Abstract
Exosomes are released membrane vesicles derived from late endosomes, which share structural and biochemical characteristics with proteasomes. Exosomes are responsible for trafficking proteins, microRNAs (miRNAs), and long non-coding RNAs (lncRNAs) among cells and regulating various cellular processes, such as differentiation, proliferation, migration, invasion, and apoptosis. Although our knowledge of the roles of exosomes in the initiation and progression of leukemia is limited, some studies have indicated that exosomes can encompass many functional factors with an appropriate sorting signal, thereby supporting the metastasis, drug resistance, and immune escape of leukemia cells. This review initially focuses on the biogenesis and composition of exosomes and then summarizes the application of exosomes as a screening biomarker and a potential therapeutic target in leukemia. Many recent reports on the functions of exosomes released from leukemia cells are also discussed, including drug resistance, immune dysfunction, and microenvironment manipulation. Given the critical roles of exosomes in leukemia, understanding the mechanisms regulating the compositions and levels of exosomes, as well as defining exosome functions, will ultimately provide additional insights into the use of exosomes as therapeutic agents for leukemia treatment.
Keywords: Exosomes, leukemia, microRNA, lncRNA, microenvironment
Introduction
Exosomes are membrane-bound vesicles that are approximately 30-100 nm in diameter and are extensively distributed in various biological fluids [1]. In 1983, Pan et al. [2] first discovered exosomes in reticulocytes as novel transfer vesicles. Exosomes are released into the extracellular space from many types of cells after fusing with the plasma membrane [3-5]. The main contents of exosomes are lipids, proteins, and various nucleic acids [6]. Although the differences between the exosomes of normal cells and those of cancer cells have been extensively studied [7-9], the biological and functional roles of exosomes in leukemia are not yet fully understood, and several regulatory mechanisms remain unclear.
To advance the field of exosomal biology and understand the roles of exosomes in leukemia, we have conducted a review regarding the concept of the exosome, as well as exosomal contents, biogenesis and functions. The application of exosomes as biomarkers for leukemia screening and the potential roles of exosomes in the development of leukemia therapeutic agents are also discussed.
Formation, isolation, detection and uptake of exosomes
Formation and secretion of exosomes
Although many types of extracellular vesicles have been identified [10], no uniform definitions of these extracellular vesicles have been established. Microvesicles, exosomes, and microparticles are terms that have been frequently used to refer to extracellular vesicles, but these terms can be confusing in many cases [11-13]. Fortunately, a new standard has been established to separate extracellular vesicles into microvesicles and exosomes by categorizing them according to how they are secreted from parent cells. Microvesicles are directly shed from the parent cell membrane, whereas exosomes are released after multivesicular bodies fuse with the plasma membrane [14]. Exosomes exhibit a cup-shaped morphology when viewed using negative staining and transmission microscopy. In addition, tetraspanin proteins, such as CD63, CD9, and CD81, can be used to identify most exosomes [3-5].
Exosome secretion via exocytosis is mediated by many factors. Ostrowski et al. [15] indicated that the secretion of exosomes was directly regulated by Rab27a and Rab27b, and that knockdown of either Rab27 or related effector molecules inhibited the secretion of exosomes from HeLa cells. Syndecan-syntenin was found to directly interact with ALIX protein to support exosome formation by promoting intraluminal budding of endosomal membranes, which was associated with heparan sulphate [16]. Additionally, exosome secretion can be affected by other factors, such as intracellular Ca2+ levels and extracellular/intracellular pH gradients [17,18]. A recent study indicated that SphK2/S1P signaling affected the contents of exosomes derived from K562 cells, and knockdown of Sphk2 significantly decreased the contents of exosomes. Furthermore, adverse contents of exosomes were enriched after inhibition of SphK using N, N-dimethylsphingosine (DMS) [19].
Isolation of exosomes
Isolating exosomes has always been challenging because exosomes are very small. Several effective methods have been established to isolate exosomes from various biological fluids, all of which have certain advantages and disadvantages, such as ultracentrifugation, ultrafiltration, chromatography, polymer-based precipitation and antibody-coupled magnetic beads [20]. Although ultracentrifugation has been extensively accepted due to its simple and easy operation, this method is time-consuming, and protein complexes, lipoproteins, and other contaminants always reduce product purity [21,22]. Ultrafiltration and chromatography produce highly purified exosomes using semipermeable polyethersulfone nanomembranes; however, the remaining proteins are very difficult to remove [23-25]. Polymeric precipitation was invented by System Biosciences to yield high-purity exosome isolates through precipitation using polymers, but this new technology can only capture and collect exosomes with sizes ranging from 60 to 150 nm [26]. Immunoaffinity purification using beads only captures exosomes with positive surface markers, including CD63, CD9, and CD81, and consequently, a large population of negative exosomes is selectively abandoned [27]. In addition, to avoid cross-contamination, serum should be depleted of exosomes by the standard ultracentrifugation method.
Transfer and uptake of exosomes
Cell-derived exosomes can facilitate gene transfer between parent and recipient cells. To date, three exosome transfer methods have been identified: (A), exosomes activate the cell surface receptors of receipt cells via transmembrane proteins [28]; (B), exosomes fuse with the plasma membranes of recipient cells and release cargo contents [24]; and (C), exosomes transform into recipient cells and merge into endosomes, mature into lysosomes, or undergo transcytosis, eventually leaving the recipient cells and being released into adjacent cells [29,30]. Thus, exosomes have great potential in research on drug delivery and therapy development [8]. One study showed that exosomes systemically administered to mice deliver siRNA to the brain, confirming the substantial therapeutic potential of exosomes [31]. However, a few studies have revealed that AnxA2 and AnxA6 promote exosome uptake in breast cancer cells, while protease K prevents exosome uptake in ovarian cancer cells, but the underlying mechanism of exosome internalization remains unclear. In addition, how exosomes travel and how long exosomes remain stable in the systemic circulation urgently need to be determined.
Contents of exosomes
MicroRNAs
Exosomal microRNAs (miRNAs) can circulate to and play functional roles in neighboring or distant recipient cells. While the impact of other types of exosomal contents on recipient cells cannot be completely excluded, miRNAs are considered pivotal regulating factors. Exosomal miRNAs serve both conventional and novel functions. Conventional miRNA functions involve negative regulation of the expression of target genes. In endothelial cells, exosomal miR-105 of breast cancer cells reduced the expression of the ZO-1 gene, thus promoting breast cancer cell metastases to the lungs and brain [32].
Exosomal miR-214, which is derived from human microvascular endothelial cell (HMEC)-1, significantly reduced the expression of the ataxia telangiectasia-mutated gene and allowed blood vessel formation, consequently stimulating migration and angiogenesis in neighboring HMEC-1 cells [33]. Novel miRNA functions have been identified in some cases where miRNAs were studied as exosomes rather than intracellular molecules. Exosomal miR-21 and miR-29a, in addition to their classic role of targeting mRNAs, were the first miRNAs discovered to have the capacity to act as toll-like receptor ligands and activate immune cells [34]. Furthermore, patients and healthy individuals exhibited different exosomal miRNA levels and compositions, suggesting the potential of these miRNAs to be used as cancer biomarkers or even as prognostic indicators. For example, let-7a, miR-1229, miR-1246, miR-150, miR-21, miR-223, and miR-23a can be used as diagnostic biomarkers of colorectal cancer [35]. In addition, miR-1290 and miR-375 can be used as prognostic markers of castration-resistant prostate cancer [36].
lncRNAs
Long non-coding RNAs (lncRNAs) are classically defined as RNA transcripts longer than 200 nucleotides that have limited to no protein-coding potential. Generally, lncRNAs have fewer exons than mRNAs, and only short open reading frames can be identified; thus, few lncRNAs are likely to encode small peptides. Recent studies have found that tumorigenesis is associated with overall deregulation of lncRNAs. This deregulation of lncRNAs derived from exosomes may be observed intracellularly, in tissue, and extracellularly in body fluids. Takahashi et al. [37]. demonstrated that the lncRNAs present in exosomes released by HepG2 hepatocellular carcinoma cells are a key factor in HepG2 cell chemoresistance to sorafenib treatment. Furthermore, chemoresistance may be transferred to other HepG2 cells via exosomes. After stimulation with TGF-β, the exosomal lncRNA content was found to be altered, further supporting the roles of exosomes as cell representations and excellent resources for the identification of potential cancer biomarkers. In addition, exosomal lncRNAs have been implicated in cancer development through modulation of the cancer microenvironment. Conigliaro et al. [38] identified exosomes from CD90+ hepatocellular cancer cells containing different types of lncRNAs, including HOTAIR, HULC, linc-ROR and H19. Upon co-culturing these cancer cells with endothelial cells in vitro, the latter cells rapidly internalized exosomes and were triggered to reorganize into tubular structures and to increase vascular endothelial growth factor (VEGF)/VEGF-R1 mRNA levels. Li et al. [39] suggested that most lncRNAs in gastric cancer patient plasma are derived from exosomes; the exosomal lncRNAs supported a gastric cancer diagnosis with a sensitivity of 48.1% and a specificity of 85.2%. Recently, exosomal lncRNAs have also been shown to have prognostic potential in cancer. lncRNAs isolated from serum exosomes of laryngeal squamous cell carcinoma patients were found to be elevated in the patients with lymph node metastases and later disease stages [40]. However, the mechanism of lncRNA secretion is not yet fully understood.
Proteins
To a certain extent, the exosomal protein content depends on the lineage, state of activation, and/or transformation of the parent cells. Proteins enriched in exosomes are likely involved in vesicle genesis or trafficking (i.e., Tsg101, ALIX, annexins, and Rab proteins) [41-43], signal transduction (i.e., kinases and G-proteins) [44], cytoskeleton organization (i.e., actin and tubulin) [44], Ag presentation or transport (i.e., MHC I and II molecules and heat-shock proteins) [45,46], vesicle targeting to recipient cells or extracellular matrix (i.e., integrins and MFG-E8/lactadherin) [47,48], protein organization in membrane microdomains (i.e., tetraspanins such as CD9, CD63, and CD81) [49], and protection from complement-mediated lysis (i.e., CD55 and CD59) [50]. Exosomes also contain enzymes required for RNA and protein synthesis and degradation [20,45]. Recently, Théry et al. [51] established the first extensive protein map of a particular exosome population in dendritic cells. Twenty-one new exosomal proteins were identified, including cytoskeletonrelated proteins, intracellular membrane transport proteins and signaling factors, as well as a novel category of apoptosis-related proteins.
Exosomes and leukemia
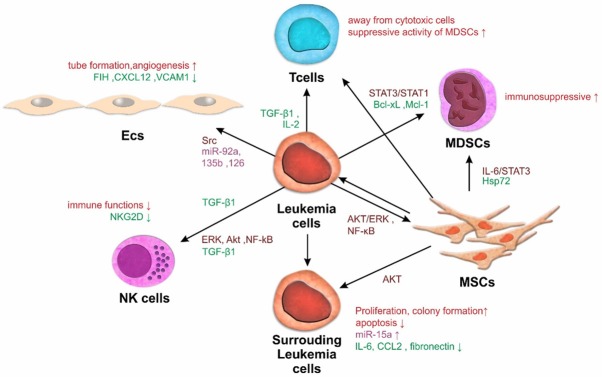
As expected, the roles of exosomes have been extensively studied in both leukemia and solid tumors. Studies examining leukemia exosomes have highlighted their possible roles in leukemia development and progression (Figure 1).
Figure 1.
The positive feedback loop favoring leukemia appears to operate in the leukemia microenvironment. Exosomes produced in the bone marrow (BM) microenvironment by leukemia or stromal cells establish bidirectional lines of communication between these cells and modify their functions: Exosomes released by leukemia cells interact with endothelial cells to promote angiogenesis. These exosomes also influence the drug resistance of leukemia cells and the development and differentiation of NK cells and T cells. Although leukemia cells in BM release exosomes that alter the functions of mesenchymal stem cells and myeloid-derived suppressor cells, the role of the exosome-mediated communication network is not well known.
Exosome-mediated cell-cell communication in leukemia
The importance of the microenvironment in leukemia progression has become widely recognized in recent years. Exosomes are small extracellular vesicles that are enriched with RNAs and proteins, released by various normal cells and leukemia cells, and are present in all body fluids. Thus, exosomes have been acknowledged as important mediators of communication among leukemia cells and in the stromal microenvironment [52-54].
Exosomes in chronic myeloid leukemia
Exosomes derived from chronic myeloid leukemia (CML) cells can be taken up by endothelial cells and promote endothelial cell tube formation. Exosomes from K562 cells induced dasatinib-sensitive Src phosphorylation and activation of downstream Src pathway proteins in endothelial cells. Dasatinib activity may be greater than imatinib (IM) activity due to the involvement of Src in both leukemia cells and the angiogenic microenvironment [55]. Furthermore, a study from Tokyo Medical University found that pre-miR-92a derived from K562 exosomes could reduce the expression of the target gene integrin a5 and then enhance endothelial cell migration and tube formation. miR-210 from CML exosomes has been shown to combine with the target gene Ephrin-A3 and play an important role in the regulation of angiogenesis and VEGF signaling [56,57]. These findings demonstrate how leukemia cells may convey signals to the microenvironment and indicate the therapeutic potential of using exosome-derived miRNAs in combination with currently available VEGF inhibitors. In addition, CML exosomes can establish an autocrine loop with their parent cells through a ligand-receptor interaction mediated by exosome-associated transforming growth factor (TGF)-β1, followed by activation of the extracellular signal-regulated kinase (ERK), protein kinase B (AKT) and nuclear factor (NF)-kB signaling pathways, leading to increased CML cell proliferation and survival [58]. These findings provide a new understanding of the roles of exosomes in cancer biology. The extensive autocrine stimulation that occurs in tumors has significant implications for innovative therapeutic and biomarker strategies for leukemia. After co-culture with bone marrow mesenchymal stem cells (BM-MSCs) and macrophages, K562-derived exosomes affect the expression of various genes, including those encoding C-X-C motif chemokine 12 (Cxcl12), Dickkopf-related protein 1 (DKK1), wnt5a, interleukin 6 (IL-6), and TNF-alpha, leading to increased production of NO and decreased production of reactive oxygen species (ROS) in a time- and concentration-dependent manner [59]. hUCMSC-derived exosomes enhanced the sensitivity of K562 cells to IM via activation of the caspase signaling pathway by increasing Bax expression and decreasing Bcl-2 expression. Therefore, combining IM with hUCMSC-derived exosomes may be a promising approach to improve the efficacy of CML treatment [60]. In addition, a BCR-ABL transcript was detected in exosomes derived from CML cell lines and the serum of CML patients, reflecting the potential of exosomes as detection targets for BCR-ABL [61].
Exosomes in acute myeloid leukemia
Compared with parent cells, miRNAs were found to be enriched in acute myeloid leukemia (AML) cell-derived exosomes. Nine hematopoiesis/leukemogenesis-related factors (i.e., GATA1, FOXP3, SHIP1, ID1, E2F1, CEBP-a/-b, MEF2C (20) and Myc) and 5 candidate biomarkers (i.e., NPM1, FLT3, CXCR4, MMP9, and IGF-1R) related to the prognosis of AML were detected in these exosomes. Kaplan-Meier analysis also revealed that a high miR-125b level was related to higher cumulative relapse and overall death rates [62]. These findings suggest potentially strong regulation of AML progression by exosomes. Moreover, significantly higher levels of miRNAs, including let-7a, miR-9, miR-99b, miR-150, miR-155, miR-191, and miR-223, have been found in AML cell-derived exosomes, ranging from 2- to 40-fold enrichment compared with the levels in parent cells [63]. Exosomes released by AML cells can be taken up by bone marrow stromal cells (BMSCs). BMSC expression of CXCR4, a target of miR-150, was markedly decreased after co-culture with AML cell-derived exosomes. As a result, the migration of AML cells toward the chemokine SDF-1a was significantly attenuated [63]. Cells of a specific AML subtype, i.e., acute promyelocytic leukemia (APL) cells, also produced considerable numbers of exosomes, which were readily taken up by cultured endothelial cells and triggered their increased survival. All-trans-retinoic acid (ATRA) treatment changes the emission profile of APL-related exosomes, resulting in a preponderance of smaller vesicles, which is an effect that occurs together with the onset of cellular differentiation. ATRA also increases IL-8 mRNA levels and protein contents in APL cells and their exosomes while decreasing VEGF and tissue factor (TF) levels. Endothelial cell uptake of NB4-derived extracellular vesicles results in these cells having higher levels of TF and procoagulant activity, and these effects are diminished by pre-treating the extracellular vesicle donor cells with ATRA [64]. These observations highlight the potential significance of changes in the angiogenic signature and activity associated with exosomes released from AML cells subjected to targeted therapy.
AML cell-derived exosomes have also been shown to participate in the suppression of residual hematopoietic function preceding widespread leukemic bone marrow invasion both directly and indirectly via stromal components. On the one hand, AML cell-derived exosomes downregulate critical retention factors (i.e., SCF and CXCL12) in stromal cells, leading to hematopoietic stem and progenitor cell (HSPC) mobilization from the bone marrow. On the other hand, they directly regulate HSPCs through reduced clonogenicity and decreased expression of CXCR4, c-Kit, and other hematopoietic transcription factors (i.e., c-M). Furthermore, exosomes released from leukemia blasts have been shown to suppress HPC functions indirectly through stromal reprogramming of niche-retention factors and to remodel the bone marrow niche into a leukemia growth-permissive microenvironment [65].
Exosomes in acute/chronic lymphocyte leukemia
Only few studies have explored the relationship between exosomes and central nervous system (CNS) leukemia after acute lymphoblastic leukemia (ALL). CNS leukemia cells have strong chemoresistance and are associated with a high risk of relapse due to penetration of the blood brain barrier (BBB), but the migration mechanism of leukemia cells remains unclear. Kinjyo et al. [5] indicated that the IL-15 containing exosomes released by B acute lymphoblastic leukemia (BCP-ALL) cells contribute to disruption of the BBB in engrafted mice. Knockdown of either IL-15 or IL-15Ra reduced the invasion of BCP-ALL cells into the CNS [66].
The pathogenesis of chronic lymphocytic leukemia (CLL) is stringently associated with a tumor-supportive microenvironment. After being taken up by stromal cells, circulating exosomes can activate AKT/ERK and NF-κB signaling and then stimulate stromal cells to induce inflammatory and protumorigenic environmental conditions, including increased angiogenesis, thus supporting the survival and outgrowth of CLL cells [67]. In addition, this is the first report describing a possible role of leukemia cell-derived exosomes in cancer-associated fibroblast (CAF) formation. Combined with the observation of exosome-mediated angiogenesis and the cross-talk among tumor cells, endothelial cells, and CAFs, tumor-derived exosomes may direct the activities of surrounding cells in hematologic neoplasms. More importantly, the exosomes control the evolution of CLL. A study on the differential contents of plasma-derived exosomes from CLL patients and Richter syndrome patients was performed using an miR direct hybridization plate array. The authors suggested that miRNA-19b was the most statistically significantly upregulated miRNA in CLL. Furthermore, miRNA-19b enhanced the proliferation and even the invasion potential of CLL by downregulating TP53 and upregulating MKI67 indirectly, leading to the evolution of therapy-resistant CLL toward RS [68].
Exosomes in drug resistance
Drug resistance is the main cause of leukemia treatment failure. Notably, stromal cell-derived exosomes also mediate leukemia cell drug resistance. Yeh et al. [69]. showed that CLL plasma-derived exosomes possess an enriched leukemia-associated miRNA signature, including the miR-29 family, miR-150, miR-155, and miR-223; this signature was associated with a poor CLL outcome. The exosome level in the plasma of CLL patients was significantly suppressed by ibrutinib treatment. Moreover, exosomes could counteract the effect of antibody-based drugs by modulating their binding to tumor cells. Lymphoma exosomes carry CD20, which binds therapeutic anti-CD20 antibodies and protects target cells from antibody attack [70].
However, chemoresistance occurs frequently in AML cells. Exosomes have also been found to participate in the development of drug resistance in myeloid neoplasms. yb, Cebp-β, and Hoxa-9) [65]. A recent study indicated that AML cells secrete VEGF/VEGFR-containing exosomes that induce glycolysis in HUVECs, leading to vascular remodeling and acquisition of chemoresistance [8]. AML-derived plasma exosomes mediate the intercellular transfer of regulatory proteins, including B-cell CLL/lymphoma 2 (BCL-2), myeloid cell leukemia 1 (MCL-1), BCL-2-like 1 isoform 1 (BCL-XL) and BCL-2-associated X protein (BAX); thus, these exosomes were identified as important determinants of therapy resistance [71]. Liu et al. [72] found that phosphoinositide 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) signaling and autophagic activity were increased significantly in the IM-resistant CML cell line K562 (K562RIMT). In addition, mTOR-independent beclin-1/Vps34 signaling was shown to be involved in exosomal release from these cells. Dasatinib can promote apoptosis through downregulation of AKT/mTOR activities while also preventing exosomal release and inhibiting autophagy by downregulating the expression of beclin-1 and Vps34 in K562RIMT cells. Min QH [73] indicated that the level of miR-365 was significantly higher in exosomes derived from drug-resistant CML cells than that in exosomes derived from sensitive cells. IM-sensitive CML cells transfected with pre-miR-365 displayed lower chemosensitivity and a decreased apoptosis rate compared with controls. Exosomal transfer of miR-365 induced drug resistance by inhibiting the expression of pro-apoptosis proteins in sensitive CML cells.
Exosomes in immune dysfunction
Understanding the interaction between immune cells and leukemia cells is critical for the development of successful immunotherapeutic strategies. Clayton et al. [74] reported that the proliferation of healthy donor peripheral blood lymphocytes in response to IL-2 was inhibited by tumor exosomes. In addition, this effect was observed in all unfractionated lymphocyte subsets. Separating CD4+ T cells, CD8+ T cells, and natural killer (NK) cells revealed that CD8+ T cell proliferation was not inhibited in the absence of CD4+ T cells and that NK cell proliferation was only slightly impaired. Other exosome effects included selective impairment of IL-2-mediated CD25 upregulation, which affected all subsets except for the CD3+CD8+ T cell subset, indicating an exosome-mediated mechanism of skewing IL-2 responsiveness in favor of regulatory T cells at the expense of cytotoxic cells. Another study showed that Hsp72 on leukemia cell-derived exosomes promotes the immunosuppressive activity of myeloid-derived suppressor cells (MDSCs) by activating the IL-6/STAT3 pathway [75]. Exosomes derived from BMSCs can also enhance the suppressive activity of MDSCs toward T cells via the STAT3/STAT1 pathways and anti-apoptosis proteins BCL-XL and Mcl-1 [76]. Recently, Szczepanskiet et al. [77] showed that AML cell-derived exosomes carried high levels of TGF-β1 and suppressed the immune functions of NK cells by reducing their cytotoxic potential and downregulating the expression of natural-killer group-2 member D (NKG2D). The NKG2D ligand was detected on Jurkat and Raji cell-derived exosomes; this ligand can downregulate NKG2D and impair NK cell function [78]. Reiners et al. [79] found that soluble CLL plasma factors suppressed NK cell cytotoxicity and downregulated the surface receptors CD16 and CD56 on NK cells from healthy donors. The inhibition of NK cell cytotoxicity was attributed to the soluble ligand BAG6/BAT3, which engages the activating receptor NKp30 expressed on NK cells. Moreover, these findings reveal potential therapeutic strategies using exosomes as immunotherapeutic agents to regulate NK cell function in leukemia patients.
Exosome enrichment in patient plasma and its potential role in leukemia therapeutics
In patients with melanoma and other solid tumors, the total protein levels of exosome fractions isolated from plasma have been reported to reflect the disease stage, tumor burden, response to therapy, and even survival, with patients characterized by high exosomal protein levels exhibiting more advanced diseases and shorter survival times [80-82]. In addition, the protein contents of exosomes isolated from the plasma of newly diagnosed leukemia patients have also been found to be enhanced compared with those found in normal control plasma. Exosomes as biomarkers have great potential for the diagnosis of cancer. However, the low concentration of cancer-derived exosomes present in biofluids complicates early diagnosis. A new technology was established using GNP-DNA-FAM conjugates to amplify MB-CD63-labeled leukemia-derived exosomes; thus, concentrations as low as 1 × 102 particles per μL of exosomes could be detected [83]. De Miguel et al. [84] found that exosomes derived from human hematologic tumor cells carried special molecular membrane markers, including TGF-β1, MHC class I polypeptide-related sequence A/B (MICA/MICB) and myeloid blast markers CD34, CD33 and CD117; furthermore, these exosomes were found to attenuate NK cell cytotoxicity. These researchers generated artificial lipid vesicles coated with bioactive Apo2 ligand/TNF-related apoptosis-inducing ligand (Apo2L/TRAIL) that resembled natural exosomes. This lipid framework (i.e., large, unilamellar vesicles (LUVs)-Apo2L/TRAIL) greatly improved Apo2L/TRAIL activity. The liposome-bound Apo2L/TRAIL overcame the resistance to soluble recombinant Apo2L/TRAIL exhibited by Jurkat cell mutants (Jurkat-shBak) and was also effective against other hematologic tumor cells. Furthermore, Szczepanski et al. [77] reported that blast-derived exosomes in sera from patients with AML have elevated protein and TGF-β1 contents and inhibit NK cell cytotoxicity. A potential role of exosomes in predicting responses to chemotherapy was evaluated in AML patients undergoing treatment. At diagnosis, protein and TGF-β1 levels were higher in AML exosomes than those in control exosomes. These values decreased after induction chemotherapy, increased during consolidation chemotherapy, and normalized during long-term, complete remission. Changes in exosomal protein and/or TGF-β1 contents may reflect responses to CT. Exosomal profiles may suggest the presence of residual disease in patients considered to have achieved complete remission [85]. CLL patients also exhibited higher levels of exosomes than healthy controls. CLL-unique microRNAs, including the miR-29 family, miR-150, miR-155 and miR-223, were also detected in plasma-derived exosomes. miR-125b in serum-derived exosomes was recently confirmed to be a potential biomaker for a poor prognosis in intermediate-risk AML patients, including individuals with the FLT3 or MLL mutation [62]. In addition, the number of exosomes was significantly decreased after ibrutinib therapy. Further studies revealed that B cell receptor (BCR) signaling was the key pathway involved in enhanced exosome secretion [69]. Overall, these studies reflect the positive correlation between plasma-derived exosomes and the leukemia cell burden, as well as the considerable potential of plasma-derived exosomes for use as predictors of AML remission after chemotherapy.
Detection of the contents of exosomes in plasma has also been used in CML therapy, especially in clinical trials for tyrosine kinase inhibitor discontinuation [86,87]. Approximately 40% of CML patients who discontinue IM therapy maintain undetectable minimal residual disease for more than one year after stopping IM (STOP-IM). Ohyashiki et al. [88] found that exosomal miR-215 and plasma miR-215 levels were downregulated in the STOP-IM group compared with those in the control group, indicating that the biological relevance of the miR-215 level in plasma and in exosomes is equivalent. Patients with low plasma miR-215 levels had significantly higher total IM intake than patients with elevated miR-215 levels. A functional annotation of miR-215 target genes estimated by the bioinformatic Database for Annotation, Visualization and Integrated Discovery (DAVID) involved the cell cycle, mitosis, DNA repair and cell cycle checkpoints. This study suggests a possible role of miR-215 in successful IM discontinuation. Further prospective studies may provide new insights into the relationship between miR-215 and therapy-free remission in CML patients.
Evidence from a preclinical myocardial infarction model showed that mesenchymal stem cell (MSC) immune-modulating factors are also secreted ex vivo and reside in supernatant fractions that are highly enriched with exosomes that bud from the plasma membrane [89]. Kordelas et al. [90] used MSC-derived exosomes to treat graft-versus-host disease (GVHD). The number of MSC exosomes obtained from the supernatant of 4 × 107 MSCs was calculated as a corresponding dosage for the bodyweight of a specific patient and was defined as 1 unit. Unit quantities were gradually increased and were administered every 2-3 days until a 4 × dosage (i.e., 4 units) was reached. Compared with the values determined before MSC-exosome therapy, the levels of IL-1, TNF-alpha, and IFN-gamma produced by PBMCs were reduced by more than 50% after the last treatment administration. Clinical GVHD symptoms improved significantly shortly after the start of MSC-exosome therapy. Due to the clinical response to MSC-exosome therapy, the steroid dosage could be reduced from 125 mg/d to 30 mg/d. The patient was stable for several months. These findings indicate that MSC-derived exosomes may serve as a new and safe tool for treating therapy-refractory GVHD and potentially for other inflammation- associated diseases. Leukemic stem cells (LSCs) are responsible for AML chemotherapy resistance and relapse. Lower expression of miR-34c-5p in LSCs was closely correlated with an adverse prognosis and poor responses to therapy in AML patients. Furthermore, miR-34c-5p can increase its intracellular level by inhibiting exosome-mediated transfer via a positive feedback loop through RAB27B, a molecule that promotes exosome shedding [91]. Recently, tumor cell-derived exosomes have attracted attention as a source of tumor antigens for use in vaccines [92]. Tumor cell-derived exosomes harbor tumor-related antigens and can induce potent antitumor immune responses [93]. Dendritic cell (DC) immunotherapy has been considered a potential maintenance therapy for leukemia due to its effective reduction of relapse rates and improvement in survival. Exosomes isolated from malignant effusions can transfer tumor antigens to dendritic cells and induce specific cytotoxic T lymphocyte (CTL) responses and antitumor immunity [94,95]. Yao et al. [96] investigated the biological characteristics of exosomes derived from K562 leukemia cells and their ability to induce anti leukemic immunity. They revealed that K562 cell-derived exosomes harbor the BCR-ABL fusion protein, which is expressed in the original K562 cell line. Furthermore, this study showed that K562 cell-derived exosomes can be taken up by DCs in vitro, and that leukemia cell-derived exosome-pulsed DCs induce a stronger antigen-specific anti leukemic CTL immune response in vivo. Vaccines based on leukemia cell-derived exosomes have been suggested as a potential therapy for prolonging the disease-free survival of leukemia patients after chemotherapy or hematopoietic stem cell transplantation. By internalizing AML-derived exosomes, the lethality of DCs was significantly reduced, which was accompanied by decreased production of INF-γ, but the expression of maturation marker CD86, which promotes T cell proliferation, did not change. Interestingly, DCs showed stronger toxicity toward target cells with increased expression of CD86 after incubation with the exosomes of K562 cells [97].
Perspectives
The current review provides a detailed description of exosomal biology and function in the context of leukemia (Table 1). In future studies, the following issues need to be addressed. (i) The isolation of exosomes is difficult because exosomes are very small. Validated isolation methods have certain disadvantages. New methods of isolating and identifying exosomes need to be established, and these methods will effectively promote the precise study of exosomes. (ii) Whether exosome compositions have interactive relationships is unclear. Establishing a nucleic acid-protein interactome map of exosomes from different cells will help to further understand the role of exosomes in angiogenesis, drug resistance and immune suppression. (iii) Studies have shown that exosomes can suppress the immune functions of NK/T cells. Although these results are preliminary, they strongly suggest that exosomes play important roles in leukemia cell immune escape. In view of the great achievements in recent years in the field of leukemia immunotherapy, the relationship between exosomes and leukemia immunity merits further investigation. (iv) The unique molecular membrane markers of exosome fractions isolated from plasma have been reported to reflect the disease stage, leukemia cell burden, response to therapy, and even survival. Thus, one can reasonably speculate that exosomes may be an important indicator of leukemia progression. However, the exact relationship between the levels of exosome protein markers and the state of leukemia has not yet been clarified.
Table 1.
Exosome mediate the cell communication in leukemia hematopoietic microenvironment
| RESOURCE | RECEIVE CELL | KEY MOLECULE | TARGET GENE | PATHWAY | FUNCTION | REFERENCE |
|---|---|---|---|---|---|---|
| AML | HUVECs | VEGF/VEGFR | --- | --- | Induce glycolysis, vascular remodeling and acquisition of chemoresistance | [8] |
| AML | BMSCs | --- | CXCR4 | Promote migration | [63] | |
| AML | BMSCs | --- | CXCR4, c-Kit | --- | Promote HSPC mobilization | [65] |
| AML | --- | --- | BCL-2 | --- | Therapy resistance | [71] |
| AML | MDSCs | --- | --- | IL-6/STAT3 signaling | Immunosuppressive activity | [75] |
| AML | NK cells | TGF-β1 | NKG2D | --- | Suppressed the immune functions | [77] |
| NB4 | ECs | --- | --- | --- | Procoagulant activity | [64] |
| drug-resistant CML | sensitive CML | miR-365 | pro-apoptosis proteins | --- | Drug resistance | [73] |
| CML | ECs | miR-210 | Ephrin-A3 | VEGF signaling | Decresed angiogenesis | [57] |
| K562 | ECs | pre-miR-92a | integrin a5 | --- | Promote cell migration, tube formation | [56] |
| K562 | BMSCs | --- | Cxcl12, DKK1, wnt5a, IL-6 | VEGF signaling | Decreased production of reactive oxygen species (ROS) | [59] |
| BCP-ALL | --- | IL-15 | --- | --- | Disruption of the blood brain barrier (BBB) | [66] |
| CLL | MSCs | --- | -- | AKT/ERK and NF-κB signaling | Induce inflammatory and protumorigenic environmental conditions | [67] |
| CLL | --- | miR-150, miR-155, and miR-223 | --- | --- | Associated with a poor CLL outcome | [69] |
| CLL | NK cells | --- | CD16, CD56 | --- | Suppressed cytotoxicity | [79] |
| APL | ECs | --- | --- | --- | Increased survival | [64] |
Note: leukemia cell derived exosmes affected the other cells of hematopoietic microenvironment via transfor a variety of content. ‘---’ means there are no detailed informations.
Acknowledgements
This research was supported by the National Key Research and Development Program of China (2016YFA0202104), the Chinese National Natural Science Foundation (No. 81370593, No. 81570131 and No. 81570097), the Research Fund from the Clinical Foundation of Army Medical University (2018JSLC0034) and Xinqiao Hospital (2018YQYLY007). We thank Dr. Shicang Yu (Institute of Pathology, Southwest Hospital, Third Military Medical University) and Dr. Xi Zhang (Medical Center of Hematology, The Xinqiao Hospital of Army Medical University) for revising the paper.
Disclosure of conflict of interest
None.
References
- 1.Harding CV, Heuser JE, Stahl PD. Exosomes: looking back three decades and into the future. J Cell Biol. 2013;200:367–371. doi: 10.1083/jcb.201212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 3.Simons M, Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Mathivanan S, Ji H, Simpson RJ. Exosomes: Extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Gross JC, Chaudhary V, Bartscherer K, Boutros M. Active Wnt proteins are secreted on exosomes. Nat Cell Biol. 2012;14:1036–1045. doi: 10.1038/ncb2574. [DOI] [PubMed] [Google Scholar]
- 6.Sato-Kuwabara Y, Melo S, Soares F, Calin G. The fusion of two worlds: non-coding RNAs and extracellular vesicles - diagnostic and therapeutic implications (Review) Int J Oncol. 2014;46:17–27. doi: 10.3892/ijo.2014.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riches A, Campbell E, Borger E, Powis S. Regulation of exosome release from mammary epithelial and breast cancer cells - a new regulatory pathway. Eur J Cancer. 2014;50:1025–1034. doi: 10.1016/j.ejca.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 8.Le MT, Hamar P, Guo C, Basar E, Perdigão-Henriques R, Balaj L, Lieberman J. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest. 2014;124:5109–5128. doi: 10.1172/JCI75695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, Lucci A, Ivan C, Calin GA, Kalluri R. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, Nana-Sinkam SP, Jarjoura D, Marsh CB. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva J, Garcia V, Zaballos A, Provencio M, Lombardia L, Almonacid L, Garcia JM, Dominguez G, Pena C, Diaz R, Herrera M, Varela A, Bonilla F. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J. 2011;37:617–623. doi: 10.1183/09031936.00029610. [DOI] [PubMed] [Google Scholar]
- 14.Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and granules. Blood. 1999;94:3791–3799. [PubMed] [Google Scholar]
- 15.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2009;12:19–30. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 16.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 17.Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, Coscia C, Iessi E, Logozzi M, Molinari A, Colone M, Tatti M, Sargiacomo M, Fais S. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. 2009;284:34211–34222. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savina A, Furlan M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278:20083–20090. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 19.Mohamed NNI, Okada T, Kajimoto T, Nakamura SI. Essential role of sphingosine kinase 2 in the regulation of Cargo contents in the Exosomes from K562 cells. Kobe J Med Sci. 2018;63:E123–E129. [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson MF, Otoc N, Sethi JK, Gupta A, Antes TJ. Integrated systems for exosome investigation. Methods. 2015;87:31–45. doi: 10.1016/j.ymeth.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 21.Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, Hoen ENN, Piper MG, Sivaraman S, Skog J, Thery C, Wauben MH, Hochberg F. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013:2. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caradec J, Kharmate G, Hosseini-Beheshti E, Adomat H, Gleave M, Guns E. Reproducibility and efficiency of serum-derived exosome extraction methods. Clin Biochem. 2014;47:1286–1292. doi: 10.1016/j.clinbiochem.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 23.Momen-Heravi F, Balaj L, Alian S, Trachtenberg AJ, Hochberg FH, Skog J, Kuo WP. Impact of biofluid viscosity on size and sedimentation efficiency of the isolated microvesicles. Front Physiol. 2012;3:162. doi: 10.3389/fphys.2012.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alvarez ML, Khosroheidari M, Ravi RK, DiStefano JK. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012;82:1024–1032. doi: 10.1038/ki.2012.256. [DOI] [PubMed] [Google Scholar]
- 25.Sabapatha A, Gercel-Taylor C, Taylor DD. Specific isolation of placenta-derived exosomes from the circulation of pregnant women and their immunoregulatory consequences. Am J Reprod Immunol. 2006;56:345–355. doi: 10.1111/j.1600-0897.2006.00435.x. [DOI] [PubMed] [Google Scholar]
- 26.Taylor DD, Zacharias W, Gercel-Taylor C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol Biol. 2011;728:235–246. doi: 10.1007/978-1-61779-068-3_15. [DOI] [PubMed] [Google Scholar]
- 27.Ibrahim AG, Cheng K, Marbán E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Reports. 2014;2:606–619. doi: 10.1016/j.stemcr.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munich S, Sobo-Vujanovic A, Buchser WJ, Beer-Stolz D, Vujanovic NL. Dendritic cell exosomes directly kill tumor cells and activate natural killer cells via TNF superfamily ligands. OncoImmunology. 2014;1:1074–1083. doi: 10.4161/onci.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014:3. doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tian T, Zhu YL, Hu FH, Wang YY, Huang NP, Xiao ZD. Dynamics of exosome internalization and trafficking. J Cell Physiol. 2013;228:1487–1495. doi: 10.1002/jcp.24304. [DOI] [PubMed] [Google Scholar]
- 31.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 32.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, Yu Y, Chow A, O’Connor ST, Chin AR, Yen Y, Wang Y, Marcusson EG, Chu P, Wu J, Wu X, Li AX, Li Z, Gao H, Ren X, Boldin MP, Lin PC, Wang SE. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Balkom BW, de Jong OG, Smits M, Brummelman J, den Ouden K, de Bree PM, van Eijndhoven MA, Pegtel DM, Stoorvogel W, Würdinger T, Verhaar MC. Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood. 2013;121:3997–4006. doi: 10.1182/blood-2013-02-478925. [DOI] [PubMed] [Google Scholar]
- 34.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akagi T, Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, Watanabe M, Nakagama H, Yokota J, Kohno T, Tsuchiya N. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, Kohli M, Thibodeau SN, Boardman L, Wang L. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conigliaro A, Costa V, Lo Dico A, Saieva L, Buccheri S, Dieli F, Manno M, Raccosta S, Mancone C, Tripodi M, De Leo G, Alessandro R. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol Cancer. 2015;14:155. doi: 10.1186/s12943-015-0426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, Wang B, Ye G, Xiao B, Guo J. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36:2007–2012. doi: 10.1007/s13277-014-2807-y. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Zhou Y, Lu J, Sun Y, Xiao H, Liu M, Tian L. Combined detection of serum exosomal miR-21 and HOTAIR as diagnostic and prognostic biomarkers for laryngeal squamous cell carcinoma. Med Oncol. 2014;31:148. doi: 10.1007/s12032-014-0148-8. [DOI] [PubMed] [Google Scholar]
- 41.Teo H, Perisic O, Gonzalez B, Williams RL. ESCRT-II, an endosome-associated complex required for protein sorting: crystal structure and interactions with ESCRT-III and membranes. Dev Cell. 2004;7:559–569. doi: 10.1016/j.devcel.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 42.Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kourembanas S. Exosomes: vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol. 2015;77:13–27. doi: 10.1146/annurev-physiol-021014-071641. [DOI] [PubMed] [Google Scholar]
- 44.Tran TH, Mattheolabakis G, Aldawsari H, Amiji M. Exosomes as nanocarriers for immunotherapy of cancer and inflammatory diseases. Clin Immunol. 2015;160:46–58. doi: 10.1016/j.clim.2015.03.021. [DOI] [PubMed] [Google Scholar]
- 45.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol. 2014;14:195–208. doi: 10.1038/nri3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segura E, Amigorena S, Théry C. Mature dendritic cells secrete exosomes with strong ability to induce antigen-specific effector immune responses. Blood Cells Mol Dis. 2005;35:89–93. doi: 10.1016/j.bcmd.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 47.Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9:871–881. doi: 10.1111/j.1600-0854.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mears R, Craven RA, Hanrahan S, Totty N, Upton C, Young SL, Patel P, Selby PJ, Banks RE. Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics. 2004;4:4019–4031. doi: 10.1002/pmic.200400876. [DOI] [PubMed] [Google Scholar]
- 49.Gupta A, Pulliam L. Exosomes as mediators of neuroinflammation. J Neuroinflammation. 2014;11:68. doi: 10.1186/1742-2094-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clayton A, Harris CL, Court J, Mason MD, Morgan BP. Antigen-presenting cell exosomes are protected from complement-mediated lysis by expression of CD55 and CD59. Eur J Immunol. 2003;33:522–531. doi: 10.1002/immu.200310028. [DOI] [PubMed] [Google Scholar]
- 51.Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 52.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 53.Méndez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, MacArthur BD, Lira SA, Scadden DT, Ma’ayan A, Enikolopov GN, Frenette PS. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohyashiki JH, Umezu T, Ohyashiki K. Exosomes promote bone marrow angiogenesis in hematologic neoplasia: the role of hypoxia. Curr Opin Hematol. 2016;23:268–273. doi: 10.1097/MOH.0000000000000235. [DOI] [PubMed] [Google Scholar]
- 55.Mineo M, Garfield SH, Taverna S, Flugy A, De Leo G, Alessandro R, Kohn EC. Exosomes released by K562 chronic myeloid leukemia cells promote angiogenesis in a src-dependent fashion. Angiogenesis. 2011;15:33–45. doi: 10.1007/s10456-011-9241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Umezu T, Ohyashiki K, Kuroda M, Ohyashiki JH. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene. 2012;32:2747–2755. doi: 10.1038/onc.2012.295. [DOI] [PubMed] [Google Scholar]
- 57.Tadokoro H, Umezu T, Ohyashiki K, Hirano T, Ohyashiki JH. Exosomes derived from hypoxic leukemia cells enhance tube formation in endothelial cells. J Biol Chem. 2013;288:34343–34351. doi: 10.1074/jbc.M113.480822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raimondo S, Saieva L, Corrado C, Fontana S, Flugy A, Rizzo A, De Leo G, Alessandro R. Chronic myeloid leukemia-derived exosomes promote tumor growth through an autocrine mechanism. Cell Commun Signal. 2015;13:8. doi: 10.1186/s12964-015-0086-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jafarzadeh N, Safari Z, Pornour M, Amirizadeh N, Forouzandeh MM, Sadeghizadeh M. Alteration of cellular and immune-related properties of bone marrow mesenchymal stem cells and macrophages by K562 chronic myeloid leukemia cell derived exosomes. J Cell Physiol. 2018;234:3697–3710. doi: 10.1002/jcp.27142. [DOI] [PubMed] [Google Scholar]
- 60.Liu Y, Song B, Wei Y, Chen F, Chi Y, Fan H, Liu N, Li Z, Han Z, Ma F. Exosomes from mesenchymal stromal cells enhance imatinib-induced apoptosis in human leukemia cells via activation of caspase signaling pathway. Cytotherapy. 2018;20:181–188. doi: 10.1016/j.jcyt.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 61.Kang KW, Jung JH, Hur W, Park J, Shin H, Choi B, Jeong H, Kim DS, Yu ES, Lee SR, Sung HJ, Kim SJ, Choi CW, Kim HK, Hong S, Park JH, Choi Y, Park Y, Kim BS. The potential of Exosomes derived from chronic myelogenous leukaemia cells as a biomarker. Anticancer Res. 2018;38:3935–3942. doi: 10.21873/anticanres.12679. [DOI] [PubMed] [Google Scholar]
- 62.Jiang L, Deng T, Wang D, Xiao Y. Elevated serum exosomal miR-125b level as a potential marker for poor prognosis in intermediate-risk acute myeloid leukemia. Acta Haematol. 2018;140:183–192. doi: 10.1159/000491584. [DOI] [PubMed] [Google Scholar]
- 63.Huan J, Hornick NI, Shurtleff MJ, Skinner AM, Goloviznina NA, Roberts CT Jr, Kurre P. RNA trafficking by acute myelogenous leukemia exosomes. Cancer Res. 2012;73:918–929. doi: 10.1158/0008-5472.CAN-12-2184. [DOI] [PubMed] [Google Scholar]
- 64.Fang Y, Garnier D, Lee TH, D’Asti E, Montermini L, Meehan B, Rak J. PML-RARa modulates the vascular signature of extracellular vesicles released by acute promyelocytic leukemia cells. Angiogenesis. 2015;19:25–38. doi: 10.1007/s10456-015-9486-1. [DOI] [PubMed] [Google Scholar]
- 65.Huan J, Hornick NI, Goloviznina NA, Kamimae-Lanning AN, David LL, Wilmarth PA, Mori T, Chevillet JR, Narla A, Roberts CT, Loriaux MM, Chang BH, Kurre P. Coordinate regulation of residual bone marrow function by paracrine trafficking of AML exosomes. Leukemia. 2015;29:2285–2295. doi: 10.1038/leu.2015.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kinjyo I, Bragin D, Grattan R, Winter SS, Wilson BS. Leukemia-derived exosomes and cytokines pave the way for entry into the brain. J Leukoc Biol. 2019;105:741–753. doi: 10.1002/JLB.3A0218-054R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paggetti J, Haderk F, Seiffert M, Janji B, Distler U, Ammerlaan W, Kim YJ, Adam J, Lichter P, Solary E, Berchem G, Moussay E. Exosomes released by chronic lymphocytic leukemia cells induce the transition of stromal cells into cancer-associated fibroblasts. Blood. 2015;126:1106–1117. doi: 10.1182/blood-2014-12-618025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jurj A, Pop L, Petrushev B, Pasca S, Dima D, Frinc I, Deak D, Desmirean M, Trifa A, Fetica B, Gafencu G, Selicean S, Moisoiu V, Micu WT, Berce C, Sacu A, Moldovan A, Colita A, Bumbea H, Tanase A, Dascalescu A, Zdrenghea M, Stiufiuc R, Leopold N, Tetean R, Burzo E, Tomuleasa C, Berindan-Neagoe I. Exosome-carried microRNA-based signature as a cellular trigger for the evolution of chronic lymphocytic leukemia into Richter syndrome. Crit Rev Clin Lab Sci. 2018;55:501–515. doi: 10.1080/10408363.2018.1499707. [DOI] [PubMed] [Google Scholar]
- 69.Yeh YY, Ozer HG, Lehman AM, Maddocks K, Yu L, Johnson AJ, Byrd JC. Characterization of CLL exosomes reveals a distinct microRNA signature and enhanced secretion by activation of BCR signaling. Blood. 2015;125:3297–3305. doi: 10.1182/blood-2014-12-618470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aung T, Chapuy B, Vogel D, Wenzel D, Oppermann M, Lahmann M, Weinhage T, Menck K, Hupfeld T, Koch R, Trümper L, Wulf GG. Exosomal evasion of humoral immunotherapy in aggressive B-cell lymphoma modulated by ATP-binding cassette transporter A3. Proc Natl Acad Sci U S A. 2011;108:15336–15341. doi: 10.1073/pnas.1102855108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wojtuszkiewicz A, Schuurhuis GJ, Kessler FL, Piersma SR, Knol JC, Pham TV, Jansen G, Musters RJ, van Meerloo J, Assaraf YG, Kaspers GJ, Zweegman S, Cloos J, Jimenez CR. Exosomes secreted by apoptosis-resistant AML blasts harbor regulatory network proteins potentially involved in antagonism of apoptosis. Mol Cell Proteomics. 2016;15:1281–1298. doi: 10.1074/mcp.M115.052944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu J, Zhang Y, Liu A, Wang J, Li L, Chen X, Gao X, Xue Y, Zhang X, Liu Y. Distinct dasatinib-induced mechanisms of apoptotic response and exosome release in imatinib-resistant human chronic myeloid leukemia cells. Int J Mol Sci. 2016;17:531. doi: 10.3390/ijms17040531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Min QH, Wang XZ, Zhang J, Chen QG, Li SQ, Liu XQ, Li J, Liu J, Yang WM, Jiang YH, Xu YM, Lin J, Gao QF, Sun F, Zhang L, Huang B. Exosomes derived from imatinib-resistant chronic myeloid leukemia cells mediate a horizontal transfer of drug-resistant trait by delivering miR-365. Exp Cell Res. 2018;362:386–393. doi: 10.1016/j.yexcr.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 74.Clayton A, Mitchell JP, Court J, Mason MD, Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 2007;67:7458–7466. doi: 10.1158/0008-5472.CAN-06-3456. [DOI] [PubMed] [Google Scholar]
- 75.Chalmin F, Ladoire S, Mignot G, Vincent J, Bruchard M, Remy-Martin JP, Boireau W, Rouleau A, Simon B, Lanneau D, De Thonel A, Multhoff G, Hamman A, Martin F, Chauffert B, Solary E, Zitvogel L, Garrido C, Ryffel B, Borg C, Apetoh L, Rébé C, Ghiringhelli F. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest. 2010;120:457–471. doi: 10.1172/JCI40483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang J, De Veirman K, De Beule N, Maes K, De Bruyne E, Van Valckenborgh E, Vanderkerken K, Menu E. The bone marrow microenvironment enhances multiple myeloma progression by exosome-mediated activation of myeloid-derived suppressor cells. Oncotarget. 2015;6:43992–44004. doi: 10.18632/oncotarget.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Szczepanski MJ, Szajnik M, Welsh A, Whiteside TL, Boyiadzis M. Blast-derived microvesicles in sera from patients with acute myeloid leukemia suppress natural killer cell function via membrane-associated transforming growth factor-1. Haematologica. 2011;96:1302–1309. doi: 10.3324/haematol.2010.039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zimmer J, Hedlund M, Nagaeva O, Kargl D, Baranov V, Mincheva-Nilsson L. Thermal- and oxidative stress causes enhanced release of NKG2D ligand-bearing immunosuppressive exosomes in leukemia/lymphoma T and B Cells. PLoS One. 2011;6:e16899. doi: 10.1371/journal.pone.0016899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reiners KS, Topolar D, Henke A, Simhadri VR, Kessler J, Sauer M, Bessler M, Hansen HP, Tawadros S, Herling M, Krönke M, Hallek M, von Strandmann EP. Soluble ligands for NK cell receptors promote evasion of chronic lymphocytic leukemia cells from NK cell anti-tumor activity. Blood. 2013;121:3658–3665. doi: 10.1182/blood-2013-01-476606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bergmann C, Strauss L, Wang Y, Szczepanski MJ, Lang S, Johnson JT, Whiteside TL. T regulatory type 1 cells in squamous cell carcinoma of the head and neck: mechanisms of suppression and expansion in advanced disease. Clin Cancer Res. 2008;14:3706–3715. doi: 10.1158/1078-0432.CCR-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Filipazzi P, Burdek M, Villa A, Rivoltini L, Huber V. Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Semin Cancer Biol. 2012;22:342–349. doi: 10.1016/j.semcancer.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 83.Huang L, Wang DB, Singh N, Yang F, Gu N, Zhang XE. A dual-signal amplification platform for sensitive fluorescence biosensing of leukemia-derived exosomes. Nanoscale. 2018;10:20289–20295. doi: 10.1039/c8nr07720g. [DOI] [PubMed] [Google Scholar]
- 84.De Miguel D, Basáñez G, Sánchez D, Malo PG, Marzo I, Larrad L, Naval J, Pardo J, Anel A, Martinez-Lostao L. Liposomes decorated with Apo2L/TRAIL overcome chemoresistance of human hematologic tumor cells. Mol Pharm. 2013;10:893–904. doi: 10.1021/mp300258c. [DOI] [PubMed] [Google Scholar]
- 85.Hong CS, Muller L, Whiteside TL, Boyiadzis M. Plasma exosomes as markers of therapeutic response in patients with acute myeloid leukemia. Front Immunol. 2014;5:160. doi: 10.3389/fimmu.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mahon FX, Rea D, Guilhot J, Guilhot F, Huguet F, Nicolini F, Legros L, Charbonnier A, Guerci A, Varet B, Etienne G, Reiffers J, Rousselot P. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre stop imatinib (STIM) trial. Lancet Oncol. 2010;11:1029–1035. doi: 10.1016/S1470-2045(10)70233-3. [DOI] [PubMed] [Google Scholar]
- 87.Ross DM, Branford S, Seymour JF, Schwarer AP, Arthur C, Yeung DT, Dang P, Goyne JM, Slader C, Filshie RJ, Mills AK, Melo JV, White DL, Grigg AP, Hughes TP. Safety and efficacy of imatinib cessation for CML patients with stable undetectable minimal residual disease: results from the TWISTER study. Blood. 2013;122:515–522. doi: 10.1182/blood-2013-02-483750. [DOI] [PubMed] [Google Scholar]
- 88.Ohyashiki K, Umezu T, Katagiri S, Kobayashi C, Azuma K, Tauchi T, Okabe S, Fukuoka Y, Ohyashiki JH. Downregulation of plasma miR-215 in chronic myeloid leukemia patients with successful discontinuation of imatinib. Int J Mol Sci. 2016;17:570. doi: 10.3390/ijms17040570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lai RC, Arslan F, Lee MM, Sze NS, Choo A, Chen TS, Salto-Tellez M, Timmers L, Lee CN, El Oakley RM, Pasterkamp G, de Kleijn DP, Lim SK. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4:214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 90.Kordelas L, Rebmann V, Ludwig AK, Radtke S, Ruesing J, Doeppner TR, Epple M, Horn PA, Beelen DW, Giebel B. MSC-derived exosomes: a novel tool to treat therapy-refractory graft-versus-host disease. Leukemia. 2014;28:970–973. doi: 10.1038/leu.2014.41. [DOI] [PubMed] [Google Scholar]
- 91.Peng D, Wang H, Li L, Ma X, Chen Y, Zhou H, Luo Y, Xiao Y, Liu L. miR-34c-5p promotes eradication of acute myeloid leukemia stem cells by inducing senescence through selective RAB27B targeting to inhibit exosome shedding. Leukemia. 2018;32:1180–1188. doi: 10.1038/s41375-018-0015-2. [DOI] [PubMed] [Google Scholar]
- 92.Kim JV, Latouche JB, Rivière I, Sadelain M. The ABCs of artificial antigen presentation. Nat Biotechnol. 2004;22:403–410. doi: 10.1038/nbt955. [DOI] [PubMed] [Google Scholar]
- 93.Thery C, Duban L, Segura E, Veron P, Lantz O, Amigorena S. Indirect activation of naive CD4+ T cells by dendritic cell-derived exosomes. Nat Immunol. 2002;3:1156–1162. doi: 10.1038/ni854. [DOI] [PubMed] [Google Scholar]
- 94.Altieri SL, Khan AN, Tomasi TB. Exosomes from plasmacytoma cells as a tumor vaccine. J Immunother. 2004;27:282–288. doi: 10.1097/00002371-200407000-00004. [DOI] [PubMed] [Google Scholar]
- 95.Hao S, Bai O, Yuan J, Qureshi M, Xiang J. Dendritic cell-derived exosomes stimulate stronger CD8+ CTL responses and antitumor immunity than tumor cell-derived exosomes. Cell Mol Immunol. 2006;3:205–211. [PubMed] [Google Scholar]
- 96.Yao Y, Wang C, Wei W, Shen C, Deng X, Chen L, Ma L, Hao S. Dendritic cells pulsed with leukemia cell-derived exosomes more efficiently induce antileukemic immunities. PLoS One. 2014;9:e91463. doi: 10.1371/journal.pone.0091463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Benites BD, da Silva Santos Duarte A, Longhini ALF, Santos I, Alvarez MC, de Morais Ribeiro LN, Paula E, Saad STO. Exosomes in the serum of acute myeloid leukemia patients induce dendritic cell tolerance: implications for immunotherapy. Vaccine. 2019;37:1377–1383. doi: 10.1016/j.vaccine.2019.01.079. [DOI] [PubMed] [Google Scholar]