Abstract
Background/Aim: Undifferentiated pleomorphic sarcoma (UPS) is an aggressive mesenchymal neoplasm characterized by chromosomal instability. The aim of this study was to identify fusion events involved in UPS. Materials and Methods: Transcriptome sequencing was performed to search for new fusion genes in 19 UPS samples, including two paired recurrent (R) and re-recurrent (RR) samples. Results: A total of 66 fusion genes were detected. Among them, 10 novel fusion genes were further confirmed by reverse transcription polymerase chain reaction (RT-PCR) and Sanger sequencing. Retinoblastoma (RB1) fusions (2 cases) were the most recurrent fusion genes. The gene fusions RB1-RNASEH2B, RB1-FGF14-AS1, and E2F6-FKBP4 were correlated with the Rb/E2F pathway. Pseudogenes were involved in the formation of the gene fusions CIC-DUX4L8 and EIF2AK4-ANXA2P2. Importantly, targetable gene fusions (PDGFRA-MACROD2 and NCOR1-MAP2K1) were detected in UPS. Conclusion: Screening for the presence of fusion transcripts will provide vital clues to the understanding of genetic alterations and the finding of new targeted therapies for UPS.
Keywords: Soft tissue sarcoma, undifferentiated pleomorphic sarcoma, transcriptome sequencing, fusion gene, RB1, pseudogene, targetable fusion
Undifferentiated pleomorphic sarcoma (UPS) is one of the most common soft tissue sarcomas, and represents an aggressive type of mesenchymal malignancy with no identifiable line of differentiation (1). Surgery, radiotherapy, and chemotherapy are the main treatments for UPS. However, UPS tends to recur and spreads to other organs (mainly the lungs), and the 5-year survival rate remains low (30-50%) (1). Therefore, investigations into the underlying mechanisms involved in UPS initiation and progression are of major importance and may lead to the development of new approaches for these patients with poor prognosis.
We and others have found that TP53, ATRX, and RB1 are highly mutated in UPS (2,3); however, no specific genetic characteristics have been identified for UPS compared with other soft tissue sarcomas[such as c-kit mutation for gastrointestinal stromal tumor (4), SS18-SSX1 fusion for synovial sarcoma and so on (5)]. Gene fusions are common in leukemia, bone and soft tissue tumors (6). Almost 300 gene fusions have been detected in mesenchymal tumors (5). Many gene fusions are driver mutations and provide new insights into the molecular mechanisms that are involved in tumorigenesis. These may serve diagnostic or risk stratification purposes and specific targets (5,7). Some gene fusions (PRDM10 and TRIO fusions) have been reported in a small number of UPS cases (8,9). However, UPS is heterogeneous at the genomic level, displaying a spectrum of genomic instability mostly driven by massive copy number changes and large structural rearrangements (10,11). Many gene fusions may represent chance events that result from chromosomal instability (12). Therefore, we hypothesized that more fusion genes may exist in UPS.
In an attempt to identify new fusion genes in UPS, we performed transcriptome sequencing on 19 UPS samples, including two paired recurrent (R) and re-recurrent (RR) samples. In this study, 66 fusion genes were found. Among them, 10 novel fusion transcripts were further verified by reverse transcription polymerase chain reaction (RT-PCR) and Sanger sequencing.
Materials and Methods
Patients. Seventeen patients diagnosed with UPS (the criteria: 1. No identifiable line of differentiation; 2. Tumor cells showing a high degree of pleomorphism) by two professional bone and soft tissue pathologists were included in this study. Four μm hematoxylin and eosin stained slides of each tumor were reviewed, a minimum tumor content of 20% was observed for all cases. Of the 17 patients, one case had a primary tumor resection three years earlier and developed lung metastasis, three cases were newly diagnosed, and 13 patients were relapsed at admission. Tumor tissues and matched adjacent normal muscle tissues were frozen and stored at -80˚C. Tumor, node, and metastasis (TNM) staging was determined using the 7th edition of the American Joint Committee on Cancer guideline of TNM classification. All patients provided written informed consent, and the study was approved by the Clinical Research Ethics Committee of Fudan University Shanghai Cancer Center (Number: 050432-4-1805C).
RNA extraction. Total RNA was extracted by TRIzol reagent (Invitrogen, Carlsbad, CA, USA) from fresh frozen UPS samples and matched adjacent normal muscle tissue samples according to the manufacturer’s instructions. The quantity and quality of RNA were evaluated by the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Samples with a RIN score >7 and RNA concentration >1000 ng were selected for library preparation.
RNA sequencing. One microgram of total RNA from UPS samples was used for library preparation, which was conducted by using the TruSeq RNA Library Prep Kit (Illumina, SanDiego, CA, USA) according to the manufacturer’s protocol. The final libraries were sequenced on an Illumina HiSeqXten platform with a paired-end 2×150 bp protocol aiming at 8 Gb per sample. To discover the fusion transcripts, the RNA-Seq data was analyzed by the STAR-Fusion software (13,14). In Star-Fusion, a built-in GRCh38 reference genome was used with Gencode v26 annotation. Fusion transcripts were found by Star-Fusion with default parameters, and at least three spanning read pairs and three split-reads were required for each fusion (14). In this study, the matched adjacent normal muscle tissue samples were not subjected to RNA sequencing.
RT-PCR and Sanger sequencing. The primers used for PCR amplification and sequencing are listed in Table I. For RT-PCR, 1 μg of total RNA was reverse-transcribed in a 20-μl reaction volume using a FastKing RT Kit (with gDNase) according to the manufacturer’s instructions (Tiangen Biotech, Beijing, China). PCR amplification was performed using PrimeSTAR Max DNA Polymerase (Takara). PCR products were analyzed by agarose gel electrophoresis. All positive RT-PCR products were confirmed by Sanger sequencing. The BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for the analysis of sequence data.
Table I. Primers used for RT-PCR of gene fusions.
F: Forward; R: reverse.
Immunohistochemical analysis. Immunohistochemical analysis was performed on 4-μm-thick tumor sections according to standard procedures. The tissue sections were dewaxed and retrieved, and endogenous peroxidase activity was blocked by hydrogen peroxide treatment. Following incubation with protein blocking solution, slides were incubated at 4˚C overnight with an RB1 antibody (1:600, 9309, against the C-Terminus of RB1, Cell Signaling Technology), PDGFRA antibody (1:400, ab203491), and MAP2K1 antibody (1:100, ab32576, against the C-Terminus of MAP2K1). Signal amplification was performed using the IHC Detection Kit (KIT-5920, Fuzhou Maixin Biotechnology, Fujian, China).
Results
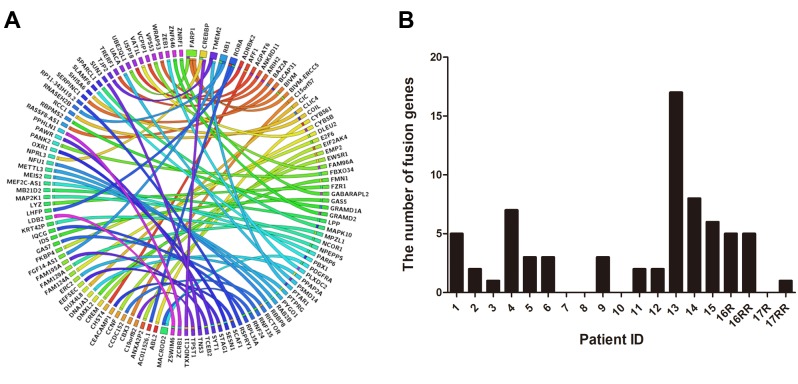
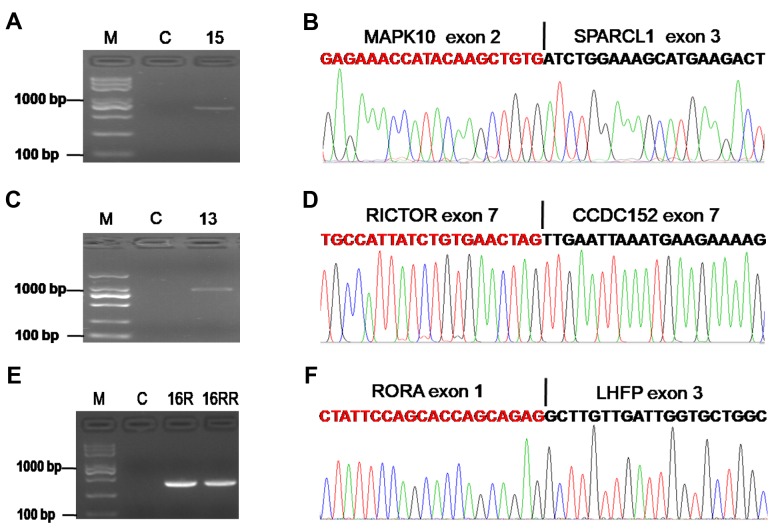
Detection of fusion genes in UPS. To identify fusion transcripts, we performed RNA-sequencing in 19 UPS samples, including two paired recurrent (R) and re-recurrent (RR) samples. The clinicopathological characteristics of patients are listed in Table II. STAR-Fusion software was used for analyzing the sequencing data, and 66 fusion genes were found. The gene fusions are shown in Figure 1A. The number of fusion genes varied in different patients (Figure 1B). Of the 17 patients, one patient (ID 13) harbored the largest number of gene fusions (n=17), and 4 patients (ID7, 8, 10, 17R) showed no fusion genes. Interestingly, the RR tumor (ID 17RR) showed a new fusion gene, ZSWIM6-LDB2. Another RR sample (ID 16RR) displayed the same number of fusion genes compared with the paired R sample (ID 16R). However, two fusion genes (C15orf57-CBX3 and RNF24-MACROD2) were newly detected, whereas two fusions (COIL-SHISA6 and PPAP2A-DNAJA3) were lost in the RR UPS (ID 16RR) compared with paired R UPS (ID 16R); three fusion genes (DLEU2-FAM124A, RB1-FGF14-AS1 and RORA-LHFP) showed no change after tumor re-recurrence. No relationship was found between gene fusions and survival in this study.
Table II. Characteristics of patients.
R: Recurrence; RR: re-recurrence.
Figure 1. Overview of fusion genes detected in UPS. A. Circle plot for fusion transcripts is presented. B. The number of fusion genes in different patients. R: Recurrence; RR: re-recurrence.
A few gene fusions (PRDM10 and TRIO fusions) have been reported in a small number of UPS cases (8,9). However, in this study, we found many fusion genes (n=66). Most of the gene fusions that were recurrent in different cases or whose gene partner affects proliferation were selected and confirmed by RT-PCR and Sanger sequencing; these may be neoplasiaassociated gene fusions. C15orf57-CBX3, RORA fusions and RB1 fusions were selected as the most recurrent fusions and were observed in 2 patients each. Twelve fusion genes (C15orf57-CBX3, RORA-AC011526.1, RORA-LHFP, RB1-RNASEH2B, RB1-FGF14-AS1, E2F6-FKBP4, CIC-DUX4L8, EIF2AK4-ANXA2P2, PDGFRA-MACROD2, NCOR1-MAP2K1, MAPK10-SPARCL1 and RICTOR-CCDC152) were further analysed in this study. Only one fusion (RORA-AC011526.1) could not be detected by RT-PCR (data not shown), although additional paired primers were used. Eleven fusions were confirmed by Sanger sequencing (Figures 2-5). The fusion genes were specifically found in UPS compared with corresponding normal tissues, except for one fusion gene (C15orf57-CBX3) that was observed both in the tumor and the corresponding normal tissue (patient ID 14, data not shown), which is consistent with a report indicating that C15orf57-CBX3 is found in healthy humans (15). To our knowledge, 10 gene fusions (RB1-RNASEH2B, RB1-FGF14-AS1, E2F6-FKBP4, CIC-DUX4L8, EIF2AK4-ANXA2P2, PDGFRA-MACROD2, NCOR1-MAP2K1, MAPK10-SPARCL1, RICTOR-CCDC152, and RORA-LHFP) were confirmed for the first time in malignant diseases. Regarding the three recurrent fusions, RORA-AC011526.1 fusion could not be verified by RT-PCR, C15orf57-CBX3 fusion was not tumor-associated as it was also detected in normal tissue, and RB1 fusions (two cases) were identified as the most recurrent fusion genes in this study.
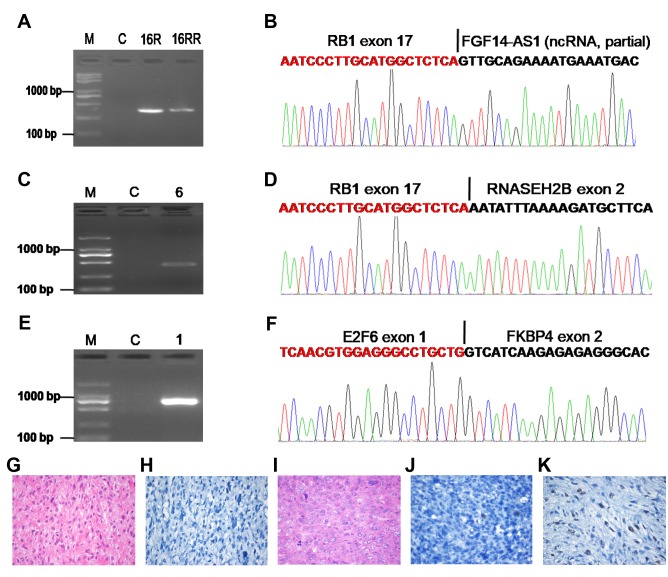
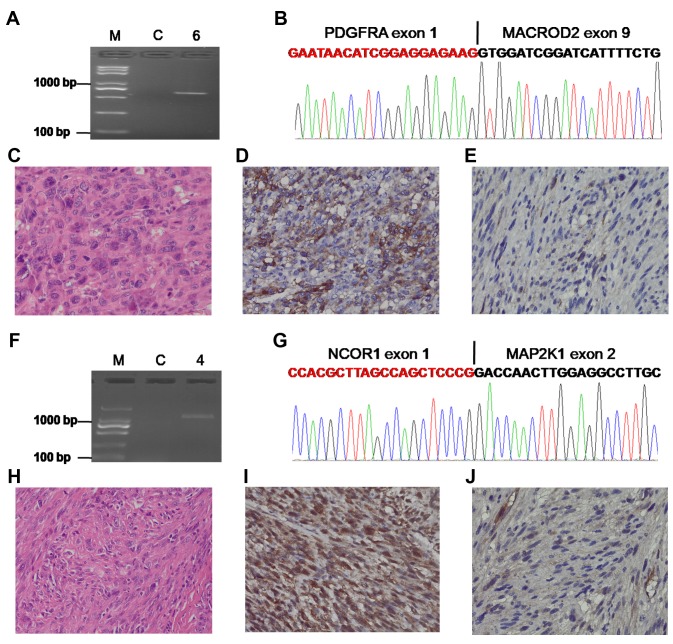
Gene fusions correlated with the Rb/E2F pathway. RB1 mutation is a common event in UPS (2,3). However, reports on RB1 fusion are scant. Interestingly, we found two patients harboring RB1 fusion genes. One case with RB1-FGF14-AS1 fusion (Figure 2A and B) had four local recurrences, and the re-recurrent tumor also showed the same fusion (Figure 2A). The patient had to undergo hindquarter amputation to control tumor recurrence, and is still alive, without tumor relapse and metastasis for three years. Another patient with the RB1-RNASEH2B fusion (Figure 2C and D) died of lung metastasis. Rb/E2F pathway dysregulation is a key factor in the pathogenesis of malignant diseases, and E2F-6 acts as a transcriptional repressor (16,17). We found a new E2F6-FKBP4 fusion transcript consisting of the first exon of the E2F6 gene fused to exon 2 of the FKBP4 gene (Figure 2E and F), which may disrupt the Rb/E2F pathway in UPS. Moreover, expression of the RB1 protein was scarcely observed or only observed in few cells of the two patients with the RB1 fusion compared with the patient with no RB1 fusion (Figure 2G-K).
Figure 2. Gene fusions correlated with the Rb/E2F pathway. A. Confirmation of the RB1-FGF14-AS1 fusion by RT-PCR for patient ID16. B. Schematic representation of the sequence around the junction point (vertical line) is shown for the RB1-FGF14-AS1 fusion. C. Confirmation of the RB1-RNASEH2B fusion by RT-PCR for patient ID 6. D. Schematic representation of the sequence around the junction point (vertical line) is shown for RB1-RNASEH2B fusion. E. Confirmation of the E2F6-FKBP4 fusion by RT-PCR for patient ID 1. F. Schematic representation of the sequence around the junction point (vertical line) is shown for the E2F6-FKBP4 fusion. G. H&E staining of UPS for patient ID16. H. Immunohistochemical analysis of RB1 expression in STS for patient ID16. I. H&E staining of UPS for patient ID 6. J. Immunohistochemical analysis of RB1 expression in STS for patient ID 6. K. Positive expression of RB1 in UPS without RB1 fusion. M: DNA marker; C: Paired normal tissue; Original magnification (G-K), ×400.
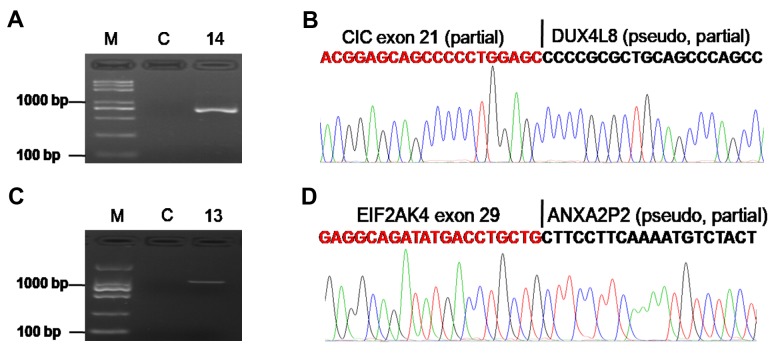
Pseudogenes are involved in formation of gene fusion. A pseudogene is a non-coding gene that is not producing protein (18). In this study, we identified two fusion genes (CIC-DUX4L8, EIF2AK4-ANXA2P2) (Figure 3) with pseudogene partners (DUX4L8, ANXA2P2).
Figure 3. Pseudogene is involved in the formation of gene fusion. A. Confirmation of the CIC-DUX4L8 fusion by RT-PCR for patient ID14. B. Schematic representation of the sequence around the junction point (vertical line) is shown for CIC-DUX4L8 fusion. C. Confirmation of the EIF2AK4-ANXA2P2 fusion by RT-PCR for patient ID 13. D. Schematic representation of the sequence around the junction point (vertical line) is shown for the EIF2AK4-ANXA2P2 fusion. M: DNA marker; C: paired normal tissue.
Targetable gene fusions. Some fusions represent a potential therapeutic target (19). Transcriptome sequencing of 19 UPS samples showed that 1 case with metastasis in the lung harboring PDGFRA-MACROD2 fusion (Figure 4A and B) could be treated with tyrosine kinase inhibitor, which has been shown to be effective in the management of patients with PDGFRA fusions (19,20). The PDGFR gene has a breakpoint between exon 1 and exon 2 (Figure 4B); however, the coding sequence for PDGFR begins at the second exon. Immunohistochemical staining was performed to detect the impact of PDGFRA-MACROD2 fusion on the expression of PDGFRA protein. High expression of PDGFRA was observed in the patient with PDGFRA-MACROD2 fusion compared with the patient without the PDGFRA-MACROD2 fusion (Figure 4C-E).
Figure 4. Targetable gene fusions. A. Confirmation of the PDGFRA-MACROD2 fusion by RT-PCR for patient ID 6. B. Schematic representation of the sequence around the junction point (vertical line) is shown for the PDGFRA-MACROD2 fusion. C. H&E staining of UPS for patient ID 6. D. High expression of PDGFRA for patient ID 6. E. Low expression of PDGFRA in a patient without PDGFRA-MACROD2 fusion. F. Confirmation of the NCOR1-MAP2K1 fusion by RT-PCR for patient ID 4. G. Schematic representation of the sequence around the junction point (vertical line) is shown for the NCOR1-MAP2K1 fusion. H. H&E staining of UPS for patient ID 4. I. Abnormally strong expression of MAP2K1 for patient ID 4. J. Low expression of MAP2K1 in a patient without the NCOR1-MAP2K1 fusion. M: DNA marker; C: paired normal tissue; Original magnification (C-E, H-J), ×400.
MAP2K1, also known as MEK1, is a key ‘gatekeeper’ of ERK signaling, making MEK inhibition a promising strategy for the treatment of various tumors (21-23). We not only found a novel NCOR1-MAP2K1 fusion gene as described in Figure 4F and 4G, but also uncovered strong diffuse immunoreactivity by immunohistochemistry using an antibody to the C terminus of MAP2K1.
Other confirmed fusions in UPS. Three other fusions were also confirmed by RT-PCR and Sanger sequencing. One case harbored a fusion between exon 2 of the MAPK10 and exon 3 of SPARCL1 gene (Figure 5A and B). Another patient displayed a RICTOR-CCDC152 fusion (Figure 5C and D). A fusion transcript RORA-LHFP composed of RORA exon 1 fused to LHFP exon 3 is shown in Figure 5E and F. The three fusion genes are speculated to be involved in tumor growth, as their partners have important roles in malignant diseases (24-26).
Figure 5. Other confirmed fusions in UPS. A. Confirmation of the MAPK10-SPARCL1 fusion by RT-PCR for patient ID 15. B. Schematic representation of the sequence around the junction point (vertical line) is shown for MAPK10-SPARCL1 fusion. C. Confirmation of the RICTOR-CCDC152 fusion by RT-PCR for patient ID 13. D. Schematic representation of the sequence around the junction point (vertical line) is shown for the RICTOR-CCDC152 fusion. E. Confirmation of the RORA-LHFP fusion by RT-PCR for patient ID 16. F. Schematic representation of the sequence around the junction point (vertical line) is shown for the RORA-LHFP fusion. M: DNA marker; C: paired normal tissue; R: recurrence; RR: re-recurrence.
Discussion
UPS is a heterogeneous disease with no definitive cell of origin or specific genetic markers. Fusion genes are considered to have one of the most cancer-specific signatures (27). However, only a few gene fusions have been reported in UPS (8,9). We identified 10 novel fusion transcripts, including RB1-RNASEH2B, RB1-FGF14-AS1, PDGFRA-MACROD2, MAPK10-SPARCL1, CIC-DUX4L8, EIF2AK4-ANXA2P2, RICTOR-CCDC152, E2F6-FKBP4, NCOR1-MAP2K1, and RORA-LHFP. RB1 fusions (2/17 cases) were the most recurrent fusion genes in this study. Pseudogenes were involved in the formation of gene fusions. The fusion transcripts were associated with the Rb/E2F pathway and may be targets for treatment. The characterization of UPS-associated gene fusions is beneficial for our general understanding of pathogenetic mechanisms and may also improve patient management.
RB1, the first tumor suppressor gene to be identified, acts as a negative regulator of E2F-regulated transcription (16). RB1 mutations at variable frequencies have been confirmed in various tumors, and RB1 loss can induce indefinite cell division (2,16). In this study, RB1 fusions (2 cases) were found to be the most recurrent fusion genes. RB1 was respectively fused to RNASEH2B and FGF14-AS1 (Figure 2A-D). Moreover, expression of RB1 protein was scarcely observed or only observed in few cells (Figure 2G-K), indicating that RB1 fusions lead to loss of RB1 expression. Our data enrich the knowledge on RB1 genetic aberration caused by gene fusion and suggest that RB gene fusion is an important event in UPS. Interestingly, in another case, a FKBP4 fusion with E2F6 was detected, which is associated with the Rb/E2F pathway. Taken together, our results suggest that gene fusions (RB1-RNASEH2B, RB1-FGF14-AS1 and E2F6-FKBP4) correlated with the Rb/E2F pathway may play important roles in sustaining tumor cell growth in UPS.
Pseudogenes, as non-coding genes, have lost the ability to produce proteins (18). Emerging evidence suggest that pseudogenes play an important role in the pathogenesis of various tumors (18,28). However, few studies have reported that pseudogenes are involved in the formation of gene fusions. Interestingly, we observed two novel pseudogene fusions (EIF2AK4-ANXA2P2 and CIC-DUX4L8) (Figure 3). EIF2AK4, also known as GCN2, a stress-activated protein kinase, has been shown to promote tumor growth and angiogenesis (29), and its function may be affected by the fusion with pseudogene ANXA2P2. Another gene, CIC, a transcriptional repressor, which has a tumor suppressive function in glioblastoma (30), was found to be fused with the pseudogene DUX4L8. These data suggest a new mechanism of genetic disorder involving pseudogenes.
Gene fusions provide important clues to the precise diagnoses and classifications (5). For example, compared with Ewing sarcoma, tumors containing CIC-rearrangements are taken as an independent tumor type with distinct clinicopathological features (31,32). One patient was found to have a fusion of CIC with a new partner (DUX4L8) (Figure 3A and B). The patient developed lung metastasis following previous surgery in her right thigh and died of disseminated disease, which is consistent with Antonescu’s study (31) indicating that tumors containing CIC rearrangements are associated with an aggressive clinical course. Therefore, tumors containing CIC rearrangements should be treated with more aggressive approaches.
Gene fusions provide important clues for targeted therapy (26). In UPS, the data on fusion gene-guided therapy are limited. Zhou et al. have reported a case with a lumbosacral UPS harboring a LMNA-NTRK1 gene fusion. The patient showed a near-complete clinical response to crizotinib (33). In this study, one case was found to harbor three fusion genes (PDGFRA-MACROD2, RB1-RNASEH2B, and PLXDC2-ZEB1) in lung metastasis. Among these, PDGFRA-MACROD2 can be a direct treatment target, as patients with PDGFRA or PDGFRB fusions often respond to tyrosine kinase inhibitor therapy (19). Unfortunately, the patient died of tumor metastasis without using targeted therapy. Another patient harboring an NCOR1-MAP2K1 fusion showed aberrant expression of MAP2K1 (Figure 4I), which can be treated with MEK inhibitors (22,23), but he showed no local recurrence and metastasis for 4 years after our extensive resection of the tumor in the right upper extremity. In this study, we selected 12 fusion transcripts for verification, and 11 gene fusions were confirmed by RT-PCR and Sanger sequencing, indicating transcriptome sequencing is a reliable approach for detecting fusion genes. Moreover, we found 66 fusion genes in 19 UPS samples, and fusion genes were positive in 14 (82%) of 17 patients, suggesting UPS is a translocation-related sarcoma. Trabectedin, a DNA-binding agent with multiple mechanisms of interfering with several transcription factors and DNA-binding proteins, has shown more clinical benefits in translocation-related sarcoma than those without translocations (34). It has also been reported that trabectedin provides a clinical benefit in UPS patients, and the disease control rate is 56% (5/9) (35). Our results indicate that targeted therapy and trabectedin may show great promise in some selected UPS patients through transcriptome sequencing.
In this study, gene fusions were frequently observed in UPS. Novel recurrent RB1 fusion genes were found that were accompanied with loss of RB1 expression. Pseudogenes were involved in gene fusions. Moreover, some fusion transcripts were associated with the Rb/E2F pathway and targeted treatment. Screening for the presence of fusion transcripts in UPS will provide vital clues to understanding genetic alterations and finding new targeted therapies.
Conflicts of Interest
The Authors disclose no potential conflicts of interest regarding this study.
Authors’ Contributions
BZ, XL, and WY designed the study. BZ, SZ, WC, JW, TW, NT, and YS carried out experiments. SZ analyzed the RNA-Seq data. BZ, XL, and WY wrote the manuscript.
Acknowledgements
Funding: This study was supported by grants from the National Natural Science Foundation of China (Grant No: 81472480, 81302103) and Research Project of Shanghai Municipal Health and Family Planning Commission (Grant No:201640261, 20164y0228).
References
- 1.Canter RJ, Beal S, Borys D, Martinez SR, Bold RJ, Robbins AS. Interaction of histologic subtype and histologic grade in predicting survival for soft-tissue sarcomas. J Am Coll Surg. 2010;210(2):191–198 e192. doi: 10.1016/j.jamcollsurg.2009.10.007. PMID: 20113939. DOI: 10.1016/ j.jamcollsurg.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell. 2017;171(4):950–965 e928. doi: 10.1016/j.cell.2017.10.014. PMID: 29100075. DOI: 10.1016/j.cell.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng B, Qu Y, Wang J, Shi Y, Yan W. Pathogenic and targetable genetic alterations in resected recurrent undifferentiated pleomorphic sarcomas identified by targeted next-generation sequencing. Cancer Genomics Proteomics. 2019;16(3):221–228. doi: 10.21873/cgp.20127. PMID: 31018952. DOI: 10.21873/cgp.20127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmirotta R, De Marchis ML, Ludovici G, Leone B, Covello R, Conti S, Costarelli L, Della-Morte D, Ferroni P, Roselli M, Guadagni F. Mutational analysis of gastrointestinal stromal tumors (gists): Procedural approach for diagnostic purposes. Cancer Genomics Proteomics. 2013;10(3):115–123. PMID: 23741027. [PubMed] [Google Scholar]
- 5.Nakano K, Takahashi S. Translocation-related sarcomas. Int J Mol Sci. 2018;19 (12) doi: 10.3390/ijms19123784. PMID: 30487384. DOI: 10.3390/ijms19123784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7(4):233–245. doi: 10.1038/nrc2091. PMID: 17361217. DOI: 10.1038/ nrc2091. [DOI] [PubMed] [Google Scholar]
- 7.Kulda V, Topolcan O, Kucera R, Kripnerova M, Srbecka K, Hora M, Hes O, Klecka J, Babuska V, Rousarova M, Benson V, Pesta M. Prognostic significance of tmprss2-erg fusion gene in prostate cancer. Anticancer Res. 2016;36(9):4787–4793. doi: 10.21873/anticanres.11037. PMID: 27630329. DOI: 10.21873/anticanres.11037. [DOI] [PubMed] [Google Scholar]
- 8.Hofvander J, Tayebwa J, Nilsson J, Magnusson L, Brosjo O, Larsson O, Vult von Steyern F, Mandahl N, Fletcher CD, Mertens F. Recurrent prdm10 gene fusions in undifferentiated pleomorphic sarcoma. Clin Cancer Res. 2015;21(4):864–869. doi: 10.1158/1078-0432.CCR-14-2399. PMID: 25516889. DOI: 10.1158/1078-0432.CCR-14-2399. [DOI] [PubMed] [Google Scholar]
- 9.Delespaul L, Lesluyes T, Perot G, Brulard C, Lartigue L, Baud J, Lagarde P, Le Guellec S, Neuville A, Terrier P, Vince-Ranchere D, Schmidt S, Debant A, Coindre JM, Chibon F. Recurrent trio fusion in nontranslocation-related sarcomas. Clin Cancer Res. 2017;23(3):857–867. doi: 10.1158/1078-0432.CCR-16-0290. PMID: 27528700. DOI: 10.1158/1078-0432.CCR-16-0290. [DOI] [PubMed] [Google Scholar]
- 10.Gibault L, Perot G, Chibon F, Bonnin S, Lagarde P, Terrier P, Coindre JM, Aurias A. New insights in sarcoma oncogenesis: A comprehensive analysis of a large series of 160 soft tissue sarcomas with complex genomics. J Pathol. 2011;223(1):64–71. doi: 10.1002/path.2787. PMID: 21125665. DOI: 10.1002/path.2787. [DOI] [PubMed] [Google Scholar]
- 11.Steele CD, Tarabichi M, Oukrif D, Webster AP, Ye H, Fittall M, Lombard P, Martincorena I, Tarpey PS, Collord G, Haase K, Strauss SJ, Berisha F, Vaikkinen H, Dhami P, Jansen M, Behjati S, Amary MF, Tirabosco R, Feber A, Campbell PJ, Alexandrov LB, Van Loo P, Flanagan AM, Pillay N. Undifferentiated sarcomas develop through distinct evolutionary pathways. Cancer Cell. 2019;35(3):441–456. doi: 10.1016/j.ccell.2019.02.002. PMID: 30889380. DOI: 10.1016/j.ccell.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshihara K, Wang Q, Torres-Garcia W, Zheng S, Vegesna R, Kim H, Verhaak RG. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34(37):4845–4854. doi: 10.1038/onc.2014.406. PMID: 25500544. DOI: 10.1038/ onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haas B, Dobin A, Stransky N, Li B, Yang X, Tickle T, Bankapur A, Ganote C, Doak T, Pochet N, Sun J, Wu C, Gingeras T, Regev A. STAR-Fusion: Fast and accurate fusion transcript detection from RNA-Seq. 2017. Available at: https://www.biorxiv.org/content/early/2017/03/24/120295.full.pdf (last accessed July 4, 2019) DOI: 10.1101/120295.
- 14.Dixon JR, Xu J, Dileep V, Zhan Y, Song F, Le VT, Yardimci GG, Chakraborty A, Bann DV, Wang Y, Clark R, Zhang L, Yang H, Liu T, Iyyanki S, An L, Pool C, Sasaki T, Rivera-Mulia JC, Ozadam H, Lajoie BR, Kaul R, Buckley M, Lee K, Diegel M, Pezic D, Ernst C, Hadjur S, Odom DT, Stamatoyannopoulos JA, Broach JR, Hardison RC, Ay F, Noble WS, Dekker J, Gilbert DM, Yue F. Integrative detection and analysis of structural variation in cancer genomes. Nat Genet. 2018;50(10):1388–1398. doi: 10.1038/s41588-018-0195-8. PMID: 30202056. DOI: 10.1038/s41588-018-0195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schrider DR, Navarro FC, Galante PA, Parmigiani RB, Camargo AA, Hahn MW, de Souza SJ. Gene copy-number polymorphism caused by retrotransposition in humans. PLoS Genet. 2013;9(1):e1003242. doi: 10.1371/journal.pgen.1003242. PMID: 23359205. DOI: 10.1371/ journal.pgen.1003242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyson NJ. Rb1: A prototype tumor suppressor and an enigma. Genes Dev. 2016;30(13):1492–1502. doi: 10.1101/gad.282145.116. PMID: 27401552. DOI: 10.1101/gad.282145.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cartwright P, Muller H, Wagener C, Holm K, Helin K. E2f-6: A novel member of the e2f family is an inhibitor of e2f-dependent transcription. Oncogene. 1998;17(5):611–623. doi: 10.1038/sj.onc.1201975. PMID: 9704927. DOI: 10.1038/sj.onc.1201975. [DOI] [PubMed] [Google Scholar]
- 18.Kong Y, Zhang L, Huang Y, He T, Zhang L, Zhao X, Zhou X, Zhou D, Yan Y, Zhou J, Xie H, Zhou L, Zheng S, Wang W. Pseudogene pdia3p1 promotes cell proliferation, migration and invasion, and suppresses apoptosis in hepatocellular carcinoma by regulating the p53 pathway. Cancer Lett. 2017;407:76–83. doi: 10.1016/j.canlet.2017.07.031. PMID: 28823960. DOI: 10.1016/j.canlet.2017.07.031. [DOI] [PubMed] [Google Scholar]
- 19.Vega F, Medeiros LJ, Bueso-Ramos CE, Arboleda P, Miranda RN. Hematolymphoid neoplasms associated with rearrangements of pdgfra, pdgfrb, and fgfr1. Am J Clin Pathol. 2015;144(3):377–392. doi: 10.1309/AJCPMORR5Z2IKCEM. PMID: 26276769. DOI: 10.1309/AJCP MORR5Z2IKCEM. [DOI] [PubMed] [Google Scholar]
- 20.David M, Cross NC, Burgstaller S, Chase A, Curtis C, Dang R, Gardembas M, Goldman JM, Grand F, Hughes G, Huguet F, Lavender L, McArthur GA, Mahon FX, Massimini G, Melo J, Rousselot P, Russell-Jones RJ, Seymour JF, Smith G, Stark A, Waghorn K, Nikolova Z, Apperley JF. Durable responses to imatinib in patients with pdgfrb fusion gene-positive and bcr-abl-negative chronic myeloproliferative disorders. Blood. 2007;109(1):61–64. doi: 10.1182/blood-2006-05-024828. PMID: 16960151. DOI: 10.1182/blood-2006-05-024828. [DOI] [PubMed] [Google Scholar]
- 21.Caunt CJ, Sale MJ, Smith PD, Cook SJ. Mek1 and mek2 inhibitors and cancer therapy: The long and winding road. Nat Rev Cancer. 2015;15(10):577–592. doi: 10.1038/nrc4000. PMID: 26399658. DOI: 10.1038/nrc4000. [DOI] [PubMed] [Google Scholar]
- 22.Grimaldi AM, Simeone E, Festino L, Vanella V, Strudel M, Ascierto PA. Mek inhibitors in the treatment of metastatic melanoma and solid tumors. Am J Clin Dermatol. 2017;18(6):745–754. doi: 10.1007/s40257-017-0292-y. PMID: 28537004. DOI: 10.1007/s40257-017-0292-y. [DOI] [PubMed] [Google Scholar]
- 23.Diamond EL, Durham BH, Ulaner GA, Drill E, Buthorn J, Ki M, Bitner L, Cho H, Young RJ, Francis JH, Rampal R, Lacouture M, Brody LA, Ozkaya N, Dogan A, Rosen N, Iasonos A, Abdel-Wahab O, Hyman DM. Efficacy of mek inhibition in patients with histiocytic neoplasms. Nature. 2019;567(7749):521–524. doi: 10.1038/s41586-019-1012-y. PMID: 30867592. DOI: 10.1038/s41586-019-1012-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao SJ, Jiang YQ, Xu NW, Li Q, Zhang Q, Wang SY, Li J, Wang YH, Zhang YL, Jiang SH, Wang YJ, Huang YJ, Zhang XX, Tian GA, Zhang CC, Lv YY, Dai M, Liu F, Zhang R, Zhou D, Zhang ZG. Sparcl1 suppresses osteosarcoma metastasis and recruits macrophages by activation of canonical wnt/beta-catenin signaling through stabilization of the wnt-receptor complex. Oncogene. 2018;37(8):1049–1061. doi: 10.1038/onc.2017.403. PMID: 29084211. DOI: 10.1038/onc.2017.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt KM, Dietrich P, Hackl C, Guenzle J, Bronsert P, Wagner C, Fichtner-Feigl S, Schlitt HJ, Geissler EK, Hellerbrand C, Lang SA. Inhibition of mtorc2/rictor impairs melanoma hepatic metastasis. Neoplasia. 2018;20(12):1198–1208. doi: 10.1016/j.neo.2018.10.001. PMID: 30404068. DOI: 10.1016/j.neo.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan J, Lv Z, Yang G, Liao TT, Xu J, Wu F, Huang Q, Guo M, Hu G, Zhou M, Duan L, Liu S, Jin Y. Retinoic acid receptor-related orphan receptors: Critical roles in tumorigenesis. Front Immunol. 2018;9:1187. doi: 10.3389/fimmu.2018.01187. PMID: 29904382. DOI: 10.3389/ fimmu.2018.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kannan K, Coarfa C, Chao PW, Luo L, Wang Y, Brinegar AE, Hawkins SM, Milosavljevic A, Matzuk MM, Yen L. Recurrent bcam-akt2 fusion gene leads to a constitutively activated akt2 fusion kinase in high-grade serous ovarian carcinoma. Proc Natl Acad Sci USA. 2015;112(11):E1272–1277. doi: 10.1073/pnas.1501735112. PMID: 25733895. DOI: 10.1073/pnas.1501735112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shang J, Wang Z, Chen W, Yang Z, Zheng L, Wang S, Li S. Pseudogene chiap2 inhibits proliferation and invasion of lung adenocarcinoma cells by means of the wnt pathway. J Cell Physiol. 2019;234(8):13735–13746. doi: 10.1002/jcp.28053. PMID: 30623445. DOI: 10.1002/jcp.28053. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Ning Y, Alam GN, Jankowski BM, Dong Z, Nor JE, Polverini PJ. Amino acid deprivation promotes tumor angiogenesis through the gcn2/atf4 pathway. Neoplasia. 2013;15(8):989–997. doi: 10.1593/neo.13262. PMID: 23908598. DOI: 10.1593/neo.13262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bunda S, Heir P, Metcalf J, Li ASC, Agnihotri S, Pusch S, Yasin M, Li M, Burrell K, Mansouri S, Singh O, Wilson M, Alamsahebpour A, Nejad R, Choi B, Kim D, von Deimling A, Zadeh G, Aldape K. Cic protein instability contributes to tumorigenesis in glioblastoma. Nat Commun. 2019;10(1):661. doi: 10.1038/s41467-018-08087-9. PMID: 30737375. DOI: 10.1038/s41467-018-08087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antonescu CR, Owosho AA, Zhang L, Chen S, Deniz K, Huryn JM, Kao YC, Huang SC, Singer S, Tap W, Schaefer IM, Fletcher CD. Sarcomas with cic-rearrangements are a distinct pathologic entity with aggressive outcome: A clinicopathologic and molecular study of 115 cases. Am J Surg Pathol. 2017;41(7):941–949. doi: 10.1097/PAS.0000000000000846. PMID: 28346326. DOI: 10.1097/PAS.00000000 00000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Loarer F, Pissaloux D, Watson S, Godfraind C, Galmiche-Rolland L, Silva K, Mayeur L, Italiano A, Michot A, Pierron G, Vasiljevic A, Ranchere-Vince D, Coindre JM, Tirode F. Clinicopathologic features of cic-nutm1 sarcomas, a new molecular variant of the family of cic-fused sarcomas. Am J Surg Pathol. 2019;43(2):268–276. doi: 10.1097/PAS.0000000000001187. PMID: 30407212. DOI: 10.1097/PAS.0000000000001187. [DOI] [PubMed] [Google Scholar]
- 33.Zhou N, Schafer R, Li T, Fang M, Liu L. A primary undifferentiated pleomorphic sarcoma of the lumbosacral region harboring a lmna-ntrk1 gene fusion with durable clinical response to crizotinib: A case report. BMC Cancer. 2018;18(1):842. doi: 10.1186/s12885-018-4749-z. PMID: 30134855. DOI: 10.1186/s12885-018-4749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Cesne A, Cresta S, Maki RG, Blay JY, Verweij J, Poveda A, Casali PG, Balana C, Schoffski P, Grosso F, Lardelli P, Nieto A, Alfaro V, Demetri GD. A retrospective analysis of antitumour activity with trabectedin in translocation-related sarcomas. Eur J Cancer. 2012;48(16):3036–3044. doi: 10.1016/j.ejca.2012.05.012. PMID: 22749255. DOI: 10.1016/j.ejca.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 35.De Sanctis R, Marrari A, Marchetti S, Mussi C, Balzarini L, Lutman FR, Daolio P, Bastoni S, Bertuzzi AF, Quagliuolo V, Santoro A. Efficacy of trabectedin in advanced soft tissue sarcoma: Beyond lipo- and leiomyosarcoma. Drug Des Devel Ther. 2015;9:5785–5791. doi: 10.2147/DDDT.S92395. PMID: 26604682. DOI: 10.2147/ DDDT.S92395. [DOI] [PMC free article] [PubMed] [Google Scholar]