Abstract
Background/Aim: Sunitinib is a tyrosine kinase inhibitor routinely used as first-line therapy in metastatic renal cell carcinoma (mRCC). Emerging evidence suggests that microRNAs (miRNAs) could be suitable biomarkers with predictive potential in mRCC. The aim of this study was to identify miRNA-based predictive biomarkers of therapy response to avoid unnecessary therapy to non-responding patients. Patients and Methods: High-throughput miRNA microarray profiling was performed on a cohort of 47 patients treated with sunitinib. Validation of candidate miRNAs was carried out on an independent cohort of 132 mRCC patients using qRT-PCR. Results: Out of 158 miRNAs (65 down-regulated, 93 up-regulated), six miRNAs were chosen for independent validation and miR-376b-3p was confirmed to be differentially expressed in tumors of patients with primary resistance versus long-term response (p<0.0002). Conclusion: A predictive miRNA associated with progression-free survival in metastatic renal cell carcinoma patients treated with sunitinib was identified.
Keywords: Renal cell carcinoma, sunitinib, microRNA, prediction
Renal cell carcinoma (RCC) accounts for more than 3% of adult solid tumors and is fatal for around 40% of patients (1). The metastatic form of this disease (mRCC) is routinely treated with tyrosine kinase inhibitors (TKIs) of the VEGF pathway to prevent formation of new blood vessels that support tumors with nutrients. Sunitinib is routinely used as a first-line therapy, followed by other therapeutic options and TKI variants in the second- and third-line treatment after first line failure, which is inevitable in most patients (2,3). However, duration of response is variable, ranging from a few months to more than two years since a proportion of the patients have primary resistance to treatment, some develop it faster than others (4). The financial burden of therapy together with many unpleasant side-effects affecting the quality of life of patients are the main drivers behind the attempts to identify predictive biomarkers reliably differentiating patients who would benefit from the first-line sunitinib from those who should be immediately redirected to other therapeutic options.
MicroRNAs (miRNAs) are a class of short non-coding RNAs, 18-25 nucleotides long, post-transcriptionally regulating gene expression of more than half of protein-coding human genes. Complementary binding to the 3’untranslated region of their target mRNAs (5), miRNAs are pivotal regulators of many vital cellular processes and their deregulation leads to over- or under-expression of proteins, causing many severe diseases, including cancer. Because of their principal role in the pathogenesis of diseases, miRNAs present potential biomarkers for diagnostic, prognostic or predictive purposes (6-8). Several studies have focused, so far, on non-coding RNAs as predictive biomarkers of TKIs. Excluding non-coding RNA biomarkers, differential levels of cytokines, some genetic variants, several blood-based molecules, circulating cancer cells and epigenetic patterns have been associated with response to various TKIs (9-15). It has been shown that the response to sunitinib in mRCC is associated with standard blood serum markers such as C-reactive protein (16) and proangiogenic and proinflammatory cytokine C-X-C motif chemokine ligand containing glutamic acid, leucine arginine motif (17,18). Significant attention has been directed on non-coding RNAs in recent years. Although several studies have been conducted, establishing the diagnostic and prognostic potential of miRNAs (19,20), their predictive value has received less attention. In addition to that, many studies have problematic validity and reproducibility due to the small number of patients analysed. There are also many discrepancies in the study design. Consequently, solitary predictive miRNAs often failed to be validated in independent cohorts. Thus, despite many efforts to identify possible biomarkers, there are currently no reliable candidates applicable in routine clinical practice.
In the present study, we present results on miRNAs as potential predictors of sunitinib response obtained on the largest cohort of mRCC published so far.
Materials and Methods
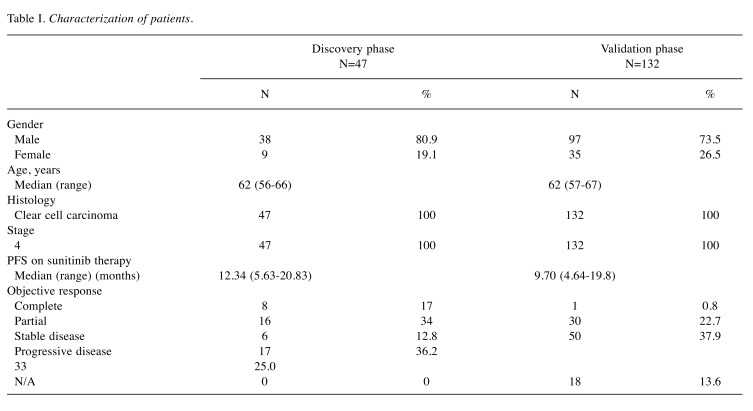
Patients, tissue samples and study design. Metastatic clear cell RCC patients treated with standard first-line sunitinib regimen between years 2008 and 2017 at four comprehensive cancer centers in the Czech Republic – Masaryk Memorial Cancer Institute in Brno, Thomayer Hospital in Prague, University Hospital Pilsen and University Hospital Hradec Králové – were included into the retrospective study. Patients characteristics and treatment outcomes were collected from the national registry RENIS and individual medical records (summarized in Table I). Local ethical committees at all centers approved the study protocol and the study was performed according to the World Medical Association Declaration of Helsinki and all patients signed an informed consent. Best response according to the Response Evaluation Criteria in Solid Tumors (RECIST) (21) and progression-free survival (PFS) interval were used for the definition of the therapeutic response. Only cases for whom there was a formalin fixed paraffin embedded (FFPE) tissue sample of primary tumor available were included into the biomarker study. Out of 179 primary tumor samples from metastatic RCC patients, 25 cases representing the good (PFS longer than 17 months, complete response, partial response or stable disease as characterized by RECIST) and 22 cases representing the poor therapeutic responses (PFS shorter than seven months, progressive disease or stable disease according to RECIST) were chosen for the purpose of the discovery phase of the study. Candidate miRNAs from the discovery phase were validated on an independent cohort of 132 patients (study design summarized in Figure 1). Contrary to the discovery phase, where only patients with very good and poor response were pre-selected, in the validation phase there was a significant proportion of patients with intermediate response. Based on this, we classified patients into three groups: (i) patients with primary resistance (PFS <5 months, progressive or stable disease according to RECIST), (ii) patients with intermediate response (PFS >5 but <12 months, progressive disease, stable disease and partial response according to RECIST) and (iii) patients with long-term response (PFS >12 months, complete response, partial response or stable disease according to RECIST).
Table I. Characterization of patients.
Figure 1. Design of the study. FFPE: Formalin-fixed paraffin embedded; RT-qPCR: reverse transcription real time quantitative polymerase chain reaction.
RNA isolation, miRNA microarrays and individual miRNA detection. Total RNA enriched with short RNA was isolated using commercial mirVana miRNA Isolation Kit (Ambion, Austin, USA). RNA concentration and purity were measured using Nanodrop 2000c (Thermo Fisher Scientific, Waltham, MA, USA). Global expression profiles of miRNAs in the discovery phase were determined using GeneChip 4.0 by Affymetrix (Thermo Fisher Scientific) according to standard protocols recommended by the manufacturer. Microarray and sample annotation data were deposited in EMBL-EBI database ArrayExpress (22) under the accession code E-MTAB-6296. For individual miRNA detection in the validation phase of the study, quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was carried out according to standard TaqMan Advanced miRNA Assay protocols (Applied Biosystems) by use of QuantStudio 12K Flex Real-Time PCR system (Applied Biosystems), TaqMan Fast Advanced Master Mix and specific miRNA probe (Thermo Fisher Scientific). PCR reactions were performed in triplicates; average threshold cycles and standard deviation values were calculated.
Data normalization and statistical analysis. Microarray data were analysed using Bioconductor package and the tool for Linear Model of Microarray Data. qPCR expression levels of each miRNA in the sample was normalized to miR-103a-3p as an endogenous control. Normalized expression data were statistically evaluated by Mann–Whitney U-test, Kruskall–Wallis test, ROC analysis and Kaplan–Meier analysis (GraphPad Prism 5, GraphPad Software, La Jolla, CA, USA). p-Values lower than 0.05 were considered statistically significant.
Results
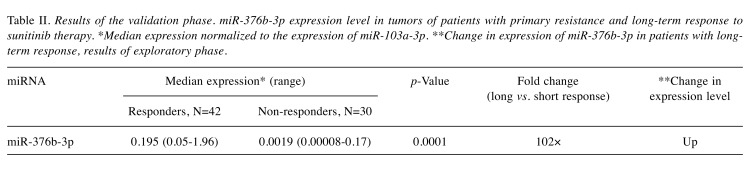
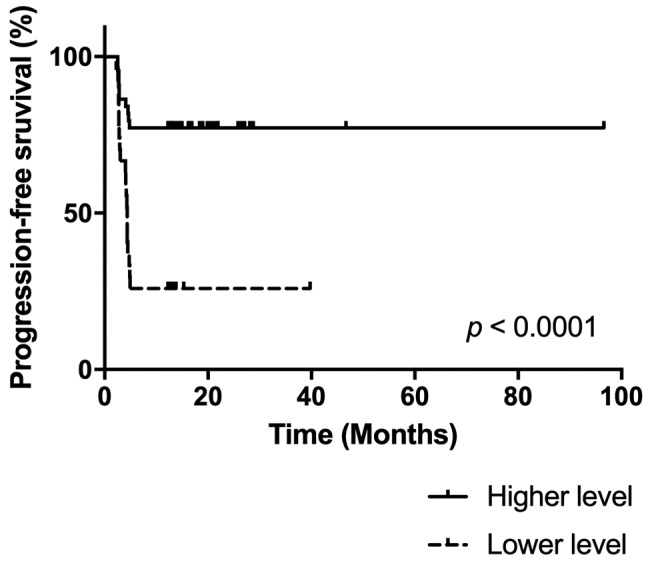
According to statistical analysis of microarray results from the discovery phase of the study, we identified 158 miRNAs (65 down-regulated, 93 up-regulated) to have significantly different expression in tumors of patients with good and poor response (p<0.05). Out of these miRNAs, miR-488-3p, miR-186-3p, miR-122-5p, miR-181d-3p, miR-615-5p and miR-376b-3p were selected for further validation based on the p-value, fold-change, average expression and available literature evidence. Six patients were excluded from the validation study due to undetectable levels of all tested miRNAs, including the reference miRNA, in qPCR measurements indicating insufficient quality of FFPE tumor tissue samples. Out of all miRNAs tested, miR-376b was successfully validated, outperforming other miRNAs in the prediction of a chosen clinical endpoint. In the validation cohort, miR-376b was down-regulated in tumors of patients with shorter PFS (Figure 2) consistently with the microarray analysis results indicating that it was lower by the factor of 0.15 in patients with shorter response. Although we observed progressively decreasing levels of miR-376b-3p in the direction from patients with long-term response to patients with intermediate response and primary resistance (Figure 2), significant results were achieved only when patients with primary resistance and long-term response where compared. In these cases, miR-376b was able to predict the response to sunitinib therapy and identify patients with long-term response with a sensitivity of 83% and specificity of 67% (p=0.0002, AUC=0,758; Figure 2, Figure 3, Table II). These results have also been confirmed using Kaplan–Meier analysis where the patients with lower expression of miR-376b-3p (median PFS=4.4 months) developed relapse significantly earlier in comparison to the patients who exhibited higher expression of miR-376b-3p (median of PFS is not reached) (p=0.0001, Log rank test; Figure 4). Other miRNAs tested in the validation phase were not subjected to statistical analysis due to their extremely low levels in FFPE tumor specimens characterized with Ct values >35 or even undetectable levels in more than 80% of samples.
Figure 2. MiR-376b-3p expression levels in tumors of patients with primary resistance (PFS=2-5 months, N=30), intermediate response (PFS 5-12 months, N=42) and long-term response to sunitinib therapy (PFS >12 months, N=42). (Kruskall–Wallis test; p=0.0003). Comparison only between patients with primary resistance and long-term response (Mann–Whitney test; ***p=0.0001).
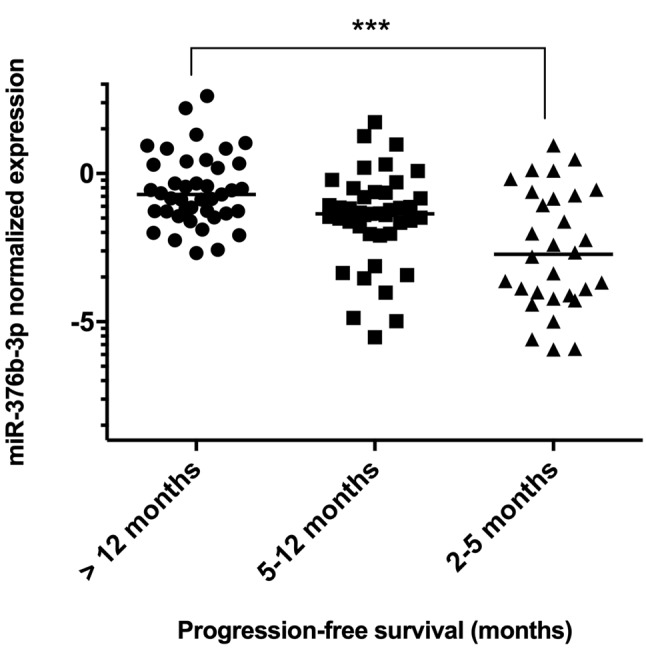
Figure 3. Receiver operating characteristic analysis of the miR-376b-3p as a discriminator between patients with primary resistance and long-term response to sunitinib treatment (p=0.0002).
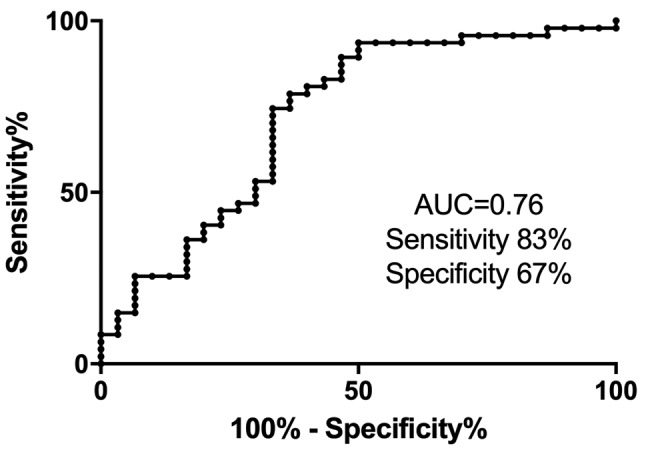
Table II. Results of the validation phase. miR-376b-3p expression level in tumors of patients with primary resistance and long-term response to sunitinib therapy. *Median expression normalized to the expression of miR-103a-3p. **Change in expression of miR-376b-3p in patients with longterm response, results of exploratory phase.
Figure 4. Survival curves of patients stratified accordingly to miR-376b-3p expression level (cut-off value=0.0244; p<0.0001).
Discussion
In the present study, we used microarray technology and identified 158 miRNAs to be differentially expressed in tumor tissues of mRCC patients with good and poor response to first-line sunitinib therapy. Based on the pre-defined criteria we selected six candidate miRNAs for validation in an independent cohort of mRCC patients treated with sunitinib. Unfortunately, five out of six validated miRNAs identified by microarray analysis were not detectable by qRT-PCR (Ct>35) in more than 80% of patients. Since biological specimens used for this biomarker study were FFPE tumor tissues, we believe, that unsatisfactory quality of the tissue specimens and consequently a low-level of RNA integrity (RNA Integrity Number ranging from 2 to 4) have not affected hybridization-based microarray experiments but significantly altered our ability to detect and quantify our miRNA candidates by highly-specific amplification-based qRT-PCR assays. Therefore, the usage of FFPE tissues is the main limitation of our study. On the other hand, in routine diagnostics, FFPE is the only widely available type of biological specimens and qRT-PCR is the most commonly used diagnostic method.
However, we successfully validated miR-376b-3p to have significantly increased levels in the tumors of patients with a long-term response (PFS >12 months). In the remaining patients, we defined two subgroups – patients with intermediate response, lasting for 5-12 months, and patients with primary resistance and PFS being 2-5 months. Further, we observed progressively decreasing levels of miR-376b-3p in the direction from patients with long-term response to patients with intermediate response and primary resistance. We believe, that the existence of these subgroups is not surprising since the response to anti-angiogenic therapy is biologically very complex and continuously variable resulting inevitably in intermediate response groups at any chosen PFS cut-off values. Twelve months present the most common cut-off for response evaluation in metastatic RCC patients; therefore, we chose it as our clinical endpoint to maintain relevance to routine clinical practice.
Only limited information about miR-376b-3p function is available in the literature. Results from Li et al. (23) suggest a role formiR-376b-3p in angiogenesis in a study focused on cerebral ischemia. These authors report that miR-376b-3p strongly inhibited angiogenesis, which corresponds with our results showing down-regulation of miR-376b-3p levels in patients with poor response to antiangiogenic treatment with sunitinib (23). Even in cases that miR-376b-3p is not involved in the development of resistance to sunitinib, its role as an inhibitor of angiogenesis could reinforce or amplify the therapeutic effect of sunitinib and therefore higher levels of this miRNA could help to achieve better therapeutic outcomes in mRCC. MiR-376b-3p was also shown to regulate autophagy by targeting key autophagy-activating proteins like Beclin 1 and Autophagy Related 4C Cystine Peptidase (24). Furthermore, miR-376b-3p is associated with modulation of mitochondrial morphology through targeting Mitochondrial Fisson Factor (25).
Concerning potential usage as a predictive biomarker in mRCC, in one study, based only on small cohorts of patients (9 non-responders vs. 11 responders), miR-376b-3p was identified to be down-regulated in tumors of patients non-responding to sunitinib therapy (p=0.032), which is in accordance with our observations (26). Importantly, definitions of the therapeutic response used in the Berkers’s study were similar to those used in our current study (PFS less than 6 months for non-responders and PFS longer than 12 months for responders was used). As a diagnostic biomarker, differential levels of miR-376b-3p have been associated also with Grave’s disease, a common autoimmune disease affecting the thyroid gland (27).
The present study has several potential limitations and many issues remain to be addressed in order to establish miR-376b-3p as a novel predictive tool. Firstly, the quality of FFPE tissue specimens could affect precision of miRNA quantification by qRT-PCR and low integrity of RNA could cause omission of other miRNA biomarkers identified by microarrays. Analytical performance of miR-376b-3p alone is not enough to enable response prediction in clinical practice (AUC<0.8). Therefore, we expect this biomarker to be used in combination with other biomarkers or predictive tools. Also, the lack of knowledge regarding the mechanism by which miR-376b-3p regulates the response to sunitinib should be addressed.
Successful independent validation of potentially predictive miRNAs remains a challenge. Overlap of results is rare in published studies of miRNAs as predictors of response to sunitinib in mRCC (8,28-30). This could be partly due to tumor heterogeneity, various technologies used for miRNA analysis, limited numbers of enrolled patients, and failure to include a validation cohort.
In conclusion, we identified miR-376b-3p as a potential predictive biomarker in mRCC patients treated with first-line sunitinib. Based on the current knowledge we believe that miR-376b-3p plays a role in angiogenesis and thus could contribute to tumor sensitivity to sunitinib treatment. miR-376b-3p could be used in future predictive models in combination with other independent biomarkers and clinical features.
Conflicts of Interest
Tomas Buchler and Alexandr Poprach have received honoraria and research support unrelated to this project from Pfizer, Roche, Ipsen, Novartis, Bristol Myers Squibb and Bayer. The remaining Authors declare no conflict of interest in regard to this study.
Authors’ Contributions
Conceptualization OS and TB; methodology OS and JKov; experimental work JKov, JJ, MS, PF; validation JKov, JJ, OF, JKop, AP and PF; formal analysis LR and TB; resources and data collection OF, JKop, AP and PF; data curation LR and PB; writing-original draft preparation JKov; writing-review and editing OS, TB, and AP; supervision OS and MS; funding acquisition OS and TB.
Acknowledgements
This research was funded by the Ministry of Health of the Czech Republic, grant no. 15-34678A.
References
- 1.Capitanio U, Bensalah K, Bex A, Boorjian SA, Bray F, Coleman J, Gore JL, Sun M, Wood C, Russo P. Epidemiology of renal cell carcinoma. Eur Urol. 2019;75(1):74–84. doi: 10.1016/j.eururo.2018.08.036. PMID: 30243799. DOI: 10.1016/j.eururo.2018.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bamias A, Escudier B, Sternberg CN, Zagouri F, Dellis A, Djavan B, Tzannis K, Kontovinis L, Stravodimos K, Papatsoris A, Mitropoulos D, Deliveliotis C, Dimopoulos MA, Constantinides CA. Current clinical practice guidelines for the treatment of renal cell carcinoma: A systematic review and critical evaluation. Oncologist. 2017;22(6):667–679. doi: 10.1634/theoncologist.2016-0435. DOI: 10.1634/theoncologist.2016-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Oudard S, Negrier S, Szczylik C, Pili R, Bjarnason GA, Garcia-del-Muro X, Sosman JA, Solska E, Wilding G, Thompson JA, Kim ST, Chen I, Huang X, Figlin RA. Overall survival and updated results for sunitinib compared with interferon alfa in patients with metastatic renal cell carcinoma. J Clin Oncol. 2009;27(22):3584–3590. doi: 10.1200/JCO.2008.20.1293. PMID: 19487381. DOI: 10.1200/JCO.2008.20.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rini BI, Atkins MB. Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol. 2009;10(10):992–1000. doi: 10.1016/S1470-2045(09)70240-2. PMID: 19796751. [DOI] [PubMed] [Google Scholar]
- 5.Friedman RC, Farh KK-H, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. PMID: 18955434. DOI: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petillo D, Kort EJ, Anema J, Furge KA, Yang XJ, Teh BT. MicroRNA profiling of human kidney cancer subtypes. Int J Oncol. 2009;35(1):109–114. doi: 10.3892/ijo_00000318. PMID: 19513557. DOI: 10.3892/ijo_00000318. [DOI] [PubMed] [Google Scholar]
- 7.Gu L, Li H, Chen L, Ma X, Gao Y, Li X, Zhang Y, Fan Y, Zhang X. MicroRNAs as prognostic molecular signatures in renal cell carcinoma: a systematic review and meta-analysis. Oncotarget. 2015;6:32545–32560. doi: 10.18632/oncotarget.5324. PMID: 26416448. DOI: 10.18632/ oncotarget.5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovacova J, Juracek J, Poprach A, Buchler T, Kopecky J, Fiala O, Svoboda M, Slaby O. Candidate microRNA biomarkers of therapeutic response to sunitinib in metastatic renal cell carcinoma: a validation study in patients with extremely good and poor response. Anticancer Res. 2018;38(5):2961–2965. doi: 10.21873/anticanres.12546. PMID: 29715124. DOI: 10.21873/anticanres.12546. [DOI] [PubMed] [Google Scholar]
- 9.Pompas-Veganzones N, Sandonis V, Perez-Lanzac A, Beltran M, Beardo P, Juárez A, Vazquez F, Cozar JM, Alvarez-Ossorio JL, Sanchez-Carbayo M. Myopodin methylation is a prognostic biomarker and predicts antiangiogenic response in advanced kidney cancer. Tumour Biol. 2016;37(10):14301–14310. doi: 10.1007/s13277-016-5267-8. PMID: 27592258. DOI: 10.1007/s13277-016-5267-8. [DOI] [PubMed] [Google Scholar]
- 10.Kim J, Hwang J, Jeong H, Song HJ, Shin J, Hur G, Park YW, Lee SH, Kim J. Promoter methylation status of VEGF receptor genes: a possible epigenetic biomarker to anticipate the efficacy of intracellular-acting VEGF-targeted drugs in cancer cells. Epigenetics. 2012;7(2):191–200. doi: 10.4161/epi.7.2.18973. PMID: 22395469. DOI: 10.4161/epi.7.2.18973. [DOI] [PubMed] [Google Scholar]
- 11.Motzer RJ, Hutson TE, Hudes GR, Figlin RA, Martini JF, English PA, Huang X, Valota O, Williams JA. Investigation of novel circulating proteins, germ line single-nucleotide polymorphisms, and molecular tumor markers as potential efficacy biomarkers of first-line sunitinib therapy for advanced renal cell carcinoma. Cancer Chemother Pharmacol. 2014;74(4):739–750. doi: 10.1007/s00280-014-2539-0. PMID: 25100134. DOI: 10.1007/s00280-014-2539-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossi E, Fassan M, Aieta M, Zilio F, Celadin R, Borin M, Grassi A, Troiani L, Basso U, Barile C, Sava T, Lanza C, Miatello L, Jirillo A, Rugge M, Indraccolo S, Cristofanilli M, Amadori A, Zamarchi R. Dynamic changes of live/apoptotic circulating tumour cells as predictive marker of response to sunitinib in metastatic renal cancer. Br J Cancer. 2012;107(8):1286–1294. doi: 10.1038/bjc.2012.388. PMID: 22955853. DOI: 10.1038/bjc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowery A, Han Z. Assessment of tumor response to tyrosine kinase inhibitors. Front Biosci (Landmark Ed) 2011;16:1996–2007. doi: 10.2741/3836. PMID: 21622159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beuselinck B, Karadimou A, Lambrechts D, Claes B, Wolter P, Couchy G, Berkers J, van Poppel H, Paridaens R, Schöffski P, Méjean A, Verkarre V, Lerut E, Joly F, Lebret T, Gravis G, Deplanque G, Descazeaud A, Leclercq NR, Molinié V, Patard JJ, Teghom C, Elaidi R, Zucman Rossi J, Oudard S. VEGFR1 single nucleotide polymorphisms associated with outcome in patients with metastatic renal cell carcinoma treated with sunitinib – a multicentric retrospective analysis. Acta Oncol. 2014;53(1):103–112. doi: 10.3109/0284186X.2013.770600. PMID: 23421954. DOI: 10.3109/ 0284186X.2013.770600. [DOI] [PubMed] [Google Scholar]
- 15.Decoster L, Vande Broek I, Neyns B, Majois F, Baurain JF, Rottey S, Rorive A, Anckaert E, De Mey J, De Brakeleer S, De Grève J. Biomarker analysis in a phase II study of sunitinib in patients with advanced melanoma. Anticancer Res. 2015;35(12):6893–6899. PMID: 26637913. [PubMed] [Google Scholar]
- 16.Pilskog M, Beisland C, Akslen LA, Bostad L, Haug Å, Heinrich D, Hjelle KM, Straume O. Predictive value of C-reactive protein in patients treated with sunitinib for metastatic clear cell renal cell carcinoma. BMC Urol. 2017;17(1):74. doi: 10.1186/s12894-017-0267-6. PMID: 28859644. DOI: 10.1186/s12894-017-0267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grépin R, Guyot M, Giuliano S, Boncompagni M, Ambrosetti D, Chamorey E, Scoazec JY, Negrier S, Simonnet H, Pagès G. The CXCL7/CXCR1/2 axis is a key driver in the growth of clear cell renal cell carcinoma. Cancer Res. 2014;74(3):873–883. doi: 10.1158/0008-5472.CAN-13-1267. PMID: 24335961. DOI: 10.1158/0008-5472.CAN-13-1267. [DOI] [PubMed] [Google Scholar]
- 18.Dufies M, Giuliano S, Viotti J, Borchiellini D, Cooley LS, Ambrosetti D, Guyot M, Ndiaye PD, Parola J, Claren A, Schiappa R, Gal J, Frangeul A, Jacquel A, Cassuto O, Grépin R, Auberger P, Bikfalvi A, Milano G, Escudier B, Rioux-Leclercq N, Porta C, Negrier S, Chamorey E, Ferrero JM, Pagès G. CXCL7 is a predictive marker of sunitinib efficacy in clear cell renal cell carcinomas. Br J Cancer. 2017;117(7):947–953. doi: 10.1038/bjc.2017.276. PMID: 28850564. DOI: 10.1038/bjc.2017.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell'Aquila ML, Alder H, Rassenti L, Kipps TJ, Bullrich F, Negrini M, Croce CM. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101(32):11755–11760. doi: 10.1073/pnas.0404432101. PMID: 15284443. DOI: 10.1073/ pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinzelmann J, Henning B, Sanjmyatav J, Posorski N, Steiner T, Wunderlich H, Gajda MR, Junker K. Specific miRNA signatures are associated with metastasis and poor prognosis in clear cell renal cell carcinoma. World J Urol. 2011;29(3):367–373. doi: 10.1007/s00345-010-0633-4. PMID: 21229250. DOI: 10.1007/s00345-010-0633-4. [DOI] [PubMed] [Google Scholar]
- 21.Litière S, Collette S, de Vries EGE, Seymour L, Bogaerts J. RECIST - learning from the past to build the future. Nat Rev Clin Oncol. 2017;14(3):187–192. doi: 10.1038/nrclinonc.2016.195. PMID: 27995946. DOI: 10.1038/nrclinonc.2016.195. [DOI] [PubMed] [Google Scholar]
- 22.Kolesnikov N, Hastings E, Keays M, Melnichuk O, Tang YA, Williams E, Dylag M, Kurbatova N, Brandizi M, Burdett T, Megy K, Pilicheva E, Rustici G, Tikhonov A, Parkinson H, Petryszak R, Sarkans U, Brazma A. ArrayExpress update-simplifying data submissions. Nucleic Acids Res. 2015;43(Database issue):D1113–1136. doi: 10.1093/nar/gku1057. PMID: 25361974. DOI: 10.1093/ nar/gku1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L-J, Huang Q, Zhang N, Wang G-B, Liu Y-H. miR-376b-5p regulates angiogenesis in cerebral ischemia. Mol Med Rep. 2014;10(1):527–535. doi: 10.3892/mmr.2014.2172. PMID: 24789343. DOI: 10.3892/mmr. 2014.2172. [DOI] [PubMed] [Google Scholar]
- 24.Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy. 2012;8(2):165–176. doi: 10.4161/auto.8.2.18351. PMID: 22248718. DOI: 10.4161/auto.8.2.18351. [DOI] [PubMed] [Google Scholar]
- 25.Sun YL, Li SH, Yang L, Wang Y. miR-376b-3p attenuates mitochondrial fission and cardiac hypertrophy by targeting mitochondrial fission factor. Clin Exp Pharmacol Physiol. 2018;45:779–787. doi: 10.1111/1440-1681.12938. PMID: 29570827. DOI: 10.1111/1440-1681. 12938. [DOI] [PubMed] [Google Scholar]
- 26.Berkers J, Govaere O, Wolter P, Beuselinck B, Schöffski P, van Kempen LC, Albersen M, Van den Oord J, Roskams T, Swinnen J, Joniau S, Van Poppel H, Lerut E. A possible role for microRNA-141 down-regulation in sunitinib resistant metastatic clear cell renal cell carcinoma through induction of epithelial-to-mesenchymal transition and hypoxia resistance. J Urol. 2013;189(5):1930–1938. doi: 10.1016/j.juro.2012.11.133. PMID: 23206420. DOI: 10.1016/ j.juro.2012.11.133. [DOI] [PubMed] [Google Scholar]
- 27.Liu R, Ma X, Xu L, Wang D, Jiang X, Zhu W, Cui B, Ning G, Lin D, Wang S. Differential microRNA expression in peripheral blood mononuclear cells from Graves’ disease patients. J Clin Endocrinol Metab. 2012;97(6):E968–972. doi: 10.1210/jc.2011-2982. PMID: 22456620. DOI: 10.1210/jc.2011-2982. [DOI] [PubMed] [Google Scholar]
- 28.Kovacova J, Poprach A, Buchler T, Cho WC, Slaby O. MicroRNAs as predictive biomarkers of response to tyrosine kinase inhibitor therapy in metastatic renal cell carcinoma. Clin Chem Lab Med. 2018;56(9):1426–1431. doi: 10.1515/cclm-2017-0861. PMID: 29451857. DOI: 10.1515/cclm-2017-0861. [DOI] [PubMed] [Google Scholar]
- 29.Lukamowicz-Rajska M, Mittmann C, Prummer M, Zhong Q, Bedke J, Hennenlotter J, Stenzl A, Mischo A, Bihr S, Schmidinger M, Vogl U, Blume I, Karlo C, Schraml P, Moch H. MiR-99b-5p expression and response to tyrosine kinase inhibitor treatment in clear cell renal cell carcinoma patients. Oncotarget. 2016;7:78433–78447. doi: 10.18632/oncotarget.12618. PMID: 27738339. DOI: 10.18632/oncotarget.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ralla B, Busch J, Flörcken A, Westermann J, Zhao Z, Kilic E, Weickmann S, Jung M, Fendler A, Jung K. miR-9-5p in nephrectomy specimens is a potential predictor of primary resistance to first-line treatment with tyrosine kinase inhibitors in patients with metastatic renal cell carcinoma. Cancers (Basel) 2018;10 doi: 10.3390/cancers10090321. PMID: 27738339. DOI: 10.18632/oncotarget.12618. [DOI] [PMC free article] [PubMed] [Google Scholar]