Abstract
Numerous studies have highlighted the self-centering activities of individual microtubule (MT) arrays in animal cells, but relatively few works address the behavior of multiple arrays that coexist in a common cytoplasm. In multinucleated Dictyostelium discoideum cells, each centrosome organizes a radial MT network, and these networks remain separate from one another. This feature offers an opportunity to reveal the mechanism(s) responsible for the positioning of multiple centrosomes. Using a laser microbeam to eliminate one of the two centrosomes in binucleate cells, we show that the unaltered array is rapidly repositioned at the cell center. This result demonstrates that each MT array is constantly subject to centering forces and infers a mechanism to balance the positions of multiple arrays. Our results address the limited actions of three kinesins and a cross-linking MAP that are known to have effects in maintaining MT organization and suggest a simple means used to keep the arrays separated.
INTRODUCTION
Interphase microtubule (MT) arrays in animal cells are typically arranged in a radial manner, with minus ends clustered near the centrosome and plus ends extending toward the cell periphery. This arrangement is simple in principle and can be recapitulated in vitro with only a nucleating center and purified tubulin (Holy et al., 1997). MT arrangements in cells are more complicated and involve prominent contributions from MT tip dynamics and dynein motors (Holy et al., 1997; Tran et al., 2001; Burakov et al., 2003; Malikov et al., 2005; Kimura and Kimura, 2011; Laan et al., 2012; Zhao et al., 2012; Koke et al., 2014; Gibeaux et al., 2017). Other motors, nonmotor cross-linkers, and even frictional components in the cytoplasm also play roles in MT arrangements (Zhu et al., 2010; De Simone and Gönczy, 2017; Howard and Garzon-Coral, 2017; Tanimoto et al., 2018). While a uniform distribution and balance of MT forces in cells likely facilitates the centering of a radial MT array, asymmetric changes to these actions (targeted alterations in MT dynamics, cross-linking by MAPs, regulation of motor activity, cell shape change, etc.) can alter this default state and move centrosomes toward the cell periphery, drive centrosome migration, or otherwise alter MT arrangement (Grill and Hyman, 2005; Subramanian and Kapoor, 2012; Tang and Marshall, 2012; Dogterom and Surrey, 2013; McNally, 2013; Letort et al., 2016; Tanimoto et al., 2016; Chaaban and Brouhard, 2017). In these ways, a cell can exert exquisite spatial control over its MT network.
When multiple MT arrays are present, the self-centering activity of each centrosome should result in convergence of the MT networks. Indeed, this is what happens in some types of cells (Brinkley et al., 1981; Quintyne et al., 2005). However, in other cases, the arrays remain separate (Telley et al., 2012; Anderson et al., 2013; Nguyen et al., 2014; Tikhonenko et al., 2016). At least two different ideas have been described that contribute to the separation of MT arrays. The first, rooted in the mitotic literature, is that MT arrays of opposite polarity actively engage one another and form a barrier that limits interdigitation (e.g., Nguyen et al., 2014, 2018). The second idea is more general; that simple pushing from MTs or motor proteins can act to set distances (e.g., Manhart et al., 2018). These are not mutually exclusive contributions and do not rule out other contributing factors. Understanding the forces acting upon the MT array is important as it lends insight into the fundamental organization of the cytoplasm. Our efforts seek to address these force balance mechanisms in a dynamic model system.
Multinucleated cells frequently occur in Dictyostelium discoideum cell culture (De Lozanne and Spudich, 1987; Knecht and Loomis, 1987; Neujahr et al., 1998); each nucleus is tightly associated with a centrosome and its corresponding MT array (Kuriyama et al., 1982; Omura and Fukui, 1985; Tikhonenko et al., 2013). A striking feature of these cells is that the multiple MT arrays generally remain separate from one another, organizing mutually exclusive territories in the common cytoplasm (Koonce and Khodjakov, 2002; Samereier et al. 2010; Tikhonenko et al., 2016; Koonce and Tikhonenko, 2018). We have previously shown that the radial interphase MT arrangement is particularly sensitive to kinesin, dynein, and MT cross-linker perturbations, suggesting that motor proteins play an integral part to the D. discoideum array dynamics (Koonce et al., 1999; Nag et al., 2008; Tikhonenko et al., 2013, 2016). In the current study, we use a laser microbeam to destroy one of the two MT arrays in binucleate cells and ask what effect this has on the remaining, unaltered array. After centrosome ablation, the unaltered array begins almost immediately to move toward the cell center, migrating into the territory previously occupied by the other MT network and establishing a central position in the cell. Further, we demonstrate that the centering forces persist in cells that lack three kinesins and a MAP that have previously been shown to impact MT organization in D. discoideum (DdKif8, 9, 10) (Nag et al., 2008; Tikhonenko et al., 2013); (DdAse1A) (Tikhonenko et al., 2016). Our results demonstrate a persistent centering force acting on MT arrays and also infer a balancing mechanism that maintains multiple MT array separation in a common cytoplasm.
RESULTS
MT organization in binucleate cells
D. discoideum centrosomes nucleate significantly fewer MTs (30–70 per centrosome; Moens 1976; Kuriyama et al., 1982) than many of the more widely studied metazoan organisms; the MT-minus ends remain strictly anchored into the centrosome corona (e.g., Brito et al., 2005), and MTs are far less dynamic. While there is evidence of growth/shrinkage at the MT plus tips (Brito et al., 2005; Samereier et al., 2011), direct observation and FRAP analyses (Samereier et al. 2010) indicate that the bulk of the MT polymer is quite stable. In an attempt to visualize potential interactions between MTs from different centrosomes, we tracked MTs in binucleate wild-type cells (Figure 1; Supplemental Figure S1). MT arrangements in the region between centrosomes are complex; however, these figures show there is neither a dedicated zone where anti-parallel MTs join to form a barrier nor extensive regions of MT interdigitation. Previous labeling does not support an actin-rich barrier that separates the multiple MT arrays (Tikhonenko et al., 2016)
FIGURE 1:
Binucleate MT arrays manage discrete territories. (A) The distance is plotted between the two centrosomes in three representative wild-type cells as a function of time. Note the spacing between centrosomes varies, indicating that they are not rigidly connected. The left column in panel B shows individual frames of each cell, taken at or very near the onset of recording. The right column shows 2D maximum intensity projections of the 121 image stacks for each cell, showing the overall range of centrosome movements during the 10 min imaging period. Panel C presents a plot of the average distance between the centrosomes for eight WT cells. Error bars are SD. Panel D shows one example of MT tracking in a fixed binucleate cell using the FilamentTracer module in Imaris. MTs are color coded either magenta or green depending on their centrosomal origin. Additional examples can be viewed in Supplemental Figure S1. Scale bars in B and D = 5 μm.
We imaged MT arrays in binucleate cells constitutively expressing α-tubulin- green fluorescent protein (GFP) (Neujahr et al., 1998) (Figure 1; Supplemental Movie 1). As all minus ends in D. discoideum converge on the centrosome, the position of this organelle is accurately defined. Our initial sequences were recorded at 5-s intervals and extended for ∼2–300 frames (∼15–20 min), in part to assess the potential for long-term photodamage and in part to establish a basal timeline for activity. Since much of the visible rearrangements post laser occurred within the first 2–3 min, we limited the bulk of our recordings to 121 frames (10 min). In control binucleate cells, each centrosome rocks back and forth in a dynamic manner, typically within a 1- to 3-μm radius. As previously noted, individual MTs show significant lateral movement and bending, with occasional arcing along the cell cortex (Koonce and Khodjakov, 2002; Brito et al., 2005). Similar rocking motions also occur in mononucleated cells. Measurements of the distance between the two centrosomes suggest that they move independent of one another and maintain some distance apart (average separation is 6 μm ± 1.2, n = 8). The two-dimensional (2D) maximum intensity projections of the image stacks also show the degree of separation over time (Figure 1).
Movie S1.
Non-irradiated control binucleate WT cell demonstrating dynamics of centrosomes and interphase MTs. Frames recorded at 5-s intervals.
Centering of the undamaged MT array upon laser ablation
To ablate the centrosome, a series of brief laser pulses were targeted at the focal point of one MT array in binucleate cells, resulting in the disappearance of the GFP fluorescence focus and followed shortly by cytoplasmic disassembly of the associated MTs (see also Brito et al., 2005). The MT array does not regenerate, indicating that the centrosome is functionally dead. Control irradiations in the cytoplasm near the centrosome had no effect on the MT arrays, and ablation of one centrosome did not noticeably affect the integrity of the second centrosome or its MT array. In some cells, it was obvious that we only partially damaged the centrosome; in these cases, most MTs disappeared in the short term, but remnants of a partial MT array persisted or were restored over time. We analyzed only cells where the irradiated array disintegrated completely.
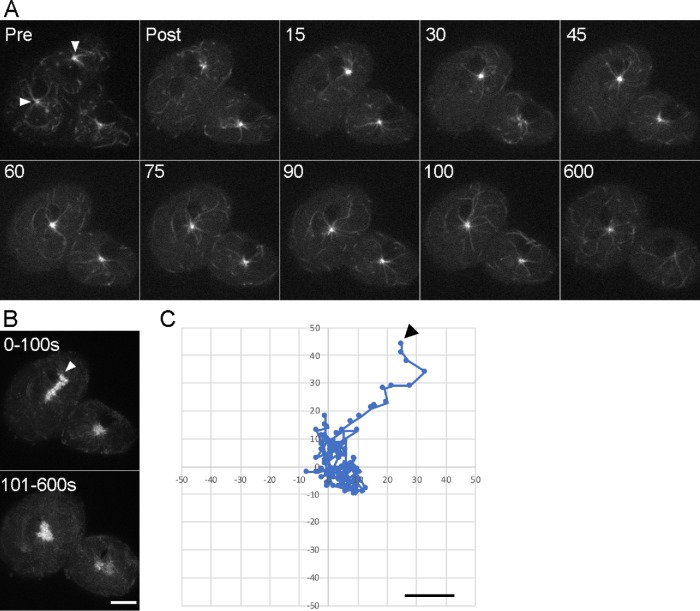
Within seconds after ablation, the undamaged centrosome began to move toward the cell center (Figure 2; Supplemental Movies 2–4). Individual MTs extended into the area previously occupied by the now disassembled MT array. These MTs were not due to new growth from the centrosome but arose from repositioning existing polymers. The 2D maximum intensity projections of the image stacks reveal an initial phase of centrosome translocation toward the cell center, generally within the first 100 s (20 image frames), followed by fluctuation around the center until the end of recording (10 min, 121 image frames). Although the average linear distance to the cell center was 3.6 μm, centrosomes moved in an irregular path (Figure 2), covering an average 7.4 μm distance at a rate of 4.6 μm/min during the initial phase. The net migration rate of the centrosome to the cell center is 1.8 μm/min. These results demonstrate that interphase MT arrays are constantly subjected to centering forces and suggest that multiple MT arrays in a common cytoplasm present an opposing influence in this model system.
FIGURE 2:
Removal of one MT array results in centering of the undamaged array. (A) Individual frames of a binucleate cell pre and post laser ablation and at intervals through the 10-min imaging period. Numbers represent seconds post ablation. The two centrosomes in the targeted cell are marked with arrowheads in the prepanel. A second, mononucleated cell is also visible in this panel. (B) Maximum intensity projections of the first 100 s post ablation (top) and of the subsequent 500 s (bottom). In the top frame, the starting point of the centrosome is indicated with the arrowhead. Note the steady initial migration of the centrosome toward the cell center in the top frame, and then movements around the center in the lower frame (scale bar = 5 μm). (C) The centrosome track at 5-s intervals. The arrowhead marks the initial time point. Axes are in pixels. Scale bar = 2 um.
Movie S2.
Examples of centrosome/MT array movement following ablation of one of two centrosomes in binucleate cells. Frames recorded at 5-s intervals.
Movie S3.
Examples of centrosome/MT array movement following ablation of one of two centrosomes in binucleate cells. Frames recorded at 5-s intervals.
Movie S4.
Examples of centrosome/MT array movement following ablation of one of two centrosomes in binucleate cells. Frames recorded at 5-s intervals.
Role of motors and MAPs in MT array centering
Dynein and MT tip dynamics are predicted to be the primary drivers of centrosome positioning and relocation. Prominent in D. discoideum are transient MT-cortical engagements that pull on the centrosome in a manner consistent with dynein-driven events (Supplemental Figure S2; Koonce et al., 1999; Hestermann and Gräf, 2004). Moreover, disruptions to dynein result in large-scale circulation of the MT array (Koonce et al., 1999; Ma et al., 1999; Rehberg et al., 2005), indicating significant forces and balance at play to maintain the radial organization.
Directly testing the role of dynein is problematic. Our unpublished observations indicate that neither ciliobrevin (Firestone et al., 2012) nor dynarrestin (Höing et al., 2018) have any impact on interphase or mitotic MT distributions in D. discoideum. Previous attempts (e.g., Brito et al., 2005) to laser-ablate motile centrosomes in dominate-negative dynein perturbations have proven challenging to completely eliminate MT nucleation activity. However, in addition to dynein, there are other motors and nonmotor MAPs in this model that impact interphase MT organization that we can test (Hestermann and Gräf, 2004; Nag et al., 2008; Tikhonenko et al., 2016). These include DdKif8 (Kinesin-4), DdKif10 (Kinesin-8), and DdAse1A (MAP65/Ase1/PRC1). Disruption of either kinesin abrogates the dynein-mediated circulation of interphase MTs; deletion of DdAse1A affects the spacing of multiple centrosomes in a common cytoplasm. To examine their potential role in centering activities, we examined centrosome movements in these null cells.
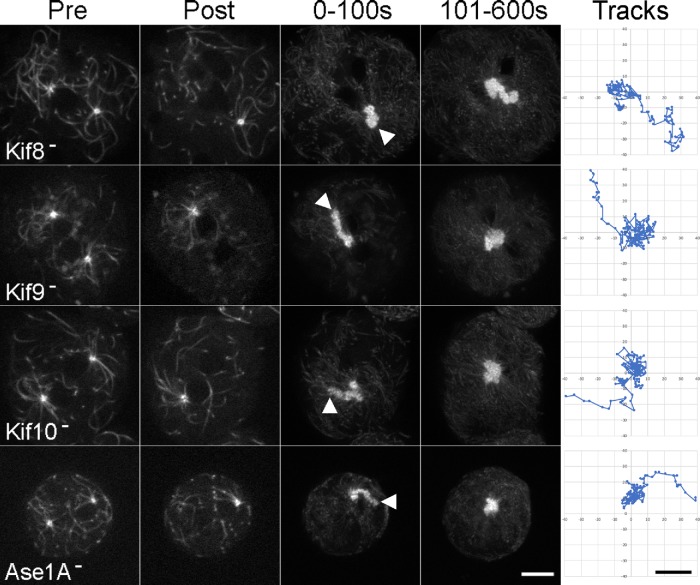
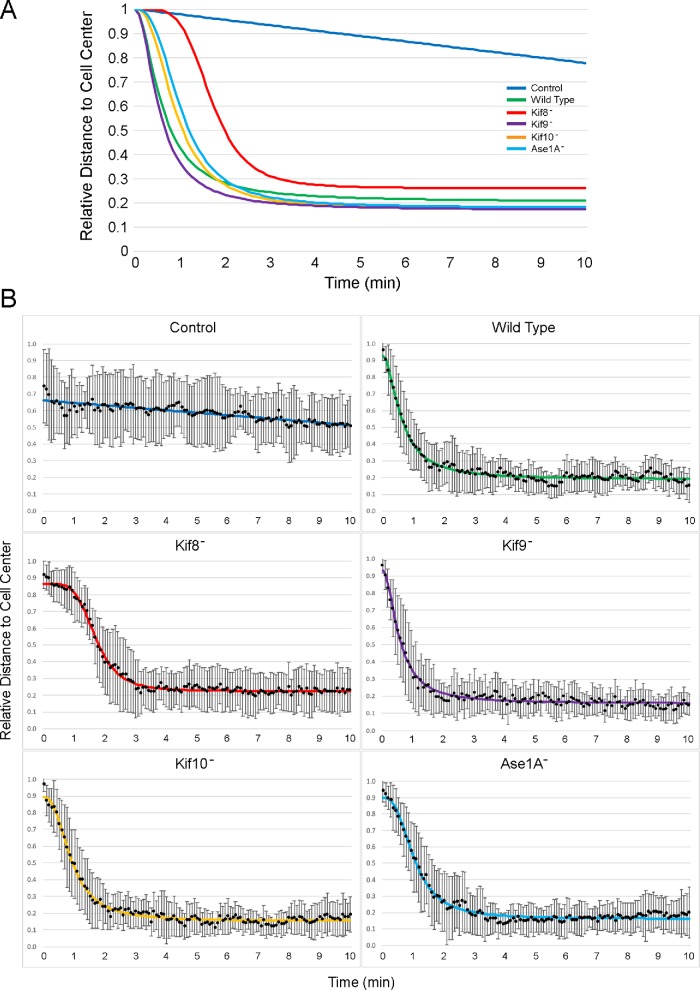
During interphase, binucleate centrosomes in each of the tested null cell lines show oscillatory dynamics that are similar to movement in wild-type cells (Supplemental Figure S3). Upon laser ablation, the undamaged centrosomes underwent a brief, ∼2-min period of migration to the cell center, followed by minor back-and-forth movements until the end of the filming period (Figure 3; Supplemental Figure S4). Detailed analyses of the movements reveal only one prominent difference: the centrosomes in the DdKif8 null cells show a brief lag period before their migration to the cell center (Figure 4). Once begun though, the directed movement in all three null cell lines occur at similar rates and persistence as in wild-type cells (Figure 5).
FIGURE 3:
Centrosome centering in mutant backgrounds. Four representative examples are shown, one each of DdKif8, Kif9, Kif10, and Ase1A null cells. Pre and post columns represent single image frames before and after ablation; the adjacent two columns show maximum intensity projections of the first 100 s post ablation (left) and of the subsequent 500 s (right). The arrowheads mark the initial position of the centrosome, before its directed movement to the center. Scale bar = 5 μm. The rightmost column (Tracks) shows the frame-by-frame movement of the undamaged centrosome. Dots are separated by 5-s intervals. Scale bar = 2 μm.
FIGURE 4:
Summary of centrosome movements. (A) Fitted plots of post ablation centrosome distance to cell center versus time for wild-type, DdKif8, Kif9, Kif10, and Ase1A null cells, and nonirradiated wild-type controls. Each curve represents the summation of 15 cells fitted to a Rodbard function using FIJI and normalized to a 1.0 starting point. (B) The raw averages, including error bars (SD) for each cell line.
FIGURE 5:
Centrosome movement rates. Box plots for each cell type shown in A, the instantaneous rate of centrosome movement (frame by frame distance covered during the 100-s movement period toward the cell center), and in B, the average rate (straight line distance to cell center over same time frame) of centrosome centering. In A, the WT instantaneous rate of movement averages 4.6 μm/min ± 1.2 (SD) (n = 16); Kif8 = 4.2 ± 1.1 (n = 18); Kif9 = 3.7 ± 1.3 (n = 16); Kif10 = 4.2 ± 0.9 (n = 15); Ase1A = 4.5 ± 1.0 (n = 15). In B, the WT centering rate averages 1.8 μm/min ± 0.5 (SD); Kif8 = 1.1 ± 0.3; Kif9 = 1.9 ± 0.6; Kif10 = 1.6 ± 0.4; Ase1A = 1.6 ± 0.5 (n = 15 in all cases).
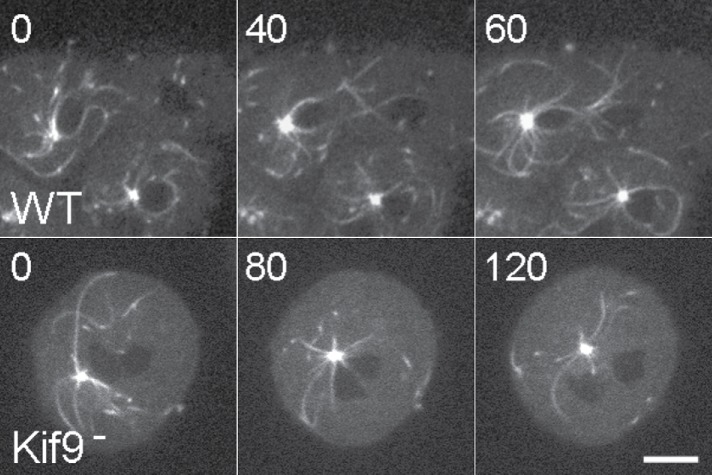
We also examined an additional kinesin knockout, DdKif9 (Figure 3). This Kin-I kinesin links centrosomes to nuclei in D. discoideum (Tikhonenko et al., 2013) and thus may indicate nuclear contributions to MT-centering activity. In some wild-type binucleate cells, MTs from the surviving array appear to establish contacts with the nucleus that lost its own MT array as a result of laser irradiation (Figure 6). These contacts may guide the movement of the nucleus toward the surviving centrosome (or vice versa). In irradiated DdKif9 null cells, centrosome-nuclear interactions are transient, but the rates and behavior of centrosome movement match those of wild-type cells. While these results are consistent with DdKif9 function as a nuclear bound motor that maintains centrosome proximity, nuclear-centrosome coupling does not appear to play a significant role in centrosome positioning.
FIGURE 6:
Nuclear engagements. Two sequences of binucleate cells, post laser ablation. The top row shows a wild-type cell where MTs from the surviving centrosome appear to engage the other nucleus for movement closer together. The bottom row is from a DdKif9 null cell where MT-nuclear engagements appear transient and not force productive. Time is in seconds; scale bar = 5 μm.
Our results suggest that none of the tested kinesins, nor the Ase1 MAP, play a substantial role in the post laser centrosome repositioning, or its back-and-forth movements about the cell center and further support a dominate role for dynein-mediated pulling motility in driving the centrosome to the cell center.
DdKif8 (Kinesin-4) motor activity
Since disruption of DdKif8 abrogates the comet-like motility of the MT array that occurs through dominate-negative dynein perturbation and shows a delay in centrosome relocation following laser ablation, we examined DdKif8 function in more detail. In mononucleates, DdKif8 null cells display a less radial, more disorganized MT array (Figure 7). In these cells the centrosome is frequently not as centered as in wild type and either there are more MTs or the MTs are longer and loop away from the cortex, adding to the array’s complexity. The DdKif8 kinesin is a member of the Kinesin-4 family (Kollmar and Glöckner, 2003); the 1874–amino acid residue sequence (209 kDa) contains an amino-terminal motor domain, a carboxy terminus with seven WD-40 repeats, and overall is most similar to the Kif21A/B subfamily (Marszalek et al., 1999).
FIGURE 7:
DdKif8 influences MT architecture, localizes to the cytoplasm and is a robust MT motor. (A) Two representative examples of interphase MT organization in mononucleate WT and in DdKif8 null cells (MTs in green, nuclei in blue). Panel B quantitates the average distance of the centrosome from the cell center in WT (1.4 μm ± 0.8 SD, n = 98) and Kif8 null (2.7 μm ± 1.2, n = 71). Panel C quantitates the number of MT segments encountered in similar line scans across cells. (WT = 18.0 ± 3.1 (SD), Kif8 null = 24.8 ± 4.9, n = 22 for both strains). (D) GFP-tagged DdKif8 distribution in the cytoplasm, forming punctate patterns during interphase and mitosis. No enrichment of the motor is seen at the MT tips, nor in the spindle midzone as has been reported for some isoforms of the Kinesin-4 family. Panels E and F illustrate DdKif8 MT binding and motility. (E) Three lanes of a Coomassie Blue stained protein gel, containing MW markers, the MT pellet, and supernatant after ATP extraction of MTs incubated with column fractions of DdKif8-GFP protein. The full-length 236 kDa fusion polypeptide is marked with an arrowhead. (F) Four sequential frames of MT gliding activity on a coverslip bound with the DdKif8 motor. Time in seconds. Scale bars in A, D, F = 5 μm.
To address DdKif8 function in greater detail, we expressed a full-length carboxy-terminal GFP-tagged version of the motor in D. discoideum and analyzed both its cytoplasmic distribution and its in vitro motility. DdKif8 protein is distributed throughout the cytoplasm and accumulates in punctate clusters (Figure 7). No enrichment of the motor is observed in the spindle overlap zones that would indicate a mitotic function, nor at the MT plus tips during interphase. When purified, the polypeptide readily cycles on and off MTs in an ATP-dependent manner and powers rapid MT gliding in simple motility assays (1.6 μm/s ± 0.15, n = 92) (Figure 7). These results suggest that DdKif8 may play a role in the rapid organelle movements seen in the cell cytoplasm, but it is not yet obvious how it might contribute to MT length control or action at the MT plus tips.
DISCUSSION
A prominent feature of the D. discoideum cytoskeleton is the spatial separation of multiple MT arrays during interphase in a common cytoplasm. Our laser ablation results directly demonstrate that this separation is actively and robustly maintained. Removal of one centrosome/MT array in a binucleate cell results in the rapid, almost immediate movement of the unaltered array toward the cell center. This result implies that there are constant centering forces acting on the MT arrays and infers the presence of a mechanism to balance this activity to ensure centrosome separation in complex multinuclear arrangements.
The dynamic behavior of MT arrays has received significant attention in the literature. Multiple models have emerged that address array spacing, including creating boundary layers between MTs of opposite polarity and MT pushing against adjacent nuclei. The first mechanism is well rooted in mitotic spindle dynamics where interdigitating MTs of opposite polarity are carefully managed by a combination of MAPs and motors to create the spindle overlap zone. Here, a combination of PRC1 (MAP65/Ase1 family member) and Kif4A (Kinesin-4 family) proteins plays a primary role in organizing MTs of opposite polarity, to regulate the degree of MT overlap, and define the local architecture of this region (Bieling et al., 2010; Subramanian et al., 2013; Wijeratne and Subramanian, 2018). Relevant not just to spindle dynamics, these proteins are also recruited to interaction zones that maintain separation of large MT asters formed in interphase Xenopus oocyte extracts (Nguyen et al., 2014, 2018). The two proteins (and Aurora Kinase B) are enriched in a narrow region of anti-parallel MTs bundles between asters where they are thought to display “anti-parallel pruning” activity to slow MT growth and encourage plus-end catastrophe to maintain aster–aster separation.
We considered this paradigm as a spacing model here; indeed, deletion of the D. discoideum homologue of PRC1 (DdAse1A) results in shrinkage of the distance between interphase centrosomes in binucleate cells and enhances a greater interdigitation of the two MT arrays (Tikhonenko et al., 2016). However, there are notable differences. First there are significantly greater numbers of MTs in the Xenopus system (hundreds vs. dozens) to enhance the likelihood of opposite polarity MT contact. DdAse1A does localize to the nucleus and the spindle overlap zone where it participates in the expected mitotic activity of holding the two spindle halves together, but we are unable to demonstrate an interphase presence of this MAP on MTs, especially on those located between the two centrosomes (Tikhonenko et al., 2016). The D. discoideum Kinesin-4 homologue does not colocalize with DdAse1A at the mitotic spindle, and gene knockouts do not yield a detectable mitotic phenotype. Instead, DdKif8 is more closely related to the Kif21A/B members of the Kinesin-4 family that have been functionally characterized in neuronal cells (Marszalek et al., 1999), in part as organelle transporters and in part to manage MT length (van der Vaart et al., 2013; Muhia et al., 2016; van Riel et al., 2017). MTs appear more numerous or longer in DdKif8 null cells, with many arcing off the cell cortex and leading back into the cell interior. The one distinctive difference in centrosome centering after laser ablation is the short delay in DdKif8 null cells. A simple explanation is that the delay may be due to hysteresis; extra polymer length may increase slack in the system that delays productive force generation. This hysteresis may also reflect the reduced centering observed in mononucleated DdKif 8 null cells (Figure 7). Ddkif8 localization illustrates a cytoplasmic distribution but does not appear enriched at the tips of interphase MTs. The Xenopus extract work also indicates a role for Aurora kinase (Nguyen et al., 2018), with localization to a narrow boundary region very similar to that of the Kif4A distribution. D. discoideum has a single Aurora kinase protein that shows mitotic activities predicted from other systems (Li et al., 2008). However, DdAurora has a diffuse cytoplasmic distribution during interphase, with no discernible enrichment on the MT system. Taken together, our results do not necessarily exclude an anti-parallel pruning model to maintain MT array separation, but do not support a simple application of this idea in our model system.
A second model addresses the idea of centrosome spacing through MT mediated pushing. A recent study describes the positioning of multiple nuclei in muscle cells (Manhart et al., 2018), but the general idea is also related to the polar ejection forces that balance chromosome-to-pole movement during the early phases of cell division (Rieder et al., 1986). The model describes pushing forces either through MT polymerization or by kinesin-generated actions at the MT plus ends that establish nuclear spacing. In this manner, nuclei repel one another, with long range forces decreasing as a function of distance. In D. discoideum, such pushing forces may contribute to close-in actions, but given the observed flexibility of MTs, it is difficult to conceive that they would be sufficiently rigid to support the longer-range centrosome separation seen in cells. Moreover, the nuclei may not play as much of a role here, as in the absence of DdKif9 or in the presence of supernumerary centrosomes, MT arrays can uncouple from nuclei and yet maintain spatial separation (Gräf et al., 2000; Tikhonenko et al., 2016; Koonce and Tikhonenko, 2018).
A third possibility is that MTs normally extend from one centrosome toward another, but these MTs fail to gain sufficient traction to exert pulling forces in this direction. The area surrounding each centrosome may form congested exclusion zones that sterically or dynamically inhibit incoming MT ends. For example, through dynein-mediated transport, the centrosomal region is well known to accumulate organelles and membrane compartments (e.g., Golgi, endosomes) (Mohrs et al., 2000; Schneider et al., 2000; Charette et al., 2006; Marko et al., 2006). In this way, the space surrounding MT asters forms a physical hindrance to MT penetration, as well as one that may also concentrate membranes rich with kinesin motors. If a MT from one centrosome extends toward the second centrosome and engages a kinesin, it could simply be moved back out of the region. This idea is different from the nuclear repulsion described above, as it does not assume a pushing force per se, just that the MT is moved out of the region to a different position and thus is unable to develop a pulling force to move one centrosome toward another. Key here is that MTs in this model organism are frequently observed to undergo bending motions indicative of motor-driven events that push the substrate instead of moving along it (e.g., Supplemental Movie S1; Koonce and Khodjakov, 2002). Several of the nonmitotic kinesins characterized in D. discoideum that could participate in MT bending are exceptionally robust, with rates from 1.6 to 2.0 μm/s (DdKif8 here; McCaffrey and Vale, 1989; Röhlk et al., 2008).
MT exclusion actions, even limited to a narrow wedge in the direction of an additional centrosome, may be sufficient to create an imbalance of forces that favor dynein-mediated cortical pulling on the opposite side of the centrosome. Laser ablation eliminates the minus-end foci of MTs and could cause the organelle-rich area to disperse, similar to what happens when cells disassemble interphase MTs and enter mitosis (e.g., Weiner et al., 1993; Schneider et al., 2000). MTs would then be able to laterally arc into this region and accumulate cortical or length-dependent pulling forces that adjust centrosome position. This idea suggests dynein as the primary driving force for centrosome centering. Additional centrosomes do not actively drive one another apart, but derive their spacing by merely inhibiting formation of dynein-mediated traction forces that would otherwise pull them together.
MATERIALS AND METHODS
Live-cell imaging and laser microsurgery
D. discoideum AX-2 cell lines (wild-type and null strains) expressing GFP-α-tubulin (Neujahr et al., 1998) were maintained on 10-cm plastic Petri dishes in HL-5 medium (Franke and Kessin, 1977) containing Pen Strep antibiotic (Sigma Chemical, P4333) and G418 (15 μg/ml; Sigma Chemical, A1720). All null strains used in this study have been previously described (Nag et al., 2008; Tikhonenko et al., 2013, 2016) and contained additional Blasticidin S selection (10 μg/ml; Sigma Chemical, 15205). For live-cell imaging, 100 μl of near confluent culture was scraped off the bottom of the Petri dish with a pipette tip, added onto a glass coverslip, and allowed to adhere for 1 h in 17 mM K/Na phosphate buffer, pH 6.0. Cells were then overlaid and flattened with small agarose sheets, as described in Yumura et al. (1984). The coverslips were assembled into Rose Chambers containing a small piece of moistened Kimwipe to maintain humidity (Brito et al., 2005).
Live-cell imaging was performed on a TE-2000E2 microscope (Nikon Instruments) in spinning-disk confocal mode (Yokogawa GSU-10). Images of single focal planes were captured with a PlanApo 100 × 1.4 NA lens on a Cascade 512B EM CCD camera (Photometrics) at 5-s intervals. Light exposure was kept to a bare minimum by hardwire synchronization of all light sources with the camera. All hardware was controlled by IPLab software (Scanalytics).
Laser microsurgery was performed with a brief (300–500 ms) series of 0.56-ns, 532-nm pulses at 250 Hz. The pulses were generated by a Q-switched Nd:YAG laser (Teem Photonics), and the beam was expanded, collimated, and steered to the back aperture of a 100 × 1.4 NA PlanApo objective lens (Nikon) as described in Magidson et al. (2007) (see also Berns et al., 1981; Khodjakov et al., 2004). For ablation, the shutter was manually opened for less than 1-s periods and repeated 2–5× until the GFP signal marking the centrosome position disappeared. Similar control exposures into the cytoplasm were also performed. Recordings of cells that showed obvious signs of photodamage (blebbing, cessation of movement in the cytoplasm) or cell death were not included in the data analysis.
Immunostaining and fixed-cell imaging
For fixed-cell imaging, coverslips containing cells were fixed/permeabilized in 50% PHEM buffer (30 mM PIPES, 12.5 mM HEPES, pH 6.9, 4 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid [EGTA], 1 mM MgCl2) containing 3.7% formaldehyde, 0.05% glutaraldehyde, 0.1% Triton X-100, and processed as described in Koonce and McIntosh (1990). In some cases, cells were preflattened as above under agarose; in others, cells were flattened by centrifugation onto coverslips (400 × g, 5 min) in phosphate buffer before fixation. Images were collected using a DeltaVision microscope workstation and softWoRx imaging package. The α-tubulin antibody was generated as described in Piperno and Fuller (1985).
DdKif8 expression and biochemistry
The full-length coding sequence for DdKif8 (5622 base pairs, 1874 amino acids) was obtained from the genome resources at DictyBase (DDB_G0284471) (Basu et al., 2013). The cDNA sequence, minus the stop codon, was commercially synthesized (GenScript) and subcloned into an expression vector containing the DdActin15 promotor, an amino terminal 8x His tag, and a G418 selectable marker. We added a 732–base pair GFP (S65T) cassette into an engineered Xho1 site at the carboxy terminus of DdKif8 and then verified all reading frames by sequencing. The final construct was introduced into DdAX-2 cells via the Ca2+PO4 precipitation method (Egelhoff et al., 1991); clones were selected for growth in 15 μg/ml G418 and screened visually for GFP fluorescence. For Kif8 protein purification, 800 ml cell cultures were grown to mid–log phase and centrifuged to pellet cells (1000 × g, 5 min). Ten grams of wet cell mass was suspended in 2× vol PMG buffer (100 mM PIPES, pH 7.2, 4 mM MgCl2, 0.9M glycerol) plus the protease inhibitors leupeptin (10 μg/ml), soybean trypsin inhibitor (0.1 mg/ml), and phenylmethylsulfonyl fluoride (1 mM). The suspension was sonicated (3 × 15 s), then clarified by centrifugation (20 min at 30,000 × g, and then 30 min at 150,000 × g [4°C]) to yield a high-speed supernatant. The supernatant was diluted in PMG buffer supplemented with Talon Resin (Clontech) (1 ml resin/50 ml), 250 mM KCL, and 30 mM imidazole (final diluted volume was 5x wet cell mass). After a 30-min binding period, the resin was washed with buffer and poured into an Econo-Pac column (Bio-Rad), and protein was eluted in PMG plus 150 mM imidazole. Fractions containing the kinesin were mixed 1:1 with sucrose freezing buffer (2.5 M sucrose, 35 mM Tris, pH 7.2, 5 mM MgSO4 [Bingham et al., 1998]); 1-ml aliquots were flash-frozen in LN2 and stored at –80°C.
For MT binding and gliding analyses, kinesin-containing aliquots were thawed, supplemented with 0.5 mg/ml paclitaxel-stabilized MTs, and brought to a final volume of 1.5 ml with BRB-80 buffer (80 mM PIPES, pH 6.8, 1 mM MgCl2, 1 mM EGTA) and 10 μM paclitaxel. After 15 min incubation at 21°C, MTs were sedimented (15 min, 150,000 × g); the pellet was resuspended in 1/10 vol buffer supplemented with paclitaxel and 10 mM Mg2+ATP, 5 min and resedimented to yield an ATP extract enriched for active motors. Simple flow cells were assembled by overlaying acid-washed coverslips onto parallel double-stick tape spacers mounted lengthwise on a glass slide. The chambers were sequentially flushed with 10 μl each of buffer (BRB-80), anti-GFP tag antibody (Life Technologies, A11120; 100 μg/ml, 5 min incubation), buffer plus casein (0.5 mg/ml), kinesin (5 min incubation), 3x buffer washes, polymerized rhodamine-labeled MTs (0.1 mg/ml, 5 min incubation), and finally buffer containing 1 mM ATP and an oxygen scavenging system (glucose, 22.5 mM; glucose oxidase 0.2 mg/ml; catalase 35 μg/ml; Mitchison Lab Protocols). The chamber was sealed with VALAP, and then observed for MT gliding activity on a DeltaVision microscopy workstation. Images were collected at 4-s intervals. MT ends were manually tracked using FIJI to determine gliding rate.
Data analyses and preparation of illustrations
Tracking of the centrosome movement was performed with the TrackMate and Manual Tracking plug-ins in FIJI (Schindelin et al., 2012; Tinevez et al., 2017). Centrosome position was defined as the centroid of the brightest part of the GFP-tubulin focus at the MT-minus ends. The X,Y coordinates were imported into Microsoft Excel for calculations and statistical analyses; box plots were generated as described in Spitzer et al. (2014). For MT number measurements in WT and DdKif8 null cells (Figure 7C), line scans across the cell axes were used to measure and count peaks that correspond to MTs, in 2D maximum intensity projections of antibody-stained cells. Curve fitting was performed in FIJI; the Rodbard function was manually selected as a best fit to the data. MT tracking was performed by importing deconvoluted image stacks obtained in fixed-cell preparations into the Imaris image analysis software (Bitplane) and following standard prompts in the FilamentTracing package. The diameter for the starting point was set at 2.5 μm, and thinnest diameter filaments were set at 0.100 μm. The final figures were assembled with Adobe Photoshop.
Supplementary Material
Acknowledgments
We appreciate the use of Wadsworth Center’s AGTC Core for DNA sequencing and are grateful for the efforts at http://dictybase.org to archive and annotate Dictyostelium genomic information ( Basu et al., 2013). Our work is supported in part by the National Science Foundation (DBI-1757170 for REU support of J.O., MCB-1510511 to M.P.K.) and the National Institutes of Health (GM-130298 to A.K.). This project was inspired by M. W. Berns from his pioneering contributions in laser microsurgery ( Berns et al., 1981).
Abbreviations used:
- GFP
green fluorescent protein
- MT
microtubule
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-01-0034) on May 8, 2019.
REFERENCES
- Anderson CA, Eser U, Korndorf T, Borsuk ME, Skotheim JM, Gladfelter AS. (2013). Nuclear repulsion enables division autonomy in a single cytoplasm. Curr Biol , 1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Fey P, Pandit Y, Dodson R, Kibbe WA, Chisholm RL. (2013). dictyBase 2013: integrating multiple Dictyostelid species. Nucleic Acids Res , D676–D683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns MW, Aist J, Edwards J, Strahs K, Girton J, McNeill P, Rattner JB, Kitzes M, Hammer-Wilson M, Liaw LH, et al (1981). Laser microsurgery in cell and developmental biology. Science , 505–513. [DOI] [PubMed] [Google Scholar]
- Bieling P, Telley IA, Surrey T. (2010). A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell , 420–432. [DOI] [PubMed] [Google Scholar]
- Bingham JB, King SJ, Schroer TA. (1998). Purification of dynactin and dynein from brain tissue. Methods Enzymol , 171–184. [DOI] [PubMed] [Google Scholar]
- Brinkley BR, Cox SM, Pepper DA, Wible L, Brenner SL, Pardue RL. (1981). Tubulin assembly sites and the organization of cytoplasmic microtubules in cultured mammalian cells. J Cell Biol , 554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito DA, Strauss J, Magidson V, Tikhonenko I, Khodjakov A, Koonce MP. (2005). Pushing forces drive the comet-like motility of microtubule arrays in Dictyostelium . Mol Biol Cell , 3334–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burakov A, Nadezhdina E, Slepchenko B, Rodionov V. (2003). Centrosome positioning in interphase cells. J Cell Biol , 963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaaban S, Brouhard GJ. (2017). A microtubule bestiary: structural diversity in tubulin polymers. Mol Biol Cell , 2924–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette SJ, Mercanti V, Letourneur F, Bennett N, Cosson P. (2006). A role for adaptor protein-3 complex in the organization of the endocytic pathway in Dictyostelium . Traffic , 1528–1538. [DOI] [PubMed] [Google Scholar]
- De Lozanne A, Spudich JA. (1987). Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science , 1086–1091. [DOI] [PubMed] [Google Scholar]
- De Simone A, Gönczy P. (2017). Computer simulations reveal mechanisms that organize nuclear dynein forces to separate centrosomes. Mol Biol Cell , 3165–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogterom M, Surrey T. (2013). Microtubule organization in vitro. Curr Opin Cell Biol , 23–29. [DOI] [PubMed] [Google Scholar]
- Egelhoff TT, Titus MA, Manstein DJ, Ruppel KM, Spudich JA. (1991). Molecular genetic tools for study of the cytoskeleton in Dictyostelium. Methods Enzymol , 319–334. [DOI] [PubMed] [Google Scholar]
- Firestone AJ, Weinger JS, Maldonado M, Barlan K, Langston LD, O’Donnell M, Gelfand VI, Kapoor TM, Chen JK. (2012). Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature , 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke J, Kessin R. (1977). A defined minimal medium for axenic strains of Dictyostelium discoideum . Proc Nat Acad Sci USA , 2157–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaux R, Politi AZ, Philippsen P, Nédélec F. (2017). Mechanism of nuclear movements in a multinucleated cell. Mol Biol Cell , 645–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräf R, Daunderer C, Schliwa M. (2000). Dictyostelium DdCP224 is a microtubule-associated protein and a permanent centrosomal resident involved in centrosome duplication. J Cell Sci , 1747–1758. [DOI] [PubMed] [Google Scholar]
- Grill SW, Hyman AA. (2005). Spindle positioning by cortical pulling forces. Dev Cell , 461–465. [DOI] [PubMed] [Google Scholar]
- Hestermann A, Gräf R. (2004). The XMAP215-family protein DdCP224 is required for cortical interactions of microtubules. BMC Cell Biol , 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höing S, Yeh TY, Baumann M, Martinez NE, Habenberger P, Kremer L, Drexler HCA, Küchler P, Reinhardt P, Choidas A, et al (2018). Dynarrestin, a novel inhibitor of cytoplasmic dynein. Cell Chem Biol , 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holy TE, Dogterom M, Yurke B, Leibler S. (1997). Assembly and positioning of microtubule asters in microfabricated chambers. Proc Natl Acad Sci USA , 6228–6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J, Garzon-Coral C. (2017). Physical limits on the precision of mitotic spindle positioining by microtubule pushing forces. Bioessays , 1700122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, La Terra S, Chang F. (2004). Laser microsurgery in fission yeast: role of the mitotic spindle midzone in anaphase B. Curr Biol , 1330–1340. [DOI] [PubMed] [Google Scholar]
- Kimura K, Kimura A. (2011). A novel mechanism of microtubule length-dependent force to pull centrosomes toward the cell center. BioArchitecture , 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht DA, Loomis WF. (1987). Antisense RNA inactivation of myosin heavy chain gene expression in Dictyostelium discoideum . Science , 1081–1086. [DOI] [PubMed] [Google Scholar]
- Koke C, Kanesaki T, Grosshans J, Schwarz US, Dunlop CM. (2014). A computational model of nuclear self-organisation in syncytial embryos. J Theoretical Biol , 92–100. [DOI] [PubMed] [Google Scholar]
- Kollmar M, Glöckner G. (2003). Identification and phylogenetic analysis of Dictyostelium discoideum kinesin proteins. BMC Genomics , 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonce MP, Khodjakov A. (2002). Dynamic microtubules in Dictyostelium . J Muscle Res Cell Motil , 613–619. [DOI] [PubMed] [Google Scholar]
- Koonce MP, Kohler J, Neujahr R, Schwartz JM, Tikhonenko I, Gerisch G. (1999). Dynein motor regulation stabilizes interphase microtubule arrays and determines centrosome position. EMBO J , 6786–6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonce MP, McIntosh JR. (1990). Identification and immunolocalization of cytoplasmic dynein in Dictyostelium . Cell Motil Cytoskel , 51–62. [DOI] [PubMed] [Google Scholar]
- Koonce MP, Tikhonenko I. (2018). Centrosome positioning in Dictyostelium: Moving beyond microtubule tip dynamics. Cells , 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R, Sato C, Fukui Y, Nishibayashi S. (1982). In vitro nucleation of microtubules from microtubule-organizing center prepared from cellular slime mold. Cell Motil , 257–272. [DOI] [PubMed] [Google Scholar]
- Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, López MP, Vale RD, Jülicher F, Reck-Peterson SL, Dogterom M. (2012). Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell , 502–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letort G, Nedelec F, Blanchoin L, Théry M. (2016). Centrosome centering and decentering by microtubule network rearrangement. Mol Biol Cell , 2833–2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chen Q, Kaller M, Nellen W, Gräf R, De Lozanne A. (2008). Dictyostelium Aurora kinase has properties of both Aurora A and Aurora B kinases. Eukaryot Cell , 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Triviños-Lagos L, Gräf R, Chisholm RL. (1999). Dynein intermediate chain mediated dynein–dynactin interaction is required for interphase microtubule organization and centrosome replication and separation in Dictyostelium . J Cell Biol , 1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V, Loncarek J, Hergert P, Rieder CL, Khodjakov A. (2007). Laser microsurgery in the GFP era: a cell biologist’s perspective. Methods Cell Biol , 239–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malikov V, Cytrynbaum EN, Kashina A, Mogilner A, Rodionov V. (2005). Centering of a radial microtubule array by translocation along microtubules spontaneously nucleated in the cytoplasm. Nat Cell Biol , 1213–1218. [DOI] [PubMed] [Google Scholar]
- Manhart A, Windner S, Baylies MK, Mogilner A. (2018). Mechanical positioning of multiple nuclei in muscle cells. PLoS Comput Biol , e1006208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko M, Prabhu Y, Müller R, Blau-Wasser R, Schleicher M, Noegel AA. (2006). The annexins of Dictyostelium . Eur J Cell Biol , 1011–1022. [DOI] [PubMed] [Google Scholar]
- Marszalek JR, Weiner JA, Farlow SJ, Chun J, Goldstein LSB. (1999). Novel dendritic kinesin sorting identified by different process targeting of two related kinesins: KIF21A and KIF21B. J Cell Biol , 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey G, Vale RD. (1989). Identification of a kinesin-like microtubule-based motor protein in Dictyostelium discoideum . EMBO J , 3229–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally FJ. (2013). Mechanisms of spindle positioning. J Cell Biol , 131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB. (1976). Spindle and kinetochore morphology of Dictyostelium discoideum . J Cell Biol , 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrs MR, Janssen K-P, Kreis T, Noegel AA, Schleicher M. (2000). Cloning and characterization of β-COP from Dictyostelium discoideum . Eur J Cell Biol , 350–357. [DOI] [PubMed] [Google Scholar]
- Muhia M, Thies E, Labonté D, Ghiretti AE, Gromova KV, Xompero F, Lappe-Siefke C, Hermans-Borgmeyer I, Kuhl D, Schweizer M, et al (2016). The kinesin KIF21B regulates microtubule dynamics and is essential for neuronal morphology, synapse function, and learning and memory. Cell Rep , 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag DK, Tikhonenko I, Soga I, Koonce MP. (2008). Disruption of four kinesin genes in Dictyostelium . BMC Cell Biol , 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neujahr R, Albrecht R, Kohler J, Matzner M, Schwartz JM, Westphal M, Gerisch G. (1998). Microtubule-mediated centrosome motility and the positioning of cleavage furrows in multinucleate myosin II-null cells. J Cell Sci (Pt 9), 1227–1240. [DOI] [PubMed] [Google Scholar]
- Nguyen PA, Field CM, Mitchison TJ. (2018). Prc1E and Kif4A control microtubule organization within and between large Xenopus egg asters. Mol Biol Cell , 304–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen PA, Groen AC, Loose M, Ishihara K, Wühr M, Field CM, Mitchison TJ. (2014). Spatial organization of cytokinesis signaling reconstituted in a cell-free system. Science , 244–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura F, Fukui Y. (1985). Dictyostelium MTOC: structure and linkage to the nucleus. Protoplasma , 212–221. [Google Scholar]
- Piperno G, Fuller MT. (1985). Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol , 2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. (2005). Spindle multipolarity is prevented by centrosomal clustering. Science , 127–129. [DOI] [PubMed] [Google Scholar]
- Rehberg M, Kleylein-Sohn J, Faix J, Ho TH, Schulz I, Gräf R. (2005). Dictyostelium LIS1 is a centrosomal protein required for microtubule/cell cortex interactions, nucleus/centrosome linkage, and actin dynamics. Mol Biol Cell , 2759–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Davison EA, Jensen LC, Cassimeris L, Salmon ED. (1986). Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J Cell Biol , 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhlk C, Rohlfs M, Leier S, Schliwa M, Liu X, Parsch J, Woehlke G. (2008). Properties of the Kinesin-1 motor DdKif3 from Dictyostelium discoideum . Eur J Cell Biol , 237–249. [DOI] [PubMed] [Google Scholar]
- Samereier M, Baumann O, Meyer I, Gräf R. (2011). Analysis of Dictyostelium TACC reveals differential interactions with CP224 and unusual dynamics of Dictyostelium microtubules. Cell Mol Life Sci , 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samereier M, Meyer I, Koonce MP, Gräf R. (2010). Live cell-imaging techniques for analyses of microtubules in Dictyostelium . Methods Cell Biol , 341–357. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider N, Schwartz J-M, Köhler J, Becker M, Schwarz H, Gerisch G. (2000). Golvesin-GFP fusions as distinct markers for Golgi and post-Golgi vesicles in Dictyostelium cells. Biol Cell , 495–511. [DOI] [PubMed] [Google Scholar]
- Spitzer M, Wildenhain J, Rappsilber J, Tyers M. (2014). BoxPlotR: a web tool for generation of box plots. Nat Methods , 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Kapoor TM. (2012). Building complexity: Insights into self-organized assembly of microtubule-based architectures. Dev Cell , 874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Ti S-C, Tan L, Darst SA, Kapoor TM. (2013). Marking and measuring single microtubules by PRC1 and kinesin-4. Cell , 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N, Marshall WF. (2012). Centrosome positioning in vertebrate development. J Cell Sci , 4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto H, Kimura AI, Minc N. (2016). Shape–motion relationships of centering microtubule asters. J Cell Biol , 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto H, Sallé J, Dodin L, Minc N. (2018). Physical forces determining the persistency and centring precision of microtubule asters. Nat Phys , 848–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley IA, Gáspár I, Ephrussi A, Surrey T. (2012). Aster migration determines the length scale of nuclear separation in the Drosophila syncytial embryo. J Cell Biol , 887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonenko I, Irizarry K, Khodjakov A, Koonce MP. (2016). Organization of microtubule assemblies in Dictyostelium synctia depends on the microtubule crosslinker, Ase1. Cell Mol Life Sci , 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonenko I, Magidson V, Gräf R, Khodjakov A, Koonce MP. (2013). A kinesin-mediated mechanism that couples centrosomes to nuclei. Cell Mol Life Sci , 1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez J-Y, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, Eliceiri KW. (2017). TrackMate: an open and extensible platform for single-particle tracking. Methods , 80–90. [DOI] [PubMed] [Google Scholar]
- Tran PT, Marsh L, Doye V, Inoue S, Chang F. (2001). A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol , 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart B, van Riel WE, Doodhi H, Kevenaar JT, Katrukha EA, Gumy L, Bouchet BP, Grigoriev I, Spangler SA, Yu KL, et al (2013). CFEOM1-associated kinesin KIF21A is a cortical microtubule growth inhibitor. Dev. Cell , 145–160. [DOI] [PubMed] [Google Scholar]
- van Riel WE, Rai A, Bianchi S, Katrukha EA, Liu Q, Heck AJR, Hoogenraad CC, Steinmetz MO, Kapitein LC, Akhmanova A. (2017). Kinesin-4 KIF21B is a potent microtubule pausing factor. eLife , e24746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner OH, Murphy J, Griffiths G, Schleicher M, Noegel AA. (1993). The actin-binding protein comitin (p24) is a component of the Golgi apparatus. J Cell Biol , 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne S, Subramanian R. (2018). Geometry of antiparallel microtubule bundles regulates relative sliding and stalling by PRC1 and Kif4A. eLife , e32595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumura S, Mori H, Fukui Y. (1984). Localization of actin and myosin for the study of amoeboid movement in Dictyostelium discoideum using improved immunofluorescence. J Cell Biol , 894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Graham OS, Raposo A, St Johnston D. (2012). Growing microtubules push the oocyte nucleus to polarize the Drosophila dorsal-ventral axis. Science , 999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Burakov A, Rodionov V, Mogilner A. (2010). Finding the cell center by a balance of dynein and myosin pulling and microtubule pushing: A computational study. Mol Biol Cell , 4418–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.