Abstract
Background/Aim: Epithelioid osteoblastoma is a rare benign tumor of the bone. Its pathogenesis is unknown and little is known regarding its genetic features. Materials and Methods: Cytogenetic, RNA sequencing, reverse transcription polymerase chain reaction (RT-PCR), genomic PCR, and Sanger sequencing analyses were performed on an epithelioid osteoblastoma. Results: G-banding analysis of short-term cultured tumor cells yielded a normal male karyotype in all examined metaphases. RNA sequencing detected a fusion of COL1A1 from 17q21 with FYN from 6q21. Both RT-PCR and genomic PCR together with Sanger sequencing verified the presence of a COL1A1-FYN fusion gene. In the COL1A1-FYN chimeric transcript, exon 43 of COL1A1 was fused to exon 2 of FYN. The genomic junction occurred in introns 43 and 1 of COL1A1 and FYN, respectively. Conclusion: A COL1A1-FYN fusion gene was found in an epithelioid osteoblastoma resulting in deregulation of FYN. Whether COL1A1-FYN represents a consistent genetic feature of epithelioid osteoblastomas, remains to be seen.
Keywords: Epithelioid osteoblastoma, COL1A1, FYN, COL1A1- FYN fusion gene, RNA sequencing
Osteoblastoma is a rare benign tumor which accounts for about 1% of all bone tumors. It is most often found in the age range of 10-30 years and is 2.5 more common in males than females (1). The tumor was first described in 1956 in two different publications, one by Jaffe and the other by Lichtenstein (2,3). In the 1970s, a more aggressive type of osteoblastoma was described with a higher recurrence rate and, upon microscopy, many epithelioid osteoblasts; various names were given to this type of tumor, such as malignant osteoblastoma (4), aggressive osteoblastoma (5), and epithelioid osteoblastoma (6,7). The initial reports emphasized that the aggressive behavior was associated with an epithelioid morphology (4,5,8); however, studying 306 osteoblastomas, Lucas et al. (9) did not find any difference in the aggressiveness between epithelioid and conventional tumors and concluded that “Aggressive behavior is within the biologic spectrum of osteoblastomas, and histopathology alone does not appear to be a reliable predictor of aggressiveness”. Examining 55 cases of osteoblastomas, Della Rocca et al. (10) also concluded that “clinically aggressive behavior of osteoblastoma is not related to particular histological features, but rather to the skeletal location”.
The cytogenetic information on osteoblastomas is limited. According to the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer, only three epithelioid (aggressive) osteoblastomas and four of the so-called conventional osteoblastomas have been karyotyped and no consistent cytogenetic pattern has emerged (http://cgap.nci.nih.gov/Chromosomes/Mitelman, database last updated on February 19, 2019). Recently, recurrent rearrangements of FOS and FOSB were found in so-called conventional osteoblastoma. Examining six tumors by whole genome and RNA sequencing, Fitall et al. (11) found FOS and FOSB rearrangements in five and one tumors, respectively. Extending the investigation to 55 additional cases using fluorescence in situ hybridization (FISH) and immunohistochemical methodologies, they found that 51 carried FOS and one FOSB rearrangements, respectively.
In the present study, we used RNA sequencing and other molecular genetic techniques to find fusion of the collagen type I alpha 1 (COL1A1) and FYN proto-oncogene, Src family tyrosine kinase (FYN) genes in an epithelioid osteoblastoma.
Materials and Methods
Ethics statement. The study was approved by the Regional Committee for Medical and Health Research Ethics, South-East Norway (REK Sør-Øst; http://helseforskning.etikkom.no) and written informed consent was obtained from the patient’s parents for publication of the case details. The ethics committee’s approval included a review of the consent procedure. All patient information has been de-identified.
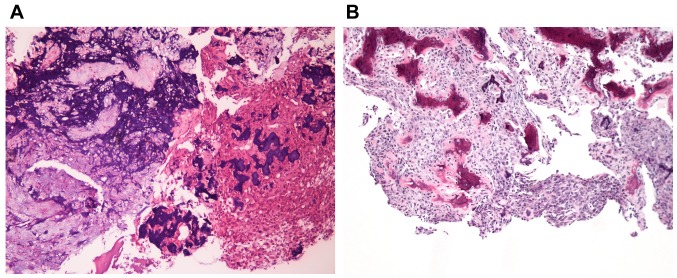
Case description. The patient was a 10-year old boy, who experienced ongoing pain in his left knee for more than 6 months. X-ray and CT scan showed an osteolytic lesion in the proximal fibula, first thought to be osteomyelitis. The local hospital performed curettage of the lesion. Histologically the lesion was bone forming, with trabeculae of osteoid, partly calcified, rimmed by osteoblasts (Figure 1A). In between the trabeculae there was fibrovascular tissue and sheets of epithelioid osteoblasts and scattered osteoclasts (Figure 1B). There was no cellular atypia and there were only few mitotic figures, none of them atypical. In addition, there were areas with “blue bone”, where the osteoid was heavily calcified and some areas were cartilage like.
Figure 1. Microscopic examination of the epithelioid osteoblastoma. A) H&E-stained section showing irregular trabeculae of woven bone with deep blue, calcified areas and areas with cartilage, ×20. B) H&E-stained section showing irregular trabeculae of woven bone rimmed by osteoblasts. In the intertrabecular space, fibrovascular tissue and sheets of epithelioid osteoblasts are shown, ×20.
G-banding and karyotyping. Fresh tissue from a representative area of the tumor was short-term cultured and analyzed cytogenetically as previously described (12).
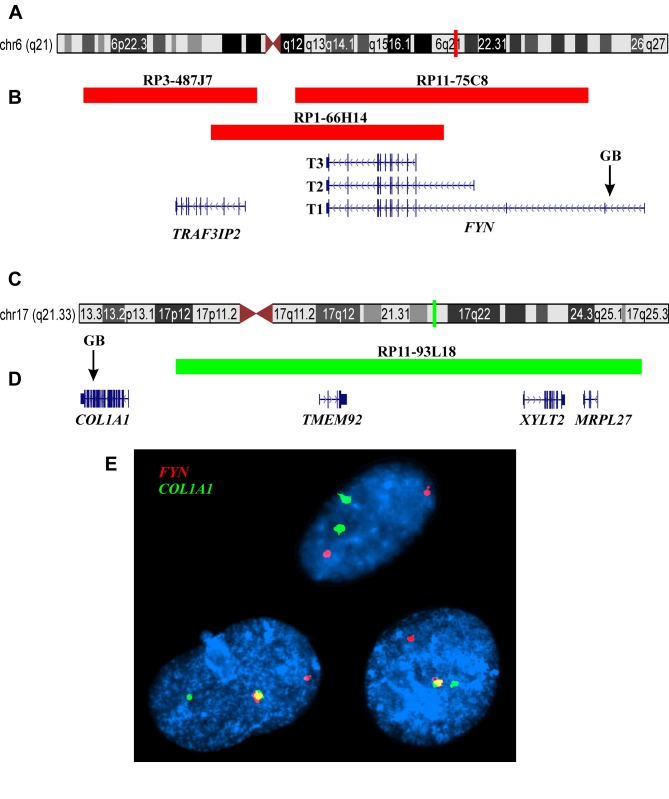
Fluorescence in situ hybridization (FISH). The BAC probes were purchased from BACPAC Resource Center located at the Children’s Hospital Oakland Research Institute (Oakland, CA, USA) (https://bacpacresources.org/). FISH analyses were performed on interphase nuclei using COL1A1 and FYN (see below) home-made dual-color single-fusion probes. Detailed information on the FISH procedure was given elsewhere (12). For the COL1A1 gene on chromosome band 17q21, the BAC clone used was RP11-93L18 (Position: chr17:50219522-50388834). For the FYN gene on chromosome band 6q21, the BAC clones used were RP11-75C8 (Position: chr6:111639316-111835441), RP1-66H14 (accession number Z97989.1; Position: chr6:111582811-111738747), and RP3-487J7 (accession number AL008730.1, Position: chr6: 111497307-111613802; Band: 6q21). The probes for COL1A1 and FYN were labelled with Fluorescein-12-dCTP (PerkinElmer, Boston, MA, USA) or Texas Red-5-dCTP (PerkinElmer) in order to obtain green and red signals, respectively. Fluorescent signals were captured and analyzed using the CytoVision system (Leica Biosystems, Newcastle, UK).
RNA sequencing. Total RNA was extracted from frozen (–80˚C) tumor tissue adjacent to that used for cytogenetic analysis and histologic examination using miRNeasy Mini Kit (Qiagen Nordic, Oslo, Norway). One μg of total RNA was sent to the Genomics Core Facility at the Norwegian Radium Hospital, Oslo University Hospital (http://genomics.no/oslo/) for high-throughput paired-end RNA-sequencing using the Illumina TruSeq Stranded mRNA protocol. The software FusionCatcher was used to find fusion transcripts (13,14).
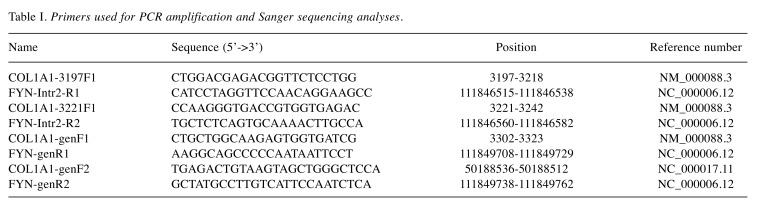
Reverse transcription (RT) and genomic PCR analyses. The primers used for PCR amplifications and Sanger sequencing analyses are shown in Table I. Genomic DNA was extracted using the Maxwell RSC Instrument and the Maxwell RSC Tissuel DNA Kit (Promega, Madison, WI, USA) and the concentration was measured using the Quantus Fluorometer and the QuantiFluor ONE dsDNA System (Promega). RT-PCR, genomic PCR, analysis of PCR products, and Sanger sequencing were performed as previously described (12).
Table I. Primers used for PCR amplification and Sanger sequencing analyses.
For amplification of the COL1A1-FYN fusion transcript, the primer combinations were the forward COL1A1-3197F1 together with the reverse FYN-Intr2R1 and COL1A1-3221F1 together with the reverse FYN-Intr2R2. For amplification of genomic COL1A1-FYN fragments, the primer combinations were COL1A1-genF1/FYN-genR1 and COL1A1-genF2/FYN-genR2. The cycling was at 94˚C for 30 sec followed by 35 cycles of 7 sec at 98˚C, 30 sec at 60˚C, 30 sec at 72˚C, and a final extension for 5 min at 72˚C. The BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for computer analysis of sequence data.
Immunohistochemistry. The FYN antibody was a mouse monoclonal antibody (2A10) purchased from ThermoFisher Scientific (Catalogue number MA5-15865) applied at 1:100 dilution. Formalin-fixed, paraffin-embedded sections from the epithelioid osteoblastoma were analyzed for FYN expression using the Dako EnVision Flex + System (K8012; Dako, Glostrup, Denmark) as previously described (15).
Results
The G-banding analysis of short-term cultured tumor cells revealed a normal karyotype, 46, XY, in all 25 examined metaphases (data not shown).
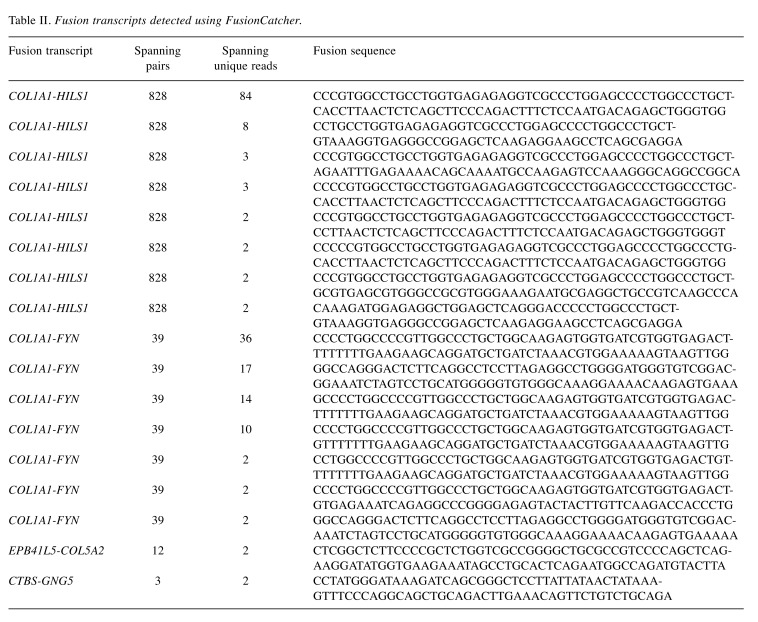
Using the FusionCatcher software with the fastq files from the RNA sequencing, four fusion genes were found with 17 fusion transcripts (Table II): a read-through COL1A1-HILS1 fusion gene with eight fusion transcripts, a COL1A1-FYN fusion gene with seven fusion transcripts, an EBP41L5-COL5A2 with one fusion transcript, and a read-through CTBS-GNG5 fusion gene with one fusion transcript. Detailed information on the fusion genes and transcripts is given in Table II. Taking into consideration that COL1A1 is fused to PDGFB in dermatofibrosarcoma protuberans (http://omim.org/ entry/120150) and FYN is a tyrosine kinase protooncogene related to SRC, FGR, and YES (http://omim.org/entry/137025) we decided to investigate further with molecular techniques the presence of COL1A1-FYN fusion gene in the tumor. No other fusion transcripts were examined.
Table II. Fusion transcripts detected using FusionCatcher.
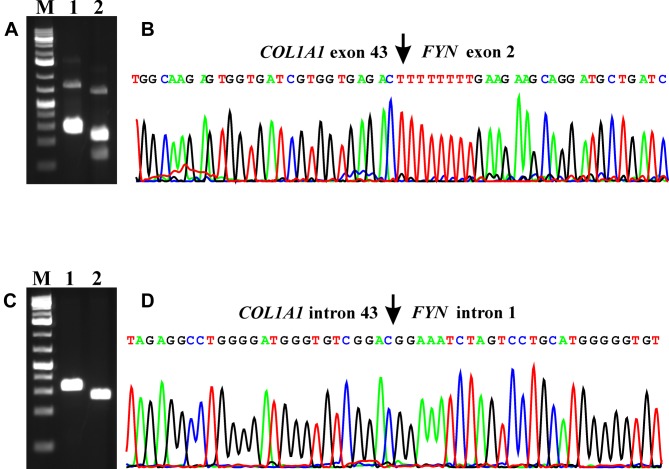
RT-PCR with the primer combinations COL1A1-3197F1/FYN-Intr2R1 and COL1A1-3221F1/FYN-Intr2R2 amplified a 252 bp fragment and a 183 bp fragment, respectively (Figure 2A). Sanger sequencing of the PCR fragments showed that they were COL1A1-FYN chimeric cDNA fragments in which exon 43 of COL1A1 (nucleotide 3333 of the COL1A1 sequence with accession number NM_000088.3) was fused to the untranslated exon 2 of FYN (nucleotide 486 of the FYN sequence with accession number NM_002037.5) (Figure 2B). The fusion point was identical to one of the 7 fusion points found by FusionCatcher analysis of the RNA sequencing data: CCCCTGGCCCCGTTGGCCCTGC TGGCAAGAGTGGTGATCGTGGTGAGACT-TTTTTTTGAAGAAGCAGGATGCTGATCTAAACGTGGAAAAAGTAAGTTGG.
Figure 2. Molecular genetic analysis of the epithelioid osteoblastoma. (A) Gel electrophoresis showing the amplified COL1A1-FYN cDNA fragment using the COL1A1-3197F1/FYN-Intr2R1 (lane 1) and COL1A1-3221F1/FYN-Intr2R2 (lane 2) primer combinations. (B) Partial sequence chromatogram of the cDNA amplified fragment showing the fusion (arrow) of exon 43 of COL1A1 with exon 2 of FYN. (C) Amplification of genomic COL1A1-FYN fragments using the primer combinations COL1A1-genF1/FYN-genR1 (lane 1) and COL1A1-genF2/FYN-genR2 (lane 2). (D) Partial sequence chromatogram of the genomic DNA amplified fragment showing the fusion (arrow) of intron 43 of COL1A1 with intron 1 of FYN. M, Thermo Scientific GeneRuler 1 kb Plus DNA Ladder.
Genomic PCR with the primer combinations COL1A1-genF1/FYN-genR1 and COL1A1-genF2/FYN-genR2 amplified a 285 bp fragment and a 230 bp fragment, respectively (Figure 2C). Direct sequencing showed that they were genomic COL1A1-FYN chimeric fragments in which a sequence from intron 43 of COL1A1 was fused to a sequence of intron 1 from FYN (Figure 2D). The genomic junction point was identical to one of the fusion points found by FusionCatcher analysis of the RNA sequencing data: GGCCAGGGACTCTTCAGGCCTCCTTAGAGGCCTGGGGATGGGTGTCGGAC-GGAAATCTAGTCCTGCATGGG GGTGTGGGCAAAGGAAAACAAGAGTGAAA.
FISH analysis, using COL1A1 and FYN home-made dual-color single-fusion probe, showed a fusion signal in 9 out of 106 (8%) examined interphase nuclei suggesting a COL1A1-FYN fusion gene in these cells (Figure 3).
Figure 3. FISH analysis of epithelioid osteoblastoma using COL1A1 and FYN home-made dual-color single-fusion probe. (A) Ideogram of chromosome 6 showing the mapping position of the FYN gene (vertical red line). (B) Diagram showing the FISH probes RP3-487J7, RP1-66H14, and RP11-75C8 for FYN. Arrow indicates the genomic breakpoint (GP) in the intron 1 of FYN. The neighbor TRAF3IP2 gene in this region is also shown. (C) Ideogram of chromosome 17 showing the mapping position of the COL1A1 gene (vertical green light). (D) Diagram showing the FISH probe RP11-93L18 for COL1A1. The TMEM92, XYLT2, and MRPL27 genes mapped in this region, distal to COL1A1, are also shown. Arrow indicates the genomic breakpoint (GP) in the intron 43 of COL1A1. (E) FISH results with the FYN (red signal) and COL1A1 (green signal) probes on three interphase nuclei (composite photo) showing a red signal, a green signal, and one yellow-fusion signal in two nuclei and two green and two red signals in one nucleus.
FYN immunohistochemical examination was uninformative (data not shown) probably due to damage done to the tissue by the decalcification procedure (16,17).
Discussion
Using the RNA sequencing methodology, we identified a COL1A1-FYN fusion gene in the cells of an epithelioid osteoblastoma. The fusion gene was further verified at both transcriptional (RNA) and genomic (DNA) levels using RT-PCR and genomic PCR together with Sanger sequencing. Interphase FISH showed that the short-term cultured cells from tumor biopsy contained a small clone of cells (8%) carrying the COL1A1-FYN fusion. The cells carrying the rearrangement most probably were not able to divide in vitro.
The FYN gene on 6q21 codes for a non-receptor tyrosine kinase, FYN, which is a member of the SRC family of kinases (18,19). The FYN protein is found in the inner layer of the cell membrane and is involved in signal transduction pathways. It phosphorylates tyrosine residues of many proteins regulating numerous functions including control of cell growth, cell-cell adhesion, and cytoskeletal remodeling (18,19). FYN is implicated in cancer, too (18,19). In vitro studies have shown that overexpression of FYN in NIH-3T3 cells induced morphologic transformation and anchorage-independent growth (20). FYN was found to be overexpressed in prostate, breast, pancreas, and thyroid cancer (2,21-24). In neuroblastoma, high expression of FYN and high FYN kinase activity was found in tumors of stage I, whereas FYN was down-regulated in stage 4 neuroblastomas (25). In chronic myeloid leukemia, FYN expression was significantly higher in blast crisis compared to chronic phase, and overexpression of FYN was an important determinant for resistance to BCR-ABL1 inhibitors (26-28).
The COL1A1 gene codes for the pro-alpha1 chain of type I collagen, a fibril-forming collagen which is abundant in bone, cornea, dermis, and tendon (29,30). Mutations in the COL1A1 gene are associated with osteogenesis imperfecta types I-IV, Ehlers-Danlos syndrome type VIIA, Ehlers-Danlos syndrome Classical type, Caffey Disease, and idiopathic osteoporosis (http://omim.org/entry/120150). Simon et al. (31) showed that in dermatofibrosarcoma protuberans, the t(17;22)(q21;q13) and supernumerary ring chromosomes characteristic of these tumors, contain a COL1A1-PDGFB fusion gene consisting of COL1A1 from 17q21 and PDGFB from 22q13 resulting in deregulation of PDGFB. In the COL1A1-PDGFB fusion gene, the exact site of the breakpoint in COL1A1 was shown to be highly variable from exons 6 to 49, whereas the PDGFB breakpoint was consistently located in intron 2 so that exon 2 of the gene was always present in the COL1A1-PDGFB fusion transcript (31-33). No correlation was found between the genomic breakpoint in COL1A1 and clinico-histopathologic dermatofibrosarcoma protuberans features (34,35).
Fusion of COL1A1 with USP6 (from 17p13) was also described in a case of aneurysmal bone cyst and in a benign bone tumor (36,37). In both instances, exon 1 of COL1A1 was fused to exon 2 of USP6 and the pathogenic consequence of the fusion gene appeared to be control of USP6 expression by the COL1A1 promoter (36,37).
In the present case of epithelioid osteoblastoma, we believe that the COL1A1-FYN fusion gene results in regulation of FYN expression by the COL1A1 promoter similar to what happens with COL1A1-PDFGB and COL1A1-USP6.
Both FYN (on chromosome band 6q21) and COL1A1 (on 17q21) are transcribed from telomere to centromere. Hence, formation of a COL1A1-FYN fusion is possible through a simple t(6;17)(q21;q21). The COL1A1-FYN is predicted to lie in the breakpoint of the putative der(6)t(6;17).
To the best of our knowledge, this is the first time that a fusion gene is described in epithelioid osteoblastoma. Whether COL1A1-FYN represents a consistent genetic feature of these tumors, and whether additional clinico-pathological features, including the aggressive behavior, may be associated with this fusion, remains to be seen.
Conflicts of Interest
No potential conflicts of interest exist regarding this study.
Authors’ Contributions
IP conceived the study, designed and performed the experiments, performed the bioinformatics analysis, and drafted the manuscript. LG performed cytogenetic analysis. IL performed pathological examination. ML-I performed pathological examination and evaluated immunohistochemical staining. KA performed cytogenetic and FISH analyses. AH performed immunohistochemistry examinations. BB performed pathological examination. SH evaluated the cytogenetic and FISH data and assisted with writing of the manuscript. All Authors read and approved the final manuscript.
Acknowledgements
This work was supported by grants from Radiumhospitalets Legater.
References
- 1.de Andrea CE, Bridge JA, Schiller A. IARC Press, Lyon. 2013. Osteoblastoma. In: World Health Organization Classification of Tumours, Volume 5. Pathology and genetics of tumours of soft tissue and bone. Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F (eds.) pp. pp. 279–280. [Google Scholar]
- 2.Jaffe HL. Benign osteoblastoma. Bull Hosp Joint Dis. 1956;17:141–151. PMID: 13413389. [PubMed] [Google Scholar]
- 3.Lichtenstein L. Benign osteoblastoma; a category of osteoid-and bone-forming tumors other than classical osteoid osteoma, which may be mistaken for giant-cell tumor or osteogenic sarcoma. Cancer. 1956;9:1044–1052. doi: 10.1002/1097-0142(195609/10)9:5<1044::aid-cncr2820090523>3.0.co;2-o. PMID: 13364889. [DOI] [PubMed] [Google Scholar]
- 4.Schajowicz F, Lemos C. Malignant osteoblastoma. J Bone Joint Surg Br. 1976;58:202–211. doi: 10.1302/0301-620X.58B2.932083. PMID: 932083. [DOI] [PubMed] [Google Scholar]
- 5.Revell PA, Scholtz CL. Aggressive osteoblastoma. J Pathol. 1979;127:195–198. doi: 10.1002/path.1711270406. PMID: 469645. DOI: 10.1002/path.17112 70406. [DOI] [PubMed] [Google Scholar]
- 6.Deyrup AT, Montag AG. Epithelioid and epithelial neoplasms of bone. Arch Pathol Lab Med. 2007;131:205–216. doi: 10.5858/2007-131-205-EAENOB. PMID: 17284104. DOI: 10.1043/1543-2165(2007)131[205:EAENOB] 2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 7.Britt JD, Murphey MD, Castle JT. Epithelioid osteoblastoma. Head Neck Pathol. 2012;6:451–454. doi: 10.1007/s12105-012-0356-5. PMID: 22528828. DOI: 10.1007/s12105-012-0356-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorfman HD, Weiss SW. Borderline osteoblastic tumors: problems in the differential diagnosis of aggressive osteoblastoma and low-grade osteosarcoma. Semin Diagn Pathol. 1984;1:215–234. PMID: 6600112. [PubMed] [Google Scholar]
- 9.Lucas DR, Unni KK, McLeod RA, O’Connor MI, Sim FH. Osteoblastoma: clinicopathologic study of 306 cases. Hum Pathol. 1994;25:117–134. doi: 10.1016/0046-8177(94)90267-4. PMID: 8119712. [DOI] [PubMed] [Google Scholar]
- 10.Della Rocca C, Huvos AG. Osteoblastoma: varied histological presentations with a benign clinical course. An analysis of 55 cases. Am J Surg Pathol. 1996;20:841–850. doi: 10.1097/00000478-199607000-00007. PMID: 8669532. [DOI] [PubMed] [Google Scholar]
- 11.Fittall MW, Mifsud W, Pillay N, Ye H, Strobl AC, Verfaillie A, Demeulemeester J, Zhang L, Berisha F, Tarabichi M, Young MD, Miranda E, Tarpey PS, Tirabosco R, Amary F, Grigoriadis AE, Stratton MR, Van Loo P, Antonescu CR, Campbell PJ, Flanagan AM, Behjati S. Recurrent rearrangements of FOS and FOSB define osteoblastoma. Nat Commun. 2018;9:2150. doi: 10.1038/s41467-018-04530-z. PMID: 29858576. DOI: 10.1038/s41467-018-04530-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panagopoulos I, Bjerkehagen B, Gorunova L, Taksdal I, Heim S. Rearrangement of chromosome bands 12q14~15 causing HMGA2-SOX5 gene fusion and HMGA2 expression in extraskeletal osteochondroma. Oncol Rep. 2015;34:577–584. doi: 10.3892/or.2015.4035. PMID: 26043835. DOI: 10.3892/or.2015.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kangaspeska S, Hultsch S, Edgren H, Nicorici D, Murumagi A, Kallioniemi O. Reanalysis of RNA-sequencing data reveals several additional fusion genes with multiple isoforms. PLoS One. 2012;7:e48745. doi: 10.1371/journal.pone.0048745. PMID: 23119097. DOI: 10.1371/journal. pone.0048745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicorici D, Satalan H, Edgren H, Kangaspeska S, Murumagi A, Kallioniemi O, Virtanen S, Kikku O. FusionCatcher – a tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 2014 DOI: 10.1101/011650. [Google Scholar]
- 15.Brunetti M, Holth A, Panagopoulos I, Staff AC, Micci F, Davidson B. Expression and clinical role of the dipeptidyl peptidases DPP8 and DPP9 in ovarian carcinoma. Virchows Arch. 2019;474:177–185. doi: 10.1007/s00428-018-2487-x. PMID: 30467600. DOI: 10.1007/ s00428-018-2487-x. [DOI] [PubMed] [Google Scholar]
- 16.Maclary SC, Mohanty SK, Bose S, Chung F, Balzer BL. Effect of hydrochloric acid decalcification on expression pattern of prognostic markers in invasive breast carcinomas. Appl Immunohistochem Mol Morphol. 2017;25:144–149. doi: 10.1097/PAI.0000000000000277. PMID: 27028239. DOI: 10.1097/PAI.0000000000000277. [DOI] [PubMed] [Google Scholar]
- 17.Clark BZ, Yoest JM, Onisko A, Dabbs DJ. Effects of hydrochloric acid and formic acid decalcification on breast tumor biomarkers and HER2 fluorescence in situ hybridization. Appl Immunohistochem Mol Morphol. 2019;27:223–230. doi: 10.1097/PAI.0000000000000564. PMID: 28877070. DOI: 10.1097/PAI.0000000000000564. [DOI] [PubMed] [Google Scholar]
- 18.Saito YD, Jensen AR, Salgia R, Posadas EM. Fyn: a novel molecular target in cancer. Cancer. 2010;116:1629–1637. doi: 10.1002/cncr.24879. PMID: 20151426. DOI: 10.1002/cncr.24879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elias D, Ditzel HJ. Fyn is an important molecule in cancer pathogenesis and drug resistance. Pharmacol Res. 2015;100:250–254. doi: 10.1016/j.phrs.2015.08.010. PMID: 26305432. DOI: 10.1016/j.phrs.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 20.Kawakami T, Kawakami Y, Aaronson SA, Robbins KC. Acquisition of transforming properties by FYN, a normal SRC-related human gene. Proc Natl Acad Sci USA. 1988;85:3870–3874. doi: 10.1073/pnas.85.11.3870. PMID: 3287380. DOI: 10.1073/pnas.85.11.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Posadas EM, Al-Ahmadie H, Robinson VL, Jagadeeswaran R, Otto K, Kasza KE, Tretiakov M, Siddiqui J, Pienta KJ, Stadler WM, Rinker-Schaeffer C, Salgia R. FYN is overexpressed in human prostate cancer. BJU Int. 2009;103:171–177. doi: 10.1111/j.1464-410X.2008.08009.x. PMID: 18990162. DOI: 10.1111/j.1464-410X.2008.08009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie YG, Yu Y, Hou LK, Wang X, Zhang B, Cao XC. FYN promotes breast cancer progression through epithelial-mesenchymal transition. Oncol Rep. 2016;36:1000–1006. doi: 10.3892/or.2016.4894. PMID: 27349276. DOI: 10.3892/or.2016.4894. [DOI] [PubMed] [Google Scholar]
- 23.Chen ZY, Cai L, Bie P, Wang SG, Jiang Y, Dong JH, Li XW. Roles of Fyn in pancreatic cancer metastasis. J Gastroenterol Hepatol. 2010;25:293–301. doi: 10.1111/j.1440-1746.2009.06021.x. PMID: 19968749. DOI: 10.1111/ j.1440-1746.2009.06021.x. [DOI] [PubMed] [Google Scholar]
- 24.Zheng J, Li H, Xu D, Zhu H. Upregulation of tyrosine kinase FYN in human thyroid carcinoma: Role in modulating tumor cell proliferation, invasion, and migration. Cancer Biother Radiopharm. 2017;32:320–326. doi: 10.1089/cbr.2017.2218. PMID: 29140740. DOI: 10.1089/cbr.2017.2218. [DOI] [PubMed] [Google Scholar]
- 25.Berwanger B, Hartmann O, Bergmann E, Bernard S, Nielsen D, Krause M, Kartal A, Flynn D, Wiedemeyer R, Schwab M, Schafer H, Christiansen H, Eilers M. Loss of a FYN-regulated differentiation and growth arrest pathway in advanced stage neuroblastoma. Cancer Cell. 2002;2:377–386. doi: 10.1016/s1535-6108(02)00179-4. PMID: 12450793. [DOI] [PubMed] [Google Scholar]
- 26.Ban K, Gao Y, Amin HM, Howard A, Miller C, Lin Q, Leng X, Munsell M, Bar-Eli M, Arlinghaus RB, Chandra J. BCR-ABL1 mediates up-regulation of Fyn in chronic myelogenous leukemia. Blood. 2008;111:2904–2908. doi: 10.1182/blood-2007-05-091769. PMID: 18180382. DOI: 10.1182/blood-2007-05-091769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grosso S, Puissant A, Dufies M, Colosetti P, Jacquel A, Lebrigand K, Barbry P, Deckert M, Cassuto JP, Mari B, Auberger P. Gene expression profiling of imatinib and PD166326-resistant CML cell lines identifies Fyn as a gene associated with resistance to BCR-ABL inhibitors. Mol Cancer Ther. 2009;8:1924–1933. doi: 10.1158/1535-7163.MCT-09-0168. PMID: 19567819. DOI: 10.1158/ 1535-7163.MCT-09-0168. [DOI] [PubMed] [Google Scholar]
- 28.Singh MM, Howard A, Irwin ME, Gao Y, Lu X, Multani A, Chandra J. Expression and activity of Fyn mediate proliferation and blastic features of chronic myelogenous leukemia. PLoS One. 2012;7:e51611. doi: 10.1371/journal.pone.0051611. PMID: 23284724. DOI: 10.1371/journal. pone.0051611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gelse K, Pöschl E, Aigner T. Collagens – structure, function, and biosynthesis. Adv Drug Deliv Rev. 2003;55:1531–1546. doi: 10.1016/j.addr.2003.08.002. PMID: 14623400. [DOI] [PubMed] [Google Scholar]
- 30.Ricard-Blum S. The collagen family. Cold Spring Harb Perspect Biol. 2011;3:a004978. doi: 10.1101/cshperspect.a004978. PMID: 21421911. DOI: 10.1101/ cshperspect.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon MP, Pedeutour F, Sirvent N, Grosgeorge J, Minoletti F, Coindre JM, Terrier-Lacombe MJ, Mandahl N, Craver RD, Blin N, Sozzi G, Turc-Carel C, O’Brien KP, Kedra D, Fransson I, Guilbaud C, Dumanski JP. Deregulation of the platelet-derived growth factor B-chain gene via fusion with collagen gene COL1A1 in dermatofibrosarcoma protuberans and giant-cell fibroblastoma. Nat Genet. 1997;15:95–98. doi: 10.1038/ng0197-95. PMID: 8988177. DOI: 10.1038/ng0197-95. [DOI] [PubMed] [Google Scholar]
- 32.O’Brien KP, Seroussi E, Dal Cin P, Sciot R, Mandahl N, Fletcher JA, Turc-Carel C, Dumanski JP. Various regions within the alpha-helical domain of the COL1A1 gene are fused to the second exon of the PDGFB gene in dermatofibrosarcomas and giant-cell fibroblastomas. Genes Chromosomes Cancer. 1998;23:187–193. PMID: 9739023. [PubMed] [Google Scholar]
- 33.Takahira T, Oda Y, Tamiya S, Higaki K, Yamamoto H, Kobayashi C, Izumi T, Tateishi N, Iwamoto Y, Tsuneyoshi M. Detection of COL1A1-PDGFB fusion transcripts and PDGFB/PDGFRB mRNA expression in dermatofibrosarcoma protuberans. Mod Pathol. 2007;20:668–675. doi: 10.1038/modpathol.3800783. PMID: 17431412. DOI: 10.1038/modpathol.3800783. [DOI] [PubMed] [Google Scholar]
- 34.Llombart B, Sanmartin O, Lopez-Guerrero JA, Monteagudo C, Serra C, Requena C, Poveda A, Vistos JL, Almenar S, Llombart-Bosch A, Guillen C. Dermatofibrosarcoma protuberans: clinical, pathological, and genetic (COL1A1-PDGFB ) study with therapeutic implications. Histopathology. 2009;54:860–872. doi: 10.1111/j.1365-2559.2009.03310.x. PMID: 19635106. DOI: 10.1111/j.1365-2559.2009. 03310.x. [DOI] [PubMed] [Google Scholar]
- 35.Giacchero D, Maire G, Nuin PA, Berthier F, Ebran N, Carlotti A, Celerier P, Coindre JM, Esteve E, Fraitag S, Guillot B, Ranchere-Vince D, Saiag P, Terrier P, Lacour JP, Pedeutour F. No correlation between the molecular subtype of COL1A1-PDGFB fusion gene and the clinico-histopathological features of dermatofibrosarcoma protuberans. J Invest Dermatol. 2010;130:904–907. doi: 10.1038/jid.2009.338. PMID: 19890351. DOI: 10.1038/jid.2009.338. [DOI] [PubMed] [Google Scholar]
- 36.Oliveira AM, Perez-Atayde AR, Dal Cin P, Gebhardt MC, Chen CJ, Neff JR, Demetri GD, Rosenberg AE, Bridge JA, Fletcher JA. Aneurysmal bone cyst variant translocations upregulate USP6 transcription by promoter swapping with the ZNF9, COL1A1, TRAP150, and OMD genes. Oncogene. 2005;24:3419–3426. doi: 10.1038/sj.onc.1208506. PMID: 15735689. DOI: 10.1038/sj.onc. 1208506. [DOI] [PubMed] [Google Scholar]
- 37.Panagopoulos I, Mertens F, Lofvenberg R, Mandahl N. Fusion of the COL1A1 and USP6 genes in a benign bone tumor. Cancer Genet Cytogenet. 2008;180:70–73. doi: 10.1016/j.cancergencyto.2007.09.017. PMID: 18068538. DOI: 10.1016/j.cancergencyto.2007.09.017. [DOI] [PubMed] [Google Scholar]