Abstract
Megalin (gp330, LRP-2) is a protein structurally related to the low-density lipoprotein receptor family that displays a large luminal domain with multiligand binding properties. Megalin localizes to the apical surface of multiple epithelia, where it participates in endocytosis of a variety of ligands performing roles important for development or homeostasis. We recently described the apical recycling pathway of megalin in Madin–Darby canine kidney (MDCK) cells and found that it is a long-lived, fast recycling receptor with a recycling turnover of 15 min and a half-life of 4.8 h. Previous work implicated clathrin and clathrin adaptors in the polarized trafficking of fast recycling basolateral receptors. Hence, here we study the role of clathrin and clathrin adaptors in megalin’s apical localization and trafficking. Targeted silencing of clathrin or the γ1 subunit of clathrin adaptor AP-1 by RNA interference in MDCK cells disrupted apical localization of megalin, causing its redistribution to the basolateral membrane. In contrast, silencing of the γ2 subunit of AP-1 had no effect on megalin polarity. Trafficking assays we developed using FM4-HA-miniMegalin-GFP, a reversible conditional endoplasmic reticulum–retained chimera, revealed that clathrin and AP-1 silencing disrupted apical sorting of megalin in both biosynthetic and recycling routes. Our experiments demonstrate that clathrin and AP-1 control the sorting of an apical transmembrane protein.
INTRODUCTION
Megalin (gp330, LRP-2) is expressed in embryonic and adult general and neuroepithelial cells, in which it mediates the endocytosis of a vast array of ligands (Kerjaschki and Farquhar, 1983; Christensen and Birn, 2002; Christensen et al., 2012). Knockout of megalin in mice causes a range of neurodevelopmental abnormalities that result in perinatal death (Willnow et al., 1996), ostensibly because megalin participates in the endocytosis and transcytosis of key differentiation factors, for example, sonic hedgehog (McCarthy and Argraves, 2003). Megalin also plays key roles in adult physiology. In the kidney a 1:1 complex of megalin and cubilin (Figure 1A) on the apical plasma membrane (PM) of proximal tubule cells binds and mediates endocytosis of a myriad of ultrafiltrate proteins (i.e., hormone, vitamin and iron carriers, enzymes, and immunoglobulin light chains; Kerjaschki and Farquhar, 1983; Christensen and Birn, 2002; Christensen et al., 2012), for subsequent lysosomal degradation and retrieval of their constituent amino acids into the blood (Maack et al., 1979). Kidney filtration of the blood is responsible for the formation of 180 l per day of glomerular ultrafiltrate containing 10–30 μg per liter of low molecular weight proteins (Maack et al., 1979; Haas et al., 1993). Therefore, megalin and cubilin are required to internalize a large amount of ultrafiltrate proteins to prevent their loss in urine (Russo et al., 2007; Christensen and Birn, 2013). Mutations in megalin or cubilin cause human genetic syndromes such as Donnai-Barrow/Facio-Oculo-Acustico-Renal Syndrome (Kantarci et al., 2007), Stickler-like syndrome (Schrauwen et al., 2013), and Imerslund-Gräsbeck disease (Aminoff et al., 1999; Densupsoontorn et al., 2012), all presenting proteinuria. Megalin-deficient mice largely recapitulate this phenotype (Nykjaer et al., 1999). Understanding how megalin is targeted to its apical location in renal epithelium is crucial for recognizing the underlying mechanisms of these diseases.
FIGURE 1:
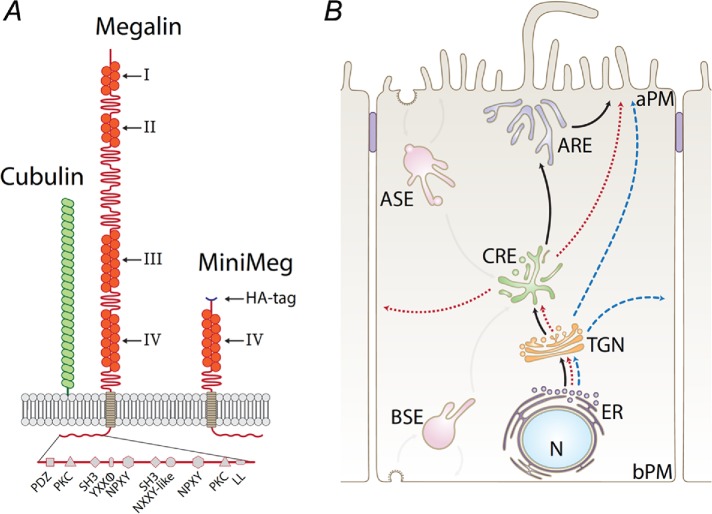
Diagrams of megalin, HA-miniMegalin, and intracellular pathways of megalin in MDCK cells. (A) Cartoon representing the structure of megalin and cubilin, as well as the structure of HA-mMeg. The various sorting motifs present in megalin’s cytoplasmic domain are indicated. (B) Megalin’s biosynthetic and recycling routes in MDCK cells. AP PM, apical plasma membrane; BL PM, basolateral plasma membrane; ER, endoplasmic reticulum; TGN, trans-Golgi network; CRE, common recycling endosome; ASE, apical sorting endosome; BSE, basal sorting endosome; ARE, apical recycling endosome.
We recently used quantitative live imaging to outline the apical recycling pathway of megalin in Madin–Darby canine kidney (MDCK) cells (Figure 1B; Perez Bay et al., 2016). We showed that megalin is a fast recycling (15 min) and long-lived (t1/2 4.8 h) receptor. Megalin is internalized from the apical PM into apical sorting endosomes (ASE) and is subsequently translocated to common recycling endosomes (CRE) where it intersects the recycling pathways of transferrin receptor (TfR) and polymeric IgA receptor (pIgR; Perez Bay et al., 2016). Although this recycling itinerary has been established, the biosynthetic route followed by megalin to the apical membrane is still unknown. After synthesis in the endoplasmic reticulum (ER) surface receptors like tfR and pIgR move to the Golgi apparatus and the trans-Golgi network (TGN). From there, they follow separate routes across endosomal compartments. The fast basolateral recycling tfR is guided to the basolateral membrane via interactions with clathrin and the clathrin adaptor AP-1 (both AP-1B and AP-1A) in both recycling and biosynthetic pathways (Gravotta et al., 2007, 2012). Could clathrin and AP-1 mediate trafficking of fast recycling megalin in the biosynthetic and/or recycling pathways? Previous observations in Caenorhabditis elegans and MDCK implicate clathrin and AP-1 in apical trafficking (see Discussion). Here, we performed silencing and targeting experiments that reveal a role of clathrin and AP-1 in the biosynthetic sorting and recycling of megalin, an apical transmembrane protein in mammalian cells.
RESULTS
Clathrin knockdown disrupts the apical localization of megalin
To study the role of clathrin in the apical localization of megalin, we utilized an MDCK cell line permanently expressing a truncated version of megalin (miniMegalin) containing only its fourth extracellular domain and an extracellular HA epitope (Figure 1A, HA-mMeg; Marzolo et al., 2003; Yuseff et al., 2007; Perez Bay et al., 2016). Silencing via small interfering RNA (siRNA) showed 99% depletion of clathrin heavy chain protein (Figure 2A). In clathrin knockdown cells, domain-selective biotinylation and quantification by Western blot of streptavidin-retrieved biotinylated proteins indicated a robust depolarization of tfR (Figure 2B), as previously reported (Deborde et al., 2008). No effect was seen on the basolateral polarity of claudin 2 (Figure 2B). Surprisingly, clathrin knockdown caused a significant (∼33%) depolarization of surface HA-mMeg to the basolateral membrane, which was also confirmed by qualitative and quantitative immunofluorescence (Figure 2C, quantification in the right panel). Altogether, these results indicate that clathrin mediates apical sorting of megalin. However, since clathrin did not fully depolarize HA-mMeg, it is likely that additional apical sorting mechanisms mediate megalin’s apical localization.
FIGURE 2:
Clathrin silencing disrupts the apical sorting of megalin. MDCK cells stably expressing HA-mMeg were transfected with siRNA targeting clathrin heavy chain and analyzed 4 d after culture on Transwells. (A) Western blot analysis of clathrin knockdown. (B) Domain-selective biotinylation (top panels) and quantification (bottom panels) showing steady-state localization of megalin, transferrin receptor (TfR), and claudin-2 in control and clathrin knockdown MDCK cells. Note the decreased polarity of HA-mMeg and TfR in clathrin KD cells. (C) Surface immunofluorescence showing steady-state HA-mMeg localization in control (left) and clathrin knockdown (right) HA-mMeg MDCK cells. Cells were incubated apically with AF488 (green) labeled anti-HA antibody or basally with AF647 (red) labeled anti-HA antibody. Right panel, integrated fluorescence quantification of images in C, in entire confocal stacks and on a pixel by pixel basis, according to Perez Bay et al. (2016).
Knockdown of γ1 and γ2 variants of clathrin adaptor AP-1
Clathrin cooperates with various adaptor protein complexes, assisting AP-2–dependent endocytosis, or AP-1–dependent vesicular trafficking from TGN and/from endosomal compartments to numerous destinations (Bonifacino and Traub, 2003; Traub, 2009; Traub and Bonifacino, 2013). AP-1 is a tetrameric complex assembled from various isoforms of heavy (γ and β), medium (μ), and small (σ) subunits (Figure 3A). Mammalian cells express the subunit isoforms μ1A, μ1B, γ1, γ2, β1, σ1A, σ1B, and σ1C and assemble them in various combinations generating a repertoire of a dozen possible AP-1 variants (Figure 3B; Mattera et al., 2011; Bonifacino, 2014). We studied the role of two major classes of AP-1 variants, by knocking down the γ1 and/or the γ2 subunits via siRNAs. In each case, half of the possible AP-1 variants (six) are knocked down (Figure 3B). Efficient knockdown of γ1 and γ2 subunits, together or individually, caused depletion of >90% of these subunits at both mRNA (quantitative real time-PCR [qRT-PCR]) and protein levels (Western blot) compared with control (luciferase siRNA) samples (Figure 3C). Although, as expected, combined or individual silencing of γ1 or/and γ2 did not affect the mRNA level of the β1 subunit, they did cause robust reductions in β1 protein level of ∼70, 60, and 30%, respectively (Figure 3C). These results are consistent with a high turnover of unassembled β1 subunits; there are equivalent experiments reporting the instability of the β2 subunit of AP-2 upon knockdown of other AP-2 subunits (Hinrichsen et al., 2003). Immunofluorescence experiments in γ1 and γ2 knockdown cells demonstrated complete depletion of γ1 or γ2 subunits, respectively (Supplemental Figure 1, A and B, and Supplemental Movies 1–9 and 10–18; rotating projection of three-dimensional [3D] reconstruction). Furthermore, knockdown of γ2 did not affect γ1 protein levels or its colocalization with the TGN marker TGN38 (Supplemental Figure 1A), or vice versa in γ2 knockdown cells (Supplemental Figure 1B). These results indicate that knockdown of γ1 or γ2 subunits causes selective depletion of the corresponding AP-1 variants in MDCK cells.
FIGURE 3:
Knockdown of clathrin adaptor AP-1 γ1 and γ2 subunits in MDCK cells. (A) Cartoon of tetrameric AP-1 displaying its assembled subunits. (B) Two-column table displaying all possible variants of AP-1. Gray-shaded and color-filled AP-1 variants represent depleted and expressed populations, respectively, after knockdown of γ1 (left column) or γ2 (right column). (C) Quantification of the depletion of AP-1 γ and β1 subunits at mRNA and protein levels after knockdown of γ1 and/or γ2 subunits. MDCK cells were transfected by electroporation three consecutive times at 3-d intervals with siRNA (Life Technologies) targeting clathrin adaptor subunits γ1 and γ2, alone or combined. The cells were then plated on Transwells and processed 4 d later. Analysis by qRT-PCR revealed an efficient and selective ∼90% silencing of γ1 and γ2 subunits, either when transfected alone or simultaneously. Western blot analysis (LI-COR, Lincoln, NE) revealed depletion levels of γ and β1 protein subunits. Values are averages ± SD from N = 3. Statistical analyses as described in Materials and Methods.
Movie S1.
Suppl. Fig.1A Movie 1. Rotating 3D‐rendered z‐stack of control MDCK cells immune‐labeled with anti‐γ1 (Green) and anti‐ZO‐1 (Magenta); (Detailed in supplementary figure 1A).
Movie S2.
Suppl. Fig. 1A Movie 2. Rotating 3D‐rendered z‐stack of control MDCK cells immune‐labeled with anti‐TGN38 (Red) and anti‐ZO‐1 (Magenta); (Detailed in supplementary figure 1A).
Movie S3.
Suppl. Fig. 1A Movie 3. Rotating 3D‐rendered z‐stack of control MDCK cells immune‐labeled with anti‐γ1 (Green), anti‐TGN38 (Red) and anti‐ZO‐1 (Magenta); (Detailed in supplementary figure 1A).
Movie S4.
Suppl. Fig. 1A Movie 4. Rotating 3D‐rendered z‐stack of γ1 knock down MDCK cells immune‐labeled with anti‐γ1 (Green) and anti‐ZO‐1 (Magenta); (Detailed in supplementary figure 1A).
Movie S5.
Suppl. Fig. 1A Movie 5. Rotating 3D‐rendered z‐stack of γ1 knock down MDCK cells immune‐labeled with anti‐TGN38 (Red) and anti‐ZO‐1(Magenta); (Detailed in supplementary figure 1A).
Movie S6.
Suppl. Fig. 1A Movie 6. Rotating 3D‐rendered z‐stack of γ1 knock down MDCK cells immune‐labeled with anti‐γ1 (Green), anti‐TGN38 (Red) and anti‐ZO‐1 (Magenta); (Detailed in supplementary figure 1A).
Movie S7.
Suppl. Fig. 1A Movie 7. Rotating 3D‐rendered z‐stack of γ2 knock down MDCK cells immune‐labeled with anti‐γ1 (Green) and anti‐ZO‐1 (Magenta); (Detailed in supplementary figure 1A).
Movie S8.
Suppl. Fig. 1A Movie 8. Rotating 3D‐rendered z‐stack of γ2 knock down MDCK cells immune‐labeled with anti‐TGN38 (Red) and anti‐ZO‐1 (Magenta); (Detailed in supplementary figure 1A).
Movie S9.
Suppl. Fig. 1A Movie 9. Rotating 3D‐rendered z‐stack of γ2 knock down MDCK cells immune‐labeled with anti‐γ1 (Green), anti‐TGN38 (Red) and anti‐ZO‐1 (Magenta); (Detailed in supplementary figure 1A).
Movie S10.
Suppl. Fig.1B Movie 10. Rotating 3D‐rendered z‐stack of control MDCK cells immune‐labeled with anti‐γ2 (Green); (Detailed in supplementary figure 1B).
Movie S11.
Suppl. Fig. 1B Movie 11. Rotating 3D‐rendered z‐stack of control MDCK cells immune‐labeled with anti‐GM‐130 (Red); (Detailed in supplementary figure 1B).
Movie S12.
Suppl. Fig. 1B Movie 12. Rotating 3D‐rendered z‐stack of control MDCK cells immune‐labeled with anti‐γ2 (Green) and anti‐GM‐130 (Red); (Detailed in supplementary figure 1B).
Movie S13.
Suppl. Fig. 1B Movie 13. Rotating 3D‐rendered z‐stack of γ1 knock down MDCK cells immune‐labeled with anti‐γ2 (Green); (Detailed in supplementary figure 1B).
Movie S14.
Suppl. Fig. 1B Movie 14. Rotating 3D‐rendered z‐stack of γ1 knock down MDCK cells immune‐labeled with anti‐GM‐130 (Red); (Detailed in supplementary figure 1B).
Movie S15.
Suppl. Fig. 1B Movie 15. Rotating 3D‐rendered z‐stack of γ1 knock down MDCK cells immune‐labeled with anti‐γ2 (Green) and anti‐GM‐130 (Red); (Detailed in supplementary figure 1B).
Movie S16.
Suppl. Fig. 1B Movie 16. Rotating 3D‐rendered z‐stack of γ2 knock down MDCK cells immune‐labeled with anti‐γ2 (Green); (Detailed in supplementary figure 1B).
Movie S17.
Suppl. Fig. 1B Movie 17. Rotating 3D‐rendered z‐stack of γ2 knock down MDCK cells immune‐labeled with anti‐GM‐130 (Red); (Detailed in supplementary figure 1B).
Movie S18.
Suppl. Fig. 1B Movie 18. Rotating 3D‐rendered z‐stack of γ2 knock down MDCK cells immune‐labeled with anti‐γ2 (Green) and anti‐GM‐130 (Red); (Detailed in supplementary figure 1B).
AP-1 silencing depolarizes apical proteins megalin, gp114, and syntaxin 3 but not gp135
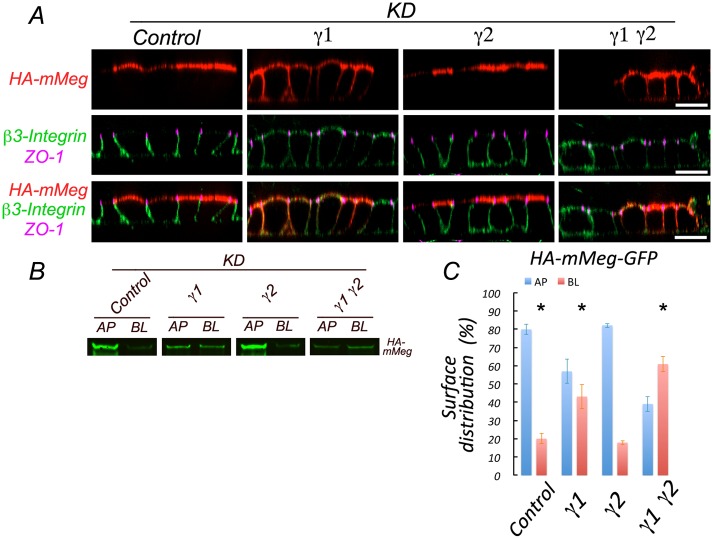
Strikingly, surface immunofluorescence labeling of MDCK cells expressing HA-mMeg subjected to combined γ1/γ2 or single γ1 knockdown revealed basolateral missorting (45%) of HA-mMeg, contrasting with its preferential apical distribution in control cells and in γ2 knockdown cells (Figure 4A). This steady-state result was confirmed biochemically by domain-selective biotinylation (Figure 4B). These data indicate that efficient apical sorting of megalin requires the set of AP-1 variants associated with the γ1 subunit. The larger depolarization observed with AP-1 relative to clathrin knockdown suggests possible clathrin-independent roles of AP-1.
FIGURE 4:
Silencing of AP-1 disrupts apical sorting of megalin and basolateral sorting of β3 integrin in MDCK cells. (A) MDCK cells expressing HA-mMeg were subjected to silencing of γ1 and/or γ2 subunits, as indicated in Figure 3. After 4 d, cells were immunolabeled at 4°C with a rabbit anti-HA antibody and a mouse monoclonal antibody against the luminal domain of β3-integrin. Cells were fixed, permeabilized, and subsequently incubated with rat anti–ZO-1. Cell-bound antibodies were visualized with Alexa-labeled secondary antibodies and analyzed by spinning disk confocal microscopy. Images are displayed as x-z sections. Bar, 20 μm. (B) Cells silenced for γ1 and/or γ2 as described above were subjected to domain-selective biotinylation, retrieval of biotinylated HA-mMeg with streptavidin, and Western blot with HA antibodies. (C) Quantification of the results in B. Values are averages ± SD from N = 3. Statistical analyses as described in Materials and Methods. * represents p < 0.01.
To extend this observation to a broader cellular context than MDCK cells, we carried out similar experiments in the thyroid epithelial cell line FRT. Thyroid cells normally utilize apical megalin to internalize thyroglobulin for degradation into lysosomes (Marino et al., 2000). Transfection of FRT cells with siRNA targeting γ1 subunits, as described for MDCK cells, caused robust, >70%, knockdown of γ1 both at mRNA (qRT-PCR) and protein levels (Western blot) compared with control (luciferase siRNA) samples (Supplemental Figure 2A). We transfected HA-mMeg into FRT cells simultaneously with siRNAs against γ1 adaptin and analyzed the cells for immunofluorescence 5 d later. FRT cells knocked down of γ1 displayed a striking depolarization of megalin, similar to what we observed in MDCK cells (Supplemental Figure 2, B and C).
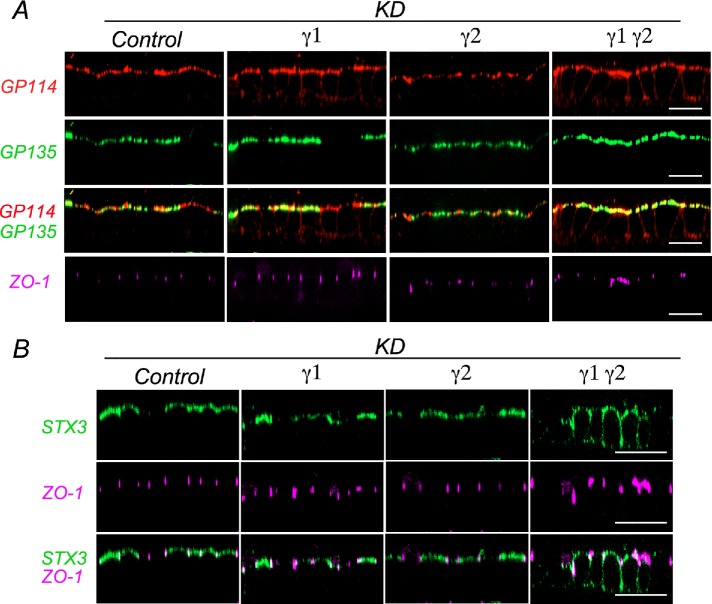
Next, we sought to extend our HA-mMeg observations to endogenous apical and basolateral proteins. Surface immunolabeling of gp114, also known as CEACAM (Fullekrug et al., 2006), revealed a partial basolateral redistribution in both combined γ1/γ2 and single γ1, but not in single γ2 knockdown cells (Figure 5A). In contrast, the apical localization of gp135, also known as podocalyxin (Cheng et al., 2005) was not disrupted by either combined γ1/γ2 or individual γ1 and γ2 knockdown cells (Figure 5A). This observation was consistent with our previous report in MDCK cells knocked down for AP-1 μ1A and μ1B subunits (Gravotta et al., 2012). On the other hand, the apical localization of syntaxin-3, a T-SNARE involved in apical delivery, was disrupted by combined γ1/γ2 knockdown but not by single γ1 or γ2 knockdown (Figure 5B). We also monitored the basolateral membrane protein β3 integrin and found apical redistribution in single γ1 and combined γ1/γ2, but not in single γ2 knockdown cells (Figure 4A).
FIGURE 5:
Silencing of AP-1 disrupts apical sorting of gp114 and syntaxin 3 proteins in MDCK cells. (A) MDCK cells expressing HA-mMeg were subjected to silencing of γ1 and/or γ2 subunits as described in Figure 3. For surface immunolabeling HA-mMeg MDCK cells were incubated at 4°C with a rabbit anti-gp114 and a mouse anti-gp135 added apically and basolaterally. Cells were fixed and permeabilized and subsequently incubated with rat anti–ZO-1. Bound antibodies were visualized with Alexa-labeled secondary antibodies and analyzed by spinning disk confocal microscopy. Images are displayed as x-z sections. Control represents luciferase KD. Bar, 20 μm. (B) MDCK cells stably expressing Myc epitope-tagged syntaxin-3 (STX3-Myc) were subjected to single or combined silencing of γ1 and/or γ2 subunits as described in A and Figures 3 and 4. Surface and total syntaxin 3 immunofluorescence distribution were revealed by staining with mouse (green) and rabbit (red) anti-Myc antibodies, on intact and subsequently permeabilized cells, respectively. Images are displayed as x-z sections. Control represents luciferase KD. Bar, 20 μm.
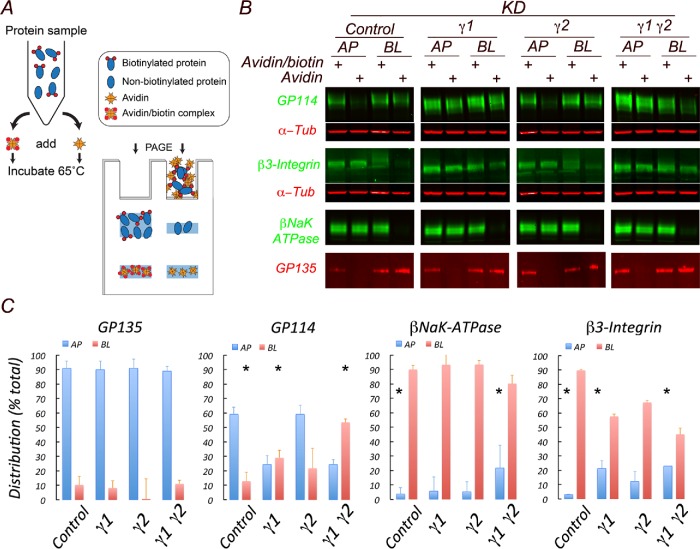
Biochemical quantification of the distribution of endogenous apical and basolateral membrane proteins in single γ1 or combined γ1/γ2 knockdown cells was carried out using a surface biotin avidin shift (SBAS) assay described earlier (Figure 6A; Gravotta et al., 2012). In this assay two samples are compared, one in which biotinylated cargo proteins are sequestered into a tight complex by the addition of avidin (lane 2) and another one exposed to avidin prebound to biotin (lane 1) representing the total, in which nonbiotinylated as well as biotinylated cargo, remain free to diffuse. The difference between the two bands reflects the amount of biotinylated protein in the mixture.
FIGURE 6:
Silencing of AP-1 in MDCK cells disrupts apical sorting of gp114 but not gp135. (A) Scheme of SBAS (surface biotinylation avidin shift) assay. MDCK cells silenced for AP-1 γ1 and/or γ2 subunits and cultured for 4 d on 24-mm Transwells are subjected to domain-selective biotinylation. The extracted samples are incubated with either avidin or avidin prebound to biotin and then analyzed by gel electrophoresis followed by Western blot. Binding of avidin to biotinylated proteins results in the formation of multimeric complexes that are excluded from the gel; the consequent decrease in the intensity of the sample containing avidin is the basis of their quantification. (B) Western blot of SBAS-treated samples. (C) Quantification of the Western blot results expressed as percent of total. Values are averages ± SD with N = 3. Statistical analyses were done as described in Materials and Methods.
These experiments revealed substantial basolateral missorting (fourfold) of apical gp114 and a substantial (sevenfold) apical redistribution of normally basolateral β3 integrin (Figure 6, B and C). On the other hand, the polarities of both apical gp135 and basolateral NaK-ATPase β subunit were only marginally affected by the same experimental conditions (Figure 6, B and C). Our biochemical observations on apical and basolateral missorting of endogenous markers in AP-1 knockdown MDCK cells supported our morphological observations indicating a robust basolateral redistribution of gp114, with minimal impact on gp135. Collectively, these findings demonstrate that apical sorting of HA-mMeg as well as of some endogenous transmembrane apical proteins is regulated by the clathrin adaptor AP-1.
Clathrin controls megalin’s apical biosynthetic and recycling routes
Is the basolateral mislocalization of HA-mMeg observed in clathrin knockdown cells caused by protein missorting in the biosynthetic and/or recycling routes? To address this question, we engineered a HA-mMeg chimera containing a C-terminal GFP and four N-terminal FM4 conditional aggregation domains followed by a furin cleavage site that results in post-Golgi removal of the FM4 domains, a strategy we previously developed to promote reversible ER retention and synchronized protein export from the ER of MDCK cells (Thuenauer et al., 2014). Experiments of surface capture with fluorescently labeled mouse anti-HA (647Ms-HA) antibody in a stable MDCK cell line expressing FM4-HA-mMeg-GFP validated our strategy. Confocal microscopy analysis of these experiments revealed negligible capture of 647Ms-HA in cells incubated in the absence of D/D solubilizer, reflecting efficient ER retention, through aggregation of FM domains. On the other hand, incubation in the presence of D/D solubilizer released FM4-HA-mMeg-GFP promoting capture of 647Ms-HA upon surface delivery (Supplemental Figure 3, A and B). The biosynthetic delivery route of HA-mMeg-GFP was monitored by surface biotinylation after release from the ER by the addition of D/D solubilizer (Figure 7A). These studies revealed a robust basolateral missorting of this protein in clathrin knockdown cells (Figure 7, B and C). A similar missorting of HA-mMeg GFP was observed when clathrin function was pharmacologically blocked by Pitstop (unpublished data).
FIGURE 7:
Clathrin regulates apical biosynthetic delivery of HA-mMeg-GFP. (A) Scheme of biotinylation assay for surface delivery. MDCK cells stably expressing ER-retained FM4-HA-mMegalin-GFP are incubated at 37°C with D/D solubilizer, to elicit synchronized export, followed by surface domain biotinylation and streptavidin retrieval of cell surface biotinylated HA-mMeg-GFP. (B) Clathrin knockdown disrupts apical delivery of HA-mMeg-GFP. Left, luciferase silencing control. Right, clathrin heavy chain silencing. (C) Quantification of the results. Note the missorting of HA-mMeg-GFP to the basolateral plasma membrane in clathrin knockdown cells. Values are averages ± SD with N = 3. Statistical analyses as described in Materials and Methods.* represents p < 0.05. ** represents p < 0.01.
AP-1 controls megalin apical biosynthetic and recycling routes
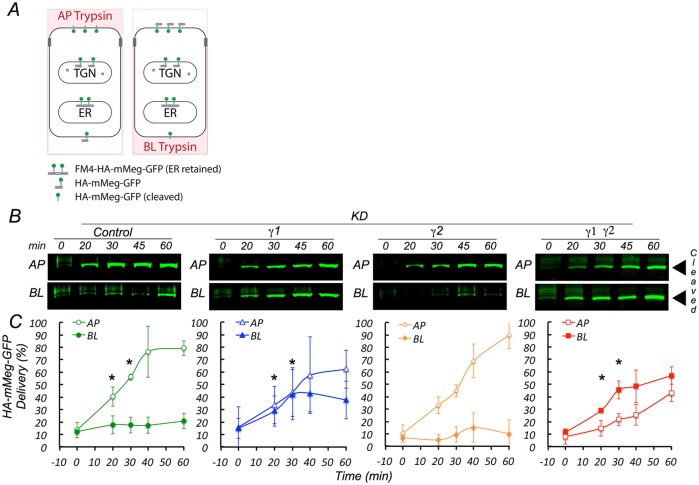
We next addressed the question of whether AP-1 regulates the biosynthetic and/or recycling pathways of megalin. To this end we used a modified assay to monitor surface arrival of HA-mMeg-GFP after its intracellular release from the ER and Golgi through disaggregation and furin cleavage of its FM4 domains. As megalin is rapidly endocytosed its apical dwelling after biosynthetic surface delivery is highly transient; hence we posited that it might be best monitored through constant polarized exposure to trypsin added apically or basolaterally during the delivery period (Figure 8A). Under these conditions the full-size 170 kDa HA-mMeg-GFP at the cell surface is cleaved by trypsin, generating a ∼130 kDa product. The uncleaved and cleaved species are easily separated by electrophoresis and easily quantified by Western blot with antibodies against GFP (Supplemental Figure 3C); for simplicity we show only the 130 kDa band (Figure 8, B and C). Control cells (luciferase siRNA) displayed preferential cleavage of HA-mMeg-GFP when trypsin was added from the apical surface; the kinetics of this process shows that >50% of HA-mMeg-GFP is delivered after 30 min of incubation and most of the trypsin cleavage occurs apically (Figure 8, B and C, control panels). However, in γ1 or combined γ1/γ2 (but not in γ2) knockdown cells HA-mMeg-GFP was cleaved from both apical and basolateral sides at early times after the addition of D/D solubilizer (20 min), consistent with missorting in its biosynthetic route (Figure 8, B and C). These experiments demonstrated that AP-1 γ1 variants regulate biosynthetic delivery of HA-mMeg-GFP.
FIGURE 8:
AP-1 controls apical biosynthetic delivery of HA-mMeg-GFP. (A) Scheme of controlled trypsin cleavage assay for biosynthetic surface delivery. MDCK cells stably expressing ER-retained FM4-HA-mMegalin-GFP are incubated at 37°C with D/D solubilizer, and with trypsin, added apically or basolaterally, to induce ER export and subsequent cleavage of HA-mMegalin-GFP upon cell surface arrival. (B) Western blotting detection with an anti-GFP antibody of cleaved ∼130 kDa HA-mMegalin-GFP. (C) Quantification of the results. Values derived from three independent experiments. *p value < 0.05. Note that single γ1 and combined γ1/γ2 knockdown but not single γ2 knockdown cause missorting of HA-mMeg-GFP.
We extended these studies to investigate the role of AP-1 on the surface delivery of HA-mMeg-GFP in the recycling route. To this end, we used a similar trypsin cleavage assay as just described for the biosynthetic route, except that the cells were first incubated with D/D solubilizer for 2 h to allow ER release, surface delivery, and endosomal equilibration of HA-mMeg-GFP (Figure 9A). Upon subsequent incubation at 37°C for short intervals in the presence of apical or basolateral trypsin, control (luciferase siRNA) samples showed preferential (85%) trypsin cleavage from the apical side (Figure 9, B and C, control panels). In contrast, single γ1 or combined γ1/γ2 knockdown cells displayed trypsin cleavage of HA-mMeg-GFP from both sides, albeit the kinetics of basolateral cleavage were somewhat slower (Figure 9, B and C). These experiments, in agreement with our morphological findings (Figure 4) demonstrated that the clathrin adaptor AP-1 variants associated with the γ1 subunit regulate surface delivery of HA-mMeg-GFP in the recycling route.
FIGURE 9:
AP-1 controls apical recycling of HA-mMeg-GFP. (A) Scheme of controlled trypsin cleavage assay for recycling. MDCK cells stably expressing ER-retained FM4-HA-mMegalin-GFP are incubated at 37°C with D/D solubilizer, for 2 h and an additional ½ h in the presence of cycloheximide, to allow HA-mMeg-GFP at the cell surface, and in the recycling compartment reach steady-state distribution. Recycling is then monitored for an additional 1 h upon trypsin addition, apically or basolaterally, in the presence of cycloheximide. (B) Trypsin cleaves HAmMeg-GFP, causing the appearance of a ∼130 kDa band that is detected by Western blot with an anti-GFP antibody (see Supplemental Figure 3C). (C) Quantification of the Western blot results. Note the polarized arrival of HA-mMeg-GFP to the cell surface in control (luciferase) and γ2 KD cells. The arrival of HA-mMeg-GFP is depolarized in γ1 and γ1/γ2 KD cells. Values are averages ± SD with N = 3. *p value < 0.05. Statistical analyses as described in Materials and Methods.
To gain some insight on which compartments might constitute the site of action of the different AP-1 variants containing γ1 and γ2 adaptins we carried out quantitative coimmunolocalization experiments of γ1 and γ2 with TGN, CRE, and apical recycling endosome (ARE) markers (Figure 10). For these experiments, we used commercial mouse antibody against γ1 and rabbit antibody against γ2. These experiments demonstrated that the largest pool of both adaptins localized to the TGN (∼40%), with smaller pools localizing at CRE (30%) and ARE (20%) (Figure 10). The distributions we found were asymmetrical as a large fraction of γ1, 56%, colocalizes with γ2; however, only 25% of γ2 colocalizes with γ1; moreover, the TGN fraction occupied by γ2 was twice (50%) the fraction occupied by γ1 (25%). These experiments suggest that distinct functions of γ1 and γ2 variants of AP-1 may be performed in the different subcellular compartments.
FIGURE 10:
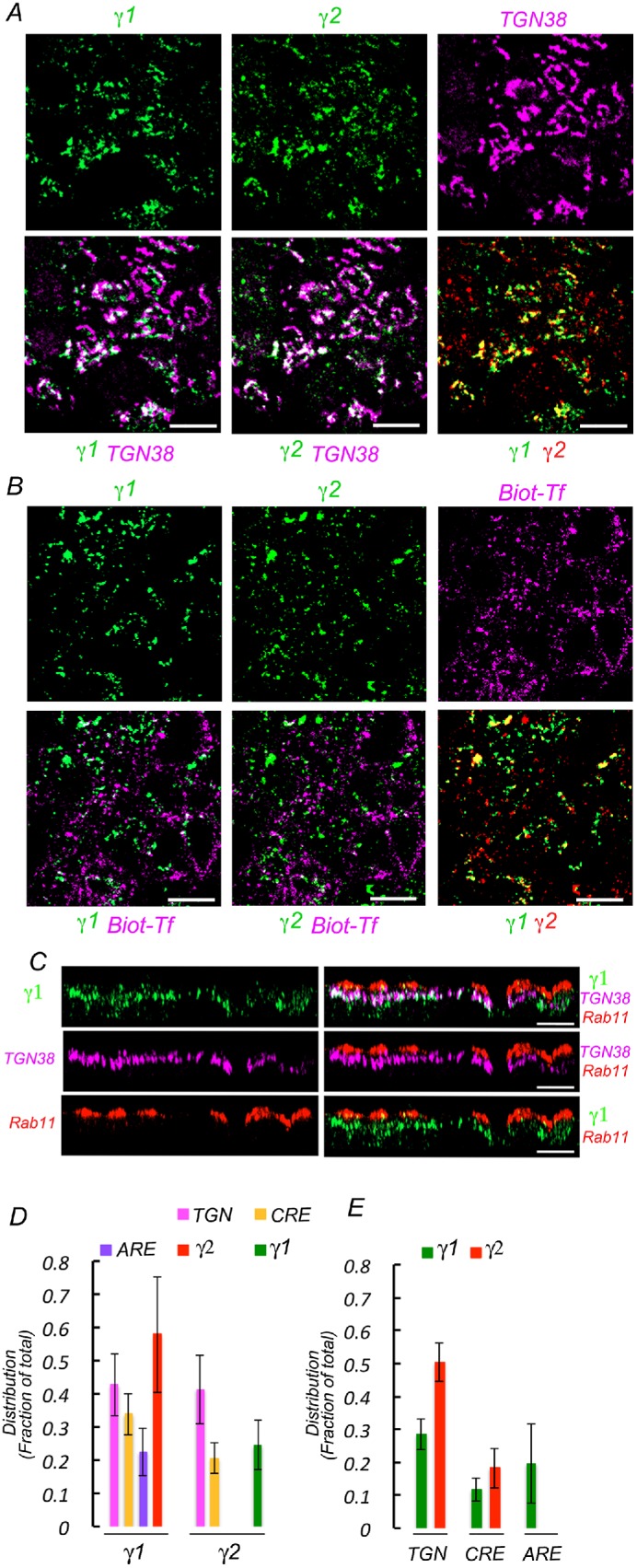
Compartmental distribution of γ1 and γ2 studied by quantitative immunofluorescence colocalization. Wild-type MDCK cells were fixed and processed for immunofluorescence using a mouse antibody against γ1 adaptin and a rabbit antibody against γ2 adaptin and different antibodies against TGN (TGN38) and ARE (Rab11) markers. Top panels display individual markers; bottom panels display pairs of colocalized markers. (A) Colocalization with TGN marker. Note areas of extensive colocalization between γ1, γ2, and TGN 38, as well as colocalization between γ1 and γ2. (B) Colocalization with CRE marker. For CRE labeling, cells were loaded with biotinylated transferrin for 90 min before fixation and processing for γ1 and γ2 immunofluorescence as described above. Subsequently, the cells are decorated with Alexa 647–streptavidin. (C) Colocalization with ARE marker. Cells are processed with antibody against γ1 adaptin and Rab 11, followed by the corresponding secondary antibodies. γ2 localization studies were prevented by lack of appropriate antibodies. Displayed images represent x-z projections of 15 slices. Note the apical localization of ARE with respect to the TGN and the extensive colocalization of γ1 and TGN. There is little colocalization of γ1 with ARE, likely reflecting projections of TGN fragments at that level. (D) Bar graph quantification of the colocalization of γ1 and γ2 with TGN, CRE, and ARE. (E) Bar graph quantification of the colocalization of TGN, CRE, and ARE with γ1 and γ2.
DISCUSSION
Megalin is apically expressed in polarized epithelia including native renal proximal tubule and cultured MDCK cells. We recently characterized megalin as a long-lived fast recycling receptor that follows the endosomal compartments associated with the apical recycling pathway (Perez Bay et al., 2016). However, very little is known about the biosynthetic pathway that megalin follows to the apical cell surface and about the molecular machinery that mediates its apical trafficking in the biosynthetic and recycling routes. Here we describe a mechanism by which clathrin and the clathrin adaptor AP-1 mediate apical sorting of megalin.
For our experiments, we used a truncated version of megalin (mMeg) for both practical and conceptual reasons. The practical reason is that, with a molecular weight of over half a million daltons, megalin is a difficult protein for performing trafficking studies. The conceptual reason is that miniMegalin (mMeg), containing just the fourth extracellular domain of megalin, appears to traffic normally along the same route followed by the full-size protein (Perez Bay et al., 2016), ostensibly because its cytoplasmic tail contains all of its known trafficking motifs (Figure 1A). Indeed, we were prompted to study the role of clathrin and AP-1 on the apical trafficking of megalin by the reported observation that its apical trafficking apparently depends on endocytic-like motifs such as NPXY contained in its cytoplasmic tail (Marzolo et al., 2003; Takeda et al., 2003). Motifs of this kind are known to participate in protein sorting via interaction with clathrin adaptors such as AP-1 and AP-2 (Bonifacino and Traub, 2003; Bonifacino, 2014; Traub and Bonifacino, 2013). Indeed, we found that that the apical sorting of megalin is modulated by clathrin and AP-1. Our experiments demonstrate that knockdown of clathrin or the gamma subunits of AP-1 decrease the apical polarization of HA-tagged miniMegalin. Our silencing experiments also demonstrate that the apical sorting of megalin may be achieved via a role of clathrin and AP-1 in the biosynthetic and recycling routes. We did not study directly which of the possible sorting determinants in the cytoplasmic tail of megalin participates in clathrin-mediated sorting. This is an important question relevant to the molecular interactions mediating apical sorting of megalin and other apical proteins, but it falls beyond the scope of the hypothesis tested here. Furthermore, as megalin remains partially apically localized after clathrin or AP-1 knockdown, it is possible that additional apical determinants are present in megalin’s transmembrane or luminal domain.
Since the discovery of AP-1B and AP-1A as regulators of epithelial polarity (Folsch et al., 1999, 2009; Ohno et al., 1999; Carvajal-Gonzalez et al., 2012; Gravotta et al., 2012; Bonifacino, 2014; Rodriguez-Boulan and Macara, 2014), they have been found to predominantly mediate basolateral protein sorting. Moreover, we previously reported that clathrin mediates preferentially sorting of basolateral proteins, based on the analysis of small subgroups of apical and basolateral proteins (Deborde et al., 2008). However, in this article we also report that, in addition to megalin, other apical transmembrane proteins also depend on AP-1 for apical sorting, for example, gp114 and syntaxin 3 but not gp135. Conversely, as previously observed, the polarity of some basolateral proteins (e.g., β-integrin, claudins, Na,K-ATPase) was variably affected by knockdown of clathrin and/or AP-1. These results highlight an emerging role of AP-1 in apical sorting and, importantly, the need to expand the repertoire of PM proteins examined in polarity studies. We have recently addressed this issue by developing a global proteomics approach that helped us identify more than 300 PM proteins in MDCK cells and demonstrate roles of AP-1 in the sorting of subgroups of both apical and basolateral PM proteins (Caceres et al., 2019).
Our findings reported here have antecedents in earlier work in C. elegans that implicated AP-1 in apical sorting (Shafaq-Zadah et al., 2012; Zhang et al., 2012) and in a recent report that implicates AP-1 and clathrin in the sorting of GPI-anchored proteins (Castillon et al., 2018), which are preferentially apically localized in most cases (Lisanti et al., 1989; Sargiacomo et al., 1989; Zurzolo and Simons, 2016). In agreement with our data, the latter group also reported an effect of AP-1 knockdown on the sorting of apical syntaxin 3; based on this finding, they proposed that the apical sorting role of the AP-1 adaptor might be through its ability to apically sort syntaxin 3. However, in our hands, knockdown of both γ1 and γ2 subunits of AP-1 is required to observe the effect on syntaxin 3 rather than single knockdown of just γ1, as reported by Castillon et al. (2018). This suggests that the mechanism through which AP-1 regulates the apical polarity of megalin or gp114 does not necessarily involve syntaxin 3. We also report here that, unlike megalin and gp114, which depend on AP-1 for trafficking, gp135/podocalyxin apical localization is independent of AP-1; this is not surprising as gp135 depends on a bipartite apical signal constituted by O-glycans and a C-terminal PDZ motif for apical sorting (Yu et al., 2007).
The AP-1 adaptor studied here is a tetramer composed of 12 possible variants resulting from different combinations of μ, γ, β, and σ subunits (Bonifacino, 2014). In epithelial cells, the μ1B variant plays a central role in basolateral trafficking because it is expressed exclusively in epithelia (Ohno et al., 1999). However, the kidney proximal tubule, which expresses megalin endogenously on the apical membrane, does not express μ1B (Schreiner et al., 2010), whereas MDCK cells do express μ1B and also direct megalin to the apical membrane; therefore, it is unlikely that the μ subunits of AP-1 have a major role in megalin apical sorting. To study a mechanism that may be more relevant to megalin sorting in the kidney proximal tubule, we decided to focus on the γ subunits of AP-1. We found an exclusive role of AP-1 variants containing γ1 in apical megalin sorting, as knockdown of γ2 had no effect and combined γ1/ γ2 knockdown did not exacerbate the effect of γ1 knockdown. This may be due to disruption of the stability of additional subunits of the AP-1 complex, highlighting that different combinations of the several subunits and isoforms may play specific sorting roles. It is not known which are the AP-1 variants that exist in native kidney proximal tubule cells.
In summary, our results describe novel roles of clathrin and AP-1 in megalin’s biosynthetic and recycling pathways to the apical PM that mediate its apical sorting. Further work is necessary to determine the sorting signals contained in the cytoplasmic tail of megalin, as well as additional components of the apical sorting machinery that mediate its apical sorting. In addition, in a more general sense, it will be necessary to develop and test specific hypotheses (e.g., different sorting signals, cargo-dependent interaction with additional sorting mechanisms) to explain how clathrin and AP-1 may participate in both apical and basolateral protein sorting.
MATERIALS AND METHODS
Reagents
Reagents used were D/D solubilizer (635054), Takara Bio, San Francisco, CA; power SYBR green RNA to Ct (4389986), Applied Biosystem, Foster City, CA; ProStar Ultra HF RT-PCR, Stratagene Cedar Creek, TX; RNeasy (74106), Qiagen, Germany.
Antibodies and sources
Antibodies used were mouse anti-HA (MMS-101P), Covance, Princeton, NJ; rat anti-HA (11867423), Roche, Indianapolis, IN; rabbit anti-GFP antibody (JM3999), MBL International, Des Plaines, IL; mouse anti-transferrin receptor (10RCD71aHU), Fitzgerald, Acton, MA; mouse anti-clathrin heavy chain (B212853), Biolegend, San Diego, CA; rabbit anti–β catenin (C2206), rabbit anti–γ2-adaptin (HPA004106), mouse anti–α-tubulin (T9026), and rabbit anti–β1-adaptin (A4450), Sigma-Aldrich, St. Louis, MO; mouse anti–Myc-tag (132500), rabbit anti-Rab11 (715300), and Alexa Fluor labeled secondary antibodies, Invitrogen, Carlsbad, CA; mouse anti–β3-integrin (555752), mouse anti–γ1-adaptin (610386), mouse anti–μ2-adaptin (611350), BD Franklin Lakes, NJ; mouse anti-CEACAM, gp114, mouse anti-gp135 (grown from a hybridoma kindly provided by J.K.O. Ojakian while at SUNY Downstate Medical Center, Brooklyn, NY), rabbit anti–claudin 2 (Thermo Fisher Scientific, Waltham, MA). Secondary antibodies for immunofluorescence, goat anti-mouse, rat, or rabbit, conjugated to either Alexa Fluor 568, 488, or 647, were from Life Technologies, Grand Island, NY. Secondary antibodies for Western blotting, goat anti-mouse, rat, or rabbit conjugated to either IRDye680 or IRDye800, were from LI-COR (Lincoln, NE). HRP-conjugated secondary antibodies were from Amersham Pharmacia Biotech, Piscataway, NJ.
The fluorescent probe mouse anti-HA (647-Ms-HA) was prepared by conjugation of monoclonal mouse-HA to the fluorophore SeTau-647 (SeTa Biomedicalos, Urbana, IL) as previously described (Perez Bay et al., 2016).
siRNA
γ1: 5′GCAUUGAUGUGGAACUCCAtt3′ (Sense, dog)
γ1: 5′UGGAGUUCCACAUCAAUGCtg3′ (Antisense, dog)
γ1: 5′GUUCCUGAACUUAUGGAGAdtdt3′ (Sense, rat)
γ1: 5′UCUCCAUAAGUUCAGGAACdtdt3′ (Antisense, rat)
γ2: 5′GCUUUGCUCUUAAUCACCAtt3′ (Sense)
γ2: 5′UGGUGAUUAAGAGCAAAGCag3′ (Antisense)
CHC: 5′GUAUGAUGCUGCUAAACUAdtdt3′ (Sense)
CHC: 5′UAGUUUAGCAGCAUCAUACdtdt3′ (Antisense)
CHC: 5′UAAUCCAAUUCGAAGACCAAdtdt3′ (Sense)
CHC: 5′UUGGUCUUCGAAUUGGAUUAdtdt3′ (Antisense)
qRT-PCR oligos
γ1: 5′AGTGGCTGCATGGTGTATAG3′ (Sense)
γ1: 5′GGTCACAGAGGTGGACATATTAG3′(Antisense)
γ2: 5′CTTCGTCTGCTTCGGATTCT3′ (Sense)
γ2: 5′GGTCAGCACCGTCTCAAATA3′ (Antisense)
β1: 5′ CTGGCAGAGCTGAAAGAGTATG3′ (Sense)
β1: 5′ GCAGATTGCTCCACCTTGAT3′ (Antisense)
Plasmids
As a strategy to study biosynthetic and recycling trafficking of megalin we generated pFM4-HAmMeg-GFP, a vector encoding a chimera of HA-mMeg flanked with the growth hormone signal sequence, four FM domains, and a furin cleavage site at its 5′ end, and by GFP at the 3′ end of HA-mMeg. We used pFM4-rhodopsin-GFP (Thuenauer et al., 2014) as a template and swapped rhodopsin for HA-mMeg.
Wild-type and permanent cell line culture
MDCK cells were grown in DMEM (Cellgro) supplemented with penicillin, streptomycin, and 10% fetal bovine serum (FBS), at 37°C, 5% CO2.
Stable MDCK cell lines expressing FM4-HA-mMegalin-GFP, were generated by transfecting MDCK cells with pFM4-HA-mMeg-GFP vector, described above, using Lipofectamine 2000, following manufacturer recommendations (Life Technologies, Grand Island, NY). Cells were selected in growth media supplemented with 0.8 mg/ml G418 (Mediatech, Manassas, VA) and subsequently selected for homogeneous expression level by fluorescence-activated cell sorting.
Stable MDCK cell lines expressing HA-mMeg, containing just the fourth domain of human megalin and an N-terminal HA epitope tag, were reported before (Perez Bay et al., 2016).
RNA interference
Canine γ1 (AP1γ1) and γ2 (AP1γ2) custom synthesized Silencing Select siRNA (SiSel-siRNA; Ambion, Life Technologies, Grand Island, NY) were tested by qRT-PCR. The most potent siRNA, with ∼90% knockdown efficiency, for either single, γ1 or γ2 or combined, γ1 and γ2, as well as any siRNA chosen against targets listed above, were transfected into MDCK cells by electroporation as previously described (Gravotta et al., 2007). Briefly, trypsinized MDCK cells suspended at 5 × 106 cells per 0.125 ml of nucleofector solution V supplemented with 160 pmol each siRNA were electroporated through a cycle run with program T-23 (Lonza Group Ltd., Allendale, NJ). This transfection step was repeated twice at 3-d intervals to produce effective depletion of targeted protein. Target knockdown mRNA and protein levels were quantified by qRT-PCR and Western blot from total RNA and cell lysates collected 4 d after the last transfection.
Surface immunofluorescence and confocal imaging
For morphological analysis, MDCK cells were plated on 12-mm polycarbonate membrane inserts (Transwells) at the density of ∼3 × 105.cm−2 and cultured for 4 d. Cell surface and intracellular labeling of HA-mMeg expressed on MDCK cells grown on Transwells was carried out in two stages. First, for surface labeling cells were rinsed twice with 37°C prewarmed Hank’s balanced salt solution (HBSS) containing Ca++ and Mg++ (HBSS-CaMg) and once with ice-chilled HBSS-CaMg. Cells were then incubated at 4°C for 4 h with rat monoclonal anti-HA 4 μg/ml in HBSS-CaMg supplemented with 1.5% bovine serum albumin (BSA). Cells were rinsed four times with HBSS-CaMg-1.5% BSA and once with HBSS-CaMg before fixation in 3.7% paraformaldehyde (PFA) for 20 min at 4°C. Excess fixative was rinsed with phosphate-buffered saline (PBS), and excess aldehyde groups were quenched with 50 mM ammonium acetate in PBS for 20 min at room temperature (RT). For intracellular antigens cells were permeabilized with 0.075% saponin in PBS at RT for 10 min and then incubated at RT for 30 min with Tris-BSA buffer (Tris-HCl 30 mM, pH 7.6, NaCl 150 mM, BSA 1.5%). Cells were incubated overnight at 4°C with mouse monoclonal anti-HA 2.5 µg/ml HA in Tris-BSA buffer followed by four rinses with Tris-BSA buffer. Bound primary antibodies were visualized with ∼2 μg/ml secondary goat antibodies conjugated to Alexa Fluor 568, 488, or 647 (Life Technologies) in Tris-BSA buffer. Cells were then rinsed four times with Tris-BSA buffer, mounted in Vectashield, and examined using a Zeiss Cell Observer SD confocal with a Yokagawa CSU-X1 spinning disk, Plan-Apochromat 63×/1.4 M27 oil-immersion objective paired with a 1.2× adapter to a Photometrics Evolve 512 EMCCD camera. Image stacks were collected by Nyquist sampling. 3D rendering rotation was generated by processing Z-stack images with Zeiss software version 3.5 (Carl Zeiss MicroImaging, LLC, Thornwood, NJ).
Colocalization and quantification of immunolabeled MDCK cells
Colocalization experiments were carried out in MDCK cells, grown on 12 mm Transwells 4 d after full polarization, as previously described (de la Fuente-Ortega et al., 2015). In studies of colocalization with CRE, cells were starved for 30 min in serum-free DMEM (SF-DMEM) before incubation at 37°C for 90 min with biotinylated transferrin (Biot-Tf) 10 μg/ml in SF-DMEM supplemented with BSA 1 mg/ml. Cells were rinsed with medium three times at 37°C in 5-min intervals to clear most surface labeling before rinsing them four times with ice-cold HBSS-CaMg at 4°C. Then all samples for colocalization experiments were rinsed, fixed, blocked, and permeabilized as described above. Visualization of the biotinylated transferrin endocytosed was carried out with streptavidin labeled with Alexa Fluor 647 included in the incubation together with the secondary antibodies. The acquired image stack (90–120 slices collected as described above) are thresholded and marked with a region of interest (ROI), containing an average of 45 cells, before being analyzed with Zen imaging software (Zeiss, Oberkochen, Germany) to generate the following data files:
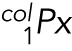 (cargo 1): Pixels within ch2 that colocalize with ch1, (cargo 2)
(cargo 1): Pixels within ch2 that colocalize with ch1, (cargo 2)
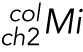 : Mean intensity of colocalizing pixels above
: Mean intensity of colocalizing pixels above  in ch2
in ch2
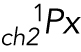 (cargo 1): Pixels within ch2 (cargo 1) that do not colocalize with ch1 (cargo 2)
(cargo 1): Pixels within ch2 (cargo 1) that do not colocalize with ch1 (cargo 2)
 : Mean intensity of the above noncolocalizing pixels
: Mean intensity of the above noncolocalizing pixels 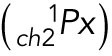 in ch2.
in ch2.
The equation described below is an operator that sums i the product of pixel (Px) number and the mean intensity (Mi) within an ROI for all z = 1 to N).
Cargo 1 (colocalized):
 |
Cargo 1 (noncolocalized):
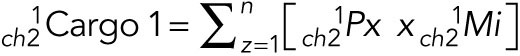 |
The values obtained represent total Cargo 1 colocalized  and total Cargo 1 noncolocalized
and total Cargo 1 noncolocalized  . The fraction of total Cargo 1 that colocalizes with Cargo 2 is calculated as follows.
. The fraction of total Cargo 1 that colocalizes with Cargo 2 is calculated as follows.
Colocalized Cargo 1 fraction =
 |
Domain-selective biotinylation and SBAS assay
For biochemical analysis MDCK cells were plated on 24-mm Transwells at a density of ∼3 × 105 cm−2 and cultured for 4 d. Cell surface biotinylation and SBAS assays were carried out as previously described (Gravotta et al., 2012). This assay, differently from the conventional surface domain biotinylation-streptavidin retrieval assay (Sargiacomo et al., 1989), quantifies the steady-state apical/basolateral surface distribution of a given protein as a fraction of the total protein pool. Briefly, polarized MDCK cells were subjected to domain-selective biotinylation using sulfo-NHS-LC-biotin (3 mg/ml), twice for 20 min at 4°C, either apically or basolaterally (Gravotta et al., 2012). The lysate from these samples (300 μl) made in 40 mM Tris-HCl, pH 7.8, 5 mM EDTA, 1% SDS, supplemented with a protease inhibitor cocktail, was sonicated for 45 s at an output power set at 25–30 (Branson Sonifier 250) and incubated at 95°C for 10 min with 25 mM dithiothreitol. Two identical (45 μl) aliquots retrieved from each lysate were mixed with 25 μl of a premixed solution containing 750 μg/ml avidin, 1.2 mM biotin, and 15 μg/ml bromophenol blue in 50% glycerol (sample ST), or 750 μg/ml avidin, 15 μg/ml bromophenol blue in 40% glycerol (sample SNB) and heated at 65°C for 10 min. The two samples were resolved side by side on PAGE and target cargo proteins processed for Western blot as described above. Sample “ST “ reflects the total, biotinylated and nonbiotinylated cargo protein, while sample “SNB“ reflects nonbiotinylated cargo proteins. The difference between these two samples calculated from values derived from quantified Western blots represents biotinylated cargo protein in the sample.
Quantitative HA-mMeg-GFP biosynthetic and recycling assays
To study the biosynthetic and recycling routes of megalin, we developed a quantitative biochemical assay using the chimera FM4-HA-mMeg-GFP (described above), which is reversibly retained at ER and can be released for synchronous export to the cell surface upon addition of D/D solubilizer. Arrival to the cell surface was monitored by surface biotinylation or by controlled trypsin cleavage from the apical or basolateral sides.
For biosynthetic delivery assays, FM4-HA-mMeg-GFP–expressing MDCK cells cultured on 12-mm Transwells) for 4 d were rinsed twice with serum-free DMEM and incubated at 37°C with 2.5 μM D/D solubilizer for indicated times in the same media supplemented with 50 μg/ml TPCK-treated trypsin added either apically or basolaterally. At the end of the incubation, cells were rinsed twice with ice-chilled HBSS-CaMg supplemented with 1 mM phenylmethylsulfonyl fluoride. Cells were lysed in sample buffer containing 5% β-mercaptoethanol and heated at 95°C for 10 min followed by a 45 s sonication at an output power set of 25–30 (Branson sonifier 250) before gel electrophoresis as described below. Western blots were developed with mouse anti-GFP to identify the intact 170 kDa, and cleaved, 130 kDa forms of HA-mMeg-GFP (for additional details, see Supplemental Figure 3C).
For recycling assays, FM4-HA-mMeg-GFP–expressing cultured MDCK cells were first incubated in serum-free DMEM at 37°C for 120 min in the presence of 2.5 μM D/D solubilizer in DMEM followed by an additional 30 min incubation with 100 μM cycloheximide. This procedure allows HA-mMeg-GFP to reach its steady-state distribution at endosomal compartments and cell surface. Recycling was then allowed to proceed by incubating cells at 37°C in serum-free DMEM in the presence of 50 μg/ml TPCK-treated trypsin added either apically or basolaterally for the indicated times. At the end of this incubation, cells were processed as described in biosynthetic delivery assays.
Biosynthetic delivery and recycling values derived from Western blot quantification of cleaved HA-mMeg-GFP (130 kDa) at different intervals were expressed as percentages of total, apical plus basolateral protein levels, determined at the latest time point, that is, 60 min for biosynthetic assays or 45 min for recycling assays.
Quantitative real-time PCR
qRT-PCR was carried out in a StepOnePlus Real-Time PCR system (Applied Biosystems). A 20 μl reaction contained 10 μl of Power SYBR Green RT-PCR premixed with 0.16 μl RT Enzyme (Applied Biosystems), 450 ng total RNA (RNeasy Mini Kit; Qiagen), and 200 nM each forward and reverse primers. Reactions were run in triplicate following the manufacturer’s standard protocol. Primer pair specificity for each target was validated through melting curves run in parallel with samples.
Gel electrophoresis and Western blot
Sample aliquots of lysates containing ∼50–125 μg of protein were added to sample buffer supplemented with β-ME and denatured at 95°C for 10 min before electrophoresis on precast 4–12% gradient polyacrylamide gels (NuPAGE; Invitrogen, Carlsbad, CA) run at 125 mV for 2 h. Protein transferred to nitrocellulose membrane (iBlot; Invitrogen, Carlsbad, CA) was incubated at RT for 30 min with Odyssey blocking buffer (LI-COR, Lincoln, NE) and subsequently incubated with targeted primary antibody. Excess antibody was removed in four consecutive washed with Tween-20 buffer (Tris-HCl 30 mM, pH 7.4, NaCl 150 mM, and Tween-20 0.1%). Bound antibody was detected with corresponding secondary antibody conjugated to either IRDye680 or IRDye800 as indicated and analyzed in an Odyssey Classic scanner (LI-COR, Lincoln, NE).
Statistical analyses
Data derived from images and biochemical quantification were processed for statistical analysis using a two-tailed t test, equal variance. Data derived from quantified Western blots were fitted to a general linear model, and a simultaneous multiple comparison procedure was conducted in R version 3.5.1.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grants no. GM-34108 and no. EY-08538 to E.R.-B. and by grants from the Research to Prevent Blindness Foundation and from the Dyson Foundation. We acknowledge the contribution of the Visual Function Core to our imaging experiments.
Abbreviations used:
- AP
apical
- AP-1
adaptor protein 1
- ARE
apical recycling endosome
- Bl
basolateral
- CRE
common recycling endosome
- GFP
green fluorescent protein
- MDCK
Madin–Darby canine kidney
- SBAS
surface biotin avidin shift
- TGN
trans-Golgi network.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-12-0811) on May 15, 2019.
REFERENCES
- Aminoff M, Carter JE, Chadwick RB, Johnson C, Grasbeck R, Abdelaal MA, Broch H, Jenner LB, Verroust PJ, Moestrup SK, et al. (1999). Mutations in CUBN, encoding the intrinsic factor-vitamin B12 receptor, cubilin, cause hereditary megaloblastic anaemia 1. Nat Genet , 309–313. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS. (2014). Adaptor proteins involved in polarized sorting. J Cell Biol , 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM. (2003). Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem , 395–447. [DOI] [PubMed] [Google Scholar]
- Caceres P, Gravotta D, Zager PJ, Dephoure N, Rodriguez-Boulan E. (2019). Quantitative surface proteomics of MDCK cells identify novel roles of clathrin adaptor AP-1 in polarized sorting of surface proteins. Proc Natl Acad Sci USA , 11796–11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal-Gonzalez JM, Gravotta D, Mattera R, Diaz F, Perez Bay A, Roman AC, Schreiner RP, Thuenauer R, Bonifacino JS, Rodriguez-Boulan E. (2012). Basolateral sorting of the coxsackie and adenovirus receptor through interaction of a canonical YXXΦ motif with the clathrin adaptors AP-1A and AP-1B. Proc Natl Acad Sci USA , 3820–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon GA, Burriat-Couleru P, Abegg D, Criado Santos N, Watanabe R. (2018). Clathrin and AP1 are required for apical sorting of glycosyl phosphatidyl inositol-anchored proteins in biosynthetic and recycling routes in Madin-Darby canine kidney cells. Traffic , 215–228. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Lin YY, Yu CY, Chen JY, Shen KF, Lin WL, Liao HK, Chen YJ, Liu CH, Pang VF, Jou TS. (2005). Molecular identification of canine podocalyxin-like protein 1 as a renal tubulogenic regulator. J Am Soc Nephrol , 1612–1622. [DOI] [PubMed] [Google Scholar]
- Christensen EI, Birn H. (2002). Megalin and cubilin: multifunctional endocytic receptors. Nat Rev Mol Cell Biol , 256–266. [DOI] [PubMed] [Google Scholar]
- Christensen EI, Birn H. (2013). Proteinuria: tubular handling of albumin-degradation or salvation? Nat Rev Nephrol , 700–702. [DOI] [PubMed] [Google Scholar]
- Christensen EI, Birn H, Storm T, Weyer K, Nielsen R. (2012). Endocytic receptors in the renal proximal tubule. Physiology (Bethesda) , 223–236. [DOI] [PubMed] [Google Scholar]
- Deborde S, Perret E, Gravotta D, Deora A, Salvarezza S, Schreiner R, Rodriguez-Boulan E. (2008). Clathrin is a key regulator of basolateral polarity. Nature , 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente-Ortega E, Gravotta D, Perez Bay A, Benedicto I, Carvajal-Gonzalez JM, Lehmann GL, Lagos CF, Rodriguez-Boulan E. (2015). Basolateral sorting of chloride channel 2 is mediated by interactions between a dileucine motif and the clathrin adaptor AP-1. Mol Biol Cell , 1728–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Densupsoontorn N, Sanpakit K, Vijarnsorn C, Pattaragarn A, Kangwanpornsiri C, Jatutipsompol C, Tirapongporn H, Jirapinyo P, Shah NP, Sturm AC, Tanner SM. (2012). Imerslund-Grasbeck syndrome: new mutation in amnionless. Pediatr Int , e19–e21. [DOI] [PubMed] [Google Scholar]
- Folsch H, Mattila PE, Weisz OA. (2009). Taking the scenic route: biosynthetic traffic to the plasma membrane in polarized epithelial cells. Traffic , 972–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsch H, Ohno H, Bonifacino JS, Mellman I. (1999). A novel clathrin adaptor complex mediates basolateral targeting in polarized epithelial cells. Cell , 189–198. [DOI] [PubMed] [Google Scholar]
- Fullekrug J, Shevchenko A, Shevchenko A, Simons K. (2006). Identification of glycosylated marker proteins of epithelial polarity in MDCK cells by homology driven proteomics. BMC Biochem , 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravotta D, Carvajal-Gonzalez JM, Mattera R, Deborde S, Banfelder JR, Bonifacino JS, Rodriguez-Boulan E. (2012). The clathrin adaptor AP-1A mediates basolateral polarity. Dev Cell , 811–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravotta D, Deora A, Perret E, Oyanadel C, Soza A, Schreiner R, Gonzalez A, Rodriguez-Boulan E. (2007). AP1B sorts basolateral proteins in recycling and biosynthetic routes of MDCK cells. Proc Natl Acad Sci USA , 1564–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas M, de Zeeuw D, van Zanten A, Meijer DK. (1993). Quantification of renal low-molecular-weight protein handling in the intact rat. Kidney Int , 949–954. [DOI] [PubMed] [Google Scholar]
- Hinrichsen L, Harborth J, Andrees L, Weber K, Ungewickell EJ. (2003). Effect of clathrin heavy chain- and α-adaptin-specific small inhibitory RNAs on endocytic accessory proteins and receptor trafficking in HeLa cells. J Biol Chem , 45160–45170. [DOI] [PubMed] [Google Scholar]
- Kantarci S, Al-Gazali L, Hill RS, Donnai D, Black GC, Bieth E, Chassaing N, Lacombe D, Devriendt K, Teebi A, et al. (2007). Mutations in LRP2, which encodes the multiligand receptor megalin, cause Donnai-Barrow and facio-oculo-acoustico-renal syndromes. Nat Genet , 957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerjaschki D, Farquhar MG. (1983). Immunocytochemical localization of the Heymann nephritis antigen (GP330) in glomerular epithelial cells of normal Lewis rats. J Exp Med , 667–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisanti MP, Caras IW, Davitz MA, Rodriguez-Boulan E. (1989). A glycophospholipid membrane anchor acts as an apical targeting signal in polarized epithelial cells. J Cell Biol , 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maack T, Johnson V, Kau ST, Figueiredo J, Sigulem D. (1979). Renal filtration, transport, and metabolism of low-molecular-weight proteins: a review. Kidney Int , 251–270. [DOI] [PubMed] [Google Scholar]
- Marino M, Zheng G, Chiovato L, Pinchera A, Brown D, Andrews D, McCluskey RT. (2000). Role of megalin (gp330) in transcytosis of thyroglobulin by thyroid cells. A novel function in the control of thyroid hormone release. J Biol Chem , 7125–7137. [DOI] [PubMed] [Google Scholar]
- Marzolo MP, Yuseff MI, Retamal C, Donoso M, Ezquer F, Farfan P, Li Y, Bu G. (2003). Differential distribution of low-density lipoprotein-receptor-related protein (LRP) and megalin in polarized epithelial cells is determined by their cytoplasmic domains. Traffic , 273–288. [DOI] [PubMed] [Google Scholar]
- Mattera R, Boehm M, Chaudhuri R, Prabhu Y, Bonifacino JS. (2011). Conservation and diversification of dileucine signal recognition by adaptor protein (AP) complex variants. J Biol Chem , 2022–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy RA, Argraves WS. (2003). Megalin and the neurodevelopmental biology of sonic hedgehog and retinol. J Cell Sci , 955–960. [DOI] [PubMed] [Google Scholar]
- Nykjaer A, Dragun D, Walther D, Vorum H, Jacobsen C, Herz J, Melsen F, Christensen EI, Willnow TE. (1999). An endocytic pathway essential for renal uptake and activation of the steroid 25-(OH) vitamin D3. Cell , 507–515. [DOI] [PubMed] [Google Scholar]
- Ohno H, Tomemori T, Nakatsu F, Okazaki Y, Aguilar RC, Foelsch H, Mellman I, Saito T, Shirasawa T, Bonifacino JS. (1999). Mu1B, a novel adaptor medium chain expressed in polarized epithelial cells. FEBS Lett , 215–220. [DOI] [PubMed] [Google Scholar]
- Perez Bay AE, Schreiner R, Benedicto I, Paz Marzolo M, Banfelder J, Weinstein AM, Rodriguez-Boulan EJ. (2016). The fast-recycling receptor Megalin defines the apical recycling pathway of epithelial cells. Nat Commun , 11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E, Macara IG. (2014). Organization and execution of the epithelial polarity programme. Nat Rev Mol Cell Biol , 225–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo LM, Sandoval RM, McKee M, Osicka TM, Collins AB, Brown D, Molitoris BA, Comper WD. (2007). The normal kidney filters nephrotic levels of albumin retrieved by proximal tubule cells: retrieval is disrupted in nephrotic states. Kidney Int , 504–513. [DOI] [PubMed] [Google Scholar]
- Sargiacomo M, Lisanti M, Graeve L, Le Bivic A, Rodriguez-Boulan E. (1989). Integral and peripheral protein composition of the apical and basolateral membrane domains in MDCK cells. J Membr Biol , 277–286. [DOI] [PubMed] [Google Scholar]
- Schrauwen I, Sommen M, Claes C, Pinner J, Flaherty M, Collins F, Van Camp G. (2013). Broadening the phenotype of LRP2 mutations: a new mutation in LRP2 causes a predominantly ocular phenotype suggestive of Stickler syndrome. Clin Genet , 282–286. [DOI] [PubMed] [Google Scholar]
- Schreiner R, Frindt G, Diaz F, Carvajal-Gonzalez JM, Perez Bay AE, Palmer LG, Marshansky V, Brown D, Philp NJ, Rodriguez-Boulan E. (2010). The absence of a clathrin adapter confers unique polarity essential to proximal tubule function. Kidney Int , 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafaq-Zadah M, Brocard L, Solari F, Michaux G. (2012). AP-1 is required for the maintenance of apico-basal polarity in the C. elegans intestine. Development , 2061–2070. [DOI] [PubMed] [Google Scholar]
- Takeda T, Yamazaki H, Farquhar MG. (2003). Identification of an apical sorting determinant in the cytoplasmic tail of megalin. Am J Physiol Cell Physiol , C1105–C1113. [DOI] [PubMed] [Google Scholar]
- Thuenauer R, Hsu YC, Carvajal-Gonzalez JM, Deborde S, Chuang JZ, Romer W, Sonnleitner A, Rodriguez-Boulan E, Sung CH. (2014). Four-dimensional live imaging of apical biosynthetic trafficking reveals a post-Golgi sorting role of apical endosomal intermediates. Proc Natl Acad Sci USA , 4127–4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub LM. (2009). Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat Rev Mol Cell Biol , 583–596. [DOI] [PubMed] [Google Scholar]
- Traub LM, Bonifacino JS. (2013). Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harb Perspect Biol , a016790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willnow TE, Hilpert J, Armstrong SA, Rohlmann A, Hammer RE, Burns DK, Herz J. (1996). Defective forebrain development in mice lacking gp330/megalin. Proc Natl Acad Sci USA , 8460–8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CY, Chen JY, Lin YY, Shen KF, Lin WL, Chien CL, ter Beest MB, Jou TS. (2007). A bipartite signal regulates the faithful delivery of apical domain marker podocalyxin/Gp135. Mol Biol Cell , 1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuseff MI, Farfan P, Bu G, Marzolo MP. (2007). A cytoplasmic PPPSP motif determines megalin’s phosphorylation and regulates receptor’s recycling and surface expression. Traffic , 1215–1230. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kim A, Abraham N, Khan LA, Hall DH, Fleming JT, Gobel V. (2012). Clathrin and AP-1 regulate apical polarity and lumen formation during C. elegans tubulogenesis. Development , 2071–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurzolo C, Simons K. (2016). Glycosylphosphatidylinositol-anchored proteins: membrane organization and transport. Biochim Biophys Acta , 632–639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.