Abstract
The relevance of the dysregulation of snoRNAs in human cancer has been widely investigated and has challenged the view that snoRNAs merely function as house-keeping genes for the posttranscriptional modification of rRNAs. Accumulating evidence has shown the intimate connection between snoRNAs and proliferation, apoptosis, invasion and migration of tumor cells via manual intervention patterns of snoRNA expression. In this review, we focused on how snoRNAs are dysregulated and its regulation of the formation and development of cancer. We summarized the non-classical functions of snoRNAs in the context of their regulations of the signaling pathways involving PI3K-AKT and K-Ras and p53-dependant manner. Under these novel functions and characteristics, snoRNAs can act as potential and feasible biomarkers for diagnosis. Simultaneously, these promising therapeutic strategies should be considered to counteract the perturbations of snoRNAs.
Keywords: SnoRNAs, dysregulation, therapeutic development, cancer
Introduction
Small nucleolar RNAs (snoRNAs), encoded in the intron of the host gene, are widely distributed in the nucleolus of eukaryotic cells. They are medium-size non-coding RNAs whose length ranges from 60-300 nucleotides (nt) [1]. Most snoRNAs are transcribed by host genes in the nucleus. Following splicing, debranching and trimming of primary transcripts containing the pre-mRNAs of introns, the mature snoRNAs are transported to the nucleolus [1-4]. Classical snoRNAs are responsible for guiding specific chemical modifications and processing ribosomal RNA (rRNA) [5,6]. They can be divided into two main categories based on their unique structural elements, which are conservative: box C/D snoRNAs (SNORDs) and box H/ACA snoRNAs (SNORAs) [7]. These two types of RNAs may have an effect based on their specific sequence, as well as their secondary structure (Figure 1). Simultaneously, their effectors and chemical modifications that they catalyzed were also indispensable in the biosynthesis of rRNAs [8].
Figure 1.
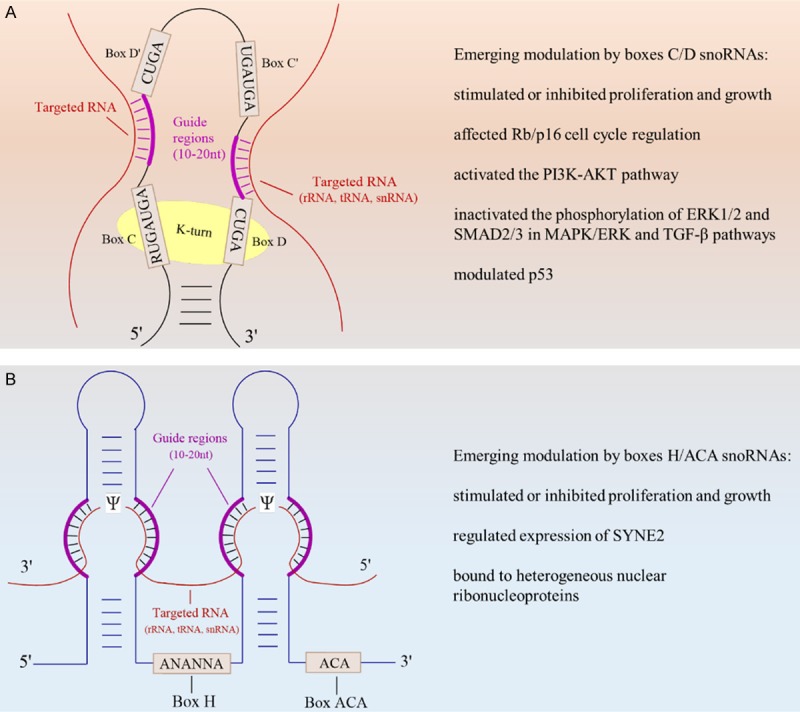
A. Box C/D snoRNAs contain two conserved elements: boxes C (RUGAUGA) and D (CUGA) located at the 5’- and 3’-termini, respectively. Usually, box C/D snoRNAs also have an additional copy of internally resided C’ or D’ boxes. A complex of box C/D snoRNA with nucleolar proteins Snu13p (15.5 kDa), fibrillarin (FBL), Nop58 and Nop56 catalyzes site-specific 2’-O-methylation of the nucleotide in targets including rRNA, tRNA, snRNA. B. Box H/ACA snoRNAs have “hairpin-hinge-hairpin-tail” structure with boxes H (ANANNA) and ACA located within the single stranded (hinge) and 3’-termini (tail) regions, respectively. Box H/ACA snoRNPs contain Nap57, Cbf5p (dyskerin) and GAR1, thereby catalyzing site-specific isomerization of uridine (U) to pseudo uridine (Ψ) in target RNA.
Typical Box C/D snoRNAs are characteristic of the 2’-O-methylation of rRNAs. They consist of two conserved motifs that are treated as recognized sequences: box C contains (RUGAUGA) and box D contains (CUGA) at 5’-termini and 3’-termini, respectively [9]. It also contains a functional structure named as a ‘K-turn’, which is formed by the non-regular base pairing (G-A) shown in boxes C and D, which are folded to form the short stem structure [3,6]. The K-turn serves as a framework and provides the possibility of combining core proteins, in order to assemble small nucleolar ribonucleoproteins (snoRNPs) [2,10]. The combination of core proteins on snoRNAs is likely to protect mature snoRNAs from exonucleolytic trimming [11]. Furthermore, the boxes C/D may contain similar C’ and D’ boxes. These two boxes are similar to their parental C/D in terms of sequence formation [9]. The 2’-O-methylation of the residues of targeted RNAs occurs when the targeted RNAs combine with the guided regions. The guided regions have been explored upstream from boxes D and/or D’, leading the strict complementarity of the featured fragment of the targeted RNAs. Experiments have unmasked that the residues correspond to the fifth nucleotide upstream from boxes D’ or D [10,12].
There is another type of modification that results from snoRNAs, which is named as ‘pseudouridylation’. The pseudouridylation of rRNAs takes place in the common secondary structure with its signature being the “hairpin-hinge-hairpin-tail” that exists in the boxes H/ACA of snoRNAs [7,9]. Considering this unique structure, the boxes H (ANANNA) are located in the hinge areas formed by single chains, and boxes ACA (ACA) are found three nucleotides upstream from the 3’ termini [13,14]. When the target RNA passes through the interior pocket of the central section of one or every hairpin, the progression of pseudouridylation is characterized by the modification of a uridine residue by the complementary base pairing of the targeted RNA and guided regions [3,6]. The guided regions of boxes H and ACA have been explored in pseudouridylation pockets of hairpins [7]. The isomerization of uridine residue in the target RNA, corresponds to the location of 14-16 nt upstream of the H boxes or the ACA boxes [3,15].
These two specific modifications and their correct locations are crucial for the processes of rRNAs. Either a defect in methylation and pseudouridylation or the modifications of other sites can result in intense interference of rRNA processing, leading to a negative effect on cell growth [16]. For instance, it has been confirmed that the pseudouridylated residues are located in the stem loop junction that stabilizes the hairpin structure of RNAs, whereas in the single-stranded loop region, they destabilize the same structure [17,18].
The assembly of snoRNPs is characterized by the combination of snoRNAs and core proteins. Recent studies have shown the significant value of the core proteins. They have reinforced the conservation of mature snoRNA ends, and to some extent, shown a positive correlation with the expression level of snoRNAs [6]. Notably, the levels of some core proteins are associated with the regulation of genes and proteins. We can distinguish snoRNPs in terms of their unique proteins and assembly methods.
The assembly of boxes C/D snoRNPs contains the following proteins: Snu13p (15.5 kDa), fibrillarin (FBL), Nop58 and Nop56 [19,20]. It is initiated by the bonding of the k-turn and Snu13p to form the secondary structure, and then, this motif attracts other core proteins involving FBL and Nop58, thereby accomplishing the assembly [21,22]. Fibrillarin is characterized as the specific structure that is same as S-adenosylmethionine (SAM)-dependent methyltransferase, and it is thus suggested that 2’-O- ribose methylation enzymes work in coordination with boxes C and D snoRNAs [5]. On the contrary, the assembly of boxes H/ACA snoRNPs combines with core proteins related to Nap57, Cbf5p (dyskerin) and GAR1 [23,24]. The snoRNPs may function in the splicing of pre-mRNAs, thereby raising the stability of mRNA transcription though extensive crosslinking between snoRNPs and mRNAs [25].
Previous studies have indicated that snoRNAs that are exclusively regarded as house-keeping genes, regulate the biogenesis of rRNA. However, this assumption has been challenged in recent years. SnoRNAs might have an effect on the regulation of proliferation and apoptosis of tumor cells, through signaling pathways and cell cycles (Summarized in Table 1). For instance, the over-expression of snoRA42 enhances the proliferation, migration, invasion, anoikis resistance and oncogenicity of cells in colorectal cancer (CRC) [26,27]. The increased expression of snoRD126 contributes to reinforce the level of fibroblast growth factor receptor 2 (FGFR2), to activate the PI3k-AKT pathway, where it facilitates hepatocellular carcinoma (HCC) and CRC cell growth [28]. SnoRD76 functions as a tumor inhibitor in glioblastoma (GBM), whereas its elevated expression induces a block on the S phase of the cell cycle, which is associated with the elevated expression of Rb and represses the proliferation and growth of cells [29]. Therefore, investigation of the mechanism leading to the dysregulation of snoRNAs is significant for carcinogenesis.
Table 1.
Summary of dysregulated snoRNAs and related cancer diseases
| SnoRNA ID | Dysregulation | Study models | Pathology | Function | Ref. |
|---|---|---|---|---|---|
| snoRA21 | ↑ | Sample from patients Cell lines: HCT116, SW480 | colorectal adenomas and cancers | proliferation, invasion, poor survival | [90] |
| snoRA23 | ↑ | Sample from patients | pancreatic ductal adenocarcinoma (PDAC) | regulates expression of SYNE2 to promote growth and metastasis | [91] |
| snoRD42 | ↑ | Sample from patients Cell lines: H226, H292, H460, A549, H1299, H1944, H358, H1792, SK-MES-1, H522 and BEAS-2B | Non-small cell lung cancer (NSCLC) | proliferation, colony formation | [26] |
| snoRD42 | ↑ | Sample from patients Cell lines: Caco2, HCT116, HT29, LoVo, SW480 and SW620 | colorectal cancer (CRC) | proliferation, migration, invasion, anoikis resistance and tumorigenicity | [27] |
| snoRD76, snoRD78 | ↑ | Sample from patients Cell lines: Caco2, HCT116, HT29, LoVo, SW480 and SW620 | CRC | [27] | |
| ACA11 | ↑ | Sample from patients Cell lines: Caco2, HCT116, HT29, LoVo, SW480 and SW622 | CRC | binds to heterogeneous nuclear ribonucleoproteins | [27] |
| snoRD33, snoRD66 | ↑ | Sample from patients | NSCLC | [36] | |
| snoRA55 | ↑ | Sample from patients Cell lines: PCa cell lines | prostate cancer (PCa) | proliferation, migration and poor prognosis | [92] |
| snoRD112-114 | ↑ | Sample from patients Cell lines: leukemic blast cells | acute promyelocytic leukemia (APL) | affects Rb/p16 cell cycle regulation to promote cell growth | [93] |
| RNU2 | ↑ | Sample from patients | pancreatic and colorectal adenocarcinoma | highly stable in serum and plasma | [37] |
| snoRD126 | ↑ | Sample from patients Cell lines: Huh-7, SW480 | hepatocellular carcinoma (HCC) and CRC | facilitates HCC and CRC cell growth via activating the PI3K-AKT pathway | [28] |
| HBII-289, U22, U3, U15b, U94, U97 | ↑ | Sample from patients Cell lines: MCF-7, MCF-10A, U-2OS, HCT-116, A549, RWPE1, Hela, U87 cells | breast and prostate cancer | modulation of p53 | [59] |
| snoRD115 | ↓ | Sample from patients | Prader-Willi syndrome (PWS) | regulates the alternative splicing of serotonin receptor 2 C | [94] |
| snoRD113-1 | ↓ | Sample from patients Cell lines: HepG2 and Huh7 | HCC | inactivates the phosphorylation of ERK1/2 and SMAD2/3 in MAPK/ERK and TGF-β pathways | [95] |
| snoRD76 | ↓ | Sample from patients Cell lines: U87-MG, LN229, U251, and SNB19c | glioblastoma (GBM) | inhibits proliferation and growth | [29] |
| snoRD50 | ↓ | Sample from patients Cell lines: Caco2-2, HCT116, HT29, LoVo, SW480 and SW620 | colon, breast and prostate cancers and B-cell lymphoma | inhibits K-Ras signaling pathway and inhibited proliferation and growth | [47] |
| snoRD123, U70c, ACA59B | ↓ | Cell lines: HCT116, normal colon mucosa cells | CRC | [43] |
Dysregulation of snoRNAs in tumor cells
The amplification and inhibition of genes may result in a dramatic transformation of genetical characteristic through the promotion of carcinogenesis. Therefore, identifying the mechanisms of those genes whose expression are increased or decreased would be conducive to discover how tumors form and develop. Interestingly, the dysregulated expression of snoRNAs appears to be unique, suggesting that these dysregulations are closely related to the occurrence of cancer.
Possible explanations of what leads to snoRNA over-expression can be divided into two main manners: increased transcription and decreased consumption. A significant example is growth arrest-specific transcript 5 (GAS5), which is regulated by the expression of p53 in CRC associated snoRNAs [30]. GAS5, located at 1q25, is a non-protein coding lncRNA encoded by the GAS5 gene and hosts several snoRNA sequences including snoRD81, snoRD47, snoRD80, snoRD79, snoRD78, snoRD44, snoRD77, snoRD76, snoRD75 and snoRD74 [31,32]. These snoRNAs along with GAS5 are frequently transcribed simultaneously, and thus the level of snoRNAs have a positive correlation with GAS5. Previous studies have supported the assumption that GAS5 and derived snoRNAs are downregulated in breast cancer [33,34], head and neck squamous cell carcinoma [33] and glioblastoma multiforme [35], whereas the overexpression of snoRD44, snoRD76 and snoRD78 has been demonstrated in non-small cell lung cancer (NSCLC) [36]. It has been suggested that the dysregulated expression level of GAS5-derived snoRNAs is associated with carcinogenesis. Furthermore, the level of p53 expression is positively correlated with the level of snoRD44 and snoRD47 in colorectal tumor samples, indicating that the level of GAS5-derived snoRNAs is regulated in a p53-dependent manner [30]. This direct regulation, which is mediated though p53 simplified the post-transcriptional modification and maturation of rRNAs, ensuring a more effective translation of genes that are needed for the stress response [30]. Acting as a well-known tumor suppressor gene, p53 deficiency has been found in tumorigenesis of gastric cancer. Further studies are needed to ascertain whether snoRNA expression conversions are related to gastric cancer.
Recent research has found that several snoRNA expression levels are not synchronized with their host genes, suggesting that their upregulation is transcribed in a host gene-independent manner or that it results from decreased consumption [26,28]. For instance, small nuclear RNAs (snRNAs) U2 are controlled by snoRNAs and they form the “vesicle” structures which are similar to microRNAs (miRNAs) [37]. Current evidence shows that extracellular transportation of microRNAs depends on the ceramide-dependent pathway, micro-vesicles or exosomes [38,39]. The specific protein Argonaute2 bind miRNAs to form these complexes, which function as a protector and allow these RNAs to escape degradation. Experiments have indicated that fragments resulting from snRNA U2 (RNU2-1f) show a cross-reaction with miR-1246 of the microRNA family [37]. For RNU2-1, corresponding vesicle-like structures may be the apoptotic bodies that contain snRNPs. The existence of apoptotic bodies may be the reason why RNU2-1f can escape from degradation [37,40]. Furthermore, other possible vesicle vectors of RNU2-1, such as exosomes, cannot be excluded. These carriers containing RNU2-1 enter the blood circulation through a tumor-specific capillary system and stabilize it at an elevated level in the serum of the patients with colorectal and pancreatic adenocarcinoma [41].
In contrast, the downregulation of snoRNAs is probably a result of reduced transcription involving the silencing of genes and elevated consumption. It has been recently reported that snoRNAs are targeted though epigenetic inactivation in tumor development [1]. Evidence supports the assumption that the transcriptional silencing of genes is associated with the hypermethylation of the CpG island [42]. Typically, snoRD123, U70c and ACA59B are progressed by their host genes using 5’-CpG islands [43]. They are members of the group of snoRNAs that undergo hypermethylation of their CpG islands in CRC cell lines, compared with that of unmethylated normal tissues [43]. The transcriptional inactivation of snoRNAs induced by CpG island-associated hypermethylation is not unique in CRC, meanwhile, this phenomenon also occurs in lung, breast, prostate and other cancers [43]. An emerging motif prompts that the repression of the expression of snoRNAs mediated by CpG island-associated hypermethylation, could be a promising approach for a tumor suppressor.
The excessive consumption of snoRNAs can be another factor that triggers the downregulation of snoRNAs. It is associated with the proliferation of tumor cells. For instance, snoRNA U50 that functions as a tumor repressor gene by binding to the K-Ras gene can further inhibit its activity [44]. It has been found that the homozygous two-base pair (TT) deletion in a stretch of four thymidines in prostate cancer, which corresponds to the heterozygous deletion in breast cancer [45,46]. snoRNA U50 mediates the post-transcriptional modification of rRNAs, and thus it is consumed within the period of this progress [47]. The requirement for more rRNAs further aggravates its consumption in the process of tumor cell proliferation [48]. The downregulation of U50 has been shown that its host gene and potential regulators do not transcribe enough to counteract its consumption. Taken together, the dysregulation of snoRNAs may function as critical mediator of cancer progression. Novel insights into the up or down-regulation and stability of snoRNAs in tumor cells, even in circulation, have begun to shed light on their feasibility of use in diagnosis and treatment.
SnoRNAs-associated oncogenesis
Discoveries indicated that the function of snoRNAs are not restricted to “house-keeping” genes are accumulation. They act as regulatory factors that affect the signaling pathway involving PI3K-AKT and K-Ras, to regulate the proliferation and apoptosis of cells during tumorigenesis (Figure 2). Recent research has indicated that snoRD126 activates the PI3K-AKT pathway by activation of fibroblast growth factor receptor (FGFR2), thereby promoting CRC cell growth [28]. Similar to members of the growth factor receptor family, FGFR2 contains tyrosine kinase residues that are auto-phosphorylated through the control of their SH2 domains [49]. The orthophosphoric acids derived from these activated receptor residues are recruited by the PI3K-AKT signaling pathway, leading to the activation of AKT [50]. The activated AKT pathway participates in the phosphorylation of several target enzymes, kinases and transcription factors though its various downstream aspects that in particular include the mTOR pathway [51-54]. The TSC1 (tuberous sclerosis complex-1)-TSC2 (tuberous sclerosis complex-2) complex acts as the GAP [55]. It functions as an inhibitor of the GTP-binding protein Rheb (Ras homolog enriched in brain). Since Rheb is essential for the activation of mTOR, the phosphorylation of TSC2 suppresses the formation of the TSC1-TSC2 complex by accepting signals from p-AKT, and as a result, both Rheb and mTOR are activated [55]. The mTOR pathway is associated with the acceleration of cell growth [56]. Additionally, current evidence indicates that mTOR pseudouridylates the 28S rRNA where growth is favorable, which is symbolic as the function of snoRNAs, suggesting that the mTOR pathway may promote cell proliferation by this method [17].
Figure 2.
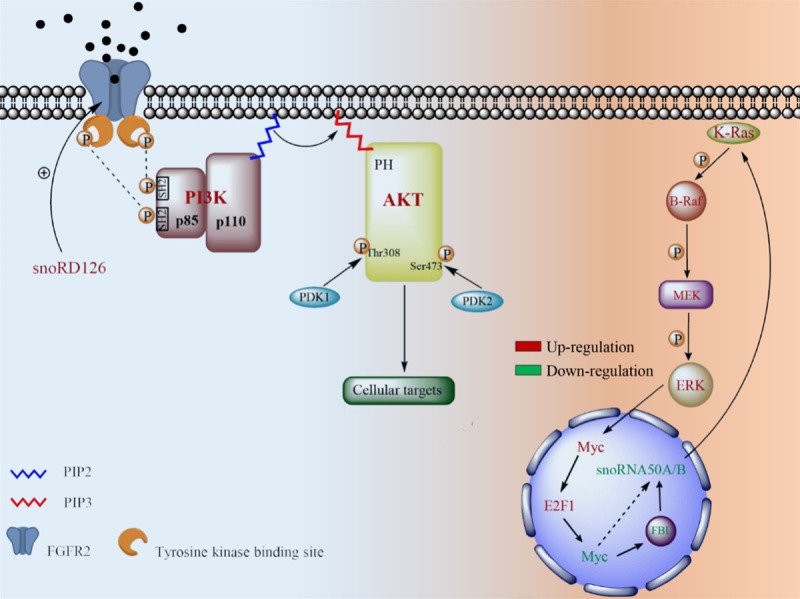
SnoRD126 activates the PI3K-AKT signaling pathway through FGFR2 and snoRD50A/B and affects the MAPK signaling cascades. The expression level of snoRD126 is positively correlated with FGFR2. It contains tyrosine kinase residues which are auto-phosphorylated through binding its ligand. Being of activated, the phosphorylated residues on tyrosine kinase binding site provide binding sites for the SH2 domain of p85. Activation of PI3K induces an alteration of PIP2 to PIP3. PIP3 functions as a second messenger when it is combined with the PH domain of AKT, leading to activation of AKT. The activation is also required members of AGC protein kinase family-PDK1 (phosphoinositide dependent kinase-1) and PDK2 (phosphoinositide dependent kinase-2) which phosphorylate residues on Thr308 and Ser473 of AKT protein, respectively. The deletion of snoRD50A/B increases the binding of GTP and K-Ras, leading to the activation of K-Ras. The activation of K-Ras activates the mitogen-activated protein kinase (MAPK) signaling cascades. It is consisted of the MAP kinase kinase kinase (MAP3K), the MAP kinase kinase (MAP2K) and MAPK. They are phosphorylated in cascades order, thereby transmitting upstream signals to downstream responders.
This effect on cells is initiated by FGFR2 and its regulatory factor snoRD126. Previous studies have demonstrated that the overexpression of snoRD126 is associated with the high protein level of FGFR2, implying the potential possibility that it promotes the translation of the FGFR2 gene [28]. However, no binding sites between snoRD126 and FGFR2 gene have been detected, indicating that the upregulation of FGFR2 might be mediated through a snoRD126-associated transcription factor [28]. This phenomenon implies that this type of snoRNA may conduct its regulation of downstream proteins by associating with other transcriptors. Furthermore, snoRD126 activation of P3K-AKT or other signaling pathways does not appear to be unique among the snoRNA family. A small part of this large family contains several similar subtypes that consist of two to dozens of analogues, leading to parallel functions [57,58]. These functions might be a result of their specific sequences, spatial structures, host genes or transcription manners, hypothesizing that other snoRNAs may function as the activation of PI3K-AKT signaling pathways in a diverse range of cells or tissues.
Downregulation of snoRNAs occurs among tumor cells by generating either the proliferation of K-Ras or apoptosis in a p53-dependent manner [44,59]. Intriguingly, these functions are at least partially motivated by the dysregulation of E2F-1. Being a member of the E2F transcription factor family, E2F-1 can stimulate resting cells to enter the S phase of the cell cycle, leading to the promotion of proliferation [60,61]. It also acts as a regulatory factor that accelerates the apoptotic process. E2F-1 retains both oncogenic and tumor suppressive functions. Previous investigations have demonstrated its crucial impact in regulating the expression of genes that participate in the synthesis of DNA and cell cycle progression [62,63]. As a transcription factor, the activation of E2F-1 is inhibited by binding to pocket proteins involving Rbs [64]. The activation of Rb is regulated by the combination of CDKs (cyclin-dependent kinases) and cyclin proteins. Additionally, this binding pattern is mainly represented by CDK4 and CDK6 combining with cyclin D, while the other model is the integration of CDK2 with cyclin E [66,67]. Dissociation of phosphorylated Rb from the Rb-E2F1 complex leads to activated E2F-1, thereby activating or repressing the expression of downstream genes [64]. Myc belongs to one of these genes whose relationship with E2F-1 remains complicated. Although previous studies have indicated that Myc acts as either an upstream or parallel regulator of E2F-1, it is simultaneously regulated by E2F-1. Its DNA-binding sites have been detected at the promoter of the c-Myc gene, and these sites are able to react with E2F-1 and E2F-dependent regulators [65-67]. It has been illustrated that the overexpression of E2F-1 is associated with the downregulation of Myc [68], whereas the inactivation of E2F-1 takes effect in cooperating with the overexpression of Myc, suggesting that it could be a downstream target of E2F-1, whereas Myc receives negative regulation from E2F-1, and thus they form the feedback loop in tumorigenesis [64].
Myc are oncogenes that encode transcription factors. It has been proved in previous studies that their dysregulation contributes to the development of cancer. Regarding E2F-1, dysregulation of Myc could function to affect both the proliferation and the apoptotic process in tumor cells [69]. Combining enhancer box sequences of DNAs when they have heterogeneous dimerization with MAX polypeptides, Myc proteins are regarded as transcription factors that activate the expression of genes and the translation of proteins [64,70]. Recent investigations have found that the overexpression of Myc induces a higher expression of FBL, which is a crucial snoRNP protein, whereas knockdown of Myc leads to the downregulation of FBL [59]. Considering that snoRNP proteins are regarded as essential elements during the accumulation of snoRNAs, the expression of snoRNAs participate in a branch of products of Myc. Moreover, the activation of RNA polymerase-III is needed for both the conversion of the downstream target of Myc and the transcription of pre-mRNAs of snoRNAs [71,72]. Thus, further evidence provides information of the feasible conditions for the formation of Myc-induced snoRNAs.
Downregulation of snoRNAs that was induced by Myc facilitates the p53-dependent apoptosis of tumor cells [59]. The expression of snoRNAs is repressed by the silencing of core proteins of the C/D box snoRNP (FBL) that leads to a significant increase in the accumulation of p53. These effects are partly associated with ribosomal proteins. Over-synthesis of ribosomes is widely observed in cancer [48,73], which is required for the formation of more ribosomal RNAs and proteins. Decreased levels of snoRNAs result in the absence of modification of rRNAs, leading to the excess of ribosomal proteins. Ribosome-free forms of some ribosomal proteins, including RPL5 and RPL11, enter the nucleoplasm, thereby suppressing the E3 ligase activity of MDM2 [74,75]. In addition, MDM2 is regarded as the E3 ligase that binds to the N-terminal trans-activation domain (TAD) of p53 [76]. The accelerated cell growth is affected by the degradation of the p53 protein, which results from p53-MDM2 interaction [76]. This interaction is inhibited by ribosomal proteins, suggesting that the accumulation of p53 is associated with the downregulation of snoRNAs in this manner [76]. On the other hand, downregulation of snoRNAs reinforces the p53 translation through a cap-independent pathway. This pathway is mediated when the activation of the p53 internal ribosome entering site (IRES) combines with the IRES-binding protein PTB, leading to an increased translation of p53 [59,77]. The arrest of cell growth, even apoptosis, induced by the downregulation of snoRNAs is displayed in tumor cells. These snoRNA deficiencies lead to p53 responses not only in colorectal cancer, but also in lung cancer, breast cancer, and osteosarcoma [59]. Conversely, the overexpression of snoRNAs through elevated levels of Myc promotes cell proliferation, which is regarded as a crucial regulator of cancer progression.
In addition to inducing apoptosis in a p53-dependent manner, the downregulation of snoRNAs may also activate the K-Ras/B-Raf-MEK-ERK pathway to facilitate the proliferation of tumor cells [44,78]. For instance, the deletion of snoRD50A and snoRD50B (snoRD50A/B) activates this pathway due to a decrease in binding with the K-Ras protein directly. These binding mainly form through the action of some potential residues--Lys5, Lys42, Arg149 and Arg161, on K-Ras proteins bound to the box C sequence of snoRD50A/B [44]. It has been recognized that binding occurs in the cytoplasm, suggesting that snoRNAs may be transferred out of the nucleus, rather than being limited to functioning in the nucleus. Furthermore, this binding inhibition as a result of hydrolysis of phosphorylate from GTP activates K-Ras at intramembrane locations [44]. Considering that GTP is essential for the activation of K-Ras, the deletion of snoRD50A/B increases the binding between GTP and K-Ras, leading to the activation of K-Ras [79]. Additionally, the oncogenic mutation of the K-Ras gene generates the mutated K-Ras protein that is found in several cancers [80,81]. The wild-type of the K-Ras protein is activated by binding to GTP when it receives the signals from the ATP-dependent phosphorylated epidermal growth factor receptor (EGFR). However, the mutant-type of K-Ras proteins can be persistently activated without the phosphorylated EGFR [82,83]. The deletion of snoRD50A/B could thus activate both the wild-type and the mutant-type of K-Ras and be synergized with the mutant in tumorigenesis. Moreover, farnesyl transferase (FTase) is indispensable to the post-translational modification of both the wild and the mutant-type of K-Ras. Deletion of snoRD50A/B increases the binding between K-Ras and FTase, thereby activating K-Ras [44].
The activation of K-Ras activates the mitogen-activated protein kinase (MAPK) signaling cascades “B-Raf-MEK-ERK”. Activated ERK then regulates the activity of several transcription factors either directly or indirectly [84,85]. These transcription factors alter the expression level of genes that are significant during the cell growth [86]. In summary, the dysregulation of snoRNAs can affect the proliferation or apoptosis of tumor cells through signaling pathways and in a p53-dependent manner. This effect is conditionally associated with E2F-1.
Molecular biological targeting of snoRNA in cancer
Introns in eukaryotes are frequently removed and degraded during gene transcription. The previous assumption was that these gene sequences in non-coding regions were afunctional. However, emerging evidence had indicated that few RNAs are in fact derived from introns identified as snoRNAs, which contribute to homeostasis. They function by guiding the post-transcriptional modification of rRNAs, transfer RNAs and small nuclear RNAs. Furthermore, the dysregulated expression of snoRNAs have been explored in several cancers.
The stability, sensitivity and specificity of snoRNAs should be considered when they are regarded as potential diagnostic markers. Dysregulation of snoRNAs have been identified in cancer, whereas, occasionally, their presence is not restricted to cancerous tissue. They can access circulation through certain carriers and have a consistent and steady expression. Considering that tumor tissue accounts for only a small fraction of weight, snoRNAs secreted by tumors are insufficient to induce such dynamic variations in their expression levels in circulation [72]. It has been demonstrated that cancer may generate systemic effects on adjacent tissues and even distant organs through cytokines or growth factors derived from cancer, stress or carcinogens [72]. Furthermore, abnormal levels of snoRNA expression have only one manifestation for a cell-type and tissue-specific modes. It has not been shown that levels are elevated or decreased simultaneously, suggesting the specificity of snoRNAs. Some snoRNAs can be repeatedly detected in cancer and manifest higher sensitivity than normal indicators. Disordering of several snoRNAs may be seen during the early stages of cancer, and even during the asymptomatic period, suggesting that they can contribute to the early detection of cancer. Moreover, dysregulation of some snoRNAs may act as predictors that are associated with the prognosis and recurrence of cancer.
We summarized that several receptors are related to the regulation of proliferation and apoptosis of tumor cells through snoRNAs, particularly involving FGFR2. FGFR2 had been widely investigated in relation with advanced gastric cancer [87,88]. Inhibition of related snoRNAs may be a feasible manner to decrease the expression of FGFR2 through the interrelation between snoRNAs and FGFR2. Moreover, the upregulation of snoRNAs can be silenced by siRNAs, and this lays the foundation for the development of novel methods of treating malignant tumors [26]. Considering the bidirectional regulation of E2F-1, its dysregulation is a result of complicated and interconnected regulatory network. In some circumstances, it participates in the signal transduction which includes snoRNAs-related pathways. The difficulties in early-stage diagnosis and the poor prognosis are shown in gastric cancer, suggesting that targeted snoRNAs might be a promising approach for the diagnosis and treatment of gastric cancer. Furthermore, negative feedback loops are involved in complex regulatory systems related to snoRNAs in vivo. It has been illustrated that the inactivation of E2F-1 results in the upregulation of Myc, and in turn, the overexpression of Myc activates E2F-1 through miRNAs [89]. These negative feedback loops can neutralize some homeostasis imbalances within a certain range. However, they may be deactivated or inhibited in cancer, being insufficient to counteract the function of over-excited carcinogenic pathways.
Conclusions and perspectives
In this review we have briefly discussed the controversy that is found in recent literature regarding the presence or absence of snoRNAs in cancer, and the assumption that snoRNAs function as critical mediators of cancer progression, which can act as an emerging field of diagnosis and treatment for cancer. Novel insights into the global roles of specific snoRNAs at phenotypic, physiological, and molecular levels may be promising approaches to solving human diseases, particularly cancers [6]. In-depth research combined with gene sequencing analyses and proteomics testing will contribute to the investigation of the non-classical effects of snoRNAs and may even be extended to ncRNAs, raising the capacity of humanity to fight against cancer up to an unprecedented level.
Acknowledgements
This work was supported by the National Science Foundation for Young Scholars of China (No. 81502120) and the National Science Foundation (No. 81560454) and Guangxi Medical University Training Program for Distinguished Young Scholars. We would like to thank Dr Zhi-Liu Cao and Si Cheng for them critical reading of this manuscript and insightful comments and the contributions from Jing Duan, Yi-Ru Wang, Ze-Feng Chen, Jing-Yu Chen.
Disclosure of conflict of interest
None.
References
- 1.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 2.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 3.Reichow SL, Hamma T, Ferre-D’Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown JW, Marshall DF, Echeverria M. Intronic noncoding RNAs and splicing. Trends Plant Sci. 2008;13:335–342. doi: 10.1016/j.tplants.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Stepanov GA, Filippova JA, Komissarov AB, Kuligina EV, Richter VA, Semenov DV. Regulatory role of small nucleolar RNAs in human diseases. Biomed Res Int. 2015;2015:206849. doi: 10.1155/2015/206849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dupuis-Sandoval F, Poirier M, Scott MS. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdiscip Rev RNA. 2015;6:381–97. doi: 10.1002/wrna.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kehr S, Bartschat S, Tafer H, Stadler PF, Hertel J. Matching of soulmates: coevolution of snoRNAs and their targets. Mol Biol Evol. 2014;31:455–67. doi: 10.1093/molbev/mst209. [DOI] [PubMed] [Google Scholar]
- 8.Cavaillé J, Nicoloso M, Bachellerie JP. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–735. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 9.Bachellerie JP, Cavaillé J, Hüttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 10.Kiss-László Z, Henry Y, Kiss T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. Embo Journal. 1998;17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darzacq X, Kiss T. Processing of intron-encoded box C/D small nucleolar RNAs lacking a 5’,3’-terminal stem structure. Mol Cell Biol. 2000;20:4522–31. doi: 10.1128/mcb.20.13.4522-4531.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kisslászló Z, Henry Y, Bachellerie JP, Caizerguesferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 13.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 14.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. Embo Journal. 2001;20:3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lui L, Lowe T. Small nucleolar RNAs and RNA-guided post-transcriptional modification. Essays Biochem. 2013;54:53–77. doi: 10.1042/bse0540053. [DOI] [PubMed] [Google Scholar]
- 16.Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009;15:1716–1728. doi: 10.1261/rna.1724409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Courtes FC, Gu C, Wong NS, Dedon PC, Yap MG, Lee DY. 28S rRNA is inducibly pseudouridylated by the mTOR pathway translational control in CHO cell cultures. J Biotechnol. 2014;174:16–21. doi: 10.1016/j.jbiotec.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Badis G. A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast. RNA. 2003;9:771–779. doi: 10.1261/rna.5240503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kishore S, Gruber AR, Jedlinski DJ, Syed AP, Jorjani H, Zavolan M. Insights into snoRNA biogenesis and processing from PAR-CLIP of snoRNA core proteins and small RNA sequencing. Genome Biol. 2013;14:R45. doi: 10.1186/gb-2013-14-5-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMahon M, Contreras A, Ruggero D. Small RNAs with big implications: new insights into H/ACA snoRNA function and their role in human disease. Wiley Interdiscip Rev RNA. 2015;6:173–189. doi: 10.1002/wrna.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watkins NJ, Dickmanns A, Luhrmann R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the Box C/D snoRNP. Mol Cell Biol. 2002;22:8342–52. doi: 10.1128/MCB.22.23.8342-8352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiss T, Fayet E, Jady BE, Richard P, Weber M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- 23.Walbott H, Machado-Pinilla R, Liger D, Blaud M, Rety S, Grozdanov PN, Godin K, van Tilbeurgh H, Varani G, Meier UT, Leulliot N. The H/ACA RNP assembly factor SHQ1 functions as an RNA mimic. Genes Dev. 2011;25:2398–2408. doi: 10.1101/gad.176834.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Duan J, Li D, Ma S, Ye K. Structure of the Shq1-Cbf5-Nop10-Gar1 complex and implications for H/ACA RNP biogenesis and dyskeratosis congenita. EMBO J. 2011;30:5010–5020. doi: 10.1038/emboj.2011.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kishore S, Gruber AR, Jedlinski DJ, Syed AP, Jorjani H, Zavolan M. Insights into snoRNA biogenesis and processing from PAR-CLIP of snoRNA core proteins and small RNA sequencing. Genome Biol. 2013;14:R45. doi: 10.1186/gb-2013-14-5-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mei YP, Liao JP, Shen J, Yu L, Liu BL, Liu L, Li RY, Ji L, Dorsey SG, Jiang ZR, Katz RL, Wang JY, Jiang F. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2012;31:2794–2804. doi: 10.1038/onc.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okugawa Y, Toiyama Y, Toden S, Mitoma H, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut. 2017;66:107–117. doi: 10.1136/gutjnl-2015-309359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang X, Yang D, Luo H, Wu S, Dong W, Xiao J, Yuan S, Ni A, Zhang KJ, Liu XY, Chu L. SNORD126 promotes HCC and CRC cell growth by activating the PI3K-AKT pathway through FGFR2. J Mol Cell Biol. 2017;9:243–255. doi: 10.1093/jmcb/mjw048. [DOI] [PubMed] [Google Scholar]
- 29.Chen L, Han L, Wei J, Zhang K, Shi Z, Duan R, Li S, Zhou X, Pu P, Zhang J, Kang C. SNORD76, a box C/D snoRNA, acts as a tumor suppressor in glioblastoma. Sci Rep. 2015;5:8588. doi: 10.1038/srep08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krell J, Frampton AE, Mirnezami R, Harding V, De Giorgio A, Roca Alonso L, Cohen P, Ottaviani S, Colombo T, Jacob J, Pellegrino L, Buchanan G, Stebbing J, Castellano L. Growth arrest-specific transcript 5 associated snoRNA levels are related to p53 expression and DNA damage in colorectal cancer. PLoS One. 2014;9:e98561. doi: 10.1371/journal.pone.0098561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith C, Steitz J. Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5’-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol. 1998;18:6897–909. doi: 10.1128/mcb.18.12.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller AJ, Chatterjee S, Teresky A, Levine AJ. The gas5 gene is disrupted by a frameshift mutation within its longest open reading frame in several inbred mouse strains and maps to murine chromosome 1. Mamm Genome. 1998;9:773–4. doi: 10.1007/s003359900862. [DOI] [PubMed] [Google Scholar]
- 33.Gee HE, Buffa FM, Camps C, Ramachandran A, Leek R, Taylor M, Patil M, Sheldon H, Betts G, Homer J. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br J Cancer. 2011;104:1168–77. doi: 10.1038/sj.bjc.6606076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mourtadamaarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 35.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 36.Liao J, Lei Y, Mei Y, Guarnera M, Shen J, Li R, Liu Z, Feng J. Small nucleolar RNA signatures as biomarkers for non-small-cell lung cancer. Mol Cancer. 2010;9:1–10. doi: 10.1186/1476-4598-9-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baraniskin A, Nopel-Dunnebacke S, Ahrens M, Jensen SG, Zollner H, Maghnouj A, Wos A, Mayerle J, Munding J, Kost D, Reinacher-Schick A, Liffers S, Schroers R, Chromik AM, Meyer HE, Uhl W, Klein-Scory S, Weiss FU, Stephan C, Schwarte-Waldhoff I, Lerch MM, Tannapfel A, Schmiegel W, Andersen CL, Hahn SA. Circulating U2 small nuclear RNA fragments as a novel diagnostic biomarker for pancreatic and colorectal adenocarcinoma. Int J Cancer. 2013;132:E48–57. doi: 10.1002/ijc.27791. [DOI] [PubMed] [Google Scholar]
- 38.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 39.Arroyo JD, Eisenman RN. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biggiogera M, Bottone MG, Martin TE, Uchiumi T, Pellicciari C. Still immunodetectable nuclear RNPs are extruded from the cytoplasm of spontaneously apoptotic thymocytes. Exp Cell Res. 1997;234:512–20. doi: 10.1006/excr.1997.3657. [DOI] [PubMed] [Google Scholar]
- 41.Halicka HD, Bedner E, Darzynkiewicz Z. Segregation of RNA and separate packaging of DNA and RNA in apoptotic bodies during apoptosis. Exp Cell Res. 2000;260:248–56. doi: 10.1006/excr.2000.5027. [DOI] [PubMed] [Google Scholar]
- 42.Lopezserra P, Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. 2012;31:1609–1622. doi: 10.1038/onc.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferreira HJ, Heyn H, Moutinho C, Esteller M. CpG island hypermethylation-associated silencing of small nucleolar RNAs in human cancer. RNA Biol. 2012;9:881–890. doi: 10.4161/rna.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siprashvili Z, Webster DE, Johnston D, Shenoy RM, Ungewickell AJ, Bhaduri A, Flockhart R, Zarnegar BJ, Che Y, Meschi F, Puglisi JD, Khavari PA. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat Genet. 2016;48:53–58. doi: 10.1038/ng.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong XY, Rodriguez C, Guo P, Sun X, Talbot JT, Zhou W, Petros J, Li Q, Vessella RL, Kibel AS. SnoRNA U50 is a candidate tumor-suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Hum Mol Genet. 2008;17:1031–42. doi: 10.1093/hmg/ddm375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong XY, Guo P, Boyd J, Sun X, Li Q, Zhou W, Dong JT. Implication of snoRNA U50 in human breast cancer. J Genet Genomics. 2009;36:447–54. doi: 10.1016/S1673-8527(08)60134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer. 2011;11:865–78. doi: 10.1038/nrc3143. [DOI] [PubMed] [Google Scholar]
- 49.Lee MS, Jeong MH, Lee HW, Han HJ, Ko A, Hewitt SM, Kim JH, Chun KH, Chung JY, Lee C, Cho H, Song J. PI3K/AKT activation induces PTEN ubiquitination and destabilization accelerating tumourigenesis. Nat Commun. 2015;6:7769. doi: 10.1038/ncomms8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dazert E, Hall MN. mTOR signaling in disease. Curr Opin Cell Biol. 2011;23:744–55. doi: 10.1016/j.ceb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 52.Hay N. The Akt-mTOR tango and its relevance to cancer. Cancer Cell. 2005;8:179–183. doi: 10.1016/j.ccr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 53.Missiaglia E, Dalai I, Barbi S, Beghelli S, Falconi M, Peruta MD, Piemonti L, Capurso G, Florio AD, Fave GD. Pancreatic endocrine tumors: expression profiling evidences a role for AKT-mTOR pathway. J. Clin. Oncol. 2010;28:245–55. doi: 10.1200/JCO.2008.21.5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- 55.Yuan S, Wu Y, Wang Y, Chen J, Chu L. An oncolytic adenovirus expressing SNORD44 and GAS5 exhibits antitumor effect in colorectal cancer cells. Hum Gene Ther. 2017;28:690–700. doi: 10.1089/hum.2017.041. [DOI] [PubMed] [Google Scholar]
- 56.Nahkuri S, Taft RJ, Korbie DJ, Mattick JS. Molecular evolution of the HBII-52 snoRNA cluster. J Mol Biol. 2008;381:810–5. doi: 10.1016/j.jmb.2008.06.057. [DOI] [PubMed] [Google Scholar]
- 57.Schmitz J, Zemann A, Churakov G, Kuhl H, Grützner F, Reinhardt R, Brosius J. Retroposed SNOfall-A mammalian-wide comparison of platypus snoRNAs. Genome Res. 2008;18:1005–10. doi: 10.1101/gr.7177908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Su H, Xu T, Ganapathy S, Shadfan M, Long M, Huang TH, Thompson I, Yuan ZM. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene. 2013;33:1348–1358. doi: 10.1038/onc.2013.89. [DOI] [PubMed] [Google Scholar]
- 59.Johnson DG, Schwarz JK, Cress WD, Nevins JR. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 60.Degregori J, Leone G, Miron A, Jakoi L, Nevins JR. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci U S A. 1997;94:7245–50. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 62.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 63.Rounbehler RJ, Rogers PM, Conti CJ, Johnson DG. Inactivation of E2f1 enhances tumorigenesis in a myc transgenic model. Cancer Res. 2002;62:3276–81. [PubMed] [Google Scholar]
- 64.Pierce AM, Schneider-Broussard R, Philhower JL, Johnson DG. Differential activities of E2F family members: unique functions in regulating transcription. Mol Carcinog. 1998;22:190–8. doi: 10.1002/(sici)1098-2744(199807)22:3<190::aid-mc7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 65.Hiebert SW, Lipp M, Nevins JR. E1A-dependent trans-activation of the human MYC promoter is mediated by the E2F factor. Proc Natl Acad Sci U S A. 1989;86:3594–8. doi: 10.1073/pnas.86.10.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oswald F, Lovec H, Möröy T, Lipp M. E2F-dependent regulation of human MYC: trans-activation by cyclins D1 and a overrides tumour suppressor protein functions. Oncogene. 1994;9:2029–2036. [PubMed] [Google Scholar]
- 67.Yan LH, Li L, Xie YB, Xiao Q, Wang CQ. Effects of E2F-1 overexpression on apoptosis of gastric cancer cells and expressions of apoptosis-related genes. Ai Zheng. 2009;28:1176–1180. doi: 10.5732/cjc.009.10192. [DOI] [PubMed] [Google Scholar]
- 68.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Armelin HA, Armelin MC, Kelly K, Stewart T, Leder P, Cochran BH, Stiles CD. Functional role for c-myc in mitogenic response to platelet-derived growth factor. Nature. 1984;310:655–660. doi: 10.1038/310655a0. [DOI] [PubMed] [Google Scholar]
- 70.Filipowicz W, Pogacić V. Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol. 2002;14:319–27. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 71.Appaiah HN, Goswami CP, Mina LA, Badve S, Sledge GW Jr, Liu Y, Nakshatri H. Persistent upregulation of U6: SNORD44 small RNA ratio in the serum of breast cancer patients. Breast Cancer Res. 2011;13:R86. doi: 10.1186/bcr2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–92. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 73.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279:44475–82. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 74.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 75.Wade M, Wong ET, Tang M, Stommel JM, Wahl GM. Hdmx modulates the outcome of p53 activation in human tumor cells. J Biol Chem. 2006;281:33036–44. doi: 10.1074/jbc.M605405200. [DOI] [PubMed] [Google Scholar]
- 76.Takagi M, Absalon MJ, Mclure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 77.Avruch J, Khokhlatchev A, Kyriakis JM, Luo Z, Tzivion G, Vavvas D, Zhang XF. Ras activation of the Raf kinase: tyrosine kinase recruitment of the MAP kinase cascade. Recent Prog Horm Res. 2001;56:127–155. doi: 10.1210/rp.56.1.127. [DOI] [PubMed] [Google Scholar]
- 78.Schulze WX, Deng L, Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol Syst Biol. 2005;1:2005.0008. doi: 10.1038/msb4100012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bai CM. KRAS mutations and benefit from cetuximab in advanced colorectal cancer. The Journal of Evidence-Based Medicine. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 80.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baines AT, Xu D, Der CJ. Inhibition of Ras for cancer treatment: the search continues. Future Med Chem. 2011;3:1787–808. doi: 10.4155/fmc.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Herrmann C, Horn G, Spaargaren M, Wittinghofer A. Differential interaction of the ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. J Biol Chem. 1996;271:6794–800. doi: 10.1074/jbc.271.12.6794. [DOI] [PubMed] [Google Scholar]
- 83.Campisi J, Gray HE, Pardee AB, Dean M, Sonenshein GE. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell. 1984;36:241–247. doi: 10.1016/0092-8674(84)90217-4. [DOI] [PubMed] [Google Scholar]
- 84.Mannava S, Grachtchouk V, Wheeler LJ, Im M, Zhuang D, Slavina EG, Mathews CK, Shewach DS, Nikiforov MA. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle. 2008;7:2392–2400. doi: 10.4161/cc.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aughey GN, Grice SJ, Liu JL. The Interplay between Myc and CTP synthase in drosophila. PLoS Genet. 2016;12:e1005867. doi: 10.1371/journal.pgen.1005867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Katoh M, Katoh M. FGFR2 and WDR11 are neighboring oncogene and tumor suppressor gene on human chromosome 10q26. Int J Oncol. 2003;22:1155–9. [PubMed] [Google Scholar]
- 87.Kunii K, Davis L, Gorenstein J, Hatch H, Yashiro M, Bacco AD, Elbi C, Lutterbach B. FGFR2-amplified gastric cancer cell lines require FGFR2 and Erbb3 signaling for growth and survival. Cancer Res. 2008;68:2340–8. doi: 10.1158/0008-5472.CAN-07-5229. [DOI] [PubMed] [Google Scholar]
- 88.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc- regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 89.Yoshida K, Toden S, Weng W, Shigeyasu K, Miyoshi J, Turner J, Nagasaka T, Ma Y, Takayama T, Fujiwara T. SNORA21 - an oncogenic small nucleolar RNA, with a prognostic biomarker potential in human colorectal cancer. Ebiomedicine. 2017;22:68–77. doi: 10.1016/j.ebiom.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cui L, Nakano K, Obchoei S, Setoguchi K, Matsumoto M, Yamamoto T, Obika S, Shimada K, Hiraoka N. Small nucleolar noncoding RNA SNORA23, upregulated in human pancreatic ductal adenocarcinoma, regulates expression of SYNE2 to promote growth and metastasis of xenograft tumors in mice. Gastroenterology. 2017;153:292. doi: 10.1053/j.gastro.2017.03.050. [DOI] [PubMed] [Google Scholar]
- 91.Crea F, Quagliata L, Michael A, Liu HH, Frumento P, Azad AA, Xue H, Pikor L, Watahiki A, Morant R. Integrated analysis of the prostate cancer small-nucleolar transcriptome reveals SNORA55 as a driver of prostate cancer progression. Mol Oncol. 2016;10:693–703. doi: 10.1016/j.molonc.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Valleron W, Laprevotte E, Gautier EF, Quelen C, Demur C, Delabesse E, Agirre X, Prósper F, Kiss T, Brousset P. Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia. 2012;26:2052–2060. doi: 10.1038/leu.2012.111. [DOI] [PubMed] [Google Scholar]
- 93.Bortolin-Cavaillé ML, Cavaillé J. The SNORD115 (H/MBII-52) and SNORD116 (H/MBII-85) gene clusters at the imprinted Prader-Willi locus generate canonical box C/D snoRNAs. Nucleic Acids Res. 2012;40:6800–7. doi: 10.1093/nar/gks321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xu G, Yang F, Ding CL, Zhao LJ, Ren H, Zhao P, Wang W, Qi ZT. Small nucleolar RNA 113-1 suppresses tumorigenesis in hepatocellular carcinoma. Mol Cancer. 2014;13:216. doi: 10.1186/1476-4598-13-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pacilli A, Ceccarelli C, Trere D, Montanaro L. SnoRNA U50 levels are regulated by cell proliferation and rRNA transcription. Int J Mol Sci. 2013;14:14923–14935. doi: 10.3390/ijms140714923. [DOI] [PMC free article] [PubMed] [Google Scholar]


