Abstract
Poly(ethylene glycol) (PEG) polymers and PEG-conjugated lipids are widely used in bioengineering and drug transport applications. A PEG layer in a drug carrier increases hydrophilic repulsion, inhibits membrane fusion and serum opsonin interactions, and prolongs the storage and circulation time. It can also change the carrier shape and have an influence on many properties related to the content release of the carrier. In this paper, we focus on the physicochemical effects of PEGylation in the lipid bilayer. We introduce laurdanC as a fluorophore for shape recognition and phase transition detection. Together with laurdanC, cryogenic transmission electron microscopy, differential scanning calorimetry, molecular dynamics simulations, and small-angle X-ray scattering/wide-angle X-ray scattering, we acquire information of the particle/bilayer morphology and phase behavior in systems containing 1,2-dipalmitoyl-sn-glycero-3-phosphocholine:1,2-distearoyl-sn-glycero-3-phosphoethanolamine-PEG(2000) with different fractions. We find that PEGylation leads to two important and potentially usable features of the system. (1) Spherical vesicles present a window of elevated chain-melting temperatures and (2) lipid packing shape-controlled liposome-to-bicelle transition. The first finding is significant for targets requiring multiple release sequences and the second enables tuning the release by composition and the PEG polymer length. Besides drug delivery systems, the findings can be used in other smart soft materials with trigger-polymers as well.
Introduction
In aqueous solutions, phospholipids form various structures. These include liposomes and micelles. Bicelles are a specific subgroup of micelles. They contain a bilayer and are discoidal in shape. Liposomes are used in drug delivery due to their ability to encapsulate hydrophilic molecules into their aqueous core. The hydrophobic part in the bilayer can also be used since lipophilic molecules have an affinity toward the lipid tails. Hence, liposomes,1 bicelles,2 and micelles3,4 can be used in drug delivery purposes alike. In the most typical scheme, a target cell internalizes a carrier via endocytosis and a hydrophilic drug is released by some internal or external trigger. These include changes in pH,5 temperature,6 or radiation in the ultraviolet,7,8 visible,9,10 or near infrared9,11−14 region. Here, the trigger is set to alter the phase order in the lipid bilayer, resulting in an increased permeation and release of the drug14,15 or any encapsulated cargo.16
Drug delivery systems face many obstacles as they pass through the hostile environments of epithelia. Unfortunately, liposomes are usually targeted by serum opsonin and opsonized liposomes are removed from the circulation by specialized phagocytic cells in a matter of hours.17,18 Surviving this clearance is a major objective, because a longer circulation time increases chances of reaching the target cell. Opsonin targeting can be prevented by adding poly(ethylene glycol) (PEG) moieties on the surface of the carrier, usually at the concentration of ca. 5 mol %, which is a procedure sometimes portrayed as “the gold standard”.19,20 The PEG layer adds a steric barrier against opsonins and other bioactive molecules.21 It increases circulation time and provides a stealth sheath that stabilizes the drug delivery system in blood and in storage. A good example of this approach is Doxil (a liposomal drug formulation that is used to deliver chemotherapeutic doxorubicin into the cancer cells). Doxil was approved by the FDA in 1995.22
In addition to the steric barrier, PEG changes the extent of hydration of the bilayer23 and inhibits its interaction with divalent ions, most commonly Ca2+, decreasing the extent of membrane fusion.24,25 Interestingly, PEG can be solubilized in both hydrophilic and hydrophobic solvents.26 Hence, it can be used to bind not only the hydrophilic molecules, like albumin,27,28 but also more hydrophobic molecules, e.g., indocyanine green.12,29 Due to its hydrophobic moieties, it has been hypothesized that PEG may even partly penetrate into the liquid crystal or fluid phase of the bilayer found at high temperatures.25 This obviously affects the behavior of a lipid system as a drug carrier and releaser.
PEG has been studied experimentally and computationally. In coarse-grained molecular dynamics (MD) simulations, transferable PEG models have been introduced by Lee et al.,30 Rossi et al.,31 and Grunewald et al.32 These models have been utilized in simulations of PEGylated lipid bilayers and lipid aggregates.25,33−38 Experimentally, PEGylation has been shown to not only affect the phase transition behavior of the lipid systems,39 but also the particle shape. The latter was also obtained in simulations by Lee and Pastor.33
The shape of the lipid aggregate depends on its lipids. For example, pure 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) form liposomes and pure PEG-terminated 1,2-distearoyl-sn-glycero-3-phosphoethanolamine (DSPE-PEG) forms globular micelles, but with sufficient PEGylation they both form bicelles.40−43 Similarly, l-α-phosphatidylcholine (EPC) forms liposomes but the EPC:DSPE-PEG system forms long cylindrical micelles instead of bicelles.40,41,43,44 The shape is usually predicted by the spontaneous curvature45−48 (or the packing parameter49,50) of the lipid or the lipid mixture. This parameter is characteristic of a single lipid, and DPPC and EPC are not actually much different in this aspect. Segregation of the lipids can cause the formation of bicelles, whereas a nonsegregated lipid system (i.e., EPC system) form long cylindrical micelles.51 The rigidity of the bilayer plays a key role52 because it affects the segregation of lipids. For instance, EPC:DSPE-PEG with cholesterol (40 mol %) forms bicelles due to the stiffening of the tail groups.40,43
In this paper, we have studied the physicochemical effects of PEGylation on the structure, shape, and sizes of the lipid systems containing DPPC and DSPE-N-[amino(poly(ethylene glycol))-2000] (DSPE-PEG(2000)). We introduce a new method for the shape recognition using fluorescence emissions of laurdanC. This method is presented along with several other techniques, including new MD simulations that provide a pervasive view on the two effects of PEGylation. The first effect is the shift in the phase transition temperature, measured with differential scanning calorimetry (DSC) and laurdanC. This shift could be utilized in partial drug release with, e.g., photothermal agents,14 since the most effective drug release occurs in the phase transition temperature region. The transformation from liposomes to bicelles is another effect of PEGylation. Here, we show that laurdanC data correlates with the average aspect ratio of the particles obtained with cryogenic transmission electron microscopy (cryo-TEM). In this way, we have determined the shape transformation threshold at 9.6 mol %, which is also corroborated with MD simulations and small-angle X-ray scattering (SAXS) analysis. Particle sizes from SAXS analysis agree with cryo-TEM images and show with MD simulations that PEGylated lipids are relocated to higher curvature regions. Hence, laurdanC measurements combined with cryo-TEM, SAXS/wide-angle X-ray scattering (WAXS), and MD simulations show that bicelles are formed when the spontaneous curvature of the mixture enables the formation of bicelle rims. This is demonstrated here for DSPE-PEG(2000), but as the findings relate particle shape directly with the polymer packing, they are generalizable to polymer–lipid systems and could be used to design, e.g., drug delivery systems.
Materials and Methods
Materials
1,2-Dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(poly(ethylene glycol))-2000] (DSPE-PEG(2000)) were purchased from Avanti Polar lipids. Ethanol was purchased from Altia. Carboxylated laurdan (laurdanC) was synthesized using the pathway described by Cheniour et al.53 with minor changes. Rest of the chemicals were purchased from Sigma-Aldrich unless otherwise stated.
Preparation of Lipid Samples
Samples containing laurdanC were prepared according to ref (14) but using N-(2-hydroxyethyl)piperazine-N′-ethanesulfonic acid (HEPES) buffer (150 mM NaCl, 20 mM HEPES in pH 7.4) instead of Tris–HCl. Other samples were prepared as follows: 20 μmol of lipids in chloroform, containing 0, 3, 6, 9, 12, 15, or 24 mol % of DSPE-PEG(2000) were added to a round flask. Chloroform was evaporated in a nitrogen stream and the remaining lipid film was heated up to 60 °C prior to the addition of 2 mL of HEPES buffer. The dispersion was sonicated in a 60 °C bath sonicator for the duration of adding the buffer solution (≤45 s). After this, the dispersion was extruded eleven times through double-stacked polycarbonate membranes with the pore size of 200 nm. The temperature in both steps was constant, at 60 °C. After the extrusion, samples were stored in a refrigerator overnight before further studies.
Cryo-TEM
Vitrified lipid samples were prepared in FEI Vitrobot (FEI) after Kuntsche et al.54 and Iancu et al.55 Briefly, ethane gas was condensed halfway into a liquid-nitrogen-cooled Vitrobot cup following liquefaction of propane gas and resulting in a liquefied gas mixture of 1:1 ethane:propane. A plasma-treated (30 s H2–O2, Gatan Solarus 950, Gatan) QUANTIFOIL R 2/2 (or R 2/1) copper TEM grid (Electron Microscopy Sciences) was placed inside the Vitrobot chamber and the humidity was increased to 100% at 22 °C. 5 μL of the sample was pipetted onto the TEM grid, the grid was blotted with a filter paper for 1 s, and immersed into the ethane–propane mixture. The grid was then moved into the cryo-TEM sample holder under liquid nitrogen. Micrographs were taken with an FEI Tecnai T12 (FEI) transmission electron microscope (TEM) with 120 kV accelerating voltage.
The size distribution and the number of distinctive particles were determined with a self-made Matlab script (Supporting Information). Each particle type was classified to groups of “liposomal”, “bicellar edge” (bicelle edge toward the microscope), “bicellar face” (bicelle face toward the microscope), and “unknown” (irregular shapes) structures. Structures at the vicinity of 5 nm in diameter were excluded from the image analyses since they are difficult to distinguish from the background. The number fraction of bicelles xbicelleTEM,num was determined from the number of bicellar shapes vs the total number of particles. However, the signal intensity in, e.g., SAXS, DSC, and dynamic light scattering does not follow the number of particles but the number and the molar mass of monomers in the particles. The number of surfactant molecules is related to the surface area of the particles. Hence, the monomeric bicelle fraction can be determined as
| 1 |
where τ is the area-scaling factor. It is a function of the average area of liposomes and bicelles
| 2 |
where d is the thickness of the bilayer, and rliposome and rbicelle are the average radii of the corresponding particle.
Differential Scanning Calorimetry and Fluorescence Spectroscopy
DSC thermograms were measured with a MicroCal VP-DSC calorimeter (MicroCal Inc., Northampton, MA) between 10 and 60 °C, starting at 10 °C. Heating and cooling rates were 1 and 0.5 °C min–1, respectively, to obtain similar peak positions and shapes, considering the differences in the phase separation kinetics. Both scan directions were measured twice to ensure the reversibility of the system.
LaurdanC measurements were performed with a PerkinElmer LS5 fluorescence spectrometer in a temperature controlled four-window cuvette. Each measurement was started after stabilization for 3 min at the given temperature between 25 and 70 °C. Smaller temperature intervals (0.5 °C) were used near the phase transition region (i.e., 34–45 °C). The lipid order was calculated with the laurdan generalized polarization (GP)
| 3 |
where Io and Id represent the emission intensities at the wavelengths corresponding to the ordered (λem = 430 nm) and disordered state (λem = 510 nm) of the lipid bilayer. In addition to GP, the derivative ∂GP/∂T was determined to portray the data similar to the DSC results.
The deconvolution of the DSC data and ∂GP/∂T peaks was calculated using the Voigt profile analysis package by Ruzi,56 using a Faddeeva function algorithm.57 The Matlab script fits multiple Voigt profiles, i.e., the convolution of Gaussian distribution and Cauchy–Lorentz distribution, simultaneously. The number of fitted Voigt profiles was set to 7 in DSC data and 5 in the GP differentials. Signals that were clearly part of the baseline, had zero intensity, or were part of the pre-phase transition were subtracted. Typically, the DSC fits found 3 distinctive Voigt profiles below the main phase transition peaks in DSC but, e.g., the PEG9a sample contained 5 distributions in the upward scan and 4 distributions in the downward scan. The peaks were labeled in the order of their position and compared with the like measurement (e.g., downward scan of PEG9 with the other downward scans).
Small/Wide-Angle X-ray Scattering
The SAXS/WAXS measurements were performed using a SAXSLab Ganesha 300XL instrument (SAXSLAB ApS, Skovlunde, Denmark), a pinhole collimated system equipped with a Genix 3D X-ray source (Xenocs SA, Sassenage, France). The scattered intensity was recorded with the detector placed at three sample-to-detector distances yielding a range of the scattering vector q of 0.05–20.0 nm–1. The samples were sealed in a 1.5 mm diameter quartz capillary (Hilgenberg GmbH, Malsfeld, Germany), and the measurements were performed at 25, 41, and 50 °C. In all cases, the temperature was controlled with water circulation with an accuracy of 0.2 °C. The two-dimensional scattering pattern was recorded by using a two-dimensional 300 K Pilatus detector (Dectris Ltd., Baden, Switzerland) and the radial averaging to obtain I(q) was performed with SAXSGui software. The measured scattering curves were corrected for solvent scattering.
The following analyses for the SAXS data were derived following refs (58−65). The total scattering cross-sections are the sum of the liposome and bicelle scattering (i.e., Ibicelle and Iliposome)
| 4 |
where the fraction of bicelles xbicelles was determined with cryo-TEM. In this form, we assume no interaction between particles. The starting point of Ibicelle and Iliposome is the corresponding core–shell structure. These are detailed in Supporting Information. Briefly, the form factor of the liposome is
| 5 |
where each component layer follows the Rayleigh form factor amplitude,66 Δρi = ρi–1 – ρi is the scattering length contrast between the component layers, ri,j is the radius, and Vi,j is the volume of the component layer, which is deflected as a function of the polydispersity index (see the Supporting Information). For the bicelle, the form factor is67
| 6 |
where each component layer follows cylindrical symmetry according to Fournet,67 α is the angle between the incident beam and the cylinder normal, J1 is the modified Bessel function of order 1, and li is the half-thickness of the component layer. When the particle surface is covered with PEG, the total scattering cross-section is given as44,58
| 7 |
where, in the order of appearance, Ps and Pc are the self-correlation terms of the lipid core and the PEG chain, respectively, Psc and Pcc are the cross-correlation terms of the core-chain and the chain–chain interactions, respectively, n is the macroscopic number density of the particles, and Nc is the number of polymer chains on a single particle. Since PEG is essentially a Gaussian random coil with the characteristics detailed by Hammouda,68,69 the self-correlation term is the well-known Debye function70
| 8 |
where ΔρPEGW = ρPEG – ρ0 is the scattering length density contrast of the PEG chain, vPEG is the partial specific molecular volume of PEG, and RG is its radius of gyration. The form factor amplitude times ΔρPEGVPEG is given as68,69
| 9 |
There is some irregularity in the presentation of fc in the literature. Most authors (e.g., refs (58−60) and (65)) follow Hammouda,68,69 but for instance, refs (44)(61), and (64) have also used fc = ΔρPEGWVPEG(1 – exp(−qRG))(qRG)−1. The expression Pc = 2(ΔρPEGVPEG)2(exp[−(qRG)2] + (qRG)2 – 1)(qRG)−2 that is found in ref (65) may also cause confusion. Since there should be a general proportionality Pc ∝ fc2, the use of eqs 8 and 9 is well justified since fc, with a Taylor approximation exp[−2(qRG)2] ≈ 1 – 2(qRG)2, results in Pc.
The cross-correlation terms of eq 7 with no azimuth angle dependency can be obtained by combining eqs 5 or 6 with eq 9
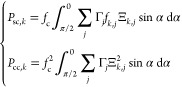 |
10 |
where Ξk,j is the form factor amplitude of an infinitely thin shell that connects the polymer with the surface of the underlying core structure.44 For a liposome with a PEG fraction xout on its outer leaflet and xin = 1 – xout on its inner leaflet
| 11 |
Since liposomes are relatively large, a good approximation is to let xout → 0.5. In this case, xin and xout can be omitted from eq 11 and the multipliers in eq 7 can be effectively substituted with Nc → Nc/2, which is the approach presented by Arleth et al.64 The form factor amplitude of a cylindrical shell is59,61
| 12 |
where the former term describes the fraction xface of the form factor amplitude with PEG only on the face of the bicelle and the latter describes the fraction xrim on the rim region. Uniform polymer coating is obtained when xrim,uniform = 2l4(r4 + 2l4)−1.61
Computational Methods
Molecular dynamics (MD) simulations of pure DPPC and mixtures of DPPC and DSPE-PEG(2000) in water were performed to support the experimental observations. The model used here is based on the implicit-solvent version of the CG MARTINI model,71,72 called DRY-MARTINI.73 Similar to the standard MARTINI PEG models,30,31 a 3-to-1 CG mapping for the PEG is used, whereas the lipids are treated with the 4-to-1 DRY-MARTINI CG mapping scheme. Additional model details are provided in the Supporting Information.
The simulations were run with Gromacs 5.1.174 in the NVT ensemble. Temperature was maintained at T = 323 K by the stochastic velocity rescaling thermostat of Bussi et al.75 with the stochastic friction time constant τt = 4.0 ps. The equations of motion were integrated by the second-order stochastic dynamics integrator using a time step of 40 fs. A cut-off of 1.1 nm was used for both the Lennard-Jones (LJ) and electrostatic interactions with the LJ interaction potential shifted to zero at the cut-off. As for the reaction-field electrostatics, relative permittivity εr = 15 was used beyond the cut-off distance. In total, the simulation protocol follows the standard DRY-MARTINI protocol73 augmented by the improved Verlet integration protocol of ref (76).
Following the experimental setup, mixtures of DPPC and DSPE-PEG(2000), i.e., DSPE lipids with 45 CH2CH2O monomers, are examined. The structures and the construction of the DSPE-PEG model can be found in our earlier publications.36,77 Four different molecular compositions were studied. These contained (1) 10 500 DPPC molecules (PEG0), (2) 10 269 DPPC and 231 DSPE-PEG molecules (PEG2.2), (3) 9400 DPPC and 1100 DSPE-PEG molecules (PEG10.5), and (4) 5250 DPPC and 5250 DSPE-PEG molecules (PEG50). Each lipid was also associated with a Na+ counter-ion to maintain the electroneutrality in the systems. The water interaction in the system is given implicitly by the DRY-MARTINI model.
Initial configurations were constructed by distributing the lipids randomly in the simulation box. The PEG chains were set to an extended coil state. A simulation box of 50 nm × 50 nm × 50 nm was used with PEG0, PEG2.2, and PEG10.5 systems and 100 nm × 100 nm × 100 nm for the PEG50 system. For assessing lipid segregation and bicelle thickness, 30 nm × 30 nm × 30 nm systems of self-assembled single bicelles of size 1050 lipids, with the corresponding DSPE-PEG2000 fractions, were analyzed. All systems were initially energy minimized by using the steepest descent algorithm after which an MD run was performed in the NVT ensemble with the total duration of 0.5 μs.
Results and Discussion
Microscopic Characteristics of PEGylated lipid systems
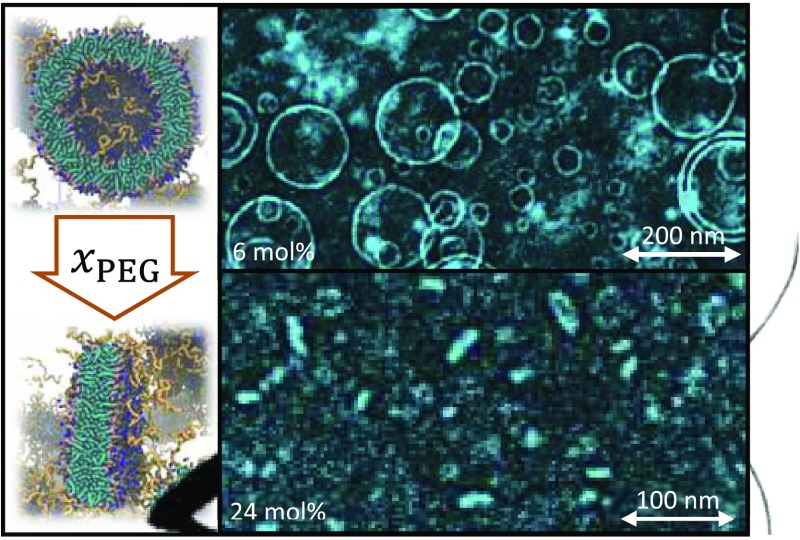
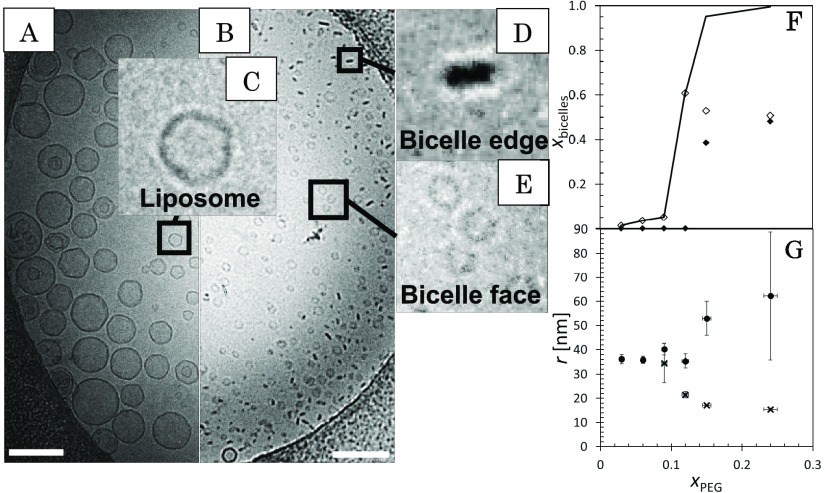
Typical cryo-TEM image of the PEG3 and PEG24 samples are shown in Figure 1A,B. In all PEGylated samples, only a few types of particles were observed. These were liposomes (Figure 1C) and bicelles (Figure 1D,E). The PEG24 sample also contained some objects close to the resolution limit of cryo-TEM (5 nm).78 These particles are difficult to qualify from the background. Hence, they were disregarded. As depicted in refs (40−43), bicelles are typically orientated with their edge (Figure 1D) or their facet (Figure 1E) facing the microscope. Intermediate orientations were also found and counted in as face oriented bicelles. The orientation is influenced by the nearby surface and the thickness of the sample film that is biconcave across the sample hole.78 Larger liposomes and bicelles with edge orientation prefer the perimeter, where the solvent layer is thicker and the aperture wall closer. Smaller particles, including face orientated bicelles, occupy the center of the image increasingly at xPEG > 0.12. In some rare occasions, seemingly saddle-shaped (in PEG12) or folded bicelles (in PEG9) could be observed. The fractions of the bicellar units are shown in Figure 1F. As can be seen, bicellar units start to emerge rapidly at xPEG > 0.09 and bicellar faces appear in the sample with 15 mol % of PEGylation. Thus, the system undergoes a transformation from liposomes to bicelles at ca. 10 mol % and the orientation becomes more random when the bicelle size decreases,78 which is evident from the cryo-TEM data in Figure 1G. Although the number of bicelles increases, their average size decreases and the size of the remaining liposomes increases. This marks a clear separation between the shapes and it shows that the smallest units have a strong affinity to produce disks.
Figure 1.
Typical cryo-TEM image of PEG3 (A) and PEG24 (B). Scale bar is 200 nm. (C), (D), and (E) display liposomes, edge bicelles, and face bicelles, respectively. The total fraction and the subset bicelle fraction are depicted in (F) with bicelle face and edge as filled diamonds and unfilled diamonds, respectively. The size of the liposomes (dots) and bicelles (crosses) is shown in (G).
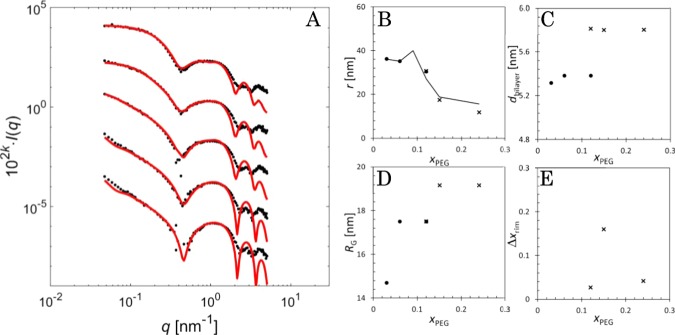
The SAXS patterns (Figure 2A) of the low PEGylation resembles those of Pili et al.,79 who measured similar systems with synchrotron-based SAXS. The SAXS data were fitted (red lines) with parameters in Table S1 of the Supporting Information. The particle radius (Figure 2B) was determined as the sum of the core radius and the half of the bilayer thickness (Figure 2C) that increases when DPPC is coupled with longer lipids (i.e., DSPE-PEG). The radii match well with the cryo-TEM results; black line in Figure 2B shows the average radii from Figure 1G.
Figure 2.
A) SAXS fits (red lines). Black dots represent the SAXS measurements of PEG3 (k = −2), PEG6 (k = −1), PEG12 (k = 0), PEG15 (k = 1), and PEG24 (k = 2). Fit parameters for liposomes (dots) and bicelles (crosses): (B) radius (the line is the cryo-TEM average), (C) bilayer thickness, (D) radius of gyration of PEG, and (E) excess amount of PEG on the rim region.
The radius of gyration RG showed a gradual increase (Figure 2D) when PEG was added. With low PEGylation (PEG3), the effect of RG is very small and more accurate estimation is difficult to obtain. At high PEGylation, RG goes to 1.75 nm and increases to 1.92 nm in samples containing only bicelles. These values are reasonable for PEG with a molar mass of 2 kDa.80,81
The location of the polymers was also determined. On PEG3 liposomes, the PEG moieties have no preference, but in PEG6, the proportion (31:69) favors the outer leaflet. In bicelles, the PEG units are more readily found on the edge, as can be seen in Figure 2E, where the ordinate displays the difference between the fitted value and the uniform polymer coating (xrim,uniform = 2l4(r4 + 2l4)−1).61
The fitting parameters differ slightly from those in the literature. The SANS data of Arleth et al.64 is equivalent to ρPEG = 1.11 × 1011 cm–2, ρtail = 0.83 × 1011 cm–2, and ρhead = 1.45 × 1011 cm–2. Quite close to this, we obtained ρPEG = 1.08 × 1011 cm–2 and ρtail = 0.87 × 1011 cm–2 for liposomes and ρtail = 0.85 × 1011 cm–2 for bicelles. The head group scattering length densities had more variation, stemming partially from the difference of D2O buffer and water. This is somewhat expected; Pedersen and Gerstenberg58 have shown that SAXS measurements of polymer micelles in H2O cannot be readily fitted with parameters obtained from SANS measurements in D2O.
The SAXS fits in Figure 2A are accurate in the q range 0.1–2 nm–1 for all PEGylated samples, and the fits of PEG12, PEG15, and PEG24 are accurate in the entire lower q range. The intermediate range provides the shape and size of the particles. Thus, the small divergence of the PEG3 and PEG6 fits in the very low q range is not significant, and the presence of spherical and discoid structures is consolidated with SAXS. The radii (Figure 2C) are almost equal to those measured with cryo-TEM (Figure 1G) although the radius of the PEG24 bicelle was smaller than that in the cryo-TEM images. This may result from the appearance of particles smaller than the ones quantified in cryo-TEM.
The fits in Figure 2A deviate from the measurements in the high q range. For simplicity, the fitting function did not contain a constant effective background to account for scattering associated with the internal fluid structure of the bilayer. Thus, the fit decays faster at high q than the experimental data. Nevertheless, the fit clearly captures the form factor oscillations, proving that the thicknesses in Figure 2C are in the correct size range. Yet, the determination of the bilayer thickness may not be accurate because the high q region is susceptible to errors in solvent subtraction and the measured intensities do not fall between the behavior of a centrosymmetric sphere model (q–4)61 and a Gaussian random coil model (q–2)61 in either the liposome or bicelle system.
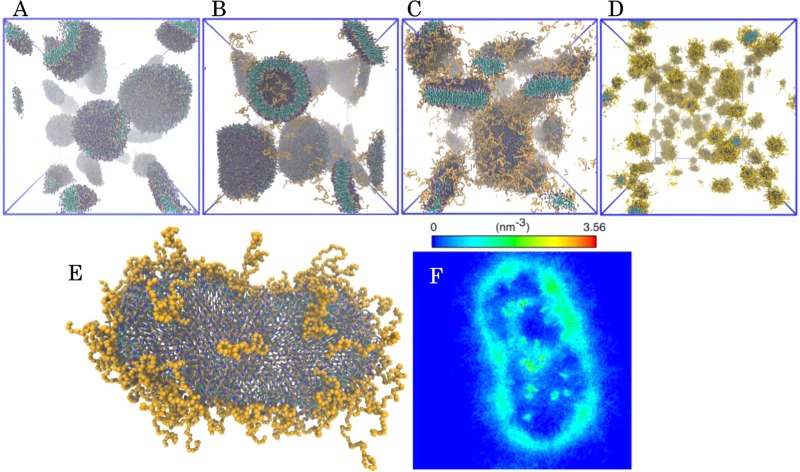
The shape transformation was also verified with MD simulations in Figure 3. Figure 3A shows that liposomes are formed from pure DPPC. This is consistent with the DRY-MARTINI model.73 Similar to Lee and Pastor33 (with DPPE-PEG), the liposome structure is also favored in PEG2.2 (Figure 3B). In PEG10.5 (Figure 3C), only bicelles are formed, whereas the PEG50 system (Figure 3D) produces slightly elongated micelles. Thus, the transition observed in the molecular modeling is in very good agreement with the cryo-TEM and SAXS results. In addition to larger simulations, Figure 3E shows a PEG10.5 bicelle of 1050 molecules. Here, the PEGylated lipids prefer the edge more likely than the bicelle face, in accordance with the SAXS result in Figure 2E and earlier computational works by Lee and Pastor33 and Shinoda et al.,35 who showed that the translocation decreases the line tension at the bicelle edge. This effect is also depicted in Figure 3F, where the density profile of DSPE lipids in bicelle (Figure 3E) is presented. These simulations represent systems with ca. 140 mM of lipids, which is 14-fold the experimental value in the equilibrium but realistic as a local concentration during the self-assembly. The concentration should not have a significant effect on the aggregate shape, because it is mainly controlled by molecular packing.
Figure 3.
Snapshots of the aggregates in 0.5 μs molecular dynamics simulations. Panel (A) shows pure DPPC, panel (B) DPPC with 2.2 mol % DSPE-PEG, panel (C) DPPC and 10.5 mol % DSPE-PEG, and panel (D) 50 mol % PEGylated lipid. Panel (E) shows a detailed structure of a bicelle of 1050 molecules containing 10.5 mol % DSPE-PEG, and panel (F) is the DSPE lipid density profile of the same bicelle.
The simulations in Figure 3A–D contain 10 500 lipids. Due to the system size and finite simulation time, the formed aggregates are smaller than in the experiments.82 For instance, the 2.2 mol % DSPE-PEG system (Figure 3B) produces liposomes with a mean radius of 7.5 nm, whereas cryo-TEM (Figure 1H) and SAXS analysis (Figure 2B) result in a radius closer to 35 nm. On the other hand, the simulation with 10.5 mol % DSPE-PEG (Figure 3C) leads to the formation of a set of bicelles with the largest ones between 9.5 and 11.5 nm in radius. These values are close to those obtained in the experiments. The PEG chain radius of gyration was 1.54 nm in all simulated samples. The value is in good agreement with the hydrodynamic radii obtained with SAXS (Figure 2D), especially as the radius of gyration calculated from the simulation does not encompass the hydration shell. Finally, in the simulations, the thickness of the bilayer corresponding to PEG3 is 5.0 ± 0.1 nm and increases for PEG11.1 to 5.2 ± 0.5 nm (see the Supporting Information). This trend is in line with the SAXS results, even though the SAXS fits are less sufficient in the high q region and the fitted head group is more involved with the water barrier than the simulations.
Thermal Analyses and the Phase Transition in the Lipid Bilayer
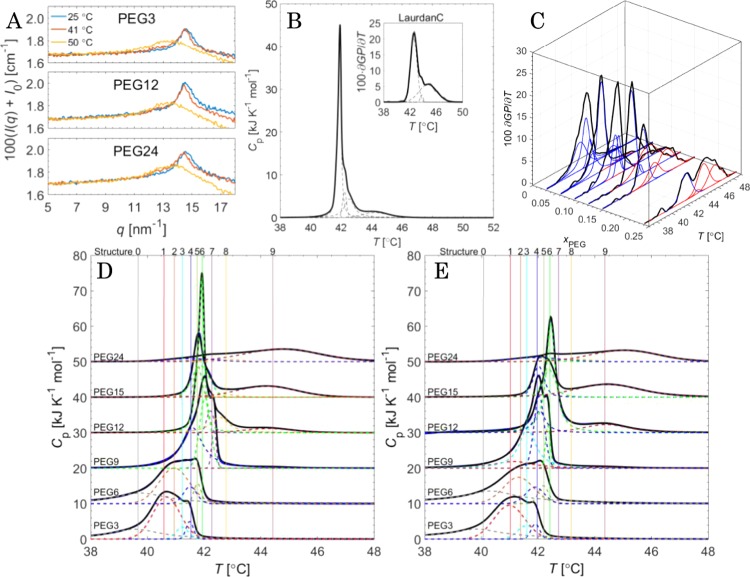
The structures of lipid bilayers showed no significant differences in the WAXS measurements of PEG3, PEG12, and PEG24 (Figure 4A, up-to-down), or in the samples shown in Figure S1. The lipid bilayer portrays near hexagonal packing or gel phase at 25 °C since the Bragg peak is found at ca. 14.5 nm–1 (blue line).83 At 50 °C (orange line), the bilayer is in the fluid phase and only a broad peak is observed just below 14 nm–1. At 41 °C (red line), DPPC is in the ripple phase and the Bragg peak is found at ca. 14.4 nm–1. However, the overall shape is almost equal to that at 25 °C.
Figure 4.
(A) WAXS intensity (I0 is the average scattering intensity of water) vs scattering vector of PEG3, PEG12, and PEG24. (B) Deconvolutions (dashed lines) of the PEG12 sample measured with DSC and laurdanC (inset) (black lines). (C) LaurdanC GP differentials of PEGylated samples with color coding liposomal (blue) and “bicellar” (red). (D) DSC down-scan and (E) up-scan with colors marking the deconvoluted subgroups.
The downward DSC scan of a PEG12 sample and the corresponding laurdanC generalized polarization differential are shown in Figure 4B. As can be seen, the enthalpy can be deconvoluted to four contributions. Similarly, GP differentials (Figure 4C), downward (Figure 4D), and upward (Figure 4E) DSC peaks are composed of multiple peaks. The emergence of particulate subgroups stems mainly from the uneven PEGylation. The average melting temperature increases as a function of PEG fraction. This can be expected since pure DPPC bilayers melt at ca. 41 °C, whereas a DSPE bilayer undergoes a phase transition at ca. 74 °C.84 The increase in the phase transition temperature is also connected with the decrease in the overall lateral pressure as the fatty acid chains of DPPC and DSPE-PEG become increasingly mismatched as the membrane enriches with DSPE-PEG.85,86 In addition to the increase in the melting temperature as shown in Figure 4D,E, broad phase transition peaks, extending below 41 °C, are obtained in the liposomal samples. This is caused by the existence of smaller liposomes (<80 nm in diameter) with wide size distribution.87
The DSC deconvolution peaks were also color coded as displayed in Figure 4D,E. In the case of laurdanC measurements (Figure 4C), only liposomal (blue) and bicellar (red) phase transition peaks were extracted because the signal contains much less data points than the DSC measurements. Both the DSC upward and downward scans of the entire sample set exhibited ten deconvolution peaks. The phase transition temperatures are obviously not the same because the kinetics of freezing and melting are slightly different.
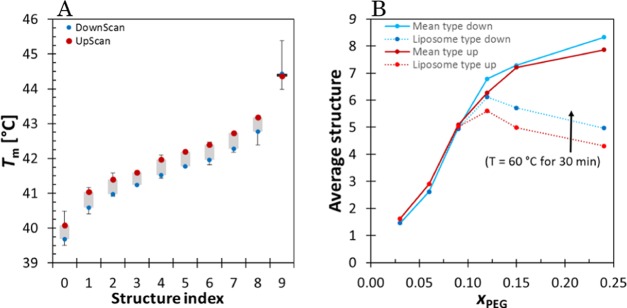
Figure 5A shows the deconvoluted subgroup peaks from Figure 4D,E with indexes from 0 to 9 in the order of their phase transition temperature. The error bars in Figure 5A mark the 95% confidence interval. The average structure in each sample is shown in Figure 5B. It shows that when the samples are kept for 30 min at 60 °C, the melting–freezing process is not completely reversible. Instead, a shift toward higher phase transition temperatures is observed in the PEG-rich samples, proving again that liposome structures in PEG12 and PEG15 are less stable than bicellar structures. The bicelle fraction was also calculated as xbicelleDSC = ΔHbicelle/ΔHtot, where the enthalpy of bicelles was calculated as the sum of the enthalpies of structures 8 and 9. This value is rather similar to that obtained with cryo-TEM (Table 1). In addition, the molar enthalpy change ΔHbicelle and ΔHliposome was calculated using xbicelle values.
Figure 5.
(A) Phase transition temperature from Figure 4D,E in the order of increasing temperature. (B) Average structure in the system vs xPEG.
Table 1. Fraction of Bicelles in DSC Samples xbicelleDSC Compared to Cryo-TEM (xbicelle) and the Molar Enthalpy Change ΔHliposome and ΔHbicelle.
| xbicelleDSC (xbicelle) (%) | ΔHliposome (kJ mol–1) | ΔHbicelle (kJ mol–1) | ||||
|---|---|---|---|---|---|---|
| xPEG | 0.09 | 0.12 | 0.15 | 0.24 | ||
| DSC up | 1.72 | 19.97 | 55.45 | 77.64 | 29.49 | 12.51 |
| DSC down | 0.97 | 30.61 | 47.82 | 83.93 | 27.75 | 13.82 |
| TEM | 1.26 ± 0.14 | 17.38 ± 0.01 | 43.97 ± 0.85 | 83.44 ± 5.96 | ||
The enthalpies in Table 1 represent only the main phase transition; the pre-phase transition was eliminated from the deconvolution for the sake of simplicity. Hence, liposomal enthalpies are ca. 2–4 kJ mol–1 lower than in the raw data. Table 1 shows a noticeable decrease in the average enthalpy when the system transforms from spherical liposomes to discoid bicelles.
Effect of Spontaneous Curvature
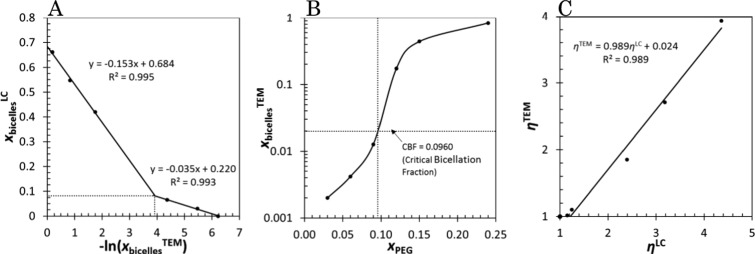
Preceding chapters imply that the onset of bicellation takes place between 9 and 12 mol % of PEG in the formulation. According to the laurdanC measurements, this transition is quite sharp. Comparison of xbicelleLC (calculated from Figure 4C) with the logarithm of xbicelle shows a change in the slope at xPEGc = 0.096 (see Figure 6A,B). We interpret this point as the critical bicellation fraction (CBF).
Figure 6.
A) Log-linear relationship between xbicelleTEM and xbicelle. The change in the slope occurs at CBF, which is converted to xPEG in (B) (see the cross-section of the dotted lines in (A) and (B)). (C) Aspect ratio ηLC vs ηTEM. ηLC values are obtained from measured xbicelleLC by using the eq 13.
LaurdanC measures electric polarity in its proximity.88,89 Hence, its response in the bilayer is related to the area of the lipid–water boundary.90,91 In the preliminary MD simulations, we found that laurdanC has lower mobility in the bilayer than the regular laurdan molecule because of its way of making hydrogen bonds with the lipid headgroups. This leads to assume that laurdanC resides in the facet of the bicelle rather than in the rims. This assumption is also supported by the fact that fraction xbicelleLC does not approach unity when xbicelle → 1 (Figure 6A). Hence, we can derive an equation for the laurdanC GP fraction
| 13 |
where Aflat and Acurved are the total areas of the flat and curved
surfaces and 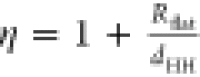 is the aspect ratio. Here, the shape of
the bicelle is the same as presented by Bergström,52 with Rflat being
the radius of the facet and dHH = 4.4
nm being the approximate bilayer thickness from the lipid head group
to another. When the measured xbicelleLC are used to calculate
ηLC with eq 13, an excellent correlation with ηTEM (determined
from the micrographs) is obtained as shown in Figure 6C. LaurdanC measurement can thus determine
the shape (aspect ratio) of the lipid system directly.
is the aspect ratio. Here, the shape of
the bicelle is the same as presented by Bergström,52 with Rflat being
the radius of the facet and dHH = 4.4
nm being the approximate bilayer thickness from the lipid head group
to another. When the measured xbicelleLC are used to calculate
ηLC with eq 13, an excellent correlation with ηTEM (determined
from the micrographs) is obtained as shown in Figure 6C. LaurdanC measurement can thus determine
the shape (aspect ratio) of the lipid system directly.
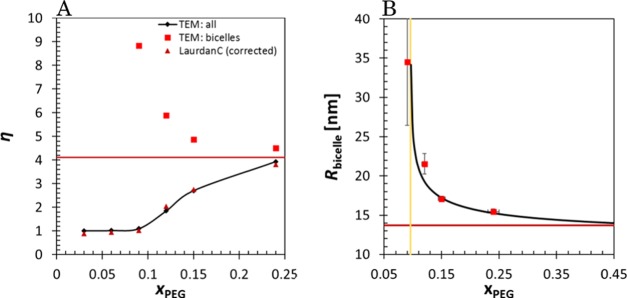
The limit radius Rflat limit = dHH(ηlimit – 1) of bicelles can be obtained by calculating the aspect ratio of the limit xbicelleLC → 0.684 (Figure 6A) with eq 13. With ηLC, this radius limit goes to Rflat limit = 16.2 nm, which is presumably the hydrodynamic radius. When the ηLC values are corrected to the TEM aspect ratio scale with the trend line in Figure 6C, an aspect ratio limit of 4.69 is reached (red line in Figure 7A), that is equivalent with the disc radius Rflat limitTEM = 13.7 nm (red line in Figure 7B). With the two critical parameters, CBF (or xPEG) and Rflat limitTEM, we can also set an empirical relationship for the bicelle radii as
| 14 |
where <A>DPPC = 52 Å2 and <A>DSPE-PEG ≈ 91 Å2. This fit is shown in Figure 7B.
Figure 7.
(A) Aspect ratio of bicelles and for the whole system in TEM (red squares and black diamonds) and the aspect ratio of the whole system with laurdanC (corrected to TEM scale, dark red triangles). The limit aspect ratio is shown as a red line. (B) The size of the bicelles, fitted with an empirical eq 14 based on CBF (light gold line) and the limit radius (red line).
The shape of a lipid particulate comes from the conditions of minimum curvature energy.45,46 Thus, it connects the critical parameters xPEGc and Rflat limit in eq 14. The connection is evident by a simple continuum mechanics examination of the system energetics. The key contributor is the bending energy
| 15 |
where κ is the bending
modulus, 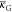 is the Gaussian
curvature modulus, K1 and K2 are the
two principal total curvatures of the bilayer, and K0 is the spontaneous total curvature. Together with the
surface tension, minimizing the surface area, the bending contribution
aims to minimize the difference K1 + K2 – K0 throughout
the bilayer. The surface stress can be written by combining eq 15 with the total curvature K12 = K1 + K2 and the surface tension γ
is the Gaussian
curvature modulus, K1 and K2 are the
two principal total curvatures of the bilayer, and K0 is the spontaneous total curvature. Together with the
surface tension, minimizing the surface area, the bending contribution
aims to minimize the difference K1 + K2 – K0 throughout
the bilayer. The surface stress can be written by combining eq 15 with the total curvature K12 = K1 + K2 and the surface tension γ
| 16 |
Even though the terms |κ| and 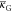 are of the same magnitude in eq 15, the latter term has little effect
on the shape (
are of the same magnitude in eq 15, the latter term has little effect
on the shape (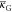 is more important
when membrane fusion
and fission are in question).52,92 Hence, the Gaussian
curvature contribution can be omitted. In addition, it should be noted
that the spontaneous total curvature K0 is double of the commonly used spontaneous curvature with the usual
notations c0 or H0.92 The minimum of eq 16 is the minimum of the free energy
and therefore one can obtain a prediction for the shape transformation
(see, e.g., ref (37)).
is more important
when membrane fusion
and fission are in question).52,92 Hence, the Gaussian
curvature contribution can be omitted. In addition, it should be noted
that the spontaneous total curvature K0 is double of the commonly used spontaneous curvature with the usual
notations c0 or H0.92 The minimum of eq 16 is the minimum of the free energy
and therefore one can obtain a prediction for the shape transformation
(see, e.g., ref (37)).
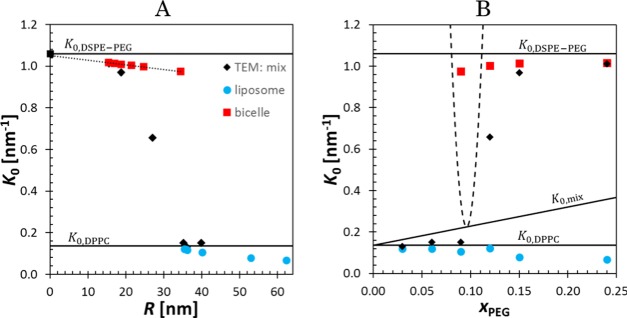
Let us assume that the spontaneous total curvature of the mixture K0,mix follows simple additivity,93K0,mix = xPEGK0,DSPE-PEG + (1 – xPEG)K0,DPPC and the surface tension is a constant. The transition from liposomes to bicelles occurs at the concentration in which the lipid mixture is capable of making the bicelle edge. This condition applies also to tubule-forming systems.40,41,43,44 The transition occurs when the spontaneous total curvature matches the curvature K12 = 1/dHH. We have used the value dHH = 4.4(5) to minimize eq 16 at xPEG = 0.096. The spontaneous total curvature values of individual lipids, K0,DPPC = 0.136 nm–1 93 and = 1.06 nm–1,94 can be extrapolated from the particle curvature vs particle radius plot, as shown in Figure 8A, where the lipid spontaneous curvatures are shown as black lines. The transition region is illustrated in Figure 8B, where the form of eq 16 b is added to the spontaneous total curvature of the mixture (rising line). The bicelle transformation occurs at the minimum of the dashed parabola. Thereafter, the bicelles are decreased in size and eventually transform into spherical micelles. This transformation occurs when the spontaneous total curvature matches that of a sphere at K12 = 4/dHH, which takes place at ca. 84 mol %. This is in perfect agreement with Ashok et al.,4 who have reported a value of 85 mol % at the final transition point; their lipid, EPC, slightly differ in spontaneous curvature and thickness from DPPC.
Figure 8.
A) Mean curvature of liposomes (blue circles), bicelles (red squares), and system average (black diamond). (B) The observed mean curvature average (black triangles) as a function of PEG fraction. The critical bicelle fraction is found at the bending energy minimum at 9.6 mol % (with a bilayer thickness of ca. 4.45 nm). The bending energy is depicted as dashed parabola on the spontaneous curvature K0,mix line.
Conclusions
PEGylation is the current state-of-the-art method to prolong circulation times of liposomal drug delivery systems in blood and in storage. Therefore, understanding the effects of PEGylation on the physicochemical properties of lipid particulates is important. In this paper, the effect of PEGylation in DPPC:DSPE-PEG(2000) systems was studied with cryo-TEM, DSC, SAXS, and fluorescence spectroscopy of laurdanC, as well as with molecular dynamics simulations. It was found that increasing the degree of PEGylation alters the fluidity of the bilayer and the shape of the lipid particulates. As demonstrated here, the shape-shift can be monitored with laurdanC. With the combination of laurdanC emission spectra and cryo-TEM, it was shown that the most visible transformation was from liposomes to bicelles when the fraction of the DSPE-PEG(2000) lipids reached the critical bicellation fraction (CBF), 9.6 mol %. We identified the greatest contribution to be the spontaneous curvature of the lipid components that is linked with the steric repulsion between PEG units in the lipid mixture. This means that lipids must relocate themselves to a region of curvature that matches their own.
The lipid curvature sets a limit to the sizes of units in the system. Liposome units have a limit size of ca. 30 nm which is characteristic to DPPC. Bicelle is characterized by the DSPE-PEG lipid that has an equivalent curvature of a micelle core 3.8 nm.94 Bicelles are formed when the lipid mixture is capable of making bicelle edges with curvature K12 = 1/dHH. The bicelle radius, in this case, was also effectively limited to the radius of ca. 14 nm.
Although the transformation to bicelles from liposomes is the most prominent phenomenon of the increased PEG fraction, we also found that other kinds of lipid particulates exist at the PEGylation fraction below bicellation. This is evident from the phase transition peak shifts of both DSC and laurdanC. Each melting temperature represents an individual PEG-layered lipid particulate. The driving force of the increasing melting temperature comes from the PEGylated lipids that were shown to affect membrane thickness and decrease the effective lateral pressure prior to bicellation.
The shift in the phase transition temperatures of spherical vesicles is connected to an increased bilayer thickness and improved shielding due to the PEG layer that evidently changes the surface pressure of the bilayer. This effect could be used to improve drug delivery systems. Since drug release occurs most effectively at the main phase transition temperature of the lipid, a mixture of liposomes with a range of phase transition temperatures can enable partial drug release by an accurate temperature control. This can be achieved with, e.g., photothermal agents.14 Liposomes transform into bicelles when the spontaneous curvature of the mixture starts to favor the latter shape. The mechanism of action is a simple one. A lipid with a sizable polymer moiety on its head group has a positive curvature that controls the transformation. Hence, with a suitable polymer, it could be possible to trigger the transformation and activate the drug release.
Acknowledgments
TEKES is acknowledged for funding via the “Light Activated Drug Delivery Systems” project (LADDS project). Academy of Finland is acknowledged for funding via grant 309324 (M.S.). Dr Dat Pham is acknowledged for the help in the DSC measurements. L.V. wishes to thank the Swedish-Finnish Cultural Foundation for a seven-day visiting grant to Lund University. This work made use of the Aalto University Nanomicroscopy Center (Aalto-NMC) premises. Computational resources from the CSC IT Center for Science, Finland, and RAMI RawMatTERS Finland Infrastructure are gratefully acknowledged.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.langmuir.8b03829.
Small/wide-angle X-ray scattering; molecular dynamics simulations: additional details; Matlab script for the Cryo-TEM analyses (PDF)
The authors declare no competing financial interest.
Footnotes
From here on, the number after “PEG” indicates its mol % in the formulation.
Bending modulus is replaced with κ → 2000 nm to better illustrate the parabolic minimum.
Supplementary Material
References
- Ali M. H.; Moghaddam B.; Kirby D. J.; Mohammed A. R.; Perrie Y. The Role of Lipid Geometry in Designing Liposomes for the Solubilisation of Poorly Water Soluble Drugs. Int. J. Pharm. 2013, 453, 225–232. 10.1016/j.ijpharm.2012.06.056. [DOI] [PubMed] [Google Scholar]
- Lin L.; Wang X.; Guo Y.; Ren K.; Li X.; Jing L.; Yue X.; Zhang Q.; Dai Z. Hybrid Bicelles as a pH-Sensitive Nanocarrier for Hydrophobic Drug Delivery. RSC Adv. 2016, 6, 79811–79821. 10.1039/C6RA18112K. [DOI] [Google Scholar]
- Han S.; Li Z.; Zhu J.; Han K.; Zeng Z.; Hong W.; Li W.; Jia H.; Liu Y.; Zhuo R. Dual-pH Sensitive Charge-Reversal Polypeptide Micelles for Tumor-Triggered Targeting Uptake and Nuclear Drug Delivery. Small 2015, 11, 2543–2554. 10.1002/smll.201402865. [DOI] [PubMed] [Google Scholar]
- Ashok B.; Arleth L.; Hjelm R. P.; Rubinstein I.; Önyüksel H. In Vitro Characterization of PEGylated Phospholipid Micelles for Improved Drug Solubilization: Effects of PEG Chain Length and PC Incorporation. J. Pharm. Sci. 2004, 93, 2476–2487. 10.1002/jps.20150. [DOI] [PubMed] [Google Scholar]
- Ferreira D. S.; Lopes S.; Franco M. S.; Oliveira M. pH-Sensitive Liposomes for Drug Delivery in Cancer Treatment. Ther. Delivery 2013, 4, 1099–1123. 10.4155/tde.13.80. [DOI] [PubMed] [Google Scholar]
- Paasonen L.; Romberg B.; Storm G.; Yliperttula M.; Urtti A.; Hennink W. E. Temperature-Sensitive Poly(N-(2-Hydroxypropyl)Methacrylamide Mono/Dilactate)-Coated Liposomes for Triggered Contents Release. Bioconjugate Chem. 2007, 18, 2131–2136. 10.1021/bc700245p. [DOI] [PubMed] [Google Scholar]
- Paasonen L.; Laaksonen T.; Johans C.; Yliperttula M.; Kontturi K.; Urtti A. Gold Nanoparticles Enable Selective Light-Induced Contents Release from Liposomes. J. Controlled Release 2007, 122, 86–93. 10.1016/j.jconrel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Paasonen L.; Sipilä T.; Subrizi A.; Laurinmäki P.; Butcher S. J.; Rappolt M.; Yaghmur A.; Urtti A.; Yliperttula M. Gold-Embedded Photosensitive Liposomes for Drug Delivery: Triggering Mechanism and Intracellular Release. J. Controlled Release 2010, 147, 136–143. 10.1016/j.jconrel.2010.07.095. [DOI] [PubMed] [Google Scholar]
- Lajunen T.; Viitala L.; Kontturi L.; Laaksonen T.; Liang H.; Vuorimaa-Laukkanen E.; Viitala T.; Le Guével X.; Yliperttula M.; Murtomäki L.; Urtti A. Light Induced Cytosolic Drug Delivery from Liposomes with Gold Nanoparticles. J. Controlled Release 2015, 203, 85–98. 10.1016/j.jconrel.2015.02.028. [DOI] [PubMed] [Google Scholar]
- Bouchaala R.; Anton N.; Anton H.; Vandamme T.; Vermot J.; Smail D.; Mély Y.; Klymchenko A. S. Light-Triggered Release from Dye-Loaded Fluorescent Lipid Nanocarriers in Vitro and in Vivo. Colloids Surf., B 2017, 156, 414–421. 10.1016/j.colsurfb.2017.05.035. [DOI] [PubMed] [Google Scholar]
- Troutman T. S.; Leung S. J.; Romanowski M. Light-Induced Content Release from Plasmon-Resonant Liposomes. Adv. Mater. 2009, 21, 2334–2338. 10.1002/adma.200900018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajunen T.; Kontturi L.; Viitala L.; Manna M.; Cramariuc O.; Róg T.; Bunker A.; Róg T.; Viitala T.; Murtomäki L.; Urtti A. Indocyanine Green-Loaded Liposomes for Light-Triggered Drug Release. Mol. Pharm. 2016, 13, 2095–2107. 10.1021/acs.molpharmaceut.6b00207. [DOI] [PubMed] [Google Scholar]
- Lajunen T.; Nurmi R.; Kontturi L.; Viitala L.; Yliperttula M.; Murtomäki L.; Urtti A. Light Activated Liposomes: Functionality and Prospects in Ocular Drug Delivery. J. Controlled Release 2016, 244, 157–166. 10.1016/j.jconrel.2016.08.024. [DOI] [PubMed] [Google Scholar]
- Viitala L.; Pajari S.; Lajunen T.; Kontturi L.; Laaksonen T.; Kuosmanen P.; Viitala T.; Urtti A.; Murtomäki L. Photothermally Triggered Lipid Bilayer Phase Transition and Drug Release from Gold Nanorod and Indocyanine Green Encapsulated Liposomes. Langmuir 2016, 32, 4554–4563. 10.1021/acs.langmuir.6b00716. [DOI] [PubMed] [Google Scholar]
- Mura S.; Nicolas J.; Couvreur P. Stimuli-Responsive Nanocarriers for Drug Delivery. Nat. Mater. 2013, 12, 991–1003. 10.1038/nmat3776. [DOI] [PubMed] [Google Scholar]
- Jansen M.; Blume A. A Comparative Study of Diffusive and Osmotic Water Permeation Across Bilayers Composed of Phospholipids with Different Head Groups and Fatty Acyl Chains. Biophys. J. 1995, 68, 997–1008. 10.1016/S0006-3495(95)80275-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima H.; Sakata K.; Funato K.; Kiwada H. Enhanced Hepatic Uptake of Liposomes through Complement Activation Depending on the Size of Liposomes. Pharm. Res. 1994, 11, 402–406. 10.1023/A:1018965121222. [DOI] [PubMed] [Google Scholar]
- Kari O. K.; Rojalin T.; Salmaso S.; Barattin M.; Jarva H.; Meri S.; Yliperttula M.; Viitala T.; Urtti A. Multi-Parametric Surface Plasmon Resonance Platform for Studying Liposome-Serum Interactions and Protein Corona Formation. Drug Delivery Transl. Res. 2017, 7, 228–240. 10.1007/s13346-016-0320-0. [DOI] [PubMed] [Google Scholar]
- Klibanov A. L.; Maruyama K.; Torchilin V. P.; Huang L. Amphipathic Polyethyleneglycols Effectively Prolong the Circulation Time of Liposomes. FEBS Lett. 1990, 268, 235–237. 10.1016/0014-5793(90)81016-H. [DOI] [PubMed] [Google Scholar]
- Bunker A.; Magarkar A.; Viitala T. Rational Design of Liposomal Drug Delivery Systems, a Review: Combined Experimental and Computational Studies of Lipid Membranes, Liposomes and their PEGylation. Biochim. Biophys. Acta, Biomembr. 2016, 1858, 2334–2352. 10.1016/j.bbamem.2016.02.025. [DOI] [PubMed] [Google Scholar]
- Peracchia M. T.; Vauthier C.; Passirani C.; Couvreur P.; Labarre D. Complement Consumption by Poly(Ethylene Glycol) in Different Conformations Chemically Coupled to Poly(Isobutyl 2-Cyanoacrylate) Nanoparticles. Life Sci. 1997, 61, 749–761. 10.1016/S0024-3205(97)00539-0. [DOI] [PubMed] [Google Scholar]
- Barenholz Y. Doxil—the First FDA-Approved Nano-Drug: Lessons Learned. J. Controlled Release 2012, 160, 117–134. 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- Tirosh O.; Barenholz Y.; Katzhendler J.; Priev A. Hydration of Poly(ethylene glycol)-Grafted Liposomes. Biophys. J. 1998, 74, 1371–1379. 10.1016/S0006-3495(98)77849-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland J. W.; Hui C.; Cullis P. R.; Madden T. D. Poly(ethylene glycol)-Lipid Conjugates Regulate the Calcium-Induced Fusion of Liposomes Composed of Phosphatidylethanolamine and Phosphatidylserine. Biochemistry 1996, 35, 2618–2624. 10.1021/bi952000v. [DOI] [PubMed] [Google Scholar]
- Magarkar A.; Karakas E.; Stepniewski M.; Róg T.; Bunker A. Molecular Dynamics Simulation of PEGylated Bilayer Interacting with Salt Ions: A Model of the Liposome Surface in the Bloodstream. J. Phys. Chem. B 2012, 116, 4212–4219. 10.1021/jp300184z. [DOI] [PubMed] [Google Scholar]
- Dinç C. Ö.; Kibarer G.; Güner A. Solubility Profiles of Poly(ethylene glycol)/Solvent Systems. II. Comparison of Thermodynamic Parameters from Viscosity Measurements. J. Appl. Polym. Sci. 2010, 117, 1100–1119. 10.1002/app.31829. [DOI] [Google Scholar]
- Furumoto K.; Yokoe J.; Ogawara K.; Amano S.; Takaguchi M.; Higaki K.; Kai T.; Kimura T. Effect of Coupling of Albumin Onto Surface of PEG Liposome on its in Vivo Disposition. Int. J. Pharm. 2007, 329, 110–116. 10.1016/j.ijpharm.2006.08.026. [DOI] [PubMed] [Google Scholar]
- Lee H.; Larson R. G. Adsorption of Plasma Proteins Onto PEGylated Lipid Bilayers: The Effect of PEG Size and Grafting Density. Biomacromolecules 2016, 17, 1757–1765. 10.1021/acs.biomac.6b00146. [DOI] [PubMed] [Google Scholar]
- Lajunen T.; Nurmi R.; Wilbie D.; Ruoslahti T.; Johansson N. G.; Korhonen O.; Rog T.; Bunker A.; Ruponen M.; Urtti A. The Effect of Light Sensitizer Localization on the Stability of Indocyanine Green Liposomes. J. Controlled Release 2018, 284, 213–223. 10.1016/j.jconrel.2018.06.029. [DOI] [PubMed] [Google Scholar]
- Lee H.; de Vries A. H.; Marrink S.; Pastor R. W. A Coarse-Grained Model for Polyethylene Oxide and Poly(ethylene glycol): Conformation and Hydrodynamics. J. Phys. Chem. B 2009, 113, 13186–13194. 10.1021/jp9058966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G.; Fuchs P.; Barnoud J.; Monticelli L. A Coarse-Grained MARTINI Model of Poly(ethylene glycol) and of Polyoxyethylene Alkyl Ether Surfactants. J. Phys. Chem. B 2012, 116, 14353–14362. 10.1021/jp3095165. [DOI] [PubMed] [Google Scholar]
- Grunewald F.; Rossi G.; de Vries A. H.; Marrink S. J.; Monticelli L. Transferable MARTINI Model of Poly(Ethylene Oxide). J. Phys. Chem. B 2018, 122, 7436–7449. 10.1021/acs.jpcb.8b04760. [DOI] [PubMed] [Google Scholar]
- Lee H.; Pastor R. W. Coarse-Grained Model for PEGylated Lipids: Effect of PEGylation on the Size and Shape of Self-Assembled Structures. J. Phys. Chem. B 2011, 115, 7830–7837. 10.1021/jp2020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.; Faller R. Pressure and Surface Tension Control Self-Assembled Structures in Mixtures of Pegylated and Non-Pegylated Lipids. Langmuir 2012, 28, 2275–2280. 10.1021/la203850z. [DOI] [PubMed] [Google Scholar]
- Shinoda W.; Discher D. E.; Klein M. L.; Loverde S. M. Probing the Structure of PEGylated-Lipid Assemblies by Coarse-Grained Molecular Dynamics. Soft Matter 2013, 9, 11549–11556. 10.1039/c3sm52290c. [DOI] [Google Scholar]
- Määttä J.; Vierros S.; Van Tassel P. R.; Sammalkorpi M. Size-Selective, Noncovalent Dispersion of Carbon Nanotubes by PEGylated Lipids: A Coarse-Grained Molecular Dynamics Study. J. Chem. Eng. Data 2014, 59, 3080–3089. 10.1021/je500157b. [DOI] [Google Scholar]
- Määttä J.; Vierros S.; Sammalkorpi M. Controlling Carbon-Nanotube-Phospholipid Solubility by Curvature-Dependent Self-Assembly. J. Phys. Chem. B 2015, 119, 4020–4032. 10.1021/jp5128173. [DOI] [PubMed] [Google Scholar]
- Vuorte M.; Määttä J.; Sammalkorpi M. Simulations Study of Single Component and Mixed N-Alkyl-PEG Micelles. J. Phys. Chem. B 2018, 122, 4851–4860. 10.1021/acs.jpcb.8b00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baekmark T. R.; Pedersen S.; Jørgensen K.; Mouritsen O. G. The Effects of Ethylene Oxide Containing Lipopolymers and Tri-Block Copolymers on Lipid Bilayers of Dipalmitoylphosphatidylcholine. Biophys. J. 1997, 73, 1479–1491. 10.1016/S0006-3495(97)78180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K.; Johnsson M.; Karlsson G.; Silvander M. Effect of Polyethyleneglycol-Phospholipids on Aggregate Structure in Preparations of Small Unilamellar Liposomes. Biophys. J. 1997, 73, 258–266. 10.1016/S0006-3495(97)78066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson M.; Edwards K. Liposomes, Disks, and Spherical Micelles: Aggregate Structure in Mixtures of Gel Phase Phosphatidylcholines and Poly(ethylene glycol)-Phospholipids. Biophys. J. 2003, 85, 3839–3847. 10.1016/S0006-3495(03)74798-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ickenstein L. M.; Sandström M. C.; Mayer L. D.; Edwards K. Effects of Phospholipid Hydrolysis on the Aggregate Structure in DPPC/DSPE-PEG 2000 Liposome Preparations After Gel to Liquid Crystalline Phase Transition. Biochim. Biophys. Acta, Biomembr. 2006, 1758, 171–180. 10.1016/j.bbamem.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Sandström M. C.; Johansson E.; Edwards K. Influence of Preparation Path on the Formation of Discs and Threadlike Micelles in DSPE-PEG2000/Lipid Systems. Biophys. Chem. 2008, 132, 97–103. 10.1016/j.bpc.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Arleth L.; Ashok B.; Onyuksel H.; Thiyagarajan P.; Jacob J.; Hjelm R. P. Detailed Structure of Hairy Mixed Micelles Formed by Phosphatidylcholine and PEGylated Phospholipids in Aqueous Media. Langmuir 2005, 21, 3279–3290. 10.1021/la047588y. [DOI] [PubMed] [Google Scholar]
- Helfrich W. Elastic Properties of Lipid Bilayers: Theory and Possible Experiments. Z. Naturforsch. C 1973, 28, 693–703. 10.1515/znc-1973-11-1209. [DOI] [PubMed] [Google Scholar]
- Deuling H.; Helfrich W. The Curvature Elasticity of Fluid Membranes: A Catalogue of Vesicle Shapes. J. Phys. 1976, 37, 1335–1345. 10.1051/jphys:0197600370110133500. [DOI] [Google Scholar]
- Marsh D. Intrinsic Curvature in Normal and Inverted Lipid Structures and in Membranes. Biophys. J. 1996, 70, 2248–2255. 10.1016/S0006-3495(96)79790-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz W.; Testard F.; Zemb T. Correspondence between Curvature, Packing Parameter, and Hydrophilicâ̂’Lipophilic Deviation Scales Around the Phase-Inversion Temperature. Langmuir 2009, 25, 112–115. 10.1021/la8028879. [DOI] [PubMed] [Google Scholar]
- Tanford C. Micelle Shape and Size. J. Phys. Chem. 1972, 76, 3020–3024. 10.1021/j100665a018. [DOI] [Google Scholar]
- Israelachvili J. N.; Mitchell D. J.; Ninham B. W. Theory of Self-Assembly of Hydrocarbon Amphiphiles into Micelles and Bilayers. J. Chem. Soc., Faraday Trans. 2 1976, 72, 1525–1568. 10.1039/f29767201525. [DOI] [Google Scholar]
- Langevin D. Micelles and Microemulsions. Annu. Rev. Phys. Chem 1992, 43, 341–369. 10.1146/annurev.pc.43.100192.002013. [DOI] [Google Scholar]
- Bergström L. M. Bending Energetics of Tablet-Shaped Micelles: A Novel Approach to Rationalize Micellar Systems. ChemPhysChem 2007, 8, 462–472. 10.1002/cphc.200600692. [DOI] [PubMed] [Google Scholar]
- Cheniour M.; Gueyrard D.; Goekjian P. G.; Granjon T.; Marcillat O. A Convenient and Versatile Synthesis of Laurdan-Like Fluorescent Membrane Probes: Characterization of their Fluorescence Properties. RSC Adv. 2016, 6, 5547–5557. 10.1039/C5RA20369D. [DOI] [Google Scholar]
- Kuntsche J.; Horst J. C.; Bunjes H. Cryogenic Transmission Electron Microscopy (Cryo-TEM) for Studying the Morphology of Colloidal Drug Delivery Systems. Int. J. Pharm. 2011, 417, 120–137. 10.1016/j.ijpharm.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Iancu C. V.; Tivol W. F.; Schooler J. B.; Dias D. P.; Henderson G. P.; Murphy G. E.; Wright E. R.; Li Z.; Yu Z.; Briegel A.; et al. Electron Cryotomography Sample Preparation using the Vitrobot. Nat. Protoc. 2006, 1, 2813–2819. 10.1038/nprot.2006.432. [DOI] [PubMed] [Google Scholar]
- Ruzi M.Voigt Line Shape Fit. https://se.mathworks.com/matlabcentral/fileexchange/57603-voigt-line-shape-fit (accessed May 21, 2017).
- Abrarov S. M.; Quine B. M.. The Voigt/Complex Error Function. https://se.mathworks.com/matlabcentral/fileexchange/47801-the-voigt-complex-error-function--second-version (accessed May 21, 2017).
- Pedersen J. S.; Gerstenberg M. C. Scattering Form Factor of Block Copolymer Micelles. Macromolecules 1996, 29, 1363–1365. 10.1021/ma9512115. [DOI] [Google Scholar]
- Pedersen J. S. Analysis of Small-Angle Scattering Data from Colloids and Polymer Solutions: Modeling and Least-Squares Fitting. Adv. Colloid Interface Sci. 1997, 70, 171–210. 10.1016/S0001-8686(97)00312-6. [DOI] [Google Scholar]
- Derici L.; Ledger S.; Mai S.; Booth C.; Hamley I. W.; Pedersen J. S. Micelles and Gels of Oxyethylene–oxybutylene Diblock Copolymers in Aqueous Solution: The Effect of Oxyethylene-Block Length. Phys. Chem. Chem. Phys. 1999, 1, 2773–2785. 10.1039/a901108k. [DOI] [Google Scholar]
- Pedersen J. S. Form Factors of Block Copolymer Micelles with Spherical, Ellipsoidal and Cylindrical Cores. J. Appl. Crystallogr. 2000, 33, 637–640. 10.1107/S0021889899012248. [DOI] [Google Scholar]
- Kučerka N.; Kiselev M. A.; Balgavý P. Determination of Bilayer Thickness and Lipid Surface Area in Unilamellar Dimyristoylphosphatidylcholine Vesicles from Small-Angle Neutron Scattering Curves: A Comparison of Evaluation Methods. Eur. Biophys. J. 2004, 33, 328–334. 10.1007/s00249-003-0349-0. [DOI] [PubMed] [Google Scholar]
- Singh D.Small Angle Scatting Studies of Self Assembly in Lipid Mixtures; The Johns Hopkins University, 2009. [Google Scholar]
- Arleth L.; Vermehren C. An Analytical Model for the Small-Angle Scattering of Poly(ethylene glycol)-Modified Liposomes. J. Appl. Crystallogr. 2010, 43, 1084–1091. 10.1107/S0021889810026257. [DOI] [Google Scholar]
- Skar-Gislinge N.; Arleth L. Small-Angle Scattering from Phospholipid Nanodiscs: Derivation and Refinement of a Molecular Constrained Analytical Model Form Factor. Phys. Chem. Chem. Phys. 2011, 13, 3161–3170. 10.1039/C0CP01074J. [DOI] [PubMed] [Google Scholar]
- Rayleigh L. The Incidence of Light upon a Transparent Sphere of Dimensions Comparable with the Wave-Length. Proc. R. Soc. A 1910, 84, 25–46. 10.1098/rspa.1910.0054. [DOI] [Google Scholar]
- Fournet G. Scattering Functions for Geometrical Forms. Bull. Soc. Fr. Mineral. Cristallogr. 1951, 74, 39–113. [Google Scholar]
- Hammouda B. Structure Factor for Starburst Dendrimers. J. Polym. Sci., Part B: Polym. Phys. 1992, 30, 1387–1390. 10.1002/polb.1992.090301209. [DOI] [Google Scholar]
- Hammouda B.SANS from Homogeneous Polymer Mixtures: A Unified Overview. In Polymer Characteristics; Springer, 1993; pp 87–133. [Google Scholar]
- Debye P. Molecular-Weight Determination by Light Scattering. J. Phys. Colloid Chem. 1947, 51, 18–32. 10.1021/j150451a002. [DOI] [PubMed] [Google Scholar]
- Marrink S. J.; de Vries A. H.; Mark A. E. Coarse Grained Model for Semiquantitative Lipid Simulations. J. Phys. Chem. B 2004, 108, 750–760. 10.1021/jp036508g. [DOI] [Google Scholar]
- Marrink S. J.; Risselada H. J.; Yefimov S.; Tieleman D. P.; De Vries A. H. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B 2007, 111, 7812–7824. 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- Arnarez C.; Uusitalo J. J.; Masman M. F.; Ingólfsson H. I.; de Jong D. H.; Melo M. N.; Periole X.; de Vries A. H.; Marrink S. J. Dry Martini, a Coarse-Grained Force Field for Lipid Membrane Simulations with Implicit Solvent. J. Chem. Theory Comput. 2015, 11, 260–275. 10.1021/ct500477k. [DOI] [PubMed] [Google Scholar]
- Pronk S.; Páll S.; Schulz R.; Larsson P.; Bjelkmar P.; Apostolov R.; Shirts M. R.; Smith J. C.; Kasson P. M.; van der Spoel D.; Hess B.; Lindahl E. GROMACS 4.5: A High-Throughput and Highly Parallel Open Source Molecular Simulation Toolkit. Bioinformatics 2013, 29, 845–854. 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G.; Donadio D.; Parrinello M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- de Jong D. H.; Baoukina S.; Ingólfsson H. I.; Marrink S. J. Martini Straight: Boosting Performance using a Shorter Cutoff and GPUs. Comput. Phys. Commun. 2016, 199, 1–7. 10.1016/j.cpc.2015.09.014. [DOI] [Google Scholar]
- Aslan S.; Määttä J.; Haznedaroglu B. Z.; Goodman J. P.; Pfefferle L. D.; Elimelech M.; Pauthe E.; Sammalkorpi M.; Van Tassel P. R. Carbon Nanotube Bundling: Influence on Layer-by-Layer Assembly and Antimicrobial Activity. Soft Matter 2013, 9, 2136–2144. 10.1039/c2sm27444b. [DOI] [Google Scholar]
- Almgren M.; Edwards K.; Karlsson G. Cryo Transmission Electron Microscopy of Liposomes and Related Structures. Colloids Surf., A 2000, 174, 3–21. 10.1016/S0927-7757(00)00516-1. [DOI] [Google Scholar]
- Pili B.; Reddy L. H.; Bourgaux C.; Lepêtre-Mouelhi S.; Desmaële D.; Couvreur P. Liposomal Squalenoyl-Gemcitabine: Formulation, Characterization and Anticancer Activity Evaluation. Nanoscale 2010, 2, 1521–1526. 10.1039/c0nr00132e. [DOI] [PubMed] [Google Scholar]
- Bhat R.; Timasheff S. N. Steric Exclusion is the Principal Source of the Preferential Hydration of Proteins in the Presence of Poly(ethylene glycol). Protein Sci. 1992, 1, 1133–1143. 10.1002/pro.5560010907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbuzenko O.; Barenholz Y.; Priev A. Effect of Grafted PEG on Liposome Size and on Compressibility and Packing of Lipid Bilayer. Chem. Phys. Lipids 2005, 135, 117–129. 10.1016/j.chemphyslip.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Sammalkorpi M.; Karttunen M.; Haataja M. Structural Properties of Ionic Detergent Aggregates: A Large-Scale Molecular Dynamics Study of Sodium Dodecyl Sulfate. J. Phys. Chem. B 2007, 111, 11722–11733. 10.1021/jp072587a. [DOI] [PubMed] [Google Scholar]
- Saveyn P.; Van der Meeren P.; Zackrisson M.; Narayanan T.; Olsson U. Subgel Transition in Diluted Vesicular DODAB Dispersions. Soft Matter 2009, 5, 1735–1742. 10.1039/b821387a. [DOI] [Google Scholar]
- Kastantin M.; Ananthanarayanan B.; Karmali P.; Ruoslahti E.; Tirrell M. Effect of the Lipid Chain Melting Transition on the Stability of DSPE-PEG(2000) Micelles. Langmuir 2009, 25, 7279–7286. 10.1021/la900310k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh D.; Bartucci R.; Sportelli L. Lipid Membranes with Grafted Polymers: Physicochemical Aspects. Biochim. Biophys. Acta, Biomembr. 2003, 1615, 33–59. 10.1016/S0005-2736(03)00197-4. [DOI] [PubMed] [Google Scholar]
- Pantusa M.; Bartucci R.; Marsh D.; Sportelli L. Shifts in Chain-Melting Transition Temperature of Liposomal Membranes by Polymer-Grafted Lipids. Biochim. Biophys. Acta, Biomembr. 2003, 1614, 165–170. 10.1016/S0005-2736(03)00171-8. [DOI] [PubMed] [Google Scholar]
- Biltonen R. L.; Lichtenberg D. The use of Differential Scanning Calorimetry as a Tool to Characterize Liposome Preparations. Chem. Phys. Lipids 1993, 64, 129–142. 10.1016/0009-3084(93)90062-8. [DOI] [Google Scholar]
- Parasassi T.; Krasnowska E.; Bagatolli L.; Gratton E. Laurdan and Prodan as Polarity-Sensitive Fluorescent Membrane Probes. J. Fluoresc. 1998, 8, 365–373. 10.1023/A:1020528716621. [DOI] [Google Scholar]
- Kawski A.; Kukliński B.; Bojarski P.; Diehla H. Ground and Excited State Dipole Moments of LAURDAN Determined from Solvatochromic and Thermochromic Shifts of Absorption and Fluorescence Spectra. Z. Naturforsch., A 2000, 55, 817–822. 10.1515/zna-2000-9-1011. [DOI] [Google Scholar]
- Vogel V.; Möbius D. Hydrated Polar Groups in Lipid Monolayers: Effective Local Dipole Moments and Dielectric Properties. Thin Solid Films 1988, 159, 73–81. 10.1016/0040-6090(88)90618-9. [DOI] [Google Scholar]
- Vogel V.; Möbius D. Local Surface Potentials and Electric Dipole Moments of Lipid Monolayers: Contributions of the Water/Lipid and the Lipid/Air Interfaces. J. Colloid Interface Sci. 1988, 126, 408–420. 10.1016/0021-9797(88)90140-3. [DOI] [Google Scholar]
- Deserno M. Fluid Lipid Membranes: From Differential Geometry to Curvature Stresses. Chem. Phys. Lipids 2015, 185, 11–45. 10.1016/j.chemphyslip.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Kollmitzer B.; Heftberger P.; Rappolt M.; Pabst G. Monolayer Spontaneous Curvature of Raft-Forming Membrane Lipids. Soft Matter 2013, 9, 10877–10884. 10.1039/c3sm51829a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuković L.; Khatib F. A.; Drake S. P.; Madriaga A.; Brandenburg K. S.; Král P.; Onyuksel H. Structure and Dynamics of Highly PEG-Ylated Sterically Stabilized Micelles in Aqueous Media. J. Am. Chem. Soc. 2011, 133, 13481–13488. 10.1021/ja204043b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.