.F., Stollar V., Li M.L. hnRNP A1 interacts with the 5' untranslated regions of enterovirus 71 and Sindbis virus RNA and is required for viral replication. J. Virol. 2009;83:6106–6114. doi: 10.1128/JVI.02476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., He Y.Q., Yi L.N., Zan H., Kung H.F., He M.L. Viral kinetics of enterovirus 71 in human abdomyosarcoma cells. World J. Gastroenterol. 2011;17:4135–4142. doi: 10.3748/wjg.v17.i36.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Yi L., Zhao J., Yu J., Chen Y., Lin M.C., Kung H.F., He M.L. Enterovirus 71 disrupts interferon signaling by reducing the level of interferon receptor 1. J. Virol. 2012;86:3767–3776. doi: 10.1128/JVI.06687-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manki A., Oda M., Seino Y. [Neurologic diseases of enterovirus infections: polioviruses, coxsackieviruses, echoviruses, and enteroviruses type 68-72] Nihon Rinsho. 1997;55:849–854. [PubMed] [Google Scholar]
- Nawijn M.C., Alendar A., Berns A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat. Rev. Cancer. 2011;11:23–34. doi: 10.1038/nrc2986. [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Shimojima M., Tano Y., Miyamura T., Wakita T., Shimizu H. Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat. Med. 2009;15:794–797. doi: 10.1038/nm.1961. [DOI] [PubMed] [Google Scholar]
- Ohlmann T., Rau M., Pain V.M., Morley S.J. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- Park C., Min S., Park E.M., Lim Y.S., Kang S., Suzuki T., Shin E.C., Hwang S.B. Pim kinase interacts with nonstructural 5A protein and regulates hepatitis C virus entry. J. Virol. 2015;89:10073–10086. doi: 10.1128/JVI.01707-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggli E., Gattesco S., Pautz A., Regazzi R. Involvement of the RNA-binding protein ARE/poly(U)-binding factor 1 (AUF1) in the cytotoxic effects of proinflammatory cytokines on pancreatic beta cells. Diabetologia. 2012;55:1699–1708. doi: 10.1007/s00125-011-2399-7. [DOI] [PubMed] [Google Scholar]
- Schmidt N.J., Lennette E.H., Ho H.H. An apparently new enterovirus isolated from patients with disease of the central nervous system. J. Infect. Dis. 1974;129:304–309. doi: 10.1093/infdis/129.3.304. [DOI] [PubMed] [Google Scholar]
- Selten G., Cuypers H.T., Berns A. Proviral activation of the putative oncogene Pim-1 in MuLV induced T-cell lymphomas. EMBO J. 1985;4:1793–1798. doi: 10.1002/j.1460-2075.1985.tb03852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih S.R., Stollar V., Lin J.Y., Chang S.C., Chen G.W., Li M.L. Identification of genes involved in the host response to enterovirus 71 infection. J. Neurovirol. 2004;10:293–304. doi: 10.1080/13550280490499551. [DOI] [PubMed] [Google Scholar]
- Sun J., Niu Y., Wang C., Zhang H., Xie B., Xu F., Jin H., Peng Y., Liang L., Xu P. Discovery of 3-benzyl-1,3-benzoxazine-2,4-dione analogues as allosteric mitogen-activated kinase kinase (MEK) inhibitors and anti-enterovirus 71 (EV71) agents. Bioorg. Med. Chem. 2016;24:3472–3482. doi: 10.1016/j.bmc.2016.05.055. [DOI] [PubMed] [Google Scholar]
- Tan C.W., Poh C.L., Sam I.C., Chan Y.F. Enterovirus 71 uses cell surface heparan sulfate glycosaminoglycan as an attachment receptor. J. Virol. 2013;87:611–620. doi: 10.1128/JVI.02226-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S.R., Sarnow P. Enterovirus 71 contains a type I IRES element that functions when eukaryotic initiation factor eIF4G is cleaved. Virology. 2003;315:259–266. doi: 10.1016/s0042-6822(03)00544-0. [DOI] [PubMed] [Google Scholar]
- Vries M., Bedke N., Smithers N.P., Loxham M., Howarth P.H., Nawijn M.C., Davies D.E. Inhibition of Pim1 kinase, new therapeutic approach in virus-induced asthma exacerbations. Eur. Respir. J. 2016;47:783–791. doi: 10.1183/13993003.00171-2015. [DOI] [PubMed] [Google Scholar]
- Wang H., Li K., Ma L., Wu S., Hu J., Yan H., Jiang J., Li Y. Berberine inhibits enterovirus 71 replication by downregulating the MEK/ERK signaling pathway and autophagy. Virol. J. 2017;14:2. doi: 10.1186/s12985-016-0674-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Dong Q., Wang H., He Y., Chen Y., Zhang H., Wu R., Chen X., Zhou B., He J. Oblongifolin M, an active compound isolated from a Chinese medical herb Garcinia oblongifolia, potently inhibits enterovirus 71 reproduction through downregulation of ERp57. Oncotarget. 2016;7:8797–8808. doi: 10.18632/oncotarget.7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W.R., Chen Y.Y., Yang S.M., Chen Y.L., Horng J.T. Phosphorylation of PI3K/Akt and MAPK/ERK in an early entry step of enterovirus 71. Life Sci. 2005;78:82–90. doi: 10.1016/j.lfs.2005.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.X., Phuektes P., Kumar P., Goh G.Y., Moreau D., Chow V.T., Bard F., Chu J.J. Human genome-wide RNAi screen reveals host factors required for enterovirus 71 replication. Nat. Commun. 2016;7:13150. doi: 10.1038/ncomms13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L., Lu J., Kung H.F., He M.L. The virology and developments toward control of human enterovirus 71. Crit. Rev. Microbiol. 2011;37:313–327. doi: 10.3109/1040841X.2011.580723. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Zhao B., Chen X., Song N., Wu J., Li G., Yu P., Han Y., Liu J., Qin C. GS-9620 inhibits enterovirus 71 replication mainly through the NF-kappaB and PI3K-AKT signaling pathways. Antiviral. Res. 2018;153:39–48. doi: 10.1016/j.antiviral.2018.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.