Abstract
Focal segmental glomerulosclerosis (FSGS) is a histological pattern of podocyte and glomerulus injury. FSGS can be primary and secondary to other diseases or due to a genetic cause. Strikingly, genetic causes for adult-onset FSGS are often overlooked, likely because identifying patients with genetic forms of FSGS based on clinical presentation and histopathology is difficult. Yet diagnosing genetic FSGS does not only have implications for prognostication and therapy but also for family and family planning. In this case series, we present 3 adult patients who presented with advanced renal disease with the histological picture of FSGS and proved to have a genetic cause of the disease, namely, variants in INF2, COL4A4 and HNF1B, respectively. We show the possibilities of identifying genetic FSGS based on clinical clues of a positive family history, early age at onset of disease, and/or severe therapy-resistant disease. We discuss ways to select the method of genetic testing for individual patients. Finally, we examine how the judicious use of genetic investigations can obviate potential harmful diagnostic procedures and direct clinical decisions in patients and their relatives.
Keywords: Focal segmental glomerulosclerosis, Kidney biopsy, Genetics, Gene panel, INF2, COL4A4, HNF1B
Background
With the advances in genetic testing methods, genetic analysis is an increasingly important diagnostic tool in nephrology [1]. This is also the case for genetic focal segmental glomerulosclerosis (FSGS), which is the focus of this paper.
FSGS is a histological pattern of podocyte loss and glomerular injury. It is characterized in a renal biopsy, by segmental sclerotic lesions in at least one glomerulus (observed with light microscopy) and effacement of the podocyte foot processes (observed with electron microscopy [EM]) [2, 3]. The underlying causes for FSGS are heterogeneous [4, 5].
FSGS is traditionally categorized according to those underlying causes, namely, primary (often involves a circulating factor causing podocyte dysfunction) and secondary to a nonrenal disease and genetic FSGS [4, 6]. Depending on the underlying cause, the patients can present with proteinuria, or nephrotic syndrome (most in primary FSGS), and end-stage renal disease (ESRD), or progress to ESRD over the course of 5–10 years [7].
There are no clear-cut clinical or histopathological findings to distinguish genetic FSGS from other types [8]. However, there are several hallmarks of genetic disease. Namely a positive family history, early age at onset of disease (~30% of FSGS with an onset before 25 years of age is genetic), and uncharacteristically severe and/or steroid-resistant disease [8, 9, 10, 11]. Conversely, because genetic disease often presents at a young age, it is often unjustly overlooked in adult-onset FSGS patients [11].
With the advances of genetic testing, however, diagnosing genetic FSGS has become much more feasible over the past few years. Not only because over 50 genes are currently known to be involved in FSGS, but also since the costs and turn-around time for genetic tests are continuously dropping, increasing their availability in daily clinical practice [8, 11, 12, 13, 14, 15].
The technique most frequently used for genetic testing is next-generation sequencing (NGS) [8, 11, 12, 13, 14]. NGS can identify disease-causing mutations in the entire genome (whole-genome sequencing), the protein-coding regions (whole-exome sequencing), or a specific set of genes of interest (targeted gene panel [TGP]) [16]. For instance, the TGP on FSGS in online supplemental Table 1 (for all online suppl. material, see www.karger.com/doi/10.1159/000499937) contains the classic FSGS genes NPHS1 and NPHS2 as well as genes recently associated with FSGS such as the COL4A3–5 genes (the Alport syndrome genes) and PAX2 (involved in nephrogenesis). Selecting the right NGS test is essential, to be able to come to a diagnosis with limited risk of the incidental findings that testing many genes (e.g., whole-exome sequencing) can bring.
Despite the abovementioned challenges, considering a genetic cause in adult-onset FSGS patients is important as it can have a large impact on the patient and his/her family members. Here, we present 3 patients with adult-onset chronic kidney disease who were clinically and histopathologically diagnosed with FSGS and were shown to carry a genetic cause thanks to a close collaboration between nephrologists, pathologists, and clinical geneticists. We use these cases to discuss the expanding possibilities of diagnosing genetic FSGS and the clinical implications of such a diagnosis.
Case 1: FSGS with ESRD at a Young Age
A 30-year-old man with asymptomatic 2 g/day proteinuria at age 20 and ESRD at age 29 (no signs of nephrotic syndrome, Table 1) was referred to our nephrogenetics out-patient clinic. There was no family history of renal disease. Renal biopsy at age 29, when the patient developed ESRD, showed FSGS (Fig. 1a), with 80% globally sclerosed glomeruli and partial podocyte foot process effacement (Fig. 1d) [17]. The patient was referred because he was planned to undergo a kidney transplant from a family member.
Table 1.
Age at first presentation, laboratory findings, and morphological findings per case
| Case number | Age at first presentation, years | Positive family history | Clinical diagnosis | eGFR at presentation (CKD-EPI) [47], mL/min/ 1.73m2 | Laboratory analysis at presentation | Renal ultrasound results | Light microscopy | Immunofluorescence microscopy | Electron microscopy | Histological classification [17] |
|---|---|---|---|---|---|---|---|---|---|---|
| Case 1 UMCU_ NG_ 012_01 | 20 | No | Secondary FSGS | <20 | Blood Albumin normal Lipids normal PT and APTT normal | Echodense kidneys, otherwise no abnormalities. Length 9.9 and 9.8 cm (normal). Changes likely due to CKD | FSGS with 80% glomerulosclerosis | No immunoreactivity | Partial podocyte effacement | FSGS NOS |
| Urine Protein (2 g/day) | ||||||||||
| Case 2 UMCU_ NG_ 044_01 | 50 | Yes | Secondary FSGS | 90 | Blood Albumin normal Triglycerides high PT and APTT normal | No abnormalities. Length 12.5 and 11.6 cm (normal) | FSGS with 50% glomerulosclerosis | A specific immunoreactivity for IgA and IgM | Partial podocyte effacement Thin basement membrane (mean 172 nm) |
FSGS NOS |
| Urine Protein (1.6 g/day) 30 erythrocytes/μL | ||||||||||
| Case 3 UMCU_ NG_ 100_01 | 33 | Yes | FSGS, etiology unknown | 39 | Blood Albumin normal Triglycerides high PT and APTT normal | No abnormalities. Length 10.2 and 10.5 cm (normal) | FSGS with 45% glomerulosclerosis | No immunoreactivity | No material | FSGS NOS |
| Urine Protein (0.6 g/day) | ||||||||||
APTT, activated partial thromboplastin time; CKD, chronic kidney disease; eGFR, electronic glomerular filtration rate; FSGS, focal segmental glomerulosclerosis; Ig, immunoglobulin; NOS, not otherwise specified; PT, prothrombin time; SRNS, steroid-resistant nephrotic syndrome.
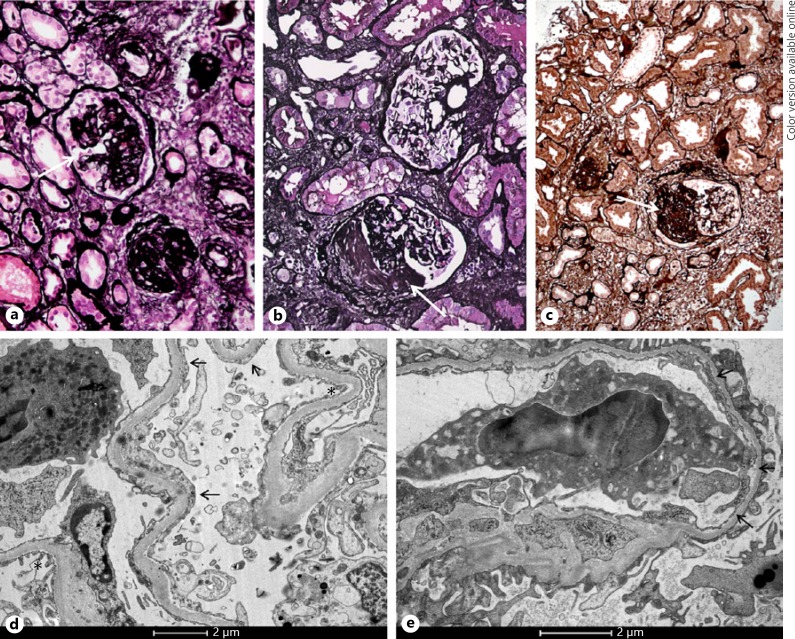
Fig. 1.
Kidney biopsy images in the 3 cases. Light microscopy (Jones staining) showed glomeruli with segmental sclerosis (arrows) in case 1 (a), case 2 (b), and case 3 (c). Electron microscopy of case 1 showed partial foot process effacement, with areas of intact foot processes (*) alternating with areas with foot process effacement (arrows, d). In addition to partial foot process effacement, EM of case 2 also showed a thin GBM thickness with a mean of 252 nm (arrows, e).
Due to the young age of onset of proteinuria in this patient, there was a marked probability of genetic FSGS, and a diagnostic TGP analysis for FSGS was performed (online suppl. Methods 1 and Table 1). This revealed a heterozygous known pathogenic mutation in the INF2 gene (OMIM610982, Table 2) [18, 19, 20, 21]. The mutation had been previously detected in FSGS patients, though one should note that no functional assessment of that specific mutation was performed [18]. Mutations in INF2 are known to be a major cause for autosomal dominant FSGS [22, 23, 24].
Table 2.
Molecular diagnosis, including the performed genetic testing and information on the genetic variant, per case
| Case number | Genetic testing performed | HGNC-approved gene name (transcript number) | OMIM number | Variant | Homozygous or heterozygous | Variant type | Reference/in silico predictions [18, 19, 20, 21] |
|---|---|---|---|---|---|---|---|
| Case 1 UMCU_ NG_ 012_01 | FSGS | INF2 (NM_022489.3) | 610982 | c.217G>A p.(Gly73Ser) | Heterozygous | Pathogenic | Barua et al. [18] (no functional analysis of this variant) PolyPhen HumDiv score 1.000, sensitivity 0.00, specificity 1.00 |
| Polyphen HumVar score 1.000, sensitivity 0.00, specificity 1.00 | |||||||
| SIFT score 0.13 (tolerated) Not present in the gnomAD database | |||||||
| Case 2 UMCU_ | FSGS | COL4A4 (NM 000092.4) | 12131 | c.2038G>C p.(Gly680Arg) | Heterozygous | Likely pathogenic | PolyPhen HumDiv score 1.000, sensitivity 0.00, specificity 1.00 |
| NG_ 044_01 | Polyphen HumVar score 1.000, sensitivity 0.00, specificity 1.00 | ||||||
| SIFT score 0.00 (deleterious) Not present in the gnomAD database | |||||||
| Case 3 UMCU_ | FSGS | HNF1B (NM 000458.3) | 189907 | c.908G>A p.(Arg303His) | Heterozygous | VUS | PolyPhen HumDiv score 0.998, sensitivity 0.27, specificity 0.99 |
| NG_ 100_01 | PAX2 Sanger sequencing | PolyPhen HumVar score 0.877, sensitivity 0.71, specificity 0.89 | |||||
| Full diagnostic renal diseases (‘RENome') | SIFT score 0.04 (deleterious) Not present in the gnomAD database | ||||||
Arg, arginine; del, deletion; FSGS, focal segmental glomerulosclerosis; Glu, glutamic acid; Gly, glycine; HGNC, HUGO gene nomenclature committee; His, histidine; OMIM, online Mendelian inheritance in man®; Ser, serine; VUS, variant of unknown significance.
To adequately counsel family members, segregation analysis was performed in the patient's healthy parents. The father did not carry the mutation and later successfully donated a kidney to our patient. In the otherwise healthy mother, a 20% mosaicism for the INF2 mutation was detected in DNA from peripheral blood. The mother was referred for extensive health screening, which revealed no abnormalities. Since she had had a son with INF2 mutation, it must therefore be present in the germline and thus possibly have been passed down to the patient's siblings. One sibling decided on testing (revealing no INF2 mutation) and one decided to undergo periodic evaluation of renal function. The patient's young child will be counseled regarding presymptomatic genetic testing when it is of age. As the earliest presentation reported in literature is at 7 years of age, the child will undergo proteinuria screening [25].
Next to the implications for family members, the molecular diagnosis impacted the patient's care directly. Mutations in INF2 can also be associated with dominant intermediate Charcot-Marie-Tooth disease, thus the patient was neurologically evaluated, showing no abnormalities [26]. Additionally, the patient and his partner wanted to have more children. After counseling, they opted to try to conceive via preimplantation genetic diagnostics, an in vitro fertilization procedure where an embryo without the INF2 mutation is transferred into the uterus [27]. At time of this publication, this has not yet led to an ongoing pregnancy.
Case 2: FSGS with a Family History of ESRD
A 50-year-old obese woman (BMI 34) of Hindustani Surinam descent (Table 1) presented in the referring hospital with mild chronic kidney disease (eGFR = 90), distinct proteinuria (1.6 g/day, no signs of nephrotic syndrome), and erythrocyturia of 30 cells/µL. Her parents had ESRD, both with an age of onset around 60 years, of which the father was diagnosed as having diabetic nephropathy. In our patient, renal biopsy displayed FSGS (Fig. 1b), with 50% globally sclerosed glomeruli, thought to be secondary to a metabolic syndrome. However, because of the erythrocyturia, the referring nephrologist wondered if COL4A3–5-related disease (mutations in these genes are detected in patients with thin basement membrane nephropathy and classical Alport syndrome) might play a role in this patient's phenotype.
To assess this possibility, the renal biopsy was revised with EM. This showed a thin GBM with a mean thickness of 172 nm, (Fig. 1e), which was well below the lower limit of 252 nm determined in our center for normal GBM thickness for females and also below the lower limit of 215 nm for the normal thickness for females reported in literature, further pointing toward COL4A3–5-related disease [28, 29]. Therefore, the diagnostic TGP analysis on FSGS was performed (online suppl. Methods and Table 1). This analysis includes the COL4A genes, since mutations in these genes have been shown to cause a histological FSGS phenotype in some cases [28, 30, 31, 32, 33, 34]. The TGP analysis showed a heterozygous likely pathogenic mutation in the COL4A4 gene (OMIM120131, Table 2), with no variants in other FSGS-linked genes [19, 20, 21].
COL4A4 codes for the type IV collagen alpha-4 chain, a protein essential to the GBM [35]. Heterozygous mutations in COL4A4 have been associated with familial hematuria [36]. There are reports suggesting that specific mutations in COL4A4 or unknown genetic modifiers might cause FSGS lesions in heterozygous carriers, while others suggest that heterozygous COL4A3–5 mutations are the most frequent underlying cause in patients with FSGS on biopsy [37, 38, 39, 40]. It is clear that the penetrance of renal disease in carriers of heterozygous COL4A3–4 mutations is far from complete [37, 38, 39, 40]. There is debate over whether this is best called autosomal dominant Alport syndrome, or for example, COL4A3–4-related disease [37, 38, 39, 40].
The specific mutation detected in our patient has not been described as pathogenic before. However, the variant causes the substitution of a highly conserved glycine residue in the collagen triple-helix repeat by a more bulky amino acid (Table 2). Based on the fact that most known pathogenic mutations in COL4A4 lead to similar substitutions, the mutation was classified as “likely pathogenic.” Segregation analysis was performed, and the mother (no diabetes) proved to be a carrier for the same mutation. The presence of the COL4A4 variant in 2 affected family members, along with erythrocyturia and a thin GBM, likely explains at least a part of our patient's COL4A3–5-related disease phenotype. With this, it is important to note that people of Hindustani Surinam descent are known to have higher risk of metabolic syndrome, which likely also played a role in this family's renal phenotype(s) [37, 41].
Genetic counseling was offered to the patient's children. Furthermore, the finding of a COL4A4 likely pathogenic variant triggered the referring nephrologist to prescribe Lisinopril, as the patient needed antihypertensive medication and ACE-inhibition is also used to attenuate renal function decline in Alport syndrome [42].
Case 3: “IgA-Related FSGS” with a Family History of ESRD
An otherwise healthy 33-year-old man presented with an eGFR of 39 and proteinuria (0.6 g/day, no signs of nephrotic syndrome). The family history revealed that the mother had died with ESRD at age 50, most likely due to hypodysplastic kidneys. Renal ultrasound in the patient showed no abnormalities and normal sized kidneys (Table 1). In the referring hospital, renal biopsy was classified as FSGS secondary to IgA depositions. The patient wondered if he could pass on the disease to his children.
Biopsy revision at our facility showed FSGS (Fig. 1c) with 45% of glomeruli globally sclerosed, but no immunoreactivity for IgA. There was not enough material to perform EM. Since the diagnosis of IgA nephropathy was doubtful, genetic diagnostics using the FSGS TGP analysis was performed (online suppl. Methods 1 and Table 1). This did not lead to a molecular diagnosis. Due to the high clinical suspicion, the analysis was expanded to a larger panel of ~225 published renal genes. This revealed a heterozygous variant of unknown significance in the HNF1B gene (OMIM189907, Table 2) [19, 20, 21, 43].
The variant had not been observed before in patients or large healthy control populations, in silico predictions suggest a possible pathogenic effect (Table 2), and the variant segregated in the patient's deceased parent. Laboratory work-up in our patient for glucose, electrolyte, and liver enzyme imbalances associated with HNF1B-related disease showed no clear abnormalities; however, genotype-phenotype correlations can be unclear [44, 45]. The HNF1B variant might thus be causal in our patient's disease and the mother's renal hypodysplasia. This is underscored by studies showing that HNF1B works as a modifier on PAX2, in which gene mutations are known to cause both isolated congenital anomalies of the kidney and urinary tract (CAKUT, such as hypodysplasia) as well as FSGS [46, 47, 48]. Also, mutations in HNF1B sometimes cause a CAKUT phenotype without abnormalities in other organs [46, 47]. Hence, it could be that mutations in HNF1B also lead to FSGS. Publication of this, to our knowledge first ever, case will hopefully stimulate further research into the HNF1B-FSGS relationship.
Though the patient cannot be conclusively diagnosed, the combination of the variant and the positive family history has led to all at-risk family members receiving advice for periodic evaluation of renal function.
Discussion
The cases presented in this paper show that although the identification of a genetic cause for FSGS presenting at an adult age can be complex, an adequate diagnosis can have far-reaching implications. That the cases were rediagnosed as genetic FSGS is due to the multidisciplinary approach with input from a nephrologist, pathologist, and clinical geneticist. These specialists discussed the possibility of genetic disease and the appropriate application of genetic testing for each patient individually. We discuss the items at the core of this discussion in detail below.
First, it is vital to recognize that though patient characteristics can give clues on patients with high risk of a genetic disease, not all patients display those hallmarks of genetic disease [8, 9, 11]. Similar to the INF2 case we presented, a family history might be absent due to germline mosaicism, or mutations that are recessive, de novo or incompletely penetrant [14]. Additionally, though a young age at presentation is an indication of genetic disease, our COL4A4 patient presented at 50 years of age [9]. The notion that genetic renal disease can present later in life is underscored by our recent finding that the classic pediatric disease nephronophthisis actually can present with ESRD to up to 61 years [49].
Second, one should consider the appropriate NGS scale for each patient. In order to test a sufficient number of genes without risk of incidental findings, we apply a tiered approach, starting with the analysis of TGPs that are limited to strictly FSGS-associated genes. If a limited TGP does not yield a diagnosis, one can opt to analyze a larger panel (as we did for our HNF1B case), or to perform whole-exome sequencing to look for variants in genes not yet associated with the patient's phenotype. To make such a step-up process even easier, we decided in 2017 to derive all TGP analyses from whole-exome sequencing data. Adequate pre- and posttest counseling (described by our group previously [38]) regarding analyses of the whole-exome data should be offered to patients, as these can reveal incidental findings.
With the continuous decrease in cost and turn-around time of NGS, the precise selection of patients and a step-up NGS method will likely become less of a question [15]. However, genetic testing should always be applied after consideration of the prognostic and therapeutic implications of finding a genetic variant for the patient and his/her family members.
For the patient, it can provide information on useful treatment strategies. Though genetic FSGS generally does not respond to corticosteroid treatment, other drugs might be beneficial, such as ACE-inhibition in COL4A-related disease [42, 50, 51]. Furthermore, a molecular diagnosis is relevant when deliberating on a renal transplantation. First, because it usually offers a favorable prognosis with respect to recurrence in a renal graft, since chances of this are very low in genetic FSGS [52]. Second, if living-related transplantation is considered, it is safest to have a genetically unaffected family member donate [53]. For this reason, we tested the INF2 patient's parent before proceeding to donation.
Family members are impacted, as they are at risk of also developing FSGS. Those at risk should be offered counseling on genetic testing and/or (presymptomatic) evaluation of renal function [53]. Likewise, future children of a genetic FSGS patient could inherit the disease. It is our experience that the knowledge that the disease is genetic is very important for patients when contemplating how to establish their family. As we saw in our INF2 case, the options for not passing the disease on not only include having less or no children but also advanced techniques such as preimplantation genetic diagnostic, when locally available [54].
In conclusion, the cases presented in this paper show that a genetic diagnosis in adult-onset FSGS can have far-reaching consequences not only for the patient but also for his/her family (planning). Identification of patients with a higher likelihood of a genetic FSGS often proves challenging, though there are several hallmarks of genetic disease. Currently, we apply a tiered method to genetic testing, to limit incidental findings. In the future, a genetic-first approach could obviate invasive renal biopsies [55]. The probability of a monogenic disease and the potential impact of a genetic diagnosis should be considered in the diagnostic work-up of all adult-onset FSGS cases.
Statement of Ethics
The patients described in this paper have given their informed written consent for their anonymized data to be included in this study. In the Netherlands, there is no need for Institutional Review Board permission to publish anonymized, retrospective patient data; therefore, no such permission was sought.
Disclosure Statement
The authors declare no conflicts of interest, financial or otherwise.
Supplementary Material
Supplementary data
References
- 1.Groopman EE, Rasouly HM, Gharavi AG. Genomic medicine for kidney disease. Nat Rev Nephrol. 2018 Feb;14((2)):83–104. doi: 10.1038/nrneph.2017.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Agati V. Pathologic classification of focal segmental glomerulosclerosis. Semin Nephrol. 2003 Mar;23((2)):117–134. doi: 10.1053/snep.2003.50012. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz MM, Korbet SM. Primary focal segmental glomerulosclerosis pathology, histological variants, and pathogenesis. Am J Kidney Dis. 1993 Dec;22((6)):874–83. doi: 10.1016/s0272-6386(12)70349-9. [DOI] [PubMed] [Google Scholar]
- 4.Fogo AB. Causes and pathogenesis of focal segmental glomerulosclerosis. Nat Rev Nephrol. 2015 Feb;11((2)):76–87. doi: 10.1038/nrneph.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiggins RC. The spectrum of podocytopathies a unifying view of glomerular diseases. Kidney Int. 2007 Jun;71((12)):1205–14. doi: 10.1038/sj.ki.5002222. [DOI] [PubMed] [Google Scholar]
- 6.Yang HC, Fogo AB. ‘Idiopathic' FSGS an increasingly obsolete diagnosis? Nephrol Dial Transplant. 2010 Mar;25((3)):654–6. doi: 10.1093/ndt/gfp750. [DOI] [PubMed] [Google Scholar]
- 7.Korbet SM. Treatment of primary FSGS in adults. J Am Soc Nephrol. 2012 Nov;23((11)):1769–76. doi: 10.1681/ASN.2012040389. [DOI] [PubMed] [Google Scholar]
- 8.De Vriese AS, Sethi S, Nath KA, Glassock RJ, Fervenza FC. Differentiating Primary, and Secondary. FSGS in Adults A Clinicopathologic Approach. J Am Soc Nephrol. 2018 Mar;29((3)):759–74. doi: 10.1681/ASN.2017090958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vivante A, Hildebrandt F. Exploring the genetic basis of early-onset chronic kidney disease. Nat Rev Nephrol. 2016 Mar;12((3)):133–46. doi: 10.1038/nrneph.2015.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollak MR. Familial FSGS. Adv Chronic Kidney Dis. 2014 Sep;21((5)):422–5. doi: 10.1053/j.ackd.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lepori N, Zand L, Sethi S, Fernandez-Juarez G, Fervenza FC. Clinical and pathological phenotype of genetic causes of focal segmental glomerulosclerosis in adults. Clin Kidney J. 2018 Apr;11((2)):179–90. doi: 10.1093/ckj/sfx143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sprangers B, Meijers B, Appel G., FSGS Diagnosis and Diagnostic Work-Up. BioMed Res Int. 2016;2016:4632768. doi: 10.1155/2016/4632768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woroniecki RP, Kopp JB. Genetics of focal segmental glomerulosclerosis. Pediatr Nephrol. 2007 May;22((5)):638–44. doi: 10.1007/s00467-007-0445-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hildebrandt F. Genetic kidney diseases. Lancet. 2010 Apr;375((9722)):1287–95. doi: 10.1016/S0140-6736(10)60236-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Nimwegen KJ, van Soest RA, Veltman JA, Nelen MR, van der Wilt GJ, Vissers LE, et al. Is the $1000 Genome as Near as We Think? A Cost Analysis of Next-Generation Sequencing. Clin Chem. 2016 Nov;62((11)):1458–64. doi: 10.1373/clinchem.2016.258632. [DOI] [PubMed] [Google Scholar]
- 16.Muzzey D, Evans EA, Lieber C. Understanding the Basics of NGS From Mechanism to Variant Calling. Curr Genet Med Rep. 2015;3((4)):158–65. doi: 10.1007/s40142-015-0076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D'Agati VD, Fogo AB, Bruijn JA, Jennette JC. Pathologic classification of focal segmental glomerulosclerosis a working proposal. Am J Kidney Dis. 2004 Feb;43((2)):368–82. doi: 10.1053/j.ajkd.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 18.Barua M, Brown EJ, Charoonratana VT, Genovese G, Sun H, Pollak MR. Mutations in the INF2 gene account for a significant proportion of familial but not sporadic focal and segmental glomerulosclerosis. Kidney Int. 2013 Feb;83((2)):316–22. doi: 10.1038/ki.2012.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60, 706 humans. Nature. 2016 Aug;536((7616)):285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng PC, Henikoff S. SIFT predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003 Jul;31((13)):3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010 Apr;7((4)):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown EJ, Schlöndorff JS, Becker DJ, Tsukaguchi H, Tonna SJ, Uscinski AL, et al. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet. 2010 Jan;42((1)):72–6. doi: 10.1038/ng.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyer O, Benoit G, Gribouval O, Nevo F, Tête MJ, Dantal J, et al. Mutations in INF2 are a major cause of autosomal dominant focal segmental glomerulosclerosis. J Am Soc Nephrol. 2011 Feb;22((2)):239–45. doi: 10.1681/ASN.2010050518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian B, Sun H, Yan P, Charoonratana VT, Higgs HN, Wang F, et al. Mice with mutant Inf2 show impaired podocyte and slit diaphragm integrity in response to protamine-induced kidney injury. Kidney Int. 2016 Aug;90((2)):363–72. doi: 10.1016/j.kint.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee HK, Han KH, Jung YH, Kang HG, Moon KC, Ha IS, et al. Variable renal phenotype in a family with an INF2 mutation. Pediatr Nephrol. 2011 Jan;26((1)):73–6. doi: 10.1007/s00467-010-1644-5. [DOI] [PubMed] [Google Scholar]
- 26.Mathis S, Funalot B, Boyer O, Lacroix C, Marcorelles P, Magy L, et al. Neuropathologic characterization of INF2-related Charcot-Marie-Tooth disease evidence for a Schwann cell actinopathy. J Neuropathol Exp Neurol. 2014 Mar;73((3)):223–33. doi: 10.1097/NEN.0000000000000047. [DOI] [PubMed] [Google Scholar]
- 27.Traeger-Synodinos J. Pre-implantation genetic diagnosis. Best Pract Res Clin Obstet Gynaecol. 2017 Feb;39:74–88. doi: 10.1016/j.bpobgyn.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 28.Miner JH. Pathology vs. molecular genetics (re)defining the spectrum of Alport syndrome. Kidney Int. 2014 Dec;86((6)):1081–3. doi: 10.1038/ki.2014.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haas M. Alport syndrome and thin glomerular basement membrane nephropathy a practical approach to diagnosis. Arch Pathol Lab Med. 2009 Feb;133((2)):224–32. doi: 10.5858/133.2.224. [DOI] [PubMed] [Google Scholar]
- 30.Malone AF, Phelan PJ, Hall G, Cetincelik U, Homstad A, Alonso AS, et al. Rare hereditary COL4A3/COL4A4 variants may be mistaken for familial focal segmental glomerulosclerosis. Kidney Int. 2014 Dec;86((6)):1253–9. doi: 10.1038/ki.2014.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatterjee R, Hoffman M, Cliften P, Seshan S, Liapis H, Jain S. Targeted exome sequencing integrated with clinicopathological information reveals novel and rare mutations in atypical suspected and unknown cases of Alport syndrome or proteinuria. PLoS One. 2013 Oct;8((10)):e76360. doi: 10.1371/journal.pone.0076360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson J, Gilbert RD, Bunyan DJ, Angus EM, Fowler DJ, Ennis S. Exome analysis resolves differential diagnosis of familial kidney disease and uncovers a potential confounding variant. Genet Res. 2013 Dec;95((6)):165–73. doi: 10.1017/S0016672313000220. [DOI] [PubMed] [Google Scholar]
- 33.McCarthy HJ, Bierzynska A, Wherlock M, Ognjanovic M, Kerecuk L, Hegde S, et al. RADAR the UK SRNS Study Group. Simultaneous sequencing of 24 genes associated with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol. 2013 Apr;8((4)):637–48. doi: 10.2215/CJN.07200712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gast C, Pengelly RJ, Lyon M, Bunyan DJ, Seaby EG, Graham N, et al. Collagen (COL4A) mutations are the most frequent mutations underlying adult focal segmental glomerulosclerosis. Nephrol Dial Transplant. 2016 Jun;31((6)):961–70. doi: 10.1093/ndt/gfv325. [DOI] [PubMed] [Google Scholar]
- 35.Thorner PS, Zheng K, Kalluri R, Jacobs R, Hudson BG. Coordinate gene expression of the alpha3, and alpha5. chains of collagen type IV. Evidence from a canine model of X-linked nephritis with a COL4A5 gene mutation. J Biol Chem. 1996 Jun;271((23)):13821–8. doi: 10.1074/jbc.271.23.13821. [DOI] [PubMed] [Google Scholar]
- 36.Badenas C, Praga M, Tazón B, Heidet L, Arrondel C, Armengol A, et al. Mutations in the COL4A4 and COL4A3 genes cause familial benign hematuria. J Am Soc Nephrol. 2002 May;13((5)):1248–54. doi: 10.1681/ASN.V1351248. [DOI] [PubMed] [Google Scholar]
- 37.Savige J. Should We Diagnose Autosomal Dominant Alport Syndrome When There Is a Pathogenic Heterozygous COL4A3 or COL4A4 Variant? Kidney Int Rep. 2018 Aug;3((6)):1239–41. doi: 10.1016/j.ekir.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stokman MF, Renkema KY, Giles RH, Schaefer F, Knoers NV, van Eerde. AM. The expanding phenotypic spectra of kidney diseases insights from genetic studies. Nat Rev Nephrol. 2016 Aug;12((8)):472–83. doi: 10.1038/nrneph.2016.87. [DOI] [PubMed] [Google Scholar]
- 39.Voskarides K, Damianou L, Neocleous V, Zouvani I, Christodoulidou S, Hadjiconstantinou V, et al. COL4A3/COL4A4 mutations producing focal segmental glomerulosclerosis and renal failure in thin basement membrane nephropathy. J Am Soc Nephrol. 2007 Nov;18((11)):3004–16. doi: 10.1681/ASN.2007040444. [DOI] [PubMed] [Google Scholar]
- 40.Pierides A, Voskarides K, Athanasiou Y, Ioannou K, Damianou L, Arsali M, et al. Clinico-pathological correlations in 127 patients in 11 large pedigrees segregating one of three heterozygous mutations in the COL4A3/ COL4A4 genes associated with familial haematuria and significant late progression to proteinuria and chronic kidney disease from focal segmental glomerulosclerosis. Nephrol Dial Transplant. 2009 Sep;24((9)):2721–9. doi: 10.1093/ndt/gfp158. [DOI] [PubMed] [Google Scholar]
- 41.Krishnadath IS, Toelsie JR, Hofman A, Jaddoe VW. Ethnic disparities in the prevalence of metabolic syndrome and its risk factors in the Suriname Health Study a cross-sectional population study. BMJ Open. 2016 Dec;6((12)):e013183. doi: 10.1136/bmjopen-2016-013183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gross O, Licht C, Anders HJ, Hoppe B, Beck B, Tönshoff B, et al. Study Group Members of the Gesellschaft für Pädiatrische Nephrologie Early angiotensin-converting enzyme inhibition in Alport syndrome delays renal failure and improves life expectancy. Kidney Int. 2012 Mar;81((5)):494–501. doi: 10.1038/ki.2011.407. [DOI] [PubMed] [Google Scholar]
- 43.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 May;17((5)):405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verhave JC, Bech AP, Wetzels JF, Nijenhuis T., Hepatocyte Nuclear Factor 1β-Associated Kidney Disease More than Renal Cysts and Diabetes. J Am Soc Nephrol. 2016 Feb;27((2)):345–53. doi: 10.1681/ASN.2015050544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clissold RL, Hamilton AJ, Hattersley AT, Ellard S, Bingham C. HNF1B-associated renal and extra-renal disease-an expanding clinical spectrum. Nat Rev Nephrol. 2015 Feb;11((2)):102–12. doi: 10.1038/nrneph.2014.232. [DOI] [PubMed] [Google Scholar]
- 46.Nakayama M, Nozu K, Goto Y, Kamei K, Ito S, Sato H, et al. HNF1B alterations associated with congenital anomalies of the kidney and urinary tract. Pediatr Nephrol. 2010 Jun;25((6)):1073–9. doi: 10.1007/s00467-010-1454-9. [DOI] [PubMed] [Google Scholar]
- 47.Paces-Fessy M, Fabre M, Lesaulnier C, Cereghini S. Hnf1b and Pax2 cooperate to control different pathways in kidney and ureter morphogenesis. Hum Mol Genet. 2012 Jul;21((14)):3143–55. doi: 10.1093/hmg/dds141. [DOI] [PubMed] [Google Scholar]
- 48.Barua M, Stellacci E, Stella L, Weins A, Genovese G, Muto V, et al. Mutations in PAX2 associate with adult-onset FSGS. J Am Soc Nephrol. 2014 Sep;25((9)):1942–53. doi: 10.1681/ASN.2013070686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snoek R, van Setten J, Keating BJ, Israni AK, Jacobson PA, Oetting WS, et al. NPHP1 (Nephrocystin-1) Gene Deletions Cause Adult-Onset ESRD. J Am Soc Nephrol. 2018 Jun;29((6)):1772–9. doi: 10.1681/ASN.2017111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antignac C. Molecular basis of steroid-resistant nephrotic syndrome. Nefrologia. 2005;25(Suppl 2):25–28. [PubMed] [Google Scholar]
- 51.Hermle T, Schneider R, Schapiro D, Braun DA, van der Ven AT, Warejko JK, et al. GAPVD1 and ANKFY1 Mutations Implicate RAB5 Regulation in Nephrotic Syndrome. J Am Soc Nephrol. 2018 Aug;29((8)):2123–38. doi: 10.1681/ASN.2017121312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jungraithmayr TC, Hofer K, Cochat P, Chernin G, Cortina G, Fargue S, et al. Screening for NPHS2 mutations may help predict FSGS recurrence after transplantation. J Am Soc Nephrol. 2011 Mar;22((3)):579–85. doi: 10.1681/ASN.2010010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rood IM, Deegens JK, Wetzels JF. Genetic causes of focal segmental glomerulosclerosis implications for clinical practice. Nephrol Dial Transplant. 2012 Mar;27((3)):882–90. doi: 10.1093/ndt/gfr771. [DOI] [PubMed] [Google Scholar]
- 54.Swift O, Vilar E, Rahman B, Side L, Gale DP. Attitudes in Patients with Autosomal Dominant Polycystic Kidney Disease Toward Prenatal Diagnosis and Preimplantation Genetic Diagnosis. Genet Test Mol Biomarkers. 2016 Dec;20((12)):741–6. doi: 10.1089/gtmb.2016.0050. [DOI] [PubMed] [Google Scholar]
- 55.Münch J, Grohmann M, Lindner TH, Bergmann C, Halbritter J. Diagnosing FSGS without kidney biopsy - a novel INF2-mutation in a family with ESRD of unknown origin. BMC Med Genet. 2016 Oct;17((1)):73. doi: 10.1186/s12881-016-0336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data