Abstract
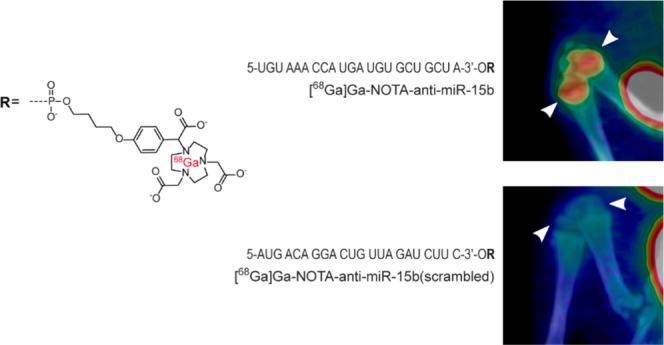
MicroRNAs (miRNAs) are endogenous, small, noncoding ribonucleic acids (RNAs) that bind to the 3′ untranslated regions of messenger RNAs (mRNAs) and induce translational repression or mRNA degradation. Although numerous studies have reported that miRNAs are of potential use for disease diagnostics and gene therapy, little is known about their fates in vivo. This study elucidated the whole-body distributions and kinetics of intravenously administered miRNA-targeting molecules in vivo by positron emission tomography (PET) imaging. A 22-mer sequence targeting miR-15b was conjugated with three different chelators and labeled with gallium-68 (68Ga). These tracers were compared with a scrambled 22-mer sequence; 22-mer with two single base substitutions; anti-miR-34 22-mer; hexathymidylate (T6), a 6-mer sequence; and an unconjugated chelator. miR-15b was chosen as a target because it is important for bone remodeling. All three 68Ga-labeled anti-miR-15b molecules had similar biodistributions and kinetics, and they all accumulated in the bones, kidneys, and liver. The bone accumulation of these tracers was the highest in the epiphyses of long tubular bones, maxilla, and mandible. By contrast, the scrambled 22-mer sequence, the 6-mer, and the unconjugated chelator did not accumulate in bones. PET imaging successfully elucidated the distributions and kinetics of 68Ga-labeled chelated miRNA-targeting molecules in vivo. This approach is potentially useful to evaluate new miRNA-based drugs.
Keywords: biodistribution, gallium-68, microRNA, PET
Introduction
Ribonucleic acid interference is a post-transcriptional gene silencing mechanism discovered by Fire and coworkers in 1998.1 In this mechanism, an exogenous double-stranded RNA silences the corresponding messenger RNA (mRNA). These exogenous RNA molecules are called short interfering RNAs. microRNAs (miRNAs) function in the same way but are endogenously expressed. miRNAs are transcribed from exonic or intronic deoxyribonucleic acid and then exported from the nucleus. The enzyme Dicer digests pre-miRNAs into fragments containing 16–23 nucleotides in the cytoplasm. These short mature miRNA fragments are incorporated into the RNA-induced silencing complex and recognize complementary mRNAs. A component of this complex, termed argonaut 2, cleaves mRNAs and thus prevents subsequent translation.2 miRNA regulation is an important mechanism in various cells and tissues. For example, blockade of the miRNA maturation pathway via tissue-specific knockout of Dicer elicits a multitude of effects in animal models, ranging from impaired organ function to embryonic lethality.3−6 Complete knockout of an miRNA is not necessary to elicit effects and may have unwanted consequences. A specific miRNA can be overexpressed by transfection or inhibited by various antisense miRNAs (anti-miRs).7−10 The main problems associated with the use of short interfering RNAs and miRNAs as therapeutic agents are their limited stabilities in vivo, difficulties in targeting their delivery to a desired tissue, their modest cellular uptake, and possible off-target effects. Several strategies have been developed to overcome these difficulties, including the use of viral vectors, liposomes, chemical modifications, and nanoparticles, as reviewed by Gao and coworkers.11 The concentrations of injected nonmodified oligonucleotides are the highest in the liver and kidneys, followed by the bone marrow, adipocytes, and lymph nodes. Cellular uptake of these oligonucleotides is mediated by endocytosis.12
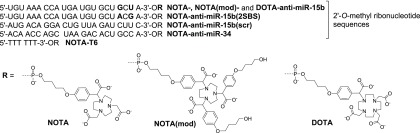
This study examined three chemically modified antisense oligonucleotides targeting miR-15b. miR-15b belongs to the miR-15 family and plays important roles in various tissues, including the bones.13−16 The novel anti-miR-15b oligonucleotides were conjugated with three different chelators to evaluate the chelator effect on biodistributions and biokinetics of the target oligonucleotide (anti-miR-15b). These chelators were chosen because they bind the positron emitter gallium-68 (68Ga) and allow straightforward synthesis of the oligonucleotide conjugates entirely on a solid phase. In previous positron emission tomography (PET) studies, oligonucleotides were usually conjugated with the macrocyclic chelator 4-(4-hydroxybut-1-oxy)phenyl 1,4,7,10-tetraazacyclododecane-N′,N″,N‴,N′′′′-tetraacetic acid (DOTA).17−19 In addition to DOTA, we used two triazacyclononane-based chelators, i.e., 4-(4-hydroxybut-1-oxy)phenyl 1,4,7-triazacyclononane-N,N′,N″-triacetic acid (NOTA) and tris[4-(4-hydroxybut-1-oxy)phenyl] 1,4,7-triazacyclononane-N,N′,N″-triacetic acid (NOTA(mod)) (Figure 1). NOTA has been previously used to label oligonucleotides.19−24 As mentioned in a review by Gao and coworkers,15 oligonucleotide stability is a major challenge when designing novel oligonucleotide tracers. Consequently, oligonucleotides used in the current study were 2′-O-methylated to increase their stabilities.
Figure 1.
Structures of the oligonucleotides.
Experimental Section
Animals
Thirty-three wild-type (Sprague-Dawley) adult rats (29 males and 4 females) weighing 240–570 g were used to evaluate the whole-body biodistributions of conjugated, 68Ga-labeled RNA oligonucleotides by PET. Eight smaller and younger animals (all males) weighing 64–97 g were used for PET/computed tomography (CT) imaging. Animals were housed at the Central Animal Laboratory of the University of Turku and had ad libitum access to food and water. All animal experiments were approved by the Lab-Animal Care & Use Committee of the State Provincial Office of Southern Finland (license number ESAVI/4835/04.10.03/2011) and were carried out in compliance with national and EU legislation related to the conduct of animal experimentation.
68Ga-RNA Molecules
Three conjugated, 68Ga-labeled RNA molecules ([68Ga]Ga-NOTA-anti-miR-15b, [68Ga]Ga-NOTA(mod)-anti-miR-15b, and [68Ga]Ga-DOTA-anti-miR-15b) were used in the study, all of which contained an anti-miR-15b sequence (5′-UGU AAA CCA UGA UGU GCU GCU A-3′). All nucleotides in the 22-mer anti-miR-15b sequence were 2′-O-methylated. In [68Ga]Ga-NOTA-anti-miR-15b, the 3′-end of the RNA sequence was conjugated with NOTA to facilitate labeling with 68Ga (t1/2 = 68 min, β+ = 89%, and EC = 11%). [68Ga]Ga-NOTA(mod)-anti-miR-15b was identical to [68Ga]Ga-NOTA-anti-miR-15b, except that the RNA sequence was conjugated with NOTA(mod) instead of NOTA. DOTA was conjugated to the 3′-end of the RNA sequence in [68Ga]Ga-DOTA-anti-miR-15b. 68Ga-labeled NOTA, [68Ga]Ga-NOTA-T6 (six thymines conjugated with 4-(4-butoxy)phenyl NOTA), [68Ga]Ga-NOTA-anti-miR-15b(scr), [68Ga]Ga-NOTA-anti-miR-15b(2SBS) with two single base substitutions, and [68Ga]Ga-NOTA-anti-miR-34 were used as controls. [68Ga]Ga-NOTA-anti-miR-15b(scr) contained the same nucleotides as [68Ga]Ga-NOTA-anti-miR-15b but in different order (5′-AUG ACA GGA CUG UUA GAU CUU C-3′) and lacked a target in the rat genome according to a BLAST search. The RNA sequence in [68Ga]Ga-NOTA-anti-miR-15b(2SBS) was 5′-UGU AAA CCA UGA UGU GCU ACG A-3′. One nucleotide was changed from guanine to adenine and another was changed from uracil to guanine at the 3′-end of this sequence. The RNA sequence in [68Ga]Ga-NOTA-anti-miR-34 was 5′-ACA ACC AGC UAA GAC ACU GCC A-3′. The effect of the 3′-68Ga-NOTA/DOTA chelate to the RNA duplex is generally slightly stabilizing as we have previously determined by Tm (UV) measurements (cold Ga3+ used as a ligand, the stabilization is of unspecific origin).23 It may be emphasized that the oligonucleotides in this study were 2′-O-methyl oligoribonucleotides without cell/tissue-specific targeting agents. While modest cellular uptake may be expected, the imaging data may more reliably demonstrate the importance of the sequence (i.e., anti-miR-15b vs anti-miR-15b(2SBS), anti-miR-15b(scr), and anti-miR-34) and plausible chelator effect (NOTA vs NOTA(mod) and DOTA) on the biodistribution and kinetics. The structures of the studied molecules are presented in Figure 1.
The oligonucleotides were synthesized by an automatic DNA/RNA synthesizer using solid-supported DOTA, NOTA, and NOTA(mod) chelators as we previously reported.19 Thereafter, the oligonucleotides were released from the supports by an optimized two-step cleavage protocol (treatment with 0.1 mol/L aqueous NaOH for 3 h at 55 °C and then incubation with concentrated ammonia overnight at 55 °C), purified by reversed-phase high-performance liquid chromatography (RP-HPLC), and lyophilized to dryness. The authenticity of the RNAs was verified by electrospray ionization time-of-flight mass spectroscopy [MS (ESI-TOF)] (Figure S1 and Table S1).
68Ga was obtained in the form of [68Ga]GaCl3 using a 68Ge/68Ga generator (Eckert & Ziegler, Valencia, CA) by elution with 0.1 M HCl composed of 30% ultrapure HCl and ultrapure water. Sodium acetate (18 mg) was added to 500 μL of [68Ga]GaCl3 (or 3 mg was added to 100 μL) to yield a final concentration of 0.4 M sodium acetate. The pH was adjusted to approximately 3.5 with 2 M HCl. Thereafter, a NOTA/DOTA-conjugated oligonucleotide (5–12 nmol, dissolved in deionized water to yield a stock solution of 0.4, 0.5, or 1 mM) was added and the reaction mixture was incubated at 95 °C for 10–15 min. Radiochemical purity was determined by reversed-phase radio-HPLC using a Jupiter C18 column (300 Å, 5 μm, 4.6 × 150 mm2; Phenomenex, Torrance, CA). The HPLC conditions were as follows for [68Ga]Ga-NOTA-anti-miR-15b, [68Ga]Ga-NOTA(mod)-anti-miR-15b, [68Ga]Ga-DOTA-anti-miR-15b, [68Ga]Ga-NOTA-anti-miR-15b(2SBS), [68Ga]Ga-NOTA-anti-miR-34, and [68Ga]Ga-NOTA-anti-miR-15b(scr): flow rate, 1 mL/min; buffer A, 0.1 M triethylammonium acetate; buffer B, acetonitrile; and buffer C, 50 mM phosphoric acid (H3PO4). The linear buffer A/B/C gradient was as follows: 0–13 min from 90/10/0 to 70/30/0, 13–14 min from 70/30/0 to 0/0/100, and 14–20 min at 0/0/100, λ = 260 nm. The HPLC conditions were as follows for 68Ga-NOTA and 68Ga-NOTA-T6: flow rate, 1 mL/min; buffer A, 0.1 M triethylammonium acetate; and buffer B, acetonitrile. The linear buffer A/B gradient was as follows: 0–22 min from 100/0 to 73/27, λ = 260 nm. The HPLC system consisted of LaChrom Instruments (Hitachi; Merck, Darmstadt, Germany) and a Radiomatic 150TR radioisotope detector (Packard, Meriden, CT).
Dynamic Whole-Body PET and CT Imaging
The biodistributions of the tracers were evaluated by performing dynamic PET imaging with a high-resolution research tomograph (HRRT) camera (Siemens Medical Solutions, Knoxville, TN) cross-calibrated with a dose calibrator (VDC-202; Veenstra Instruments, Joure, Netherlands) and a 1480 Wizard 3″ γ counter (PerkinElmer, Turku, Finland). Two rats were imaged simultaneously. For PET imaging, rats were anesthetized with isoflurane (Baxter, 3–4% with an airflow of 600–700 mL/min for induction and 1.5–2.5% with an airflow of 400–500 mL/min for maintenance) and placed on a warm pallet. A catheter was inserted into the tail vein (24-gauge cannula; BD Neoflon, Becton Dickinson Infusion Therapy AB, Helsingborg, Sweden). After a 6 min transmission scan using a 137Cs point source for attenuation correction, 68Ga tracer (19.1 ± 4.8 MBq) was intravenously injected as a bolus. Dynamic imaging was started at the time of injection and continued for 60 min.
PET data acquired in list mode were reconstructed into 5 × 10, 1 × 15, 7 × 20, 1 × 40, 2 × 60, 1 × 120, 4 × 180, 1 × 240, 5 × 300, and 1 × 400 s time frames using the ordinary Poisson three-dimensional ordered-subset expectation maximization algorithm (with 8 iterations; 16 subsets; and attenuation, random, and scatter corrections). Each reconstructed image consisted of 29 frames and a 256 × 256 × 207 voxel matrix (voxel size, 1.22 × 1.22 × 1.22 mm3).
Dynamic PET/CT (Inveon Multimodality scanner, Siemens Medical Solutions) scanning was performed on eight animals, each of which was injected with a different tracer [[68Ga]Ga-NOTA-anti-miR-15b, [68Ga]Ga-NOTA(mod)-anti-miR-15b, [68Ga]Ga-DOTA-anti-miR-15b, [68Ga]Ga-NOTA-anti-miR-15b(2SBS), [68Ga]Ga-NOTA-anti-miR-15b(scr), [68Ga]Ga-NOTA-anti-miR-34, [68Ga]Ga-NOTA-T6, or [68Ga]Ga-NOTA]. These rats were smaller than those imaged with an HRRT camera due to the smaller imaging field-of-view of the Inveon scanner (12.5 cm). Anesthesia was induced and maintained with ∼4 and ∼2% isoflurane, respectively, for tail vein cannulation and imaging. After CT for 10 min, 10.8 ± 0.9 MBq of the tracer was injected and PET data were acquired for 60 min in list mode. CT data were acquired using 121 projections with an exposure time of 330 ms, an X-ray voltage of 80 kVp, and an anode current of 500 μA for full 360° rotation. CT images were reconstructed with filtered back projection and a voxel size of 0.22 × 0.22 × 0.22 mm3. PET images were reconstructed into 28 time frames (5 × 10, 1 × 15, 7 × 20, 1 × 40, 2 × 60, 1 × 120, 4 × 180, 1 × 240, 5 × 300, 1 × 400 s) with an ordered-subset expectation maximization algorithm followed by fastMAP reconstruction with 16 subsets and 18 iterations. The matrix size was 128 × 128 × 159 and voxel size was 0.8 × 0.8 × 0.8 mm3.
Image Analysis
All reconstructed images were converted to standardized uptake value (SUV) images using the Imgsuv 0.1.0 program (Turku PET Centre, Finland, http://www.turkupetcentre.net/software/list.php). Volumes of interest were defined for the brain, salivary glands, heart, liver, kidney medulla and cortex, knees, spleen, and urinary bladder using Carimas 2.0.2 software (Turku PET Centre, Finland, http://www.turkupetcentre.net/carimasturku/). Time-activity curves (TACs) were extracted from the data accordingly.
The PET images presented in this study represent the mean of all time frames. To create an average image of the 30 middle planes, the sum plane and arithmetic functions of Vinci 2.54 software (Max-Planck-Institut für neurologische Forschung, Cologne, Germany) were used.
PET/CT images were processed with Inveon Research Workplace 4.0 software (Siemens), fused, transformed into an maximum a posterior (MAP) projection, scaled, and rotated into the same position.
Pharmacokinetic parameters were calculated using plasma TACs extracted from SUV-normalized PET images. Plasma TACs were extracted by drawing a spheroid volume of interest on the most radioactive area of the heart in the first frames where blood radioactivity was the highest. At this point, the TAC represented the whole-blood radioactivity. To generate plasma TACs, the whole-blood curve was multiplied by 1.80 for correction. This estimation was based on the plasma/blood radioactivity ratio measured ex vivo (Table S2). The area under the curve (AUC0→∞) from 0 to infinity, total clearance (ClT), elimination rate constant (kel), and plasma half-life (t1/2) were calculated using Paucinf 1.4.2 (Turku PET Centre, Finland, http://www.turkupetcentre.net/software/list.php). The Paucinf program calculates the AUC between 0 and 55.8 min using the linear trapezoidal rule. Remaining AUCs (between 55.8 min and infinity) were calculated with the Paucinf program using log-linear regression followed by the linear trapezoidal rule. The maximum concentration (Cmax) and time of maximum concentration (Tmax) were defined from plasma TACs.
Ex Vivo Analysis
After PET imaging, the rats were euthanized and blood (by cardiac puncture), urine (by bladder puncture), and tissue samples were excised and weighed. Total radioactivity was measured using a γ counter (1480 Wizard 3″, PerkinElmer, Turku, Finland). Measurements were converted from counts per minute to kilobecquerel using the calibration constant of 68Ga. All radioactivity values were decay-corrected to the time of injection.
Statistical Analysis
Differences between tracers in ex vivo biodistribution and TAC results were analyzed by the unpaired Student t-test using the GraphPad online calculator (https://www.graphpad.com/quickcalcs/). When determining differences between TACs, only the last measurement points were used in the comparison. P values of less than 0.05 were considered statistically significant.
Results
Radiolabeling
According to radio-HPLC analysis, the radiochemical yields and purities of the 68Ga-labeled oligonucleotides were 96 ± 3.1% (range, 86–98%, n = 15) throughout the study. The specific molar activity was 7.4 ± 3.5 MBq/nmol (range 2–12 MBq/nmol, n = 15).
In Vivo Imaging
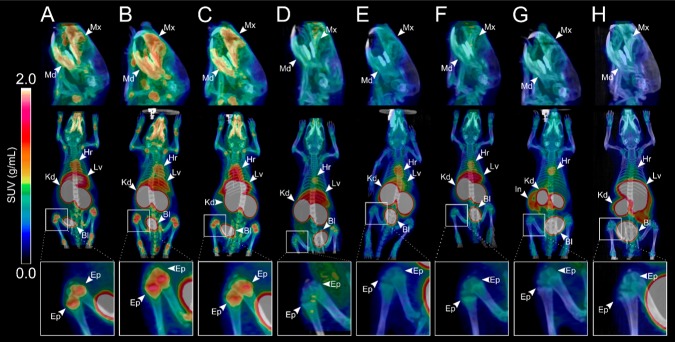
[68Ga]Ga-NOTA-anti-miR-15b, [68Ga]Ga-NOTA(mod)-anti-miR-15b, and [68Ga]Ga-DOTA-anti-miR-15b had a similar pattern of distribution with some differences (Figures 2A–C and 3A–C, Table S3), which demonstrated that the chelator had no significant effect on the distribution. Radioactivity accumulated most in the kidneys, urinary bladder, and liver. Some radioactivity was also observed in the salivary glands. [68Ga]Ga-NOTA-anti-miR-15b(2SBS) and [68Ga]Ga-NOTA-anti-miR-34 also had similar distribution in kidneys, liver, and urine. All three tracers tended to accumulate in the epiphyses of long bones (Figures 2A–C and 3A–C). Radioactivity was the highest in the distal femur and proximal tibia at the knee joint. In PET/CT imaging of small animals, radioactivity was clearly visible in the epiphyses of long bones at the ankle, hip, wrist, elbow, and shoulder, as well as in the pelvic bone, lumbar vertebra, maxilla, and mandibula (Figure 3A–C). [68Ga]Ga-NOTA-anti-miR-15b(2SBS) had lower activity in bone than [68Ga]Ga-NOTA-anti-miR-15b but higher than scrambled (Figure 3A,D,E). [68Ga]Ga-NOTA-anti-miR-34 had similar levels to scrambled (Figure 4A). [68Ga]Ga-NOTA-T6 and [68Ga]Ga-NOTA had similar distributions to each other. Injection with these two controls resulted in lower radioactivity in kidneys and higher in urinary bladder in comparison with the three oligonucleotides. These two controls did not accumulate in bones. PET/CT imaging demonstrated that [68Ga]Ga-NOTA-T6 accumulated in the intestine (Figures 2G and 3G, Table S3).
Figure 2.
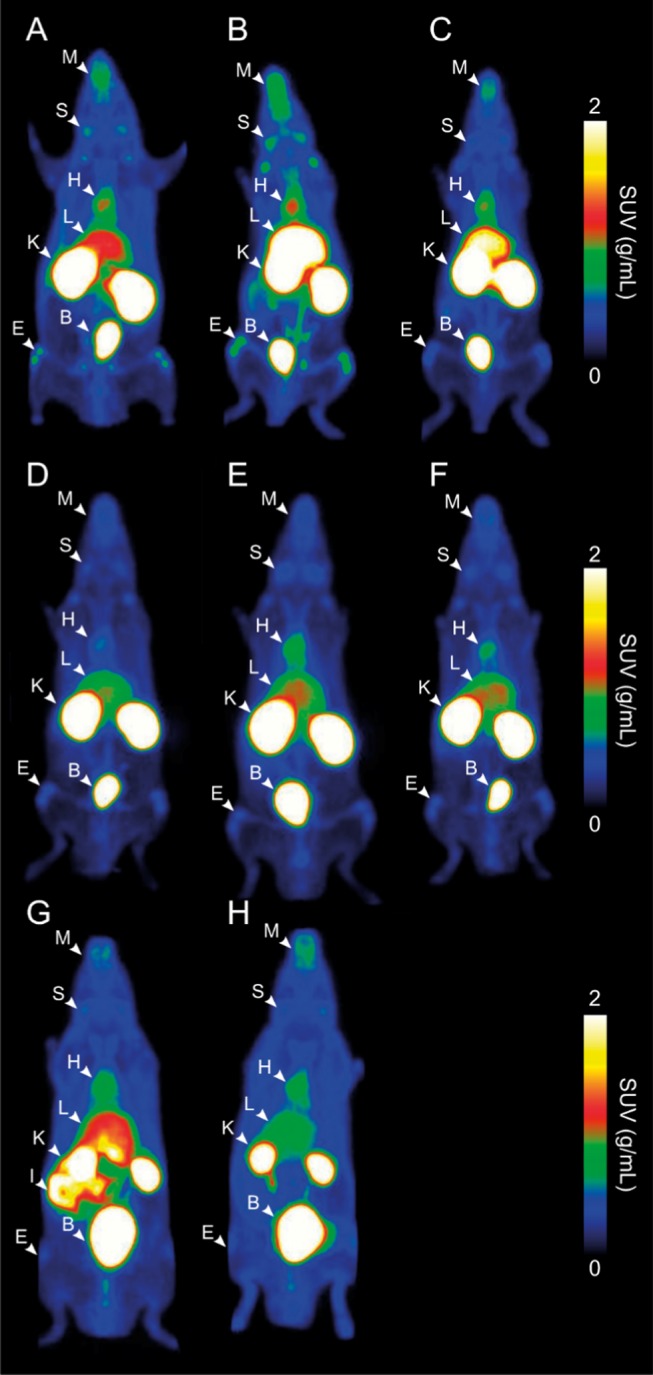
Representative coronal whole-body PET images showing the distributions of radioactivity in rats intravenously injected with 68Ga-labeled anti-miR-15b oligonucleotides and control tracers. (A) [68Ga]Ga-NOTA-anti-miR-15b, (B) [68Ga]Ga-NOTA(mod)-anti-miR-15b, (C) [68Ga]Ga-DOTA-anti-miR-15b, (D) [68Ga]Ga-NOTA-anti-miR-15b(2SBS), (E) [68Ga]Ga-NOTA-anti-miR-15b(scr), (F) [68Ga]Ga-NOTA-anti-miR-34, (G) [68Ga]Ga-NOTA-T6, and (H) [68Ga]Ga-NOTA. Images are maximum-intensity projections, the mean of all time frames, and presented using the same color scale as SUVs. Arrowheads denote organs in which the radioactivity concentration was the highest. B = urinary bladder, E = epiphyses, H = heart, I = intestine, K = kidney, L = liver, S = salivary glands, and M = maxilla and mandibula.
Figure 3.
Representative PET/CT images of the biodistributions of (A) [68Ga]Ga-NOTA-anti-miR-15b, (B) [68Ga]Ga-NOTA(mod)-anti-miR-15b, (C) [68Ga]Ga-DOTA-anti-miR-15b, (D) [68Ga]Ga-NOTA-anti-miR-15b(2SBS), (E) [68Ga]Ga-NOTA-anti-miR-15b(scr), (F) [68Ga]Ga-NOTA-anti-miR-34, (G) [68Ga]Ga-NOTA-T6, and (H) [68Ga]Ga-NOTA in male rats. Upper images are in the sagittal plane at 2× magnification and show accumulation of radioactivity in the maxilla (Mx) and mandible (Mn). Middle images are in the coronal plane and show whole-body distributions. Radioactivity was clearly observed in the liver (Lv), urinary bladder (Bl), kidneys (Kd), spleen (Sp), heart (Hr), and intestine (In) (arrowheads). Lower images are of the knee joint at 3× magnification and show accumulation of radioactivity in epiphyses (Ep). All images represent the mean at 0–60 min after tracer injection and are presented in the same color scale as SUVs.
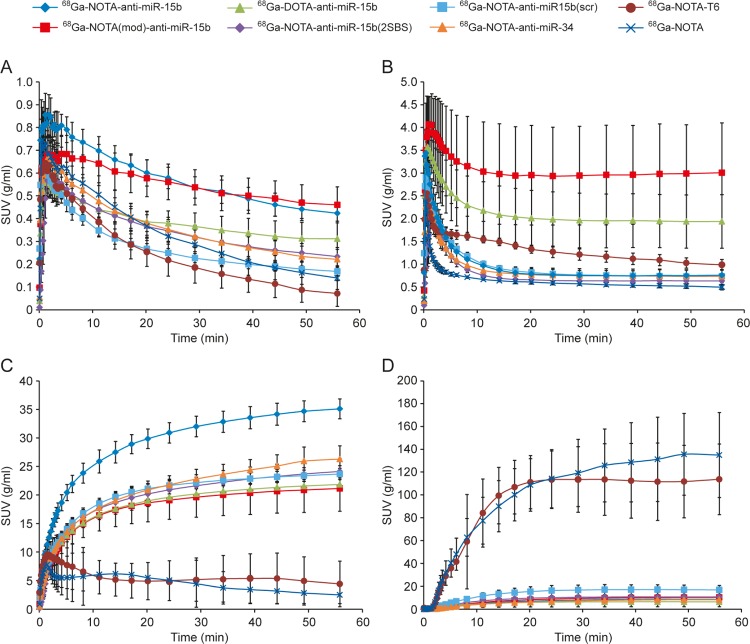
Figure 4.
TACs of the (A) epiphysis of the femur, (B) liver, (D) kidney cortex, and (D) urinary bladder in rats intravenously injected with 68Ga-labeled anti-miR-15b oligonucleotides and control tracers. Lines represent mean SUVs, and error bars denote SD values.
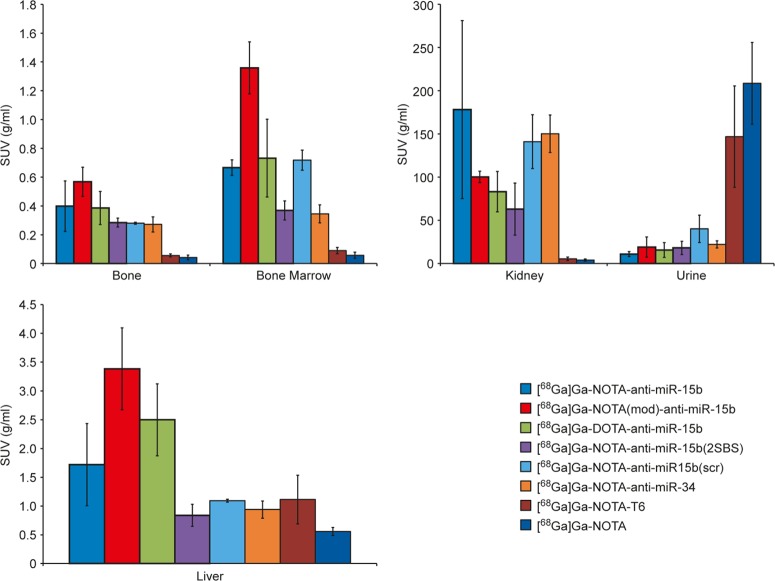
Ex Vivo Measurements
Ex vivo measurements were consistent with the PET images. Radioactivity in the kidneys, bones, and bone marrow was much higher in rats injected with [68Ga]Ga-NOTA-anti-miR-15b, [68Ga]Ga-NOTA(mod)-anti-miR-15b, and [68Ga]Ga-DOTA-anti-miR-15b than in those injected with [68Ga]Ga-NOTA-T6 and [68Ga]Ga-NOTA (Table 1 and Figure 5). The ex vivo measurements of [68Ga]Ga-NOTA-anti-miR-15b(2SBS), [68Ga]Ga-NOTA-anti-miR-15b(scr), and [68Ga]Ga-NOTA-anti-miR-34 were similar to those of [68Ga]Ga-NOTA-anti-miR-15b. The level of radioactivity in bones was only marginally lower in rats injected with [68Ga]Ga-NOTA-anti-miR-15b(scr) than in those injected with [68Ga]Ga-NOTA-anti-miR-15b. The control tracers [68Ga]Ga-NOTA-T6 and [68Ga]Ga-NOTA did not accumulate in the kidneys, bones, or bone marrow but were excreted in urine (Figure 5). [68Ga]Ga-NOTA-T6 tended to accumulate in the intestine (Figures 2G and 3G, Table S3).
Table 1. Ratios of Radioactivity Levels between 68Ga-Labeled Anti-miR-15b RNA Oligonucleotides and Control Tracers.
| bone | |||||
|---|---|---|---|---|---|
| [68Ga]Ga-NOTA-anti-miR-15b(2SBS) | [68Ga]Ga-NOTA-anti-miR-15b(scr) | [68Ga]Ga-NOTA-anti-miR-34 | [68Ga]Ga-NOTA-T6 | [68Ga]Ga-NOTA | |
| [68Ga]Ga-NOTA-anti-miR-15b | <2 | <2 | <2 | 7a | 9a |
| [68Ga]Ga-NOTA(mod)-anti-miR-15b | 10a | 13a | |||
| [68Ga]Ga-DOTA-anti-miR-15b | 7a | 9a | |||
| bone marrow | |||||
|---|---|---|---|---|---|
| [68Ga]Ga-NOTA-anti-miR-15b(2SBS) | [68Ga]Ga-NOTA-anti-miR-15b(scr) | [68Ga]Ga-NOTA-anti-miR-34 | [68Ga]Ga-NOTA-T6 | [68Ga]Ga-NOTA | |
| [68Ga]Ga-NOTA-anti-miR-15b | 2 | 1 | 2 | 7a | 12a |
| [68Ga]Ga-NOTA(mod)-anti-miR-15b | 15a | 24a | |||
| [68Ga]Ga-DOTA-anti-miR-15b | 8a | 13a | |||
| kidneys | |||||
|---|---|---|---|---|---|
| [68Ga]Ga-NOTA-anti-miR-15b(2SBS) | [68Ga]Ga-NOTA-anti-miR-15b(scr) | [68Ga]Ga-NOTA-anti-miR-34 | [68Ga]Ga-NOTA-T6 | [68Ga]Ga-NOTA | |
| [68Ga]Ga-NOTA-anti-miR-15b | 3 | 1 | <2 | 33a | 45a |
| [68Ga]Ga-NOTA(mod)-anti-miR-15b | 18a | 25a | |||
| [68Ga]Ga-DOTA-anti-miR-15b | 15a | 21a | |||
| urine | |||||
|---|---|---|---|---|---|
| [68Ga]Ga-NOTA-anti-miR-15b(2SBS) | [68Ga]Ga-NOTA-anti-miR-15b(scr) | [68Ga]Ga-NOTA-anti-miR-34 | [68Ga]Ga-NOTA-T6 | [68Ga]Ga-NOTA | |
| [68Ga]Ga-NOTA-anti-miR-15b | 0.60 (2) | 0.27 (4)a | 0.49 (2) | 0.07 (13)a | 0.05 (19)a |
| [68Ga]Ga-NOTA(mod)-anti-miR-15b | 0.13 (8)a | 0.09 (11)a | |||
| [68Ga]Ga-DOTA-anti-miR-15b | 0.11 (9)a | 0.08 (13)a | |||
Inverted ratios are shown in parentheses (P < 0.05).
Figure 5.
Ex vivo radioactivity measurements of excised tissue samples obtained from rats intravenously injected with 68Ga-labeled tracers. Results are expressed as mean SUVs. Error bars denote SD values.
Pharmacokinetics
[68Ga]Ga-NOTA-anti-miR-15b, [68Ga]Ga-NOTA(mod)-anti-miR-15b, [68Ga]Ga-DOTA-anti-miR-15b, [68Ga]Ga-NOTA-anti-miR-15b(2SBS), [68Ga]Ga-NOTA-anti-miR-15b(scr), and [68Ga]Ga-NOTA-anti-miR-34 had similar total clearances and elimination rate constants (Table 2). However, the maximum concentration and time of the maximum concentration varied greatly between the tracers. The total clearances and elimination rate constants of [68Ga]Ga-NOTA-T6 and [68Ga]Ga-NOTA were higher than those of the 22-mers.
Table 2. Pharmacokinetic Parameters of Intravenously Administered 68Ga-labeled Anti-miR-15b RNA Oligonucleotides and Control Tracers in Healthy Adult Ratsa.
| tracer | AUC0→∞ (min × g/mL) | ClT (mL/min × g) | kel (mL/min × g) | t1/2 (min) | Cmax (mL/g) | Tmax (s) |
|---|---|---|---|---|---|---|
| [68Ga]Ga-NOTA-anti-miR-15b | 120.0 ± 3.6 | 0.0083 ± 0.0003 | 0.0231 ± 0.0030 | 30.4 ± 3.9 | 23.6 ± 1.6 | 5.0 ± 0.0 |
| [68Ga]Ga-NOTA(mod)-anti-miR-15b | 125.6 ± 14.9 | 0.0080 ± 0.0010 | 0.0209 ± 0.0031 | 33.7 ± 5.5 | 18.3 ± 6.8 | 25.0 ± 26.5 |
| [68Ga]Ga-DOTA-anti-miR-15b | 229.7 ± 128.1 | 0.0057 ± 0.0032 | 0.0155 ± 0.0056 | 48.7 ± 15.2 | 16.8 ± 3.1 | 17.5 ± 12.6 |
| [68Ga]Ga-NOTA-anti-miR-15b(scr) | 101.8 ± 5.6 | 0.0098 ± 0.0005 | 0.0257 ± 0.0021 | 27.1 ± 2.2 | 15.4 ± 1.7 | 10.0 ± 5.8 |
| [68Ga]Ga-NOTA-anti-miR-15b(2SBS) | 68.6 ± 6.6 | 0.0147 ± 0.0015 | 0.0220 ± 0.0016 | 31.7 ± 2.2 | 8.6 ± 0.9 | 79.5 ± 17.5 |
| [68Ga]Ga-NOTA-anti-miR-34 | 92.3 ± 21.1 | 0.0112 ± 0.0024 | 0.0201 ± 0.0024 | 34.9 ± 4.5 | 14.4 ± 5.8 | 30.5 ± 6.0 |
| [68Ga]Ga-NOTA-T6 | 100.2 ± 13.7 | 0.0101 ± 0.0013 | 0.0329 ± 0.0066 | 21.7 ± 4.4 | 17.3 ± 3.9 | 7.5 ± 5.0 |
| [68Ga]Ga-NOTA | 100.0 ± 15.7 | 0.0102 ± 0.0017 | 0.0323 ± 0.0083 | 22.4 ± 5.3 | 18.6 ± 2.4 | 12.5 ± 5.0 |
AUC0→∞ = area under the curve; ClT = total clearance; kel = elimination rate constant; t1/2 = plasma half-life; Cmax = maximum concentration; Tmax = time of maximum concentration.
Discussion
The present study evaluated the biodistributions of intravenously administered conjugated anti-miR-15b in rats. miR-15b functions in various tissues including bones and is expressed in mesenchymal stem-cell-derived osteogenic cells.15,16 Although 68Ga-labeled anti-miR-15b has been previously investigated, this study evaluated the fate of anti-miR-15b tracers in more detail, with a particular focus on blood pharmacokinetics and the specificity of bone uptake.19,22
In vivo PET imaging and ex vivo analysis of excised tissue samples demonstrated that bone and kidney uptake of [68Ga]Ga-NOTA-anti-miR-15b markedly differed from that of [68Ga]Ga-NOTA-T6 and 68Ga-NOTA. However, it is unclear whether the RNA sequence itself or the length of the RNA oligonucleotide is responsible for these differences. Short RNA molecules accumulate in the kidneys and liver.25 Nephrons filter short [68Ga]Ga-NOTA-T6 molecules (2.3 kDa), whereas larger 22-mer sequences (7.9–8.2 kDa) are probably filtered and reabsorbed later in the proximal tubuli. The renal filtration cutoff molecular weight for globular proteins is about 70 kDa.26 Wu and coworkers studied the biodistributions of phosphorothioate oligonucleotides complementary to rat chromogranin A with various sequence lengths (30-, 20-, 12-, and 6-mer). The 20-mer in their study accumulates in the kidneys and liver, similar to the 22-mers used in the present study; however, the levels of 6-mers differed between the previous study and the present study. In contrast with the findings of Wu et al., we demonstrated that [68Ga]Ga-NOTA-T6 was not present at high levels in the kidneys but accumulated in the intestines (Figures 2G, 3G, Table S3).
The question remains whether the bone accumulation observed is due to the anti-miR-15b sequence or the length of the RNA oligonucleotide and the properties of the chelator and 68Ga. The control measurement using three different chelators for anti-miR-15b confirmed that the chelator played a minor role in the distribution, which is in the line with our previous studies.19 However, 68Ga accumulates in bone and consequently the radioactivity detected in bones may correspond to free 68Ga detached from the chelator, rather than to the oligonucleotide itself. To exclude this possibility, quality control for radiochemical purity was performed for all tracers using radio-HPLC. Moreover, bone radioactivity was not observed in rats injected with [68Ga]Ga-NOTA-T6 and [68Ga]Ga-NOTA, which harbored the same chelator and labeled [68Ga]Ga-NOTA-anti-miR-15b.
A scrambled sequence was synthesized to determine if the bone accumulation is due to the anti-miR-15b sequence. This scrambled sequence contained the same nucleotides as the anti-miR-15b sequence; however, the order of the nucleotides was randomized and this sequence lacked a specific target in the rat genome according to a BLAST search. PET/CT imaging revealed that the level of [68Ga]Ga-NOTA-anti-miR-15b(scr) in the epiphyses was much lower than that of [68Ga]Ga-NOTA-anti-miR-15b. However, ex vivo analyses of larger rats using an HRRT camera showed that the level of [68Ga]Ga-NOTA-anti-miR-15b(scr) was only slightly lower than that of [68Ga]Ga-NOTA-anti-miR-15b in bones. This discrepancy is because the excised bone samples and bone ROIs in PET/CT analyses did not correspond to the same region. In vivo PET/CT imaging indicated that radioactivity was mainly present in the epiphyses of long bones. The ex vivo samples consisted of the entire tibia, not just the epiphyses. TACs can be used to objectively compare the level of radioactivity between samples. The SUVs, which take into account the injected radioactivity dose and animal weight, of [68Ga]Ga-NOTA-anti-miR-15b(scr) and [68Ga]Ga-NOTA-anti-miR-15b significantly differed (P < 0.0001, Figure 4A).
The quantities of synthesized RNA conjugates were limited; therefore, we were unable to perform in vivo stability analyses using rat plasma samples. This would have required larger quantities of the injected tracers because the sensitivity of radio-HPLC is limited. Instead, we modeled pharmacokinetics using plasma radioactivity curves derived from dynamic PET data, which represented the total radioactivity concentration as a function of time after injection. The sample size in each group was limited to 4–6 animals. Both female and male rats were analyzed, and the results were pooled because no differences were observed between genders.
In conclusion, PET imaging successfully elucidated the distributions and kinetics of 68Ga-labeled chelated miRNA-targeting molecules in vivo. Our results revealed that intravenously injected 68Ga-labeled anti-miR-15b is specifically accumulated in the bone. This study confirms that PET imaging is a useful approach to evaluate new miRNA-based drugs.
Acknowledgments
The authors thank Aake Honkaniemi and Timo Kattelus for technical assistance. This work was financially supported by the Academy of Finland (grant numbers 308931, 251539, and 252097) and Jane and Aatos Erkko Foundation.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.molpharmaceut.8b01169.
MS (ESI-TOF) data and representative RP-HPLC chromatograms of the oligonucleotides; radioactivity concentration of 68Ga-labeled molecules in rat blood and plasma (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Fire A.; Xu S.; Montgomery M. K.; Kostas S. A.; Driver S. E.; Mello C. C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Bushati N.; Cohen S. M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Mizoguchi F.; Izu Y.; Hayata T.; Hemmi H.; Nakashima K.; Nakamura T.; Kato S.; Miyasaka N.; Ezura Y.; Noda M. Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J. Cell. Biochem. 2010, 109, 866–875. 10.1002/jcb.22228. [DOI] [PubMed] [Google Scholar]
- Sugatani T.; Hruska K. Impaired micro-RNA pathways diminish osteoclast differentiation and function. J. Biol. Chem. 2009, 284, 4667–4678. 10.1074/jbc.M805777200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur T.; Hussain S.; Mudhasani R.; Parulkar I.; Colby J. L.; Frederick D.; Kream B. E.; van Wijnen A. J.; Stein J. L.; Stein G. S.; Jones S. N.; Lian J. B. Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse. Dev. Biol. 2010, 340, 10–21. 10.1016/j.ydbio.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T.; Lu J.; Cobb B. S.; Rodda S. J.; McMahon A. P.; Schipani E.; Merkenschlager M.; Kronenberg H. M. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 1949–1954. 10.1073/pnas.0707900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri A. A.; So A. Y.-L.; Sinha N.; Gibson W. S. J.; Taganov K. D.; O’Connell R. M.; Baltimore D. MicroRNA-125b potentiates macrophage activation. J. Immunol. 2011, 187, 5062–5068. 10.4049/jimmunol.1102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E. A.; Kong L.; Bai X. H.; Luan Y.; Liu C. J. miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J. Biol. Chem. 2009, 284, 11326–11335. 10.1074/jbc.M807709200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau B. N.; Xin C.; Hartner J.; Ren S.; Castano A. P.; Linn G.; Li J.; Tran P. T.; Kaimal V.; Huang X.; Chang A. N.; Li S.; Kalra A.; Grafals M.; Portilla D.; MacKenna D. A.; Orkin S. H.; Duffield J. S. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci. Transl. Med. 2012, 4, 121ra18 10.1126/scitranslmed.3003205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang C.; Lu Y. M.; Meng L. R. MicroRNA-125b down-regulation mediates endometrial cancer invasion by targeting ERBB2. Med. Sci. Monit. 2012, 18, BR149–BR155. 10.12659/MSM.882617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Liu X. L.; Li X. R. Research progress on siRNA delivery with nonviral carriers. Int. J. Nanomed. 2011, 6, 1017–1025. 10.2147/IJN.S17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geary R. S.; Norris D.; Yu R.; Bennett C. F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Delivery Rev. 2015, 87, 46–51. 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Porrello E. R.; Johnson B. A.; Aurora A. B.; Simpson E.; Nam Y. J.; Matkovich S. J.; Dorn G. 2nd; van Rooij E.; Olson E. N. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ. Res. 2011, 109, 670–679. 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An F.; Gong B.; Wang H.; Yu D.; Zhao G.; Lin L.; Tang W.; Yu H.; Bao S.; Xie Q. miR-15b and miR-16 regulate TNF mediated hepatocyte apoptosis via BCL2 in acute liver failure. Apoptosis 2012, 17, 702–716. 10.1007/s10495-012-0704-7. [DOI] [PubMed] [Google Scholar]
- Gao J.; Yang T.; Han J.; Yan K.; Qiu X.; Zhou Y.; Fan Q.; Ma B. MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. J. Cell. Biochem. 2011, 112, 1844–1856. 10.1002/jcb.23106. [DOI] [PubMed] [Google Scholar]
- Palmieri A.; Pezzetti F.; Brunelli G.; Martinelli M.; Lo Muzio L.; Scarano A.; Degidi M.; Piattelli A.; Carinci F. Peptide-15 changes miRNA expression in osteoblast-like cells. Implant Dent. 2008, 17, 100–108. 10.1097/ID.0b013e318166d182. [DOI] [PubMed] [Google Scholar]
- Lendvai G.; Velikyan I.; Bergström M.; Estrada S.; Laryea D.; Välilä M.; Salomäki S.; Långström B.; Roivainen A. Biodistribution of 68Ga-labelled phosphodiester, phosphorothioate, and 2′-O-methyl phosphodiester oligonucleotides in normal rats. Eur. J. Pharm. Sci. 2005, 26, 26–38. 10.1016/j.ejps.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Roivainen A.; Tolvanen T.; Salomäki S.; Lendvai G.; Velikyan I.; Numminen P.; Välilä M.; Sipilä H.; Bergström M.; Härkönen P.; Lönnberg H.; Långström B. 68Ga-labeled oligonucleotides for in vivo imaging with PET. J. Nucl. Med. 2004, 45, 347–355. [PubMed] [Google Scholar]
- Kiviniemi A.; Mäkelä J.; Mäkilä J.; Saanijoki T.; Liljenbäck H.; Poijärvi-Virta P.; Lönnberg H.; Laitala-Leinonen T.; Roivainen A.; Virta P. Solid-supported NOTA and DOTA chelators useful for the synthesis of 3′-radiometalated oligonucleotides. Bioconjugate Chem. 2012, 23, 1981–1988. 10.1021/bc300253t. [DOI] [PubMed] [Google Scholar]
- Hwang D. W.; Ko H. Y.; Lee J. H.; Kang H.; Ryu S. H.; Song I. C.; Lee D. S.; Kim S. A nucleolin-targeted multimodal nanoparticle imaging probe for tracking cancer cells using an aptamer. J. Nucl. Med. 2010, 51, 98–105. 10.2967/jnumed.109.069880. [DOI] [PubMed] [Google Scholar]
- Rockey W. M.; Huang L.; Kloepping K. C.; Baumhover N. J.; Giangrande P. H.; Schultz M. K. Synthesis and radiolabeling of chelator-RNA aptamer bioconjugates with copper-64 for targeted molecular imaging. Bioorg. Med. Chem. 2011, 19, 4080–4090. 10.1016/j.bmc.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäkilä J.; Jadhav S.; Kiviniemi A.; Käkelä M.; Liljenbäck H.; Poijärvi-Virta P.; Laitala-Leinonen T.; Lönnberg H.; Roivainen A.; Virta P. Synthesis of multi-galactose-conjugated 2′-O-methyl oligoribonucleotides and their in vivo imaging with positron emission tomography. Bioorg. Med. Chem. 2014, 22, 6806–6813. 10.1016/j.bmc.2014.10.034. [DOI] [PubMed] [Google Scholar]
- Jadhav S.; Käkelä M.; Bourgery M.; Rimpilä K.; Liljenbäck H.; Siitonen R.; Mäkilä J.; Laitala-Leinonen T.; Poijärvi-Virta P.; Lönnberg H.; Roivainen A.; Virta P. In vivo bone-targeting of bis(phosphonate)-conjugated double helical RNA monitored by positron emission tomography. Mol. Pharmacol. 2016, 13, 2588–2595. 10.1021/acs.molpharmaceut.6b00261. [DOI] [PubMed] [Google Scholar]
- Jadhav S.; Käkelä M.; Mäkilä J.; Kiugel M.; Liljenbäck H.; Virta J.; Poijärvi-Virta P.; Laitala-Leinonen T.; Kytö V.; Jalkanen S.; Saraste A.; Roivainen A.; Lönnberg H.; Virta P. Synthesis and in vivo PET imaging of hyaluronan conjugates of oligonucleotides. Bioconjugate Chem. 2016, 27, 391–403. 10.1021/acs.bioconjchem.5b00477. [DOI] [PubMed] [Google Scholar]
- Wu F.; Yngve U.; Hedberg E.; Honda M.; Lu L.; Eriksson B.; Watanabe Y.; Bergström M.; Långström B. Distribution of 76Br-labeled antisense oligonucleotides of different length determined ex vivo in rats. Eur. J. Pharm. Sci. 2000, 10, 179–186. 10.1016/S0928-0987(00)00067-1. [DOI] [PubMed] [Google Scholar]
- Vegt E.; de Jong M.; Wetzels J. F.; Masereeuw R.; Melis M.; Oyen W. J.; Gotthardt M.; Boerman O. C. Renal toxicity of radiolabeled peptides and antibody fragments: mechanisms, impact on radionuclide therapy, and strategies for prevention. J. Nucl. Med. 2010, 51, 1049–1058. 10.2967/jnumed.110.075101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.