Summary
Background
Pneumonia is the leading cause of death among children younger than 5 years. In this study, we estimated causes of pneumonia in young African and Asian children, using novel analytical methods applied to clinical and microbiological findings.
Methods
We did a multi-site, international case-control study in nine study sites in seven countries: Bangladesh, The Gambia, Kenya, Mali, South Africa, Thailand, and Zambia. All sites enrolled in the study for 24 months. Cases were children aged 1–59 months admitted to hospital with severe pneumonia. Controls were age-group-matched children randomly selected from communities surrounding study sites. Nasopharyngeal and oropharyngeal (NP-OP), urine, blood, induced sputum, lung aspirate, pleural fluid, and gastric aspirates were tested with cultures, multiplex PCR, or both. Primary analyses were restricted to cases without HIV infection and with abnormal chest x-rays and to controls without HIV infection. We applied a Bayesian, partial latent class analysis to estimate probabilities of aetiological agents at the individual and population level, incorporating case and control data.
Findings
Between Aug 15, 2011, and Jan 30, 2014, we enrolled 4232 cases and 5119 community controls. The primary analysis group was comprised of 1769 (41·8% of 4232) cases without HIV infection and with positive chest x-rays and 5102 (99·7% of 5119) community controls without HIV infection. Wheezing was present in 555 (31·7%) of 1752 cases (range by site 10·6–97·3%). 30-day case-fatality ratio was 6·4% (114 of 1769 cases). Blood cultures were positive in 56 (3·2%) of 1749 cases, and Streptococcus pneumoniae was the most common bacteria isolated (19 [33·9%] of 56). Almost all cases (98·9%) and controls (98·0%) had at least one pathogen detected by PCR in the NP-OP specimen. The detection of respiratory syncytial virus (RSV), parainfluenza virus, human metapneumovirus, influenza virus, S pneumoniae, Haemophilus influenzae type b (Hib), H influenzae non-type b, and Pneumocystis jirovecii in NP-OP specimens was associated with case status. The aetiology analysis estimated that viruses accounted for 61·4% (95% credible interval [CrI] 57·3–65·6) of causes, whereas bacteria accounted for 27·3% (23·3–31·6) and Mycobacterium tuberculosis for 5·9% (3·9–8·3). Viruses were less common (54·5%, 95% CrI 47·4–61·5 vs 68·0%, 62·7–72·7) and bacteria more common (33·7%, 27·2–40·8 vs 22·8%, 18·3–27·6) in very severe pneumonia cases than in severe cases. RSV had the greatest aetiological fraction (31·1%, 95% CrI 28·4–34·2) of all pathogens. Human rhinovirus, human metapneumovirus A or B, human parainfluenza virus, S pneumoniae, M tuberculosis, and H influenzae each accounted for 5% or more of the aetiological distribution. We observed differences in aetiological fraction by age for Bordetella pertussis, parainfluenza types 1 and 3, parechovirus–enterovirus, P jirovecii, RSV, rhinovirus, Staphylococcus aureus, and S pneumoniae, and differences by severity for RSV, S aureus, S pneumoniae, and parainfluenza type 3. The leading ten pathogens of each site accounted for 79% or more of the site's aetiological fraction.
Interpretation
In our study, a small set of pathogens accounted for most cases of pneumonia requiring hospital admission. Preventing and treating a subset of pathogens could substantially affect childhood pneumonia outcomes.
Funding
Bill & Melinda Gates Foundation.
Introduction
Pneumonia remains the greatest cause of death globally in children younger than 5 years, accounting for an estimated 12·8% of annual deaths beyond the neonatal period.1 A substantial reduction in the past few decades of estimated pneumonia deaths (0·9 million in 2015 vs 1·7 million deaths in 1990) reflects not only economic development, improved nutrition, and reduced household crowding, but also the use of pneumonia-specific interventions such as improved case management—including empirical antibiotic treatment—and effective vaccines against leading pneumonia pathogens.2, 3 Despite these advances, continued progress to reduce pneumonia mortality is constrained by the absence of vaccines against the remaining common pathogens.
The Pneumonia Etiology Research for Child Health (PERCH) study sought to characterise the causes of severe childhood pneumonia requiring hospital admission among children living in high-burden, low-resource regions in the era of routine use of vaccines against pertussis, selected pneumococcal serotypes, Haemophilus influenzae type b (Hib), and measles.4 The PERCH study sought to provide rigorous data that would inform future prevention and treatment strategies in low-income and lower-middle-income settings and to address challenges faced by previous pneumonia aetiology studies, such as imperfect standardisation of methods and analyses, narrow generalisability, and absence of an analytical method to provide their primary output of interest, an aetiological distribution.5, 6
Research in context.
Evidence before this study
Before designing and doing the PERCH study, we did a detailed review to assess the body of evidence on pneumonia aetiology and the epidemiological, laboratory, and analytical methods used to quantify causes of pneumonia in young children. Additionally, we established an expert panel, the Pneumonia Methods Working Group (PMWG), to provide additional advice on the key epidemiological, laboratory, study design, and analytical issues that the PERCH study needed to optimise. The search dates, databases used, search terms, and restrictions are detailed in a separate publication (Zunera Gilani and colleagues, 2012). The results of our reviews of the pre-existing evidence and the decisions reached by the PMWG have been published in a set of methods papers and in the protocol. Since this review, the descriptive results of three large paediatric pneumonia aetiology studies in children younger than 5 years have been published—from the USA (EPIC), South Africa (Drakenstein), and a multicountry study (GABRIEL)—and were considered in our analysis.
Added value of this study
The PERCH study is a comprehensive, systematic, multisite study of the causes of severe and very severe pneumonia with hospital admission among children aged 1–59 months in the context of routine use of Haemophilus influenzae type b and pneumococcal conjugate vaccine. The emphasis on a single protocol with highly standardised implementation and quality control steps throughout the study conduct, the inclusion of diverse sites but with routine implementation of existing pneumonia vaccines, the duration of enrolment to capture seasonal trends, the use of a 33-target multiplex quantitative PCR, the inclusion of multiple body fluid specimens, the inclusion of an appropriate control group, the adjustment for the sensitivity of the laboratory tests, and the large size of the study all provided strength to the inferential value of the results. Our study provides results for over 30 pathogens and allows for cross-site comparisons of aetiological distribution because of its highly standardised methodology. It is the first infectious aetiology study to integrate the results of testing of multiple pathogens from multiple body fluid specimens, by using a range of laboratory assays, and to provide an aetiological distribution of the pathogens, with uncertainty intervals, both by individual and in the population. Our results showed that ten pathogens were responsible for 79–90% of cases with severe pneumonia requiring hospital admission in children younger than 5 years across a wide range of geographical and epidemiological settings. Causes of pneumonia varied by age strata and severity status and, to a lesser degree, by geography.
Implications of all the available evidence
From a policy perspective, the low number of pathogens that are common causes of pneumonia requiring hospital admission in children younger than 5 years across all sites could allow for targeted development of pneumonia interventions. Respiratory syncytial virus was the most common cause of disease and should be a primary target for dedicated prevention and treatment efforts, especially for children younger than 6 months. Bacterial pathogens, cumulatively, caused a substantial proportion of disease and, being treatable and commonly fatal, should remain a target for early access to treatment. Our study clearly shows that the inferential value of nasopharyngeal specimens varies by pathogen, with only a low number of pathogens providing evidence of causality. The PERCH Integrated Analysis shows the value of analytical approaches that account for pathogen prevalence, test performance, multiple specimens, and multiple results for an individual pathogen. The methods developed by the PERCH study for integrating such data, in a principled approach, offer the opportunity for other aetiology studies to also adapt and use these methods.
Here, we report the descriptive clinical and microbiological findings of the PERCH study using a traditional analytical approach, so that results can be compared with those of other pneumonia studies. Acknowledging that aetiology cannot be inferred from these results alone, we also developed a new analytical approach to assess the aetiological distribution of pneumonia.7, 8, 9 This approach integrates microbiology results from multiple specimens types and multiple results for one pathogen from an individual; this approach also accounts for the sensitivity and specificity of the individual diagnostic tests used. We report the probability distributions of the microbial aetiology in the study population, and in individual cases.
Methods
Study design and participants
The PERCH case-control study design has been previously described.4, 10, 11 Briefly, we assessed the causes of WHO-defined12 severe or very severe pneumonia (pre-2013 definitions, originally presented in 2005) among children aged 1–59 months presented and admitted to a hospital with this condition. The nine study locations in seven countries (Basse, The Gambia; Bamako, Mali; Lusaka, Zambia; Soweto, South Africa; Kilifi, Kenya; Dhaka and Matlab, Bangladesh; and Nakhon Phanom and Sa Kaeo, Thailand) were selected to represent diverse epidemiological conditions through an open call for applications.4 Site characteristics, vaccination schedules for each country, and screening and enrolment procedures are described in the appendix.4 All sites, except those in Thailand, were routinely using Hib vaccine; pneumococcal conjugate vaccine (PCV) was in routine use in The Gambia, Kenya, Mali, and South Africa and was in routine use in Zambia for the last few months of the PERCH study. Influenza vaccine was not in routine use at any site.
Cases were enrolled at the time of presentation to the hospital. Severe pneumonia was defined as cough or difficulty breathing with lower chest wall indrawing; very severe pneumonia was defined as cough or difficulty breathing and at least one of the following signs: central cyanosis, difficulty breastfeeding or drinking, vomiting everything, convulsions, lethargy, unconsciousness, or head nodding.12 Elevated respiratory rate was not part of case definitions (appendix). Exclusion criteria for cases and controls were hospitalisation within the preceding 14 days, having been discharged as a PERCH case within the preceding 30 days, and residence outside the study catchment area. Cases had the additional exclusion criterion of resolution of lower chest wall indrawing after bronchodilator therapy for children with wheeze. At all sites, case assessment occurred within 24 h of admission. In Bangladesh, children from the Dhaka site that met the case definition were identified at an outpatient clinic and referred for hospital admission; children whose parents refused hospital admission were still enrolled if they met all other inclusion criteria (n=33). Each site enrolled participants over a 24-month period (appendix). Case screening was done 24 h per day and 7 days per week at four sites (Kilifi, Sa Kaeo, Nakhon Phanom, and Matlab), during which all eligible consenting cases were enrolled. At the remaining sites, screening was done during established hours; all eligible consenting cases presenting during predefined screening hours were enrolled, except for Mali, where a systematic sampling process was used (appendix).
Controls were randomly selected from residents of the same catchment area as cases and frequency matched to cases by age group (1 to <6 months, 6 to <12 months, 12 to <24 months, and 24–59 months of age), as previously described.11 Sites aimed to enrol a minimum of 25 controls monthly or matched to case counts when they had enrolled more than 25 cases in a month. Controls were enrolled regardless of respiratory symptoms to provide the least biased comparison for estimating pneumonia causes, but were ineligible if they met the PERCH case definition.11, 13
All enrolled children were tested for HIV, except those in Bangladesh and controls in The Gambia and Thailand, because these were low HIV-prevalence settings. HIV-positive controls were oversampled in sites with high HIV prevalence (Zambia and South Africa) by recruitment at HIV treatment clinics.11
The study protocol was approved by the Institutional Review Boards or Ethical Review Committees for each of the seven institutions and at The Johns Hopkins School of Public Health.14 Parents or guardians of participants provided written informed consent. We followed standardised procedures for assessment of enrolment criteria, clinical assessment, specimen collection, data collection, and laboratory testing.10, 15, 16, 17 The study protocol, data collection forms, standard operating procedures, and clinical training videos are publicly available.18
Procedures
Cases underwent clinical examination at admission, at 24 h, and at 48 h (if the child was still hospitalised) including assessments of respiratory signs, anthropomorphic measurements, and peripheral oxygen saturation (on room air whenever possible).15, 18 We assessed the vital status of cases during a follow-up visit or telephone interview done 30 days after admission (with a window of 21–90 days). Controls were similarly assessed for clinical findings at enrolment but had no follow-up assessment.18 Clinical definitions used in analyses are described in the appendix.
At enrolment, we collected nasopharyngeal and oropharyngeal (NP-OP), urine, and blood specimens from cases and controls and blood cultures, induced sputum, lung aspirate, pleural fluid, and gastric aspirates from cases only. Results of bacterial and tuberculosis cultures of blood, lung aspirate, and pleural fluid were immediately available to the treating physicians. PCR results were batched and not available for clinical management. Chest x-rays were obtained from cases at enrolment and interpreted by two members of the PERCH Chest Radiograph Reading Panel trained in the WHO method;19 discordant readings were arbitrated as described elsewhere.20 Readers were masked to site and clinical factors. Chest x-rays were classified as consolidation, other infiltrate, both, normal, or uninterpretable by standardised WHO criteria.19, 20, 21 We defined x-rays as abnormal (positive chest x-ray) if they showed either consolidation, other infiltrate, or both.
Specimen collection procedures, laboratory testing methods, and determination of pathogen-specific PCR density thresholds used in analyses have been described separately elsewhere (appendix).17, 22, 23, 24, 25, 26, 27, 28 Briefly, we used a 33-pathogen multiplex quantitative PCR (FTD Resp-33, Fast Track Diagnostics, Sliema, Malta) and cultures to test NP-OP swabs (of cases and controls; cultures were done for Streptococcus pneumoniae alone), induced sputum (of cases), lung aspirates (of eligible cases in The Gambia, Bangladesh, Mali, and South Africa), and pleural fluid (at the discretion of the treating paediatrician in cases with pleural effusions on chest x-ray); gastric aspirates from cases were cultured for mycobacteria alone. We tested blood samples for S pneumoniae by PCR for cases and controls and with cultures for cases only. Pleural fluid specimens that signalled positive by automated culture but yielded no pathogen on subculture were also tested for pneumococcal antigen (BinaxNOW, Alere, Orlando, FL, USA; appendix). We tested serum specimens from cases and controls for the presence of antimicrobial activity; specimens from cases were also tested for C-reactive protein.
All pathogens detected by blood culture, except for contaminants, were included as potential pathogens. We excluded PCR results for Klebsiella pneumoniae because of poor assay specificity, which was identified after our testing protocol was established.29 For Moraxella catarrhalis, NP-OP PCR results are described in this study, but were excluded from the integrated aetiology analysis; these results did not provide useful diagnostic evidence because control prevalence of M catarrhalis was substantially higher than case prevalence (appendix). We implemented confirmatory uniplex PCR testing for specimens that tested positive on the multiplex assay for Legionella spp, pertussis, and Hib (appendix). For several pathogens with similar prevalence in cases and controls (ie, S pneumoniae, H influenzae, cytomegalovirus, and P jirovecii from NP-OP specimens and S pneumoniae from whole blood specimens),24, 25, 27 positivity was redefined by use of quantitative PCR density thresholds based on our analyses. For all remaining pathogens, the standard assay thresholds were used. Blood culture contaminants were defined by standard criteria (appendix) and these results were excluded from the aetiology analysis.
We obtained a single induced sputum or gastric aspirate specimen from each case, except in South Africa, where two specimens were routinely collected for increased tuberculosis detection, or at physicians' discretion for cases with suspected tuberculosis diagnosis. The first specimen alone was used to define a positive result for Mycobacterium tuberculosis to ensure cross-site comparability; the lower sensitivity of a single specimen was accounted for in the aetiology analysis. With the exception of tuberculosis, we did not use the induced sputum results in the aetiology analysis because the microbiological findings from sputum culture and PCR did not add information beyond that added by the NP-OP specimens, which had the analytical advantage of availability from both cases and controls.30, 31, 32
Pneumococcal serotypes were assessed by Quellung reaction or PCR; for NP-OP specimens that were culture negative but PCR positive and met the density threshold criteria for positivity and adequate specimen volume, we used a microarray assay to assess serotype (n=52; appendix).17, 33 These serotypes were grouped into vaccine-type or non-vaccine type according to the 13-valent PCV (PCV13) formulation. The Gambia, Mali, South Africa, and Zambia (for part of the study period) used PCV13, whereas Kenya used the ten-valent PCV (PCV10). The multiplex PCR panel included a target for all H influenzae and for H influenzae type b; cultured isolates were serotyped by slide agglutination or PCR. NP-OP specimens from cases who met study defined clinical criteria for measles (n=33) were tested for measles by PCR (appendix). Antibiotic exposure at the time of specimen collection was defined as either documented administration of antibiotics in the facility or antibiotic activity in the serum (measured by Staphylococcus aureus bioassay), collected at the same time as the specimen.28
Statistical analysis
Data were single-data entered into a centralised electronic data capture system (Emmes Corporation, Rockville, MD, USA);16 the Kenya site extracted data from its existing electronic data capture system, which conformed to the study data entry standards.
The number of sites and the number of participants enrolled per site were based on a combination of feasibility within budget and the case prevalence at each site. Site eligibility included, among other criteria, a minimum of 100 cases per year of severe pneumonia requiring hospital admission in children aged 1–59 months. Seven sites were selected so that results would represent diverse epidemiological settings in Asia and Africa, and sites were enrolled for 2 years rather than 1 year to enable assessment of seasonality, annual variations, and outbreaks in aetiology.
Unless otherwise specified, we restricted analyses to cases and controls without HIV infection because the aetiological distribution was expected to vary substantially according to HIV infection status (to be reported in a separate publication). The primary analysis was to estimate the aetiological fraction of severe and very severe pneumonia among cases with a positive chest x-ray; cases were restricted to positive results in chest x-rays because the analytical method for assessing causes, the PERCH Integrated Analysis (PIA) model, assumed that all cases had a lung infection. A secondary analysis of all cases without HIV infection, regardless of chest x-ray, was done and is reported in the appendix.34 Children missing all laboratory measurements used in the integrated aetiology analysis (blood cultures, NP-OP PCR, whole blood PCR, and tuberculosis cultures for cases; NP-OP PCR and whole blood PCR for controls) were excluded from the analyses. For children with at least one of these measurements, any missing data for the other measurements were imputed during model estimation by use of standard Bayesian methods.35
Statistical comparisons of demographic and clinical characteristics between sites and case-control groups were done with logistic regression that adjusted for age in months, with or without site as appropriate. We compared the prevalence of organisms measured in both cases and controls using odds ratios (ORs) with 95% CIs calculated by logistic regression, adjusting for site, age in months, and the other pathogens detected (this adjustment for NP-OP PCR results alone); we could not adjust for previous antibiotic use because it was rare among controls. Except for selected site-stratified analyses, results are for all sites combined; comprehensive site-specific results will be published separately. We used elevated C-reactive protein concentrations in the case descriptive analysis, but this was not a variable in the PIA model.
We developed and used the PIA model, a novel application of Bayesian latent class analysis that integrates the case and control data across multiple specimens and tests, resulting in an estimate of the aetiology distribution for each individual case and for the population of cases.7, 8, 9, 36 This method assumes that infections at body sites peripheral to the lung are indicative of the cause of the putative lung infection and that measurements are independent of one another. This method was tested with use of simulated datasets comprising cases with known aetiology who had a mix of measurements that varied in their sensitivity and specificity, which were prespecified, and it was shown to give accurate estimates of the aetiological fraction, even under mild violations of the independence assumption.9 The PIA model uses a Markov Chain Monte Carlo algorithm that, on convergence, samples from the posterior distribution of the aetiology estimates, given the observed measurements (appendix). The population-level estimate for each pathogen is the average of the individual case probabilities and has a 95% credible interval (95% CrI), the Bayesian analogue of the confidence interval. Sensitivity analyses were done to assess the influence of the values used for aetiology and sensitivity priors (appendix). To allow for the possibility that the pneumonia event was caused by a pathogen not tested for, we included a pathogen category termed not otherwise specified (NOS). Cases who had negative tests for all pathogens or positive results only in measurements with poor specificity had a higher individual-level probability to be included in NOS category than other cases.
The PIA model assumes that a pneumonia event is caused by a single pathogen. For cases with multiple pathogens detected in silver standard measurements (eg, blood cultures, lung aspirates, or pleural fluid), the model cannot distinguish which one is the dominant cause. For these cases, the model distributes the aetiology probability equally across the pathogens detected. This distribution leads to an underestimation of the contribution of the true cause of the disease because the sum of the pathogen probabilities for an individual case in this primary pathogen model must be 100%. For cases who are negative by silver standard measurements, the cause is distributed across multiple pathogens according to the strength of evidence for each pathogen. This evidence includes whether the pathogen was detected in multiple samples from the individual, the pathogen's prevalence among cases, the pathogen's odds ratio, and a-priori assumptions regarding sensitivity and aetiology.
As a Bayesian analysis, we assigned a-priori estimates for both the aetiology distribution and the sensitivity of each laboratory measurement. These estimates quantify the preceding uncertainty about the model parameters. The a-priori aetiology distribution favoured no pathogen over another (uninformed) because we wanted to estimate the causes using the evidence in the PERCH study alone. The a-priori sensitivity distribution, selected by an internal working group using available information external to the PERCH study, varied by laboratory test method and pathogen (table 1 , appendix). We classified specimens according to their estimated measurement error as either bronze (ie, imperfect sensitivity and specificity) or silver standard (ie, imperfect sensitivity, but 100% specificity); no test was categorised as gold standard (ie, 100% sensitivity and specificity; table 1, a-priori estimates of measurement-specific sensitivity are detailed in the appendix).9 Measurements designated as silver standard were those of blood cultures, of cultures or PCR of percutaneous lung aspirate and pleural fluid specimens, and of M tuberculosis cultures of induced sputum (or gastric aspirate). PCR results from the NP-OP swab and whole blood (S pneumoniae alone) specimens were included as bronze standard measurements, with controls providing the data to estimate specificity. We reduced the sensitivity for detecting bacterial pathogens in NP-OP PCR (S pneumoniae and H influenzae only) and blood culture specimens for individuals whose specimens were collected after documented antibiotic exposure and for blood culture specimens with low blood volume (appendix).
Table 1.
Integrated aetiology analysis input values for sensitivity and specificity of laboratory test measures
| Base sensitivity priors | Reduced sensitivity priors | Specificity | |
|---|---|---|---|
| Blood cultures*† | |||
| Streptococcus pneumoniae | 5–20% | 1–13% | 100% |
| Haemophilus influenzae | 5–20% | 1–13% | 100% |
| Moraxella catarrhalis | 5–15% | 1–10% | 100% |
| Staphylococcus aureus | 5–15% | 1–10% | 100% |
| Non-fermentative Gram-negative rods | 5–15% | 1–10% | 100% |
| Candida spp | 5–15% | 1–10% | 100% |
| Non-pneumococcal streptococci, including enterococci | 5–15% | 1–10% | 100% |
| Salmonella spp | 10–50% | 1–34% | 100% |
| Enterobacteriaceae | 10–50% | 1–34% | 100% |
| Neisseria meningitidis | 10–50% | 1–34% | 100% |
| NP-OP PCR | |||
| S pneumoniae | 50–90%† | 15–55% | 1–control prevalence‡ |
| H influenzae | 50–90%† | 15–55% | 1–control prevalence‡ |
| Salmonella spp | 0·5–90% | 0·5–90% | 1–control prevalence‡ |
| Legionella spp | 0·5–90% | 0·5–90% | 1–control prevalence‡ |
| All other PCR targets | 50–90% | 50–90% | 1–control prevalence‡ |
| Whole blood PCR | |||
| S pneumoniae | 12–65% | 12–65% | 1–control prevalence‡ |
| Induced sputum | |||
| Mycobacterium tuberculosis | 10–30% | 10–30% | 100% |
| Lung aspirate | |||
| All pathogens by culture or PCR | NA§ | NA§ | 100% |
| Pleural fluid | |||
| All pathogens by culture or PCR | NA§ | NA§ | 100% |
Background information supporting choice of sensitivity priors is provided in the appendix. For base priors, specimens had blood culture volume greater than 1·5 mL (blood culture only) and no evidence of previous antibiotic exposure; for reduced priors, specimens had blood culture volume lower than 1·5 mL or evidence of previous antibiotic exposure. These criteria were not applicable for some pathogen measurements (more details in the appendix). NP-OP=nasopharyngeal and oropharyngeal. NA=Not applicable.
Direct evidence of the diagnostic sensitivity of blood cultures for S pneumoniae and H influenzae obtained from vaccine probe studies; for all other pathogens, we set the base blood culture sensitivity prior to 5–15%, with the exception of Salmonella spp, Enterobacteriaceae, and N meningitidis, for which we selected wider priors (10–50%) to reflect their greater uncertainty.
Adjusted (reduced) for previous antibiotic exposure and low blood volume (blood culture only; see appendix for more details).
See appendix for control prevalence.
Results of lung aspirate and pleural fluid were used to update the aetiology prior distribution that was applied to cases with similar clinical characteristics.
The PIA model integrated the a-priori distributions with the observed data, changing (updating) both the a-priori distribution and sensitivity estimates with contributions of the study data, to produce the aetiology estimates.9 The positive lung aspirate results were used to update the a-priori aetiology distribution for cases with similar chest x-ray characteristics (appendix). Intrinsic within the PIA model, the sensitivity of the bronze standard measurements (eg, NP-OP) for individual bacteria was updated by use of the results of cases with positive blood cultures.
The PIA model was applied on data from all sites combined and assumed that the measurement sensitivities were common across sites, while allowing the aetiological distributions and control prevalences to vary across sites (appendix). We did analyses stratified by age (<1 year and ≥1 year) and WHO pneumonia severity (severe and very severe). The all-site results were based on cases with a positive chest x-ray, wherein each case, without regard to site, contributed equally to the estimated aetiology distribution, and based on all controls. To assess differences in the aetiological distribution related to site, we standardised the age and severity of case distributions at each site to that of all sites combined (appendix). We compared the age and severity-standardised results across sites for ten focus pathogens (H influenzae, M tuberculosis, S aureus, S pneumoniae, human metapneumovirus [HMPV], influenza, parainfluenza, rhinovirus, respiratory syncytial virus [RSV], and P jirovecii). We defined focus pathogens on the basis of results of the all-site aetiological analysis. Focus pathogens were those with aetiology estimates higher than 5% (n=7) or higher than 2% that were of epidemiological interest (defined as treatable by antibiotics [P jirovecii and S aureus] or having an available vaccine [influenza virus]). The 95% CrI around the aetiological fraction of these three pathogens overlap with some non-focus pathogens; therefore, we used the term focus pathogen rather than labelling these ten as the most common pathogens. In this study, we also report the list of the site-specific ten most common pathogens and the overall aetiological fraction of those pathogens (ie, the ten pathogens with the highest aetiological fraction at a given site).
We did statistical analyses with SAS, version 9.4, and with Bayesian inference software JAGS 4.2.0; NP-OP PCR figures were created with R, version 3.3.1. The PIA model used an open-source R software package that we developed called the Bayesian Analysis Kit for Etiology Research.
Role of the funding source
Representatives from the Bill & Melinda Gates Foundation participated in site selection and in Pneumonia Methods Working Group meetings, which informed the study design. They had no role in the data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
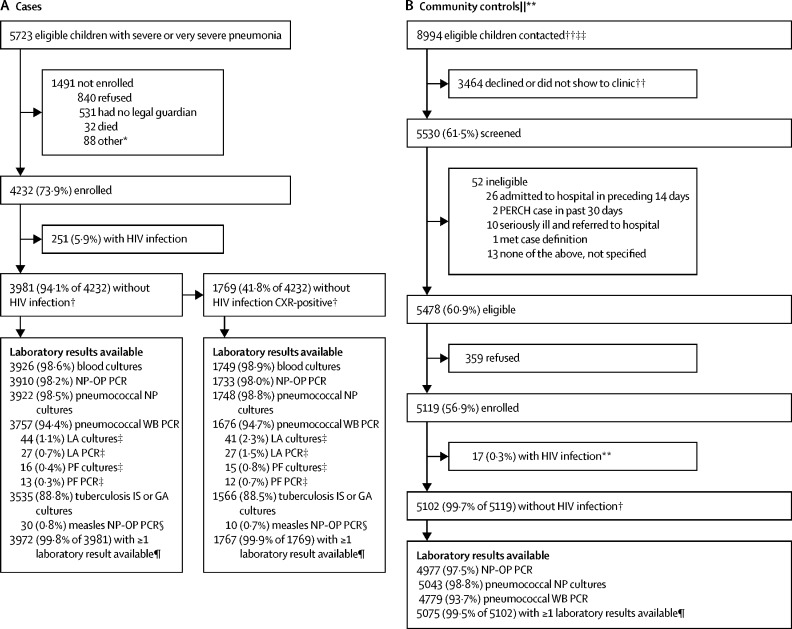
Between Aug 15, 2011, and Jan 30, 2014, we enrolled 4232 (73·9%) of 5723 eligible children with severe or very severe pneumonia as cases and 5119 (56·9%) of 8994 eligible children in the study catchment areas contacted as community controls; an additional 206 controls with HIV infection were enrolled from HIV clinics in South Africa and Zambia for HIV-specific analyses to be reported separately (figure 1 ). Enrolment of cases and controls varied by season (appendix). Approximately 90% of the planned specimens (ranging from 88·8% for tuberculosis cultures to 98·6% for blood cultures) were collected and tested (figure 1); reasons for missing specimens included specimens not collected because of physician discretion or contraindication (particularly for induced sputum), specimens lost in transport, and parental refusal.16 Cases (251 [5·9%] of 4232) and controls (17 [0·3%] of 5119) with HIV infection were otherwise excluded from analyses in this report, leaving 3981 (94·1%) of 4232 cases and 5102 (99·7%) of 5119 controls categorised as without HIV infection (figure 1, appendix). The primary analysis further restricted cases to those with positive chest x-rays (1769 [52·1%] of 3396 who had an interpretable x-ray; table 2 , 3 appendix). The descriptive laboratory analysis and the primary aetiology analysis excluded an additional two cases and 27 controls missing all microbiology test results. The secondary all-case aetiology analysis (ie, regardless of x-ray findings) included 3968 cases and 5075 controls with at least one microbiology test result (figure 1); of the 3972 cases with at least one test result, four cases who tested positive for measles by NP-OP PCR (all without an infiltrate on their chest x-rays) were excluded from the all-case analysis.
Figure 1.
Case (A) and control (B) enrolment and specimen availability profile
CXR=chest x-ray. NP=nasopharyngeal. OP=oropharyngeal. WB=whole blood. LA=lung aspirate. PF=pleural fluid. IS=induced sputum. GA=gastric aspirate. *Of the 88 children not enrolled because of other reasons, 45 were not enrolled because of an enrolment cap at the Mali site, ten because of political unrest in Bangladesh, and 24 in Kenya and nine in Zambia because of reasons not stated. †Included in clinical descriptive analysis. ‡Lung aspirate and pleural fluid specimens were collected on a subset of cases eligible for the procedures; for samples with low volumes, only culture was done. §Measles testing was done on a subset of cases who met the study defined clinical criteria for measles. ¶Included in laboratory descriptive analysis and aetiology analysis; at least one of the following specimens was required for a child to be included in the aetiology analysis: blood culture, NP-OP PCR, WB PCR, or tuberculosis culture for cases; and NP-OP PCR or WB PCR for controls. ||Number contacted, screened, and eligible includes some extrapolated data for the Zambia and South Africa sites; data were available for 16 of 24 months for Zambia and 10·5 of 24 months for South Africa; for each of these sites, available data were used to extrapolate numbers for the months with missing data assuming that contact, participation, and eligibility rates were constant over time.**Not shown here are an additional 206 controls with HIV infection enrolled from HIV clinics at the South Africa and Zambia sites to ensure adequate sample size of children with HIV infection; these children will be described in forthcoming manuscripts devoted to the causes of severe and very severe pneumonia in children with HIV infection. ††Data for total number of children contacted and number of children who declined or did not show to clinic were not available for the Mali site. ‡‡Among sites with available data for total number of children contacted (ie, all sites except Mali), 8149 (73·9%) of 11 033 randomly selected children or households were contacted; the number of children or guardians contacted is used as the denominator for the percentage of children screened, eligible, and enrolled; because the denominator excludes Mali but the numerator does not, the percentages for screened, eligible, and enrolled are overestimated.
Table 2.
Demographic and clinical characteristics of cases without HIV infection and controls for all sites
| All cases | Cases with a positive x-ray | Controls | p value | |||
|---|---|---|---|---|---|---|
| Total | 3981 | 1769 | 5102 | .. | ||
| Age, months | .. | .. | .. | <0·0001 | ||
| 28 days–5 | 1619 (40·7%) | 691 (39·1%) | 1598 (31·3%) | .. | ||
| 6–11 | 908 (22·8%) | 425 (24·0%) | 1210 (23·7%) | .. | ||
| 12–23 | 903 (22·7%) | 436 (24·6%) | 1262 (24·7%) | .. | ||
| 24–59 | 551 (13·8%) | 217 (12·3%) | 1032 (20·2%) | .. | ||
| Sex | ||||||
| Girls | 1684 (42·3%) | 778 (44·0%) | 2533 (49·7%) | <0·0001 | ||
| Respiratory tract illness (controls only) | .. | .. | 1206 (23·6%) | .. | ||
| Positive malaria smear* | 91 (3·9%) | 19 (1·9%) | 35 (1·3%) | 0·1480 | ||
| DTP vaccine | ||||||
| Number of doses (regardless of age) | .. | .. | .. | <0·0001 | ||
| 0 | 480 (12·5%) | 201 (11·8%) | 249 (5·0%) | .. | ||
| 1–2 | 1086 (28·3%) | 484 (28·4%) | 1060 (21·1%) | .. | ||
| ≥3 | 2267 (59·1%) | 1017 (60·0%) | 3708 (73·9%) | .. | ||
| Fully vaccinated for age† | ||||||
| Age <1 year | 1724 (70·3%) | 751 (69·5%) | 2258 (81·0%) | <0·0001 | ||
| Age ≥ 1 year | 1264 (91·5%) | 566 (91·1%) | 2083 (93·5%) | 0·1407 | ||
| PCV | ||||||
| Number of doses (regardless of age) | .. | .. | .. | 0·0014 | ||
| 0 | 1690 (43·8%) | 701 (40·8%) | 2404 (47·9%) | .. | ||
| 1–2 | 1021 (26·4%) | 480 (28·0%) | 1056 (21·0%) | .. | ||
| ≥3 | 1151 (29·8%) | 536 (31·2%) | 1561 (31·1%) | .. | ||
| Fully vaccinated for age‡ | ||||||
| Age <1 year | 1226 (49·7%) | 566 (52·0%) | 1384 (49·6%) | 0·0004 | ||
| Age ≥1 year | 640 (45·9%) | 291 (46·3%) | 867 (38·8%) | 0·7019 | ||
| Full vaccination for measles§ | 1453 (86·8%) | 646 (85·3%) | 2459 (90·6%) | 0·0135 | ||
| Mid-upper arm circumference <11·5 cm¶ | 171 (7·4%) | 93 (8·8%) | 29 (0·8%) | <0·0001 | ||
| Weight-for-age Z score | .. | .. | .. | <0·0001 | ||
| Severe (< −3) | 631 (15·9%) | 338 (19·2%) | 196 (3·9%) | .. | ||
| Moderate (≥ −3 to < −2) | 754 (19·0%) | 361 (20·5%) | 515 (10·1%) | .. | ||
| Normal (≥ −2) | 2584 (65·1%) | 1062 (60·3%) | 4368 (86·0%) | .. | ||
| Antibiotic activity detected in serum | 918 (24·5%) | 425 (25·3%) | 85 (1·8%) | <0·0001 | ||
| Previous exposure to antibiotics‖ | 1530 (38·9%) | 704 (40·1%) | 85 (1·7%) | <0·0001 | ||
| Median duration of illness (days) | 3·0 (2·0–5·0) | 3·0 (2·0–5·0) | .. | .. | ||
| Very severe pneumonia | 1279 (32·1%) | 519 (29·3%) | .. | .. | ||
| Wheeze on auscultation | 1340 (33·9%) | 555 (31·7%) | .. | .. | ||
| Tachypnoea | 3256 (82·4%) | 1492 (85·3%) | .. | .. | ||
| Hypoxaemia | 1423 (35·8%) | 748 (42·3%) | .. | .. | ||
| Oxygen use at admission** | 1243 (37·4%) | 671 (43·9%) | .. | .. | ||
| Temperature ≥38·0°C | 1213 (30·5%) | 591 (33·5%) | .. | .. | ||
| CRP ≥40 mg/L | 925 (27·6%) | 526 (34·9%) | .. | .. | ||
| Elevated WBC count | 1620 (43·4%) | 793 (47·8%) | .. | .. | ||
| Chest x-ray findings | ||||||
| Positive | 1769 (44·4%) | 1769 (100%) | .. | .. | ||
| Consolidation | 841 (21·1%) | 841 (47·5%) | .. | .. | ||
| Other infiltrate | 928 (23·3%) | 928 (52·5%) | .. | .. | ||
| Negative | 1627 (40·9%) | .. | .. | .. | ||
| Uninterpretable | 351 (8·8%) | .. | .. | .. | ||
| Missing | 234 (5·9%) | .. | .. | .. | ||
| Blood culture volume, mL†† | ||||||
| 1 to <2 | 1649 (48·7%) | 753 (51·4%) | .. | .. | ||
| ≥2 to <3 | 1313 (38·8%) | 537 (36·7%) | .. | .. | ||
| ≥3 | 425 (12·6%) | 175 (12·0%) | .. | .. | ||
| Died in hospital or within 30 days of admission | 292 (7·3%) | 114 (6·4%) | .. | .. | ||
| Died in hospital | 251 (6·3%) | 95 (5·4%) | .. | .. | ||
| Died post discharge, within 30 days of admission‡‡ | 41 (1·0%) | 19 (1·1%) | .. | .. | ||
| Missing 30-day vital status | 346 (8·7%) | 146 (8·3%) | .. | .. | ||
Data are n (%) or median (IQR). Positive cases are those with a positive chest x-ray. p values obtained from logistic regression models adjusted for age in months (all variables expect age category) and site for cases with a positive x-ray versus controls. During the study, pneumococcal conjugate vaccine (PCV) was in routine use in Kenya (introduced February, 2011), The Gambia (August, 2009), Mali (March, 2011), and South Africa (April, 2009); PCV was introduced in Zambia in July, 2013 (Lusaka), 3 months before the end of study enrolment. Duration of illness was defined as duration (in days) of cough, wheeze, fever, or difficulty breathing, whichever was longest. Tachypnoea was defined as 60 or more breaths per min (<2 months), 50 or more breaths per min (2–11 months), and 40 or more breaths per min (12–59 months). Hypoxaemia was defined as oxygen saturation lower than 92% (or <90% for sites at elevation above 1200 m: Zambia and South Africa), or supplemental oxygen use if a room air oxygen saturation reading was not available; a room air oxygen saturation reading was available for 3514 (88·3%) children; the South African site, at an altitude of 1600 m above sea level, had a standard clinical practice to administer supplemental oxygen for all children admitted to hospital with a diagnosis of severe pneumonia. Elevated white blood cell (WBC) count was defined as greater than 15 × 109 cells per L for children aged 1–11 months and greater than 13 × 109 cells per L for children aged 12–59 months. CRP=C-reactive protein.
Restricted to endemic sites (Kenya, Gambia, Mali, and Zambia).
For children younger than 1 year, full vaccination was defined as having received at least one dose and being up to date for age on the basis of the child's age at enrolment, doses received, and country schedule (allowing a 4-week window for each dose); for children aged 1 year or older, full vaccination was defined as having received three or more doses.
For children younger than 1 year, full vaccination was defined as having received at least one dose and being up to date for age on the basis of the child's age at enrolment, doses received, and country schedule (allowing a 4-week window for each dose); for children aged 1 year or older in all sites except Kenya, full vaccination was defined as having received three or more doses; for children older than 1 year in Kenya (which introduced PCV with catch-up campaign), full vaccination was defined as having received three or more doses, two doses if given at least 8 weeks apart and the child was older than 1 year of age at first dose, and one dose if the child was older than 2 years at any dose or at introduction.
Data restricted to those age-eligible; at least one dose was restricted to children older than 10 months (>10·5 months in Bangladesh).
Restricted to children aged 6 months or older.
Defined as serum bioassay positive (cases and controls), antibiotics administered at the referral facility, or antibiotic administration before the collection of whole blood specimens at the study facility (cases only); restricted to children with blood culture (cases only) or whole blood (cases or controls) collected.
Data was recorded in the Mali site on the basis of time at presentation, not time at admission, as was done at all other sites; Mali is excluded from the all site summary.
13·7% of cases with a blood culture collected were missing blood culture volume data.29
Restricted to children discharged alive who had vital status data obtained 21 days or longer after admission.
Demographic and clinical characteristics are presented in table 2, table 3 , and the appendix. Cases were unevenly distributed across the seven sites; of all cases with a positive chest x-ray (1769), Thailand made up the lowest percentage (5·5%), whereas South Africa made up the highest (24·6%; appendix) of the seven countries. For every site except Thailand, more than 75% of cases with a positive x-ray were younger than 24 months, with the majority being younger than 12 months (appendix). Controls were older than cases in The Gambia, Mali, South Africa, and Dhaka (Bangladesh) sites (p<0·020; appendix). The proportion of cases with very severe pneumonia was lowest in Bangladesh and The Gambia and highest in Mali and Kenya (p<0·0001; table 2, 3, appendix). 30-day case-fatality ratio of cases with a positive x-ray was 6·4% (114 of 1769 cases), which varied markedly by site (p<0·0001) and was higher in cases with very severe pneumonia (13·3% of 519 very severe cases without HIV infection and with a positive x-ray; range by site 3·7–28·2%) than in cases with severe pneumonia (3·6% of 1250 severe cases without HIV infection and with a positive x-ray; range by site 0·5–9·7%; appendix).
Table 3.
Demographic and clinical characteristics of cases with a positive chest x-ray and without HIV infection and controls for each PERCH site
|
The Gambia |
Mali |
Kenya |
Zambia |
South Africa |
Bangladesh |
Thailand |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases with a positive x-ray | Controls | Cases with a positive x-ray | Controls | Cases with a positive x-ray | Controls | Cases with a positive x-ray | Controls | Cases with a positive x-ray | Controls | Cases with a positive x-ray | Controls | Cases with a positive x-ray | Controls | |||
| Total | 286 | 654 | 241 | 725 | 282 | 863 | 208 | 601 | 435 | 828 | 219 | 772 | 98 | 659 | ||
| Age, months | ||||||||||||||||
| 28 days–5 | 106 (37·1%) | 199 (30·4%) | 98 (40·7%) | 247 (34·1%) | 90 (31·9%) | 234 (27·1%) | 110 (52·9%) | 286 (47·6%) | 219 (50·3%) | 320 (38·6%) | 50 (22·8%) | 221 (28·6%) | 18 (18·4%) | 91 (13·8%) | ||
| 6–11 | 71 (24·8%) | 133 (20·3%%) | 59 (24·5%) | 188 (25·9%) | 66 (23·4%) | 190 (22·0%) | 49 (23·6%) | 150 (25·0%) | 111 (25·5%) | 228 (27·5%) | 50 (22·8%) | 168 (21·8%) | 19 (19·4%) | 153 (23·2%) | ||
| 12–23 | 70 (24·5%) | 181 (27·7%) | 61 (25·3%) | 165 (22·8%) | 79 (28·0%) | 247 (28·6%) | 37 (17·8%) | 108 (18·0%) | 78 (17·9%) | 148 (17·9%) | 81 (37·0%) | 196 (25·4%) | 30 (30·6%) | 217 (32·9%) | ||
| 24–59 | 39 (13·6%) | 141 (21·6%) | 23 (9·5%) | 125 (17·2%) | 47 (16·7%) | 192 (22·2%) | 12 (5·8%) | 57 (9·5%) | 27 (6·2%) | 132 (15·9%) | 38 (17·4%) | 187 (24·2%) | 31 (31·6%) | 198 (30·0%) | ||
| Sex | ||||||||||||||||
| Girls | 108 (37·8%) | 308 (47·1%) | 97 (40·2%) | 366 (50·5%) | 130 (46·1%) | 410 (47·6%) | 91 (43·8%) | 300 (49·9%) | 222 (51·0%) | 424 (51·2%) | 87 (39·7%) | 402 (52·1%) | 43 (43·9%) | 323 (49·0%) | ||
| Respiratory tract illness (controls only) | .. | 159 (24·3%) | .. | 299 (41·2%) | .. | 211 (24·4%) | .. | 69 (11·5%) | .. | 45 (5·4%) | .. | 169 (21·9%) | .. | 254 (38·5%) | ||
| Positive malaria smear* | 3 (1·2%) | 7 (1·1%) | 6 (2·5%) | 8 (1·1%) | 9 (3·2%) | 15 (1·8%) | 1 (0·5%) | 5 (0·8%) | .. | .. | · | .. | .. | .. | ||
| DTP vaccine | ||||||||||||||||
| Number of doses (regardless of age) | ||||||||||||||||
| 0 | 41 (15·0%) | 32 (5·2%) | 29 (12·3%) | 51 (7·0%) | 11 (4·0%) | 13 (1·5%) | 23 (11·9%) | 35 (5·8%) | 79 (19·4%) | 89 (11·0%) | 8 (3·7%) | 12 (1·6%) | 10 (10·4%) | 17 (2·6%) | ||
| 1–2 | 68 (24·8%) | 166 (26·9%) | 75 (31·9%) | 153 (21·1%) | 55 (19·8%) | 119 (14·1%) | 72 (37·3%) | 175 (29·2%) | 163 (40·0%) | 240 (29·6%) | 32 (14·7%) | 104 (13·6%) | 19 (19·8%) | 103 (15·7%) | ||
| ≥3 | 165 (60·2%) | 419 (67·9%) | 131 (55·7%) | 521 (71·9%) | 212 (76·3%) | 712 (84·4%) | 98 (50·8%) | 389 (64·9%) | 166 (40·7%) | 482 (59·4%) | 178 (81·7%) | 650 (84·9%) | 67 (69·8%) | 535 (81·7%) | ||
| Fully vaccinated for age† | ||||||||||||||||
| Age <1 year | 96 (56·1%) | 218 (67·1%) | 112 (72·7%) | 346 (79·5%) | 133 (85·8%) | 387 (91·5%) | 113 (74·8%) | 368 (84·8%) | 189 (60·2%) | 378 (70·0%) | 85 (85·9%) | 346 (88·9%) | 23 (62·2%) | 215 (88·5%) | ||
| Age ≥ 1 year | 97 (94·2%) | 271 (92·8%) | 69 (85·2%) | 278 (95·9%) | 117 (95·1%) | 405 (96·2%) | 40 (95·2%) | 152 (92·1%) | 75 (79·8%) | 208 (76·8%) | 114 (95·8%) | 364 (96·6%) | 54 (91·5%) | 405 (98·3%) | ||
| PCV | ||||||||||||||||
| Number of doses (regardless of age) | ||||||||||||||||
| 0 | 48 (17·5%) | 66 (10·7%) | 53 (22·6%) | 212 (29·3%) | 14 (5·0%) | 34 (4·0%) | 199 (96·6%) | 584 (97·2%) | 74 (18·1%) | 81 (10·0%) | 218 (100%) | 772 (100%) | 95 (99·0%) | 655 (100%) | ||
| 1–2 | 63 (23·0%) | 169 (27·5%) | 71 (30·2%) | 146 (20·2%) | 87 (31·1%) | 236 (27·9%) | 7 (3·4%) | 17 (2·8%) | 252 (61·8%) | 488 (60·2%) | 0 (0·0%) | 0 (0·0%) | 0 (0·0%) | 0 (0·0%) | ||
| ≥3 | 163 (59·5%) | 379 (61·7%) | 111 (47·2%) | 365 (50·5%) | 179 (63·9%) | 575 (68·0%) | 0 (0·0%) | 0 (0·0%) | 82 (20·1%) | 242 (29·8%) | 0 (0·0%) | 0 (0·0%) | 1 (1·0%) | 0 (0·0%) | ||
| Fully vaccinated for age‡ | ||||||||||||||||
| Age <1 year | 95 (55·6%) | 214 (66·0%) | 112 (72·7%) | 332 (76·5%) | 136 (87·2%) | 392 (92·7%) | 6 (3·8%) | 15 (3·4%) | 217 (69·1%) | 431 (79·8%) | 0 (0·0%) | 0 (0·0%) | 0 (0·0%) | 0 (0·0%) | ||
| Age ≥1 year | 94 (91·3%) | 232 (80·0%) | 48 (59·3%) | 134 (46·4%) | 93 (75·0%) | 330 (78·2%) | 0 (0·0%) | 0 (0·0%) | 55 (58·5%) | 171 (63·1%) | 0 (0·0%) | 0 (0·0%) | 1 (1·7%) | 0 (0·0%) | ||
| Full vaccination for measles§ | 119 (91·5%) | 303 (89·9%) | 78 (78·0%) | 326 (88·1%) | 131 (90·3%) | 458 (92·0%) | 42 (77·8%) | 188 (85·5%) | 106 (84·1%) | 345 (92·0%) | 110 (80·9%) | 384 (88·1%) | 60 (90·9%) | 455 (95·0%) | ||
| Mid-upper arm circumference <11·5 cm¶ | 9 (5·0%) | 3 (0·7%) | 36 (25·2%) | 1 (0·2%) | 24 (12·6%) | 8 (1·3%) | 10 (10·3%) | 9 (2·9%) | 9 (4·6%) | 7 (1·5%) | 3 (1·8%) | 1 (0·2%) | 2 (2·5%) | 0 (0·0%) | ||
| Weight-for-age Z score | ||||||||||||||||
| Severe (< −3) | 34 (11·9%) | 35 (5·4%) | 71 (29·5%) | 27 (3·7%) | 68 (24·4%) | 38 (4·4%) | 35 (16·8%) | 28 (4·7%) | 81 (18·8%) | 20 (2·4%) | 31 (14·2%) | 43 (5·6%) | 18 (18·4%) | 5 (0·8%) | ||
| Moderate (≥ −3 to < −2) | 64 (22·4%) | 83 (12·8%) | 44 (18·3%) | 66 (9·2%) | 65 (23·3%) | 109 (12·7%) | 31 (14·9%) | 47 (7·8%) | 71 (16·5%) | 31 (3·8%) | 70 (32·0%) | 138 (17·9%) | 16 (16·3%) | 41 (6·2%) | ||
| Normal (≥ −2) | 188 (65·7%) | 531 (81·8%) | 126 (52·3%) | 628 (87·1%) | 146 (52·3%) | 710 (82·8%) | 142 (68·3%) | 525 (87·5%) | 278 (64·7%) | 770 (93·8%) | 118 (53·9%) | 591 (76·6%) | 64 (65·3%) | 613 (93·0%) | ||
| Antibiotic activity detected in serum | 20 (7·6%) | 1 (0·2%) | 41 (17·1%) | 19 (2·7%) | 26 (10·0%) | 21 (2·7%) | 50 (25·3%) | 21 (3·9%) | 224 (54·2%) | 8 (1·0%) | 45 (21·6%) | 10 (1·4%) | 19 (19·4%) | 5 (0·9%) | ||
| Previous exposure to antibiotics‖ | 30 (10·6%) | 1 (0·2%) | 54 (22·4%) | 19 (2·6%) | 99 (35·1%) | 21 (2·6%) | 189 (92·2%) | 21 (3·6%) | 249 (57·2%) | 8 (1·0%) | 52 (24·6%) | 10 (1·4%) | 30 (30·6%) | 5 (0·8%) | ||
| Median duration of illness (days) | 3 (2–4) | .. | 5 (3–7) | .. | 3 (2–4) | .. | 3 (2–6) | .. | 3 (2–5) | .. | 3 (2–5) | .. | 3 (2–4) | .. | ||
| Very severe pneumonia | 39 (13·6%) | .. | 96 (39·8%) | .. | 114 (40·4%) | .. | 70 (33·7%) | .. | 153 (35·2%) | .. | 27 (12·3%) | .. | 20 (20·4%) | .. | ||
| Wheeze on auscultation | 84 (29·6%) | .. | 35 (14·5%) | .. | 41 (14·6%) | .. | 22 (10·6%) | .. | 122 (29·0%) | .. | 213 (97·3%) | .. | 38 (38·8%) | .. | ||
| Tachypnoea | 260 (90·9%) | .. | 205 (85·1%) | .. | 224 (79·4%) | .. | 188 (90·8%) | .. | 331 (78·3%) | .. | 210 (95·9%) | .. | 74 (80·4%) | .. | ||
| Hypoxaemia | 33 (11·5%) | .. | 133 (55·2%) | .. | 106 (37·7%) | .. | 97 (46·6%) | .. | 329 (75·8%) | .. | 22 (10·0%) | .. | 28 (28·6%) | .. | ||
| Oxygen use at admission** | 34 (11·9%) | .. | .. | .. | 75 (26·6%) | .. | 122 (58·7%) | .. | 392 (90·1%) | .. | 12 (5·5%) | .. | 36 (36·7%) | .. | ||
| Temperature ≥38·0 °C | 117 (40·9%) | .. | 89 (36·9%) | .. | 128 (45·4%) | .. | 122 (58·9%) | .. | 42 (9·7%) | .. | 38 (17·4%) | .. | 55 (56·1%) | .. | ||
| CRP ≥40 mg/L | 86 (48·9%) | .. | 89 (43·0%) | .. | 78 (35·9%) | .. | 91 (48·4%) | .. | 134 (31·4%) | .. | 26 (12·9%) | .. | 22 (24·2%) | .. | ||
| Elevated WBC count | 100 (49·8%) | .. | 86 (35·8%) | .. | 145 (52·0%) | .. | 97 (47·8%) | .. | 194 (44·7%) | .. | 123 (60·0%) | .. | 48 (49·5%) | .. | ||
| Chest x-ray findings | ||||||||||||||||
| Consolidation | 101 (35·3%) | .. | 136 (56·4%) | .. | 119 (42·2%) | .. | 135 (64·9%) | .. | 250 (57·5%) | .. | 58 (26·5%) | .. | 42 (42·9%) | .. | ||
| Other infiltrate | 185 (64·7%) | .. | 105 (43·6%) | .. | 163 (57·8%) | .. | 73 (35·1%) | .. | 185 (42·5%) | .. | 161 (73·5%) | .. | 56 (57·1%) | .. | ||
| Blood culture volume, mL†† | ||||||||||||||||
| 1 to <2 | 101 (39·5%) | .. | 87 (36·6%) | .. | 180 (64·1%) | .. | 104 (52·0%) | .. | 154 (85·1%) | .. | 90 (42·7%) | .. | 37 (37·8%) | .. | ||
| 2 to <3 | 119 (46·5%) | .. | 73 (30·7%) | .. | 82 (29·2%) | .. | 83 (41·5%) | .. | 19 (10·5%) | .. | 119 (56·4%) | .. | 42 (42·9%) | .. | ||
| ≥3 | 36 (14·1%) | .. | 78 (32·8%) | .. | 19 (6·8%) | .. | 13 (6·5%) | .. | 8 (4·4%) | .. | 2 (1·0%) | .. | 19 (19·4%) | .. | ||
| Died in hospital or within 30 days of admission | 16 (5·6%) | .. | 31 (12·9%) | .. | 17 (6·0%) | .. | 25 (12·0%) | .. | 18 (4·1%) | .. | 2 (0·9%) | .. | 5 (5·1%) | .. | ||
| Died in hospital | 12 (4·2%) | .. | 25 (10·4%) | .. | 17 (6·0%) | .. | 23 (11·1%) | .. | 16 (3·7%) | .. | 0 (0·0%) | .. | 2 (2·0%) | .. | ||
| Died post-discharge, within 30 days of admission‡‡ | 4 (1·5%) | .. | 6 (2·9%) | .. | 0 (0·0%) | .. | 1 (1·0%) | .. | 2 (0·5%) | .. | 2 (0·9%) | .. | 3 (3·2%) | .. | ||
| Missing 30-day vital status | 2 (0·7%) | .. | 8 (3·3%) | .. | 2 (0·7%) | .. | 82 (39·4%) | .. | 48 (11·0%) | .. | 3 (1·4%) | .. | 1 (1·0%) | .. | ||
Data are n (%) or median (IQR). Positive cases are those with a positive chest x-ray. p values obtained from logistic regression models adjusted for age in months (all variables expect age category) and site for cases with a positive x-ray versus controls. Diphtheria-tetanus-pertussis (DTP) vaccine formulation varied by site: pentavalent vaccine (DTP-Hib-HepB) was used in Kenya, Gambia, Mali, Zambia, and Bangladesh; DTP-only and DTP-HepB was used in Thailand; and pentaxim (DTaP-Hib-IPV) was used in South Africa. During the study, pneumococcal conjugate vaccine (PCV) was in routine use in Kenya (introduced February, 2011), The Gambia (August, 2009), Mali (March, 2011), and South Africa (April, 2009); PCV was introduced in Zambia in July, 2013 (Lusaka), 3 months before the end of study enrolment. Duration of illness was defined as duration (in days) of cough, wheeze, fever, or difficulty breathing, whichever was longest. Tachypnoea was defined as 60 or more breaths per min (<2 months), 50 or more breaths per min (2–11 months), and 40 or more breaths per min (12–59 months). Hypoxaemia was defined as oxygen saturation lower than 92% (or <90% for sites at elevation above 1200 m: Zambia and South Africa), or supplemental oxygen use if a room air oxygen saturation reading was not available; a room air oxygen saturation reading was available for 3514 (88·3%) children; the South African site, at an altitude of 1600 m above sea level, had a standard clinical practice to administer supplemental oxygen for all children admitted to hospital with a diagnosis of severe pneumonia. Elevated white blood cell (WBC) count was defined as greater than 15 × 109 cells per L for children aged 1–11 months and greater than 13 × 109 cells per L for children aged 12–59 months. CRP=C-reactive protein.
Restricted to endemic sites (Kenya, The Gambia, Mali, and Zambia).
For children younger than 1 year, full vaccination was defined as having received at least one dose and being up to date for age on the basis of the child's age at enrolment, doses received, and country schedule (allowing a 4-week window for each dose); for children aged 1 year or older, full vaccination was defined as having received three or more doses.
For children younger than 1 year, full vaccination was defined as having received at least one dose and being up to date for age on the basis of the child's age at enrolment, doses received, and country schedule (allowing a 4-week window for each dose); for children aged 1 year or older in all sites except Kenya, full vaccination was defined as having received three or more doses; for children older than 1 year in Kenya (which introduced PCV with catch-up campaign), full vaccination was defined as having received three or more doses, two doses if given at least 8 weeks apart and the child was older than 1 year of age at first dose, and one dose if the child was older than 2 years at any dose or at introduction.
Data restricted to those age-eligible; at least one dose was restricted to children older than 10 months (>10·5 months in Bangladesh).
Restricted to children aged 6 months or older.
Defined as serum bioassay positive (cases and controls), antibiotics administered at the referral facility, or antibiotic administration before the collection of whole blood specimens at the study facility (cases only); restricted to children with blood culture (cases only) or whole blood (cases or controls) collected.
Data was recorded in the Mali site on the basis of time at presentation, not time at admission, as was done at all other sites; in The Gambia, on the basis of evidence from a concurrent project of clinical care, all children received oxygen if they had hypoxaemia and were coded as such for this study.
13·7% of cases with a blood culture collected were missing blood culture volume data (0·0% in Bangladesh and Thailand, 0·2% in Kenya, 1·8% in Zambia and Mali, 7·0% in The Gambia, and 59·1% in South Africa).29
Restricted to children discharged alive who had vital status data obtained 21 days or longer after admission.
Clinical characteristics at admission of cases with a positive chest x-ray varied substantially by site (Table 2, Table 3, appendix), particularly for wheezing (range by site 10·6–97·3%), fever (9·7–58·9%), hypoxaemia (10·0–75·8%), and oxygen administration (5·5–90·1%; p<0·0001 for all). It should be noted that the South African site, at an altitude of 1600 m above sea level, had a standard clinical practice to administer supplemental oxygen at admission and often before recording oxygen saturation for all children admitted to hospital with a diagnosis of severe pneumonia. Additionally, Mali oxygen administration data were excluded from analysis because of inadequate standardisation of data recording. Tachypnoea was commonly observed at all sites (85·3% of cases). Malnutrition, defined as WHO weight-for-age Z score37 lower than −2, was found in 39·7% of cases with a positive x-ray and without HIV infection, with significant variability by site (p<0·0001 for all); malnutrition was more common in cases than in controls at all sites (appendix). The prevalence of any respiratory symptom among controls without HIV infection was 23·6% (range by site 5·4–41·2%, appendix).
Previous exposure to antibiotics varied by site and case or control status, as did coverage with vaccines against pertussis, Hib, S pneumoniae, and measles (appendix; p<0·050 for all). Enrolment of cases varied by season at all sites, except in Mali and Zambia (appendix).
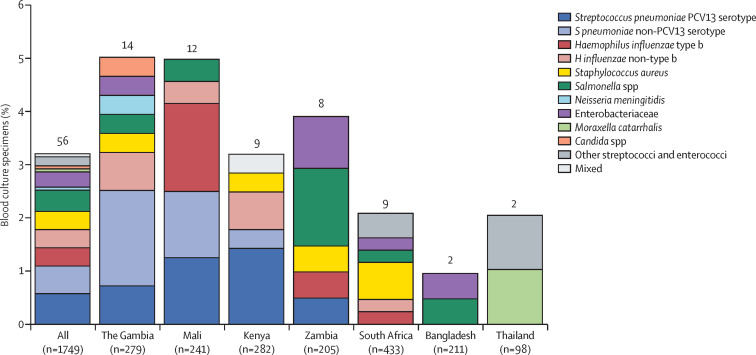
Blood cultures were positive for a pathogen in 56 (3·2%) of 1749 cases with positive chest x-rays and available blood cultures (percentage range by site 1·0–5·0%; figure 2 , appendix). Isolation rates did not vary significantly by age strata (p=0·21), but did vary by case severity (p=0·014) and were 45% lower among cases with previous antibiotic exposure or with blood volume lower than 1·0 mL (appendix).28 The bacterial species most commonly isolated from blood cultures was S pneumoniae (19 [34%] of 56 positive blood cultures); however, no pneumococcal isolates were cultured from sites in South Africa, Thailand, or Bangladesh. Although not part of the core methods, South Africa had two blood cultures that were flagged as positive and tested positive for pneumococcal antigen, but were negative for pneumococcal culture (appendix).
Figure 2.
Blood culture results by study site in cases with positive chest x-ray and without HIV infection
Enterobacteriaceae includes Escherichia coli, Enterobacter spp, and Klebsiella spp, excluding mixed Gram-negative rods. Other streptopcocci and enterococci include Streptococcus pyogenes and Enterococcus faecium. Mixed label includes Salmonella spp and other streptopcocci and enterococci. Contaminants, including those organisms deemed to be contaminants after clinical review, were excluded from the analysis. Figure is restricted to cases with available blood culture results. The numbers on the top of the bars refer to the total number of positive blood cultures. Two of the cases positive for pneumococcus in Kenya were pneumococcal conjugate vaccine (PCV) 13-type but not PCV10-type (serotypes 19A and 6A). Antibiotic pretreatment (defined as having a positive serum bioassay result, antibiotics administered at the referral facility, or antibiotic administration before whole-blood specimen collection at the study facility) varied by site: The Gambia (composite 10·6%, bioassay 7·6%), Mali (22·4%, 17·1%), Kenya (35·1%, 10·0%), Zambia (92·2%, 25·3%), South Africa (57·2%, 54·2%), Bangladesh (24·6%, 21·6%), and Thailand (30·6%, 19·4%).
M tuberculosis was cultured from 24 (1·5%) of 1571 cases with positive x-rays (23 from induced sputum and one from gastric aspirate; appendix). 11 (29%) of 37 lung aspirates and 12 (80%) of 15 pleural fluid specimens tested by culture, PCR, or both were positive for a pathogen; bacterial pathogens (most often S pneumoniae [lung aspirate] and S aureus [pleural fluid]) were more commonly detected (in 23 cases) than viral pathogens (three cases), and all viral pathogens were co-detected with bacteria (table 4 , appendix). Multiple pathogens were observed in six (55%) of 11 lung aspirates that yielded at least one pathogen by culture or PCR.
Table 4.
Organisms identified by culture and PCR of lung aspirate or pleural fluid specimens in cases with a positive chest x-ray and without HIV infection
| Culture | PCR | Culture, PCR, or both | ||
|---|---|---|---|---|
| Lung aspirate | ||||
| Cases with available results (n) | 37 | 25 | 37 | |
| Any positive* | 5 (13·5%) | 9 (36·0%) | 11 (29·7%) | |
| Streptococcus pneumoniae† | 5 (13·5%) | 6 (24·0%) | 8 (21·6%) | |
| Haemophilus influenzae non-b‡ | 1 (2·7%) | 4 (16·0%) | 4 (10·8%) | |
| Chlamydophila pneumoniae | 0 | 1 (4·0%) | 1 (2·7%) | |
| Moraxella catarrhalis | 0 | 4 (16·0%) | 4 (10·8%) | |
| Human metapneumovirus | NA | 1 (4·0%) | 1 (2·7%) | |
| Adenovirus | NA | 1 (4·0%) | 1 (2·7%) | |
| Combinations | ||||
| S pneumoniae + H influenzae | 1 (2·7%)§ | 1 (4·0%) | 1 (2·7%) | |
| S pneumoniae + M catarrhalis | 0 | 2 (8·0%) | 2 (5·4%) | |
| Adenovirus + C pneumoniae | NA | 1 (4·0%) | 1 (2·7%) | |
| H influenzae + M catarrhalis + S pneumoniae | NA | 1 (4·0%) | 1 (2·7%)§ | |
| H influenzae + M catarrhalis + human metapneumovirus | NA | 1 (4·0%) | 1 (2·7%) | |
| Negative | 32 (86·5%) | 16 (64·0%) | 26 (70·3%) | |
| Pleural fluid | ||||
| Cases with available results (n) | 15 | 12 | 15 | |
| Any positive* | 9 (60·0%) | 7 (58·3%) | 12 (80·0%) | |
| S pneumoniae† | 1 (6·7%) | 4 (33·3%) | 5 (33·3%)¶ | |
| Staphylococcus aureus | 7 (46·7%) | 4 (33·3%) | 7 (46·7%) | |
| Escherichia coli | 1 (6·7%) | 0 | 1 (6·7%) | |
| Streptococcus group F | 1 (6·7%) | 0 | 1 (6·7%) | |
| Human bocavirus | NA | 1 (8·3%) | 1 (6·7%) | |
| Combinations | ||||
| E coli + streptococcus group F | 1 (6·7%) | NA | 1 (6·7%) | |
| S aureus + human bocavirus | NA | 1 (8·3%) | 1 (6·7%) | |
| S pneumoniae + S aureus | 0 | 0 | 1 (6·7%)¶ | |
| Negative | 6 (40·0%) | 5 (41·7%) | 3 (20·0%) | |
Data are n (%). Results were restricted to specimens obtained within 3 days of enrolment and to those pathogens determined by the clinical review team to be non-contaminants. NA=not applicable.
Total number of cases with a positive culture result; does not equal the number of organisms identified because some cases appear in multiple rows (any positive and in combination).
For S pneumoniae in lung aspirates, PCV13 types were identified in three cases, non-PCV13 types were identified in two cases, and three cases did not have serotyping available (PCR-positive alone); for S pneumoniae in pleural fluid, PCV13 type was identified in one case and four cases did not have serotyping available (positive result only for PCR test, antigen test, or both).
One case positive for H influenzae by lung aspirate was missing serotyping data for the culture isolate but was negative for H influenzae type b by lung aspirate PCR.
This case was positive for S pneumoniae and H influenzae by lung aspirate culture and positive for H influenzae, M catarrhalis, and S pneumoniae by lung aspirate PCR; therefore, it is reported twice in this table.
Three cases were positive in pleural fluid for S pneumoniae by antigen testing; two of these were also positive for S pneumoniae in culture, PCR, or both, and one was positive only for S pneumoniae in antigen testing.
None of the ten cases with positive chest x-rays who met the clinical screening definition for measles testing were positive for measles (appendix). Legionella longbeachae infection was identified in one case, a 19-month-old Zambian child without HIV infection with very severe pneumonia, in both NP-OP and induced sputum specimens by multiplex PCR and confirmatory uniplex PCR.
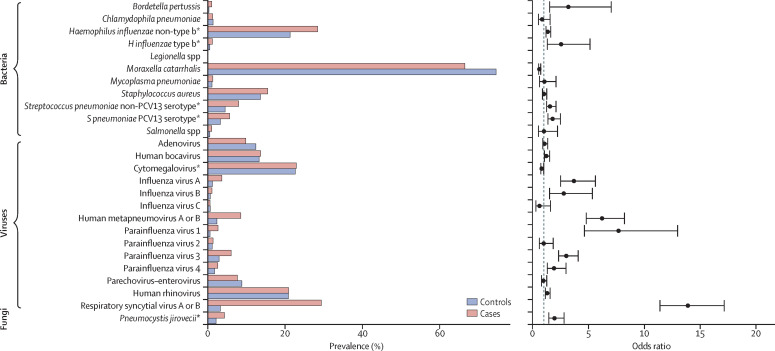
Almost all cases (3867 [98·9%] of 3910) and controls (4886 [98·0%] of 4984) had at least one pathogen detected (ie, ignoring density threshold) by PCR in the NP-OP specimen; most cases (2313 [59·2%]) and controls (2667 [53·5%]) had four or more pathogens detected (appendix). The mean number of potential pathogens detected in NP-OP specimens was 3·8 (SD 1·5) among cases and 3·6 (SD 1·5) among controls. Among cases with multiple pathogens detected in NP-OP specimens, 3219 (88·4%) of 3642 had both viral and bacterial detections; no pathogen was commonly found alone (appendix). In cases with a positive chest x-ray, the most common bacteria detected in NP-OP specimens were S pneumoniae (1265 [72·8%] of 1737), M catarrhalis (1153 [66·4%]), H influenzae (1004 [57·8%]), and S aureus (268 [15·4%]), whereas cytomegalovirus (890 [51·2%]) and RSV (506 [29·1%]) were the most common viruses (appendix). Influenza virus was rarely detected (influenza A 62 [3·6%]; influenza B 18 [1·0%]; influenza C 10 [0·6%]). S pneumoniae was more commonly detected in controls (3846 [77·2%] of 4984) than in cases with a positive chest x-ray (p=0·0001). After applying PCR density thresholds that best distinguished cases from controls,24, 25 the proportion of positive results in cases with a positive x-ray changed for S pneumoniae (from 72·8% to 13·5%), H influenzae (from 57·8% to 29·5%), P jirovecii (from 8·9% to 4·3%), and cytomegalovirus (from 51·2% to 22·7%). This changed the rank order of most commonly detected bacteria in the NP-OP specimens to M catarrhalis, H influenzae, S aureus, and S pneumoniae (appendix). Compared with controls, having a positive chest x-ray in cases was significantly associated with the presence of 15 different pathogens in NP-OP specimens (figure 3 ). The adjusted ORs for NP-OP PCR results ranged widely across pathogens, with RSV (OR 14·0, 95% CI 11·4–17·1), parainfluenza 1 (7·7, 4·6–13·0), and human metapneumovirus A or B (6·3, 4·8–8·2) having the highest adjusted ORs, followed by influenza A (3·6, 2·4–5·3) and Bordetella pertussis (3·3, 1·6–7·2); parainfluenza 1, influenza A, and B pertussis were infrequently detected in cases (figure 3, appendix). M catarrhalis was the only pathogen significantly negatively associated with case status (OR 0·6, 95% CI 0·5–0·7). We observed only minor changes in the magnitude and direction of the ORs after removing the adjustment for detection of the other pathogens (appendix).
Figure 3.
Nasopharyngeal-oropharyngeal (NP-OP) pathogen prevalence* and adjusted odds ratios (OR) in cases with positive chest x-ray and without HIV infection and in controls without HIV infection
Pathogens are ordered alphabetically among bacteria, followed by viruses and fungi. ORs adjusted for age (months), site, and presence of other pathogens detected by NP-OP PCR, but not adjusted for previous antibiotic use, which is known to influence bacterial positivity. *Prevalence defined by use of NP-OP PCR density thresholds for four pathogens: Pneumocystis jirovecii, 4 log10 copies per mL; Haemophilus influenzae, 5·9 log10 copies per mL; cytomegalovirus, 4·9 log10 copies per mL; Streptococcus pneumoniae, 6·9 log10 copies per mL; NP-OP PCR results based on positivity are in the appendix. PCV=pneumococcal conjugate vaccine.
Whole blood PCR was positive for S pneumoniae in cases with a positive x-ray (128 [7·6%] of 1676) at nearly the same prevalence as in controls (255 [5·3%] of 4779; OR 1·2, 95% CI 1·0–1·6); after applying a PCR density threshold (≥2·2 log10 copies per mL) to enhance differentiation of cases and controls,26, 27 prevalence was 91 (5·4%) of 1676 cases with a positive chest x-ray versus 144 (3·0%) of 4779 controls (1·6, 1·2–2·1; appendix).
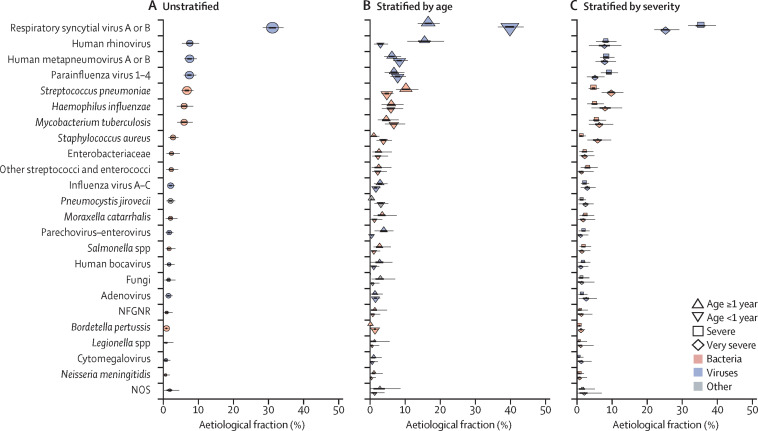
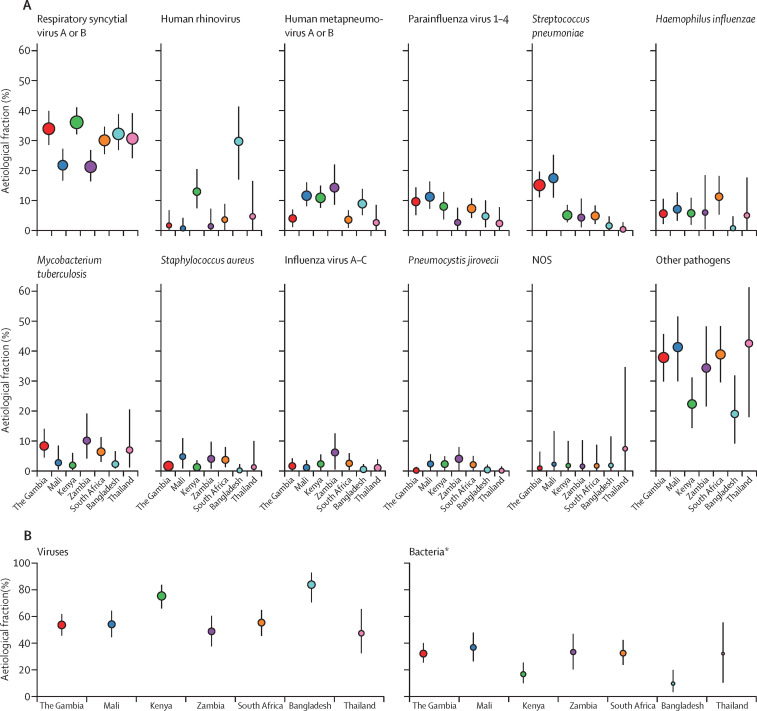
Integrating the microbiology results from the multiple specimens among the 1767 cases with a positive chest x-ray and the 5075 controls, we estimated that more disease was due to viral pathogens (61·4%, 95% CrI 57·3–65·6) than to bacterial pathogens (other than M tuberculosis; 27·3%, 23·3–31·6); the remaining aetiological fractions were attributed to M tuberculosis, Candida spp, P jirovecii, and causes not otherwise specified (figure 4A , appendix). RSV was the most common cause (31·1%, 95% CrI 28·4–34·2), and other common pathogens (each accounting for 5% or more of the aetiological distribution) included human rhinovirus, HMPV A or B, human parainfluenza virus (types 1–4 combined), S pneumoniae, M tuberculosis, and H influenzae. Unknown causes (NOS category) accounted for 1·8% (95% CrI 0·2–4·5) of the pneumonia aetiology distribution. The fraction of pneumonia attributable to pathogens targeted by currently licensed vaccines (Hib, vaccine-serotype S pneumoniae, pertussis, M tuberculosis, and influenza A or B) was 14·0% (11·3–17·0).
Figure 4.
Aetiological fraction unstratified (A), stratified by age (B), and stratified by severity (C) for cases with a positive chest x-ray and without HIV infection from all PERCH sites combined
Lines represent 95% credible interval; the darker region of the line represents the IQR. The size of the symbol is scaled on the basis of the ratio of the estimated aetiological fraction to its SE. Of two identical aetiological fraction estimates, the estimate associated with a larger symbol is more informed by the data than the priors. Positive chest x-rays defined as consolidation or other infiltrate on the x-ray. The following pathogens contributed less than 1% to the aetiological fraction (overall and after stratifying by age and severity) and were excluded from the figure: Coronavirus, Chlamydophila pneumoniae, and Mycoplasma pneumoniae. Other streptococci and enterococci includes Streptococcus pyogenes and Enterococcus faecium. Non-fermentative Gram-negative rods (NFGNR) includes Acinetobacter spp and Pseudomonas spp. Enterobacteriaceae includes Escherichia coli, Enterobacter spp, and Klebsiella spp, excluding mixed Gram-negative rods. Pathogens that were estimated at the subspecies level, but grouped to the species level for display include parainfluenza virus type 1, 2, 3 and 4; Streptococcus pneumoniae PCV 13 and S pneumoniae non-PCV 13 types; Haemophilus influenzae type b and H influenzae non-type b; and influenza A, B, and C. Exact figures, including subspecies and serotype disaggregation (eg, PCV13 type and non-PCV13 type), are given in the appendix. NOS=not otherwise specified (ie, pathogens we did not test for).
Some pathogens differed in their aetiological fraction by age strata. B pertussis, parainfluenza type 3, S aureus, P jirovecii, and RSV had a greater aetiological fraction in children younger than 1 year than in those aged 1 year or older, whereas parechovirus–enterovirus, parainfluenza type 1, rhinovirus, and S pneumoniae were more common causes in children aged 1 year or older than in those younger than 1 year (figure 4B; appendix). We found seven pathogens in the top ten pathogens of both age strata (RSV, rhinovirus, HMPV, parainfluenza, S pneumoniae, M tuberculosis, and H influenzae). The aetiological distribution by finer age strata is provided in the appendix. We observed differences in aetiological fraction by pneumonia severity for RSV, S aureus, S pneumoniae, parainfluenza type 3, and all viral causes combined (figure 4C, appendix).
In analyses of all cases regardless of chest x-ray findings compared with cases with a positive x-ray, a greater aetiological fraction was attributed to rhinovirus (11·4% in all cases vs 7·5% in cases with a positive x-ray), parechovirus–enterovirus (4·6% vs 1·6%) and non-fermenting Gram-negative bacteria (2·8% vs 0·9%) in all cases, but otherwise, the analyses did not differ substantively (appendix). Aetiology by chest x-ray findings will be reported in a separate manuscript.
The site-specific aetiology results standardised for age and severity of the ten focus pathogens showed consistency across the sites for most pathogens (figure 5 , appendix), including for influenza, which did not exceed 3% in any site except Zambia (6·1%). Exceptions to this consistency included the following: rhinovirus was substantially more common in Bangladesh (29·7%) and Kenya (12·9%); H influenzae was extremely uncommon in Bangladesh (0·7%); H influenzae type b in Zambia (3·4%) and Mali (4·2%) and non-type b H influenzae in South Africa (10·0%) were disproportionately higher than in other sites; and S pneumoniae, both vaccine-types and non-vaccine-types, was higher in The Gambia (15·1%) and Mali (17·4%) than in other sites. The fraction of cases attributed to bacteria was lower in Bangladesh (10·4%) and Kenya (17·5%) than in other sites (range 32·7–37·6). M tuberculosis had a larger aetiological fraction in Zambia, The Gambia, Thailand, and South Africa (6·4–10·2) than in other sites (<3%). The NOS contribution was 2% or less in all sites except Thailand (7·4%). The results standardised for age and severity (appendix) did not differ from the primary, non-standardised, site-specific results (ie, based on the mix of cases enrolled at each site).
Figure 5.
Site-specific aetiology results for ten focus pathogens in cases with a positive chest x-ray and without HIV infection
The size of the symbol is scaled on the basis of the ratio of the estimated aetiological fraction to its SE. Of two identical aetiological fraction estimates, the estimate associated with a larger symbol is more informed by the data than the priors. Positive chest x-rays defined as consolidation or other infiltrate on the x-ray. Graph restricted to the ten focus pathogens from the all-site analysis, which include those with aetiology estimate higher than 5% (n=7) or higher than 2% that were of epidemiological interest (defined as treatable by antibiotics [Pneumocystis jirovecii and Staphylococcus aureus] or having an available vaccine [influenza virus]). The 95% credibility intervals for the aetiological fractions of these three pathogens overlap with some non-focus pathogens, hence our use of the term focus pathogen rather than labelling these ten as the most common pathogens. Other pathogens category represents the sum of the aetiological fraction for all remaining pathogens tested for, but not presented in this figure. Site-specific results were standardised to the following case mix: 40% younger than 1 year with severe pneumonia, 20% younger than 1 year with very severe pneumonia, 30% aged 1 year or older with severe pneumonia, and 10% aged 1 year or older with very severe pneumonia. Pathogens estimated at the subspecies level, but grouped to the species level for display include parainfluenza virus types 1, 2, 3, and 4; Streptococcus pneumoniae PCV13 and S pneumoniae non-PCV13 types; Haemophilus influenzae type b and H influenzae non-type b; and influenza virus A, B, and C. Exact figures are given in the appendix. NOS=not otherwise specified (ie, pathogens we did not test for). *The summary for bacteria excludes Mycobacterium tuberculosis.
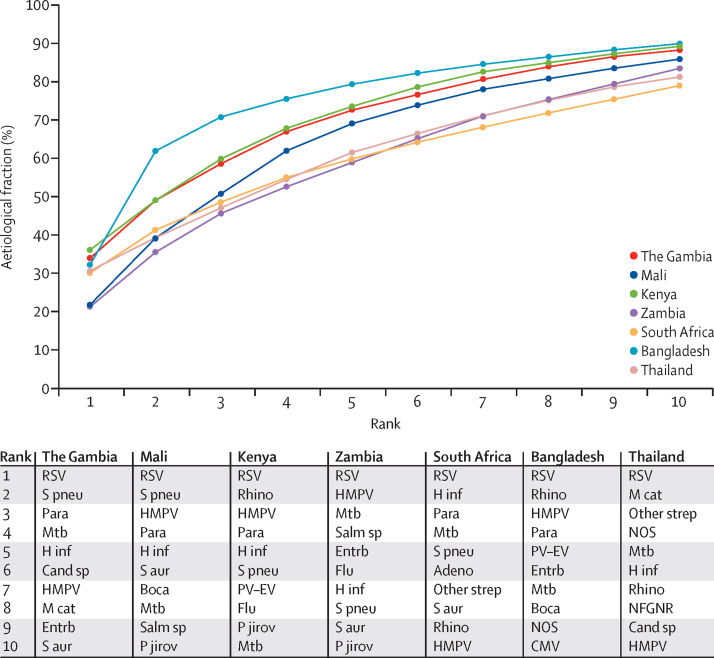
The cumulative aetiological fraction of the standardised and site-specific ten most common pathogens accounted for between 79·0% and 90·0% of the aetiological fraction at every site (figure 6 ). By contrast, the cumulative aetiological fraction of the ten focus pathogens ranged by site from 55·1% to 86·6%, reflecting heterogeneity across the sites in the rank order of each pathogen (appendix). The distribution of the individual case aetiological probability for each pathogen, for the case-specific leading pathogen, and for any virus by site are provided in the appendix.
Figure 6.
Cumulative contribution of site-specific ten most common pathogens in cases with a positive chest x-ray and without HIV infection
Positive chest x-rays defined as consolidation or other infiltrate on the x-ray. Site-specific results were standardised to the following case mix: 40% younger than 1 year with severe pneumonia, 20% younger than 1 year with very severe pneumonia, 30% aged 1 year or older with severe pneumonia, and 10% aged 1 year or older with very severe pneumonia. Ranks correspond to the site-specific rank from the top ten pathogens of each site; the pathogen corresponding to each rank varies by site (see inset panel). Other strep category includes Streptococcus pyogenes and Enterococcus faecium. Non-fermentative Gram-negative rods (NFGNR) includes Acinetobacter spp and Pseudomonas spp. Enterobacteriaceae category (Entrb) includes Escherichia coli, Enterobacteriaceae spp, and Klebsiella spp, excluding mixed Gram-negative rods. Pathogens estimated at the subspecies level, but grouped to the species level for display include parainfluenza virus types 1, 2, 3, and 4; Streptococcus pneumoniae PCV13 and S pneumoniae non-PCV13 types; Haemophilus influenzae type b and H influenzae non-type b; and influenza virus A, B, and C. Boca=human bocavirus. Cand sp=Candida spp. CMV=cytomegalovirus. Flu=influenza virus A, B and C. H inf=H influenzae. HMPV=human metapneumovirus A or B. Mtb=Mycobacterium tuberculosis. M cat=Moraxella catarrhalis. NOS=not otherwise specified (ie, pathogens we did not test for). P jirov=Pneumocystis jirovecii. Para=parainfluenza virus type 1, 2, 3 and 4. PV-EV=parechovirus–enterovirus. Rhino=human rhinovirus. RSV=respiratory syncytial virus A or B. S aur=Staphylococcus aureus. S pneu=S pneumoniae. Salm sp=Salmonella spp.
Sensitivity analyses were done to assess the effect of the assumptions of PIA model sensitivity and aetiology priors on the aetiology findings. Lowering and narrowing the pneumococcal blood culture sensitivity prior, or increasing its aetiology prior, had little effect on the S pneumoniae aetiology estimate, probably because of the strength of evidence regarding S pneumoniae from the other measurements (NP-OP PCR and whole blood PCR; appendix). Increasing the aetiology prior of NOS from 3% (base case) to 25% resulted in an increase in the NOS aetiology estimate from lower than 8% at all sites to 8–25% across sites (appendix).
Discussion
The principal finding of the PERCH study was that, in low-income and low-middle-income settings with widespread uptake of Hib vaccine and PCV, viruses, especially RSV, were the predominant cause of pneumonia requiring hospital admission in children younger than 5 years at all sites, but bacteria continued to cause from a quarter to a third of all cases. In each site, a small number of pathogens accounted for most cases admitted to hospital with radiographically confirmed pneumonia; the ten most common pathogens at a given site accounted for 79% or more of cases of disease at that site. Furthermore, most cases at every site were caused by a handful of pathogens that were commonly found in all sites. Nonetheless, the relative contribution of these pathogens varied by age and, to a lesser degree, by severity of the disease.
Approximately half of children admitted to hospital defined by WHO as having severe or very severe pneumonia had an infiltrate on chest x-rays, and among these cases, clinical characteristics varied substantially across all sites, including case severity at presentation, presence of wheeze, and case-fatality ratios. Nevertheless, we found little variability in the pathogens commonly detected from body fluid specimens of cases across study sites, except for the low recovery of bacteria from blood specimens in the Asian sites, or the pathogens accounting for the majority of aetiology distribution.
RSV was the dominant pathogen in all sites, accounting for 31·1% of the aetiology distribution, more than three times greater than the next leading pathogen. RSV was notable not only for its frequent detection in the NP-OP of cases with a positive chest x-ray, but also for its uncommon detection in controls, making it one of a few pathogens (including parainfluenza 1 and HMPV) for which detection had high predictive value for aetiological attribution. RSV was a common pathogen across age strata and pneumonia severities, although the aetiological fraction was notably higher in infants than in older children. The predominance of RSV emphasises its importance as a prevention and therapeutic target. In the past decade, pneumonia aetiology studies29, 38 in the USA and South Africa and the GABRIEL multisite study39 also identified RSV as an important target.
The contribution of parainfluenza viruses and HMPV to the aetiological distribution varied little by age strata or disease severity but did vary by site. We recorded a low prevalence (<5%) of influenza A or B in all sites and the aetiological contribution of influenza A or B did not exceed 3% in any site except Zambia (6·1%). Influenza circulation varies annually, and the 2 years of enrolment in the PERCH study might have been low circulation years.
Of the tested pathogens, human rhinovirus was associated with the most geographical heterogeneity in aetiological fraction, ranging from 30% in Bangladesh and 13% in Kenya, to less than 2% in Zambia, The Gambia, and Mali. Human rhinovirus predominance in Kenya was not explained by an overrepresentation of respiratory infection seasons (three compared with two seasons in all other African sites; appendix), on the basis of an analysis of the Kenya data that excluded the third respiratory season (data not shown). Although well described as a cause of upper respiratory tract infection, the role of human rhinovirus as a cause of pneumonia is uncertain.40, 41, 42 It is possible that human rhinovirus, as several other respiratory viruses, might be incorrectly attributed as the true cause of pneumonia despite all methodological and analytical efforts to address this. This general limitation extends beyond this study to most case-control studies for pneumonia aetiology, leading to an exaggeration of the contribution of viruses to pneumonia when the primary specimen used to assess causality is from the upper respiratory tract. However, the OR and the significant association with wheeze (data not shown) argues against this.
For bacteria that commonly colonise the upper respiratory tract, such as S pneumoniae and H influenzae, the inferential role of NP-OP PCR for estimating aetiology in a case-control study has inherent uncertainties. To mitigate the risk of underestimating the attributable aetiology of these bacterial pathogens, analyses incorporated pathogen density measures from the NP-OP specimens to distinguish cases from controls, adjustment for antibiotic exposure that lowers detection in the NP-OP PCR, and blood PCR for S pneumoniae. Additionally, for these and all bacterial pathogens, we directly sampled from the lung (albeit in few cases), optimised and adjusted for the volume of blood for culture, accounted for the effect of previous antibiotic exposure on the sensitivity of bacterial pathogen detection for blood cultures, and accounted for the low sensitivity of blood cultures.
About 3% of cases in the PERCH study had a non-contaminant organisms isolated from blood culture. S pneumoniae was the most common isolate, consistent with other childhood pneumonia aetiology studies.29, 38 Although we attempted to improve identification of cases with true pneumococcal pneumonia using whole blood PCR,26, 27 the specificity was poor because prevalence was similar between cases and controls (aOR 1·2); this improved somewhat when bacterial load was considered (aOR 1·6). Perhaps the poor specificity of blood PCR is due to low level seeding, swallowed sputum, or gastrointestinal tract absorption of pneumococcal antigens from common colonisation with S pneumoniae in the nasopharynx.43 This mechanism is also believed to be the source of poor pneumococcal urine antigen specificity.44 Despite the high prevalence of pneumococcal NP-OP colonisation among children in every site,25 the prevalence of positive blood PCR for S pneumoniae was lower in Asian sites26 and in the Zambia site, which had the highest prevalence of antibiotic pretreatment. Neither the Asian sites nor the South Africa site had a positive blood culture for S pneumoniae, unlike the other four African sites. We investigated the site performance of blood collection and culture and found no methodological explanation for the inability to isolate S pneumoniae from blood cultures at these three sites, nor for the lower prevalence of positive pneumococcal results in the Asian sites compared with that of other sites. However, the South Africa site had two blood culture samples that were alarm positive, culture negative, with pneumococci in chains detected on Gram stain, and Binax positive, and all three sites isolated pneumococci from blood in patients who were not enrolled in the PERCH study during the same period. Antibiotic treatment before specimen collection is known to influence the isolation of bacteria from blood and NP, as we have previously described,25, 28 but does not substantially influence pneumococcal detection by whole blood PCR.26, 27 The prevalence of antibiotic pretreatment was particularly high in Zambia (92%), South Africa (57%), and Kenya (35%); this was accounted for, as described previously, by reducing the sensitivity.
There were reasons to expect that, in our study, there would be a lower burden of pneumococcal disease than in earlier studies. By design, sites with PCV in routine use were targeted for participation in the PERCH study (all except Zambia, Thailand, and Bangladesh used PCV), which could have reduced the pneumococcal pneumonia disease burden by approximately 70% at those sites, if the serotype distribution matched that of invasive pneumococcal disease.45 The low pneumococcal aetiological attribution is supported by the low prevalence of radiographic consolidation that has been shown to be associated with bacterial pneumonia,46, 47, 48 which comprised only 25% of all PERCH cases and half of the cases with a positive chest x-ray. Furthermore, 60% of cases in the PERCH study were younger than 1 year, with many younger than 6 months, a group often excluded from the analyses of clinical trials of pneumococcal vaccine that partly formed the basis for our expectations about pneumococcal burden. Economic development and improved access to care might also have reduced hospital admissions for pneumococcal pneumonia through effective early outpatient treatment. In some sites, such as Thailand, the absence of any detected cases of pneumococcal bacteraemia might simply reflect low enrolment numbers, given that invasive pneumococcal disease has been observed at this site in other studies.49, 50, 51
Among other vaccine-preventable pathogens, M tuberculosis burden varied by site, accounting for 6–10% of the aetiology in four sites (The Gambia, Zambia, South Africa, and Thailand), but less than 3% elsewhere. M tuberculosis was rarely cultured from induced sputum or gastric aspirates and, in the South Africa site, testing more than one specimen increased the yield.52 The sensitivity of M tuberculosis culture is low when testing a single induced sputum specimen (30%) and remains low even when multiple samples were taken from children with pulmonary tuberculosis;53 this was accounted for in the aetiology model. Perhaps owing to high vaccination rates, B pertussis was rarely identified but, when present, was most often found in children younger than 12 months. Detection of B pertussis by PCR of NP-OP specimens was highly predictive of case status, with very few positives identified in controls.54 We saw few cases that were clinically compatible with measles, therefore it is unlikely that we missed many measles-related cases of pneumonia, highlighting the efforts of measles control at the sites during the study. Hib was an uncommon cause for pneumonia in our study and was less common than non-type b H influenzae. The longstanding use of Hib vaccine in several study sites could have accounted for the low contribution of Hib to the aetiological distribution. However, the two sites where the contribution of Hib exceeded the contribution of non-b H influenzae (Mali and Zambia) did not have the lowest Hib vaccination rates. Thailand was the only site not using Hib vaccine; the reason for the small contribution of Hib to pneumonia aetiology in Thailand is unclear, but might relate to access to outpatient antibiotics.
P jirovecii is a prominent cause of pneumonia among children with HIV infection. The PERCH study has shown that it is also a relevant cause of pneumonia in a subset of children without HIV infection. Among cases, those who had P jirovecii detected in the NP-OP specimens were more likely than other cases to be younger than 6 months and malnourished. To the best of our knowledge, the single case of legionellosis (in the Zambia site) that we identified is the first reported case of childhood infection due to L longbeachae in Africa.
The aetiological contribution of non-tested pathogens (ie, the NOS category) was approximately 2% in the all-site analysis. Sensitivity analyses that assumed a higher fraction for the NOS category (aetiology prior set at 25% vs 3%) concluded that the NOS category would probably not account for more than 15% of cases across the sites, except in Thailand (25%). This emphasises that strong evidence exists for the aetiological contribution of pathogens tested for in our study, unless there is substantial confounding of the true cause by these pathogens. Because the value chosen for the aetiology prior influences the fraction attributed to the NOS category, our analysis cannot be used to estimate precisely the size of the NOS category. However, it does affirm that NOS accounts for a small fraction of pneumonia causes.
The contribution of NP-OP data for assessing aetiological probability depends on differences in frequency of detection between cases and controls. The validity of NP-OP infection as a source of aetiological inference for a selected group of pneumonia pathogens is supported by the finding that clear distinctions in prevalence by case or control status were found consistently across sites. However, NP-OP specimens provide little inferential value to the cause of pneumonia at the individual case level, except for a small group of pathogens, such as RSV and B pertussis. Collection of induced sputum added little value beyond that of the NP-OP specimen, despite the high quality of the lower respiratory tract specimens.30, 32 For those pathogens with NP-OP OR lower than 1, ruling out their role in the causal chain was not possible with this study design; probe or longitudinal study designs are needed to better understand the contribution of these pathogens to pneumonia aetiology and better explore the sequence from NP-OP infection to lung infection. Despite their limitations, NP-OP specimens remain of substantial importance for pneumonia aetiology studies because they are easily collected, can be subjected to molecular diagnostic testing, and can provide valid information about infection of certain pathogens in the respiratory tract.
Four sites (The Gambia, South Africa, Mali, and Bangladesh) included lung aspirate specimen collection as part of the study protocol. Because of the clinical criteria for specimen collection (ie, cases with anterior, large, and right-sided consolidation), hesitation from clinicians and families, and delays in ethical approvals and training at some sites, only 37 (6·8%) of 545 cases with radiographic consolidation enrolled at these four sites had a lung aspirate specimen collected. Pathogen detection from lung aspirates was lower (29% overall, 41% when both culture and PCR were undertaken) than that reported in previous studies, possibly because the timing of lung aspirates occurred after antibiotic administration.55 Pleural fluid specimens, included in the study protocol in all sites at the discretion of the treating clinicians, were only available from 15 (8%) of 180 cases in whom at least one standardised chest x-ray reader identified pleural fluid; however, a pathogen was detected in 80% of these specimens. Bacteria were more commonly detected on these specimens than viruses, with the most common being S pneumoniae in lung aspirates and S aureus in pleural fluid. These specimens, although most directly reflective of the true cause of pneumonia, were collected in a highly selected group of cases and the results are unlikely to be generalisable.
Clinical characteristics and case outcomes varied substantially by site. The high variation in wheezing prevalence among cases, especially the extremely high prevalence in Bangladesh, might be related to poor air quality, infection, or underlying propensity for bronchospasm in the children presenting for care.56, 57, 58 Sites also differed in the mix of case severity, which might reflect underlying host factors (eg, nutritional status), other concurrent illnesses (eg, malaria), or patient propensity for seeking care and ability to access to care. Because pathogen findings differed by case severity, it is possible that the stratification of cases on finer or differently defined severity indicators might control for some confounding by site. Case-fatality ratios varied by site, reflecting differences in case severity and clinical care. Among all cases who died in the 30 days after hospital admission, only a small proportion of deaths (41 [14%] of 292) occurred after discharge. This is similar to the proportion of mortality (16% of deaths) observed to occur after discharge in a study of severe pneumonia requiring hospital admission in children younger than 5 years in Kilifi,59 but contrasts with the substantial proportion of mortality (74% of deaths) observed to occur in the 90 days after enrolment for diarrhoeal illness in the GEMS study,60 although those children were followed for a longer period.
We did not obtain chest x-rays from a small proportion of cases (6%, n=234), many (33%) because of death soon after enrolment, limiting the representation of microbiological findings in immediately fatal cases in the primary analysis. Among cases with a chest x-ray, 53% had either a normal or uninterpretable finding; by design, the WHO clinical pneumonia case definitions are highly sensitive, but non-specific. We focused on the epidemiological and laboratory characteristics of cases with pneumonia with a positive x-ray to enhance the specificity of the clinical case definition for pneumonia, recognising that this might introduce bias by excluding cases with pneumonia whose x-ray findings lagged behind the clinical findings or cases with lower tract disease caused by pathogens who did not have chest x-ray infiltrates (eg, pertussis). We assessed this bias by providing analyses of all cases, regardless of x-ray findings, and observed little difference in the findings. Notably, among cases without a positive chest x-ray, 26% either had a missing or uninterpretable x-ray.
We did the PERCH study over a 24-month period at each site to obtain more reliable data on annual seasonality patterns and to increase the probability of detecting pathogens that exhibit multi-year epidemic cycles. We were unlikely to miss seasonal outbreaks; however, some pathogens (eg, B pertussis, influenza subtypes, parainfluenza viruses, and Mycoplasma pneumoniae) have periodicities that are longer than 24 months and, therefore, we might have observed quiescent periods.61, 62, 63, 64
Among the ten focus pathogens of our study, a subset (HMPV, parainfluenza, S pneumoniae, human rhinovirus, H influenzae, and M tuberculosis) differed in their importance across sites. The estimated contributions of some pathogens varied by age strata (RSV, pertussis, parainfluenza types 1 and 3, P jirovecii, S aureus, S pneumoniae, parechovirus-enterovirus, and rhinovirus) and by disease severity (all viral causes combined, RSV, S aureus, S pneumoniae, and parainfluenza type 3), which might have implications for treatment guideline revisions. Because preventive measures against current pathogens are increasingly implemented, the proportional aetiological distribution will continue to change over time.
The PERCH study results can be compared with other childhood pneumonia aetiology studies.29, 39, 65, 66 The multisite BOSTID study66, 67 from the 1980s, an important historical reference, detected viruses and bacteria from upper respiratory specimens and bacterial antigens from urine specimens and found high prevalence of RSV, S pneumoniae, and Hib. The PERCH study differed substantially from BOSTID in study design through the inclusion of controls, in strict standardisation of case definitions and methods, and in the breadth and types of laboratory testing. The multicountry GABRIEL pneumonia case-control aetiology study,65 done in low-income settings and published in 2017, also highlighted RSV as a major causative pathogen. However, the aetiological attribution for RSV was substantially lower in the GABRIEL study than in the PERCH study (18% vs 31%).65 This discordance appears mainly attributable to differences in the prevalence of RSV in GABRIEL (20% of cases) compared with that in the PERCH study (29% of cases), rather than a result of different analytical approaches. Study design and methodological differences probably affected the rate of RSV detection; specifically, the age distribution of cases enrolled in GABRIEL was older than that of cases in PERCH, and GABRIEL controls were enrolled from facilities, rather than from the community. Although S pneumoniae detection in NP specimens in cases was similar between the two studies (68% in GABRIEL vs 73% in PERCH [any detection]), the control prevalence was considerably lower in GABRIEL (48% vs 77%), resulting in a high OR and attributable fraction in the GABRIEL study. The difference in pneumococcal findings between the PERCH and GABRIEL studies is unlikely to lie in the case definitions used, which were similar between studies. Instead, notable differences existed in PCV use (nearly absent in GABRIEL, but widely used in PERCH) and in control eligibility (facility-based recruitment and no respiratory symptoms in GABRIEL vs community controls regardless of respiratory symptoms, if not case-defining, in PERCH). Additionally, the proportion of cases and controls who had detection of both viruses and bacteria was substantially higher in PERCH (83·5% cases and 75·8% of controls had mixed bacterial-viral detection) than in GABRIEL (59·6% cases and 36·1% controls), suggesting that other differences in the selection of participants or in clinical or laboratory methods might have also contributed. In the EPIC study38 of childhood pneumonia aetiology from the USA, RSV and human rhinovirus were the most commonly detected pathogens from the upper respiratory tract among young children hospitalised with radiographic pneumonia, and a larger proportion of children had no pathogen detected compared with that of PERCH (18·9% in EPIC vs 0·9% in PERCH). In the South African Drakenstein case-control substudy29 of pneumonia aetiology (using the same multiplex PCR assay as PERCH), cases were predominantly children with non-severe pneumonia, managed in outpatient settings, and with a median age of 5 months. Nevertheless, a range of pathogens similar to that of the PERCH study was associated with pneumonia case status, but the prevalence of influenza was higher. Unlike the PERCH study of severe pneumonia with hospital admission, the Drakenstein substudy found that cytomegalovirus, bocavirus, and adenovirus were associated with case status.29
Our study has several limitations, particularly related to viral pathogens as confounders of pneumonia aetiology, to the copathogen interpretation, and to bacterial aetiology. Our descriptive analysis underscored limitations inherent to nearly every pneumonia aetiology study—multiple specimens, multiple tests with varying sensitivity and specificity, and the absence of an established and principled method to integrate the results in a single aetiology analysis—and justifies the novel analytical approach we have taken. As in all case-control studies, residual confounding cannot be excluded. Despite matching controls by community of residence, age, and season of cases, there are likely to be factors that influence nasopharyngeal infection and independently influence infection with significant lung pathogens (eg, crowding); this could overestimate the role of viruses and underestimate the role of bacteria. The PIA model integrates the multiple observed laboratory results, acknowledging the inherent error in laboratory measurements, the reduced sensitivity of assays because of previous antibiotic use or low blood culture volume, and the indirect nature of data obtained from sampling body fluids distant from the lung. The model assumes each case has a primary pathogen as the cause and does not attempt to estimate whether specific pathogen combinations are necessary for pneumonia. This approach has been described in detail elsewhere9 and has been contrasted with the standard attributable fraction method. The PIA model includes assumptions on the sensitivity of laboratory measures and aetiology priors, each with a range provided. The validity of the assumptions regarding sensitivity is limited by the availability of data on diagnostic test performance. Despite the potential misspecification of these assumptions, sensitivity analyses showed that the aetiology results were robust to changes in these values, except the aetiology prior for NOS.9 Furthermore, the inherent ability of an observational study to accurately define the contribution of bacteria to pneumonia is uncertain. To advance this problem will require better diagnostics (such as discriminating PCR in blood samples or other pathogen-specific, infection-specific biomarkers), specimens other than the NP-OP swabs (such as lung aspirates, which are very challenging to obtain), and the sampling of patients before they have had antibiotics, which, in the era of community case management, is increasingly unlikely. A vaccine probe study is the optimal way to reveal the contribution of a pathogen to pneumonia,68 but this can only be undertaken for one or two pathogens at a time. Finally, the all-site PERCH study pneumonia aetiological distribution reflected estimates of the distribution of cases with pneumonia who were admitted to a hospital, outside of the neonatal period, across the PERCH study sites during the period of observation. We recognise that pneumonia is an important illness in neonates. Because there was a sister study, ANISA,69 assessing the causes of neonatal disease during the same period, the PERCH study focused on children outside the neonatal period.
The results from the PERCH study should not be extrapolated as the global aetiological distribution of severe childhood pneumonia or interpreted as the aetiological distribution at the national level for each of the participating sites. At the time of study initiation (2011), 88% of countries eligible for programmes of Gavi, the Vaccine Alliance, were using Hib vaccine and 21% were using PCV in their routine immunisation programmes.70 However, the PERCH study was intentionally designed to reflect the aetiology in the setting of high PCV and Hib vaccine use. By 2020, these vaccines are expected to be used in 100% (Hib vaccine) and 93% (PCV vaccine) of Gavi-eligible countries.70 Furthermore, the aetiological distribution among the cases does not necessarily reflect the aetiological distribution among pneumonia deaths, especially in children who died in the community without the provision of curative care efforts.
The PERCH study was designed to update the evidence base on causes of serious pneumonia with use of the most advanced clinical, microbiological, and analytical methods, deployed across geographically and epidemiologically diverse study sites in Africa and south Asia, where the pneumonia burden is greatest. Beyond comparing the clinical and microbiological findings of cases and controls, as has traditionally been done in aetiology studies, we have used a novel analytical method to overcome some of the limitations of this traditional approach and estimate the aetiology distribution, with 95% CrI, in both the population and each individual. Previously, infectious aetiology studies were unable to integrate the results from multiple specimen types for multiple pathogens.
Our results highlight the consistency of the pneumonia aetiology findings across sites, disease severity, and age strata, providing evidence to target the handful of pathogens that are consistently identified as important for prevention and treatment. RSV is the most common cause of serious pneumonia with hospital admission in these settings and should be a target for dedicated prevention and treatment efforts. Bacterial pathogens cumulatively cause a substantial proportion of disease and, because they are treatable and commonly fatal, should remain as targets for early access to appropriate supportive and curative treatment. The PERCH study estimated that 61% of pneumonia requiring hospital admission in children younger than 5 years and without HIV infection had a primary viral cause; however, we cannot firmly exclude the presence of bacterial co-infection, as discussed above. Nevertheless, this finding strengthens the need to adjust pneumonia treatment algorithms to emphasise supportive care and identify children for whom antibiotics are unlikely to be therapeutic, thereby reducing the pressure for antimicrobial resistance. Future pneumonia research needs to address the conundrum of multiple pathogens and the sequence of their role in pneumonia pathogenesis. The findings from the PERCH study should prompt additional investments in pneumonia prevention, testing, triage, and treatment and should encourage the use of innovative design and analyses in aetiology studies, especially those that can further assess pneumonia as a multipathogen disease.
Correspondence to: Dr Katherine L O'Brien, Department of International Health, International Vaccine Access Center, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD 21231, USA obrienk@who.int
This online publication has been corrected. The corrected version first appeared at thelancet.com on August 29, 2019
Data sharing
We are in the process of making the PERCH dataset publicly available in ClinEpiDB (University of Pennsylvania, PA, USA; https://clinepidb.org; we estimate availability in 2020).
Acknowledgments
Acknowledgments
The PERCH study was supported by grant 48968 from the Bill & Melinda Gates Foundation to the International Vaccine Access Center, Department of International Health, Johns Hopkins Bloomberg School of Public Health (Baltimore, MD, USA). JAGS was supported by a clinical fellowship from the Wellcome Trust (UK; 098532). We gratefully acknowledge the substantial contributions to the design of the study by the Pneumonia Methods Working Group (Zulfiqar A Bhutta, Robert E Black, Harry Campbell, Thomas Cherian, Derrick Crook, Menno D de Jong, Scott F Dowell, Stephen M Graham, Keith P Klugman, Claudio F Lanata, Shabir A Madhi, Paul Martin, James Nataro, Franco Piazza, Shamim Ahmed Qazi, Heather Zar) and the PERCH Site Selection Committee (David L Blazes, Robert Bollinger, Claire V Broome, John A Crump, Olivier Fontaine, Robert Gie, David Goldblatt, Patrick W Kelley, Sharon J Peacock, Mark S Riddle, Montse Soriano-Gabarró, Annelies Van Rie, Christopher W Woods). The PERCH Expert Group (William C Blackwelder, Harry Campbell, John A Crump, Menno D de Jong, Adegoke Falade, Claudio F Lanata, Kim Mulholland, Shamim Ahmed Qazi, Cynthia G Whitney) provided valuable advice during the conduct of the study and the analysis of the results for which we are indebted. We also recognise the support provided by the Institutional Review Boards for study oversight. We appreciate the helpful discussions with our many colleagues who were conducting aetiology studies of pneumonia, neonatal sepsis, and diarrhoea during the same period, including the EPIC study, the GABRIEL study, the ANISA study, and the GEMS study. We acknowledge the work of all PERCH contributors who were involved in data collection at the local sites and central laboratories, members of the PERCH Chest Radiograph Reading Panel (see appendix for the list of members in each group), and Shalika Jayawardena and Rose Watt from Canterbury Health Laboratories. Finally, we gratefully recognise the willingness and confidence in the study by the parents of the participating children and express our gratitude for their commitment to the advancement of knowledge toward better health for children of their community and those of other communities. This paper is published with the permission of the Director of the Kenya Medical Research Institute. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the US Centers for Disease Control and Prevention, US Department of Health and Human Services, or the US Government.
Contributors
KLO, OSL, HCB, WAB, DRF, LLH, SRCH, MDK, KLK, SAMad, DRM, JAGS, DMT, JCh, JCr, AND, AJD, NF, RAK, SAMal, and SLZ designed the study. Principal investigators of the study were KLO and OSL (overall); WAB, DG, and KZ (Bangladesh); SRCH and SMAZ (The Gambia); JAGS and JOA (Kenya); KLK, SOS, and MDT (Mali); SAMad and DPM (South Africa); HCB, SAMal, and PA (Thailand); and DMT, JCh, LM, and PSe (Zambia). Team leads included DRF (clinical and epidemiological), LLH (clinical and epidemiological), MDK (data analysis), and DRM (laboratory). KLO, OSL, HCB, WAB, DRF, LLH, SRCH, MDK, KLK, SAMad, DRM, JAGS, DMT, PVA, PA, TPA, MA, JOA, VLB, CB, JCh, MJC, JCr, AND, AJD, BEE, HPE, NF, DG, MJG, MMH, LH, YJ, EWK, AK, RAK, SK, NK, LK, GK, EMM, GM, NM, SAMal, JLMc, JLMi, DPM, SCM, AM, LM, JM, MSO, UO, DEP, CP, JR, PSa, PSe, AS, EAFS, SS, SWS, SOS, MS, BT, MDT, ST, AT, NLW, KZ, and SMAZ collected the data. MDK, SLZ, ZW, QS, WF, ML, CP, DEP, MMH, MH, and NLW supervised or did the analysis. KLO, OSL, HCB, WAB, DRF, LLH, SRCH, MDK, KLK, SAMad, DRM, JAGS, DMT, SLZ, ZW, QS, WF, ML, CP, DEP, MMH, MH, NLW interpreted the data. KLO, DRM, MDK, and CP drafted the paper. OSL, HCB, WAB, DRF, LLH, SRCH, KLK, SAMad, JAGS, DMT, SLZ, and ZW provided substantial contributions to the review and revision of the text. CP drafted the descriptive appendix. MDK, SLZ, ZW, CP, and QS drafted the analytical methods appendix. All authors reviewed the draft. KLO had final responsibility for the decision to submit for publication.
The PERCH Study Group
Katherine L O'Brien, Orin S Levine, Maria Deloria Knoll, Daniel R Feikin, Andrea N DeLuca, Amanda J Driscoll, Nicholas Fancourt, Wei Fu, Meredith Haddix, Laura L Hammitt, Melissa M Higdon, E Wangeci Kagucia, Ruth A Karron, Mengying Li, Daniel E Park, Christine Prosperi, Qiyuan Shi, Zhenke Wu, Scott L Zeger, Nora L Watson, Jane Crawley, David R Murdoch, W Abdullah Brooks, Hubert P Endtz, Khalequ Zaman, Doli Goswami, Lokman Hossain, Yasmin Jahan, Mohammod Jobayer Chisti, Stephen R C Howie, Bernard E Ebruke, Martin Antonio, Jessica L McLellan, Eunice M Machuka, Arifin Shamsul, Syed M A Zaman, Grant Mackenzie, J Anthony G Scott, Juliet O Awori, Susan C Morpeth, Alice Kamau, Sidi Kazungu, Micah Silaba Ominde, Karen L Kotloff, Milagritos D Tapia, Samba O Sow, Mamadou Sylla, Boubou Tamboura, Uma Onwuchekwa, Nana Kourouma, Aliou Toure, Seydou Sissoko, Shabir A Madhi, David P Moore, Peter V Adrian, Vicky L Baillie, Locadiah Kuwanda, Azwifarwi Mudau, Michelle J Groome, Nasreen Mahomed, Eric A F Simões, Henry C Baggett, Somsak Thamthitiwat, Susan A Maloney, Charatdao Bunthi, Julia Rhodes, Pongpun Sawatwong, Pasakorn Akarasewi, Donald M Thea, Lawrence Mwananyanda, James Chipeta, Phil Seidenberg, James Mwansa, Somwe Wa Somwe, Geoffrey Kwenda, Trevor P Anderson, Joanne L Mitchell.
Affiliations
Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA (Katherine L O'Brien, Orin S Levine [currently affiliated with Bill & Melinda Gates Foundation, Seattle, WA, USA], Maria Deloria Knoll, Daniel R Feikin [also affiliated with the US Centers for Disease Control and Prevention, Atlanta, GA, USA], Andrea N DeLuca, Amanda J Driscoll, Nicholas Fancourt, Wei Fu, Meredith Haddix, Laura L Hammitt, Melissa M Higdon, E Wangeci Kagucia, Ruth A Karron, Mengying Li, Daniel E Park, Christine Prosperi, Qiyuan Shi, Zhenke Wu, and Scott L Zeger); The Emmes Corporation, Rockville, MD, USA (Nora L Watson); Nuffield Department of Clinical Medicine, University of Oxford, Oxford, UK (Jane Crawley); University of Otago, Christchurch, New Zealand (David R Murdoch); International Centre for Diarrhoeal Disease Research, Bangladesh, (W Abdullah Brooks, Hubert P Endtz, Khalequ Zaman, Doli Goswami, Lokman Hossain, Yasmin Jahan, and Mohammod Jobayer Chisti); Medical Research Council, Basse, The Gambia (Stephen R C Howie, Bernard E Ebruke, Martin Antonio, Jessica L McLellan, Eunice M Machuka, Arifin Shamsul, Syed M A Zaman, Grant Mackenzie); KEMRI–Wellcome Trust Research Programme, Kilifi, Kenya (J Anthony G Scott [also affiliated with London School of Hygiene and Tropical Medicine, London, UK], Juliet O Awori, Susan C Morpeth, Alice Kamau, Sidi Kazungu, and Micah Silaba Ominde); Division of Infectious Disease and Tropical Pediatrics, Department of Pediatrics, Center for Vaccine Development, University of Maryland School of Medicine, Baltimore, MD, USA (Karen L Kotloff, Milagritos D Tapia, Samba O Sow, Mamadou Sylla, Boubou Tamboura, Uma Onwuchekwa, Nana Kourouma, Aliou Toure, and Seydou Sissoko); Centre pour le Développement des Vaccins (CVD-Mali), Bamako, Mali (Karen L Kotloff, Milagritos D Tapia, Samba O Sow, Mamadou Sylla, Boubou Tamboura, Uma Onwuchekwa, Nana Kourouma, Aliou Toure, Seydou Sissoko); Medical Research Council, Respiratory and Meningeal Pathogens Research Unit and Department of Science and Technology–National Research Foundation, Vaccine Preventable Diseases, University of the Witwatersrand, Johannesburg, South Africa (Shabir A Madhi, David P Moore, Peter V Adrian, Vicky L Baillie, Locadiah Kuwanda, Azwifarwi Mudau, Michelle J Groome, Nasreen Mahomed, and Eric A F Simões); Thailand Ministry of Public Health–US Centers for Disease Control and Prevention Collaboration, Nonthaburi, Thailand (Henry C Baggett, Somsak Thamthitiwat, Susan A Maloney, Charatdao Bunthi, Julia Rhodes, Pongpun Sawatwong, and Pasakorn Akarasewi); Boston University School of Public Health, Boston, MA, USA (Donald M Thea, Lawrence Mwananyanda, James Chipeta, Phil Seidenberg, James Mwansa, Somwe Wa Somwe, and Geoffrey Kwenda); University Teaching Hospital, Lusaka, Zambia (Donald M Thea, Lawrence Mwananyanda, James Chipeta, Phil Seidenberg, James Mwansa, Somwe Wa Somwe, and Geoffrey Kwenda); Canterbury Health Laboratories, Christchurch, New Zealand (Trevor P Anderson and Joanne L Mitchell).
Declaration of interests
KLO has received grant funding from GlaxoSmithKline and Pfizer and participates on technical advisory boards for Merck, Sanofi-Pasteur, PATH, Affinivax, and ClearPath. MDK has received funding for consultancies from Merck, Pfizer, and Novartis and grant funding from Merck. LLH has received grant funding from Pfizer and GlaxoSmithKline. KLK has received grant funding from Merck Sharp & Dohme. SAMad has received honorarium for advisory board from the Bill & Melinda Gates Foundation, Pfizer, Medimmune, and Novartis; institutional grants from GlaxoSmithKline, Novartis, Pfizer, Minervax, and Bill & Melinda Gates Foundation; and speakers bureau for Sanofi Pasteur and GlaxoSmithKline. WAB reported funding from Sanofi, PATH, Bill & Melinda Gates Foundation, and contributions to contemporaneous studies from Serum Institute of India, Roche, and Sanofi. All other authors declare no competing interests.
Contributor Information
The Pneumonia Etiology Research for Child Health (PERCH) Study Group:
Katherine L. O'Brien, Henry C. Baggett, W. Abdullah Brooks, Daniel R. Feikin, Laura L. Hammitt, Melissa M. Higdon, Stephen R.C. Howie, Maria Deloria Knoll, Karen L. Kotloff, Orin S. Levine, Shabir A. Madhi, David R. Murdoch, Christine Prosperi, J. Anthony G. Scott, Qiyuan Shi, Donald M. Thea, Zhenke Wu, Scott L. Zeger, Peter V. Adrian, Pasakorn Akarasewi, Trevor P. Anderson, Martin Antonio, Juliet O. Awori, Vicky L. Baillie, Charatdao Bunthi, James Chipeta, Mohammod Jobayer Chisti, Jane Crawley, Andrea N. DeLuca, Amanda J. Driscoll, Bernard E. Ebruke, Hubert P. Endtz, Nicholas Fancourt, Wei Fu, Doli Goswami, Michelle J. Groome, Meredith Haddix, Lokman Hossain, Yasmin Jahan, E. Wangeci Kagucia, Alice Kamau, Ruth A. Karron, Sidi Kazungu, Nana Kourouma, Locadiah Kuwanda, Geoffrey Kwenda, Mengying Li, Eunice M. Machuka, Grant Mackenzie, Nasreen Mahomed, Susan A. Maloney, Jessica L. McLellan, Joanne L. Mitchell, David P. Moore, Susan C. Morpeth, Azwifarwi Mudau, Lawrence Mwananyanda, James Mwansa, Micah Silaba Ominde, Uma Onwuchekwa, Daniel E. Park, Julia Rhodes, Pongpun Sawatwong, Phil Seidenberg, Arifin Shamsul, Eric A.F. Simões, Seydou Sissoko, Somwe Wa Somwe, Samba O. Sow, Mamadou Sylla, Boubou Tamboura, Milagritos D. Tapia, Somsak Thamthitiwat, Aliou Toure, Nora L. Watson, Khalequ Zaman, and Syed M.A. Zaman
Supplementary Material
References
- 1.Liu L, Oza S, Hogan D. Global, regional, and national causes of under-5 mortality in 2000–15: an updated systematic analysis with implications for the Sustainable Development Goals. Lancet. 2016;388:3027–3035. doi: 10.1016/S0140-6736(16)31593-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feikin D, Flannery B, Hamel M, Stack M, Hansen P. Disease control priorities: reproductive, maternal, newborn, and child health. 3rd. World Bank; Washington DC: 2016. Vaccines for children in low- and middle-income countries; pp. 187–204. [PubMed] [Google Scholar]
- 3.Lee LA, Franzel L, Atwell J. The estimated mortality impact of vaccinations forecast to be administered during 2011–2020 in 73 countries supported by the GAVI Alliance. Vaccine. 2013;31:B61–B72. doi: 10.1016/j.vaccine.2012.11.035. [DOI] [PubMed] [Google Scholar]
- 4.Levine OS, O’Brien KL, Deloria-Knoll M. The Pneumonia Etiology Research for Child Health Project: a 21st century childhood pneumonia etiology study. Clin Infect Dis. 2012;54:S93–S101. doi: 10.1093/cid/cir1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilani Z, Kwong YD, Levine OS. A literature review and survey of childhood pneumonia etiology studies: 2000–2010. Clin Infect Dis. 2012;54(suppl 2):S102–S108. doi: 10.1093/cid/cir1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feikin DR, Hammitt LL, Murdoch DR, O’Brien KL, Scott JAG. The enduring challenge of determining pneumonia etiology in children: considerations for future research priorities. Clin Infect Dis. 2017;64:S188–S196. doi: 10.1093/cid/cix143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Z, Deloria-Knoll M, Hammitt LL, Zeger SL. Partially latent class models for case-control studies of childhood pneumonia aetiology. J R Stat Soc Ser C Appl Stat. 2016;65:97–114. doi: 10.1111/rssc.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Z, Deloria-Knoll M, Zeger S. Nested partially-latent class models for dependent binary data; estimating disease etiology. Biostatistics. 2017;18:200–213. doi: 10.1093/biostatistics/kxw037. [DOI] [PubMed] [Google Scholar]
- 9.Deloria Knoll M, Fu W, Shi Q. Bayesian estimation of pneumonia etiology: epidemiologic considerations and applications to the Pneumonia Etiology Research for Child Health Study. Clin Infect Dis. 2017;64:S213–S227. doi: 10.1093/cid/cix144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott JAG, Wonodi C, Moïsi JC. The definition of pneumonia, the assessment of severity, and clinical standardization in the Pneumonia Etiology Research for Child Health study. Clin Infect Dis. 2012;54(suppl 2):S109–S116. doi: 10.1093/cid/cir1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deloria-Knoll M, Feikin DR, Scott JAG. Identification and selection of cases and controls in the Pneumonia Etiology Research for Child Health Project. Clin Infect Dis. 2012;54:S117–S123. doi: 10.1093/cid/cir1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.WHO Pocket book of hospital care for children: guidelines for the management of common illnesses with limited resources. 2005 http://whqlibdoc.who.int/publications/2005/9241546700.pdf [PubMed] [Google Scholar]
- 13.Higdon MM, Hammitt LL, Deloria Knoll M. Should controls with respiratory symptoms be excluded from case-control studies of pneumonia etiology? Reflections from the PERCH study. Clin Infect Dis. 2017;64:S205–S212. doi: 10.1093/cid/cix076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLuca AN, Regenberg A, Sugarman J, Murdoch DR, Levine O. Bioethical considerations in developing a biorepository for the Pneumonia Etiology Research for Child Health Project. Clin Infect Dis. 2012;54:S172–S179. doi: 10.1093/cid/cir1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawley J, Prosperi C, Baggett HC. Standardization of clinical assessment and sample collection across all PERCH study sites. Clin Infect Dis. 2017;64:S228–S237. doi: 10.1093/cid/cix077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watson NL, Prosperi C, Driscoll AJ. Data management and data quality in PERCH, a large international case-control study of severe childhood pneumonia. Clin Infect Dis. 2017;64:S238–S244. doi: 10.1093/cid/cix080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Driscoll AJ, Karron RA, Morpeth SC. Standardization of laboratory methods for the PERCH study. Clin Infect Dis. 2017;64:S245–S252. doi: 10.1093/cid/cix081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The PERCH Study Group The Pneumonia Etiology Research for Child Health Project (PERCH) study materials. 2012 https://www.jhsph.edu/ivac/resources/perch-background-and-methods/ [Google Scholar]
- 19.Cherian T, Mulholland EK, Carlin JB. Standardized interpretation of paediatric chest radiographs for the diagnosis of pneumonia in epidemiological studies. Bull World Health Organ. 2005;83:353–359. [PMC free article] [PubMed] [Google Scholar]
- 20.Fancourt N, Deloria Knoll M, Barger-Kamate B. Standardized interpretation of chest radiographs in cases of pediatric pneumonia from the PERCH study. Clin Infect Dis. 2017;64:S253–S261. doi: 10.1093/cid/cix082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fancourt N, Deloria-Knoll M, Baggett HC. Chest radiograph findings in childhood pneumonia cases from the multi-site PERCH study. Clin Infect Dis. 2017;64(suppl 3):S262–S270. doi: 10.1093/cid/cix089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murdoch DR, O’Brien KL, Driscoll AJ, Karron RA, Bhat N. Laboratory methods for determining pneumonia etiology in children. Clin Infect Dis. 2012;54(suppl 2):S146–S152. doi: 10.1093/cid/cir1073. [DOI] [PubMed] [Google Scholar]
- 23.Feikin DR, Fu W, Park DE. Is Higher viral load in the upper respiratory tract associated with severe pneumonia? Findings from the PERCH study. Clin Infect Dis. 2017;64:S337–S346. doi: 10.1093/cid/cix148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park DE, Baggett HC, Howie SRC. Colonization density of the upper respiratory tract as a predictor of pneumonia—Haemophilus influenzae, Moraxella catarrhalis, Staphylococcus aureus, and Pneumocystis jirovecii. Clin Infect Dis. 2017;64:S328–S336. doi: 10.1093/cid/cix104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baggett HC, Watson NL, Deloria Knoll M. Density of upper respiratory colonization with Streptococcus pneumoniae and its role in the diagnosis of pneumococcal pneumonia among children aged <5 years in the PERCH study. Clin Infect Dis. 2017;64:S317–S327. doi: 10.1093/cid/cix100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morpeth SC, Deloria Knoll M, Scott JAG. Detection of pneumococcal DNA in blood by polymerase chain reaction for diagnosing pneumococcal pneumonia in young children from low- and middle-income countries. Clin Infect Dis. 2017;64:S347–S356. doi: 10.1093/cid/cix145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deloria Knoll M, Morpeth SC, Scott JAG. Evaluation of pneumococcal load in blood by polymerase chain reaction for the diagnosis of pneumococcal pneumonia in young children in the PERCH study. Clin Infect Dis. 2017;64:S357–S367. doi: 10.1093/cid/cix149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Driscoll AJ, Deloria Knoll M, Hammitt LL. The effect of antibiotic exposure and specimen volume on the detection of bacterial pathogens in children with pneumonia. Clin Infect Dis. 2017;64:S368–S377. doi: 10.1093/cid/cix101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zar HJ, Barnett W, Stadler A, Gardner-Lubbe S, Myer L, Nicol MP. Aetiology of childhood pneumonia in a well vaccinated South African birth cohort: a nested case-control study of the Drakenstein Child Health Study. Lancet Respir Med. 2016;4:463–472. doi: 10.1016/S2213-2600(16)00096-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murdoch DR, Morpeth SC, Hammitt LL. Microscopic analysis and quality assessment of induced sputum from children with pneumonia in the PERCH study. Clin Infect Dis. 2017;64:S271–S279. doi: 10.1093/cid/cix083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murdoch DR, Morpeth SC, Hammitt LL. The diagnostic utility of induced sputum microscopy and culture in childhood pneumonia. Clin Infect Dis. 2017;64:S280–S288. doi: 10.1093/cid/cix090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thea DM, Seidenberg P, Park DE. Limited utility of polymerase chain reaction in induced sputum specimens for determining the causes of childhood pneumonia in resource-poor settings: findings from the Pneumonia Etiology Research for Child Health (PERCH) study. Clin Infect Dis. 2017;64:S289–S300. doi: 10.1093/cid/cix098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner P, Hinds J, Turner C. Improved detection of nasopharyngeal cocolonization by multiple pneumococcal serotypes by use of latex agglutination or molecular serotyping by microarray. J Clin Microbiol. 2011;49:1784–1789. doi: 10.1128/JCM.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.WHO . World health Organization; Geneva: 1991. Technical bases for the WHO recommendations on the management of pneumonia in children at first-level health facilities: programme for the control of acute respiratory infections. [Google Scholar]
- 35.Gelman A, Carlin JB, Stern HS, Dunson DB, Vehtari A, Rubin DB. 3rd. Taylor & Francis Group; Boca Raton, FL: 2014. Bayesian data analysis. [Google Scholar]
- 36.Hammitt LL, Feikin DR, Scott JAG. Addressing the analytic challenges of cross-sectional pediatric pneumonia etiology data. Clin Infect Dis. 2017;64:S197–S204. doi: 10.1093/cid/cix147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.WHO WHO AnthroPlus software. 2009 http://www.who.int/growthref/tools/en/ [Google Scholar]
- 38.Jain S, Williams DJ, Arnold SR. Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med. 2015;372:835–845. doi: 10.1056/NEJMoa1405870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bénet T, Picot VS, Awasthi S. Severity of pneumonia in under 5-year-old children from developing countries: a multicenter, prospective, observational study. Am J Trop Med Hyg. 2017;97:68–76. doi: 10.4269/ajtmh.16-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang JW, Lam TT, Zaraket H. Global epidemiology of non-influenza RNA respiratory viruses: data gaps and a growing need for surveillance. Lancet Infect Dis. 2017;17:e320–e326. doi: 10.1016/S1473-3099(17)30238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spichak TV, Yatsyshina SB, Katosova LK, Kim SS, Korppi MO. Is the role of rhinoviruses as causative agents of pediatric community-acquired pneumonia over-estimated? Eur J Pediatr. 2016;175:1951–1958. doi: 10.1007/s00431-016-2791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pretorius MA, Tempia S, Walaza S. The role of influenza, RSV and other common respiratory viruses in severe acute respiratory infections and influenza-like illness in a population with a high HIV sero-prevalence, South Africa 2012–2015. J Clin Virol. 2016;75:21–26. doi: 10.1016/j.jcv.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dagan R, Shriker O, Hazan I. Prospective study to determine clinical relevance of detection of pneumococcal DNA in sera of children by PCR. J Clin Microbiol. 1998;36:669–673. doi: 10.1128/jcm.36.3.669-673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dowell SF, Garman RL, Liu G, Levine OS, Yang YH. Evaluation of Binax NOW, an assay for the detection of pneumococcal antigen in urine samples, performed among pediatric patients. Clin Infect Dis. 2001;32:824–825. doi: 10.1086/319205. [DOI] [PubMed] [Google Scholar]
- 45.Johnson HL, Deloria-Knoll M, Levine OS. Systematic evaluation of serotypes causing invasive pneumococcal disease among children under five: the pneumococcal global serotype project. PLoS Med. 2010;7:e1000348. doi: 10.1371/journal.pmed.1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cutts FT, Zaman SMA, Enwere G. Efficacy of nine-valent pneumococcal conjugate vaccine against pneumonia and invasive pneumococcal disease in The Gambia: randomised, double-blind, placebo-controlled trial. Lancet. 2005;365:1139–1146. doi: 10.1016/S0140-6736(05)71876-6. [DOI] [PubMed] [Google Scholar]
- 47.Klugman KP, Madhi S a, Huebner R. A trial of a 9-valent pneumococcal conjugate vaccine in children with and those without HIV infection. N Engl J Med. 2003;349:1341–1348. doi: 10.1056/NEJMoa035060. [DOI] [PubMed] [Google Scholar]
- 48.Tregnaghi MW, Sáez-Llorens X, López P. Efficacy of pneumococcal nontypable Haemophilus influenzae protein D conjugate vaccine (PHiD-CV) in young Latin American children: a double-blind randomized controlled trial. PLoS Med. 2014;11:e1001657. doi: 10.1371/journal.pmed.1001657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piralam B, Tomczyk SM, Rhodes JC. Incidence of pneumococcal pneumonia among adults in rural Thailand, 2006–2011: Implications for pneumococcal vaccine considerations. Am J Trop Med Hyg. 2015;93:1140–1147. doi: 10.4269/ajtmh.15-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhodes J, Dejsirilert S, Maloney SA. Pneumococcal bacteremia requiring hospitalization in rural Thailand: an update on incidence, clinical characteristics, serotype distribution, and antimicrobial susceptibility, 2005–2010. PLoS One. 2013;8:e66038. doi: 10.1371/journal.pone.0066038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baggett HC, Peruski LF, Olsen SJ. Incidence of pneumococcal bacteremia requiring hospitalization in rural Thailand. Clin Infect Dis. 2009;48:S65–S74. doi: 10.1086/596484. [DOI] [PubMed] [Google Scholar]
- 52.Moore DP, Higdon MM, Hammitt LL. The incremental value of repeated induced sputum and gastric aspirate samples for the diagnosis of pulmonary tuberculosis in young children with acute community-acquired pneumonia. Clin Infect Dis. 2017;64:S309–S316. doi: 10.1093/cid/cix099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicol MP, Zar HJ. New specimens and laboratory diagnostics for childhood pulmonary TB: progress and prospects. Paediatr Respir Rev. 2011;12:16–21. doi: 10.1016/j.prrv.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barger-Kamate B, Deloria Knoll M, Kagucia EW. Pertussis-associated pneumonia in infants and children from low- and middle-income countries participating in the PERCH Study. Clin Infect Dis. 2016;63:S187–S196. doi: 10.1093/cid/ciw546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ideh RC, Howie SRC, Ebruke B. Transthoracic lung aspiration for the aetiological diagnosis of pneumonia: 25 years of experience from The Gambia. Int J Tuberc Lung Dis. 2011;15:729–735. doi: 10.5588/ijtld.10.0468. [DOI] [PubMed] [Google Scholar]
- 56.Dawood FS, Fry AM, Goswami D. Incidence and characteristics of early childhood wheezing, Dhaka, Bangladesh, 2004–2010. Pediatr Pulmonol. 2016;51:588–595. doi: 10.1002/ppul.23343. [DOI] [PubMed] [Google Scholar]
- 57.Murray EL, Brondi L, Kleinbaum D. Cooking fuel type, household ventilation, and the risk of acute lower respiratory illness in urban Bangladeshi children: a longitudinal study. Indoor Air. 2012;22:132–139. doi: 10.1111/j.1600-0668.2011.00754.x. [DOI] [PubMed] [Google Scholar]
- 58.Silk BJ, Cohen AL, Abedin J. Household air quality risk factors associated with childhood pneumonia in urban Dhaka, Bangladesh. Am J Trop Med Hyg. 2014;90:968–975. doi: 10.4269/ajtmh.13-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ngari MM, Fegan G, Mwangome MK. Mortality after inpatient treatment for severe pneumonia in children: a cohort study. Paediatr Perinat Epidemiol. 2017;31:233–242. doi: 10.1111/ppe.12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kotloff KL, Nataro JP, Blackwelder WC. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 61.Fry AM, Curns AT, Harbour K, Hutwagner L, Holman RC, Anderson LJ. Seasonal trends of human parainfluenza viral infections: United States, 1990–2004. Clin Infect Dis. 2006;43:1016–1022. doi: 10.1086/507638. [DOI] [PubMed] [Google Scholar]
- 62.Cox N. Influenza seasonality: timing and formulation of vaccines. Bull World Health Organ. 2014;92:311–411. doi: 10.2471/BLT.14.139428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Broutin H, Guégan J-F, Elguero E, Simondon F, Cazelles B. Large-scale comparative analysis of pertussis population dynamics: periodicity, synchrony, and impact of vaccination. Am J Epidemiol. 2005;161:1159–1167. doi: 10.1093/aje/kwi141. [DOI] [PubMed] [Google Scholar]
- 64.Omori R, Nakata Y, Tessmer HL, Suzuki S, Shibayama K. The determinant of periodicity in Mycoplasma pneumoniae incidence: an insight from mathematical modelling. Sci Rep. 2015;5:14473. doi: 10.1038/srep14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bénet T, Sánchez Picot V, Messaoudi M. Microorganisms associated with pneumonia in children <5 years of age in developing and emerging countries: the GABRIEL pneumonia multicenter, prospective, case-control study. Clin Infect Dis. 2017;54:S93–S101. doi: 10.1093/cid/cix378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Selwyn BJ, Coordinated Data Group of BOSTID Researchers The epidemiology of acute respiratory tract infection in young children: comparison of findings from several developing countries. Rev Infect Dis. 1990;12:S870–S888. doi: 10.1093/clinids/12.supplement_s870. [DOI] [PubMed] [Google Scholar]
- 67.Bale JR. Creation of a research program to determine the etiology and epidemiology of acute respiratory tract infection among children in developing countries. Rev Infect Dis. 1990;12(suppl 8):S861–S866. doi: 10.1093/clinids/12.supplement_8.s861. [DOI] [PubMed] [Google Scholar]
- 68.Feikin D, Scott J, Gessner B. Use of vaccines as probes to define disease burden. Lancet. 2014;383:1762–1770. doi: 10.1016/S0140-6736(13)61682-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saha SK, Schrag SJ, El Arifeen S. Causes and incidence of community-acquired serious infections among young children in south Asia (ANISA): an observational cohort study. Lancet. 2018;392:145–159. doi: 10.1016/S0140-6736(18)31127-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.International Vaccine Access Center VIEW-hub: Data visualization platform. http://www.view-hub.org
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We are in the process of making the PERCH dataset publicly available in ClinEpiDB (University of Pennsylvania, PA, USA; https://clinepidb.org; we estimate availability in 2020).