Abstract.
Leishmaniasis is a vector-borne disease caused by protozoan parasites of the Leishmania genus. In Australia, leishmaniasis is an imported disease that is presenting itself at increased rates because of international travel, the influx of immigrants, and deployment of military operations to endemic regions. Although Leishmania species are morphologically indistinguishable, there is a strong correlation between some causative species of leishmaniasis and the subsequent response to the treatments available and patient outcome. Consequently, identification of the infective species is imperative as misidentification can result in the administering of an ineffective drug. The aim of this study was to develop a simple diagnostic tool with high sensitivity and specificity, which is capable of detecting the presence of the parasite and accurately differentiating the causative species in question. Using the advantageous properties of the maxi-circle kinetoplast DNA, a polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) targeting the ND7 gene was developed for the analysis of imported cases of human leishmaniasis in Australia. Designed as a dual analysis, concurrent PCR of Leishmania maxi-circle DNA and digestion with two separate enzymes (NlaIII and HpyCH4IV), this study provides an appraisal on 24 imported cases of leishmaniasis between 2008 and 2017. Five Leishmania species were reported, with members of the Viannia subgenus being the most common. The implementation of novel diagnostic procedures for leishmaniasis such as the one reported here is needed to establish a gold standard practice for the diagnosis and treatment of leishmaniasis.
INTRODUCTION
Leishmaniasis is a complex disease with a global distribution, affecting approximately 12 million people and a further 350 million at risk of contracting the disease.1 Leishmania infections manifest as three distinct clinical forms of leishmaniasis depending on the species in question and location of infection: cutaneous leishmaniasis (CL), mucocutaneous leishmaniasis (MCL), and visceral leishmaniasis (VL or kala-azar). This broad-spectrum disease can manifest from either zoonotic or anthroponotic transmission. Zoonotic leishmaniasis tends to occur in rural settings across endemic regions, whereas anthroponotic leishmaniasis tends to occur in densely populated urban regions.1,2 Species of the Leishmania (Viannia) (L. [V.]) subgenus are restricted to the Neotropics (New World), whereas the subgenus Leishmania (Leishmania) (L. [L.]) occurs in both the New World and Old World (Figure 1). Representing a major public health problem, in the absence of a “gold standard” or reference biomarker, a wide array of laboratory methods has been developed over the last 20 years for the diagnosis of leishmaniasis.3–10 Traditionally, diagnosis was based primarily on clinical manifestations of the disease and the microscopic detection of Leishmania parasites.11 The obvious drawback to this technique is that the sensitivity is dependent on the skill of the technician with reported sensitivities ranging from 43% to 85%.12–14 As such, conventional methods involving parasitological and serological detection are now routinely used for the examination of stained biopsy smears and identification of infected reservoir hosts, respectively.15 Although useful for patient diagnosis, limitations surrounding these conventional methods and their ability to definitively identify the species causing the infection are unfavorable and detrimental to disease treatment and patient outcome.
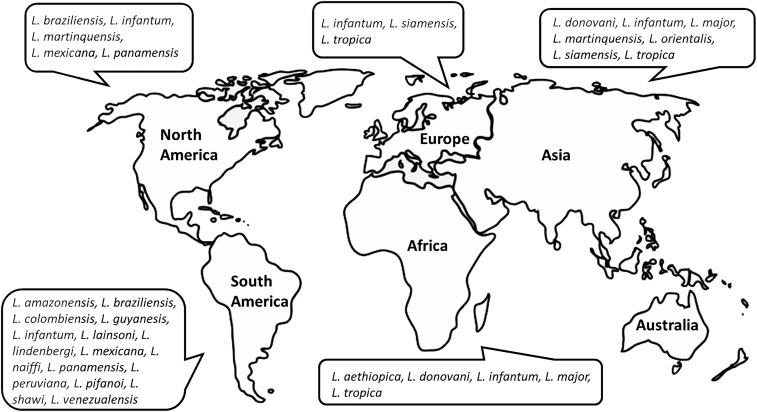
Figure 1.
World map of geographical distribution of clinically important, human-infecting Leishmania species. Travel history is an important factor in diagnosing leishmaniasis. This diagram illustrates the epidemiological pattern of various New World (the Americas) and Old World (Africa, Asia, and Europe) Leishmania species. This is particular important in non-endemic areas where any species of Leishmania can be imported from areas where more than one species is found.
Accurate speciation in clinical cases of leishmaniasis is crucial for an effective diagnosis of the disease. Furthermore, research has revealed a link between some Leishmania spp. and 1) severity and clinical outcome of the disease and 2) treatment outcome.16 Consequently, knowledge of the aetiological agent causing an infection is necessary to choose the correct course of chemotherapeutic drugs. A focused treatment chosen as a result of species identification will work more effectively in the first instance, avoiding the administering of an ineffective drug, which are also highly toxic substances. Diagnostics based on the analyses of DNA sequences offers the most direct and accurate approach for the identification and discrimination between species at the taxonomic level.11 Advancements in molecular biology have revolutionized the genetic approach to trypanosomatid identification, allowing relatively straightforward and simple analyses to reveal the true genotypic relationships between closely related organisms.17
The polymerase chain reaction (PCR) allows rapid isolation, amplification, and sequencing of DNA samples.11 In recent years, the conventional PCR has been widely applied for the diagnosis of diseases caused by the Trypanosomatidae including Chagas disease, human African trypanosomiasis, and leishmaniasis. Copious gene markers including the small subunit (SSU) rDNA, internal transcribed spacer 1 (ITS1), heat-shock protein 70 (Hsp70), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) genes have been routinely targeted for the identification of Leishmania and closely related trypanosomatid species.18 Furthermore, PCR assays have been developed for the specific diagnosis of all three clinical manifestations of leishmaniasis, including CL, MCL, and VL.19 Polymerase chain reaction–based diagnostics from CL or MCL samples have high sensitivity (87–98%) and specificity (≥ 84%), enabling rapid detection and the potential for speciation.15 This technique allows for the diagnosis and identification of the infective species before the administration of therapeutic drugs and has shown to be a superior method in sensitivity and specificity than the conventional diagnostics involving microscopy and parasite culture.16 It is of paramount importance to equip health systems with a simple diagnostic tool with high sensitivity and specificity, which is capable of detecting the presence of the parasite and accurately differentiating the causative Leishmania spp. in question. However, assays that allow simultaneous multispecies testing are perhaps the most applicable in a laboratory setting and is a current trend appearing in diagnostic laboratories.
Residing in the class Kinetoplastida, species of the genus Leishmania are characterized by the presence of a large, catenated network of DNA circles that are organized into a disk-shaped structure known as the kinetoplast.20,21 It is composed of approximately 10,000 minicircles and 20–50 maxicircles that form the intricate network of the kinetoplast DNA (kDNA).22–25 In a previous publication, we demonstrated the importance of the maxicircle for the taxonomic and phylogenetic analyses of Leishmania spp., providing a superior model to improve the resolution of trypanosomatid systematics.26 Thus, using the advantageous properties of the maxi-circle (i.e., potential to discriminate between closely related species) in the development, a quick and reliable test in the form of a PCR-RFLP has a clear potential in a laboratory setting. PCR-RFLP detects the variation between the band patterns of the DNA fragments produced through the digestion of PCR amplicons with specific restriction enzymes.27 Universal primers targeting a specific marker for all Leishmania spp. and an assay that does not require prior gel purification are the ideal requirements of any future diagnostic technique. The PCR-RFLP molecular method combines the sensitivity of PCR with the means of species differentiation without additional sequencing for the diagnosis and identification of Leishmania species. In addition, when RFLP is linked with PCR, only nanogram volumes of parasite DNA are required for analysis compared with that of conventional RFLP methods requiring micrograms of relatively high-quality DNA.11 Despite the development of more updated methods in a diagnostic laboratory setting and in the absence of a cost-effective alternative that can offer an efficient turn-around time from collection to diagnosis, the PCR-RFLP remains a useful method for the detection and identification of Leishmania species. Presently, the protocol described by Schönian et al.8 targeting the ITS region is the most frequented and cited PCR-RFLP used in the diagnosis of leishmaniasis. Through RFLP, the amplicon is digested without prior purification and is able to identify almost all pathogenic Leishmania species. This is extremely useful in a diagnostic setting, allowing rapid and direct identification of the infection-causing species.28 Despite its practical usefulness, the key phrase “almost all” is problematic, demonstrating the limitations of ITS-1 as a genetic marker for the determination of the pathogenic Leishmania species. This PCR-RFLP cannot differentiate between closely related species within the (L. [V.]) braziliensis complex (L. [V.] and braziliensis/L. [V.] guyanensis/L. [V.] panamensis) and difficulties arise with the L. (L.) donovani complex (L. [L.] donovani/L. [L.] infantum).29 When analyzing L. (L.) donovani and L. (L.) infantum using the ITS1-PCR, the restriction pattern is extremely similar, with only minor variations seen when comparing the two species alongside one another. Subsequently, without the concurrent digestion of these two species for reference, they cannot be differentiated. Furthermore, as with minicircles of the kDNA, the ITS-1 of Leishmania is a heterogeneous multi-copy gene, which varies greatly in copy number.29 Because of its heterogeneous nature, sequencing of the ITS1 to correctly delineate between species that cannot be differentiated by RFLP is complicated, requiring laborious and time-consuming molecular techniques such as cloning of the PCR products. Thus, although conducive to a high degree of sensitivity, the heterogeneous nature is not favorable as a target for DNA-based molecular detection and identification.
In Australia, cases of imported human leishmaniasis are increasing at an alarming rate because of international travel, the influx of immigrants, and military operations.29,30 Cutaneous leishmaniasis is the most common disease encountered, with 93% of cases reported since 2005, presenting with severe lesions affecting the skin. Visceral leishmaniasis was reported in the remaining 7% of patients, with no cases of MCL yet encountered.29,30 This influx highlights the need to provide diagnostic services and the difficulties present, particularly with imported cases in Australia in distinguishing between Leishmania and closely related species. In this study, we describe a universal PCR assay targeting the ND7 gene of the kDNA for the accurate diagnosis and identification of the principal Leishmania spp. associated with clinical leishmaniasis.
MATERIALS AND METHODS
Subjects and sample collection.
Cultured promastigotes of L. (L.) tropica, L. (L.) major, and L. (V.) braziliensis were used for the preliminary analyses in this study. Extracted DNA from the biopsies of positive cases of imported leishmaniasis were provided by the Microbiology Department of St. Vincent’s Hospital, Sydney. All positive cases had been previously diagnosed using the ITS1 PCR-RFLP described by Schönian et al.8 Samples from 24 Leishmania-infected individuals were used to assess the performance of the PCR-RFLP, targeting the kDNA in the diagnosis of leishmaniasis. This panel was composed of samples from across the globe where leishmaniasis is endemic.
DNA extraction of Leishmania cultures.
Cultures of Leishmania were first cultured on Novy-MacNeal-Nicolle (NNN) slopes and subsequently transferred to Minimum Essential Medium with 20% fetal calm serum.31 Parasite cultures were centrifuged at 4,000 × g for 15 minutes to pellet the parasite cells. The supernatant was removed and cell pellets were resuspended in 1,000 μL DNA extraction buffer (0.2 M Tris-HCL, 0.025 M ethylenediaminetetraacetic acid (EDTA), 0.5% EDTA, 0.25 M NaCl, and 0.3 mg/mL proteinase K) and followed by incubation overnight at 55°C. Samples were spun at 4,000 × g for 3 minutes and the supernatant was removed with care to a new tube so as to not disrupt the pellet. The cells were then subject to DNA extraction using a phenol–chloroform extraction. Briefly, 500 μL of phenol was added to the lysate and vortexed for approximately 1 minute. Next, 500 μL of chloroform was added and vortexed for an additional minute. The mixture was then centrifuged at 13,000 × g for 1 minute. The aqueous layer was carefully removed and extracted twice more with phenol and chloroform, as previously described. This was followed by a final extraction of the aqueous phase once more with 500 μL of chloroform. DNA was then precipitated overnight at −20°C with the addition of 8 μL 5 M NaCl and 1× volume isopropanol. The tubes were centrifuged at 13,000 × g for 15 minutes to pellet the DNA. Once the supernatant was removed, the pellet was rinsed 3× with 1 mL of 70% ethanol. Once decanted, the DNA pellet was air-dried for 10 minutes at room temperature and eluted in a total volume of 50 μL ddH2O. All DNA templates were stored at−20°C until assayed.
Variant calling.
To assess the suitability of the maxicircle coding region, a Burrows-Wheeler Alignment tool, was used to align whole genome sequence reads of various Leishmania spp. against the reference of each respective species.32 SAMtools and VarScan were then used to identify and filter variants in the form of single nucleotide polymorphisms (SNPs) and indels.33,34 The location of SNPs between the various Leishmania spp. was plotted to investigate the distribution of polymorphisms between species and to determine if a particular gene might represent an ideal candidate for the discrimination of Leishmania through an RFLP analysis. A detailed description of the SNPs distribution throughout the maxicircle can be found in Supplemental File 1. Species of the Viannia subgenus: L. (V.) braziliensis/L. (V.) peruviana and L. (V.) guyanensis/L. (V.) panamensis, and the Leishmania subgenus: L. (L.) infantum/L. (L.) donovani were chosen as they show very little genetic distance between species within their respective pairs. These species were hypothesized to exhibit very little variation in their restriction patterns and so variant calling was performed to identify “hot-spots” that could discriminate between these closely related species. Leishmania major, Leishmania amazonensis, and Leishmania mexicana were also selected as they represent well-defined species, whose taxonomic validity has not come under scrutiny.
Polymerase chain reaction amplification and sensitivity.
Primers were designed to amplify the ND7 gene of Leishmania kDNA producing fragments of approximately 800 bp. The PCR mixtures were prepared in a final volume of 50 µL with ND7-F (GTGCATTTATGCGTTTATTAATGTG) and ND7-R (ACAACATCAACATTACCAATAACTGC). The reaction was carried out in a final volume of 50 μL using reagents provided in the MyTaq PCR Kit (Bioline, London, UK) in the following quantities: 10 μL 5 × MyTaq reaction buffer, 1 μL 10 µM each primer, 1 μL MyTaq DNA Polymerase, 1 μL bovine serum albumin (BSA), and 10–100 ng of DNA. Reactions were carried out with an initial denaturation step at 95°C for 5 minutes, followed by 39 cycles at 95°C for 1 minute, 59°C for 1 minute, 72°C for 1 minute, and a final extension step at 72°C for 5 minutes. To confirm successful amplification, 4 μL of the PCR product was subjected to electrophoresis on 2% agarose gel stained with GelRed™ Nucleic Acid Gel Stain Biotium (Fremont, CA). The ND7 PCR was performed on the DNA extracted directly from the patient material (i.e., skin biopsies) of 24 previously identified cases. Positive controls containing DNA from L. (L.) major (culture) and a negative control void of DNA were included in each assay reaction. Specificity was tested using a negative control group consisting of 30 tissue-negative, six bacteria-positive, seven fungi-positive, and two yeast-positive clinical samples previously submitted to St. Vincent’s Hospital (Table 1). Sensitivity was estimated using a series of 10-fold dilutions of DNA from extracted L. (L.) major cultures to assess the lowest detection threshold of each PCR assay. Reaction templates corresponded to decreasing concentrations from 102 to 10−3 ng/μL DNA per PCR tube. The final concentrations of the samples were 100 ng/μL, 20 ng/μL, 4 ng/μL, 0.8 ng/μL, 0.16 ng/μL, 0.032 ng/μL, 0.0064 ng/μL, and 0.00128 ng/μL.
Table 1.
List of bacterial and fungal species tested as a negative control group
| Sample type | Clinical sample (number of samples tested) |
|---|---|
| Negative | Negative tissue samples (n = 30) |
| Bacteria | Staphylococcus aureus (n = 1) |
| Staphylococcus epidermidis (n = 1) | |
| Micrococcus luteus (n = 1) | |
| Pseudomonas aeruginosa (n = 1) | |
| Bacillus cereus (n = 1) | |
| Escherichia coli (n = 1) | |
| Yeast | Candida albicans (n = 1) |
| Candida glabrata (n = 1) | |
| Fungi | Sporothrix schenckii (n = 1) |
| Fusarium sp. (n = 1) | |
| Trichophyton mentagrophytes (n = 2) | |
| Epidermophyton floccosum (n = 1) | |
| Trichophyton rubrum (n = 1) | |
| Aspergillus fumigatus (n = 1) |
Sequencing.
For the purposes of sequencing, the entire PCR product of 21 selected clinical samples was loaded onto gels across multiple wells so that the maximum mass of PCR product could be obtained for sequencing. The PCR products of the correct size were excised from the agarose gel using a sterile scalpel blade. The PCR product was extracted from gel slices using a QIAquick® Gel Extraction Kit (Cat. No. 28706; QIAGEN, Hilden, Germany) following the manufacturer’s protocol. A slight modification was made to the protocol at the DNA elution stage in step nine of the manufacturer’s kit insert, where 33 μL of ddH2O was used instead of the recommended 50 μL. The concentration of DNA obtained from the gel slices was quantitated using a Nanodrop® Spectrophotometer ND-1000 (Thermo Fisher Scientific, Waltham, MA). All purified PCR products were stored at −20°C until they were shipped to the service provider Macrogen (Seoul, South Korea), for sequencing. Sequencing was performed on an ABI 3730XL capillary sequencer, twice in each forward and reverse direction. An NCBI BLAST database was created on command line using every available ND7 sequence as the query.
Virtual digestion of amplified ND7 gene sequences.
In silico modeling of the ND7 gene products was used to generate the predicted DNA fragments that would result from an RFLP digestion. Additional sequences obtained from the NCBI GenBank were included to generate a model containing every available Leishmania ND7 gene sequence. Virtual digests were performed using the Geneious software package, version 11.0.2. Single digestions were performed with every commercially available enzyme to assess the suitability for certain restriction enzymes.
RFLP analysis of amplified ND7.
Two restriction endonucleases: HpyCH4IV and NlaIII (New England Biolabs, Hertfordshire, United Kingdom) were used to individually digest the ND7 kDNA fragments. Polymerase chain reaction products (10 μL) were digested separately for each enzyme in a final volume of 25 µL with the corresponding reaction buffers and conditions summarized in Table 2. The restriction fragments were subjected to electrophoresis in 3% agarose gel containing GelRed Nucleic Acid Gel Stain Biotium for 2–3 hours and visualized under UV light using a LKB Bromma 2011 Macrovue Transilluminator.
Table 2.
Reaction conditions for each restriction enzyme used for the ND7 PCR-RFLP
| Enzyme (µL) | Product source | Recognition site | Reaction buffer (µL) | Reaction conditions |
|---|---|---|---|---|
| HpyCH4IV (1 μL) | Helicobacter pylori CH4 | 5′… A/C G T …3′.3′… T G C/A …5′ | 1 × CutSmart® Buffer (2.5 μL) | 2 hours at 37°C |
| NlaIII (1 μL) | Neisseria lactamica | 5′… C A T G/ …3′ 3′… /G T A C …5′ | 1 × CutSmart Buffer (2.5 μL) | 2 hours at 37°C |
Phylogenetic analysis.
Phylogenetic trees were constructed from the DNA sequences to infer the evolutionary relationships between the 21 clinical samples of Leishmania. Multiple alignments were performed using the MUSCLE algorithmic approach implemented in the Seaview software package.35 Phylogenetic trees were constructed using PAUP* version 4.0.36 Phylogenies were inferred using a heuristic search under the parsimony criterion, which involved random stepwise addition with tree bisection and reconnection (TBR) branch swapping and 1,000 random replicates.37 Bootstrap support for clade topologies was estimated following the analysis of 1,000 pseudo-replicate datasets. Inter- and intraspecies similarities of the ND7 region were calculated for 21 clinical isolates using MEGA software v.7.0.21.38
RESULTS
Variant calling.
The number of SNPs and indels identified between the various Leishmania species can be seen in Table 3. As expected, species of the Viannia subgenus and L. (L.) infantum and L. (L.) donovani exhibited very few polymorphisms when compared with one another. The L. (V.) braziliensis and the L. (V.) peruviana maxicircle coding region differed by 25 SNPs and three indels. L. (V.) guyanensis and L. (V.) panamensis differed by 19 SNPs and seven indels, whereas L. (L.) infantum and L. (L.) donovani differed by 125 SNPs and 13 indels. The distribution of variants called along the maxicircle coding region identified ND7 gene as an ideal candidate for the RFLP analysis of Leishmania. Supplemental File 1 contains a list of the distribution and gene location of SNPs when comparing the various Leishmania spp.
Table 3.
Number of single nucleotide polymorphisms (above diagonal) and indels (below diagonal) identified between the various Leishmania (L.) species
| L. braziliensis | L. peruviana | L. guyanensis | L. panamensis | L. infantum | L. donovani | L. major | L. amazonensis | L. mexicana | |
|---|---|---|---|---|---|---|---|---|---|
| L. braziliensis | – | 27 | 218 | 215 | 1,179 | 1,366 | 1,243 | 1,680 | 1,753 |
| L. peruviana | 3 | – | 226 | 226 | 1,178 | 1,365 | 1,253 | 1,675 | 1,753 |
| L. guyanensis | 25 | 27 | – | 21 | 1,425 | 1,444 | 1,268 | 1,402 | 1,381 |
| L. panamensis | 25 | 25 | 7 | – | 1,228 | 1,372 | 1,292 | 1,683 | 1,738 |
| L. infantum | 29 | 30 | 40 | 31 | – | 133 | 1,241 | 1,528 | 1,584 |
| L. donovani | 41 | 41 | 42 | 45 | 13 | – | 1,227 | 1,510 | 1,567 |
| L. major | 39 | 41 | 42 | 43 | 69 | 67 | – | 1,382 | 1,430 |
| L. amazonensis | 72 | 73 | 41 | 80 | 94 | 83 | 63 | – | 712 |
| L. mexicana | 76 | 81 | 42 | 77 | 101 | 108 | 57 | 71 | – |
Application of the ND7 PCR assay to clinical samples.
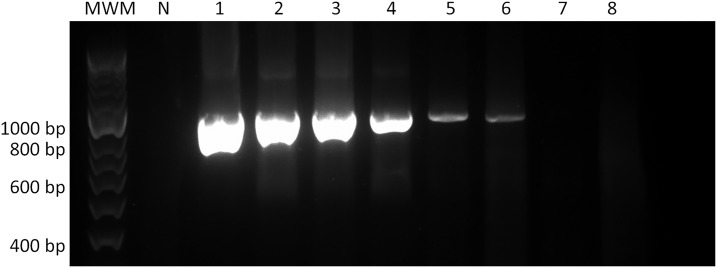
The ability of the PCR assays to detect Leishmania infection in clinical materials was tested with 24 positive samples submitted to the Department of Microbiology, St. Vincent’s Hospital, Sydney, between 2004 and 2017 (Table 4). Among all clinical isolates and cultured reference strains, specific amplification of an 800-bp fragment of the ND7 region was observed. The lowest detection threshold of each PCR method was equivalent to 0.032 ng/μL (Figure 2). No parasite DNA nor PCR amplification was detected by the PCR assay in neither the negative control where only water was used as the template nor the negative control group of 45 tissues samples containing commonly reported bacteria, fungi, and yeast species.
Table 4.
Patient details, associated risk factor, clinical presentation, and species identified
| Case Study | Age (years) | Gender | Risk factor | Country | Clinical presentation | Leishmania species identification | |||
|---|---|---|---|---|---|---|---|---|---|
| Previous ITS-1 RFLP29 | ND7 NlaIII RFLP | ND7 HpyCH4IV RFLP | Sequencing (BLAST identity) | ||||||
| 1 | 19 | F | Travel | South America | CL (finger) | L. (Viannia) spp. | L. (Viannia) spp. | L. (V.) guyanensis/L. (V.) panamensis | L. (V.) panamensis (100%) |
| 2 | 69 | M | Travel | South America | CL (forehead) | L. (Viannia) spp. | L. (Viannia) spp. | L. (V.) guyanensis/L. panamensis | L. (V.) panamensis (98%) |
| 3 | 26 | F | Travel | Peru | CL (arm) | No speciation | L. (Viannia) spp. | L. (V.) braziliensis | L. (V.) braziliensis (99.7%) |
| 4 | 25 | M | Army | Iraq | CL | L. (L.) major | L. (L.) major | L. (L.) major | L. (L.) major (100%) |
| 5 | 31 | M | Travel | South America | CL (arm) | L. (Viannia) spp. | L. (Viannia) spp. | L. (V.) braziliensis | L. (V.) braziliensis (98%) |
| 6 | 36 | M | Travel | Middle East | CL | L. (L.) tropica | L. (L.) tropica | L. (L.) tropica | L. (L.) tropica (100%) |
| 7 | 9 | M | Travel | – | CL (cheek) | L. (L.) tropica | L. (L.) tropica | L. (L.) tropica | L. (L.) tropica (100%) |
| 8 | – | M | Travel | – | CL (leg) | L. (Viannia) spp. | L. (Viannia) spp. | L. (V.) braziliensis | L. (V.) braziliensis (99.8%) |
| 9 | 54 | M | Travel | Worldwide | CL (leg) | L. (L.) donovani complex | L. (L.) donovani complex | L. (L.) donovani complex | L. (L.) infantum (100%) |
| 10 | 48 | M | Travel | South Spain | CL (spleen) | L. (L.) donovani complex | L. (L.) donovani complex | L. (L.) donovani complex | L. (L.) infantum (100%) |
| 11 | 67 | M | Travel | Peru | CL (chest) | L. (L.) donovani complex | L. (L.) donovani complex | L. (L.) donovani complex | L. (L.) infantum (100%) |
| 12 | 53 | M | Travel | Italy | CL (leg) | L. (L.) donovani complex | L. (L.) donovani complex | L. (L.) donovani complex | L. (L.) infantum (100%) |
| 13 | 35 | F | Travel | Costa Rica | CL (arm) | L. (Viannia) spp. | L. (Viannia) spp. | L. (V.) guyanensis/L. panamensis | L. (V.) panamensis (100%) |
| 14 | 73 | M | Travel | Brazil | CL (chest) | L. (Viannia) spp. | L. (Viannia) spp. | L. (V.) braziliensis | L. (V.) braziliensis (100%) |
| 15 | 27 | M | Travel | Bolivia | CL (leg) | L. (Viannia) spp. | L. (Viannia) spp. | L. (V.) braziliensis | * |
| 16 | 30 | F | Migrant | Middle East | CL (arm) | L. (L.) tropica | L. (L.) tropica | L. (L.) tropica | L. (L.) tropica (100%) |
| 17 | 20 | F | Travel | Egypt | CL | L. (L.) major | L. (L.) major | L. (L.) major | L. (L.) major (100%) |
| 18 | 34 | M | Migrant | Middle East | CL (lip) | L. (L.) tropica | L. (L.) tropica | L. (L.) tropica | L. (L.) tropica (100%) |
| 19 | 21 | F | Travel | Panama | CL (leg) | L. (Viannia) spp. | L. (Viannia) spp. | L. (V.) guyanensis/L. (V.) panamensis | * |
| 20 | – | F | Travel | South America | CL (nose) | L. (Viannia) spp. | L. (Viannia) spp. | L. (V.) guyanensis/L. (V.) panamensis | * |
| 21 | 8 | M | Migrant | Middle East | CL (cheek) | L. (L.) tropica | L. (L.) tropica | L. (L.) tropica | L. (L.) tropica (100%) |
| 22 | 2 | M | Migrant | Afghanistan | CL (forehead) | L. (L.) tropica | L. (L.) tropica | L. (L.) tropica | L. (L.) tropica (100%) |
| 23 | 24 | M | Travel | Peru | CL | L. (Viannia) spp. | L. (Viannia) spp. | L. (V.) braziliensis | L. (V.) braziliensis (99.9%) |
| 24 | 27 | M | Travel | Bolivia | CL (leg) | L. (Viannia) spp. | L. (Viannia) spp. | L. (V.) braziliensis | L. (V.) braziliensis (99.7%) |
CL = cutaneous leishmaniasis; Leishmania (Leishmania) = L. (L.); Leishmania (Viannia) = L. (V.).
* Clinical specimen not sequenced because of lack of sample volume.
Figure 2.
DNA electrophoresis of sensitivity test of ND7-PCR of Leishmania (Leishmania) major. Samples were run alongside a 50-bp molecular weight marker (Bioline). A negative control, void of DNA is marked by the “N” lane. The lanes labeled 1–8 correspond to the concentrations 100 ng/μL, 20 ng/μL, 4 ng/μL, 0.8 ng/μL, 0.16 ng/μL, 0.032 ng/μL, 0.0064 ng/μL, and 0.00128 ng/μL, respectively. The lowest detection threshold of the ND7-PCR was equivalent to 0.032 ng/μl (lane 6).
In silico digestion and species identification by PCR-RFLP of the amplified ND7 region in clinical samples.
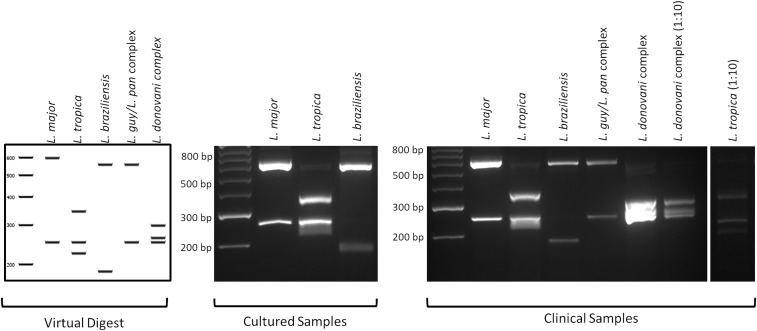
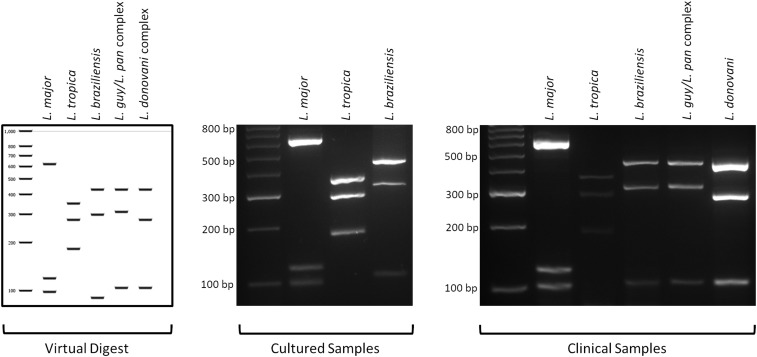
Six hundred twenty-five commercially available enzymes were assessed for their ability to discriminate between species of the Leishmania genus. Following the analyses using the software application Geneious, version 11.0.2, two enzymes, NlaIII and HpyCH4IV, were selected for the subsequent digestion of the ND7 amplicon. All 24 clinical samples found positive by PCR amplification of the ND7 were further tested by digestion with HpyCH4IV and NlaIII. The ND7 PCR-RFLP was designed as a dual diagnostic assay, performing two separate digestions simultaneously, using different enzymes for each assay on a single clinical isolate. The major advantage of digestion of the ND7-PCR with the restriction enzyme NlaIII is that L. (V.) braziliensis can be differentiated from the L. (V.) guyanensis/L. (V.) panamensis complex of the Viannia subgenus (Figure 3). The differentiation of L. (L.) major (Old World) and L. (V.) guyanensis/L. (V.) panamensis complex (New World) may cause difficulties if travel history is unknown, but the concurrent digestion with the restriction enzyme HpyCH4IV solves this problem (Figure 4). Digestion with NlaIII gave the following fragments: L. (L.) major (598 and 251 bp), L. (L.) tropica (344, 251, and 225 bp), L. (V.) braziliensis (557 and 186 bp), L. (V.) guyanensis/L. (V.) panamensis complex (557 and 251 bp), and L. (L.) donovani complex (305, 263, and 251 bp) (Figure 3). Digestion with HpyCH4IV gave the following fragments: L. (L.) major (620, 119, and 98 bp), L. (L.) tropica (350, 278, and 182 bp), L. (V.) braziliensis (428, 301, and 90 bp), L. (V.) guyanensis/L. (V.) panamensis complex (428, 311, and 104 bp), and L. (L.) donovani complex (428, 278, and 104 bp) (Figure 4). Both NlaIII and HpyCH4IV were unable to differentiate between L. (V.) guyanensis/L. (V.) panamensis and L. (L.) donovani/L. (L.) infantum. However, subsequent sequencing of the ND7 gene successfully resolved identification at the species level between L. (L.) donovani and L. (L.) infantum.
Figure 3.
PCR-RFLP analysis of Leishmania species with the restriction enzyme NlaIII. Samples were run alongside a 50-bp molecular weight marker. Comparison of the band patterns of the DNA fragments generated from the NlaIII digestion of in silico, culture, and clinical isolates. Digestion with NlaIII can successfully differentiate between the extremely closely related Leishmania (Viannia) (L. [V.]) braziliensis and L. (V.) guyanensis/L. (V.) panamensis complex of the Viannia subgenus, which is extremely important in a clinical setting where treatment outcome can vary greatly between these two species.
Figure 4.
PCR-RFLP analysis of Leishmania species with the restriction enzyme HpyCH4IV. Samples were run alongside a 50-bp molecular weight marker. Comparison of the band patterns of the DNA fragments generated from the HpyCH4IV digestion of in silico, culture, and clinical isolates. Digestion with HpyCH4IV can successfully differentiate between New World and Old World Leishmania species isolated from both culture and clinical specimens.
Sequencing and phylogenetic tree analysis.
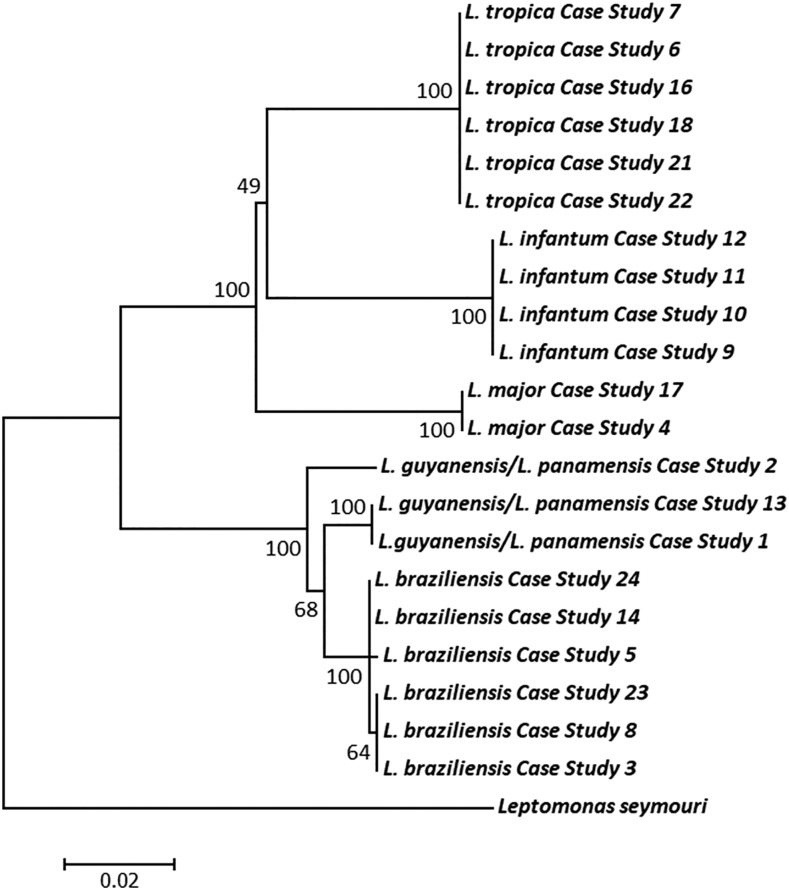
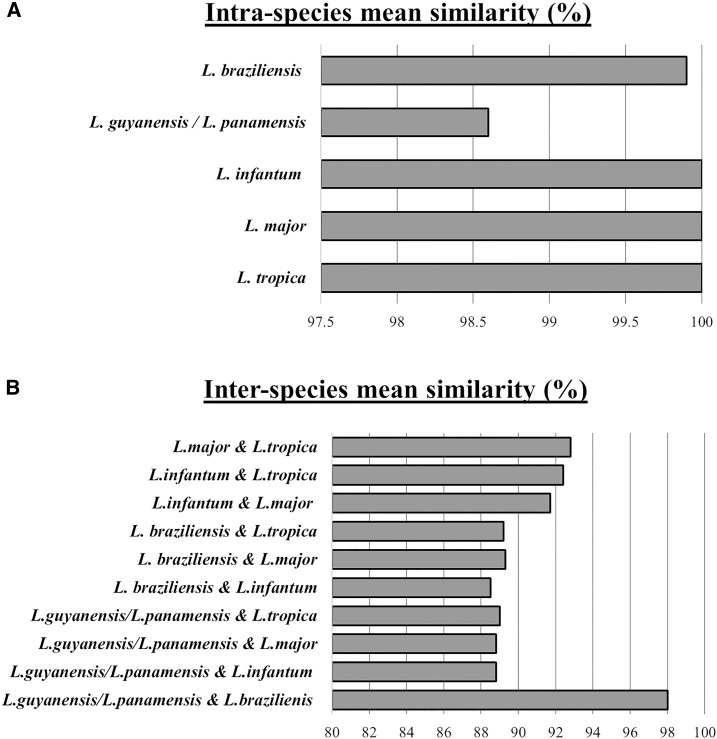
Of the 21 samples selected for sequencing, five species of the L. (L.) and L. (V.) were identified: L. (L.) major, L. (L.) tropica, L. (V.) braziliensis, L. (L.) infantum, and L. (V.) guyanensis/L. (V.) panamensis complex (Table 4). The ND7 sequence was unable to discriminate between L. (V.) guyanensis/L. (V.) panamensis and is hereafter referred to as the L. (V.) guyanensis/L. (V.) panamensis complex. From the heuristic search of the ND7, the most parsimonious tree was found to be 304. Of the 844 characters analyzed under the parsimony optimality criterion, 634 characters were constant, 64 variable characters were parsimony-uninformative, and a final 146 characters were considered parsimony-informative. The evolutionary relationships showing the genetic distance between clinical isolates of Leishmania is shown in Figure 5. In the inferred phylogeny, all Leishmania clades corresponded to previous classifications of the Leishmaniinae subfamily.18,39,40 Each clinical isolate clustered according to their species-specific identification, forming five distinct, monophyletic clades. Intraspecies similarities of the ND7 region in isolates of L. (L.) major, L. (L.) tropica, L. (V.) braziliensis, L. (L.) infantum, and L. (V.) guyanensis/L. (V.) panamensis complex were 100%, 100%, 99.9%, 100%, and 98.6%, respectively (Figure 6). The highest inter-species similarity was observed between the L. (Viannia) subgenera, with 98% similarity between L. (V.) braziliensis and the L. (V.) guyanensis/L. (V.) panamensis complex.
Figure 5.
Inferred evolutionary relationships showing genetic distance between clinical isolate of imported cases of leishmaniasis. The structure of this tree was inferred from DNA sequence data using the parsimony criterion with 1,000 bootstrap replicates. The scale bar represents the number of nucleotide substitutions per site. Each clinical isolate clustered according to their species-specific identification, forming five distinct, monophyletic clades.
Figure 6.
Levels of intra- and inter-species mean similarities of Leishmania spp. (A) Intraspecies mean similarities for Leishmania species from leishmaniasis cases based on the ND7 region sequence. Intraspecies similarities of the ND7 region in isolates of Leishmania (Viannia) (L. [V.]) guyanensis/L. (V.) panamensis complex, L. (V.) braziliensis, Leishmania (Leishmania) (L. [L.]) infantum, L. (L.) major, and L. (L.) tropica were 98.6%, 99.9%, 100%, 100%, and 100%, respectively. (B) Inter-species mean similarities for Leishmania species from leishmaniasis cases based on the ND7 region sequence. The greatest inter-species similarity was seen between L. (V.) braziliensis and the L. (V.) guyanensis/L. (V.) panamensis complex with 98% similarity.
DISCUSSION
Whereas previous cases of imported leishmaniasis into Australia have been described (case studies 1–14)29,30, this study reports an additional 10 cases imported throughout 2017 (case studies 15–24). Over the last two decades, a multitude of diagnostic assays targeting various genes including the ITS-1, Hsp70, antigen-encoding genes gp63 and cpb, RNA polymerase II, and the N-acetylglucosamine-1-phosphate transferase have been designed for the detection and identification of Leishmania species.41–45 In the absence of a globally accepted “gold standard” for diagnosis, the downside of such an immense range of tests is that many of these methods are obscurely used in very few or individual laboratories throughout the world. Cited more than 185 times in PubMed (viewed March 23, 2019), and used for the detection and identification of various Leishmania species in 63 articles (Supplemental File 2), the ITS-1 remains the most extensively used tool for the diagnosis of leishmaniasis. The main drawback of the ITS-1 as a diagnostic target is its inability to distinguish between important species of the Viannia subgenus. The main aim of this study was to address the limitations of the widely used ITS-1 PCR-RFLP and establish a diagnostic PCR-RFLP assay that is capable of not only identifying the principal Leishmania spp. associated with clinical leishmaniasis but also discriminating between species that the ITS-1 cannot. The design of a PCR-RFLP targeting the ND7 is a reliable and cost- and time-effective method that does not require prior gel purification, making it an ideal candidate for the diagnosis and identification of the principal Leishmania species. Using the ND7 PCR-RFLP, we provide an appraisal on the molecular epidemiology of previously imported cases of leishmaniasis in Australia.
This study was limited by the spectrum of species that are imported into Australia. Despite being only one of two continents free of endemic, clinically important Leishmania spp., cases of imported human leishmaniasis are rising at an ever-increasing rate because of international travel, the influx of immigrants, and military operations.29,30 This trend highlights the difficulties present in diagnostics, particularly in non-endemic areas where any species of Leishmania can be imported, often from endemic areas where more than one species is present (Figure 1).8 In addition, diagnosis based on clinical symptoms in conjunction with area of travel is difficult because several species can cause a wide spectrum of clinical manifestations such as localized CL and MCL.46 The etiological agents primarily responsible for the imported cases of human leishmaniasis in Australia are typically members of the species that either cannot be differentiated by the ITS-1 PCR-RFLP reported in Schönian et al.8 This includes the L. (V.) braziliensis/L. (V.) panamensis/L. (V.) guyanensis complex or those that can cause difficulties if not run simultaneously alongside one another (L. [L.] donovani/L. [L.] infantum). As a result of this epidemiological trend, we set out to design a universal diagnostic PCR method that combines sensitivity and is capable of species-specific differentiation through a combination of both RFLP and sanger sequencing.
Over the last 10 years, PCR-RFLP as a diagnostic method has become the method of choice to detect and identify the leishmaniasis infective species from imported cases in Australia. Important advantages of PCR-RFLP for genetic analysis and species identification include its relative inexpensiveness and less laborious method with greater sensitivity than that of the traditional microscopic detection of parasites in culture. The ND7 PCR-RFLP offers a highly sensitive technique, allowing the characterization of Old World and New World Leishmania species in clinical infections. This is crucial to not only inform the most effective treatment but also provide valuable epidemiology on the distribution of Leishmania species being imported into Australia. In a clinical setting, the disadvantage of RFLP analysis is the risk of contamination by amplicons resulting in false-positive results.10 However, the use of two separate enzymes (NlaIII and HpyCH4IV) that require identical assay conditions including reagents, reaction volume, digestion temperature, and time addresses this drawback. These enzymes were chosen for their ability to generate an easily distinguishable restriction pattern, low cost, and availability, and that they can be run simultaneously, requiring identical assay conditions. In addition, NlaIII was chosen because of its specificity for L. (V.) braziliensis and HpyCH4IV was chosen for its ability to distinguish between Old World and New World species of the subgenus L. (L.). By ensuring that the same fingerprinting identification is reached from two separate digestions run concurrently, the restriction digest of ND7-PCR amplicons with NlaIII and HpyCH4IV provide a robust, 2-fold diagnosis. Thus, in a clinical setting, it requires minimal effort to perform a simultaneous dual restriction analysis that is sufficient to distinguish between the medically relevant Leishmania species being imported into Australia. In addition, the ND7 as a biomarker allows for additional species-specific identification through sequencing, providing greater support for the classification of clinical isolates. If a PCR product can be obtained from either the ITS1-PCR or ND7-PCR, the whole PCR diagnosis (DNA extraction, PCR and RFLP) can be performed in 2–2.5 days. However, if identification cannot be resolved at a species level, sequencing of the ND7-PCR (purification, transportation, and Sanger sequencing) will require an additional 3–4 days compared with ITS1 sequencing (molecular cloning, transportation, and Sanger sequencing) that requires an additional 11–14 days.
In the present study, CL was the most common clinical manifestation described (n = 23) (Table 4). As previously reported by Roberts et al.,29 lesions on the legs and arms were the most common presentation in patients. No cases of MCL were observed in the 10 cases reported in 2017. Five species/complexes of Leishmania were identified; two L. (L.) major, six L. (L.) tropica, seven L. (V.) braziliensis, five L. (V.) guyanensis/L. (V.) panamensis complex and four L. (L.) infantum. From our analyses, the infective species of five case studies previously identified as belonging to the generic L. (V.) braziliensis complex have now been correctly reclassified as L. (V.) guyanensis/L. (V.) panamensis complex. Despite the ND7 PCR-RFLP diagnostic assay being insufficient to discriminate between L. (L.) donovani and L. (L.) infantum, species identity was successfully resolved through sequencing and the four isolates previously identified as belonging to the L. (L.) donovani complex revealed a 100% BLAST identity with L. (L.) infantum. One specimen (case study 3) that could not be specifically identified because of the presence of a very weak positive PCR product resulting in an ambiguous restriction pattern was identified as L. (V.) braziliensis. From the 2017 specimens, L. (L.) tropica was the most common species identified, which coincides with the patients’ travel history where all L. (L.) tropica-infected patients were civilian immigrants from Afghanistan or the Middle East. The vast majority of travelers from the 2017 cohort examined in this study were infected with species of the Leishmania (Viannia) subgenus, coinciding with most patients having reported previous travel to Leishmania endemic regions of South America.
The taxonomic status of L. (V.) guyanensis and L. (V.) panamensis has often been debated, with reports questioning the validity of the division of these complexes into two distinct species.47–49 Together both PCR-RFLP and sequencing of the ND7 gene do not possess the discriminatory power to discern between species of the L. (V.) guyanensis/L. (V.) panamensis complex. Variant calling identified only 26 variants between the two species (Table 3) and from our previous analyses on the coding region of the Leishmaniinae maxicircle, we demonstrated that L. (V.) guyanensis/L. (V.) panamensis are separated by a genetic distance of only 0.001. Considering their extremely close genetic relatedness in context of the previously published concerns surrounding these species, it is important to acknowledge the outstanding questions surrounding the validity of these Viannia parasites and whether these parasites warrant speciation as distinct organisms or is merely genomic variability within a sole species. However, the greatest intraspecies divergence was seen within species of the Viannia subgenus (Figure 6). This diversity is reflected in Figure 5, where the phylogenetic analysis revealed two clusters within the L. (V.) guyanensis/L. (V.) panamensis complex isolates and two within the L. (V.) braziliensis isolates. No correlations were seen between the intraspecies diversity through SNPs and geographic distribution of the species. Clinical isolates of the L. (L.) infantum, L. (L.) major, and L. (L.) tropica case studies showed no diversity and formed independent, monophyletic clades with 100% bootstrap confidence.
Despite the recent appraisal, in which Leishmania (L.) infantum and Leishmania (L.) donovani were recognized as the only species of the L. (L.) donovani complex,50 issues still surround the taxonomic status of this complex. The Leishmania donovani complex have a largely discrete geographic distribution; L. (L.) donovani in the Old World is predominately found in East Africa, India, and parts of the Middle East, and L. (L.) infantum in both the Old World and New World, which is found in Europe, North Africa, and South and Central America.50,51 However, the genetic diversity of these two species are the cause for complications regarding the classification of these parasites. In the past, phylogenetic analyses based on insufficient taxonomic markers that were unable to detect the diversity (if any) between these extremely similar species have been the main source for confusion. Nevertheless, the use of a large dataset based on the entire coding region of the maxicircle only detected few DNA polymorphisms (Table 3) between L. (L.) donovani and L. (L.) infantum, separated by a genetic divergence of only 0.007.26 As such, the ND7 PCR-RFLP does not have the discriminatory power to delineate the boundary between these two species. However, subsequent sequencing of the ND7 amplicons did show that the ND7-PCR was capable of resolving these species, with 100% BLAST identity of four clinical samples aligning with L. (L.) infantum. Despite this, treatment for L. (L.) donovani and L. (L.) infantum is the same, with amphotericin B and miltefosine being the first-line treatment option of choice.52,53 Thus, the ability of the ND7-RFLP to identify the L. (L.) donovani complex is sufficient for the administration of correct treatment.
The need for accurate speciation in clinical cases of leishmaniasis is of paramount importance, particularly when dealing with species of the Viannia subgenus. Leishmania (Viannia) species of the New World can lead to diverse clinical presentations and as such often have a diverse and unpredictable response to treatment.54 Previous reports have demonstrated a strong correlation between inappropriately treated CL forms of the disease and the subsequent occurrence of MCL for both L. (V.) braziliensis and L. (V.) guyanensis.55,56 Pentavalent antimonials are the most frequently used first-line treatment for both CL and VL.52 However, the therapeutic response to antimonials in patients with CL is highly dependent on the L. (Viannia) etiological agent causing the infection.57 Romero et al.57 demonstrated that patients infected with L. (V.) guyanensis had a higher failure rate in response to treatment with meglumine antimoniate than L. (V.) braziliensis, and Francesconi et al.54 also showed that fluconazole was not efficacious against CL caused by L. (V.) guyanensis in adult men. In addition, the Viannia species are susceptible not only to the drug of choice but also to the route of administration, with Christen et al.58 demonstrating that an intramuscular dosage of pentamidine isethionate in L. (V.) guyanensis–infected patients increased the risk of treatment failure. Thus, it is essential for clinicians to have the tools available that allow the correct differentiation between L. (V.) braziliensis and L. (V.) guyanensis, ensuring the administration of the most effective first-line treatment with the correct course of chemotherapeutic drugs. The ND7 PCR-RFLP provides a species-specific approach that can not only identify the various causative species across the Leishmania genus but also differentiate L. (V.) braziliensis from the L. (V.) guyanensis/L. (V.) panamensis complex. Use of this fingerprinting technology in clinical settings may therefore ultimately lead to higher cure rates and lower costs due to side effects from the administration of highly toxic ineffective drugs.
To conclude, we describe a dual-analysis PCR-RFLP capable of not only identifying the principal species of both New World and Old World Leishmania but also possessing the discriminatory power to differentiate between the extremely closely related species of the L. (Viannia) subgenus. Thus, superior to the extensively used ITS1 PCR-RFLP described by Schönian et al.,8 the PCR-RFLP targeting the ND7 gene of the kDNA can be applied to not only imported cases of leishmaniasis in Australia but also in other endemic and non-endemic areas where more than one specie is most likely to be present. The diagnostic method combines the sensitivity to detect Leishmania in extracted clinical materials and the ability to fingerprint and identify the clinically important species. In this study, we provide an appraisal on 14 previously reported case studies between 2008 and 2014 and present data on the molecular identification and epidemiology of an additional 10 cases of imported leishmaniasis in Australia. Over a 1-year period in 2017, L. (L.) tropica was the most common species identified from civilian immigrants and L. (Viannia) species were the infective species for the vast majority or travelers. These results demonstrate the continuing trend in risk factor, country of infection, clinical presentation, and the predominant causative species in imported cases of leishmaniasis. Although leishmaniasis is not presently a notifiable disease in Australia, the number of new cases reported at SydPath has increased considerably over the last 10 years at an alarming rate. The results highlight the need for providing Australian clinicians with the tools specific for diagnosing and determining the causative Leishmania species.
Supplementary Material
Acknowledgments:
We would like to thank Rogan Lee for providing the L. (L.) tropica samples. This study was completed by A. K. in partial fulfillment of the degree at UTS.
Note: Supplemental files appear at www.ajtmh.org.
REFERENCES
- 1.World Health Organization , 2018. Leishmaniasis. Available at: http://www.who.int/gho/neglected_diseases/leishmaniasis/en/. Accessed July 16, 2018. [Google Scholar]
- 2.Faulde M, Schrader J, Heyl G, Amirih M, 2008. Differences in transmission seasons as an epidemiological tool for characterization of anthroponotic and zoonotic cutaneous leishmaniasis in northern Afghanistan. Acta Trop 105: 131–138. [DOI] [PubMed] [Google Scholar]
- 3.Akhoundi M, Kuhls K, Cannet A, Votýpka J, Marty P, Delaunay P, Sereno D, 2016. A historical overview of the classification, evolution, and dispersion of Leishmania parasites and sandflies. PLoS Negl Trop Dis 10: e0004349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ceccarelli M, Diotallevi A, Andreoni F, Vitale F, Galluzzi L, Magnani M, 2018. Exploiting genetic polymorphisms in metabolic enzymes for rapid screening of Leishmania infantum genotypes. Parasit Vectors 11: 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Morais RC, da Costa Oliveira CN, de Albuquerque Sda C, Mendonca Trajano Silva LA, Pessoa ESR, Alves da Cruz HL, de Brito ME, de Paiva Cavalcanti M, 2016. Real-time PCR for Leishmania species identification: evaluation and comparison with classical techniques. Exp Parasitol 165: 43–50. [DOI] [PubMed] [Google Scholar]
- 6.Hernández C, Alvarez C, González C, Ayala MS, León CM, Ramírez JD, 2014. Identification of six New World Leishmania species through the implementation of a high-resolution melting (HRM) genotyping assay. Parasit Vectors 7: 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrera G, Hernandez C, Ayala MS, Florez C, Teheran AA, Ramirez JD, 2017. Evaluation of a multilocus sequence typing (MLST) scheme for Leishmania (Viannia) braziliensis and Leishmania (Viannia) panamensis in Colombia. Parasit Vectors 10: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schönian G, Nasereddin A, Dinse N, Schweynoch C, Schallig H, Presber W, Jaffe CL, 2003. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis 47: 349–358. [DOI] [PubMed] [Google Scholar]
- 9.Van der Auwera G, Ravel C, Verweij JJ, Bart A, Schonian G, Felger I, 2014. Evaluation of four single-locus markers for Leishmania species discrimination by sequencing. J Clin Microbiol 52: 1098–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sagi O, Berkowitz A, Codish S, Novack V, Rashti A, Akad F, Shemer-Avni Y, 2017. Sensitive molecular diagnostics for cutaneous leishmaniasis. Open Forum Infect Dis 4: ofx037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McManus DP, Bowles J, 1996. Molecular genetic approaches to parasite identification: their value in diagnostic parasitology and systematics. Int J Parasitol 26: 687–704. [DOI] [PubMed] [Google Scholar]
- 12.Mouttaki T, Morales-Yuste M, Merino-Espinosa G, Chiheb S, Fellah H, Martin-Sanchez J, Riyad M, 2014. Molecular diagnosis of cutaneous leishmaniasis and identification of the causative Leishmania species in Morocco by using three PCR-based assays. Parasit Vectors 7: 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pourmohammadi B, Motazedian M, Hatam G, Kalantari M, Habibi P, Sarkari B, 2010. Comparison of three methods for diagnosis of cutaneous leishmaniasis. Iran J Parasitol 5: 1–8. [PMC free article] [PubMed] [Google Scholar]
- 14.Sundar S, Rai M, 2002. Laboratory diagnosis of visceral leishmaniasis. Clin Diagn Lab Immunol 9: 951–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson SM, 1995. DNA-based methods in the detection of Leishmania parasites: field applications and practicalities. Ann Trop Med Parasitol 89: 95–100. [PubMed] [Google Scholar]
- 16.Reithinger R, Dujardin JC, 2007. Molecular diagnosis of leishmaniasis: current status and future applications. J Clin Microbiol 45: 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaufer A, Ellis J, Stark D, Barratt J, 2017. The evolution of trypanosomatid taxonomy. Parasit Vectors 10: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barratt J, Kaufer A, Peters B, Craig D, Lawrence A, Roberts T, Lee R, McAuliffe G, Stark D, Ellis J, 2017. Isolation of novel trypanosomatid, Zelonia australiensis sp. nov. (Kinetoplastida: Trypanosomatidae) provides support for a Gondwanan origin of dixenous parasitism in the Leishmaniinae. PLoS Negl Trop Dis 11: e0005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mugasa CM, Laurent T, Schoone GJ, Basiye FL, Saad AA, el Safi S, Kager PA, Schallig H, 2010. Simplified molecular detection of Leishmania parasites in various clinical samples from patients with leishmaniasis. Parasit Vectors 3: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin RH, Lai DH, Zheng LL, Wu J, Lukes J, Hide G, Lun ZR, 2015. Analysis of the mitochondrial maxicircle of Trypanosoma lewisi, a neglected human pathogen. Parasit Vectors 8: 665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Telleria J, Lafay B, Virreira M, Barnabe C, Tibayrenc M, Svoboda M, 2006. Trypanosoma cruzi: sequence analysis of the variable region of kinetoplast minicircles. Exp Parasitol 114: 279–288. [DOI] [PubMed] [Google Scholar]
- 22.Flegontov PN, Zhirenkina EN, Gerasimov ES, Ponirovsky EN, Strelkova MV, Kolesnikov AA, 2009. Selective amplification of maxicircle classes during the life cycle of Leishmania major. Mol Biochem Parasitol 165: 142–152. [DOI] [PubMed] [Google Scholar]
- 23.Gerasimov ES, Gasparyan AA, Litus IA, Logacheva MD, Kolesnikov AA, 2017. Minicircle kinetoplast genome of insect trypanosomatid Leptomonas pyrrhocoris. Biochemistry (Mosc) 82: 572–578. [DOI] [PubMed] [Google Scholar]
- 24.Simpson L, Douglass SM, Lake JA, Pellegrini M, Li F, 2015. Comparison of the mitochondrial genomes and steady state transcriptomes of two strains of the trypanosomatid parasite, Leishmania tarentolae. PLoS Negl Trop Dis 9: e0003841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yatawara L, Le TH, Wickramasinghe S, Agatsuma T, 2008. Maxicircle (mitochondrial) genome sequence (partial) of Leishmania major: gene content, arrangement and composition compared with Leishmania tarentolae. Gene 424: 80–86. [DOI] [PubMed] [Google Scholar]
- 26.Kaufer A, Barratt J, Stark D, Ellis J, 2019. The complete coding region of the maxicircle as a superior phylogenetic marker for exploring evolutionary relationships between members of the Leishmaniinae. Infect Genet Evol 70: 90–100. [DOI] [PubMed] [Google Scholar]
- 27.Akhoundi M, et al. 2017. Leishmania infections: molecular targets and diagnosis. Mol Asp Med 57: 1–29. [DOI] [PubMed] [Google Scholar]
- 28.Bensoussan E, Nasereddin A, Jonas F, Schnur LF, Jaffe CL, 2006. Comparison of PCR assays for diagnosis of cutaneous leishmaniasis. J Clin Microbiol 44: 1435–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts T, Barratt J, Sandaradura I, Lee R, Harkness J, Marriott D, Ellis J, Stark D, 2015. Molecular epidemiology of imported cases of leishmaniasis in Australia from 2008 to 2014. PLoS One 10: e0119212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stark D, van Hal S, Lee R, Marriott D, Harkness J, 2008. Leishmaniasis, an emerging imported infection: report of 20 cases from Australia. J Travel Med 15: 351–354. [DOI] [PubMed] [Google Scholar]
- 31.Chouihi E, Amri F, Bouslimi N, Siala E, Selmi K, Zallagua N, Ben Abdallah R, Bouratbine A, Aoun K, 2009. Cultures on NNN medium for the diagnosis of leishmaniasis. Pathol Biol (Paris) 57: 219–224. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Durbin R, 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L, 2009. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25: 2283–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gouy M, Guindon S, Gascuel O, 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224. [DOI] [PubMed] [Google Scholar]
- 36.Swofford DL, 1993. PAUP–a computer-program for phylogenetic inference using maximum parsiomy. J Gen Physiol 102: A9. [Google Scholar]
- 37.Swofford DL, Bell CD, 2017. PAUP Manual Available at: http://phylosolutions.com/paup-documentation/paupmanual.pdf. Accessed July 17, 2018. [Google Scholar]
- 38.Kumar S, Stecher G, Tamura K, 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Espinosa OA, Serrano MG, Camargo EP, Teixeira MMG, Shaw JJ, 2018. An appraisal of the taxonomy and nomenclature of trypanosomatids presently classified as Leishmania and Endotrypanum. Parasitology 145: 430–442. [DOI] [PubMed] [Google Scholar]
- 40.Jirků M, Yurchenko VY, Lukeš J, Maslov DA, 2012. New species of insect trypanosomatids from Costa Rica and the proposal for a new subfamily within the trypanosomatidae. J Eukaryot Microbiol 59: 537–547. [DOI] [PubMed] [Google Scholar]
- 41.El Hamouchi A, El Kacem S, Ejghal R, Lemrani M, 2018. Genetic polymorphism in Leishmania infantum isolates from human and animals determined by nagt PCR-RFLP. Infect Dis Poverty 7: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Espada CR, Ortiz PA, Shaw JJ, Barral AMP, Costa JML, Uliana SRB, Coelho AC, 2018. Identification of Leishmania (Viannia) species and clinical isolates of Leishmania (Leishmania) amazonensis from Brazil using PCR-RFLP of the heat-shock protein 70 gene reveals some unexpected observations. Diagn Microbiol Infect Dis 91: 312–318. [DOI] [PubMed] [Google Scholar]
- 43.Mauricio IL, Gaunt MW, Stothard JR, Miles MA, 2001. Genetic typing and phylogeny of the Leishmania donovani complex by restriction analysis of PCR amplified gp63 intergenic regions. Parasitology 122: 393–403. [DOI] [PubMed] [Google Scholar]
- 44.Pratlong F, Lami P, Ravel C, Balard Y, Dereure J, Serres G, Baidouri FE, Dedet JP, 2013. Geographical distribution and epidemiological features of old world Leishmania infantum and Leishmania donovani foci, based on the isoenzyme analysis of 2277 strains. Parasitology 140: 423–434. [DOI] [PubMed] [Google Scholar]
- 45.Simon S, Veron V, Carme B, 2010. Leishmania spp. identification by polymerase chain reaction-restriction fragment length polymorphism analysis and its applications in French Guiana. Diagn Microbiol Infect Dis 66: 175–180. [DOI] [PubMed] [Google Scholar]
- 46.Monroy-Ostria A, Nasereddin A, Monteon VM, Guzman-Bracho C, Jaffe CL, 2014. ITS1 PCR-RFLP diagnosis and characterization of Leishmania in clinical samples and strains from cases of human cutaneous leishmaniasis in states of the Mexican southeast. Interdiscip Perspect Infect Dis 2014: 607287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bañuls AL, Guerrini F, LePont F, Barrera C, Espinel I, Guderian R, Echeverria R, Tibayrenc M, 1997. Evidence for hybridization by multilocus enzyme electrophoresis and random amplified polymorphic DNA between Leishmania braziliensis and Leishmania panamensis/guyanensis in Ecuador. J Eukaryot Microbiol 44: 408–411. [DOI] [PubMed] [Google Scholar]
- 48.Fraga J, Montalvo AM, Van der Auwera G, Maes I, Dujardin JC, Requena JM, 2013. Evolution and species discrimination according to the Leishmania heat-shock protein 20 gene. Infect Genet Evol 18: 229–237. [DOI] [PubMed] [Google Scholar]
- 49.Garcia AL, Kindt A, Quispe-Tintaya KW, Bermudez H, Llanos A, Arevalo J, Banuls AL, De Doncker S, Le Ray D, Dujardin JC, 2005. American tegumentary leishmaniasis: antigen-gene polymorphism, taxonomy and clinical pleomorphism. Infect Genet Evol 5: 109–116. [DOI] [PubMed] [Google Scholar]
- 50.Lukeš J, et al. 2007. Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc Natl Acad Sci USA 104: 9375–9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jamjoom MB, Ashford RW, Bates PA, Chance ML, Kemp SJ, Watts PC, Noyes HA, 2004. Leishmania donovani is the only cause of visceral leishmaniasis in east Africa; previous descriptions of L. infantum and “L. archibaldi” from this region are a consequence of convergent evolution in the isoenzyme data. Parasitology 129: 399–409. [DOI] [PubMed] [Google Scholar]
- 52.de Menezes JP, Guedes CE, Petersen AL, Fraga DB, Veras PS, 2015. Advances in development of new treatment for leishmaniasis. Biomed Res Int 2015: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hendrickx S, Van den Kerkhof M, Mabille D, Cos P, Delputte P, Maes L, Caljon G, 2017. Combined treatment of miltefosine and paromomycin delays the onset of experimental drug resistance in Leishmania infantum. PLoS Negl Trop Dis 11: e0005620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Francesconi VA, Francesconi F, Ramasawmy R, Romero GAS, Alecrim M, 2018. Failure of fluconazole in treating cutaneous leishmaniasis caused by Leishmania guyanensis in the Brazilian Amazon: an open, nonrandomized phase 2 trial. PLoS Negl Trop Dis 12: e0006225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guerra JA, Prestes SR, Silveira H, Coelho LI, Gama P, Moura A, Amato V, Barbosa M, Ferreira LC, 2011. Mucosal leishmaniasis caused by Leishmania (Viannia) braziliensis and Leishmania (Viannia) guyanensis in the Brazilian Amazon. PLoS Negl Trop Dis 5: e980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Achtman JC, Ellis DL, Saylors B, Boh EE, 2016. Cutaneous leishmaniasis caused by Leishmania (Viannia) panamensis in 2 travelers. JAAD Case Rep 2: 95–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Romero GA, Guerra MV, Paes MG, Macedo VO, 2001. Comparison of cutaneous leishmaniasis due to Leishmania (Viannia) braziliensis and L. (V.) guyanensis in Brazil: therapeutic response to meglumine antimoniate. Am J Trop Med Hyg 65: 456–465. [DOI] [PubMed] [Google Scholar]
- 58.Christen JR, et al. 2018. Use of the intramuscular route to administer pentamidine isethionate in Leishmania guyanensis cutaneous leishmaniasis increases the risk of treatment failure. Trav Med Infect Dis 24: 31–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.