Abstract
Rheumatoid arthritis (RA) is the most common inflammatory arthritis and exhibits genetic overlap with other autoimmune and inflammatory disorders. Although predominant associations with the HLA-DRB1 locus have been known for decades, recent data have revealed additional insight into the likely causative variants within HLA-DRB1 as well as within other HLA loci that contribute to disease risk. In addition, more than 100 common variants in non-HLA loci have been implicated in disease susceptibility. Genetic factors are involved not only in the development of RA, but also with various disease subphenotypes, including production and circulating levels of autoantibodies and joint destruction. The major current challenge is to integrate these new data into a precise understanding of disease pathogenesis, including the critical cell types and molecular networks involved as well as interactions with environmental factors. We predict that delineating the functional effects of genetic variants is likely to drive new diagnostic and therapeutic approaches to the disease.
Keywords: HLA, endophenotype
1. INTRODUCTION
Rheumatoid arthritis (RA) is the most common immune-mediated arthritis in adults. The disease is female predominant, with a 3:1 to 4:1 female:male ratio (15) and a worldwide prevalence of 0.5–0.8% (92, 184). Inflammatory processes in RA lead to bony destruction of the joints (33) and can involve the lungs, skin, and other organs (33). Because of the clinical overlap with other forms of arthritis and the heterogeneity of the associated antibody serologies, the definition of RA continues to evolve. Indeed, these serological differences between patients have been important for understanding the underlying genetics as well as the role of environmental factors in RA. The overlap of risk alleles in RA with alleles for many other autoimmune disorders suggests shared mechanistic pathways between these conditions. A major challenge for all autoimmune disorders is to understand genetic variations in the context of a deeper functional understanding of disease pathogenesis and then to apply this information to improve disease management. Our discussion in this review attempts to put the current knowledge of RA genetics into this context.
2. PHENOTYPIC AND SEROLOGICAL DIVERSITY IN RHEUMATOID ARTHRITIS
Since the seminal discovery of rheumatoid factor (RF) in the late 1940s, serological studies have played a critical role in diagnosing and defining RA (117). The presence or absence of a specific serology has long been used to define seropositive and seronegative RA. But RF was quickly recognized to be nonspecific for RA; it may be present in patients with other autoimmune diseases, infections, or liver diseases (131). Despite early evidence of additional informative serological specificities (94), it was not until 50 years later that serological reactivity to citrullinated peptides was identified as a marker for seropositive RA (123). Anti-citrullinated protein/peptide antibodies (ACPAs) are now recognized as more highly specific for RA than RF (123, 124), and ACPAs are now part of the official diagnostic criteria for the disease (1). These antibodies have provided a powerful method of identifying a phenotypic subgroup, both for characterizing disease populations and for performing genetic analysis. The anti-cyclic citrullinated peptide (CCP) antibody is a widely used clinical test to quantify ACPAs. Meta-analysis has shown that 67% of patients with RA are positive for anti-CCP antibodies (95). Positivity of ACPA correlates with that of RF; more than 90% of ACPA(+) RA patients are positive for RF, and approximately half of ACPA(−) RA patients are positive for RF (155). Strictly speaking, seronegative RA should be negative for both ACPA and RF, although in practice seropositive disease is often defined with either specificity. It is important to note that methods for detecting ACPA(+) are rapidly evolving, and mixtures of different autoantigens are detected by the various methods. This can also influence the definition of seropositive and may impact genetic studies of this serological subgroup.
3. FAMILIAL AGGREGATION AND HERITABILITY OF RHEUMATOID ARTHRITIS
Evidence for familial aggregation of RA was reported shortly after the discovery of RF (75, 79). Familial studies revealed that individuals with affected first-degree relatives have a risk of developing RA that is two to four times the risk of those without such relatives. Frisell et al. (35) examined Swedish patient registries with more than 80,000 subjects and observed an increased risk of seropositive RA in individuals with a first-degree relative with seropositive RA [odds ratio (OR) = 3.7−4.2]. They observed a similar but weaker effect in seronegative RA (OR = 2.3−3.4). The risk was independent of gender. This study showed potential heterogeneity in the heritability of seropositive and seronegative RA. A related study by Hemminki et al. (48) used the Multigeneration Register in Sweden and reported that the standardized incidence ratio of RA was 3.02 for offspring, 4.64 in siblings, 9.31 in multiplex families, 6.48 in twins, and 1.17 in spouses. A study using the Danish nationwide registry reported that the hazard ratio for RA in offspring is 2.6−2.9 regardless of parental and offspring gender (135).
Twin studies have also been informative for estimating the heritability of RA. Most published results are from European populations. The proportion of variance in developing the disease that is attributable to genetic factors is called broad-sense heritability; the proportion of variance explained by additive effects of genetic components is called narrow-sense heritability. MacGregor et al. (85) studied a total of 185 and 231 twins in the United Kingdom and Finland, respectively, and estimated the narrow-sense heritability of RA to be 53−65%. A Dutch twin study using 148 RA twins estimated the heritability of RA to be 66% (169). A Swedish study using a total of 12,590 twins estimated the heritability of RA to be 39% (49). A recent study reported from Demark that recruited 162 twins reported a much lower heritability estimate of 12% (148). In addition to these twin studies, a Swedish family-based study estimated the heritability of RA as 40% (35), and calculation from a genome-wide association study (GWAS) estimated the heritability as 52% (136). Although twin studies outside European countries have not yet been reported, a study of Japanese twins has estimated the heritability of RAin the Japanese population to be approximately 62% (150), which is comparable to reports in European populations. Thus, with a few exceptions, the estimated heritability of RA seems to fall consistently in the range of 40–60%.
As discussed below, major histocompatibility complex (MHC) associations explain approximately 12% of total heritability. Recent GWAS investigations have identified a total of 100 non-human leukocyte antigen (HLA) susceptibility genes, yet these variants together explain only approximately 5.5% of the total variance of RA susceptibility (101), although a somewhat higher fraction of variance could be explained by adjusting for poor tagging of causal variants by the GWAS markers (44). Stahl et al. (138) estimated that approximately 18% of heritability could be explained by additional polygenic signals, capturing undiscovered common variant associations. Therefore, in aggregate, the known alleles and polygenic signals explain 3 5 % of the total liability, which falls short of known heritability estimates of approximately 50%. This discrepancy may reflect the heterogeneity of phenotypic definitions used in twin studies compared with population-based studies, gene-gene interactions, gene-environment interactions, or undiscovered alleles.
4. ENVIRONMENTAL FACTORS
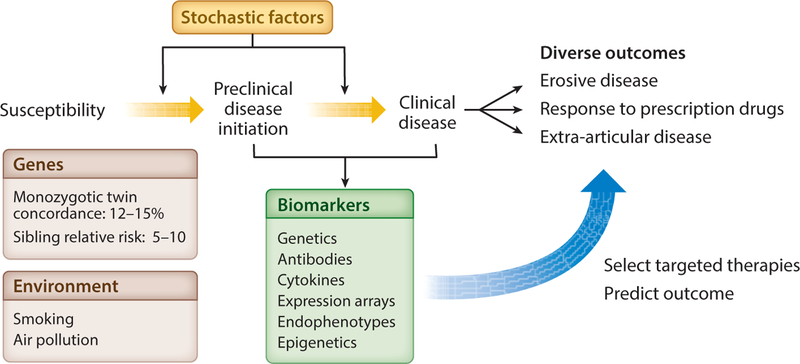
The development of RA occurs after a prolonged preclinical period in which autoantibodies develop in susceptible individuals, which is followed by the appearance of clinical disease (Figure 1). Several environmental risk factors for RA have been identified, including smoking, obesity (82), and exposure to air pollution (28), which likely act during the preclinical period. Smoking is the most well-established risk factor, increasing risk of disease 1.3- and 2.0-fold for women and men, respectively (143). Smoking is a particularly important consideration in the context of this review because it has been shown to interact with genotype, particularly in northern European populations (62, 84, 103). This has led to a model of pathogenesis in which smoking exposure may lead to the presence of citrullinated proteins in the lung, which is followed by the development of ACPAs in susceptible individuals (12). Data showing production of anti-citrulline antibodies in the inflamed rheumatoid joint reinforce the importance of anti-citrulline specificity in disease pathogenesis (2).
Figure 1.
The stages of development of rheumatoid arthritis. Genetic susceptibility can interact with environmental factors, which together with chance (stochastic factors) can lead to the initiation of autoimmune and inflammatory subclinical phenotypes in a subset of individuals. These phenotypes may in turn progress to overt clinical disease in some individuals. It is likely that a combination of genetics and other endophenotypes (detectable with appropriate cellular assays or biomarkers) can influence disease heterogeneity and longer-term clinical outcomes.
Inasmuch as it likely reflects the action of an environmental exposure, the presence of ACPA antibodies in an unaffected individual is clearly a risk factor for the future development of RA. Johansson et al. (59) further demonstrated that a combination of smoking, the presence of HLA risk alleles, and the presence of PTPN22 risk alleles (discussed below) leads to very high risk for the development of RA within a period of years, at least in some populations. This observation is one of the best examples of gene-environment interactions in the field of autoimmunity and emphasizes that consideration of environmental factors is important for the discovery and understanding of genetic contributions to disease pathogenesis.
Including environmental measurements in GWAS analysis is challenging because of the difficulty of collecting accurate data. However, doing so will likely be important for integrating genetic data into models of RA patho genesis.
5. GENETIC DISCOVERY
5.1. Refining HLA Associations with Rheumatoid Arthritis
Polymorphisms in the HLA loci within the MHC are by far the strongest source of genetic susceptibility to RA (140), with a subgroup of DRB1*04 alleles conferring greatest risk of disease (38, 141). The MHC molecules are critical for the presentation of self and foreign antigens. The MHC class I proteins are encoded by HLA-A, -B, and -C genes; they are expressed in almost all nucleated cells and function to present antigen to CD8+ T cells. The MHC class II molecules are encoded by HLA-DR, -DP, and -DQ genes. These proteins present peptide antigens to CD4+ T cells and are expressed mainly on professional antigen-presenting cells, such as dendritic cells, B cells, and macrophages. The HLA genes encoding both class I and class II molecules are located in a single region on chromosome 6. The region is under a high degree of selection and is thus highly polymorphic, with complex patterns of linkage disequilibrium structure (162). This makes it extremely challenging to sort out the independent effects of risk loci in this region.
5.1.1. HLA alleles.
The first evidence for an association of RA with HLA class II alleles dates back to the cellular and serological studies by Stastny (140, 141) and colleagues in the 1970s. Careful examination of individual alleles in patient cohorts revealed associations among some but not all alleles corresponding to the DR4 serotype. For example, studies demonstrated case associations with DRB1*04:01 (141), DRB1*04:04 (93), and DRB1*04:05 but failed to show associations between DRB1*04:02 or DRB1*04:03 and RA (180). Other associations with non-DR4 serotypes were also recognized, including associations with DRB1*01:01, DRB1*10:01 (183), and DRB1*14:02 (91).
The advent of molecular cloning and sequencing of HLA alleles in the 1980s led to the definition of the shared epitope (SE), a common peptide sequence (positions 70–74) shared among many different HLA class II alleles at the DRB1 locus (43). This provided a structural rationale for the diverse patterns of DRB1 allelic associations. However, some variation was observed among different populations. For example, DRB1*09:01 in Asian populations confers strong susceptibility to RA (97), yet it differs at position 74 from other SE alleles while sharing a triple arginine sequence with the DRB1*10:01 susceptibility alleles found in Caucasians. Thus, subtle structural differences among DRB1 risk alleles required explanation.
5.1.2. Fine mapping HLA alleles to individual amino acid sites.
More recent studies have taken advantage of large-scale single-nucleotide polymorphism (SNP) data collected for GWAS and have utilized HLA imputation to infer HLA alleles (24, 56, 185). This has been an important methodological advance because direct HLA typing remains costly, even with next-generation sequencing approaches; the highly polymorphic nature of these genes makes even targeted sequencing difficult without very long reads. The availability of highly dense GWAS data in reference populations with known HLA typing allows for the construction of haplotypes and imputation of specific HLA alleles in individuals with GWAS data. Based on the combination of amino acid residues encoded by the genotyped or imputed alleles, HLA alleles or amino acid residues can be inferred and tested for associations (56). Construction of ethnicity-specific reference panels leads to more accurate imputation results (104).
Using this approach with 5,018 cases and 14,974 controls, Raychaudhuri et al. (112) refined and extended the SE hypothesis and highlighted the predominant importance of amino acid position 13 in the DRB1 molecule, at the base of the antigen-binding cleft, with secondary independent associations at positions 71 and 74. These three positions together explain 9.7% of the liability of seropositive RA, corresponding to approximately one-fifth of the total heritability of RA. Together, these positions define the P4 binding pocket in DRB1, suggesting that this pocket plays a critical role in antigen binding and triggering of a critical immune response. Indeed, this structural feature of the DRB1 protein may be important for binding of citrullinated antigenic peptides that either initiate or perpetuate the disease (122).
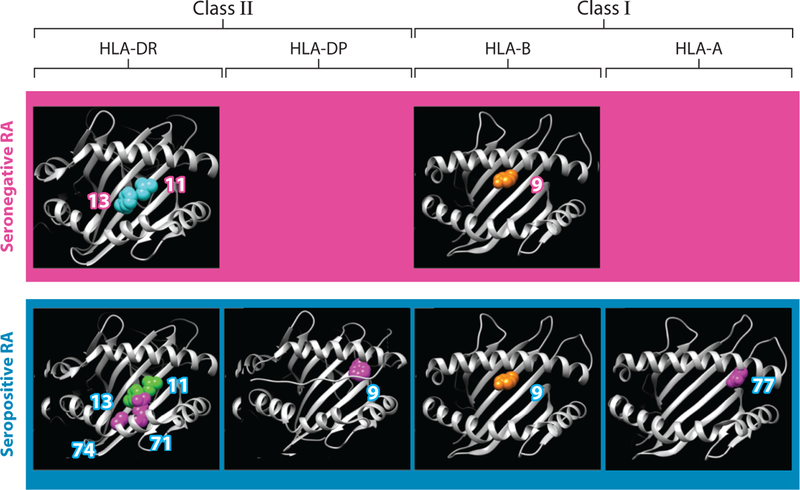
Using detailed conditional analysis, these studies have also implicated additional loci within the MHC that influence disease susceptibility, including position 9 in HLA-B, position 9 in HLA-DPB1, and position 77 in HLA-A (46, 112) (Figure 2). All of these amino acid sites are in peptide-binding grooves that are critical for antigen recognition to discern self and nonself peptides. Okada et al. (98) applied a similar approach in Asian populations and demonstrated that ACPA(+) RA shows associations with HLA amino acid residues and positions in these populations that are similar to those in European populations. Although positions 11 and 13 are tightly linked in European populations, this study identified position 13 as the strongest position in Asian populations, and the difference from European populations was contributed mainly by DRB1*09:01. Intriguingly, the authors argued for the potential importance of amino acid position of 57 in HLA-DRβ1, possibly reflecting cross-ethnic differences of DRB1*04:05 in Asian populations. Reynolds et al. (115) genotyped 561 African American cases for HLA-DRβ1 and showed that amino acid position 13 conferred most of the susceptibility associations. In conditional analysis, they found an additional significant signal in amino acid position of 57, similar to that in Asians.
Figure 2.
Three-dimensional structures of amino acid positions in HLA class I and II molecules that affect susceptibility to seropositive and seronegative rheumatoid arthritis (RA). Modified from Reference 46 with permission.
It should be noted that the HLA region includes the other proteins with multiple polymorphisms that were not imputed by the method, including nonclassical HLA alleles. Thus, there is potential for additional discoveries within the MHC region as more sophisticated sequencing-based approaches are developed and implemented.
5.1.3. HLA interactions.
Investigators have reported excess risk for heterozygous genotypes composed of certain combinations of HLA-DRB1 alleles. These results have suggested that HLA alleles interact with each other to confer the risk of developing RA (55, 181). These interactions might be mediated, for example, by complementary binding of pathogenic antigens. A challenge in this area is understanding whether the increased risk is due to partial dominance effects of individual alleles or to interactions between allelic combinations. Hall et al. (45) reported that subjects carrying a combination of DRB1*04:01 and DRB1*04:04 had a risk of developing RA that was thirty times that of subjects carrying no risk alleles. Fries et al. (34) confirmed the increased risk of this heterozygote combination in 2002.
In the largest examination of HLA allelic interactions so far, Lenz et al. (78) systematically analyzed interactive effects for RA, psoriasis, celiac disease, type I diabetes, and achalasia. They observed that HLA-DRB1 alleles showed significant interactive effects for RA susceptibility. The HLA alleles showing significant interactive effects for RA susceptibility included combinations of DRB1*01:01 and *04:01, DRB1*03:01 and *04:01, DRB1*01:01 and *15:01, and DRB1*01:01 and *07:01. Although the heterozygote of DRB1*04:01 and *04:04 showed higher odds ratios than homozygotes of DRB1*04:01 or *04:04, with an overlap of confidence intervals, the interaction was not significant. They found a suggestive dominant effect of DRB1*04:01. Collectively, the combination of partial dominance effects and interaction effects could explain 1.4% of the variance of RA development.
5.2. Genetic Mapping of Non-MHC Loci
Although the MHC contains the strongest individual risk alleles for RA, a much larger number of risk alleles have been defined outside of the MHC region. Most of these risk alleles are fairly common, although an increasing number of rare susceptibility variants are also being identified.
5.2.1. Common variants.
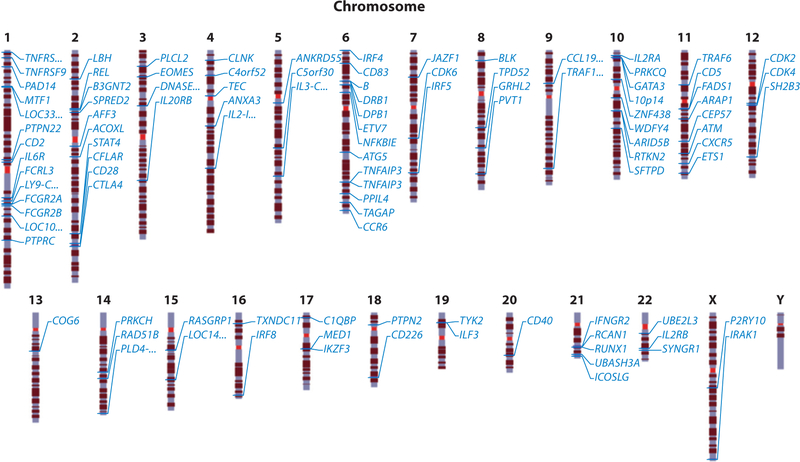
Major research efforts to identify genes outside of the MHC have been carried out over nearly two decades; these efforts have included linkage analysis, candidate gene studies, and most recently GWAS. Although linkage analysis was largely unsuccessful, researchers were able to identify PTPN22 and CTLA4 via a candidate approach that prioritized genes with specific immune functions (6, 127). Similarly, candidate gene studies in Japanese cases identified PADI4, encoding a citrullination enzyme (144); FCRL3 (67); SLC22A4 (161); and CD244 (145). Technical advances have enabled the performance of GWAS by genotyping hundreds of thousands to millions of SNPs. The first GWAS reports for RA were made in 2007, resulting in the identification of TRAF1/C5 (107) and STAT4 (114) as susceptibility loci for RA in Swedish and American patient cohorts. Another group contemporaneously reported TRAF1/C5 in a study that used a candidate gene approach (73). Since then, many RA GWAS and their meta-analyses have identified various non-HLA susceptibility genes not only in European populations but also in Asian populations (29, 40, 57, 66, 87, 100, 101, 106, 111, 113, 137, 153, 157, 160) (Figure 3, Table 1). These variants implicate a wide range of candidate pathways, including pathways important for activation and development of T and B cells, regulation and synthesis of immunoglobulins and cytokines, cell signaling, innate immunity, apoptosis, ubiquitination, and citrullination. It is important to note that the major findings from these GWAS results focused mainly on ACPA(+) RA.
Figure 3.
Non-HLA susceptibility loci for rheumatoid arthritis. The chromosomal positions of markers outside of the HLA region that are associated with susceptibility to rheumatoid arthritis are indicated. Table 1 lists the leading candidate genes associated with these marker regions. Ellipses indicate uncertainty about the actual locus with respect to the gene name.
Table 1.
Non-HLA loci found in genome-wide association studies of rheumatoid arthritis, grouped by putative functional relationships
| Chr | SNP | Gene | Allele (A1/A2) | Asian | European | ||||
|---|---|---|---|---|---|---|---|---|---|
| Frequency of A1 | OR (95% CI) | Frequency of A1 | OR (95% CI) | ||||||
| Case | Control | Control | Case | ||||||
| T cell | |||||||||
| 1 | rs624988 | CD2 | T/C | 0.43 | 0.43 | 0.96 (0.92–1.01) | 0.42 | 0.40 | 1.09(1.06–1.12) |
| rs2476601 | PTPN22 | A/G | NA | NA | NA | 0.16 | 0.095 | 1.80 (1.73–1.88) | |
| 2 | rs1980422 | CD28 | C/T | 0.083 | 0.067 | 1.10(1.00–1.20) | 0.25 | 0.24 | 1.13 (1.09–1.17) |
| rs3087243 | CTLA4 | G/A | 0.78 | 0.74 | 1.12 (1.06–1.18) | 0.59 | 0.55 | 1.15 (1.12–1.18) | |
| rs11889341 | STAT4 | T/C | 0.34 | 0.30 | 1.16(1.10–1.22) | 0.25 | 0.23 | 1.12 (1.09–1.16) | |
| 4 | rs45475795 | 1L2-1L21 | G/A | NA | NA | NA | 0.085 | 0.074 | 1.14(1.08–1.20) |
| 6 | rs2451258 | TAGAP | T/C | 0.97 | 0.98 | 1.17(1.01–1.37) | 0.67 | 0.64 | 1.10(1.07–1.13) |
| 10 | rs706778 | 1L2RA | T/C | 0.57 | 0.56 | 1.04 (0.99–1.09) | 0.43 | 0.40 | 1.12 (1.09–1.15) |
| rs3824660 | GATA3 | C/T | 0.36 | 0.35 | 1.03 (0.98–1.08) | 0.41 | 0.39 | 1.10(1.06–1.13) | |
| 12 | rs10774624 | SH2B3-PTPN11 | G/A | NA | NA | NA | 0.51 | 0.50 | 1.09 (1.06–1.13) |
| 18 | rs8083786 | PTPN2 | G/A | 0.37 | 0.34 | 1.18(1.13–1.24) | 0.17 | 0.16 | 1.12 (1.08–1.16) |
| 21 | rs2236668 | 1COSLG-A1RE | C/T | 0.68 | 0.66 | 1.09(1.03–1.16) | 0.73 | 0.71 | 1.07 (1.03–1.10) |
| 22 | rs3218251 | 1L2RB | A/T | 0.076 | 0.072 | 0.99 (0.90–1.09) | 0.28 | 0.27 | 1.08 (1.04–1.11) |
| B cell | |||||||||
| 2 | rs34695944 | REL | C/T | 0.033 | 0.038 | 1.00 (0.88–1.14) | 0.39 | 0.36 | 1.13 (1.09–1.16) |
| 4 | rs13142500 | CLNK | C/T | 0.58 | 0.56 | 1.10(1.04–1.15) | 0.47 | 0.45 | 1.10(1.06–1.15) |
| 8 | rs2736337 | BLK | C/T | 0.71 | 0.67 | 1.15 (1.09–1.22) | 0.26 | 0.25 | 1.09 (1.06–1.13) |
| 10 | rs71508903 | AR1D5B | T/C | 0.25 | 0.24 | 1.18(1.12–1.25) | 0.22 | 0.21 | 1.15 (1.11–1.20) |
| 11 | rs508970 | CD5 | A/G | NA | NA | NA | 0.50 | 0.49 | 1.07 (1.04–1.10) |
| 20 | rs4239702 | CD40 | C/T | 0.65 | 0.62 | 1.08 (1.03–1.14) | 0.75 | 0.72 | 1.14(1.11–1.18) |
| Immunoglobulin related | |||||||||
| 1 | rs2317230 | FCRL3 | T/G | 0.41 | 0.39 | 1.10(1.05–1.16) | 0.44 | 0.43 | 1.06(1.03–1.09) |
| rs72717009 | FCGR2A | T/C | NA | NA | NA | 0.12 | 0.12 | 1.12 (1.07–1.18) | |
| chr1:161644258 | FCGR2B | C/G | 0.29 | 0.29 | 1.12 (1.06–1.19) | NA | NA | NA | |
| 6 | chr6:14103212 | CD83 | T/C | 0.18 | 0.15 | 1.16(1.09–1.24) | 0.056 | 0.053 | 1.10(1.02–1.18) |
| 18 | rs2469434 | CD226 | C/T | 0.39 | 0.37 | 1.11 (1.07–1.15) | 0.42 | 0.41 | 1.05 (1.02–1.08) |
| Cytokine | |||||||||
| 1 | chr1:2523811 | TNFRSF14-MMEL1 | G/A | 0.61 | 0.58 | 1.16(1.09–1.23) | 0.72 | 0.70 | 1.10(1.07–1.14) |
| rs227163 | TNFRSF9 | C/T | 0.44 | 0.42 | 1.11 (1.08–1.16) | 0.42 | 0.42 | 1.00 (0.97–1.03) | |
| rs2228145 | 1L6R | A/C | 0.61 | 0.60 | 1.10(1.05–1.15) | 0.62 | 0.60 | 1.07 (1.04–1.10) | |
| 3 | rs9826828 | 1L20RB | A/G | NA | NA | NA | 0.023 | 0.018 | 1.44(1.28–1.61) |
| 5 | rs657075 | 1L3-CSF2 | A/G | 0.35 | 0.35 | 1.12 (1.07–1.18) | 0.10 | 0.097 | 1.07 (1.01–1.12) |
| 6 | rs9378815 | 1RF4 | C/G | 0.38 | 0.38 | 1.10(1.04–1.15) | 0.67 | 0.66 | 1.09 (1.05–1.12) |
| rs1571878 | CCR6 | C/T | 0.53 | 0.48 | 1.28 (1.22–1.35) | 0.47 | 0.44 | 1.13 (1.10–1.17) | |
| 7 | chr7:128580042 | 1RF5 | G/A | 0.25 | 0.23 | 1.11 (1.05–1.18) | 0.48 | 0.46 | 1.12 (1.08–1.15) |
| 9 | rs11574914 | CCL19-CCL21 | A/G | 0.052 | 0.046 | 1.05 (0.92–1.19) | 0.36 | 0.33 | 1.13 (1.09–1.16) |
| rs10985070 | TRAF1-C5 | C/A | 0.53 | 0.52 | 1.08 (1.03–1.13) | 0.44 | 0.42 | 1.09(1.06–1.12) | |
| 11 | rs331463 | TRAF6-RAG1/2 | T/A | 0.93 | 0.92 | 1.03 (0.94–1.12) | 0.86 | 0.85 | 1.12 (1.07–1.16) |
| rs10790268 | CXCR5 | G/A | 0.85 | 0.85 | 1.03 (0.96–1.11) | 0.81 | 0.79 | 1.17(1.13–1.22) | |
| 16 | rs13330176 | 1RF8 | A/T | 0.45 | 0.44 | 1.12 (1.06–1.17) | 0.25 | 0.23 | 1.12 (1.08–1.17) |
| 17 | chr17:38031857 | 1KZF3-CSF3 | G/T | 0.28 | 0.26 | 1.09(1.04–1.15) | 0.47 | 0.46 | 1.09(1.06–1.12) |
| 19 | chr19:10771941 | 1LF3 | C/T | NA | NA | NA | 0.98 | 0.97 | 1.47 (1.30–1.67) |
| rs34536443 | TYK2 | G/C | NA | NA | NA | 0.97 | 0.95 | 1.46(1.33–1.60) | |
| 21 | rs73194058 | 1FNGR2 | C/A | 0.50 | 0.48 | 1.03 (0.98–1.08) | 0.89 | 0.87 | 1.13 (1.08–1.18) |
| Cell signaling | |||||||||
| 1 | rs28411352 | MTF1-1NPP5B | T/C | 0.25 | 0.23 | 1.12 (1.06–1.19) | 0.28 | 0.25 | 1.10(1.07–1.14) |
| rs17668708 | PTPRC | C/T | NA | NA | NA | 0.91 | 0.89 | 1.12 (1.06–1.18) | |
| 2 | rs10175798 | LBH | A/G | 0.36 | 0.33 | 1.07 (1.02–1.13) | 0.64 | 0.62 | 1.09(1.06–1.12) |
| rs1858037 | SPRED2 | T/A | 0.20 | 0.16 | 1.19(1.12–1.26) | 0.68 | 0.65 | 1.09(1.06–1.13) | |
| rs9653442 | AFF3 | C/T | 0.48 | 0.47 | 1.10(1.05–1.16) | 0.49 | 0.46 | 1.12 (1.09–1.15) | |
| 3 | rs3806624 | EOMES | G/A | 0.84 | 0.84 | 1.06 (0.99–1.14) | 0.47 | 0.46 | 1.08 (1.05–1.12) |
| 4 | rs2664035 | TEC | A/G | 0.30 | 0.29 | 1.03 (0.97–1.08) | 0.42 | 0.40 | 1.08 (1.05–1.11) |
| rs10028001 | ANXA3 | T/A | 0.46 | 0.44 | 1.11 (1.06–1.17) | 0.38 | 0.37 | 1.02 (0.98–1.06) | |
| 6 | rs7752903 | TNFA1P3 | G/T | 0.083 | 0.067 | 1.34(1.23–1.46) | 0.041 | 0.031 | 1.41 (1.31–1.52) |
| rs17264332 | TNFA1P3 | G/A | NA | NA | NA | 0.24 | 0.21 | 1.17(1.13–1.21) | |
| rs2234067 | ETV7 | C/A | 0.97 | 0.96 | 1.22 (1.06–1.41) | 0.88 | 0.87 | 1.14(1.09–1.19) | |
| rs2233424 | NFKB1E | T/C | 0.24 | 0.21 | 1.24(1.17–1.31) | 0.052 | 0.043 | 1.33 (1.20–1.47) | |
| 7 | rs67250450 | JAZF1 | T/C | 0.017 | 0.014 | 1.02 (0.84–1.23) | 0.80 | 0.79 | 1.11 (1.07–1.14) |
| rs4272 | CDK6 | G/A | 0.12 | 0.11 | 1.06 (0.98–1.15) | 0.23 | 0.21 | 1.10(1.07–1.14) | |
| 8 | rs998731 | TPD52 | T/C | 0.85 | 0.85 | 1.02 (0.96–1.10) | 0.45 | 0.44 | 1.09(1.06–1.12) |
| rs678347 | GRHL2 | G/A | 0.80 | 0.80 | 1.03 (0.98–1.10) | 0.29 | 0.27 | 1.10(1.06–1.13) | |
| 10 | rs947474 | PRKCQ | A/G | 0.89 | 0.88 | 1.05 (0.98–1.13) | 0.83 | 0.81 | 1.12 (1.08–1.16) |
| rs6479800 | RTKN2 | C/G | 0.11 | 0.10 | 1.19(1.11–1.29) | 0.24 | 0.23 | 1.08 (1.04–1.13) | |
| 11 | rs4409785 | CEP57 | C/T | 0.089 | 0.076 | 1.16 (1.07–1.27) | 0.19 | 0.17 | 1.12 (1.08–1.16) |
| chr11:107967350 | ATM | A/G | NA | NA | NA | 0.93 | 0.92 | 1.21 (1.13–1.29) | |
| rs73013527 | ETS1 | C/T | 0.74 | 0.72 | 1.14(1.08–1.21) | 0.51 | 0.50 | 1.08 (1.05–1.11) | |
| 12 | rs773125 | CDK2 | A/G | 0.80 | 0.79 | 1.10(1.04–1.17) | 0.62 | 0.60 | 1.09(1.06–1.12) |
| rs1633360 | CDK4 | T/C | 0.27 | 0.24 | 1.03 (0.98–1.09) | 0.60 | 0.58 | 1.08 (1.05–1.11) | |
| 14 | rs3783782 | PRKCH | A/G | 0.24 | 0.22 | 1.14(1.09–1.19) | 0.013 | 0.011 | 1.12 (0.96–1.31) |
| 15 | rs8032939 | RASGRP1 | C/T | 0.62 | 0.60 | 1.12 (1.06–1.17) | 0.28 | 0.25 | 1.13 (1.09–1.17) |
| 17 | rs1877030 | MED1 | C/T | 0.83 | 0.83 | 1.09 (1.04–1.14) | 0.85 | 0.84 | 1.09(1.05–1.13) |
| 21 | chr21:35928240 | RCAN1 | C/T | 0.96 | 0.94 | 1.08 (0.97–1.21) | 0.89 | 0.88 | 1.12 (1.07–1.17) |
| rs8133843 | RUNX1-LOC100506403 | A/G | 0.52 | 0.50 | 1.06(1.01–1.11) | 0.64 | 0.62 | 1.09(1.06–1.12) | |
| X | chrX:78464616 | P2RY10 | A/C | 0.48 | 0.44 | 1.11 (1.07–1.15) | 0.008 | 0.008] | 1.16 (0.78–1.75) |
| rs5987194 | 1RAK1 | C/G | 0.78 | 0.75 | 1.15 (1.08–1.22) | 0.15 | 0.13 | 1.16(1.12–1.21) | |
| Innate immunity | |||||||||
| 1 | rs4656942 | LY9-CD244 | G/A | 0.71 | 0.70 | 1.05 (1.00–1.11) | 0.77 | 0.77 | 1.01 (0.98–1.05) |
| 10 | rs726288 | SFTPD | T/C | 0.21 | 0.18 | 1.22 (1.14–1.31) | 0.028 | 0.028 | 0.96 (0.86–1.06) |
| 17 | rs72634030 | C1QBP | A/C | 0.67 | 0.65 | 1.12 (1.07–1.18) | 0.073 | 0.066 | 1.12 (1.06–1.19) |
| Apoptosis | |||||||||
| 2 | rs6715284 | CFLAR-CASP8 | G/C | NA | NA | NA | 0.11 | 0.097 | 1.15 (1.10–1.20) |
| 3 | rs73081554 | DNASE1L3-ABHD6-PXK | T/C | NA | NA | NA | 0.082 | 0.073 | 1.18 (1.11–1.25) |
| 11 | rs11605042 | ARAP1 | G/A | 0.56 | 0.51 | 1.09(1.04–1.14) | 0.52 | 0.51 | 1.05 (1.01–1.09) |
| Ubiquitination | |||||||||
| 21 | rs1893592 | UBASH3A | A/C | 0.77 | 0.75 | 1.11 (1.05–1.18) | 0.73 | 0.72 | 1.11 (1.07–1.15) |
| 22 | rs11089637 | UBE2L3-YDJC | C/T | 0.48 | 0.46 | 1.06(1.02–1.10) | 0.17 | 0.16 | 1.10(1.06–1.15) |
| Citrullination | |||||||||
| 1 | rs2301888 | PAD14 | G/A | 0.46 | 0.41 | 1.19(1.14–1.25) | 0.67 | 0.65 | 1.11 (1.07–1.14) |
| Others | |||||||||
| 1 | rs12140275 | LOC339442 | A/T | NA | NA | NA | 0.77 | 0.75 | 1.11 (1.07–1.14) |
| rs2105325 | LOC100506023 | C/A | 0.93 | 0.92 | 1.13 (1.04–1.23) | 0.77 | 0.74 | 1.12 (1.08–1.15) | |
| 2 | rs13385025 | B3GNT2 | A/G | 0.27 | 0.26 | 1.14(1.07–1.20) | 0.13 | 0.12 | 1.08 (1.02–1.15) |
| rs6732565 | ACOXL | A/G | 0.61 | 0.59 | 1.04(1.00–1.08) | 0.64 | 0.62 | 1.10(1.07–1.14) | |
| 3 | rs4452313 | PLCL2 | T/A | 0.54 | 0.53 | 1.04 (0.99–1.09) | 0.31 | 0.29 | 1.11 (1.08–1.15) |
| 4 | rs11933540 | C4orf52 | C/T | NA | NA | NA | 0.34 | 0.31 | 1.15 (1.11–1.19) |
| 5 | rs7731626 | ANKRD55 | G/A | NA | NA | NA | 0.67 | 0.63 | 1.21 (1.17–1.26) |
| rs2561477 | C5orf30 | G/A | 0.73 | 0.72 | 1.04 (0.98–1.09) | 0.70 | 0.68 | 1.11 (1.08–1.14) | |
| 6 | rs9372120 | ATG5 | G/T | 0.058 | 0.054 | 1.16(1.05–1.28) | 0.22 | 0.20 | 1.10(1.06–1.14) |
| rs9373594 | PP1L4 | T/C | 0.34 | 0.33 | 1.11 (1.07–1.15) | 0.85 | 0.85 | 1.07 (1.02–1.12) | |
| 8 | rs1516971 | PVT1 | T/C | NA | NA | NA | 0.88 | 0.87 | 1.16(1.11–1.21) |
| 10 | rs12413578 | 10p14 | C/T | NA | NA | NA | 0.91 | 0.90 | 1.20 (1.12–1.29) |
| rs793108 | ZNF438 | T/C | 0.49 | 0.48 | 1.09(1.04–1.14) | 0.52 | 0.50 | 1.07 (1.04–1.10) | |
| rs2671692 | WDFY4 | A/G | 0.31 | 0.29 | 1.10(1.05–1.14) | 0.65 | 0.63 | 1.06(1.03–1.09) | |
| 11 | rs968567 | FADS1-FADS2-FADS3 | C/T | NA | NA | NA | 0.84 | 0.83 | 1.12 (1.07–1.16) |
| 13 | rs9603616 | COG6 | C/T | 0.77 | 0.75 | 1.08(1.02–1.14) | 0.68 | 0.66 | 1.11 (1.07–1.14) |
| 14 | rs1950897 | RAD51B | T/C | 0.89 | 0.88 | 1.16(1.08–1.25) | 0.73 | 0.71 | 1.09(1.06–1.12) |
| rs2582532 | PLD4-AHNAK2 | C/T | 0.75 | 0.73 | 1.18(1.11–1.25) | 0.99 | 0.99 | 0.93 (0.72–1.21) | |
| 15 | rs8026898 | LOC145837 | A/G | 0.067 | 0.052 | 1.14(1.02–1.27) | 0.29 | 0.27 | 1.15 (1.11–1.18) |
| 16 | rs4780401 | TXNDC11 | T/G | 0.48 | 0.49 | 1.03 (0.98–1.08) | 0.60 | 0.59 | 1.09(1.06–1.13) |
| 22 | rs909685 | SYNGR1 | A/T | 0.86 | 0.85 | 1.23 (1.14–1.33) | 0.32 | 0.30 | 1.11 (1.08–1.15) |
Abbreviations: Chr, chromosome; CI, confidence interval; NA, not applicable; OR, odds ratio; SNP, single-nucleotide polymorphism.
5.2.2. Rare genetic variants.
Following the success in identifying common risk alleles, researchers have now focused their attention on the discovery of rare variants that may have high effect sizes and penetrance. Although rare variants are unlikely to explain substantial heritability in RA, they may lead to a deeper understanding of the disease mechanisms because the discovery of such variants might clearly pinpoint causal genes in loci and derive alleles that are more amenable to functional follow-up. The effect of PCSK9 variants in cholesterol regulation is perhaps the most compelling example in which uncommon or rare variant analysis has led to drug discovery (17, 142). This approach has been successfully applied to discover rare alleles in RA and other autoimmune diseases. For example, Nejentsev et al. (90) identified four rare protective coding alleles of IFIH1 associated with type I diabetes, and Rivas et al. (116) identified multiple rare variants, including four independent variants in NOD2, associated with inflammatory bowel diseases. This paradigm could well lead to similar insights that affect the treatment of autoimmune diseases and therefore justifies further efforts in this area.
In 2011, Eyre et al. (30) took advantage of the Wellcome Trust Case Control Consortium genotyping data to address enrichment of rare variant signals in the regions showing linkage signals. However, they found that enrichment of the signals was driven by the MHC region and failed to identify independent signals of the MHC. To address rare variants that affect RA susceptibility, Diogo et al. (26) focused on the 25 genes identified as RA susceptibility genes up to 2012 and performed deep exon sequencing using 500 cases and 650 controls, followed by a replication study consisting of 10,609 cases and 35,605 controls. They found a suggestive trend of enrichment of rare CD2 coding alleles in RA, but no replication studies have confirmed these findings.
In 2014, Bang et al. (5) used a multifaceted approach to select a total of 398 genes and performed exon sequencing for 1,217 cases and 717 controls. They also used GWAS data on 4,799 individuals and immunochip data on 4,722 individuals. However, they failed to identify any single rare variants associated with RA beyond the GWAS significant level or a significant burden signal in any gene. A subsequent study by Diogo et al. (25) integrated immunochip dense genotyping, exome chip genotyping, and targeted exon sequencing for a total of 13,066 cases and 30,671 controls and identified a total of three protein-coding variants in TYK2 as independent susceptibility variants with RA. Intriguingly, one study has examined the role of a rare deletion on chromosome 12 in risk for RA, with the conclusion that loss of one copy of SLC2A3, encoding the glucose receptor GLUT3, may confer protection for RA (172). Collectively, these results suggest that very large sample sizes will be needed to definitively identify rare variant signals. Whole-genome sequencing using a large number of RA patients has not been reported to date but could deepen our understanding of RA genetics, especially for noncoding regions.
5.3. Seronegative Rheumatoid Arthritis
Many consider seronegative RA to be a distinct subphenotype of RA that has a different genetic basis and clinical course from seropositive RA (72, 97, 102). There is substantial evidence that seronegative RA has a heritable component, although it may be less heritable than seropositive RA (35). The study of seronegative RA has been complicated by potential clinical heterogeneity; other seronegative inflammatory arthritides with atypical presentations can be misclassified as seronegative RA. For example, individuals with psoriatic arthritis presenting before the appearance of psoriasis can be classified as having seronegative RA, as might individuals who are negative for anti-CCP antibodies but positive for other ACPA serologies not utilized in a clinical setting. This heterogeneity has been seen in genetic studies of seronegative RA (46,108). It was recognized early on that HLA associations with seronegative RA and seropositive RA are quite distinct from one another. Verpoort et al. (175) reported an association between DRB1*03 and ACPA(−) RA using 171 ACPA(−) RA and 423 healthy subjects. Irigoyen et al. (54) contemporaneously reported the association between DRB1*03 and ACPA(−) RA. Ohmura et al. (97) studied 185 Japanese ACPA(−) cases and 1,508 controls and demonstrated different genetic associations with the HLA alleles. In an expanded study of 869 Japanese ACPA(−) cases and 2,008 controls, Terao et al. (154) identified three distinct susceptibility alleles for ACPA(−) RA: DRB1*12:01, DRB1*14:03, and homozygotes of DRB1*08. HLA differences between ACPA(−) RF(+) RA and ACPA(−) RF(−) RA have also been observed (152).
Han et al. (46) comprehensively fine mapped the HLA alleles in seronegative RA. They took advantage of 2,406 ACPA(−) RA cases and 13,930 controls and revealed that ACPA(−) RA is associated with DRB1*03, which is characterized by a serine residue at amino acid position 13, and HLA-B*08, which is characterized by an aspartate residue at amino acid position 9; both of these positions are associated with ACPA(+) RA. The serine residue at position 13 showed opposing effects between ACPA(−) RA and ACPA(+) RA, indicating that this position plays a critical role in the production of ACPAs in RA. Critical to the success of this large study was the recognition of clinical heterogeneity, in particular the understanding that misclassified seropositive cases constitute a substantial proportion of seronegative RA cohorts. Successful fine mapping requires the application of statistical methods to correct for this heterogeneity and the use of highly sensitive ACPA assays to validate findings.
For non-HLA regions, identification of novel alleles has been challenging. Using a candidate gene approach, Viatte et al. (176) genotyped 2,040 ACPA(−) RA cases and 13,009 controls for a total of 36 SNPs and identified the SE, ANKRD55/IL6ST, BLK, and PTPN22 as susceptibility genes in European populations. In an unbiased approach, Padyukov et al. (102) performed a GWAS using 774 ACPA(−) patients and 1,079 controls from the Swedish population and failed to identify any loci with genome-wide significance. Similarly, Bossini-Castillo et al. (8) analyzed a total of 1,922 patients with ACPA(−) RA and 7,087 controls and did not find any significant associations in the non-HLA region. Terao et al. (155) reported a GWAS of 670 ACPA(−) RA cases and 16,891 controls in Japanese subjects followed by a replication study of 916 ACPA(−) RA cases and 3,764 controls, and also failed to identify signals with genome-wide significance. A recent Chinese GWAS also did not identify alleles beyond the GWAS significant level outside of the HLA region in ACPA(−) RA patients (57).
5.4. Gene-Environment and Gene-Gene Interactions in Rheumatoid Arthritis
Gene-environment and gene-gene interactions have been investigated in RA, especially in the context of ACPA(+) subsets. Interactions between ACPA production and genetic factors suggest alleles that are specific to seropositive RA. For example, a study in the Dutch population reported that smoking and DRB1*01:01, *01:02, or *10:01 showed interaction with ACPA(+) RA development (167). In 2006, Linn-Rasker et al. (80) reported that smoking is associated with ACPAs only in subjects carrying the SE. Pedersen et al. (103) carried out a nationwide study in Denmark and reported a strong gene-environment effect on ACPA(+) RA development, namely, the combination of the SE and smoking, coffee consumption, or use of an oral contraceptive. A recent report described an association of HLA risk alleles and smoking with specific ACPA isotypes in a Swedish cohort (147). Another study of Dutch populations reported that smoking increased the number of ACPA isotypes even in SE-negative subjects (174); further studies are necessary to clarify the entire picture of interaction among the SE, smoking, and ACPAs.
PTPN22 is also associated with ACPA production (88), and an interaction between PTPN22 and smoking in production of anti-α-enolase CEP1 antibodies has been reported (86). Källberg et al. (60) reported an interaction between the SE and PTPN22 using three different cohorts comprising 1,977 subjects.
Although multiple studies have reported interactive effects between HLA alleles, smoking, and PTPN22, these studies did not evaluate interaction effects in the same way. In the US cohorts, they did not find interactions defined by effects beyond the multiplicative model between the SE and smoking (76) or among the SE, PTPN22, and smoking that affected the production of ACPAs in RA cases (88). Some studies have utilized a multiplicative model and found a significant interaction beyond multiplicative effects for part of the results (60, 86), whereas others have detected significance only for additive interactions (88). In general, the evidence for multiplicative effects is more modest than the evidence for additive effects (60, 86).
6. FUNCTIONAL IMPLICATIONS OF GENETIC FINDINGS
Functional follow-up on disease alleles is crucial if genetic discoveries are to yield mechanistic insights. However, functional investigation of disease alleles has been challenging in RA, and indeed in many other complex diseases. A common approach is to consider the current biological knowledge surrounding the genes in each associated region and attempt to develop hypotheses. This method is hampered by incomplete knowledge of the biology of candidate genes as well as uncertainty about exactly which gene is causative within a given associated haplotype. Ideally, one variant or combination of variants would lead to an RA-like disease in a mouse model. In practice, however, it is not clear that these models can translate directly to functions for specific candidate genes across species. For example, the SKG mouse carrying the Zap70 mutation has altered T cell signaling and develops arthritis, and is regarded as a good mouse model of RA (119). However, genetic investigation of RA in humans has failed to identify disease-associated variants of ZAP70 in humans. Investigation of TRAF1, the susceptibility gene first identified by an RA GWAS, in the KRN/I-Ag7 mouse model by knockout demonstrated no altered arthritis phenotype (14).
6.1. Implicated Genome Elements, eQTLs, Pathways, and Cell Types
An alternative approach is to begin by understanding what cell types and biological pathways are implicated by the expression of candidate genes. Hu et al. (52) took advantage of a compendium of gene expression data in different immune and nonimmune cell types and assessed whether genes within GWAS loci were specifically expressed within any of these cell types. They found that effector memory CD4+ T cells showed enrichment for expression of genes within RA susceptibility loci, suggesting the important involvement of effector memory T cells in the initial stages of RA pathogenesis. In a subsequent study, Hu et al. (51) quantified gene expression of effector memory CD4+ T cells before and after the stimulation of T cell receptors. They used an expanded list of SNPs associated with susceptibility to RA and other immunological diseases. They observed expression quantitative trait locus (eQTL) signals specific to cell state or specific to effector memory CD4+ T cells undetected in peripheral blood mononuclear cells and found several variants that completely explained eQTL signals in the regions.
Using a complementary approach, Trynka et al. (163) focused on cell type-specific histone marks and overlap between cell-specific peaks of histone marks and susceptibility SNPs. They found that trimethylation of histone H3 on lysine 4 (H3K4me3) was the most cell type-specific mark and found significant overlap between RA and histone marks in CD4+ regulatory T cells. Finucane et al. (32) partitioned RA heritability and analyzed the polygenic contributions of cell type-specific functional annotations. They found the strongest enrichment of H3K4me1 in stimulated Th17 cells among cell type-specific elements.
Okada et al. (101) expanded the functional analysis using 101 susceptibility SNPs from a multiple-ethnicity GWAS meta-analysis and found enrichment in regulatory T cells in eQTLs and histone marks. Based on the mega-GWAS of RA, the 44% of SNPs showing the strongest association signals in the regions have cis-eQTL effects, indicating that alteration of the expression of RA-related genes is critical for the development of RA. They also showed that there is an overlap of susceptibility genes between RA and primary immune deficiency or hematopoietic cell malignancies. Their genetic findings also suggest the importance of B cell pathways and cytokine signal pathways as well as CD4+ T cell pathways. Finally, a recent attempt to integrate genome annotations to identify causal variants for autoimmunity implicated both CD4+ T cell and B cell involvement in RA pathogenesis (31).
6.2. Quantitative Immune Traits
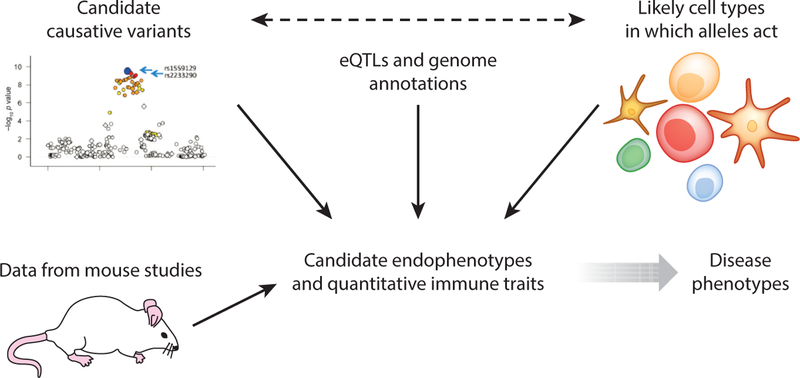
The identification of disease-relevant quantitative immune traits is likely to be essential in order to fully understand the role of genetics in RA and other immune disorders (41). Quantitative immune traits, or endophenotypes, are more directly regulated by risk alleles than are highly complex disease phenotypes. We take the view that defining the factors involved in relevant endophenotypes provides a more tractable approach to defining the components of risk for disease (Figure 4). The selection of endophenotypes for study is driven by accumulating immunobiological knowledge, observing phenotypes in mice, and integrating data on cell-specific gene regulation and annotation as well as evolving gene network analysis. Although the current genetic data are extensive, they are not complete, particularly for uncommon or rare variants, and thus future discovery will be an iterative process, as depicted in Figure 1. Because some murine quantitative traits do not translate directly to humans, direct human studies are essential to confirm observations. Therefore, genetically characterized human volunteer populations have emerged as an important resource for research (22, 41). For example, Hu et al. (51) identified a SNP that altered CD4+ effector memory T cell proliferation in response to nonantigenic bead stimulation.
Figure 4.
Schematic representation of an iterative approach to establishing the identity and function of rheumatoid arthritis risk alleles. Further population studies will be needed to identify all statistically convincing risk alleles. These alleles in turn will be integrated with expression quantitative trait locus (eQTL) analysis and genomic annotation to determine likely cell types in which these alleles act. The results may drive further examination of genetic variation, which can in turn be combined with functional information from mouse studies to drive the examination of candidate immune endophenotypes in human populations, often using a living biobank of normal subjects who are characterized for the presence of risk alleles. The identification of disease-relevant endophenotypes will lead to a deeper understanding of disease pathogenesis.
Although quantitative effects on gene expression are a common feature of disease risk alleles for RA and other autoimmune disorders, relatively few autoimmune disease risk alleles have been directly related to an immune endophenotype. With regard to RA-associated alleles, the intracellular phosphatase gene PTPN22 has been of major interest because it is the most strongly associated locus after the MHC (allelic OR of approximately 1.8) (6). Risk for RA is conferred by a single amino acid change, R620W, in a region of the molecule that disrupts binding to the intracellular kinase CSK. Studies in cell lines, mice, and humans are consistent with the risk allele leading to a change in the signaling threshold for T cell receptors (9). However, the data are complex and generally conflicting with respect to findings in mouse and human systems. Thus, a lower threshold for T cell receptor signaling is generally observed in mice, with expansion and activation of memory T cells, in both knockout and knock-in animals (47, 110). By contrast, human carriers of the PTPN22 620W allele appear to have reduced signaling activity in many studies (110). In addition to effects on T cell receptor signaling, PTPN22 has been implicated in B cell receptor signaling as well as Toll-like receptor signaling, leading to reduced production of interferon in myeloid cells (179). In the latter context, differences in binding of PTPN22 to TRAF3 are observed with the disease allele. More recently, Chang et al. (13) reported that PTPN22 variants can bind to PADI4 and affect citrullination activity in myeloid cells.
Although the PTPN22 620W allele is also a risk factor for many other autoimmune diseases, such as type 1 diabetes, systemic lupus erythematosus, and myasthenia gravis, the patterns of association are quite different when considering the larger universe of autoimmune disorders (139). In particular, PTPN22 has no impact on risk for multiple sclerosis but is protective for risk of inflammatory bowel disease (139). Interestingly, PTPN22 has been associated with protection against some infectious diseases (7). Thus, it is clear that more work on PTPN22 function needs to be completed in order to understand its role in disease susceptibility.
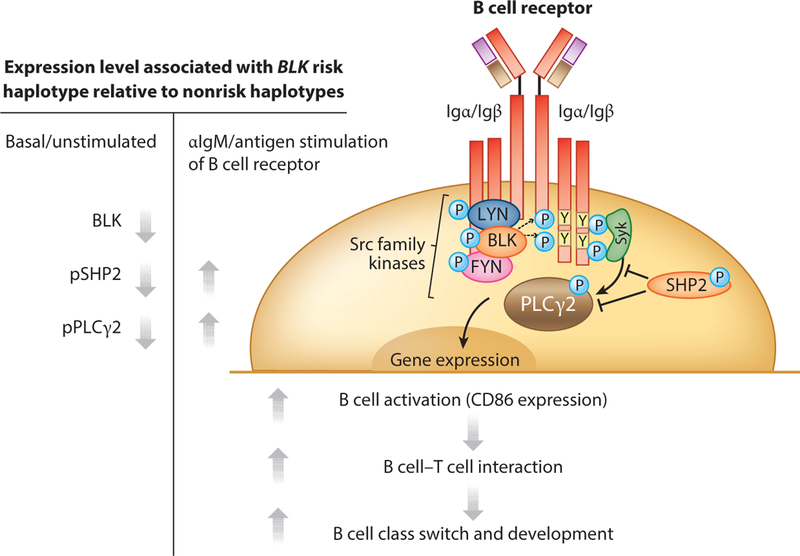
In contrast to PTPN22, BLK offers an example of an RA risk gene in which quantitative functional effects in mice and humans appear to be well aligned. BLK has been associated with RA (40) and other autoimmune disorders (39, 50, 96). The risk haplotype is associated with reduced expression of BLK in B cell lines, native B cells (133), and resting and stimulated CD4+ T cells (51). Previous studies in Blk knockout mice showed no obvious phenotypes (159), and this was presumed to be secondary to redundant functions with other Src kinases involved in B cell receptor signaling, such as Lyn and Fyn (118). However, more recent studies have shown subtle effects on B cell function in hemizygous mice, including enhanced activation after B cell receptor triggering, as well as changes in B cell subsets and some susceptibility to lupus-like phenotypes (120, 121). Consistent with these effects of quantitative expression, it is now apparent that the BLK risk haplotype is also associated with enhanced B cell responsiveness in humans, relatively increased CD86 expression, and increased ability to activate T cell responses. This also translates into increased numbers of switched memory B cells and changes in the B cell repertoire (132). Overall, these data are consistent with a negative regulatory function of BLK (Figure 5).
Figure 5.
B cell receptor signaling pathways and B cell–related endophenotypes mediated by BLK risk alleles. Blk seems to act mainly as a negative regulator of B cell activation, leading to increased B cell–T cell interactions and enhanced immunoglobulin class switching, even in normal carriers of the rheumatoid arthritis risk allele. Abbreviations: pPLCγ2, phospholipase Cγ2; pSHP2, SH2 domain–containing phosphatase 2; P, phosphorylated; Y, tyrosine. Modified from Reference 132 with permission. Copyright © 2015, American College of Rheumatology.
These are some of the first examples of specific quantitative immune traits involving RA risk loci, and these data indicate that primary endophenotypes in both B and T cells are likely to be involved in disease susceptibility. Similar examples of quantitative immune traits have been reported for other autoimmunity risk alleles, such as alleles in PRDM1 (61) and IL23R (23). Given that more than 100 common risk alleles in a variety of putative risk genes have been reported for RA (see Table 1), there is clearly much more to be done, and we expect that the paradigm illustrated in Figure 4 will be useful in focusing on relevant endophenotypes to explore.
7. CLINICAL SUBPHENOTYPES IN RHEUMATOID ARTHRITIS: ADDITIONAL CONSIDERATIONS
7.1. Autoantibody Reactivity and Titers
As discussed above, the genetic factors underlying ACPA(+) and ACPA(−) RA as defined by standard clinical testing are clearly distinct. However, recent data suggest that the fine specificities of ACPA are also under genetic control. Lundberg et al. (83) showed that the associations among the SE, PTPN22, and smoking are different among fine-specific ACPAs. A study in a Dutch population reported that the SE is associated with citrullinated vimentin antibody and not with citrullinated fibrinogen antibody (173). Additional refinements in the genetic influence on ACPA specificities have emerged from studies considering combinations of HLA and non-HLA risk loci (134).
In addition to specificity, the titers of autoantibodies need to be considered as a separate trait. Previous studies have revealed that ACPA titers are strongly affected by genetic factors, and in particular by the HLA alleles. In addition to the HLA-DRB1 SE alleles, Irigoyen et al. (54) reported that DRB1*03 showed a significant association with decreasing ACPA levels. Cui et al. (19) reported that the heritability of ACPA levels is 44% based on a calculation by the Genome-Wide Complex Trait Analysis (GCTA) tool (182) using GWAS data from 1,975 RA subjects. They found strong association signals in the HLA region, all of which are in linkage disequilibrium with DRB1*03. Balsa et al. (4) also found suggestive evidence of an independent signal outside of the HLA genes. Laid et al. (74) reported that DRB1*15 is associated with high levels of ACPA in European populations.
In Japanese populations, Okada et al. (99) performed a study of 1,883 ACPA(+) subjects and found that DRB1*09:01, a frequent allele in Asians, is associated with decreasing ACPA levels. Terao et al. (151) replicated this finding in a study of 2,457 independent ACPA(+) subjects and additionally found an association with DRB1*15:01. A meta-analysis of these the two studies demonstrated associations with DRB1*09:01, *15:01, *15:02, *14:06, and *08:03 as well as involvement of amino acid positions 74, 60, or 57 in HLA-DRB1 (156).
Anti-carbamylated protein antibodies (anti-CarPs) are newly identified autoantibodies in patients with RA (130). Carbamylation is a nonenzymatic posttranslational modification in which isocyanic acid is bound to the amino terminus residues. Anti-CarPs are also present in patients with other diseases, including chronic kidney disease, cardiovascular disease, and juvenile idiopathic arthritis (130). There are not yet clear data to evaluate the presence of anti-CarPs in patients with other autoimmune diseases. Shi et al. (128, 130) observed anti-CarPs in 30% of ACPA(−) patients and 70% of ACPA(+) patients, but genetic influences on this reactivity have not been reported. A recent study reported that anti-CarP positivity was not associated with the HLA-DRB1 SE alleles, PTPN22, or smoking in two cohorts comprising a total of 2,831 Swedish subjects with RA (58).
7.2. Clinical Outcome
As indicated in Figure 1, the rate of progression and clinical outcome vary among patients with RA. The development of bony erosions and joint destruction is a major marker of progressive disease, and there is evidence for a genetic influence on this phenotype. Van der Helm-van Mil et al. (166) reported a smaller variation of joint destruction in monozygotic twins compared with dizygotic twins and uncorrelated pairs, indicating some heritability of the degree of joint destruction in RA. In a study of Icelandic individuals affected by joint destruction, Knevel et al. (64) estimated that 45–58% of the joint destruction was attributable to genetic components.
Many studies have reported strong associations of HLA-DRB1 risk alleles, such as SE alleles, with joint destruction (146, 149). However, ACPA positivity and RF positivity are also strongly associated with these alleles and with joint destruction. As a result, separating causality from joint damage, serologies, and HLA alleles is challenging. A majority of recent studies in European populations indicate that the SE alone has a negligible effect on progression to erosive disease independent of ACPA status (125, 149, 170). By contrast, Suzuki et al. (146) reported that, when they analyzed a total of 865 patients for erosions using the total Sharp score (TSS) stratified by ACPA positivity, they still found a significant association between TSS and the SE. Another Japanese study of 861 patients with ACPA(+) RA analyzed TSS data at year 5 after RA onset (158). The component of the association between TSS and the SE independent of ACPA status was largely explained by HLA-DRB1*04:05. Overall, these data suggest that, in some patient populations, HLA background may contribute to bony erosion independent of the patient’s serological status. This conclusion is supported by an analysis of HLA polymorphisms in a large longitudinal cohort of RA (177) in which haplotypes of amino acid positions 11, 71, and 74 of HLA-DRB1 were significantly associated with the Larsen score independent of ACPA status.
Genetic variants outside of the MHC have also been implicated in the development of bony erosions in RA. The degree of erosive disease is highly dependent on the duration of the disease because the erosions develop over time. Therefore, the most convincing studies involve observations in longitudinal cohorts, preferably with serial radiographic data. At a minimum, assessment of erosive disease must take disease duration into account. A recent study implicated PADI4 polymorphisms in disease outcome independent of ACPA status (146). Van der Linden et al. (168) reported that rs4810485 in CD40 was associated with TSS in two different cohorts as well. SNP variants in MMP9 (20), IL2RA (63), IL4, and IL4R (70) as well as other loci (65) have also been reported to be associated with progression to erosive disease. In general, these effects are rather modest. FOXO3 has been implicated in the outcome of several inflammatory and infectious diseases, including Crohn’s disease, malaria, and RA (77). These data were initially not replicated in a mixture of several populations (171), but a more recent large cohort study of RA in the United Kingdom has confirmed this association (K. Smith, personal communication). This association is of interest because it implicates a TGF-β pathway in monocytes involved in regulating both pro-and anti-inflammatory cytokines (42).
7.3. Genetic Regulation of Response to Therapy
The introduction of biological therapies such as tumor necrosis factor (TNF) inhibitors has brought about a paradigm shift in the treatment of RA, with dramatic improvements in long-term erosive outcome. TNF inhibition leads to changes in peripheral blood gene expression pattern as well as regulatory T cell subsets (11, 27, 89). Nevertheless, there are no established predictors of response to anti-TNF therapy, and although some studies have identified putative biomarkers of response to biological therapy, these have not been validated. In fact, there is no direct evidence of heritability of drug response to these agents, and many candidate gene studies of this issue (71, 81, 105, 164) have not been convincingly replicated. A large GWAS has suggested a very modest effect of CD84 on response (18). However, it is important to emphasize that the phenotype of drug response is extremely difficult to measure reliably, particularly in retrospective analysis. Genetic predictors of response to biological drugs targeted to other pathways, such as IL6 (178), have been at best suggestive with modest effects, and have not yet been replicated.
8. CLINICAL IMPLICATIONS OF RHEUMATOID ARTHRITIS GENETICS
8.1. Clarifying Ambiguous Diagnoses
The definition of genetic risk alleles for RA and other diseases may benefit the management of patients in clinical settings. For patients with no symptoms, the clinical predictive value of these alleles is very limited. However, there is a potential application for patients with undefined inflammatory arthritides. We anticipate that, ultimately, many of these patients will be genotyped to enhance diagnostic accuracy. In these instances, genotypic data might help to clarify ambiguous diagnoses. A genetic risk assessment for RA, gout, psoriatic arthritis, ankylosing spondylitis, and other conditions can be used to define probabilities of likely diseases. The integration of epidemiological information and autoantibody status with genetic data (126) should lead to better accuracy in supporting ambiguous diagnoses by applying prior probabilities.
8.2. Intermediate Biomarkers That Predict Disease
Because ACPA is often found in at-risk populations before RA develops, further clarification of pre-RA status could lead to potential preventive therapies. The establishment of the Studies of the Etiology of Rheumatoid Arthritis (SERA) study has been a major step forward in addressing this issue by recruiting and following first-degree relatives of patients with RA (53).
Detailed information about ACPA or other autoantibodies and inflammatory biomarkers may provide useful information regarding the risk of developing RA (68, 69). Demoruelle et al. (21) reported that pre-RA patients may have a different ACPA repertoire in comparison with non-pre-RA subjects positive for ACPA. Van de Stadt et al. (165) reported that they did not find skewing antigen preferentially recognized by ACPA in pre-RA patients and that epitope spreading occurred during pre-RA without specific patterns. Arkema et al. (3) reported that monocyte chemotactic protein 1 was elevated in both seropositive and seronegative pre-RA subjects. Several studies have reported that anti-CarPs could be detected in pre-RA patients and were associated with RA development independent of ACPA status (36, 129). MicroRNAs have been explored as biomarkers, but their clinical utility remains to be established (16). Recent data suggest that assessment of novel environmental exposures may add to the predictive power of these immune biomarkers (37). The integration of genetic information and intermediate biomarkers may lead to more efficient interpretation of pre-RA status, offering opportunities for early intervention.
9. FUTURE DIRECTIONS OF RHEUMATOID ARTHRITIS GENETICS
Over the last decade, RA research efforts have benefited from the remarkable technological progress that has enabled genome-wide association mapping. However, we are still in the early stages of understanding the functional significance of the many risk variants that have been identified. For many associations, the actual causative alleles have not been defined, implying the need for even greater efforts to fine map all risk loci. This will require even larger data sets of patients and controls, including analyses across the major population groups. The Rheumatoid Arthritis Consortium International will likely be a leader in this effort, bringing together most of the world’s major research groups and facilitating sharing and analysis of data in a single location at the New York Genome Center. Integration of genetic data with genome annotation will clearly be important for progress, and this information then needs to inform targeted functional studies of disease-relevant endophenotypes.
In addition to these basic studies to elucidate disease pathogenesis, the integration of genetic information and novel biomarkers into clinical practice should be achievable, even if it has so far been elusive. This will clearly require the development of longitudinal cohorts of patients, an effort that is well established in the European research community. We believe that progress will also depend on the application of techniques to query the noncoding genome, such as the assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) method (10, 109), which can be applied to small numbers of cells and will likely reflect both genetic and environmental factors that affect disease risk and expression. In addition, most functional and biomarker studies have focused on peripheral blood analysis, which may not capture important aspects of disease heterogeneity at the tissue level. The Accelerating Medicines Partnership is an international collaborative effort to address this issue by examining the biological diversity of synovial tissue, the major site of inflammatory lesions in RA.
Just as the extraordinary technological advances of the last decade have enabled a rapid expansion of the list of risk loci for all common human diseases, we expect that further technological advances will lead to new insights into disease heterogeneity and pathogenesis. These advances will yield more specific correlations between genotype and phenotype, leading in turn to more precisely targeted treatment and management of RA.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, et al. 2010. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum 62:2569–81 [DOI] [PubMed] [Google Scholar]
- 2.Amara K, Steen J, Murray F, Morbach H, Fernandez-Rodriguez BM, et al. 2013. Monoclonal IgG antibodies generated from joint-derived B cells of RA patients have a strong bias toward citrullinated autoantigen recognition. J. Exp. Med 210:445–55 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Arkema EV, Lu B, Malspeis S, Karlson EW, Costenbader KH. 2015. Monocyte chemotactic protein-1 elevation prior to the onset of rheumatoid arthritis among women. Biomark. Med 9:723–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balsa A, Cabezon A, Orozco G, Cobo T, Miranda-Cams E, et al. 2010. Influence of HLA DRB1 alleles in the susceptibility of rheumatoid arthritis and the regulation of antibodies against citrullinated proteins and rheumatoid factor. Arthritis Res. Ther 12:R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bang SY, Na YJ, Kim K, Joo YB, Park Y, et al. 2014. Targeted exon sequencing fails to identify rare coding variants with large effect in rheumatoid arthritis. Arthritis Res. Ther 16:447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, et al. 2004. A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am. J. Hum. Genet 75:330–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boechat AL, Ogusku MM, Sadahiro A, dos Santos MC. 2013. Association between the PTPN22 1858C/T gene polymorphism and tuberculosis resistance. Infect. Genet. Evol 16:310–13 [DOI] [PubMed] [Google Scholar]
- 8.Bossini-Castillo L, de Kovel C, Källberg H, van ‘t Slot R, Italiaander A, et al. 2015. A genome-wide association study of rheumatoid arthritis without antibodies against citrullinated peptides. Ann. Rheum. Dis 74:e15. [DOI] [PubMed] [Google Scholar]
- 9.Bottini N, Peterson EJ. 2014. Tyrosine phosphatase PTPN22: multifunctional regulator of immune signaling, development, and disease. Annu. Rev. Immunol 32:83–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. 2015. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol 109:21.29. 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byng-Maddick R, Ehrenstein MR. 2015. The impact of biological therapy on regulatory T cells in rheumatoid arthritis. Rheumatology 54:768–75 [DOI] [PubMed] [Google Scholar]
- 12.Catrina AI, Ytterberg AJ, Reynisdottir G, Malmström V, Klareskog L. 2014. Lungs, joints and immunity against citrullinated proteins in rheumatoid arthritis. Nat. Rev. Rheumatol 10:645–53 [DOI] [PubMed] [Google Scholar]
- 13.Chang HH, Dwivedi N, Nicholas AP, Ho IC. 2015. The W620 polymorphism in PTPN22 disrupts its interaction with peptidylarginine deiminase type 4 and enhances citrullination and NETosis. Arthritis Rheumatol 67:2323–34 [DOI] [PubMed] [Google Scholar]
- 14.Cheng T, Choi Y, Finkel TH, Tsao PY, Ji MQ, Eisenberg RA. 2013. Tumor necrosis factor receptor-associated factor 1 influences KRN/I-Ag7 mouse arthritis autoantibody production. J. Clin. Immunol 33:759–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung WS, Peng CL, Lin CL, Chang YJ, Chen YF, et al. 2014. Rheumatoid arthritis increases the risk of deep vein thrombosis and pulmonary thromboembolism: a nationwide cohort study. Ann. Rheum. Dis 73:1774–80 [DOI] [PubMed] [Google Scholar]
- 16.Churov AV, Oleinik EK, Knip M. 2015. MicroRNAs in rheumatoid arthritis: altered expression and diagnostic potential. Autoimmun. Rev 14:1029–37 [DOI] [PubMed] [Google Scholar]
- 17.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. 2005. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet 37:161–65 [DOI] [PubMed] [Google Scholar]
- 18.Cui J, Stahl EA, Saevarsdottir S, Miceli C, Diogo D, et al. 2013. Genome-wide association study and gene expression analysis identifies CD84 as a predictor of response to etanercept therapy in rheumatoid arthritis. PLOS Genet 9:e1003394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui J, Taylor KE, Lee YC, Källberg H, Weinblatt ME, et al. 2014. The influence of polygenic risk scores on heritability of anti-CCP level in RA. Genes Immun 15:107–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Rooy DP, Zhernakova A, Tsonaka R, Willemze A, Kurreeman BA, et al. 2014. A genetic variant in the region of MMP-9 is associated with serum levels and progression of joint damage in rheumatoid arthritis. Ann. Rheum. Dis 73:1163–69 [DOI] [PubMed] [Google Scholar]
- 21.Demoruelle MK, Parish MC, Derber LA, Kolfenbach JR, Hughes-Austin JM, et al. 2013. Performance of anti-cyclic citrullinated peptide assays differs in subjects at increased risk of rheumatoid arthritis and subjects with established disease. Arthritis Rheum 65:2243–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dendrou CA, Plagnol V, Fung E, Yang JH, Downes K, et al. 2009. Cell-specific protein phenotypes for the autoimmune locus IL2RA using a genotype-selectable human bioresource. Nat. Genet 41:1011–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Meglio P, Di Cesare A, Laggner U, Chu CC, Napolitano L, et al. 2011. The IL23R R381Q gene variant protects against immune-mediated diseases by impairing IL-23-induced Th17 effector response in humans. PLOS ONE 6:e17160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dilthey AT, Moutsianas L, Leslie S, McVean G. 2011. HLA*IMP—an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics 27:968–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diogo D, Bastarache L, Liao KP, Graham RR, Fulton RS, et al. 2015. TYK2 protein-coding variants protect against rheumatoid arthritis and autoimmunity, with no evidence of major pleiotropic effects on non-autoimmune complex traits. PLOS ONE 10:e0122271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diogo D, Kurreeman F, Stahl EA, Liao KP, Gupta N, et al. 2013. Rare, low-frequency, and common variants in the protein-coding sequence of biological candidate genes from GWASs contribute to risk of rheumatoid arthritis. Am. J. Hum. Genet 92:15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehrenstein MR, Evans JG, Singh A, Moore S, Warnes G, et al. 2004. Compromised function of regulatory T cells in rheumatoid arthritis and reversal by anti-TNFα therapy. J. Exp. Med 200:277–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Essouma M, Noubiap JJ. 2015. Is air pollution a risk factor for rheumatoid arthritis? J. Inflamm 12:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eyre S, Bowes J, Diogo D, Lee A, Barton A, et al. 2012. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat. Genet 44:1336–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eyre S, Ke X, Lawrence R, Bowes J, Panoutsopoulou K, et al. 2011. Examining the overlap between genome-wide rare variant association signals and linkage peaks in rheumatoid arthritis. Arthritis Rheum 63:1522–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, et al. 2015. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518:337–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, et al. 2015. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet 47:1228–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Firestein GS. 2003. Evolving concepts of rheumatoid arthritis. Nature 423:356–61 [DOI] [PubMed] [Google Scholar]
- 34.Fries JF, Wolfe F, Apple R, Erlich H, Bugawan T, et al. 2002. HLA-DRB1 genotype associations in 793 white patients from a rheumatoid arthritis inception cohort: frequency, severity, and treatment bias. Arthritis Rheum 46:2320–29 [DOI] [PubMed] [Google Scholar]
- 35.Frisell T, Holmqvist M, Kallberg H, Klareskog L, Alfredsson L, Askling J. 2013. Familial risks and heritability of rheumatoid arthritis: role of rheumatoid factor/anti-citrullinated protein antibody status, number and type of affected relatives, sex, and age. Arthritis Rheum 65:2773–82 [DOI] [PubMed] [Google Scholar]
- 36.Gan RW, Trouw LA, Shi J, Toes RE, Huizinga TW, et al. 2015. Anti-carbamylated protein antibodies are present prior to rheumatoid arthritis and are associated with its future diagnosis. J. Rheumatol 42:572–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gan RW, Young KA, Zerbe GO, Demoruelle MK, Weisman MH, et al. 2016. Lower omega-3 fatty acids are associated with the presence of anti-cyclic citrullinated peptide autoantibodies in a population at risk for future rheumatoid arthritis: a nested case-control study. Rheumatology 55:367–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibofsky A, Winchester RJ, Patarroyo M, Fotino M, Kunkel HG. 1978. Disease associations of the Ia-like human alloantigens. Contrasting patterns in rheumatoid arthritis and systemic lupus erythematosus. J.Exp.Med 148:1728–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gourh P, Agarwal SK, Martin E, Divecha D, Rueda B, et al. 2010. Association of the C8orf13-BLK region with systemic sclerosis in North-American and European populations. J. Autoimmun 34:155–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gregersen PK, Amos CI, Lee AT, Lu Y, Remmers EF, et al. 2009. REL, encoding a member of the NF-κB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat. Genet 41:820–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregersen PK, Klein G, Keogh M, Kern M, DeFranco M, et al. 2015. The Genotype and Phenotype (GaP) registry: a living biobank for the analysis of quantitative traits. Immunol. Res 1:107–12 [DOI] [PubMed] [Google Scholar]
- 42.Gregersen PK, Manjarrez-Orduño N. 2013. FOXO in the hole: leveraging GWAS for outcome and function. Cell 155:11–12 [DOI] [PubMed] [Google Scholar]
- 43.Gregersen PK, Silver J, Winchester RJ. 1987. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum 30:1205–13 [DOI] [PubMed] [Google Scholar]
- 44.Gusev A, Bhatia G, Zaitlen N, Vilhjalmsson BJ, Diogo D, et al. 2013. Quantifying missing heritability at known GWAS loci. PLOS Genet 9:e1003993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hall FC, Weeks DE, Camilleri JP, Williams LA, Amos N, et al. 1996. Influence of the HLA-DRB1 locus on susceptibility and severity in rheumatoid arthritis. QJM 89:821–29 [DOI] [PubMed] [Google Scholar]
- 46.Han B, Diogo D, Eyre S, Källberg H, Zhernakova A, et al. 2014. Fine mapping seronegative and seropositive rheumatoid arthritis to shared and distinct HLA alleles by adjusting for the effects of heterogeneity. Am.J. Hum. Genet 94:522–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hasegawa K, Martin F, Huang G, Tumas D, Diehl L, Chan AC. 2004. PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science 303:685–89 [DOI] [PubMed] [Google Scholar]
- 48.Hemminki K, Li X, Sundquist J, Sundquist K. 2009. Familial associations of rheumatoid arthritis with autoimmune diseases and related conditions. Arthritis Rheum 60:661–68 [DOI] [PubMed] [Google Scholar]
- 49.Hensvold AH, Magnusson PK, Joshua V, Hansson M, Israelsson L, et al. 2013. Environmental and genetic factors in the development of anticitrullinated protein antibodies (ACPAs) and ACPA-positive rheumatoid arthritis: an epidemiological investigation in twins. Ann. Rheum. Dis 74:375–80 [DOI] [PubMed] [Google Scholar]
- 50.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, et al. 2008. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med 358:900–9 [DOI] [PubMed] [Google Scholar]
- 51.Hu X, Kim H, Raj T, Brennan PJ, Trynka G, et al. 2014. Regulation of gene expression in autoimmune disease loci and the genetic basis of proliferation in CD4+ effector memory T cells. PLOS Genet 10:e1004404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu X, Kim H, Stahl E, Plenge R, Daly M, Raychaudhuri S. 2011. Integrating autoimmune risk loci with gene-expression data identifies specific pathogenic immune cell subsets. Am. J. Hum. Genet 89:496–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hughes-Austin JM, Deane KD, Derber LA, Kolfenbach JR, Zerbe GO, et al. 2013. Multiple cytokines and chemokines are associated with rheumatoid arthritis-related autoimmunity in first-degree relatives without rheumatoid arthritis: Studies of the Aetiology of Rheumatoid Arthritis (SERA). Ann. Rheum. Dis 72:901–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Irigoyen P, Lee AT, Wener MH, Li W, Kern M, et al. 2005. Regulation of anti-cyclic citrullinated peptide antibodies in rheumatoid arthritis: contrasting effects of HLA-DR3 and the shared epitope alleles. Arthritis Rheum 52:3813–18 [DOI] [PubMed] [Google Scholar]
- 55.Jawaheer D, Li W, Graham RR, Chen W, Damle A, et al. 2002. Dissecting the genetic complexity of the association between human leukocyte antigens and rheumatoid arthritis. Am. J. Hum. Genet 71:585–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jia X, Han B, Onengut-Gumuscu S, Chen WM, Concannon PJ, et al. 2013. Imputing amino acid polymorphisms in human leukocyte antigens. PLOS ONE 8:e64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang L, Yin J, Ye L, Yang J, Hemani G, et al. 2014. Novel risk loci for rheumatoid arthritis in Han Chinese and congruence with risk variants in Europeans. Arthritis Rheumatol 66:1121–32 [DOI] [PubMed] [Google Scholar]
- 58.Jiang X, Trouw LA, van Wesemael TJ, Shi J, Bengtsson C, et al. 2014. Anti-CarP antibodies in two large cohorts of patients with rheumatoid arthritis and their relationship to genetic risk factors, cigarette smoking and other autoantibodies. Ann. Rheum. Dis 73:1761–68 [DOI] [PubMed] [Google Scholar]
- 59.Johansson M, Arlestig L, Hallmans G, Rantapaa-Dahlqvist S. 2006. PTPN22 polymorphism and anti-cyclic citrullinated peptide antibodies in combination strongly predicts future onset of rheumatoid arthritis and has a specificity of 100% for the disease. Arthritis Res. Ther 8:R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Källberg H, Padyukov L, Plenge RM, Rönnelid J, Gregersen PK, et al. 2007. Gene-gene and gene-environment interactions involving HLA-DRB1, PTPN22, and smoking in two subsets of rheumatoid arthritis. Am. J. Hum. Genet 80:867–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim SJ, Gregersen PK, Diamond B. 2013. Regulation of dendritic cell activation by microRNA let-7c and BLIMP1. J. Clin. Investig 123:823–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klareskog L, Stolt P, Lundberg K, Källberg H, Bengtsson C, et al. 2006. A new model for an etiology of rheumatoid arthritis: Smoking may trigger HLA-DR (shared epitope)-restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum 54:38–46 [DOI] [PubMed] [Google Scholar]
- 63.Knevel R, de Rooy DPC, Zhernakova A, Grondal G, Krabben A, et al. 2013. Association of variants in IL2RA with progression of joint destruction in rheumatoid arthritis. Arthritis Rheum 65:1684–93 [DOI] [PubMed] [Google Scholar]
- 64.Knevel R, Grondal G, Huizinga TW, Visser AW, Jonsson H, et al. 2012. Genetic predisposition of the severity of joint destruction in rheumatoid arthritis: a population-based study. Ann. Rheum. Dis 71:707–9 [DOI] [PubMed] [Google Scholar]
- 65.Knevel R, Klein K, Somers K, Ospelt C, Houwing-Duistermaat JJ, et al. 2014. Identification of a genetic variant for joint damage progression in autoantibody-positive rheumatoid arthritis. Ann. Rheum. Dis 73:2038–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kochi Y, Okada Y, Suzuki A, Ikari K, Terao C, et al. 2010. A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nat. Genet 42:515–19 [DOI] [PubMed] [Google Scholar]
- 67.Kochi Y, Yamada R, Suzuki A, Harley JB, Shirasawa S, et al. 2005. A functional variant in FCRL3, encoding Fc receptor-like 3, is associated with rheumatoid arthritis and several autoimmunities. Nat. Genet 37:478–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kokkonen H, Mullazehi M, Berglin E, Hallmans G, Wadell G, et al. 2011. Antibodies of IgG, IgA and IgM isotypes against cyclic citrullinated peptide precede the development of rheumatoid arthritis. Arthritis Res. Ther 13:R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kolfenbach JR, Deane KD, Derber LA, O’Donnell CI, Gilliland WR, et al. 2010. Autoimmunity to peptidyl arginine deiminase type 4 precedes clinical onset of rheumatoid arthritis. Arthritis Rheum 62:2633–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krabben A, Wilson AG, de Rooy DP, Zhernakova A, Brouwer E, et al. 2013. Association of genetic variants in the IL4 and IL4R genes with the severity of joint damage in rheumatoid arthritis: a study in seven cohorts. Arthritis Rheum 65:3051–57 [DOI] [PubMed] [Google Scholar]
- 71.Krintel SB, Palermo G, Johansen JS, Germer S, Essioux L, et al. 2012. Investigation of single nucleotide polymorphisms and biological pathways associated with response to TNFα inhibitors in patients with rheumatoid arthritis. Pharmacogenet. Genom 22:577–89 [DOI] [PubMed] [Google Scholar]
- 72.Kroot EJ, de Jong BA, van Leeuwen MA, Swinkels H, van den Hoogen FH, et al. 2000. The prognostic value of anti-cyclic citrullinated peptide antibody in patients with recent-onset rheumatoid arthritis. Arthritis Rheum 43:1831–35 [DOI] [PubMed] [Google Scholar]
- 73.Kurreeman FA, Padyukov L, Marques RB, Schrodi SJ, Seddighzadeh M, et al. 2007. A candidate gene approach identifies the TRAF1/C5 region as a risk factor for rheumatoid arthritis. PLOS Med 4:e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Laki J, Lundström E, Snir O, Rönnelid J, Ganji I, et al. 2012. Very high levels of anti-citrullinated protein antibodies are associated with HLA-DRB1*15 non-shared epitope allele in patients with rheumatoid arthritis. Arthritis Rheum 64:2078–84 [DOI] [PubMed] [Google Scholar]
- 75.Lawrence JS. 1970. Heberden Oration, 1969. Rheumatoid arthritis—nature or nurture? Ann. Rheum. Dis 29:357–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee HS, Irigoyen P, Kern M, Lee A, Batliwalla F, et al. 2007. Interaction between smoking, the shared epitope, and anti-cyclic citrullinated peptide: a mixed picture in three large North American rheumatoid arthritis cohorts. Arthritis Rheum 56:1745–53 [DOI] [PubMed] [Google Scholar]
- 77.Lee JC, Espeli M, Anderson CA, Linterman MA, Pocock JM, et al. 2013. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell 155:57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lenz TL, Deutsch AJ, Han B, Hu X, Okada Y, et al. 2015. Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat. Genet 47:1085–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lewis-Faning E 1950. Report on an enquiry into the aetiological factors associated with rheumatoid arthritis. Ann. Rheum. Dis 9(Suppl.):1–9418623830 [Google Scholar]
- 80.Linn-Rasker SP, van der Helm-van Mil AH, van Gaalen FA, Kloppenburg M, de Vries RR, et al. 2006. Smoking is a risk factor for anti-CCP antibodies only in rheumatoid arthritis patients who carry HLA-DRB1 shared epitope alleles. Ann. Rheum. Dis 65:366–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu C, Batliwalla F, Li W, Lee A, Roubenoff R, et al. 2008. Genome-wide association scan identifies candidate polymorphisms associated with differential response to anti-TNF treatment in rheumatoid arthritis. Mol. Med 14:575–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu B, Hiraki LT, Sparks JA, Malspeis S, Chen CY, et al. 2014. Being overweight or obese and risk of developing rheumatoid arthritis among women: a prospective cohort study. Ann. Rheum. Dis 73:1914–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lundberg K, Bengtsson C, Kharlamova N, Reed E, Jiang X, et al. 2012. Genetic and environmental determinants for disease risk in subsets of rheumatoid arthritis defined by the anticitrullinated protein/peptide antibody fine specificity profile. Ann. Rheum. Dis 72:652–58 [DOI] [PubMed] [Google Scholar]
- 84.Lundström E, Källberg H, Alfredsson L, Klareskog L, Padyukov L. 2009. Gene-environment interaction between the DRB1 shared epitope and smoking in the risk of anti-citrullinated protein antibody-positive rheumatoid arthritis: All alleles are important. Arthritis Rheum 60:1597–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.MacGregor AJ, Snieder H, Rigby AS, Koskenvuo M, Kaprio J, et al. 2000. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum 43:30–37 [DOI] [PubMed] [Google Scholar]
- 86.Mahdi H, Fisher BA, Källberg H, Plant D, Malmström V, et al. 2009. Specific interaction between genotype, smoking and autoimmunity to citrullinated α-enolase in the etiology of rheumatoid arthritis. Nat. Genet 41:1319–24 [DOI] [PubMed] [Google Scholar]
- 87.McAllister K, Yarwood A, Bowes J, Orozco G, Viatte S, et al. 2013. Identification of BACH2 and RAD51B as rheumatoid arthritis susceptibility loci in a meta-analysis of genome-wide data. Arthritis Rheum 65:3058–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morgan AW, Thomson W, Martin SG, Carter AM, Erlich HA, et al. 2009. Reevaluation of the interaction between HLA-DRB1 shared epitope alleles, PTPN22, and smoking in determining susceptibility to autoantibody-positive and autoantibody-negative rheumatoid arthritis in a large UK Caucasian population. Arthritis Rheum 60:2565–76 [DOI] [PubMed] [Google Scholar]
- 89.Nadkarni S, Mauri C, Ehrenstein MR. 2007. Anti-TNF-α therapy induces a distinct regulatory T cell population in patients with rheumatoid arthritis via TGF-β. J. Exp. Med 204:33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. 2009. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science 324:387–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nelson JL, Mickelson E, Masewicz S, Barrington R, Dugowson C, et al. 1991. Dw14(DRB1*0404) is a Dw4-dependent risk factor for rheumatoid arthritis. Rethinking the “shared epitope” hypothesis. Tissue Antigens 38:145–51 [DOI] [PubMed] [Google Scholar]
- 92.Neovius M, Simard JF, Askling J, group As. 2011. Nationwide prevalence of rheumatoid arthritis and penetration of disease-modifying drugs in Sweden. Ann. Rheum. Dis 70:624–29 [DOI] [PubMed] [Google Scholar]
- 93.Nepom GT, Holbeck SL, Seyfried CE, Wilske KR, Nepom BS. 1986. Identification of HLA-Dw14 genes in DR4+ rheumatoid arthritis. Lancet 328:1002–5 [DOI] [PubMed] [Google Scholar]
- 94.Nienhuis RL, Mandema E. 1964. A new serum factor in patients with rheumatoid arthritis: the antiper-inuclear factor. Ann. Rheum. Dis 23:302–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nishimura K, Sugiyama D, Kogata Y, Tsuji G, Nakazawa T, et al. 2007. Meta-analysis: diagnostic accuracy of anti-cyclic citrullinated peptide antibody and rheumatoid factor for rheumatoid arthritis. Ann. Intern. Med 146: 797–808 [DOI] [PubMed] [Google Scholar]
- 96.Nordmark G, Kristjansdottir G, Theander E, Appel S, Eriksson P, et al. 2011. Association of EBF1, FAM167A(C8orf13)-BLK and TNFSF4 gene variants with primary Sjogren’s syndrome. Genes Immun 12:100–9 [DOI] [PubMed] [Google Scholar]
- 97.Ohmura K, Terao C, Maruya E, Katayama M, Matoba K, et al. 2010. Anti-citrallinated peptide antibody-negative RA is a genetically distinct subset: a definitive study using only bone-erosive ACPA-negative rheumatoid arthritis. Rheumatology 49:2298–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Okada Y, Kim K, Han B, Pillai NE, Ong RT, et al. 2014. Risk for ACPA-positive rheumatoid arthritis is driven by shared HLA amino acid polymorphisms in Asian and European populations. Hum. Mol. Genet 23:6916–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Okada Y, Suzuki A, Yamada R, Kochi Y, Shimane K, et al. 2010. HLA-DRB1*0901 lowers anti-cyclic citrullinated peptide antibody levels in Japanese patients with rheumatoid arthritis. Ann. Rheum. Dis 69:1569–70 [DOI] [PubMed] [Google Scholar]
- 100.Okada Y, Terao C, Ikari K, Kochi Y, Ohmura K, et al. 2012. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat. Genet 44:511–16 [DOI] [PubMed] [Google Scholar]
- 101.Okada Y, Wu D, Trynka G, Raj T, Terao C, et al. 2014. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506:376–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Padyukov L, Seielstad M, Ong RT, Ding B, Rönnelid J, et al. 2011. A genome-wide association study suggests contrasting associations in ACPA-positive versus ACPA-negative rheumatoid arthritis. Ann. Rheum. Dis 70:259–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pedersen M, Jacobsen S, Garred P, Madsen HO, Klarlund M, et al. 2007. Strong combined gene-environment effects in anti-cyclic citrullinated peptide-positive rheumatoid arthritis: a nationwide case-control study in Denmark. Arthritis Rheum 56:1446–53 [DOI] [PubMed] [Google Scholar]
- 104.Pillai NE, Okada Y, Saw WY, Ong RT, Wang X, et al. 2014. Predicting HLA alleles from high-resolution SNP data in three Southeast Asian populations. Hum. Mol. Genet 23:4443–51 [DOI] [PubMed] [Google Scholar]
- 105.Plant D, Bowes J, Potter C, Hyrich KL, Morgan AW, et al. 2011. Genome-wide association study of genetic predictors of anti-tumor necrosis factor treatment efficacy in rheumatoid arthritis identifies associations with polymorphisms at seven loci. Arthritis Rheum 63:645–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Plenge RM, Cotsapas C, Davies L, Price AL, de Bakker PI, et al. 2007. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat. Genet 39:1477–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, et al. 2007. TRAF1-C5 as a risk locus for rheumatoid arthritis—a genomewide study. N. Engl. J. Med 357:1199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pouget JG, Gonçalves VF, Schizophr. Work. Group Psychiatr. Genom. Consort., Spain SL, Finucane HK, et al. 2015. Genome-wide association studies suggest limited immune gene enrichment in schizophrenia compared to six immune diseases. BioRxiv. doi: 10.1101/030411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Qu K, Zaba LC, Giresi PG, Li R, Longmire M, et al. 2015. Individuality and variation of personal regulomes in primary human T cells. Cell Syst 1:51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rawlings DJ, Dai X, Buckner JH. 2015. The role of PTPN22 risk variant in the development of autoimmunity: finding common ground between mouse and human. J. Immunol 194:2977–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Raychaudhuri S, Remmers EF, Lee AT, Hackett R, Guiducci C, et al. 2008. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat. Genet 40:1216–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, et al. 2012. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet 44:291–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Raychaudhuri S, Thomson BP, Remmers EF, Eyre S, Hinks A, et al. 2009. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat. Genet 41:1313–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, et al. 2007. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N. Engl. J. Med 357:977–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Reynolds RJ, Ahmed AF, Danila MI, Hughes LB, Consort. Longitud. Eval. Afr. Am. Early Rheum. Arthritis Investig., et al. 2014. HLA-DRB1-associated rheumatoid arthritis risk at multiple levels in African Americans: hierarchical classification systems, amino acid positions, and residues. Arthritis Rheumatol 66:3274–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, et al. 2011. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet 43:1066–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rose HM, Ragan C, Pearce E, Lipman MO. 1948. Differential agglutination of normal and sensitized sheep erythrocytes by sera of patients with rheumatoid arthritis. Proc. Soc. Exp. Biol. Med 68:1–6 [DOI] [PubMed] [Google Scholar]
- 118.Saijo K, Schmedt C, Su IH, Karasuyama H, Lowell CA, et al. 2003. Essential role of Src-family protein tyrosine kinases in NF-κB activation during B cell development. Nat. Immunol 4:274–79 [DOI] [PubMed] [Google Scholar]
- 119.Sakaguchi N, Takahashi T, Hata H, Nomura T, Tagami T, et al. 2003. Altered thymic T-cell selection due to a mutation of the ZAP-70 gene causes autoimmune arthritis in mice. Nature 426:454–60 [DOI] [PubMed] [Google Scholar]
- 120.Samuelson EM, Laird RM, Maue AC, Rochford R, Hayes SM. 2012. Blk haploinsufficiency impairs the development, but enhances the functional responses, of MZ B cells. Immunol. Cell Biol 90:620–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Samuelson EM, Laird RM, Papillion AM, Tatum AH, Princiotta MF, Hayes SM. 2014. Reduced B lymphoid kinase (Blk) expression enhances proinflammatory cytokine production and induces nephrosis in C57BL/6-lpr/lpr mice. PLOS ONE 9:e92054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Scally SW, Petersen J, Law SC, Dudek NL, Nel HJ, et al. 2013. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J. Exp. Med 210:2569–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schellekens GA, de Jong BA, van den Hoogen FH, van de Putte LB, van Venrooij WJ. 1998. Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis-specific autoantibodies. J. Clin. Investig 101:273–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schellekens GA, Visser H, de Jong BA, van den Hoogen FH, Hazes JM, et al. 2000. The diagnostic properties of rheumatoid arthritis antibodies recognizing a cyclic citrullinated peptide. Arthritis Rheum 43:155–63 [DOI] [PubMed] [Google Scholar]
- 125.Scherer HU, van der Woude D, Willemze A, Trouw LA, Knevel R, et al. 2011. Distinct ACPA fine specificities, formed under the influence of HLA shared epitope alleles, have no effect on radiographic joint damage in rheumatoid arthritis. Ann. Rheum. Dis 70:1461–64 [DOI] [PubMed] [Google Scholar]
- 126.Scott IC, Seegobin SD, Steer S, Tan R, Forabosco P, et al. 2013. Predicting the risk of rheumatoid arthritis and its age of onset through modelling genetic risk variants with smoking. PLOS Genet 9:e1003808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Seidl C, Donner H, Fischer B, Usadel KH, Seifried E, et al. 1998. CTLA4 codon 17 dimorphism in patients with rheumatoid arthritis. Tissue Antigens 51:62–66 [DOI] [PubMed] [Google Scholar]
- 128.Shi J, Knevel R, Suwannalai P, van der Linden MP, Janssen GM, et al. 2011. Autoantibodies recognizing carbamylated proteins are present in sera of patients with rheumatoid arthritis and predict joint damage. PNAS 108:17372–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Shi J, van de Stadt LA, Levarht EW, Huizinga TW, Toes RE, et al. 2013. Anti-carbamylated protein antibodies are present in arthralgia patients and predict the development of rheumatoid arthritis. Arthritis Rheum 65:911–15 [DOI] [PubMed] [Google Scholar]
- 130.Shi J, van Veelen PA, Mahler M, Janssen GM, Drijfhout JW, et al. 2014. Carbamylation and antibodies against carbamylated proteins in autoimmunity and other pathologies. Autoimmun. Rev 13:225–30 [DOI] [PubMed] [Google Scholar]
- 131.Shmerling RH, Delbanco TL. 1991. The rheumatoid factor: an analysis of clinical utility. Am. J. Med 91:528–34 [DOI] [PubMed] [Google Scholar]
- 132.Simpfendorfer KR, Armstead BE, Shih A, Li W, Curran M, et al. 2015. Autoimmune disease-associated haplotypes of BLK exhibit lowered thresholds for B cell activation and expansion of Ig class-switched B cells. Arthritis Rheumatol 67:2866–76 [DOI] [PubMed] [Google Scholar]
- 133.Simpfendorfer KR, Olsson LM, Manjarrez-Orduño N, Khalili H, Simeone AM, et al. 2012. The autoimmunity-associated BLK haplotype exhibits cis-regulatory effects on mRNA and protein expression that are prominently observed in B cells early in development. Hum. Mol. Genet 21:3918–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Snir O, Gomez-Cabrero D, Montes A, Perez-Pampin E, Gómez-Reino JJ, et al. 2014. Non-HLA genes PTPN22, CDK6 and PADI4 are associated with specific autoantibodies in HLA-defined subgroups of rheumatoid arthritis. Arthritis Res. Ther 16:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Somers EC, Antonsen S, Pedersen L, Sorensen HT. 2013. Parental history of lupus and rheumatoid arthritis and risk in offspring in a nationwide cohort study: Does sex matter? Ann. Rheum. Dis 72:525–29 [DOI] [PubMed] [Google Scholar]
- 136.Speed D, Hemani G, Johnson MR, Balding DJ. 2012. Improved heritability estimation from genome-wide SNPs. Am. J. Hum. Genet 91:1011–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, et al. 2010. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet 42:508–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Stahl EA, Wegmann D, Trynka G, Gutierrez-Achury J, Do R, et al. 2012. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nat. Genet 44:483–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stanford SM, Bottini N. 2014. PTPN22: the archetypal non-HLA autoimmunity gene. Nat. Rev. Rheumatol 10:602–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Stastny P 1976. Mixed lymphocyte cultures in rheumatoid arthritis. J. Clin. Investig 57:1148–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Stastny P 1978. Association of the B-cell alloantigen DRw4 with rheumatoid arthritis. N. Engl. J. Med 298:869–71 [DOI] [PubMed] [Google Scholar]
- 142.Stein EA, Mellis S, Yancopoulos GD, Stahl N, Logan D, et al. 2012. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N. Engl. J. Med 366:1108–18 [DOI] [PubMed] [Google Scholar]
- 143.Sugiyama D, Nishimura K, Tamaki K, Tsuji G, Nakazawa T, et al. 2010. Impact of smoking as a risk factor for developing rheumatoid arthritis: a meta-analysis of observational studies. Ann. Rheum. Dis 69:70–81 [DOI] [PubMed] [Google Scholar]
- 144.Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, et al. 2003. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat. Genet 34:395–402 [DOI] [PubMed] [Google Scholar]
- 145.Suzuki A, Yamada R, Kochi Y, Sawada T, Okada Y, et al. 2008. Functional SNPs in CD244 increase the risk of rheumatoid arthritis in a Japanese population. Nat. Genet 40:1224–29 [DOI] [PubMed] [Google Scholar]
- 146.Suzuki T, Ikari K, Yano K, Inoue E, Toyama Y, et al. 2013. PADI4 and HLA-DRB1 are genetic risks for radiographic progression in RA patients, independent of ACPA status: results from the IORRA cohort study. PLOS ONE 8:e61045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Svard A, Skogh T, Alfredsson L, Ilar A, Klareskog L, et al. 2015. Associations with smoking and shared epitope differ between IgA- and IgG-class antibodies to cyclic citrullinated peptides in early rheumatoid arthritis. Arthritis Rheumatol 67:2032–37 [DOI] [PubMed] [Google Scholar]
- 148.Svendsen AJ, Kyvik KO, Houen G, Junker P, Christensen K, et al. 2013. On the origin of rheumatoid arthritis: the impact of environment and genes—a population based twin study. PLOS ONE 8:e57304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Syversen SW, Goll GL, van der Heijde D, Landewe R, Lie BA, et al. 2010. Prediction of radiographic progression in rheumatoid arthritis and the role of antibodies against mutated citrullinated vimentin: results from a 10-year prospective study. Ann. Rheum. Dis 69:345–51 [DOI] [PubMed] [Google Scholar]
- 150.Terao C, Ikari K, Nakayamada S, Takahashi Y, Yamada R, et al. 2016. A twin study of rheumatoid arthritis in the Japanese population. Mod. Rheumatol In press, doi: 10.3109/14397595.2015.1135856 [DOI] [PubMed] [Google Scholar]
- 151.Terao C, Ikari K, Ohmura K, Suzuki T, Iwamoto T, et al. 2012. Quantitative effect of HLA-DRB1 alleles to ACPA levels in Japanese rheumatoid arthritis: no strong genetic impact of shared epitope to ACPA levels after stratification of HLA-DRB1*09:01. Ann. Rheum. Dis 71:1095–97 [DOI] [PubMed] [Google Scholar]
- 152.Terao C, Ohmura K, Ikari K, Kochi Y, Maruya E, et al. 2012. ACPA-negative RA consists of two genetically distinct subsets based on RF positivity in Japanese. PLOS ONE 7:e40067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Terao C, Ohmura K, Katayama M, Takahashi M, Kokubo M, et al. 2011. Myelin basic protein as a novel genetic risk factor in rheumatoid arthritis—a genome-wide study combined with immunological analyses. PLOS ONE 6:e20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Terao C, Ohmura K, Kochi Y, Ikari K, Maruya E, et al. 2011. A large-scale association study identified multiple HLA-DRB1 alleles associated with ACPA-negative rheumatoid arthritis in Japanese subjects. Ann. Rheum. Dis 70:2134–39 [DOI] [PubMed] [Google Scholar]
- 155.Terao C, Ohmura K, Kochi Y, Ikari K, Okada Y, et al. 2015. Anti-citrullinated peptide/protein antibody (ACPA)-negative RA shares a large proportion of susceptibility loci with ACPA-positive RA: a meta-analysis of genome-wide association study in a Japanese population. Arthritis Res. Ther 17:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Terao C, Suzuki A, Ikari K, Kochi Y, Ohmura K, et al. 2015. An association between the 74th amino acid position of HLA-DRB1 and ACPA levels of Japanese ACPA-positive RA. Arthritis Rheumatol 67:2038–45 [DOI] [PubMed] [Google Scholar]
- 157.Terao C, Yamada R, Ohmura K, Takahashi M, Kawaguchi T, et al. 2011. The human AIRE gene at chromosome 21q22 is a genetic determinant for the predisposition to rheumatoid arthritis in Japanese population. Hum. Mol. Genet 20:2680–85 [DOI] [PubMed] [Google Scholar]
- 158.Terao C, Yano K, Ikari K, Furu M, Yamakawa N, et al. 2015. Main contribution of DRB1*04:05 among the shared epitope alleles and involvement of DRB1 amino acid position 57 in association with joint destruction in anti-citrullinated protein antibody-positive rheumatoid arthritis. Arthritis Rheumatol 67:1744–50 [DOI] [PubMed] [Google Scholar]
- 159.Texido G, Su IH, Mecklenbrauker I, Saijo K, Malek SN, et al. 2000. The B-cell-specirlc Src-family kinase Blk is dispensable for B-cell development and activation. Mol. Cell. Biol 20:1227–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Thomson W, Barton A, Ke X, Eyre S, Hinks A, et al. 2007. Rheumatoid arthritis association at 6q23. Nat. Genet 39:1431–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tokuhiro S, Yamada R, Chang X, Suzuki A, Kochi Y, et al. 2003. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat. Genet 35:341–48 [DOI] [PubMed] [Google Scholar]
- 162.Trowsdale J, Knight JC. 2013. Major histocompatibility complex genomics and human disease. Annu. Rev. Genom. Hum. Genet 14:301–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Trynka G, Sandor C, Han B, Xu H, Stranger BE, et al. 2013. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet 45:124–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mirkov Umiċeviċ, Mirkov M, Cui J, Vermeulen SH, Stahl EA, Toonen EJ, et al. 2013. Genome-wide association analysis of anti-TNF drug response in patients with rheumatoid arthritis. Ann. Rheum. Dis 72:1375–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.van de Stadt LA, de Koning MH, van de Stadt RJ, Wolbink G, Dijkmans BA, et al. 2011. Development of the anti-citrullinated protein antibody repertoire prior to the onset of rheumatoid arthritis. Arthritis Rheum 63:3226–33 [DOI] [PubMed] [Google Scholar]
- 166.van der Helm-van Mil AH, Kern M, Gregersen PK, Huizinga TW. 2006. Variation in radiologic joint destruction in rheumatoid arthritis differs between monozygotic and dizygotic twins and pairs of unrelated patients. Arthritis Rheum 54:2028–30 [DOI] [PubMed] [Google Scholar]
- 167.van der Helm-van Mil AH, Verpoort KN, le Cessie S, Huizinga TW, de Vries RR, Toes RE. 2007. The HLA-DRB1 shared epitope alleles differ in the interaction with smoking and predisposition to antibodies to cyclic citrullinated peptide. Arthritis Rheum 56:425–32 [DOI] [PubMed] [Google Scholar]
- 168.van der Linden MP, Feitsma AL, le Cessie S, Kern M, Olsson LM, et al. 2009. Association of a single-nucleotide polymorphism in CD40 with the rate of joint destruction in rheumatoid arthritis. Arthritis Rheum 60:2242–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.van der Woude D, Houwing-Duistermaat JJ, Toes RE, Huizinga TW, Thomson W, et al. 2009. Quantitative heritability of anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis. Arthritis Rheum 60:916–23 [DOI] [PubMed] [Google Scholar]
- 170.van Gaalen FA, van Aken J, Huizinga TW, Schreuder GM, Breedveld FC, et al. 2004. Association between HLA class II genes and autoantibodies to cyclic citrullinated peptides (CCPs) influences the severity of rheumatoid arthritis. Arthritis Rheum 50:2113–21 [DOI] [PubMed] [Google Scholar]
- 171.van Steenbergen HW, Rantapaa-Dahlqvist S, van Nies JA, Berglin E, Huizinga TW, et al. 2014. Does a genetic variant in FOXO3A predict a milder course of rheumatoid arthritis? Arthritis Rheumatol 66:1678–81 [DOI] [PubMed] [Google Scholar]
- 172.Veal CD, Reekie KE, Lorentzen JC, Gregersen PK, Padyukov L, Brookes AJ. 2014. A 129-kb deletion on chromosome 12 confers substantial protection against rheumatoid arthritis, implicating the gene SLC2A3. Hum. Mutat 35:248–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Verpoort KN, Cheung K, Ioan-Facsinay A, van der Helm-van Mil AH, de Vries-Bouwstra JK, et al. 2007. Fine specificity of the anti-citrullinated protein antibody response is influenced by the shared epitope alleles. Arthritis Rheum 56:3949–52 [DOI] [PubMed] [Google Scholar]
- 174.Verpoort KN, Papendrecht-van der Voort EA, van der Helm-van Mil AH, Jol-van der Zijde CM, van Tol MJ, et al. 2007. Association of smoking with the constitution of the anti-cyclic citrullinated peptide response in the absence of HLA-DRB1 shared epitope alleles. Arthritis Rheum 56:2913–18 [DOI] [PubMed] [Google Scholar]
- 175.Verpoort KN, van Gaalen FA, van der Helm-van Mil AH, Schreuder GM, Breedveld FC, et al. 2005. Association of HLA-DR3 with anti-cyclic citrullinated peptide antibody-negative rheumatoid arthritis. Arthritis Rheum 52:3058–62 [DOI] [PubMed] [Google Scholar]
- 176.Viatte S, Plant D, Bowes J, Lunt M, Eyre S, et al. 2012. Genetic markers of rheumatoid arthritis susceptibility in anti-citrullinated peptide antibody negative patients. Ann. Rheum. Dis 71:1984–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Viatte S, Plant D, Han B, Fu B, Yarwood A, et al. 2015. Association of HLA-DRB1 haplotypes with rheumatoid arthritis severity, mortality, and treatment response. JAMA 313:1645–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wang J, Bansal AT, Martin M, Germer S, Benayed R, et al. 2013. Genome-wide association analysis implicates the involvement of eight loci with response to tocilizumab for the treatment of rheumatoid arthritis. Pharmacogenom. J 13:235–41 [DOI] [PubMed] [Google Scholar]
- 179.Wang Y, Shaked I, Stanford SM, Zhou W, Curtsinger JM, et al. 2013. The autoimmunity-associated gene PTPN22 potentiates toll-like receptor-driven, type 1 interferon-dependent immunity. Immunity 39:111–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wordsworth P, Lanchbury JS, Sakkas LI, Welsh KI, Panayi GS, Bell JI. 1989. HLA-DR4 subtype frequencies in rheumatoid arthritis indicate that DRB1 is the major susceptibility locus within the HLA class II region. PNAS 86:10049–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wordsworth P, Pile KD, Buckely JD, Lanchbury JS, Oilier B, et al. 1992. HLA heterozygosity contributes to susceptibility to rheumatoid arthritis. Am. J. Hum. Genet 51:585–91 [PMC free article] [PubMed] [Google Scholar]
- 182.Yang J, Lee SH, Goddard ME, Visscher PM. 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet 88:76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yelamos J, Garcia-Lozano JR, Moreno I, Aguilera I, Gonzalez MF, et al. 1993. Association of HLA-DR4-Dw15 (DRB1*0405) and DR10 with rheumatoid arthritis in a Spanish population. Arthritis Rheum 36:811–14 [DOI] [PubMed] [Google Scholar]
- 184.Yu KH, See LC, Kuo CF, Chou IJ, Chou MJ. 2013. Prevalence and incidence in patients with autoimmune rheumatic diseases: a nationwide population-based study in Taiwan. Arthritis Care Res 65:244–50 [DOI] [PubMed] [Google Scholar]
- 185.Zheng X, Shen J, Cox C, Wakefield JC, Ehm MG, et al. 2014. HIBAG—HLA genotype imputation with attribute bagging. Pharmacogenom. J 14:192–200 [DOI] [PMC free article] [PubMed] [Google Scholar]