Abstract
Many neurological and psychiatric diseases in humans are caused by disruptions to large-scale functional properties of the brain, including functional connectivity. There has been growing interest in discovering the functional organization of brain networks in larger animal models. As a result, the use of translational pig models in neuroscience has significantly increased in the past decades. The gyrencephalic pig brain resembles the human brain more in anatomy, growth, and development than the brains of commonly used small laboratory animals such as rodents. In this work, resting-state functional magnetic resonance imaging (rs-fMRI) and diffusion tensor imaging (DTI) data were acquired from a group of pigs (n = 12). rs-fMRI data were analyzed for resting-state networks (RSNs) by using independent component analysis and sparse dictionary learning. Six RSNs (executive control, cerebellar, sensorimotor, visual, auditory, and default mode) were detected that resemble their counterparts in human brains, as measured by Pearson spatial correlations and mean ratios. Supporting evidence of the validity of these RSNs was provided through the evaluation and quantification of structural connectivity measures (mean diffusivity, fractional anisotropy, fiber length, and fiber density) estimated from the DTI data. This study shows that as a translational, large animal model, pigs demonstrate great potential for mapping connectome-scale functional connectivity in experimental modeling of human brain disorders.
Keywords: independent component analysis, MRI, pigs, resting-state networks, sparse dictionary learning
Introduction
Resting-state functional magnetic resonance imaging (rs-fMRI) is used to examine functional connectivity in the brain through the recording and analysis of blood-oxygenation-level dependent (BOLD) oscillations originating from synchronized intrinsic neuronal activity in the absence of any explicit task (Biswal et al., 1995; Chang et al., 2018; Fox et al., 2005). In rs-fMRI analysis, functional connectivity is quantified by using various temporal and/or spatial metrics, such as correlation, covariance, and mutual information between the BOLD time series signals collected from different brain regions. This quantification of functional connectivity can highlight sets of distinct cortical and subcortical areas that form integrated information processing networks and can be used to construct connectivity maps.
Numerous prior rs-fMRI studies have reported a number of functional connectivity networks, commonly referred to as resting-state networks (RSNs) or intrinsic connectivity networks (ICNs; Hutchison et al., 2013) in the human brain. Atlases of the most common human brain RSNs or ICNs have been developed (Laird et al., 2011; Smith et al., 2009) and have been used to quantify fMRI activation patterns in healthy subjects.
In addition to functional connectivity, structural connectivity has also been studied by using diffusion tensor imaging (DTI) to measure the diffusion of water molecules in the brain. Many studies have reported associations between functional and structural connectivity (Honey et al., 2009), for example in the default mode network (DMN) (Greicius et al., 2008) and in the aging population (Damoiseaux, 2017). Some studies have shown that the strength of resting-state functional connectivity is positively correlated with that of structural connectivity (Damoiseaux and Greicius, 2009). However, other studies have also reported functional connectivity detected between regions with little or no structural connectivity (Zimmermann et al., 2016), suggesting that functional connectivity may also be mediated by indirect structural connections.
Inspired by the discovery of RSNs in the human brain from rs-fMRI analysis, there has been a growing interest in finding similar networks of functional connectivity in animal models. The ability to map homologous connectome-scale functional connectivity in a highly translational animal model will improve characterization of the effects of neural diseases and brain injuries on major neural functions and allow such findings to have meaningful translational implications in humans.
Rodents and nonhuman primates have been the most commonly used animal models (Lu et al., 2007; Vincent et al., 2007). However, these animal models are nonideal due to a number of limitations: the rodent brain is lissencephalic (lacks gyri and sulci) and has significantly different brain anatomy relative to the human brain, and economic considerations hinder the use of nonhuman primates in brain research. Because of this, there is a growing need for the establishment of alternative non-primate, large animal models.
Use of the pig brain as a model has been rapidly increasing in neuroscience research due to its similarities in size, structure, composition, and development to that of the human brain. The percentage of brain weight at birth, brain growth spurts, and prenatal and postnatal brain maturation patterns in humans are also more similar to pigs relative to other animal models (Dobbing and Sands, 1979). The pig brain is larger in volume and weight (∼180 g) than that of a rodent (∼10 g) and is comparable in size to that of a nonhuman primate (∼300 g), while being much less expensive (Sauleau et al., 2009).
The pig brain, similar to the human brain, is also gyrencephalic and follows a similar gyral pattern (Gieling et al., 2011). The volume of the prefrontal cortex in the pig constitutes 10% of total brain volume, which is comparable to humans whose prefrontal cortex constitutes ∼12.5% of total brain volume (Jelsing et al., 2006; McBride et al., 1999). The dorsal striatum of the pig brain is split by the internal capsule into two distinct structures: the caudate nucleus and the putamen; whereas in comparison, the rodent brain only contains a single caudate-putamen structure (Felix et al., 1999; Matsas et al., 1986). The pig hippocampus has been well described and found to be structurally more similar to the human hippocampus than the rodent, having a degree of encephalization that lies between rodent and primate (Holm and West, 1994).
A number of descriptive, comparative anatomical studies have been performed for the pig brain thalamus, hypothalamus, hypothalamic nuclei (Campos-Ortega, 1970; Junge, 1977; Szteyn et al., 1980), brainstem structures (Freund, 1969; Otabe and Horowitz, 1970), and cerebellum (Larsell, 1954). Similar to humans, sensory cortices such as the motor cortex (Breazile et al., 1966) and the somatosensory cortex (Craner and Ray, 1991) are arranged somatotopically.
In establishing a translational animal model for brain injury and neurological disease research, it is important that the brain of the animal has similar anatomy and organization to the human brain, as it impacts the brain regions injured, vascular responses, short- and long-term effects of the injury, and, ultimately, whether the findings are translational to humans (Duhaime, 2006). Pig models of stroke and traumatic brain injury (TBI) have recently been developed and show comparable anatomical damage and physiological responses as would be expected in human patients (Baker et al., 2017; Platt et al., 2014). However, groundwork studies to characterize functional connectivity in the pig animal model have yet to be conducted.
To model rs-fMRI data and determine brain functional connectivity, a variety of data-driven approaches have been proposed over the past decades. Independent component analysis (ICA) is one method that has been very successful (Calhoun and Adali, 2004). Alternatively, sparse dictionary learning (sDL; Mairal et al., 2010) is another data-driven approach that has been successfully applied to reconstruct brain functional connectivity.
In sDL-based methods, fMRI data is decomposed into two matrices: a dictionary basis matrix (which represents the time-domain activity of functional networks) and a reference weight matrix (which represents the spatial distributions of the functional networks within the brain). Generally, the sDL approach provides a compact, high-fidelity representation of the fMRI data and discovers spatial patterns representing functional connectivity (Lv et al., 2015; Zhang et al., 2018).
In this article, rs-fMRI and DTI data were acquired from a group of 3-week-old piglets. From the rs-fMRI data, both ICA and sDL were employed to empirically detect RSNs in the pig brain. To quantify the degree of homology between the detected pig RSNs and human RSNs, a reference pig RSN atlas was created by combining anatomies from a standard pig brain atlas (Saikali et al., 2010) that are known to be associated with human RSNs. Using the reference atlas and empirically detected pig RSNs, Pearson spatial correlation coefficients and mean ratio metrics were calculated and used to determine the degree of homology between the detected pig RSNs and human RSNs. To the best knowledge of the authors, this is the first study to attempt to characterize RSNs in the pig brain to date.
From acquired DTI data, structural connectivity measures (mean diffusivity [MD], fractional anisotropy [FA], fiber length [FL], and fiber density [FD]) were quantified for each detected pig RSN to provide supporting evidence of their validity.
Materials and Methods
Subjects
Three-week-old Landrace-cross pigs (n = 12) were used in this study. The pigs were initially sedated through an intramuscular injection of xylazine (7 mg/kg), butorphanol (0.3 mg/kg), and midazolam (0.3 mg/kg). Then, mild anesthesia was maintained with 1.5% inhalational isoflurane in oxygen to keep the pigs sedated while reducing anesthetic agents from interfering with pig brain neural activity and neurovascular coupling. All experimental procedures were approved by the Institutional Animal Care and Use Committee (University of Georgia).
Data acquisition
Using a GE Signa HDx 3T scanner and an HD quadrature knee coil, rs-fMRI, T1-weighted anatomical, and DTI data were acquired by using the following three sequences: (1) rs-fMRI: gradient echo-planar imaging (EPI) sequence (repetition time [TR] = 3 s, TE = 30 ms, FA = 80°, field of view [FOV] = 12.8 × 12.8 × 6.4 cm, a matrix size of 96 × 96 × 32, 300 total volumes, and an acquisition time of 15 min), (2) T1-weighted anatomical: three-dimensional (3D) fast spoiled gradient echo sequence (TR = 5.5 s, TE = 2.1 ms, FA = 9°, FOV = 12.8 × 12.8 × 6.4 cm, slice thickness = 1 mm, and a matrix size of 256 × 256 × 112), and (3) DTI: spin echo EPI sequence (TR = 15.5 s, TE = min-full, FOV = 12.8 × 12.8 × 6.4 cm, a matrix size of 64 × 64 × 32, 3 b = 0 images, and 30 diffusion weighted images using b = 1000 s/mm2).
Data preprocessing
Pig rs-fMRI data were preprocessed to realign images to correct for motion, perform slice-timing correction, and execute spatial normalization by using the Realign and Unwarp, slice timing, and Old Normalize Statistical Parametric Mapping software algorithms, respectively (Ashburner et al., 2016; Penny et al., 2011). One pig was chosen as the template, and the rs-fMRI datasets of the other 11 pigs were spatially normalized to the template pig fMRI space. The first volume from each rs-fMRI dataset was used to calculate a spatial transformation, and the transformation was then applied to the rest of the volumes. All spatial transformations in this work were performed by first applying a 12-parameter affine transformation, followed by a nonlinear deformation transformation (Ashburner and Friston, 1999; Ashburner et al., 1997).
Next, brain tissue was separated from the skull and other surrounding tissues by manual slice-by-slice segmentation of the images, and the rs-fMRI time series from the 12 pigs were temporally concatenated into a group dataset. Due to severe movement that occurred for one of the pigs toward the end of its rs-fMRI data acquisition, eight volumes could not be corrected for motion and were removed; therefore, the total time series length of the group dataset was 3592 (total number of volumes).
Data analysis
The group dataset was analyzed by using FastICA and sDL, respectively. ICA attempts to extract independent signal components from a mixed signal through matrix decomposition. In fMRI studies, spatial ICA is typically used, which extracts independent spatial components representing proposed functional networks. In this study, spatial ICA was performed by using the Group ICA of fMRI Toolbox's (GIFT; Calhoun and Adali, 2004) FastICA algorithm, which is a commonly used iterative algorithm for doing approximate ICA efficiently on large datasets. FastICA first performs a whitening process on the input data matrix before iteratively calculating a series of components through maximization of non-Gaussianity of the projection of the whitened input data matrix along the component.
An automated process to estimate the number of independent components for ICA decomposition from the group dataset was performed by using the GIFT Toolbox, and it was estimated that the data were composed of nine independent components. To reduce error in this estimation, ICA decomposition was performed by using 20 independent components, which doubles the estimated component number and is also commonly used throughout the literature (Smith et al., 2009).
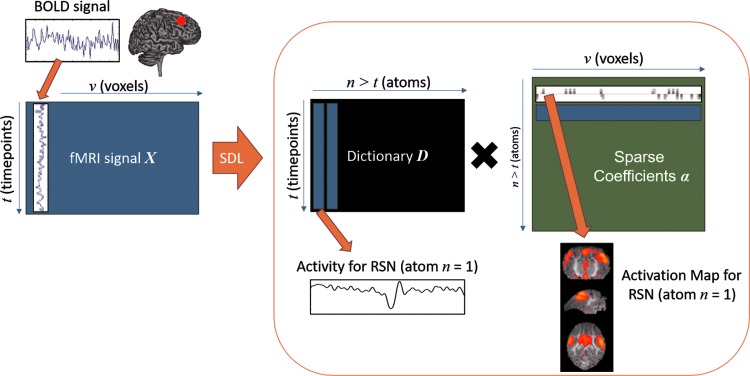
sDL is an iterative algorithm for learning a sparse representation of the input data as a linear combination of a typically complete or over-complete dictionary of elements, or “atoms” (Mairal et al., 2010). In fMRI analysis, the entire signal volume of v voxels over t time points is arranged as a two-dimensional signal matrix 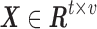 , where the time course data of each of the v voxels are represented as a column with t rows.
, where the time course data of each of the v voxels are represented as a column with t rows.
Given a desired n number of atoms, sDL iteratively decomposes matrix 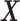 into a dictionary matrix
into a dictionary matrix 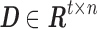 and a sparse coefficient matrix
and a sparse coefficient matrix 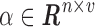 , such that
, such that 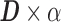 closely approximates the signal matrix
closely approximates the signal matrix 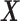 through the minimization of the mean squared error between
through the minimization of the mean squared error between 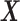 and
and 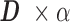 , while fulfilling an L1-regularization sparsity constraint of λ on
, while fulfilling an L1-regularization sparsity constraint of λ on 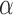 , as summarized in Equation (1). Again,
, as summarized in Equation (1). Again, 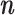 is typically selected so that the dictionary matrix
is typically selected so that the dictionary matrix 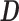 is complete or over-complete
is complete or over-complete 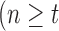 ), allowing the
), allowing the 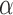 matrix to be sparse.
matrix to be sparse.
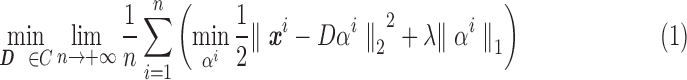 |
Through this decomposition, the original signal time series of each voxel is represented as a linear combination of n dictionary atoms of length t, where each atom corresponds to the time series of a potential functional network detected by the decomposition. The corresponding row in 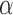 represents the strength of involvement of each of v voxels in the detected functional network, which can be mapped back to a brain volume to generate a map of each of the n potential functional networks, as illustrated in Figure 1.
represents the strength of involvement of each of v voxels in the detected functional network, which can be mapped back to a brain volume to generate a map of each of the n potential functional networks, as illustrated in Figure 1.
FIG. 1.
Illustrated representation of the decomposition process of sDL. A matrix representing the complete volume of v voxel signals acquired from a scan of time t is arranged into a signal matrix 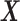 . Through sDL, the matrix is decomposed into a dictionary matrix
. Through sDL, the matrix is decomposed into a dictionary matrix 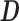 and
and 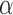 , such that
, such that 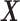 approximates
approximates 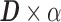 . Dictionary
. Dictionary 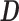 contains activity for n number of atoms, or detected RSNs, whereas
contains activity for n number of atoms, or detected RSNs, whereas  contains representations of the detect RSN maps that can be mapped back to a brain volume. BOLD, blood-oxygenation-level dependent; fMRI, functional magnetic resonance imaging; RSNs, resting-state networks; sDL, sparse dictionary learning.
contains representations of the detect RSN maps that can be mapped back to a brain volume. BOLD, blood-oxygenation-level dependent; fMRI, functional magnetic resonance imaging; RSNs, resting-state networks; sDL, sparse dictionary learning.
In this study, the SPArse Modeling Software (SPAMS) toolbox (Mairal et al., 2010) was used to implement the Online Dictionary Learning (ODL) algorithm. ODL trains a dictionary by iteratively using each sample of an input data matrix to calculate a sparse coding using least angle regression, and then using the calculated sparse coding to update the dictionary (Mairal et al., 2010). As an iterative algorithm, ODL is suitable for performing sDL on large datasets that are too large to fit into memory (Lv et al., 2015). When performing sDL, a complete dictionary (n = 292) and a sparsity parameter of λ = 0.15 were used. For details regarding the optimization of n and λ, see the Supplementary Data.
Pig reference RSN analysis
To quantify the degree of homology between the detected pig RSNs and established human RSNs, a reference pig RSN atlas was created by first spatially normalizing a standard pig brain atlas (Saikali et al., 2010) to the anatomical space of the template pig. Spatial normalization was accomplished by calculating a spatial transformation using anatomical data provided in the same space as, and associated with, the standard pig brain atlas and the anatomical data of the template pig. The calculated transformation was then applied to the atlas.
The anatomically homologous pig brain regions associated with six major human RSNs (Laird et al., 2011; Smith et al., 2009) were manually selected and combined to construct the following six pig RSNs: executive control (EX), cerebellar (CERE), visual (VIS), sensorimotor (SM), auditory (AUD), and DMN. These six manually constructed pig RSNs formed a reference atlas that was assumed to be perfectly homologous to the six human RSNs and subsequently used as a reference to calculate the degree of homology of the empirically detected RSNs found by ICA and sDL. Table 1 lists the anatomies and corresponding atlas labels from the standard pig brain atlas (Saikali et al., 2010) that were used to construct each of the six reference pig RSNs.
Table 1.
Reference Pig Resting-State Network (RSN) Atlas and Human RSN Anatomies
| Pig brain | Human brain |
|---|---|
| Executive control | Executive control |
| Primary somatosensory cortex (101, 201) | Primary somatosensory cortex |
| Dorsolateral prefrontal cortex (109, 209) | Prefrontal cortex |
| Anterior prefrontal cortex (210, 211) | |
| Orbitofrontal cortex (111, 211) | Orbitofrontal cortex |
| Insular cortex (113, 213) | Insular cortex |
| Ventral anterior cingulate cortex (124, 224) | Anterior cingulate cortex |
| Dorsal anterior cingulate cortex (132, 232) | |
| Cerebellar | Cerebellar |
| Cerebellum (160–193) | Cerebellum |
| Visual | Visual |
| Primary visual cortex (117, 217) | Visual cortices (V1–V6) |
| Secondary visual cortex (118, 218) | |
| Associative visual cortex (119, 219) | |
| Sensorimotor | Sensorimotor |
| Primary motor cortex (104, 204) | Primary motor cortex |
| Somatosensory associative cortex (105, 205) | Secondary somatosensory cortex |
| Premotor cortex (106, 206) | Premotor cortex |
| Auditory | Auditory |
| Superior temporal gyrus (122, 222) | Superior temporal gyrus |
| Auditory cortex (141, 241) | Primary auditory cortex |
| Associative auditory cortex | |
| Default mode | Default mode |
| Hippocampus (36, 37) | Hippocampus |
| Anterior prefrontal cortex (110, 210) | Dorsal medial prefrontal cortex |
| Ventral medial prefrontal cortex | |
| Orbitofrontal cortex (111, 211) | Orbitofrontal cortex |
| Inferior temporal gyrus (120, 220) | Inferior temporal gyrus |
| Ventral posterior cingulate cortex (123, 223) | Posterior cingulate cortex |
| Dorsal posterior cingulate cortex (131,231) | |
| Retrosplenial cingular cortex (129, 229) | Retrosplenial cortex |
| Anterior entorhinal cortex (134, 234) | Entorhinal cortex |
| Parahippocampal cortex (136, 236) | Parahippocampal cortex |
| Inferior parietal lobule |
Anatomies from a standard pig brain atlas (Saikali et al., 2010; atlas labels in parentheses) that were combined to create six reference pig RSNs (left column) and corresponding human anatomies known to be associated with the same six RSNs (right column).
RSNs, resting-state networks.
To determine which brain connectivity decomposition method best identified the six reference RSNs, the functional activation maps generated by each method were thresholded by using a z-score of 1, normalized between 0 and 1, and then spatially normalized to the anatomical space of the template pig. Spatial normalization was accomplished by calculating a spatial transformation using the template pigs' fMRI data and anatomical data; then applying the calculated transformation to the activation maps.
The activation maps that produced the maximum Pearson spatial correlation coefficients for each of the six reference RSNs were then determined for each method, and mean ratio values were calculated for the selected activation maps. The Pearson spatial correlation coefficient was defined as
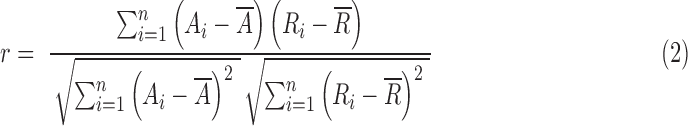 |
where n is the number of voxels within the brain, A is the activation value or sparse coefficient, R is the atlas value (which is binary), and an overbar denotes the arithmetic mean. The mean ratio was defined as
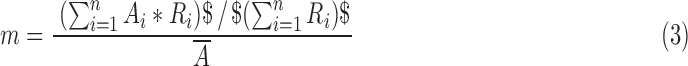 |
or simply stated, the mean activation value within a given RSN atlas divided by the mean activation value of the entire brain. Rose maps of the Pearson and mean ratio values were used to evaluate the effectiveness of each method at generating activation maps that correlate well with the reference pig RSN atlas.
DTI analysis
Group tractography was performed by using an early registration and superset method (Vo et al., 2013), which included two spatial transformations. First, the DTI dataset of each pig was spatially normalized to the DTI space of the template pig. The spatial transformation was calculated by using the DTI volumes of each pig that were acquired when no diffusion gradient (b = 0) was applied, and the transformation was then applied to all DTI volumes. Next, the volumes from all of the spatially normalized datasets were combined into a group superset, giving a total of 396 (33 diffusion gradient directions per pig × 12 pigs) DTI volumes, and brain tissue was separated from surrounding tissue by manual segmentation.
The DTI superset was then spatially normalized to the anatomical space. The second spatial transformation was calculated by using the DTI volume of the template pig acquired with no diffusion gradient applied (b = 0) and the anatomical data, and the transformation was then applied to all volumes within the superset. During this second transformation, voxel size was forced to be 0.75 mm cubed, opposed to the 0.5 mm cubed voxels of anatomical space, to reduce the computational size of the dataset. The DTI gradient vectors were corrected accordingly based on each spatial transformation applied to the DTI data.
Tractography was performed on the spatially normalized DTI superset by using Medinria (Toussaint et al., 2007). The Tensor ToolKit's (TTK) tensor estimation algorithm was used to determine the DTI tensors, and from the estimated tensors, MD and FA maps were created. Tractography was also performed by using the TTK's tensor tractography algorithm. Whole-brain tractography was performed with fibers seeding from voxels with FA greater than approximately double the whole-brain average, fibers stopping at voxels with FA less than approximately two-thirds of the whole-brain average, and the fibers were not restricted to any minimum FL.
In addition, tractography of just the major fibers was performed with fibers seeding from voxels with FA greater than approximately double the whole-brain average, fibers stopping at voxels with FA less than approximately four-thirds of the whole-brain average, and a minimum FL of 10 mm.
Before making quantitative measurements, the activation maps determined by the pig RSN analysis described earlier were corrected by removing any regions of the activation volume that were not overlapping with gray matter structures. Using the whole-brain tractography result, measurements of MD, FA, and FL were then obtained for the fibers intersecting with the remaining activation volume. The FD, defined as the number of fibers intersecting with an activation volume divided by the activation volume, was also determined. All measurements of MD, FA, FL, and FD were normalized by their respective whole-brain averages, calculated from all brain structures for comparison.
Results
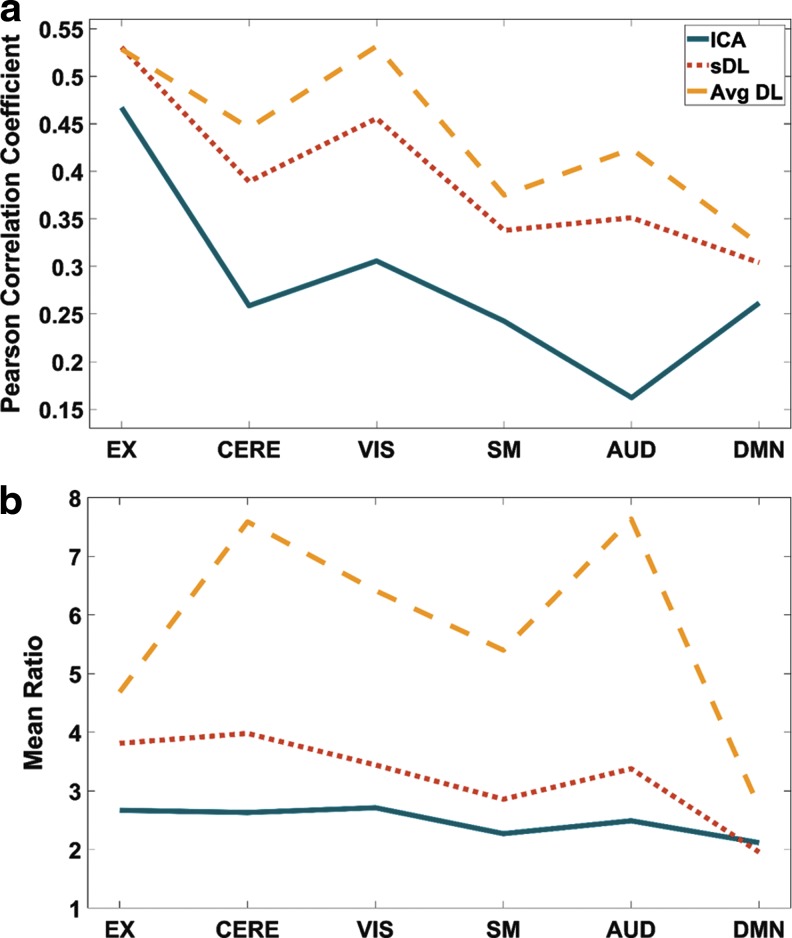
Using the Pearson correlation analysis described earlier, the maximum Pearson values obtained from the 20 activation maps generated by using ICA and the 292 maps generated by using sDL for each reference RSN are given in Figure 2a, and Figure 2b gives the associated mean ratio values for these maps. The activation maps produced by sDL show stronger Pearson correlations with the atlas than the maps produced by ICA (values in the range of 0.30–0.53 and 0.16–0.47, respectively), as well as greater mean ratio values (range of 2.00–3.98 and 2.12–2.71, respectively).
FIG. 2.
Maximum Pearson spatial correlation coefficient values (a) and corresponding mean ratio values (b) from activation maps generated by ICA (solid line), sDL (dotted line), and averaged sDL (dashed line) for six RSNs defined by the reference pig RSN atlas (EX, CERE, VIS, SM, AUD, and DMN). AUD, auditory; CERE, cerebellar; DMN, default mode; EX, executive control; ICA, independent component analysis; sDL, sparse dictionary learning; SM, sensorimotor; VIS, visual.
It was observed that sDL tends to produce multiple maps with similar activation patterns and similarly high Pearson values for each RSN from the atlas. When using a complete or over-complete dictionary to achieve sparseness, redundant representations of the same signal are possible. Therefore, averaged sDL activation maps were created by averaging the top three maps with the highest Pearson values for each reference RSN. These averaged maps produce even stronger Pearson correlations with the reference atlas (range of 0.32–0.53) and even higher mean ratio values (range of 2.72–7.64) in comparison to values obtained by ICA and non-averaged sDL (Fig. 2).
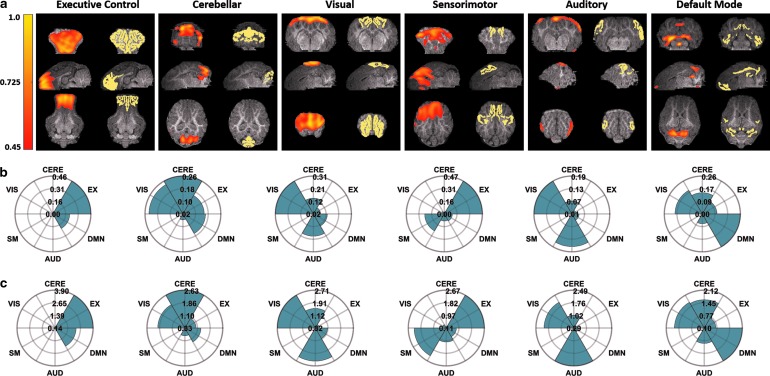
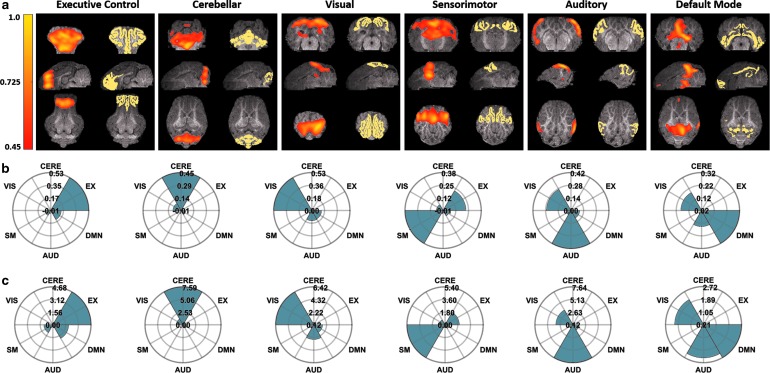
Cross-sectional images of the activation maps associated with the maximum Pearson values generated by ICA for each reference RSN are displayed in Figure 3a, as well as cross-sectional images of the reference RSN itself. All cross-sectional images are overlaid on the corresponding anatomical images. Rose maps giving the associated Pearson values (Fig. 3b) and mean ratio values (Fig. 3c) for the displayed activation maps are also presented.
FIG. 3.
(a) Representative cross-sectional images of activation maps generated by using ICA (left) for six RSNs and the corresponding reference pig RSN atlas (right). All activation map and atlas cross-sections are overlaid on the corresponding anatomical images. (b, c) Rose maps of the corresponding Pearson spatial correlation coefficient values (b) and mean ratio values (c) for the displayed activation maps.
The activation map generated by ICA that gives the maximum Pearson value for the EX RSN shows a strong correlation with this RSN exclusively, as evidenced by its low correlations with the other five RSNs within the reference atlas (Fig. 3b, c, first column). However, this exclusivity is not consistent for all RSNs. For the CERE, VIS, and DMN RSNs, the maximally correlated activation maps simultaneously exhibit comparatively strong correlations with their respective RSNs and other RSNs as well (Fig. 3b, c, second, third, and sixth column); for the SM and AUD RSNs, the maximally correlated activation maps show stronger correlations with other RSNs within the atlas than with their respective RSN (Fig. 3b, c, fourth and fifth column).
The maximally correlated activation maps generated by sDL for the EX, CERE, and VIS RSNs all show strong and exclusive correlations with their respective RSNs (Fig. 4b, c, first–third columns). However, for the SM, AUD, and DMN RSNs, sDL produced maximally correlated activation maps with comparatively strong correlations with other RSNs as with their respective RSNs (Fig. 4b, c, fourth–sixth columns). In comparison to ICA, sDL tends to produce maps that correlate better with an exclusive RSN.
FIG. 4.
(a) Representative cross-sectional images of activation maps generated by using sDL (left) for six RSNs and the corresponding reference pig RSN atlas (right). All activation map and atlas cross-sections are overlaid on the corresponding anatomical images. (b, c) Rose maps of the corresponding Pearson spatial correlation coefficient values (b) and mean ratio values (c) for the displayed activation maps.
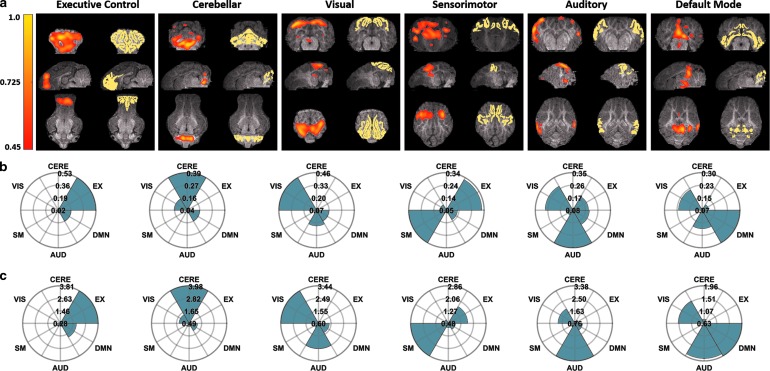
For the averaged sDL activation maps, strong and exclusive correlations between each maximally correlated activation map and its respective reference RSN are observed (Fig. 5b, c), except for the mean ratio of the DMN RSN. In comparison to the maps generated by ICA and sDL, the averaged sDL maps tend to correlate more strongly with an exclusive RSN. Whole brain 3D projections of each of the averaged sDL activation maps, as well as each of the reference atlases, can be found in Supplementary Figures S4 and S5.
FIG. 5.
(a) Representative cross-sectional images of the averaged sDL activation maps (left) for six RSNs and the corresponding reference pig RSN atlas (right). All activation map and atlas cross-sections are overlaid on the corresponding anatomical images. (b, c) Rose maps of the corresponding Pearson spatial correlation coefficient values (b) and mean ratio values (c) for the displayed activation maps.
Correlations between the averaged sDL activation maps and the individual anatomical components of each reference RSN given in Table 1 were further explored by using spatial Pearson correlations, mean ratios, and the volumetric percentage of each anatomical component in relation to its corresponding RSN volume, as determined from the reference pig RSN atlas (Supplementary Table S1). Good correlations and high mean ratios were observed for the majority of individual anatomical components in each RSN. However, poor correlations were observed between the EX network and the ventral anterior cingulate cortex and between the DMN and the anterior prefrontal, orbitofrontal, and parahippocampal cortices.
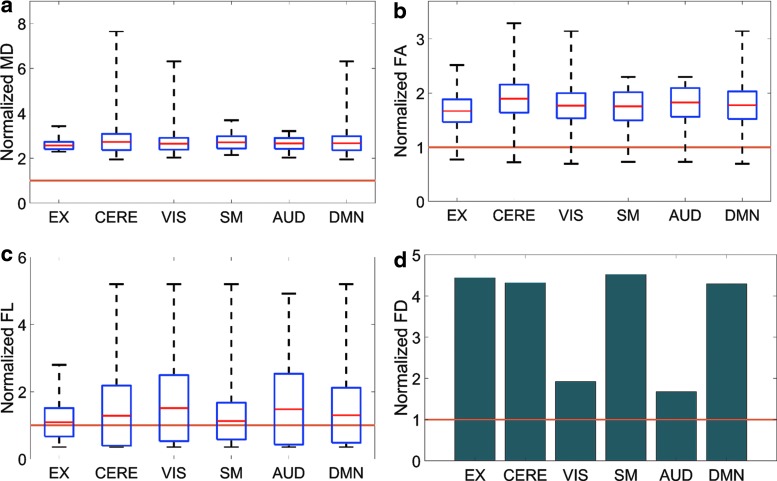
The averaged sDL activation maps were also further evaluated by examining the normalized MD, FA, FL, and FD measurements determined for the fibers intersecting with each activation volume (Fig. 6). The overwhelming majority of the MD and FA values for the fibers intersecting with each activation volume are greater than the average value of the whole brain (Fig. 6a, b), whereas the average FL of these fibers only tends to be slightly greater than the average FL in the whole brain (Fig. 6c). The EX, CERE, SM, and DMN RSNs all have FD values of approximately four times greater than the average FD of the whole brain; whereas the VIS and AUD RSNs only have FD values of approximately two times greater than the average FD of the whole brain (Fig. 6d).
FIG. 6.
Normalized MD (a), FA (b), FL (c), and FD (d) measurements for the fibers intersecting with each activation volume of the averaged sDL activation maps. The lower and upper whiskers of the boxplots represent the minimum and maximum measurements, respectively. The central red line in each box represents the mean of the measurements, and the box represents one standard deviation from the mean. All measurements were normalized by their respective whole-brain averages, represented as the brown line across all six RSNs. FA, fractional anisotropy; FD, fiber density; FL, fiber length; MD, mean diffusivity.
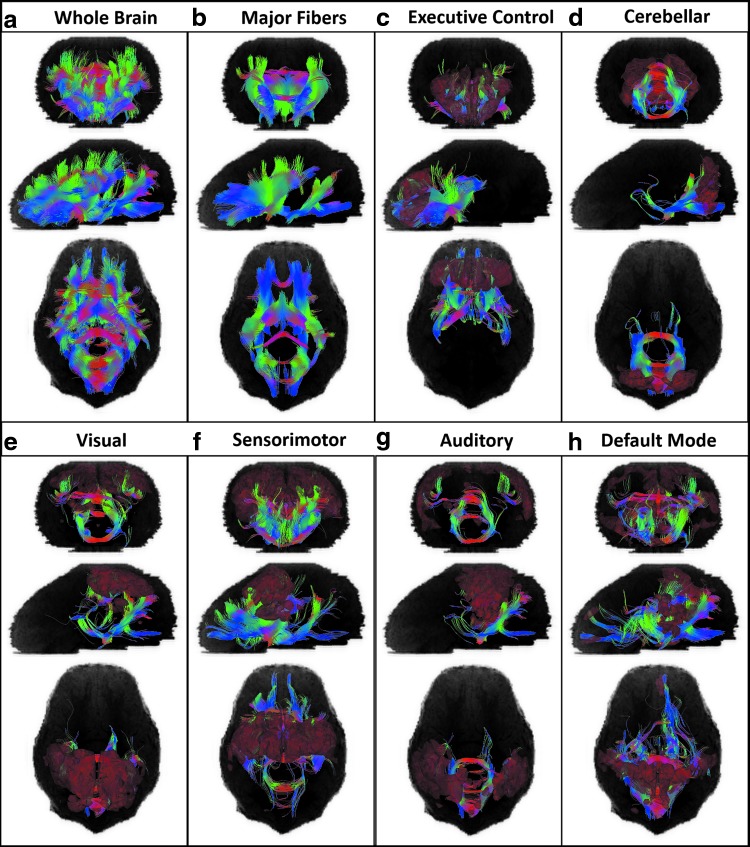
To better demonstrate how structural fibers interact with each individual RSN, tractography of the whole brain, major fibers, and the fibers intersecting with the activation volume for each RSN determined from the averaged sDL activation maps are shown in Figure 7. The activation maps were corrected by removing any regions of the activation volume that were not overlapping with gray matter structures.
FIG. 7.
Group tractography of the whole brain (a) and major fibers (b) obtained by using an early registration and superset method, and the fibers intersecting with the activation volume (shown in red) for each RSN determined from the averaged sDL activation maps (c–h). The activation maps were corrected by removing any regions of the activation volume that were not overlapping with gray matter structures. Coronal, sagittal, and axial views are projections from the front, left, and top of the brain, respectively.
Discussion
Using ICA and sDL, the described empirical analysis generated six activation maps that have high degrees of correlation with the six reference pig RSNs (EX, CERE, SM, VIS, AUD, and DMN), which were derived from well-established human RSNs, indicating a high degree of homology. This finding suggests that there are RSNs in the translational pig brain model that may be homologous to RSNs found in the human brain. To the best of the authors' knowledge, this is the first study to identify RSNs in the pig brain and draw comparisons between those RSNs experimentally found in pigs and those previously established in humans.
In this work, the use of ICA and sDL to detect RSNs produces activation maps with Pearson values that fall within the typical range reported in the literature (Brookes et al., 2011; Lois et al., 2014; Smith et al., 2009); by averaging the top three sDL maps with the highest Pearson values, an averaged map can be generated that produces even stronger Pearson correlations in the range of 0.32–0.53 (Fig. 2). These maps demonstrate six well-defined RSNs within the pig brain (Fig. 5).
The detection of these RSNs using two separate methods, as well as evidence provided from structural connectivity analysis, further supports the validity of these RSNs, as functional and structural connectivity have been shown to be positively correlated (Greicius et al., 2008; Honey et al., 2009). The MD and FA values of the fibers intersecting with each RSN show much greater diffusion and directionality of the diffusion along structural pathways both through and within the RSNs in comparison to the average diffusion and directionality of the diffusion throughout the whole brain (Fig. 6a, b). The fibers intersecting with each RSN also show greater average FL than the average FL found throughout the whole brain (Fig. 6c), and each RSN's FD is greater than the FD found throughout the whole brain (Fig. 6d).
The discovery of the six RSNs defined in this work may help to establish an experimental RSN atlas of a healthy pig brain, which will allow for future investigation of the effects of neural diseases and disorders on major neural connections and functions within the pig brain model. There are a number of important pig neural injury, disease, and developmental models that cause functional changes closely associated with the six RSNs identified in this study.
One such example is TBI. Multiple types of TBI in both pigs and humans have previously been shown to cause changes in activity in the SM network, as well as the EX network and DMN network, resulting in a variety of symptoms, including deficits in motor coordination, proprioception, problem-solving, cognitive flexibility, and regular modulation of day and night activity patterns (Etkin and Wager, 2007; Iandolo et al., 2018; Jak et al., 2018; Tanaka et al., 2008; Ustinova et al., 2015).
Stroke, the number one cause of long-term disability in the world, is another example of a major neurological injury successfully modeled in pigs and may cause changes in RSN activity (Baker et al., 2017; Duberstein et al., 2014; Platt et al., 2014; Tanaka et al., 2008; Webb et al., 2018). In the pig model, studies found that ischemic stroke causes lesioning of the temporal and piriform lobes in regions housing the insular and AUD cortices, which are associated with the EX and AUD networks, respectively (Platt et al., 2014).
Middle cerebral artery occlusion stroke in pigs can lead to atrophy in the hippocampus, parahippocampal cortex, and inferior temporal gyrus regions, all of which are associated with the DMN network (Platt et al., 2014). The high amount (>60%) of white matter in the pig brain currently also makes it a suitable model for studying lucunar infarctions, small areas of necrosis in deep cerebral white matter occurring in 25% of ischemic stroke patients that can affect the activation of the primary motor cortex and SM network (Izquierdo et al., 2017; Swindale, 1998).
By identifying pig RSNs homologous to the human SM, AUD, EX, and DMN networks, which are associated with TBI and multiple types of ischemic stroke, the findings presented here further support the extended use of pigs as a translational model for characterizing the effects of TBI and stroke on functional network disruption and brain injury. Many previous pig studies of TBI, stroke, and nutrition (Mudd et al., 2017, 2018a,b) have heavily relied on observed tissue or whole animal function (e.g., behavior and cognitive tests, such as open field) changes; however, most were not capable of studying changes in brain functional networks due, in part, to the absence of developed pig functional network maps. This further highlights the importance of the pig RSNs reported in this study.
Despite the good correlations between the averaged sDL activation maps and most of the individual anatomical components of each RSN given in Supplementary Table S1, some poor correlations for specific RSN anatomies were observed, including: the ventral anterior cingulate cortex of the EX network and the anterior prefrontal, orbitofrontal, and parahippocampal cortices of the DMN. These poor correlations are possibly due to one or more of three reasons: (1) the anatomical region is not associated with the RSN, (2) anesthetic agents suppressed neural activity in the anatomical region, and/or (3) only a sub-region or fractional volume of the anatomical region is involved in the RSN.
Although minimal, feasible amounts of anesthetic agents were used to minimize interference with brain activity, the use of anesthesia is known to have potentially confounding effects on functional connectivity, as many anesthetic agents are known to interfere with neural activity and neurovascular coupling (Hamilton et al., 2017; Liang et al., 2012). It is possible that the RSNs reported in this study may have been affected by the anesthesia protocol, and specific anatomical regions may have been affected more than others.
This study is also potentially limited by the creation of the reference RSN atlas from a pre-existing standard pig brain atlas and the assumption that the reference atlas is perfectly homologous with the human brain. In humans, certain anatomies, such as the anterior prefrontal cortex, are known to consist of sub-regions that may be functionally activated separately from the rest of the region. For example, only the medial area of the prefrontal cortex is involved in the human DMN (Buckner et al., 2008), and this is also observed in the pig brain (Fig. 5a, sixth column). However, the anterior prefrontal cortex listed in the standard pig brain atlas (Saikali et al., 2010) was not segmented into smaller regions, and the medial area of the pig brain was unable to be differentiated from the rest of the region.
Among the four anatomical regions that were identified as having poor correlations with their corresponding functional activation maps, the orbitofrontal cortex is likely not associated with the DMN RSN. Since this anatomical region was also included in the reference atlas for the EX network and shows good correlation with the functional activation map for this network, significant interference from anesthesia is unlikely, and the volume of this region is relatively small, making fractional volume effects also unlikely.
Although anesthesia can have potentially confounding effects on functional connectivity, it should not interfere with structural connectivity. Therefore, to better determine which of the three reasons may have caused the poor correlations observed in the other three anatomical regions yet discussed (the ventral anterior cingulate, anterior prefrontal, and parahippocampal cortices), structural connectivity measures were evaluated for these regions (Supplementary Figs. S2 and S3).
For the ventral anterior cingulate cortex of the EX network, high structural connectivity measures were observed in comparison to the whole-brain values (Supplementary Fig. S2). Since structural connectivity and functional connectivity have been shown to be positively correlated (Greicius et al., 2008; Honey et al., 2009), it is hypothesized that interference from anesthesia may have possibly affected this region. Since the volume of this region is relatively small, fractional volume effects are considered less likely.
For the anterior prefrontal and parahippocampal cortices of the DMN, high structural connectivity measures were also observed in comparison to the whole-brain values (Supplementary Fig. S2). However, when comparing the measures of structural connectivity for the anatomical region defined by the reference atlas with the measures obtained for just the functionally activated sub-region from the averaged sDL activation maps of each anatomical region, increased FL and FD were observed within the sub-regions that were empirically determined to be activated (Supplementary Fig. S3). This observation, as well as the fact that these two anatomical regions are relatively large, leads to the hypothesis that these two regions only have fractional volumes that are involved in the DMN RSN.
As a whole, the DMN shows lower correlation values compared with the other five RSNs (Fig. 2). This is possibly due to these fractional volume activations and may also be due, in part, to the young age of the pigs used in this study. Studies have shown that in the human infant brain, there is limited evidence of DMN activation; whereas functional connectivity is more consistent in children aged 9–12 years and older, suggesting that the DMN undergoes developmental change (Broyd et al., 2009). A possible explanation for the lower correlation values observed for the DMN in this work is that the pigs used are still young (3 weeks old), and the DMN may not be fully developed at such a young age.
One other limitation of this study that the authors intend to improve on in future work is the relatively small sample size (n = 12). For larger sample sizes, better-defined RSNs may be obtained, possibly leading to stronger correlations with the reference pig RSN atlas and/or better determination of noninvolved anatomies contained within the atlas and fractional volumes may be feasible.
The results presented in this study provide a groundwork for the development of a standard pig brain functional network and RSN atlas, which can be used to support future translational pig model studies characterizing functional network disruption caused by disease and injury. Over the years, many studies have reported associations between functional and structural connectivity (Greicius, 2008; Honey et al., 2009), and some studies have shown that the strength of resting-state functional connectivity is positively correlated with that of structural connectivity (Damoiseaux and Greicius, 2009). However, other studies have detected functional connectivity between regions with little or no structural connectivity (Zimmermann et al., 2016), suggesting that functional connectivity is likely mediated by indirect structural connections.
In future work, the authors intend to continue exploring the relationship between functional and structural networks in the pig brain at multiple time points during brain development by using advanced deep learning methods such as convolutional neural networks and deep generative models (Shen et al., 2017), and they intend to explore how these networks are effected by acute ischemic stroke and potential treatments of stroke.
Conclusion
The use of the pig model in neuroscience has significantly increased in the past two decades. The pig brain, which is gyrencephalic, resembles the human brain more in anatomy, growth, and development than the brains of commonly used small laboratory animals such as rodents. Using ICA and sDL, six RSNs (EX, CERE, SM, VIS, AUD, and DMN) were detected in the pig brain that resemble their counterparts in humans, as measured by Pearson spatial correlations and mean ratios, and supporting evidence of the validity of these RSNs was provided through the evaluation of structural connectivity. This study shows that as a translational, large animal model, pigs demonstrate great potential for mapping connectome-scale functional connectivity in experimental modeling of human brain disorders.
Supplementary Material
Acknowledgments
This work was supported in part by NIH RO1 grant RO1NS093314, faculty research grant from Office of Research at the University of Georgia, and Georgia Experimental Agricultural Station, HATCH No. GEO00795.
Author Disclosure Statement
No competing financial interests exist.
Supplementary Material
References
- Ashburner J, Barnes G, Chen C, Daunizeau J, Flandin G, Friston K, et al. 2016. SPM12 manual. www.fil.ion.ucl.ac.uk/spm/doc/spm12_manual.pdf Last accessed April16, 2019
- Ashburner J, Friston KJ. 1999. Nonlinear spatial normalization using basis functions. Hum Brain Mapp 7:254–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Neelin P, Collins DL, Evans A, Friston K. 1997. Incorporating prior knowledge into image registration. Neuroimage 6:344–352 [DOI] [PubMed] [Google Scholar]
- Baker EW, Platt SR, Lau VW, Grace HE, Holmes SP, Wang L, et al. 2017. Induced pluripotent stem cell-derived neural stem cell therapy enhances recovery in an ischemic stroke pig model. Sci Rep 7:10075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal BF, Yetkin Z, Haughton VM, Hyde JS. 1995. Functional connectivity in the motor cortex of resting human brain using echo‐planar MRI. Magn Reson Med 34:537–541 [DOI] [PubMed] [Google Scholar]
- Breazile JE, Swafford BC, Thompson WD. 1966. Study of the motor cortex of the domestic pig. Am J Veterinary Res 27:1369–1373 [Google Scholar]
- Brookes MJ, Woolrich M, Luckhoo H, Price D, Hale JR, Stephenson MC, et al. 2011. Investigating the electrophysiological basis of resting state networks using magnetoencephalography. Proc Natl Acad Sci U S A 108:16783–16788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyd SJ, Demanuele C, Debener S, Helps SK, James CJ, Sonuga-Barke EJS. 2009. Default-mode brain dysfunction in mental disorders: a systematic review. Neurosci Biobehav Rev 33:279–296 [DOI] [PubMed] [Google Scholar]
- Buckner RL, Andrews‐Hanna JR, Schacter DL. 2008. The brain's default network. Ann N Y Acad Sci 1124:1–38 [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T. 2004. Group ICA of fMRI toolbox (GIFT). http://icatb.sourceforge.net Last accessed October17, 2018
- Campos-Ortega JA. 1970. The distribution of retinal fibres in the brain of the pig. Brain Res 19:306–312 [DOI] [PubMed] [Google Scholar]
- Chang C, Keilholz S, Miller R, Woolrich M. 2018. Mapping and interpreting the dynamic connectivity of the brain. Neuroimage 180:335–336 [DOI] [PubMed] [Google Scholar]
- Craner SL, Ray RH. 1991. Somatosensory cortex of the neonatal pig: I. Topographic organization of the primary somatosensory cortex (SI). J Comp Neurol 306:24–38 [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS. 2017. Effects of aging on functional and structural brain connectivity. Neuroimage 160:32–40 [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Greicius MD. 2009. Greater than the sum of its parts: a review of studies combining structural connectivity and resting-state functional connectivity. Brain Struct Funct 213:525–533 [DOI] [PubMed] [Google Scholar]
- Dobbing J, Sands J. 1979. Comparative aspects of the brain growth spurt. Early Hum Dev 3:79–83 [DOI] [PubMed] [Google Scholar]
- Duberstein KJ, Platt SR, Holmes SP, Dove CR, Howerth EW, Kent M, et al. 2014. Gait analysis in a pre- and post-ischemic stroke biomedical pig model. Physiol Behav 125:8–16 [DOI] [PubMed] [Google Scholar]
- Duhaime AC. 2006. Large animal models of traumatic injury to the immature brain. Dev Neurosci 28:380–387 [DOI] [PubMed] [Google Scholar]
- Etkin A, Wager TD. 2007. Functional neuroimaging of anxiety: A meta-analysis of emotional processing in PTSD, social anxiety disorder, and specific phobia. Am J Psychiatry 164:1476–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix B, Leger ME, Albe-Fessard D, Marcilloux JC, Rampin O, Laplace JP. 1999. Stereotaxic atlas of the pig brain. Brain Res Bull 49:1–137 [DOI] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC. Raichle ME. 2005. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A 102:9673–9678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund E. 1969. [Cytoarchitectonics of the mesencephalon and pons in the domestic pig (Sus scrofa domestica)]. Anat Anz 125:345–362 [PubMed] [Google Scholar]
- Gieling ET, Schuurman T, Nordquist RE, van der Staay FJ. 2011. The pig as a model animal for studying cognition and neurobehavioral disorders. Curr Top Behav Neurosci 7:359–383 [DOI] [PubMed] [Google Scholar]
- Greicius M. 2008. Resting-state functional connectivity in neuropsychiatric disorders. Curr Opin Neurol 21:424–430 [DOI] [PubMed] [Google Scholar]
- Greicius MD, Menon V, Supekar K, Dougherty RF. 2008. Resting-state functional connectivity reflects structural connectivity in the default mode network. Cereb Cortex 19:72–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton C, Ma Y, Zhang N. 2017. Global reduction of information exchange during anesthetic-induced unconsciousness. Brain Struct Funct 222:3205–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm IE, West MJ. 1994. Hippocampus of the domestic pig: a stereological study of subdivisional volumes and neuron numbers. Hippocampus 4:15–125 [DOI] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. 2009. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci U S A 106:2035–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, et al. 2013. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage 80:360–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iandolo R, Bellini A, Saiote C, Marre I, Bommarito G, Oesingmann N, et al. 2018. Neural correlates of lower limbs proprioception: an fMRI study of foot position matching. Hum Brain Mapp 39:1929–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo A, Brigman JL, Radke AK, Rudebeck PH, Holmes A. 2017. The neural basis of reversal learning: an updated perspective. Neuroscience 345:12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jak AJ, Jurick S, Crocker LD, Sanderson-Cimino M, Aupperle R, Rodgers CS, et al. 2018. SMART-CPT for veterans with comorbid post-traumatic stress disorder and history of traumatic brain injury: a randomised controlled trial. J Neurol Neurosurg Psychiatry 90:333–341 [DOI] [PubMed] [Google Scholar]
- Jelsing J, Hay-Schmidt A, Dyrby T, Hemmingsen T, Uylings HB, Pakkenberg B. 2006. The prefrontal cortex in the Gottingen minipig brain defined by neural projection criteria and cytoarchitecture. Brain Res Bull 70:322–336 [DOI] [PubMed] [Google Scholar]
- Junge D. 1977. [The topography and cytoarchitectonic of the diencephalon of the cow (Bos taurus var. domesticus L.). II. The internal structure of the diencephalon of the cow (Bos taurus var. domesticus L.) (author's transl)]. Anat Anz 141:478–497 [PubMed] [Google Scholar]
- Laird AR, Fox PM, Eickhoff SB, Turner JA, Ray KL, McKay DR, et al. 2011. Behavioral interpretations of intrinsic connectivity networks. J Cogn Neurosci 23:4022–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsell O. 1954. The development of the cerebellum of the pig. Anat Rec 118:73–107 [DOI] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N. 2012. Intrinsic organization of the anesthetized brain. J Neurosci 32:10183–10191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois G, Linke J, Wessa M. 2014. Altered functional connectivity between emotional and cognitive resting state networks in euthymic bipolar I disorder patients. PLoS One 9:e107829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zuo Y, Gu H, Waltz JA, Zhan W, Scholl CA, et al. 2007. Synchronized delta oscillations correlate with the resting-state functional MRI signal. Proc Natl Acad Sci U S A 104:18265–18269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J, Jiang X, Li X, Zhu D, Chen H, Zhang T, et al. 2015. Sparse representation of whole-brain fMRI signals for identification of functional networks. Med Image Anal 20:112–134 [DOI] [PubMed] [Google Scholar]
- Mairal J, Bach F, Ponce J, Sapiro G. 2010. Online learning for matrix factorization and sparse coding. J Mach Learn Res 11:19–60 [Google Scholar]
- Matsas R, Kenny AJ, Turner AJ. 1986. An immunohistochemical study of endopeptidase-24.11 (“enkephalinase”) in the pig nervous system. Neuroscience 18:991–1012 [DOI] [PubMed] [Google Scholar]
- McBride T, Arnold SE, Gur RC. 1999. A comparative volumetric analysis of the prefrontal cortex in human and baboon MRI. Brain Behav Evol 54:159–166 [DOI] [PubMed] [Google Scholar]
- Mudd A, Fil J, Knight L, Lam F, Liang ZP, Dilger R. 2018a. Early-life iron deficiency reduces brain iron content and alters brain tissue composition despite iron repletion: a neuroimaging assessment. Nutrients 10:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd A, Fleming S, Labhart B, Chichlowski M, Berg B, Donovan S, Dilger R. 2017. Dietary sialyllactose influences sialic acid concentrations in the prefrontal cortex and magnetic resonance imaging measures in corpus callosum of young pigs. Nutrients 9:1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd AT, Getty CM, Dilger RN. 2018b. Maternal dietary choline status influences brain gray and white matter development in young pigs. Curr Dev Nutr 2:nzy015-nzy15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otabe JS, Horowitz A. 1970. Morphology and cytoarchitecture of the red nucleus of the domestic pig (Sus scrofa). J Comp Neurol 138:373–389 [DOI] [PubMed] [Google Scholar]
- Penny WD, Friston KJ, Ashburner JT, Kiebel SJ, Nichols TE. 2011. Statistical Parametric Mapping: The Analysis of Functional Brain Images. Cambridge, MA: Elsevier [Google Scholar]
- Platt SR, Holmes SP, Howerth EW, Duberstein KJJ, Dove CR, Kinder HA, et al. 2014. Development and characterization of a Yucatan miniature biomedical pig permanent middle cerebral artery occlusion stroke model. Exp Transl Stroke Med 6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikali S, Meurice P, Sauleau P, Eliat PA, Bellaud P, Randuineau G, et al. 2010. A three-dimensional digital segmented and deformable brain atlas of the domestic pig. J Neurosci Methods 192:102–109 [DOI] [PubMed] [Google Scholar]
- Sauleau P, Lapouble E, Val-Laillet D, Malbert C. 2009. The pig model in brain imaging and neurosurgery. Animal 3:1138–1151 [DOI] [PubMed] [Google Scholar]
- Shen D, Wu G, Suk HI. 2017. Deep learning in medical image analysis. Ann Rev Biomed Eng 19:221–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, et al. 2009. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci U S A 106:13040–13045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindale NV. 1998. Orientation tuning curves: empirical description and estimation of parameters. Biol Cybern 78:45–56 [DOI] [PubMed] [Google Scholar]
- Szteyn S, Galert D, Dynowski J, Hoczyk W. 1980. The stereotaxic configuration of hypothalamus nerve centres in the pig. Anat Anz 147:12–32 [PubMed] [Google Scholar]
- Tanaka Y, Imai H, Konno K, Miyagishima T, Kubota C, Puentes S, et al. 2008. Experimental model of lacunar infarction in the gyrencephalic brain of the miniature pig. Stroke 39:205–212 [DOI] [PubMed] [Google Scholar]
- Toussaint N, Souplet J-C, Fillard P. MedINRIA: Medical Image Navigation and Research Tool by INRIA. In Proceedings of MICCAI, Brisbane, Australia, 2007, p. 280 [Google Scholar]
- Ustinova KI, Chernikova LA, Dull A, Perkins J. 2015. Physical therapy for correcting postural and coordination deficits in patients with mild-to-moderate traumatic brain injury. Physiother Theory Pract 31:1–7 [DOI] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, et al. 2007. Intrinsic functional architecture in the anaesthetized monkey brain. Nature 447:83. [DOI] [PubMed] [Google Scholar]
- Vo A, Argyelan M, Eidelberg D, Uluğ AM. 2013. Early registration of diffusion tensor images for group tractography of dystonia patients. J Magn Reson Imaging 37:67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb RL, Kaiser EE, Jurgielewicz BJ, Spellicy S, Scoville SL, Thompson TA, et al. 2018. Human neural stem cell extracellular vesicles improve recovery in a porcine model of ischemic stroke. Stroke 49:1248–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Lv J, Li X, Zhu D, Jiang X, Zhang S, et al. 2018. Experimental comparisons of sparse dictionary learning and independent component analysis for brain network inference from fMRI data. IEEE Trans Biomed Eng 66:289–299 [DOI] [PubMed] [Google Scholar]
- Zimmermann J, Ritter P, Shen K, Rothmeier S, Schirner M, McIntosh AR. 2016. Structural architecture supports functional organization in the human aging brain at a regionwise and network level. Hum Brain Mapp 37:2645–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.