Abstract
Oocytes of many species lack centrioles and therefore form acentriolar spindles. Despite the necessity of oocyte meiosis for successful reproduction, how these spindles mediate accurate chromosome segregation is poorly understood. We have gained insight into this process through studies of the kinesin-4 family member Kif4 in mouse oocytes. We found that Kif4 localizes to chromosomes through metaphase and then largely redistributes to the spindle midzone during anaphase, transitioning from stretches along microtubules to distinct ring-like structures; these structures then appear to fuse together by telophase. Kif4’s binding partner PRC1 and MgcRacGAP, a component of the centralspindlin complex, have a similar localization pattern, demonstrating dynamic spindle midzone organization in oocytes. Kif4 knockdown results in defective midzone formation and longer spindles, revealing new anaphase roles for Kif4 in mouse oocytes. Moreover, inhibition of Aurora B/C kinases results in Kif4 mislocalization and causes anaphase defects. Taken together, our work reveals essential roles for Kif4 during the meiotic divisions, furthering our understanding of mechanisms promoting accurate chromosome segregation in acentriolar oocytes.
INTRODUCTION
Meiosis is an important type of cell division that is essential for sexual reproduction. During meiosis, cells undergo one round of DNA replication followed by two rounds of division, reducing their chromosome number by half to form haploid gametes. Female meiosis in many species is unique because oocytes lack centriole-containing centrosomes, which typically serve to nucleate microtubules and organize spindle poles in mitotic and male meiotic spindles (Szollosi et al., 1972; Prosser and Pelletier, 2017). Thus, acentriolar spindles are morphologically distinct from centriolar spindles and employ unique mechanisms for bipolar spindle assembly (Schuh and Ellenberg, 2007; Wolff et al., 2016; Mullen and Wignall, 2017; Radford et al., 2017). Despite the importance of oocyte meiosis for the propagation of sexually reproducing species, mechanisms responsible for ensuring accurate chromosome partitioning on these acentriolar spindles are poorly understood.
Microtubule-based motors (including dynein and multiple families of kinesins) play essential roles in cell division and have diverse functions in spindle assembly and maintenance as well as in facilitating chromosome movements (Wordeman, 2010). Kinesins that bind chromosomes, termed chromokinesins, have been well studied in mitosis in a variety of systems. Xenopus laevis Xklp1, the first-characterized member of the kinesin-4 family of chromokinesins, was shown to play a role in both spindle bipolarity and chromosome congression in egg extracts, which is indicative of its ability to bind both chromosomes and microtubules (Vernos et al., 1995). Moreover, mammalian kinesin-4 (Kif4) has been extensively studied. Human Kif4 is a plus-end–directed motor with an N-terminal motor domain and a C-terminal DNA-binding region. Additionally, Kif4 contains a nuclear localization sequence and Aurora kinase (AURK) phosphorylation sites in its coiled-coil region (Lee et al., 2001; Samejima et al., 2012; Nunes Bastos et al., 2013). In mitosis, Kif4 localizes to chromosome arms during metaphase and to the midzone in anaphase and has roles in spindle bipolarity and in chromosome structure, congression, and segregation (Mazumdar et al., 2004; Hu et al., 2011; Samejima et al., 2012). Moreover, the nonmotile microtubule cross-linker PRC1 (protein regulator of cytokinesis 1) binds Kif4’s C-terminus in vitro, and both Kif4 and PRC1 localize to and are required to build a proper anaphase spindle midzone, thus preventing overelongation of the spindle during chromosome segregation (Kurasawa et al., 2004; Lee and Kim, 2004; Zhu and Jiang, 2005; Zhu et al., 2005; Gruneberg et al., 2006; Bieling et al., 2010; Subramanian et al., 2010; Hu et al., 2011; Nunes Bastos et al., 2013). However, despite the extensive research that has been done on human Kif4 in mitosis, little is known about its role in meiosis.
Mouse and human Kif4 are nearly identical, sharing major domains and conserved sites (Oh et al., 2000). Recent studies of Kif4 in mouse oocyte meiosis showed that Kif4 displays both kinetochore and diffuse spindle localization, and demonstrated that knockdown causes abnormal spindle morphology and defective polar body (PB) extrusion (Camlin et al., 2017; Tang et al., 2018). However, Kif4 was not reported to have broad chromosomal localization in either study, raising the question of whether it acts as a chromokinesin in oocytes. Additionally, little is known about whether Kif4 has roles in anaphase. In particular, because the composition and organization of the anaphase spindle midzone has not been extensively studied in mouse oocytes, whether Kif4 has roles in midzone formation, as it does in mitosis, is not known.
In this study, we revisit the role of Kif4 in mouse oocytes, providing insight into these questions. We find that Kif4 has dynamic localization during the cell cycle: it is nuclear localized before germinal vesicle breakdown (GVBD), then localizes to chromosomes through metaphase and transitions to the spindle midzone during anaphase; this pattern is similar to its reported mitotic localization. Moreover, we present the first high-resolution study of the anaphase spindle midzone in mouse oocytes, characterizing the localization of Kif4, PRC1, and components of the centralspindlin and chromosomal passenger complexes. We also describe new anaphase roles for Kif4 in midzone formation and spindle length regulation in oocytes. Finally, we demonstrate that Kif4 is regulated by Aurora kinases, as AURKB/C inhibition prevents Kif4 chromosomal targeting and causes anaphase defects. Together, these studies provide new insights into the organization of the anaphase spindle midzone in mouse oocytes and reveal roles for Kif4 in ensuring the integrity of the meiotic divisions on acentriolar spindles.
RESULTS
Chromokinesin Kif4 has a dynamic localization pattern during oocyte meiosis
Previous work by ourselves and others showed that a Kif4 homologue in Caenorhabditis elegans (KLP-19) is part of a protein complex that plays important roles in chromosome congression and segregation in oocytes (Wignall and Villeneuve, 2009; Dumont et al., 2010; Muscat et al., 2015; Pelisch et al., 2017; Davis-Roca et al., 2018), and other studies have revealed important roles for mammalian Kif4 during mitosis (Mazumdar et al., 2004; Samejima et al., 2012; Nunes Bastos et al., 2013). However, Kif4’s functions during oocyte meiosis are not as well understood. Therefore, we investigated the role of this kinesin in mouse oocytes, beginning by assessing its localization at each of the major stages of meiosis (Figure 1).
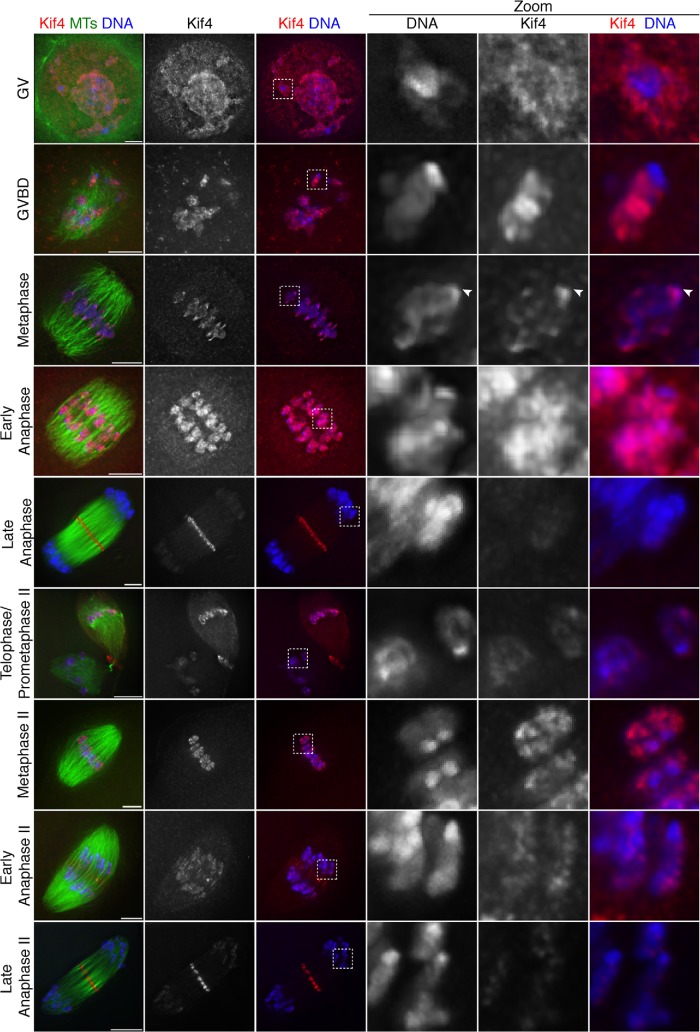
FIGURE 1:
Dynamic Kif4 localization in mouse oocytes. Representative deconvolved images of the major stages of mouse oocyte meiosis I and II. Shown are Kif4 (red), microtubules (green), and DNA (blue). Kif4 is chromosome-associated through early anaphase and then largely leaves the chromosomes, which is highlighted by the zoomed regions. Early in prometaphase, Kif4 appears excluded from the region near the ends of bivalents, where kinetochores are located, but spreads to this region by metaphase (arrowhead). All images are partial projections chosen to highlight chromosomal staining. Bars = 5 µm.
Previous studies reported that Kif4 localizes diffusely to the spindle (Camlin et al., 2017; Tang et al., 2018) and to kinetochores (Camlin et al., 2017) during metaphase in mouse oocytes. In contrast, we observed a different localization pattern. Consistent with previous studies of this protein in mitosis (Lee et al., 2001), we found that Kif4 appears to be a chromokinesin, localizing broadly to chromosomes in prometaphase/metaphase, and then mostly relocalizing from the chromosomes to the central spindle during anaphase (Figure 1). Before GVBD, Kif4 localizes throughout the germinal vesicle, and is somewhat enriched on the chromatin. Following GVBD, during the stage in which bivalents undergo individualization (Schuh and Ellenberg, 2007), Kif4 begins to localize more strongly to chromosomes and remains chromosome-associated through metaphase. Interestingly, at GVBD, Kif4 appeared excluded from the ends of chromosomes, where kinetochores are located, but then spread to this region in metaphase (Figure 1, arrowheads); because this is the stage at which kinetochore fibers (K-fibers) are formed (Brunet et al., 1999), the change in localization could indicate that Kif4 achieves kinetochore localization once microtubule attachments are made. During anaphase, the majority of Kif4 localizes to the spindle midzone with a small population remaining on segregating chromosomes, similar to mitosis. In meiosis II, Kif4 reloads onto the chromosomes during prometaphase II, and then follows a similar localization pattern as in meiosis I, leaving the chromosomes during anaphase II.
These findings differ substantially from previous reports of Kif4 localization in oocytes (Camlin et al., 2017; Tang et al., 2018). One major difference between our studies and previous reports is that we used methanol fixation, whereas other studies used formaldehyde. Notably, when we used formaldehyde fixation, our staining appeared similar to the other studies, with Kif4 localized weakly in the vicinity of the spindle (Supplemental Figure S1). Therefore, the differences in fixation conditions likely led to the reported discrepancies in localization; because the pattern we report more closely mimics the localization of Kif4 in mitotic cells (Kurasawa et al., 2004), we propose that it more accurately represents Kif4’s meiotic localization.
Aurora kinase B/C regulate Kif4 localization and inhibition of these kinases causes a range of meiotic defects
Next, we sought to assess how Kif4 localization is regulated. In C. elegans oocyte meiosis, the Aurora kinase B homologue (AURKBAIR-2) is required for targeting Kif4KLP-19 to chromosomes (Wignall and Villeneuve, 2009). Additionally, mammalian Kif4 is regulated by AURKB in mitosis (Tipton et al., 2017) and the faint Kif4 kinetochore localization observed in a previous study of mouse oocytes was reduced following Aurora kinase inhibition (Camlin et al., 2017). Given that we observed broad chromosomal Kif4 localization (Figure 1), and AURKC is localized to chromosome arms at this stage (Balboula and Schindler, 2014), we asked whether the Aurora kinase pathway regulates this Kif4 population.
For these experiments we used the Aurora kinase inhibitor AZD1152 at concentrations that have been reported to inhibit the enzymatic activity of both AURKB and the related meiosis-specific kinase AURKC (Sharif et al., 2010). To validate this inhibitor, we assessed phospho-histone H3 serine 10 (pH3S10) staining during metaphase, because this mark has been previously used to assess Aurora kinase B/C activity (Balboula and Schindler, 2014), and we also stained with an antibody that recognizes phospho-Aurora kinase A/B/C (pAURK). We found that both marks were reduced with our inhibitor conditions (Figure 2A and Supplemental Figure S2, A and B). Although almost all control spindles showed strong chromosomal pH3S10 staining (30/32), most AZD1152-treated spindles lost this signal (5/47 had signal; Supplemental Figure S2A). Similarly, for pAURK, all control oocytes displayed clear chromosomal staining (27/27), whereas most AZD1152-treated oocytes lost pAURK signal in this location (1/29 oocytes had signal; Figure 2A). In contrast, AZD1152 treatment did not affect pAURK staining at the spindle poles (Figure 2A) where AURKA predominantly localizes (Saskova et al., 2008). Therefore, our inhibitor conditions do not appear to significantly affect AURKA, validating our use of AZD1152 to inhibit AURKB/C activity in mouse oocytes.
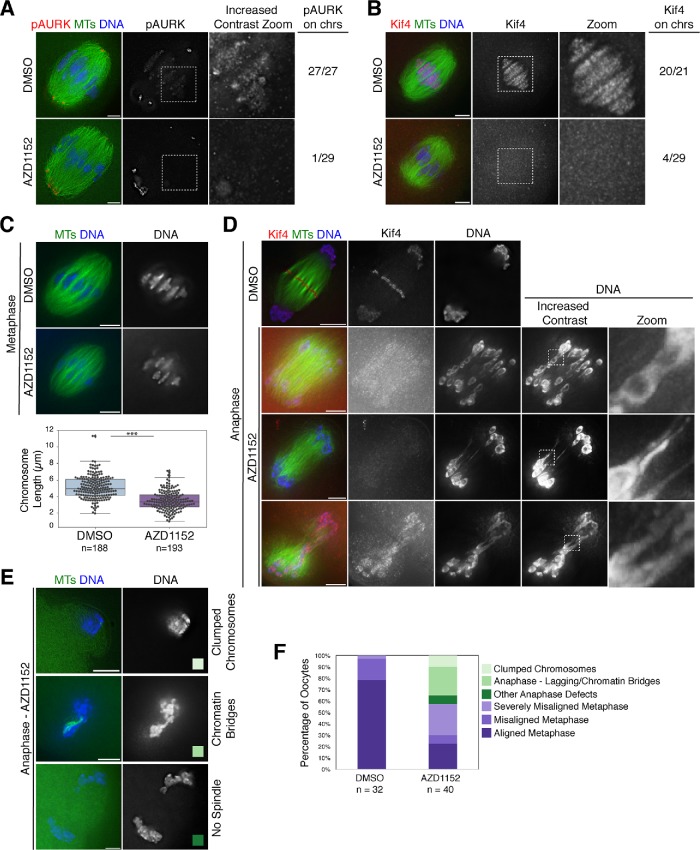
FIGURE 2:
Aurora B/C regulate Kif4’s localization and inhibition of these kinases causes a variety of meiotic defects. (A) Metaphase I spindles stained for phospho-Aurora kinase A/B/C (pAURK; red), microtubules (green), and DNA (blue) in DMSO control and 100 nM AZD1152-treated oocytes. Although pAURK pole staining (primarily Aurora kinase A) is unaffected, pAURK fluorescence on the chromosomes is reduced. Although the chromosomal staining appears dim in the middle column, given the brighter pole staining, this difference is more apparent in the zoom of the chromosomal region where we increased contrast (right column). (B) Metaphase I spindles stained for Kif4 (red), microtubules (green), and DNA (blue) in DMSO control and 100 nM AZD1152-treated oocytes. Kif4 fluorescence is reduced following AZD1152 treatment, highlighted in the zoom. For A and B, numbers at the right indicate the number of images examined that showed substantial pAURK or Kif4 chromosomal staining. (C) Metaphase spindles stained for microtubules (green) and DNA (blue) showing change in chromosome alignment and bivalent structure following AZD1152 treatment. Quantification of chromosome lengths is shown below the images. Average chromosome length is 5.2 µm ± 0.1 (n = 188 chromosomes from 11 DMSO-treated spindles) and 3.5 µm ± 0.1 (n = 193 chromosomes from 10 AZD1152-treated spindles), demonstrating that chromosomes are shorter following AZD1152 treatment. Asterisks denote p < 0.05. (D) Anaphase I spindles stained for Kif4 (red), microtubules (green), and DNA (blue). AZD1152 treatment causes mislocalization of Kif4 in anaphase and chromosome segregation errors with lagging chromosomes and chromatin bridges; images in the right two columns have increased contrast to highlight these features. (E) Examples of other anaphase defects, stained for microtubules (green) and DNA (blue), following AZD1152 treatment, including clumped chromosomes (top row) and chromatin bridges and/or aberrant spindles (bottom two rows) color-coded to reflect the graph in F. (F) Quantification of spindles from two independent experiments; oocytes were matured for 6 h, at which time most control oocytes are in metaphase with mild to no chromosome misalignment. AZD1152-treated oocytes show increased chromosome congression defects (categories displayed in light purple; misaligned metaphase with one to two chromosomes misaligned or severely misaligned with more than three chromosomes misaligned), enter anaphase prematurely (categories displayed in green), and show a variety of anaphase defects with examples shown in E. Bars = 5 µm.
Importantly, under these inhibitor conditions, we found that Kif4 was mislocalized; although Kif4 displayed strong chromosomal localization in control metaphase oocytes (20/21), this localization was lost in most AZD1152-treated oocytes (4/29 metaphase spindles had signal; Figure 2B). Therefore, AURKB/C regulate Kif4 chromosomal localization in mouse oocytes.
Given this finding, we next asked whether AURKB/C inhibition led to other phenotypes during meiosis. Following AZD1152 treatment, we observed defects in metaphase chromosome alignment (Figure 2C), and we also found that some oocytes entered anaphase at a timepoint where most control oocytes were still in metaphase (6 h following release of the oocytes from prophase arrest; Figure 2F), consistent with previous studies (Shuda et al., 2009; Sharif et al., 2010; Schindler et al., 2012). However, unlike these reports, we imaged these precocious anaphase spindles, which revealed a range of anaphase defects (Figure 2, D and E). In some spindles (10/17 anaphase oocytes), we observed lagging chromosomes that had chromatin bridges connecting them to their homologue (Figure 2, D and E). We also observed some oocytes (4/17 anaphase oocytes) with either segregated chromatin masses or a combined chromatin mass, primarily within the oocyte, but also sometimes in a PB-like structure (Figure 2E).
In some of these AZD1152-treated spindles, the chromosomes in the severe chromatin bridges appeared to be unraveling, suggesting that there were defects in bivalent structure. To investigate this, we assessed metaphase I chromosome structure. Although we did not observe gross defects in bivalent organization, chromosomes were significantly shorter in AZD1152-treated oocytes compared with the control group (Figure 2C), suggesting that there are alterations in compaction. This difference in chromosome length is not likely caused by a lack of tension on the chromosomes from reduced spindle forces, as the metaphase I spindle lengths were not significantly different (Supplemental Figure S2C). Therefore, AURKB/C regulate Kif4 localization and are also essential for proper chromosome structure and segregation in oocytes.
Kif4 regulates anaphase spindle length in mammalian oocytes
Next, we wanted to investigate the function of Kif4. Therefore, we knocked down Kif4 using morpholino (MO) microinjection and then assessed effects on meiotic progression. First, we confirmed that Kif4 levels were reduced using Western blotting and immunofluorescence imaging (Figure 3, A and B). Notably, chromosomal Kif4 staining was reduced in 17 out of 19 spindles following morpholino injection, demonstrating that our depletion conditions were largely effective and also validating the specificity of the chromosomal localization that we observed with the Kif4 antibody.
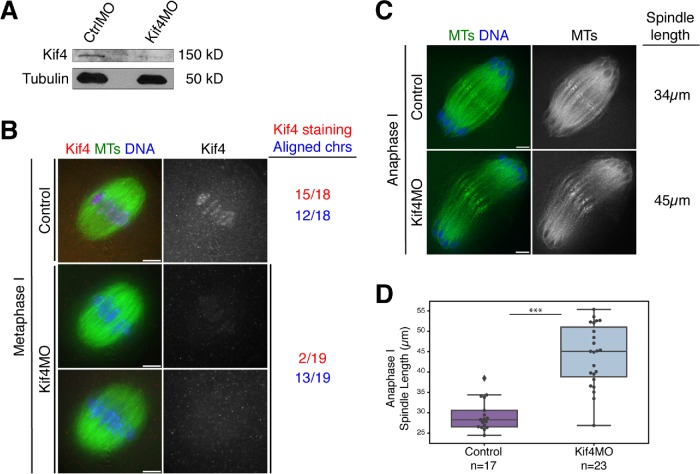
FIGURE 3:
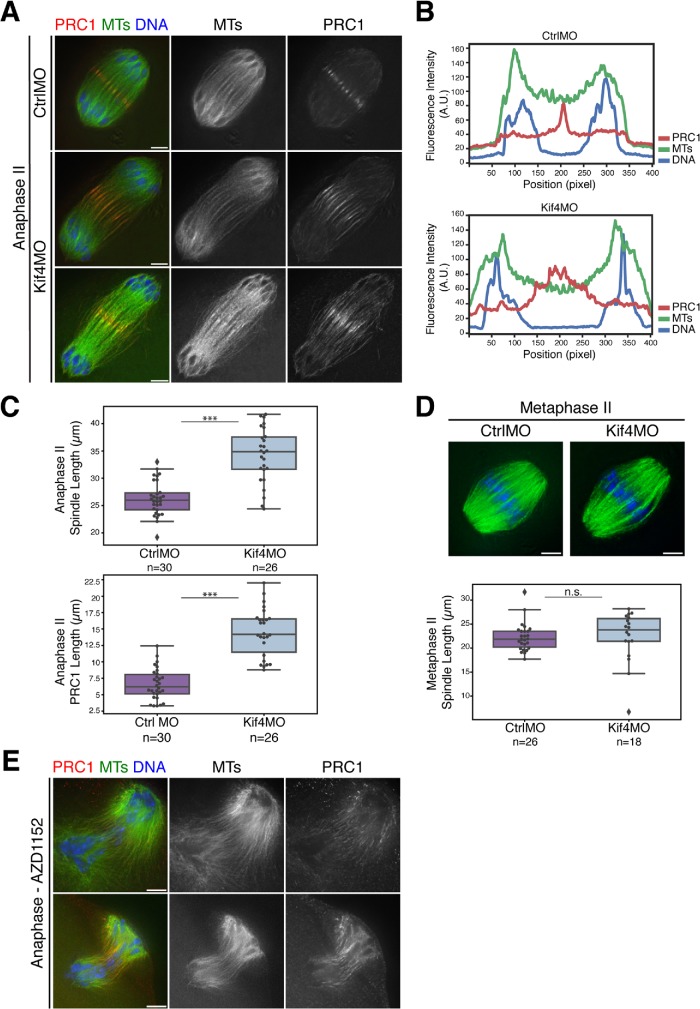
Kif4 knockdown causes anaphase spindle defects. (A) Western blot showing reduced Kif4 protein levels after microinjection of Kif4 MO. Tubulin was used as a loading control. Each lane contains 25 GV oocytes. (B) Metaphase I oocytes stained for Kif4 (red), microtubules (green), and DNA (blue). Kif4 fluorescence is reduced after microinjection of Kif4 morpholino (Kif4 MO) in most oocytes, but there were no obvious congression defects. Quantification of 18 control and 19 Kif4 MO metaphase I spindles for Kif4 staining levels (red) and chromosome alignment (blue) is shown to the right of the images. Images are single z-slices from the center of the spindle stack. (C) Anaphase I spindles stained for microtubules (green) and DNA (blue). Kif4 knockdown causes elongated anaphase spindles. The lengths of the representative spindles shown are noted on the right of the images. (D) Quantification of anaphase I spindle lengths in control compared with Kif4-depleted oocytes. The control category represents a mixture of both uninjected oocytes (n = 8) and oocytes injected with a control MO (n = 9); we did not detect a significant difference (by the Kolmogorov–Smirnov test) in the spindle lengths in these two control categories, so they were pooled in the graph. For this analysis, only spindles where chromosomes were near the ends of spindle microtubules were used for consistency in staging. Average anaphase I spindle lengths were 29.8 µm ± 1.1 (n = 17) for control oocytes and 46.8 µm ± 1.3 (n = 23) for Kif4MO-injected oocytes. Scatter plot overlaid on box plot shows individual spindle length measurements. Three asterisks denote p < 0.0005. Bars = 5 µm.
Although Kif4 is a chromokinesin, our experiments did not point to a major role for this motor in either chromosome congression or segregation. In metaphase I there was not a higher incidence of polar chromosomes or expanded metaphase plates (13/19 Kif4 MO injected oocytes had normal metaphase alignment, compared with 12/18 in controls; Figure 3B). Moreover, in 32 out of 34 Kif4-depleted oocytes, chromosome segregation appeared normal. Curiously, the other two oocytes had severe defects; in those spindles, the segregating chromosome masses appeared to be connected by strings of chromatin and chromosomes lagged in the anaphase spindle midzone (Supplemental Figure S3A). Although this phenotype resembled the severe chromosome segregation defects observed following AURKB/C inhibition (Figure 2D), we observed them in only two oocytes and we also did not detect major changes in metaphase chromosome lengths following Kif4 depletion (Supplemental Figure S3B), as we did following AZD1152 treatment (Figure 2C). Therefore, future studies would be needed to determine whether Kif4 plays a significant role in chromosome segregation in oocytes.
Nevertheless, our depletion experiments uncovered another potential role for Kif4 during anaphase. Specifically, we found that anaphase I spindles were significantly longer in the absence of Kif4 (44.0 µm ± 1.6 average length for Kif4 depletion (n = 23), compared with 29.3 µm ± 0.9 for control spindles (n = 17) (Figure 3, C and D). Therefore, Kif4 may play a role in organizing the anaphase spindle.
Midzone components organize into ring-like structures in the anaphase spindle
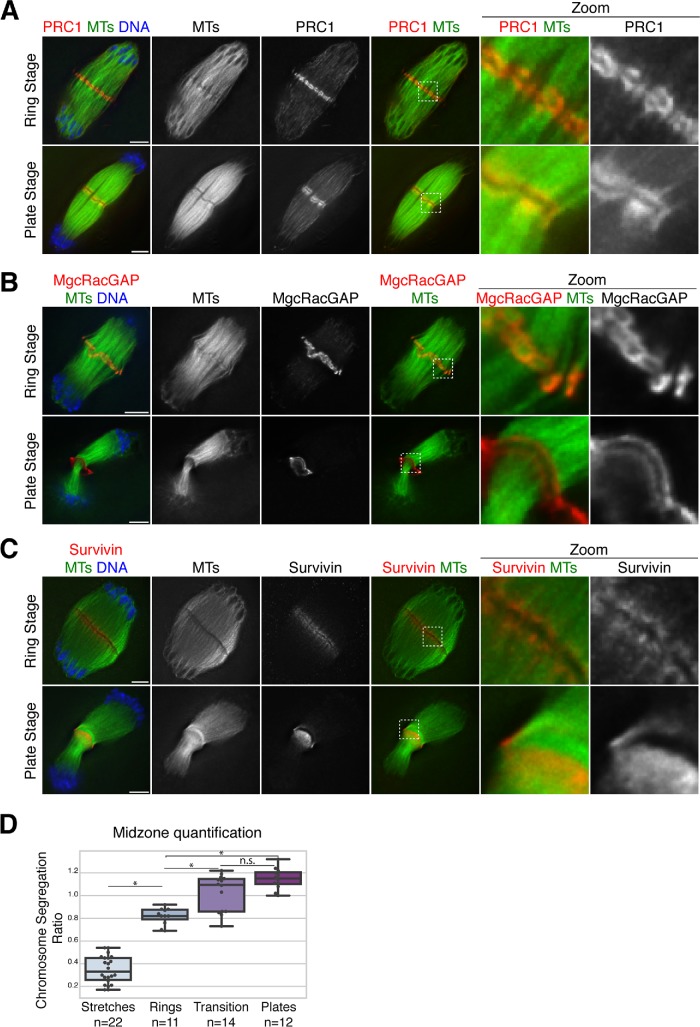
Given this phenotype, we next sought to investigate a potential role for Kif4 in anaphase spindle organization. Because we had found that Kif4 largely leaves the chromosomes in anaphase and relocalizes to the center of the spindle (Figure 1), we set out to more carefully analyze this localization pattern. This analysis revealed that although in early anaphase Kif4 is chromosome-associated (Figure 1), by mid-to-late anaphase it relocalizes to the midzone, where it forms ring-like structures that associate with distinct bundles of microtubules (Figure 4A, “Ring Stage,” and Supplemental Movies S1 and S2). Moreover, we also observed some late anaphase spindles where Kif4 appeared to form two lines (or “plates”) at the midzone, which are most clearly apparent when looking through the z-stacks of these images (Figure 4A, “Plate Stage,” and Supplemental Movies S3 and S4). This suggests that the rings may fuse and transition into plates as anaphase progresses toward telophase, a hypothesis supported by the fact that we observed some spindles that appeared to be at an intermediate stage, containing both individual rings and short plate-like structures (Figure 4B and Supplemental Movie S5).
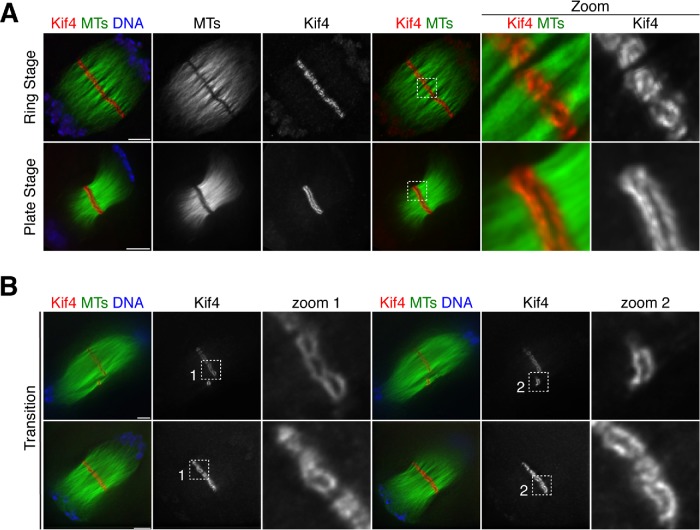
FIGURE 4:
Dynamic reorganization of Kif4 during anaphase progression. (A) Anaphase spindles stained for Kif4 (red), microtubules (green), DNA (blue). Kif4 forms ring-like structures that associate with microtubule bundles in the spindle midzone (“ring stage”) and then transitions to a plate-like formation (“plate stage”). Images are partial projections, chosen to highlight midzone features. (B) Anaphase I spindles stained for Kif4 (red), microtubules (green), and DNA (blue) showing examples of intermediates between the ring stage and the plate stage. Images in columns 1–3 and 4–6 are different partial projections of the same spindle, chosen to highlight different spindle features. Note how in row 1, the rings appear large in zoom 1 and the second zoom shows a plate-like structure that does not stretch across the entire spindle. Bars = 5 µm.
Movie S1.
Movie S1 is a z‐stack animation of an Anaphase I spindle in side view, with a midzone in the “ring stage” labeled with Kif4 (red), microtubules (green), DNA (blue) and corresponds to Figure 4A, row 1. Microtubule channel blinks in and out for clarity. Bar = 5 μm.
Movie S2.
Movie S2 is a z‐stack animation of an Anaphase II spindle in end‐on view, with a midzone in the “ring stage” labeled with Kif4 (red), microtubules (green), and DNA (blue). Bar = 5 μm.
Movie S3.
Movie S3 is a z‐stack animation of an Anaphase I spindle in side view, with a midzone in the “plate stage” labeled with Kif4 (red), microtubules (green), and DNA (blue) and corresponds to Figure 4A, row 2. Bar = 5 μm.
Movie S4.
Movie S4 is a z‐stack animation of an Anaphase I spindle in an angled view, with a midzone in the “plate stage” labeled with Kif4 (red), microtubules (green), and DNA (blue). Bar = 5 μm.
Movie S5.
Movie S5 is a z‐stack animation of an Anaphase I spindle in side view, with a midzone in the “transition stage” labeled with Kif4 (red), microtubules (green), and DNA (blue) and corresponds to Figure 4B. Microtubule channel blinks in and out for clarity. Bar = 5 μm.
Given Kif4’s relocalization from chromosomes to the midzone (Figure 1) and its plus-end–directed motor activity (Sekine et al., 1994), we next hypothesized that it might utilize the microtubules between segregating chromosomes to reach the midzone, where the plus ends reside. Consistent with this idea, we found that Kif4 localizes to microtubule bundles in mid anaphase I and II, between segregating chromosomes (Figure 1 and Supplemental Figure S4A). Although this localization could also be explained in other ways, for example if Kif4 has an affinity for anti-parallel microtubules, our data raise the possibility that Kif4 uses its plus-end–directed motor activity to walk along microtubules to reach the anaphase midzone, where it assembles into distinct ring-like structures.
Next, we characterized two other midzone proteins that had not previously been studied in mouse oocytes and found that they behave similarly to Kif4. Both PRC1, a Kif4-binding partner that bundles anti-parallel microtubules, and MgcRacGAP, a member of the centralspindlin complex, localize to similar ring and plate-like structures during anaphase (Figure 5, A and B, Supplemental Figure S5A, and Supplemental Movies S6 and S7), and in mid-anaphase PRC1 has a similar pattern to Kif4, and localizes to microtubule bundles between segregating chromosomes (Supplemental Figure S4B). Although we were not able to directly evaluate colocalization of these proteins for technical reasons, the high similarity of their localization patterns suggests that PRC1, Kif4, and MgcRacGAP may all be components of the same structures at the midzone. To quantitatively assess changes in midzone organization during anaphase progression, we measured the extent of chromosome segregation in each spindle (i.e., the distance the chromosomes had segregated divided by the total spindle length), and classified the localization of midzone proteins based on morphology as either “stretches along microtubules,” “rings,” “intermediate/transition,” or “plates” (Figure 5D). We found that these stages correlate with chromosome segregation progression. This analysis reinforced the view that these stages are sequential and demonstrated that the midzone undergoes dynamic rearrangements as anaphase progresses.
FIGURE 5:
Midzone organization in anaphase oocyte spindles. (A, B) Anaphase spindles stained for microtubules (green), DNA (blue), and PRC1 (red, panel A) or centralspindlin complex component MgcRacGAP (red, panel B). Both PRC1 and MgcRacGAP have similar localization to Kif4, forming ring-like structures and transitioning to a plate stage. (C) Anaphase spindles stained for the CPC component survivin (red), microtubules (green), and DNA (blue). In contrast to Kif4/PRC1/MgcRacGAP, survivin does not appear to form distinct structures and instead localizes adjacent to the dark zone region where these other proteins reside. Note that even though we were not able to costain with an antibody that would mark the rings, we were able to categorize the top spindle as “ring stage” by measuring the extent of chromosome segregation and confirming that this spindle was at a similar stage of anaphase as those that contain rings (analysis shown in Supplemental Figure S5B). Images in all panels are partial projections, chosen to highlight midzone features. (D) Quantification of midzone changes during anaphase progression. For each spindle, midzone organization was classified (i.e., whether midzone proteins Kif4, PRC1, or MgcRacGAP were in stretches on microtubules, in rings, in plates, or with a mixture of rings and plates in “transition”) and the stage of anaphase was determined by measuring the chromosome segregation ratio (calculated by dividing chromosome segregation distance by spindle length). Average segregation ratio for stretches is 0.35 ± 0.03 (n = 22), for rings is 0.82 ± 0.02 (n = 11), for transition is 1.02 ± 0.04 (n = 14), and for plates is 1.15 ± 0.03 (n = 12); this trend suggests that these stages are sequential and the segregation ratio can distinguish between stages (p = 1.6e-26, one-way ANOVA). Bonferroni adjusted p < 0.05 has a single asterisk; adjusted p > 0.05 denoted n.s.
Movie S6.
Movie S6 is a z‐stack animation of an Anaphase I spindle in end‐on view, with a midzone in the “ring stage” labeled with MgcRacGAP (red), microtubules (green), and DNA (blue) and corresponds to Figure 5B, row 1. Microtubule channel blinks in and out for clarity. Bar = 5 μm.
Movie S7.
Movie S7 is a z‐stack animation of an Anaphase II spindle in side view, with a midzone in the “transition stage” labeled with PRC1 (red), microtubules (green), DNA (blue) and corresponds to Supplemental Figure 5A, row 1. Microtubule channel blinks in and out for clarity. Bar = 5 μm.
In some of our images of anaphase spindles, it appeared as if the microtubules had a “break” in the center of the spindle, with the midzone rings localized in between, raising the possibility that these structures serve to bridge two separate spindle halves. However, this “dark zone” was not apparent in all of our images (Figure 5), and following careful examination we observed that the midzone ring structures often appeared to localize along or encircle individual microtubule bundles, a feature that was more clear when the bundles were splayed away from the dense midzone region (Supplemental Figure S6, A and B). Therefore, we favor the idea that the break is instead caused by exclusion of the tubulin antibody from this region, as has been reported in studies of the mitotic spindle midzone (Hu et al., 2012; D’Avino and Capalbo, 2016), and that these midzone structures therefore associate with continuous microtubule bundles rather than bridging separate spindle halves.
To determine whether all midzone components localize to the ring structures, we next assessed survivin, a member of the chromosomal passenger complex (CPC). In contrast to Kif4, PRC1, and MgcRacGAP, survivin did not form distinct ring structures, and instead localized on anaphase microtubule bundles, in regions on both sides of the “dark zone” in the microtubule staining (Figure 5C), where the rings reside (Figures 4A and 5, A and B). Although we were not able to image survivin in combination with other ring components to directly show that they do not colocalize due to technical restrictions from reagents, the spindles displaying this survivin staining pattern were all at a similar stage of anaphase to those that had the Kif4/PRC1/MgcRacGAP rings, judged by the extent of chromosome segregation (Supplemental Figure S5B). Therefore, the fact that survivin did not display this ring pattern suggests that not every midzone component is part of these structures and supports the view that the acentriolar spindle midzone has distinct domains.
Kif4 helps organize the anaphase spindle midzone
Given this interesting anaphase midzone organization (Figures 4 and 5 and Supplemental Figure S4), and the finding that Kif4 depletion resulted in longer anaphase I spindles (Figure 3), we next sought to determine whether Kif4 helps to organize the anaphase spindle midzone. To more rigorously and quantitatively investigate this possible role in anaphase spindle regulation, we knocked down Kif4 and assessed anaphase II. Anaphase II can be synchronized by parthenogenetically activating metaphase II–arrested oocytes and then fixing at defined times to allow a more precise comparison of spindle lengths during anaphase progression. Our studies revealed that, similar to anaphase I, anaphase II spindles were significantly longer in the absence of Kif4 (34.2 µm ± 1.0 in Kif4 depletion [n = 26] compared with 26.3 µm ± 0.6 in injected controls [n = 30]), and that their midzone microtubules appeared disorganized and/or less bundled (Figure 6, A–C). Additionally, PRC1 distribution at the midzone was broader in Kif4-depleted anaphase II spindles (14.5 µm ± 0.7 in Kif4 depletion (n = 26) compared with 6.6 µm ± 0.4 in injected controls (n = 30), indicating that aspects of midzone establishment are defective in the absence of Kif4 (Figure 6, A–C).
FIGURE 6:
Anaphase spindle length and midzone organization is regulated by Kif4 in oocytes. (A) Anaphase II spindles from oocytes injected with either control or Kif4 morpholinos stained for PRC1 (red), microtubules (green), and DNA (blue). Following Kif4 depletion, PRC1 does not form distinct structures and instead is spread on an expanded region of the anaphase spindle. (B) Line scans of the top two images from panel A, showing a tight peak of PRC1 in the control compared with a broader peak following Kif4 depletion. (C) Quantification of anaphase II spindle lengths (top) and PRC1 midzone spread (bottom); spindles are longer following Kif4 depletion and PRC1 localizes to a broader domain. Scatter plot overlaid on box plot shows individual spindle or PRC1 length measurements. Average anaphase II spindle lengths were 26.2 µm ± 0.6 (n = 30) for control-injected oocytes and 34.2 µm ± 1.0 (n = 26) for Kif4MO-injected oocytes. Average length of the PRC1 midzone is 6.6 µm ± 0.4 (n = 30) for the control and 14.5 µm ± 0.7 (n = 26) for Kif4MO. Conditions are significantly different (p < 0.0005, denoted by three asterisks). (D) Metaphase II spindles stained for microtubules (green) and DNA (blue). No major congression errors or spindle abnormalities were observed in 26 control and 18 Kif4-depleted spindles. Quantification of metaphase II spindle lengths are below the images; Kif4 depletion does not significantly affect metaphase II spindle length. As in C, box plots with overlaid scatter plots show individual spindle length measurements. No significant difference (p > 0.05, denoted by n.s.) between the two conditions. (E) AZD1152-treated anaphase I spindles stained for PRC1 (red), microtubules (green), and DNA (blue). In 10/10 aberrant anaphase spindles, there were disorganized midzones and mislocalized PRC1 with AURKB/C inhibition (two representative images shown). Bars = 5 µm.
To ensure that the longer anaphase II spindles were not a result of previous spindle defects, we measured the lengths of metaphase II spindles and found no significant difference between control and Kif4 morpholino-injected oocytes; control spindles averaged 22.3 µm ± 0.6 (n = 26) and the Kif4-depleted spindles averaged 22.4 µm ± 1.3 (n = 18; Figure 6D). Together, these results indicate that Kif4 plays an anaphase-specific role in spindle length regulation, likely through its role in establishing the spindle midzone.
Finally, because we had found that AURKB/C inhibition caused mislocalization of chromosomal Kif4 (Figure 2B), we assessed whether inhibition of these kinases also affected midzone formation. Notably, Kif4 was not targeted properly to anaphase midzone microtubules following AZD1152 treatment, and instead localized faintly to the chromosomes (Figure 2D); 6/6 AZD1152-treated anaphase spindles lacked a significant microtubule-associated population. Moreover, PRC1 localized broadly to the disorganized anaphase spindle microtubules and did not form distinct midzone structures in 10/10 AZD1152-treated anaphase spindles (Figure 6E). These findings likely explain the aberrant anaphase spindle phenotypes we observed following AZD1152 treatment (Figure 2, E and F), and demonstrate that AURKB/C kinases play a role in setting up the spindle midzone, potentially at least in part through their role in targeting Kif4.
DISCUSSION
Kif4 contributes to anaphase spindle organization
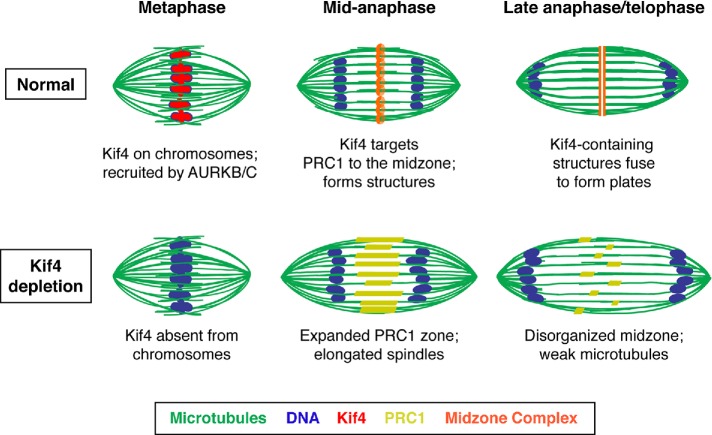
Taken together, our studies support a model in which Kif4 contributes to the fidelity of the meiotic divisions in mouse oocytes (Figure 7). First, Kif4 is targeted to chromosomes following GVBD. During this time Kif4 transitions from being excluded from the ends of bivalents, where kinetochores are located, to covering the entire chromosome, suggesting a dynamic regulation of this protein during this period. Then, during anaphase, Kif4 progressively relocalizes to the anaphase spindle midzone, where it forms distinct structures that likely also contain PRC1 and MgcRacGAP. Kif4 appears to be essential for proper anaphase spindle organization, as Kif4 depletion causes PRC1 mislocalization and results in longer anaphase spindles with poorly defined midzones. Altogether, our studies suggest that Kif4 aids in the organization of the anaphase midzone in acentriolar oocyte spindles.
FIGURE 7:
Model for Kif4’s roles in oocyte meiosis. Under normal conditions, Kif4 is chromosome-associated and then relocalizes to form ring-shaped structures in the midzone that contain PRC1 and MgcRacGAP1, and that stabilize the anaphase spindle (top). In the absence of Kif4 (bottom), PRC1 no longer concentrates into midzone structures, leading to elongated anaphase spindles with a disorganized central spindle. Kif4 (red), microtubules (green), DNA (blue), midzone complex (orange), and PRC1 (yellow).
Our studies also implicate Kif4 in anaphase spindle length regulation. This proposed function is consistent with previous studies, which demonstrated that Kif4 can suppress microtubule dynamic instability in vitro (Bringmann et al., 2004; Bieling et al., 2010; Nunes Bastos et al., 2013). Therefore, one role for Kif4 at the midzone of oocyte spindles may be to suppress microtubule plus-end dynamics, preventing overgrowth of microtubules and thus constraining anaphase spindle length, similar to its role in mitotic cells. In both anaphase I and II, the midzone microtubules do not appear to be as strongly bundled as they are in controls, a phenotype that could be a result of the overelongation of midzone microtubules.
Importantly, our studies also revealed that Kif4 is regulated by AURKB/C in mouse oocytes. Inhibition of these kinases prevented Kif4 metaphase chromosomal localization, affected bivalent organization, and led to anaphase bridges. Moreover, AURKB/C inhibition also caused defective midzone organization, reflected by splayed and disorganized spindle microtubules and improper Kif4 and PRC1 targeting. Interestingly, phosphorylation of conserved Kif4 residues (T799 and S801) by AURKB increases Kif4’s processivity and allows Kif4 to halt microtubule polymerization in vitro (Nunes Bastos et al., 2013). This regulation is necessary in mitosis for normal midzone formation, and given our findings, it is possible that this site contributes to Kif4 regulation in oocytes as well. However, given that the defects observed following AURKB/C inhibition are much more severe than those seen with Kif4 depletion alone, AURKB/C inhibition likely results in the misregulation of multiple proteins; future studies aimed at identifying other targets of AURKB/C in mouse oocytes would therefore be valuable.
Organization and dynamics of the acentriolar anaphase spindle midzone
Our studies also yielded new insight into the dynamic reorganization of acentriolar spindles during chromosome segregation. We found that Kif4 forms distinct ring-like structures at the anaphase spindle midzone; each ring interacts with a prominent microtubule bundle that runs through the midzone. These rings then appear to fuse into a plate-like structure before cytokinesis. Moreover, we found that Kif4’s binding partner PRC1 and the centralspindlin component MgcRacGAP form similar ring and plate structures, likely colocalizing with Kif4. In contrast, survivin, a CPC component, does not localize to these structures and is instead adjacent to them. Therefore, the midzone of acentriolar oocyte spindles is highly organized, comprising multiple domains. In the future it will be interesting to determine whether similar ring structures are also present in mitosis; previous studies have reported that Kif4 and PRC1 localize in foci at the spindle midzone in mitosis (Hu et al., 2011; Nunes Bastos et al., 2013), but whether they form rings is not clear.
The discovery of these midzone domains leads to the important question of how this organization is established and maintained. Our studies have revealed that Kif4 is an important player in midzone formation, as Kif4 depletion resulted in a disorganized anaphase central spindle. Although PRC1 was still able to localize to microtubules and sometimes form structures under these depletion conditions, it displayed broader localization. Because both Kif4 and PRC1 normally localize along anaphase spindle microtubules in mid-anaphase (Supplemental Figure S4), one possibility is that Kif4 helps build the midzone by transiting from the chromosomes to the central spindle using plus-end–directed microtubule movement, transporting PRC1 with it. This would facilitate the restriction of PRC1 to the midzone structures, where it could then cross-link anti-parallel microtubules, helping to stabilize the anaphase central spindle. This idea has been previously proposed to explain PRC1 targeting in mitosis, because Kif4 can walk to the plus ends of microtubules with PRC1 in vitro, forming end tags that are dependent on microtubule length (Subramanian et al., 2013). However, the extent to which Kif4 carries PRC1 in vivo is debated, because PRC1 has a nanomolar affinity for overlapping microtubules, which might be sufficient for its midzone localization (Bieling et al., 2010). Therefore, an alternative possibility is that PRC1 mistargeting in the oocyte following Kif4 depletion is a downstream consequence of an extended overlap zone due to loss of Kif4’s repression of microtubule dynamics; in this scenario Kif4 would not directly carry PRC1 but would establish a restricted overlap zone that would facilitate its proper localization. This idea is consistent with a previous mitosis study that showed that PRC1 localizes normally in the presence of a mutant Kif4 that is incapable of binding PRC1 (Hu et al., 2011), indicating that PRC1 is capable of localizing to a midzone that is correctly established by Kif4 without directly binding to Kif4. These ideas are consistent with our finding that PRC1 has a broadened midzone localization and ultimately forms disorganized structures in the absence of Kif4. Future studies in mouse oocytes will be important to better distinguish between these different models and to reveal additional principles underlying midzone establishment in acentriolar spindles.
Looking forward, it will also be interesting to assess the localization of other potential midzone components in relation to the ones described here. A good candidate is the kinase Plk1, which has been reported to form dot-like particles at the anaphase spindle midzone (Otsuki et al., 2009); we therefore predict that this protein colocalizes with the structures we describe. Moreover, in addition to looking at other midzone components, further high-resolution studies of mouse oocytes may provide insight into the general organization of anaphase spindle midzones in acentriolar spindles, revealing the nature of the midzone structures’ interaction with anaphase microtubule bundles and how this changes as the cells progress to telophase.
MATERIALS AND METHODS
Oocyte collection and handling
CD-1 strain Mus musculus (Envigo) were kept in compliance with Institutional Animal Care and Use Committee (IACUC) policies and provided food and water ad libitum. Female animals aged 5–13 wk were peritoneally injected with 5IU pregnant mare serum gonadotropin (PMSG; Millipore) 42–46 h before sacrificing and dissecting ovaries. For each experiment, multiple animals were used, and oocytes were mixed. Oocytes were collected by ovarian puncture and manipulated outside the incubator in L15 (Life Technologies), 3 mg/ml polyvinylpyrrolidone (PVP; Sigma), 50 U/ml Pen, 50 µg/ml Strep (Life Technologies). Oocytes were held and/or matured in a cell-culture incubator at 37°C with 5% CO2 in air, in α-MEM GlutaMax (Life Technologies), 3 mg/ml bovine serum albumin (BSA; Sigma), 50 U/ml Pen, 50 µg/ml Strep (Life Technologies), in center-well organ dishes (Nunc). The germinal vesicle (GV) arrest stage was maintained with 1 µM milrinone (Sigma), which was washed out to allow meiotic resumption. Oocytes that remained at the GV stage after 90 min of release were counted and removed.
Oocytes were treated with 100 nM AZD1152-HQPA (Sigma) in α-MEM following milrinone washout, with dimethyl sulfoxide (DMSO) as a vehicle control. Time of incubation/maturation is from milrinone washout and treatment initiation. Anaphase II parthenogenetic activation was carried out by treatment of metaphase-II–arrested oocytes either with 7% EtOH (FitzHarris, 2012) in L15 PVP media at 25°C for 7 min, 10 mM SrCl2, and 2 mM EGTA (ethylene glycol tetraacetic acid) (Kishigami and Wakayama, 2007) in L15 PVP media at 25°C for 10 min, or with 10 mM SrCl2 in homemade Ca2+-free KSOM (potassium-supplemented simplex optimized media, Cold Spring Harbor Protocol; gift of O’Halloran lab, Northwestern University).
Immunofluorescence microscopy
For formaldehyde fixation, oocytes were fixed at the given stages in HEM buffer (100 nM HEPES, pH 7, with KOH, 50 mM EGTA, 1 mM MgCl2) with 2% PFA (MeOH Free; Electron Microscopy Sciences) and 0.2% Triton X-100 for 30–60 min at 37°C. For MeOH fixation (Kif4 visualization), the zona pellucida was removed using warmed Acidic Tyrode’s Solution (Sigma), recovered in the incubator for 15 min, and fixed for 5 min in −20°C MeOH. Fixed oocytes were extracted overnight in PBS with 0.2% Triton X-100 (PBST) at 4°C and were then blocked in 3% BSA-PBST (blocking solution) for at least 1 h at room temperature and placed in primary antibody diluted in blocking solution overnight at 4°C. Oocytes were washed three times for 5 min in PBST on a rocker at room temperature between primary and secondary antibodies and before Hoechst staining. Alexa Fluor Secondaries (Molecular Probes), diluted 1:500 in blocking solution, were applied to oocytes for 1–3 h at room temperature.
All primary antibodies were raised in rabbit unless otherwise noted and all primary and secondary antibodies were used at a 1:500 dilution. Primary antibodies used were α-Kif4 (sc-134593 [raised against amino acids 361–660 of the human Kif4]; Santa Cruz Biotechnology, Dallas, TX), α-survivin (#2808; Cell Signaling Technologies [CST], Danvers, MA), α-pH3S10 (#53348; CST), α-pAURK (#2914; CST), α-PRC1 (#15617-1-AP; Proteintech, Chicago, IL), and α-RacGAP (#13739-1-AP; Proteintech). Directly conjugated mouse FITC-α-tubulin (MA1-19581; Invitrogen/Molecular Probes [MP]) was used to visualize tubulin and added at the same time as Alexa Fluor 555 α-rabbit (A-21428; MP).
Chromosome spreads
Chromosome spreading was done as described previously (Chambon et al., 2013). Briefly, the zona pellucida was removed from oocytes using prewarmed Acidic Tyrode’s Solution (Sigma) by washing through drops under oil and gentle pipetting. Oocytes were recovered in the incubator for at least 15 min. Oocytes were placed in 10 µl drops of fixation/spreading solution (1% PFA, 0.15% Triton X-100, 3 mM dithiothreitol, in H2O, pH 9.2) in the wells of a Teflon-coated slide and left to dry undisturbed at room temperature. Slides were stored at −20°C until processing for immunofluorescence. Immunofluorescence was carried out as described above, but slides were washed with PBST before blocking and all solutions were pipetted onto the slide and removed with an aspirator.
Microscopy
Fixed immunofluorescence images were acquired on a DeltaVision Core deconvolution microscope with a 100× objective (NA = 1.4; Applied Precision). The DeltaVision microscope is housed by Northwestern’s Biological Imaging Facility supported by the Northwestern University Office for Research. Image stacks were taken with a 0.2-µm step size and deconvolved using SoftWoRx (Applied Precision). The images in this study were deconvolved (unless otherwise noted) and represent maximum projections of full or partial spindles as noted in the figures and legends.
Microinjection of morpholinos
GV stage oocytes were held at GV stage with milrinone and held in drops of L15 5% fetal bovine serum (FBS) Pen Strep media for injecting on an inverted Olympus IX53 microscope fitted with Eppendorf micromanipulators and a PLI-100A Picoliter Microinjector (Warner Instruments) with a foot pedal. The microinjection setup and cell-culture incubator are housed in the Quantitative Biological Imaging Center and maintained by the Biological Imaging Facility. Oocytes were injected with either control MO (standard control oligo from GeneTools) or distilled water as a control. Oocytes were held at the GV stage in an incubator in α-MEM 10% FBS PenStrep for 24 h to allow for protein turnover and washed in drops of fresh L15 PVP three times for 2 min at 37°C to remove milrinone and returned to the incubator to mature. GVBD was monitored at 90 min postrelease and GV oocytes were removed. Morpholinos were designed by and obtained from GeneTools. Sequence for Kif4MO: GAA TCC CCT TCA CCT CTT CTT TCA T. Sequence for CtrlMO: CCT CTT ACC TCA GTT ACA ATT TAT A.
Immunoblot
Twenty-five GV oocytes from each injection condition were collected after a 24-h holding period and washed through warmed PBS to remove BSA and residual media. Oocytes were transferred to a centrifuge tube in a minimal amount of buffer and snap frozen in liquid nitrogen. Samples were boiled in Laemmli sample buffer at 95°C for 5 min and loaded on a 15-well SDS–PAGE gel. Primary α-Kif4 antibody (Santa Cruz) was used at 1:1000, HRP α-rabbit (BioRad) was used at 1:5000, and blots were processed with SuperSignal West Femto Maximum Sensitivity Substrate ECL (Thermo Scientific).
Data visualization
All box plots show the inner quartile range represented by a colored box, the minimum and maximum shown with whiskers, and outliers as diamonds, with an overlaid scatter plot of each individual measurement reported. Plots were generated using Seaborn in Jupyter notebook.
Quantification of specific figures
Figure 1: Kif4 localization was assessed by eye. The number of images that showed the same staining pattern as the representative image displayed in the figure are as follows. For GV stage, 15/15 images from two experiments. For GVBD, 25/28 images from three experiments. For metaphase I, 27/28 images from three experiments. For early anaphase I, 3/3 images from two experiments. For anaphase I, 20/20 images from four experiments. For telophase/prometaphase II, 2/2 images from one experiment. For metaphase II, 16/16 images from four experiments. For early anaphase II, 7/7 images from two experiments. For late anaphase II, 18/18 images from four experiments.
Figure 2: Kif4, pH3S10, and pAURK levels were assessed by eye and with the “Data Inspector” tool on SoftWoRx image software (Applied Precision) to differentiate background levels and chromosome levels, by drawing a box around the chromosomes and assessing levels in chromosome areas versus background using the three-dimensional graph view. Proteins were judged to be depleted if they were below the intensity range of the control experiment from the same day. This analysis was performed on two independent experiments for each group. Chromosome lengths were measured in Imaris (Bitplane) using “Spots” tool and measuring the length of 15–20 (>75%) chromosomes for each spindle. Errors in AZD1152 treatment conditions versus the control were categorized by eye from two independent experiments.
Figure 3: Kif4 staining levels were assessed by eye and with the SoftWoRx “Data Inspector” tool, as described for Figure 2. Metaphase chromosome alignment at 6 h and anaphase errors were assessed by eye. A spindle was categorized as having misaligned chromosomes if at least one chromosome was separated from the metaphase plate by more than 50% of its length. Spindle lengths were measured using Imaris “Measurement Points” tool. A surface was created for the spindle using the “Surfaces” tool and then the ends of the microtubule density at each pole was determined and used to measure spindle length. Spindles were excluded from analysis if they were bent and could not be precisely measured.
Figures 4 and 5: The localization and morphology of midzone components was assessed by eye. The number of images that showed the same staining pattern as the representative images displayed in the figures are as follows. For Kif4, 38 ring stage from eight experiments and 28 plate stage from seven experiments. For PRC1, 17 ring stage from four experiments and 9 plate stage from three experiments. For MgcRacGAP, 9 ring stage from five experiments and 4 plate stage from two experiments. For survivin, 8 “ring” stage and 3 plate stage, both from one experiment. For anaphase midzone characterization (Figure 4C), anaphase spindles from multiple experiments were measured using Imaris Measurement Points (method adapted from Davis-Roca et al., 2017). Surfaces were created for both the chromosomes and the spindle. The center of each set of segregating chromosomes was determined and then the distance between them was measured, to calculate the chromosome segregation distance. The ends of the microtubule density at each pole was determined and used to measure spindle length. Spindles were excluded from analysis if they were bent or if midzone staining was not optimal (as double staining midzone components leads to punctate patterns). Transition category was scored by the presence of fused rings or extended line structures that did not span the entire width of the spindle. “Segregation Ratio” is calculated by dividing chromosome segregation distance by spindle length and plotted in box plots with overlaid scatter plots.
Figure 6: Line scans were done in ImageJ with a 25-pixel-wide line of the same length for each image to account for different spindle lengths. Data from ImageJ were analyzed using Python in Jupyter notebook. Late anaphase I spindle lengths (staged by only assessing spindles where the chromosomes were near the ends of microtubules at the spindle poles) were measured using Imaris (Bitplane) software using the surfaces tool on the microtubule channel and measuring pole to pole distance using “Measurement Points” tool by marking a point near the center of each pole. Only spindles where the resulting line coincided with the spindle midzone were used, to avoid measurement errors from assessing bent spindles. Anaphase II spindle lengths were measured in Imaris in the same manner.
PRC1 midzone spread was measured using the LineProbe module in Amira (Thermo). Points were placed at the poles of each spindle, and the values were averaged with a radius of 3. Data were exported and plotted in Microsoft Excel to measure peaks and determine the length of the PRC1 midzone. The Amira workstation is housed in the Center for Advanced Molecular Imaging core at Northwestern. Metaphase spindle lengths were measured in Imaris by creating a surface around the tubulin intensity and measuring distances between the two pole surfaces.
Statistics
All p values were calculated using a two-tailed Student’s t test, unless otherwise noted in the figure legends. Standard error is reported with length measurement averages. For one-way analysis of variance (ANOVA) in Figure 5D, p values were adjusted using a Bonferroni correction of six pairwise comparisons.
Supplementary Material
Acknowledgments
We thank Teresa Woodruff, members of the Wignall and Woodruff labs, and the WiLa Cell Biology Group for their help and support; Amanda Davis-Roca, Nikita Divekar, Jeremy Hollis, Rachel Kadzik, Tim Mullen, and Ian Wolff for critical reading of the manuscript; and Joseph Draut for initial anaphase spindle characterization work. This work was supported by a March of Dimes Research Grant and by National Institutes of Health R01GM124354 (to S.M.W.) and by the Cell and Molecular Basis of Disease Training Program at Northwestern University, Grant no. T32GM008061 (to C.M.H.).
Abbreviations used:
- AURK
Aurora kinase
- CPC
chromosomal passenger complex
- GV
germinal vesicle
- GVBD
germinal vesicle breakdown
- K-fibers
kinetochore fibers
- MO
morpholino
- MT
microtubule
- PAURK
phospho-Aurora kinase
- PB
polar body
- pH3S10
phospho-histone H3 serine 10
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-10-0666) on May 8, 2019.
REFERENCES
- Balboula AZ, Schindler K. (2014). Selective disruption of aurora C kinase reveals distinct functions from aurora B kinase during meiosis in mouse oocytes. PLoS Genet , e1004194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Telley IA, Surrey T. (2010). A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell , 420–432. [DOI] [PubMed] [Google Scholar]
- Bringmann H, Skiniotis G, Spilker A, Kandels-Lewis S, Vernos I, Surrey T. (2004). A kinesin-like motor inhibits microtubule dynamic instability. Science , 1519–1522. [DOI] [PubMed] [Google Scholar]
- Brunet S, Maria AS, Guillaud P, Dujardin D, Kubiak JZ, Maro B. (1999). Kinetochore fibers are not involved in the formation of the first meiotic spindle in mouse oocytes, but control the exit from the first meiotic M phase. J Cell Biol , 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camlin NJ, McLaughlin EA, Holt JE. (2017). Kif4 is essential for mouse oocyte meiosis. PLoS One , e0170650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon JP, Hached K, Wassmann K. (2013). Chromosome spreads with centromere staining in mouse oocytes. Methods Mol Biol , 203–212. [DOI] [PubMed] [Google Scholar]
- D’Avino PP, Capalbo L. (2016). Regulation of midbody formation and function by mitotic kinases. Semin Cell Dev Biol , 57–63. [DOI] [PubMed] [Google Scholar]
- Davis-Roca AC, Divekar NS, Ng RK, Wignall SM. (2018). Dynamic SUMO remodeling drives a series of critical events during the meiotic divisions in Caenorhabditis elegans. PLoS Genet , e1007626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Roca AC, Muscat CC, Wignall SM. (2017). Caenorhabditis elegans oocytes detect meiotic errors in the absence of canonical end-on kinetochore attachments. J Cell Biol , 1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J, Oegema K, Desai A. (2010). A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat Cell Biol , 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzHarris G. (2012). Anaphase B precedes anaphase A in the mouse egg. Curr Biol , 437–444. [DOI] [PubMed] [Google Scholar]
- Gruneberg U, Neef R, Li X, Chan EH, Chalamalasetty RB, Nigg EA, Barr FA. (2006). KIF14 and citron kinase act together to promote efficient cytokinesis. J Cell Biol , 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CK, Coughlin M, Field CM, Mitchison TJ. (2011). KIF4 regulates midzone length during cytokinesis. Curr Biol , 815–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CK, Coughlin M, Mitchison TJ. (2012). Midbody assembly and its regulation during cytokinesis. Mol Biol Cell , 1024–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishigami S, Wakayama T. (2007). Efficient strontium-induced activation of mouse oocytes in standard culture media by chelating calcium. J Reprod Dev , 1207–1215. [DOI] [PubMed] [Google Scholar]
- Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K. (2004). Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J , 3237–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YM, Kim W. (2004). Kinesin superfamily protein member 4 (KIF4) is localized to midzone and midbody in dividing cells. Exp Mol Med , 93–97. [DOI] [PubMed] [Google Scholar]
- Lee YM, Lee S, Lee E, Shin H, Hahn H, Choi W, Kim W. (2001). Human kinesin superfamily member 4 is dominantly localized in the nuclear matrix and is associated with chromosomes during mitosis. Biochem J , 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar M, Sundareshan S, Misteli T. (2004). Human chromokinesin KIF4A functions in chromosome condensation and segregation. J Cell Biol , 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen TJ, Wignall SM. (2017). Interplay between microtubule bundling and sorting factors ensures acentriolar spindle stability during C. elegans oocyte meiosis. PLoS Genet , e1006986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscat CC, Torre-Santiago KM, Tran MV, Powers JA, Wignall SM. (2015). Kinetochore-independent chromosome segregation driven by lateral microtubule bundles. eLife , e06462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes Bastos R, Gandhi SR, Baron RD, Gruneberg U, Nigg EA, Barr FA. (2013). Aurora B suppresses microtubule dynamics and limits central spindle size by locally activating KIF4A. J Cell Biol , 605–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Hahn H, Torrey TA, Shin H, Choi W, Lee YM, Morse HC, Kim W. (2000). Identification of the human homologue of mouse KIF4, a kinesin superfamily motor protein. Biochim Biophys Acta , 219–224. [DOI] [PubMed] [Google Scholar]
- Otsuki J, Nagai Y, Chiba K. (2009). Association of spindle midzone particles with polo-like kinase 1 during meiosis in mouse and human oocytes. Reprod Biomed Online , 522–528. [DOI] [PubMed] [Google Scholar]
- Pelisch F, Tammsalu T, Wang B, Jaffray EG, Gartner A, Hay RT. (2017). A SUMO-dependent protein network regulates chromosome congression during oocyte meiosis. Mol Cell , 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser SL, Pelletier L. (2017). Mitotic spindle assembly in animal cells: a fine balancing act. Nat Rev Mol Cell Biol , 187–201. [DOI] [PubMed] [Google Scholar]
- Radford SJ, Nguyen AL, Schindler K, McKim KS. (2017). The chromosomal basis of meiotic acentrosomal spindle assembly and function in oocytes. Chromosoma , 351–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima K, Samejima I, Vagnarelli P, Ogawa H, Vargiu G, Kelly DA, de Lima Alves F, Kerr A, Green LC, Hudson DF, et al (2012). Mitotic chromosomes are compacted laterally by KIF4 and condensin and axially by topoisomerase IIα. J Cell Biol , 755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saskova A, Solc P, Baran V, Kubelka M, Schultz RM, Motlik J. (2008). Aurora kinase A controls meiosis I progression in mouse oocytes. Cell Cycle , 2368–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler K, Davydenko O, Fram B, Lampson MA, Schultz RM. (2012). Maternally recruited Aurora C kinase is more stable than Aurora B to support mouse oocyte maturation and early development. Proc Natl Acad Sci USA , E2215–E2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Ellenberg J. (2007). Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell , 484–498. [DOI] [PubMed] [Google Scholar]
- Sekine Y, Okada Y, Noda Y, Kondo S, Aizawa H, Takemura R, Hirokawa N. (1994). A novel microtubule-based motor protein (KIF4) for organelle transports, whose expression is regulated developmentally. J Cell Biol , 187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif B, Na J, Lykke-Hartmann K, McLaughlin SH, Laue E, Glover DM, Zernicka-Goetz M. (2010). The chromosome passenger complex is required for fidelity of chromosome transmission and cytokinesis in meiosis of mouse oocytes. J Cell Sci , 4292–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda K, Schindler K, Ma J, Schultz RM, Donovan PJ. (2009). Aurora kinase B modulates chromosome alignment in mouse oocytes. Mol Reprod Dev , 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Ti SC, Tan L, Darst SA, Kapoor TM. (2013). Marking and measuring single microtubules by PRC1 and kinesin-4. Cell , 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Wilson-Kubalek EM, Arthur CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, Kapoor TM. (2010). Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell , 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szollosi D, Calarco P, Donahue RP. (1972). Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J Cell Sci , 521–541. [DOI] [PubMed] [Google Scholar]
- Tang F, Pan MH, Lu Y, Wan X, Zhang Y, Sun SC. (2018). Involvement of Kif4a in spindle formation and chromosome segregation in mouse oocytes. Aging Dis , 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton AR, Wren JD, Daum JR, Siefert JC, Gorbsky GJ. (2017). GTSE1 regulates spindle microtubule dynamics to control Aurora B kinase and Kif4A chromokinesin on chromosome arms. J Cell Biol , 3117–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernos I, Raats J, Hirano T, Heasman J, Karsenti E, Wylie C. (1995). Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell , 117–127. [DOI] [PubMed] [Google Scholar]
- Wignall SM, Villeneuve AM. (2009). Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat Cell Biol , 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff ID, Tran MV, Mullen TJ, Villeneuve AM, Wignall SM. (2016). Assembly of C. elegans acentrosomal spindles occurs without evident MTOCs and requires microtubule sorting by KLP-18/kinesin-12 and MESP-1. Mol Biol Cell , 3122–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L. (2010). How kinesin motor proteins drive mitotic spindle function: lessons from molecular assays. Semin Cell Dev Biol , 260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Jiang W. (2005). Cell cycle-dependent translocation of PRC1 on the spindle by Kif4 is essential for midzone formation and cytokinesis. Proc Natl Acad Sci USA , 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, Abraham RT, Jiang W. (2005). Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell , 3187–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.