SUMMARY
CRISPR-Cas9-mediated gene editing has enabled the direct manipulation of gene function in many species. However, the reproductive biology of reptiles presents unique barriers for the use of this technology, and there are no reptiles with effective methods for targeted mutagenesis. Here, we demonstrate that the microinjection of immature oocytes within the ovaries of Anolis sagrei females enables the production of CRISPR-Cas9-induced mutations. This method is capable of producing F0 embryos and hatchlings with monoallelic or biallelic mutations. We demonstrate that these mutations can be transmitted through the germline to establish genetically modified strains of lizards. Direct tests of gene function can now be performed in Anolis lizards, an important model for studies of reptile evolution and development.
In Brief
The reproductive biology of reptiles makes accessing single-cell embryos difficult and presents a major barrier for deploying gene-editing technologies in these species. Rasys et al. report that the microinjection of Cas9 RNP into immature lizard oocytes enables the production of lizards with targeted mutations.
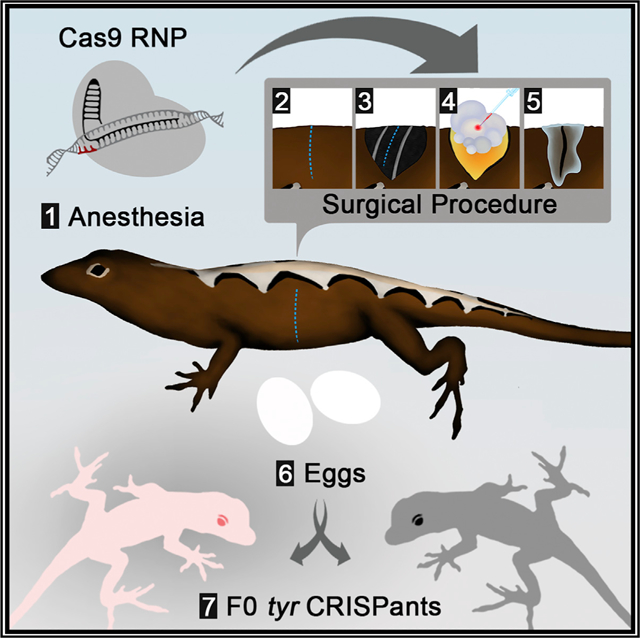
Graphical Abstract
INTRODUCTION
Squamates (lizards and snakes) comprise a diverse group of reptiles represented by >10,000 recognized species (Uetz and Stylianou, 2018). However, mechanistic studies of gene function in squamates and other reptiles lag behind other major vertebrate groups. While the adoption of CRISPR-Cas9-mediated gene editing has enabled the direct manipulation of gene function in many fish (Ansai and Kinoshita, 2014; Hwang et al., 2013), amphibian (Blitz et al., 2013; Flowers et al., 2014), avian (Oishi et al., 2016), and mammalian species (Honda et al., 2015; Wang et al., 2013), there remain no reptilian model systems with established methods for the production of targeted sequence alterations. A major barrier for the production of genetically modified reptiles is accessing zygotes. Since the reproductive biology of reptiles makes the microinjection of single-cell embryos impractical, attempts to manipulate gene function in reptiles have been limited to a small number of studies using a whole-embryo culture coupled to viral- or electroporation-based methods to introduce transgenes (Nomura et al., 2015; Tschopp et al., 2014). These methods produce transient, localized, and highly mosaic patterns of transgenesis. Moreover, these techniques have not been used to engineer targeted gene modifications in any reptile species.
Among squamates, Anolis lizards are compelling candidates for the establishment of gene-editing methods. Over the past 50 years, anoles have become one of the central model systems for studies of reptile evolution, physiology, and development (Sanger and Kircher, 2017). This group has experienced an extensive adaptive radiation in the Caribbean, with hundreds of described species that display a wide range of morphological, behavioral, and physiological differences. Studies of the convergent evolution of similar sets of Anolis “ecomorphs,” or habitat specialists, on different Caribbean islands have produced a rich literature on the biology of Anolis lizards. Here, we demonstrate that the microinjection of CRISPR-Cas9 into unfertilized oocytes is an effective method to produce targeted mutations in the brown anole lizard Anolis sagrei. We anticipate that this approach can be applied to many species of reptiles.
RESULTS
Gene-Editing Strategy
CRISPR-Cas9-mediated genome editing is an effective method for producing genetically modified vertebrates (Komor et al., 2017). In the most common approach, CRISPR-Cas9 components are microinjected into vertebrate embryos at the one-cell stage to generate individuals potentially harboring alterations at the locus of interest. However, there are significant challenges associated with the microinjection of Anolis zygotes. These challenges include internal fertilization and the long-term storage of sperm within the oviducts of adult females, which makes timing the microinjection of single-cell embryos extremely difficult. At the time of ovulation, Anolis eggs are also quite large (~8 mm in length) and are filled with substantial amounts of yolk; these oocytes are fragile and are difficult to manipulate without rupturing. Furthermore, after fertilization, the eggshell must be deposited around the egg, and embryonic development is initiated before the egg is laid (Sanger et al., 2008b). Finally, unlike the hard shells of birds, the eggshells that enclose Anolis embryos are pliable, and no air space is present within the egg, presenting obstacles for embryo manipulation within the eggshell. Most of these reproductive challenges for microinjection are not unique to anoles, but are features that are typical of many reptiles.
To circumvent the challenges associated with accessing Anolis zygotes, we developed an approach in which CRISPR-Cas9 is injected into unfertilized oocytes. Although many Anolis species can be successfully raised in the lab, we chose to develop our genome-editing method in the brown anole lizard, A. sagrei. This invasive lizard is now found far beyond its native Caribbean range and is ideal for genetic studies due to its small size, ease of husbandry, long breeding season, and relatively short generation time. Reproductively active A. sagrei females lay ~1 egg every week, similar to other anole species (Andrews, 1985). Each ovary contains a series of ~10 maturing ovarian follicles arranged by size, with the smallest follicle closest to the germinal bed and the largest vitellogenic follicle positioned distally (Figure S1). With the exception of the largest follicle, the oocytes within the developing follicles are previtellogenic or in the early stages of vitellogenesis and display a similar range of sizes in the left and right ovaries.
In our approach, female lizards are anesthetized (Rasys et al., 2019) and are placed on a surgical platform underneath a standard dissecting scope. Left and right ovaries are separately accessed via vertical incisions positioned along the left or right flank, respectively (Figure 1). During surgery, the ovary can be gently moved to allow easy observation and injection of the oocytes under the dissecting microscope. Oocytes that are 0.75–5 mm in diameter are microinjected with Cas9 ribonucleoprotein complex (Cas9 RNP) while remaining associated with the ovary (Figures 1 and S1; Video S1). A typical anole ovary has 4–6 oocytes in this range. Therefore, ~10 oocytes in this size range can be injected per animal. Oocytes >5 mm in diameter are not injected due to the increased risk of rupturing these large, yolk-filled oocytes. After the microinjection of oocytes is completed on one side, the incision is sealed with veterinary glue. The procedure is then repeated on the opposite side. Following recovery, the microinjected oocytes continue to mature within the female and are eventually ovulated and fertilized through natural mating with an introduced male or via stored sperm from previous matings.
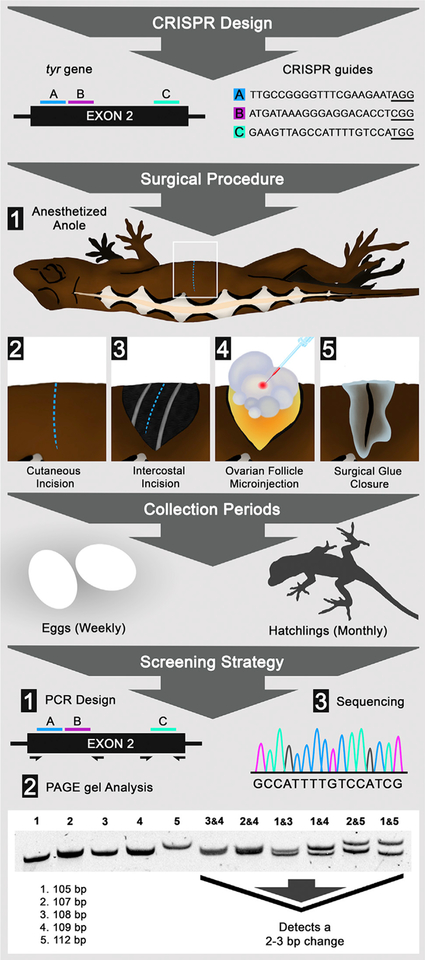
Figure 1. Gene Editing in Lizards through Microinjection of Ovarian Follicles.
Flow diagram detailing CRISPR design, surgical procedure, collection periods, and screening strategy. CRISPR design shows the placement and sequence of CRISPR guides A (blue), B (pink), and C (cyan) within exon 2 of the tyr gene; protospacer adjacent motif (PAM) sites are underlined. Surgical procedure: lizard anesthesia and the surgical steps to access and microinject ovary follicles. Collection periods: the time between gathering eggs and raising hatchlings. Screening strategy: the steps used to detect tyr crispants, including (1) PCR primer design; (2) PAGE analysis, which can reliably detect 2–3 bp changes; and (3) Sanger sequencing.
CRISPR-Induced Mutations at the tyr Locus
To assess the effectiveness of our approach, we targeted the second exon of the tyrosinase (tyr) gene. Tyrosinase was chosen for this study because loss-of-function mutations are viable in a wide range of vertebrates, the resulting pigmentation phenotypes are readily detected, and we wished to develop a new Anolis model to investigate eye defects associated with human albinism. Cas9 protein coupled to a mixture of three different synthetic tyr guide RNAs was injected into immature oocytes. The decision to simultaneously inject three guide RNAs was motivated by the presence of a number of single-nucleotide polymorphisms (SNPs) in tyr exon 2 within the population of lizards used for these experiments. For these experiments, a total of 146 oocytes from 21 reproductively active females were microinjected over the course of 8 surgical sessions.
We obtained 9 F0 animals harboring mutations in tyr exon 2 (Figure 2). Four of these animals were phenotypically albino and harbored loss-of-function mutations at both tyr alleles. Five animals carried heterozygous loss-of-function mutations and exhibited normal pigmentation. Mutant alleles could be visualized by PAGE after PCR amplification across tyr exon 2 using genomic DNA prepared from F0 embryos or hatchlings as a template. The changes associated with each of the CRISPR-Cas9-induced mutations were determined by sequencing the amplicons from each animal and comparing the sequence to that of wild-type lizards in the colony. The overall mutation frequency in terms of mutant lizards per follicle injected was 6.2%. A mutation frequency of 9.7% was obtained from microinjected follicles that were 1.5–2.5 mm in diameter, while follicles 0.75–1.0 mm and 3–5 mm in diameter yielded frequencies of 9.3% and 5.6%, respectively (Figure S2). No mutations were obtained from microinjection into follicles <0.5 mm in diameter. Consistent with the results in other vertebrates, CRISPR-Cas9 genome editing in lizards resulted in indels that ranged in size from 3–17 bp.
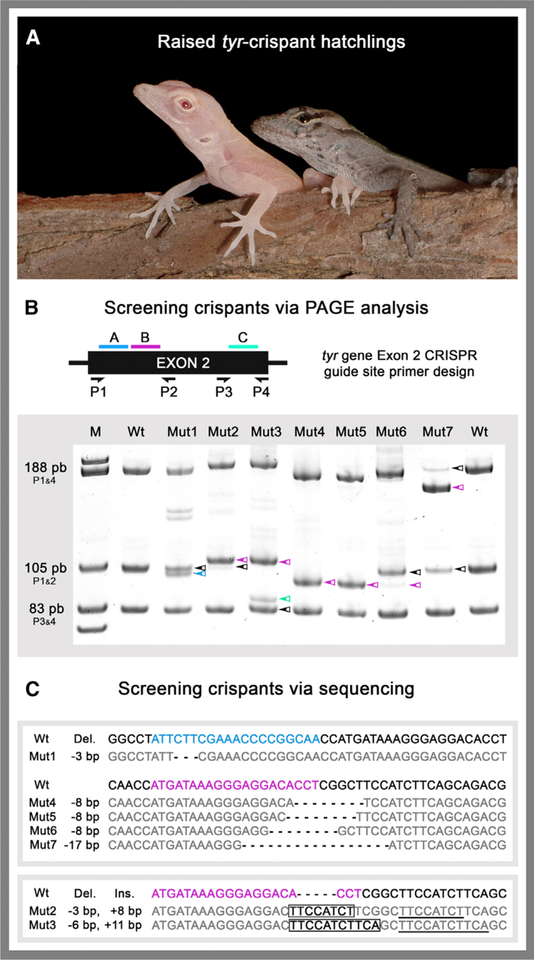
Figure 2. Detection and Sequencing of tyr Crispants.
(A) Albino tyr crispant (left) and wild-type (right) age-matched hatchlings.
(B) PCR primer placement (P1–P4) relative to CRISPR target sites A (blue), B (pink), and C (cyan). Representative PAGE results are shown for seven of the mutant lizards. Colored arrows denote bands with altered mobility relative to wild type (WT).
(C) Sequences of CRISPR-Cas9-induced indels from representative tyr crispants. (Top) Mut1 and Mut4–7 sequences with deletions. (Bottom) Mut2 and Mut3 sequences with insertions. Targeted guide sites A (blue), B (pink), and C (cyan) are highlighted in WT reference sequences. tyr crispants deletions are indicated in bold, insertions boxed, and sequences matching WT in gray.
See also Figure S2.
Germline Transmission of a tyr Mutation
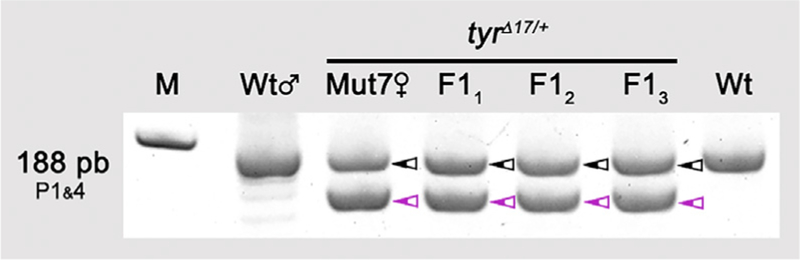
To establish stable lines of genetically modified anoles, it is necessary to transmit mutations through the germline. Therefore, we mated a wild-type male lizard to an F0 female lizard that was heterozygous for a 17-bp deletion in tyr exon 2 (Figures 2 B and 2C; F0 female is designated Mut7). PAGE genotyping revealed that the first three offspring from this mating pair inherited the tyrΔ17 allele from the mother (Figure 3). Thus, stable lines can be established from brown anole lizards that carry CRISPR-induced mutations generated via the microinjection of oocytes.
Figure 3. Germline Transmission of a CRISPR-Induced tyr Mutation.
PCR genotyping for the presence of WT and Δ17 tyr alleles. M, marker; Wt♂, tyr+/+ father; Mut7♀, tyrΔ17/+ F0 mother; F11–3, tyrΔ17/+ F1 offspring; Wt, tyr+/+ control; black arrowhead, tyr+ allele, magenta arrowhead, tyrΔ17 allele.
DISCUSSION
While CRISPR-based gene editing has been reported in a host of different species, the generation of non-mosaic mutant animals or the germline transmission of mutations requires that CRISPR components be introduced into the appropriate cell type. Adoption of CRISPR technology in mouse, chick, Xenopus, and zebrafish has been rapid, in part due to the existence of well-established methods for the manipulation of fertilized eggs, early-stage embryos, and germ cells of these species. Experience in these canonical model systems has clearly aided the expansion of CRISPR gene editing to other, related species. In contrast, there has historically been much less work on methods to culture or manipulate reptilian eggs and embryos. Moreover, there is no reptile model around which a large community of developmental biologists has coalesced.
Many of the reproductive features that pose challenges for gene editing in reptiles are also characteristics that make developmental studies less convenient to perform in these animals (e.g., seasonal breeding, opaque eggs shells, and an inability to precisely time fertilization). Our results demonstrate that microinjection of CRISPR-Cas9 into immature oocytes can generate F0 lizards carrying monoallelic or biallelic mutations in a targeted gene without the need to manipulate embryos. Thus, our approach offers an effective path for directly testing gene requirement in anoles. The production of F0 offspring with biallelic mutations was somewhat unexpected because it may take several days or weeks for microinjected oocytes to mature and be fertilized by sperm. Our results suggest that active Cas9 RNP can perdure long enough to target the paternal allele after fertilization occurs. For genes in which loss-of-function mutations are homozygous lethal or for studies requiring heterozygous animals, we expect that naturally occurring SNPs can be used to facilitate the targeting of one allele only. In our research colony, female A. sagrei generally reach reproductive age by 5 months and males by 6–7 months. This generation time is short enough that it should be feasible for mutations of interest to be maintained through breeding, allowing detailed investigations of mutant phenotypes.
We anticipate that the gene-editing strategy we have used in anoles can also be successfully applied to many other squamates. Moreover, the microinjection of immature oocytes may also provide a viable approach for gene editing in avians. As with squamates, accessing and injecting avian zygotes is technically challenging. A small number of studies in chickens have shown that targeted mutations can be introduced in chicken primordial germ cells (PGCs) grown in culture, circumventing the need to manipulate zygotes (Dimitrov et al., 2016; Oishi et al., 2016; Park et al., 2014). In this alternative approach, the modified PGCs can be introduced into the bloodstream of a host chicken embryo, where they migrate into the gonads of the host, contribute to the germ cell population, and can eventually transmit mutations by breeding the host animal. In comparison, the microinjection of CRISPR reagents into immature oocytes may provide a more direct method to create genetically modified strains of chickens. Furthermore, since optimal PGC culture conditions differ between species, the injection of immature oocytes may prove to be a more expedient approach for gene editing in other, less studied avian species in which PGC culture conditions have not been established.
The editing efficiency that we observed at the A. sagrei tyr locus (6%) is relatively low compared to CRISPR efficiencies that have been reported in other vertebrates (Burger et al., 2016; Flowers et al., 2014). However, all of our crispants were obtained from Anolis oocytes >0.75 mm in diameter, while no CRISPR-induced mutations were generated from injections of smaller oocytes. Therefore, focusing injection efforts on larger oocytes, which are also easier to inject, may be a simple way to improve gene-editing efficiency in Anolis lizards. While sequencing control animals and F0 animals from CRISPR injections, we also noted that some animals carried alleles with naturally occurring SNPs at tyr guide sites A and B. These variants are likely to be resistant to the guide RNAs that we used and may have reduced our editing efficiency. Therefore, we recommend surveying target sites for polymorphisms before selecting target sites or designing PCR primers for screening F0 offspring. When targeting the tyr locus, we simultaneously injected three guide RNAs in an attempt to increase the probability of inducing targeted mutations. However, this may also increase the chance of off-target events. Although we did not assess the frequency of off-target mutations in this study, in silico selection of guide sites with the fewest predicted off-target matches and the use of high-fidelity Cas9 variants should reduce the chances of inducing unintended mutations (Haeussler et al., 2016; Vakulskas et al., 2018).
Even with the modest efficiencies we obtained in this study, 1 day of microinjection surgeries can be expected to yield 2–4 mutants (surgeries on 5 females can be completed in a typical day, with 10 oocytes injected per female). As injection volumes and reagent concentrations are further optimized, it is reasonable to expect that gene-editing efficiency will improve. Moreover, mutations carried by F0 animals can be transmitted through the germline to establish strains of lizards that carry defined mutations. The establishment of CRISPR-Cas9 editing in this inexpensive reptilian system will finally permit mechanistic studies of gene function to be performed in reptiles.
STAR⋆METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Douglas Menke (dmenke@uga.edu). Anolis tyr mutants generated in this study will be made available upon request, subject to the successful establishment of breeding stocks at the University of Georgia.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Animals used in this study were wild-caught Anolis sagrei from Orlando, FL. Lizards were housed at University of Georgia following published guidelines (Sanger et al., 2008a). Breeding cages housed up to 4 females and 1 male together. Twenty-one adult females from cages that consistently produced eggs were selected for this study. All experiments followed the National Research Council’s Guide for the Care and Use of Laboratory Animals and were performed with the approval and oversight of the University of Georgia Institutional Animal Care and Use Committee (A2016 09-008-Y2-A3).
METHOD DETAILS
Selection of crRNA guide sequences and preparation of Cas9 RNP
The CRISPOR target selection tool (version 4.4) was used to select target regions with efficiency scores of 50% or greater within the second exon of the A. sagrei tyr gene (Haeussler et al., 2016). The tyr gene reference sequence was obtained from a draft genome assembly of Anolis sagrei. Alt-R CRISPR-Cas9 crRNAs, tracrRNA, and Cas9 V3 were purchased from Integrated DNA Technologies, Inc. The crRNA target sequences were as follows:
AsagTyrEx2A: 5′-TTGCCGGGGTTTCGAAGAAT-3′
AsagTyrEx2B: 5′-ATGATAAAGGGAGGACACCT-3′
AsagTyrEx2C: 5′-GAAGTTAGCCATTTTGTCCA-3′
Cas9 RNP was prepared by following manufacturer recommendations. A 5 μM injection solution was made using a standard microinjection solution (10 mM Tris-HCl, pH 7.4) containing phenol red to help verify that injected solutions entered the oocytes.
Anesthesia and analgesia
Lizards were anesthetized by administering 30 mg/kg of Alfaxalone (Alfaxan, 10 mg/mL, Jurox) in combination with 0.1 mg/kg of dexmedetomidine (Dexdomitor 5 mg/10 mL, Zoetis/Orion) (Rasys et al., 2019). To ensure accurate dosing, these drugs were administered by subcutaneous injection in the cervical area as an Alfaxalone/Dexmedetomidine (A/D) mixture. Preoperative analgesia was obtained by subcutaneous injection of meloxicam (0.3 mg/kg, Loxiject, 5 mg/mL, Henry Schein) in the dorsal epaxial area just above the shoulder and topical application of lidocaine (2.0 mg/kg, Lidocaine HCL, 2%, Hospira) over the surgical site. See video for injection sites. The anesthetic combination A/D provided approximately 30 min of surgical anesthesia time. Lizards typically recovered about 40–45 min following A/D administration. If a longer anesthesia time was required, a second dose of 30 mg/kg of alfaxalone alone was administered 25–30 min post induction dose, providing an additional 30 min of surgical anesthesia.
The method and location where injections were made were specifically chosen to avoid some of the challenges with administering drugs to reptiles. One such issue to be aware of is the hepatic-first pass effect which is a phenomenon found in many reptiles where, drugs, if administered in hindlimb or caudal regions, are rapidly cleared by the ventral abdominal and hepatic portal veins and metabolized by the liver, inhibiting wide systemic circulation. We have found that administering A/D subcutaneously in the dorsal epaxial area just above the shoulder region results in only moderate to light levels of anesthesia. Contrary to this, A/D administered subcutaneously in the cervical area are rapidly induced and reach a surgical plane of anesthesia within 1 min. Because injection volumes can be large, a subcutaneous method was also preferred over an intramuscular route of injection. For these reasons, the cervical area and a subcutaneous route were used in this study.
Another important factor that can influence drug metabolism in reptiles is body temperature. Lizards are ectothermic and are dependent upon the environmental temperature to regulate their body warmth which in turn impacts their metabolic rate. A decrease in body temperature will lead to a decrease in drug metabolism potentially resulting in a persistence of circulating drugs. In such an instance, this can result in an animal responding poorly, prolongment of anesthesia recovery time, or in some cases lead to death. The converse is also true. Animals maintained at too high a temperature may metabolize and clear drugs quickly resulting in insufficient anesthesia time. To avoid this, anesthetized anoles should be maintained on a heating source at around 32°C until fully recovered.
Surgery and microinjection
After successful anesthesia induction, the lizard becomes non-responsive to any noxious stimuli (i.e., an absence of response to a cloacal/tail clamp that normally induces severe discomfort). The anesthetized lizard was placed into right lateral recumbency and the left flank was aseptically prepared by alternating disinfection with 70% ethanol and 7.5% povidone-iodine (Surgical Scrub Solution, 16 fl. oz. 473 mL, Dynarex) wipes for 5 minutes.
Following standard surgical practices, sterile iris scissors (FST, item 15023–10) were used to make an 8–10 mm vertical cutaneous incision on the left side, in the mid-coelom region. A second incision between the ribs was made through the musculature (i.e., internal/external intercostal and pigmented coelom muscle layers) to enter the coelom. The ovary can be found dorsally in the midcoelom region and was easily accessible by shifting intestines gently aside using blunt forceps (FST: 45° angled forceps, item 00649–11; FST: strait forceps, item 00632–11). Once located, the ovary was carefully rotated and repositioned to expose immature follicles ranging anywhere from 0.25 mm to 5 mm in size.
Using the blunt forceps to clasp and hold the ovary in place, a microinjection needle was visually guided into the follicle center at an angle between 35–45° degrees relative to horizontal. 5 μM Cas9 RNP solution was then injected into follicles at differing volumes ranging from as little as 15 nL to as much as 575 nL which was dependent upon needle and follicle size. Retrospectively, ideal injection volumes were determined (3 ≤ 5 mm, 300–500 nL; 2 ≤ 2.5 mm, 200–250 nL; 1 ≤ 1.5 mm, 100–150 nL; and 0.75 mm, 25 nL) based on surgical sessions that produced mutants. Large yolky follicles greater than 5 mm in diameter, and eggs already present in the oviduct, were not injected. Sterile drops of P-Lytes solution (Veterinary Plasma-Lyte A Injection pH 7.4) were applied directly on the ovary or in coeliotomy opening throughout the procedure to prevent tissue dehydration.
After injection, the ovary was gently returned into the coelom and overlying musculature and skin was lightly pulled together to close the cavity. Tissue adhesive (3M Vetbond, #1469SB) was carefully applied only to only the external surface of the skin, avoiding the underlying musculature. Once the tissue adhesive was dry, the lizard was re-positioned into left lateral recumbency and the procedure was repeated for a right coeliotomy.
During recovery, triple antibiotic ointment (Bacitracin Zinc, Neomycin Sulfate, Polymyxin B Sulfate) was applied topically to the surgical wounds. Lizards were monitored daily for 1 week for any signs of infection, pain, or inflammation. After recovery from anesthesia, females were housed together with their previous female mates and allowed to recover for 7 days prior to reintroducing the male.
All surgeries were performed using sterilized equipment and tools (i.e., forceps and iris scissors) under a dissecting scope (Zeiss Stemi SV11) with a top light (AmScope 80-LED illuminator). Body temperature was maintained throughout the procedure by placing lizards on a heating platform (Fisher Scientific: model 77, serial # 802N0041CAT 12–594) with surgical towels draped between the heat source and the lizards to provide a barrier. The contact surface temperature was held at 32°C and readings continuously taken using thermometer strips. Each laparotomy was performed within 10–12 minutes. Follicle injections were carried out using a standard zebrafish/Xenopus microinjection rig (Harvard Apparatus PLI-100 Pico-Injector) set at 20 PSI with an injection time of 50–60 msecs. Initially, a manual micromanipulator (Märzhaüser Wetzlar; MMJ-rechts: 00-42-107-0000) was used to perform steady needle injections. However, use of this limited the degrees of freedom to inject the ovary from multiple directions and angles, therefore, a simple hand-guided technique using no micromanipulator was ultimately preferred and proved to be more efficient. Injection needles with a gradual taper typically used for zebrafish microinjection were made following the Sutter Instrument Company Pipette Cookbook guidelines using a Flaming/Brown Micropipette Puller (Model P-97) and cut to have a 20 to 40 μm diameter opening.
Mutation screening
Cages were monitored for a specified number of weeks following surgery which was based on the highest number of follicles injected in an ovary (e.g., if 8 and 5 follicles were injected in the right and left ovaries of one lizard, respectively, a cage housing this lizard would be monitored for n = 16 weeks). Because these females often had 1 or 2 eggs in the oviduct as well as 2 large (> 5mm) un-injected large follicles, cages were monitored for an additional 3–4 weeks following surgery.
Embryos and hatchlings from surgery cages were screened via PCR PAGE analysis under conditions that reliably detect a 2–3bp change (VanLeuven et al., 2018). DNA was extracted from tail clips from hatchlings or from tissue collected from embryos following standard protocols. PCR was performed using the following primers: P1, 5-CAAGAACTTTGCAATGGAACAAATG-3′; P2, 5′-GAATTCAACGTCTGCTGAAGATG-3′; P3, 5′-TGTTTAAGTCTGACTCAGTACGAAG-3′; P4, 5′-GGATTACCTTCCAAAGTATTCCTG-3′. See Figure 2 for primer location relative to targeting sites. Sanger sequencing was performed on PCR products to fully characterize mutations.
QUANTIFICATION AND STATISTICAL ANALYSES
Follicle train assignment for quantification of gene targeting efficiencies
Targeting efficiencies were calculated based on the number of follicles injected over a given size range and the number of resulting mutants arising from those injections. To determine what size of injected follicle a mutant likely originated from, we had to use the timing that egg lay occurred relative to when the microinjection procedure occurred (using the logic that larger follicles will be laid sooner than smaller follicles). Follicles were ranked by size and by alternating left and right ovarian contributions to infer a probable timeline of egg lay for a given follicle size at time of injection. This method of follicle train assignment assumes 1) eggs present in the oviduct will be laid within one week, 2) follicles greater than 8–10 mm in 2 weeks, and 3) follicles less than or equal to 5mm in diameter will be laid no sooner than 3 weeks. Our reason for including these 3 underlying assumptions derives from the observation that females who had 1–2 eggs present in their oviduct, also had a follicle greater than 8–10 mm in diameter followed by a follicle between 3–5 mm in each ovary, suggesting at least a week interval between these sizes. As each lizard possesses a “leading” ovary and “lagging” ovary in follicle sizes, the leading ovary is given preferential ordering in train position. It is important to note that this method of ordering does not account for any potential loss of follicles accidently destroyed in the microinjection process and assumes that if such an event occurred, the follicle developmental timeline of that ovary is unaffected. See Figure S2 for a graphical depiction of follicle train assignment.
DATA AND CODE AVAILABILITY
The published article includes all data analyzed for this study.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides, and Recombinant Proteins | ||
| Alt-R® S.p. Cas9 Nuclease V3, 500 μg | Integrated DNA Technologies | Cat#1081059 |
| Experimental Models: Organisms/Strains | ||
| Anolis sagrei - wild caught | Orlando, FL | N/A |
| Oligonucleotides | ||
| Alt-R® CRISPR-Cas9 tracrRNA, 20 nmol | Integrated DNA Technologies | Cat#1072533 |
| Alt-R® CRISPR-Cas9 crRNA, 2 nmol Target: AsagTyrEx2A: 5′-TTGCCGGG GTTTCGAAGAAT-3′ | Integrated DNA Technologies | N/A |
| Alt-R® CRISPR-Cas9 crRNA, 2 nmol Target: AsagTyrEx2B: 5′-ATGATAAAGG GAGGACACCT-3′ | Integrated DNA Technologies | N/A |
| Alt-R® CRISPR-Cas9 crRNA, 2 nmol Target: AsagTyrEx2C: 5′-GAAGTTAGCC ATTTTGTCCA-3′ | Integrated DNA Technologies | N/A |
| Primer P1, 5-CAAGAACTTTGCAATGG AACAAATG-3′ | Integrated DNA Technologies | N/A |
| Primer P2, 5′-GAATTCAACGTCTGCTG AAGATG-3′ | Integrated DNA Technologies | N/A |
| Primer P3, 5′-TGTTTAAGTCTGACTCAGT ACGAAG-3′ | Integrated DNA Technologies | N/A |
| Primer P4, 5′-GGATTACCTTCCAAAGTAT TCCTG-3′ | Integrated DNA Technologies | N/A |
Highlights.
Microinjection of Anolis oocytes with Cas9 RNP can induce targeted mutations
F0 lizards from microinjected oocytes can carry monoallelic or biallelic mutations
CRISPR-Cas9-induced mutations can be transmitted through the lizard germline
ACKNOWLEDGMENTS
We thank S. Divers for guidance on reptile anesthesia and surgical techniques and J. Eggenschwiler and C. Sabin for comments on this manuscript. This work was funded by National Science Foundation awards 1149453 to D.B.M. and 1827647 to D.B.M. and J.D.L. and a Society for Developmental Biology Emerging Models grant to A.M.R. A.M.R. was supported by NIH training grant T32GM007103 and by an ARCS Foundation Scholarship.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.07.089.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Andrews RM (1985). Oviposition frequency of Anolis carolinensis. Copeia 1, 259–262. [Google Scholar]
- Ansai S, and Kinoshita M (2014). Targeted mutagenesis using CRISPR/Cas system in medaka. Biol. Open 3, 362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz IL, Biesinger J, Xie X, and Cho KWY (2013). Biallelic genome modification in F(0) Xenopus tropicalis embryos using the CRISPR/Cas system. Genesis 51, 827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger A, Lindsay H, Felker A, Hess C, Anders C, Chiavacci E, Zaugg J, Weber LM, Catena R, Jinek M, et al. (2016). Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development 143, 2025–2037. [DOI] [PubMed] [Google Scholar]
- Dimitrov L, Pedersen D, Ching KH, Yi H, Collarini EJ, Izquierdo S, van de Lavoir M-C, and Leighton PA (2016). Germline gene editing in chickens by efficient CRISPR-mediated homologous recombination in primordial germ cells. PLoS One 11, e0154303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers GP, Timberlake AT, McLean KC, Monaghan JR, and Crews CM (2014). Highly efficient targeted mutagenesis in axolotl using Cas9 RNA-guided nuclease. Development 141, 2165–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud J-B, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda A, Hirose M, Sankai T, Yasmin L, Yuzawa K, Honsho K, Izu H, Iguchi A, Ikawa M, and Ogura A (2015). Single-step generation of rabbits carrying a targeted allele of the tyrosinase gene using CRISPR/Cas9. Exp. Anim 64, 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh J-RJ, and Joung JK (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol 31, 227–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Badran AH, and Liu DR (2017). CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168, 20–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T, Yamashita W, Gotoh H, and Ono K (2015). Genetic manipulation of reptilian embryos: toward an understanding of cortical development and evolution. Front. Neurosci 9, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi I, Yoshii K, Miyahara D, Kagami H, and Tagami T (2016). Targeted mutagenesis in chicken using CRISPR/Cas9 system. Sci. Rep 6, 23980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TS, Lee HJ, Kim KH, Kim J-S, and Han JY (2014). Targeted gene knockout in chickens mediated by TALENs. Proc. Natl. Acad. Sci. USA 111, 12716–12721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasys AM, Divers SJ, Lauderdale JD, and Menke DB (2019). A systematic study of injectable anesthetic agents in the brown anole lizard (Anolis sagrei). Lab. Anim Published online July 26, 2019. 10.1177/0023677219862841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger TJ, and Kircher BK (2017). Model clades versus model species: Anolis lizards as an integrative model of anatomical evolution. Methods Mol. Biol 1650, 285–297. [DOI] [PubMed] [Google Scholar]
- Sanger TJ, Hime PM, Johnson MA, Diani J, and Losos JB (2008a). Laboratory protocols for husbandry and embryo collection of Anolis lizards. Herpetol. Rev 39, 58–63. [Google Scholar]
- Sanger TJ, Losos JB, and Gibson-Brown JJ (2008b). A developmental staging series for the lizard genus Anolis: a new system for the integration of evolution, development, and ecology. J. Morphol 269, 129–137. [DOI] [PubMed] [Google Scholar]
- Tschopp P, Sherratt E, Sanger TJ, Groner AC, Aspiras AC, Hu JK, Pourquié O, Gros J, and Tabin CJ (2014). A relative shift in cloacal location repositions external genitalia in amniote evolution. Nature 516, 391–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P, and Stylianou A (2018). The original descriptions of reptiles and their subspecies. Zootaxa 4375, 257–264. [DOI] [PubMed] [Google Scholar]
- Vakulskas CA, Dever DP, Rettig GR, Turk R, Jacobi AM, Collingwood MA, Bode NM, McNeill MS, Yan S, Camarena J, et al. (2018). A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med 24, 1216–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanLeuven AJ, Park S, Menke DB, and Lauderdale JD (2018). A PAGE screening approach for identifying CRISPR-Cas9-induced mutations in zebrafish. Biotechniques 64, 275–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, and Jaenisch R (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all data analyzed for this study.