In the title compound, intermolecular N—H⋯O hydrogen bonds link the molecules into a three-dimensional supramolecular network.
Keywords: crystal structure, Hirshfeld surface analysis, pyridazine, pyridazine derivative, pyridazinone
Abstract
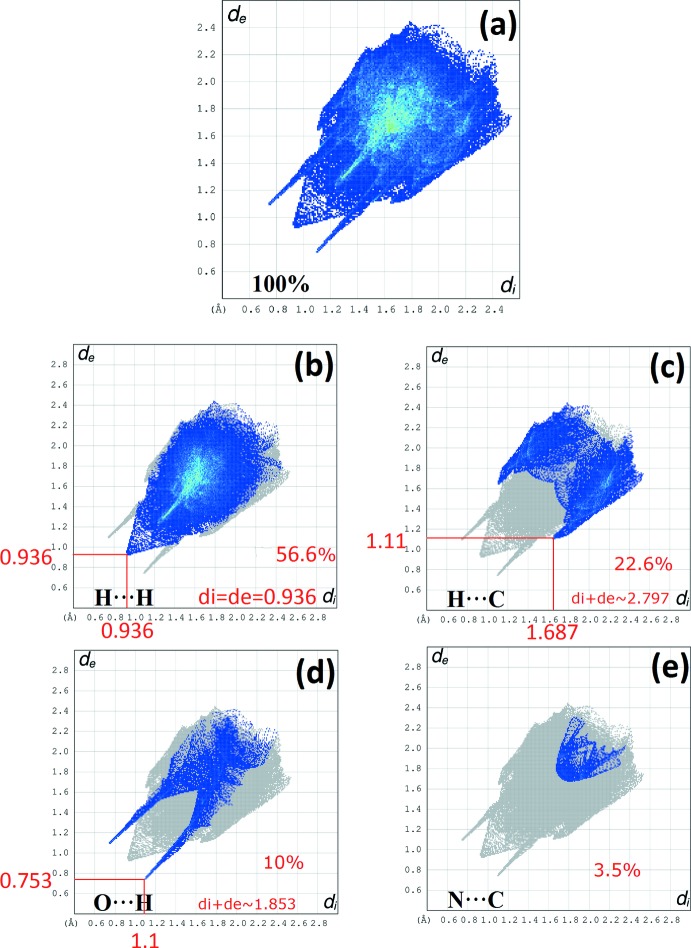
In this paper, we describe the synthesis of a new dihydro-2H-pyridazin-3-one derivative. The molecule, C18H16N2O, is not planar; the benzene and pyridazine rings are twisted with respect to each other, making a dihedral angle of 11.47 (2)°, and the toluene ring is nearly perpendicular to the pyridazine ring, with a dihedral angle of 89.624 (1)°. The molecular conformation is stabilized by weak intramolecular C—H⋯N contacts. In the crystal, pairs of N—H⋯O hydrogen bonds link the molecules into inversion dimers with an R 2 2(8) ring motif. The intermolecular interactions were investigated using Hirshfeld surface analysis and two-dimensional (2D) fingerprint plots, revealing that the most important contributions for the crystal packing are from H⋯H (56.6%), H⋯C/C⋯H (22.6%), O⋯H/H⋯O (10.0%) and N⋯C/C⋯N (3.5%) interactions.
Chemical context
Pyridazines are an important family of six-membered aromatic heterocycles containing two N atoms. Pyridazinone is an important pharmacophore possessing a wide range of biological applications (Asif, 2014 ▸; Akhtar et al., 2016 ▸). The chemistry of pyridazinones has been an interesting field of study for decades and this nitrogen heterocycle has become a scaffold of choice for the development of potential drug candidates (Dubey & Bhosle, 2015 ▸; Thakur et al., 2010 ▸). A review of the literature has revealed that substituted pyridazinones have received a lot of attention in recent years because of their significant potential as antimicrobial (Sönmez et al., 2006 ▸), antidepressant (Boukharsa et al., 2016 ▸), anti-inflammatory (Barberot et al., 2018 ▸), antihypertensive (Siddiqui et al., 2011 ▸), analgesic (Gökçe et al., 2009 ▸), anti-HIV (Livermore et al., 1993 ▸), anticonvulsant (Partap et al., 2018 ▸; Sharma et al., 2014 ▸), cardiotonic (Wang et al., 2008 ▸), antihistaminic (Tao et al., 2012 ▸), glucan synthase inhibitors (Zhou et al., 2011 ▸), phosphodiesterase (PDE) inhibitors (Ochiai et al., 2012 ▸) and herbicidal agents (Asif, 2013 ▸). In continuation of our work in this field (El Kali et al., 2019 ▸; Chkirate et al., 2019a ▸,b ▸; Karrouchi et al., 2015 ▸, 2016a ▸,b ▸), we report the synthesis and the crystal and molecular structures of the title compound, as well as an analysis of its Hirshfeld surface.
Structural commentary
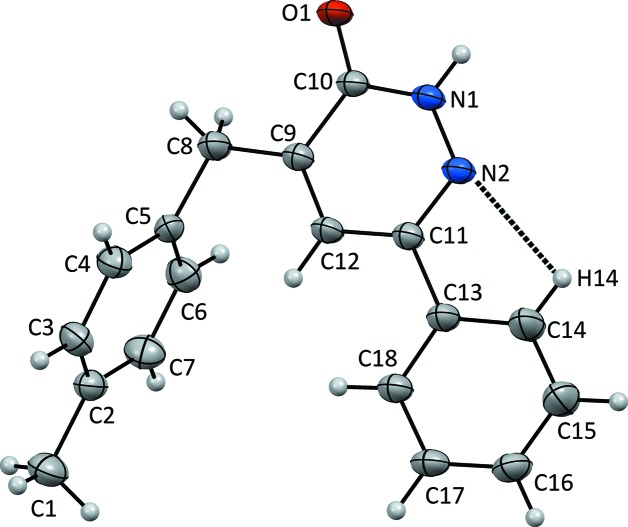
In the title molecule (Fig. 1 ▸), the C10=O1 bond length is 1.241 (3) Å while the N1—N2 and C11=N2 bond lengths are 1.347 (3) and 1.311 (4) Å, respectively (Table 1 ▸). The C9—C8—C5 bond angle is 113.7 (2)°, while the C4—C5—C8—C9, C6—C5—C8—C9 and C10—C9—C8—C5 torsion angles are 90.0 (3), −87.1 (3) and 169.1 (3)°, respectively. The molecule is not planar as the benzene and pyridazine rings are twisted with respect to each other, making a dihedral angle of 11.469 (2)°. The toluene ring is nearly perpendicular to the pyridazine ring, with a dihedral angle of 89.624 (1)°.
Figure 1.
The molecular structure of the title compound, with the atom labelling. Displacement elipsoids are drawn at the 20% probability level.
Table 1. Selected geometric parameters (Å, °).
| O1—C10 | 1.241 (3) | N1—C10 | 1.352 (4) |
| N1—N2 | 1.347 (3) | N2—C11 | 1.311 (4) |
| O1—C10—N1 | 120.9 (3) | C10—C9—C8 | 117.5 (2) |
| O1—C10—C9 | 123.9 (3) | C9—C8—C5 | 113.7 (2) |
| N1—N2—C11—C13 | 177.4 (3) | C4—C5—C8—C9 | 90.3 (4) |
| C10—C9—C8—C5 | 169.2 (3) | C6—C5—C8—C9 | −86.8 (4) |
Supramolecular features
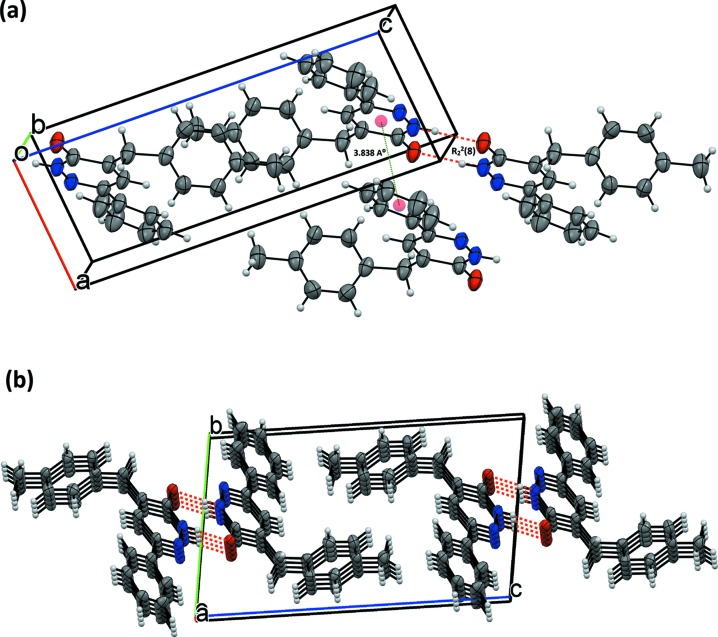
The molecules are connected two-by-two through N1—H1⋯O1 hydrogen bonds (Table 2 ▸), with a  (8) graph-set motif (Bernstein et al., 1995 ▸), and form inversion dimers (Fig. 2 ▸
a). Weak C—H⋯O hydrogen bonds and weak off-set π-stacking stabilize the packing. In the crystal, hydrogen bonds link the chains into a two-dimensional (2D) network parallel to (011) (Fig. 2 ▸
b and Table 2 ▸). The stacking occurs between the pyridazine rings of inversion-related molecules [Cg1⋯Cg3 (at x − 1, y, z)], with a centroid-to-centroid distance of 3.8333 (18) Å and a slippage of 1.460 Å (Cg1 is the centroid of the C9–C11/N1/N2 ring and Cg3 is the centroid of the C13–C18 ring) (Fig. 2 ▸
a).
(8) graph-set motif (Bernstein et al., 1995 ▸), and form inversion dimers (Fig. 2 ▸
a). Weak C—H⋯O hydrogen bonds and weak off-set π-stacking stabilize the packing. In the crystal, hydrogen bonds link the chains into a two-dimensional (2D) network parallel to (011) (Fig. 2 ▸
b and Table 2 ▸). The stacking occurs between the pyridazine rings of inversion-related molecules [Cg1⋯Cg3 (at x − 1, y, z)], with a centroid-to-centroid distance of 3.8333 (18) Å and a slippage of 1.460 Å (Cg1 is the centroid of the C9–C11/N1/N2 ring and Cg3 is the centroid of the C13–C18 ring) (Fig. 2 ▸
a).
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1i | 0.86 | 1.98 | 2.836 (3) | 175 |
| C14—H14⋯N2 | 0.93 | 2.43 | 2.764 (3) | 101 |
Symmetry code: (i) 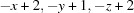 .
.
Figure 2.
(a) A view along the c-axis direction of the title structure. Red dashed lines denote N—H⋯O hydrogen bonds. (b) A view along the a-axis direction of the title compound (Xu et al., 2005 ▸).
Database survey
A search of the Cambridge Structural Database (CSD, Version 5.40, update of November 2018; Groom et al., 2016 ▸) using 4-ethyl-6-methylpyridazin-3(2H)-one (see A in Scheme) as the main skeleton revealed the presence of two structures containing the pyridazine moiety with different substituents similar to the title compound in this study. The structures are 4-benzyl-6-p-tolylpyridazin-3(2H)-one (CSD refcode YOTVIN; Oubair et al., 2009 ▸) and 4-aryl-2,5-dioxo-8-phenylpyrido[2,3-d]pyridazines (BARQUG; Pita et al., 2000 ▸). In YOTVIN, the molecules are connected two-by-two through N—H⋯O hydrogen bonds, with an 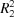 (8) graph-set motif, building a pseudo-dimer arranged around the inversion centre. Weak C—H⋯O hydrogen bonds and weak off-set π–π stacking stabilize the packing. In BARQUG, the dihedral angle between the least-squares planes of the substituted phenyl and pyridone rings is 79.78 (2)° and between the pyridazinone ring and the unsubstitued phenyl ring is 57.37 (2)°.
(8) graph-set motif, building a pseudo-dimer arranged around the inversion centre. Weak C—H⋯O hydrogen bonds and weak off-set π–π stacking stabilize the packing. In BARQUG, the dihedral angle between the least-squares planes of the substituted phenyl and pyridone rings is 79.78 (2)° and between the pyridazinone ring and the unsubstitued phenyl ring is 57.37 (2)°.
Hirshfeld surface (HS) analysis
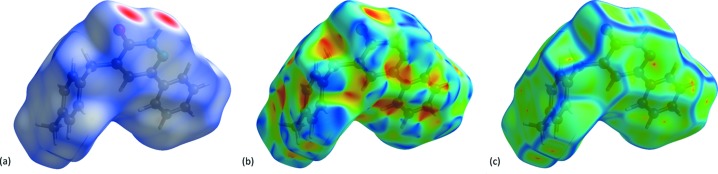
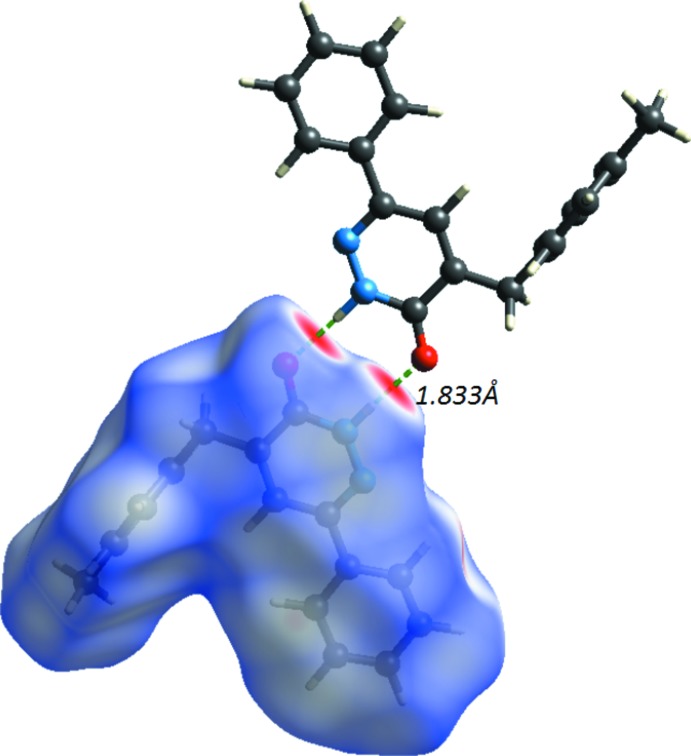
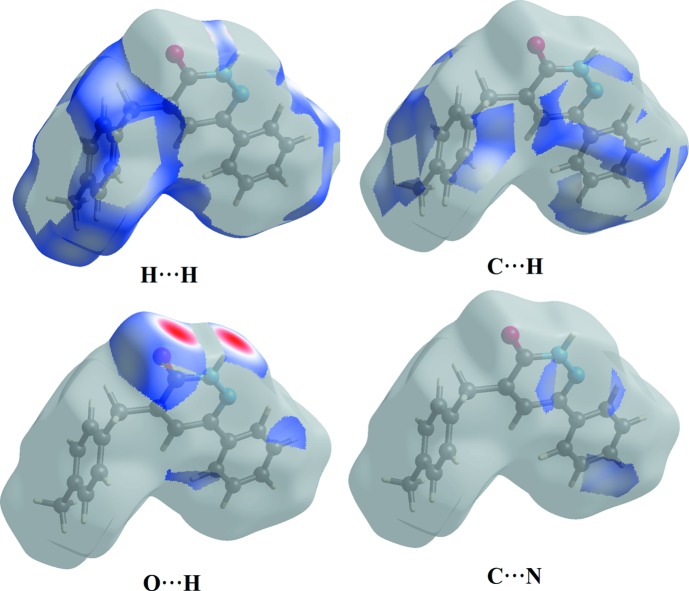
The Hirshfeld surface analysis (Spackman & Jayatilaka, 2009 ▸) and the associated 2D fingerprint plots (McKinnon et al., 2007 ▸) were performed with CrystalExplorer17 (Turner et al., 2017 ▸). The Hirshfeld surface was calculated using a standard (high) surface resolution with the three-dimensional (3D) d norm surface plotted over a fixed colour scale of −0.6048 (red) to 1.4188 a.u. (blue). The 3D d norm surface of the title complex is illustrated in Figs. 3 ▸(a) and 4 ▸. The pale-red spots symbolize short contacts and negative d norm values on the surface correspond to the N—H⋯O interactions (Table 2 ▸). The overall 2D fingerprint plot and the 2D fingerprint plots for the H⋯H, H⋯C/C⋯H, H⋯O/O⋯H and N⋯C/C⋯N contacts are shown in Fig. 5 ▸ (McKinnon et al., 2007 ▸), respectively, associated with their relative contributions to the Hirshfeld surface. The largest interaction is H⋯H, contributing 56.6% to the overall crystal packing. In the fingerprint plot representing H⋯H contacts, the 56.6% contribution to the overall crystal packing, is reflected by widely scattered points of high density due to the large hydrogen content of the molecule. The single spike in the centre at d e = d i = 0.936 Å in Fig. 5 ▸(b) is due to short interatomic H⋯H contacts. In the absence of C—H⋯π interactions in the crystal, the pair of characteristic wings in the fingerprint plot representing H⋯C/C⋯H contacts (22.6% contribution to the HS) have a symmetrical distribution of points (Fig. 5 ▸ c), with the tips at d e + d i = 2.797 Å. The O⋯H (Fig. 5 ▸ d) contacts contribute 10% to the HS and have a symmetrical distribution of points, with the tips at d e + d i = 1.853 Å. The contribution of the other contact to the Hirshfeld surface is N⋯C/C⋯N (3.5%). The Hirshfeld surface representations with the function d norm plotted on the surface are shown for the H⋯H, H⋯C/C⋯H, H⋯O/O⋯H, C⋯C and H⋯N/N⋯H interactions in Figs. 6 ▸. The Hirshfeld surface analysis confirms the importance of H-atom contacts in establishing the packing. The large number of H⋯H, H⋯C/C⋯H, H⋯O/O⋯H, C⋯C and H⋯N/N⋯H interactions suggest that van der Waals interactions and hydrogen bonding play the major roles in the crystal packing (Hathwar et al., 2015 ▸).
Figure 3.
(a) d norm mapped on the Hirshfeld surface for visualizing the intermolecular interactions; (b) shape-index map; (c) curvedness map of the title compound.
Figure 4.
d norm mapped on the Hirshfeld surface for visualizing the intermolecular interactions and showing the dimer formed by inversion-related N—H⋯O hydrogen bonds.
Figure 5.
(a) The overall 2D fingerprint plot and (b) H⋯H, (c) C⋯H, (d) O⋯H and (e) N⋯C interactions are shown.
Figure 6.
Hirshfeld surface representation with the function d norm plotted on the surface for H⋯H, C⋯H, O⋯H and N⋯C interactions.
A shape-index map of the title compound was generated in the range −1 to 1 Å (Fig. 3 ▸ b). The convex blue regions on the shape-index symbolize hydrogen-donor groups and the concave red regions symbolize hydrogen-acceptor groups. The π–π interactions on the shape-index map of the Hirshfeld surface are generally indicated by adjacent red and blue triangles.
A curvedness map of the title compound was generated in the range −4 to 0.4 Å (Fig. 3 ▸ c). This shows large regions of green indicating a relatively flat surface area (planar), while the blue regions indicate areas of curvature. The presence of π–π stacking interactions is also evident in the flat regions around the rings on the Hirshfeld surface plotted over curvedness (see the Supramolecular features section above).
Synthesis and crystallization
To a solution (0.15 g, 1 mmol) of 6-phenyl-4,5-dihydropyridazin-3(2H)-one and (0.12 g, 1 mmol) of 4-methylbenzaldehyde in ethanol (30 ml), sodium hydroxide (10%, 0.5 g, 3.5 mmol) was added. The solvent was evaporated under vacuum and the residue was purified through silica-gel column chromatography using hexane/ethyl acetate (7:3 v/v). Slow evaporation at room temperature leads to single crystals.
Refinement
H atoms were fixed geometrically and treated as riding, with C—H = 0.97 Å and U iso(H) = 1.5U eq(C) for methyl, C—H = 0.96 Å and U iso(H) = 1.2U eq(C) for methylene, C—H = 0.93 Å and U iso(H) = 1.2U eq(C) for aromatic and C—H = 0.98 Å and U iso(H) = 1.2U eq(C) for methine H atoms. Crystal data, data collection and structure refinement details are summarized in Table 3 ▸.
Table 3. Experimental details.
| Crystal data | |
| Chemical formula | C18H16N2O |
| M r | 276.33 |
| Crystal system, space group | Triclinic, P
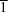
|
| Temperature (K) | 296 |
| a, b, c (Å) | 5.8479 (5), 8.5738 (7), 15.2439 (12) |
| α, β, γ (°) | 80.693 (6), 83.147 (7), 78.164 (7) |
| V (Å3) | 735.27 (11) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.08 |
| Crystal size (mm) | 0.27 × 0.20 × 0.06 |
| Data collection | |
| Diffractometer | Stoe IPDS 2 |
| Absorption correction | Integration (X-RED32; Stoe & Cie, 2002 ▸) |
| T min, T max | 0.966, 0.996 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 9453, 2887, 1471 |
| R int | 0.086 |
| (sin θ/λ)max (Å−1) | 0.617 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.068, 0.208, 1.05 |
| No. of reflections | 2887 |
| No. of parameters | 191 |
| No. of restraints | 84 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.30, −0.32 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989019011551/mw2146sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989019011551/mw2146Isup3.hkl
CCDC reference: 1947718
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors acknowledge the Faculty of Arts and Sciences, Ondokuz Mayıs University, Turkey, for the use of the Stoe IPDS 2 diffractometer (purchased under grant F.279 of the University Research Fund).
supplementary crystallographic information
Crystal data
| C18H16N2O | Z = 2 |
| Mr = 276.33 | F(000) = 292 |
| Triclinic, P1 | Dx = 1.248 Mg m−3 |
| a = 5.8479 (5) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 8.5738 (7) Å | Cell parameters from 8216 reflections |
| c = 15.2439 (12) Å | θ = 2.5–30.7° |
| α = 80.693 (6)° | µ = 0.08 mm−1 |
| β = 83.147 (7)° | T = 296 K |
| γ = 78.164 (7)° | Prism, colorless |
| V = 735.27 (11) Å3 | 0.27 × 0.20 × 0.06 mm |
Data collection
| Stoe IPDS 2 diffractometer | 2887 independent reflections |
| Radiation source: sealed X-ray tube, 12 x 0.4 mm long-fine focus | 1471 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.086 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 26.0°, θmin = 2.5° |
| rotation method scans | h = −7→7 |
| Absorption correction: integration (X-RED32; Stoe & Cie, 2002) | k = −10→10 |
| Tmin = 0.966, Tmax = 0.996 | l = −18→18 |
| 9453 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: dual space |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.068 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.208 | H-atom parameters constrained |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0939P)2] where P = (Fo2 + 2Fc2)/3 |
| 2887 reflections | (Δ/σ)max < 0.001 |
| 191 parameters | Δρmax = 0.30 e Å−3 |
| 84 restraints | Δρmin = −0.32 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.9692 (4) | 0.6487 (3) | 0.90252 (15) | 0.0776 (7) | |
| N1 | 0.7762 (4) | 0.4423 (3) | 0.94429 (17) | 0.0653 (7) | |
| H1 | 0.848007 | 0.419860 | 0.992137 | 0.078* | |
| N2 | 0.6315 (4) | 0.3435 (3) | 0.93530 (17) | 0.0621 (7) | |
| C10 | 0.8227 (5) | 0.5725 (4) | 0.8874 (2) | 0.0627 (8) | |
| C13 | 0.3636 (6) | 0.2661 (4) | 0.8525 (2) | 0.0728 (8) | |
| C11 | 0.5151 (5) | 0.3799 (4) | 0.8641 (2) | 0.0594 (8) | |
| C5 | 0.6414 (6) | 0.7618 (4) | 0.6536 (2) | 0.0668 (9) | |
| C9 | 0.6964 (5) | 0.6101 (4) | 0.8080 (2) | 0.0630 (8) | |
| C12 | 0.5470 (5) | 0.5134 (4) | 0.7992 (2) | 0.0671 (9) | |
| H12 | 0.463050 | 0.535285 | 0.749060 | 0.080* | |
| C2 | 0.4368 (7) | 0.7711 (4) | 0.4949 (2) | 0.0738 (10) | |
| C18 | 0.2108 (6) | 0.2984 (4) | 0.7881 (2) | 0.0750 (8) | |
| H18 | 0.200948 | 0.394224 | 0.748873 | 0.090* | |
| C3 | 0.3212 (7) | 0.8506 (5) | 0.5622 (3) | 0.0806 (11) | |
| H3 | 0.170292 | 0.909110 | 0.555296 | 0.097* | |
| C17 | 0.0695 (6) | 0.1901 (5) | 0.7802 (3) | 0.0812 (9) | |
| H17 | −0.034226 | 0.214814 | 0.736029 | 0.097* | |
| C8 | 0.7452 (7) | 0.7504 (4) | 0.7414 (2) | 0.0793 (11) | |
| H8A | 0.913650 | 0.742443 | 0.729872 | 0.095* | |
| H8B | 0.682622 | 0.848692 | 0.766918 | 0.095* | |
| C16 | 0.0804 (7) | 0.0510 (5) | 0.8351 (3) | 0.0895 (10) | |
| H16 | −0.016404 | −0.019967 | 0.829942 | 0.107* | |
| C4 | 0.4200 (7) | 0.8474 (4) | 0.6402 (2) | 0.0770 (10) | |
| H4 | 0.335386 | 0.904240 | 0.684478 | 0.092* | |
| C6 | 0.7593 (6) | 0.6817 (5) | 0.5855 (3) | 0.0819 (11) | |
| H6 | 0.910392 | 0.623176 | 0.591920 | 0.098* | |
| C7 | 0.6572 (7) | 0.6867 (5) | 0.5077 (3) | 0.0870 (11) | |
| H7 | 0.741083 | 0.631025 | 0.462885 | 0.104* | |
| C1 | 0.3250 (8) | 0.7769 (6) | 0.4096 (3) | 0.1041 (14) | |
| H1A | 0.164486 | 0.831458 | 0.415749 | 0.156* | |
| H1B | 0.330986 | 0.669253 | 0.398093 | 0.156* | |
| H1C | 0.408494 | 0.833710 | 0.360869 | 0.156* | |
| C14 | 0.3757 (8) | 0.1221 (5) | 0.9057 (3) | 0.1055 (11) | |
| H14 | 0.482967 | 0.094249 | 0.948657 | 0.127* | |
| C15 | 0.2333 (8) | 0.0152 (6) | 0.8979 (3) | 0.1111 (11) | |
| H15 | 0.243396 | −0.081609 | 0.936290 | 0.133* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0901 (16) | 0.0843 (16) | 0.0741 (15) | −0.0425 (13) | −0.0351 (12) | −0.0038 (12) |
| N1 | 0.0704 (16) | 0.0782 (18) | 0.0577 (16) | −0.0304 (14) | −0.0247 (13) | −0.0051 (14) |
| N2 | 0.0635 (15) | 0.0736 (18) | 0.0579 (16) | −0.0271 (13) | −0.0179 (12) | −0.0074 (13) |
| C10 | 0.0695 (19) | 0.069 (2) | 0.059 (2) | −0.0260 (16) | −0.0192 (15) | −0.0100 (16) |
| C13 | 0.0799 (18) | 0.0761 (18) | 0.0732 (19) | −0.0356 (16) | −0.0284 (15) | −0.0002 (15) |
| C11 | 0.0597 (18) | 0.066 (2) | 0.0576 (19) | −0.0200 (15) | −0.0160 (15) | −0.0061 (16) |
| C5 | 0.078 (2) | 0.0570 (19) | 0.073 (2) | −0.0278 (17) | −0.0288 (18) | 0.0040 (17) |
| C9 | 0.0674 (19) | 0.066 (2) | 0.062 (2) | −0.0214 (16) | −0.0219 (15) | −0.0037 (16) |
| C12 | 0.0689 (19) | 0.074 (2) | 0.067 (2) | −0.0271 (17) | −0.0306 (16) | −0.0010 (17) |
| C2 | 0.092 (3) | 0.077 (2) | 0.061 (2) | −0.036 (2) | −0.0167 (19) | −0.0020 (18) |
| C18 | 0.0798 (18) | 0.0817 (18) | 0.0737 (18) | −0.0301 (15) | −0.0258 (15) | −0.0083 (15) |
| C3 | 0.078 (2) | 0.086 (3) | 0.078 (3) | −0.008 (2) | −0.028 (2) | −0.007 (2) |
| C17 | 0.0827 (18) | 0.093 (2) | 0.083 (2) | −0.0327 (16) | −0.0299 (16) | −0.0198 (16) |
| C8 | 0.096 (3) | 0.079 (2) | 0.075 (2) | −0.043 (2) | −0.034 (2) | 0.0059 (19) |
| C16 | 0.098 (2) | 0.093 (2) | 0.095 (2) | −0.0501 (18) | −0.0263 (17) | −0.0138 (17) |
| C4 | 0.087 (3) | 0.080 (2) | 0.068 (2) | −0.012 (2) | −0.0180 (19) | −0.0154 (19) |
| C6 | 0.068 (2) | 0.083 (3) | 0.095 (3) | −0.0052 (19) | −0.020 (2) | −0.015 (2) |
| C7 | 0.090 (3) | 0.102 (3) | 0.073 (3) | −0.017 (2) | −0.006 (2) | −0.026 (2) |
| C1 | 0.132 (4) | 0.117 (3) | 0.077 (3) | −0.044 (3) | −0.038 (2) | −0.006 (2) |
| C14 | 0.121 (2) | 0.101 (2) | 0.110 (2) | −0.0590 (19) | −0.0567 (19) | 0.0217 (18) |
| C15 | 0.128 (2) | 0.102 (2) | 0.119 (2) | −0.0644 (19) | −0.0481 (19) | 0.0192 (19) |
Geometric parameters (Å, º)
| O1—C10 | 1.241 (3) | C18—H18 | 0.9300 |
| N1—N2 | 1.347 (3) | C3—C4 | 1.378 (5) |
| N1—C10 | 1.352 (4) | C3—H3 | 0.9300 |
| N1—H1 | 0.8600 | C17—C16 | 1.336 (5) |
| N2—C11 | 1.311 (4) | C17—H17 | 0.9300 |
| C10—C9 | 1.449 (4) | C8—H8A | 0.9700 |
| C13—C14 | 1.357 (5) | C8—H8B | 0.9700 |
| C13—C18 | 1.364 (4) | C16—C15 | 1.344 (6) |
| C13—C11 | 1.488 (4) | C16—H16 | 0.9300 |
| C11—C12 | 1.415 (4) | C4—H4 | 0.9300 |
| C5—C4 | 1.372 (5) | C6—C7 | 1.381 (5) |
| C5—C6 | 1.375 (5) | C6—H6 | 0.9300 |
| C5—C8 | 1.515 (4) | C7—H7 | 0.9300 |
| C9—C12 | 1.353 (4) | C1—H1A | 0.9600 |
| C9—C8 | 1.496 (4) | C1—H1B | 0.9600 |
| C12—H12 | 0.9300 | C1—H1C | 0.9600 |
| C2—C3 | 1.359 (5) | C14—C15 | 1.386 (5) |
| C2—C7 | 1.361 (5) | C14—H14 | 0.9300 |
| C2—C1 | 1.512 (5) | C15—H15 | 0.9300 |
| C18—C17 | 1.391 (4) | ||
| N2—N1—C10 | 128.0 (2) | C16—C17—H17 | 119.5 |
| N2—N1—H1 | 116.0 | C18—C17—H17 | 119.5 |
| C10—N1—H1 | 116.0 | C9—C8—C5 | 113.7 (2) |
| C11—N2—N1 | 116.5 (3) | C9—C8—H8A | 108.8 |
| O1—C10—N1 | 120.9 (3) | C5—C8—H8A | 108.8 |
| O1—C10—C9 | 123.9 (3) | C9—C8—H8B | 108.8 |
| N1—C10—C9 | 115.1 (2) | C5—C8—H8B | 108.8 |
| C14—C13—C18 | 116.8 (3) | H8A—C8—H8B | 107.7 |
| C14—C13—C11 | 120.7 (3) | C17—C16—C15 | 119.1 (3) |
| C18—C13—C11 | 122.5 (3) | C17—C16—H16 | 120.5 |
| N2—C11—C12 | 121.3 (3) | C15—C16—H16 | 120.5 |
| N2—C11—C13 | 116.2 (3) | C5—C4—C3 | 121.0 (4) |
| C12—C11—C13 | 122.4 (3) | C5—C4—H4 | 119.5 |
| C4—C5—C6 | 117.0 (3) | C3—C4—H4 | 119.5 |
| C4—C5—C8 | 121.1 (4) | C5—C6—C7 | 121.1 (4) |
| C6—C5—C8 | 121.9 (3) | C5—C6—H6 | 119.4 |
| C12—C9—C10 | 117.4 (3) | C7—C6—H6 | 119.4 |
| C12—C9—C8 | 125.0 (3) | C2—C7—C6 | 121.7 (4) |
| C10—C9—C8 | 117.5 (2) | C2—C7—H7 | 119.1 |
| C9—C12—C11 | 121.7 (3) | C6—C7—H7 | 119.1 |
| C9—C12—H12 | 119.1 | C2—C1—H1A | 109.5 |
| C11—C12—H12 | 119.1 | C2—C1—H1B | 109.5 |
| C3—C2—C7 | 117.0 (3) | H1A—C1—H1B | 109.5 |
| C3—C2—C1 | 121.2 (4) | C2—C1—H1C | 109.5 |
| C7—C2—C1 | 121.8 (4) | H1A—C1—H1C | 109.5 |
| C13—C18—C17 | 120.9 (4) | H1B—C1—H1C | 109.5 |
| C13—C18—H18 | 119.6 | C13—C14—C15 | 122.0 (4) |
| C17—C18—H18 | 119.6 | C13—C14—H14 | 119.0 |
| C2—C3—C4 | 122.2 (4) | C15—C14—H14 | 119.0 |
| C2—C3—H3 | 118.9 | C16—C15—C14 | 120.1 (4) |
| C4—C3—H3 | 118.9 | C16—C15—H15 | 119.9 |
| C16—C17—C18 | 121.0 (3) | C14—C15—H15 | 119.9 |
| C10—N1—N2—C11 | −2.3 (5) | C1—C2—C3—C4 | −179.7 (4) |
| N2—N1—C10—O1 | −176.7 (3) | C13—C18—C17—C16 | −0.4 (6) |
| N2—N1—C10—C9 | 1.2 (5) | C12—C9—C8—C5 | −9.4 (6) |
| N1—N2—C11—C12 | 1.9 (5) | C10—C9—C8—C5 | 169.2 (3) |
| N1—N2—C11—C13 | 177.4 (3) | C4—C5—C8—C9 | 90.3 (4) |
| C14—C13—C11—N2 | −9.6 (5) | C6—C5—C8—C9 | −86.8 (4) |
| C18—C13—C11—N2 | 171.6 (3) | C18—C17—C16—C15 | −1.0 (7) |
| C14—C13—C11—C12 | 165.7 (4) | C6—C5—C4—C3 | 0.7 (5) |
| C18—C13—C11—C12 | −13.0 (5) | C8—C5—C4—C3 | −176.5 (3) |
| O1—C10—C9—C12 | 178.0 (3) | C2—C3—C4—C5 | −0.5 (6) |
| N1—C10—C9—C12 | 0.2 (5) | C4—C5—C6—C7 | −0.6 (5) |
| O1—C10—C9—C8 | −0.7 (5) | C8—C5—C6—C7 | 176.6 (3) |
| N1—C10—C9—C8 | −178.5 (3) | C3—C2—C7—C6 | 0.0 (6) |
| C10—C9—C12—C11 | −0.4 (5) | C1—C2—C7—C6 | 179.9 (4) |
| C8—C9—C12—C11 | 178.2 (3) | C5—C6—C7—C2 | 0.2 (6) |
| N2—C11—C12—C9 | −0.7 (5) | C18—C13—C14—C15 | −2.7 (7) |
| C13—C11—C12—C9 | −175.9 (3) | C11—C13—C14—C15 | 178.5 (4) |
| C14—C13—C18—C17 | 2.2 (6) | C17—C16—C15—C14 | 0.5 (7) |
| C11—C13—C18—C17 | −179.0 (3) | C13—C14—C15—C16 | 1.4 (8) |
| C7—C2—C3—C4 | 0.1 (6) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1i | 0.86 | 1.98 | 2.836 (3) | 175 |
| C14—H14···N2 | 0.93 | 2.43 | 2.764 (3) | 101 |
Symmetry code: (i) −x+2, −y+1, −z+2.
References
- Akhtar, W., Shaquiquzzaman, M., Akhter, M., Verma, G., Khan, M. F. & Alam, M. M. (2016). Eur. J. Med. Chem. 123, 256–281. [DOI] [PubMed]
- Asif, M. (2013). Mini-Rev. Org. Chem. 10, 113–122.
- Asif, M. (2014). Mini Rev. Med. Chem. 14, 1093–1103. [DOI] [PubMed]
- Barberot, C., Moniot, A., Allart-Simon, I., Malleret, L., Yegorova, T., Laronze-Cochard, M. & SAPI, J. (2018). Eur. J. Med. Chem. 146, 139–146. [DOI] [PubMed]
- Bernstein, J., Davis, R. E., Shimoni, L. & Chang, N.-L. (1995). Angew. Chem. Int. Ed. Engl. 34, 1555–1573.
- Boukharsa, Y., Meddah, B., Tiendrebeogo, R. Y., Ibrahimi, A., Taoufik, J. & Cherrah, Y. (2016). Med. Chem. Res. 25, 494–500.
- Chkirate, K., Kansiz, S., Karrouchi, K., Mague, J. T., Dege, N. & Essassi, E. M. (2019a). Acta Cryst. E75, 154–158. [DOI] [PMC free article] [PubMed]
- Chkirate, K., Kansiz, S., Karrouchi, K., Mague, J. T., Dege, N. & Essassi, E. M. (2019b). Acta Cryst. E75, 33–37. [DOI] [PMC free article] [PubMed]
- Dubey, S. & Bhosle, P. A. (2015). Med. Chem. Res. 24, 3579–3598.
- El Kali, F., Kansiz, S., Daoui, S., Saddik, R., Dege, N., Karrouchi, K. & Benchat, N. (2019). Acta Cryst. E75, 650–654. [DOI] [PMC free article] [PubMed]
- Gökçe, M., Utku, S. & Küpeli, E. (2009). Eur. J. Med. Chem. 44, 3760–3764. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hathwar, V. R., Sist, M., Jørgensen, M. R. V., Mamakhel, A. H., Wang, X., Hoffmann, C. M., Sugimoto, K., Overgaard, J. & Iversen, B. B. (2015). IUCrJ, 2, 563–574. [DOI] [PMC free article] [PubMed]
- Karrouchi, K., Ansar, M., Radi, S., Saadi, M. & El Ammari, L. (2015). Acta Cryst. E71, o890–o891. [DOI] [PMC free article] [PubMed]
- Karrouchi, K., Radi, S., Ansar, M. H., Taoufik, J., Ghabbour, H. A. & Mabkhot, Y. N. (2016a). Z. Kristallogr. New Cryst. Struct. 231, 883–886.
- Karrouchi, K., Radi, S., Ansar, M. H., Taoufik, J., Ghabbour, H. A. & Mabkhot, Y. N. (2016b). Z. Kristallogr. New Cryst. Struct. 231, 839–841.
- Livermore, D., Bethell, R. C., Cammack, N., Hancock, A. P., Hann, M. M. & Green, D. (1993). J. Med. Chem. 36, 3784–3794. [DOI] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- McKinnon, J. J., Jayatilaka, D. & Spackman, M. A. (2007). Chem. Commun. pp. 3814–3816. [DOI] [PubMed]
- Ochiai, K., Takita, S., Eiraku, T., Kojima, A., Iwase, K. & Kishi, T. (2012). Bioorg. Med. Chem. 20, 1644–1658. [DOI] [PubMed]
- Oubair, A., Daran, J.-C., Fihi, R., Majidi, L. & Azrour, M. (2009). Acta Cryst. E65, o1350–o1351. [DOI] [PMC free article] [PubMed]
- Partap, S., Akhtar, M. J., Yar, M. S., Hassan, M. Z. & Siddiqui, A. A. (2018). Bioorg. Chem. 77, 74–83. [DOI] [PubMed]
- Pita, B., Sotelo, E., Suarez, M., Ravina, E., Ochoa, E., Verdecia, Y., Novoa, H., Blaton, N., Ranter, C. & Peeters, O. M. (2000). Tetrahedron, 56, 2473–2479.
- Sharma, B., Verma, A., Sharma, U. K. & Prajapati, S. (2014). Med. Chem. Res. 23, 146–157.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Siddiqui, A. A., Mishra, R., Shaharyar, M., Husain, A., Rashid, M. & Pal, P. (2011). Bioorg. Med. Chem. Lett. 21, 1023–1026. [DOI] [PubMed]
- Sönmez, M., Berber, İ. & Akbaş, E. (2006). Eur. J. Med. Chem. 41, 101–105. [DOI] [PubMed]
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Stoe & Cie (2002). X-AREA and X-RED32. Stoe & Cie GmbH, Darmstadt, Germany.
- Tao, M., Aimone, L. D., Gruner, J. A., Mathiasen, J. R., Huang, Z. & Lyons, J. (2012). Bioorg. Med. Chem. Lett. 22, 1073–1077. [DOI] [PubMed]
- Thakur, A. S., Verma, P. & Chandy, A. (2010). Asian J. Res. Chem. 3, 265–271.
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17. University of Western Australia. http://hirshfeldsurface.net.
- Wang, T., Dong, Y., Wang, L.-C., Xiang, B.-R., Chen, Z. & Qu, L.-B. (2008). Arzneimittelforschung, 58, 569–573. [DOI] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
- Xu, H., Song, H.-B., Yao, C.-S., Zhu, Y.-Q., Hu, F.-Z., Zou, X.-M. & Yang, H.-Z. (2005). Acta Cryst. E61, o1561–o1563.
- Zhou, G., Ting, P. C., Aslanian, R., Cao, J., Kim, D. W., Kuang, R. & Zych, A. J. (2011). Bioorg. Med. Chem. Lett. 21, 2890–2893. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989019011551/mw2146sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989019011551/mw2146Isup3.hkl
CCDC reference: 1947718
Additional supporting information: crystallographic information; 3D view; checkCIF report