Abstract
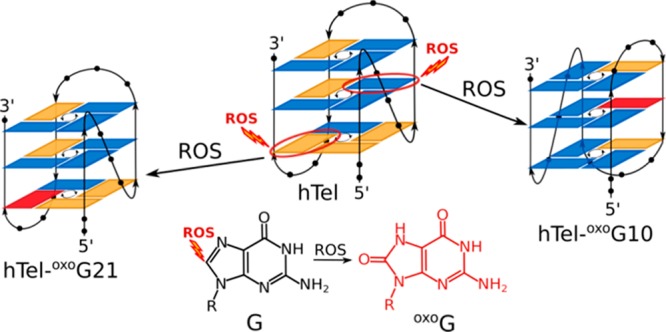
Telomere attrition is closely associated with cell aging and exposure to reactive oxygen species (ROS). While oxidation products of nucleotides have been studied extensively in the past, the underlying secondary/tertiary structural changes in DNA remain poorly understood. In this work, we systematically probed guanine positions in the human telomeric oligonucleotide sequence (hTel) by substitutions with the major product of ROS, 8-oxo-7,8-dihydroguanine (oxoG), and evaluated the G-quadruplex forming ability of such oligonucleotides. Due to reduced hydrogen-bonding capability caused by oxoG, a loss of G-quadruplex structure was observed for most oligonucleotides containing oxidative lesions. However, some positions in the hTel sequence were found to tolerate substitutions with oxoG. Due to oxoG’s preference for the syn conformation, distinct responses were observed when replacing guanines with different glycosidic conformations. Accommodation of oxoG at sites originally in syn or anti in nonsubstituted hTel G-quadruplex requires a minor structural rearrangement or a major conformational shift, respectively. The system responds by retaining or switching to a fold where oxoG is in syn conformation. Most importantly, these G-quadruplex structures are still stable at physiological temperatures and should be considered detrimental in higher-order telomere structures.
Introduction
Reactive oxygen species (ROS) are a byproduct of aerobic cellular metabolism in all living organisms. Cells possess enzymatic and non-enzymatic antioxidant mechanisms, which are able to manage normal levels of endogenous ROS. However, environmental factors such as pollutants and (non)ionizing radiation can cause spikes in ROS levels. The inability to neutralize excessive ROS results in oxidative stress, which has a damaging effect on cellular structures. All macro-molecular classes are affected by ROS and the major detrimental effects include lipid peroxidation, protein oxidation, and DNA damage.1,2
Oxidative stress causes many types of DNA lesions, and cells respond by activating relevant DNA repair pathways or apoptosis, if repair is unsuccessful. Most common DNA damages caused by ROS include single- and double-strand breaks, formation of apurinic/apyrimidinic sites, and DNA base substitutions. Among the four DNA nucleobases, guanine has the lowest redox potential and is therefore the easiest to oxidize.3,4 Furthermore, guanine tracts are even more susceptible to oxidation than isolated guanines.5,6 It has been suggested that G-rich regions serve as trapping sites for oxidative damage caused by one-electron oxidations via radical cation migration through the DNA duplex.7 Consequently, noncoding G-rich regions could protect the rest of the genome from mutagenesis induced by ROS. Numerous products resulting from guanine oxidation have been reported thus far of which 8-oxo-7,8-dihydroguanine (oxoG) is commonly used as a biomarker of oxidative stress.8,9 Due to its redox potential, which is lower than for the parent guanine, oxoG is preferentially oxidized even in the presence of a large excess of guanine.10
Telomeric regions are found at ends of chromosomes and protect the chromosome termini from deterioration or from fusion with neighboring chromosomes.11,12 Their length was found to diminish under conditions of high oxidative stress, for example, in diabetes patients.13,14 Due to their high G content, repetitive sequences in telomeres are able to fold into short four-stranded structures called G-quadruplexes. The integral part of a G-quadruplex is the G-quartet, which is a planar arrangement of four guanine nucleobases connected with Hoogsteen hydrogen bonds. Several G-quartets stack in order to form the G-quadruplex core. Contiguous guanine segments, called G-tracts, are connected by (usually) non-G nucleotides forming loops of different orientations contributing to tremendous structural polymorphism of G-quadruplexes.15,16
NMR and X-ray crystallographic studies found that oligonucleotide sequences containing four human telomeric repeats, d(T2AG3), lead to several distinct G-quadruplex topologies.17−26 In solutions containing K+ ions, these oligonucleotides were found to adopt so-called hybrid-1 and hybrid-2 folds, which feature three G-quartet planes with mixed parallel/antiparallel G-tract directionalities. In these (3 + 1) topologies, G-tracts are connected by two edgewise loops. The remaining first or third loop is of a double-chain-reversal type in hybrid-1 or hybrid-2, respectively. A 24-nt d[T2G3(T2AG3)3A] oligonucleotide (hTel) adopts a hybrid-1 type G-quadruplex fold in solutions containing K+ ions.21 In the current study, hTel was selected as a model system for a systematic survey of oxidation-related changes in G-quadruplex structure and stability.
Compared to regular G, the Watson–Crick edge of oxoG is unaltered, while N7 on its Hoogsteen edge is protonated and is a hydrogen-bond donor (Figure S1). Due to different hydrogen-bonding capabilities, replacing G with oxoG results in reduced stability of a G-quartet and consequently the G-quadruplex structure.27,28 Furthermore, incorporation of oxoG into a planar G-quartet is a potential source of steric clashes of H7 with the amino group of adjacent guanine. Several recent studies showed that it is favorable to exclude the lesion-containing G-tract from a G-quadruplex fold and replace it with a nearby lesion-free G-tract, if one is available in the same strand.29−32 Similarly, we have shown that a G-quadruplex structure adopted by the vascular endothelial growth factor (VEGF) promoter sequence is disrupted with the introduction of an oxoG lesion. However, the G-quadruplex fold can be recovered by adding a short pyrene conjugated lesion-free G-rich oligonucleotide.33
While reduced stabilities of G-quadruplex structures containing oxidative lesions are most likely observed due to alterations in the hydrogen-bonding network, the underlying structural changes have not been explored by high-resolution techniques to date. It has been shown that oxoG prefers the syn conformation in double-stranded DNA.34,35 We expect the same preference for syn conformation for oxoG in G-quadruplex structures. Furthermore, we hypothesize that the structural changes and consequent (de)stabilizing effects on G-quadruplex structures will depend on the position of oxoG substitution in the oligonucleotide sequence and whether a specific site corresponds to a guanine in anti or syn conformation in the parent G-quadruplex structure.
In the current study, we show that single substitutions of G with oxoG at distinct sites in the hTel sequence result in changes of the G-quadruplex structure. In the resulting structures, oxoG nucleotides were always found to exhibit a syn conformation and their corresponding G-quadruplexes conformed to hybrid-1 or hybrid-2 like topologies. Importantly, several structures based on the parent hTel and related oligonucleotides containing oxoG were found to be stable at physiological temperatures and could have implications for oxidative stress related changes in human telomeres.
Results
Analogues of hTel Containing oxoG Can Form G-Quadruplex Structures
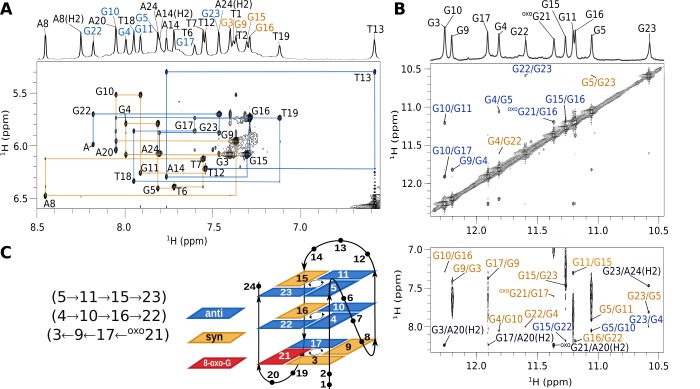
Guanines were individually substituted with oxoG at all 12 positions in the hTel sequence, and G-quadruplex forming ability was evaluated through examination of one-dimensional (1D) 1H NMR spectra (Figure S2). Well-resolved 1H signals were obtained when oxoG was substituted at positions 10 and 21. The spectrum of hTel-oxoG10 exhibits 12 guanine imino proton resonances in the region from δ 10.70 to 11.70 ppm and two additional broader resonances at δ 12.45 and 13.90 ppm, which were identified as imino signals of thymines (Figure 1). Eleven well-resolved imino proton resonances in the region from δ 10.61 to 12.29 ppm were observed in the spectrum of hTel-oxoG21. While chemical shift dispersion of imino proton resonances of hTel-oxoG21 is comparable to that of hTel, spectrum of hTel-oxoG10 shows a different chemical shift pattern. Spectra of both oligonucleotides with oxoG substitutions contain a set of low intensity peaks corresponding to minor species.
Figure 1.
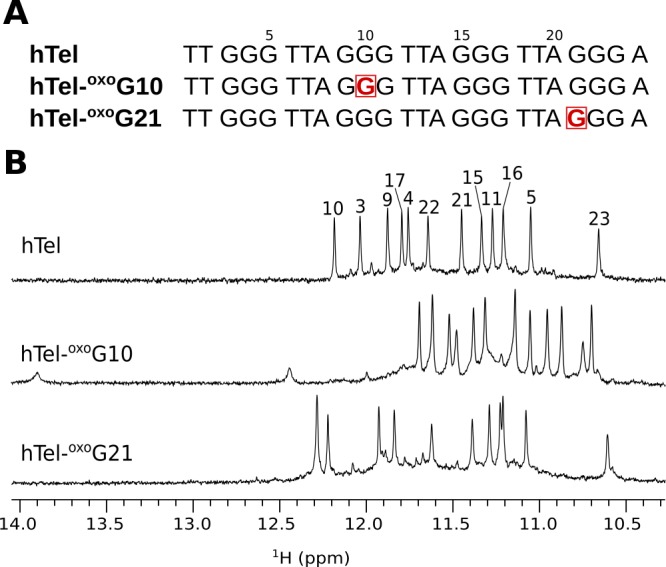
(A) Oligonucleotide sequences of hTel and its analogues with oxoG substitutions at positions 10 and 21 (shown squared in red color). (B) Imino regions of 1D 1H NMR spectra of hTel, hTel-oxoG10 and hTel-oxoG21. Imino proton assignment of hTel is shown on the top of the spectrum.21 Spectra were acquired in 90% H2O and 10% 2H2O, 70 mM KCl, and 20 mM K-phosphate buffer, pH 7, on an 800 MHz NMR spectrometer. Oligonucleotide concentrations were ∼0.5 mM.
Spectra of oligonucleotides with oxoG substitutions at the remaining 10 guanine positions exhibit broader resonances in imino and aromatic regions hindering a detailed structural analysis (Figure S2). However, an evaluation of imino regions of 1H spectra suggests that hTel-oxoG23 forms a single major species, while hTel-oxoG3, hTel-oxoG9, hTel-oxoG11, and hTel-oxoG17 form more than one structure in solution. Spectra of hTel-oxoG5 and hTel-oxoG15 exhibit several sharp imino resonances, but their low number suggests that in the major species, one of the G-quartets is destabilized. Spectra of remaining hTel-oxoG4, hTel-oxoG16 and hTel-oxoG22 containing broad resonances indicate destabilization of the G-quadruplex structure.
hTel-oxoG21 Retains the Parent G-Quadruplex Fold
Position 21 in the hTel sequence is located in the outer G-quartet of the parent G-quadruplex and contains a G in syn conformation along its glycosidic bond. Comparison of cross-peak fingerprints between NOESY spectra of hTel and hTel-oxoG21 revealed a great degree of similarity (Figure S3). All imino, aromatic and anomeric resonances of hTel-oxoG21 could be assigned by analyzing NOESY spectra (Figure 2 and Figure S3). Several nucleotides were residue-specifically 13C, 15N labeled and imino, and aromatic assignments were confirmed through acquisition of 15N and 13C-edited HSQC spectra (Figure S4A). Interestingly, H1 of oxoG21 gives a sharp NMR signal at δ 11.39 ppm, while its H7 resonance is not observed. Four intense H8–H1′ cross-peaks in NOESY spectrum acquired with a short mixing time of 80 ms were identified as G3, G9, G15, and G16 and were assigned a syn conformation along their glycosidic bonds. The complete sequential walk could not be traced throughout the oligonucleotide sequence due to large interproton distances at the following anti–syn steps: T2 (anti)–G3 (syn), A8 (anti)–G9 (syn), and A14 (anti)–G15 (syn). Furthermore, the lack of H8 resonance of oxoG21 interrupts the sequential walk at the A20 (anti)–oxoG21 (syn) step. However, oxoG21H1′-G22H8 cross-peak has been observed for this syn–anti step. Additionally, intense NOESY cross-peaks between H2′/H2′′ of oxoG21 and H8 of G22 indicating short distances correspond to a structure with oxoG21 in syn conformation. Furthermore, chemical shifts of H2′ and H2′′ of oxoG21 are δ 3.51 and 2.80 ppm, respectively. Such downfield chemical shift values are characteristic for H2′/H2′′ in syn nucleotides.36
Figure 2.
hTel-oxoG21 assignment of NOESY spectra and G-quadruplex topology. (A) Aromatic–anomeric region of a NOESY spectrum (τm = 250 ms). Assignments are shown next to H6/H8n-H1n′ cross-peaks. Orange lines connect cross-peaks from G3 to T12 and blue lines from T12 to A24. Letter “A” indicates the G22H8-oxoG21H1′ cross-peak. 1D trace of the NOESY spectrum is shown on the top with signal assignments, where assignments corresponding to syn and anti guanine nucleotides are shown in orange and blue, respectively. Adenine and thymine resonances are in black. (B) Imino–imino (top) and imino–aromatic (bottom) regions of the NOESY spectrum. Assignments in blue and orange represent cross-peaks between guanines of the inner and outer G-quartets, respectively. Assignments in black correspond to cross-peaks between H1 protons of guanines and aromatic protons of adenines. (C) hTel-oxoG21 G-quadruplex topology and hydrogen-bond directionality in G-quartets.
NOE connectivities between imino–imino and imino–aromatic protons were used to determine the fold of the G-quadruplex structure, which is comprised of G3·G9·G17·oxoG21, G4·G10·G16·G22, and G5·G11·G15·G23 quartets (Figure 2C). Central and top G-quartets exhibit anti–anti–syn–anti conformations as well as counterclockwise hydrogen-bond donor–acceptor directionalities. On the other hand, the bottom G-quartet exhibits syn–syn–anti–syn conformations and clockwise hydrogen-bond directionality. G-tracts are connected by TTA loops of which the first one (T6-T7-A8) adopts a double-chain-reversal topology, while the following two (T12-T13-A14 and T18-T19-A20) form edgewise loops. These structural features are consistent with the hybrid-1 topology adopted by the parent hTel G-quadruplex.
A solution-state structure of hTel-oxoG21 was determined on the basis of NMR data using a restrained simulated annealing (SA) protocol. Simulations utilized 287 NOE distance and 140 backbone torsion angle restraints. An additional 22 hydrogen-bond restraints were used for Hoogsteen base-pairs for all Gs involved in G-quartets. Only hydrogen bonds, where oxoG21 is the donor, were restrained, while hydrogen-bond restraints between G3 and oxoG21 were omitted. Eleven χ angle restraints for Gs (excluding oxoG21) were also used in SA simulations. A good convergence was achieved by running a series of 100 SA simulations. Twelve structures with the lowest energy of hTel-oxoG21 exhibit an RMSD of 0.26 Å (Table 1 and Figure 3A).
Table 1. Statistics of Structures of hTel-oxoG21 and hTel-oxoG10 G-Quadruplexes.
| hTel-oxoG21 | hTel-oxoG10 | |
|---|---|---|
| NMR Restrains | ||
| total NOE distance restraints | 287 | 329 |
| intranucleotide | 141 | 173 |
| internucleotide | 146 | 156 |
| sequential | 85 | 83 |
| long-range | 61 | 73 |
| hydrogen-bond restraints | 22 | 24 |
| glycosidic torsion angle restraints | 11 | 11 |
| backbone torsion angles restraints | 140 | 140 |
| Structure Statistics | ||
| violations | ||
| mean NOE restraint (Å) | 0.11 | 0.10 |
| max. NOE restraint (Å) | 0.21 | 0.17 |
| Deviations from the Idealized Geometry | ||
| bond lengths (Å) | 0.01 | 0.01 |
| torsion angles (deg) | 2.47 | 2.44 |
| Pairwise Heavy Atom RMSD (Å) | ||
| G-quartets | 0.26 | 0.56 |
| G-quartets and T6-A8 | 0.24 | 0.53 |
| G-quartets and T12-A14 | 0.25 | 0.63 |
| G-quartets and T18-A20 | 0.28 | 0.67 |
| G-quartets and T1-T2, A24 | 0.25 | 0.53 |
| all heavy atoms | 0.26 | 0.66 |
Figure 3.
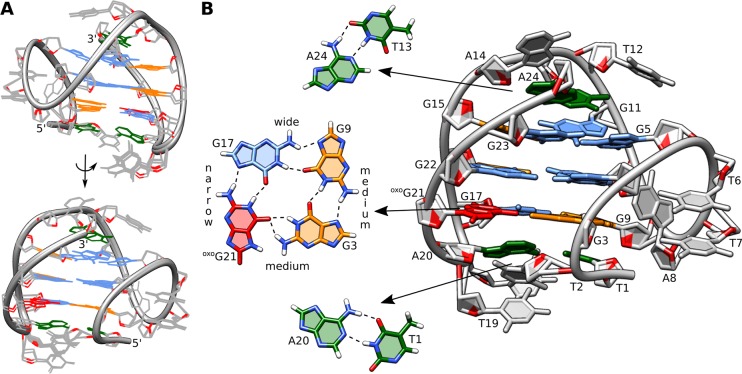
Details of the structure of hTel-oxoG21. (A) Superposition of 12 lowest energy structures. View into the wide and medium grooves. (B) Enlarged view into the medium groove. G-quartet with oxoG substitution, T13·A24 reversed Watson–Crick pair, and T1·A20 Watson–Crick pair are shown separately.
Our structure of hTel-oxoG21 reveals that oxoG21 is well tolerated in the G3·G9·G17·oxoG21 quartet (Figure 3B). Three of oxoG21’s atoms are involved in four hydrogen bonds with neighboring in-plane guanines. Two are with G17 (oxoG21H21-G17N7 and oxoG21H1-G17O6), typical for a Hoogsteen base-pair. Additionally, O6 of oxoG21 is a bifurcated hydrogen-bond acceptor for adjacent G3 (oxoG21O6-G3H1, 2.0 Å and oxoG21O6-G3H21, 1.9 Å). H7 of oxoG21 is slightly shifted from the G-quartet center and in close proximity (∼2.6 Å) to the amino group of G3. Since H7 is not involved in hydrogen bonding and is located in the outer G-quartet, the lack of its NMR resonance is likely due to efficient exchange with solvent.
Nucleobases T6, T7, and A8 are part of a double-chain-reversal loop with T6 and T7 stacking on each other, hiding their hydrophobic methyl groups from solvent. A8 nucleobase is oriented perpendicularly to the thymines. Nucleobases T12, T13, A14 and T18, T19, A20 form edgewise loops, which exhibit efficient stacking on the outer G-quartets (Figure 3). SA simulations suggest formation of T1·A20 and T13·A24 Watson–Crick base-pairs (Figure 3B), however, no imino resonance could be observed in NMR spectra to experimentally support this. While the existence of these base-pairs is probably only transient, they could contribute to stabilization of the G-quadruplex structure.
Introduction of oxoG at Position 10 Results in a Structural Rearrangement
Substitution of G10, which exhibits an anti conformation in the parent G-quadruplex structure, with oxoG10 results in NMR spectra with a distinct chemical shift pattern compared to hTel or hTel-oxoG21 (Figure 1B). Although the hTel-oxoG10 G-quadruplex is stable at 25 °C, all 2D NMR data were collected at 5 °C due to favorable spectral properties and reduced resonance overlap. Imino and aromatic proton assignment was aided by acquisition of 13C and 15N-edited HSQC spectra of 13C, 15N residue-specifically labeled oligonucleotides (Figure S4). Remaining resonances could be unambiguously assigned through sequential connectivities. Four intense H8/H6–H1′ cross-peaks can be observed in a NOESY spectrum acquired with a mixing time of 80 ms, however, only two were assigned to guanines in syn conformation (G9 and G11) with the remaining two cross-peaks belonging to T12 and A24 nucleotides. The sequential walk could be traced throughout the oligonucleotide sequence with an expected interruption at A8 (anti)–G9 (syn) step due to a large interproton distance (Figure 4A). oxoG10 (syn)–G11 (syn) step is not observable due to the lack of H8 resonance in oxoG10, while the weak G9H8–oxoG10H1′ cross-peak is observable in NOESY spectrum despite the inherent syn–syn step.
Figure 4.
hTel-oxoG10 assignment of NOESY spectra and G-quadruplex topology. (A) Aromatic–anomeric region of a NOESY spectrum (τm = 250 ms). Assignments are shown next to H6/H8n-H1n′ cross-peaks. Orange lines connect cross-peaks from T1 to T12 and blue lines from T12 to A24. Letter “A” indicates the oxoG10H1′-G11H8 cross-peak, “B” is A8H2-G17H1′, and “C” is G11H8-G4H21. 1D trace of the NOESY spectrum is shown on the top with signal assignments, where assignments corresponding to syn and anti guanine nucleotides are shown in orange and blue, respectively. Adenine and thymine resonances are in black. (B) Imino–imino (top) and imino–aromatic (bottom) regions of the NOESY spectrum. Assignments in orange and blue represent cross-peaks between guanines of the inner and outer G-quartets, respectively. Assignments in black correspond to cross-peaks between H1 protons of guanines and aromatic protons of adenines. (C) hTel-oxoG10 topology and hydrogen-bond directionality in G-quartets.
It is worth noting that H2′ and H2′′ resonances of oxoG10 were observed at δ 2.75 and 2.44 ppm, respectively. These chemical shifts are similar to those found for G9 and G11, while a more upfield chemical shift is expected for H2′/H2′′ in guanines in an anti conformation.
Examination of internucleotide NOE connectivities (Figure 4B) revealed that the G-quadruplex is comprised of G3·G11·G15·G21, G4·oxoG10·G16·G22, and G5·G9·G17·G23 quartets. Guanines in all G-quartets exhibit anti–syn–anti–anti conformations, and hydrogen-bond donor–acceptor directionalities follow a counterclockwise arrangement (Figure 4C). The first two loops (T6-T7-A8 and T12-T13-A14) are edgewise, while the last one (T18-T19-A20) forms a double-chain-reversal topology. The two broader imino signals at δ 13.85 and 12.39 ppm were assigned to T2 and T7, respectively (Figure S4C). NOE cross-peaks between T2 and T7 imino protons and A14 and A24 aromatic protons (Figure S5) are indicative of T2·A14 Watson–Crick and T7·A24 Hoogsteen base-pairs, which are stacked on the bottom and top G-quartets, respectively (Figure 5). This topology is a deviation from the hTel G-quadruplex structure.
Figure 5.
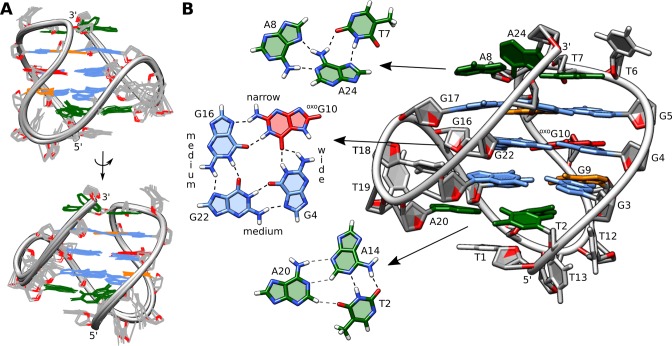
Details of the structure of hTel-oxoG10. (A) Superposition of 12 lowest energy structures. View into the narrow and medium grooves. (B) Enlarged view into the medium groove. G-quartet with oxoG substitution, T7·A8·A24 base triplet, and T2·A14 reversed Watson–Crick pair, which forms a transient base triplet with A20, are showed separately.
A SA protocol was used to determine the structure of hTel-oxoG10 G-quadruplex. Apart from the G-quartet hydrogen-bond restraints and backbone torsion angles, 329 NOE derived distance restrains were included in the simulations. A series of 100 simulations resulted in a well converged set of structures. Twelve lowest energy structures of hTel-oxoG10 with an overall pairwise heavy atoms RMSD of 0.66 Å (Table 1) were selected for further analysis (Figure 5A).
The oxidative lesion in the hTel-oxoG10 G-quadruplex is located in the central G4·oxoG10·G16·G22 quartet. Three atoms of oxoG10 are involved in four hydrogen bonds with adjacent guanines. Two typical Hoogsteen hydrogen bonds are formed with G16 (oxoG10H21-G16N7 and oxoG10H1-G16O6), while O6 of oxoG10 is a bifurcated hydrogen-bond acceptor for G4 (oxoG10O6-G4H1, 1.9 Å and oxoG10O6-G4H21, 2.0 Å) (Figure 5B).
H7 of oxoG10 is shifted from the G-quartet center and positioned further away (∼3.0 Å) from amino group of residue G4. However, due to its location in the central G-quartet, it is protected from solvent exchange processes therefore exhibiting a weak imino signal at δ 11.10 ppm at 5 °C. Amino protons of G4 exhibit resolved signals (δ 5.4 and 8.7 ppm) observable even at 25 °C, suggesting that both are protected from exchange with solvent despite only one being hydrogen bonded.
Due to the lack of H8 resonance, we were unable to directly evaluate the χ torsion angle of oxoG10, and therefore no restraint was used in SA simulations. However, a syn conformation along the glycosidic bond of oxoG10 was found to be favorable. Perusal of 2D NOESY spectra revealed cross-peaks of H7 of oxoG10 with G4 and G5 amino protons and with G11 and G9 aromatic H8 protons. Cross-peak intensities are in agreement with the syn conformation of oxoG10, since the anti conformation would result in a different NOE pattern. Nevertheless, we performed a SA simulation with the χ torsion angle of oxoG10 restrained in anti (1000 kcal/mol·rad2), which still resulted in a syn conformation, albeit with a high restraint penalty.
Loop nucleotides of hTel-oxoG10 are involved in stable structural elements. The double-chain-reversal loop (T18-T19-A20) is stabilized by favorable stacking interactions between T18 and A20, while T19 is flexible and rotated away from the G-quadruplex core. Nucleobases belonging to the two edgewise loops (T6-T7-A8 and T12-T13-A14) of hTel-oxoG10 exhibit efficient stacking on the outer G-quartets (Figure 6). Additionally, A24 forms a Hoogsteen base-pair with T7 resulting in an observable imino signal at δ 12.45 ppm. Furthermore, T7·A24 interacts with A8 to form a base triplet, where A24’s Watson–Crick edge is involved in hydrogen bonding with the Hoogsteen edge of A8. A T2·A14·A20 base triplet was observed in the final set of structures in which the Watson–Crick edge of A20 forms hydrogen bonds with H2 and N3 atoms of A14 and also with O4 of T2. While T2·A14 pairing is stable with an observable imino signal at δ 13.90 ppm, no NOE’s with A20 could be observed, suggesting a transient nature of the base triplet (Figure 5B). It is noteworthy that T2 and T7 imino protons involved in Watson–Crick and Hoogsteen base-pairs give well observable signals at 25 °C, suggesting their stable nature (Figure 1B).
Figure 6.
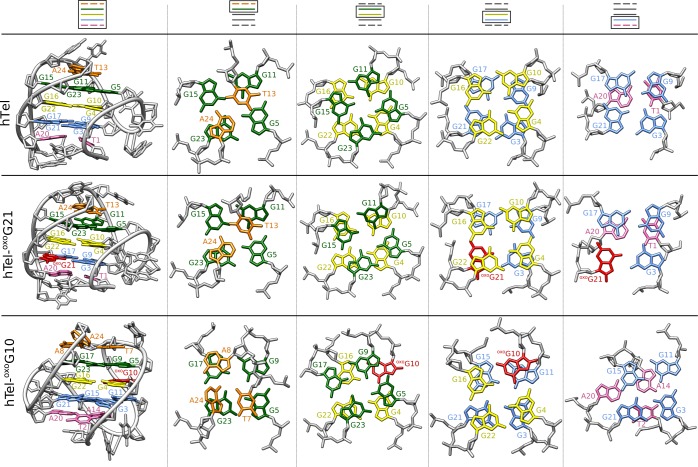
Comparison of stacking between different layers in the hTel,21 hTel-oxoG10, and hTel-oxoG21. Bottom, central and top G-quartets are in blue, yellow, and green, respectively. Stacked loop nucleotides are in pink and orange. oxoG is shown in red.
Oxidative Lesions Reduce G-Quadruplex Stability
The parent hTel G-quadruplex is relatively stable and exhibits a melting temperature of 65 °C, while hTel-oxoG10 and hTel-oxoG21 melt at 44 and 49 °C, respectively (Figure S6). Also, a strong hysteresis is observed in the melting curve of hTel-oxoG10, which suggests it is a slow system to fold into a G-quadruplex structure.
Discussion
Oligonucleotides originating from the human telomeric repeat sequence can fold into G-quadruplex structures even when their sequences contain oxidative lesions in the form of oxoG. While the most obvious implication of oxoG substitutions is a considerable reduction in thermal stability compared to the parent hTel, these structures are stable at physiological temperatures and may be considered relevant.
Substitution with oxoG nucleotides changes the hydrogen-bonding network in a G-quartet due to O6 being the sole hydrogen-bond acceptor on its Hoogsteen edge. As a result oxoG can form stable G-quartets when the adjacent G is substituted with xanthine.37−39 However, O6 of oxoG can form two hydrogen bonds with amino and imino protons when positioned next to a G. This causes a slight shift of the oxoG nucleobase out of the center of G-quartet. Consequently, the distance of the metal–oxygen bond between O6 of oxoG and the K+ cation(s) is increased, which could contribute to decreased stability of such structures.40
The oxoG’s preference for the syn conformation is important for the magnitude of structural rearrangements in lesion-containing G-quadruplexes. Our data demonstrate that either a minor structural adjustment or a major conformational shift is required to accommodate a oxoG nucleotide. G21 adopts a syn conformation in the parent hTel G-quadruplex, and its H8 atom is located in the medium groove, where it is exposed to solvent. Consequently, oxoG is a well-tolerated substitution at position 21 of the hTel sequence and results in only minor structural differences between hTel and hTel-oxoG21 G-quadruplexes. Solvent exposure of H8 also makes position G21 a likely site for nucleobase oxidation by ROS.
On the other hand, G10 has an anti conformation in hTel G-quadruplex, and substitution with oxoG causes a major conformational shift. A similar conformational rearrangement has already been observed in d[G3ATG3ACACAG4ACG3], when three syn guanines were replaced by 2′-fluoro-2′deoxyribo guanines, which favor the anti conformation.41 The original structure changed into a G-quadruplex with one purely syn G-tract and three purely anti G-tracts. However, in the case of hTel-oxoG10, we observed the overturning of G-tracts G9-G11 and G15-G17, which results in loop rearrangement. Considering loop orientations with respect to the G-quartet core, hTel-oxoG10 adopts the hybrid-2 form.23 However, differences can be found in the G3·G11·G15·G21 quartet, which contains only one syn guanine (G11) and in reversal of G-quartet’s hydrogen-bond directionality. Furthermore, the T7·A24·A8 base triplet is observed in the hTel-oxoG10 structure, which involves the terminal 3′ nucleotide. Similarly, the hybrid-2 form of the human telomeric G-quadruplex exhibits a T·T·A base triplet, which also involves equivalent T and A loop nucleotides as well as the terminal 3′ nucleotide.23 This base triplet in the hybrid-2 form has an important role in stabilizing the G-quadruplex structure, especially the 3′-end, which is less stable than the 5′-end.42
MD simulations showed, that syn–anti and anti–anti steps are the most stable dinucleotide fragments in antiparallel and parallel G-quadruplex structures, while the syn–syn fragments are the least favorable.43 Interestingly, in the hTel-oxoG10 structure, the unfavorable all-syn G-tract is counterbalanced by three all-anti G-tracts, and together they form a stable G-quadruplex structure. This G-tract arrangement also results in favorable positioning of the O8 atom of the lesion in the wide groove where it is exposed to solvent.
Telomeres are repetitive regions, which are able to form higher-order G-quadruplex structures. Individual G-quadruplex units either have no mutual interactions and conform to the “beads on a string” model or they interact with each other via G-quartets or loops.44−47 The 5′- and 3′-ends of hybrid-1 and hybrid-2 types of G-quadruplexes are found on opposite sides of the G-quadruplex core, which allows these structures to be folded and to stack on each other in long human telomeric segments. Both hTel-oxoG10 and hTel-oxoG21 also adopt hybrid-type structures, which could allow them to fold and to stack with adjacent nonoxidized G-quadruplex structures. Apart from the lower thermal stability of lesion-containing G-quadruplexes, the higher-order structure of telomeres should be unaffected.
Interestingly, the low-energy barrier between hybrid-1 and hybrid-2 type structures of human telomeric G-quadruplexes allows interconversions between the two, which are influenced by different factors such as temperature, cation concentration, or protein binding.23 The conformational shift following oxidation at position G10 could therefore be a part of normal human telomere metabolism.
Oligonucleotides with oxoG substitutions at remaining ten G positions in the hTel sequence were found to form multiple structures in solutions, or the G-quadruplex structures were destabilized as indicated by broad NMR resonances. Substitutions in the central G-quartet, excluding hTel-oxoG10, are destabilizing. On the other hand, oligonucleotides with oxoG substitutions in outer G-quartets exhibit mostly destabilization of the affected G-quartet or form multiple structures. Other positions where the substituted G is in syn conformation in the parent structure should be able to tolerate oxoG residues, but this is not the case. We hypothesize that some oxoG containing structures are stabilized through base pairing and/or stacking of adjacent loop nucleotides, even if only transiently as is the case with hTel-oxoG21.
Generation and repair of oxidative lesions are dynamic processes in vivo. Following consequent DNA (re)folding in real-time is currently subjected to technical limitations. We believe the ability to follow oxidative structural changes at an atomic level could prove to be in the future an invaluable tool in our understanding of such processes. Nevertheless, due to undisputed presence of oxoG nucleotides in the human genome and stability of some structures described in this study, G-quadruplex structures containing oxoG are very relevant. Identification of some oxidation products and their effect on the structural landscape contributes important data toward understanding this highly dynamic system and widens the extent of structural polymorphism of human telomeric DNA. Our structural characterization shows that the effect of oxidative stress on the telomeric region and its G-quadruplex building blocks is mostly detrimental.
In summary, this work explores the behavior of G-quadruplex structures originating from the human telomeric region under conditions of oxidative stress. To the best of our knowledge, this is the first report of human telomeric G-quadruplex structures containing single oxoG lesions. Several positions in the hTel oligonucleotide sequence can tolerate the major DNA product of oxidative stress - oxoG. Depending on the site of oxoG substitution, accommodation of the lesion can be achieved by a minor structural adjustment or a major conformational shift. However, resulting structures conform to either hybrid-1-or hybrid-2-like topologies and apart from a stability decrease should have little influence on higher-order telomere structure. Telomere length is associated with age, and studies found that ROS are one of the main causes for telomere shortening in humans.48−50 Our work offers a detailed insight into ROS-induced alterations in G-quadruplex structures and could prove significant for detection and mitigation of oxidative lesions in human telomeres and related diseases.
Experimental Section
Samples Preparation
All oligonucleotides were synthesized on K&A Laborgeraete DNA/RNA Synthesizer H-8 using standard phosphoramidite chemistry. Residue-specifically labeled samples contained 10% 13C, 15N enriched guanine and thymine nucleotides. Oligonucleotides were deprotected with AMA (1:1 mixture of aqueous ammonium hydroxide and aqueous methylamine) at 65 °C for 30 min. Samples were purified using reverse-phase HPLC chromatography, followed by removal of the DMT group with 80% AcOH for 30 min. Then the oligonucleotide was transferred to a pure water phase by ether extraction and desalted using FPLC and a Sephadex G25 column. DNA solutions were dried on a vacuum centrifuge and redissolved in 9:1 H2O/2H2O. All samples included 20 mM potassium phosphate buffer, pH 7, and 70 mM KCl. DNA concentrations were determined by measuring UV absorption at 260 nm. An extinction coefficient of 244300 M–1 cm–1 was calculated with the nearest-neighbor method for nonsubstituted hTel and was used for all samples. The final oligonucleotide concentrations were in the range from 0.5 to 1.0 mM.
UV Melting
UV melting experiments were performed on a Varian CARY-100 BIO UV–vis spectrophotometer using 1 cm path length cells. Samples were heated/cooled at a rate of 0.1 °C/min in the range of 15–90 °C and absorbance at 295 nm was measured. Tm was determined from the first derivative of A295 versus temperature plot.
NMR Experiments
All NMR data were collected on Agilent/Varian 600 and 800 MHz NMR spectrometers at 5 or 25 °C. All homonuclear spectra were acquired with DPFGSE solvent suppression. 2D NOESY experiments (τm = 80, 150, 250, 300 ms) were utilized for 1H resonance assignment. Identification of guanine and thymine imino proton resonances was aided by 1D 15N-edited HSQC experiments, while H6/H8 aromatic proton resonances were assigned with the help of 13C-edited HSQC experiments using 10% 13C, 15N site-specifically labeled oligonucleotides. 2D TOCSY experiments (τm = 80 ms) gave information on sugar puckering, and 2D 1H–13C HSQC spectra were used to assign H2 aromatic resonances of adenines. Data were analyzed with VNMRJ, NMRpipe, CcpNmr, and Origin.51,52
Distance and Dihedral Angle Restrains
Interproton distances were calculated from 250 ms NOESY spectra, which were chosen from NOE buildup curves. The average of thymine aromatic-methyl proton (H6-Me) distances was used as a reference (2.99 Å). Restraint bounds for cross-peaks in aromatic and anomeric regions were set at ±0.3 Å for dij < 3 Å, ±0.4 Å for 3 Å < dij < 4 Å, ±0.5 Å for 4 Å < dij < 4.5 Å, and ±0.6 Å for dij > 4.5 Å. For some restraints corresponding to overlapping cross-peaks, larger distance ranges were used (±0.7–1.5 Å). All cross-peaks in imino–aromatic, imino–imino, amino–imino and amino–aromatic regions were classified as strong (2.5 Å), medium (3.5 Å), or weak (4.5 Å) with ±1–1.5 Å restraint ranges.
Dihedral angle χ was restrained to a range between −90° and 90° for syn and between 90° and 270° for anti nucleotides, and oxoG was left unrestrained. Restraints for backbone dihedral angles included α (−120° to 120°), β (150° to 210°), γ (30° to 90°), δ (130°–190°), ε (170°–300°), and ζ (−120°–120°). All sugar puckers were determined to be of S-type (C2′-endo) based on 2D TOCSY NMR spectra.
Structural Calculations
Molecular dynamics calculations were performed with AMBER 14 software using the ff99bsc0 force field and ε/ζOL1 and χOL4 modifications.53,54 Force field parameters for oxoG nucleotides were derived from the RESP ESP charge Derive (R.E.D.) Server.55
Calculations were started from initial linear structures of the oligonucleotides, created with the LEAP module of AMBER 14. A total of 100 structures were obtained in 1 ns restrained SA simulations using the Born implicit solvent model with random starting velocities. In the first 200 ms of SA, the temperature was kept at 1000 K, followed by slow cooling in the next 600 ms down to 300 K and to 0 K in the last 200 ms. Force constants were 100 kcal·mol–1 Å–2 for hydrogen bonds, 20 kcal·mol–1 Å–2 for NOE distance restrains, 200 kcal·mol–1 rad–2 for backbone, and 50 kcal·mol–1 rad–2 for the χ torsion angle. Twelve structures were selected based on the lowest energy and subjected to energy minimization with a maximum of 10000 steps.
Acknowledgments
This work was supported by the Slovenian Research Agency (ARRS, grants J1-6733, J3-6800 and P1-0242).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b12748.
Schematic representation of a substituted G-quartet, 1H NMR spectra of all hTel analogues, 2D NOESY spectra, 15N-edited HSQC spectra, UV melting curves (PDF)
Accession Codes
The coordinates for the 12 lowest energy structures of hTel-oxoG10 and hTel-oxoG21 have been deposited in the Protein Data Bank with accession codes 6IA0 and 6IA4, respectively. Chemical shifts have been deposited in the Biological Magnetic Resonance Data Bank as entries 34331 and 34332.
The authors declare no competing financial interest.
Supplementary Material
References
- Apel K.; Hirt H. REACTIVE OXYGEN SPECIES: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55 (1), 373–399. 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Halliwell B. Reactive Oxygen Species in Living Systems: Source, Biochemistry, and Role in Human Disease. Am. J. Med. 1991, 91 (S3), S14–S22. 10.1016/0002-9343(91)90279-7. [DOI] [PubMed] [Google Scholar]
- Steenken S.; Jovanovic S. V. How Easily Oxidizable Is DNA? One-Electron Reduction Potentials of Adenosine and Guanosine Radicals in Aqueous Solution. J. Am. Chem. Soc. 1997, 119 (3), 617–618. 10.1021/ja962255b. [DOI] [Google Scholar]
- Prat F.; Houk K. N.; Foote C. S. Effect of Guanine Stacking on the Oxidation of 8-Oxoguanine in B-DNA. J. Am. Chem. Soc. 1998, 120 (4), 845–846. 10.1021/ja972331q. [DOI] [Google Scholar]
- Sugiyama H.; Saito I. Theoretical Studies of GG-Specific Photocleavage of DNA via Electron Transfer: Significant Lowering of Ionization Potential and 5′-Localization of HOMO of Stacked GG Bases in B-Form DNA. J. Am. Chem. Soc. 1996, 118 (30), 7063–7068. 10.1021/ja9609821. [DOI] [Google Scholar]
- Kino K.; Saito I.; Sugiyama H. Product Analysis of GG-Specific Photooxidation of DNA via Electron Transfer: 2-Aminoimidazolone as a Major Guanine Oxidation Product. J. Am. Chem. Soc. 1998, 120 (29), 7373–7374. 10.1021/ja980763a. [DOI] [Google Scholar]
- Saito I.; Nakamura T.; Nakatani K.; Yoshioka Y.; Yamaguchi K.; Sugiyama H. Mapping of the Hot Spots for DNA Damage by One-Electron Oxidation: Efficacy of GG Doublets and GGG Triplets as a Trap in Long-Range Hole Migration. J. Am. Chem. Soc. 1998, 120 (48), 12686–12687. 10.1021/ja981888i. [DOI] [Google Scholar]
- Burrows C. J.; Muller J. G. Oxidative Nucleobase Modifications Leading to Strand Scission. Chem. Rev. 1998, 98 (3), 1109–1152. 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- Neeley W. L.; Essigmann J. M. Mechanisms of Formation, Genotoxicity, and Mutation of Guanine Oxidation Products. Chem. Res. Toxicol. 2006, 19 (4), 491–505. 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- Hickerson R. P.; Prat F.; Muller J. G.; Foote C. S.; Burrows C. J. Sequence and Stacking Dependence of 8-Oxoguanine Oxidation: Comparison of One-Electron vs Singlet Oxygen Mechanisms. J. Am. Chem. Soc. 1999, 121 (40), 9423–9428. 10.1021/ja991929q. [DOI] [Google Scholar]
- Wright W. E.; Tesmer V. M.; Huffman K. E.; Levene S. D.; Shay J. W. Normal Human Chromosomes Have Long G-Rich Telomeric Overhangs at One End. Genes Dev. 1997, 11 (21), 2801–2809. 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E. H.; Epel E. S.; Lin J. Human Telomere Biology: A Contributory and Interactive Factor in Aging, Disease Risks, and Protection. Science 2015, 350 (6265), 1193–1198. 10.1126/science.aab3389. [DOI] [PubMed] [Google Scholar]
- Ma D.; Zhu W.; Hu S.; Yu X.; Yang Y. Association between Oxidative Stress and Telomere Length in Type 1 and Type 2 Diabetic Patients. J. Endocrinol. Invest. 2013, 36 (11), 1032–1037. [DOI] [PubMed] [Google Scholar]
- Tesovnik T.; Kovac J.; Hovnik T.; Dovc K.; Bratina N.; Battelino T.; Trebušak Podkrajšek K. Association of Glycemic Control and Cell Stress With Telomere Attrition in Type 1 Diabetes. JAMA Pediatr. 2018, 172, 879–881. 10.1001/jamapediatrics.2018.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webba Da Silva M. Geometric Formalism for DNA Quadruplex Folding. Chem. - Eur. J. 2007, 13 (35), 9738–9745. 10.1002/chem.200701255. [DOI] [PubMed] [Google Scholar]
- Karsisiotis A. I.; O’Kane C.; Webba da Silva M. DNA Quadruplex Folding Formalism - A Tutorial on Quadruplex Topologies. Methods 2013, 64 (1), 28–35. 10.1016/j.ymeth.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Patel D. J. Solution Structure of the Human Telomeric Repeat d[AG3(T2AG3)3] G-Tetraplex. Structure 1993, 1 (4), 263–282. 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- Parkinson G. N.; Lee M. P. H.; Neidle S. Crystal Structure of Parallel Quadruplexes from Human Telomeric DNA. Nature 2002, 417, 876–880. 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- Ambrus A.; Chen D.; Dai J.; Bialis T.; Jones R. A.; Yang D. Human Telomeric Sequence Forms a Hybrid-Type Intramolecular G-Quadruplex Structure with Mixed Parallel/Antiparallel Strands in Potassium Solution. Nucleic Acids Res. 2006, 34 (9), 2723–2735. 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A. T.; Luu K. N.; Patel D. J. Different Loop Arrangements of Intramolecular Human Telomeric (3 + 1) G-Quadruplexes in K+ Solution. Nucleic Acids Res. 2006, 34 (19), 5715–5719. 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu K. N.; Phan A. T.; Kuryavyi V.; Lacroix L.; Patel D. J. Structure of the Human Telomere in K+ Solution: An Intramolecular (3 + 1) G-Quadruplex Scaffold. J. Am. Chem. Soc. 2006, 128 (30), 9963–9970. 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J.; Punchihewa C.; Ambrus A.; Chen D.; Jones R. A.; Yang D. Structure of the Intramolecular Human Telomeric G-Quadruplex in Potassium Solution: A Novel Adenine Triple Formation. Nucleic Acids Res. 2007, 35 (7), 2440–2450. 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J.; Carver M.; Punchihewa C.; Jones R. A.; Yang D. Structure of the Hybrid-2 Type Intramolecular Human Telomeric G-Quadruplex in K+ Solution: Insights into Structure Polymorphism of the Human Telomeric Sequence. Nucleic Acids Res. 2007, 35 (15), 4927–4940. 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A. T.; Kuryavyi V.; Luu K. N.; Patel D. J. Structure of Two Intramolecular G-Quadruplexes Formed by Natural Human Telomere Sequences in K+ Solution. Nucleic Acids Res. 2007, 35 (19), 6517–6525. 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim K. W.; Amrane S.; Bouaziz S.; Xu W.; Mu Y.; Patel D. J.; Luu K. N.; Phan A. T. Structure of the Human Telomere in K+ Solution: A Stable Basket-Type G-Quadruplex with Only Two G-Tetrad Layers. J. Am. Chem. Soc. 2009, 131 (12), 4301–4309. 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A. T. Human Telomeric G-Quadruplex: Structures of DNA and RNA Sequences. FEBS J. 2010, 277 (5), 1107–1117. 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- Lech C. J.; Cheow Lim J. K.; Wen Lim J. M.; Amrane S.; Heddi B.; Phan A. T. Effects of Site-Specific Guanine C8-Modifications on an Intramolecular DNA G-Quadruplex. Biophys. J. 2011, 101 (8), 1987–1998. 10.1016/j.bpj.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorlícková M.; Tomasko M.; Sagi A. J.; Bednarova K.; Sagi J. 8-Oxoguanine in a Quadruplex of the Human Telomere DNA Sequence. FEBS J. 2012, 279 (1), 29–39. 10.1111/j.1742-4658.2011.08396.x. [DOI] [PubMed] [Google Scholar]
- Fleming A. M.; Zhou J.; Wallace S. S.; Burrows C. J. A Role for the Fifth G-Track in G-Quadruplex Forming Oncogene Promoter Sequences during Oxidative Stress: Do These “Spare Tires” Have an Evolved Function?. ACS Cent. Sci. 2015, 1 (5), 226–233. 10.1021/acscentsci.5b00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. M.; Ding Y.; Burrows C. J. Oxidative DNA Damage Is Epigenetic by Regulating Gene Transcription via Base Excision Repair. Proc. Natl. Acad. Sci. U. S. A. 2017, 114 (10), 2604–2609. 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omaga C. A.; Fleming A. M.; Burrows C. J. The Fifth Domain in the G-Quadruplex-Forming Sequence of the Human NEIL3 Promoter Locks DNA Folding in Response to Oxidative Damage. Biochemistry 2018, 57 (20), 2958–2970. 10.1021/acs.biochem.8b00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoi S.; Ferino A.; Miglietta G.; Pedersen E. B.; Xodo L. E. The Regulatory G4Motif of the Kirsten Ras (KRAS) Gene Is Sensitive to Guanine Oxidation: Implications on Transcription. Nucleic Acids Res. 2018, 46 (2), 661–676. 10.1093/nar/gkx1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S.; Kim K. T.; Podbevšek P.; Plavec J.; Kim B. H.; Sugimoto N. Recovery of the Formation and Function of Oxidized G-Quadruplexes by a Pyrene-Modified Guanine Tract. J. Am. Chem. Soc. 2018, 140 (17), 5774–5783. 10.1021/jacs.8b01577. [DOI] [PubMed] [Google Scholar]
- Kouchakdjian M.; Bodepudi V.; Shibutani S.; Eisenberg M.; Johnson F.; Grollman A. P.; Patel D. J. NMR Structural Studies of the Ionizing Radiation Adduct 7-Hydro-8-Oxodeoxyguanosine (8-Oxo-7H-DG) Opposite Deoxyadenosine in a DNA Duplex. 8-Oxo-7H-DG(Syn)•dA(Anti) Alignment at Lesion Site. Biochemistry 1991, 30 (5), 1403–1412. 10.1021/bi00219a034. [DOI] [PubMed] [Google Scholar]
- Fujimoto H.; Pinak M.; Nemoto T.; Bunta J. K. Structural Analysis of Base Mispairing in DNA Containing Oxidative Guanine Lesion. Cent. Eur. J. Phys. 2007, 5 (1), 49–61. 10.2478/s11534-006-0044-8. [DOI] [Google Scholar]
- Norman D.; Abuaf P.; Hingerty B. E.; Live D.; Grunberger D.; Broyde S.; Patel D. J. NMR and Computational Characterization of the IV-(Deoxyguanosin-8-Yl)Aminofluorene Adduct [(AF)G] Opposite Adenosine in DNA: (AF)G[Syn]·A[Anti] Pair Formation and Its PH Dependence. Biochemistry 1989, 28 (18), 7462–7476. 10.1021/bi00444a046. [DOI] [PubMed] [Google Scholar]
- Fujii T.; Podbevšek P.; Plavec J.; Sugimoto N. Effects of Metal Ions and Cosolutes on G-Quadruplex Topology. J. Inorg. Biochem. 2017, 166, 190–198. 10.1016/j.jinorgbio.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Cheong V. V.; Heddi B.; Lech C. J.; Phan A. T. Xanthine and 8-Oxoguanine in G-Quadruplexes: Formation of a G·G·X·O Tetrad. Nucleic Acids Res. 2015, 43 (21), 10506–10514. 10.1093/nar/gkv826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong V. V.; Lech C. J.; Heddi B.; Phan A. T. Inverting the G-Tetrad Polarity of a G-Quadruplex by Using Xanthine and 8-Oxoguanine. Angew. Chem., Int. Ed. 2016, 55 (1), 160–163. 10.1002/anie.201507034. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya D.; Mirihana Arachchilage G.; Basu S. Metal Cations in G-Quadruplex Folding and Stability. Front. Chem. 2016, 4, 1–14. 10.3389/fchem.2016.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerhoff J.; Weisz K. Flipping a G-Tetrad in a Unimolecular Quadruplex without Affecting Its Global Fold. Angew. Chem., Int. Ed. 2015, 54 (19), 5588–5591. 10.1002/anie.201411887. [DOI] [PubMed] [Google Scholar]
- Bugaut A.; Alberti P. Understanding the Stability of DNA G-Quadruplex Units in Long Human Telomeric Strands. Biochimie 2015, 113, 125–133. 10.1016/j.biochi.2015.04.003. [DOI] [PubMed] [Google Scholar]
- Cang X.; Šponer J.; Cheatham T. E. Explaining the Varied Glycosidic Conformational, G-Tract Length and Sequence Preferences for Anti-Parallel G-Quadruplexes. Nucleic Acids Res. 2011, 39 (10), 4499–4512. 10.1093/nar/gkr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.; Ishizuka T.; Kurabayashi K.; Komiyama M. Consecutive Formation of G-Quadruplexes in Human Telomerieoverhang DNA: A Protective Capping Structure for Telomere Ends. Angew. Chem., Int. Ed. 2009, 48 (42), 7833–7836. 10.1002/anie.200903858. [DOI] [PubMed] [Google Scholar]
- Yu H. Q.; Miyoshi D.; Sugimoto N. Characterization of Structure and Stability of Long Telomeric DNA G-Quadruplexes. J. Am. Chem. Soc. 2006, 128 (48), 15461–15468. 10.1021/ja064536h. [DOI] [PubMed] [Google Scholar]
- Petraccone L.; Spink C.; Trent J. O.; Garbett N. C.; Mekmaysy C. S.; Giancola C.; Chaires J. B. Structure and Stability of Higher-Order Human Telomeric Quadruplexes. J. Am. Chem. Soc. 2011, 133 (51), 20951–20961. 10.1021/ja209192a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petraccone L.; Trent J. O.; Chaires J. B. The Tail of the Telomere. J. Am. Chem. Soc. 2008, 130 (49), 16530–16532. 10.1021/ja8075567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samassekou O.; Gadji M.; Drouin R.; Yan J. Sizing the Ends: Normal Length of Human Telomeres. Ann. Anat. 2010, 192 (5), 284–291. 10.1016/j.aanat.2010.07.005. [DOI] [PubMed] [Google Scholar]
- Epel E. S.; Blackburn E. H.; Lin J.; Dhabhar F. S.; Adler N. E.; Morrow J. D.; Cawthon R. M. Accelerated Telomere Shortening in Response to Life Stress. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (49), 17312–17315. 10.1073/pnas.0407162101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkowitz O. M.; Mellon S. H.; Epel E. S.; Lin J.; Dhabhar F. S.; Su Y.; Reus V. I.; Rosser R.; Burke H. M.; Kupferman E.; Compagnone M.; Nelson J. C.; Blackburn E. H. Leukocyte Telomere Length in Major Depression: Correlations with Chronicity, Inflammation and Oxidative Stress - Preliminary Findings. PLoS One 2011, 6 (3), e17837. 10.1371/journal.pone.0017837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F.; Grzesiek S.; Vuister G.; Zhu G.; Pfeifer J.; Bax A. NMRPipe: A Multidimensional Spectral Processing System Based on UNIX Pipes. J. Biomol. NMR 1995, 6 (3), 277–293. 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Vranken W. F.; Boucher W.; Stevens T. J.; Fogh R. H.; Pajon A.; Llinas M.; Ulrich E. L.; Markley J. L.; Ionides J.; Laue E. D. The CCPN Data Model for NMR Spectroscopy: Development of a Software Pipeline. Proteins: Struct., Funct., Genet. 2005, 59 (4), 687–696. 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Krepl M.; Zgarbová M.; Stadlbauer P.; Otyepka M.; Banáš P.; Koča J.; Cheatham T. E.; Jurečka P.; Šponer J. Reference Simulations of Noncanonical Nucleic Acids with Different χ Variants of the AMBER Force Field: Quadruplex DNA, Quadruplex RNA, and Z-DNA. J. Chem. Theory Comput. 2012, 8 (7), 2506–2520. 10.1021/ct300275s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgarbová M.; Luque F. J.; Šponer J.; Cheatham T. E.; Otyepka M.; Jurečka P. Toward Improved Description of DNA Backbone: Revisiting Epsilon and Zeta Torsion Force Field Parameters. J. Chem. Theory Comput. 2013, 9 (5), 2339–2354. 10.1021/ct400154j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanquelef E.; Simon S.; Marquant G.; Garcia E.; Klimerak G.; Delepine J. C.; Cieplak P.; Dupradeau F.-Y. R.E.D. Server: A Web Service for Deriving RESP and ESP Charges and Building Force Field Libraries for New Molecules and Molecular Fragments. Nucleic Acids Res. 2011, 39 (S2), W511–W517. 10.1093/nar/gkr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.