Abstract
The microtubule-severing enzyme katanin (KTN1) regulates the organization and turnover of microtubule arrays by the localized breakdown of microtubule polymers. In land plants, KTN1 activity is essential for the formation of linearly organized cortical microtubule arrays that determine the axis of cell expansion. Cell biological studies have shown that even though KTN1 binds to the sidewalls of single and bundled microtubules, severing activity is restricted to microtubule cross-over and nucleation sites, indicating that cells contain protective mechanisms to prevent indiscriminate microtubule severing. Here, we show that the microtubule-bundling protein MAP65-1 inhibits KTN1-mediated microtubule severing in vitro. Severing is inhibited at bundled microtubule segments and the severing rate of nonbundled microtubules is reduced by MAP65-1 in a concentration-dependent manner. Using various MAP65-1 mutant proteins, we demonstrate that efficient cross-linking of microtubules is crucial for this protective effect and that microtubule binding alone is not sufficient. Reduced severing due to microtubule bundling by MAP65-1 correlated to decreased binding of KTN1 to these microtubules. Taken together, our work reveals that cross-linking of microtubules by MAP65-1 confers resistance to severing by inhibiting the binding of KTN1 and identifies the structural features of MAP65-1 that are important for this activity.
INTRODUCTION
The microtubule cytoskeleton is a multipurpose structure whose configuration dynamically changes depending on the needs of the cell. Microtubule-severing enzymes have emerged as key players for the formation, remodeling, and breakdown of microtubule arrays in eukaryotes (McNally and Roll-Mecak, 2018). Katanin was the first identified microtubule-severing enzyme and consists of a catalytic p60 subunit and a regulatory p80 subunit (McNally and Vale, 1993; Hartman et al., 1998). The p60 subunit belongs to the ATPases associated with diverse cellular activities superfamily and is sufficient for microtubule severing in an ATP-dependent manner (McNally and Vale, 1993; Hartman et al., 1998). The p80 subunit confers targeting and some degree of regulation of the p60 catalytic activity (Hartman et al., 1998; McNally et al., 2014; Wang et al., 2017). Biochemical and structural studies have shown that the p60 and p80 subunits assemble into hexameric ring structures that interact with the C-terminal tails of tubulin and remove tubulin dimers from microtubule sidewalls to promote severing (McNally and Vale, 1993; Hartman and Vale 1999; Zehr et al., 2017, Nithianantham et al., 2018; Vemu, Szczesna, et al., 2018).
In the land plant Arabidopsis thaliana, the p60 subunit of katanin is encoded by a single gene called KTN1 (Burk et al., 2001; Stoppin-Mellet et al., 2002) and null mutants exhibit developmental defects that are associated with aberrant organization of interphase and mitotic microtubule arrays (Bichet et al., 2001; Burk and Ye, 2002; Bouquin et al., 2003; Komis, Luptovcˇiak, Ovecˇka, et al., 2017). Live-imaging studies of the interphase cortical microtubule array revealed that KTN1 severs microtubules almost exclusively at microtubule cross-over junctions and at nucleation sites to generate coaligned microtubule organization and to drive light-induced array reorientation (Nakamura et al., 2010; Lindeboom et al., 2013; Wightman, Chomicki, et al., 2013; Zhang et al., 2013; Fan et al., 2018). Interestingly, green fluorescent protein (GFP)-labeled KTN1 localizes not only to microtubule cross-over and nucleation sites in a p80-dependent manner (Wang et al., 2017) but also extensively along the shaft of cortical microtubules (Lindeboom et al., 2013; Zhang et al., 2013). The lack of severing along the cortical microtubule shaft suggests that off-target severing is inhibited; however, the underlying mechanism remains unknown.
The neuronal microtubule-associated proteins (MAPs) tau and MAP2, which bundle and stabilize microtubules in axons and dendrites, respectively, have been shown to protect microtubules against KTN1 activity when expressed in fibroblasts (Qiang, Yu, et al., 2006; Qiang et al., 2010). In addition, depletion of tau or phosphorylation-induced dissociation of tau from microtubules has been correlated with increased KTN1-based microtubule severing in neurons (Qiang, Yu, et al., 2006). These data suggest that microtubule-cross-linking proteins inhibit microtubule severing by KTN1.
Microtubule bundling is also a prominent feature of the plant cortical microtubule cytoskeleton and is thought to be important for the construction of higher-order microtubule structures (Shaw et al., 2003; Dixit and Cyr, 2004). Therefore, protecting cortical microtubule bundles against severing is likely to be important to maintain array integrity. Plant cortical microtubule bundling is mediated by the evolutionarily conserved MAP65 family of microtubule cross-linking proteins (Smertenko et al., 2004; Smertenko, Chang, et al., 2006; Chang, Smertenko, et al., 2005; Lucas et al., 2011; Lucas and Shaw 2012). Similar to the mammalian protein regulator of cytokinesis 1 and fungal anaphase spindle elongation protein 1 homologues, plant MAP65 cross-links microtubules and dynamically labels the bundled regions of microtubules both in vivo (Lucas et al., 2011) and in vitro (Tulin et al., 2012), making it ideally suited to protect bundled regions of cortical microtubules against severing.
Here, we report that purified MAP65-1 protein inhibits microtubule severing in vitro in a concentration-dependent manner and that this effect is primarily due to the cross-linking of microtubules by MAP65-1 rather than just the binding of MAP65-1 to microtubules. In addition, we show that the inhibitory effect of MAP65-1 correlates with reduced binding of KTN1 to bundled microtubules. Together, our findings provide a mechanistic model for why bundled microtubules are not prone to severing in cells.
RESULTS AND DISCUSSION
GFP-KTN1 dwell time is reduced on bundled microtubules
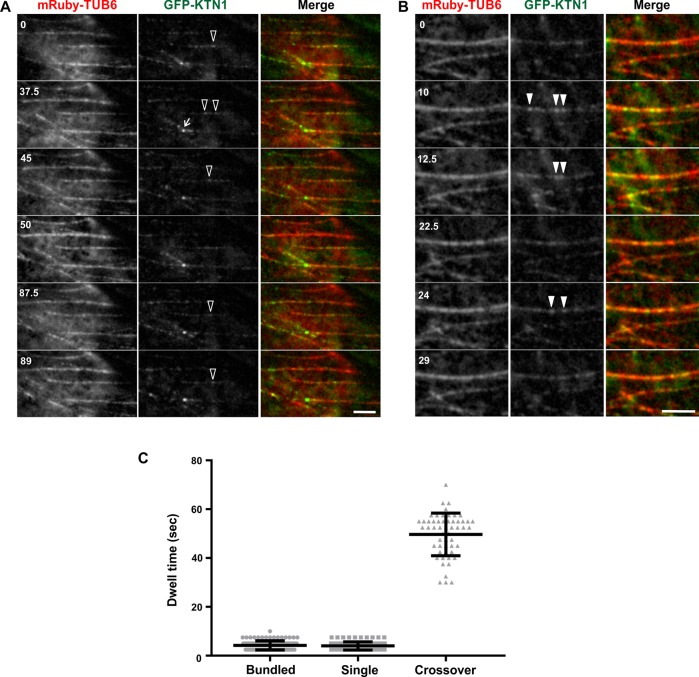
KTN1 has been reported to bind to cortical microtubule bundles without detectably severing them (Lindeboom et al., 2013; Zhang et al., 2013). Since the likelihood of severing correlates to the dwell time of KTN1 on a microtubule (Zhang et al., 2013), we wondered whether microtubule bundles impair the binding of KTN1. To determine the dwell time of KTN1 on bundled microtubules, we conducted time-lapse imaging of A. thaliana ktn1-2 mutant plants coexpressing GFP-KTN1 (Lindeboom et al., 2013) and an mRuby-TUB6 microtubule marker. From these time series, we measured the duration of individual GFP-KTN1 puncta on single cortical microtubules, bundled cortical microtubules, and at microtubule cross-over sites (Figure 1, A and B). We found that the dwell time of GFP-KTN1 on bundled microtubules (4.2 ± 1.9 s) is dramatically lower than at cross-over sites that are severed (49.7 ± 8.7 s) (Figure 1C). In addition, the dwell time of GFP-KTN1 on single microtubules that are not severed (4.0 ± 1.7 s) is comparable to that of bundled microtubules (Figure 1C).
FIGURE 1:
GFP-KTN1 binds more transiently to bundled cortical microtubules than to cross-over sites. (A, B) Time course of GFP-KTN1 binding to single (A) and bundled (B) cortical microtubules in vivo. Empty arrowheads indicate GFP-KTN1 particles on single microtubules. Solid arrowheads indicate GFP-KTN1 particles on bundled microtubules. Arrow points to a cross-over site. Scale bar is 5 µm. Numbers indicate time in seconds. (C) Dwell times of individual GFP-KTN1 particles on single microtubules (n = 96), bundled microtubules (n = 109), and at cross-over sites (n = 54). Values are means ± SD.
The MAP65-1 protein inhibits KTN1-mediated microtubule severing
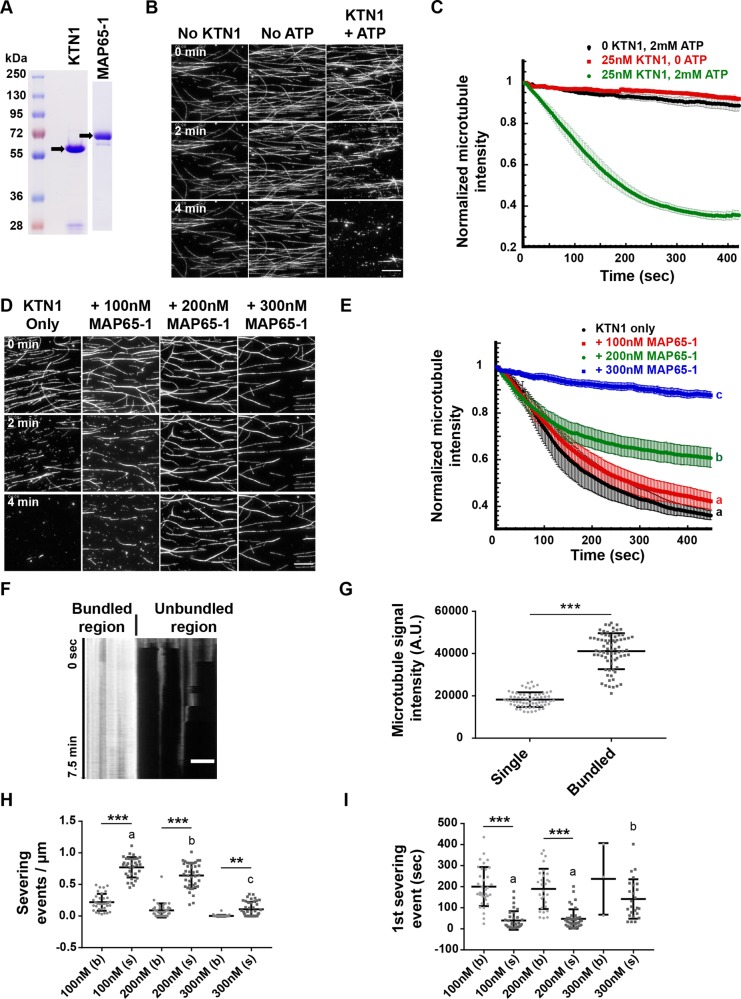
Proteins of the MAP65 family mediate bundling of cortical microtubules. In this study, we selected the MAP65-1 isoform because it abundantly binds to and bundles cortical microtubules in vivo (Smertenko et al., 2004; Gaillard, Neumann, et al., 2008; Lucas et al., 2011) and also bundles microtubules in vitro with similar characteristics (Tulin et al., 2012). To determine whether microtubule bundles created by MAP65-1 are resistant to KTN1 activity, we used an in vitro microtubule severing assay with purified recombinant KTN1 and MAP65-1 proteins (Figure 2A). Our purified KTN1 protein severs taxol-stabilized microtubules in an ATP-dependent manner (Figure 2B and Supplemental Movie 1) as reported previously (McNally and Vale, 1993; Stoppin-Mellet et al., 2002). We measured the microtubule fluorescence intensity over time in these assays and found that KTN1 severs essentially all microtubules by 6 min under our experimental conditions (Figure 2C). Next, we preincubated microtubules with MAP65-1 for 10 min to induce bundling followed by the addition of KTN1 (Figure 2D; Supplemental Movie 2). We found that microtubule severing was inhibited by MAP65-1 in a concentration-dependent manner (Figure 2E). Bundled microtubule regions were particularly resistant to severing compared with unbundled regions (Figure 2F), similar to tobacco NtMAP65-1c inhibiting severing of microtubule bundles in vitro (Meng et al., 2010). To quantify this observation, we examined the frequency of severing and the time to first observed severing for both single and bundled microtubules at different MAP65-1 concentrations. Bundled microtubules were defined by the merging of at least two distinct microtubules and having an intensity value at least twice that of nearby apparent single microtubules (Figure 2G). At all MAP65-1 concentrations tested, the severing frequency of single microtubules was significantly greater than that of the corresponding bundled microtubules (Figure 2H) and severing of single microtubules occurred earlier than in bundled microtubules (Figure 2I). In addition, increasing MAP65-1 concentrations progressively enhanced the resistance of single microtubules to severing (Figure 2, H and I). These findings suggest that MAP65-1 can protect both single and bundled microtubules in a concentration-dependent manner, although the efficacy of protecting bundled microtubules is higher at any given concentration.
FIGURE 2:
Microtubule bundling by MAP65-1 inhibits KTN1-mediated severing in a dose-dependent manner. (A) Coomassie Blue-stained SDS–PAGE gel loaded with purified KTN1 or MAP65-1 proteins. Arrows indicate the expected bands for KTN1 and MAP65-1. (B) Time course of rhodamine-labeled, taxol-stabilized microtubules in the presence of 2 mM ATP, 25 nM KTN1, or 25 nM KTN1 and 2 mM ATP. Scale bar is 10 µm. See also Supplemental Movie 1. (C) Plot of microtubule signal intensity over time from experiments shown in B. Each image in a series is normalized to the fluorescence intensity of the first frame of that series. Error bars represent SEM of at least four separate protein preparations (0 KTN1, 2 mM ATP, n = 12 movies; 25 nM KTN1, 0 ATP, n = 12 movies; 25 nM KTN1, 2 mM ATP, n = 18 movies). (D) Time course of rhodamine-labeled, taxol-stabilized microtubules in the presence of either 25 nM KTN1 alone or with a 10-min preincubation with 100, 200, or 300 nM of MAP65-1. See also Supplemental Movie 2. (E) Plot of microtubule signal intensity over time from experiments shown in D. Each image in a series is normalized to the fluorescence intensity of the first frame of that series. Error bars represent SEM (25 nM KTN1 alone, n = 8 movies; +100 nM MAP65-1, n = 10 movies; + 200 nM MAP65-1, n = 16 movies; + 300 nM MAP65-1, n = 10 movies). Letters at the end of the curves indicate statistical groups of the end-point signal intensities with p < 0.001 using analysis of variance (ANOVA). (F) Kymograph showing differences in signal intensity and severing frequency of bundled and unbundled regions of a microtubule after a 10-min preincubation with 200 nM MAP65-1 followed by the addition of 25 nM KTN1. (G) Dot-plots showing the quantification of microtubule signal intensities for single and bundled microtubules after a 10-min incubation with 200 nM MAP65-1. Bars represent mean ± SD (single microtubules, n = 65, bundled microtubules, n = 74). Asterisks indicate statistical difference with p < 0.001 using Student's t test. (H) Dot-plot showing quantification of severing events/µm of bundled (b) and single (s) microtubules from the experiments in D. Bars represent mean ± SD (100 nM MAP65-1; bundled microtubules, n = 34, single microtubules, n = 34, 200 nM MAP65-1; bundled microtubules, n = 40, single microtubules, n = 40, 300 nM MAP65-1; bundled microtubules, n = 40, single microtubules, n = 40). Statistically significant differences between single and bundled microtubules determined by ANOVA are indicated by asterisks, ***p < 0.001, **p < 0.01. Letters indicate statistical groups of single microtubule fluorescence intensities with p < 0.001 using ANOVA. (I) Dot-plot showing quantification of the time until the first severing event is observed for bundled (b) and single (s) microtubules from the experiments in D. Bars represent mean ± SD (100 nM MAP65-1; bundled microtubules, n = 34, single microtubules, n = 34, 200 nM MAP65-1; bundled microtubules, n = 28, single microtubules, n = 40, 300 nM MAP65-1; bundled microtubules, n = 3, single microtubules, n = 25). Statistically significant differences between single and bundled microtubules determined by ANOVA are indicated by asterisks, ***p < 0.001. Letters indicate statistical groups of single microtubule severing times with p < 0.001 using ANOVA.
Movie S1.
Microtubule severing by purified KTN1 protein.
Movie S2.
MAP65-1 protects microtubules against severing in a concentration-dependent manner.
Streptavidin binding and cross-linking of microtubules weakly inhibits severing activity
To determine whether microtubule severing can be inhibited by cross-linking microtubules by other means, we used streptavidin to artificially cross-link biotinylated microtubules. Using microtubule cosedimentation assays, we established that streptavidin bound well to microtubules containing 48% of biotin-labeled tubulin (Supplemental Figure S1A). We then conducted in vitro microtubule severing assays using microtubules containing 48% biotin-labeled tubulin either in the absence of streptavidin or with 900 nM streptavidin, a concentration sufficient to saturate the available biotin residues. We found that streptavidin-bound single and optically bundled microtubules were marginally protected against severing at early time points but were essentially completely severed over the course of 6 min (Supplemental Figure S1, B and C). Therefore, the protective role of MAP65-1 is a specific effect and not a general outcome of microtubule cross-linking.
Microtubule cross-linking by MAP65-1 is required for efficient inhibition of severing activity
One possible mechanism by which MAP65-1 inhibits microtubule severing is by competing for the same binding site on the microtubule as KTN1. Alternatively, the cross-bridges formed by MAP65-1 between microtubules might sterically block KTN1 from accessing the microtubule. The first mechanism predicts that the microtubule-binding domain of MAP65-1 should be sufficient to inhibit microtubule severing. The second mechanism predicts that the cross-bridge length of MAP65-1 would impact severing activity.
To test these predictions, we generated a novel MAP65-1 mutant protein that we refer to as MAP65-1 ND, which consists only of the microtubule-binding domain and therefore cannot homodimerize to form microtubule bundles (Figure 3, A and B). To study the contribution of the cross-bridge length, we took advantage of previously characterized MAP65-1 mutants (Tulin et al., 2012) that either decrease the spacing between microtubules (MAP65-1 ∆R1) or increase the spacing between microtubules (MAP65-1 R1R4) compared with wild-type MAP65-1 (WT MAP65-1), respectively (Figure 3, A and B). To confirm that MAP65-1 ND does not bundle microtubules, we incubated WT MAP65-1, MAP65-1 ∆R1, MAP65-1 R1R4 and MAP65-1 ND with 500 nM of rhodamine-labeled, taxol-stabilized microtubules for 10 min before imaging. We observed microtubule bundling with the wild type, ∆R1, and R1R4 mutants but not with MAP65-1 ND mutant (Figure 3C). We also found that the MAP65-1 ∆R1 and MAP65-1 R1R4 mutants exhibited weaker and stronger microtubule bundling compared with WT MAP65-1, respectively (Figure 3C), probably because the longer rod domain of R1R4 bundles microtubules more efficiently compared with the shorter rod domain of ∆R1 (Tulin et al., 2012).
FIGURE 3:
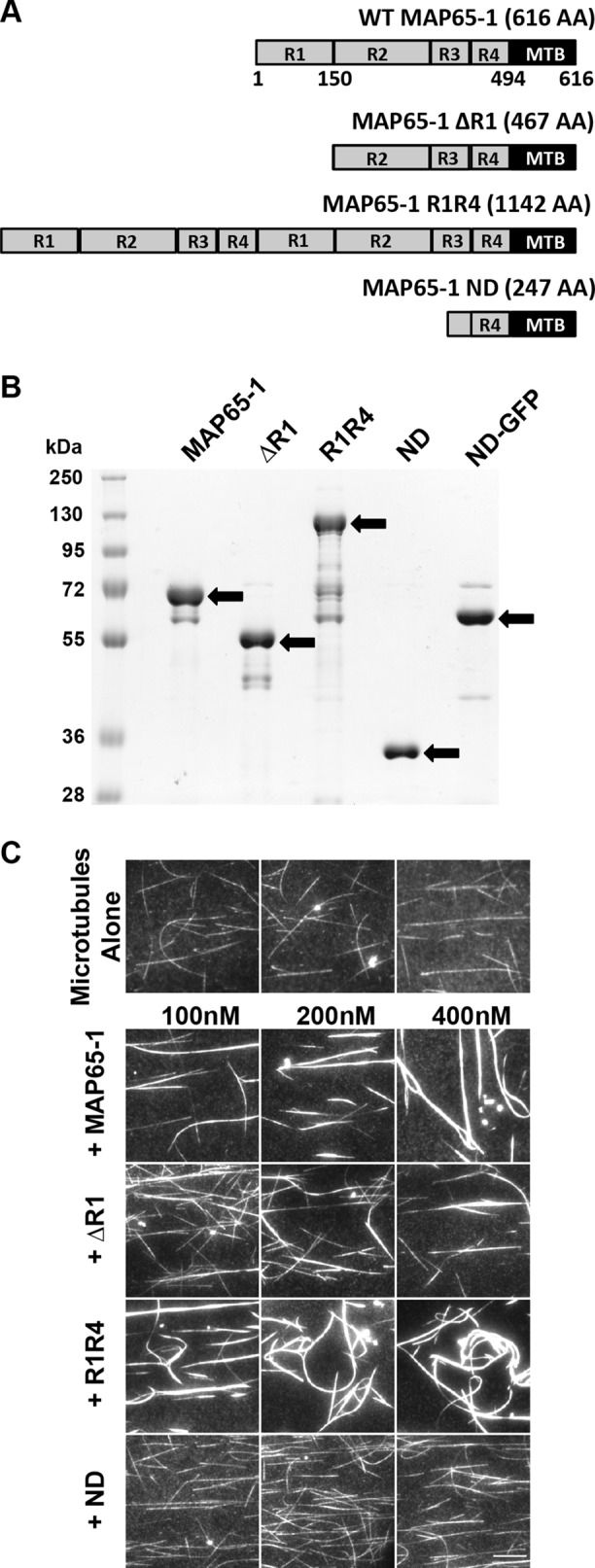
The MAP65-1 ND mutant does not bundle microtubules. (A) Schematic diagram of wild-type MAP65-1 and mutants used in this study. (B) Coomassie Blue-stained SDS–PAGE gel loaded with wild-type and mutant versions of MAP65-1 protein. Arrows indicate the expected bands for each protein. (C) Fluorescence micrographs of rhodamine-labeled, taxol-stabilized microtubules alone or in the presence of various concentrations of wild-type or mutant MAP65-1 proteins. All images were acquired under identical conditions. Scale bar is 10 µm.
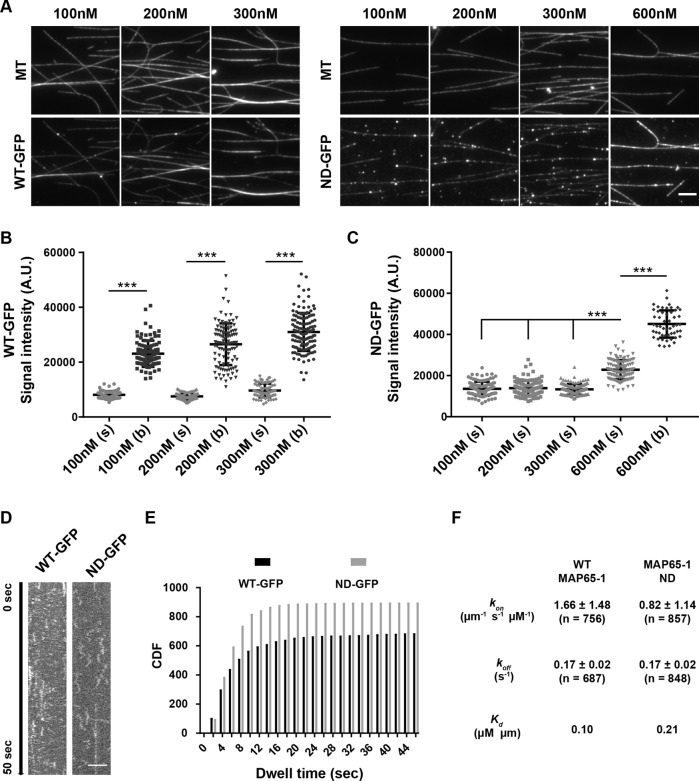
Since we did not observe microtubule bundling with the MAP65-1 ND mutant, we wanted to confirm that this mutant protein binds to microtubules. We generated a MAP65-1 ND-GFP fusion protein and compared its microtubule binding to WT MAP65-1-GFP. Consistent with previous findings (Tulin et al., 2012), WT MAP65-1-GFP binds more extensively to bundled microtubule regions than unbundled regions at all concentrations that we tested (Figure 4, A and B). At 300 nM WT MAP65-1-GFP, most of the microtubules were extensively bundled and there was more MAP65-1-GFP bound to single microtubules than at lower MAP65-1-GFP concentrations (Figure 4, A and B), which might explain why single microtubules were protected to a greater degree by 300 nM MAP65-1 (Figure 2, H and I). In contrast to WT MAP65-1-GFP, 100–300 nM ND-GFP bound relatively uniformly along the length of microtubules (Figure 4, A and C). Interestingly, at 600 nM ND-GFP, there was a sharp increase in microtubule binding and microtubules also appeared to be bundled (Figure 4, A and C). The apparent bundling activity at high ND-GFP concentrations could be due to masking of the negatively charged C-terminal regions of tubulin by bound ND-GFP, leading to microtubule association. Alternatively, the short N-terminal region of MAP65-1 ND (Figure 3A) might mediate microtubule interactions at high concentrations.
FIGURE 4:
The MAP65-1 ND mutant binds more weakly to microtubules compared with WT MAP65-1. (A) Fluorescence micrographs of rhodamine-labeled, taxol-stabilized microtubules incubated for 10 min with 100, 200, or 300 nM of WT MAP65-1 GFP or 100, 200, 300, or 600 nM MAP65-1 ND GFP. Scale bar is 10µm. (B) Dot-plot showing the quantification of WT MAP65-1 GFP signal intensities on single (s) and bundled (b) microtubules from A. Bars represent mean ± SD (100 nM [s], n = 100; 100 nM [b], n = 104; 200 nM [s], n = 95; 200 nM [b], n = 109; 300 nM [s], n = 84; 300 nM [b], n = 130). Values were compared between single and bundled microtubules within a treatment using ANOVA. Asterisks indicate statistical groups with p < 0.001. (C) Dot-plot showing the quantification of MAP65-1 ND GFP signal intensities on single (s) and bundled (b) microtubules from A. Bars represent mean ± SD (100 nM [s], n = 105; 200 nM [s], n = 111; 300 nM [s], n = 113; 600 nM [s], n = 112; 600 nM [b], n = 54). Values were compared between single microtubules as well as single and bundled microtubules at 600 nM MAP65-1 ND GFP. Asterisks indicate statistical groups with p < 0.001 using ANOVA. (D) Kymographs showing the behavior of WT MAP65-1-GFP and MAP65-1 ND-GFP on single microtubules. The brightness and contrast of the images were adjusted to better visualize the individual particles. Scale bar is 4 µm. (E) Cumulative distribution frequency plots of the dwell times of single WT MAP65-1-GFP (black) and MAP65-1 ND-GFP (gray) molecules. (F) Binding rate constant (kon), dissociation rate constant (koff), the apparent dissociation constant (Kd) of WT MAP65-1-GFP and MAP65-1 ND-GFP determined from kymograph analysis of single molecules. Values represent mean ± SD (n = number of molecules).
Using single-molecule imaging conditions to compare the behavior of WT MAP65-1-GFP and ND-GFP on unbundled regions of microtubules (Figure 4, D and E), we found that the binding rate constant (kon) of MAP65-1 ND was approximately twofold lower than that of WT MAP65-1 (Figure 4F), whereas the dissociation rate constant (koff) of MAP65-1 ND was similar to that of WT MAP65-1 (Figure 4F). Therefore, the apparent dissociation constant, Kd, of MAP65-1 ND is about twofold lower than that of WT MAP65-1. On the basis of these data, we conclude that MAP65-1 ND binds to microtubules but with a slightly lower affinity compared with WT MAP65-1.
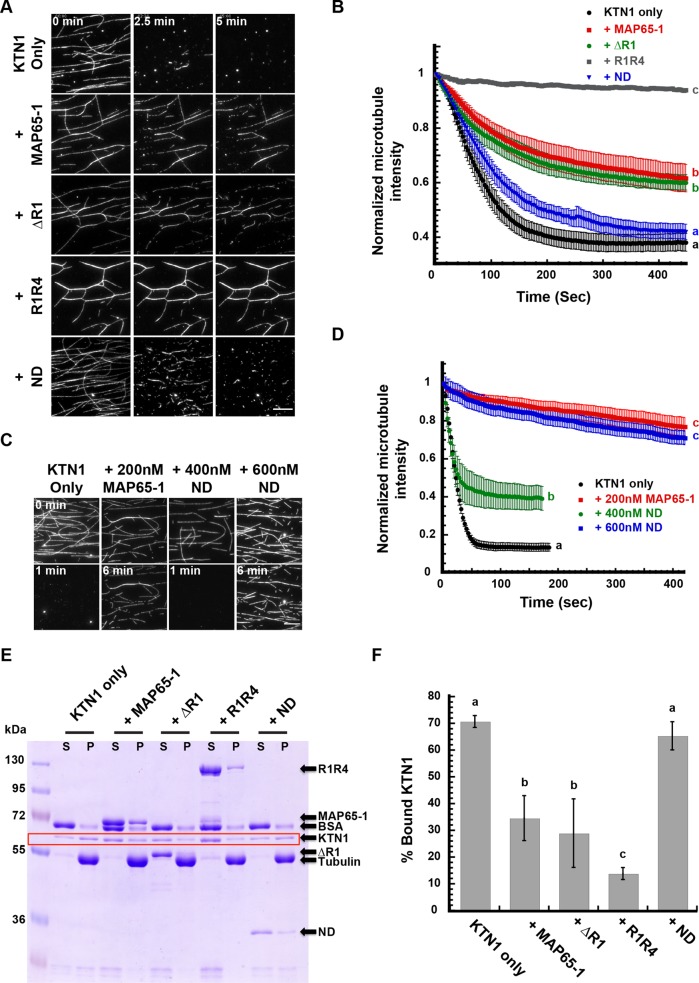
To examine the impact of the various MAP65-1 mutant proteins on KTN1-mediated microtubule severing, we preincubated microtubules for 10 min with 200 nM of WT or mutant MAP65-1 proteins before introducing KTN1. We found that the MAP65-1 ND mutant had a minimal inhibitory effect on microtubule severing compared with WT MAP65-1 (Figure 5, A and B; Supplemental Movie 3). To compensate for the lower binding affinity of the MAP65-1 ND mutant, we doubled the concentration of MAP65-1 ND to 400 nM and found that it still does not protect microtubules to the level of 200 nM WT MAP65-1 (Figure 5, C and D). However, 600 nM MAP65-1 ND, which densely binds to and bundles microtubules (Figure 4A), reduced microtubule severing similar to 200 nM WT MAP65-1 (Figure 5, C and D). In these experiments, both MAP65-1 ∆R1 and MAP65-1 R1R4 reduced microtubule severing and their degree of protection correlated with their strength of microtubule bundling (Figure 5, A and B). Together, these data indicate that bundling offers the strongest protection to microtubules against severing and that dense binding of MAP65-1 can protect single microtubules against severing to a certain degree.
FIGURE 5:
Bundling of microtubules by MAP65-1 inhibits severing by decreasing the binding of KTN1 to microtubules. (A) Time course of rhodamine-labeled, taxol-stabilized microtubules in the presence of 25 nM KTN1 alone or with a 10-min preincubation with 200 nM of wild-type MAP65-1, MAP65-1 ∆R1, MAP65-1 R1R4, or MAP65-1 ND proteins. Scale bar is 10 µm. See also Supplemental Movie 3. (B) Plot of microtubule signal intensity over time from experiments shown in A. Each image in a series is normalized to the fluorescence intensity of the first frame of that series. Error bars represent SEM (25 nM KTN1, n = 10 movies; wild-type MAP65-1, n = 10 movies; MAP65-1 ∆R1, n = 12 movies; MAP65-1 R1R4, n = 10 movies; MAP65-1 ND, n = 10 movies). Letters at the end of the curves indicate statistical groups of the end-point signal intensities with p < 0.001 using ANOVA. (C) Time course of rhodamine-labeled, taxol-stabilized microtubules in the presence of 25 nM KTN1 alone or with a 10-min preincubation with 200 nM MAP65-1 or 400 or 600 nM MAP65-1 ND proteins. Scale bar is 10 µm. (D) Plot of microtubule signal intensity over time from experiments in C. Each image in a series is normalized to the fluorescence intensity of the first frame of that series. Error bars represent SEM (KTN1 only, n = 5 movies, 200 nM MAP65-1, n = 6 movies, 400 nM ND, n = 5 movies, 600 nM ND, n = 6 movies). Letters at the end of the curves indicate statistical groups of the end-point signal intensities with p < 0.001 using ANOVA. (E) Coomassie Blue-stained SDS–PAGE gel of a microtubule cosedimentation assay. 1µM rhodamine-labeled, taxol-stabilized microtubules were incubated with either 1 µM KTN1 alone or were preincubated for 10 min with 2 µM of the indicated MAP65-1 proteins before adding KTN1. S, supernatant; P, pellet. Arrows identify the different protein bands. The KTN1 bands are boxed in red. (F) Quantification of the microtubule-bound KTN1 fraction for the experiments shown in E. The bars represent the mean ± SD (n = 6 for each condition). Letters above the bars indicate statistical groups with α < 0.05 using ANOVA.
Movie S3.
Comparison of the protective activity of wild-type and mutant MAP65-1 proteins.
Microtubule bundling by MAP65-1 inhibits KTN1 binding
On the basis of our live imaging observation of reduced GFP-KTN1 dwell time on cortical microtubule bundles and our finding that microtubule bundling inhibits severing in vitro, we hypothesized that microtubule bundling by MAP65-1 inhibits the binding of KTN1 to microtubules. To test this, we performed microtubule cosedimentation experiments. Microtubules were preincubated with WT MAP65-1, MAP65-1 mutants, or buffer alone for 10 min before introducing KTN1 (Figure 5E). We conducted these assays using the nonhydrolyzable ATP analogue, AMPPNP, to stabilize the binding of KTN1 to microtubules without severing them. We found that the microtubule-bound fraction of KTN1 decreased by twofold when microtubules were preincubated with either WT MAP65-1 or the MAP65-1 ∆R1 mutant (Figure 5F). In contrast, the MAP65-1 R1R4 mutant inhibited KTN1 binding by approximately fivefold compared with the control. Notably, the MAP65-1 ND mutant did not significantly inhibit KTN1 binding to microtubules (Figure 5F). On the basis of these results, we conclude that microtubule bundling inhibits severing by decreasing the binding of KTN1 to those microtubules.
Together, our findings indicate that bundling of microtubules by MAP65-1 is an effective protective mechanism to prevent unwanted microtubule severing by KTN1. The microtubule-binding domain of MAP65-1 by itself offers modest protection against severing, which requires much higher concentrations than full-length MAP65-1. One reason for this could be that the affinity of the microtubule-binding domain alone for microtubules is less compared with full-length MAP65-1. In addition, full-length MAP65-1 stays bound to microtubules for a significantly longer duration upon a bundling reaction (Tulin et al., 2012), whereas the microtubule-binding domain alone lacks this ability because it cannot engage in a bivalent interaction with two microtubules. In any case, our results reveal that the microtubule-binding domain alone is not sufficient to maximally inhibit severing, consistent with our in vitro observation that single microtubules bound by MAP65-1 are more prone to severing than microtubules bundled by MAP65-1. Our examination of the impact of the rod domain on severing activity demonstrated a strong correlation between the strength of microtubule bundling and the degree of inhibition of microtubule severing. Therefore, the ability to cross-link microtubules is a major determinant of the protective effect of MAP65-1. In addition to MAP65-1 reducing microtubule severing by inhibiting KTN1 binding to cross-linked microtubules, microtubule bundling by MAP65-1 would also limit the number of cross-over events in vivo, thus further reducing the substrates for KTN1 binding.
Recent work has discovered that at short timescales, KTN1 damages microtubules by removing tubulin dimers from microtubule sidewalls (Vemu, Szczesna, et al., 2018). These injury sites can be healed by the incorporation of fresh GTP-tubulin dimers. However, prolonged incubation with KTN1 tips the balance toward damage and leads to microtubule severing (Vemu, Szczesna, et al., 2018). Since the residence time of KTN1 is lower on bundled microtubules, one potential mechanism for why bundled microtubules are more resistant to severing is a reduction in the rate of removal of tubulin dimers from these microtubules. If the dimer removal rate is slower than the rate of insertion of new GTP-tubulin dimers, then the microtubule lattice is more likely to be repaired than get severed. Another potential mechanism that is not mutually exclusive is that MAP65-1 cross-links brace-damaged microtubules and thus inhibit complete severing from occurring in these regions.
Our finding that MAP65-1 inhibits severing in a concentration-dependent manner suggests a potential mechanism for cells to tune the level of protection against KTN1 activity. For example, cells can regulate the amount of MAP65-1 protein to globally modulate the extent of microtubule severing. Furthermore, phosphorylation of MAP65-1, which is known to decrease its microtubule binding (Smertenko, Chang, et al., 2006), could serve as a mechanism to locally control the extent of microtubule severing. Therefore, the balance between bundling and severing provides a potential mechanism to control the architecture and stability of microtubule arrays.
The MAP65 gene family has expanded considerably in land plants compared with mammals and yeast (Hussey et al., 2002). Plant MAP65 isoforms differ considerably in polypeptide length and in their preference for polarity of microtubule bundling (Gaillard, Neumann, et al., 2008; Smertenko et al., 2008; Fache et al., 2010; Ho et al., 2012). Certain plant MAP65 isoforms also localize to specific microtubule arrays during the cell cycle or even to specific zones of particular microtubule arrays (Van Damme et al., 2004; Li, Sun, et al., 2017). Future work will determine whether different MAP65 isoforms differ in their ability to inhibit severing, which would provide another mechanism for plants to differentially protect microtubule arrays from severing. Additional work is also needed to determine whether the mammalian and yeast homologues of MAP65-1 inhibit microtubule severing in the mitotic microtubule arrays to which they localize.
MATERIALS AND METHODS
Plant materials, growth conditions, and imaging
We generated a pGAG UBQ10promoter::mRuby-TUB6-KanR plasmid using multisite-Gateway recombination and introduced it into A. thaliana Col-0 plants via Agrobacterium tumefaciens–mediated transformation. Homozygous transformants were crossed to the ktn1-2 mutant expressing GFP-KTN1. Seeds from plants homozygous for the ktn1-2 mutation and at least heterozygous for GFP-KTN1 and mRuby-TUB6 were surface-sterilized, plated on 1/2× Murashige and Skoog with 1% agar, and stratified in the dark for 2 d at 4°C. Seedlings were grown at 22°C in a 16-h/8-h light/dark cycle for 3–4 d before observation. Seedlings were imaged by VAEM microscopy (Konopka and Bednarek, 2008) using 488- and 514-nm laser lines for excitation and 500–550 and 582–636 nm emission filter sets for GFP and mRuby imaging, respectively. Two-color, time-lapse images were collected every 2.5 s. Image analysis was conducted using the Fiji ImageJ package (Schindelin et al., 2012).
Constructs for recombinant protein expression
pMAL C5-X (New England Biolabs) was modified by using PCR to add a 6×-histidine tag in frame upstream of the maltose-binding protein (MBP) coding sequence and a tobacco etch virus protease (TEV) cleavage site in frame downstream of the MBP coding sequence, generating the expression vector pHis-MAL-TEV (pHMT). The cDNA sequence of KTN1 was introduced into pHMT using the NotI and SalI restriction sites.
The C-terminal microtubule-binding domain of MAP65-1 (amino acids 369–616) was cloned by PCR from the pTEV-MAP65-1 construct (Tulin et al., 2012) with primers CATATGTCTGCTCGTGAGAGAATCATGT and GGATCCTCATGGTGAAGCTGGAAC introducing NdeI and BamHI restriction sites for cloning into pTEV. To generate the ND-GFP construct, primers CATATGTCTGCTCGTGAGAGAATCATGT and GGATCCTGGTGAAGCTGGAACTTG were used to amplify the MAP65-1 ND fragment, and primers GGATCCATGGTGAGCAAGGGCGAG and GAGCTCTTACTTGTACAGCTCGTC were used to amplify mEGFP. The PCR products were both digested with BamHI and ligated together. The ligation was then used as a template for PCR using primers CATATGTCTGCTCGTGAGAGAATCATGT and GAGCTCTTACTTGTACAGCTCGTC. This PCR product was cloned into pTEV using NdeI and SacI. All constructs were confirmed by DNA sequencing.
Expression and purification of recombinant KTN1 and MAP65-1 proteins
The pHMT-KTN1 plasmid was transformed into BL21-CodonPlus (DE3)-RIPL competent cells (Agilent Technologies). Protein was affinity-purified using Ni-NTA agarose (Qiagen #301210) and exchanged into severing buffer (SB) I (pH 7.0, 20 mM HEPES, 50 mM NaCl, 75 mM MgSO4, 2 mM MgCl2, 10% sucrose, 5 mM dithiothreitol [DTT], 50 µM ATP) using PD-10 desalting columns (GE Healthcare #17085101). Protein was then digested with TEV protease and run through a subtractive Ni-NTA column to remove TEV protease and uncut protein. MAP65-1 proteins were purified from Rosetta DE3 cells as described previously (Tulin et al., 2012). Protein aliquots were snap-frozen in liquid nitrogen and stored at –80°C until use.
Microtubule-severing assays
Imaging chambers were prepared by affixing silanized coverslips to slides using strips of double-sided tape. A solution of 0.8% anti-tubulin antibody (Sigma #T4026) in SBII (pH 7.0, 20 mM HEPES, 3 mM MgCl2, 10% sucrose) was coated on the coverslip and then blocked with a 5% pluronic F-127 (Sigma #P2443) solution in SBII. A 1:100 dilution of 5 mg/ml taxol-stabilized 1:25 rhodamine-labeled porcine microtubules was then flowed in and allowed to bind to the antibody for 5 min. Excess microtubules were then washed out with two washes of 20 µM taxol in SBII. Severing mix (25 nM KTN1, 2 mM ATP, 50 mM DTT, 800 µg/ml glucose oxidase, 175 µg/ml catalase, 22.5 mg/ml glucose, 20 µM taxol in SBII) was then flowed in and the imaging chamber was immediately mounted and imaged by total internal reflection fluorescence microscopy. Time-lapse images were acquired every 3 s for 7.5 min using a back-illuminated electron-multiplying CCD camera (ImageEM; Hamamatsu, Bridgewater, NJ) and a 582–636 nm emission filter set. Image analysis was performed using the Fiji ImageJ package (Schindelin et al., 2012).
For severing assays conducted with MAP65-1 proteins, SBII with 20 µM taxol and the indicated amount of MAP65-1 protein were flowed in following the microtubule washout step and incubated for 10 min before the severing mix (supplemented with the same concentration of MAP65-1 protein used for the incubation step) was flowed in, and then the chamber was mounted and imaged as described above. For severing assays conducted with streptavidin, a 1:100 dilution of 5 mg/ml taxol-stabilized 1:25 rhodamine-labeled microtubules containing 48% biotin-labeled tubulin was used. After the microtubule washout step, 900 nM streptavidin (estimated to be at least a twofold higher concentration than the bound microtubules) was flowed in and allowed to bind to microtubules for 5 min. Unbound streptavidin was washed out with two washes of 20 µM taxol in SBII, and then 1:100 diluted rhodamine-labeled microtubules containing 48% biotin-labeled tubulin was flowed in to promote cross-linking to the affixed microtubules bound to streptavidin. Unbound microtubules were washed out with two washes of 20 µM taxol in SBII, and then the severing mix was flowed in and the chamber was imaged as described above.
Well-separated single microtubules and microtubule bundles consisting of at least two microtubules that were visibly merging were used to analyze their signal intensities. The mean gray values for regions of single and bundled microtubules were measured using the Fiji ImageJ package (Schindelin et al., 2012). Statistical analysis was performed using Kaliedagraph 4.1 (Synergy Software).
Imaging of WT MAP65-1 GFP and MAP65-1 ND GFP
Imaging chambers were prepared as for the severing assays. After a 10-min incubation with the indicated concentrations of WT MAP65-1-GFP and MAP65-1 ND-GFP, the imaging chambers were mounted and imaged using the 582–636 and 500–550 nm emission filter sets. All images were acquired with identical settings to be able to quantitatively compare signal intensities. The mean GFP signal intensities were measured along single and bundled microtubules using the Fiji ImageJ package (Schindelin et al., 2012). Statistical analysis was performed using Kaliedagraph 4.1 (Synergy Software).
Single molecule imaging
Imaging chambers were prepared as for the microtubule severing assays. Following the microtubule washout step, 2.5 nM MAP65-1 or 10 nM MAP65-1 ND-GFP was then flowed in and immediately imaged. An initial snapshot of the microtubules was taken using the 582–636 nm emission filter set. Streaming-mode images were acquired with the 500–550 nm filter set every 100 ms for 50 s. Kymographs were generated along the microtubules and analyzed using the Fiji ImageJ package (Schindelin et al., 2012). Bright, static particles were considered to be protein aggregates and were not included in our analysis.
Microtubule bundling assays
The 500 nM taxol-stabilized, 1:25 rhodamine-labeled microtubules were incubated for 10 min with the indicated concentrations of MAP65-1 proteins, then flowed into an imaging chamber and imaged using the 582–636 nm emission filter set. All images were acquired with identical settings so that signal intensity reflects the degree of microtubule bundling.
Microtubule cosedimentation assays
To assess KTN1 binding to microtubules in the presence of MAP65-1 mutant proteins, 1 µM of rhodamine-labeled, taxol-stabilized microtubules was coincubated with 2 µM of MAP65-1 proteins for 10 min in SBII supplemented with 50 µM DTT, 40 µM taxol, 0.1 mg/ml bovine serum albumin, and 2 mM AMPPNP before adding 1 µM KTN1 to the mixture. This mixture was then incubated at room temperature for 30 min before centrifugation at 15°C for 25 min. The supernatant and pellet fractions were run out on SDS–PAGE and Coomassie-stained. Band densitometry was performed using Fiji software (Schindelin et al., 2012). Statistical analysis was performed using Kaliedagraph 4.1 (Synergy Software).
Supplementary Material
Acknowledgments
We thank M. Bezanilla (Dartmouth College) for providing the cDNA for mRuby and the pDonor plasmid and D. Ehrhardt (Carnegie Institute for Science, Stanford University) for providing the GFP-KTN1 complemented ktn1-2 mutant. This work was supported by National Institutes of Health Grant R01 GM-114678.
Abbreviations used:
- GFP
green fluorescent protein
- KTN1
katanin
- MAP
microtubule-associated protein
- MBP
maltose-binding protein
- SB
severing buffer
- TEV
tobacco etch virus protease.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-12-0776) on April 24, 2019.
REFERENCES
Boldface names denote co–first authors.
- Bichet A, Desnos T, Turner S, Grandjean O, Höfte H. (2001). BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J , 137–148. [DOI] [PubMed] [Google Scholar]
- Bouquin T, Mattsson O, Naested H, Foster R, Mundy J. (2003). The Arabidopsis lue1 mutant defines a katanin p60 ortholog involved in hormonal control of microtubule orientation during cell growth. J Cell Sci , 791–801. [DOI] [PubMed] [Google Scholar]
- Burk DH, Liu B, Zhong R, Morrison WH, Ye ZH. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell (4), 807–827. [PMC free article] [PubMed] [Google Scholar]
- Burk DH, Ye ZH. (2002). Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule-severing protein. Plant Cell , 2145–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HY, Smertenko AP, Igarashi H, Dixon DP, Hussey PJ. (2005). Dynamic interaction of NtMAP65–1a with microtubules in vivo. J Cell Sci , 3195–3201. [DOI] [PubMed] [Google Scholar]
- Dixit R, Cyr R. (2004). Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell , 3274–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fache V, Gaillard J, Van Damme D, Geelen D, Neumann E, Stoppin-Mellet V, Vantard M. (2010). Arabidopsis kinetochore fiber-associated MAP65–4 cross-links microtubules and promotes microtubule bundle elongation. Plant Cell , 3804–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Burkart GM, Dixit R. (2018). The Arabidopsis SPIRAL2 protein targets and stabilizes microtubule minus ends. Curr Biol (6), 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard J, Neumann E, Van Damme D, Stoppin-Mellet V, Ebel C, Barbier E, Geelen D, Vantard M, Drubin DG. (2008). Two microtubule-associated proteins of arabidopsis map65s promote antiparallel microtubule bundling. Mol Biol Cell , 4534–4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman JJ, Vale RD. (1999). Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science , 782–785. [DOI] [PubMed] [Google Scholar]
- Hartman JJ, Mahr J, McNally K, Okawa K, Iwamatsu A, Thomas S, Cheesman S, Heuser J, Vale RD, McNally FJ. (1998). Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell , 277–287. [DOI] [PubMed] [Google Scholar]
- Ho CMK, Lee YRJ, Kiyama LD, Dinesh-Kumar SP, Liu B. (2012). Arabidopsis microtubule-associated protein MAP65–3 cross-links antiparallel microtubules toward their plus ends in the phragmoplast via its distinct C-terminal microtubule binding domain. Plant Cell , 2071–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey PJ, Hawkins TJ, Igarashi H, Kaloriti D, Smertenko A. (2002). The plant cytoskeleton: recent advances in the study of the plant microtubule-associated proteins MAP-65, MAP-190 and the Xenopus MAP215-like protein, MOR1. Plant Mol Biol , 915. [DOI] [PubMed] [Google Scholar]
- Komis G, Luptovcˇiak I, Ovecˇka M, Samakovli D, Šamajová O, Šamaj J. (2017). Katanin effects on dynamics of cortical microtubules and mitotic arrays in arabidopsis thaliana revealed by advanced live-cell imaging. Front Plant Sci . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka CA, Bednarek SY. (2008). Variable-angle epifluorescence microscopy: a new way to look at protein dynamics in the plant cell cortex. Plant J , 186–196. [DOI] [PubMed] [Google Scholar]
- Li H, Sun B, Sasabe M, Deng X, Machida Y, Lin H, Lee YRJ, Liu B. (2017). Arabidopsis MAP65–4 plays a role in phragmoplast microtubule organization and marks the cortical cell division site. New Phytol , 187–201. [DOI] [PubMed] [Google Scholar]
- Lindeboom J.J, Nakamura M, Hibbel A, Shundyak K, Gutierrez R, Ketelaar T, Emons AMC, Mulder BM, Kirik V, Ehrhardt DW. (2013). A mechanism for reorientation of cortical microtubule arrays driven by microtubule severing. Science , 1245533. [DOI] [PubMed] [Google Scholar]
- Lucas JR, Courtney S, Hassfurder M, Dhingra S, Bryant A, Shaw SL. (2011). Microtubule-associated proteins MAP65-1 and MAP65-2 positively regulate axial cell growth in etiolated Arabidopsis hypocotyls. Plant Cell , 1889–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas JR, Shaw SL. (2012). MAP65-1 and MAP65-2 promote cell proliferation and axial growth in Arabidopsis roots. Plant J , 454–463. [DOI] [PubMed] [Google Scholar]
- McNally FJ, Roll-Mecak A. (2018). Microtubule-severing enzymes: from cellular functions to molecular mechanism. J Cell Biol , 4057–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally FJ, Vale RD. (1993). Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell , 419–429. [DOI] [PubMed] [Google Scholar]
- McNally K, Berg E, Cortes DB, Hernandez V, Mains PE, McNally FJ, Doxsey S. (2014). Katanin maintains meiotic metaphase chromosome alignment and spindle structure in vivo and has multiple effects on microtubules in vitro. Mol Biol Cell , 1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Du J, Li J, Lu X, Zeng X, Yuan M, Mao T. (2010). Tobacco microtubule-associated protein, MAP65-1c, bundles and stabilizes microtubules. Plant Mol Biol , 537–47. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Ehrhardt DW, Hashimoto T. (2010). Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat Cell Biol , 1064. [DOI] [PubMed] [Google Scholar]
- Nithianantham S, McNally FJ, Al-Bassam J. (2018). Structural basis for disassembly of katanin heterododecamers. J Biol Chem , 10590–10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L, Yu W, Andreadis A, Luo M, Baas PW. (2006). Tau protects microtubules in the axon from severing by katanin. J Neurosci , 3120–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L, Yu W, Liu M, Solowska JM, Baas PW. (2010). Basic fibroblast growth factor elicits formation of interstitial axonal branches via enhanced severing of microtubules. Mol Biol Cell , 334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Kamyar R, Ehrhardt DW. (2003). Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science , 1715–1718. [DOI] [PubMed] [Google Scholar]
- Smertenko AP, Chang H-Y, Sonobe S, Fenyk SI, Weingartner M, Bögre L, Hussey PJ. (2006). Control of the AtMAP65-1 interaction with microtubules through the cell cycle. J Cell Sci , 3227–3237. [DOI] [PubMed] [Google Scholar]
- Smertenko AP, Chang HY, Wagner V, Kaloriti D, Fenyk S, Sonobe S, Lloyd C, Hauser MT, Hussey PJ. (2004). The arabidopsis microtubule-associated protein AtMAP65-1: molecular analysis of its microtubule bundling activity. Plant Cell , 2035–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko AP, Kaloriti D, Chang H-Y, Fiserova J, Opatrny Z, Hussey PJ. (2008). The C-terminal variable region specifies the dynamic properties of Arabidopsis microtubule-associated protein MAP65 isotypes. Plant Cell , 3346–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppin-Mellet V, Gaillard J, Vantard M. (2002). Functional evidence for in vitro microtubule severing by the plant katanin homologue. Biochem J , 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A, McClerklin S, Huang Y, Dixit R.(2012). Single-molecule analysis of the microtubule cross-linking protein MAP65-1 reveals a molecular mechanism for contact-angle-dependent microtubule bundling. Biophys J , 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D, Van Poucke K, Boutant E, Ritzenthaler C, Inzé D, Geelen D. (2004). In Vivo dynamics and differential microtubule-binding activities of MAP65 proteins. Plant Physiol , 3956–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemu A, Szczesna E, Zehr EA, Spector JO, Grigorieff N, Deaconescu AM, Roll-Mecak A. (2018). Severing enzymes amplify microtubule arrays through lattice GTP-tubulin incorporation. Science , eaau1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Liu W, Wang G, Li J, Dong L, Han L, Wang Q, Tian J, Yu Y, Gao C, Kong Z. (2017). KTN80 confers precision to microtubule severing by specific targeting of katanin complexes in plant cells. EMBO J , 3435–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman R, Chomicki G, Kumar M, Carr P, Turner SR. (2013). SPIRAL2 determines plant microtubule organization by modulating microtubule severing. Curr Biol , 1902–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr E, Szyk A, Piszczek G, Szczesna E, Zuo X, Roll-Mecak A. (2017). Katanin spiral and ring structures shed light on power stroke for microtubule severing. Nat Struct Mol Biol , 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Fishel E, Bertroche T, Dixit R. (2013). Microtubule severing at crossover sites by katanin generates ordered cortical microtubule arrays in Arabidopsis . Curr Biol , 2191–2195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.