Abstract
αT (Testes)-catenin, a critical factor regulating cell–cell adhesion in the heart, directly couples the cadherin-catenin complex to the actin cytoskeleton at the intercalated disk (ICD), a unique cell–cell junction that couples cardiomyocytes. Loss of αT-catenin in mice reduces plakophilin2 and connexin 43 recruitment to the ICD. Since αT-catenin is subjected to mechanical stretch during actomyosin contraction in cardiomyocytes, its activity could be regulated by mechanical force. To provide insight in how force regulates αT-catenin function, we investigated the mechanical stability of the putative, force-sensing middle (M) domain of αT-catenin and determined how force impacts vinculin binding to αT-catenin. We show that 1) physiological levels of force, <15 pN, are sufficient to unfold the three M domains; 2) the M1 domain that harbors the vinculin-binding site is unfolded at ∼6 pN; and 3) unfolding of the M1 domain is necessary for high-affinity vinculin binding. In addition, we quantified the binding kinetics and affinity of vinculin to the mechanically exposed binding site in M1 and observed that αT-catenin binds vinculin with low nanomolar affinity. These results provide important new insights into the mechanosensing properties of αT-catenin and how αT-catenin regulates cell–cell adhesion at the cardiomyocyte ICD.
INTRODUCTION
Cell–cell adhesion formation and function is essential for many fundamental physiological processes including morphogenesis during embryogenesis, tissue development during fetal life, and tissue maintenance during adulthood (Edelman, 1986). This is particularly true for the heart which is under constant mechanical load. In cardiac muscle tissue, cardiomyocytes are connected to each other at the intercalated disk (ICD), a specialized cell–cell junction found only in cardiac muscle tissue that mechanically and electrically couples cells (Sheikh et al., 2009; Vermij et al., 2017). A primary component of the ICD is the adherens junction that connects actin filaments across adjacent cardiomyocytes, thereby allowing cells to retain their shape upon mechanical stress (Bays et al., 2014). In addition, the adherens junction is also involved in force transmission and sensing (Sheikh et al., 2009; Wang et al., 2012). The core of the cardiac adherens junction is the transmembrane protein N-cadherin, and its extracellular domain homodimerizes with N-cadherin molecules from neighboring cells. The N-cadherin cytosolic tail binds p120-catenin and β-catenin (Noorman et al., 2009; Brasch et al., 2012). β-catenin, in turn, recruits the actin-binding protein α-catenin, forming the cadherin-catenin complex. This complex links to the actin filament network either directly via α-catenin or through the recruitment of actin-binding proteins like vinculin (Watabe-Uchida et al., 1998; Weiss et al., 1998). In addition to the adherens junction, the ICD also contains the desmosome and gap junction that together establish mechanoelectrical coupling in heart muscle tissue (Vermij et al., 2017).
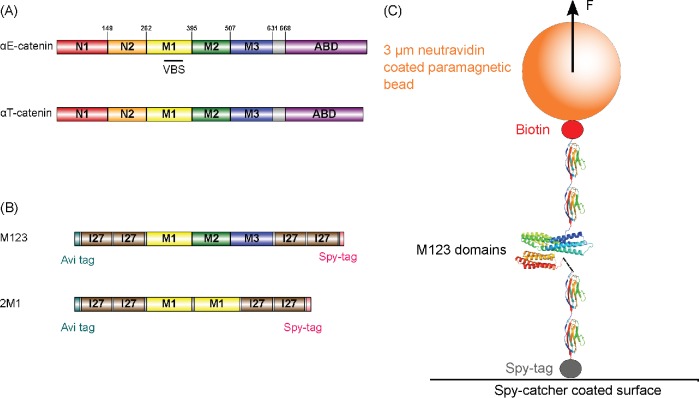
As one of the primary components of the cadherin-catenin complex, the α-catenin family of proteins consists of three known members in mammals: αE (Epithelial)-catenin, αT (Testes)-catenin, and αN (Neuronal)-catenin, with αE-catenin the best studied. All three α-catenins are highly similar, with amino acid sequence identity greater than 57%. Based on sequence homology, it can be assumed that αT-catenin and αN-catenin share a similar domain organization with αE-catenin. The structure of αE-catenin is comprised of a series of helical bundles (Pokutta and Weis, 2000; Pokutta et al., 2002, 2014; Ishiyama et al., 2013; Rangarajan and Izard, 2013). At the N-terminus of αE-catenin, there are two N domains (N1 and N2), each formed by a four-helix bundle and capable of binding β-catenin and mediating homodimerization (Pokutta and Weis, 2000). The actin-binding domain (ABD), located at the C-terminus, is a five-helix bundle (Imamura et al., 1999; Pokutta et al., 2008). The middle (M) domain, an adhesion modulation domain (Imamura et al., 1999; Yang et al., 2001), is composed of three four-helix bundles (M1, M2, and M3), with a vinculin-binding site (VBS) in M1 (Choi et al., 2012; Li et al., 2015) (Figure 1A).
FIGURE 1:
Protein constructs and experimental design. (A) Domain organization of αE-catenin and αT-catenin. The VBS from amino acid 300–360 is indicated. (B) Protein constructs for single-molecule stretching experiments used in this study. (C) Schematic of the M123 construct tethered between a coverslip and a superparamagnetic bead under an applied force F.
Of the three isoforms of α-catenin, only αE-catenin and αT-catenin are expressed in cardiomyocytes, and both are localized primarily at the ICD (Janssens et al., 2001; Wickline et al., 2016). It has been found recently that at the ICD, adherens junction and desmosome proteins intermingle to form the area composite, a junction unique to heart muscle tissue (Borrmann et al., 2006; Pieperhoff and Franke, 2007). A critical component of the ICD is αT-catenin, which mainly exists as a monomer in solution and is capable of binding to F-actin, thereby directly linking the cadherin-catenin complex to the actin cytoskeleton (Wickline et al., 2016). αT-catenin binds β-catenin through its N1-2 domains and also the desmosomal protein plakophilin2 via its M3 domain, thereby potentially acting like a “bridge” that connects adherens junction and desmosome proteins (Janssens et al., 2001; Goossens et al., 2007). αT-catenin is unique to amniotes and mainly expressed in cardiomyocytes and testis (Janssens et al., 2001; Folmsbee et al., 2014). Loss of αT-catenin in mice causes earlier onset of dilated cardiomyopathy (as compared with cardiac-specific αE-catenin conditional knockout), altered plakophilin2 distribution, and gap junction remodeling (Li et al., 2012).
In addition to the cadherin-catenin family of proteins, vinculin localizes to the ICD (Tokuyasu et al., 1981; Pardo et al., 1983). Vinculin is a compact globular protein that consists of a series of four helical bundles, which is divided into a head domain (D1–D4) connected to a tail domain by a proline-rich linker (Peng et al., 2011). Vinculin binds to αE-catenin through its head domain (Weiss et al., 1998; Choi et al., 2012) and actin filaments through its tail domain (Menkel et al., 1994; Johnson and Craig, 1995) at cell–cell adhesions, implying a potential role as a bridge in connecting actin to mature cell–cell adhesions. In cardiomyocytes, vinculin plays important roles in cell–cell adhesion, as heterozygous vinculin knockout mice exhibit abnormal ICD structure and cardiac function either at baseline or upon increased hemodynamic stress imposed by transverse aortic constriction (Zemljic-Harpf et al., 2004, 2007). Vinculin is auto-inhibited: the head and tail domains interact with each other to blocking the binding sites for α-catenin proteins and for actin (Johnson and Craig, 1995). Auto-inhibition is relieved by combinatorial ligand binding (Hino et al., 2019).
Biochemical and structural data indicate that two α-helices (residues 304–316 and 328–353) within αE-catenin M1 forms the core VBS (residues 300–360) that interacts with vinculin D1 (Yonemura et al., 2010; Choi et al., 2012). The α-helix of residues 328–353 inserts into the first vinculin D1 α-helix bundle, while the α-helix of residues 304–316 interacts with the second vinculin D1 α-helix bundle (Yonemura et al., 2010; Choi et al., 2012). Structural data further indicate that αE-catenin can adopt an auto-inhibitory conformation (Rangarajan and Izard, 2012; Li et al., 2015) whereby the VBS is buried in the folded M1 domain. A recent single-molecule study shows that mechanical unfolding of the M1 is needed to create a high-affinity binding site for the vinculin D1 domain (Yao et al., 2014b). Interactions between the M1 domain and both the M2 and M3 helix bundles are believed to further enhance the auto-inhibition against vinculin binding (Yonemura et al., 2010; Li et al., 2015). However, the interaction of vinculin with these two isoforms (αT-catenin and αN-catenin) has not been confirmed experimentally as they are much less understood compared with αE-catenin.
Recent studies have shown that adherens junction-related proteins in cardiomyocytes are able to sense and respond to mechanical cues. N-cadherin, the sole cadherin expressed in cardiomyocytes (Noorman et al., 2009), is up-regulated in response to applied mechanical stretch in neonatal ventricular rat myocytes (Chopra et al., 2011). In addition, neonatal ventricular rat myocytes exhibit cytoskeletal-mediated mechanical remodeling, including changes in cell shape and myofibrillar organization, when plated on N-cadherin–coated substrates of varying stiffness that mimic changes in cell–cell adhesion strength (Chopra et al., 2011). Vinculin also responds to cell–cell junction formation, as demonstrated when myoblasts plated on N-cadherin–coated surfaces were found to recruit β-catenin as well as p120, vinculin, and αE-catenin to the lamellipodium (Gavard et al., 2004). As a component of the cadherin-catenin complex in cardiomyocytes, αT-catenin is under constant mechanical load; however, its mechanosensing properties are not defined.
To investigate the mechanosensing and vinculin-binding functions of αT-catenin, we used magnetic tweezers to apply forces between 1 and 40 pN to various αT-catenin M domain constructs. We found that all three M domains, M1–M3, unfold within a physiological force range of less than 15 pN. The M1 domain that contains a putative VBS unfolds at near 6 pN and is insensitive to pulling rate. The unfolding of M1 is necessary for high-affinity vinculin binding. In addition, we quantified the binding kinetics and affinity of vinculin to the mechanically exposed binding site of αT-catenin. Together, these results provide important new insights into the potential mechanosensing function of αT-catenin.
RESULTS
Mechanical response of αT-catenin M1–M3 domains
Our experiments were carried out using in-house developed magnetic tweezers (Chen et al., 2011) that allowed us to stretch a single protein at low forces within a physiological range over multiple hours without significant mechanical drift (Chen et al., 2015; Pang et al., 2018). Based on I-TASSER structure prediction, the αT-catenin M domain is formed by three four-helix bundles, similar to αE-catenin (Zhang, 2008; Roy et al., 2010; Wickline et al., 2016). The 61 amino acid sequence in the VBS is largely conserved across all three α-catenin isoforms, with 82% identity with αT-catenin and 95% identity with αN-catenin (Figure 1A; Supplemental Figure S1), indicating that vinculin likely interacts with αT-catenin and αN-catenin. To investigate the force response of the three M domains of αT-catenin, we expressed the following two constructs: M123 that comprised all three M domains and 2M1 that comprised two repeats of the M1 domain (Figure 1B). A pair of the titin I27 immunoglobulin-like domain were included at both ends to serve as a spacer handle and as a positive control using its highly characteristic unfolding transition signals at high force (Chen et al., 2015; Yuan et al., 2017) (Figure 1C; Supplemental Figure S2).
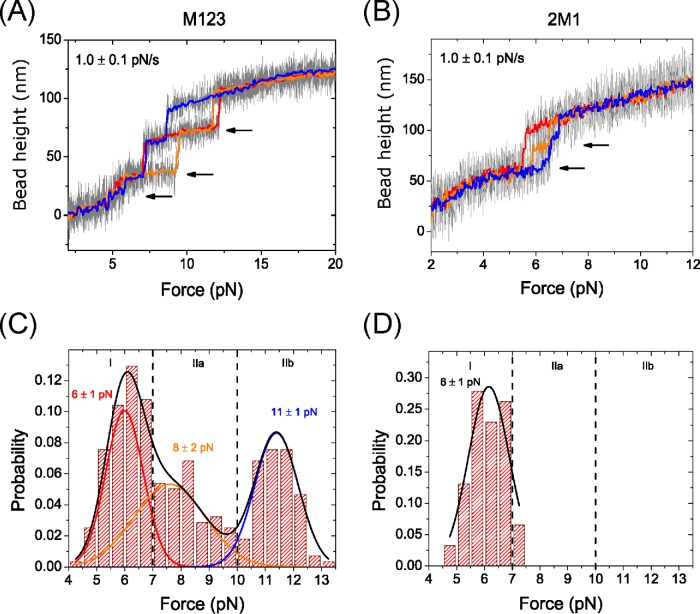
In linearly increasing force loading rate experiments, a linearly increasing force with a loading rate of 1.0 ± 0.1 pN/s was applied to the M123 construct at a force range between 2 and 20 pN (Figure 2A). Three unfolding transitions were observed, one occurring near 6 pN and the other two occurring at higher forces of 7–13 pN, as indicated by the stepwise bead height increase (Figure 2A, arrows). Each of these unfolding transitions corresponds to one M domain, indicating that all three M domains are well structured though expected to have different mechanical properties. Analysis of the histogram of the forces where unfolding events occurred revealed three distinct mechanical stability groups defined as I (4–7 pN), IIa (7–10 pN), and IIb (10–13.5 pN). These events can be fit to a triple-Gaussian distribution that peaks at 6 ± 1 pN, 8 ± 2 pN, and 11 ± 1 pN, with error reported as the width of the Gaussian (Figure 2C).
FIGURE 2:
Force-response of αT-catenin middle domains. Representative changes of the height of the bead tethered to an M123 construct (A) and a 2M1 construct (B) when subjected to a linearly increasing force loading rate of 1.0 ± 0.1 pN/s. The arrows indicate the domain unfolding events accompanied by a stepwise height increase. (C, D) The unfolding force distribution obtained from the M123 construct (C) and the 2M1 construct (D). The vertical dashed lines indicate the boundary between different mechanical stability groups indicated by I, IIa, and IIb. The black line in C is three-peak Gaussian fitting to the unfolding force histogram. The colored lines are Gaussian distributions to individual groups plotted based on the fitted parameters. The line in D is the single-peak Gaussian fitting to the unfolding force histogram of the 2M1 construct.
To identify which unfolding step corresponds to the unfolding of the VBS-bearing M1, a similar linearly increasing force loading rate experiment was performed on the 2M1 construct. Two unfolding events were observed at low force ranging from 4.5–7.5 pN (Figure 2B, arrows). The histogram of the unfolding forces was similar to that of mechanical stability group I in (Figure 2, C and D). Therefore, the M1 domain is the mechanically weakest domain of the three M domains.
In addition, when the 2M1 construct was subjected to a constant force in the range of 5–7 pN, the M1 domains underwent a folding-unfolding fluctuation (Figure 3A). This further supports that the unfolding transition at ∼6 pN observed in the M123 construct (Figure 2A) is caused by the unfolding of M1. To further investigate the timescale of M1 unfolding at such forces, the force-dependent unfolding and folding rates in the force range of 5–7 pN were measured using the 2M1 construct. As there are two repeats of the same M1 domain, transition rates were obtained based on pseudo dwell time analysis of the lifetimes of the folding/unfolding states during constant force experiments (Supplemental Methods). The unfolding and folding rates obtained at different forces are shown in Figure 3B on a logarithmic scale.
FIGURE 3:
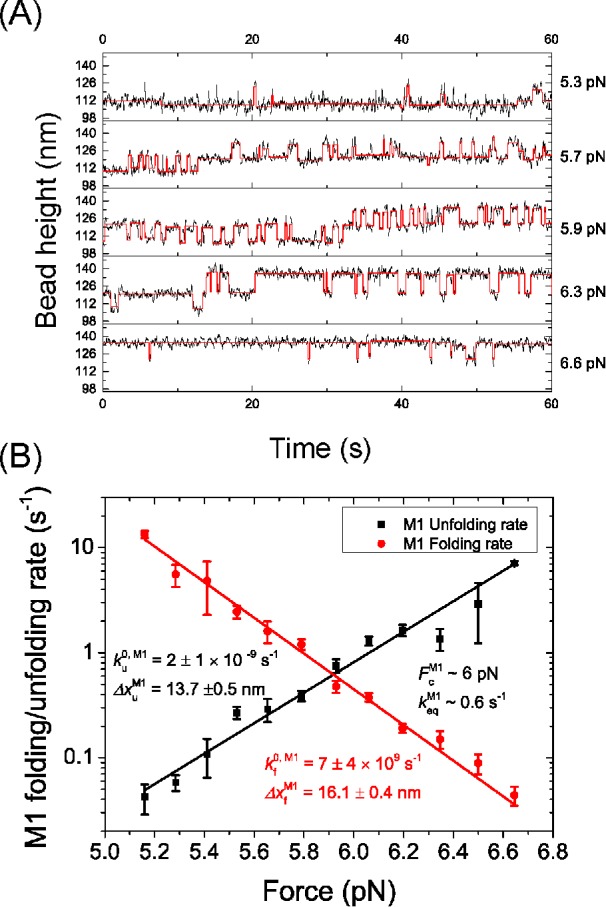
Mechanical stability of the M1 domain. (A) Representative time traces of the height of the bead tethered to a 2M1 construct at various constant forces over a duration of 60 s and smoothed over 0.1 s (black). The digitized stepwise changes in bead height were determined based on hidden Markov analysis using stepfit1 software (red) (Aggarwal et al., 2012). (B) The force-dependent unfolding (black) and folding (red) rates of the M1 domain calculated from the dwell times of the folded/unfolded states obtained from A. The errors are estimated using Bootstrap analysis (Supplemental Methods). The rates are fitted with Bell’s model where the best fitting parameters are indicated in the figure panel.
Within the force range of 5–7 pN, both the force-dependent unfolding rate, 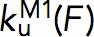 , and the folding rate,
, and the folding rate, 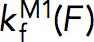 , had a linear profile on the logarithm scale. Therefore, they were fit with Bell’s model,
, had a linear profile on the logarithm scale. Therefore, they were fit with Bell’s model, 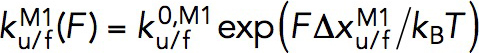 , where
, where 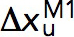 is the transition distance during unfolding transitions that typically has a positive value,
is the transition distance during unfolding transitions that typically has a positive value, 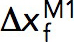 is the transition distance during folding transitions that typically has a negative value, and
is the transition distance during folding transitions that typically has a negative value, and 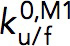 is the extrapolated unfolding/folding rate at no force, so as to identify the critical force, Fc, where unfolding and folding rates are equal. From such analysis, a critical force of Fc ∼6 pN was obtained at which the equilibrium transition rate was ∼0.6 s–1.
is the extrapolated unfolding/folding rate at no force, so as to identify the critical force, Fc, where unfolding and folding rates are equal. From such analysis, a critical force of Fc ∼6 pN was obtained at which the equilibrium transition rate was ∼0.6 s–1.
The folding energy was estimated based on the critical force of the equilibrium two-state transition of the M1 domain. The calculation of the folding energy depends on the force-extension curves of the domain in the folded and unfolded states. For the folded state, its force-extension curve,  , can be modeled as that of a freely rotatable rigid rod of a certain length, which has an analytical solution:
, can be modeled as that of a freely rotatable rigid rod of a certain length, which has an analytical solution: 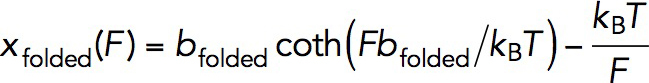 , where
, where  = 5.8 nm is the length of the folded state estimated based on I-TASSER predicted structure as well as that of αE-catenin M1 domain (PDBID: 4IGG). For the unfolded state, the force-extension curve depends on whether the unfolded polypeptide is a disordered polymer, a chain of four α-helices, or a mixture of them. The folding energy was estimated based on two extreme cases, assuming the unfolded polypeptide was a disordered polymer or a chain of four α-helices, which yielded an estimated folding energy of ∼–16 kBT and ∼–12 kBT, respectively. Based on the unfolding/folding step size around the critical force, it is consistent with the assumption of the unfolded polypeptide as a chain of four α-helices (Supplemental Methods and Supplemental Figure S3). Therefore, we estimated that the folding energy was ∼–12 kBT. More details of the estimation can be found in the Supplemental Methods.
= 5.8 nm is the length of the folded state estimated based on I-TASSER predicted structure as well as that of αE-catenin M1 domain (PDBID: 4IGG). For the unfolded state, the force-extension curve depends on whether the unfolded polypeptide is a disordered polymer, a chain of four α-helices, or a mixture of them. The folding energy was estimated based on two extreme cases, assuming the unfolded polypeptide was a disordered polymer or a chain of four α-helices, which yielded an estimated folding energy of ∼–16 kBT and ∼–12 kBT, respectively. Based on the unfolding/folding step size around the critical force, it is consistent with the assumption of the unfolded polypeptide as a chain of four α-helices (Supplemental Methods and Supplemental Figure S3). Therefore, we estimated that the folding energy was ∼–12 kBT. More details of the estimation can be found in the Supplemental Methods.
Force activates vinculin binding onto M1
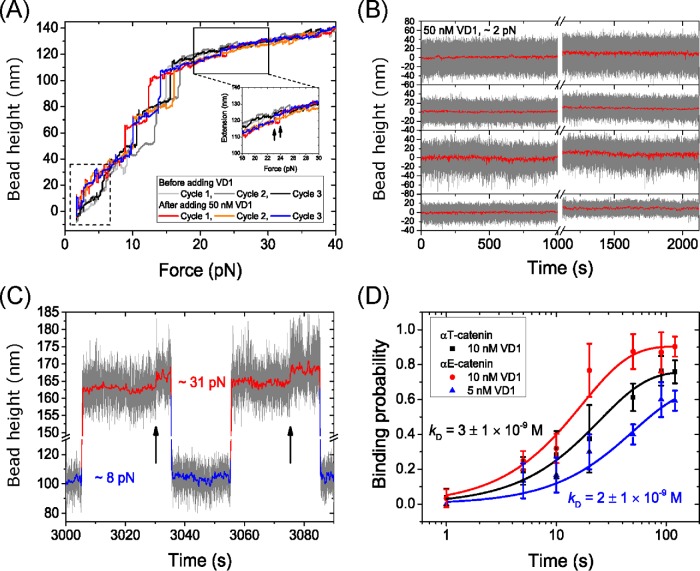
We next investigated whether the third α−helix in M1 is indeed a VBS as predicted based on sequence homology (Supplemental Figure S1) and whether force was needed to expose it to allow vinculin binding. A loading rate of 3.5 ± 0.4 pN/s was applied to the M123 construct before (three curves in gray) and after (three curves in color) introducing 50 nM of vinculin head domain 1 (VD1) (Figure 4A). As shown by the data, before VD1 was introduced, three characteristic unfolding steps were obtained in each force-increase scan. In contrast, after VD1 was introduced, there were only two unfolding steps that correspond to the M2-3 domains. The loss of the characteristic M1 unfolding event indicates that it did not refold when the tether was held at ∼2 pN for 30 s, suggesting that it was bound by VD1 and blocked from refolding. Consistently, at forces less than 7 pN, the bead height was higher than that of the tether when all the domains are folded (Figure 4A, dashed box). Blocking of M1 domain refolding by bound VD1 was also observed in the 2M1 construct (Supplemental Figure S4).
FIGURE 4:
Force-dependent VD1 binding. (A) Representative changes in the height of the bead tethered to a M123 construct when subjected to a linearly increasing (3.5 ± 0.4 pN/s) force loading rate before (gray traces) and after the addition of 50 nM VD1 (colored traces), smoothed over 0.1 s. In the presence of VD1, the bead height is higher than that before introducing VD1 at low forces (dashed box), indicating VD1 binding inhibits M1 refolding. At forces >20 pN, there is an additional ∼3- to 4-nm step indicating the unbinding of VD1 from M1 (insert, arrow showing two of the steps). (B) Four representative time traces of the height change of a bead tethered to an originally folded M123 construct in 50 nM VD1 held at a constant force of ∼2 pN for 1000 s (before the break) and those of the same tethers for another 1000 s (after the break) at the same force after unfolding the M1 domain by transiently holding the tethers at ∼8 pN for 2 min. (C) A representative time trace of the height of a bead tethered to a M123 construct during force jumping between ∼8 pN and ∼31 pN. The two 3- to 4-nm stepwise bead height increases at ∼31 pN indicate VD1 dissociation events (black arrows). (D) The time evolution of the binding probability of VD1 onto mechanically exposed VBS in the αT-catenin M123 construct (black) and in the αE-catenin M123 construct (red and blue). The corresponding concentrations of VD1 are indicated in the figure panel. Error bars show SD estimated from Bootstrap analysis (Supplemental Methods). The probabilities were fit to an exponential function, from which kon, koff and KD were determined.
In our previous studies, we found that the VD1-bound VBSs of talin and αE-catenin became unstable at forces higher than 20 pN (Yao et al., 2014a,b). This was also true for the αT-catenin VBS, as shown by a small ∼3–4 nm step at forces higher than 20 pN (Figure 4A, inset, arrows). After this step, the bead height of the tethered M123 construct returned to the same position as before the VD1 was added. This suggests that the unfolding step corresponds to the dissociation of the bound VD1, concurrent with further unfolding of the α-helix VBS into a disordered peptide chain. Indeed, the ∼3- to 4-nm step is consistent with the extension difference between a flexible polypeptide conformation and the α-helix conformation of the VBS (Supplemental Methods and Supplemental Figure S5).
To study the effect of force on the interaction between the VBS in M1 and VD1, 50 nM of VD1 was introduced to the M123 construct held at a low force of ∼2 pN that is well below the critical force, Fc ∼6 pN, of M1. At this force, M1 was predominantly in the folded state. Over a long duration of 1000 s, the bead height of the tethered M123 construct remained unchanged in four out of five independent experiments, indicating that the no binding of VD1 had occurred (Figure 4B). Following this, the M1 domain was unfolded by applying a force of ∼8 pN for 120 s, and the force was then reduced back to ∼2 pN. The bead height increased by ∼10 nm, consistent with the extension change of the ∼130 amino acid M1 domain and indicating that VD1 bound to the mechanically exposed VBS and prevented M1 domain refolding after the force was reduced to ∼2 pN (Figure 4B). In only one experiment was VD1 binding to VBS observed at ∼2 pN (without facilitated unfolding by transient force jumping to ∼8 pN) after the tether was held for ∼960 s (Supplemental Figure S6). Taken together, these observations indicate the importance of force in unfolding αT-catenin M1 and exposing the VBS for high-affinity VD1 binding.
The ∼10 nm increase in bead height was maintained in all five experiments over a long duration of 1000 s, indicating that M1 refolding at ∼2 pN had not occurred (Figure 4B and Supplemental Figure S6). As M1 has a folding rate of >13 s–1 at 5.1 ± 0.5 pN (Figure 3B), at a lower force of ∼2 pN the folding rate of M1 is ≫13 s–1 and folding should occur in less than 1 s. The lack of M1 refolding indicates that VD1 remained bound to M1 over this timeframe.
The association and dissociation rates, kon and koff, of VD1 binding were determined using a force jumping’ assay. In this assay, the M123 construct was exposed to 10 nM of VD1 and held at ∼8 pN to expose the VBS in M1 for a time interval between 1 and 120 s before "force jumping" to ∼31 pN for 30 s (Figure 4C). If VD1 binding had occurred within the time interval at ∼8 pN, force jumping will produce the characteristic ∼3- to 4-nm dissociation step at ∼31 pN (Figure 4C, arrow), which therefore can serve to detect if the exposed VBS was bound with a vinculin D1 domain when it was held at ∼8 pN. The force is then jumped back to ∼8 pN to begin the next cycle. This was repeated on five independent tethers until more than 40 cycles were obtained for each time interval. At each time interval, t, the binding probability, P(t), was obtained by calculating the ratio of the number of cycles with observed VD1 binding to the total number of cycles (Figure 4D, black square). The time evolution of the binding probability from 1 to 120 s was then fitted with an exponential curve, 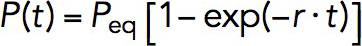 , where Peq is the equilibrium probability of binding, and r is the relaxation rate, both of which were treated as fitting parameters. The Peq and r are related to kon and koff as
, where Peq is the equilibrium probability of binding, and r is the relaxation rate, both of which were treated as fitting parameters. The Peq and r are related to kon and koff as 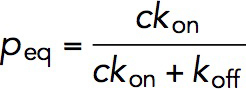 and
and 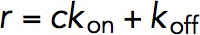 , where c is the concentration of VD1. By solving these two equations, both kon, koff, and hence KD, which is the ratio of koff to kon, can be obtained.
, where c is the concentration of VD1. By solving these two equations, both kon, koff, and hence KD, which is the ratio of koff to kon, can be obtained.
It was determined that the Peq of VD1 binding to αT-catenin was 0.76 ± 0.03, and r was 4.0 ± 0.6 × 10–2 s–1 (Figure 4D, black square and line). From these, it was determined that the kon = 3.0 ± 0.5 × 106 M–1 s–1 and koff = 9 ± 4 × 10–3 s–1, and hence KD = 3 ± 1 × 10–9 M. For comparison, the same experiment was repeated for an αE-catenin construct containing the M123 domain in 10 nM of VD1. In this case, Peq was 0.91 ± 0.03 and r was 0.058 ± 0.008 s–1 (Figure 4D, red circle and line). To reduce the error for koff calculation due to the high Peq, this experiment was repeated at a lower VD1 concentration of 5 nM (Figure 4D, blue triangle and line). Taking the average, the affinity rates for VD1 were determined to be kon = 4.8 ± 0.7 × 106 M–1 s–1 and koff = 8 ± 5 × 10–3 s–1, and hence KD = 2 ± 1 × 10–9 M. Thus, comparing αT-catenin and αE-catenin revealed that both α-catenins have a similar strong binding affinity for vinculin VD1.
DISCUSSION
In this study, we systematically studied the mechanical properties of αT-catenin, a protein highly expressed in cardiomyocytes, using magnetic tweezers. We found that all the three M domains can be unfolded at physiological levels of force, <15 pN (Figure 2, A and B). In particular, the M1 domain that contains a VBS (Figure 4A; Supplemental Figure S4) is unfolded at ∼6 pN (Figures 2, C and D, and 3, A and B). In addition, the force-dependent interaction between both αT-catenin and αE-catenin and vinculin were investigated. We observed that vinculin interacts with the M1 domain in αT-catenin and a force of more than ∼6 pN is required to mechanically expose the VBS in M1 for high-affinity VD1 binding (Figure 4, A and B). Importantly, we report here a label-free single-molecule methodology used to measure the binding and unbinding rate and affinity of VD1 for αT-catenin and αE-catenin (Figure 4, C and D). From this, we obtained KD = 3 ± 1 × 10–9 M (αT-catenin) and 2 ± 1 × 10–9 M (αE-catenin).
Our data show that a force of ∼6 pN is necessary to mechanically unfold the M1 domain, exposing the buried VBS for high-affinity VD1 binding. M1 was estimated to have a folding energy of ∼–12 kBT, far greater than the average thermal energy of ∼1 kBT, implying that it is unlikely for M1 to be unfolded by thermal fluctuation alone (Supplemental Methods). In addition, at a low force of ∼2 pN and 50 nM of VD1, the binding of VD1 to a mechanically exposed VBS of M1 should take place in <10 s, according to the kon (3.0 ± 0.5 × 106 M–1 s–1). This is in sharp contrast to the observation that, when the M1 domain was in the folded state at ∼2 pN, no binding was observed for more than 10 min in the presence of 50 nM VD1 (Figure 4B). Together, these results strongly suggest that mechanical unfolding of αT-catenin M1 is required for high-affinity vinculin binding.
In comparing αT-catenin with αE-catenin, it was observed that both α-catenins have similar mechanical stabilities. Our results show that αT-catenin M domains unfold in the range of <15 pN with the M1 domain unfolds at ∼6 pN and the M2 and M3 domains unfold at ∼8-11 pN, when subjected to a linearly increasing force loading rate of 1.0 ± 0.1 pN/s. The force-responses of the M domains in αT-catenin are similar to that of αE-catenin measured in previous kinetic experiments (Yao et al., 2014b), where the unfolding of M1 domain was observed at ∼5 pN, the M2 and M3 domains were observed at ∼12 pN with a force loading rate of ∼4 pN/s. In addition, our results show that both α-catenins have similar vinculin-binding affinities in the mechanically exposed state, with a KD of 3 ± 1 × 10–9 M for αT-catenin and a KD of 2 ± 1 × 10–9 M for αE-catenin. Importantly, our calculated affinity of αE-catenin for vinculin is similar to the previously reported affinity of 5.2 × 10–9 M between the exposed VBS in αE-catenin M1-M2 (amino acids 273–510) and vinculin D1 as measured by isothermal titration calorimetry (Choi et al., 2012). The high similarity in KD between αE-catenin and αT-catenin can be explained by the high amino acid identity (82%) of the VBS between them (Supplemental Figure S1).
Nonetheless, αT-catenin is known to play a unique role in the heart, as αT-catenin knockout in mice leads to dilated cardiomyopathy despite the presence of αE-catenin (Li et al., 2012). We speculate that molecular differences between the two α-catenin isoforms exist in other domains. For example, the M3 domain of αT-catenin bears a unique binding site for plakophilin2 and loss of αT-catenin in the heart reduces plakophilin2 recruitment to the ICD (Goossens et al., 2007). Likewise, the αT-catenin N2 domain is only 39% identical to αE-catenin, and αT-catenin homodimerization properties (mediated by N1-2) in solution are distinct from αE-catenin (Wickline et al., 2016). These molecular differences could contribute to αT-catenin’s unique function in cell–cell adhesion at the ICD.
MATERIALS AND METHODS
Protein expression
Two fragments of the αT-catenin middle domains (M123: 259–667; M1: 259–295) were synthesized by PCR using plasmid template containing the full-length αT-catenin sequence. DNA fragments were then subcloned into expression vector pET151/D-TOPO with an N-terminal avi-tag and two I27 domain, and a C-terminal spy-tag and two I27 domains, using HiFi DNA Assembly (NEBuilder). Each of the resulting plasmids were cotransformed with a BirA plasmid (Kay et al., 2009). Subsequently, the proteins of interest were expressed in Escherichia coli BL21 (DE3) cultured in Luria-Bertani media with D-Biotin (Sigma-Aldrich) and affinity purified using the His-tag.
Single-molecule manipulation
The single-molecule manipulation experiments were carried out on a custom-built magnetic tweezers platform that can exert forces up to 100 pN with ∼1 nm extension resolution for tethered bead at a 200 Hz sampling rate (Chen et al., 2011; Le et al., 2016). For the given magnets and bead, the force is solely dependent on the distance, d, between the magnet and the bead, F(d), which can be calibrated based on its fluctuation at low force, as described in a previous publication, with ∼10% uncertainty due to the heterogeneous manufactured bead sizes (Chen et al., 2011). For the magnetic tweezers experiments, the protein of interest was immobilized onto the glass coverslip of a laminar flow chamber and to a 3-μm (Dynabeads M270-epoxy) paramagnetic bead coated with neutravidin using specific tethering through Spy-tag/Spy-catcher and biotin/neutravidin chemistry. All experiments were carried out in 1 × phosphate-buffered saline, 1% bovine serum albumin, 1 mM dithiothreitol, and 10 mM sodium l-ascorbate at room temperature of 21 ± 1°C. More details of the magnetic tweezers setup and protein manipulation can be found in previous publications (Chen et al., 2015; Yuan et al., 2017; Pang et al., 2018).
Unfolding/folding rates of αT-catenin M123 domains
The force-dependent unfolding/folding rates of the M123 domains were determined from constant force measurements of folded/unfolded state lifetime. For the M1 domain, due to its short lifetime at near critical force, the M1-2R construct was subjected to various constant forces between 5 and 7 pN near its critical force for 60 s, and the smoothed bead heights were fitted with the stepfit1 function (Aggarwal et al., 2012) from which the lifetimes of the folded/unfolded state were obtained. This was repeated multiple times until more than 60 lifetimes were obtained at each force. As there are two repeats of the same M1 domain, pseudo dwell time analysis was performed on the lifetimes of the folding/unfolding states to obtain the pseudo dwell times (Cao et al., 2008) (Supplemental Methods). The pseudo dwell times were fitted with an exponential decay function to obtain the folding/unfolding rates at each force. Bootstrap analysis was used to obtain the statistical error for the unfolding/folding rates (Supplemental Methods).
Unfolding/folding kinetics parameters
Within the force range at which the force-dependent unfolding/folding rates of M1 domains were obtained (5–7 pN), the force-dependent unfolding/folding rates at force F, 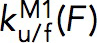 were fitted directly to the Bell’s model,
were fitted directly to the Bell’s model, 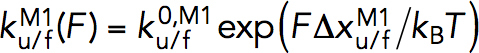 ,
,
where 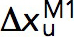 is the transition distance during unfolding transition which typically has a positive value,
is the transition distance during unfolding transition which typically has a positive value, 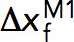 is the transition distance during folding transition which typically has a negative value, and
is the transition distance during folding transition which typically has a negative value, and 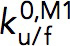 are the extrapolated unfolding/folding rates at no force. The
are the extrapolated unfolding/folding rates at no force. The 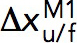 and
and 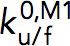 are treated as fitting parameters. From the Bell’s model fitting, the critical force, Fc, where unfolding and folding rates are equal, and the corresponding equilibrium transition rate was determined.
are treated as fitting parameters. From the Bell’s model fitting, the critical force, Fc, where unfolding and folding rates are equal, and the corresponding equilibrium transition rate was determined.
VD1 binding affinity rate calculation
The M123-I27 construct, in the presence of 5 or 10 nM of VD1, was first subjected to a force of ∼8 pN in order to expose the VBS. This was maintained for a duration of between 1 and 120 s before jumping the force immediately to ∼31 pN for 30 s to determine if any VD1 binding has occurred, as can be detected by the characteristic ∼3- to 4-nm dissociation step at this force (Supplemental Methods; Supplemental Figure S5). This was then repeated on five independent tethers until more than 40 times cycles were obtained for each time interval. At each time interval, the probability of binding can be calculated by dividing the total number of binding events observed over the total number of cycles. Bootstrap analysis was performed to obtain statistical error for the binding probability by taking the SD of the results from the 1000 data sets generated (Supplemental Methods). P was then plotted against t and fit using 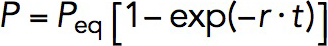 , where Peq is the equilibrium probability, r is the relaxation rate. The kon and koff are related to the Peq and r in these two equations:
, where Peq is the equilibrium probability, r is the relaxation rate. The kon and koff are related to the Peq and r in these two equations: 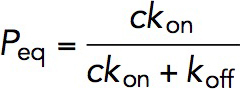 , and
, and 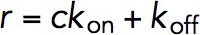 , where c is the concentration of VD1. By solving these two equations, both kon and koff and hence KD, which is equal to koff divided by kon, was obtained.
, where c is the concentration of VD1. By solving these two equations, both kon and koff and hence KD, which is equal to koff divided by kon, was obtained.
Supplementary Material
Acknowledgments
We thank the Mechanobiology Institute protein expression facility for protein purification. The research was funded by the Singapore Ministry of Education Academic Research Fund Tier 3 (MOE2016-T3-1-002); the National Research Foundation (NRF), Prime Minister’s Office, Singapore, under its NRF Investigatorship Programme (NRF Investigatorship Award No. NRF-NRFI2016-03); the NRF, Prime Minister’s Office, Singapore, and the Ministry of Education under the Research Centres of Excellence programme; and a Human Frontier Science Program RGP00001/2016 grant to J.Y. This work was also supported by National Institutes of Health R01 HL-127711 to A.V.K.
Abbreviations used:
- αE
αEpithelial
- αN
αNeuronal
- αT
αTestes
- Fc
critical force
- ICD
intercalated disk
- koff
dissociation rate
- kon
association rate
- M
middle
- VBS
vinculin-binding site
- VD1
vinculin head domain 1.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-02-0102) on July 17, 2019.
REFERENCES
- Aggarwal T, Materassi D, Davison R, Hays T, Salapaka M. (2012). Detection of steps in single molecule data. Cell Mol Bioeng , 14–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays JL, Peng X, Tolbert CE, Guilluy C, Angell AE, Pan Y, Superfine R, Burridge K, DeMali KA. (2014). Vinculin phosphorylation differentially regulates mechanotransduction at cell–cell and cell–matrix adhesions. J Cell Biol , 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrmann CM, Grund C, Kuhn C, Hofmann I, Pieperhoff S, Franke WW. (2006). The area composita of adhering junctions connecting heart muscle cells of vertebrates. II. Colocalizations of desmosomal and fascia adhaerens molecules in the intercalated disk. Eur J Cell Biol , 469–485. [DOI] [PubMed] [Google Scholar]
- Brasch J, Harrison OJ, Honig B, Shapiro L. (2012). Thinking outside the cell: how cadherins drive adhesion. Trends Cell Biol , 299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Kuske R, Li H. (2008). Direct observation of Markovian behavior of the mechanical unfolding of individual proteins. Biophys J , 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Fu H, Zhu X, Cong P, Nakamura F, Yan J. (2011). Improved high-force magnetic tweezers for stretching and refolding of proteins and short DNA. Biophys J , 517–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Yuan G, Winardhi RS, Yao M, Popa I, Fernandez JM, Yan J. (2015). Dynamics of equilibrium folding and unfolding transitions of titin immunoglobulin domain under constant forces. J Am Chem Soc , 3540–3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H-J, Pokutta S, Cadwell GW, Bobkov AA, Bankston LA, Liddington RC, Weis WI. (2012). αE-catenin is an autoinhibited molecule that coactivates vinculin. Proc Natl Acad Sci USA , 8576–8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra A, Tabdanov E, Patel H, Janmey PA, Kresh JY. (2011). Cardiac myocyte remodeling mediated by N-cadherin-dependent mechanosensing. Am J Physiol Heart Circ Physiol , H1252–H1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman GM. (1986). Cell adhesion molecules in the regulation of animal form and tissue pattern. Annu Rev Cell Biol , 81–116. [DOI] [PubMed] [Google Scholar]
- Folmsbee SS, Morales-Nebreda L, Van Hengel J, Tyberghein K, Van Roy F, Budinger GS, Bryce PJ, Gottardi CJ. (2014). The cardiac protein αT-catenin contributes to chemical-induced asthma. Am J Physiol Lung Cell Mol Physiol , L253–L258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavard J, Lambert M, Grosheva I, Marthiens V, Irinopoulou T, Riou J-F, Bershadsky A, Mège R-M. (2004). Lamellipodium extension and cadherin adhesion: two cell responses to cadherin activation relying on distinct signalling pathways. J Cell Sci , 257–270. [DOI] [PubMed] [Google Scholar]
- Goossens S, Janssens B, Bonné S, De Rycke R, Braet F, van Hengel J, Van Roy F. (2007). A unique and specific interaction between αT-catenin and plakophilin-2 in the area composita, the mixed-type junctional structure of cardiac intercalated discs. J Cell Sci , 2126–2136. [DOI] [PubMed] [Google Scholar]
- Hino N, Ichikawa T, Kimura Y, Matsuda M, Ueda K, Kioka N. (2019). An amphipathic helix of vinexin α is necessary for substrate stiffness-dependent conformational change in vinculin. J Cell Sci , 217349. [DOI] [PubMed] [Google Scholar]
- Imamura Y, Itoh M, Maeno Y, Tsukita S, Nagafuchi A. (1999). Functional domains of α-catenin required for the strong state of cadherin-based cell adhesion. J Cell Biol , 1311–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama N, Tanaka N, Abe K, Yang YJ, Abbas YM, Umitsu M, Nagar B, Bueler SA, Rubinstein JL, Takeichi M. (2013). An autoinhibited structure of α-catenin and its implications for vinculin recruitment to adherens junctions. J Biol Chem , 15913–15925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens B, Goossens S, Staes K, Gilbert B, van Hengel J, Colpaert C, Bruyneel E, Mareel M, Van Roy F. (2001). αT-catenin: a novel tissue-specific β-catenin-binding protein mediating strong cell-cell adhesion. J Cell Sci , 3177–3188. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Craig SW. (1995). F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature , 261. [DOI] [PubMed] [Google Scholar]
- Kay BK, Thai S, Volgina VV. (2009). High-throughput biotinylation of proteins. In: High Throughput Protein Expression and Purification, New York: Springer, 185–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S, Liu R, Lim CT, Yan J. (2016). Uncovering mechanosensing mechanisms at the single protein level using magnetic tweezers. Methods , 13–18. [DOI] [PubMed] [Google Scholar]
- Li J, Goossens S, van Hengel J, Gao E, Cheng L, Tyberghein K, Shang X, De Rycke R, Van Roy F, Radice GL. (2012). Loss of αT-catenin alters the hybrid adhering junctions in the heart and leads to dilated cardiomyopathy and ventricular arrhythmia following acute ischemia. J Cell Sci , 1058–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Newhall J, Ishiyama N, Gottardi C, Ikura M, Leckband DE, Tajkhorshid E. (2015). Structural determinants of the mechanical stability of α-catenin. J Biol Chem , 18890–18903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkel AR, Kroemker M, Bubeck P, Ronsiek M, Nikolai G, Jockusch BM. (1994). Characterization of an F-actin-binding domain in the cytoskeletal protein vinculin. J Cell Biol , 1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noorman M, van der Heyden MA, van Veen TA, Cox MG, Hauer RN, de Bakker JM, van Rijen HV. (2009). Cardiac cell–cell junctions in health and disease: electrical versus mechanical coupling. J Mol Cell Cardiol , 23–31. [DOI] [PubMed] [Google Scholar]
- Pang SM, Le S, Yan J. (2018). Mechanical responses of the mechanosensitive unstructured domains in cardiac titin. Biol Cell , 65–76. [DOI] [PubMed] [Google Scholar]
- Pardo JV, Siliciano JDA, Craig SW. (1983). Vinculin is a component of an extensive network of myofibril-sarcolemma attachment regions in cardiac muscle fibers. J Cell Biol , 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Nelson ES, Maiers JL, DeMali KA. (2011). New insights into vinculin function and regulation. Int Rev Cell Mol Biol , 191–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieperhoff S, Franke WW. (2007). The area composita of adhering junctions connecting heart muscle cells of vertebrates–IV: coalescence and amalgamation of desmosomal and adhaerens junction components–late processes in mammalian heart development. Eur J Cell Biol , 377–391. [DOI] [PubMed] [Google Scholar]
- Pokutta S, Choi H-J, Ahlsen G, Hansen SD, Weis WI. (2014). Structural and thermodynamic characterization of cadherin· β-catenin· α-catenin complex formation. J Biol Chem , 13589–13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S, Drees F, Takai Y, Nelson WJ, Weis WI. (2002). Biochemical and structural definition of the l-afadin-and actin-binding sites of α-catenin. J Biol Chem , 18868–18874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S, Drees F, Yamada S, Nelson WJ, Weis WI. (2008). Biochemical and Structural Analysis of α-Catenin in Cell–Cell Contacts, London, UK: Portland Press Limited. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokutta S, Weis WI. (2000). Structure of the dimerization and β-catenin-binding region of α-catenin. Mol Cell , 533–543. [DOI] [PubMed] [Google Scholar]
- Rangarajan ES, Izard T. (2012). The cytoskeletal protein α-catenin unfurls upon binding to vinculin. J Biol Chem , 18492–18499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan ES, Izard T. (2013). Dimer asymmetry defines α-catenin interactions. Nat Struct Mol Biol , 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc , 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh F, Ross RS, Chen J. (2009). Cell-cell connection to cardiac disease. Trends Cardiovasc Med , 182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu K, Dutton AH, Geiger B, Singer S. (1981). Ultrastructure of chicken cardiac muscle as studied by double immunolabeling in electron microscopy. Proc Natl Acad Sci USA , 7619–7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermij SH, Abriel H, van Veen TA. (2017). Refining the molecular organization of the cardiac intercalated disc. Cardiovasc Res , 259–275. [DOI] [PubMed] [Google Scholar]
- Wang Q, Lin JL-C, Wu K-H, Wang D-Z, Reiter RS, Sinn HW, Lin C-I, Lin JJ-C. (2012). Xin proteins and intercalated disc maturation, signaling and diseases. Front Biosci , 2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M, Uchida N, Imamura Y, Nagafuchi A, Fujimoto K, Uemura T, Vermeulen S, Van Roy F, Adamson ED, Takeichi M. (1998). α-Catenin-vinculin interaction functions to organize the apical junctional complex in epithelial cells. J Cell Biol , 847–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EE, Kroemker M, Rüdiger A-H, Jockusch BM, Rüdiger M. (1998). Vinculin is part of the cadherin–catenin junctional complex: complex formation between α-catenin and vinculin. J Cell Biol , 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickline ED, Dale IW, Merkel CD, Heier JA, Stolz DB, Kwiatkowski AV. (2016). αT-Catenin is a constitutive actin-binding α-catenin that directly couples the cadherin· catenin complex to actin filaments. J Biol Chem , 15687–15699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Dokurno P, Tonks NK, Barford D. (2001). Crystal structure of the M‐fragment of α‐catenin: implications for modulation of cell adhesion. EMBO J , 3645–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Goult BT, Chen H, Cong P, Sheetz MP, Yan J. (2014a). Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci Rep , 4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M, Qiu W, Liu R, Efremov AK, Cong P, Seddiki R, Payre M, Lim CT, Ladoux B, Mege R-M. (2014b). Force-dependent conformational switch of α-catenin controls vinculin binding. Nat Commun , 4525. [DOI] [PubMed] [Google Scholar]
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. (2010). α-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol , 533. [DOI] [PubMed] [Google Scholar]
- Yuan G, Le S, Yao M, Qian H, Zhou X, Yan J, Chen H. (2017). Elasticity of the transition state leading to an unexpected mechanical stabilization of titin immunoglobulin domains. Angew Chem , 5582–5585. [DOI] [PubMed] [Google Scholar]
- Zemljic-Harpf AE, Miller JC, Henderson SA, Wright AT, Manso AM, Elsherif L, Dalton ND, Thor AK, Perkins GA, McCulloch AD. (2007). Cardiac-myocyte-specific excision of the vinculin gene disrupts cellular junctions, causing sudden death or dilated cardiomyopathy. Mol Cell Biol , 7522–7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemljic-Harpf AE, Ponrartana S, Avalos RT, Jordan MC, Roos KP, Dalton ND, Phan VQ, Adamson ED, Ross RS. (2004). Heterozygous inactivation of the vinculin gene predisposes to stress-induced cardiomyopathy. Am J Pathol , 1033–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics , 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.