Abstract
Background
Over the last decade, reports about dengue cases have increase worldwide, which is particularly worrisome in South America due to the historic record of dengue outbreaks from the seventeenth century until the first half of the twentieth century. Dengue is a viral disease that involves insect vectors, namely Aedes aegypti and Ae. albopictus, which implies that, to prevent and combat outbreaks, it is necessary to understand the set of ecological and biogeographical factors affecting both the vector species and the virus.
Methods
We contribute with a methodology based on fuzzy logic that is helpful to disentangle the main factors that determine favorable environmental conditions for vectors and diseases. Using favorability functions as fuzzy logic modelling technique and the fuzzy intersection, union and inclusion as fuzzy operators, we were able to specify the territories at biogeographical risk of dengue outbreaks in South America.
Results
Our results indicate that the distribution of Ae. aegypti mostly encompasses the biogeographical framework of dengue in South America, which suggests that this species is the principal vector responsible for the geographical extent of dengue cases in the continent. Nevertheless, the intersection between the favorability for dengue cases and the union of the favorability for any of the vector species provided a comprehensive map of the biogeographical risk for dengue.
Conclusions
Fuzzy logic is an appropriate conceptual and operational tool to tackle the nuances of the vector-illness biogeographical interaction. The application of fuzzy logic may be useful in decision-making by the public health authorities to prevent, control and mitigate vector-borne diseases.
Keywords: Aedes aegypti, Ae. albopictus, Favorability function, Fuzzy operators, Vector-illness interaction
Background
Dengue is one of the diseases with most epidemiological global relevance in the last decades [1–9]. Over this century, dengue has become a growing public health problem and about half of the world’s population is currently at risk of dengue infection [7, 8, 10–12]. This is especially a concern in South America, where historical records of dengue epidemic outbreaks report upsurges every three or five years from the seventeenth century until the first half of the twentieth century [13, 14].
Aedes mosquitoes, namely the yellow fever mosquito (Aedes aegypti) and the Asian tiger mosquito (Ae. albopictus), are the most important dengue vectors in the world [15–17]. The number of studies about the mosquitoes of the genus Aedes as transmission vectors of human infectious diseases has recently increased remarkably [7, 10, 18–21]. Different authors have studied relevant aspects of mosquito-dengue relationships from phylogenetic [22], ecological [17–19], physicochemical [20, 23], genetic [21] and biogeographical perspectives [7, 10, 24, 25].
A biogeographical approach to the study of zoonotic diseases, known as pathogeography, has contributed with relevant advances in the knowledge of infectious disease macroecology and distribution [26–29]. It has also provided a proper analytical framework for the study of vector-illness interaction useful for management or surveillance. Species distribution models (hereinafter SDM) have been particularly used to investigate the environmental drivers for the distribution of Aedes species [25], to map the global distribution of the Aedes species according to the effect of temperature [10, 25], precipitation, and some land cover variables [7, 10], or to forecast the possible effects of climate change scenarios for Aedes species distributions [10]. Other studies also took into account economic information [4], or focused on predicting and determining the global burden of dengue [4, 24]. However, the biogeographical framework of vector-illness interaction that could reveal the large-scale risk of dengue occurrence remains poorly understood.
The current range occupied by Aedes mosquitoes (Ae. aegypti and Ae. albopictus) in South America is wider than the known dengue cases. For some reason not yet fully elucidated, there are territories occupied by Aedes vectors with and without dengue cases. This suggests that the relationship between the occurrence of Aedes mosquito populations and cases of dengue is not clear-cut, and that a fuzzy-logic approach is worth considering. In contrast to crisp logic, Zadeh [30] proposed the fuzzy set theory, which avoids the use of discrete true-or-false syllogisms, thus conferring a conceptual malleability suitable for real-life situations. Salski and Kandzia [31] emphasized the continuous character of nature, which implies that living beings are distributed in time and space essentially in a gradual and fuzzy manner. A fuzzy logic approach is consequently useful for processing and modelling environmental data [32]. Thus, the application of fuzzy logic could be helpful to recognize the biogeographical vector-illness interaction and the dynamism in the risk of dengue occurrence, and to establish the biogeographical framework in which the disease occurs.
Fuzzy logic led to the notion of environmental favorability, a concept related to, but different from, probability of occurrence [33]. Favorability functions can be used in SDM, and are particularly helpful when models of several species are involved in the study, as they allow the comparison between models for species or cases differing in prevalence, using fuzzy logic tools [28, 34–38].
In this study, our aims were to establish the biogeographical context in which dengue cases occur in South America and to map the areas favourable for new cases to occur. We assessed vector-illness biogeographical relationship using fuzzy logic to determine the different environmental drivers that favor the occurrence of both Aedes vectors and of dengue cases. We aimed to identify the territories more at biogeographical risk of dengue outbreaks, which may be helpful in order to apply measures for the management and control of this recurrent disease.
Methods
Study area and species range
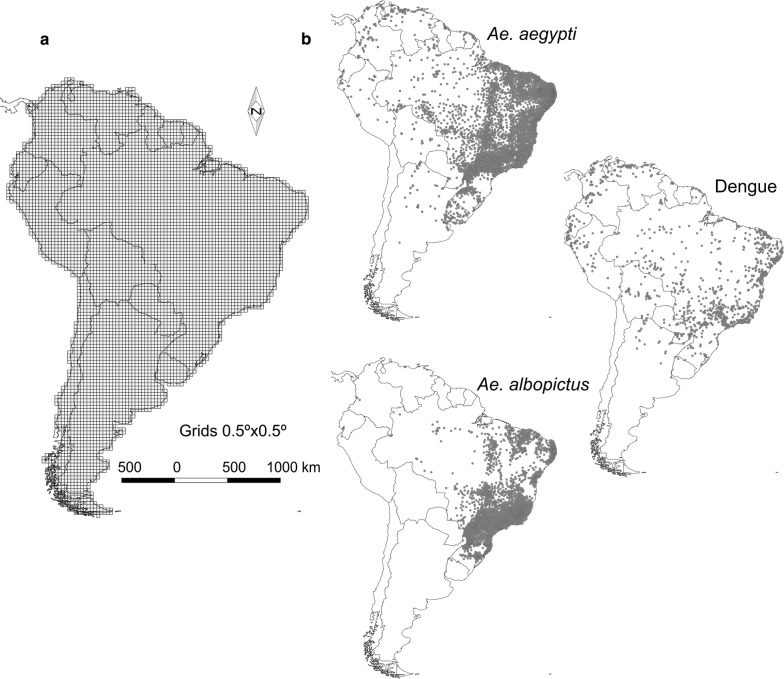
We analyzed Ae. aegypti, Ae. albopictus and dengue virus occurrences on a 0.5° × 0.5° grid (6430 cells of approximately 50 km × 50 km at the equator) to identify the biogeographical relationship between Aedes vectors and dengue cases. We used grids instead of geographical locations, thus solving a large part of the spatial autocorrelation problems derived from sampling bias or observation spatial clustering. Dengue virus occurrences were obtained from global occurrence records published from 1960 to 2012 in Messina et al. [5], with 731 grid cells with confirmed cases, which cover 11.37% of South America (Fig. 1). Aedes aegypti and Ae. albopictus occurrences were obtained from the global compendium of Aedes aegypti and Ae. albopictus occurrence [8] and from the Faculty of Science of the Republic University of Uruguay (inbuy.fcien.edu.uy, accessed in May 2017), with data spanning from 1960 to 2013, and from 1986 to 2014, respectively. Aedes aegypti was confirmed to occur in 1688 cells whereas Ae. albopictus presence was confirmed in 957 cells, covering 26.25 and 14.88% of South America, respectively (Fig. 1).
Fig. 1.
Study area and distribution data: a the grid of 0.5° latitude × 0.5° longitude squares in which the study area was divided to represent the occurrence data; b occurrence data of vectors and dengue infection cases. The grid layer was created with the tool “Create grid” of the software QGIS (www.qgis.org). The country layer was obtained from https://www.naturalearthdata.com and licensed CC BY. The maps were developed using QGIS in the composer tool. The final composition was created using CorelDRAW X8
Environment predictors and distribution modelling
We modelled the distribution of the two vector species and of dengue virus occurrence (the target variables) on the basis of a set of explanatory variables that could potentially affect them at the spatial resolution here applied [39, 40] (Table 1). The explanatory variables were related to different environmental factors that could determine the area occupied by both Aedes species and the extent of dengue virus occurrence in South America: spatial configuration, topography, climate (rainfall and temperature), hydrology, land use and other human activities (Table 1).
Table 1.
Explanatory variables used in Ae. aegypti, Ae. albopictus and dengue virus models in South America. Climate variables which do not have a pairwise correlation value above 0.80 according to Spearmanʼs test are shown in bolditalic
| Abbreviation | Variable | Abbreviation | Variable |
|---|---|---|---|
| SP | Spatial lineal combination (ysp)a | ||
| Topography | |||
| A | Mean altitude (m)b | S | Slope (◦) (calculated from altitude) |
| DA | Difference altitude (m) (calculated from altitude) | ON/S | Orientation N/S (calculated from slope) |
| Climatic variables | |||
| BIO1 | Mean annual temperature (°C) c | BIO11 | Mean annual temperatures of the coldest quarter (°C)c |
| BIO2 | Mean diurnal range temperatures (°C)c | BIO 12 | Annual precipitation (mm) c |
| BIO3 | Isotermality (BIO2/BIO17)(*100) (°C)c | BIO13 | Precipitation of the wettest month (mm)c |
| BIO4 | Seasonal temperatures (°C)c | BIO14 | Precipitation of the driest month (mm)c |
| BIO5 | Maximum temperatures of the warmest month (°C)c | BIO 15 | Seasonal precipitation (coeficiente de variación) (mm) c |
| BIO6 | Minimum temperatures of the coldest month (°C)c | BIO16 | Precipitation of wettest quarter (mm)c |
| BIO7 | Annual temperatures range (BIO5–BIO6)c | BIO17 | Precipitation of dry quarterc |
| BIO8 | Mean annual temperatures of the wetter quarterc | BIO 18 | Precipitation of warmest quarter c |
| BIO9 | Mean annual temperatures of the dry quarterc | BIO 19 | Precipitation of coldest quarter c |
| BIO10 | Mean annual temperatures of the warmest quarterc | ||
| Hydrology | |||
| DistRiver | Minimum distance to rivers (km)d | SumRiver | Sum of km of rivers per grid (km)d |
| Land use | |||
| Forests | Forests (%)e | Crops | Crops (%)e |
| NatField | Natural field (%)e | BareSoil | Bare soil (%)e |
| FlooVeg | Flooding vegetation (%)e | ||
| Human activities | |||
| PopDen | Population densityf | DistRoad | Minimum distance to paved roads (km)h |
| DistUrban | Minimum distance to urban centers (km)g | ||
aSpatial variables, latitude and longitude, were generated using QGIS (www.qgis.org) according to the vector geometry tools: (i) with “centroids of polygons” the centroid of each grid was calculated, and (ii) with “Export/Add columns of geometry” values of length and latitude expressed in the 1984 World Geodetic System were assigned to each centroid (WGS84). The spatial variable used in the multivariate modelling procedure is the linear polynomial combination (ysp) resulting from a spatial logistic regression
bUnited States Geological Survey. GTOPO30. Land Processes Distributed Active Archive Center. EROS Data Center, https://www.usgs.gov/centers/eros/science/usgs-eros-archive-digital-elevation-global-30-arc-second-elevation-gtopo30?qt-science_center_objects=0#qt-science_center_objects. 1996 (Accessed April 2016)
cWorldClim—Global Climate Data available. Described in: Fick, S. E. and R. J. Hijmans. Worldclim 2: New 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology. 2017. In: http://www.worldclim.org/ (Accessed May 2016)
dUnited States Geological Survey. HydroShed. Hydrological data and maps based on SHuttle Elevation Derivatives at multiple Scales. Available in: http://hydrosheds.cr.usgs.gov/index.php/ (Accessed May 2016)
eGlobCover 2009. Global land cover map. 2006. Avalaible at: http://due.esrin.esa.int/page_globcover.php (Accessed April 2016)
fGridded Population of the World (GPW), v4. Socioeconomic Data and Applications Center (SEDAC). A Data Center in NASA’s Earth Observing System Data and Information System (EOSDIS). Hosted by CIESIN at the Columbia University. 2010. (Accessed June 2016)
gNatural Earth Data. North American Cartographic Information Society (NACIS). Available at: http://www.naturalearthdata.com/ (Accessed April 2016)
hDiva-Gis 1.4, Plant Genetic Resources Newsletter. Available in: http://www.diva-gis.org/ (Accesed April 2016)
To define the spatial structure of each distribution, we considered a polynomial trend-surface analysis [41] that included a quadratic and cube effect of latitude and longitude and interactions between them. Spatial structure is known to be functional in biogeography, as purely spatial trends derive from biological processes such as history, spatial ecology and population dynamics [42]. Spatial structure is known to be functional in biogeography, as purely spatial trends derive from biological processes such as history, spatial ecology and population dynamics [42]. To define the spatial structure of each distribution, we considered a polynomial trend surface analysis [41] that included a quadratic and cube effect of latitude and longitude and interactions between them. For this, we performed a logistic regression of Ae. aegypti, Ae. albopictus and dengue virus distribution data on latitude (Lat), longitude (Lon), Lat2, Lon2, Lat3, Lon3, Lat × Lon, Lat2 × Lon and Lat × Lon2. Specifically, we performed a backward stepwise logistic regression with each event (both Aedes vectors and dengue cases) and those nine spatial terms as predictor variables in order to remove the non-significant spatial terms from models [41]. In this way, in the modelling procedure we included the resulting lineal combinations (ysp) as the spatial variable without non-significant spatial terms.
We used this spatial lineal combination (ysp) and the rest of variables listed in Table 1 (environmental factors) to produce distribution models according to all the explanatory factors together. To do this, we first analysed the effect of each explanatory variable on each target variable on a bivariate basis, by performing a logistic regression of each target variable on each explanatory variable separately. So, as Miller et al. [43] indicated, including the variation of the response variables separated into environmental and spatial components (represented by a trend surface of geographical coordinates) is a way to quantify the spatial dependence in distribution models ([44, 45], among others). We controlled the false discovery rate (FDR) with the aim of avoiding the increase in type I errors arising from the number of variables used in the analyses [46]. An explanatory variable was selected only when it was significantly related to the target variable (P < 0.05) under a FDR of q < 0.05, with q being the false discovery rate. Then, we calculated Spearman correlation coefficients to control multicollinearity between the selected explanatory variables. Out of any group of explanatory variables whose pairwise correlation value was higher than 0.80, we retained the variable most significantly related with the distribution of the target variable. In this way we obtained a filtered set of potentially explanatory variables for each target variable.
Finally, we performed a forward-backward stepwise logistic regression of the target variable on the polynomial combination of the spatial structure (ysp) and the filtered set of environmental variables, which produced increasingly more complex multivariate models while avoiding the inclusion of redundant variables. We used Akaike’s information criterion (AIC) to select the multivariate model that best-balanced information and parsimony (AIC; [47]). All analyses mentioned so far were performed with the fuzzySim R package [38]. Then, we evaluated the relative weight of each variable included in the models through the Wald parameter [48] using the survey package [49, 50]. Variables with non-significant coefficients left in the model (Chi-square test, P < 0.05) were eliminated until we obtained a model with all the coefficients significantly different from zero according to Crawley’s [51] procedure.
Then we used the Favorability Function according to Real et al. [33] and Acevedo and Real [52].
where F is the environmental favorability (ranging between 0 and 1), P is the probability of occurrence obtained from the multivariate logistic regression performed for each target variable, n1 is the number of presences and n0 in the number of absences, in each case. This analysis was carried out with the fuzzySim R package [38].
Favorability values factor out the weight of the initial species presence/absences ratio, inherent to any probability function [33, 52] and, thus, depend exclusively on the effect of the environmental conditions of the territory on the distribution under analysis. In addition, local favorability reflects the degree of membership of the locality in the fuzzy set of areas favorable for the occurrence of the event, so allowing the comparison between models through fuzzy logic tools [36, 52].
In this way, we obtained favorability models for the occurrence of the two Aedes vectors (Ae. aegypti and Ae. albopictus) and of dengue in South America, F-Ae. aegypti, F-Ae. albopictus and F-dengue, respectively. We evaluated the discrimination and classification capacity of the models with the modEva R package [53]. The discrimination ability of the models was evaluated using the area under the curve (AUC) [54], and the classification capacity was estimated through the model sensitivity, specificity, kappa and correct classification rate (CCR), using the value of F = 0.5 as classification threshold. We checked the autocorrelation spatial using the Moran’s I spatial autocorrelation statistic from the residuals of the models [55].
According to the thresholds proposed by Muñoz and Real [56], we calculated the number of grid cells in each South American country classified as highly favorable (F ≥ 0.8), for which the favorability odds are more than 4:1 in favor, hereinafter at high risk, and of intermediate favorability (0.2 < F < 0.8), which odds are under 4:1 and above 1:4 in favor, hereinafter vulnerable, for Aedes vectors, for dengue cases, and for vector-dengue cases simultaneously (see below).
Biogeographical vector-dengue relationships and dengue risk maps
We used the fuzzy modelling approach to assess the vector-dengue biogeographical interaction in South America. The logic underlying fuzzy sets was applied to the favorability function to indicate to what degree each grid cell belongs to the set of favorable areas for the presence of each species [52]. Then we used fuzzy logic tools to analyze the fuzzy vector-dengue biogeographical interactions and to detect the territories at high risk or vulnerable to new dengue cases.
Based on the values of F-Ae. aegypti, F-Ae. albopictus and F-dengue models, we calculated the fuzzy intersection (minimum favorability value for two events at a given location) [30] to identify the fuzzy set of areas simultaneously favorable for dengue outbreaks and for any of the two species separately (i.e. F-Ae. aegypti ∩ F-dengue and F-Ae. albopictus ∩ F-dengue). Then, we analyzed how the favorability for each vector presence and for the occurrence of dengue cases changed along the gradient of favorability intersection (i.e. of shared favorability for both vector and disease). To this aim, we established 10 bins of equal-range F-Ae. aegypti ∩ F-dengue values and F-Ae. albopictus ∩ F-dengue values and calculated in each bin the average favorability values for the corresponding vector species and for dengue virus. If the vector species is a limiting factor in the distribution of the disease, then the favorability for dengue should be equal to or lower than that for the mosquito along the shared favorability range.
We also calculated to what extent the favorable areas for dengue (F-dengue) are contained in those for F-Ae. aegypti and for F-Ae. albopictus models, by applying the fuzzy inclusion equation [57]:
which indicates how much the set A is included in the set B. In this way, we calculated the inclusion of one into the other for the models F-Ae. aegypti, F-Ae. albopictus and F-dengue, and also for vector-dengue intersections (i.e. F-Ae. aegypti ∩ F-dengue and F-Ae. albopictus ∩ F-dengue). Those fuzzy inclusion operations are defined in terms of the cardinal of each fuzzy set (i.e. the sum of the favorabilities values of all the grids). Thus, for example, the cardinal of F-Ae. aegypti ∩ F-dengue divided by the cardinal of F-dengue indicates the degree of inclusion of the distribution of dengue into that of Aedes aegypti.
To obtain the comprehensive biogeographical risk map for dengue in South America in the current context of vector-dengue biogeographical relationship, we identified the fuzzy set of areas simultaneously favorable for dengue outbreaks and for any of the two vector species. To do this we first calculated the fuzzy union of the favorability for any vector species, F-Ae. aegypti ∪ F-Ae. albopictus (or maximum favorability value for any of them), which can identify the fuzzy set of areas favorable to either vector species [30]. Then, we calculated the fuzzy intersection between F-Ae. aegypti ∪ F-Ae. albopictus and F-dengue [(F-Ae. aegypti ∪ F-Ae. albopictus) ∩ F-dengue].
Results
Favorable conditions for vectors and dengue cases
The variables that were significantly associated with the occurrence of each vector species and with dengue cases are shown in Table 2 (Additional file 1: Table S1). All the factors explained to some extent the occurrence of both vectors and dengue, with the exception of the hydrology for Ae. albopictus and hydrology and land use for dengue.
Table 2.
Predictor variables included in Ae. aegypti, Ae. albopictus and dengue cases favorability models. Signs in brackets show the positive or negative relationship between favorability and the variables in the models. The Wald parameter indicates the relative weight of every variable in each model. Variable abbreviations are given in Table 1
| Environmental factor | Ae. aegypti | Wald | Ae. albopictus | Wald | Dengue cases | Wald |
|---|---|---|---|---|---|---|
| Spatial situation | Sp | 1221.597 | Sp | 770.6308 | Sp | 252.8375 |
| Topography | A (−) | 15.03676 | A (+) | 30.7381 | ||
| O N/S (+) | 21.09944 | ON/S (+) | 65.61643 | ON/S (+) | 13.52308 | |
| S (+) | 11.75523 | S (+) | 8.108446 | |||
| Climatic | BIO1 (+) | 73.15295 | BIO1 (+) | 24.7797 | BIO1 (+) | 83.51969 |
| BIO7 (+) | 7.520641 | BIO7 (+) | 18.91607 | |||
| BIO12 (+) | 8.464809 | BIO12 (+) | 32.34416 | |||
| BIO15 (−) | 11.84533 | |||||
| BIO19 (−) | 17.86841 | BIO19 (−) | 25.6279 | |||
| Hydrology | SumRiver (−) | 10.33488 | ||||
| Land use | Crops (+) | 19.87312 | Crops (−) | 6.809295 | ||
| NatFields (−) | 6.809295 | |||||
| Human activities | PopDen (+) | 21.1916 | PopDen (+) | 7.74626 | PopDen (+) | 33.15417 |
| DistUrban (−) | 148.1178 | DistUrban (−) | 43.05502 | DistUrban (−) | 173.8293 | |
| DistRoad (−) | 7.287445 |
The distribution of dengue was favored in territories of a certain elevation (435.06 m.a.s.l. on average), of predominant orientation towards the south, of high mean annual temperatures (23.13 °C on average), with low precipitation in the colder months (185.86 mm on average), few differences between maximum and minimum precipitations, high population density (221 inhabitants/km2 on average) and moderate distance to urban centers (7750 m on average).
The distribution of both Aedes vectors was favored by similar variables with similar effect (positive or negative), except for the land use factor. A high proportion of crops was favorable for Ae. aegypti while it was unfavorable for Ae. albopictus. According to the Wald test, in both Ae. aegypti and dengue models, the three most explanatory variables were the spatial structure, closeness to urban centers and mean annual temperature (Table 2). The spatial structure, North-South orientation and proximity to urban centers were the three most explanatory variables for Ae. albopictus.
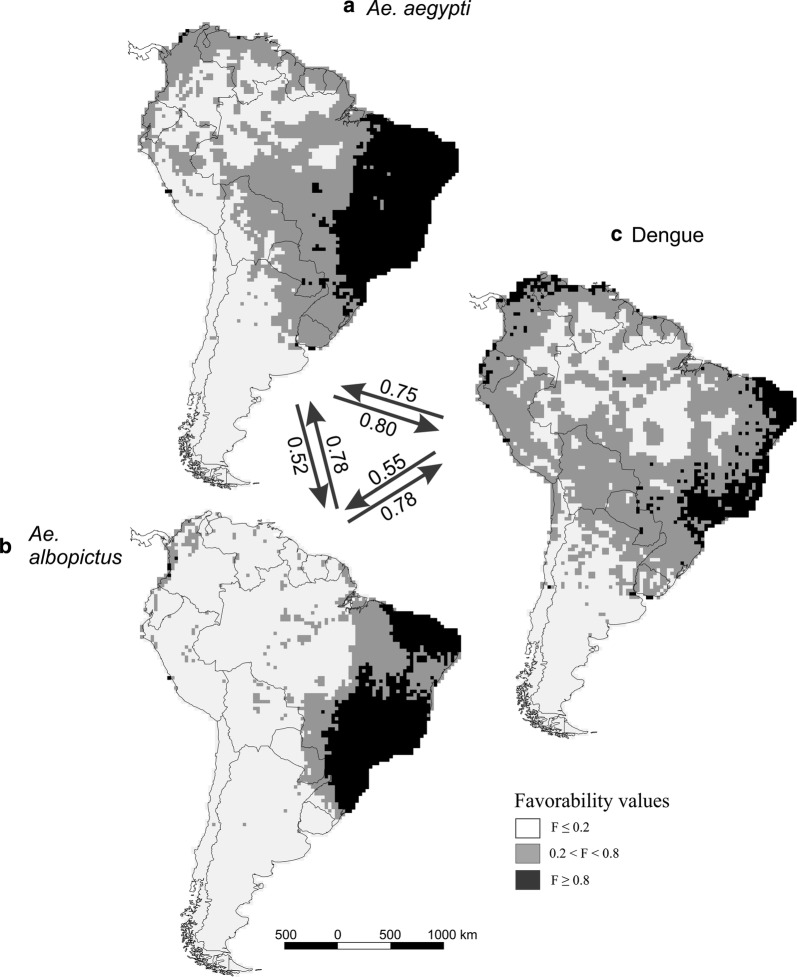
In Fig. 2 we show the cartographic favorability models for the vector species and for dengue (F-Ae. aegypti, F-Ae. albopictus and F-dengue) separately, with values grouped in three favorability classes: F values lower than 0.2 indicate low favorability, values between 0.2 and 0.8 indicate vulnerable areas, and values higher than 0.8 indicate areas at high risk [58]. The F-Ae. aegypti model depicted a large principal nucleus of high risk in Brazil, and some dispersed high-risk cells in Venezuela, Colombia, Peru, Paraguay, Argentina and Uruguay. The F-Ae. albopictus map revealed a main high-risk nucleus in Brazil, and some dispersed high-risk cells in Colombia and Peru. The F-dengue model detected two main nuclei of high risk for the occurrence of dengue cases, one in Brazil and another, more dispersed, in Venezuela, Colombia and Ecuador. Some dispersed high-risk cells are also found in Peru, Guyana, Surinam, Paraguay and Uruguay. The F-dengue model detected at least one vulnerable grid cell (0.2 < F < 0.8) in every South American country. Although only 11.37% (731 squares) of the total analyzed squares (n = 6430) have recorded dengue cases, 60.14% (3867) of the squares showed at least vulnerable conditions (F > 0.2) according the F-dengue model, while 8.94% of the cells (575) were at high risk (F ≥ 0.8).
Fig. 2.
Favorability models of: a Aedes aegypti, b Ae. albopictus and c dengue cases. Favorable areas are shown in black (favorability values or F ≥ 0.8), grey (0.2 < F < 0.8) and white (F ≤ 0.2). The arrows show inclusion values between the different models, one into the other. The maps were developed using QGIS (www.qgis.org) in the composer tool. The final composition was created using CorelDRAW X8
Both Aedes and dengue favorability models attained general acceptable scores according to the parameters considered to assess discrimination and classification capacities (Table 3). Discrimination (AUC) was always higher than 0.86, which is “excellent” according to Hosmer and Lemeshow [59]. Sensitivity values were always higher than 0.79, specificity was always higher than 0.74 and CCR was higher than 0.75. Kappa was higher than 0.6 for both Aedes vectors, which is “substantial” according to Landis and Kock [60]; it was 0.31, which is “fair”, for dengue cases. On the other hand, according to the analysis of residuals, we detected a minor autocorrelation (Moran’s I < 0.019), or approximately zero, below 1600 km and only in the Ae. albopictus model. These results indicate that there is no relevant spatial autocorrelation resulting from sampling bias with the grid system employed [55]. None of the Moran’s I-values were significant in the Ae. aegypti or the dengue models. The residuals did not show problems of spatial autocorrelation in our models and therefore we did not find relevant effects of spatial autocorrelation that invalidate our results.
Table 3.
Comparative assessment of models for Aedes aegypti, Ae. albopictus and dengue cases, as well as the fuzzy intersection between the vector species and dengue cases, according to their discrimination and classification capacity
| Evaluation indices | Favorability models | Vector-dengue favorability intersection | ||||
|---|---|---|---|---|---|---|
| Ae. aegypti | Ae. albopictus | Dengue cases | Ae. aegypti ∩ dengue | Ae. albopictus ∩ dengue | ||
| Discrimination | ||||||
| AUC | 0.914 | 0.966 | 0.862 | 0.844 | 0.794 | |
| Classification | ||||||
| Sensitivity | 0.791 | 0.918 | 0.819 | 0.640 | 0.511 | |
| Specificity | 0.871 | 0.901 | 0.742 | 0.848 | 0.881 | |
| CCR | 0.850 | 0.903 | 0.751 | 0.824 | 0.839 | |
| Kappa | 0.630 | 0.682 | 0.312 | 0.358 | 0.329 | |
Abbreviations: AUC, area under the ROC (receiving operating characteristic) curve; CCR, correct classification rate
Vector-dengue biogeographical interactions
Compared to the F-dengue model, both vector-disease intersections improved classification capacity according to kappa, CCR and specificity, whereas sensitivity and discrimination capacity decreased (Table 3).
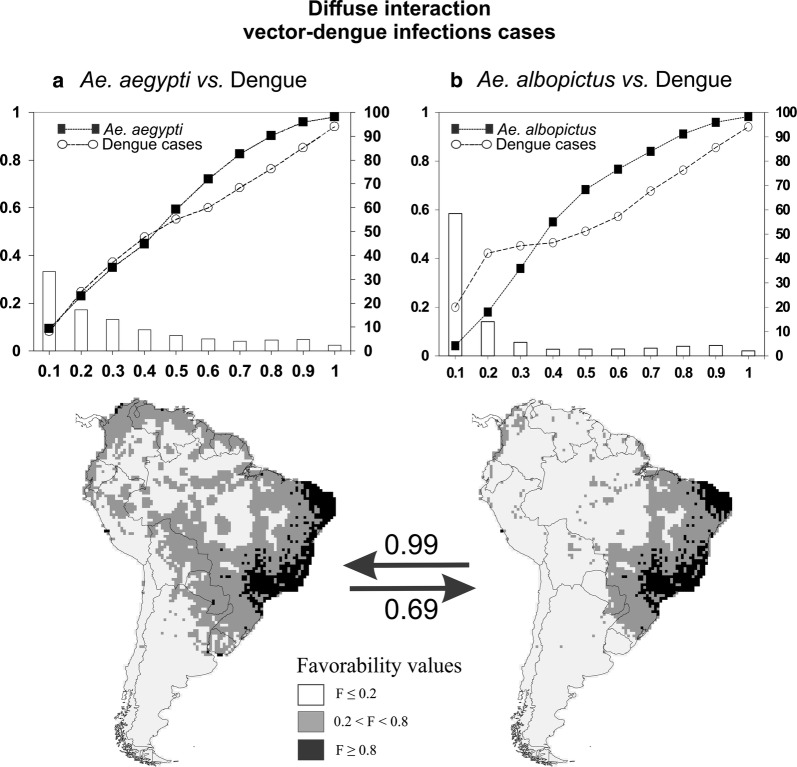
In Fig. 3 we show the fuzzy intersection between the favorability for dengue and vector species for both Ae. aegypti and Ae. albopictus in South America. Aedes aegypti and dengue favorability values increased together until a fuzzy intersection of 0.5 was reached; then, both continued to increase with higher favorability values for the mosquito (Fig. 3a). The intersection between Ae. albopictus and dengue favorability values indicated that dengue cases had higher favorability values than the mosquito up to fuzzy intersection = 0.3; after that point, the vector showed higher favorability values than the disease.
Fig. 3.
Plots and maps show the fuzzy intersection (simultaneous favorability) between the favorability for: a Ae. aegypti and dengue infection cases; and b favorability for Ae. albopictus and dengue infection cases. Fuzzy intersection values are shown on the horizontal axes (ranging from 0.1 to 1), grouped in 10 bins of values of equal favorability range. The average favorability values for both mosquito vectors in each bin are represented by solid lines and filled squares, and for dengue infection cases by dashed lines and blank circles, (on the left vertical axes ranging from 0 to 1). Columns represent the percentage of grid cells at each fuzzy intersection bin (on the right vertical axes). On maps, the arrows show inclusion values between both fuzzy intersection models (ranging from 0 to 1). The graphics were made using LibreOffice (https://es.libreoffice.org). The maps were developed using QGIS (www.qgis.org) in the composer tool. The final composition was created using CorelDRAW X8
According to the intersection between F-Ae. aegypti and F-dengue (Table 4), nine of the 14 South American countries have more than 50% of the country surface area at least vulnerable (F > 0.2) to dengue-cases occurrence transmitted by Ae. aegypti. In contrast, only Brazil has more than 50% of the country at least vulnerable (F > 0.2) to dengue-cases occurrence transmitted by Ae. albopictus. Seven South American countries have some locations at high risk (F ≥ 0.8) based on the intersection between F-Ae. aegypti and F-dengue. Two countries, Brazil and Peru, have locations at high risk of dengue occurrence due to Ae. albopictus, based on the intersection between the F-Ae. albopictus and F-dengue models (Table 4).
Table 4.
Percentages of the country surface with intermediate and high risk (F > 0.2, and F ≥ 0.8, respectively) of both vectors (Aedes aegypti and Ae. albopictus), of dengue cases, and of vector-dengue favorability intersection (with respect to the total number of grid cells per country in the leftmost column). Countries were ordered from highest to lowest percentage of the country surface of dengue cases detected in Messina et al. [5]
| Country | Cells by country | % of risk for Ae. aegypti | % of risk for Ae. albopictus | % of risk for dengue cases | % of risk intersection F-Ae. aegypti-F-dengue | % of risk intersection F-Ae. albopictus-F-dengue | % of the country with dengue cases | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F > 0.2 | F ≥ 0.8 | F > 0.2 | F ≥ 0.8 | F > 0.2 | F ≥ 0.8 | F > 0.2 | F ≥ 0.8 | F > 0.2 | F ≥ 0.8 | |||
| Brazil | 2860 | 84.056 | 42.343 | 58.776 | 33.741 | 69.720 | 16.049 | 67.552 | 15.839 | 53.497 | 14.196 | 16.958 |
| Colombia | 349 | 48.424 | 1.146 | 19.198 | 1.146 | 70.201 | 11.461 | 57.593 | 1.146 | 21.490 | 0.000 | 13.181 |
| Venezuela | 365 | 60.274 | 0.274 | 4.932 | 0.000 | 64.932 | 11.781 | 47.397 | 0.274 | 2.740 | 0.000 | 11.507 |
| Peru | 509 | 29.666 | 0.393 | 4.322 | 0.196 | 79.371 | 0.786 | 25.344 | 0.393 | 4.322 | 0.196 | 8.251 |
| Bolivia | 325 | 60.000 | 0.000 | 5.538 | 0.000 | 91.385 | 0.000 | 76.615 | 0.000 | 8.000 | 0.000 | 11.692 |
| Paraguay | 182 | 89.560 | 4.396 | 40.659 | 2.747 | 97.253 | 3.846 | 87.363 | 1.099 | 40.659 | 0.000 | 19.780 |
| Argentina | 1062 | 13.653 | 0.094 | 0.659 | 0.094 | 19.115 | 0.188 | 10.829 | 0.094 | 0.659 | 0.000 | 1.507 |
| Ecuador | 82 | 46.341 | 0.000 | 12.195 | 0.000 | 96.341 | 15.854 | 46.341 | 0.000 | 12.195 | 0.000 | 18.293 |
| French Guyana | 32 | 78.125 | 0.000 | 15.625 | 0.000 | 62.500 | 0.000 | 53.125 | 0.000 | 9.375 | 0.000 | 12.500 |
| Guyana | 72 | 58.333 | 0.000 | 4.167 | 0.000 | 58.333 | 1.389 | 61.111 | 0.000 | 5.556 | 0.000 | 5.556 |
| Surinam | 65 | 73.846 | 3.077 | 7.692 | 0.000 | 63.077 | 1.538 | 63.077 | 0.000 | 7.692 | 0.000 | 4.615 |
| Chile | 423 | 1.182 | 0.000 | 0.236 | 0.000 | 12.530 | 0.236 | 1.182 | 0.000 | 0.236 | 0.000 | 0.000 |
| Uruguay | 93 | 93.548 | 2.151 | 1.075 | 0.000 | 68.817 | 2.151 | 67.742 | 2.151 | 0.000 | 0.000 | 0.000 |
| Panamá | 11 | 100.000 | 0.000 | 27.273 | 0.000 | 100.000 | 18.182 | 54.545 | 0.000 | 9.091 | 0.000 | 0.000 |
Fuzzy-inclusion relationships between models
In Fig. 2 we show the values for F-Ae. aegypti, F-Ae. albopictus and F-dengue inclusion into one another and in Fig. 3 the values of the inclusion of the two vector-dengue intersections one into the other. The main results were that F-dengue was included in a higher proportion into F-Ae. aegypti (0.75) than into F-Ae. albopictus (0.55), and that the intersection F-Ae. albopictus ∩ F-dengue was more included into the F-Ae. aegypti ∩ F-dengue (0.99) model than vice versa (0.69).
Dengue risk map in the current biogeographical context of vector-dengue interaction
In Fig. 4, we show the comprehensive map of areas at high risk and vulnerable to dengue cases due to the two vector species combined, resulting from F-dengue ∩ (F-Ae. aegypti ∪ F-Ae. albopictus).
Fig. 4.
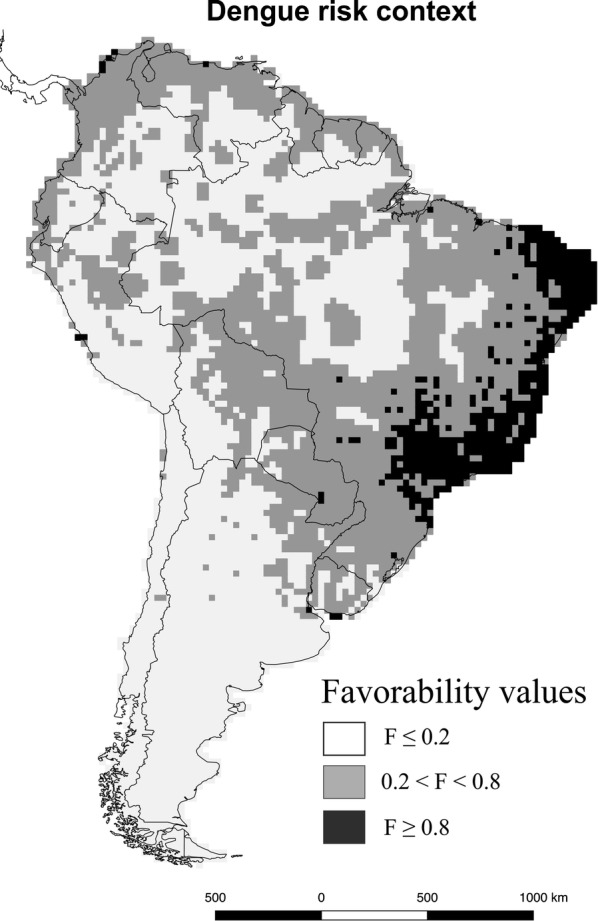
Dengue risk map in South America in the current biogeographical context of vector-dengue interaction. The map shows the intersection favorability values between the union of the favorability for the two mosquito species with the favorability for dengue (F-Ae. aegypti ∪ F-Ae. albopictus) ∩ F-dengue. The maps were developed using QGIS (www.qgis.org) in the composer tool. The final composition was created using CorelDRAW X8
Discussion
Environmental drivers of Aedes vectors and dengue cases in South America
Many studies have explained the occurrence of both Aedes species in South America in terms of only climate [10, 25], climate and some land cover variables [7], or climate and economic information [4]. In general, they detected that temperature was the main factor limiting the distribution of the two Aedes species. In contrast, our favorability models detected a more complex pattern of drivers for the presence of these vectors (Table 2). Spatial structure and closeness to urban centers were among the most relevant variables for both Aedes species, while mean annual temperature was more important for Ae. aegypti and topography was more relevant for Ae. albopictus.
The most important drivers of dengue cases, according to our model, are the same as those favoring Ae. aegypti, including temperature (Table 2), which coincides with the conclusion of Capinha et al. [61]. As Campbell et al. [10] suspected, requirements for the presence of Ae. aegypti in South of America better reflect the risky environmental conditions for dengue occurrence than those for Ae. albopictus. Our results also concur with what Messina et al. [62] and Brady et al. [25] suggested, that the distribution of dengue occurrences is better modelled by incorporating drivers of different nature, such as climate, topography and human activities.
Distribution of favourable areas
Although our explanatory models were more complex than those previously described, we detected favorable regions for both Aedes species coarsely similar to those described by other authors [7, 10, 25]. Areas highly favorable for Ae. albopictus were mostly located in Brazil and Paraguay (Fig. 2). High-favorability territories for Ae. aegypti were more concentrated in eastern South America, in Brazil, and some high-favorability territories reached further south than those indicated by Kraemer et al. [7], particularly in Uruguay (Fig. 2).
The dengue favorability model got lower discrimination and classification scores than the vector models (Table 3). This may result from the fact that, as other authors have pointed out [63, 64], accuracy of distribution models gets worse when a distribution is more poorly known. The published distribution data of this disease shows a scattered pattern that point to some possible bias in the quality of dengue-virus infection reports, despite the effort of Messina et al. [14]. Nevertheless, Bhatt et al. [24], by using descriptors based on climate, vegetation and human variables, described a pattern of dengue risk in South America similar to our F-dengue model (Fig. 2). However, they did not define risk areas in southern countries such as Chile and Uruguay, while we obtained areas vulnerable or at high risk in these countries. These areas represent a risk for dengue that was hidden up to now (Fig. 2). In the case of Chile, the vulnerable zones are restricted to a few low-altitude grids that were also favorable for Ae. aegypti. It should be noted that we found areas at high risk and vulnerable in many squares neighboring those with reported cases. This suggests that, although these areas are apparently dengue-free, they are in fact at high risk, and extreme precautions and management, control and prevention plans should be applied there.
The greatest risk for the disease in South America may be considered to occur in areas favourable to dengue (F-dengue) with reported presence of vectors Aedes aegypti and/or Ae. albopictus (Figs. 2, 4): much of Brazil and scattered regions of Venezuela, Colombia, Peru and Paraguay for Ae. aegypti; and much of Brazil for Ae. albopictus. The case of Uruguay is particularly interesting. In this country, the F-dengue model detected vulnerable locations in areas where no dengue cases had been reported for a century [11]. Uruguay was classified by Brady et al. [65] as with complete or good evidence consensus on dengue absence. However, in the summer of 2016, about 20 cases of indigenous dengue occurred in Montevideo city [11], specifically where our F-dengue model indicated a high risk of dengue occurrence (Fig. 4). Taking into account that these cases have not been considered as presences for model training in this work, this supports the predictive capacity of our model. According to Real et al. [66], the favorability function may be considered to be, for the distribution of species, analogous to what the wave function is for the distribution of quantum particles, a mathematical conceptualization of the forces that are behind the distribution of the species. This being the case, favorability values could be a better representation of the distribution of a species than the dataset of specific observations, which are always incomplete and contingent on the observation effort.
The biogeographical context of dengue risk
In agreement with the proposal of Messina et al. [62], our approach of biogeographical modelling and fuzzy logic applied to the interaction with Aedes vectors has proved to be a useful method for unravelling the biogeographical context of dengue cases. The maximum simultaneous vector-dengue favorability occurs in much of Brazil for both species (Fig. 3). Additionally, some scattered areas of Colombia, Venezuela, Paraguay, Peru and Uruguay are simultaneously favorable for Aedes aegypti and dengue occurrence, all of them areas where disease cases have been recorded.
The F-dengue model was more included into the F-Ae. aegypti model than into F-Ae. albopictus (Fig. 2), which indicates that the favorability for Ae. aegypti explained to a higher extent than that for Ae. albopictus the dengue cases in South America. Coinciding with Campbell et al. [10], these results indicate that the distribution of Ae. aegypti mostly encompasses the biogeographical framework of dengue in South America, which also suggests that this species is the principal vector responsible for the dengue cases in the continent. Some authors already reported that the increase in the cases of dengue in Brazil and Argentina, for example, was directly linked to the expansion of Ae. aegypti [67–69]. Brathwaite et al. [13] also found a relationship between an increase in dispersion of Ae. aegypti between 2001 and 2010 in America and a corresponding increase in dengue virus circulation. In our analyses, compared to the model built on dengue cases alone, the model based on the intersection between dengue and Ae. aegypti included 26% more no-case-record locations within areas of low risk (F ≤ 0.2), i.e. had a higher specificity (Table 3). This corroborates that incorporating vector information in the biogeographical analysis of disease drivers provides a more plausible explanation about the pattern of cases occurrence [28, 29], which was previously suggested specifically for dengue as well [10, 62].
Although both mosquito species are known to act as vectors of dengue, 99% of the F-Ae. albopictus ∩ F-dengue model was included in the F-Ae. aegypti ∩ F-dengue model (Fig. 3). In addition, while the favorability for Ae. aegypti seems to effectively limit that for dengue (Fig. 3a), this does not happen with the favorability for Ae. albopictus, particularly when the favorability for the mosquito is 0.3 or lower (Fig. 3b). Consequently, in South America, in order to manage the epidemiological risk of new dengue cases, the intersection between Ae. aegypti and dengue favorability should be used as a the most parsimonious map of dengue risk. Nevertheless, a few territories at risk of dengue were attributed exclusively to the F-Ae. albopictus model. Therefore, the most appropriate risk map should include the interaction of all vectors and cases of dengue, which can be readily obtained using fuzzy logic. The fuzzy intersection between the favorability for dengue cases and the fuzzy union of the favorability for any of the vector species provided a comprehensive map of the biogeographical risk for dengue (Fig. 4). This proposal of a risk map for dengue in South America is based on the geographical-environmental, disease-trait and human profiling that constitute the starting point for risk assessments in pathogeography [29].
Conclusions
Our results corroborate that incorporating vector information in the biogeographical analysis of disease drivers provides a more plausible explanation about the pattern of cases occurrence, and confirm that fuzzy logic is an appropriate conceptual and operational tool to deal with the nuances of the vector-illness biogeographical interactions. Thus, the application of fuzzy logic may help health authorities to better prevent, control and mitigate vector-borne diseases.
Supplementary information
Additional file 1: Table S1. Dengue cases, Aedes aegypti and Ae. albopictus occurrences (presences as 1, and absences as 0 in each South America grid), values of the significant variables in the models by South America grid, and favorability values for each model by grid (F-dengue; F-Ae. aegypti and F-Ae. albopictus). Variable abbreviations are given in Table 1.
Acknowledgements
Not applicable.
Abbreviations
- SDM
species distribution models
- AIC
Akaike’s information criterion
- F
environmental favorability
- AUC
area under the curve
- CCR
correct classification rate
Authors’ contributions
DR processed and analyzed part of the data, participated in and designed the field work and wrote the first draft. JO worked on the discussion section and made revisions to the manuscript. RR participated in the design of the field work and revised the last version of the manuscript. JCG elaborated the database, analyzed part of the data and defined the study aims. All authors read and approved the final manuscript.
Funding
DR was supported by a grant from the Agencia Nacional de Investigación e Innovación de Uruguay (ANII) (2016–2018), and from the Graduate Academic Commission (CAP, from Spanish acronym Comisión Académica de Posgrado) of the Universidad de la República (2018–2020). This study was supported by the project CGL2016-76747-R (Ministerio de Economía y Competitividad, España, y el Fondo Europeo de Desarrollo Regional, Unión Europea).
Availability of data and materials
We have included the data table used as Additional file 1: Table S1.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jesús Olivero, Raimundo Real and José Carlos Guerrero contributed equally to this work
Contributor Information
David Romero, Email: davidrpbio@fcien.edu.uy.
Jesús Olivero, Email: jesusolivero@uma.es.
Raimundo Real, Email: rrgimenez@uma.es.
José Carlos Guerrero, Email: jguerrero@fcien.edu.uy.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13071-019-3691-5.
References
- 1.Gubler DJ. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 2002;10:100–103. doi: 10.1016/S0966-842X(01)02288-0. [DOI] [PubMed] [Google Scholar]
- 2.Gubler DJ. The changing epidemiology of yellow fever and dengue, 1900 to 2003: full circle? Comp Immunol Microbiol Infect Dis. 2004;27:319–330. doi: 10.1016/j.cimid.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Delatte H, Gimonneau G, Triboire A, Fontenille D. Influence of temperature on immature development, survival, longevity, fecundity, and gonotrophic cycles of Aedes albopictus, vector of chikungunya and dengue in the Indian Ocean. J Med Entomol. 2009;46:33–41. doi: 10.1603/033.046.0105. [DOI] [PubMed] [Google Scholar]
- 4.Aström C, Rocklöv J, Hales S, Béguin A, Louis V, Sauerborn R. Potential distribution of dengue fever under scenarios of climate change and economic development. EcoHealth. 2013;9:448–454. doi: 10.1007/s10393-012-0808-0. [DOI] [PubMed] [Google Scholar]
- 5.Messina JP, Brady OJ, Pigott DM, Brownstein JS, Hoen AG, Hay SI. 2014. Global dengue occurrence database: 1960–2012. [DOI] [PMC free article] [PubMed]
- 6.Morin CW, Comrie AC, Ernst K. Climate and dengue transmission: evidence and implications. Environ Health Perspect. 2013;121:1264–1272. doi: 10.1289/ehp.1306556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraemer MU, Sinka ME, Duda KA, Mylne AQ, Shearer FM, Barker CM, et al. The global distribution of the arbovirus vectors Aedes aegypti and A. albopictus. eLife. 2015;4:e08347. doi: 10.7554/eLife.08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kraemer MUG, Sinka ME, Duda KA, Mylne A, Shearer FM, Barker CM, et al. The global compendium of Aedes aegypti and Ae. albopictus occurrence. Sci Data. 2015;2:150035. doi: 10.1038/sdata.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeller H, Van Bortel W, Sudre B. Chikungunya: its history in Africa and Asia and its spread to new regions in 2013–2014. J Infect Dis. 2016;214:436–440. doi: 10.1093/infdis/jiw391. [DOI] [PubMed] [Google Scholar]
- 10.Campbell LP, Luther C, Moo-Llanes D, Ramsey JM, Danis-Lozano R, Peterson AT. Climate change influences on global distributions of dengue and chikungunya virus vector. Philos Trans R Soc London B Biol Sci. 2015;370:20140135. doi: 10.1098/rstb.2014.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smink V. Cómo llegó el dengue a Uruguay después de 100 años sin el virus. 2016. https://www.bbc.com/mundo/noticias/2016/02/160225_uruguay_dengue_brote_vs. Accessed 1 June 2016.
- 12.Gubler DJ, Vasilakis N, Musso D. History and emergence of Zika virus. J Infect Dis. 2017;216:860–867. doi: 10.1093/infdis/jix451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brathwaite DO, San Martín JL, Montoya RH, del Diego J, Zambrano B, Dayan GH. Review: the history of dengue outbreaks in the Americas. Am J Trop Med Hyg. 2012;87:584–593. doi: 10.4269/ajtmh.2012.11-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messina JP, Bradu OJ, Scott TW, Zou C, Pigott DM, Duda KA, et al. Global spread of dengue virus types: mapping the 70 year history. Trends Microbiol. 2014;22:138–146. doi: 10.1016/j.tim.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powell JR, Tabachnick WJ. Genetics and the origin of a vector population: Aedes aegypti, a case study. Science. 1980;208:1385–1387. doi: 10.1126/science.7375945. [DOI] [PubMed] [Google Scholar]
- 16.Braks MAH, Honório N, Lounibos L, Lourenço-de-Oliveira R, Juliano S. Interspecific competition between two invasive species of container mosquitoes, Aedes aegypti and Aedes albopictus (Diptera: Culicidae), in Brazil. Ann Entomol Soc Am. 2004;97:130–139. doi: 10.1603/0013-8746(2004)097[0130:ICBTIS]2.0.CO;2. [DOI] [Google Scholar]
- 17.Ferguson NM, Kien DT, Clapham H, Aguas R, Trung VT, Chau TN, et al. Modeling the impact on virus transmission of Wolbachia-mediated blocking of dengue virus infection of Aedes aegypti. Sci Transl Med. 2015;7:279ra37. doi: 10.1126/scitranslmed.3010370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brady OJ, Johansson MA, Guerra CA, Bhatt S, Golding N, Pigott DM, et al. Modelling adult Aedes aegypti and Aedes albopictus survival at different temperatures in laboratory and field settings. Parasites Vectors. 2013;6:351. doi: 10.1186/1756-3305-6-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gama ZP, Nakagoshi N, Islamiyah M. Distribution patterns and relationship between elevation and the abundance of Aedes aegypti in Mojokerto city 2012. Open J Anim Sci. 2013;3:11–16. doi: 10.4236/ojas.2013.34A1003. [DOI] [Google Scholar]
- 20.Goindin D, Delannay C, Ramdini C, Gustave J, Fouque F. Parity and longevity of Aedes aegypti according to temperatures in controlled conditions and consequences on dengue transmission risks. PLoS ONE. 2015;10:e0135489. doi: 10.1371/journal.pone.0135489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rašić G, Endersby-Harshman N, Tantowijoyo W, Goundar A, White V, Yang Q, et al. Aedes aegypti has spatially structured and seasonally stable populations in Yogyakarta, Indonesia. Parasites Vectors. 2015;8:610. doi: 10.1186/s13071-015-1230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown JE, Evans BR, Zheng W, Obas V, Barrera-Martinez L, Egizi A, et al. Human impacts have shaped historical and recent evolution in Aedes aegypti, the dengue and yellow fever mosquito. Evolution. 2014;68:14–25. doi: 10.1111/evo.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatterjee S, Chakraborty A, Sinha SK. Spatial distribution and physicochemical characterization of the breeding habitats of Aedes aegypti in and around Kolkata, West Bengal, India. Indian J Med Res. 2015;142:79–86. doi: 10.4103/0971-5916.176631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brady OJ, Golding N, Pigott DM, Kraemer MU, Messina JP, Reiner RC, et al. Global temperature constraints on Aedes aegypti and Ae. albopictus persistence and competence for dengue virus transmission. Parasites Vectors. 2014;7:338. doi: 10.1186/1756-3305-7-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cliff AD, Haggett P. The epidemiological significance of islands. Health Place. 1995;1:199–209. doi: 10.1016/1353-8292(95)00029-1. [DOI] [Google Scholar]
- 27.Smith KF, Guégan J-F. Changing geographic distributions of human pathogens. Annu Rev Ecol Evol Syst. 2010;41:231–250. doi: 10.1146/annurev-ecolsys-102209-144634. [DOI] [Google Scholar]
- 28.Olivero J, Fa JE, Real R, Farfán MA, Márquez AL, Vargas JM, et al. Mammalian biogeography and the Ebola virus in Africa. Mammal Rev. 2017;47:24–37. doi: 10.1111/mam.12074. [DOI] [Google Scholar]
- 29.Murray KA, Olivero J, Roche B, Tiedt S, Guégan JF. Pathogeography: leveraging the biogeography of human infectious diseases for global health management. Ecography. 2018;41:1411–1427. doi: 10.1111/ecog.03625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zadeh LA. Fuzzy sets. Inform Control. 1965;8:338–353. doi: 10.1016/S0019-9958(65)90241-X. [DOI] [Google Scholar]
- 31.Salski A, Kandzia P. Fuzzy sets and fuzzy logic in ecological modelling. EcoSys. 1996;4:85–97. [Google Scholar]
- 32.Salski A. Ecological applications of fuzzy logic. In: Recknagel F, editor. Ecological informatics: scope, techniques and applications. Berlin: Springer; 2006. pp. 3–14. [Google Scholar]
- 33.Real R, Barbosa AM, Vargas JM. Obtaining environmental favourability functions from logistic regression. Environ Ecol Stat. 2006;13:237–245. doi: 10.1007/s10651-005-0003-3. [DOI] [Google Scholar]
- 34.Acevedo P, Ward AI, Real R, Smith GC. Assessing biogeographical relationships of ecologically related species using favourability functions: a case study on British deer. Divers Distrib. 2010;16:515–528. doi: 10.1111/j.1472-4642.2010.00662.x. [DOI] [Google Scholar]
- 35.Romero D, Báez JC, Ferri-Yáñez F, Bellido JJ, Real R. Modelling favourability for invasive species encroachment to identify areas of native species vulnerability. Sci World J. 2014;2014:519710. doi: 10.1155/2014/519710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Romero D, Olivero J, Brito JC, Real R. Comparison of approaches to combine species distribution models based on different sets of predictors. Ecography. 2016;39:561–571. doi: 10.1111/ecog.01477. [DOI] [Google Scholar]
- 37.Romo H, García-Barros E, Márquez AL, Moreno JC, Real R. Effects of climate change on the distribution of ecologically interacting species: butterflies and their main food plants in Spain. Ecography. 2014;37:1063–1072. [Google Scholar]
- 38.Barbosa AM. fuzzySim: applying fuzzy logic to binary similarity indices in ecology. Methods Ecol Evol. 2015;6:853–858. doi: 10.1111/2041-210X.12372. [DOI] [Google Scholar]
- 39.Willis KJ, Whittaker RJ. Species diversity—scale matters. Science. 2002;295:1245–1248. doi: 10.1126/science.1067335. [DOI] [PubMed] [Google Scholar]
- 40.Pearson RG, Dawson TP. Predicting the impacts of climate change on the distribution of species: are bioclimate envelope models useful? Glob Ecol Biogeogr. 2003;12:361–371. doi: 10.1046/j.1466-822X.2003.00042.x. [DOI] [Google Scholar]
- 41.Legendre P, Legendre L. Numerical ecology. 2. Amsterdam: Elsevier Science; 1998. [Google Scholar]
- 42.Legendre P. Spatial autocorrelation: trouble or new paradigm? Ecology. 1993;74:1659–1673. doi: 10.2307/1939924. [DOI] [Google Scholar]
- 43.Miller J, Franklin J, Aspinall R. Incorporating spatial dependence in predictive vegetation models. Ecol Model. 2007;202:225–242. doi: 10.1016/j.ecolmodel.2006.12.012. [DOI] [Google Scholar]
- 44.Lobo JM, Lumaret J-P, Jay-Robert P. Modelling the species richness distribution of French dung beetles (Coleoptera, Scarabaeidae) and delimiting the predictive capacity of different groups of explanatory variables. Glob Ecol Biogeogr. 2002;11:265–277. doi: 10.1046/j.1466-822X.2002.00291.x. [DOI] [Google Scholar]
- 45.Nogués-Bravo D, Martinez-Rica JP. Factors controlling the spatial species richness pattern of four groups of terrestrial vertebrates in an area between two different biogeographic regions in northern Spain. J Biogeogr. 2004;31:629–640. doi: 10.1046/j.1365-2699.2003.01041.x. [DOI] [Google Scholar]
- 46.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B. 1995;57:289–300. [Google Scholar]
- 47.Akaike H. Information theory and an extension of the maximum likelihood principle. In: Petrov BN, Csaki F, editors. Proceedings of the second international symposium on information theory. Budapest, Hungary: Akademiai Kiado; 1973. p. 267–81.
- 48.Wald A. Tests of statistical hypotheses concerning several parameters with applications to problems of estimation. Trans Am Math Soc. 1943;54:426–482. doi: 10.1090/S0002-9947-1943-0012401-3. [DOI] [Google Scholar]
- 49.Lumley T. Analysis of complex survey samples. J Stat Softw. 2004;9:1–19. doi: 10.18637/jss.v009.i08. [DOI] [Google Scholar]
- 50.Lumley T. Survey: analysis of complex survey samples. R package version 3.35. 2018.
- 51.Crawley MJ. The R book. Chichester: Wiley; 2007. [Google Scholar]
- 52.Acevedo P, Real R. Favourability: concept, distinctive characteristics and potential usefulness. Naturwissenschaften. 2012;99:515–522. doi: 10.1007/s00114-012-0926-0. [DOI] [PubMed] [Google Scholar]
- 53.Barbosa AM, Brown JA, Jimenez-Valverde A, Real R. modEvA: model evaluation and analysis. R package version 1.3.2. 2016. https://CRAN.R-project.org/package=modEvA.
- 54.Lobo J, Jiménez-Valverde A, Real R. AUC: a misleading measure of the performance of predictive distribution models. Glob Ecol Biogeogr. 2008;17:145–151. doi: 10.1111/j.1466-8238.2007.00358.x. [DOI] [Google Scholar]
- 55.Cliff AD, Ord JK. Spatial processes: models and applications. London: Pion; 1981. pp. 2–10. [Google Scholar]
- 56.Muñoz AR, Real R. Assessing the potential range expansion of the exotic monk parakeet in Spain. Divers Distrib. 2006;12:656–665. doi: 10.1111/j.1472-4642.2006.00272.x. [DOI] [Google Scholar]
- 57.Kunchenva LI. Using measures of similarity and inclusion for multiple classifier fusion by decision templates. Fuzzy Set Syst. 2001;122:401–407. doi: 10.1016/S0165-0114(99)00161-X. [DOI] [Google Scholar]
- 58.Muñoz AR, Real R, Barbosa AM, Vargas JM. Modelling the distribution of Bonelli’s eagle in Spain: implications for conservation planning. Divers Distrib. 2005;11:477–486. doi: 10.1111/j.1366-9516.2005.00188.x. [DOI] [Google Scholar]
- 59.Hosmer DW, Lemeshow S. Applied logistic regression. 2. New York: Wiley; 2000. [Google Scholar]
- 60.Landis JR, Koch GC. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 61.Capinha C, Rocha J, Sousa CA. Macroclimate determines the global range limit of Aedes aegypti. EcoHealth. 2014;11:420–428. doi: 10.1007/s10393-014-0918-y. [DOI] [PubMed] [Google Scholar]
- 62.Messina JP, Brady OJ, Pigott DM, Golding N, Kraemer MUG, Scott TW, et al. The many projected futures of dengue. Nat Rev Microbiol. 2015;13:230–239. doi: 10.1038/nrmicro3430. [DOI] [PubMed] [Google Scholar]
- 63.Stockwell DRB, Peterson AT. Effects of sample size on accuracy of species distribution models. Ecol Model. 2002;148:1–13. doi: 10.1016/S0304-3800(01)00388-X. [DOI] [Google Scholar]
- 64.Gurutzeta GA. Modelling of species distributions, range dynamics and communities under imperfect detection: advances, challenges and opportunities. Ecography. 2017;40:281–295. doi: 10.1111/ecog.02445. [DOI] [Google Scholar]
- 65.Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, Hoen AG, et al. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl Trop Dis. 2012;6:e1760. doi: 10.1371/journal.pntd.0001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Real R, Barbosa AM, Bull JW. Species distributions, quantum theory, and the enhancement of biodiversity measures. Syst Biol. 2017;66:453–462. doi: 10.1093/sysbio/syw072. [DOI] [PubMed] [Google Scholar]
- 67.Vezzani D, Carbajo AE. Aedes aegypti, Aedes albopictus, and dengue in Argentina: current knowledge and future directions. Mem Inst Oswaldo Cruz. 2008;103:66–74. doi: 10.1590/S0074-02762008005000003. [DOI] [PubMed] [Google Scholar]
- 68.Serpa LLN, Marques GRAM, de Lima AP, Voltolini JC, de Brito Arduino M, Barbosa GL, et al. Study of the distribution and abundance of the eggs of Aedes aegypti and Aedes albopictus according to the habitat and meteorological variables, municipality of São Sebastião, São Paulo State, Brazil. Parasites Vectors. 2013;6:321. doi: 10.1186/1756-3305-6-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.da Rocha Taranto MF, Pessanha JE, dos Santos M, dos Santos Pereira Andrade AC, Camargos VN, Alves SN, et al. Dengue outbreaks in Divinopolis, south-eastern Brazil and the geographic and climatic distribution of Aedes albopictus and Aedes aegypti in 2011–2012. Trop Med Int Health. 2015;20:77–88. doi: 10.1111/tmi.12402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Messina JP, Brady OJ, Pigott DM, Brownstein JS, Hoen AG, Hay SI. 2014. Global dengue occurrence database: 1960–2012. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Additional file 1: Table S1. Dengue cases, Aedes aegypti and Ae. albopictus occurrences (presences as 1, and absences as 0 in each South America grid), values of the significant variables in the models by South America grid, and favorability values for each model by grid (F-dengue; F-Ae. aegypti and F-Ae. albopictus). Variable abbreviations are given in Table 1.
Data Availability Statement
We have included the data table used as Additional file 1: Table S1.