Abstract
The mitochondrial calcium uniporter (MCU) complex mediates the uptake of Ca2+ into mitochondria. Its activity is regulated by a heterodimer of MICU1 and MICU2, two EF‐hand‐containing proteins that act as the main gatekeeper of the uniporter. Herein we report the crystal structure of human MICU2 at 1.96 Å resolution. Our structure reveals a dimeric architecture of MICU2, in which each monomer adopts the canonical two‐lobe structure with a pair of EF‐hands in each lobe. Both Ca2+‐bound and Ca2+‐free EF‐hands are observed in our structure. Moreover, we characterize the interaction sites within the MICU2 homodimer, as well as the MICU1–MICU2 heterodimer in both Ca2+‐free and Ca2+‐bound conditions. Glu242 in MICU1 and Arg352 in MICU2 are crucial for apo heterodimer formation, while Phe383 in MICU1 and Glu196 in MICU2 significantly contribute to the interaction in the Ca2+‐bound state. Based on our structural and biochemical analyses, we propose a model for MICU1–MICU2 heterodimer formation and its conformational transition from apo to a more compact Ca2+‐bound state, which expands our understanding of this co‐regulatory mechanism critical for MCU's mitochondrial calcium uptake function.
Keywords: EF‐hands, MICU1–MICU2 complex, MICU2, mitochondrial calcium uniporter
Subject Categories: Membrane & Intracellular Transport, Structural Biology
Introduction
Mitochondrial calcium uniporter (MCU) located in the inner mitochondrial membrane (IMM) is an ion channel with high Ca2+ selectivity and affinity (K d ≤ 2 nM) 1, which regulates and maintains mitochondrial calcium homeostasis. The uniporter calcium uptake is driven by electrochemical potential (Δψm) of IMM 2, 3, which can be inhibited by ruthenium red 4, 5. Various components of uniporter holocomplex (uniplex) have been identified by comparative proteomics and genomics studies, including pore‐forming MCU 4, 5, 6, metazoan‐specific essential MCU regulator (EMRE) 7, EF‐hand calcium‐binding proteins mitochondrial calcium uptake 1 and 2 (MICU1 and MICU2) 8, 9, and MCU regulatory subunit b (MCUb) 10. Although the architectures of MCU from Caenorhabditis elegans and fungi have been unequivocally established 11, 12, 13, 14, 15, the fundamental Ca2+ uptake and Ca2+‐dependent gating mechanisms of the metazoan uniporter are still ambiguous.
Regarded as MCU's gatekeepers to modulate Ca2+ uptake 16, 17, 18, MICU1 and MICU2 are regulatory subunits located in the intermembrane space (IMS) 17, 19, 20, 21 and present in most eukaryotic organisms except in fungi 22. It was demonstrated that EMRE mediates the interaction between MCU and MICU1–MICU2 7. The interaction between MICU1–MICU2 and MCU inhibited Ca2+ uptake in resting condition; the interaction was abolished in the presence of high cytosolic Ca2+ concentrations ([Ca2+]c) level to result in the loss of MCU inhibition 23. Recently, two articles reported that MICU1 can directly interact with the conserved D‐ring of MCU to regulate the Ca2+ uptake 24, 25. MICU1–MICU2 complex has been shown to exert regulatory function for MCU's activity 7, 17, 23, 26, 27, 28, 29, although how MICU1 interacts with MICU2 to regulate Ca2+ uptake is not clear.
Crystal structures of human MICU1 revealed that MICU1 forms a hexamer (trimer of dimer) in the apo state to inhibit MCU and rearranges to form multiple oligomers upon Ca2+ binding to EF‐hands, resulting in MCU activation 30. Analogous to the dependence of EMRE expression on MCU 7, 31, MICU2 requires MICU1 to become stable 9, 17, 23, 26. Furthermore, it has been suggested that disulfide formation between MICU2 and MICU1 which is facilitated by oxidoreductase Mia40 stabilizes the heterodimer in physiological condition 23. Distinct from MICU1, only MICU1–MICU2 heterodimer but not MICU2 alone was detected in cells 18, 23, 26, 28. Recent studies revealed that MICU2 spatially limited crosstalk between inositol 1,4,5‐trisphosphate receptor and the uniporter 18 and mediated the cardiovascular homeostasis 32, along with implications in neurodevelopmental disorder 33. Additionally, in most EF‐hand‐containing proteins, EF‐hands typically pack in pairs with four amphipathic helixes bundled together to make a hydrophobic core 34, 35. When Ca2+ ions are bound, the hydrophobic core which is buried in the apo state exposes to the solvent and creates a target protein interaction site 34, 35, 36.
Three main models of MICU1–MICU2 complex were proposed to regulate the MCU activities 17, 18, 26, 37, 38. In one model, MICU1 and MICU2 were suggested to play the distinct roles in modulating MCU activities 26. MICU2 inhibited MCU activity in low [Ca2+]c preventing matrix overload, and MICU1 exerted stimulatory function to facilitate accumulation of mitochondrial Ca2+ rapidly in high [Ca2+]c 26. The second model demonstrated that MICU1 and MICU2 would play nonredundant roles and set the [Ca2+]c threshold collectively to regulate Ca2+ uptake 17, 27, 38. The authors pointed out that MICU1–MICU2 complex inhibited MCU when [Ca2+]c was lower than the threshold (600–800 nM) and eliminated the inhibition of the channel when [Ca2+]c was high 17, 27. In this model, Ca2+‐induced MICU1–MICU2 heterodimer conformational change was the key point of the close‐to‐open functional transition 27, 30, 39. Recently, it was shown that Mn2+ ions were transported in MICU1 KO cells but failed in WT since Mn2+ would not be able to induce conformational change in MICU1–MICU2 complex 39. The third model is the combination of the first two models which suggests that MICU1 and MICU2 inhibit MCU activity at low Ca2+ level and MICU1 activates MCU at high Ca2+ level 37.
Although there have been various studies focusing on MICU1 or MICU2 individually, many results obtained under different experimental conditions are nonconclusive or even controversial. This is not surprising since it is not easy to isolate one's function from another to “untangle” the two proteins because MICU1 and MICU2 form a tight functional complex. As such, experiments aiming to understand the complex and its role are timely and important. Indeed, increasing evidences have shown that although MICU1 and MICU2 can form homodimers respectively in vitro 27, 40, MICU1 and MICU2 mainly work together as a heterodimer to function as MCU's gatekeeper 17, 18, 26, 37, 38. Previous studies have demonstrated that MICU1 and MICU2 formed stable complex not only in the absence of disulfide bond but also independent of Ca2+, implying a robust MICU1–MICU2 interaction 27, 40. However, since there has been no detailed investigation of MICU1–MICU2 interaction and there is no complex structure available, the interaction modes of MICU1–MICU2 for both states (Ca2+‐free and Ca2+‐bound) and its functional implication remain poorly understood. Additionally, Ca2+‐induced conformational change of MICU1–MICU2 complex has not been evaluated.
Herein we report the MICU2 structure which reveals both Ca2+‐bound and Ca2+‐free EF‐hands in one structure at the resolution of 1.96 Å by X‐ray crystallography. The overall MICU2 structure is similar to MICU1, although some local structural differences are observed. These findings have provided the structural basis for probing the interactions between MICU1 and MICU2 and for establishing a heterodimer model. Using glutathione S‐transferase (GST) pull‐down experiments and various truncation derivatives and point mutants, we investigated MICU1–MICU2 interactions in both apo and Ca2+‐bound states. In the apo form, Glu242 in MICU1 and Arg352 in MICU2 are critical residues for MICU1–MICU2 complex formation. In Ca2+‐bound from, EF‐hand 1 in MICU2 exposed a hydrophobic core and attracted Phe383 in MICU1 via hydrophobic interaction. We have also investigated binding affinities between MICU1 and MICU2 which yielded K d of 224 ± 46 nM and 134 ± 52 nM in the apo and Ca2+‐bound states by isothermal calorimetry (ITC), respectively. Our data have allowed us to establish a plausible MICU1–MICU2 heterodimer model and understand its transitioning between Ca2+‐free state and Ca2+‐bound state, providing the structural basis for understanding the gatekeeping activity of MICU1–MICU2 in response to Ca2+ fluctuation.
Results
Comparison of mitochondrial calcium regulator structures
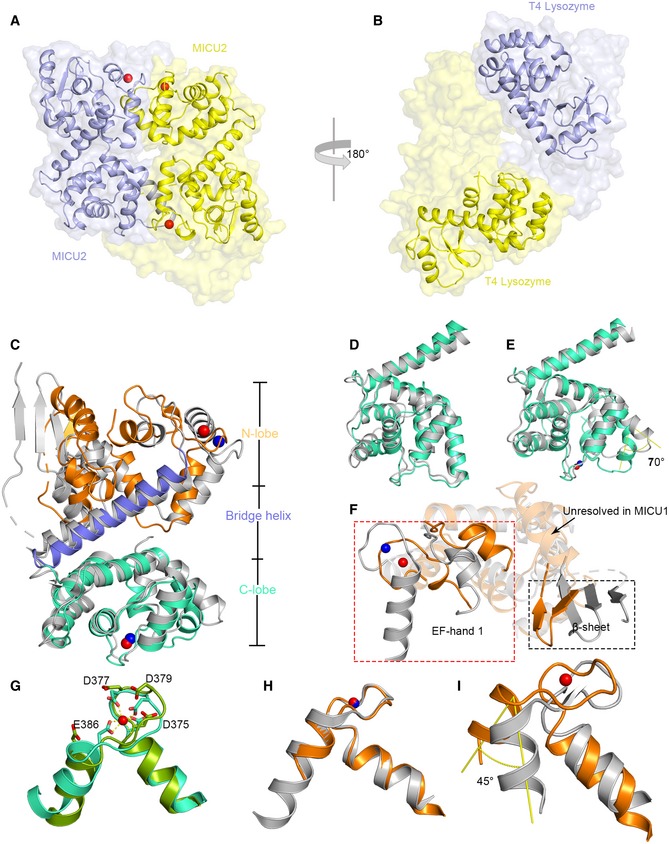
To determine the structure of MICU2, we employed the construct (85–406 aa) which was used in the previous report 40. To circumvent the difficulty of crystallization optimization, we fused the bacteriophage T4 lysozyme (T4L) to the N‐terminus of the MICU2 construct. We subsequently solved the structure by single‐wavelength anomalous dispersion. The structure was refined to final Rwork = 18.65% and Rfree = 22.67%, which contains two T4L‐MICU2 molecules in the asymmetric unit. The two MICU2 molecules are related by twofold symmetry and form an intimate dimer which is packed in an antiparallel mode, rather than head‐to‐head (Fig 1A). Interestingly, two T4L molecules (1–161 aa) are not related by symmetry and positioned in different orientations (Fig 1B). Unambiguous electron densities were observed for three Ca2+ ions (Fig EV1). EF‐hand 1 Ca2+ ions were observed in both monomers whereas EF‐hand 2 Ca2+ ion was seen in only one of the monomers (molecule 2, chain B; Fig EV1), although Ca2+ ions were not only absent in both purification and crystallization procedures, but the chelating reagent (EGTA) was also added.
Figure 1. Overall structure of T4L‐MICU2 and structural comparison of MICU1 and MICU2.
-
A, BSpace filling and cartoon representation of two MICU2 (A) and T4L molecules (B). The protein chain with two calcium ions bound is colored in yellow, and the protein chain with one calcium ion bound is colored in purple. Calcium ions are shown as red spheres.
-
CThe structure of Ca2+‐bound MICU2 (molecule 2) is superimposed with Ca2+‐bound MICU1 (PDB code: 4NSD). MICU1 is colored in gray; MICU2 is colored in orange, marine, and green‐cyan for different domains. Calcium ions are shown as blue and red spheres in MICU1 and MICU2, respectively.
-
D, EComparison of C‐lobes of MICU2 (green‐cyan) and MICU1 (gray) in the Ca2+‐free (D) and Ca2+‐bound (E) states.
-
FComparison of N‐lobes of MICU1 (gray) and MICU2 (orange) shows large divergence in the EF‐hand 1 (box in red dotted line) and β‐sheet (box in black dotted line). Calcium ions are shown as blue and red spheres in MICU1 and MICU2, respectively.
-
GSuperimposition of MICU2 EF‐hand 2 in Ca2+‐bound (green‐cyan) form and Ca2+‐free form (bright green). Calcium ion is shown as a red sphere.
-
HSuperimposition of MICU2 EF‐hand 1(with Ca2+‐bound) and MICU1 Ca2+‐bound EF‐hand 1. Calcium ions are shown as blue and red spheres in MICU1 and MICU2, respectively.
-
ISuperimposition of MICU2 EF‐hand 1 (with Ca2+‐bound) and MICU1 Ca2+‐free EF‐hand 1. The angle (yellow) represents the conformational difference. The calcium ion in MICU2 is shown as a red sphere.
Figure EV1. Electron density maps of EF‐hand loops.
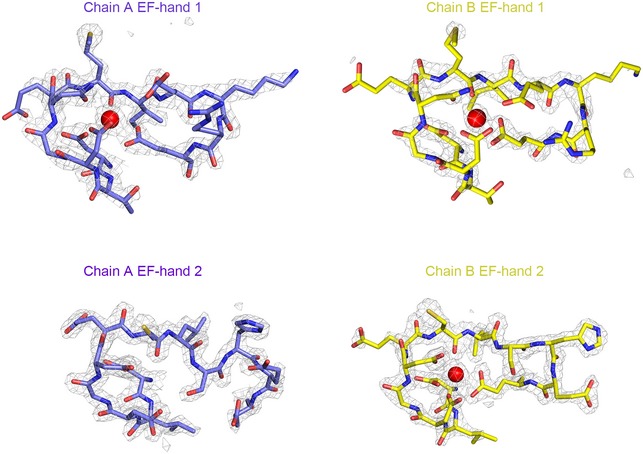
2Fo‐Fc electron density maps (gray mesh) of Ca2+ ions and EF‐hands under the sigma level of 2. EF‐hands are shown as sticks and colored in purple and yellow in both molecules 1 and 2, respectively. The calcium ions are shown as red spheres.
Analogous to most EF‐hand Ca2+‐binding proteins such as calmodulin, MICU2 structure possesses N‐lobe and C‐lobe, bridged by a long oblique α‐helix (Fig 1C). Each lobe of MICU2 contains an EF‐hand pair which consists of one canonical EF‐hand with the capacity to chelate Ca2+ and one pseudo‐EF‐hand incompetent to bind Ca2+ owing to the lack of key acidic residues (Appendix Fig S1). We shall refer to the competent EF‐hands in N‐lobe and C‐lobe as EF‐hand 1 and EF‐hand 2, respectively. Two MICU2 chains are superimposed well except for small conformational differences mainly in EF‐hand 2 (Appendix Fig S2). The bi‐lobe monomer structure of MICU2 is similar to MICU1 (Fig 1C). The packing mode of the MICU2 dimer exhibits more resemblance to the MICU1 dimer in the Ca2+‐free form than the Ca2+‐bound form (Appendix Fig S3), even though three Ca2+ ions are bound in the MICU2 structure. This observation is also supported by buried surface analysis. The buried surface of MICU2 dimer is ~ 1,390 Å2, much larger than the MICU1 Ca2+‐bound structure (~ 434 Å), but closer to the MICU1 Ca2+‐free form (~ 1,122 Å) 30.
Despite the overall structural similarity between MICU1 and MICU2, significant local differences exist. While the C‐lobe and the bridging α‐helix overlap reasonably well, the N‐lobe does not (Fig 1C–F). A detailed comparison shows that the C‐lobes of MICU2 molecule 1 (Ca2+‐free) and MICU1 in Ca2+‐free state, together with the long oblique helix, superimpose well with only small differences (Fig 1D). However, when comparing the Ca2+‐bound state of both MICU1 and MICU2 (molecule 2), the bent α‐helix displays a rotation of ~ 70° in MICU2 (Fig 1E). Additionally, there is a considerable conformational difference between the N‐lobes of MICU1 and MICU2 in Ca2+‐bound form, rendering superimposition challenging. For example, the EF‐hand pair and β‐sheet rotate toward each other in MICU2, an arrangement that makes MICU2 a more compact structure (Fig 1F). Moreover, an α‐helix is seen to follow the β‐sheet in MICU2 but unresolved in the MICU1 structure likely owing to flexibility (Fig 1F).
Changes in EF‐hand pair upon Ca2+ binding
Like other canonical EF‐hands, Ca2+ ions are coordinated by side chains of acidic groups or backbone of the EF‐hands in MICU2. For EF‐hand 1, these residues are Asp185, Asp187, Asn189, Met191, Glu193, and Glu196, while EF‐hand 2 features Asp375, Asp377, Asp379, Ser383, Glu386, and one water molecule (Fig EV2). The EF‐hand pair is positioned in a face‐to‐face manner, packed through approximate twofold symmetry with the four α‐helixes bundling together (Appendix Fig S1). In general, the coordinating groups of EF‐hand loop point toward the center in the Ca2+‐bound form to form a coordination sphere (Figs 1G and EV2). The side chain of Glu386 (at the loop position 12) moves away in the absence of Ca2+ (Fig 1G). As mentioned above, we obtained both Ca2+‐free and Ca2+‐bound forms of EF‐hand 2 in MICU2 structure which have allowed comparison of EF‐hands in the two different states. The EF‐hand 2 displays negligible conformational change in interhelical angle in both states (Fig 1G). To understand the conformational change process of EF‐hand 1 in MICU2 from apo form to Ca2+‐bound form, we used EF‐hands 1 of MICU1 in both states as references. The superimposition reveals that Ca2+‐bound EF‐hands 1 in both MICU1 and MICU2 align near perfectly (Fig 1H). Next, we superimposed MICU2 EF‐hand 1 in Ca2+‐bound form with that of MICU1 in Ca2+‐free state. The exiting α‐helix of MICU2 EF‐hand 1 shows a significant rotation of ~ 45° (Fig 1I). This conformational change would increase the interhelical angle in EF‐hand pair and expose a hydrophobic core (Appendix Fig S4). A similar change has been observed for many proteins in the calmodulin family 34, but has not been discovered in EF‐hand in C‐lobe (Appendix Fig S4).
Figure EV2. Ca2+ coordination in EF‐hand 1 (left) and in EF‐hand 2 (right) of MICU2.
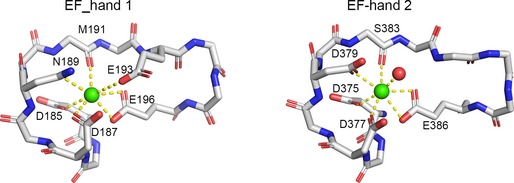
The main chain of EF‐hand loops and Ca2+‐bound residues are shown as sticks. Calcium ions and water molecules are shown as green and red spheres, respectively.
Interaction of MICU2 homodimer in both states
Since MICU2 can dimerize in solution in vitro 27, we wondered whether T4L affects the oligomerization state of MICU2 in solution and carried out multi‐angle laser light scattering (MALLS) experiments. The results revealed monodisperse T4L‐MICU2 with molecular weights ~ 106 and ~ 120 kDa in EGTA and CaCl2 conditions (Appendix Fig S5A and B), in agreement with the calculated molecular weight of the T4L‐MICU2 dimer (116 kDa). These findings are consistent with our T4L‐MICU2 dimer structure, indicating that T4 lysozyme did not affect the MICU2 oligomerization state.
In our MICU2 structure, the homodimer interface is stabilized by hydrophobic and electrostatic interactions largely contributed by EF‐hand 1 and the bent helix. The hydrophobic interactions are mediated by several hydrophobic residues (Ile333, Met337 in bent helix) and the hydrophobic core created by EF‐hand 1 (Figs 2A right and EV3 left). The electrostatic interactions comprise many hydrogen bonds which include (i) the side chain of R352 and the main chains of K182, M183, and D185, (ii) the side chain of R343 and the main chain of K199, (iii) the side chain of E349 and the side chain of K199, and (iv) the side chain of E329 and the side chain of K172 (Figs 2A left and EV3 right). To probe whether the hydrophobic core is generated by EF‐hand 1 upon Ca2+ binding, we created EF1mut (D185A & E196K) and EF2mut (D375A & E386K) double mutants and two single mutants (D185A, E196K; Appendix Table S1). To investigate the electrostatic interactions, we created three mutants including MICU2_R352A, MICU2_E329A, and MICU2_R343A (Appendix Table S1). In our previous report, D330 contributed to MICU1 and MICU2 interaction 40. To test whether it also disrupts the MICU2 homodimer in both states, we also created the MICU2_D330A mutant. The SEC results show that MICU2_EF1mut and MICU2_E196K mutation disrupted homodimer formation in the presence of 2 mM CaCl2, while MICU2_EF2mut mutation did not (Fig 2B). In comparison, MICU2_R352A, MICU2_E329A, MICU2_R343A, and MICU2_D330A mutations did not affect the homodimer in the presence of Ca2+ (Fig 2B). Interestingly, the retention volume of MICU2_D185A was between monomer and dimer in the Ca2+ condition, probably due to the coexistence of monomer and dimer (Fig 2B). Although MICU2_R352A mutation failed to disturb the homodimer in the presence of Ca2+, it did in the absence of Ca2+ (Fig 2C). Other MICU2 mutations did not change the dimer state in the EGTA condition (Fig 2C). Compared with MICU1, MICU2 has the same interaction modes in both states which are hydrophobic interaction in Ca2+‐bound form and electrostatic interaction in the Ca2+‐free form 30. MALLS results show that the molecular weights of MICU2_R352A in EGTA condition and MICU2_E196K in Ca2+ condition are ~ 50 kDa, close to the monomer's theoretical molecular weight of 40 kDa (Appendix Fig S5C and D). MICU1 and MICU2 possess the similar interaction modes in both states, highlighting the general structural conservation between MICU1 and MICU2 despite the differences mentioned above.
Figure 2. Interaction interface of the MICU2 homodimer in both states.
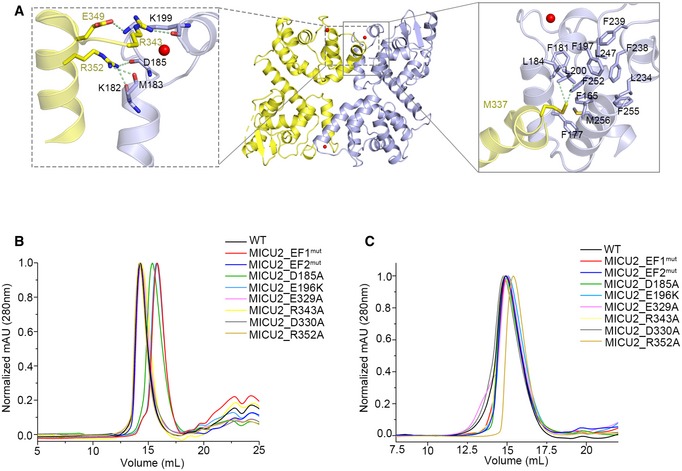
- Interaction interface of the MICU2 homodimer in the Ca2+‐bound state. The left box (gray dotted line) shows residues involved in electrostatic interactions and the right box (gray line) residues involved in hydrophobic interactions. Molecule 1 and molecule 2 in the dimer are colored in purple and yellow, respectively. Side chains of residues that make crucial interactions are shown in stick representation.
- Size exclusion chromatography (SEC) results for MICU2_WT and a set of mutants in the presence of Ca2+ monitored at 280 nm absorption wavelength.
- Size exclusion chromatography (SEC) results for MICU2_WT and a set of mutants in the presence of EGTA monitored at 280 nm absorption wavelength.
Figure EV3. Interaction interface of the MICU2 homodimer in Ca2+‐bound state.
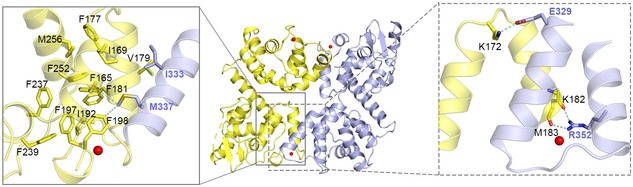
The expanded boxes show the detailed atomic hydrophobic interactions (gray boxes) and electrostatic interactions (gray dotted boxes) in MICU2. Molecule 1 and molecule 2 in the dimer are colored in purple and yellow, respectively. The crucial residues are shown as sticks.
Interactions of the MICU1–MICU2 complex in both states
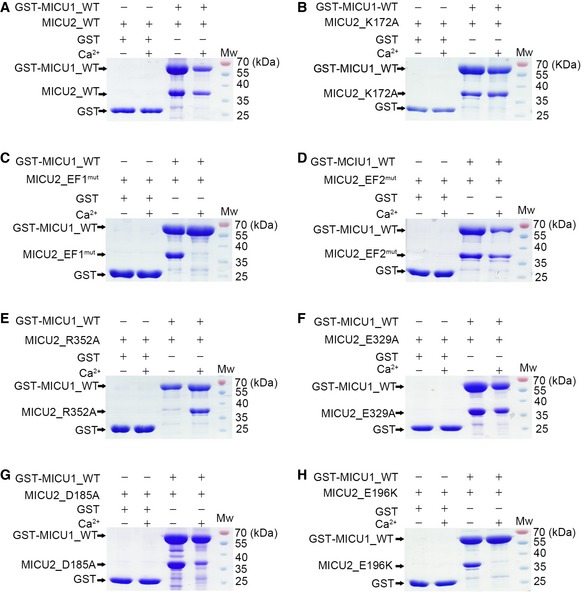
Structural conservation and similar interaction modes in MICU1 and MICU2 prompted us to investigate the interaction between MICU1 and MICU2. Based on the MICU2 homodimer electrostatic and hydrophobic interactions, we prepared a series of MICU2 mutants including EF1mut (D185A, E196K), EF2mut (D375A & E386K), R352A, K172A, and E329A (Appendix Table S1). Next, we employed GST pull‐down experiments using GST‐MICU1 as bait to pull down MICU2 wild‐type (WT) and various mutants in the presence or absence of Ca2+. Our results reveal that MICU1 and MICU2_WT interacted strongly in both states (Fig 3A). MICU2_EF2mut, MICU2_K172A, and MICU2_E329A mutants were all able to interact with MICU2 in both states (Fig 3B, D and F). In comparison, MICU2_EF1mut and MICU2_R352A mutants were not pulled down in the presence and absence of Ca2+, respectively (Fig 3C and E). To evaluate relative contribution of Asp185 or Glu196 in EF‐hand 1 of MICU2, we used the MICU2_D185A and MICU2_E196K single mutants for pull‐down assays. As expected, MICU2_D185A mutant displayed weakened interaction with MICU1 in Ca2+ condition, whereas MICU2_E196K mutant abolished the interaction completely (Fig 3G and H). Thus, Glu196 in MICU2 is a critical residue involved in MICU1–MICU2 heterodimer interaction and MICU2 homodimer in the presence of Ca2+, whereas Arg352 is important in the absence of Ca2.
Figure 3. Pull‐down experiments to test the contribution of individual MICU2 residues to the interaction with MICU1 in the apo and Ca2+‐bound states.
-
A, BMICU2_WT (A) and MICU2_K172A (B) were pulled down by GST‐MICU1 in both the absence and presence of 2 mM Ca2+.
-
C, DMICU2_EF1mut was not pulled down by GST‐MICU1 in the presence of 2 mM Ca2+ (C), while MICU2_EF2mut was pulled down by GST‐MICU1 in both the absence and presence of 2 mM Ca2+ (D).
-
E, FMICU2_R352A was not pulled down by GST‐MICU1 in the absence of 2 mM Ca2+ (E), while MICU2_E329A was pulled down by GST‐MICU1 in both the absence and presence of 2 mM Ca2+ (F).
-
GMICU2_D185A was partially pulled down by GST‐MICU1 in the presence of Ca2+, while they showed a strong interaction in the absence of 2 mM Ca2+.
-
HMICU2_E196K was not pulled down by GST‐MICU1 in the presence of 2 mM Ca2+, while a strong interaction was observed in the absence of 2 mM Ca2+.
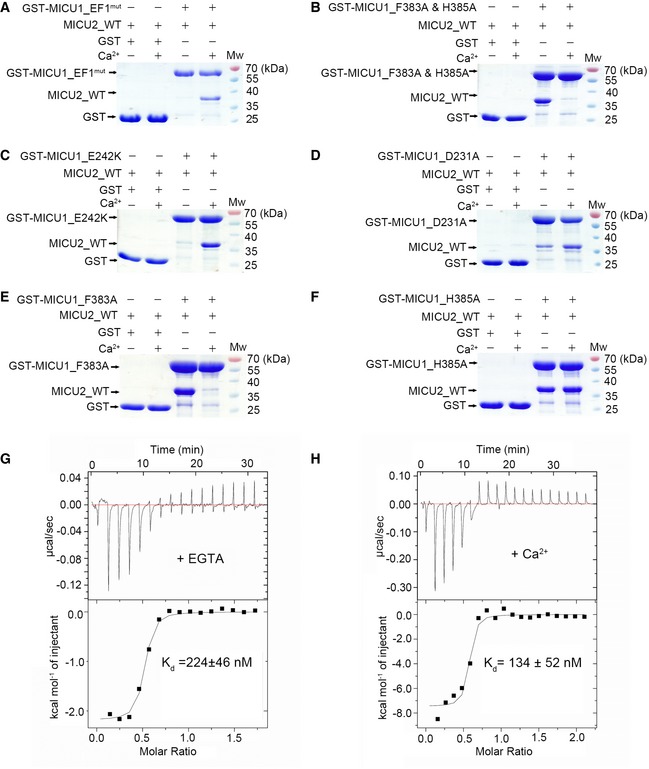
We next investigated which residues in MICU1 participate in the MICU1–MICU2 interactions. Based on the MICU1 homodimer electrostatic and hydrophobic interactions 30, we created MICU1 mutants including EF1mut (D231A & E242K), F383A & H385A double mutant, and D376A single mutant (Appendix Table S1). Here, we used GST‐MICU1 mutants as baits to pull down MICU2_WT, and in reverse, GST‐MICU2_WT as bait to pull down MICU1 mutants. The results indicate that MICU1_EF1mut and MICU1_F383A & H385A double mutants abrogated the interaction with MICU2_WT in the absence and presence of Ca2+, respectively (Figs 4A and B, and EV4B). In comparison, the mutation of Asp376 in MICU1 did not affect the interaction in both conditions (Fig EV4A). To unequivocally confirm the residues which play significant roles in MICU1–MICU2 interaction, we produced MICU1_D231A, MICU1_E242K, MICU1_F383A, and MICU1_H385A single mutants for further studies. The results reveal that MICU1_E242K mutant destroyed MICU1–MICU2 complex in the absence of Ca2+, while MICU1_D231A still showed interactions with MICU2_WT (Figs 4C and D, and EV4C and D). MICU1_F383A mutant destroyed MICU1–MICU2 complex in the presence of Ca2+, while MICU1_H383A still showed interactions with MICU2_WT in both conditions (Figs 4E and F, and EV4E and F). Additionally, we semi‐quantitatively analyzed the pull‐down SDS–PAGE grayscales of critical residues (Fig EV5). In conclusion, Glu242 and Phe383 in MICU1 and Arg352 and Glu196 in MICU2 are crucial for heterodimer formation in the absence and presence of Ca2+, respectively.
Figure 4. Characterization of MICU1 and MICU2 interactions in both apo and Ca2+‐bound states.
-
AGST‐MICU1_EF1mut failed to pull down MICU2_WT in the absence of 2 mM Ca2+, whereas they displayed a strong interaction in the presence of 2 mM Ca2+.
-
BGST‐MICU1_F383A & H385A failed to pull down MICU2_WT in the presence of Ca2+, whereas they displayed a strong interaction in the absence of 2 mM Ca2+.
-
CGST‐MICU1_E242K failed to pull down MICU2_WT in the absence of Ca2+, whereas they displayed a strong interaction in the presence of 2 mM Ca2+.
-
DGST‐MICU1_D231A partly pulled down MICU2_WT in the absence of Ca2+, whereas they displayed a strong interaction in the presence of 2 mM Ca2+.
-
EGST‐MICU1_F383A failed to pull down MICU2_WT in the presence of Ca2+, whereas they displayed a strong interaction in the absence of 2 mM Ca2+.
-
FGST‐MICU1_H385A exhibited a strong interaction with MICU2 in both the absence and presence of 2 mM Ca2+.
-
G, HITC analyses of MICU1 and MICU2 binding properties in the presence of 2 mM EGTA (G) and CaCl2 (H).
Figure EV4. Pull‐down experiment to assess the interactions between MICU1 and MICU2 in the apo and Ca2+‐bound states.
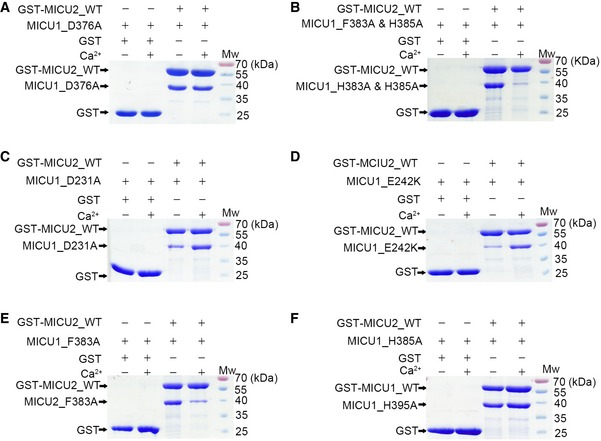
- MICU1_D376A exhibited a strong interaction with GST‐MICU2_WT in both the absence and presence of 2 mM Ca2+.
- MICU1_F383A & H385A was not pulled down by GST‐MICU2_WT in the presence of Ca2+, whereas they displayed a strong interaction in the absence of 2 mM Ca2+.
- MICU1_D231A was partially pulled down by GST‐MICU1_WT in the absence of Ca2+, while they showed a strong interaction in the presence of 2 mM Ca2+.
- MICU2_E242K was not pulled down by GST‐MICU1_WT in the absence of Ca2+, whereas they displayed a strong interaction in the presence of 2 mM Ca2+.
- MICU2_F383A was not pulled down by GST‐MICU1_WT in the presence of 2 mM Ca2+, while they showed a strong interaction in the absence of 2 mM Ca2+.
- MICU2_H385A exhibited a strong interaction in both the absence and presence of 2 mM Ca2+.
Figure EV5. Semi‐quantitative analyses of pull‐down results.
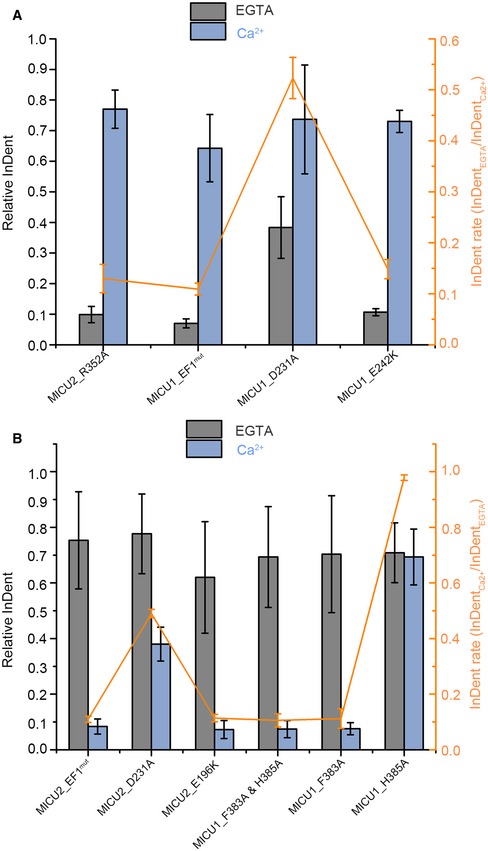
- Semi‐quantification of crucial residues which contribute to MICU1–MICU2 heterodimer formation in the absence of Ca2+. Relative InDent values (referred to as Ix) represent the ratio of grayscales (referred to as Gx) of GST‐tag‐free protein and GST‐tag‐containing protein in the same condition. For example, relative InDent of MICU2_R352A in the absence of Ca2+ represents the ratio of grayscales of MICU2_R352A and GST‐MICU1 in the absence of Ca2+(GR352A/GGST). The orange broken line represents the ratios (referred to as Px) of . Taking MICU2_R352A as an example, PR352 = .
- Semi‐quantification of crucial residues which contribute to MICU1–MICU2 heterodimer in the presence of Ca2+. Relative InDent is the same as (A). The orange broken line represents the ratios (Px) of . For example, PEF1 = .
To quantitatively estimate the binding affinity between MICU1 and MICU2, we fused maltose‐binding protein (MBP) to MICU2 to enhance protein solubility for ITC experiments. MICU1 interacted with MICU2 yielding K d value of 224 ± 46 nM in the presence of EGTA and 134 ± 52 nM in the presence of Ca2+ (Fig 4G and H). These results demonstrate that Ca2+ binding to EF‐hands promotes MICU1–MICU2 interaction and stability, consistent with our structural analyses and previous reports 27. Moreover, we tested the interactions between MBP‐MICU1 and MICU2_E196K and MICU2_R352A in Ca2+ and EGTA conditions using ITC, respectively. The results show that MICU2_R352A mutation reduced the K d value (14.9 ± 2.3 mM; Appendix Fig S6A) by about five orders of magnitude in EGTA condition compared with WT, while MICU2_E196K and MBP‐MICU1 had no obvious interaction in Ca2+ condition (Appendix Fig S6B). Similarly, we also detected the interactions of MBP_MICU2 and MICU1_F383A and MICU1_E242K in Ca2+ and EGTA conditions, respectively (Appendix Fig S6C and D). The results demonstrate that both mutants of MICU1 did not interact with MBP‐MICU2, consistent with our pull‐down assays.
Discussion
Although many studies investigated the regulatory functions of MICU1 and MICU2, the underlying mechanism is still ambiguous. It is important to note that they function in the context of a heterodimer 17, 18, 26, 37, 38 and help to maintain MCU's normal Ca2+ uptake function. Structural differences between MICU1 and MICU2, such as the different orientation of EF‐hand 1 and β‐sheet (Fig 1F), result in a more compact structure in MICU2, which might be the basis for their different roles in regulating MCU's Ca2+ uptake activities.
Calcium ions in MICU2 homodimer
We observed strong and unambiguous electron densities encompassed by the EF‐hand loop, which guided us to fill the metal ions. Although EF‐hands can coordinate other divalent metal ions (Mg2+, Mn2+), Ca2+ is the optimal candidate due to the following reasons: (i) The affinity of EF‐hand for Ca2+ is higher than that for Mg2+ or Mn2+ 41, 42. (ii) Ca2+ preferentially adopts a pentagonal bipyramidal coordination geometry with seven ligands, whereas the coordination geometry of Mg2+ or Mn2+ ions is octahedral with six ligands 34, 42. (iii) The bidentate coordination to metal ion provided by Glu12 (at position 12 of the EF‐loop) is the characteristic symbol of Ca2+‐ligand complex 42. Mg2+‐ or Mn2+‐binding EF‐hands would maintain the apo‐like conformation since the Glu12 does not participate in coordinating metal ions or only contributes one ligand 41. It is worth noting that our structure is in accordance with all Ca2+‐binding properties, although we cannot completely rule out that the observed density is due to the presence of other divalent metal ions.
Potential interaction sites between MICU1 and MICU3 in both states
We aligned the MICU1, MICU2, and MICU3 protein sequences to observe whether the critical residues in our pull‐down assays are conserved (Appendix Fig S7). MICU3, the paralog of MICU1 and MICU2, is highly expressed in the central nervous system (CNS) 9, 28, and was observed to interact with MICU1, but not MICU2 28. The crucial glutamine residue in EF‐hand 1 at loop position 12 is conserved in all three paralogous proteins (Glu242 in MICU1, Glu196 in MICU2, and Glu256 in MICU3; Appendix Fig S7). As for Phe383, although it is not conserved in MICU2 and MICU3, the corresponding sites are both hydrophobic methionine residues (Met337 in MICU2, Met445 in MICU3; Appendix Fig S7). Interestingly, Met337 in the MICU2 homodimer structure interacts with the EF‐hand 1 hydrophobic core (Figs 2A and EV3) and also appears to interact with the hydrophobic core of MICU1 EF‐hand 1 in the putative Ca2+‐bound heterodimer, similar to Phe383 in MICU1. Additionally, Arg352 in MICU2 is highly conserved in MICU3 (the corresponding residue is Arg460), but not in MICU1 (the corresponding residue is Gln398; Appendix Fig S7). This is very important because MICU1 can heterodimerize with MICU2 and MICU3, but MICU2 and MICU3 cannot interact with each other 28. Moreover, Arg352 contributes to the MICU1–MICU2 interaction in the absence of Ca2+. Arg460 and Met445 in MICU3 may be the crucial residues in MICU1–MICU3 interactions in Ca2+‐free and Ca2+‐bound conditions, respectively. According to the characteristics and conservation of Glu242 and Phe383 in MICU1 and Glu196 and Arg352 in MICU2, we speculate that Glu256, Met445, and Arg460 in MICU3 are likely crucial residues in the MICU1–MICU3 heterodimer.
Heterodimer conformational transition from apo to Ca2+‐bound state
In most EF‐hand‐containing proteins, EF‐hands are typically packed in pairs with four amphipathic helixes bundled together to make a hydrophobic core. When Ca2+ ions are bound, the hydrophobic core buried in the apo state is exposed to the solvent and creates a hydrophobic area. The hydrophobic cores are observed in both Ca2+‐bound MICU1 and MICU2 homodimer. EF‐hands 1 play significant roles in conformational transition by exposing hydrophobic cores upon Ca2+ binding. Overexpression of both MICU1_EF1mut in MICU1 KO and MICU2_EF1mut in MICU2 KO cells led to severe inhibition of Ca2+ uptake at high Ca2+ concentrations 17, 18, consistent with our structural analysis.
Fortunately, in our MICU2 structure, we obtained both Ca2+‐bound and Ca2+‐free EF‐hand conformations, which helped us to understand the conformational transition from the apo to the Ca2+‐bound state. Considering the identical interaction sites and modes (hydrophobic interaction in the apo state, electrostatic in the Ca2+‐bound state) observed in the MICU2 homodimer and the MICU1–MICU2 heterodimer, we propose a conformational transition process of the MICU1–MICU2 heterodimer from apo to Ca2+‐bound state. In the apo state, MICU1 and MICU2 interact with each other via electrostatic interaction mediated by Glu242 in MICU1 and Arg352 in MICU2. When EF‐hands coordinate Ca2+ ions, the exiting helixes of EF‐hands 1 in MICU1/2 rotate outside and expose a hydrophobic core. Subsequently, the hydrophobic residues (Phe383 in MICU1, Ile333, Met337 in MICU2) in bent helixes of MICU1/2 are attracted and form a more compact heterodimer.
Putative complex structure models of MICU1 and MICU2 in both states
In our model, the MICU1–MICU2 complex would form a more compact structure in the Ca2+‐bound than in the apo form, which may have significant implications in regulating uniporter Ca2+ uptake. Based on our structural and interaction studies, we attempted to build MICU1–MICU2 heterodimer models based on the fact that MICU2 homodimer and MICU1–MICU2 heterodimer possess similar interaction sites in both conditions. We used MICU2 homodimer as a heterodimer model at the beginning, but severe clashes were observed in the model. Additionally, the packing mode of MICU2 dimer exhibits more resemblance to the MICU1 dimer in the Ca2+‐free form than the Ca2+‐bound form (Appendix Fig S3). Thus, we elected to use MICU1 Ca2+‐free structure as a heterodimer model in the end. The calculated buried surface values of heterodimer in the apo and Ca2+‐bound states are ~ 724 and ~ 927 Å2 (Fig 5A and B), supported by our experiment results (Fig 4G and H).
Figure 5. Working model of the MICU1–MICU2 heterodimer and the regulation of Ca2+ uptake.
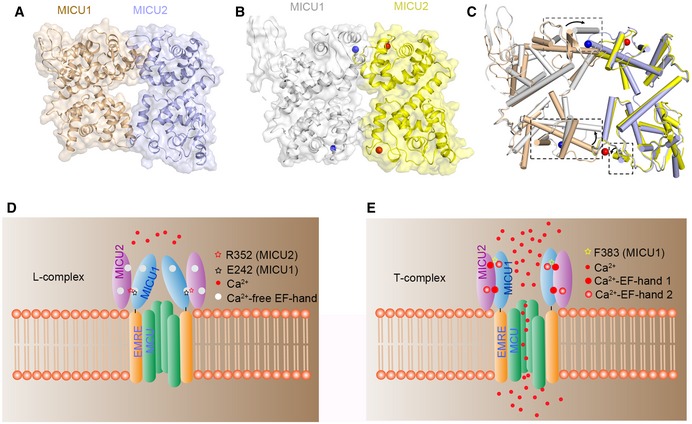
-
A, BSpace filling and cartoon representation of heterodimer structures in the Ca2+‐free (A) and Ca2+‐bound states (B). Both structures are built based on the MICU1 homodimer structure (PDB code: 4NSC) in the apo state. Ca2+‐free and Ca2+‐bound MICU1 were colored in wheat and gray, respectively. Ca2+‐free and Ca2+‐bound MICU2 were colored in purple and yellow, respectively. Calcium ions are shown as blue and red spheres in MICU1 and MICU2, respectively.
-
CComparison of MICU1–MICU2 structures in the apo (wheat and purple) and Ca2+‐bound states (gray and yellow). The conformational differences are highlighted by boxes (black dotted lines). The black arrows show the direction of the conformational change from apo to Ca2+‐bound state.
-
D, EProposed model for mitochondrial Ca2+ uptake regulated by MICU1–MICU2. In the apo state, MICU1–MICU2 form a loose complex which inhibits the uniporter. Upon Ca2+ binding, the conformational changes of the EF‐hands promote a transition to a more compact state, which activates Ca2+ uptake.
Potential reasons why EF‐hands 2 have smaller conformational change than EF‐hands 1
EF‐hands 2 in MICU1/2 display smaller conformational change than EF‐hands 1 in MICU1/2 when Ca2+ is present. We speculate that this is determined by the antiparallel nature of the MICU1–MICU2 heterodimer (or individual homodimer) and the positions of the EF‐hands. Both EF‐hands 1 in MICU1 and MICU2 are located in the homodimer interface region, while both EF‐hands 2 are located in the periphery far away from the interface. In our putative structural model of the complex, both EF‐hands 1 are close to the bent helixes. EF‐hand 1 would expose the hydrophobic core in the presence of Ca2+ ions, and the bent helix would be attracted to stabilize the structure. In contrast, there is no α‐helix or hydrophobic residue to stabilize the MICU2 pair when EF‐hand pair 2 exposes the hydrophobic core. Obviously, the hydrophobic area cannot be directly exposed to the solution.
Working model for the heterodimer of MICU1–MICU2 regulates Ca2+ uptake
Previous work indicated that the association between the MICU1–MICU2 complex and MCU is Ca2+‐dependent. At low Ca2+ concentrations, the MICU1–MICU2 complex binds to the D‐ring of MCU 24, 25. Increasing Ca2+ concentrations induce a conformational change in the MICU1–MICU2 heterodimer and its release from MCU 23. However, the exact nature of the Ca2+‐induced conformational changes is still unclear. Based on previous models, we attempted to improve the working model of MICU1–MICU2 complex formation to illuminate the structural changes and the regulatory mechanism of uniporter (Fig 5D and E). In the resting condition, we suggest that MICU1 associates with MICU2 to form a loose or less compact structure to prevent Ca2+ uptake (Fig 5C and D). The conformation of MICU1–MICU2 complex changes from loose state (L) to tight state (T) upon Ca2+ binding to EF‐hands (Fig 5C and E) 27. In this scenario, MICU2 pulls MICU1 to form a more compact complex resulting in the loss of direct interactions between the highly conserved arginine of MICU1 and the D‐ring of MCU 24, 25, relieving the inhibition of the Ca2+ channel. EMRE would tether the MICU1–MICU2 complex at the opening position of the pore to prevent waggling in the IMS under condition of high Ca2+ level 7. The two MICU1–MICU2 forms would switch back and forth in response to changes in Ca2+ levels.
Recently, Kamer et al and Xing et al published mouse and human Ca2+‐free MICU2 structures, respectively 43, 44. Intriguingly, there are considerable differences between the two publications. Xing et al 43 proposed a back‐to‐back dimer arrangement for human MICU2, while Kamer et al 44 reported a face‐to‐face dimer involving the C‐terminus of mouse MICU2. Our structure with three Ca2+ ions bound assembles in a face‐to‐face manner, similar to the mouse Ca2+‐free MICU2 homodimer reported by Kamer et al The two dimerization modes are both possible as a result of crystal packing. In fact, both face‐to‐face and back‐to‐back modes can be observed in all three structures depending on the crystallographic symmetry (Appendix Fig S8). For our Ca2+‐bound structure, the face‐to‐face mode is reasonable because of canonical hydrophobic interactions (Fig 2A). For the two apo structures reported in the literature, both packing modes seem justified with their functional analyses, but the face‐to‐face manner would be more appropriate since this packing mode promotes C‐terminal domain bundling, which generates a more compact dimer. EF‐hand 1 and the bent helix (also referred to as helix of EF‐hand 3) contribute to the homodimer interaction of the mouse Ca2+‐free and our human Ca2+‐bound MICU2 structure, although the interaction mode differs. In the apo mouse structure, EF‐hand 1 and EF‐hand 3 helixes interact with each other via surface hydrophobic interaction 44, whereas the bent helix inserts into the hydrophobic core of EF‐hand 1 pair in our Ca2+‐bound structure (Fig 2A). Moreover, within the Ca2+‐free mouse MICU2 homodimer interface, we also observe that R352 (R349 in mouse MICU2) participates in the Ca2+‐free homodimer interaction (Appendix Fig S9). As reported in the literature, MICU1 and MICU2 were shown to form stable disulfide bonds in cells through cysteine residues in C‐terminal domains 23, 26. In our pull‐down experiments, we deleted the C‐terminal domains of MICU2 and MICU1 in order to explore other interactions sites independent of the disulfide bond. Our findings that the constructs lacking the cysteine residue can still robustly form the complex clearly demonstrate that interactions other than disulfide bond are also important for heterodimer formation. However, we believe that the C‐terminal domain is crucial for MICU2 function and that our work lacking the C‐terminal domain may raise some ambiguities. For example, is the dimer assembly mode of Ca2+‐bound MICU2 containing C‐terminal domain similar to Ca2+‐free or arranged in a more complicated stacking manner? Would the orientation of the C‐terminal domain in the Ca2+‐bound state change when Ca2+ ions bind EF‐hands?
It is important to note that in order to study individual MICUs in the cell, one of them needs to be knocked out. The results from such studies, for example, cells lacking MICU2 showing lower threshold than WT cells 17, 18, would not necessarily mean that MICU1 can actually function by its own. Current evidence suggests that under physiological conditions where both MICU1 and MICU2 exist, their heterodimer is likely the main structure that helps modulate mitochondrial Ca2+ uptake 17, 18, 26, 37, 38. In summary, our structural and biochemical analyses have allowed us to establish a working model for MICU1–MICU2 complex formation and helped to analyze Ca2+‐induced conformational changes. These results provide insight into the regulatory mechanisms of MICU1–MICU2, in addition to establishing a structural framework for understanding mitochondrial calcium uptake.
Materials and Methods
Protein expression
T4L‐MICU2 (BC031089.1, residues 85–406), MICU1 (BC004190.2, residues 97–444), and individual mutants were cloned into pET‐28b(+) plasmid with an N‐terminal His‐tag. The T4 lysozyme was designed with triple mutation (C54T, C97A, D20N) to prevent cysteine oxidation. GST‐MICU1 (residues 97–444) and GST‐MICU2 (residues 85–406) and their individual mutants were cloned into pGEX‐6p‐1 containing GST sequence in N‐terminus. The recombinant plasmids were expressed in Escherichia coli BL21 (DE3). The cells were grown to OD600 ~ 0.8 at 37°C, and the temperature was lowered to 16°C. Then, the bacteria were induced by isopropyl β‐D‐1‐thiogalactopyranoside (IPTG, Amresco, Cat. No. 0487‐100g) at the final concentration of 5 mM. After incubation for 20 h, the cells were harvested and stored at −40°C. The Se‐Met T4L‐MICU2 recombinant plasmids were expressed in E. coli strain BL21 (DL41), which is unable to synthesize methionine and requires methionine or Se‐methionine in the growth media. The cells were grown in M9 media (Molecular Dimensions, Cat. No. MD12‐501 and MD12‐502) with addition of 30 μg/l Se‐methionine at 37°C. Other procedures are similar to the native protein.
Protein purification
The harvested cells were lysed in buffer I containing 20 mM HEPES pH 8.0, 300 mM NaCl, 20 mM imidazole, 0.1% protease inhibitor (phenylmethylsulfonyl fluoride, PMSF, Amresco, Cat. No. 97064‐898‐EA), and 0.1% Triton X‐100 (Amresco, Cat. No. 0694‐1L) by sonication. Insoluble cell debris were removed by centrifugation at 18,000 rpm (39,000 × g) for 30 min. The supernatant was loaded to nickel–nitrilotriacetic acid (Ni‐NTA; GE) gravity column pre‐equilibrated with buffer I. The gravity column was washed with 100 ml buffer II containing 20 mM HEPES pH 8.0, 300 mM NaCl, and 30 mM imidazole. The protein was eluted with 20 ml buffer III containing 20 mM HEPES pH 8.0, 300 mM NaCl, and 500 mM imidazole. The eluted protein was concentrated with 10‐kDa concentrator and further purified by SEC on a HiLoad 16/600 Superdex 200 pg column (GE), pre‐equilibrated with buffer IV containing 20 mM HEPES pH 8.0, 300 mM NaCl, 2 mM EGTA, and 1 mM DTT. The purification protocol of Se‐Met protein was identical to the native protein.
Protein crystallization and X‐ray data collection
Crystallization trials were performed using the hanging drop vapor diffusion method. T4L‐MICU2 protein (containing 2 mM EGTA) was concentrated to 4 mg/ml and mixed with reservoir solution composed of 100 mM sodium malonate pH 5.0 and 5% w/v PEG 3350 at a 2 μl:2 μl ratio. The crystals were grown in 1 week at 20°C. Crystals were dipped in the cryoprotectant consisting of reservoir solution and the addition of 30% glycerol and then flash‐frozen in liquid nitrogen. The Se‐Met crystals were obtained in the same crystallization condition as the native protein. The native and anomalous diffraction data were collected at the wavelength of 0.979 Å in beamline BL19U at Shanghai Synchrotron Radiation Facility (SSRF).
Structural determination
We first solved the T4L‐MICU2 structure by single anomalous dispersion (SAD) at the lower resolution (2.55 Å) and then improved this structure using the native dataset at a higher resolution (1.96 Å). Both the native and Se‐Met X‐ray diffraction datasets of T4L‐MICU2 fusion protein were processed and scaled by HKL3000R 45 suit. The initial model was built by SHARP/autoSHARP 46 software, which was subsequently refined using iterative model‐building by phenix.refine in PHENIX 47 suit and COOT 48 program. X‐ray data, structure, and validation statistics are listed in Appendix Table S2.
GST pull‐down experiments
Pull‐down assay protocol followed the previous report 49. GST‐MICU1 or GST‐MICU2 pellet was suspended in lysis buffer containing 20 mM HEPES pH 8.0, 300 mM NaCl, 1 mM PMSF, 0.1% Triton X‐100, and 2 mM EGTA or 2 mM CaCl2. The resuspended cells were sonicated with pulse of 10 s on/50 s off at 40% amplitude for 15 min on the ice. The supernatant was incubated with GST resin pre‐equilibrated with lysis buffer for 2 h at 4°C. Then, the GST resin was washed three times with washing buffer containing HEPES pH 8.0, 300 mM NaCl, and 2 mM EGTA or 2 mM CaCl2. MICU1 or MICU2 (without GST‐tag) pellet was split into two equal parts and sonicated as aforementioned. The supernatants were mixed with GST‐MICU1 or GST‐MICU2‐bound resin and blank resin (negative control), respectively. After binding for 2 h at 4°C, the resins were washed as aforementioned. Finally, the GST resins were resuspended with 1× loading buffer and analyzed by SDS–PAGE.
Isothermal titration calorimetry
All ITC experiments were performed using a VP‐ITC instrument (GE MicroCal) at 20°C. For determining the binding properties of MICU1 and MICU2, all the proteins were purified with the buffer containing 20 mM HEPES pH 8.0, 300 mM NaCl, 2 mM DTT, and 2 mM EGTA or CaCl2. The MBP‐fused proteins and titrated sample proteins were concentrated to 400 and 40 μM, respectively. 2 μl 400 μM MBP‐fused protein was injected into 200 μl 40 μM of the second sample (non‐MBP‐fused protein) for 19 times, with time intervals of 300 s. The titration procedure was the same as the calcium‐binding assays. The binding affinity and thermodynamic parameter were generated using the MicroCal Origin package.
Multi‐angle laser light scattering
Multi‐angle laser light scattering measurements were performed as previously described 40, using MALLS detector (Wyatt Technology) equipped with an in‐line SEC instrument. The protein samples were purified in buffer containing 20 mM HEPES pH 8.0, 300 mM NaCl, 2 mM DTT, and 2 mM EGTA or CaCl2 and concentrated to 2–4 mg/ml. The samples were injected into a chromatographic column with the running rate of 0.5 ml/min. The molecular weights were calculated by peaks integration using ASTRA software.
Complex structure modeling
The L‐complex and T‐complex were built based on the Ca2+‐free MICU1 dimer structure (PDB code: 4NSC; the dimer was extracted from the hexamer which is a trimer of dimer). For L‐complex, the chain A (one Ca2+ ion in the EF‐hand) of MICU2 (PDB code: 6IIH) was aligned with Ca2+‐free MICU1 dimer. The superimposed chain of MICU1 was replaced by MICU2 chain A. For T‐complex, the chain A of Ca2+‐bound MICU1 (PDB code: 4NSD) and the chain B (two Ca2+ ions in the EF‐hands) of MICU2 were aligned with Ca2+‐free MICU1 dimer, respectively. The superimposed chains of model were replaced by chain A of Ca2+‐bound MICU1 and chain B of MICU2.
Author contributions
WW and QS performed the experiments. QS and ZL determined the structure WW, QS, ZL, ZQ, DL, HP, JZ, and ZJ analyzed the data. WW, JZ, and ZJ designed the project and wrote the article.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We would like to thank the staff at the BL19U of Shanghai Synchrotron Radiation Facility (SSRF) for X‐ray data collection. This work was supported by grants from the National Natural Science Foundation of China (No. 21773014), as well as Natural Sciences and Engineering Research Council of Canada (No. RGPIN‐2018‐04427).
EMBO Reports (2019) 20: e47488
Contributor Information
Jimin Zheng, Email: jimin_z@bnu.edu.cn.
Zongchao Jia, Email: jia@queensu.ca.
Data availability
The structures produced in this study are available in the following database: protein atomic coordinates data: PDB 6IIH (https://www.rcsb.org/structure/6iih).
References
- 1. Kirichok Y, Krapivinsky G, Clapham DE (2004) The mitochondrial calcium uniporter is a highly selective ion channel. Nature 427: 360–364 [DOI] [PubMed] [Google Scholar]
- 2. Santo‐Domingo J, Demaurex N (2010) Calcium uptake mechanisms of mitochondria. Biochem Biophys Acta 1797: 907–912 [DOI] [PubMed] [Google Scholar]
- 3. Drago I, Pizzo P, Pozzan T (2011) After half a century mitochondrial calcium in‐ and efflux machineries reveal themselves. EMBO J 30: 4119–4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher‐Timme CA, Sancak Y, Bao XR, Strittmatter L, Goldberger O, Bogorad RL et al (2011) Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature 476: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Stefani D, Raffaello A, Teardo E, Szabo I, Rizzuto R (2011) A forty‐kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 476: 336–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chaudhuri D, Sancak Y, Mootha VK, Clapham DE (2013) MCU encodes the pore conducting mitochondrial calcium currents. Elife 2: e00704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sancak Y, Markhard AL, Kitami T, Kovacs‐Bogdan E, Kamer KJ, Udeshi ND, Carr SA, Chaudhuri D, Clapham DE, Li AA et al (2013) EMRE is an essential component of the mitochondrial calcium uniporter complex. Science 342: 1379–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perocchi F, Gohil VM, Girgis HS, Bao XR, McCombs JE, Palmer AE, Mootha VK (2010) MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature 467: 291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Plovanich M, Bogorad RL, Sancak Y, Kamer KJ, Strittmatter L, Li AA, Girgis HS, Kuchimanchi S, De Groot J, Speciner L et al (2013) MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS One 8: e55785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raffaello A, De Stefani D, Sabbadin D, Teardo E, Merli G, Picard A, Checchetto V, Moro S, Szabo I, Rizzuto R (2013) The mitochondrial calcium uniporter is a multimer that can include a dominant‐negative pore‐forming subunit. EMBO J 32: 2362–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoo J, Wu M, Yin Y, Herzik MA Jr, Lander GC, Lee SY (2018) Cryo‐EM structure of a mitochondrial calcium uniporter. Science 361: 506–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fan C, Fan M, Orlando BJ, Fastman NM, Zhang J, Xu Y, Chambers MG, Xu X, Perry K, Liao M et al (2018) X‐ray and cryo‐EM structures of the mitochondrial calcium uniporter. Nature 559: 575–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baradaran R, Wang C, Siliciano AF, Long SB (2018) Cryo‐EM structures of fungal and metazoan mitochondrial calcium uniporters. Nature 559: 580–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nguyen NX, Armache JP, Lee C, Yang Y, Zeng W, Mootha VK, Cheng Y, Bai XC, Jiang Y (2018) Cryo‐EM structure of a fungal mitochondrial calcium uniporter. Nature 559: 570–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Oxenoid K, Dong Y, Cao C, Cui T, Sancak Y, Markhard AL, Grabarek Z, Kong L, Liu Z, Ouyang B et al (2016) Architecture of the mitochondrial calcium uniporter. Nature 533: 269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mallilankaraman K, Doonan P, Cardenas C, Chandramoorthy HC, Muller M, Miller R, Hoffman NE, Gandhirajan RK, Molgo J, Birnbaum MJ et al (2012) MICU1 is an essential gatekeeper for MCU‐mediated mitochondrial Ca(2+) uptake that regulates cell survival. Cell 151: 630–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kamer KJ, Mootha VK (2014) MICU1 and MICU2 play nonredundant roles in the regulation of the mitochondrial calcium uniporter. EMBO Rep 15: 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Payne R, Hoff H, Roskowski A, Foskett JK (2017) MICU2 restricts spatial crosstalk between InsP3R and MCU channels by regulating threshold and gain of MICU1‐mediated inhibition and activation of MCU. Cell Rep 21: 3141–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tsai MF, Phillips CB, Ranaghan M, Tsai CW, Wu Y, Willliams C, Miller C (2016) Dual functions of a small regulatory subunit in the mitochondrial calcium uniporter complex. Elife 5: e15545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Csordas G, Golenar T, Seifert EL, Kamer KJ, Sancak Y, Perocchi F, Moffat C, Weaver D, de la Fuente Perez S, Bogorad R et al (2013) MICU1 controls both the threshold and cooperative activation of the mitochondrial Ca(2)(+) uniporter. Cell Metab 17: 976–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vais H, Mallilankaraman K, Mak DD, Hoff H, Payne R, Tanis JE, Foskett JK (2016) EMRE is a matrix Ca(2+) sensor that governs gatekeeping of the mitochondrial Ca(2+) uniporter. Cell Rep 14: 403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bick AG, Calvo SE, Mootha VK (2012) Evolutionary diversity of the mitochondrial calcium uniporter. Science 336: 886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petrungaro C, Zimmermann KM, Kuttner V, Fischer M, Dengjel J, Bogeski I, Riemer J (2015) The Ca(2+)‐dependent release of the Mia40‐induced MICU1‐MICU2 dimer from MCU regulates mitochondrial Ca(2+) uptake. Cell Metab 22: 721–733 [DOI] [PubMed] [Google Scholar]
- 24. Phillips CB, Tsai CW, Tsai MF (2019) The conserved aspartate ring of MCU mediates MICU1 binding and regulation in the mitochondrial calcium uniporter complex. Elife 8: e41112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paillard M, Csordas G, Huang KT, Varnai P, Joseph SK, Hajnoczky G (2018) MICU1 interacts with the D‐ring of the MCU pore to control its Ca(2+) flux and sensitivity to Ru360. Mol Cell 72: 778–785 e773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patron M, Checchetto V, Raffaello A, Teardo E, Vecellio Reane D, Mantoan M, Granatiero V, Szabo I, De Stefani D, Rizzuto R (2014) MICU1 and MICU2 finely tune the mitochondrial Ca2+ uniporter by exerting opposite effects on MCU activity. Mol Cell 53: 726–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kamer KJ, Grabarek Z, Mootha VK (2017) High‐affinity cooperative Ca(2+) binding by MICU1‐MICU2 serves as an on‐off switch for the uniporter. EMBO Rep 18: 1397–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Patron M, Granatiero V, Espino J, Rizzuto R, De Stefani D (2018) MICU3 is a tissue‐specific enhancer of mitochondrial calcium uptake. Cell Death Differ 26: 179–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vecellio Reane D, Vallese F, Checchetto V, Acquasaliente L, Butera G, De Filippis V, Szabo I, Zanotti G, Rizzuto R, Raffaello A (2016) A MICU1 splice variant confers high sensitivity to the mitochondrial Ca(2+) uptake machinery of skeletal muscle. Mol Cell 64: 760–773 [DOI] [PubMed] [Google Scholar]
- 30. Wang L, Yang X, Li S, Wang Z, Liu Y, Feng J, Zhu Y, Shen Y (2014) Structural and mechanistic insights into MICU1 regulation of mitochondrial calcium uptake. EMBO J 33: 594–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsai CW, Wu Y, Pao PC, Phillips CB, Williams C, Miller C, Ranaghan M, Tsai MF (2017) Proteolytic control of the mitochondrial calcium uniporter complex. Proc Natl Acad Sci USA 114: 4388–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bick AG, Wakimoto H, Kamer KJ, Sancak Y, Goldberger O, Axelsson A, DeLaughter DM, Gorham JM, Mootha VK, Seidman JG et al (2017) Cardiovascular homeostasis dependence on MICU2, a regulatory subunit of the mitochondrial calcium uniporter. Proc Natl Acad Sci USA 114: E9096–E9104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shamseldin HE, Alasmari A, Salih MA, Samman MM, Mian SA, Alshidi T, Ibrahim N, Hashem M, Faqeih E, Al‐Mohanna F et al (2017) A null mutation in MICU2 causes abnormal mitochondrial calcium homeostasis and a severe neurodevelopmental disorder. Brain 140: 2806–2813 [DOI] [PubMed] [Google Scholar]
- 34. Gifford JL, Walsh MP, Vogel HJ (2007) Structures and metal‐ion‐binding properties of the Ca2+‐binding helix‐loop‐helix EF‐hand motifs. Biochem J 405: 199–221 [DOI] [PubMed] [Google Scholar]
- 35. Ikura M (1996) Calcium binding and conformational response in EF‐hand proteins. Trends Biochem Sci 21: 14–17 [PubMed] [Google Scholar]
- 36. Lewit‐Bentley A, Rety S (2000) EF‐hand calcium‐binding proteins. Curr Opin Struct Biol 10: 637–643 [DOI] [PubMed] [Google Scholar]
- 37. Matesanz‐Isabel J, Arias‐del‐Val J, Alvarez‐Illera P, Fonteriz RI, Montero M, Alvarez J (2016) Functional roles of MICU1 and MICU2 in mitochondrial Ca(2+) uptake. Biochem Biophys Acta 1858: 1110–1117 [DOI] [PubMed] [Google Scholar]
- 38. Ahuja M, Muallem S (2014) The gatekeepers of mitochondrial calcium influx: MICU1 and MICU2. EMBO Rep 15: 205–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kamer KJ, Sancak Y, Fomina Y, Meisel JD, Chaudhuri D, Grabarek Z, Mootha VK (2018) MICU1 imparts the mitochondrial uniporter with the ability to discriminate between Ca(2+) and Mn(2). Proc Natl Acad Sci USA 115: E7960–E7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li D, Wu W, Pei H, Wei Q, Yang Q, Zheng J, Jia Z (2016) Expression and preliminary characterization of human MICU2. Biol Open 5: 962–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Senguen FT, Grabarek Z (2012) X‐ray structures of magnesium and manganese complexes with the N‐terminal domain of calmodulin: insights into the mechanism and specificity of metal ion binding to an EF‐hand. Biochemistry 51: 6182–6194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grabarek Z (2011) Insights into modulation of calcium signaling by magnesium in calmodulin, troponin C and related EF‐hand proteins. Biochem Biophys Acta 1813: 913–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xing Y, Wang M, Wang J, Nie Z, Wu G, Yang X, Shen Y (2019) Dimerization of MICU proteins controls Ca(2+) influx through the mitochondrial Ca(2+) uniporter. Cell Rep 26: 1203–1212 e1204 [DOI] [PubMed] [Google Scholar]
- 44. Kamer KJ, Jiang W, Kaushik VK, Mootha VK, Grabarek Z (2019) Crystal structure of MICU2 and comparison with MICU1 reveal insights into the uniporter gating mechanism. Proc Natl Acad Sci USA 116: 3546–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Minor W, Cymborowski M, Otwinowski Z, Chruszcz M (2006) HKL‐3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62: 859–866 [DOI] [PubMed] [Google Scholar]
- 46. Vonrhein C, Blanc E, Roversi P, Bricogne G (2007) Automated structure solution with autoSHARP. Methods Mol Biol 364: 215–230 [DOI] [PubMed] [Google Scholar]
- 47. Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW et al (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu W, Zheng J, Jia Z (2019) Expression and characterization of MICU2, a Ca(2+) sensor protein. Methods Mol Biol 1929: 261–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File
Data Availability Statement
The structures produced in this study are available in the following database: protein atomic coordinates data: PDB 6IIH (https://www.rcsb.org/structure/6iih).


