Abstract
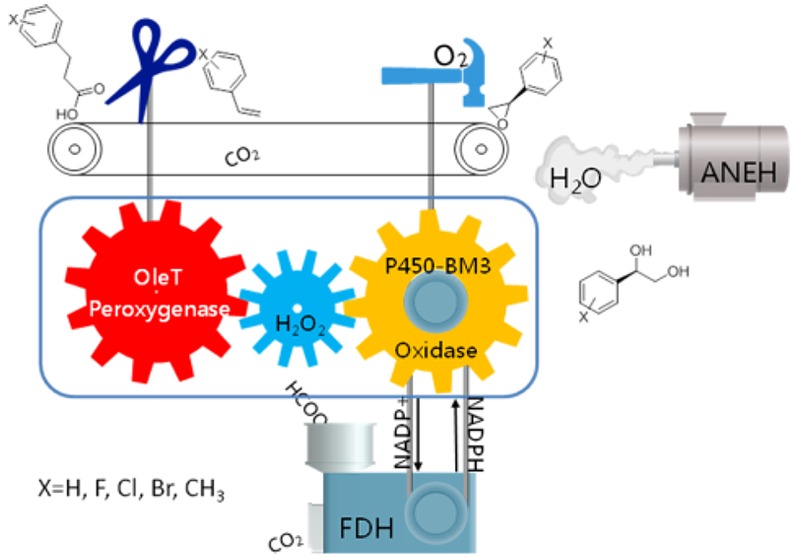
A unique P450 monooxygenase–peroxygenase mutual benefit system was designed as the core element in the construction of a biocatalytic cascade reaction sequence leading from 3-phenyl propionic acid to (R)-phenyl glycol. In this system, P450 monooxygenase (P450-BM3) and P450 peroxygenase (OleTJE) not only function as catalysts for the crucial initial reactions, they also ensure an internal in situ H2O2 recycle mechanism that avoids its accumulation and thus prevents possible toxic effects. By directed evolution of P450-BM3 as the catalyst in the enantioselective epoxidation of the styrene-intermediate, formed from 3-phenyl propionic acid, and the epoxide hydrolase ANEH for final hydrolytic ring opening, (R)-phenyl glycol and 9 derivatives thereof were synthesized from the respective carboxylic acids in one-pot processes with high enantioselectivity.
Oxygen is a ubiquitous oxidant which exists in a variety of life processes, such as respiration, metabolism, and nutrition.1 Inevitably, when it is used by an oxidase to perform an oxidative reaction, hydrogen peroxide (H2O2) will be generated in the redox process.2−5 H2O2 is a potentially toxic agent for the cells, because it injures proteins and damages nucleic acids in many ways.6 For example, it can inhibit cytoplasmic enzymes which use Fe2+ ion or [4Fe–4S] as cofactors7−11 and induce the Fenton reaction with generation of DNA-damaging hydroxyl radicals.12 To relieve oxidative stress caused by H2O2, organisms have developed versatile approaches to degrade this toxicant.13,14 In these processes, H2O2 is decomposed into water and oxygen by catalase or it is utilized by peroxidases or peroxygenases as an oxidant to generate new oxidative products.15,16 Thus, based on the activities of oxidases and peroxidases/peroxygenases, cascade reactions with high economic efficiency can be created. This is achieved upon combining oxygen-dependent oxidases with peroxidases/peroxygenases. In these reactions, the only oxidative source is oxygen (O2), and H2O2 is produced slowly as a direct product or byproduct when oxygen is reduced by oxidases. Peroxidases/peroxygenases accept H2O2 to synthesize new compounds. The two types of enzymes can benefit from each other in what can be called “oxidase–peroxidase/peroxygenase mutual benefit systems” (OPMBS) (Figure 1).
Figure 1.
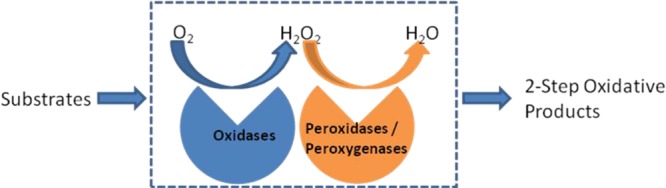
Concept of oxidase–peroxidase/peroxygenase mutual benefit systems.
In previous work, oxidases–peroxidases/peroxygenases fusion proteins or oxidases–peroxidases/peroxygenases in mixtures were reported, as in the synthesis of divanillin, lignin oligmers and α-ketoacids.17−20 However, to the best of our knowledge, such systems have not been exploited to perform asymmetric synthesis, which would be of notable interest in synthetic organic chemistry. Moreover, most studies utilize external H2O2 to switch on the reaction, specifically when peroxidases/peroxygenases function in the first catalytic step, which may have significant disadvantages.
Here, we demonstrate a biocatalytic one-pot cascade reaction sequence in which the core steps are catalyzed by a unique oxidase–peroxygenase mutual benefit system, thereby enabling the enantioselective production of (R)-phenyl glycols from 3-phenyl propionic acid or derivatives thereof. In this approach, the initial 2-step oxidative reaction sequence is triggered by the peroxygenase without any external H2O2. Furthermore, all of the enzymes involved in this reaction can be used in their cell-free lysate form, which catalyzes the target reactions with high conversion and excellent enantioselectivity.
The catalytic system introduced here is composed of two P450 enzymes, P450-BM3 (a P450 monoxygenase)21 and OleTJE (a P450 peroxygenase)22 for the first and second steps, respectively (Scheme 1). The final step in the overall cascade reaction sequence is the selective hydrolytic ring-opening of the respective epoxide catalyzed by the epoxide hydrolase from Aspergillus niger (ANEH).23 To regenerate the cofactor NADPH needed for P450-BM3 catalysis, a formate dehydrogenase mutant (FDH) from Candida bodinii was applied.24 Notice that this cascade starts from 3-phenyl propionic acid and is therefore different from the elegant one-pot sequence starting from styrene, catalyzed by styrene monooxygenase in combination with the epoxide hydrolase from Sphingomonas sp. HXN-200, as reported by Li et al.25
Scheme 1. Designed Cascade Sequence Leading from 3-Phenyl Propionic Acid to the Asymmetric Production of (R)-Phenyl Glycol Based on an Oxidase–Peroxygenase Mutual Benefit System.
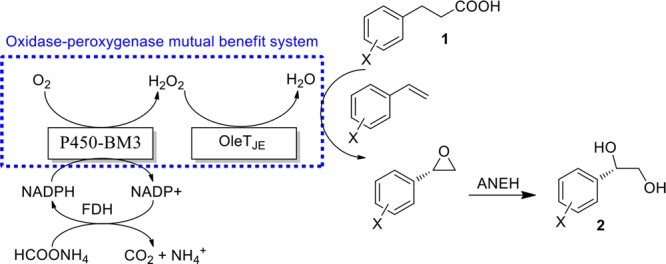
The first step in the overall sequence in which chirality is created is the P450-BM3 catalyzed epoxidation of styrene. In this model reaction, WT P450-BM3 leads to the formation of (R)-styrene oxide with an enantioselectivity of only 20% ee.26 In order to enhance (R)-selectivity, we applied a special form of iterative saturation mutagenesis at the active site (CAST/ISM)27 which we had used previously to improve and to reverse the enantioselectivity of P450-BM3-catalyzed sulfoxidation of thiochroman-4-one.28 Briefly, on the basis of earlier experience, we first screened a small previous mutant library and used the best (S)-selective mutants with high activity (56% and 63% ee) as templates for further focused ISM employing highly reduced amino acid alphabets. After 3 cycles, the best mutant, SO5, improved (R)-selectivity to ee = 96% (Table S2).
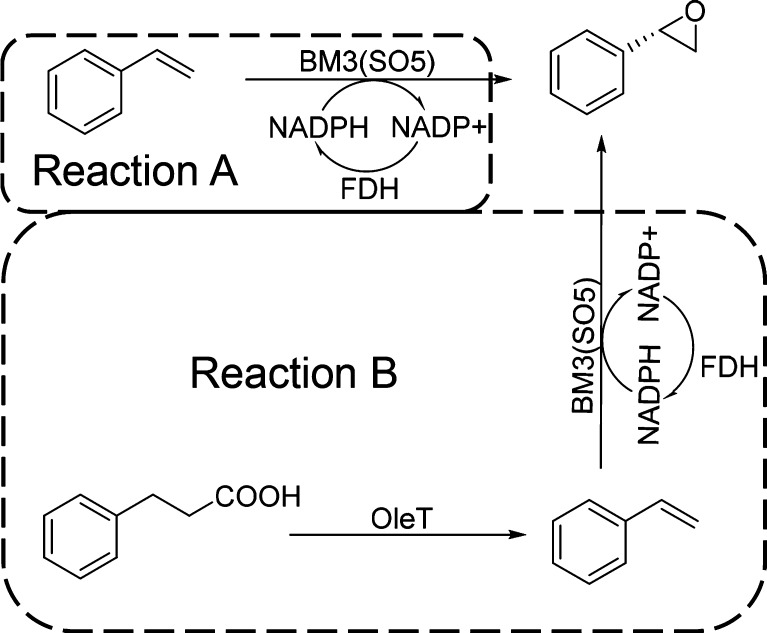
In order to confirm the first two steps of the overall cascade reaction sequence, purified OleTJE was mixed with the purified SO5 variant, and tested in the model reaction using 3-phenyl propionic acid as the starting substrate (Scheme 2). The reaction was monitored by gas chromatography (GC). As shown in Figure 2, styrene oxide was produced successfully with high conversion, demonstrating that OleTJE was indeed activated successfully to switch on the cascade reaction. To confirm that the reaction was switched on by H2O2, a control reaction with catalase was performed in the desaturase transformation (Reaction B). As expected, the reaction was inhibited almost completely, essentially no styrene epoxide being detected in the GC run (Figure 2.).
Scheme 2. In Reaction A, P450-BM3 Mutant SO5 Transforms Styrene into Styrene Oxide, while the OleT-Catalyzed Reaction B Supplies the Necessary Styrene from 3-Phenyl Propionic Acid.
Figure 2.
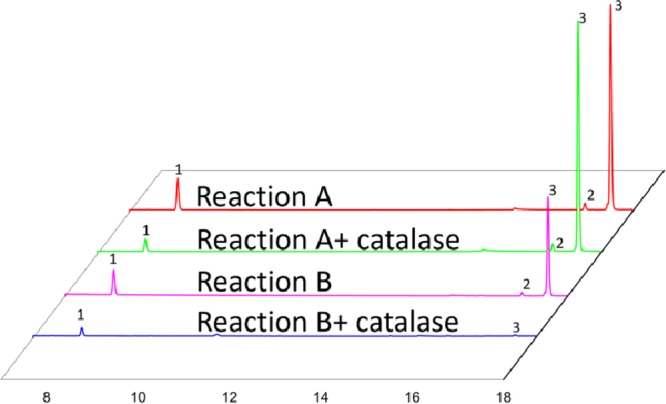
Different responses to the influence of catalase in Reaction A and Reaction B as measured by GC. Peak 1, styrene; peak 2, (S)-styrene oxide; peak 3, (R)-styrene oxide.
H2O2 is usually particularly toxic for enzymes when it accumulates.29,30 To verify whether it can inhibit the activity of variant SO5, an enzyme assay was designed with purified enzyme (Reaction A, Scheme 2/Figure 2). The results indicate that the addition of catalase actually increases the SO5-activity by about 10% (Figure 2). Since H2O2 usually results from the uncoupling pathway, we measured the uncoupling efficiency by comparing the consumption of NADPH with the production of styrene oxide in reaction A, which was found to account for 43% of the result. Therefore, it can be concluded that in the SO5-catalyzed epoxidation, H2O2 causes enzyme stress. Consequently, combining OleTJE with mutant SO5 not only enables the cascade reaction, it also relieves oxidative stress originating from H2O2. This is the reason why we call the overall technique “oxidase–peroxygenase mutual benefit system”.
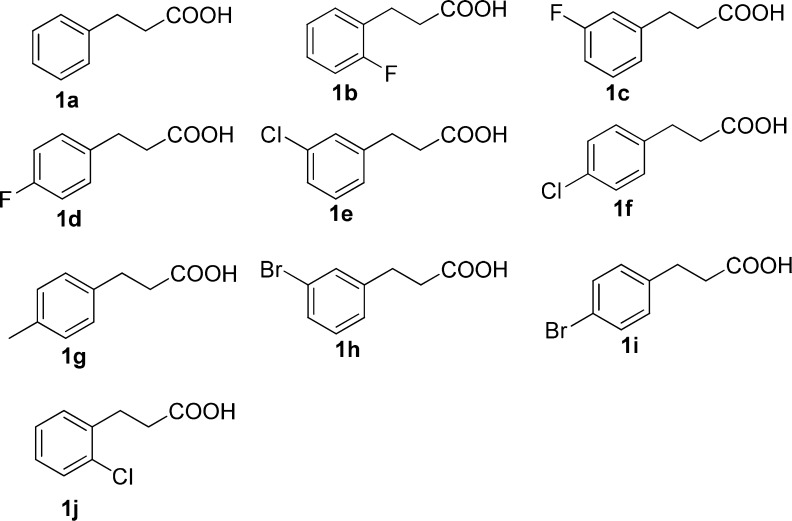
In order to synthesize enantiopure phenyl glycols, we cloned ANEH, following SO5, under the same T7 promoter, and expressed them in a single strain. OleTJE was expressed separately, then both of them were subjected to sonification, and the respective supernatants were lyophilized into cell-free powders. The overall cascade reaction sequence was performed with these powders as biocatalysts, specifically by adjusting the ratio of the SO5-ANEH powder and the OleTJE powder. When the ratio of SO5-ANEH to OleTJE was adjusted to 10 g/L: 20 g/L (enzymes’ concentration ratio is 5.6 μM: 4.5 μM), the conversion of 3-phenyl propionic acid to (R)-phenyl glycol amounted to only 31% with an ee-value of 97%. Since OleTJE is known to favor high salt concentrations,31 300 mM of NaCl were added to the system, which increased conversion to >92%. With the aim of testing the substrate scope of our catalytic system, 9 different 3-phenyl propionic acid derivatives were tested. As summarized in Table 1, generally excellent results were obtained, exceptions being the two ortho-substituted derivatives.
Table 1. Transformation of 3-Phenyl Propionic Acid and Derivatives Thereof Using the Designed Oxidase–Peroxygenase Mutual Benefit System.
| substrate | conv. (%) | %-ee(R) of diol 2 |
|---|---|---|
| 1a | 92 | 97 |
| 1b | 74 | 59 |
| 1c | 93 | 98 |
| 1d | 76 | 92 |
| 1e | 94 | 98 |
| 1f | 95 | 95 |
| 1g | 79 | 95 |
| 1h | 97 | 93 |
| 1i | 88 | 91 |
| 1j | 36 | 25 |
Standard reaction condition: 50 mM PBS buffer (pH = 8.0), ammonium formate (100 mM), NADP+ (0.8 mM), NaCl (300 mM), substrate (5 mM), OleTJE (20 mg/mL), BM3-ANEH (10 mg/mL); total volume: 500 μL.
In summary, the designed “oxidase–peroxygenase mutual benefit system” described herein constitutes a synthetically attractive way to combine a monooxidase with a peroxygenase, thereby avoiding the destructive use of toxic H2O2. The concept extends the utility of biocatalytic cascade reactions considerably.5,31−33 The results also shed light on the mechanism of the involved processes. We expect that our study will accelerate more research based on this approach for accessing useful products in an ecologically and economically viable manner.2,16−19,32−35
Acknowledgments
J.-b.W. thanks Hunan Normal University for the start-up funding. M.T.R. is grateful to the Max-Planck-Society for generous support.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.9b01939.
Experimental materials and procedure details concerning mutagenesis, screening, plasmid construction and reaction details (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Lyons T. W.; Reinhard C. T.; Planavsky N. J. The rise of oxygen in Earth’s early ocean and atmosphere. Nature 2014, 506, 307–315. 10.1038/nature13068. [DOI] [PubMed] [Google Scholar]
- Holtmann D.; Hollmann F. The Oxygen Dilemma: A Severe Challenge for the Application of Monooxygenases?. ChemBioChem 2016, 17, 1391–1398. 10.1002/cbic.201600176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmann D.; Fraaije M. W.; Arends I. W.; Opperman D. J.; Hollmann F. The Taming of Oxygen: Biocatalytic Oxyfunctionalisations. Chem. Commun. 2014, 50, 13180–13200. 10.1039/C3CC49747J. [DOI] [PubMed] [Google Scholar]
- Mifsud M.; Gargiulo S.; Iborra S.; Arends I. W.; Hollmann F.; Corma A. Photobiocatalytic chemistry of oxidoreductases using water as the electron donor. Nat. Commun. 2014, 5, 3145. 10.1038/ncomms4145. [DOI] [PubMed] [Google Scholar]
- Dong J.; Fernandez-Fueyo E.; Hollmann F.; Paul C. E.; Pesic M.; Schmidt S.; Wang Y.; Younes S.; Zhang W. Biocatalytic Oxidation Reactions: A Chemist’s Perspective. Angew. Chem., Int. Ed. 2018, 57, 9238–9261. 10.1002/anie.201800343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T.; Holbrook N. J. Oxidants, Oxidative Stress and the Biology of Ageing. Nature 2000, 408, 239–247. 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Kuo C. F.; Mashino T.; Fridovich I. Alpha, beta-dihydroxyisovalerate dehydratase. A superoxide-sensitive enzyme. J. Biol. Chem. 1987, 262, 4724–4727. [PubMed] [Google Scholar]
- Flint D. H.; Tuminello J. F.; Emptage M. H. The Inactivation of Fe-S Cluster Containing Hydro-lyases by Superoxide. J. Biol. Chem. 1993, 268, 22369–22376. [PubMed] [Google Scholar]
- Jang S.; Imlay J. A. Micromolar Intracellular Hydrogen Peroxide Disrupts Metabolism by Damaging Iron-sulfur Enzymes. J. Biol. Chem. 2007, 282, 929–937. 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobota J. M.; Imlay J. A. Iron Enzyme Ribulose-5-phosphate 3-epimerase in Escherichia coli is Rapidly Damaged by Hydrogen Peroxide but can be Protected by Manganese. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 5402–5407. 10.1073/pnas.1100410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M.; Imlay J. A. Superoxide Poisons Mononuclear Iron Enzymes by Causing Mismetallation. Mol. Microbiol. 2013, 89, 123–134. 10.1111/mmi.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay j. A.; Chin S. M.; Linn S. Toxic DNA Damage by Hydrogen Peroxide Through the Fenton Reaction in Vivo and in Vitro. Science 1988, 240, 640–642. 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- Khademian M.; Imlay J. A. Escherichia coli Cytochrome c Peroxidase is a Respiratory Oxidase that Enables the Use of Hydrogen Peroxide as a Terminal Electron Acceptor. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E6922–E6931. 10.1073/pnas.1701587114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray H. B.; Winkler J. R. Hole Hopping Through Tyrosine/Tryptophan Chains Protects Proteins from Oxidative Damage. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 10920–10925. 10.1073/pnas.1512704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire A. H.; Carey L. M.; de Serrano V.; Dali S.; Ghiladi R. A. Peroxidase versus Peroxygenase Activity: Substrate Substituent Effects as Modulators of Enzyme Function in the Multifunctional Catalytic Globin Dehaloperoxidase. Biochemistry 2018, 57, 4455–4468. 10.1021/acs.biochem.8b00540. [DOI] [PubMed] [Google Scholar]
- Shoji O.; Fujishiro T.; Nishio K.; Kano Y.; Kimoto H.; Chien S.-C.; Onoda H.; Muramatsu A.; Tanaka S.; Hori A.; Sugimoto H.; Shiro Y.; Watanabe Y. A Substrate-binding-state Mimic of H2O2-dependent Cytochrome P450 Produced By One-point Mutagenesis and Peroxygenation of Non-native Substrates. Catal. Sci. Technol. 2016, 6, 5806–5811. 10.1039/C6CY00630B. [DOI] [Google Scholar]
- Habib M. H. M.; Deuss P. J.; Lončar N.; Trajkovic M.; Fraaije M. W. A Biocatalytic One-Pot Approach for the Preparation of Lignin Oligomers Using an Oxidase/Peroxidase Cascade Enzyme System. Adv. Synth. Catal. 2017, 359, 3354–3361. 10.1002/adsc.201700650. [DOI] [Google Scholar]
- Colpa D. I.; Loncar N.; Schmidt M.; Fraaije M. W. Creating Oxidase-Peroxidase Fusion Enzymes as a Toolbox for Cascade Reactions. ChemBioChem 2017, 18, 2226–2230. 10.1002/cbic.201700478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews S.; Tee K. L.; Rattray N. J.; McLean K. J.; Leys D.; Parker D. A.; Blankley R. T.; Munro A. W. Production of alkenes and novel secondary products by P450 OleTJE using novel H2O2 -generating fusion protein systems. FEBS Lett. 2017, 591, 737–750. 10.1002/1873-3468.12581. [DOI] [PubMed] [Google Scholar]
- Gandomkar S.; Dennig A.; Dordic A.; Hammerer L.; Pickl M.; Haas T.; Hall M.; Faber K. Biocatalytic Oxidative Cascade for the Conversion of Fatty Acids to alpha-Ketoacids via Internal H2O2 Recycling. Angew. Chem., Int. Ed. 2018, 57, 427–430. 10.1002/anie.201710227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse C. J.; Bell S. G.; Wong L. L. P450(BM3) (CYP102A1): Connecting the Dots. Chem. Soc. Rev. 2012, 41, 1218–1260. 10.1039/C1CS15192D. [DOI] [PubMed] [Google Scholar]
- Rude M. A.; Baron T. S.; Brubaker S.; Alibhai M.; Del Cardayre S. B.; Schirmer A. Terminal Olefin (1-alkene) Biosynthesis by a Novel p450 Fatty Acid Decarboxylase from Jeotgalicoccus species. Appl. Environ. Microbiol. 2011, 77, 1718–27. 10.1128/AEM.02580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J.; Hallberg B. M.; Bergfors T.; Oesch F.; Arand M.; Mowbray S. L.; Jones T. A. Structure of Aspergillus niger Epoxide Hydrolase at 1.8 Å Resolution: Implications for the Structure and Function of the Mammalian Microsomal Class of Epoxide Hydrolases. Structure 2000, 8, 111–122. 10.1016/S0969-2126(00)00087-3. [DOI] [PubMed] [Google Scholar]
- Wu W.; Zhu D.; Hua L. Site-saturation Mutagenesis of Formate Dehydrogenase from Candida bodinii Creating Effective NADP+-dependent FDH enzymes. J. Mol. Catal. B: Enzym. 2009, 61, 157–161. 10.1016/j.molcatb.2009.06.005. [DOI] [Google Scholar]
- Wu S.; Zhou Y.; Wang T.; Too H. P.; Wang D. I. C.; Li Z. Highly regio- and enantioselective multiple oxy- and amino-functionalizations of alkenes by modular cascade biocatalysis. Nat. Commun. 2016, 7, 11917. 10.1038/ncomms11917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee K. L.; Schwaneberg U. A Screening System for the Directed Evolution of Epoxygenases: Importance of Position 184 in P450 BM3 for Stereoselective Styrene Epoxidation. Angew. Chem., Int. Ed. 2006, 45, 5380–5383. 10.1002/anie.200600255. [DOI] [PubMed] [Google Scholar]
- Reetz M. T. Laboratory Evolution of Stereoselective Enzymes: a Prolific Source of Catalysts for Asymmetric Reactions. Angew. Chem., Int. Ed. 2011, 50, 138–174. 10.1002/anie.201000826. [DOI] [PubMed] [Google Scholar]
- Wang J.; Ilie A.; Reetz M. T. Chemo- and Stereoselective Cytochrome P450-BM3-Catalyzed Sulfoxidation of 1-Thiochroman-4-ones Enabled by Directed Evolution. Adv. Synth. Catal. 2017, 359, 2056–2060. 10.1002/adsc.201700414. [DOI] [Google Scholar]
- Andre C.; Kim S. W.; Yu X. H.; Shanklin J. Fusing Catalase to an Alkane-producing Enzyme Maintains Enzymatic Activity by Converting the Inhibitory Byproduct H2O2 to the Cosubstrate O2. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 3191–3196. 10.1073/pnas.1218769110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Ren H.; Zhao H. Expanding the Boundary of Biocatalysis: Design and Optimization of in vitro Tandem Catalytic Reactions for Biochemical Production. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 115–129. 10.1080/10409238.2018.1431201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcher J.; McLean K. J.; Matthews S.; Woodward L. S.; Fisher K.; Rigby S. E.; Nelson D. R.; Potts D.; Baynham M. T.; Parker D. A.; Leys D.; Munro A. W. Structure and Biochemical Properties of the Alkene Producing Cytochrome P450 OleTJE (CYP152L1) from the Jeotgalicoccus sp. 8456 Bacterium. J. Biol. Chem. 2014, 289, 6535–6550. 10.1074/jbc.M113.527325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrittwieser J. H.; Velikogne S.; Hall M.; Kroutil W. Artificial Biocatalytic Linear Cascades for Preparation of Organic Molecules. Chem. Rev. 2018, 118, 270–348. 10.1021/acs.chemrev.7b00033. [DOI] [PubMed] [Google Scholar]
- France S. P.; Hepworth L. J.; Turner N. J.; Flitsch S. L. Constructing Biocatalytic Cascades: In Vitro and in Vivo Approaches to de Novo Multi-Enzyme Pathways. ACS Catal. 2017, 7, 710–724. 10.1021/acscatal.6b02979. [DOI] [Google Scholar]
- Sperl J. M.; Sieber V. Multienzyme Cascade Reactions—Status and Recent Advances. ACS Catal. 2018, 8, 2385–2396. 10.1021/acscatal.7b03440. [DOI] [Google Scholar]
- Wu S.; Zhou Y.; Li Z. Biocatalytic Selective Functionalisation of Alkenes via Single-step and One-pot Multi-step Reactions. Chem. Commun. 2019, 55, 883–896. 10.1039/C8CC07828A. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.