Abstract
Selenium deficiency has been proven to induce calcium disorders in the chicken heart. However, detailed regulatory mechanisms, e.g., the long noncoding RNA (lncRNA)-microRNA (miRNA)-mRNA regulatory axis, have not yet been described. Here, we point out lnc-2315, miR-1594, and Troponin T (TNNT2) based on the results of lncRNA and miRNA comparative genomics group analysis of Se-deficient chicken hearts compared with control hearts. We employed lnc-3215 and TNNT2 knockdown, miR-1594 knockdown, and overexpression models in the chicken embryos in vivo, and lnc-3215, miR-1594, and TNNT2 knockdown and overexpression models in cardiomyocytes in vitro. The dual-luciferase reporter assay and quantitative real-time PCR were used to confirm the relationships between miR-1594 and TNNT2, lnc-3215, and miR-1594 in cardiomyocytes. Our results revealed that TNNT2 suppression induced cardiac calcium overload in vivo and in vitro. miR-1594 activates cardiac calcium overload by targeting TNNT2. Moreover, we found that lnc-3215 regulates miR-1594, and thus influences the TNNT2 expression in vivo and in vitro; these conclusions were verified by gene knockdown in chicken embryos. Our present study revealed a novel regulatory model of a calcium program, which comprises lnc-3215, miR-1594, and TNNT2 in the chicken heart. Our conclusions may provide a feasible diagnostic tool for Se-deficient cardiomyocytes injury.
Keywords: selenium deficiency, calcium overload, chicken heart, lnc3-215-miR1594-TNNT2 axis
Introduction
Selenium (Se) is a pivotal trace element in the body; Se is involved in various cellular processes and signaling pathways through the synthesis of selenoproteins. Se deficiency is involved in Keshan disease in humans,1 white muscle disease and exudative diathesis in chicken,2, 3 liver necrosis and immune system disease,4, 5, 6 and mulberry heart disease.7, 8 The heart is one of the main targets of Se deficiency; in our previous study, we have determined that Se deficiency caused cardiomyocyte damage through activation of apoptosis9 and necrosis.10 Troponin (Tn) is a regulatory protein of muscle contraction located on the thin filament of contractile proteins and plays an important regulatory role in muscle contraction and relaxation. Tn is the main regulatory protein of the myocardium and is a diagnostic maker of myocardial damage more sensitive than creatine kinase-myoglobin (CK-MB). Troponin T (TNNT), Troponin I (TNNI), and Troponin C (TNNC) are different subunits and are the key factors determining the characteristic sensitivity for tension control in the myocardium.11 Calcium (Ca2+), an important second messenger in the body, participates in almost all physiological activities. Horner et al.12 found that the Ca2+ homeostasis is associated with metabolic diseases of the bone. The disorder of intracellular Ca2+ concentration can trigger apoptosis through the mitochondrial pathway,13 and intracellular Ca2+ is related to the rate of force development in twitch contraction of the heart.14 In our previous study, we found that Se deficiency significantly decreased the expression of TNNT2 in the chicken heart, and that Se-deficient chicken heart undergoes Ca2+ overload in the cytoplasm. However, there are few reports about the relationship between Ca2+ and TNNT2.
MicroRNAs (miRNAs) are approximately 20–25 nt long and are a class of noncoding single-stranded RNA molecules encoded by the endogenous genes. miRNAs participate in the modulation of silencing complex degradation or repression of the target mRNA translation through recognition of the target mRNA by base pairing. Until now, 28,645 miRNAs molecules have been found in plants, animals, and viruses.15 Most of the miRNAs exist in the genome in the form of single copies, multiple copies, or clusters.16 miRNAs are involved in various physiological processes as shown since the first discovery of miRNAs in 1983. Zhao et al.17 found that miRNA-155 ortholog is encoded by the oncogenic Marek’s disease virus. Serum miRNA-155 is a potential biomarker of breast cancer.18 Insulin growth factor 1 (IGF-1) deficiency resists cardiac hypertrophy and myocardial contractile dysfunction via alleviated downregulation of miR-1 and miR-133a.19 miR-185 plays an anti-hypertrophic role in the heart via multiple targets in the Ca2+-signaling pathways.20 In our previous study, we have shown that miR-1954 induces autophagy and apoptosis in Se-deficient myocardium by targeting phosphatidylinositol 3-kinase (PI3K).21 miR-200a-5p mediates myocardial necroptosis induced by Se deficiency by targeting RNF11. However, the mechanism of regulation of Ca2+ homeostasis by miRNAs in chicken cardiomyocytes remains largely unknown.
Long noncoding RNAs (lncRNAs) are a class of RNA that is not translated into a polypeptide, and lncRNAs are longer than 200 nt. lncRNAs play an important role in numerous cellular processes such as dose compensation effect, epigenetic regulation, cell-cycle regulation, and cell differentiation regulation.22 The hypothesis of competing endogenous RNAs (ceRNAs) is a new mechanism for interaction between RNAs, which includes lncRNAs, miRNAs, and mRNA. miRNAs are known to induce target gene silencing by binding to mRNA, whereas lncRNAs regulate gene expression by competitively binding to miRNAs. lncRNAs can bind miRNAs through miRNA response elements to influence miRNA-induced target gene silence. In recent years, a number of studies demonstrated that ceRNAs are involved in various diseases. Fu et al.23 found that the lncRNA-ATB/miRNA-200a/β-catenin regulatory axis is involved in the progression of HCV-related hepatic fibrosis. LINC00520 promotes acute kidney injury development through the PI3K/AKT signaling pathway by targeting miR-27b-3p to regulate the OSMR expression level.24 KCNQ1OT1 facilitates progression of non-small-cell lung carcinoma via modulating the miRNA-27b-3p/HSP90AA1 axis.25 However, it is not known whether ceRNA participates in the regulation of Ca2+ disorder induced by Se deficiency.
Results
Se Deficiency Induces TNNT2 Suppression and Ca2+ Overload in the Chicken Heart
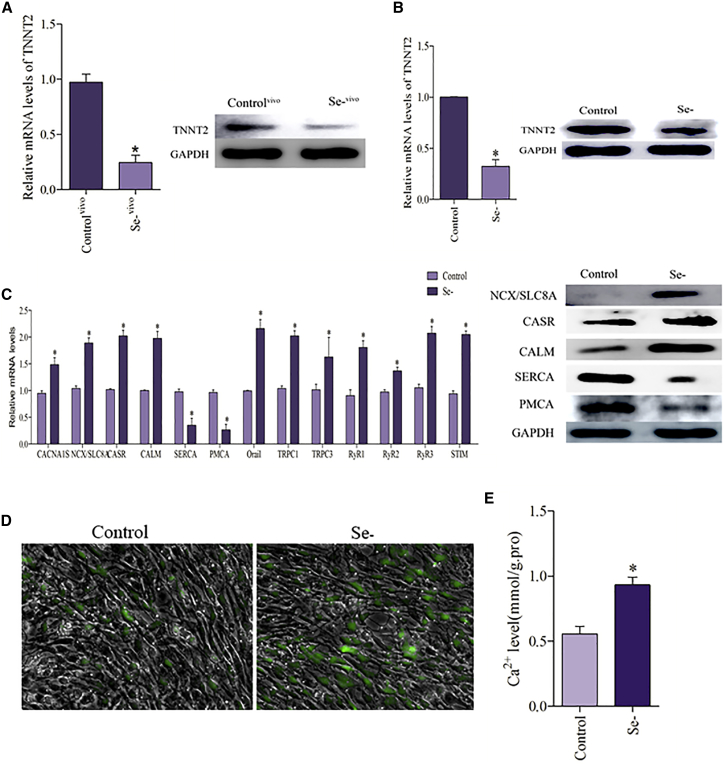
The expression of TNNT2 in the heart tissues was shown in Figure 1A; the results indicated that Se deficiency significantly reduced the expression of TNNT2 at the mRNA and protein levels. Our previous study has proved that Se deficiency induced heart injury via modulation of the Ca2+ homeostasis-related genes and induced subsequent Ca2+ overload in the heart of broilers.26
Figure 1.
Se Deficiency Induces TNNT2 Suppression and Ca2+ Overload in the Chicken Heart
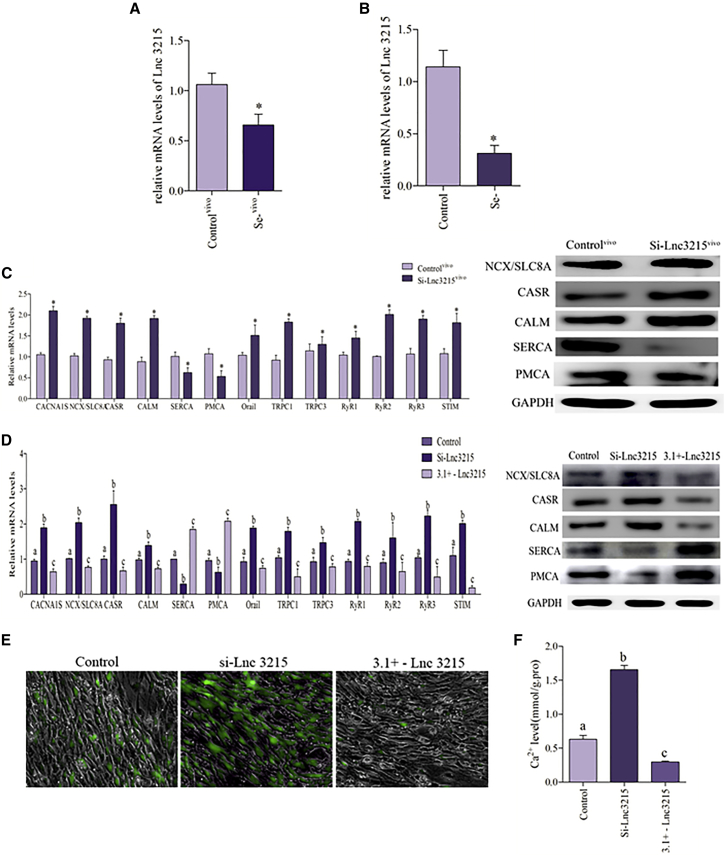
(A) The mRNA and protein levels of TNNT2 in the chicken heart. The results were calculated from at least three independent experiments (n = 3). The data are expressed as the mean ± SD. (B) The mRNA and protein levels of TNNT2 in the cardiomyocytes. (C) The mRNA levels of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, SERCA, PMCA, RyR 1, RyR 2, RyR 3, and STIM and the protein levels of NCX, CASR, CALM SERCA, and PMCA were detected in Se-deficient cardiomyocytes. (D) Cardiomyocytes were measured by the fluorescence intensity of Fluo-4 AM (green fluorescence, 5 mM). Cardiomyocytes were visualized using fluorescence microscopy; the greener the fluorescence, the greater was the concentration of Ca2+. (E) Cardiomyocytes were measured by a fluorescent microplate reader. Asterisks indicate significant difference from the corresponding normal (*p < 0.05). Control, normal cardiomyocytes; Controlvivo, control myocardial tissue; Se−, Se-deficient cardiomyocytes; Se−vivo, Se-deficient myocardial tissue.
Se-deficient cardiomyocytes were tested to validate these results. Se deficiency significantly decreased the expression of TNNT2 in cardiomyocytes at the mRNA and protein levels (Figure 1B). Then we examined the Ca2+-related pathway genes, and the results are shown in Figure 1C; the mRNA expression levels of CANA1S, Na+/Ca2+ exchanger (NCX), calcium-sensing receptor (CASR), calmodulin (CALM), calcium release-activated calcium channel protein 1 (Orai1), transient receptor potential cation (TRPC) 1, TRPC 3, RyR 1, RyR 2, RyR 3, and stromal interaction molecule (STIM) were significantly elevated; however, Se deficiency significantly decreased the sarcoplasmic reticulum calcium ATPase (SERCA) and plasma membrane Ca2+-ATPases (PMCA) expression in cardiomyocytes. A similar significant effect on the protein expression of NCX, CASR, CALM, SERCA, and PMCA was observed. The results of Ca2+ concentration were shown in Figures 1D and 1E; our results revealed that the concentration of Ca2+ in the cytoplasm of Se-deficient cardiomyocytes was significantly higher than that of control cells. These results suggested that Se deficiency caused Ca2+ overload in the chicken heart.
TNNT2 Is Involved in Maintaining the Balance of Ca2+
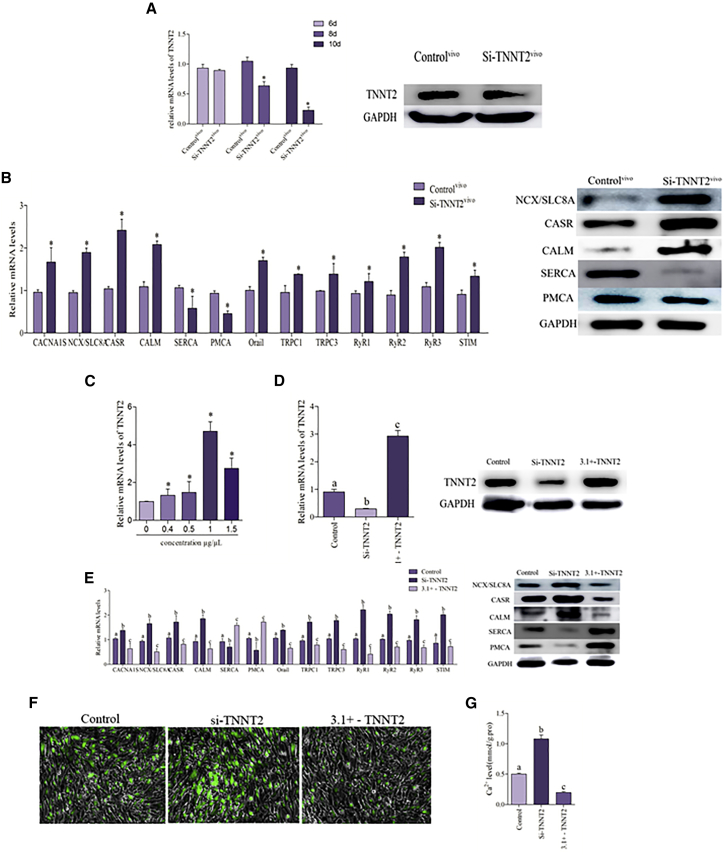
To explore the role of TNNT2 in Ca2+ homeostasis, we employed the TNNT2 knockdown model in chicken embryos. As shown in Figure 2A, we sampled the chicken embryo heart at various time points for detection of TNNT2 mRNA levels; the results revealed that the mRNA expression of TNNT2 was significantly decreased in 10-day chicken embryos. The results of assay of TNNT2 protein levels in 10-day chicken embryos indicate that the TNNT2 knockdown model was successfully established. Several classical Ca2+ channel-related genes were detected, and data are shown in Figure 2B. TNNT2 suppression significantly increased the expression of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, RyR 1, RyR 2, RyR 3, and STIM at the mRNA level, and significantly decreased the expression of SERCA and PMCA at the mRNA level. The same trend was observed in the case of the protein expression of NCX, CASR, CALM, SERCA, and PMCA.
Figure 2.
TNNT2 Is Involved in Maintaining the Balance of Ca2+
(A) To investigate the most appropriate transfection time in the chicken embryo model with transfecting 1 μg Si-TNNT2 on different days, mRNA and protein levels of TNNT2 are determined by real-time PCR and western blot. The optimum time of transfecting Si-TNNT2 is day 10 of chicken embryo. The results were calculated from at least three independent experiments (n = 3). The data are expressed as the mean ± SD. (B) The mRNA levels of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, SERCA, PMCA RyR 1, RyR 2, RyR 3, and STIM and the protein levels of NCX, CASR, CALM, SERCA, and PMCA were detected in the chicken embryo heart that normal and transfect with Si-TNNT2. (C) The most appropriate transfection concentration in cardiomyocytes with transfecting different plasmid concentrations of pcDNA3.1+ - TNNT2 for 24 h. mRNA levels of TNNT2 were determined by real-time PCR. The optimum plasmid concentration of pcDNA3.1+ - TNNT2 used in this paper is always 1 μg/μL. (D) The models of TNNT2 knockdown and TNNT2 overexpression were employed in cardiomyocytes; mRNA and protein levels of TNNT2 were detected by real-time PCR and western blot. Bars that do not share the same letters are significantly different (p < 0.05) from each other. (E) The mRNA levels of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, SERCA, PMCA, RyR 1, RyR 2, RyR 3, and STIM and the protein levels of NCX, CASR, CALM SERCA, and PMCA were detected in cardiomyocytes. (F and G) Cardiomyocytes were measured by the fluorescence intensity of Fluo-4 AM (green fluorescence, 5 mM). Cardiomyocytes were visualized using fluorescence microscopy and the fluorescent microplate reader; the greener the fluorescence, the greater was the concentration of Ca2+. Asterisks indicate significant difference from the corresponding normal (*p < 0.05). 3.1+ - TNNT2, cardiomyocytes that transfect with pcDNA3.1+ - TNNT2; Control, normal cardiomyocytes; Controlvivo, normal chicken embryo heart; Se−, Se-deficient cardiomyocytes; Si-TNNT2, cardiomyocytes that transfect with Si-TNNT2; Si-TNNT2vivo, chicken embryo heart that transfected with Si-TNNT2.
Primary cultures of cardiomyocytes treated with Si-TNNT2 and TNNT2-pcDNA3.1 were used to verify the results. The effect of the plasmid concentration of TNNT2-pcDNA3.1 was shown in Figure 2C; the result revealed that 1 μg/μL was the optimal concentration of the plasmid. The mRNA and protein levels of TNNT2 in the models of TNNT2 knockdown and TNNT2 overexpression were shown in Figure 2E. The results revealed that the expression of TNNT2 was significantly decreased in cardiomyocytes that were transfected with Si-TNNT2 and significantly increased in cardiomyocytes that transfected with TNNT2-pcDNA3.1 at the mRNA and protein levels, indicating that the models of TNNT2 knockdown and TNNT2 overexpression were successfully established. Ca2+ channel-related genes were detected, and the data are shown in Figure 2E. TNNT2 knockdown significantly increased the expression of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, RyR 1, RyR 2, RyR 3, and STIM and significantly decreased the expression of SERCA and PMCA in cardiomyocytes at the mRNA levels. Similar results were obtained in the case of protein levels of NCX, CASR, CALM, SERCA, and PMCA. However, the changes observed in the 3.1+ - TNNT2 group were opposite to those in the Si-TNNT2 group. As shown in Figures 2F and 2G, the Ca2+ concentration was significantly increased in cardiomyocytes that were transfected with Si-TNNT2. Conversely, the cytoplasmic Ca2+ concentration of cardiomyocytes that were transfected with 3.1+ - TNNT2 was significantly reduced. These data demonstrated that TNTN2 is involved in maintaining the balance of Ca2+ in the chicken heart.
miR-1594 Participates in the Regulation of TNNT2 Expression In Vivo and In Vitro
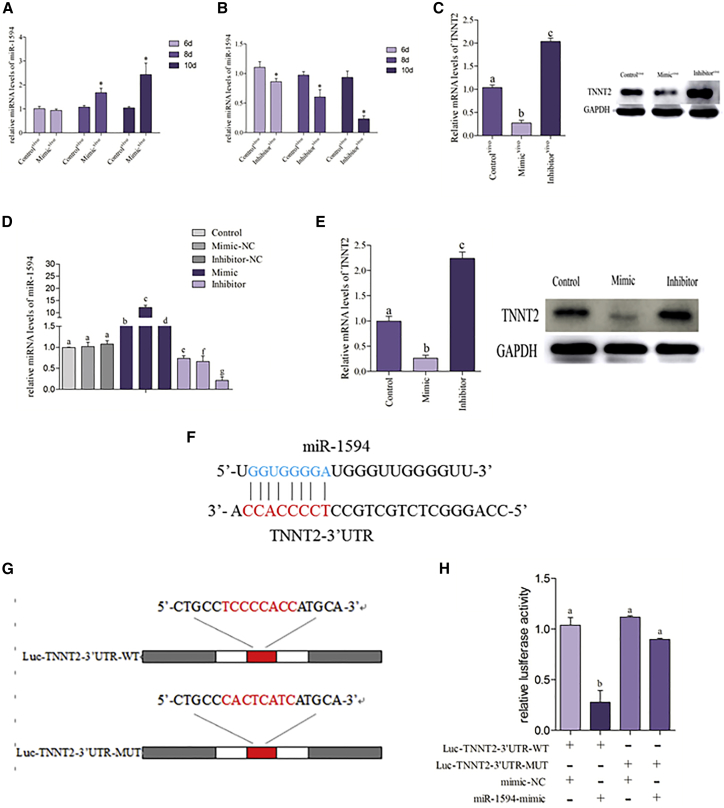
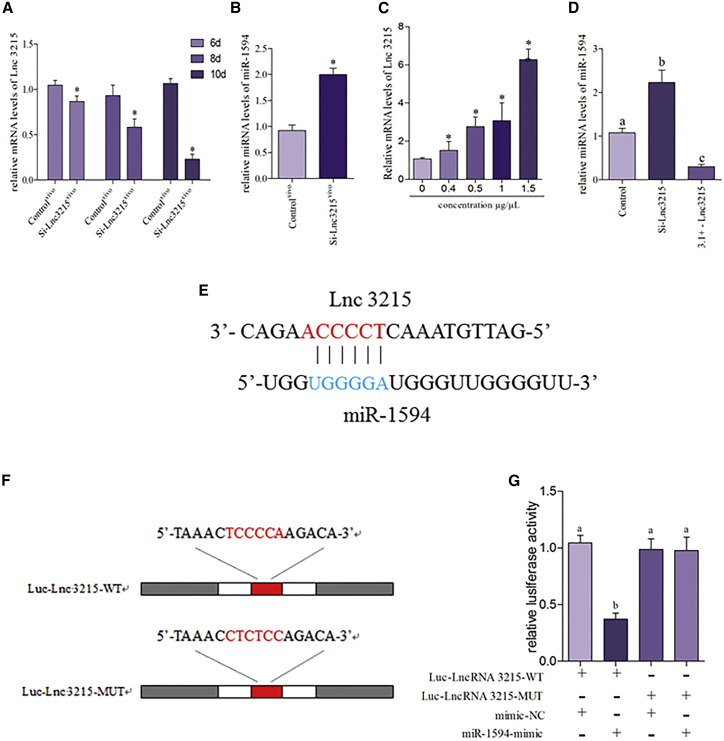
To investigate whether miRNAs participate in the regulation of TNNT2, we established the models of miR-1594 knockdown and overexpression in chicken embryos. The results of time selection of in vivo transfection were shown in Figures 3A and 3B; we found significant differential expression of miR-1594 in 10th day chicken embryo, indicating that day of the chicken embryo was optimal for transfection with a mimic and an inhibitor. As shown in Figure 3C, miR-1594 overexpression in the chicken embryo significantly reduced the expression of TNNT2 at the mRNA and protein levels, but opposite results were observed in the case of miR-1594 knockdown in chicken embryos.
Figure 3.
miR-1594 Participates in the Regulation of TNNT2 Expression In Vivo and In Vitro
(A and B) Different transfection times with 1 μg miRNA-1594 mimic (A) and inhibitor (B) were applied to the selection of the optimum transfection times; the expression of miRNA-1594 is determined by real-time PCR. The optimum time of transfect mimic and inhibitor is day 10 of chicken embryo. The results were calculated from at least three independent experiments (n = 3). The data are expressed as the mean ± SD. (C) The mRNA and protein levels of TNNT2 were detected in the chicken embryo heart that transfects with miR-1594-mimic and miR-1594-inhibitor. Bars that do not share the same letters are significantly different (p < 0.05) from each other. (D) To investigate the most appropriate transfection concentration in the cardiomyocytes model with transfecting different concentrations of miR-1594-mimic, miR-1594-inhibitor, mimic negative control, and inhibitor negative control for 24 h. miRNA levels of miR-1594 were determined by real-time PCR. The optimum concentrations of miR-1594-mimic and miR-1594-inhibitor used in this paper were always 100 nM and 200 nM. (E) The mRNA and protein levels of TNNT2 were detected in cardiomyocytes that transfect with miR-1594-mimic and miR-1594-inhibitor. (F) Putative miR-1594 binding site in the 3′ UTR region of TNNT2. (G) TNNT2 wild-type (WT) 3′ UTR and a mutated 3′ UTR in the miR-1594-binding site are shown. (H) miR-1594-mimic inhibits TNNT2-WT expression, but not mutant TNNT2-MUT expression. Asterisks indicate significant difference from the corresponding normal (*p < 0.05). Control, normal cardiomyocytes; Controlvivo, normal chicken embryo heart; Inhibitor, chicken embryo heart that transfects with miRNA-1594 inhibitor; Inhibitorvivo, chicken embryo heart that transfects with miRNA-1594 inhibitor; Mimic, cardiomyocytes that transfect with miRNA-1594 mimic; Mimicvivo, chicken embryo heart that transfects with miRNA-1594 mimic.
The models of miR-1594 knockdown and overexpression in the cardiomyocytes were used to confirm this observation in vitro. As shown in Figure 3D, different transfection concentrations of the mimic and inhibitor were used for selection of an optimal concentration. The results revealed that the optimal transfection concentration of mimic and inhibitor are 100 and 200 nM, respectively. The expression of TNNT2 was detected, and the data are shown in Figure 3E; the mRNA and protein expression levels of TNNT2 expression were decreased in response to the increasing level of miR-1594 and elevated in response to the decreasing levels of miR-1594 in cardiomyocytes. Then we performed a luciferase assay to explore whether miR-1594 can directly bind to TNNT2. As shown in Figures 3F–3H, we used the 3′ UTR-mediated luciferase activity assay and found that the miR-1594 mimic markedly modulates the luciferase activity driven by TNNT2 mRNA 3′ UTR plasmids. However, the miR-1594 mimic failed to inhibit the luciferase activity driven by the target sequences in the mutated 3′ UTR plasmids. These data suggested that TNNT2 was a specific target of miR-1594.
miR-1594 Regulates Ca2+ Homeostasis in the Chicken Heart
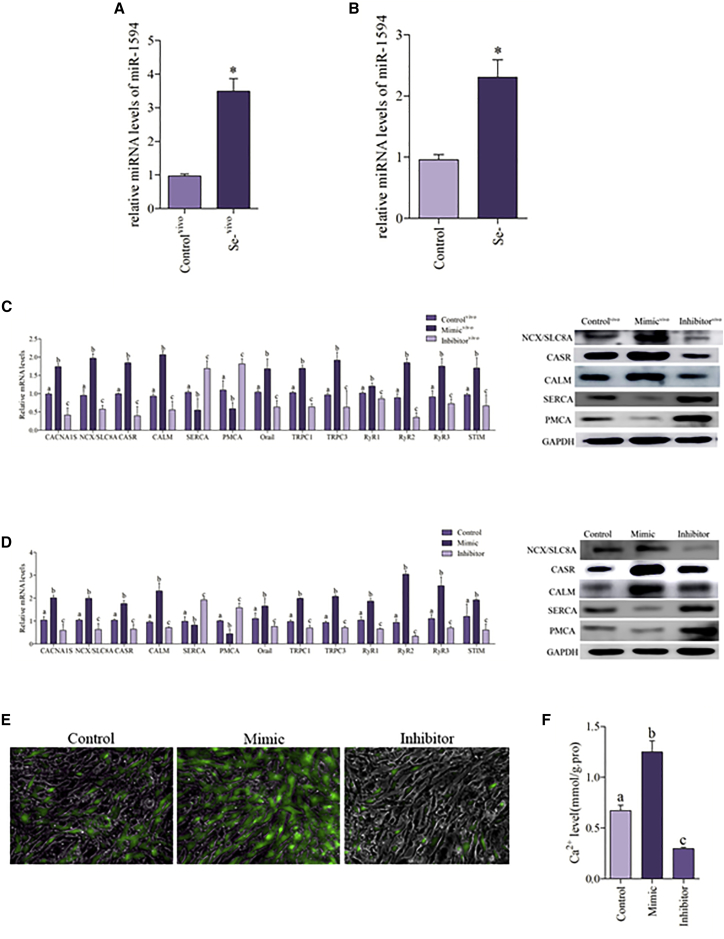
miR-1594 was selected in the chicken heart based on miRNA genomics group analysis (Figure S1A and Table S1). Quantitative real-time PCR was used to confirm the results of the genomics group analysis. The expression of miR-1594 was detected in Se-deficient myocardial tissue (Figure 4A) and Se-deficient cardiomyocytes (Figure 4B); we found that the expression of miR-1594 was substantially elevated in Se-deficient myocardium and Se-deficient cardiomyocytes. As shown in Figure 4C, overexpression of the miR-1594 in the chicken embryo significantly increased the mRNA expression of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, RyR 1, RyR 2, RyR 3, and STIM, and significantly decreased the mRNA expression of SERCA and PMCA. Similar results were obtained at the protein level in the case of NCX, CASR, CALM, SERCA, and PMCA.
Figure 4.
miR-1594 Regulates Ca2+ Homeostasis in the Chicken Heart
(A) miRNA levels of miR-1594 were detected in the chicken heart. Controlvivo, control myocardial tissue. The results were calculated from at least three independent experiments (n = 3). The data are expressed as the mean ± SD. (B) The miRNA levels of miR-1594 were detected in Se-deficient cardiomyocytes. (C) The mRNA levels of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, SERCA, PMCA, RyR 1, RyR 2, RyR 3, and STIM and the protein levels of NCX, CASR, CALM SERCA, and PMCA were detected in the chicken embryo heart. Controlvivo, normal chicken embryo heart. Bars that do not share the same letters are significantly different (p < 0.05) from each other. (D) The mRNA levels of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, SERCA, PMCA, RyR 1, RyR 2, RyR 3, and STIM and the protein levels of NCX, CASR, CALM SERCA, and PMCA were detected in cardiomyocytes. (E and F) Cardiomyocytes were measured by the fluorescence intensity of Fluo-4 AM (green fluorescence, 5 mM). Cardiomyocytes were visualized using fluorescence microscopy and the fluorescent microplate reader; the greener the fluorescence, the greater was the concentration of Ca2+. Asterisks indicate significant difference from the corresponding control (*p < 0.05). Control, normal cardiomyocytes; Inhibitor, chicken embryo heart that transfects with miRNA-1594 inhibitor; Inhibitorvivo, chicken embryo heart that transfects with miRNA-1594 inhibitor; Mimic, cardiomyocytes that transfect with miRNA-1594 mimic; Mimicvivo, chicken embryo heart that transfects with miRNA-1594 mimic; Se−, Se-deficient cardiomyocytes; Se−vivo, Se-deficient myocardial tissue.
The cardiomyocytes that were transfected with miR-1594 mimic and inhibitor were used for verification. As shown in Figure 4D, the cardiomyocytes with miR-1594 overexpression had significantly increased expression of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, RyR 1, RyR 2, RyR 3, and STIM, and significantly reduced expression of SERCA and PMCA in the mRNA levels. Similar results in the case of NCX, CASR, CALM, SERCA, and PMCA were observed in protein levels. The Ca2+ concentration was detected by fluorescence staining and is shown in Figures 4E and 4F; we found that miR-1594 overexpression increased the cytosolic Ca2+ concentration in cardiomyocytes; conversely, inhibition of miR-1594 reduced Ca2+ concentration in the cytoplasm of cardiomyocytes. These findings suggest that miR-1594 participates in the regulation of Ca2+ balance.
lnc-3215 Is Able to Directly Bind to miR-1594 and Regulate miR-1594 Activity
To explore the underlying mechanism that regulates the expression of miR-1594, we detected the expression of miR-1594 in the lnc-3215 knockdown model of chicken embryos. The results of time selection of in vivo transfection were shown in Figure 5A; the expression of lnc-3215 was significantly decreased in 10-day chicken embryos, indicating that day 10 chicken embryo was the optimal option for transfection of Si-lnc-3215. The expression of miR-1594 was detected, and the data are shown in Figure 5B; we found that lnc-3215 knockdown significantly increased the expression of miR-1594 in the chicken embryos.
Figure 5.
lnc-3215 Is Able to Directly Bind to miR-1594 and Regulate miR-1594 Activity
(A) Different transfection times with 1 μg miRNA-1594 mimic and inhibitor were applied to the selection of the optimum transfection times, and the expression of lnc-3215 was determined by real-time PCR. The optimum time of transfect mimic and inhibitor was day 10 of chicken embryo. The results were calculated from at least three independent experiments (n = 3). The data were expressed as the mean ± SD. (B) The expression of lnc-3215 was determined in the lnc-3215-deficient chicken embryo heart. (C) The most appropriate transfection concentration in cardiomyocytes with transfecting different plasmid concentrations of pcDNA3.1+ - lnc-3215 for 24 h. The expression of lnc-3215 is determined by real-time PCR. The optimum plasmid concentration of pcDNA3.1+ - lnc-3215 used in this paper was always 1.5 μg/μL. (D) The models of lnc-3215 knockdown and lnc-3215 overexpression are employed in cardiomyocytes; miR-1594 was detected by quantitative real-time PCR. The results were calculated from at least three independent experiments (n = 3). Bars that do not share the same letters were significantly different (p < 0.05) from each other. (E) Putative lnc-3215 binding site in the miR-1594. (F) lnc-3215 wild-type (WT) binding site and a mutated binding site in the miR-1594 were shown. (G) miR-1594-mimic inhibits lnc-3215-WT expression, but not mutant lnc-3215-MUT expression. Asterisks indicate significant difference from the corresponding control (*p < 0.05). 3.1+ - TNNT2, cardiomyocytes that transfect with pcDNA3.1+ - lnc-3215; Control, normal cardiomyocytes; Controlvivo, normal chicken embryo heart; Si-lnc-3215, cardiomyocytes that transfect with Si-lnc-3215; Si-lnc-3215vivo, chicken embryo heart that transfects with Si-lnc-3215.
The models of lnc-3215 knockdown and overexpression in cardiomyocytes were used to verify the associations between lnc-3215 and miR-1594. The effect of plasmid concentration of lnc-3215-pcDNA3.1 was shown in Figure 5C; the result revealed that 1.5 μg/μL is the optimal concentration. As shown in Figure 5D, the expression of miR-1594 was upregulated in cardiomyocytes after transfection with Si-lnc-3215, and the opposite result was found in cardiomyocytes that were treated with 3.1+ - lnc-3215. Luciferase assay was performed to confirm this relationship. As shown in Figures 5E–5G, our results revealed that the miR-1594 mimic markedly modulated the luciferase activity driven by lnc-3215 plasmids. However, the miR-1594 mimic failed to inhibit the luciferase activity driven by the lnc-3215 plasmids with mutated binding sites. Based on our results, we concluded that lnc-3215 is able to directly bind to miR-1594 and regulate miR-1594 activity.
lnc-3215 Was Involved in the Regulation of Ca2+ Homeostasis
lnc-3215 was identified by screening based on the results of lncRNA genomics group analysis (Figure S1B and Table S2). Then we carried out quantitative real-time PCR to detect the lnc-3215 levels in Se-deficient heart and cardiomyocytes. Our results revealed that the expression of lnc-3215 was significantly decreased in Se-deficient myocardial tissue and Se-deficient cardiomyocytes (Figures 6A and 6B). The key Ca2+ channel-related genes were detected in the lnc-3215 suppression chicken embryo and shown in Figure 6C; our results indicate that the mRNA expression of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, RyR 1, RyR 2, RyR 3, and STIM was activated by lnc-3215 suppression. At the same time, lnc-3215 knockdown inhibited the mRNA expression of SERCA and PMCA. Similar results were obtained at the protein levels in the case of NCX, CASR, CALM, SERCA, and PMCA.
Figure 6.
lnc-3215 Was Involved in the Regulation of Ca2+ Homeostasis
(A) lncRNA levels of lnc-3215 were detected in the chicken heart. Controlvivo indicates control myocardial tissue. The results were calculated from at least three independent experiments (n = 3). The data are expressed as the mean ± SD. (B) The LncRNA levels of lnc-3215 were detected in Se-deficient cardiomyocytes. (C) The mRNA levels of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, SERCA, PMCA RyR 1, RyR 2, RyR 3, and STIM and the protein levels of NCX, CASR, CALM SERCA, and PMCA were detected in the chicken embryo heart. Controlvivo indicates the normal chicken embryo heart. (D) The mRNA levels of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, SERCA, PMCA, RyR 1, RyR 2, RyR 3, and STIM and the protein levels of NCX, CASR, CALM SERCA, and PMCA were detected in cardiomyocytes. Bars that do not share the same letters are significantly different (p < 0.05) from each other. (E and F) Cardiomyocytes were measured by the fluorescence intensity of Fluo-4 AM (green fluorescence, 5 mM). Cardiomyocytes were visualized using fluorescence microscopy and the fluorescent microplate reader; the greener the fluorescence, the greater was the concentration of Ca2+. Asterisk indicates significant difference from the corresponding control (*p < 0.05). Control, normal cardiomyocytes; Se−, Se-deficient cardiomyocytes; Se−vivo, Se-deficient myocardial tissue; Si-lnc-3215, cardiomyocytes that transfect with Si-lnc-3215; Si-lnc-3215vivo, chicken embryo heart that transfects with Si-lnc-3215.
We used the cardiomyocytes that were transfected with Si-lnc-3215 and 3.1+ - lnc-3215 for verification. The results of assay of Ca2+-related genes were shown in Figure 6D; the results revealed that the cardiomyocytes that transfected with Si-lnc-3215 have significantly increased mRNA expression of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, RyR 1, RyR 2, RyR 3, and STIM, while having significantly reduced the mRNA expression of SERCA and PMCA. The results of protein level assay of NCX, CASR, CALM, SERCA, and PMCA were similar. The Ca2+ concentration was detected, and the dates are shown in Figures 6E and 6F; we found that lnc-3215 knockdown increased the cytosolic Ca2+ concentration in cardiomyocytes, and overexpression of lnc-3215 reduces the Ca2+ concentration in the cytoplasm of cardiomyocytes. Overall, these findings collectively suggested that lnc-3215 was involved in the regulation of Ca2+ homeostasis.
Discussion
Tn is a marker of myocardial injury and necrosis use for diagnosis of myocardial damage and is more sensitive than CK-MB.27 Tn conformation is changed when Ca2+ concentrations fluctuate in the cytoplasm of cardiomyocytes. Tn includes three subtypes, TNNT, TNNI, and TNNC. TNNT2 is a specific myocardial Tn. TNNT2 mutations are involved in various diseases. Hanson et al.28 found deletion of TNNT2 lysine 210 in a family with dilated cardiomyopathy. TNNT2 mutation can be associated with a predisposition to prominent left ventricular hypertrophy.29 The Ca2+ channels are the molecules predominantly located in the endoplasmic reticulum (ER) and cytoplasmic membranes. Ca2+ channels are the main tools for regulation of the balance of intracellular and extracellular Ca2+. Several types of Ca2+ channels have been found in cardiomyocytes; intracellular calcium release channels include: ryanodine receptor (RyR), L-type voltage-dependent Ca2+ channel dihydropyridine receptors (CACNA1S), inositol trisphosphate receptors (IP3Rs), STIM1, and Ca2+ pump (SERCA). Extracellular Ca2+ channels include PMCA, transient receptor potential channels (TRPC, calcium release activates calcium [CRAC]), plasma membrane NCX, and Orai1. Ca2+ channels are activated upon stimulation, which indicate that Ca2+ plays the role of the second messenger.30
Ca2+-ATPase is a class of enzymes located in the cytoplasmic and the ER membranes, which include SERCA and PMCA. Ca2+-ATPase can transfer Ca2+ across the membrane to the extracellular space.31 Cytoplasmic Ca2+ can enter the ER with the assistance of SERCA; ER Ca2+ inflow into the cytoplasm is assisted by RyR and IP3R.32 RyR2 is a specific Ca2+ channel in cardiomyocytes that triggers the release and accumulation of intracellular Ca2+. STIM1 and Orai1 are present on the ER and cytoplasmic membranes, respectively. The amount of Orai1 on the plasma membrane is increased based on STIM1 aggregation to the ER membrane upon intracellular Ca2+ depletion. Orai1 interacts with TRPC1 of the TRP family and finally led to the formation of functional ORCA, causing extracellular Ca2+ influx.33 NCX acts as a two-way ion transporter that transfers Na+ into the cell and transfers Ca2+ out of the cell, which is important for Ca2+ excretion in the diastolic phase of cardiomyocytes.34 The signal transduction function of Ca2+ depends on CALM, a Ca2+ binding protein. CALM is an activator of certain specific enzymes and undergoes structural changes when it binds Ca2+. CALM-Ca2+ enzyme activation is an indispensable component in the regulation of enzyme function.35 Mifflin et al.36 found that transgenic mice expressing mutated TNNT2 had greater Ca2+ sensitivity of myofilaments and higher incidence of ventricular tachycardia after isoproterenol administration. However, there are only a few reports about the direct relationship between TNNT2 and Ca2+. In our present study, we found that Se deficiency significantly decreased the expression of TNNT2 in cardiomyocytes. The cytoplasmic Ca2+ concentration was significantly elevated in Se-deficient cardiomyocytes through activating the expression of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, RyR 1, RyR 2, RyR 3, and STIM, and inhibiting the expression of SERCA and PMCA. Similar results were obtained in the TNNT2 knockdown chicken embryos and cardiomyocytes; the results were opposite in the cardiomyocytes that overexpress TNNT2, indicating that Se deficiency induced Ca2+ overload in the chicken heart through inhibition of TNNT2.
miRNAs are the short noncoding RNAs that participate in various diseases and cellular activities. miRNAs can induce target mRNA degradation or translational inhibition by binding to the 3′ UTR of the target mRNA through base complementation at the posttranscriptional level. Additional miRNA functions have been revealed in recent years; however, a few studies identified the regulation mechanism of these miRNAs in Se-deficient Ca2+ imbalance. Zhang et al.37 found that miRNA-367 can influence breast cancer risk and calcification by binding to the 3′ UTR of RYR3. Suppression of miRNA-708 promotes metastases by Ca2+-induced cell migration.38 miR-34c-5p mediate pain response by targeting Cav2.3 containing Ca2+ channels.39 In the present study, miR-1594 was selected because of its differential expression in the control and Se-deficient chicken heart based on miRNA genomics group analysis. Then we used miR-1594 knockdown and overexpression models in vivo and in vitro. Our results revealed that miR-1594 overexpression significantly decreased the expression of TNNT2, whereas the cytoplasmic Ca2+ concentration was significantly elevated by miR-1594 overexpression. miR-1594 overexpression significantly increased the expression of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, RyR 1, RyR 2, RyR 3, and STIM, and significantly decreased the expression of SERCA and PMCA. Opposite results were obtained in miR-1594 knockdown chicken embryo and cardiomyocytes, indicating that miR-1594 induced Ca2+ overload in the chicken heart by targeting TNNT2.
lncRNAs are a class of RNA that is longer than 200 bp and does not participate in protein encoding. The ceRNA hypothesis suggests that lncRNAs influence the regulation of miRNAs on target mRNAs by binding to miRNAs, thereby regulating the expression of the related target genes. The role of the lncRNA-miRNA-mRNA network in diseases such as rheumatoid arthritis,40 pancreatic cancer,41 and breast cancer42 has been described. The study of lncRNA function in cardiac diseases has only just begun, although lncRNAs are involved in various diseases gradually discovered by researchers. In the present study, lnc-3215 was selected because of its differential expression in the control and Se-deficient chicken heart based on lncRNA genomics group analysis. Transcriptional levels of lnc-3215 are inhibited by Se deficiency. lnc-3215 knockdown and overexpression were performed to determine the potential adjustment mechanism of Se-deficient Ca2+ overload. We found that lnc-3215 knockdown significantly increased the expression of miR-1594, and that lnc-3215 knockdown causes significant elevation of the cytoplasmic Ca2+ concentration. lnc-3215 knockdown significantly increased the expression of CANA1S, NCX, CASR, CALM, Orai1, TRPC 1, TRPC 3, RyR 1, RyR 2, RyR 3, and STIM, and significantly decreased the expression of SERCA and PMCA; opposite results were obtained in the miR-1594 knockdown chicken embryos and cardiomyocytes. In addition, we found that lnc-3215 can repair miR-1594-induced TNNT2 reduction (Figures S1C and S1D). Thus, lnc-3215 is accessible to miR-1594, and the interaction between lnc-3215 and miR-1594 is effective.
In conclusion, we found that Se deficiency inhibited the expression of TNNT2, and TNNT2 suppression activated Ca2+ channels that flow outward in cells and inhibited Ca2+ channels in the cell’s inward flow. miRNA-1594 can specifically bind to TNNT2 to regulate the function and activity. lnc-3215 as a competing endogenous RNA regulated TNNT2 expression by inhibiting miRNA-1594. In a word, we concluded that lnc-3215 regulates Se-deficient myocardial calcium overload by targeting miR-1594/TNNT2.
Materials and Methods
Treatment of Experimental Animals
All procedures used in this study were approved by the Institutional Animal Care and Use Committee of Northeast Agricultural University (SRM-11).
The Model of Se-Deficient Chicken
One hundred eighty 1-day-old chickens (Weiwei, Harbin, China) were randomly divided into two groups, Controlvivo and Se−vivo. Treatment groups were maintained on a Se-sufficient diet (Controlvivo) containing 0.23 mg/kg Se or a Se-deficient diet (Se−vivo) containing 0.033 mg/kg Se for 25 days. Composition of Se-deficient feed and a detailed process to establish the Se-deficiency model have been described by our group.9, 43
The Models of TNNT2 Knockdown, miR-1594 Knockdown and Overexpression, and lnc-3215 Knockdown in the Chicken Embryo
Ninety 0-day-old chicken embryos were randomly divided into two groups, including the Controlvivo and Si-TNNT2vivo groups. The chicken embryos in Controlvivo were injected with 1 μL entranster-in vivo (Engreen Biosystem, Beijing, China) in the subgerminal cavity, and the chicken embryos in the Si-TNNT2vivo group were injected with 1 μL entranster-in vivo+Si-TNNT2 in the subgerminal cavity. The details of the concentrations of the mixture are as follows (the total volume is 200 μL):
The chicken embryos were incubated in a constant temperature incubator. The hearts were taken at 6, 8, and 10 days and used in subsequent experiments. The models of lnc-3215 knockdown and miR-1594 knockdown and overexpression in the chicken embryos were established in a similar manner.
Cardiomyocytes Culture and Treatment
Twelve-day-old chicken embryos were used to obtain primary cardiomyocytes. Subsequent to surface disinfection (using 75% alcohol), the chest was dissected to collect the apical portion of the pericardium (approximately one-third of the heart), which was immediately transferred to PBS (4°C) and washed to remove fat, connective tissue, and blood clots. Then the myocardial tissue was cut into small pieces and washed three times using PBS. After enzymatic digestion with collagenase II (0.1 g %) for 15 min with a constant temperature magnetic stirrer (37°C, 100 r/min) collection and centrifugation, an equal volume of DMEM/F12 containing 10% fetal bovine serum (FBS) and 1× mycillin was added to terminate the digestion. The pellet was redigested until the small tissue fragments are completely digested. All supernatant was collected using sequential 300 and 500 mesh filters. The cell suspension was centrifuged at 600 rpm for 5 min, resuspended twice in DMEM/F12w, and placed in disposable Petri dishes for differential adhesion (the first round of adhesion is 1 h and the second is 1.5 h). Non-adherent cells (cardiomyocytes) were collected, centrifuged at 600 rpm, counted, plated in six-well plates at 3 × 105, and incubated at 37°C, with 5% CO2 in adherent culture box for 48 h; at this time point, cardiomyocytes attain 80% confluence. All cells were randomly divided into various groups according to the specific experimental requirements. Approximately 48 h after the transfection, the cells were harvested for analysis.
Se-deficient medium was used for the cardiomyocytes model of Se deficiency; the cells were preconditioned with DMEM/F12 medium supplemented with 1% FBS, 10 μg/mL insulin, and 5 μg/mL transferrin for 5 days. The Se-deficient medium is configured according to the method of Yan et al.44
Transfection of the Si-TNNT2 and pcDNA3.1 + - TNNT2 in Cardiomyocytes
Synthetic, chemically modified short RNA oligonucleotides were purchased from Suzhou Ribo Life Science. The Si-TNNT2 siRNA sequences were sense, 5′-GGGCAGAGCAGCAGCGCAT-3′, and anti-sense, 5′-ATGCGCTGCTGCTCTGCCC-3′, respectively. Cells were transfected with the Si-TNNT2 at 100 nM. The transfection reactions were performed using the RNAiMAX (Invitrogen) in Opti-MEM medium (pre-warming at 37°C) according to the manufacturer’s instructions.
Cells were transfected with the recombinant plasmid of pcDNA3.1 + - TNNT2 (synthesizing in Sangon, Shanghai, China) at various concentrations. The transfection reactions were performed using the Lipofectamine 2000 reagent (Invitrogen) in Opti-MEM medium (pre-warming at 37°C) according to the manufacturer’s instructions.
Transfection of the miR-1594-Mimic and miR-1594-Inhibitor in Cardiomyocytes
Synthetic, chemically modified short RNA oligonucleotides were purchased from Suzhou Ribo Life Science. The miR-1594 mimic and mimic negative control (mimic-NC) sequences were 5′- UGGUGGGGAUGGGUUGGGGUU-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′, respectively. The miR-1594 inhibitor and inhibitor negative control (inhibitor-NC) sequences are 5′- mUmGmCmUmAmGmGmAmGmUmGmGmAmAmUmGmGmGmGmAmUmG-3′ (m represents -Ome) and 5′-CAGUACUUUUGUGUAGUACAA-3′, respectively. Cells were transfected with the miR-1594-mimic and mimic-NC at 100 nM, and with the miR-1594-inhibitor and inhibitor-NC at 200 nM. The transfection reactions were performed using the Lipofectamine 2000 reagent (Invitrogen) in Opti-MEM medium (pre-warmed to 37°C) according to the manufacturer’s instructions.
Transfection of the Si-lnc-3215 and pcDNA3.1 + - lnc-3215 in Cardiomyocytes
Synthetic, chemically modified short RNA oligonucleotides were purchased from Suzhou Ribo Life Science. The Si-lnc-3215 sequences were sense, 5′-GTCATCATAGCACAAACAA-3′, and anti-sense, 5′- TTGTTTGTGCTATGATGAC-3′. Cells were transfected with the Si-lnc-3215 at 100 nM. The transfection reactions were performed using the RNAiMAX (Invitrogen) in Opti-MEM medium (pre-warmed to 37°C) according to the manufacturer’s instructions.
Cells were transfected with recombinant plasmid pcDNA3.1 + - lnc-3215 (synthesized by Sangon, Shanghai, China) at various concentrations. The transfection reactions were performed using the Lipofectamine 2000 reagent (Invitrogen) in Opti-MEM medium (pre-warming at 37°C) according to the manufacturer’s instructions.
Preparation of the Luciferase Construct of TNNT2 3′ UTR, lnc-3215-miR-1594 Binding Sites, and Luciferase Activity Assay
The pMIR-REPORT plasmids for the miR-1594 target TNNT2 3′ UTR were constructed including wild-type (WT) pMIR-TNNT2 containing two tandem repeats of miR-17-5p response elements based on the TNNT2 3′ UTR and mutant (MUT) pMIR-TNNT2. The sequences of the single-stranded oligo pairs were used to generate the pMIR- TNNT2 constructs (WT and MUT). The oligonucleotides were then annealed and inserted into the pMIR-REPORT vector (Thermo Fisher). The empty vector of pMIR-REPORT was used as the negative control. Cells were co-transfected with the plasmid constructs of pMIR-TNNT2-WT or pMIR-TNNT2-MUT and miR-mimic using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Forty-eight hours after the transfection, luciferase activity was assayed using a Dual-Luciferase Reporter Assay System (Promega). The activity of the Renilla luciferase was normalized to the activity of Firefly luciferase (Renilla LUC/Firefly LUC).
The preparation of luciferase constructs of lnc-3215-miR-1594 binding sites was performed in a similar manner based on the sequence of lnc-3215.
Intracellular Ca2+ Concentration Detection
Intracellular Ca2+ concentration was detected using a Fluo-4 AM assay kit (Beyotime Biotechnology Institute, China). The stock of Fluo-4 AM was diluted to 0.5–5 μM with PBS, culture medium was discarded from the cell samples to be tested, and the cells were washed three times using PBS (37°C preheated). After that, 1,000 μL working solution was added to six-well plates and incubated in a constant temperature incubator (37°C) for 45 min. The working solution was discarded, and PBS (37°C preheated) was used to wash the cells three times. Finally, the cells were collected for detection of the concentration of intracellular Ca2+ using a fluorescence microscope and a fluorescence microplate reader.
Western Blot Analysis
For total protein extraction, a solution of protein lysate was subjected to 15% SDS-PAGE under the reducing conditions. The separated proteins were then transferred to nitrocellulose membrane for 2 h at 100 mA in a tank transfer apparatus containing Tris-glycine buffer and 20% methanol. The membrane was blocked with 5% skim milk for 24 h and incubated overnight with diluted primary antibodies against TNNT2 (1:1,000; Abclonal, Wuhan, China), NCX (1:1500; Santa Cruz Biotechnology, USA), CASR (1:1,000; Santa Cruz Biotechnology, USA), CALM (1:1,000; Santa Cruz Biotechnology, USA), SERCA (1:500; Proteintech, China), and PMCA (1:1,000; Proteintech, China). To verify equal loading of the samples, the membrane was incubated with a monoclonal glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (1:1,500; Santa Cruz Biotechnology, USA), followed by a horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG; 1:3,000) secondary antibody. The signal was detected with an X-ray film (TransGen Biotech, Beijing, China). The optical density (OD) of each band was determined using an Image VCD gel imaging system, and the relative abundance of the proteins was calculated and presented as the ratios of OD of each protein to that of GAPDH.
Quantitative Real-Time PCR Analysis on mRNA, miRNA, and lncRNA Levels
For the quantification of miR-1594 and mRNAs of target genes by real-time PCR, total miRNA and total RNA were isolated from the myocardial tissues and cultured cardiomyocytes. Reverse transcription was performed using the miRcute miRNA First-Strand cDNA Synthesis Kit (Tiangen Biotech, Beijing) according to the manufacturer’s instructions (Roche). The primers for the detection of lnc-3215, miR-1594, and target mRNAs by real-time PCR are shown in Table 1. GAPDH or U6 were used as an internal reference. Quantitative real-time PCR was performed using a LightCycler 480 System (Roche, Basel, Switzerland) and Fast Universal SYBR Green Master Mix (Roche, Basel, Switzerland). Only a single peak for each PCR product was present in the melting curve analysis. The relative abundance of mRNA was calculated according to the 2−ΔΔCt method, accounting for gene-specific efficiencies, and was normalized to the mean expression index as described above. The sequences of miR-1594 and lnc-3215 are provided in Table 2.
Table 1.
The Primers Used in the Present Study
| Gene | Serial No. | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|---|
| TNNT2 | NM_205449.1 | GAGTATGTGGAAGAAGAAGAG | CTGCTCAATCCTGTCCTT |
| SERCA | NM_204891.1 | CCCTGGTCACAACTCTGCTG | GTCAGTGGAACCTTGGCTGT |
| NCX | XM_015283450.1 | ACCGCTTCATGTCCTCCA | ATGAGCCCAAAGCCATCA |
| PMCA | NM_001168002.3 | GATGGAAGGCTCTGGAAGG | TTCAGTGGCTGCATTTCC |
| STIM | NM_001030838.1 | CGGCTTCCAGATCGTCAA | CATCCAGGTCATCCACGTC |
| CACNA1S | XM_015298692.1 | CGAGGCCATGCTCAAGAT | CCAGGGAAACGATGGAGA |
| TRPC 1 | XM_015276873.1 | CGCTACCTCCACCTTTCAAT | CGTTTCACTTTGCCACTCG |
| TRPC 3 | XM_015276103.1 | GCAATCAGCAAGGGCTACAT | GTGCCGTCTTCGTCATAGG |
| Orai1 | NM_001030658.1 | GAGGTGGTGTTGCTGTGTTG | CTGCCTGTCCTGATGTGATG |
| CALM | NM_205005.1 | GATGGAGTTGGTAAAATGAGGGAA | ACGCACTGGAAAACTAGGGTCA |
| CASR | AB175677.1 | GGAACACTGCTGCCTATG | CAAGAGACGACTGGATGAG |
| TRP 1 | NM_001174059.1 | GGTGGCAGAACATTCCTCA | TTGACTGAAGTGCCGGAAG |
| TRP 2 | XM_015291434.1 | ATAACAGCACCAGCCTCCAG | GCTTTCTGAGTTGGGTTTGG |
| TRP 3 | XM_015298987.1 | TGTTCCGACTGTGCTACCG | AAGGCTGTGATGGTGTCCTC |
| RyR 1 | XM_015272496.1 | AGCCGAGCGTGGTCTATTAC | GAGGCAGTTGTAGCCGATGA |
| RyR 2 | XM_015284420.1 | AGAGGAGAAGGCAGAAGG | TTAGAAGCAGGATGGATAGTC |
| RyR 3 | XM_015287292.1 | TGGTTGAGGTAATGGCAGAA | TCTCCTTGGCTGTGAGTGTG |
| GAPDH | NM_204305.1 | AGAACATCATCCCAGCGT | AGCCTTCACTACCCTCTTG |
Table 2.
Sequence of lnc-3215 and miRNA-1594
| Sequence Name | 5ʹ–3ʹ |
|---|---|
| miRNA-1594 | 5′-UGGUGGGGAUGGGUUGGGGUU-3′ |
| lnc-3215 | 5′-GCAGUACUGAGAUCCAAACAGGCAAAAUCCAAUUUCAACGUCAGAUCUCAAAGAUUGUAAACUCCCCAAGACAAGAUCGAUCCCU AUGCAGACUGCCUUGCACAACAAGAGUCCAGAUCAAGGCUUCCAGAUGUCAUCAUAGCACAAACAAAACAGAACAUUCAUGAAUAU AAACGUUUGCACUGAUAUGGAACUAACAACUGAGCAGAUGAAUCAGCUCUCAAGAUGGUAAAGGGCCUUUCAUGUGCAAACUUUCU UCUCAUAUUCAGAUGAAAAAACUUCUGCACUUCCAGCUGUAAACCUUAAAUAAGUGACAGUAUCCCUGCAAAUUGCUUGAAGACUC AAGAGUAGCAAUUUGCACAUUCAAAGCUUUACAGCUGUCAAUUGACAUUUUCCUAGCUAAUUAGGACACAUACAACCAACGUAAAU AAAA-3′ |
Statistical Analysis
Statistical analyses of the data were performed using GraphPad Prism (version 5.0; GraphPad Software, San Diego, CA, USA). All data were analyzed by Student’s t test or one-way ANOVA, had a normal distribution, and passed equal variance testing. Quantitative data are presented as the mean ± SD. Samples with different superscript letters represented statistically significant differences (p < 0.05).
Author Contributions
Z.Z. conceived and designed the experiments. J.Y., Y.G., J.C., and Q.L. performed the experiments. J.Y. analyzed the data and wrote the paper. Z.Z. assisted in critically revising the manuscript. All authors have read the manuscript and agreed to submit it in its current form for consideration for publication.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (grant 31872531); Earmarked Fund for China Agriculture Research System (grant CARS 35-04); Merit-based Funding for Returned Oversea Student of Heilongjiang Province (grant 2018QD0005); State Key Laboratory of Animal Nutrition (grant 2004DA125184F1725); Postdoctoral Scientific Research Developmental Fund of Heilongjiang Province (grant LBH-Q18018); and the “Academic backbone” Project of Northeast Agricultural University (grant 17XG11).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.08.003.
Supplemental Information
References
- 1.Yang J., Wang T., Wu C., Liu C. Selenium level surveillance for the year 2007 of Keshan disease in endemic areas and analysis on surveillance results between 2003 and 2007. Biol. Trace Elem. Res. 2010;138:53–59. doi: 10.1007/s12011-010-8609-1. [DOI] [PubMed] [Google Scholar]
- 2.Ishihara H., Kanda F., Matsushita T., ChiharaK, Itoh K. White muscle disease in humans: myopathy caused by selenium deficiency in anorexia nervosa under long term total parenteral nutrition. J. Neurol. Neurosurg. Psychiatry. 1999;67:829–830. doi: 10.1136/jnnp.67.6.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y., Zhao H., Shao Y., Liu J., Li J., Luo L., Xing M. Copper (II) and/or arsenite-induced oxidative stress cascades apoptosis and autophagy in the skeletal muscles of chicken. Chemosphere. 2018;206:597–605. doi: 10.1016/j.chemosphere.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 4.Sun Z., Xu Z., Wang D., Yao H., Li S. Selenium deficiency inhibits differentiation and immune function and imbalances the Th1/Th2 of dendritic cells. Metallomics. 2018;10:759–767. doi: 10.1039/c8mt00039e. [DOI] [PubMed] [Google Scholar]
- 5.Wang S., Zheng S., Zhang Q., Yang Z., Yin K., Xu S. Atrazine hinders PMA-induced neutrophil extracellular traps in carp via the promotion of apoptosis and inhibition of ROS burst, autophagy and glycolysis. Environ. Pollut. 2018;243(Pt A):282–291. doi: 10.1016/j.envpol.2018.08.070. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y., Zhao H., Liu J., Shao Y., Li J., Luo L., Xing M. Copper and arsenic-induced oxidative stress and immune imbalance are associated with activation of heat shock proteins in chicken intestines. Int. Immunopharmacol. 2018;60:64–75. doi: 10.1016/j.intimp.2018.04.038. [DOI] [PubMed] [Google Scholar]
- 7.Nielsen T.K., Wolstrup C., Schirmer A.L., Jensen P.T. Mulberry heart disease in young pigs without vitamin E and selenium deficiency. Vet. Rec. 1989;124:535–537. doi: 10.1136/vr.124.20.535. [DOI] [PubMed] [Google Scholar]
- 8.Gong Z.G., Wang X.Y., Wang J.H., Fan R.F., Wang L. Trehalose prevents cadmium-induced hepatotoxicity by blocking Nrf2 pathway, restoring autophagy and inhibiting apoptosis. J. Inorg. Biochem. 2019;192:62–71. doi: 10.1016/j.jinorgbio.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Yang J., Zhang Y., Hamid S., Cai J., Liu Q., Li H., Zhao R., Wang H., Xu S., Zhang Z. Interplay between autophagy and apoptosis in selenium deficient cardiomyocytes in chicken. J. Inorg. Biochem. 2017;170:17–25. doi: 10.1016/j.jinorgbio.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Yang T., Cao C., Yang J., Liu T., Lei X.G., Zhang Z., Xu S. miR-200a-5p regulates myocardial necroptosis induced by Se deficiency via targeting RNF11. Redox Biol. 2018;15:159–169. doi: 10.1016/j.redox.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Babu A., Scordilis S.P., Sonnenblick E.H., Gulati J. The control of myocardial contraction with skeletal fast muscle troponin C. J. Biol. Chem. 1987;262:5815–5822. [PubMed] [Google Scholar]
- 12.Horner J. Basic science of disorders of calcium metabolism and metabolic diseases of bone. Surgery (Oxford) 2006;24:215–219. [Google Scholar]
- 13.Wang M., Ruan Y., Chen Q., Li S., Wang Q., Cai J. Curcumin induced HepG2 cell apoptosis-associated mitochondrial membrane potential and intracellular free Ca(2+) concentration. Eur. J. Pharmacol. 2011;650:41–47. doi: 10.1016/j.ejphar.2010.09.049. [DOI] [PubMed] [Google Scholar]
- 14.Yue D.T. Intracellular [Ca2+] related to rate of force development in twitch contraction of heart. Am. J. Physiol. 1987;252:H760–H770. doi: 10.1152/ajpheart.1987.252.4.H760. [DOI] [PubMed] [Google Scholar]
- 15.Laxman N., Rubin C.J., Mallmin H., Nilsson O., Pastinen T., Grundberg E., Kindmark A. Global miRNA expression and correlation with mRNA levels in primary human bone cells. RNA. 2015;21:1433–1443. doi: 10.1261/rna.049148.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathews D.H. Using an RNA secondary structure partition function to determine confidence in base pairs predicted by free energy minimization. RNA. 2004;10:1178–1190. doi: 10.1261/rna.7650904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y., Yao Y., Xu H., Lambeth L., Smith L.P., Kgosana L., Wang X., Nair V. A functional MicroRNA-155 ortholog encoded by the oncogenic Marek’s disease virus. J. Virol. 2009;83:489–492. doi: 10.1128/JVI.01166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y., Wang M., Lin G., Sun S., Li X., Qi J., Li J. Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS ONE. 2012;7:e47003. doi: 10.1371/journal.pone.0047003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua Y., Zhang Y., Ren J. IGF-1 deficiency resists cardiac hypertrophy and myocardial contractile dysfunction: role of microRNA-1 and microRNA-133a. J. Cell. Mol. Med. 2012;16:83–95. doi: 10.1111/j.1582-4934.2011.01307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J.O., Song D.W., Kwon E.J., Hong S.E., Song H.K., Min C.K., Kim D.H. miR-185 plays an anti-hypertrophic role in the heart via multiple targets in the calcium-signaling pathways. PLoS ONE. 2015;10:e0122509. doi: 10.1371/journal.pone.0122509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q., Cai J., Gao Y., Yang J., Gong Y., Zhang Z. miR-2954 Inhibits PI3K Signaling and Induces Autophagy and Apoptosis in Myocardium Selenium Deficiency. Cell. Physiol. Biochem. 2018;51:778–792. doi: 10.1159/000495332. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Liang D., Jin J., Liu C., He Y. [Progress of Long Non-coding RNA in Non-small Cell Lung Cancer] Zhongguo Fei Ai Za Zhi. 2018;21:43–49. doi: 10.3779/j.issn.1009-3419.2018.01.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu N., Zhao S.X., Kong L.B., Du J.H., Ren W.G., Han F., Zhang Q.S., Li W.C., Cui P., Wang R.Q. LncRNA-ATB/microRNA-200a/β-catenin regulatory axis involved in the progression of HCV-related hepatic fibrosis. Gene. 2017;618:1–7. doi: 10.1016/j.gene.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Tian X., Ji Y., Liang Y., Zhang J., Guan L., Wang C. LINC00520 targeting miR-27b-3p regulates OSMR expression level to promote acute kidney injury development through the PI3K/AKT signaling pathway. J. Cell. Physiol. 2019;234:14221–14233. doi: 10.1002/jcp.28118. [DOI] [PubMed] [Google Scholar]
- 25.Dong Z., Yang P., Qiu X., Liang S., Guan B., Yang H., Li F., Sun L., Liu H., Zou G., Zhao K. KCNQ1OT1 facilitates progression of non-small-cell lung carcinoma via modulating miRNA-27b-3p/HSP90AA1 axis. J. Cell. Physiol. 2019;234:11304–11314. doi: 10.1002/jcp.27788. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z., Liu M., Guan Z., Yang J., Liu Z., Xu S. Disbalance of calcium regulation-related genes in broiler hearts induced by selenium deficiency. Avian Pathol. 2017;46:265–271. doi: 10.1080/03079457.2016.1259528. [DOI] [PubMed] [Google Scholar]
- 27.Nageh T., Sherwood R.A., Harris B.M., Byrne J.A., Thomas M.R. Cardiac troponin T and I and creatine kinase-MB as markers of myocardial injury and predictors of outcome following percutaneous coronary intervention. Int. J. Cardiol. 2003;92:285–293. doi: 10.1016/s0167-5273(03)00105-0. [DOI] [PubMed] [Google Scholar]
- 28.Hanson E.L., Jakobs P.M., Keegan H., Coates K., Bousman S., Dienel N.H., Litt M., Hershberger R.E. Cardiac troponin T lysine 210 deletion in a family with dilated cardiomyopathy. J. Card. Fail. 2002;8:28–32. doi: 10.1054/jcaf.2002.31157. [DOI] [PubMed] [Google Scholar]
- 29.Komamura K., Iwai N., Kokame K., Yasumura Y., Kim J., Yamagishi M., Morisaki T., Kimura A., Tomoike H., Kitakaze M., Miyatake K. The role of a common TNNT2 polymorphism in cardiac hypertrophy. J. Hum. Genet. 2004;49:129–133. doi: 10.1007/s10038-003-0121-4. [DOI] [PubMed] [Google Scholar]
- 30.Yang J., Gong Y., Liu Q., Cai J., Zhang B., Zhang Z. Thioredoxin silencing-induced cardiac supercontraction occurs through endoplasmic reticulum stress and calcium overload in chicken. Metallomics. 2018;10:1667–1677. doi: 10.1039/c8mt00206a. [DOI] [PubMed] [Google Scholar]
- 31.Dally S., Bredoux R., Corvazier E., Andersen J.P., Clausen J.D., Dode L., Fanchaouy M., Gelebart P., Monceau V., Del Monte F. Ca2+-ATPases in non-failing and failing heart: evidence for a novel cardiac sarco/endoplasmic reticulum Ca2+-ATPase 2 isoform (SERCA2c) Biochem. J. 2006;395:249–258. doi: 10.1042/BJ20051427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowycky M.C., Fox A.P., Tsien R.W. Three types of neuronal calcium channel with different calcium agonist sensitivity. Nature. 1985;316:440–443. doi: 10.1038/316440a0. [DOI] [PubMed] [Google Scholar]
- 33.Luik R.M., Wang B., Prakriya M., Wu M.M., Lewis R.S. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature. 2008;454:538–542. doi: 10.1038/nature07065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Wagoner D.R., Bond M. Reperfusion Arrhythmias: New Insights into the Role of the Na(+)/Ca(2+) Exchanger. J. Mol. Cell. Cardiol. 2001;33:2071–2074. doi: 10.1006/jmcc.2001.1470. [DOI] [PubMed] [Google Scholar]
- 35.Cheung W.Y. Calmodulin plays a pivotal role in cellular regulation. Science. 1980;207:19–27. doi: 10.1126/science.6243188. [DOI] [PubMed] [Google Scholar]
- 36.Mifflin J.J., Dupuis L.E., Alcala N.E., Russell L.G., Kern C.B. Intercalated cushion cells within the cardiac outflow tract are derived from the myocardial troponin T type 2 (Tnnt2) Cre lineage. Dev. Dyn. 2018;247:1005–1017. doi: 10.1002/dvdy.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L., Liu Y., Song F., Zheng H., Hu L., Lu H., Liu P., Hao X., Zhang W., Chen K. Functional SNP in the microRNA-367 binding site in the 3'UTR of the calcium channel ryanodine receptor gene 3 (RYR3) affects breast cancer risk and calcification. Proc. Natl. Acad. Sci. USA. 2011;108:13653–13658. doi: 10.1073/pnas.1103360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryu S., McDonnell K., Choi H., Gao D., Hahn M., Joshi N., Park S.M., Catena R., Do Y., Brazin J. Suppression of miRNA-708 by polycomb group promotes metastases by calcium-induced cell migration. Cancer Cell. 2013;23:63–76. doi: 10.1016/j.ccr.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 39.Gandla J., Lomada S.K., Lu J., Kuner R., Bali K.K. miR-34c-5p functions as pronociceptive microRNA in cancer pain by targeting Cav2.3 containing calcium channels. Pain. 2017;158:1765–1779. doi: 10.1097/j.pain.0000000000000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang H., Ma R., Zou S., Wang Y., Li Z., Li W. Reconstruction and analysis of the lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveal functional lncRNAs in rheumatoid arthritis. Mol. Biosyst. 2017;13:1182–1192. doi: 10.1039/c7mb00094d. [DOI] [PubMed] [Google Scholar]
- 41.Ye S., Yang L., Zhao X., Song W., Wang W., Zheng S. Bioinformatics method to predict two regulation mechanism: TF-miRNA-mRNA and lncRNA-miRNA-mRNA in pancreatic cancer. Cell Biochem. Biophys. 2014;70:1849–1858. doi: 10.1007/s12013-014-0142-y. [DOI] [PubMed] [Google Scholar]
- 42.Davoren P.A., McNeill R.E., Lowery A.J., Kerin M.J., Miller N. Identification of suitable endogenous control genes for microRNA gene expression analysis in human breast cancer. BMC Mol. Biol. 2008;9:76. doi: 10.1186/1471-2199-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W., Chen M., Jin X., Li X., Yang Z., Lin H., Xu S. H2S induces Th1/Th2 imbalance with triggered NF-κB pathway to exacerbate LPS-induce chicken pneumonia response. Chemosphere. 2018;208:241–246. doi: 10.1016/j.chemosphere.2018.05.152. [DOI] [PubMed] [Google Scholar]
- 44.Yan J., Zheng Y., Min Z., Ning Q., Lu S. Selenium effect on selenoprotein transcriptome in chondrocytes. Biometals. 2013;26:285–296. doi: 10.1007/s10534-013-9610-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.