Abstract
Guanine nucleotide exchange factors (GEFs) are the initiators of signaling by every regulatory GTPase, which in turn act to regulate a wide array of essential cellular processes. To date, each family of GTPases is activated by distinct families of GEFs. Bidirectional membrane trafficking is regulated by ADP-ribosylation factor (ARF) GTPases and the development throughout eukaryotic evolution of increasingly complex systems of such traffic required the acquisition of a functionally diverse cohort of ARF GEFs to control it. We performed phylogenetic analyses of ARF GEFs in eukaryotes, defined by the presence of the Sec7 domain, and found three subfamilies (BIG, GBF1, and cytohesins) to have been present in the ancestor of all eukaryotes. The four other subfamilies (EFA6/PSD, IQSEC7/BRAG, FBX8, and TBS) are opisthokont, holozoan, metazoan, and alveolate/haptophyte specific, respectively, and each is derived from cytohesins. We also identified a cytohesin-derived subfamily, termed ankyrin repeat-containing cytohesin, that independently evolved in amoebozoans and members of the SAR and haptophyte clades. Building on evolutionary data for the ARF family GTPases and their GTPase-activating proteins allowed the generation of hypotheses about ARF GEF protein function(s) as well as a better understanding of the origins and evolution of cellular complexity in eukaryotes.
INTRODUCTION
The eukaryotic membrane trafficking system is a network of interconnected organelles, each maintaining distinct protein and lipid compositions. Bidirectional membrane trafficking in all eukaryotic cells can be thought of as originating either at the endoplasmic reticulum (ER), for anterograde or secretory traffic, or at the plasma membrane for retrograde or endocytic traffic (Bonifacino, 2014). Whereas some other eukaryotic organelles are derived from ancient symbioses (e.g., mitochondria and plastids, with α-proteobacteria and cyanobacteria, respectively), the membrane trafficking system is believed to have arisen autogenously (Keeling and Koonin, 2014). How a complex network of organelles can arise from a cellular configuration with little to no subcompartmentalization is one of the largest remaining questions in evolutionary biology.
The organelle paralogy hypothesis (OPH) proposes an evolutionary mechanism to explain the complexity of membrane-trafficking organelles (Dacks and Field, 2007). The process of membrane trafficking is divided into two essential phases: vesicle formation, whereby cargo destined for transport are selected and packaged into membrane-bound vesicular carriers, and vesicle fusion, whereby the vesicles fuse and deliver their cargo to target organelles (Springer et al., 1999). Each of these processes depends on molecular components that are increasingly well understood, although far more complicated than originally envisioned. Although first described in animal and fungal model organisms, comparative genomic and phylogenetic analyses have demonstrated the conservation of much of the molecular machinery responsible for vesicle formation and fusion in diverse eukaryotes and by deduction in the last eukaryotic common ancestor (LECA) (Dacks and Field, 2018). The basis of the OPH is the observation that a limited number of paralogous protein families govern the major steps of membrane trafficking, with different paralogues from each family carrying out essentially the same functions at different steps along the route. The OPH postulates that gene duplication, followed by sequence divergence and coevolution of interacting members of different families to create preferential partnerships between organelle-specific paralogues, would have facilitated the emergence of novel organelles (Dacks and Field, 2007).
Regulatory GTPases control or modulate a wide array of cellular systems and are central to cellular responses to extracellular stimuli, maintenance of homeostasis, and communication between different parts of the eukaryotic cell (Cherfils and Zeghouf, 2013). A critical element of the vesicle formation process is the action of ADP-ribosylation factors (ARFs) and their regulators (Kahn, 2009; Donaldson and Jackson, 2011). ARFs are ∼21 kDa, monomeric GTPases that nucleate vesicle formation on organellar membranes (Kahn et al., 2006). Like all regulatory GTPases, ARFs cycle between active GTP-bound and inactive GDP-bound states. This cycling is mediated by two classes of regulatory proteins. ARF guanine nucleotide exchange factors (ARF GEFs) activate ARF signaling by increasing the rate of release of bound GDP from its target GTPase, resulting in a GEF-(apo) GTPase intermediate that dissociates on the binding of GTP (Shin and Nakayama, 2004; Zeghouf et al., 2005; Casanova, 2007). ARF GTPase-activating proteins (ARF GAPs) bind specifically to the activated (GTP-bound) form of ARFs, increase the rate of GTP hydrolysis, and thereby can terminate ARF signaling (Kahn et al., 2008; Spang et al., 2010; Vitali et al., 2017). GAPs and GEFs are important components of ARF signaling pathways as ARFs release GDP very slowly in the absence of a GEF and hydrolyze them either not at all or very slowly in the absence of a GAP. GEFs are the initiators of this essential signaling and as such play fundamental roles in an equally large fraction of all cell biology (Cherfils and Zeghouf, 2013). GEFs act on specific subsets or families of regulatory GTPases and themselves comprise several families and subfamilies. ARF GAPs also, somewhat paradoxically, typically serve as effectors as well as signal terminators (Zhang et al., 1998; East and Kahn, 2011). Thus, these modulators are essential components that provide temporal and spatial resolution to signaling by this essential family of regulatory GTPases.
In animals, there are six ARF paralogues (ARF1-6) that share a high degree (>65%) of primary sequence identity. The LECA has been reconstructed to have possessed only a single ARF (Li et al., 2004). Similarly, comparative genomic analyses sampling the breadth of eukaryotes allowed the discrimination of 11 subfamilies of ARF GAPs, of which six were present in the LECA, two were animal-specific, one was opisthokont-specific, and one was lost in opisthokonts (Schlacht et al., 2013). To gain a more complete understanding of the evolution of the ARF system, a complementary analysis of the ARF GEFs is needed.
ARF GEFs have been identified, and in fact are defined, by the presence of the Sec7 domain (Chardin et al., 1996; Cherfils et al., 1998). Note that, while we use the terms ARF GEF and Sec7 domain-containing proteins interchangeably herein, only a very few of the proteins analyzed have been shown to possess ARF GEF activity, and the specificities of even those for different GTPases are incompletely characterized. Given the size and complexity in domain organization among the ARF GEFs, it is quite likely that at least some have other activities and functions in cells, although this issue is not explored further here. In addition, the presence (or absence) of domains outside Sec7 have been shown to play critical roles in regulating the GEF activity, in recruitment of the protein to its site of action, and in binding to lipids and other proteins (Chantalat et al., 2004; Casanova, 2007; Malaby et al., 2013; Wright et al., 2014). There remains much unexplored diversity in domain organization and this comparative genomics analysis is the first step to uncovering novel ARF GEF function(s).
The Sec7 domain is ∼200 amino acid residues in length, encodes the nucleotide exchange activity, and is the target of the fungal toxin brefeldin A (BFA) (Peyroche et al., 1996). BFA is used extensively in the functional dissection of ARF pathways as it is membrane-permeant, rapidly acting, readily reversible, and appears to be a specific inhibitor of a subset of ARF GEFs that act at the Golgi (Fujiwara et al., 1988). The mechanism of BFA action involves its stabilization of an ARF-GDP-Sec7 domain complex and structural studies identified the key residues within the Sec7 domain that determine its sensitivity or resistance to BFA (Peyroche et al., 1999), a basis on which the six animal ARF GEFs have been classified.
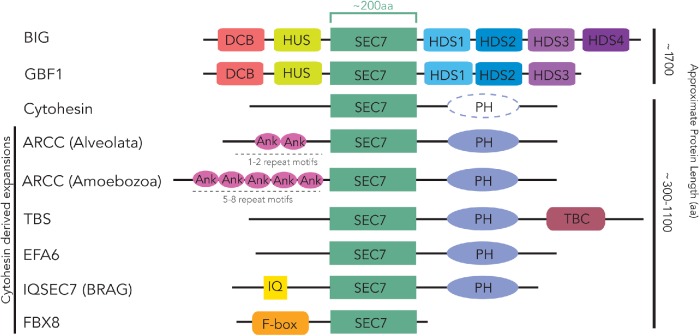
The paradoxically named Golgi BFA-resistant factor 1 (GBF1) is in fact sensitive to BFA but was first cloned in a screen that overexpressed the protein and conferred BFA resistance to the cells (Claude et al., 1999). GBF1 has previously been shown to be pan-eukaryotic (Cox et al., 2004; Bui et al., 2009). GBF1 is involved in the ARF-dependent recruitment of COPI to the cis-Golgi and the ERGIC (ER-Golgi intermediate compartment) and is able to interact with both class I (ARF1-3) and class II (ARF4, 5) ARFs in mammalian cells (Zhao et al., 2006; Bouvet et al., 2013; Jackson, 2014). In Arabidopsis thaliana, the GBF1 homologue GNOM localizes to endosomes (Geldner et al., 2003). Rather than this representing differences in GBF1 functions between organisms, it is likely that GBF1 (and perhaps all ARF GEFs) localizes to multiple sites in cells and that the fractional occupancy at any one site differs between organisms or cell types. The other subfamily of BFA-sensitive ARF GEFs is the aptly named BFA-inhibited GEFs (BIGs). BIGs have also been found in diverse eukaryotic taxa (Cox et al., 2004; Bui et al., 2009; Mouratou et al., 2005) and are involved in regulating ARF-dependent trafficking at the trans-Golgi network (TGN) and at recycling endosomes (Shinotsuka et al., 2002a,b). GBF1 and BIG proteins in animals possess characteristic domain organizations (Figure 1). Dimerization and cyclophilin binding (DCB) and homology upstream of Sec7 (HUS) domains are found upstream of the Sec7 domain, and homology downstream of Sec7 (HDS) 1, 2, and 3 are downstream of the Sec7 domain in both proteins, while an additional HDS (HDS4) is present in BIGs (Mouratou et al., 2005).
FIGURE 1:
Ancestral configuration of domains in the ARF GEF subfamilies. Conserved domains present in and that help define each ARF GEF subfamily are shown. These represent the configurations likely found in the ancestral sequence of each subfamily. The domain organization of ARF GEF proteins found in the human proteins is conserved across the entire distribution of each subfamily and represents the domains present in the earliest ancestor of each subfamily. Dotted representation of the cytohesin PH domain indicates frequent independent loss of this domain. Non-opisthokonta ARF GEFs, TBS and ARCC, are also shown, based on their consistent distribution in multiple amoebozoan and SAR lineages. Ank, Ankyrin repeat; DCB, dimerization and cyclophilin-binding domain; F-box, F-box domain; HDS1, homology downstream of Sec7 1; HDS2, homology downstream of Sec7 2; HDS3, homology downstream of Sec7 3; HDS4, homology downstream of Sec7 4; HUS, homology upstream of Sec7; IQ, IQ motif; PH, pleckstrin homology domain; Sec7, Sec7/ARF GEF catalytic domain; TBC, Tre-2/Bub2/Cdc16 Rab GTPase domain. Linear depictions of each subfamily and domains are not drawn to scale, with approximate lengths indicated on the right. The number of Ank repeats varies, as indicated.
The BFA-resistant subfamilies of GEFs are the cytohesins, the BFA-resistant ARF GEFs (BRAGs, also known as IQSEC7 [IQ and Sec7 domain-containing]), exchange factor for ARF6 (EFA6), and F-box only protein 8 (FBX8). Some cytohesins have previously been named ARF guanine nucleotide site opener (Chardin et al., 1996) or general receptors for phosphoinositides 1; herein, we exclusively use the term cytohesin. Because the BRAG terminology is more prominent in the literature than is IQSEC7, we will occasionally use both (BRAG/IQSEC7) for clarity. Similarly, we prefer the EFA6 nomenclature to that of PSDs (PH and Sec7 Domain), as it is far more common in the ARF GEF field, and the acronym PSD is also used for the unrelated protein phosphatidylserine decarboxylase. Cytohesins localize primarily to the cell periphery, where they act as GEFs to activate several ARFs, most notably perhaps ARF6, but also others by recruitment of the ARFs to the plasma membrane via their PH domains (Macia et al., 2001; Cohen et al., 2007). At the plasma membrane they are involved in the docking and fusion of secretory granules, the endocytosis of G-protein coupled receptors, and have important roles in integrin-mediated cell adhesion and movement (Claing et al., 2000; Geiger et al., 2000; Liu et al., 2006). Cytohesins and IQSEC7s have each been identified in metazoa with the latter only found there (Cox et al., 2004). IQSEC7s localize to the plasma membrane where they regulate the endocytosis of specific cargo and interact with ARF6 (e.g., β-integrins; Someya et al., 2001; Dunphy et al., 2006). EFA6 proteins are also localized to the plasma membrane and interact with both ARF1 and ARF6, serving to coordinate endocytosis, cytoskeletal dynamics, and maintenance of cellular junctions (Frank et al., 1998; Franco et al., 1999; Macia et al., 2001; Klein et al., 2008; Padovani et al., 2014). Cytohesins, EFA6, and IQSEC7s share the domain architecture of a Sec7 domain followed by a C-terminal PH domain, with IQSEC7s also possessing an N-terminal characteristic IQ domain (Figure 1). FBX8 is probably the least understood ARF GEF. It functions as part of a multisubunit ubiquitin-ligase complex, resulting in the suppression of ARF6 activity through its ubiquitination and subsequent degradation (Kipreos and Pagano, 2000; Cox et al., 2004; Yano et al., 2008). It is also unique in possessing an F-box domain N-terminal to the Sec7 domain, but no additional C-terminal domains (Figure 1).
A phylogenetic analysis of ARF GEFs was published previously (Cox et al., 2004), although at that time far fewer genomes had been sequenced and thus those analyses were performed on a far smaller collection of proteins from fewer species. Here, we have taken advantage of the large number of sequenced eukaryotic genomes to carry out detailed comparative genomic and phylogenetic analyses of Sec7 domain-containing proteins.
RESULTS
Comparative genomic analysis of ARF GEF proteins identifies three subfamilies present in the LECA
Previous phylogenetic analyses of the ARF GEF family were largely restricted to model organisms belonging to the supergroup opisthokonta (Cox et al., 2004) or to a subset of the ARF GEFs known at that time (Bui et al., 2009). To establish the range of eukaryotes in which each ARF GEF subfamily is found, we searched for proteins carrying the Sec7 domain in a representative set of 77 eukaryotic genomes from organisms that span the full taxonomic breadth of eukaryotes (Supplemental Table S1). The phylogenetic placement of these lineages on the eukaryotic tree of life is supported by numerous robust phylogenomic studies and previously described, unique lineage-specific morphological and molecular adaptions exist which define each major lineage (Adl et al., 2019).
We identified a total of 506 Sec7 domain-containing proteins (Supplemental Table S2). These proteins were initially classified based on the reciprocal best hit (RBH) method using the five-order and two-order e-value criteria for classification as reported in our previous analysis of ARF GAPs (Schlacht et al., 2013). These criteria require that a BLAST query retrieve a member of one of the human subfamilies as its top hit with an e-value at least two orders, or more stringently five orders, of magnitude better than the top sequence of the next best subfamily. This adds a level of additional rigor to the standard RBH protocol that is based solely on top reciprocal hit. Initial classification was further aided by additional domain analyses using the full-length sequences. For example, this allowed for further classification of BIG and GBF1 family members based on the presence of DCB, HUS, and HDS domains, or of cytohesins if the protein contained one or more C-terminal PH domains. Other domains were diagnostic for ARF GEFs IQSEC7 or FBX8: if a C-terminal PH and N-terminal IQ motif or F-box domains were present, respectively. C-terminal PH or Tre-2/Bub2/Cdc16 (TBC) domains sorted proteins into EFA6 or TBS (TBC-Sec7) proteins, respectively (Mouratou et al., 2005). This sorting into the previously described subfamilies proved to be unambiguous, because although there were instances where domains were absent, no Sec7-domain-containing proteins were found to have different combinations of these other domains, with the exception of the ankyrin (Ank) repeat-containing cytohesin (ARCC) subfamily described below.
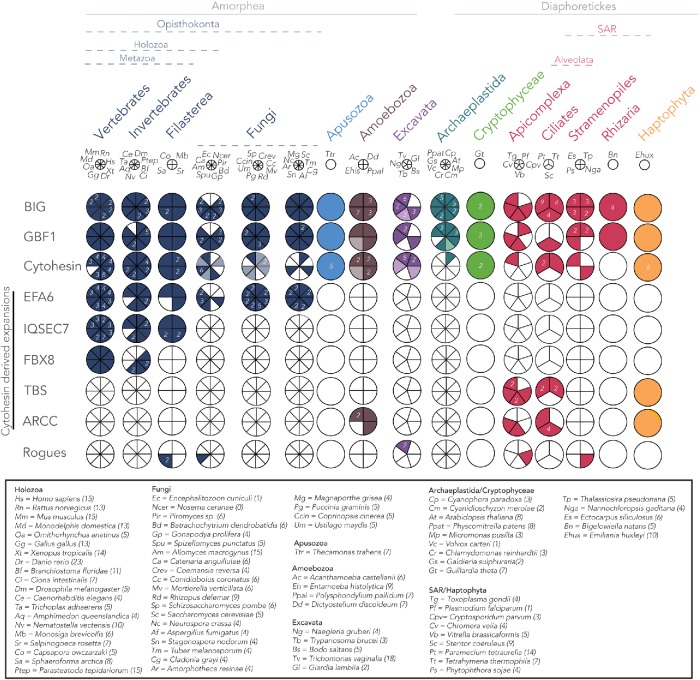
Of the 506 Sec7 domain-containing sequences identified, 480 were readily classified into one of the six ARF GEF subfamilies found in humans based on the criteria just described. The remaining 26 Sec7 domain-containing sequences could not be confidently ascribed to a specific subfamily in this way. To mitigate the evolutionary distance between the taxa in question and Homo sapiens, subsequent analysis using the same criteria but using the non-redundant (nr) database at the National Center for Biotechnology Information (NCBI) as the database for the reverse BLAST search allowed classification of 16 additional proteins as putative BIG, GBF1, or cytohesin, leaving the 10 remaining proteins as unclassified “Rogues” or as hypothetical proteins within the genomes. Subsequent phylogenetic analyses, based on RBH top hit retrieval, allowed resolution of three more proteins that were eventually determined to be fungal Syt proteins (see below), leaving only seven Sec7 domain proteins that failed to reciprocally retrieve any ARF GEF from H. sapiens or lineages deposited in the NCBI nr database. The presence or absence of ARF GEF subfamilies found across eukaryotes is shown in Figure 2.
FIGURE 2:
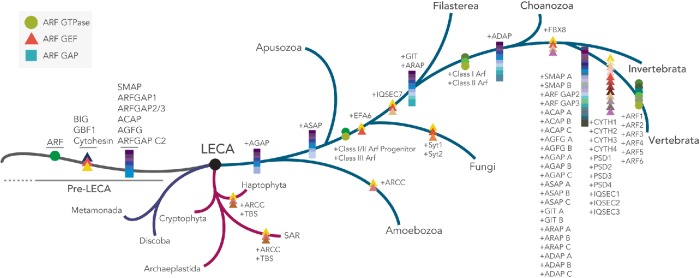
Coulson plot depicting distribution of ARF GEF subfamilies across eukaryotic taxa. Three ARF GEF subfamilies are present in at least four eukaryotic supergroups, indicating their presence in the LECA. Opisthokonta-specific families include EFA6, IQSEC7, and FBX8. EFA6 is found only in holozoa and fungi, with numerous paralogues present in both lineages. FBX8 is specific to the metazoa but is missing from some early-diverging animals. Large taxonomic groups are color coded. Sectors with solid colors indicate orthology based on a combination of RBH, domain structure, and phylogenetic analyses while light colors indicate putative orthology. The presence of multiple paralogues are indicated by numbers within each sector. The key for the species name abbreviations is boxed. The total number of Sec7 domain-containing proteins identified in each organism is indicated in brackets. Eukaryotic classification is based on Adl et al. (2019).
Representatives of BIG, GBF1, and cytohesin subfamilies were found across a wide array of eukaryotes. BIGs and GBF1s were found in all major eukaryotic lineages, whereas cytohesins were also widely distributed but slightly less universal in their distribution and with independent losses frequently occurring at various points within individual lineages (Figure 3). These results demonstrate that at least three subfamilies (GBF1, BIG, and cytohesin) were present in the LECA.
FIGURE 3:
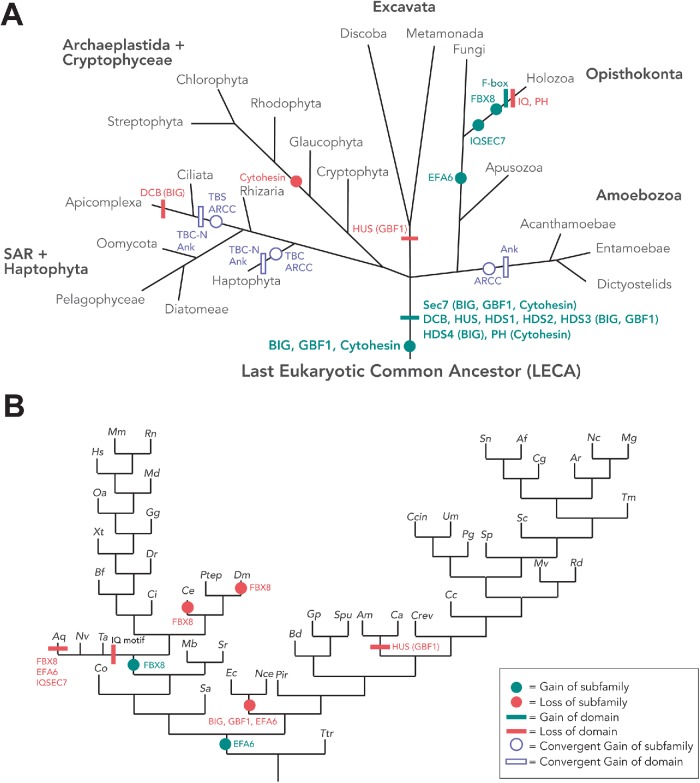
Gain and loss of other domains among the Sec7 domain-containing subfamilies in eukaryotes. (A) The tree of eukaryotes depicting the ARF GEF subfamilies and domains present in the LECA, as well as gains and losses of additional domains and subfamilies. To ensure confidence in the predictions, losses are only proposed when two instances of loss have occurred in the relevant lineage. The open symbol depiction of the ARCC and TBS and their corresponding Ank-repeat and TBC-N domains, respectively, depicts putative origins of these families in the haptophyte E. huxleyi, alveolata, and amoebozoa. Phylogenetic analyses point toward the convergent evolution of these families with the acquisition of Ank repeats in amoebozoa, haptophytes and alveolates, and TBC-N domain in haptophytes and alveolates independently. (B) Gain and loss of ARF GEF subfamilies and domains in opisthokonts. The symbol legend is shown in the inset in B.
Intrasubfamily duplications occurring prior to the LECA still remained a possibility, meaning that more than one paralogue of these subfamilies might have been present in the LECA. In an attempt to determine how many BIG, GBF1, and cytohesin proteins were present in the LECA, separate phylogenetic analyses of each family were carried out. Initially, a Sec7-only phylogeny with both GBF1 and BIG was carried out. However, no backbone resolution was observed (data available on request) and tree building for BIG and GBF1 sequences were carried out separately, again using the Sec7 domain (Supplemental Figure S1). Our analyses found no evidence of pre-LECA gene duplications for BIG (Supplemental Figure S1A), GBF1 (Supplemental Figure S1B), or cytohesin (Supplemental Figure S2), suggesting that only one copy of each of these genes was present in the LECA. Interestingly, we also observed vertebrate-specific expansions in both the BIG (Supplemental Figure S1A) and cytohesin subfamilies (Supplemental Figure S2B), whereas GBF1 (Supplemental Figure S1B) has remained as the sole member of its subfamily. Notably, we also observed that cytohesins were lost near the base of the archaeplastida and are found only in the basal archaeplastidan lineage Cyanophora (a glaucophyte algae), as well as the cryptophyte algae Guillardia theta, a representative from a major eukaryotic lineage which may share a relationship with the archaeplastids (Figure 2).
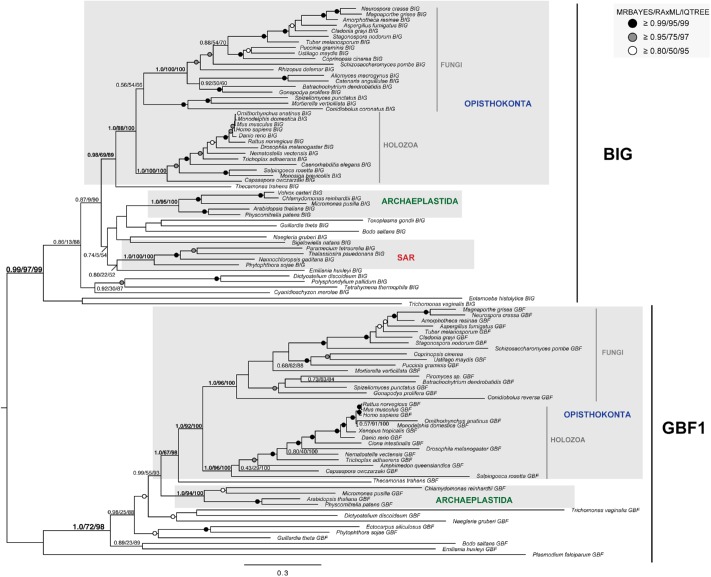
We also performed a phylogenetic analysis of full-length sequences of BIG and GBF1 together in order to validate the orthologue classifications as the proteins share predominantly common domain architectures. The results from these analyses showed two separate and well-supported clades (clade support 0.99/97/99 by MRBAYES, RAxML, and IQTREE) (Figure 4). This also clearly implies that there are distinct residues in each of the non-SEC7 domains that distinguish GBF1 and BIGs from one another (alignment available on request). Given that the functions of these domains are unknown, the distinguishing residues might be a jump-off point for future molecular cell biological analysis. We observed expansions in GBF1 within embryophytes (Supplemental Figure S1B) and in BIGs in the Viridiplantae (Supplemental Figure S1A; Figure 5). Together with the loss of cytohesins in the archaeplastida, these expansions represent an evolutionary remodeling of the ARF GEF signaling system in this major lineage of eukaryotes.
FIGURE 4:
Phylogenetic resolution of pan-eukaryotic BIG and GBF1 families revealed that a single paralogue of each was present in the LECA. The BIG and GBF1 clades were well resolved as separate but without internal structure, suggesting the presence of a single BIG and a single GBF1 pre-LECA. Notably, single well-supported clades of major eukaryotic lineages were found for both BIG and GBF1. The best Bayesian topology is shown. Numerical values represent Bayesian posterior probabilities (MRBAYES)/maximum-likelihood non-parametric bootstrap values (RAxML)/ultrafast bootstrap values (IQTREE). Nodes of interest are in bold. Values for other supported nodes have been replaced with symbols: closed dark circle ≥0.99/95/99, closed light circles ≥0.95/75/97, and open circles ≥0.80/50/95.
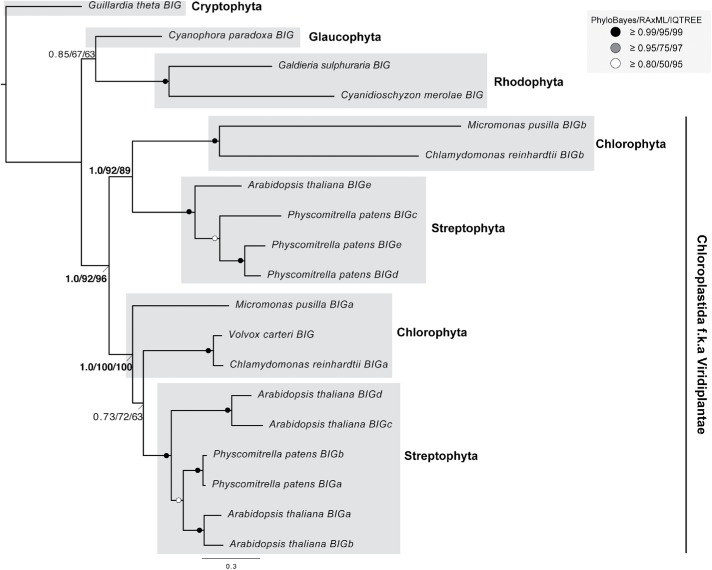
FIGURE 5:
Phylogenetic analysis of BIG proteins in archaeplastida. Phylogenetic analysis of identified archaeplastid BIG paralogues robustly supports a duplication of BIG at the base of the Viridiplantae. The best Bayesian topology is shown. Numerical values represent Bayesian posterior probabilities (PhyloBayes)/maximum-likelihood non-parametric bootstrap values (RAxML)/ultrafast bootstrap values (IQTREE). Abbreviation: f.k.a., formerly known as. Nodes of interest are in bold. Values for other supported nodes have been replaced with symbols: closed dark circles ≥0.99/95/99, closed light circles ≥0.95/75/97, and open circles ≥0.80/50/95.
From our analyses of GBF1 and BIG proteins, including our manual inspection of alignments containing both BIG and GBF1 sequences (Figure 4), we found their domain organization to be highly conserved across eukaryotes, suggesting its presence in the LECA (Supplemental Table S2; Figure 1). The only notable exception is the loss of the HUS domain in GBF1 proteins from the excavates (Figure 3A). The only significant and conserved difference in domain organization between the two ARF GEF subfamilies is that all identified BIG orthologues possess the characteristic HDS4 domain with no lineage-specific losses of this domain while the HDS4 domain was absent in all pan-eukaryotic GBF1 paralogues identified (Supplemental Table S2). Of note, BLAST searches could not identify the HUS or HDS domains in any protein families other than the GBF1 and BIGs. The conservation of the non-SEC7 domains of these two proteins subfamilies, in both their organization and their near ubiquity, raises questions about their origins and function in eukaryotes that are beyond the scope of this work, but certainly of interest.
This conservation is in striking contrast to the domains found in the cytohesin-derived ARF GEFs. Cytohesins are characterized by a PH domain C-terminal to the Sec7 domain (Supplemental Table S2; Figure 1). This organization is observed in the span of identified cytohesin proteins, albeit with more sporadic conservation of the PH domain in many organisms (Supplemental Table S2; Figure 1), as compared with the strict conservation of the PH domain when present in other subfamilies (EFA6, TBS, and ARCC). In cytohesins, this PH domain is critical for recruitment to membranes and binding to specific phospholipids so one might speculate that differences in the lipid composition of membranes among species could contribute to this less stringent conservation of the PH domain in the cytohesins (Klarlund et al., 1997; Paris et al., 1997; Klarlund et al., 2000; Cronin et al., 2004; Oh and Santy, 2012).
Phylogenetic analyses of the opisthokonta-restricted ARF GEFs reveal multiple cytohesin-derived expansions
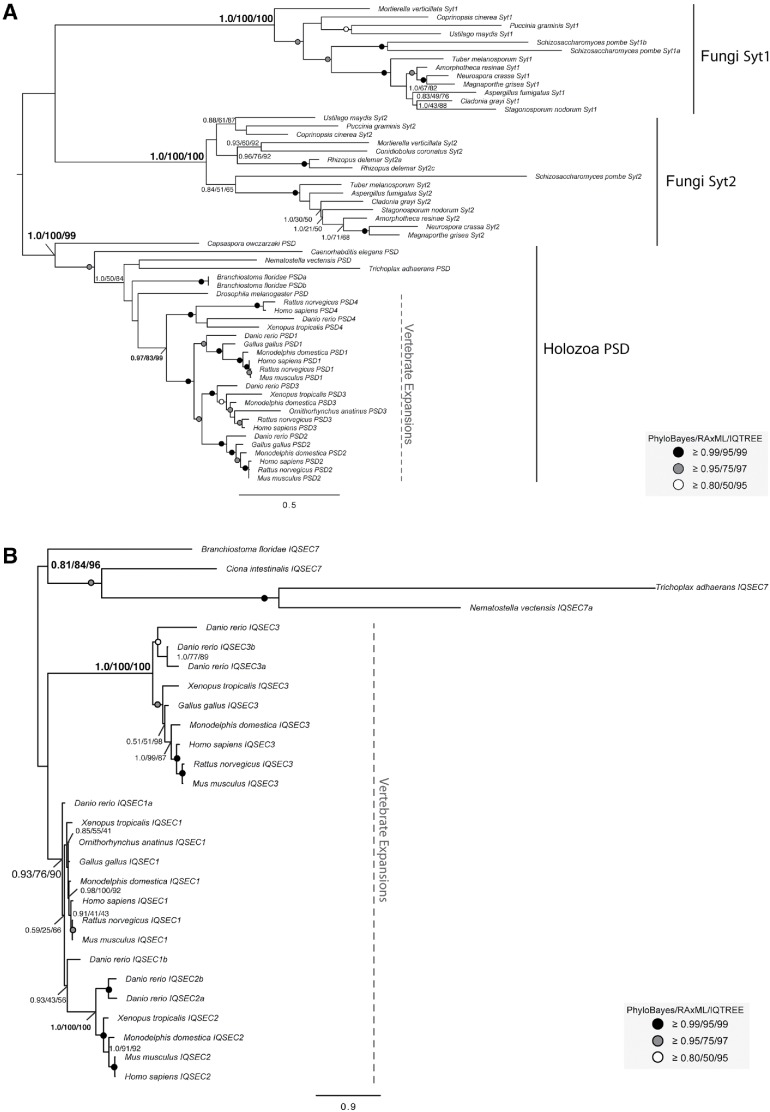
In contrast to the three most broadly conserved subfamilies (BIG, GBF1, and cytohesin), we observed only proteins from the other three previously described subfamilies of ARF GEFs (EFA6, IQSEC7/BRAGs, and FBX8) in genomes from the opisthokonta supergroup, the eukaryotic supergroup consisting of animal and fungal lineages. Previous studies have proposed EFA6 to be present only in metazoa (vertebrates and invertebrates) (Cox et al., 2004). However, in that same analysis, the Saccharomyces cerevisiae Syt1p and Yel1p proteins were also speculated to be fungal EFA6 homologues because of a region of high sequence similarity and overall domain architecture shared with EFA6 (Cox et al., 2004; Gillingham and Munro, 2007). Our homology searches using EFA6 queries found orthologues in animals and in fungi (Syt1 and 2), the latter regularly retrieved H. sapiens EFA6/PSD proteins as top reciprocal BLASTp hits (Supplemental Table S2) supporting the previously hinted at orthology between these two protein families (Cox et al., 2004). This result was further validated by our phylogenetic analysis (Figure 6A). Therefore, we hereby demonstrate that the EFA6 subfamily encompasses PSDs in metazoa and Syts in fungi as well as orthologous sequences from opisthokont taxa and that the EFA6 subfamily likely originated at the base of the opisthokonta (Figure 3A). Subsequent independent duplications in the metazoa and fungi gave rise to PSD 1–4 and Syt 1–2, respectively (Figure 6A; see below).
FIGURE 6:
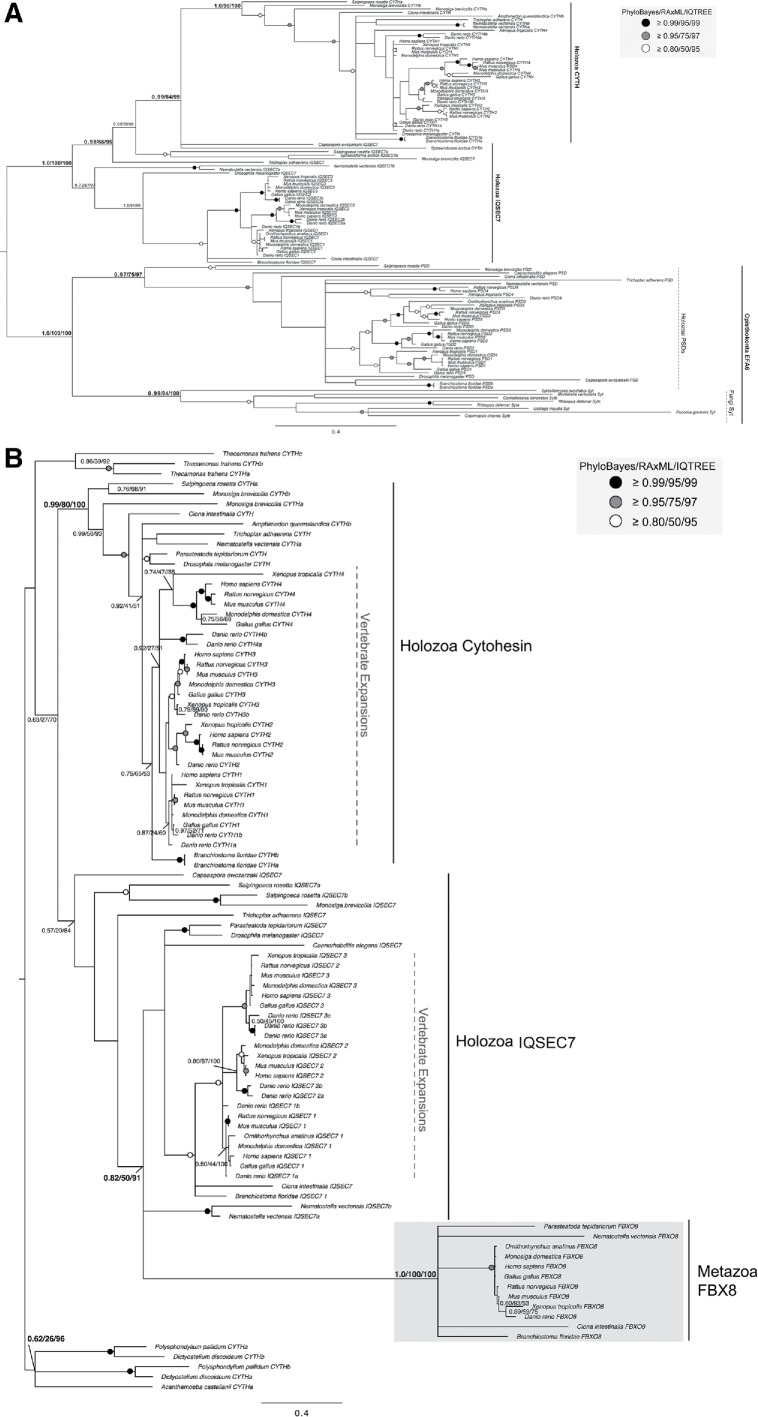
Phylogenetic analyses of the opisthokont-specific cytohesin-derived proteins. (A) Phylogeny of the opisthokonta EFA6/PSD, IQSEC7, and cytohesin (CYTH) proteins reveal evolutionary origins of the IQSEC7 subfamily from cytohesins at the base of filasterea. Additionally, the tree shows support for the monophyly of the holozoan EFA6/PSD proteins and Fungal Syt proteins suggesting common evolutionary origins of the EFA6 family at the base of opisthokonta. (B) Phylogeny of the amorphea (Amoebozoa and Opisthokonta) IQSEC7, cytohesin, and FBX8 proteins analysis supports the conclusion that FBX8 proteins evolved from IQSEC7s in holozoa. The best Bayesian topology is shown. Numerical values represent Bayesian probabilities (PhyloBayes)/maximum-likelihood non-parametric bootstrap values (RAxML)/ultrafast bootstrap values (IQTREE). In both instances, nodes of interest are in bold. Values for other supported nodes have been replaced with symbols: closed dark circle ≥0.99/95/99, closed light circles ≥0.95/75/97, and open circles ≥0.80/50/95.
By contrast, our comparative genomic analyses of the IQSEC7/BRAG proteins showed a more restricted distribution within holozoa, a lineage consisting of filasterea (single-celled, basal branching, relatives of animals), invertebrates, and vertebrates, suggesting origins of this subfamily prior to filasterea but following the split from fungi (Figures 2 and 3). Last, the FBX8 subfamily is the most restricted in its distribution of the identified ARF GEFs, being found only in vertebrates and a subset of invertebrates, while it is missing from others (Trichoplax adhaerens, Amphimedon queenslandica, Caenorhabditis elegans, and Drosophila melanogaster) (Figures 2 and 3B). Notably, the presence of an FBX8 orthologue in spiders (Parasteatoda tepidariorum) confirms the independent loss in D. melanogaster.
To determine from which of the pan-eukaryotic subfamilies each of these opisthokont or metazoan-specific proteins arose, we relied on information derived from the RBH analysis, as described under Materials and Methods. Specifically, we investigated which subfamily was returned as the next best hit after the orthologous protein was retrieved. In all cases, human cytohesins were the top scoring candidates retrieved, consistent with the cytohesins as the precursor to these other subfamilies. This is also consistent with the similarity in domain architecture between EFA6, IQSEC7 and cytohesins in all possessing C-terminal PH domains (Figure 1). This was not the case for FBX8, as it shares no domains other than Sec7 with cytohesin, BIG, or GBF1 (Figure 1). Thus, our data indicate not only that cytohesins were present in the LECA but also that they are the likely progenitors of three other subfamilies that emerged in opisthokonts (IQSEC7/BRAG, EFA6, and FBX8) and share similarities in domain organization, while being clearly distinct in origin and domain organization from GBF1 and BIGs.
To further investigate the timing and provenance of these putative cytohesin-derived subfamilies, we undertook additional phylogenetic analyses. The distribution of EFA6 proteins in both holozoa (filasterea and metazoa) and fungi, as well as the presence of multiple paralogues in each of these lineages, prompted further investigations to determine when these duplications took place. Our phylogenetic analyses show that the fungal and the holozoan EFA6 proteins are related to one another forming a separate monophyletic clade (clade support 1.0/100/100 by PhyloBayes, RAxML, and IQTREE; Figure 6A). This suggests that an ancestral duplication event from an opisthokont cytohesin protein took place to yield the subfamily, EFA6. Additionally, we found that fungal EFA6 underwent a duplication to yield proteins Syt1 and Syt2 and an expansion within the holozoa (Figure 7A). The EFA6 subfamily (called PSD in animals) underwent duplications to yield paralogues PSD1–4 in vertebrates, including in humans (Figures 6A and 7A).
FIGURE 7:
Phylogenetic examination of holozoan cytohesin-derived family paralogues reveals multiple expansion events. (A) Phylogeny of the holozoan and fungal EFA6 proteins identify two separate clades. A duplication event in the EFA6 proteins at the base of opisthokonta yields holozoan PSD proteins and fungal Syt proteins. Further duplications in the EFA6/PSD subfamily have occurred with the transition into metazoa to yield PSD1-4 and in the fungal Syt to yield Syt1 and Syt2. (B) Phylogenetic analysis of IQSEC7 paralogues from holozoa was carried out to determine when the duplication events producing the three vertebrate paralogues occurred. The analysis reveals two duplications at the base of vertebrates, resulting in three paralogues, with IQSEC7 3/BRAG3 diverging first, followed by IQSEC7 1/BRAG1 and IQSEC7 2/BRAG2. The best Bayesian topology is shown. In both instances, numerical values represent Bayesian probabilities (PhyloBayes)/maximum-likelihood non-parametric bootstrap values (RAxML)/ultrafast bootstrap values (IQTREE). Nodes of interest are in bold. Values for other supported nodes have been replaced by symbols: closed dark circle ≥0.99/95/99, closed light circles ≥0.95/75/97, and open circles ≥0.80/50/95. The tree was rooted at filasterean IQSEC7 sequences.
Based on its taxonomic distribution and domain structure, the IQSEC7/BRAG subfamily could have been derived from either cytohesin or EFA6. However, our phylogenetic analyses reveal that IQSEC7 originates as a single orthologue at the base of holozoa, giving rise to the IQSEC7 family, forming a sister clade within the tree to the clade of opisthokont EFA6 sequences (Figure 6A). This means that it is derived from cytohesin directly and not from holozoan EFA6. Cytohesins were also separately expanded to yield cytohesins 1–4. Additionally, an expansion within the IQSEC7 subfamily in metazoan lineages yields three paralogues, IQSEC1, IQSEC2, and IQSEC3 (Figures 6B and 7B). Finally, we show that FBX8 arose from a duplication event within IQSEC7s at the base of invertebrates (tree support of 0.82/50/92 by PhyloBayes, RAxML and IQTREE, respectively; Figure 6B). Our RBH analyses of FBX8 proteins consistently retrieve H. sapiens IQSEC7s as top non-orthologous hits (Supplemental Table S2), despite lacking shared domains other than Sec7. The duplication event that gave rise to FBX8 was accompanied by significant domain modification whereby a loss of the canonical IQSEC7 IQ domain was replaced with an F-box domain. The acquisition of this domain is correlated with a distinctive gain of function, as FBX8 is thought to function in ubiquitination of ARF6 via the interaction of its F-box domain with the SCF ubiquitin E3 ligase complex (Yano et al., 2008). Unlike the EFA6/PSD and IQSEC7 proteins, which underwent duplications to yield multiple paralogues, only single copies of the FBX8 proteins were identified in vertebrates. Thus, the FBX8 subfamily provides yet another example of flexibility within the cytohesin family to undergo modifications and acquire new functions.
In analyzing the EFA6 homologues, the PH domain was included in the sequences used as search queries to help with the classification, potentially biasing our analysis against observing EFA6 sequences lacking the PH domain. Acknowledging this caveat to the analysis, nonetheless, we did not find any candidate EFA6 orthologues that did not possess a PH domain, even at a low e-value cutoff, which we did observe for cytohesin. By contrast, only the Sec7 and PH domains were used to classify the IQSEC7 and FBX8 proteins, allowing for an analysis of other domain architectures within these proteins without this potential bias. We found that the majority of the proteins classified as IQSEC7s possess identifiable N-terminal IQ- motif and C-terminal PH domains. Similarly, all FBX8 orthologues also possess F-box domains N-terminal to the Sec7 domain (Supplemental Table S2).
Comparative genomics of the unclassified ARF GEF proteins identify two subfamilies of cytohesin-derived proteins found exclusively in lineages outside of the animal and fungal model systems
The identification of clear lineage-specific ARF GEF subfamilies in the opisthokonta and holozoa, as well as the presence of Sec7 domain-containing proteins within the amoebozoa and SAR lineages that remained unclassified, prompted us to question whether these might represent additional lineage-specific subfamilies. Investigation of domain architecture in this collection identified additional domains that served as distinguishing hallmarks allowing further analyses and classification into two subfamilies, the previously described TBS and the newly proposed ARCC subfamilies (Figures 1 and 2).
The TBS subfamily (Mouratou et al., 2005) is characterized by a Sec7 domain followed by a PH and TBC Rab GAP domain (Figure 1). The TBC Rab GAPs regulate the activity of Rab GTPases and at least 10 TBC subfamilies were present in the LECA (Gabernet-Castello et al., 2013). The TBS ARF GEF subfamily was first identified in the ciliates Paramecium tetraurelia and Tetrahymena thermophila, as well as the apicomplexan Cryptosporidium, and consequently predicted as alveolate-specific (Mouratou et al., 2005). Our data confirm this prediction with orthologues in all sampled apicomplexan except Plasmodium and report a similar domain fusion in the haptophyte, Emiliania huxleyi. We also supplemented our sampling of alveolates for this protein only and were able to identify clear TBS proteins in the genome of the dinoflagellate Perkinsus marinus. Examination of the Toxoplasma gondii (ToxoDB) and Cryptosporidium (CryptoDB) databases found transcriptomic and proteomic support for these proteins being expressed in those organisms. Top-next BLAST hit analysis shows that TBS proteins are derived from cytohesins and phylogenetic analysis of the Sec7 domain shows that these have arisen at least twice independently, once in the alveolates and once in E. huxleyi (Supplemental Figure S3A). To pinpoint the origin of the TBC domain in the TBS protein, phylogenetic analysis of the TBC domain was also performed and showed that the fused domain is derived from a duplication of TBC-N, one of the 10 previously described LECA-specific TBC proteins (Gabernet-Castello et al., 2013). This analysis was also consistent with the suggestion that there were at least two independent fusions of a Sec7 and TBC-N domain as the TBC portion of TBS proteins from alveolates grouped together and to the exclusion of E. huxleyi (Supplemental Figure S3B).
Finally, our searches using protein queries with the Sec7 domain identified an additional set of ARF GEFs, which RBH analysis classified as cytohesins. This subset, however, possessed a novel domain architecture, consisting of N-terminal Ank repeat domains the Sec7 domain and a C-terminal PH domain and were found only in amoebozoa, alveolata, and the haptophyte E. huxleyi (Figures 1 and 2). We designate this subset of proteins as ARCCs. The presence of Ank repeats in these proteins, identified in these evolutionarily distant lineages, prompted an in-depth analysis of those repeats. We identified amoebozoan lineages to possess five to seven Ank repeat domains while E. huxleyi and alveolate lineages possessed a maximum of two Ank repeat domains (Supplemental Table S2), clearly distinguishing these proteins. To further investigate whether these are orthologous proteins with subsequent losses occurring in other supergroups, we carried out additional analyses. Alveolate and haptophyte ARCCs failed to retrieve amoebozoan ARCCs as reciprocal hits and vice versa in BLASTp searches, suggesting independent emergence of these proteins. Furthermore, analysis of cytohesin and ARCC proteins shows moderate support separating these into two independent clades of alveolate and haptophyte sequences (Supplemental Figure S4). Hence, while it is still technically possible that these proteins in alveolates and haptophytes could be the result of a fused precursor that was lost in stramenopiles and rhizarians, by far the most parsimonious explanation for the origins of these families is independent innovations at the base of amoebozoa, alveolates, and haptophytes, respectively (Figure 3A). Investigation of the relevant public databases (e.g., CryptoDB, CilliateDB, and dictyBase) also showed transcriptomic and proteomic support for ARCC proteins (Supplemental Table S1). On the basis of the presence of the canonical C-terminal PH domain and the RBH analyses, as well as phylogenetic analyses, we conclude that the ARCC proteins represent a novel category of cytohesin-derived ARF GEFs present in diverse eukaryotic lineages (Figure 1).
DISCUSSION
Here we have presented a comprehensive molecular evolutionary analysis of the Sec7 domain-containing subfamilies of ARF GEFs across eukaryotes. We were able to elucidate the encoded ARF GEF complement in an extensive array of 77 eukaryotic genomes and from this deduce the points in eukaryotic evolution at which the various subfamilies arose. In the case of the lineage-specific subfamilies, we were also able to determine their relationships to the pan-eukaryotic ones. We find that GBF1, BIG, and cytohesin ARF GEFs were present in the LECA, while EFA6, IQSEC7, and FBX8 are opisthokont-, holozoan-, and metazoan-specific, respectively, and derived from cytohesins. We further characterize additional cytohesin-derived subfamilies specific to amoebozoans and members of the SAR plus haptophyte clades. Our analyses have yielded insight not only into the evolution of the individual ARF GEF subfamilies but also that of the ARF GEF system as a whole. These, and previously published data on the ARF GAPs and the ARF GTPases themselves, then allow for broad evolutionary perspective on the three classical components of this GTPase system.
Previous molecular evolutionary analyses of ARF GEF proteins, using the set of eukaryotic genomes available at that time (eight eukaryotic genomes), identified GBF1 and BIG as likely present in the LECA (Cox et al., 2004). Here we confirm and extend this analysis using more advanced methodology to enumerate the encoded complement of Sec7 domain-containing proteins in 77 organisms. This complement of over 500 robustly classified proteins provides a jumping-off point for molecular cell biological investigations to better understand organisms that include parasites with major global health relevance (e.g., the causative agents of giardiasis, malaria, and amoebic dysentery), as well as free-living organisms, both single and multicellular, of agricultural and environmental importance. In several of these organisms (e.g., apicomplexans parasites, ciliates), we observed losses of one ARF GEF subfamily but retention or expansion of another, including novel lineage-specific paralogues. Given the utility of BFA and golgicide A as specific inhibitors of a subset of ARF GEFs, it could be attractive to target inhibitor screens for ARF GEFs in these or other pathogens.
Ancient eukaryotic versus lineage-specific ARF GEFs
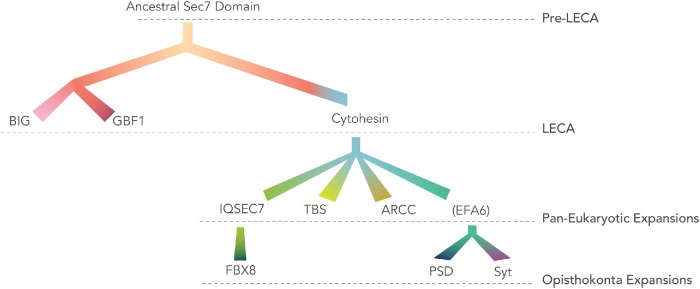
Our analyses have enabled us to resolve the relationships between the Sec7 domain-containing ARF GEFs in eukaryotes (Figure 8). Previous analyses had identified GBF1 and BIG as present in taxa across eukaryotes and consequently as likely present in the LECA. From our analyses, we can conclusively deduce that cytohesin must also have been present in the LECA, along with GBF1 and BIG (Figure 8). While the LECA possessed three major subfamilies, notable plasticity was observed in the complement of the descendent lineages. The archaeplastid complement of ARF GEFs appears to have undergone modification with an early loss of cytohesin after the divergence of glaucophytes from red and green algae, with later duplications of BIGs in the common ancestor of green algae and land plants, and of GBF1 in the ancestor of embryophytes. Because these duplications took place well after the loss of cytohesin, the additional paralogues are unlikely to have provided compensatory activity allowing for the losses, as has been used to explain gene losses in other systems (Sparvoli et al., 2018). However, once the loss of cytohesin was tolerated, possibly in response to the shift from heterotrophy to photoautotrophy, the duplications of the other ARF GEFs could provide additional material for later elaboration of ARF regulatory systems in archaeplastid evolution. This pattern of loss of canonical eukaryotic machinery in basal archaeplastid lineages, with expansion of machinery later in the land plants, echoes even more extreme examples such as those described within the Rab GTPase family (Petrželková and Elias, 2014) and other membrane traffic machinery (Barlow and Dacks, 2018).
FIGURE 8:
Relationships between the ARF GEF subfamilies deduced from domain composition, BLAST, and phylogenetic analyses. All ARF GEFs are derived from an ancestral Sec7 domain-containing protein. BIG and GBF1 form a clade based on RBHs and domain conservation. Domain composition, RBH, and phylogenetics suggest that IQSEC7, EFA6, ARCC, and TBS are derived from cytohesin. Additional derivations from the holozoan IQSEC7 proteins have resulted in FBX8 and while holozoan and fungal PSD/Syt proteins were derived from a common EFA6 family, indicated in brackets to denote this as a term for the clade deduced as present in the last opisthokont ancestor and now encompassing the animal (PSD) and fungal (Syt) names.
The loss of cytohesin in the archaeplastid lineage stands in contrast to the observed diversity and prevalence of cytohesin-derived subfamilies in several other eukaryotic lineages. For example, the TBS subfamily has been previously reported (Mouratou et al., 2005) and here we confirm and extend these findings with the report of TBS proteins in a much larger array of ciliates and apicomplexans, as well as new reports in dinoflagellates and haptophytes (Figures 1 and 3A). The existence of these proteins, at least the apicomplexan complement, is supported by publicly available proteomic and transcriptomic data (CryptoDB and ToxoDB). The observed taxonomic distribution of TBS proteins can be explained most simply by independent fusion events in the alveolates and haptophytes. We were also able to identify for the first time that TBS is definitely the product of the fusion of a Sec7 domain derived from cytohesin and the TBC domain from the TBC-N family of RabGAPs. The presence of these two domains, which both act in the endosomal system, within the same protein, leads us to predict that the TBS proteins act at endolysosomally derived organelles, potentially rhoptries and micronemes in the apicomplexans. Furthermore, the convergent fusion of these domains, each with well-defined functions, leads us to hypothesize that these may serve as sites of crossalk between ARF and Rab GTPase regulated pathways in alveolates and haptophytes in ways that warrant exploration. Previous data from yeast have also argued for such crosstalk between ARF and Rab signaling (e.g., see Jones et al., 1999; McDonold and Fromme, 2014; D’Souza et al., 2014). Indeed, we speculate that the convergent fusion of cytohesin and TBC-N proteins could be hinting at crosstalk between these two proteins more generally, even in organisms where the fusion has not taken place. Crosstalk between ARF GEFs and Rabs was also seen in studies demonstrating roles for a Rab (Rab4) to recruit BIGs (via ARL1) onto early endosomes, where they act on ARF1 and ARF3 (D’Souza et al., 2014), further highlighting both the complexity and the integration of these regulatory GTPase networks.
We were also able to identify a novel set of cytohesin-derived ARF GEFs containing Ank repeats as well as the C-terminal PH domain, termed the ARCCs. This domain organization has most likely evolved convergently on at least three occasions, based on the large evolutionary distance between the two major groups of taxa in which these proteins are found (i.e., the amoebozoans, alveolates, and haptophytes), and the fact that our phylogenetic analyses of the ARCCs in alveolates and the haptophyte, E. huxleyi, showed these as separate clades (Supplemental Figure S4). Ank repeats are ancestral short stretches of alpha helices that have been identified in a large and diverse array of eukaryotic, archaeal, and bacterial proteins (Al-Khodor et al., 2010). Biochemical and biophysical characterization of these repeats suggests their role in protein–protein interactions (Mosavi et al., 2002). The repeated appearance of Ank repeats in ARF GEFs as well as in multiple ARF GAP subfamilies might suggest the possibility that these different types of ARF regulators share common regulatory systems or possibly even partners, although Ank repeats are also present in GAPs and GEFs acting on other families of GTPase, including the Rabs (Schlacht et al., 2013; Herman et al., 2018).
While GBF1, BIG, and cytohesins are found across eukaryotes, the other three ARF GEF subfamilies found in mammals (EFA6/PSD, IQSEC7, and FBX8) are much more restricted in their phylogenetic distribution, all three in fact being lineage-specific duplications of cytohesins just like the TBS and ARCC proteins (Figure 8). We are able to show that EFA6/PSD proteins in animals are orthologous to Syt proteins in fungi, all within the EFA6 subfamily. This means that this protein evolved in the common ancestor of opisthokonts and also that experimental work from Syt1p in yeast is likely relevant to EFA6/PSD protein functions in animals. We also were able to pinpoint IQSEC7 as a holozoan expansion of cytohesin and, surprisingly, that FBX8 was derived from IQSEC7. The latter relation is not predicted from domain composition alone.
Distinct evolutionary dynamics of GBF1/BIG versus cytohesin-derived ARF GEFs
The eukaryotic ARF GEFs appear to be following two distinctive evolutionary dynamics. The GBF1 and BIG subfamilies are highly conserved and rarely lost in eukaryotic genomes. In fact, only in the most highly reduced or divergent parasites (e.g., microsporidians, Giardia, and Plasmodium) did we fail to identify a BIG orthologue, with loss of GBF1 apparently being only slightly more frequent. Such findings point to essential, and distinct, functions for these two subfamilies, despite the sharing of multiple domains outside of the Sec7 domain. Indeed, the domains in BIG and GBF1 are numerous and highly conserved, although distinctive. And yet the domains (DCB, HUS, and HDS) are found in no other eukaryotic proteins. This highly conserved evolutionary dynamic contrasts sharply with the cytohesin family and its derivatives. Here extensive plasticity is observed with losses in some systems, but more strikingly frequent duplication. This, in some cases, yielded multiple cytohesin paralogues (e.g., metazoan, haptophytes, some excavates), but in direct contrast to GBF1 and BIG, yielded novel cytohesin-derived subfamilies through loss and acquisition of novel domains. BIG and GBF1 act in the early secretory system, while cytohesins act in the late secretory and endocytic systems. The overall plasticity of the cytohesin ARF GEFs and derived subfamilies versus the conservation of the GBF1 and BIG subfamilies fits well with observation of other membrane traffic proteins families. That is, in both general surveys and in evolutionary analyses of individual systems or complexes, the proteins that act in the late secretory and endocytic systems tend to be more patchily distributed and show greater evidence of losses and compensatory duplications (Field et al., 2007; Schlacht et al., 2014). For example, COPI and COPII coats are infrequently lost or reduced and at the same time infrequently elaborated on, as compared with the array of coats found to act toward membrane traffic systems in the cell periphery, such as AP3-5, or TSET (Field et al., 2007; Hirst et al., 2014; Schlacht and Dacks, 2015).
Reconstructing the ARF regulatory system in the LECA
The above analyses allow, for the first time, a holistic view of the evolution of an entire GEF–GTPase–GAP system and its reconstruction in the LECA. While previous analyses have pointed to the presence of a single (Li et al., 2004; Berriman et al., 2005) or more likely two (R. Petrželková and M. Eliáš, personal communication) ARF GTPases and of six ARF GAPs in the LECA, the data presented here indicate the presence of three ARF GEFs. This suggests a much simpler cellular configuration of this system in the LECA, as compared with conventionally studied systems (Cox et al., 2004; Kahn et al., 2008) (Figure 9). The presence of one or two ARF GTPase(s) would indicate that these ancestral homologue(s) would have been able to act at multiple organelles, a task that has likely been divided between up to six ARF proteins that may act in pairs or other combinations in mammals (Volpicelli-Daley et al., 2005). This would imply that ARF specificity is not encoded in the ARFs themselves, but rather by the combined actions of the ARF regulators (GAPs and GEFs) and effectors. This may still be the case in extant organisms even with larger numbers of GAPs, GEFs, and GTPases. Alternatively, any organelle specificity encoded by ARFs in extant organisms, such as mammals, may be the result of lineage-specific evolution. The finding that the GAPs outnumber both the GEFs and the GTPases suggests that it is the GAPs that drive the complexity within this system. This is also consistent with the conclusion that ARF GAPs act as both effectors (downstream mediators of the biological signal) and terminators of GTPase signaling (as a consequence of their GTPase stimulating activity) (Zhang et al., 1998; East and Kahn, 2011). Multiple GAPs would greatly increase the potential for crosstalk and integration of signals between parallel pathways but cannot be responsible for the specific targeting of ARFs to membranes, as their association with the GTPase requires the GTP-bound form. Although we cannot say with certainty whether the GAPs or the GEFs expanded first, given the comparative numbers in the LECA, it is tempting to speculate that the GAPs were the first components in this system to expand, followed by the GEFs, and eventually the ARFs themselves. This scenario would indicate a period during which early eukaryotes possessed multiple GAPs, a single GEF, and a single GTPase. Such a scenario might invoke the necessity of additional upstream regulatory components in order to recruit the GEF and the ARF to the appropriate membrane. Hints to this type of regulation have already been observed; the recruitment and activation of the S. cerevisiae Sec7p require interaction with ARL1, Ypt1, Ypt31, and ARF1 (McDonold and Fromme, 2014). Note that although there was only one, or at most two, ARFs in the LECA, there were already more than a dozen members of the ARF family of GTPases, most of which are ARF-likes (ARLs), and, like ARL1 in this example, may be acting in similar pathways to those of the ARFs, despite not being activated by the ARF GEFs. Further analyses are required to confirm the conservation of this regulation in other systems and to identify upstream mechanisms of this nature for other ARF GEFs but could nonetheless provide a basis for the targeting of a single GEF to different membranes in early eukaryotes.
FIGURE 9:
Tracing the expansion of ARF/ARF GEF/ARF GAP proteins across eukaryotes. Predicted evolutionary expansion events in the ARF GTPase and its regulators across eukaryotes with primary focus on vertebrata expansions. Of all eukaryotic lineages, the majority of the ARF GTPase, GEF, and GAP expansion events occurred within opisthokonta with the largest diversity occurring on transition into vertebrates. Schematic exclusively depicts duplication events (and not losses) traced in the current study as well as previous published and unpublished comparative analyses on ARF and ARF GAP evolution across eukaryotes (Schlacht et al., 2013; Manolea et al., 2010).
Identifying the site of action and the in vivo function of the lineage-specific, novel ARF GEFs identified here, as well as the novel ARF GAPC2 described earlier (Schlacht et al., 2013), will greatly enhance our understanding of the range of functions carried out by these ARF regulators. Similarly, further characterization of ARF GEFs in non-standard model systems should identify both novel functionality and conserved roles for ARF GEFs in membrane trafficking. The technological development of such non-standard model systems represents a tremendous opportunity to explore the diversity of membrane-trafficking biology in eukaryotes.
Evolution of ARF GAPs and ARF GEFs in vertebrates mimics the evolution of ARFs
The patterns of ARF GAP and ARF GEF duplication observed in vertebrates are quite similar to the pattern of duplications seen for the ARFs. Manolea et al. (2010) demonstrated that the ancestral opisthokont possessed two ARF proteins, progenitors of the class I/II ARFs and a class III ARF (Figure 9). The class I/II progenitor duplicated near the ancestor of metazoa and choanoflagellates to produce distinct class I and II ARFs (Figure 9) (Manolea et al., 2010). Near the base of vertebrates, these ARFs duplicated again; the class I ARF duplicated twice, producing ARFs 1–3, and the class II ARF underwent a single duplication, producing ARFs 4 and 5 (Figure 9). Although the pattern of ARF GAP subfamilies is more complex, the overall patterns are strikingly similar with all subfamilies, except for ARF GAP1, undergoing one or two gene duplications in the vertebrate ancestor (Schlacht et al. 2013) (Figure 9). Although the correlation is less strong, a similar pattern is observed for the ARF GEFs: IQSEC7 has undergone two duplications at the base of vertebrates, EFA6 and cytohesin have undergone three gene duplications, and BIG has duplicated only once in vertebrates (Figure 9). While in the case of the GEFs, this correlation does not align perfectly with experimentally established substrate preferences, it should be remembered that the majority of ARF–ARF GEF interactions are analyzed from the perspective of their interactions with ARF1 and ARF6. That is, testing of substrate specificities among those Sec7 domain proteins shown to possess ARF GEF activity is limited and quite incomplete (Sztul et al., 2019). One exception would be the interaction of GBF1 with ARF4 and ARF5 at the ERGIC (Chun et al., 2008); however, this may be a special case, as GBF1 has also been shown to localize to the TGN through interactions with ARF1 (Wright et al., 2014).
Our analyses are expected to allow researchers to further dissect the interactions of the GEF regulatory system to generate stronger, testable hypotheses regarding their functions as enzymes and in cells based on proposed evolutionary relationships. In metazoa in particular, the multiple whole genome duplications concurrent with the emergence of vertebrates would have generated multiple paralogues of each of the ARF GEF subfamilies. While there is undoubtedly some degree of contingency at play, the subsequent evolutionary fate of these paralogues could well be indicative of their cellular roles or importance. In some cases, such as the cytohesins and related proteins, the paralogues have been largely retained and cannot simply be explained by tissue-specific expression. This suggests that there may have been some selective benefit to redundancy or subtle diversity of function between the family members and that these subtle differences could be teased out by knockout or complementation approaches. By contrast, the fact that GBF1 has been retained in a single copy could suggest that there was a selection against multiple paralogues, with the protein playing a central role in trafficking that redundancy might have disrupted. This predicts that GBF1 should be very difficult to knockout or knockdown without dire consequences to the cell.
On a larger scale, our work raises predictions about the regulatory network in general. ARF GAPs and ARF GEFs may also act on other proteins, the most obvious would be the ARLs, as the animal ARLs share 40–60% primary sequence identity with animal ARFs. This promiscuity of GAP and GEF action on ARF and ARL GTPases has been suggested to be the case for the S. cerevisiae GEF Syt1p, which is proposed to mediate exchange on ARL1p and for Gcs1p (a S. cerevisiae ARF GAP) acting as a GAP for ARL1p (Liu et al., 2005; Chen et al., 2010), although the much slower turnover rates raise questions about the biological significance of this activity. Further analyses will be required to assess the biological implications of these interactions and to determine whether they represent solitary cases of ARF GAPs and GEFs able to regulate ARLs. The current lack of information regarding ARL GEFs and GAPs is clearly hampering development of stronger and more global models of signaling by the ARF family in general (Sztul et al., 2019). The only known ARL GEF activity described to date is that of ARL13B acting on ARL3 (Gotthardt et al., 2015; Ivanova et al., 2017). As ARL3 at least is predicted to have orthologues in the LECA, it will be interesting to see whether other examples of one GTPase acting directly on another may be found (Li et al., 2004). Much more analysis will be needed to integrate the other ARF-related GTPases (i.e., ARLs and Sar), as well as their regulators, into this story both from an evolutionary and a molecular cell biological perspective.
The best characterized ARL GAPs, the ELMOD proteins, were also present in the LECA and have activity as GAPs for both ARLs and ARFs despite lacking the ARF GAP domain (East et al., 2012; Ivanova et al., 2014). These results clearly demonstrate the potential for ARFs and ARLs to share far more commonality in biochemical and cellular signaling than is currently envisioned. We would be remiss in ending without pointing out the real possibility that this also raises the possibility of there being proteins with ARF GEF activity that lack the Sec7 domain. Such a finding, currently pure speculation, would obviously add greatly to the complexity of signaling by ARF family GTPases, which is already complex enough.
Conclusions
In summary, our comprehensive evolutionary analyses of the ARF GEF protein family have established a concrete dataset of proteins and novel functional hypotheses for further testing. It has established the complement of ARF GEF proteins present in the eukaryotic common ancestor as well as resolved the relationships of the lineage-specific ARF GEFs, including those of the human complement. Finally, we can present for the first time an integrated perspective on the ARF system of GEFs–GTPases–GAPs and in doing so, we believe a richer understanding of how the bidirectional membrane trafficking systems in eukaryotes have evolved and may be regulated.
MATERIALS AND METHODS
Genome and sequence collections
A total of 77 genomes were obtained from a variety of databases at the NCBI, the Joint Genomes Institute, and lineage-specific databases. These genomes were chosen due to their overall quality (depth of coverage/annotation), completeness, and ability to represent different lineages within the diversity of eukaryotes that fall within the major taxonomic supergroups. We noted that, in the case of the haptophytes, cryptophytes, and rhizarians, the taxon sampling was restricted due to genome sequence availability. These represent exciting areas for future sampling of genomic diversity. Consequently, we have been cautious in our conclusions regarding these taxa given the limited data. Owing to the pattern of duplications that we observed in opisthokonta (animal and fungal lineages), a larger number of genomes was sampled to more accurately trace their evolutionary origins. The genomes analyzed and links to databases mined are listed in Supplemental Table S1.
Comparative genomics
Searches were initiated by first obtaining and isolating the conserved Sec7 domains from all identified ARF GEFs from H. sapiens and S. cerevisiae using the extraseq program available through EMBOSS explorer (www.bioinformatics.nl/cgi-bin/emboss/extractseq). The Sec7 domains from this group of 20 proteins (15 from H. sapiens and 5 from S. cerevisiae) were subsequently aligned using the MUSCLE alignment tool (Edgar, 2004). The resulting alignment was then used to generate a seed hidden Markov model (HMM) matrix to iteratively search close relatives of H. sapiens and S. cerevisiae within the opisthokonta supergroup using the hmmsearch tool available through the HMMER3 package (http://hmmer.org) (Eddy, 1998). Hits below an e-value cutoff of 0.05 were analyzed by the RBH method (Schlacht et al., 2013). Retrieval of H. sapiens ARF GEFs as best hits were kept as candidates. These hits were screened for the Sec7 domain using the Conserved Domain Database (CDD, available through the NCBI) and a domain extraction was carried out as described above. The resulting Sec7 domains were then incorporated into the starting HMM for HMMER searches into more distantly related genomes with the iterative identification and addition of Sec7 domains to previous alignment. This process was repeated for a total of 77 genomes from the five major eukaryotic supergroups and three extra-supergroup taxa. A final search, using a global HMM matrix containing Sec7 domains from a total of 503 identified proteins, was performed in all 77 query genomes in order to identify any distantly related proteins that may have been missed in any previous searches. This resulted in identification of three more Sec7 domain-containing proteins. Given that the final HMM was built from taxa across the breadth of eukaryotes, chosen to be taxonomically balanced, this should mitigate and reduce any bias toward the model organisms of yeast or mammalian cells and should be well-suited to detect Sec7 proteins in diverse eukaryotes. A total of 506 Sec7 domain-containing proteins were identified and all full-length protein sequences were subject to RBH, as described above for preliminary orthology assignment. All identified protein sequence loci and their corresponding annotations are listed in Supplemental Table S2. Searches for TBC domain-containing proteins were carried out using an HMM derived from the seed alignment for the RabGAP-TBC (PF00566) entry in Pfam. All hits with a minimum e-value of 0.05 were subject to further reciprocal BLASTp and domain analyses, as described above. All identified sequence loci for the TBC protein searches are summarized in Supplemental Table S3. In all cases, identified ORFs with boldface indicate orthology based on meeting the RBH criteria and, when relevant, robust phylogenetic classification. H. sapiens top RBH as well as the next best non-orthologous hits, and their e-values are also listed in both Supplemental Tables S2 and S3.
Domain analyses
Further validation of orthology of all 506 identified Sec7 domain-containing proteins were subject to additional defining domain analyses flanking either upstream or downstream of the conserved Sec7 domain using InterProScan (available through European Molecular Biology Laboratory; www.ebi.ac.uk/interpro/search/sequence-search), the CDD (available through the the NCBI; www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), and Pfam (available through the European Bioinformatics Institute; https://pfam.xfam.org/). InterProScan analyses were carried out using the default parameters with all 14 algorithms selected. Pfam searches were carried out with an e-value cut-off set at 0.01 and CDD searches were carried out using the default parameters to search the CDD v3.16 database with an e-value cutoff set at 0.01. In all cases, full-length sequences were used. Sec7 domain-containing proteins lacking other defining domains were left as putative ARF GEFs and are indicated in regular font and as a lighter color in Supplemental Table S2 and the Coulson plot (Figure 2), respectively. In cases where neither domain analyses nor reciprocal BLASTp was conclusive to enable assignment to a specific protein family, proteins were termed Rogue ARF GEFs. Only seven of the 506 proteins analyzed failed to be assigned into a specific subfamily and were kept as Rogues. All domain analysis results, for Sec7 domain-containing proteins and TBC domain-containing proteins, are summarized in Supplemental Tables S2 and S3, respectively.
Phylogenetic analyses
Phylogenetic analyses were carried out on the identified Sec7 domain-containing sequences using the extracted Sec7 domains isolated using the protocol described above. Sequences for specific phylogenetic hypothesis testing were first aligned using MUSCLE v. 3.8.31 (Edgar, 2004). All alignments were subsequently inspected and manually trimmed using Mesquite v. 3.5 (www.mesquiteproject.org/) to remove heterogeneous regions lacking obvious homology and partial or misaligned sequences (Maddison and Maddison, 2018).
Maximum likelihood analysis was undertaken using two separate tools; RAxML-HPC2 on XSEDE v.8.2.10 and IQTREE v.1.6.6, with the first for non-parametric bootstrapping and the latter for ultrafast bootstrapping (Stamatakis, 2006; Minh et al., 2013; Nguyen et al., 2015). The best protein matrix model for RAxML was tested using ProtTest v.3.4.2 (Abascal et al., 2005) set to account for Gamma rate variation (+G), invariant sites (+I), and observed frequency of amino acids (+F) with the default tree. Non-parametric bootstrapping was carried out by running 100 pseudoreplicates with the default tree faster hill climbing method (-f b, -b, -N 100), and a consensus tree was obtained using the Consense package, available through the Phylip v.3.66 package (Felsenstein, 1989). RAxML analyses were performed on the Cyberinfrastructure for the Phylogenetic Research (CIPRES)webserver (Miller et al., 2010). Ultrafast bootstrapping was performed using IQTREE (Minh et al., 2013; Nguyen et al., 2015). Model testing was performed using the in-built ModelFinder program with the best model selected according to the BIC criterion, and 1000 pseudoreplicates were obtained until tree convergence reached default convergence coefficient (Kalyaanamoorthy et al., 2017). IQTREE tree building was carried out locally.
Bayesian inference was carried out using PhyloBayes v.3.3f using an empirical matrix, LG, and analyses were run using parameters specifying a sample size of 100 trees and burnin fraction of 20% until splits frequency fell below 0.1 ensuring tree convergence (Lartillot et al., 2009). PhyloBayes analyses were performed on a local computing cluster. In the few instances (pan-eukaryotic BIG and GBF1, BIG only, and cytohesin phylogenetic analyses) where PhyloBayes analyses failed to converge despite running for several thousand CPU hours, an alternate tool, MRBAYES on XSEDE v3.2.6, was used and run on CIPRES (Huelsenbeck and Ronquist, 2001). Parameters specified included 10 million Markov chain Monte Carlo generations under a mixed amino acid model with the number of gamma rate categories set at 4. Sampling frequency was set to occur every 1000 generations with a burnin fraction of 25%. Tree convergence was ensured when the average SD of split frequency values fell below 0.01. Both non-parametric and ultrafast bootstraps as well as Bayesian posterior probabilities were overlaid on the best Bayes topology with a combined value of greater than 50, 95, and 0.80, respectively, indicating node support. Tree visualization and rooting was carried out in FigTree v.1.4.4 (Rambaut, 2009). Node support value overlay and additional annotations were prepared using Adobe Illustrator CS4. All alignments available on request.
Of note, a global Sec7 phylogeny was carried out using only the Sec7 domain from all 506 identified ARF GEFs in order to confirm the classifications based on homology searching. However, the resulting phylogeny remained unresolved (data available on request).
Supplementary Material
Acknowledgments
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (RES0021028, RES0043758, RES0046091) to J.B.D. and the National Institute of General Medical Sciences (R35 GM122568) to R.A.K. J.B.D. is the CRC (Tier II) in Evolutionary Cell Biology. S.V.P. is funded by a Frederick Banting and Charles Best Canada Graduate Scholarship (Master’s). A.S. was funded by the NSERC Postgraduate Scholarship-Doctoral Program, while C.M.K. is funded by a Canada Vanier Graduate Scholarship. We thank members of the Dacks lab for careful proofreading of the manuscript and insightful discussions, as well as Cathy Jackson, Paul Melançon, Elizabeth Sztul, Lorraine Santy, and James Casanova for their expert suggestions on ARF and ARF GEF cell biology and Anna Karnkowska and Vladimir Hampl for their help with initial cytohesin analyses.
Abbreviations used:
- Ank
ankyrin
- ARCC
ankyrin repeat-containing cytohesin
- ARF
ADP-ribosylation factor
- BFA
brefeldin A
- BIG
BFA-inhibited ARF GEF
- BRAG
BFA-resistant ARF GEF (herein using IQSEC7; IQ motif and Sec7 domain-containing protein)
- DCB
dimerization and cyclophilin binding
- EFA6
exchange factor for ARF6
- ER
endoplasmic reticulum
- ERGIC
ER-Golgi intermediate compartment GAP
- FBX8
F-box only protein 8
- GAP
GTPase-activating protein
- GBF1
Golgi BFA resistant factor 1
- GEF
guanine nucleotide exchange factor
- HDS
homology downstream of Sec7
- HMM
hidden Markov model
- HUS
homology upstream of Sec7
- LECA
last eukaryotic common ancestor
- nr
nonredundant
- OPH
organelle paralogy hypothesis
- PSD
PH and Sec7 domain
- RBH
reciprocal best hit
- TBC
Tre-2/Bub2/Cdc16
- TBS
TBC and Sec7
- TGN
trans-Golgi network.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-01-0073) on May 29, 2019.
REFERENCES
- Abascal F, Zardoya R, Posada D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics , 2104–2105. [DOI] [PubMed] [Google Scholar]
- Adl SM, Bass D, Lane CE, Lukes J, Schoch CL, Smirnov A, Agatha S, Berney C, Brown MW, Burki F, et al. (2019). Revisions to the classification, nomenclature, and diversity of eukaryotes. J Euk Microbiol , 4–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khodor S, Price CT, Kalia A, Abu Kwaik Y. (2010). Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol , 132–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow LD, Dacks JB. (2018). Seeing the endomembrane system for the trees: Evolutionary analysis highlights the importance of plants as models for eukaryotic membrane-trafficking. Semin Cell Dev Biol , 142–152. [DOI] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, et al (2005). The genome of the African trypanosome Trypanosoma brucei. Science , 416–422. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS. (2014). Adaptor proteins involved in polarized sorting. J Cell Biol , 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet S, Golinelli-Cohen M-P, Contremoulins V, Jackson CL. (2013). Targeting of the Arf-GEF GBF1 to lipid droplets and Golgi membranes. J Cell Sci , 4794–4805. [DOI] [PubMed] [Google Scholar]
- Bui QT, Golinelli-Cohen M-P, Jackson CL. (2009). Large Arf1 guanine nucleotide exchange factors: evolution, domain structure, and roles in membrane trafficking and human disease. Mol Genet Genomics , 329–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova JE. (2007). Regulation of Arf activation: The Sec7 family of guanine nucleotide exchange factors. Traffic , 1476–1485. [DOI] [PubMed] [Google Scholar]
- Chantalat S, Park SK, Hua Z, Liu K, Gobin R, Peyroche A, Rambourg A, Graham TR, Jackson CL. (2004). The Arf activator Gea2p and the P-type ATPase Drs2p interact at the Golgi in Saccharomyces cerevisiae. J Cell Sci , 711–722. [DOI] [PubMed] [Google Scholar]
- Chardin P, Paris S, Antonny B, Robineau S, Béraud-Dufour S, Jackson CL, Chabre M. (1996). A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature , 481–484 [DOI] [PubMed] [Google Scholar]
- Chen K-Y, Tsai P-C, Hsu J-W, Hsu H-C, Fang C-Y, Chang L-C, Tsai Y-T, Yu C-J, Lee F-JS. (2010). Syt1p promotes activation of Arl1p at the late Golgi to recruit Imh1p. J Cell Sci , 3478–3489. [DOI] [PubMed] [Google Scholar]
- Cherfils J, Ménétrey J, Mathieu M, Le Bras G, Robineau S, Béraud-Dufour S, Antonny B, Chardin P. (1998). Structure of the Sec7 domain of the Arf exchange factor ARNO. Nature , 101. [DOI] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. (2013). Regulation of Small GTPases by GEFs, GAPs, and GDIs. Physiol Rev , 269–309. [DOI] [PubMed] [Google Scholar]
- Chun J, Shapovalova Z, Dejgaard SY, Presley JF, Melancon P. (2008). Characterization of class I and II ADP-ribosylation factors (Arfs) in live cells: GDP-bound class II Arfs associate with the ER-Golgi intermediate compartment independently of GBF1. Mol Biol Cell , 3488–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claing A, Perry SJ, Achiriloaie M, Walker JKL, Albanesi JP, Lefkowitz RJ, Premont RT. (2000). Multiple endocytic pathways of G protein-coupled receptors delineated by GIT1 sensitivity. Proc Natl Acad Sci USA , 1119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claude A, Zhao BP, Kuziemsky CE, Dahan S, Berger SJ, Yan JP, Armold AD, Sullivan EM, Melancon P. (1999). GBF1: A novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J Cell Biol , 71–84. [PMC free article] [PubMed] [Google Scholar]
- Cohen LA, Honda A, Varnai P, Brown FD, Balla T, Donaldson JG. (2007). Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell , 2244–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R, Mason-Gamer RJ, Jackson CL, Segev N. (2004). Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol Biol Cell , 1487–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin TC, DiNitto JP, Czech MP, Lambright DG. (2004). Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains. EMBO J , 3711–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacks JB, Field MC. (2007). Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. J Cell Sci 2977–2985. [DOI] [PubMed] [Google Scholar]
- Dacks JB, Field MC. (2018). Evolutionary origins and specialisation of membrane transport. Curr Opin Cell Biol , 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. (2011). ARF family G proteins and their regulators: Roles in membrane transport, development and disease. Nat Rev Mol Cell Biol , 362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza RS, Semus R, Billings EA, Meyer CB, Conger K, Casanova JE. (2014). Rab4 orcherstrates a small GTPase cascade for recruitment of adaptor proteins to early endosomes. Curr Biol , 1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy JL, Moravec R, Ly K, Lasell TK, Melancon P, Casanova JE. (2006). The Arf6 GEF GEP100/BRAG2 regulates cell adhesion by controlling endocytosis of β1 integrins. Curr Biol , 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East MP, Bowzard JB, Dacks JB, Kahn RA. (2012). ELMO domains, evolutionary and functional characterization of a novel GTPase-activating protein (GAP) domain for Arf protein family GTPases. J Biol Chem , 39538–39553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East MP, Kahn RA. (2011). Models for the functions of Arf GAPs. Sem Cell Dev Biol , 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. (1998). Profile hidden Markov models. Bioinformatics , 755–763. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res , 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. (1989). PHYLIP-Phylogeny Inference Package (Version 3.2). Cladistics , 164–166. [Google Scholar]
- Field MC, Gabernet-Castello C, Dacks JB. (2007). Reconstructing the evolution of the endocytic system: insights from genomics and molecular cell biology. Adv Exp Med Biol , 84–96. [DOI] [PubMed] [Google Scholar]
- Franco M, Peters PJ, Boretto J, van Donselaar E, Neri A, D’Souza-Schorey C, Chavrier P. (1999). EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J , 1480–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Upender S, Hansen SH, Casanova JE. (1998), ARNO is a guanine nucleotide exchange factor for ADP-ribosylation factor 6. J Biol Chem , 23–27. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Oda K, Yokota S, Takatsuki A, Ikehara Y. (1988). Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J Biol Chem ,18545–18552. [PubMed] [Google Scholar]
- Gabernet-Castello C, O’Reilly AJ, Dacks JB, Field MC. (2013). Evolution of Tre-2/Bub2/Cdc16 (TBC) Rab GTPase-activating proteins. Mol Biol Cell , 1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger C, Nagel W, Boehm T, van Kooyk Y, Figdor CG, Kremmer E, Hogg N, Zeitlmann L, Dierks H, Weber KSC, Kolanus W. (2000). Cytohesin-1 regulates β-2 integrin-mediated adhesion through both ARF-GEF function and interaction with LFA-1. EMBO J , 2525–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell , 219–230. [DOI] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. (2007). Identification of a guanine nucleotide exchange factor for Arf3, the yeast orthologue of mammalian Arf6. PloS One , e842–e842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotthardt K, Lokaj M, Koerner C, Falk N, Giessl A, Wittinghofer A. (2015). A G-protein activation cascade from Arl13B to Arl3 and implications for ciliary targeting of lipidated proteins. Elife , e11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman EK, Ali M, Field MC, Dacks JB. (2018). Regulation of early endosomes across eukaryotes: evolution and functional homology of Vps9 proteins. Traffic , 546–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Schlacht A, Norcott JP, Traynor D, Bloomfield G, Antrobus R, Kay RR, Dacks JB, Robinson MS. (2014). Characterization of TSET, an ancient and widespread membrane trafficking complex. ELife , e02866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics , 754–755. [DOI] [PubMed] [Google Scholar]
- Ivanova AA, Caspary T, Seyfried NT, Duong DM, West AB, Liu Z, Kahn RA. (2017). Biochemical characterization of purified mammalian ARL13B protein indicates that it is an atypical GTPase and ARL3 guanine nucleotide exchange factor (GEF). J Biol Chem , 11091–11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova AA, East MP, Yi SL, Kahn RA. (2014). Characterization of recombinant ELMOD (cell engulfment and motility domain) proteins as GTPase-activating proteins (GAPs) for ARF family GTPases. J Biol Chem , 11111–11121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CL. (2014). GEF-effector interactions. Cell Logist , e943616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Jedd G, Kahn RA, Franzusoff A, Bartolini F, Segev N. (1999). Genetic interactions in yeast between Ypt GTPases and Arf guanine nucleotide exchangers. Genetics , 1543–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA. (2009). Toward a model for Arf GTPases as regulators of traffic at the Golgi. FEBS Lett , 3872–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Bruford E, Inoue H, Logsdon JM, Nie Z, Premont RT, Randazzo PA, Satake M, Theibert AB, Zapp ML, Cassel D. (2008). Consensus nomenclature for the human ArfGAP domain-containing proteins. J Cell Biol , 1039–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Cherfils J, Elias M, Lovering RC, Munro S, Schurmann A. (2006). Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. J Cell Biol , 645–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods , 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P, Koonin E. (2014). The Origin and Evolution of Eukaryotes: A Subject Collection from Cold Spring Harbor Perspectives in Biology, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Kipreos ET, Pagano M. (2000). The F-box protein family. Genome Biol , REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarlund JK, Guilherme A, Holik JJ, Virbasius JV, Chawla A, Czech MP. (1997). Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and sec7 homology domains. Science , 1927–1930. [DOI] [PubMed] [Google Scholar]
- Klarlund JK, Tsiaras W, Holik JJ, Chawla A, Czech MP. (2000). Distinct polyphosphoinositide binding selectivities for pleckstrin homology domains of GRP1-like proteins based on diglycine versus triglycine motifs. J Biol Chem , 32816–32821. [DOI] [PubMed] [Google Scholar]
- Klein S, Partisani M, Franco M, Luton F. (2008). EFA6 facilitates the assembly of the tight junction by coordinating an Arf6-dependent and -independent pathway. J Biol Chem , 30129–30138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. (2009). PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics , 2286–2288. [DOI] [PubMed] [Google Scholar]
- Li Y, Kelly WG, Logsdon JMJ, Schurko AM, Harfe BD, Hill-Harfe KL, Kahn RA. (2004). Functional genomic analysis of the ADP-ribosylation factor family of GTPases: phylogeny among diverse eukaryotes and function in C. elegans. FASEB J ,1834–1850. [DOI] [PubMed] [Google Scholar]
- Liu Y-W, Huang C-F, Huang K-B, Lee F-JS. (2005). Role for Gcs1p in regulation of Arl1p at trans-Golgi compartments. Mol Biol Cell , 4024–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-W, Lee S-W, Lee F-JS. (2006). Arl1p is involved in transport of the GPI-anchored protein Gas1p from the late Golgi to the plasma membrane. J Cell Sci , 3845–3855. [DOI] [PubMed] [Google Scholar]
- Macia E, Chabre M, Franco M. (2001). Specificities for the small G proteins ARF1 and ARF6 of the guanine nucleotide exchange factors ARNO and EFA6. J Biol Chem , 24925–24930. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. (2018). Mesquite: a modular system for evolutionary analysis, Version 3.51, www.mesquiteproject.org. [Google Scholar]
- Malaby AW, van den Berg B, Lambright DG. (2013). Structural basis for membrane recruitment and allosteric activation of cytohesin family Arf GTPase exchange factors. Proc Natl Acad Sci USA , 14213–14218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolea F, Chun J, Chen DW, Clarke I, Summerfeldt N, Dacks JB, Melancon P. (2010). Arf3 is activated uniquely at the trans-Golgi network by brefeldin A-inhibited guanine nucleotide exchange factors. Mol Biol Cell , 1836–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonold CM, Fromme JC. (2014). Four GTPases differentially regulate the Sec7 Arf-GEF to direct traffic at the trans-golgi network. Dev Cell , 759–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, 1–8 . 10.1109/GCE.2010.5676129. [DOI] [Google Scholar]
- Minh BQ, Nguyen MAT, von Haeseler A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol , 1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi LK, Minor DL, Peng Z -Y. (2002). Consensus-derived structural determinants of the ankyrin repeat motif. Proc Natl Acad Sci USA , 16029–16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouratou B, Biou V, Joubert A, Cohen J, Shields DJ, Geldner N, Jürgens G, Melancon P, Cherfils J. (2005). The domain architecture of large guanine nucleotide exchange factors for the small GTP-binding protein Arf. BMC Genomics , 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol , 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SJ, Santy LC. (2012). Phosphoinositide specificity determines which cytohesins regulate β1 integrin recycling. J Cell Sci , 3195–3201. [DOI] [PubMed] [Google Scholar]
- Padovani D, Folly-Klan M, Labarde A, Boulakirba S, Campanacci V, Franco M, Zeghouf M, Cherfils J. (2014). EFA6 controls Arf1 and Arf6 activation through a negative feedback loop. Proc Natl Acad Sci USA , 12378–12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris S, Béraud-Dufour S, Robineau S, Bigay J, Antonny B, Chabre M, Chardin P. (1997). Role of protein-phospholipid interactions in the activation of ARF1 by the guanine nucleotide exchange factor Arno. J Biol Chem , 22,221–22,226. [DOI] [PubMed] [Google Scholar]
- Petrželková R, Eliáš M. (2014). Contrasting patterns in the evolution of the Rab GTPase family in Archaeplastida. Act Soc Bot Pol Tow Bot , 303–315. [Google Scholar]
- Peyroche A, Antonny B, Robineau S, Acker J, Cherfils J, Jackson CL. (1999). Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol Cell , 275–285. [DOI] [PubMed] [Google Scholar]
- Peyroche A, Paris S, Jackson CL. (1996). Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature , 479–481. [DOI] [PubMed] [Google Scholar]
- Rambaut A. (2009). FigTree version 1.4.4. Computer program distributed by the author http://tree.bio.ed.ac.uk/software/figtree/. [Google Scholar]
- Schlacht A, Dacks JB. (2015). Unexpected ancient paralogs and an evolutionary model for the COPII coat complex. Genome Evol Biol , 1098–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacht A, Herman EK, Klute MJ, Field MC, Dacks JB. (2014). Missing pieces of an ancient puzzle: evolution of the eukaryotic membrane-trafficking system. Cold Spring Harb Perspect Biol , a016048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacht A, Mowbrey K, Elias M, Kahn RA, Dacks JB. (2013). Ancient complexity, opisthokont plasticity, and discovery of the 11th subfamily of Arf GAP proteins. Traffic , 636–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H-W, Nakayama K. (2004). Guanine nucleotide-exchange factors for arf GTPases: their diverse functions in membrane traffic. J Biochem , 761–767. [DOI] [PubMed] [Google Scholar]
- Shinotsuka C, Waguri S, Wakasugi M, Uchiyama Y, Nakayama K. (2002a). Dominant-negative mutant of BIG2, an ARF-guanine nucleotide exchange factor, specifically affects membrane trafficking from the trans-Golgi network through inhibiting membrane association of AP-1 and GGA coat proteins. Biochem Biophys Res Commun , 254–260. [DOI] [PubMed] [Google Scholar]
- Shinotsuka C, Yoshida Y, Kawamoto K, Takatsu H, Nakayama K. (2002b). Overexpression of an ADP-ribosylation factor-guanine nucleotide exchange factor, BIG2, uncouples brefeldin A-induced adaptor protein-1 coat dissociation and membrane tubulation. J Biol Chem , 9468–9473. [DOI] [PubMed] [Google Scholar]
- Someya A, Sata M, Takeda K, Pacheco-Rodriguez G, Ferrans VJ, Moss J, Vaughan M. (2001). ARF-GEP(100), a guanine nucleotide-exchange protein for ADP-ribosylation factor 6. Proc Natl Acad Sci USA , 2413–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Shiba Y, Randazzo PA. (2010). Arf GAPs: gatekeepers of vesicle generation. FEBS Lett , 2646–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparvoli D, Richardson E, Osakada H, Lan X, Iwamoto M, Bowman GR, Kontur C, Bourland WA, Lynn DH, Pritchard JK, et al (2018). Remodeling the Specificity of an endosomal CORVET tether underlies formation of regulated secretory vesicles in the ciliate tetrahymena thermophila. Curr Biol , 697–710.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer S, Spang A, Schekman R. (1999). A primer on vesicle budding. Cell ,145–148. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics , 2688–2690. [DOI] [PubMed] [Google Scholar]
- Sztul E, Chen P-W, Cherfils J, Dacks J, Lambright DS, Lee F-J, Randazzo PA, Schurmann A, Wilhelmi I, Yohe ME, Kahn RA. (2019). ARF GTPases and their GEFs and GAPs: concepts and challenges. Mol Biol Cell , 1249–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali T, Girald-Berlingeri S, Randazzo PA, Chen P-W. (2017). Arf GAPs: A family of proteins with disparate functions that converge on a common structure, the integrin adhesion complex. Small GTPases , 280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli-Daley LA, Li Y, Zhang C-J, Kahn RA. (2005). Isoform-selective effects of the depletion of ADP-ribosylation factors 1-5 on membrane traffic. Mol Biol Cell , 4495–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J, Kahn RA, Sztul E. (2014). Regulating the large Sec7 ARF guanine nucleotide exchange factors: the when, where and how of activation. Cell Mol Life Sci , 3419–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano H, Kobayashi I, Onodera Y, Luton F, Franco M, Mazaki Y, Hashimoto S, Iwai K, Ronai Z, Sabe H. (2008). Fbx8 makes Arf6 refractory to function via ubiquitination. Mol Biol Cell , 822–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeghouf M, Guibert B, Zeeh J-C, Cherfils J. (2005). Arf, Sec7 and Brefeldin A: a model towards the therapeutic inhibition of guanine nucleotide-exchange factors. Biochem Soc Trans , 1265–1268. [DOI] [PubMed] [Google Scholar]
- Zhang CJ, Cavenagh MM, Kahn RA. (1998). A family of Arf effectors defined as suppressors of the loss of Arf function in the yeast Saccharomyces cerevisiae. J Biol Chem , 19792–19796. [DOI] [PubMed] [Google Scholar]
- Zhao X, Claude A, Chun J, Shields DJ, Presley JF, Melancon P. (2006). GBF1, a cis-Golgi and VTCs-localized ARF-GEF, is implicated in ER-to-Golgi protein traffic. J Cell Sci , 3743–3753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.