Abstract
WDR92 associates with a prefoldin-like cochaperone complex and known dynein assembly factors. WDR92 has been very highly conserved and has a phylogenetic signature consistent with it playing a role in motile ciliary assembly or activity. Knockdown of WDR92 expression in planaria resulted in ciliary loss, reduced beat frequency and dyskinetic motion of the remaining ventral cilia. We have now identified a Chlamydomonas wdr92 mutant that encodes a protein missing the last four WD repeats. The wdr92-1 mutant builds only ∼0.7-μm cilia lacking both inner and outer dynein arms, but with intact doublet microtubules and central pair. When cytoplasmic extracts prepared by freeze/thaw from a control strain were fractionated by gel filtration, outer arm dynein components were present in several distinct high molecular weight complexes. In contrast, wdr92-1 extracts almost completely lacked all three outer arm heavy chains, while the IFT dynein heavy chain was present in normal amounts. A wdr92-1 tpg1-2 double mutant builds ∼7-μm immotile flaccid cilia that completely lack dynein arms. These data indicate that WDR92 is a key assembly factor specifically required for the stability of axonemal dynein heavy chains in cytoplasm and suggest that cytoplasmic/IFT dynein heavy chains use a distinct folding pathway.
INTRODUCTION
Ciliary motility is required for the movement of individual cells such as mammalian sperm and various protists, as well as the transport of fluids that bathe ciliated epithelia, including in the lungs, brain ventricles, trachea, and oviduct (for recent reviews, see various chapters in Marshall and Basto, 2017; King, 2018b,c). The motile behavior of these microtubule-based organelles is driven by the axonemal dyneins that form the highly complex inner- and outer-arm arrays along doublet microtubules (Nicastro et al., 2006; Lin and Nicastro, 2018; and for a brief review of earlier work, see King and Sale, 2018). These 1- to 2-MDa multicomponent microtubule motors are built around ∼530-kDa heavy chain (HC) motor units that associate with a variety of additional components, including WD-repeat intermediate chains (ICs) and various light chains (LCs) (reviewed in King, 2018a). In outer arm dyneins, which contain two or three different HCs depending on species, two WD-repeat ICs bind each other and several LC types to form an IC/LC complex that is required for formation of the holoenzyme; this complex is stable in the absence of HCs (Tang et al., 1982; Pfister and Witman, 1984). Other LCs interact directly with the HCs and mediate regulatory effects in response to various signaling inputs including changes in Ca2+ levels and alterations in redox poise (King and Patel-King, 1995; Patel-King et al., 1996; Wakabayashi and King, 2006; Sakato et al., 2007); in addition, one highly conserved outer-arm LC (LC1/DNAL1) associates with an HC microtubule-binding domain and may modulate motility directly (Benashski et al., 1999; Baron et al., 2007; Patel-King and King, 2009; King and Patel-King, 2012; Ichikawa et al., 2015).
As cilia lack ribosomes (Pazour et al., 2005), all axonemal dyneins must be synthesized in the cytoplasm, and then transported to the ciliary base, moved into the growing cilium through the transition zone gate and ultimately docked at very precise sites within the axonemal superstructure (Takada and Kamiya, 1994; Ahmed et al., 2008; Oda et al., 2014). Failure or disruption of any of these key steps leads to immotile or dyskinetic cilia (e.g., Kamiya and Okamoto, 1985; Mitchell and Rosenbaum, 1985; Omran et al., 2008; Panizzi et al., 2012; Loges and Omran, 2018); in mammals this can cause many phenotypes, including both male and female infertility, situs inversus, chronic respiratory problems, hydrocephalus, and congenital heart defects (Ibanez-Tallon et al., 2003; Fliegauf et al., 2007; Li et al., 2016; Loges and Omran, 2018). Moreover, an axonemal dynein assembly factor mutant that exhibits disrupted cerebrospinal fluid flow has recently been shown to affect assembly of the Reissner fiber in zebrafish that is essential for normal formation and alignment of the body axis (Cantaut-Belarif et al., 2018).
Requirement for the large amounts of dynein HCs and other components needed for building axonemes places an enormous biosynthetic burden on cells. Ribosomal synthesis of each HC takes ∼15 min (at a rate of ∼5 residues/s), during which time the nascent protein must be kept in a partially folded but stable state so that aggregation does not occur; although an issue for any large protein, this is especially problematic for dynein HCs, as the circular arrangement of the motor domain demands that full stability can only occur once the final AAA domain (AAA6), and possibly the C-terminal region, have been completely synthesized. Every 10-μm cilium requires ∼16,000 HCs (with a combined mass of ∼8.5 GDa), and individual multiciliated epithelial cells and various protists (e.g., Tetrahymena and Paramecium) can contain hundreds of cilia. Thus, the folding machinery needs to be plentiful, highly efficient, and error-free to avoid causing proteotoxic cell stress through the generation of large amounts of misfolded components. Once synthesized, individual HCs must then interact with one another, specific LCs, and the IC/LC complex in a directed manner to form a functional dynein particle; whether these components can associate with partially synthesized HCs once the appropriate binding sites have been built remains unexplored.
As might be expected given the inherent complexities of axonemal dynein motors, the factors now known to be required for their stable assembly in cytoplasm are both many and varied (Table 1) (for recent review, see Mitchell, 2018). They fall into several distinct classes including 1) scaffolding components with various protein–protein interaction domains, 2) PIH domain proteins that recruit the chaperone HSP90, and 3) the RuvBL1/2 (pontin/reptin) AAA+ ATPases that form the catalytic core of the R2TP complex and presumed axonemal dynein-specific R2TP variants that have differing PIH and scaffolding components (Kakihara and Houry, 2012; Maurizy et al., 2018; Yamaguchi et al., 2018). However, how all these proteins associate, the kinetics of the interactions, their precise role in the assembly pathway, the mechanisms by which their activities are coordinated, and how R2TP variants exhibit HC specificity and why this is necessary remain very unclear.
TABLE 1:
Known cytoplasmic factors needed for axonemal dynein assembly.
| Chlamydomonas gene name | Human gene name | Other aliases | Attributes |
|---|---|---|---|
| ODA5 | ––– | –– | Coiled-coil protein; Chlamydomonas specific |
| ODA8 | LRRC56 | –– | N-terminal leucine-rich repeats |
| ODA10 | CCDC151 | –– | Coiled-coil protein |
| DAP1 | DNAAF2 | PF13, Kintoun | PIH domain protein, recruits HSP90; has C-terminal CS domain; associates with RPAP3, SPAG1, and DYX1C1 |
| PF23 | DNAAF4 | DYX1C1 | N-terminal CS domain and C-terminal TPR repeats; different isoforms bind DNAAF2 and PIH1D3. |
| TWI1 | PIH1D3 | Twister | PIH domain protein, recruits HSP90; has C-terminal CS domain; binds a DYX1C1 isoform. |
| MOT47 | LRRC6 | Seahorse | N-terminal leucine-rich repeats, C-terminal CS domain |
| CrZMYND10 | ZMYND10 | –– | MYND zinc finger domain |
| FBB18 | CFAP298 | C21orf59, kurly | DUF2870 domain |
| DAP2 | PIH1D2 | IDA10, MOT48 | PIH domain protein, recruits HSP90; C-terminal CS domain; associates with SPAG1 |
| DAU1 | DNAAF1 | ODA7 | N-terminal leucine-rich repeats |
| WDR92 | WDR92 | Monad | Highly conserved WD repeat protein; binds SPAG1, RPAP3, and prefoldins |
| DAB1 | DNAAF3 | PF22 | DUF4470/4471 domains |
| CrSPAG1 | SPAG1 | –– | Has TPR repeats and a RPAP3_C domain; binds WDR92, PIH1D2, and RUVBL1/2 |
| CrHEATR2 | DNAAF5 | HEATR2 | Contains HEAT repeats |
| RUVBL1 | RUVBL1 | Pontin | AAA ATPase; RUVBL1/2 hexamers bind RPAP3-PIH1D1 heterodimers, forming the R2TP complex; hexamers can dimerize. |
| RUVBL2 | RUVBL2 | Reptin | AAA ATPase; RUVBL1/2 hexamers bind RPAP3-PIH1D1 heterodimers, forming the R2TP complex; hexamers can dimerize. |
This table does not include factors such as ODA16/WDR69 and C11orf70, which act with the IFT machinery in the trafficking of axonemal dyneins into the cilium.
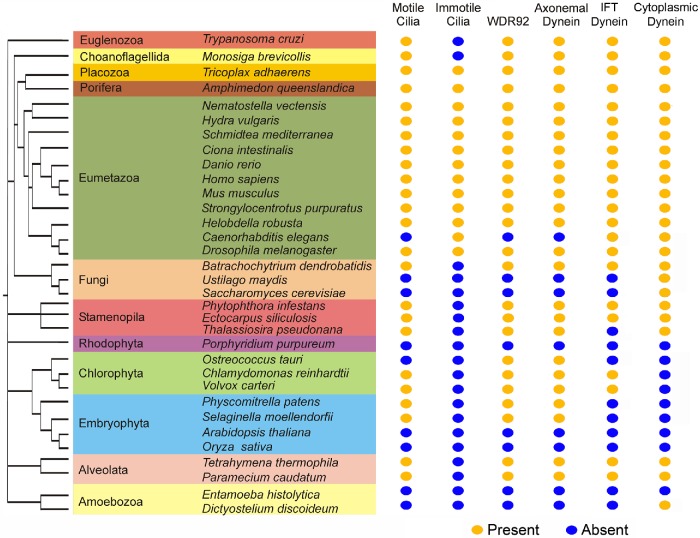
The WD-repeat protein WDR92 has been reported to associate with RPAP3_C domains (Itsuki et al., 2008; Maurizy et al., 2018) and with a prefoldin-like cochaperone complex (Boulon et al., 2010; Kakihara and Houry, 2012; Millán-Zambrano and Chávez, 2014). RPAP3_C domains are found only in RNA polymerase II–associated protein 3 (a component of the canonical R2TP complex [Kakihara and Houry, 2012] with N-terminal TPR repeats and a C-terminal RPAP3_C domain) and the dynein assembly factors SPAG1 (which has a domain organization similar to RPAP3) and CCDC103. Furthermore, WDR92 is only expressed in organisms that encode axonemal dynein HCs (Figure 1); nearly all these also have motile cilia, one known exception being the pico-chlorophyte alga Ostreococcus, which, although it encodes inner-arm I1/f HC components, appears to completely lack basal bodies, the intraflagellar transport (IFT) system, and cilia (Palenik et al., 2007). In contrast, WDR92 is absent from organisms that do not build cilia (e.g., angiosperms and ascomycetous yeasts) or that have only nonmotile sensory cilia (e.g., the nematode Caenorhabditis elegans). On the basis of this phylogenetic signature we predicted that WDR92 might be important for motile cilia function (Patel-King and King, 2016). As an initial test of this hypothesis, we previously knocked down WDR92 expression in planaria and observed pleiomorphic defects in ciliary architecture—most notably loss of dynein arms, incomplete closure of the outer-doublet microtubule B-tubules, and occasional lack of the central pair complex—as well as loss of many ventral cilia and dyskinetic motion of the remaining organelles (Patel-King and King, 2016). More recently, studies in Drosophila suggest that WDR92 associates with SPAG1 to regulate the R2TP complex (it forms part of the variant R2SP and R2SD complexes), and its loss leads to the failure of dynein arm assembly into sperm flagella and sensory neuronal cilia in insects (zur Lage et al., 2018). Based on proteomic analyses, Drosophila WDR92 has been proposed to associate with both HCs and ICs and to act at a late stage in the cytoplasmic assembly process for axonemal dyneins, perhaps by targeting partially assembled dyneins to the R2TP complex (zur Lage et al., 2018). A very recent report has also described axonemal dynein HC defects in a wdr92 Chlamydomonas mutant and its involvement with the protein-folding machinery (Liu et al., 2018).
FIGURE 1:
WDR92 is expressed only in organisms that encode axonemal dyneins. Phylogenetic analysis of the eukaryotes illustrating members of the major lineages and indicating whether they have motile and/or immotile cilia, and whether they encode axonemal dynein HCs and WDR92 (present, orange dot; absent, blue dot). There is a precise correspondence between the presence of specifically axonemal dynein HCs and WDR92. In contrast, organisms that have only immotile cilia or completely lack these organelles do not encode a WDR92 orthologue. Note that, although the pico-chlorophyte alga Ostreococcus apparently lacks all ciliary structures, it does encode the two HCs and several ICs of axonemal inner dynein arm I1/f (Palenik et al., 2007). The presence of WDR92 does not correlate with an organism encoding either cytoplasmic and/or IFT dynein HCs.
Here, we describe an insertional mutation in the Chlamydomonas WDR92 gene that disrupts the encoded protein and show that loss of WDR92 leads to very short cilia lacking dynein arms; this axonemal assembly phenotype can be rescued by mutation in a tubulin polyglutamylase, which reduces microtubule dynamic instability at the distal ciliary tip. Our biochemical data reveal that WDR92 is absolutely required for the cytoplasmic stability of outer arm dynein HCs, but not the associated ICs or LCs, and provide direct support for the idea that WDR92 is a key cytoplasmic factor needed for axonemal dynein formation. Furthermore, we find that WDR92 is required specifically for axonemal dynein HC assembly and that stability and organization of the dynein HC that powers retrograde IFT is unaffected by its loss.
RESULTS
Axonemal dynein assembly factors exhibit distinct patterns of diurnal rhythmic expression
Ciliary assembly/disassembly in Chlamydomonas exhibits a diurnal rhythm. Specifically, when grown under photoautotrophic conditions on a light/dark cycle, cells resorb cilia near the beginning of the dark phase before mitotic entry and rebuild these structures later in the dark phase following cell division. Dynein structural components and assembly factors show three general diurnal transcriptional profiles (here these are termed groups I–III and are exemplified in Figure 2 by the outer arm γ HC, LC10, and PF22/DNAAF3, respectively); the original data are from Zones et al. (2015). For group I, which includes the HCs, ODA8 (LRRC56), ODA10 (CCDC151), DAP1(DNAAF2), DYX1C1(DNAAF4), and CrHEATR2, expression is low at the onset of the light phase, but shows a peak near the start of the dark phase, and then further increases until almost the end of the dark phase ∼10 h later, when expression drops significantly. Components placed in group II, such as LC10, show a distinct expression peak near or just after the light/dark transition and a subsequent sustained reduction. WDR92 is a member of group II, exhibiting a strong peak after entry into the dark phase followed by sustained lower-level expression (Figure 2); other assembly factors, including CrZMYND10, MOT47(LRRC6), CFAP298, DAP2(IDA10), and ODA7(DNAAF1), also fall into this group. In contrast, two other cytoplasmic assembly factors PF22/DNAAF3 and CrSPAG1 (group III) show clear expression peaks in the light phase several hours before up-regulation of other dynein components and assembly proteins; indeed, these two genes show low expression at the time when transcription of all other factors and dynein components is increasing (individual profiles and group assignments for known assembly factors and representative dynein structural components are shown in Supplemental Figure S1). The very clear expression of PF22/DNAAF3 and CrSPAG1 several hours before up-regulation of dynein-encoding genes suggests that these two factors may either play key early roles in the assembly process or have other, nonaxonemal dynein-associated functions.
FIGURE 2:
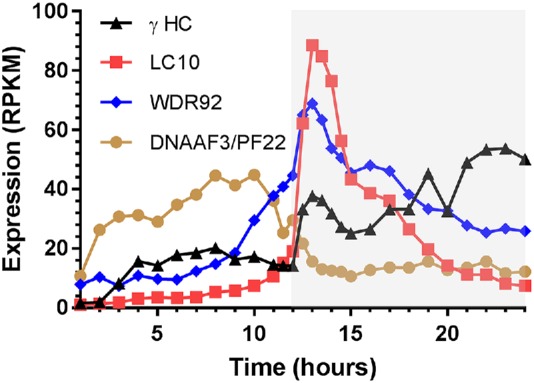
Diurnal expression of axonemal dynein components and assembly factors. When Chlamydomonas is grown on a light/dark cycle under photoautotrophic conditions, axonemal dynein components and their cytoplasmic assembly factors exhibit three broad patterns of transcription during the diurnal cycle; gray shading indicates the dark phase. These are exemplified here by the outer arm γ HC (group I; black), outer arm LC10 (group II; red), and PF22/DNAAF3 (group III; brown); WDR92 (blue) is a member of group II. The original transcriptomics data are from Zones et al. (2015), and profiles for other assembly factors and dynein components are shown in Supplemental Figure S1. RPKM, reads per kilobase per million mapped reads.
The Chlamydomonas wdr92-1 mutant
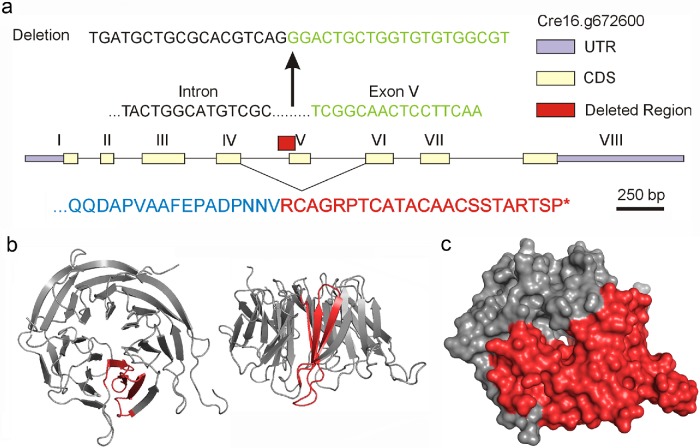
In Chlamydomonas, the 3.5-kb WDR92 gene is located at Cre16.g672600 and contains eight exons encoding a 358-residue protein consisting of seven WD repeats; WDR92 has been highly conserved, and the Chlamydomonas and human proteins are 58% identical. Examination of the CLiP mutant library database (www.chlamylibrary.org) identified a potential Chlamydomonas mutant (LMJ.RY0402.137495) with a putative C1B1 paromomycin-resistance cassette insertion within the coding region of the WDR92 gene. PCR was used to amplify the insertion boundaries using primers from within the resistance cassette paired with primers within the WDR92 gene upstream and downstream of the putative insertion site (Supplemental Table S1). Subsequent sequence analysis of the PCR products (Figure 3a) revealed that the insertion deleted a 38–base pair region that spans the junction between exon V and the immediately upstream intron, removing the splice site and thus the coding sequence for an entire blade of the WDR92 β-propeller (Figure 3b). In consequence, exon IV can only splice to exon VI, leading to a frameshift, 23 new residues, and then an in-frame stop codon (Figure 3a); thus, the encoded protein lacks the C-terminal four WD repeats (Figure 3c).
FIGURE 3:
Analysis of the wdr92-1 insertional mutant. (a) Map of the WDR92 intron/exon gene structure showing the region (red box) and sequence deleted by insertion of the C1B1 paromomycin resistance cassette. Removal of the 5′ splice site for exon V allows exon IV to splice to exon VI, leading to a frameshift, 23 new residues, and a stop codon (red in lower sequence). (b) Two views of the ribbon structure (PDB 3I2N) for human WDR92 illustrating in red the blade of the β-propeller that is encoded by exon V. (c) Surface rendering of the WDR92 structure indicating in red the C-terminal region missing in the encoded wdr92-1 mutant.
wdr92-1 cells assemble very short cilia lacking dynein arms
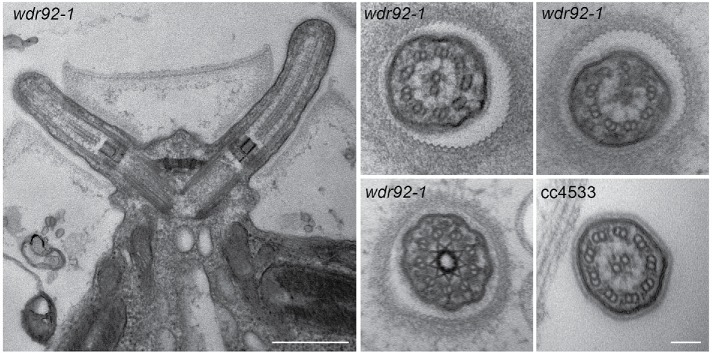
By differential interference contrast light microscopy, wdr92-1 cells appear to completely lack cilia (Figure 4, left). However, transmission electron microscopic analysis (Figure 5) revealed that they build very short ciliary stubs that barely protrude beyond the cell wall and have a length (as measured from the distal face of the transition zone to the ciliary tip) of 0.78 ± 0.21 μm (n = 15); to try and ensure that the cilium was not bent out of the plane of the ultrathin section, only cilia in which the central pair could be followed to the tip were included in the measurement statistics. Unlike cilia in wdr92-deficient planaria, the wdr92-1 Chlamydomonas cilia have both intact doublet microtubules and the central pair complex. There are minor accumulations of amorphous material between the doublet microtubules and the membrane in the very short wdr92-1 mutant cilia. Furthermore, although the transition zone appears morphologically normal, cross-sections reveal that these cilia lack both inner and outer dynein arms. Consistent with this, no immunofluorescent staining associated with ciliary stubs was observed in wdr92-1 cells probed with a monoclonal antibody (1869A) against the outer arm IC2 protein (Figure 4, right). The insertional wdr92 allele described by Liu et al. (2018) exhibits more variable ciliary length (37% aciliate, 61% very short < 2-μm stumps, 2% 2- to 4-μm cilia). This minor phenotypic difference between the two wdr92 mutants may reflect the distinct parental strains used for the original mutagenesis and/or the different culture media employed—minimal medium (used by Liu et al., 2018) versus R medium (used here), which contains acetate and allows growth to higher cell densities.
FIGURE 4:
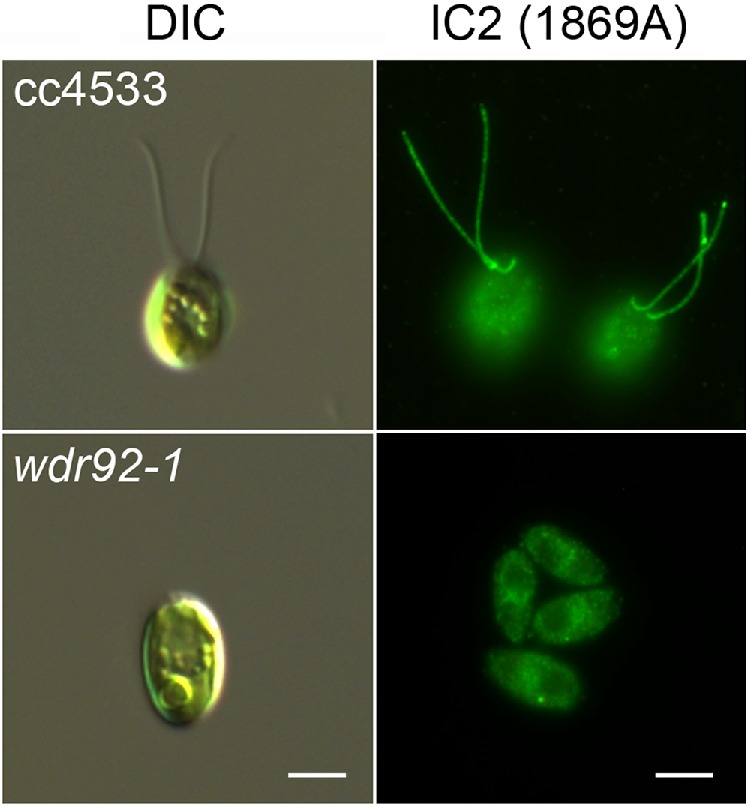
wdr92-1 mutant cells lack cilia. Left, differential interference contrast (DIC) images of control (cc4533) and wdr92-1 cells. The mutant lacks obvious ciliary structures. Right, immunofluorescence micrographs of cc4533 and wdr92-1 cells probed with monoclonal antibody 1869A that specifically recognizes outer arm dynein IC2 (King et al., 1985). No obvious staining of ciliary structures or stubs was observed. Scale bars: 5 μm.
FIGURE 5:
wdr92-1 mutant ciliary stubs are missing dynein arms. Left, electron micrograph of the basal body region of a wdr92-1 cell showing the two short ciliary structures that barely protrude beyond the cell wall. The general architecture of the transition zone and basal bodies appears normal. Scale bar: 500 nm. Right, cross-sections through wdr92-1 cilia reveal that the doublet microtubules and central pair microtubules are intact but that both inner and outer dynein arms appear to be missing; a section through a cc4533 control cilium is shown for comparison. Scale bar: 100 nm.
The ciliary length defect is rescued in a wdr92-1 tpg1-2 double mutant
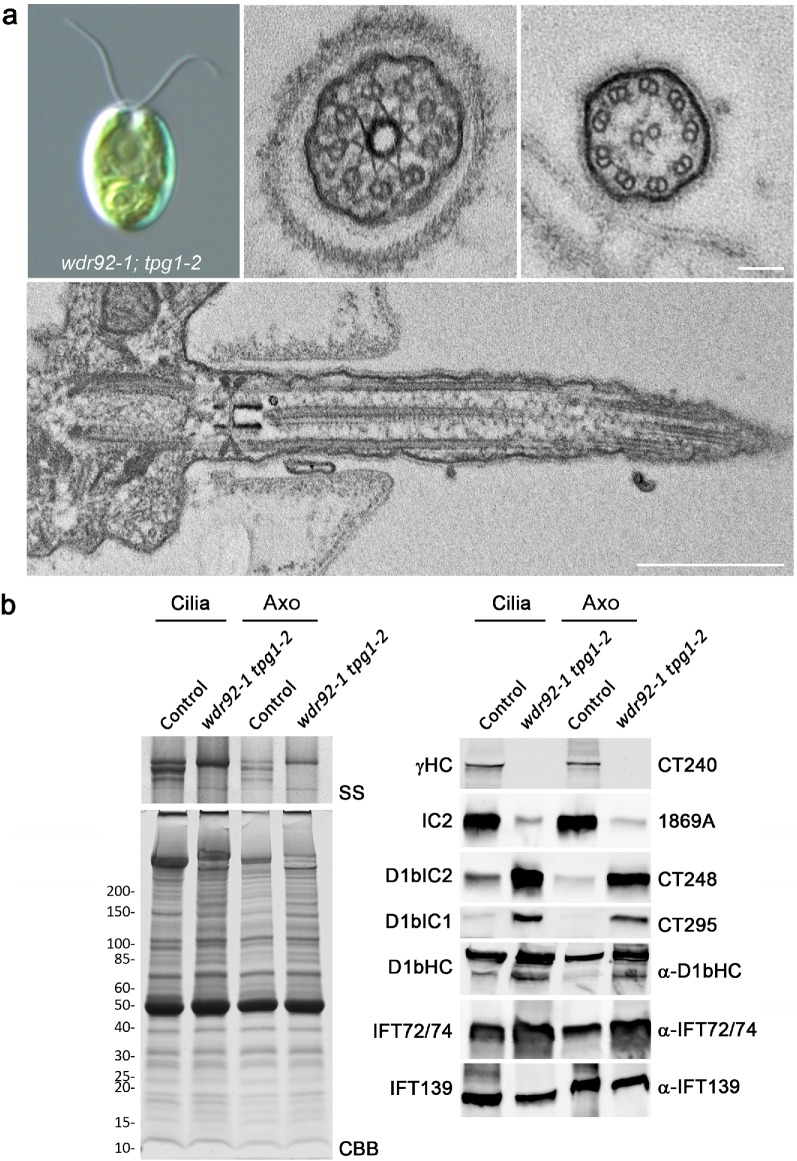
To ensure that the dynein assembly/short cilia phenotype was due to insertion of the paromomycin-resistance cassette within the WDR92 gene, the wdr92-1 insertional mutant was crossed to the tpg1-2 mutant, which is defective in a tubulin polyglutamylase; this mutation suppresses the short cilia phenotype commonly observed in strains lacking both outer and inner dynein arms by reducing microtubule dynamics at the distal ciliary tip and thereby at least partially compensating for the structural instability induced by the lack of dynein arms (Kubo et al., 2010, 2015). Thus, progeny containing the wdr92-1 mutation were predicted to be either essentially aciliate (wdr92-1 TPG1) or to have immotile cilia that are somewhat shorter than wild type (wdr92-1 tpg1-2). In three octads and 36 random paromomycin-resistant progeny, paromomycin resistance always cosegregated (n = 43) with essentially aciliate cells or those with 2- to 7-μm cilia that were completely immotile (Figure 6a) and appeared rather flaccid, as the cells drifted around before settling onto the microscope slide (see Supplemental Figure S2). In contrast, all paromomycin-sensitive progeny had motile cilia of normal length (n = 11).
FIGURE 6:
wdr92-1 tpg1-2 cells with reduced tubulin polyglutamylation build immotile cilia. (a) Differential interference contrast micrograph of a wdr92-1 tpg1-2 cell revealing that ∼7-μm-long cilia assemble in the absence of WDR92 when tubulin polyglutamylation is also defective (top left). Electron micrographs (other panels) show these cilia lack outer and inner dynein arms but retain nexin-DRC linkers, radial spokes, and the central pair complex. In addition, the transition zone and Y-links appear intact, and there is no obvious abnormal accumulation of IFT material. (b) Electrophoretic and immunoblot analysis of isolated cilia (Cilia) and demembranated axonemes (Axo) from a control tpg1-2 strain and the wdr92-1 tpg1-2 double mutant; the cilia were isolated using the pH shock method. Left, the high molecular weight region of a 4–15% acrylamide gradient gel that had been silver stained (SS) and the entire gradient gel stained with Coomassie blue (CBB). Dynein HCs are not evident, and the single prominent band represents the major membrane glycoprotein FMG-1. Right, immunoblots probed for the outer-arm γ-HC and IC2; the IFT dynein components D1bIC2, D1bIC1, and D1bHC; and the IFT72/74 (IFT complex B) and IFT139 (IFT complex A) proteins. No outer arm HC was detected in the double mutant, which surprisingly contained enhanced levels of IFT dynein components.
Cilia of the wdr92-1 tpg1-2 double mutants completely lack both inner and outer dynein arms, but otherwise appeared morphologically normal, with an intact transition zone and Y-links and no obvious accumulations of IFT material (Figure 6a). Electrophoretic and immunoblot analysis further confirmed the essentially complete absence of axonemal HCs and greatly reduced levels of the outer-arm IC2 protein (Figure 6b). In contrast, considerably enhanced levels of the IFT dynein HC and ICs (D1bIC1 and D1bIC2) were observed in both cilia and axoneme samples. As Kubo et al. (2015) demonstrated that reduced glutamylation in the tpg1 and tpg2 mutant strains did not affect ciliary levels of IFT components, this suggests that the IFT dynein increase observed in wdr92-1 tpg1-2 cilia represents a cellular response to the lack of axonemal dyneins available for ciliary assembly.
WDR92 is required for the cytoplasmic stability of axonemal dynein heavy chains
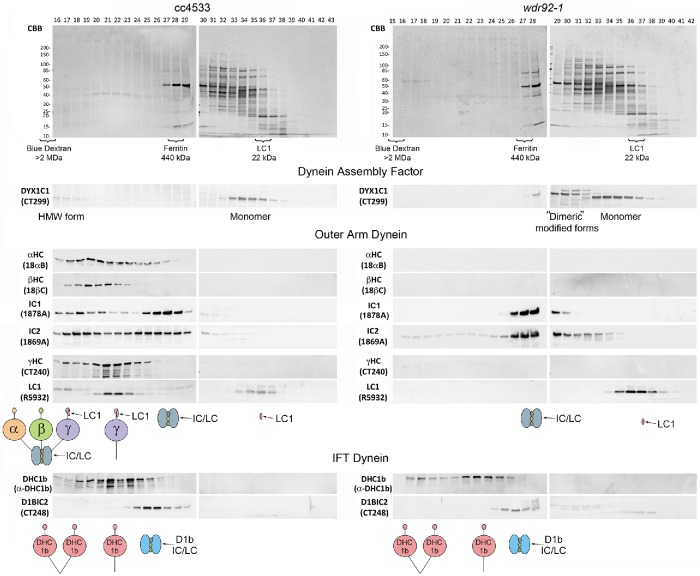
To assess the role of WDR92 in dynein preassembly, we prepared cytoplasmic extracts from cc4533 (control) and wdr92-1 cells using multiple rounds of freeze–thaw to release cellular contents. Unlike other standard protocols that involve either vortexing cells with glass beads or cell disruption by passage through a French press, this gentle method of cell breakage, which is particularly effective for “cell wall–less” strains and can likely be applied to autolysin-treated walled cells, resulted in concentrated extracts containing minimal contamination from the chloroplast; that is, the resulting supernatant solutions were clear rather than a dark green from released chlorophyll. Samples (500-μl injection volume) were then fractionated by gel-filtration chromatography using a Superose 6 10/300 column that allows for the separation of multi-megadalton complexes (Figure 7). Immunoblotting revealed that the α and β outer arm dynein HCs generally coeluted, whereas the γ HC was mostly present in a distinct peak of smaller mass that cofractionated with the γ HC–associated LC1 protein. In addition, a small amount of the γ HC comigrated with α and β HCs near the void volume and thus likely represents fully assembled outer arms containing all three HCs. In support of this interpretation, the WD-repeat IC1 and IC2 proteins were also found in two peaks: one comigrating with the three HCs and the αβ HC dimer, and a second peak of significantly smaller mass that is consistent with the intact IC/LC complex. In wdr92-1 cytoplasm, the α, β, and γ HCs were all barely detectable, suggesting that WDR92 is required for their stable formation. Both IC1 and IC2 were present only as part of the free IC/LC complex, and the 22-kDa LC1 protein that binds the γ HC microtubule-binding domain migrated solely as a monomer; neither the ICs or LC1 were present as the high molecular mass forms seen in control samples.
FIGURE 7:
Lack of outer arm dynein heavy chains in wdr92-1 cytoplasm. Freeze–thaw cytoplasmic extracts from cc4533 and wdr92-1 cells were fractionated in a Superose 6 10/300 gel-filtration column. Equal volumes of each fraction were electrophoresed in 4–15% gradient SDS gels and either stained with Coomassie blue (top) or blotted to nitrocellulose and probed with antibodies against various outer arm and IFT dynein components (bottom). The void volume (as indicated by blue dextran) eluted at fractions 15–16. Elution of other mass markers is indicated below the gels; note that, although recombinant LC1 has a mass of 22 kDa, it migrates during gel filtration with an apparent mass closer to 50–60 kDa, as it is a highly elongated molecule with a long axis approximately twice that of the orthogonal axes (Wu et al., 2003). In the absence of WDR92, outer arm HCs are almost undetectable, the IC/LC complex migrates as a single unit, and LC1 behaves solely as a monomer. Furthermore, a high molecular weight form of DYX1C1 that is likely associated with outer-arm HCs is missing from wdr92-1 cytoplasm and is replaced by a peak of appropriate mass to be a dimeric form; this altered DYX1C1 also exhibits modified electrophoretic mobility. In contrast, the IFT dynein HC is still present in the wdr92-1 mutant cytoplasm, migrating as a mix of HC dimers and monomers. Intriguingly, the IFT dynein IC/LC complex is not HC-associated in either mutant or control cytoplasm.
Intriguingly, the oligomeric status of the DYX1C1/PF23 (DNAAF4) assembly factor also revealed a dependence on WDR92 (Figure 7). Within control cytoplasm, most of this protein was present in a single peak with a mass suggestive of a monomeric form. However, a small amount was also observed at very high molecular weight in fractions containing all three outer arm HCs. In the absence of WDR92, several changes in the DYX1C1 pattern were observed. First, the high molecular weight form was completely absent, suggesting that it represents DYX1C1 associated with outer-arm HCs. It is possible that this complex represents a relatively stable intermediate in the dynein assembly pathway. Second, a new peak was observed with a mass consistent with formation of a DYX1C1 dimer or other DYX1C1-containing complexes; intriguingly, this peak consisted of two DYX1C1 immunoreactive bands, both of which exhibited altered electrophoretic mobility and were barely detectable in control samples.
IFT dynein HCs are stable in wdr92-1 cell cytoplasm
Analysis of wdr92-1 cytoplasmic extracts using antibodies against the IFT dynein HC (D1bHC) (Figure 7, bottom) demonstrated that D1bHC is stable and made in approximately wild-type amounts. Previously, we observed that the IFT dynein HCs and D1bIC2 (FAP133/WDR34) comigrate when isolated from the ciliary matrix, although an additional D1bIC2 peak of smaller mass was also evident (Rompolas et al., 2007). In contrast, the gel-filtration profile for this dynein in cytoplasm revealed that most of the IFT HCs migrate as monomers, peaking in the same fraction as the outer arm γ HC; a dimer peak is also clearly evident in the wdr92-1 extract. Furthermore, the ICs are not associated with HCs under these conditions, even those that migrate as HC dimers, suggesting that assembly of the IFT dynein holoenzyme might occur immediately before ciliary entry. Although the IFT dynein HC dimer can adopt an autoinhibited φ conformation with stacked motor domains and crossed microtubule-binding stalks (Pigino and King, 2017; Toropova et al., 2017), one possibility is that control of the HC-IC/LC complex association reflects an additional regulatory step that prevents premature loading of fully functional, albeit autoinhibited, dyneins onto IFT particles.
DISCUSSION
Role of WDR92 and R2TP variants in axonemal dynein assembly
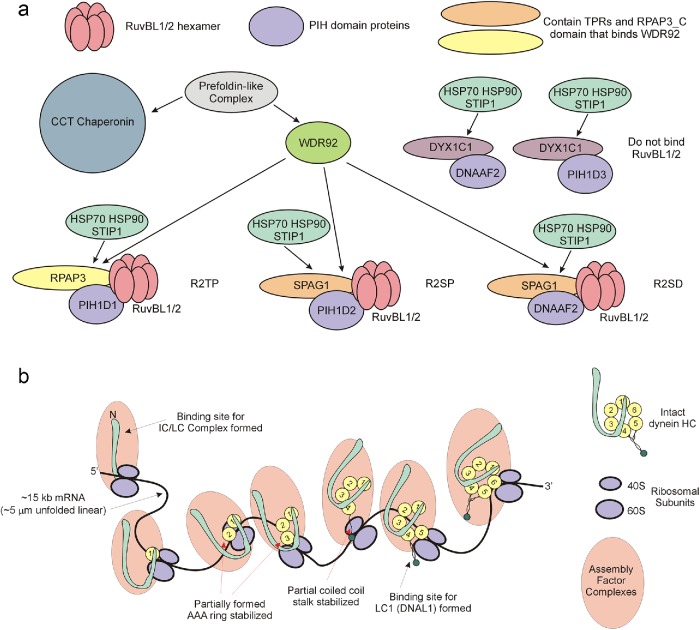
Currently, three variants of the R2TP complex (R2TP, R2SP, and R2SD) are recognized (Figure 8a) (Maurizy et al., 2018). These consist of a RuvBL1/2 (pontin/reptin) hetero-hexamer that can undergo dimerization to yield a dodecameric AAA+ domain ATPase, a scaffolding component containing both TPR and RPAP3_C domains (RPAP3 or SPAG1) associated with a PIH domain protein (PIH1D1, PIH1D2, or DNAAF2) (Yamamoto et al., 2010) that together recruit the chaperones HSP70 and HSP90, and the cochaperone STIP1. In addition, both DNAAF2 and PIH1D3 can bind DYX1C1, another TPR scaffold protein, to form additional complexes that appear to lack RuvBL1/2 but are required for axonemal dynein assembly (Tarkar et al., 2013; Yamamoto et al., 2017; Maurizy et al., 2018). It is thought that WDR92 binds the RPAP3_C domains of both RPAP3 and SPAG1 (Cloutier et al., 2017) and so can potentially participate in both canonical and variant R2TP-mediated assembly. As different axonemal dyneins require distinct PIH domain proteins for assembly (Yamaguchi et al., 2018), this observation is consistent with the loss of both inner and outer dynein arms in the wdr92 mutant.
FIGURE 8:
The role of WDR92 in the axonemal dynein assembly pathway. (a) Diagram illustrating the central role that WDR92 may play in coordinating association of R2TP (and variant) complexes with the prefoldins and the CCT chaperonin. WDR92 interacts directly with both prefoldins and the RPAP3_C domains present in RPAP3 and SPAG1. These in turn scaffold interactions with the RuvBL1/2 AAA+ ATPases, and PIH proteins that mediate recruitment of HSP70 and HSP90. As eukaryotes (such as yeast, higher plants, and nematodes) missing axonemal dyneins also lack WDR92, how prefoldins and chaperonins associate with R2TP in these organisms remains unclear. Furthermore, although clearly essential for axonemal dynein HC formation, why cytoplasmic and IFT dynein HCs, which have similar folding requirements, are unaffected remains unexplored. (b) Model illustrating the synthesis of dynein HCs. Each spliced HC mRNA is ∼15 kb, which equates to an unfolded linear molecule of ∼5 μm; at an incorporation rate of 5 residues/s, a ribosome would take ∼13–15 min to track along an mRNA of this length. Simultaneous association of numerous ribosomes would lead to a group of partially synthesized dynein HCs associated with a complex array of cytoplasmic assembly and stabilization factors. Given the circular arrangement of AAA domains within the dynein motor unit, an HC can be fully stabilized only once the terminal AAA unit has been synthesized. However, binding sites for individual dynein components such as LC1 (DNAL1), which associates near the ATP-dependent microtubule-binding domain, or the IC/LC complex, which interacts with the N-terminal region, may become available and occupied before completion of HC synthesis.
What role does WDR92 play in the R2TP complex? In addition to binding RPAP3_C domains, WDR92 also associates with a prefoldin-like cochaperone complex and might act to target that complex and thus the CCT chaperonin to dynein assembly sites (Cloutier et al., 2017). Prefoldins are of ancient origin; they are ubiquitous throughout the eukaryotes and are also present in Archaea (Vainberg et al., 1998). Chlamydomonas encodes eight prefoldin subunits (Cao, 2016)—canonical prefoldins 1–6 plus subunits related to the noncanonical human prefoldin-like proteins UXT and RPB5. These subunits form hetero-hexamers that associate with unfolded proteins and target them to chaperonins for refolding. Prefoldins do not exhibit enzymatic activity per se, but rather interact with their targets through a series of relatively weak hydrophobic interfaces on six coiled-coil “tentacles” that extend from a double β-barrel core; that is, prefoldins act as “holdases” using ATP-independent interactions to maintain unfolded protein segments in a stable nonaggregated state (Siegert et al., 2000; Comyn et al., 2016). Thus, WDR92 appears to function as a specific scaffolding hub to recruit these key cochaperone complexes to sites of active dynein HC synthesis and thereby maintain the nascent HCs in a state conducive to subsequent folding once the complete molecule has been synthesized; direct interactions with multiple highly conserved components of the folding pathway likely explain the high degree of sequence identity between Chlamydomonas and human WDR92 that covers much of the molecular surface (Patel-King and King, 2016). This model for WDR92 function makes the testable prediction that cells defective in the UXT and/or RPB5 interactor noncanonical prefoldins may exhibit ciliary dynein assembly defects by disrupting the HC folding pathway.
Using mass spectrometry of wdr92 mutant Drosophila extracts, zur Lage et al. (2018) reported a decrease in both axonemal dynein HCs and ICs and predicted WDR92 acts with the canonical R2TP complex at a mid- to late stage of dynein preassembly. In Chlamydomonas, although Liu et al. (2018) reported a decrease in outer-arm ICs, we did not observe a significant reduction in outer arm IC2 signal but merely a shift to a low molecular mass species consistent with a stable IC/LC complex not associated with HCs. Similarly, LC1, which normally binds to the MT-binding domain of the γ HC, was found only in monomeric form, but in wild-type amounts, in the absence of WDR92. Thus, our data confirm a key role for WDR92 in the formation of axonemal dyneins and furthermore reveal that WDR92 is needed specifically for the stable synthesis of dynein HCs and that other outer-arm components appear generally unaffected by its loss.
General assembly of dyneins in cytoplasm
Recent studies have suggested that axonemal dynein assembly in multiciliated cells takes place in phase-separated compartments that exhibit some fluid-like properties, including fission/coalescence and rapid intraparticle exchange of some components (Huizar et al., 2018). These studies examined the colocalization of multiple dynein assembly factors and several dynein components, including outer-arm ICs and LCs and two inner-arm components. Although no HCs were included in these analyses, the accumulations contained multiple factors known to be required for HC assembly, and their formation may, at least in part, be reflective of the sheer scale imparted by dynein HC synthesis. The mRNA for an HC is ∼15 kb; without folding, this is ∼5 μm long. When a HC mRNA is docked onto many ribosomes, each of which would have a partially synthesized and therefore unstable HC associated, addition of the needed stabilization factors would then form an enormous synthetic complex whose geometry is dictated by mRNA folding and, possibly, interactions among the assembly factors (Figure 8b). Assuming mRNAs for both key outer arm HCs (i.e., those orthologous to the β and γ HCs of Chlamydomonas) are nearby each other, this might also explain why the ICs and LCs examined by Huizar et al. (2018) did not exhibit rapid exchange, while the assembly factors did. For example, once the N-terminal HC regions have been synthesized, they could interact and thereby provide a stable template for association of the IC/LC complex. IC/LC complexes can form independently and remain stable in the absence of HCs; indeed, it has been reported that the IC/LC complex can associate with the β HC alone (Pfister and Witman, 1984), which would be sufficient to stabilize it within the synthetic complex. Similarly, once the microtubule-binding domain of the HC (DNAH5 and/or 8) equivalent to the Chlamydomonas γ HC was made, the LC1 (DNAL1) protein would be able to stably bind; these components do not generally undergo rapid dissociation or exchange under normal conditions and indeed, at least in Chlamydomonas, are stable to high ionic strength (e.g., Pfister et al., 1982; King, 2013). In contrast, cytoplasmic assembly factors must interact with nascent dyneins relatively weakly, as they need dissociate once formation of the motor is complete.
Given the inherent similarity and common origin of dynein HCs (Wickstead, 2018), the general cellular requirements for synthesis, folding, and formation of the dynein that powers retrograde IFT and for canonical cytoplasmic dynein are likely highly similar to those needed to assemble axonemal dyneins. However, it seems clear from many mutational studies in diverse organisms (including vertebrates, planarians, and algae) that these dyneins must employ an assembly pathway distinct from that used by axonemal dyneins, as their synthesis and formation appear unaffected by mutations that completely abolish axonemal HCs (e.g., Omran et al., 2008; Tarkar et al., 2013). Our studies on WDR92 further confirm this. WDR92 is present only in organisms whose genomes encode axonemal dynein HCs and is specifically missing in those such as C. elegans that only make cytoplasmic/IFT dynein motors; yeasts that have only cytoplasmic dynein; or angiosperms, amoebozoa, and red algae, which lack all dyneins. Thus, we predict that a component functionally analogous to WDR92 that recruits folding pathway components is required for cytoplasmic/IFT dynein HC formation; identifying this factor will provide further insight into the general mechanisms of HC formation.
In conclusion, it is now clear the WDR92 is a cytoplasmic factor needed specifically for the assembly and/or stability of axonemal dynein HCs. The very high degree of sequence identity among divergent species suggests that it plays a key, conserved, and central role in the assembly process potentially targeting noncanonical prefoldins to sites of dynein HC formation. Furthermore, the observation that neither the IFT dynein nor canonical cytoplasmic dynein HCs require WDR92 for their formation implies that there is a separate, currently unknown assembly pathway for these motors that likely includes a functional analogue of WDR92.
MATERIALS AND METHODS
Chlamydomonas strains and cell culture
Chlamydomonas reinhardtii strains were cultured in liquid TAP or R medium with aeration on a 12 h/12 h light/dark cycle. The paromomycin-resistant wdr92 mutant (CLiP library strain LMJ.RY0402.137495; Li et al., 2019), which we have previously termed wdr92-1 (King et al., 2018), and the control strain cc4533 were obtained from the Chlamydomonas Resource Center at the University of Minnesota. The wdr92-1 strain was crossed to the tubulin polyglutamylase–deficient strain tpg1-2 that suppresses the short cilia phenotype induced by lack of inner and outer dynein arms by Susan Dutcher (Washington University School of Medicine, St. Louis, MO). Octad and random progeny were screened for the C1B1 cassette on TAP plates containing paromomycin, and ciliary length and motility were monitored by phase-contrast microscopy.
Molecular biology
Colony PCR amplification of the C1B1 cassette insert boundaries was performed using insert and gene-specific primers (Supplemental Table S1). All DNA products were sequenced by GenScript (Piscataway, NJ).
Light and electron microscopy
Differential interference contrast micrographs of live and formaldehyde-fixed cells were taken on an Olympus BX51 microscope equipped with a ProgRes CFscan CCD camera (Jenoptik, Jena, Germany) using a 60×/1.40 oil-immersion objective lens.
For thin-section electron microscopy, log-phase cells in TAP medium were fixed by addition of an equal volume of TAP containing 5% glutaraldehyde (EM grade; EM Sciences, Hatfield, PA). After 15 min, cells were harvested by gentle centrifugation and resuspended in 2.5% glutaraldehyde in 0.1 M Na cacodylate (pH 7.4) for 45 min. Cells were then washed five times with cacodylate buffer and postfixed with 1% OsO4 and 0.8% K3Fe(CN)6 in cacodylate buffer for 60 min. Following multiple washes with distilled water, samples were stained en bloc with 1% aqueous uranyl acetate, dehydrated through an ethanol series, transitioned to propylene oxide, and embedded in Poly/Bed 812 epoxide resin (Polysciences, Warrington, PA). Ultrathin sections with a nominal thickness of 55 nm were picked up on unsupported 300-mesh copper grids, poststained with 6.25% uranyl acetate in 50% methanol, and examined in a Hitachi H-7650 transmission electron microscope operating at 80 kV.
All micrographs were cropped and adjusted for brightness/contrast using Adobe Photoshop CS4.
Fractionation of cytoplasmic extracts and cilia isolation
For examination of the status of axonemal dyneins in cytoplasm, cc4533 and wdr92-1 strains were grown to mid–log phase, and cells from ∼50 ml of culture were pelleted by low-speed centrifugation. The medium was then removed, and the pellets were frozen at −80°C. Subsequently, pellets were defrosted and resuspended in 750 μl of 20 mM Tris·Cl (pH 8.0), 150 mM NaCl containing broad-spectrum protease inhibitors (10 μl/ml P8340; Sigma, St. Louis, MO), and the samples were subjected to three rounds of freeze–thaw alternating between −80°C and room temperature. Cell remnants were removed by centrifugation in a microfuge, and the resulting clear supernatants (0.5-ml injection volume) lacking obvious chloroplast contamination were then fractionated by gel filtration in a Superose 6 10/300 column running at 0.25 ml/min on an ÄktaPurifier 100 chromatography workstation; 0.5-ml fractions were collected.
Control and wdr92-1 tpg1-2 cells were deflagellated either with dibucaine or by pH shock, and the cilia were purified by our standard methods (King, 2013). Samples were electrophoresed in 4–15% SDS–polyacrylamide gels and either stained with Coomassie blue or blotted to nitrocellulose. Blots were probed with previously described specific antibodies (Supplemental Table S2), and immunoreactive bands were detected using enhanced chemiluminescence. For more detailed analysis of dynein HCs, samples were electrophoresed in 4–15% SDS–polyacrylamide gels and silver stained.
Phylogenetic analysis and structure display
Searches of the nonredundant sequence databases with Chlamydomonas and human WDR92 sequences were performed using BLAST. Ribbon and surface diagrams of the human WDR92 crystal structure (PDB accession 3I2N) were generated and colored using the PyMOL molecular graphics system, v. 2.0 (Schrödinger).
Supplementary Material
Acknowledgments
We are very grateful to Susan Dutcher (Washington University School of Medicine) for crossing the wdr92-1 mutant to tpg1-2. We also thank David Mitchell (SUNY Upstate Medical University) for his advice concerning the DAP1/PF13 gene model, and Yuqing Hou and George Witman (University of Massachusetts Medical School) for antibody against the IFT dynein heavy chain. This study was supported by grant GM051293 from the National Institutes of Health (to S.M.K.).
Abbreviations used:
- HC
heavy chain
- IC
intermediate chain
- IFT
intraflagellar transport
- LC
light chain.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-03-0139) on May 22, 2019.
REFERENCES
- Ahmed N, Gao C, Lucker B, Cole D, Mitchell D. (2008). ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery. J Cell Biol , 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron DM, Kabututu ZP, Hill KL. (2007). Stuck in reverse: loss of LC1 in Trypanosoma brucei disrupts outer dynein arms and leads to reverse flagellar beat and backward movement. J Cell Sci , 1513–1520. [DOI] [PubMed] [Google Scholar]
- Benashski SE, Patel-King RS, King SM. (1999). Light chain 1 from the Chlamydomonas outer dynein arm is a leucine-rich repeat protein associated with the motor domain of the γ heavy chain. Biochemistry , 7253–7264. [DOI] [PubMed] [Google Scholar]
- Boulon SV, Pradet-Balade BRR, Verheggen CL, Molle DE, Boireau SP, Georgieva M, Azzag K, Robert M-CC, Ahmad Y, Neel H, et al. (2010). HSP90 and its R2TP/prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell , 912–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantaut-Belarif Y, Sternberg JR, Thouvenin O, Wyart C, Bardet P-L. (2018). The Reissner fiber in the cerebrospinal fluid controls morphogenesis of the body axis. Curr Biol , 2479–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J. (2016). Analysis of the prefoldin gene family in 14 plant species. Front Plant Sci , 317–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier P, Poitras C, Durand M, Hekmat O, Fiola-Masson É, Bouchard A, Faubert D, Chabot B, Coulombe B. (2017). R2TP/prefoldin-like component RUVBL1/RUVBL2 directly interacts with ZNHIT2 to regulate assembly of U5 small nuclear ribonucleoprotein. Nat Comm , 15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comyn SA, Young BP, Loewen CJ, Mayor T. (2016). Prefoldin promotes proteasomal degradation of cytosolic proteins with missense mutations by maintaining substrate solubility. PLoS Genet , e1006184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, Omran H. (2007). When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol , 880–893. [DOI] [PubMed] [Google Scholar]
- Huizar RL, Lee C, Boulgakov AA, Horani A, Tu F, Marcotte EM, Brody SL, Wallingford JB. (2018). A liquid-like organelle at the root of motile ciliopathy. eLife , e38497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez-Tallon I, Heintz N, Omran H. (2003). To beat or not to beat: roles of cilia in development and disease. Hum Mol Genet , R27–R35. [DOI] [PubMed] [Google Scholar]
- Ichikawa M, Saito K, Yanagisawa H-A, Yagi T, Kamiya R, Yamaguchi S, Yajima J, Kushida Y, Nakano K, Numata O, Toyoshima YY. (2015). Axonemal dynein light chain-1 locates at the microtubule-binding domain of the γ heavy chain. Mol Biol Cell , 4236–4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsuki Y, Saeki M, Nakahara H, Egusa H, Irie Y, Terao Y, Kawabata S, Yatani H, Kamisaki Y. (2008). Molecular cloning of novel Monad binding protein containing tetratricopeptide repeat domains. FEBS Lett , 2365–2370. [DOI] [PubMed] [Google Scholar]
- Kakihara Y, Houry WA. (2012). The R2TP complex: discovery and functions. Biochim Biophys Acta , 101–107. [DOI] [PubMed] [Google Scholar]
- Kamiya R, Okamoto M. (1985). A mutant of Chlamydomonas reinhardtii that lacks the flagellar outer dynein arm but can swim. J Cell Sci , 181–191. [DOI] [PubMed] [Google Scholar]
- King SM. (2013). Biochemical and physiological analysis of axonemal dyneins. Methods Enzymol , 124–145. [DOI] [PubMed] [Google Scholar]
- King SM. (2018a). Composition and assembly of axonemal dyneins. In: Dyneins: Structure, Biology and Disease, Vol. , The Biology of Dynein Motors, ed. King SM, Oxford, UK: Elsevier, Academic Press, 163–201. [Google Scholar]
- King SM. (ed.) (2018b). Dyneins: Structure, Biology and Disease, Vol. , The Biology of Dynein Motors, Oxford, UK: Elsevier, Academic Press. [Google Scholar]
- King SM. (ed.) (2018c). Dyneins: Structure, Biology and Disease, Vol. , Dynein Mechanics, Dysfunction and Disease, Oxford, UK: Elsevier, Academic Press. [Google Scholar]
- King SM, Otter T, Witman GB. (1985). Characterization of monoclonal antibodies against Chlamydomonas flagellar dyneins by high-resolution protein blotting. Proc Natl Acad Sci USA , 4717–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Patel-King RS. (1995). Identification of a Ca(2+)-binding light chain within Chlamydomonas outer arm dynein. J Cell Sci , 3757–3764. [DOI] [PubMed] [Google Scholar]
- King SM, Patel-King RS. (2012). Functional architecture of the outer arm dynein conformational switch. J Biol Chem , 3108–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Sakato-Antoku M, Patel-King RS. (2018). WDR92 is required for outer arm dynein assembly in cytoplasm. Mol Biol Cell , 3063 (abstract #P2021). [Google Scholar]
- King SM, Sale WS. (2018). Fifty years of microtubule sliding in cilia. Mol Biol Cell , 698–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Hirono M, Aikawa T, Kamiya R, Witman GB, Marshall W. (2015). Reduced tubulin polyglutamylation suppresses flagellar shortness in Chlamydomonas . Mol Biol Cell , 2810–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Yanagisawa H-A, Yagi T, Hirono M, Kamiya R. (2010). Tubulin polyglutamylation regulates axonemal motility by modulating activities of inner-arm dyneins. Curr Biol , 441–445. [DOI] [PubMed] [Google Scholar]
- Li X, Patena W, Fauser F, Jinkerson RE, Saroussi S, Meyer MT, Ivanova N, Robertson JM, Yue R, Zhang R, et al. (2019). A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis. Nat Genet , 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yagi H, Onuoha EO, Damerla RR, Francis R, Furutani Y, Tariq M, King SM, Hendricks G, Cui C, et al. (2016). DNAH6 and its interactions with PCD genes in heterotaxy and primary ciliary dyskinesia. PLoS Genet , e1005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Nicastro D. (2018). Asymmetric distribution and spatial switching of dynein activity generates ciliary motility. Science , eaar1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Wang L, Pan J. (2018). Chlamydomonas WDR92 in association with R2TP-like complex and multiple DNAAFs to regulate ciliary dynein preassembly. J Mol Cell Biol, 10.1093/jmcb/mjy067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loges NT, Omran H. (2018). Dynein dysfunction as a cause of primary ciliary dyskinesia and other ciliopathies. In: Dyneins: Structure, Biology and Disease, Vol. , Dynein Mechanics, Dysfunction and Disease, ed. King SM, Oxford, UK: Elsevier, Academic Press, 317–355. [Google Scholar]
- Marshall W, Basto R. (2017). Cilia, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Maurizy C, Quinternet M, Abel Y, Verheggen C, Santo PE, Bourguet M, Paiva ACF, Paiva A, Bragantini B, Chagot M-E, et al. (2018). The RPAP3-C terminal domain identifies R2TP-like quaternary chaperones. Nat Comm , 2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millán-Zambrano G, Chávez S. (2014). Nuclear functions of prefoldin. Open Biol , 140085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR. (2018). Cytoplasmic preassembly and trafficking of axonemal dyneins. In: Dyneins: Structure, Biology and Disease, Vol. , The Biology of Dynein Motors, ed. King SM, Oxford, UK: Elsevier, Academic Press, 141–161. [Google Scholar]
- Mitchell DR, Rosenbaum JL. (1985). A motile Chlamydomonas flagellar mutant that lacks outer dynein arms. J Cell Biol , 1228–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. (2006). The molecular architecture of axonemes revealed by cryoelectron tomography. Science , 944–948. [DOI] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Kamiya R, Kikkawa M. (2014). A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science , 857–860. [DOI] [PubMed] [Google Scholar]
- Omran H, Kobayashi D, Olbrich H, Tsukahara T, Loges NT, Hagiwara H, Zhang Q, Leblond G, O/'Toole E, Hara C, et al. (2008). Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature , 611–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenik B, Grimwood J, Aerts A, Rouzé P, Salamov A, Putnam N, Dupont C, Jorgensen R, Derelle E, Rombauts S, et al. (2007). The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci USA , 7705–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizzi J, Becker-Heck A, Castleman V, Al-Mutairi D, Liu Y, Loges NT, Pathak N, Austin-Tse C, Sheridan E, Schmidts M, et al. (2012). CCDC103 mutations cause primary ciliary dyskinesia by disrupting assembly of ciliary dynein arms. Nat Genet , 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel-King RS, Benashki SE, Harrison A, King SM. (1996). Two functional thioredoxins containing redox-sensitive vicinal dithiols from the Chlamydomonas outer dynein arm. J Biol Chem , 6283–6291. [DOI] [PubMed] [Google Scholar]
- Patel-King RS, King SM. (2009). An outer arm dynein light chain acts in a conformational switch for flagellar motility. J Cell Biol , 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel-King RS, King SM. (2016). A prefoldin-associated WD-repeat protein (WDR92) is required for the correct architectural assembly of motile cilia. Mol Biol Cell , 1204–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G, Agrin N, Leszyk J, Witman G. (2005). Proteomic analysis of a eukaryotic flagellum. J Cell Biol , 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister KK, Fay RB, Witman GB. (1982). Purification and polypeptide composition of dynein ATPases from Chlamydomonas flagella. Cell Motil , 525–547. [DOI] [PubMed] [Google Scholar]
- Pfister KK, Witman GB. (1984). Subfractionation of Chlamydomonas 18 S dynein into two unique subunits containing ATPase activity. J Biol Chem , 12072–12080. [PubMed] [Google Scholar]
- Pigino G, King SM. (2017). Switching dynein motors on and off. Nat Struct Mol Biol , 557–559. [DOI] [PubMed] [Google Scholar]
- Rompolas P, Pedersen L, Patel-King RS, King SM. (2007). Chlamydomonas FAP133 is a dynein intermediate chain associated with the retrograde intraflagellar transport motor. J Cell Sci , 3653–3665. [DOI] [PubMed] [Google Scholar]
- Sakato M, Sakakibara H, King SM. (2007). Chlamydomonas outer arm dynein alters conformation in response to Ca2+. Mol Biol Cell , 3620–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert R, Leroux MR, Scheufler C, Hartl FU, Moarefi I. (2000). Structure of the molecular chaperone prefoldin: unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell , 621–632. [DOI] [PubMed] [Google Scholar]
- Takada S, Kamiya R. (1994). Functional reconstitution of Chlamydomonas outer dynein arms from alpha-beta and gamma subunits: requirement of a third factor. J Cell Biol , 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WJ, Bell CW, Sale WS, Gibbons IR. (1982). Structure of the dynein-1 outer arm in sea urchin sperm flagella. I. Analysis by separation of subunits. J Biol Chem , 508–515. [PubMed] [Google Scholar]
- Tarkar A, Loges NT, Slagle CE, Francis R, Dougherty GW, Tamayo JV, Shook B, Cantino M, Schwartz D, Jahnke C, et al. (2013). DYX1C1 is required for axonemal dynein assembly and ciliary motility. Nat Genet , 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toropova K, Mladenov M, Roberts AJ. (2017). Intraflagellar transport dynein is autoinhibited by trapping of its mechanical and track-binding elements. Nat Struct Mol Biol , 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainberg IE, Lewis SA, Rommelaere H, Ampe C, Vandekerckhove J, Klein HL, Cowan NJ. (1998). Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell , 863–873. [DOI] [PubMed] [Google Scholar]
- Wakabayashi K, King SM. (2006). Modulation of Chlamydomonas reinhardtii flagellar motility by redox poise. J Cell Biol , 743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B. (2018). The evolutionary biology of dyneins. In: Dyneins: Structure, Biology and Disease, Vol. , The Biology of Dynein Motors, ed. King SM, Oxford, UK: Elsevier, Academic Press, 101–138. [Google Scholar]
- Wu H, Blackledge M, Maciejewski MW, Mullen GP, King SM. (2003). Relaxation-based structure refinement and backbone molecular dynamics of the dynein motor domain-associated light chain. Biochemistry , 57–71. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Oda T, Kikkawa M, Takeda H. (2018). Systematic studies of all PIH proteins in zebrafish reveal their distinct roles in axonemal dynein assembly. eLife , e36979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Hirono M, Kamiya R. (2010). Discrete PIH proteins function in the cytoplasmic preassembly of different subsets of axonemal dyneins. J Cell Biol , 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Obbineni JM, Alford LM, Ide T, Owa M, Hwang J, Kon T, Inaba K, James N, King SM, et al. (2017). Chlamydomonas DYX1C1/PF23 is essential for axonemal assembly and proper morphology of inner dynein arms. PLoS Genet , e1006996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zones JM, Blaby IK, Merchant SS, Umen JG. (2015). High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. Plant Cell , 2743–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Lage P, Stefanopoulou P, Styczynska-Soczka K, Quinn N, Mali G, von Kriegsheim A, Mill P, Jarman AP. (2018). Ciliary dynein motor preassembly is regulated by Wdr92 in association with HSP90 co-chaperone, R2TP. J Cell Biol , 2583–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.