Abstract
The linker of the nucleoskeleton and cytoskeleton (LINC) complex is formed by the conserved interactions between Sad-1 and UNC-84 (SUN) and Klarsicht, ANC-1, SYNE homology (KASH) domain proteins, providing a physical coupling between the nucleoskeleton and cytoskeleton that mediates the transfer of physical forces across the nuclear envelope. The LINC complex can perform distinct cellular functions by pairing various KASH domain proteins with the same SUN domain protein. For example, in Caenorhabditis elegans, SUN protein UNC-84 binds to two KASH proteins UNC-83 and ANC-1 to mediate nuclear migration and anchorage, respectively. In addition to distinct cytoplasmic domains, the luminal KASH domain also varies among KASH domain proteins of distinct functions. In this study, we combined in vivo C. elegans genetics and in silico molecular dynamics simulations to understand the relation between the length and amino acid composition of the luminal KASH domain, and the function of the SUN–KASH complex. We show that longer KASH domains can withstand and transfer higher forces and interact with the membrane through a conserved membrane proximal EEDY domain that is unique to longer KASH domains. In agreement with our models, our in vivo results show that swapping the KASH domains of ANC-1 and UNC-83, or shortening the KASH domain of ANC-1, both result in a nuclear anchorage defect in C. elegans.
INTRODUCTION
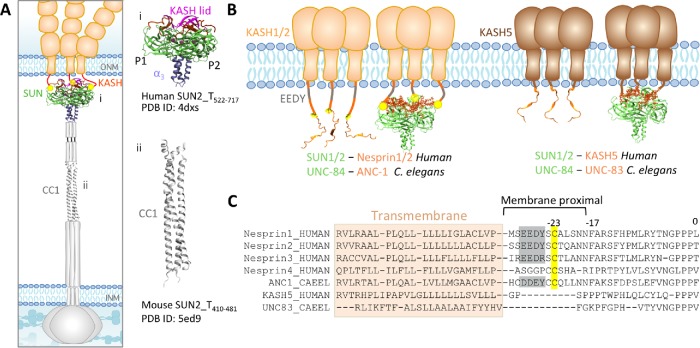
The double-layered nuclear envelope acts as a physical barrier between the constituents of the nucleus and the cytoplasm. The linker of the nucleoskeleton and cytoskeleton (LINC) complexes span this physical barrier and regulate the physical connection between the interior of the nucleus and the cytoplasm during various cellular functions. Through this physical connection, the LINC complex withstands and transfers mechanical forces across the nuclear envelope (Lombardi et al., 2011; Cain et al., 2014; Arsenovic and Conway, 2018; Jahed and Mofrad, 2018, 2019). LINC complexes are composed of Sad1/UNC-84 (SUN) proteins that are anchored to the inner nuclear membrane (INM), and Klarsicht/ANC-1/SYNE homology (KASH) proteins that are anchored to the outer nuclear membrane (ONM). The large cytoplasmic domains of KASH proteins bind to various elements of the cytoskeleton, whereas their 10–30 amino acid KASH domains reside in the perinuclear space (PNS) where they bind to SUN proteins (Figure 1). Several SUN–KASH pairs have been identified to date and each performs distinct functions within the cell (Padmakumar et al., 2005; Crisp et al., 2006; McGee et al., 2006; Kim et al., 2015; Jahed et al., 2016).
FIGURE 1:
Structure of LINC complexes at the nuclear envelope. (A) Schematic representation of LINC complex: inner nuclear membrane (INM) protein SUN, and outer nuclear membrane (ONM) protein KASH. (i) Structure of the human SUN2–KASH2 hexamer and (ii) structure of the coiled-coil (CC1) region of mouse SUN2. (B) Model of SUN–KASH interactions at the ONM. SUN1 or SUN2 in humans binds to Nesprin1, Nesprin2. In C. elegans, ANC-1 binds to UNC-84. KASH5 (human), and UNC-83 (C. elegans) are shorter in the length of their KASH domains and also bind to SUN1/2 and UUNC-84, respectively. (C) Sequences of transmembrane and KASH domains of longer KASH domains (Nesrpin1–4 in humans and ANC-1 in C. elegans), as well as shorter KASH domains (KASH5 in humans and UNC-83, KDP-1, and ZYG-12 in C. elegans). The transmembrane domains are highlighted; a conserved C-23 and EEDY motif are highlighted on the sequences of KASH domains of longer lengths, namely, Nesprin1–3 and ANC-1.
Interestingly, different KASH domain proteins can independently bind to the same SUN protein to mediate distinct cellular functions. For example, in mammals the SUN protein SUN1 can transiently associate with KASH protein KASH5 in the PNS, and with telomeres in the nucleus to mediate microtubule-dependent meiotic chromosome movement (Horn et al., 2013b). On the other hand, SUN1 can bind to Nesprin1 and Nesprin2 to mediate actin-dependent nuclear movement (Padmakumar et al., 2005; Haque et al., 2006; Yu et al., 2011; Nishioka et al., 2016). Similarly, in Caenorhabditis elegans SUN protein UNC-84 transiently binds to KASH protein UNC-83 in embryonic hypodermal cells to mediate microtubule-dependent nuclear migration during development (Starr et al., 2001; McGee et al., 2006; Fridolfsson et al., 2010; Bone et al., 2014). Later, the same SUN protein UNC-84 independently binds to a different KASH protein, ANC-1, to anchor nuclei in place for several days (Starr and Han, 2002; Cain et al., 2018).
Crystal structure of the SUN–KASH complex
The crystal structure of the conserved regions of SUN2 in complex with the KASH domain of Nesprin1/2 revealed a trimeric SUN domain that binds to three KASH peptides simultaneously (Figure 1; PDB ID: 4DXS; Sosa et al., 2012, 2013). In this structure, residues 0 to –17 of each KASH peptide of Nesprin2 bind in a groove formed by two neighboring promoters of the SUN trimer. This groove is formed by an ∼20 residue β-hairpin extending from the SUN domain of protomer 1, known as the “KASH-lid,” and the β-sandwich core of SUN protomer 2 (Figure 1; Sosa et al., 2012). After interactions with this groove, membrane proximal regions consisting of residues −18 to −23 interact exclusively with protomer 2. The SUN–KASH interaction is further enhanced by a disulfide bond formed between cysteine –23 on KASH and conserved cysteine 563 on protomer 2 of SUN2. The remaining membrane proximal residues of KASH between –23 and the transmembrane domain (residues –24 to –30), including a conserved EEDY motif are thought to not interact with SUN.
Despite being well conserved, the luminal domains of KASH5 and UNC-83 are much shorter than the luminal domains of mammalian Nesprins 1–4, C. elegans ANC-1 (Figure 1). Both UNC-83 and KASH5 lack the membrane proximal interaction domain, including the conserved cysteine at –23 (Figure 1), which is suggested to form a disulfide bond with the SUN domain (Sosa et al., 2012; Jahed et al., 2015; Cain et al., 2018). Interestingly, UNC-83 and KASH5 have relatively short-term roles in nuclear migration and meiotic chromosome movements, respectively. Herein, we combined in silico molecular dynamics (MD) simulations with in vivo C. elegans genetics to explore the role of the KASH domain length in the dynamics and function of LINC complexes. Specifically, we asked: how does the SUN–KASH complex with a shorter KASH domain withstand and transmit tensile forces compared with longer KASH domains? Does swapping long and short KASH domains between ANC-1 and UNC-83 disrupt nuclear positioning? What is the role of the membrane proximal residues in the dynamics of the LINC complex at the membrane? Our results show that the specific length of the KASH domain is important for force transmission and LINC function in vivo and in simulations. Our results also suggest that the membrane proximal EEDY motifs of longer KASH domains may play a role in anchoring the SUN–KASH complex to the ONM.
RESULTS
Longer KASH domains transfer higher forces
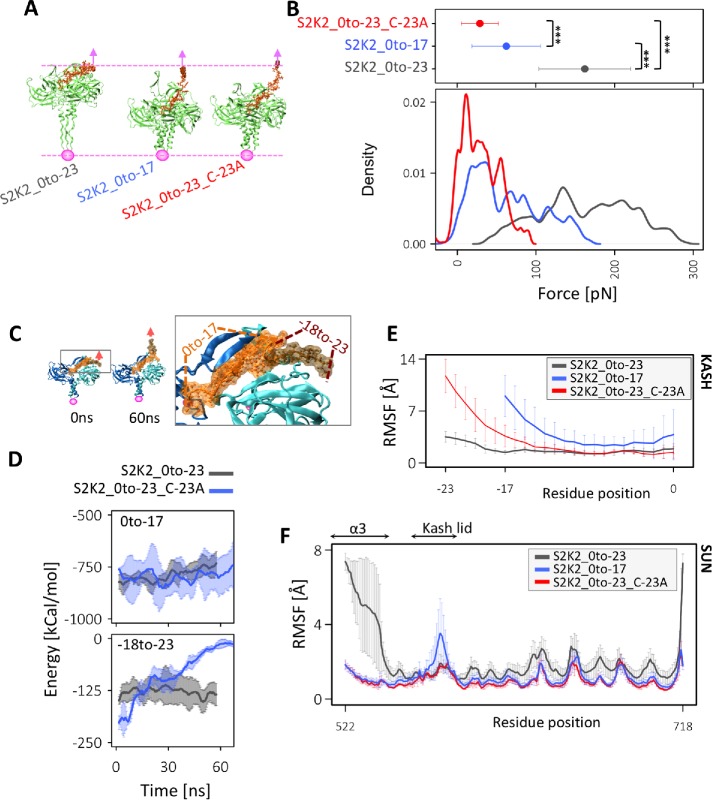
We previously developed a molecular model of the LINC complex under tensile forces and showed that the presence of a conserved cysteine residue at position –23 plays an important role in nuclear positioning, and the transmission of maximal forces across the complex (Jahed et al., 2015; Cain et al., 2018; Jahed and Mofrad, 2018). In this work, we set out to determine whether the length of the KASH domain bound to SUN also affects nuclear positioning and force transmission across LINC. To this end, we deleted residues in the membrane proximal part of KASH domains (–18 to –23) and obtained a model of SUN2 in complex with a shorter length KASH2 peptide (S2K2_0to-17). We then compared the molecular mechanisms of force transmission across this model with the previously developed models of SUN2-KASH2, namely, S2K2_0to-23 and S2K2_0to-23_C-23A by applying forces at a constant velocity of 0.5 Å/ns to the membrane proximal end residue of the KASH peptide in each model (Figure 2A). The forces required to displace the KASH peptide by 30 Å (equivalent to pulling for 60 ns at 0.5 Å/ns) were compared among the three models (Figure 2A). Note that the reported forces were averaged over the 60 ns simulation times as well as over three independent runs for each model where forces were sampled at 2000 fs (Figure 2B). These results suggest that the model with the longest KASH domain, which contains C-23, can withstand average forces that are on average 97 pN higher than S2K2_0to-17 and 131 pN higher than S2K2_0to-23_C-23A (p value < 2.2e-16; Figure 2B). Interestingly, a short KASH domain, S2K2_0to-17, endured significantly higher forces than the longer KASH with a cysteine mutation, S2K2_0to-23_C-23A (p value < 2.2e-16) suggesting that in the absence of C-23, a longer KASH domain would transmit forces less efficiently than a 17-residue-long KASH domain (Figure 2B). We therefore hypothesized that in the absence of C-23, residues –18 to –23 interfere with the stability of the SUN–KASH complex under tensile forces. This could be a reason why these residues were rapidly lost in KASH peptides that lack C-23 (i.e., mammalian KASH5 and C. elegans UNC-83; see Figure 1C). To test this, we calculated the nonbonded interaction energies between two regions of the KASH peptide (region1: residues 0 to –17, or region2: residues –18 to –23; Figure 2C), and the SUN2 trimer in the S2K2_0to-23 and S2K2_0to-23_C-23A models. Our results show that the energies of both regions are highly stable under force in the S2K2_0to-23 model (Figure 2D). Additionally, the energy between region1 and the SUN2 trimer is highly stable under force in the S2K2_0to-23_C-23A model (Figure 2D). However, once C-23 is mutated, region2 consisting of residues –18 to –23 dissociates with the SUN2 trimer under force, and the energies between this region and the SUN2 trimer abruptly reduces to zero (Figure 2, C and D). These results suggest that KASH domains that lack the cysteine residues at position –23, can transmit higher forces if residues between positions –18 and –23 are also absent.
FIGURE 2:
Force transmission from KASH to SUN. (A) Representative images of the final simulation frames of each SUN–KASH model after 60 ns of pulling. (B) Magnitude of forces required to pull on KASH for 60 ns at 0.5 Å/ns (i.e., total displacement of 30 Å), average values of force for three independent 60-ns simulation runs are shown at the top (averaged over time and simulation run; error bars correspond to 1 SD), density plots are shown at the bottom; ***, P value < 2.2e-16). (C) Conformation of S2K2_0to-23_C-23A model before and after force application showing the detachment of residues –18 to –23 and zoomed view of SUN2–KASH2 interaction showing residues 0 to –17 and residues –18 to –23. (D) Nonbonded interaction energies between residues 0 and –17 and SUN trimer (top) or residues –18 to –23 (bottom) in S2K2_0to-23_C-23A indicating the dispensability of residues –18 to –23 in the absence of C-23. (E) Average RMSF of KASH peptides under force, where average RMSF values of each KASH residue are also averaged over three independent simulation runs, and three KASH peptides of the SUN2–KASH hexamer in each run. Error bars show the range of data over three runs and three KASH peptides in each run. (F) Average RMSF of SUN under force where average RMSF values of each SUN2 residue are also averaged over three independent simulation runs, and three SUN2 protomers of the SUN2–KASH2 hexamer in each run. Error bars show the range of data over three runs and three SUN2 protomers in each run.
In our previous models of SUN2–KASH2 under tension, we had also shown that forces applied on KASH are directly transmitted to the coiled-coil regions of SUN2 formed by α3, in the S2K2_0to-23 model, but not in the S2K2_0to-23_C-23A. This was evident from the average root-mean-square fluctuations (RMSF) of KASH (Figure 2E) and SUN2 (Figure 2, E and F) in each model under tension. We computed and compared the RMSF of SUN2 and KASH in the S2K2_0to-17 model with the previous models all pulled for 60 ns (30 Å displacement; Figure 2, E and F). As shown in Figure 2E, in the S2K2_0to-23_C-23A model, the highest RMSF values are observed in residues –23 to –17, indicating that forces on residue –23 result in fluctuations in residues –23 to –17 of the KASH peptide in this model. Similarly, the average RMSF values of residues of the KASH peptide in the S2K2_0to-17 model indicate that forces on the end residue of this KASH peptide (residue –17) result in fluctuations in all residues of the KASH peptide. On the other hand, in the S2K2_0to-23 model, the RMSF values of all KASH residues are very small compared with the other two models indicating that forces on the end residue of KASH do not result in fluctuations of the KASH peptide in this model. Instead, in the S2K2_0to-23 model, forces on KASH result in fluctuations in SUN, and most prominently in the α3 regions as shown in Figure 2F. In the S2K2_0to-17, some forces are transferred to the KASH-lids of SUN, resulting in higher RMSF values for these regions. These results suggest that forces are most efficiently transmitted to the CC regions of SUN in longer KASH peptides.
Conserved EEDY motif links SUN–KASH to the membrane
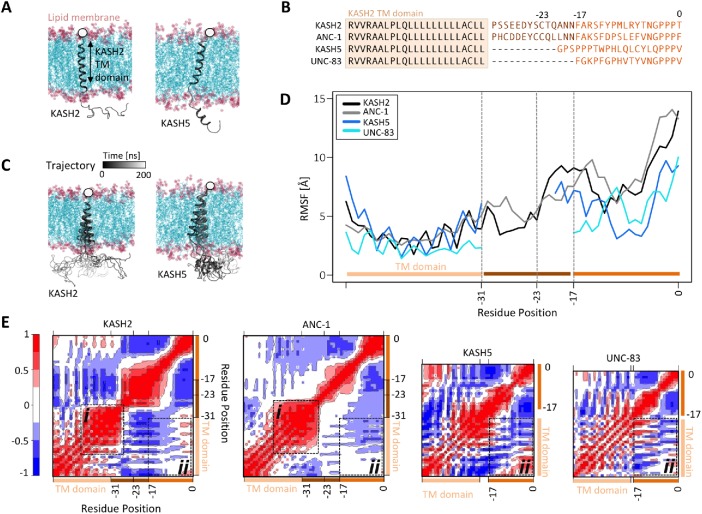
The results from our model of SUN2–KASH2 under force provided insights into the mechanism of force transfer along the LINC complex for various lengths of KASH peptides. In the next step, we asked whether the length of the KASH peptide would alter the dynamics of KASH at the membrane. To answer this question, we developed a model of luminal KASH domains of KASH2, ANC1, KASH5, and UNC-83 using homology modeling. To model the anchorage of KASH domains to the outer nuclear membrane, we also included a transmembrane (TM) domain in each model (Figure 3, A and B). For all four KASH peptides, the TM domain was modeled using the sequence of the TM domain of KASH2 (Figure 3B). All TM domains were inserted into a lipid bilayer as shown in Figure 3A. After relaxation and equilibration of the model (see Materials and Methods for more details), the position of the Cα atoms of the terminal residue on the cytoplasmic side of the TM domains of all four models were fixed in position (Figure 3A). Finally, the dynamics of the four models embedded in the membrane were monitored over 180 ns MD simulation times. Representative trajectories of the positions of KASH2 and KASH5 at the membrane are shown over the 180 ns simulation time in Figure 3C. To compare the dynamics of all four models, we computed the average RMSF of all residues in each KASH peptide (Figure 3D), as well as the average dynamical cross-correlation of residues over the simulation time (Figure 3E). To better compare the fluctuations of similar residue positions in KASH peptides of various lengths, the RMSF values shown in Figure 3D were aligned based on sequence alignment of the residues as shown in Figure 3B (hence the gap between positions –31 and –20 in KASH5, and –31 and –17 in UNC-83). For KASH2 and ANC-1, the residues immediately after the TM domain (–31 to –23) exhibited lower fluctuations compared with residues –23–0 and the RMSF increased after residue –23 in these two models. The dynamical cross-correlation analysis also revealed a strong positive correlation between the fluctuations of end residues of the TM domain, and the luminal residues immediately after the TM domain in KASH2 and ANC-1 (Figure 3Ei). On the other hand, the TM domain did not show significant correlations with residues 0 to –17 in KASH2 or ANC-1, but showed strong negative correlations between the same regions in KASH5 and UNC-83 (Figure 3Eii). These results suggest that the movement of the KASH peptide is correlated with the TM domain distinctly in the four different KASH peptides. In ANC-1 and KASH2, the residues immediately after the TM domain move in the same direction as the TM domain, in other words their movements are coupled with the movement of their TM domains, and the remaining luminal residues either move in the opposite direction (residues –27 to –14 in KASH2) or show no significant correlation with the TM domain (residues –14–0 in KASH2 and –23–0 in ANC-1). In KASH5 and UNC-83, the KASH domain residues 0 to –17 move in the opposite direction as the TM domain. This is also evident from the trajectory of KASH5 shown in Figure 3C where the luminal domain moved farther to the right as the TM domain moves to the left over the simulation time.
FIGURE 3:
Dynamics of KASH peptides at the outer nuclear membrane. (A) Molecular dynamics (MD) model of KASH domain proteins anchored to the outer nuclear membrane (KASH2 and KASH5 are shown on the left and right, respectively). Each structural model consists of the transmembrane domain of KASH2 embedded in the membrane, and the luminal domain of the corresponding KASH domain protein (i.e., KASH2, ANC-1, KASH5, or UNC-83). The position of the Cα atoms of the terminal residue on the cytoplasmic side is fixed (shown with a white circle). (B) Sequences used for modeling the KASH domain proteins KASH2, ANC-1, KASH5, and UNC-83. All models consist of the same transmembrane domain. (C) The trajectories of KASH2 (left) and KASH5 (right) over 200 ns MD simulation time. (D) Average RMSF of KASH2, ANC-1, KASH5, and UNC-83 (a gap is shown between residue –17 or –20 and the TM domain for KASH5 and UNC-83 to better compare the fluctuations of KASH domains). (E) Dynamic cross-correlation heatmap between various regions of each model averaged over MD simulation time. Positive cross-correlations represent fluctuations/displacements in the same direction, whereas negative cross-correlation values represent fluctuations/displacements in the opposite direction. (i) Dynamical cross-correlation between the TM domains and luminal domain residues immediately after the TM domain in KASH2 and ANC-1. (ii) Dynamical cross-correlation between the TM domains and residues 0 to –17 in KASH2, ANC-1, KASH5, and UNC-83.
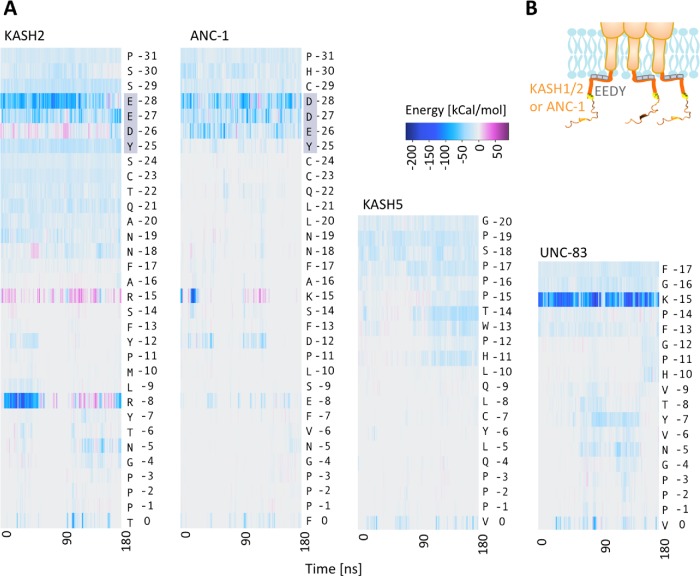
We predicted that interactions with the lipid membrane may be responsible for the reduced fluctuations in residues –31 to –23 in KASH2 and ANC-1, so we calculated the nonbonded interaction energies between each KASH peptide and the lipid membrane. The total pairwise nonbonded interaction energies were calculated between each residue in the four KASH peptides and the lipid membrane over simulation time as shown in Figure 4A. These results show that in KASH2, an EEDY motif forms long-term nonbonded interactions with the lipid membrane over the time of our simulations. ANC-1 also has a similar DDEY motif at the same position, which also interacts with the membrane (Figure 4, A and B). No such domains are present in the KASH domains of KASH5 or UNC-83; however, a lysine residue at position –15 (K-15) in UNC-83 also formed strong nonbonded interactions with the lipid membrane. The results obtained from our MD simulations suggest that the conserved EEDY in longer KASH domains may alter the dynamics of the KASH domain by binding to the lipid membrane.
FIGURE 4:
Interactions of KASH domains with the outer nuclear membrane. (A) Total nonbonded interaction energies between all residues of the luminal KASH domains of KASH2, ANC-1, KASH5, and UNC-83, and the lipid membrane over simulation time. The conserved EEDY motif in KASH2 and ANC-1 interacts with the membrane. (B) Schematic representation of EEDY motif interacting with the outer nuclear membrane.
Swapping KASH domains disrupts LINC function in vivo
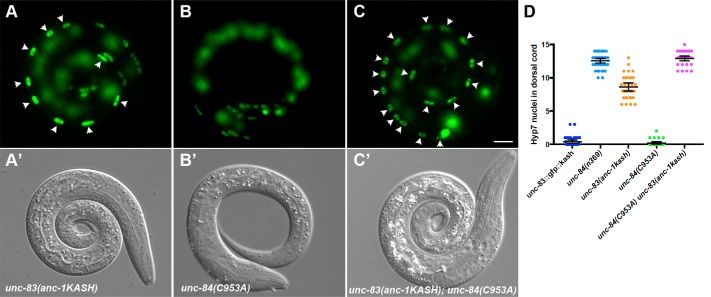
Our simulations (Figure 2, A and B) predict that shorter KASH proteins can transmit more forces across LINC complexes than longer KASH domains with mutant cysteines at –23. We set out to test this prediction in the context of a live, developing organism where nuclear positioning is easily followed—the hypodermis of C. elegans (Starr and Han, 2002; Fridolfsson et al., 2010). We investigated how changing the lengths of the UNC-83 or ANC-1 KASH domains affected their in vivo function in C. elegans developing hypodermal cells using our established nuclear migration and nuclear anchorage assays (Bone et al., 2014; Cain et al., 2018; Fridolfsson et al., 2018). We first used our nuclear migration assay to test the ability of different KASH peptides to mediate nuclear movements in embryonic hyp7 precursors. We hypothesized that longer KASH domains with a cysteine at –23 would form a stable interaction with SUN proteins and mediate nuclear migration normally. Unexpectedly, replacing the shorter KASH domain in unc-83 with the longer KASH from anc-1 to make unc-83 (anc-1KASH), caused a significant nuclear migration defect with 8.4 ± 0.9 (mean ± 95% CI) nuclei in the dorsal cord (Figure 5, A and D). This unc-83(anc-1KASH) nuclear migration defect was intermediate, significantly less severe than unc-84 or unc-83 null animals (Starr et al., 2001; Bone et al., 2014) but significantly more defective than wild type (Figure 5D; P < 0.0001). Our simulations indicated that LINC complexes with longer KASH domains including a cysteine at –23 are able to transmit more force than shorter KASH domains, but our in vivo experiments showed that the longer KASH domain with a –23 cysteine somehow interferes with nuclear migration in embryonic hyp7 precursors. Thus, the longer ANC-1 KASH domain might inhibit the migratory function of UNC-83(ANC-1 KASH) by forming overly stabilized SUN–KASH interactions containing a disulfide bond between UNC-83(ANC-1KASH) and UNC-84.
FIGURE 5:
Nuclear migration defects in KASH-swap alleles in C. elegans. (A–C) Lateral view of L1 animals in a background expressing hypodermal nuclear GFP. (A′–C′) Corresponding DIC images of the same animals. (A) unc-83(yc55[anc-1kash]); strain UD597. (B) unc-84(yc35[C953A]); strain UD605. (C) unc-83(yc55[anc-1kash]) unc-84(yc35[C953A]); strain UD604. Arrowheads mark nuclei that are abnormally in the dorsal cord, representing failed migrations. Scale bar, 10 μm. (D) Quantification of nuclear migration defects. Each point represents the number of nuclei in the dorsal cord of a single animal. Means with 95% CI error bars are shown.
We then tested the extent to which removing the possibility of a disulfide bond between UNC-83 and UNC-84 might affect nuclear migration. We previously showed that mutating of the conserved cysteine in the SUN domain of UNC-84 in unc-84(C953A) mutants had no effect on nuclear migration in the presence of the shorter, wild-type KASH domain of UNC-83 (Cain et al., 2018). We engineered a LINC complex with a long KASH domain but a mutation in the cysteine in the SUN domain so that no disulfide bond could form. unc-84(C963A); unc-83(anc-1KASH) double mutants had a completely penetrant nuclear migration defect (Figure 5, C and D). Thus, consistent with our simulations (Figure 2), in the absence of a disulfide bond between SUN and KASH in vivo, a short KASH domain appears more functional than a long one.
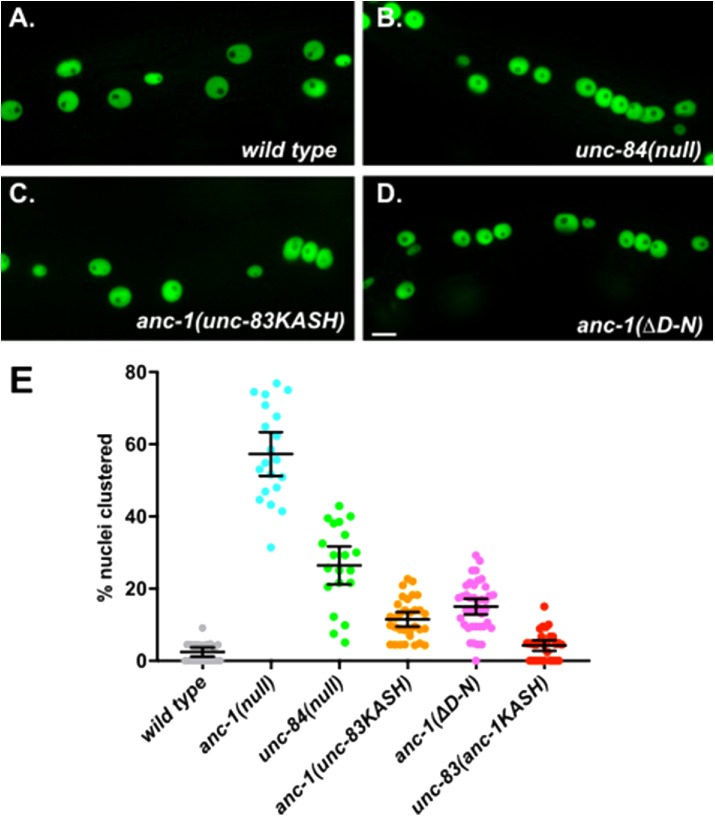
After nuclei move using the KASH protein UNC-83, they anchor in place using the giant KASH protein ANC-1 (Starr and Han, 2002). We tested the hypothesis that the longer KASH domain of ANC-1 is required for efficient nuclear anchorage. We replaced the long KASH domain of endogenous ANC-1 with the shorter UNC-83 KASH domain to make anc-1(unc-83KASH) mutant animals. We observed a significant nuclear anchorage defect with 11.5 ± 2.0 (mean ± 95% CI) percent of nuclei clustered in anc-1(unc-83KASH) animals compared with 2.4 ± 1.3 percent of nuclei clustered in wild type (P = 0.0005 in a one-way analysis of variance with multiple comparisons; Figure 6, A, C, and E). However, the anc-1(unc-83KASH) nuclear anchorage defect was not as severe as the unc-84(null) with 26.4 ± 5.3 percent clustered nuclei (P < 0.0001; Figure 6, B, C, and E), supporting our simulations that a short KASH is at least partially functional. Alternatively, we deleted 11 residues from –28 to –18 of the endogenous ANC-1 KASH domain to make anc-1(ΔD-N) mutant animals, which had 15.0 ± 2.2 percent nuclei clustered (Figure 6, D and E). Thus, both anc-1(ΔD-N) and anc-1(unc-83KASH) had similar defects (P = 0.40), suggesting that either short KASH domain is partially functional to anchor nuclei.
FIGURE 6:
Nuclear anchorage defects in KASH-swap alleles in C. elegans. (A–D) Lateral views of young adult C. elegans are shown. Hypodermal nuclei are marked with nuclear GFP from ycEx249 in the following genetic backgrounds: (A) wild type; strain UD522. (B) unc-84(n369); strain UD532. (C) unc-83(yc55[anc-1kash]); strain UD582. (D) anc-1(yc60[ΔD-N]); strain UD589. Scale bar, 10 μm. (E) Quantification of nuclear anchorage defects. Each point represents the percentage of clustered nuclei in one side of a single animal. Means with 95% CI error bars are shown.
In conclusion, our in vivo data are consistent with a model that a short KASH domain can transmit forces more efficiently than a long KASH without a disulfide bond. However, it is important to note that comparing the function of short KASH domains to long KASH domains stabilized by disulfide bonds to SUN proteins is more difficult to interpret, because other factors may be involved in vivo that are missing in our simulations.
Transfer of forces beyond the SUN domain
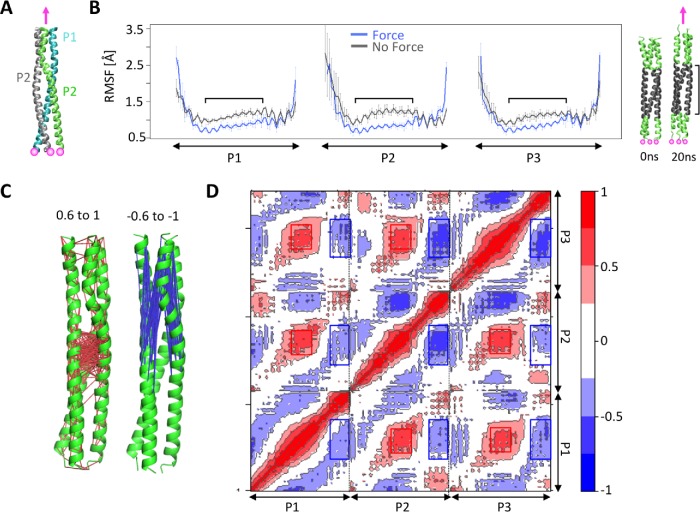
We showed that for long KASH peptides, forces on the terminal residue of the KASH domain were transferred to the trimeric CC domains of SUN (Figure 2). We next asked how tensile forces are further transmitted across the trimeric CC domains of SUN. Using the solved crystal structure of CC1 of SUN2, we applied tensile forces on the C-terminal residue of each of its three protomers, namely, P1, P2 and P3, while fixing their N-terminal residues (Figure 7A). To determine the dynamics of CC1 under force, we compared the per-residue RMSF values of P1, P2, and P3 in force and no force conditions (Figure 7B). We expected the forces applied at the C-terminus to be further transferred to the N-terminus of CC1. Indeed, the highest fluctuations were observed at the two terminal residues of each protomer when forces were applied, indicating that some of the forces at the C-terminus are directly transferred to the N-terminus. Surprisingly, the RMSF values of residues at the central core of each protomer of CC1 were significantly lower under force as compared with a no force condition (Figure 7B), indicating that the fluctuations in these regions are reduced when CC1 is under tensile forces (Figure 7B). Next, to obtain further insights into the dynamics of CC1 under force and to determine how the fluctuations of various residues across protomers are coupled, we also calculated the dynamical cross-correlations between residues on P1, P2, and P3 in our simulations (Figure 7, C and D). Our results show that the residues at the central core of CC1 exhibit strong positive correlations (i.e., move together in the same direction). On the other hand, the C-terminal residues on which forces are applied show strong negative correlations with the residues at the central core of CC1 (Figure 7, C and D). Specifically, the C-terminal residue of each protomer correlates negatively with the central residues of itself, as well the other two protomers (P2 and P3; Figure 7D), indicating that these regions move in opposite directions during our simulations. These results suggest that cytoskeletal forces may be translated to the residues at the central core of CC1, resulting in some conformational changes in these regions.
FIGURE 7:
Force transmission across the CC domain of SUN2. (A) Model of CC1 of SUN2 under force. Three protomers in the SUN2 CC1 trimer (P1, P2, and P3) were pulled at a constant velocity in the direction shown with a pink arrow. Forces were applied to the C-terminal residue of each protomer while the Cα residues of the N-terminal residues were fixed (shown with pink circles). (B) RMSF of CC1 protomers (P1, P2, and P3) under force, compared with a no force control. Average values are shown for three independent simulation runs, and error bars show data spread. Regions where the RMSF is different between the two conditions are indicated on the plot and mapped onto the structure on the left. (C, D) Dynamic cross-correlation between various regions of each model under force, averaged over MD simulation time. Positive cross-correlations (red) represent fluctuations/displacements in the same direction, whereas negative cross-correlation values (blue) represent fluctuations/displacements in the opposite direction. The residues that are positively correlated with a correlation value between 0.6 and 1 are connected with red lines and residues that are negatively cross-correlated with a correlation value between –0.6 and –1 are connected with blue lines in C, while corresponding residues are represented with red and blue boxes in D.
DISCUSSION
There is direct evidence that the LINC complex is subject to tension at the nuclear envelope, and that the transmission of forces across the LINC complex is essential for several cellular functions (Grady et al., 2005; Bone et al., 2014; Arsenovic et al., 2016; Arsenovic and Conway, 2018). Various KASH proteins pair with SUN proteins to mediate distinct functions of the cell (Jahed et al., 2016, 2018). In addition to their distinct cytoplasmic domains, the transluminal domains of KASH proteins which bind to SUN proteins are also different in length. Specifically, shorter KASH proteins lack a conserved membrane proximal domain that is responsible for additional interactions with SUN proteins (Figure 1). Our results suggest that the lack of this membrane proximal domain reduces the amount of force that the SUN–KASH complex can withstand. Furthermore, our MD simulations show that an EEDY motif, conserved in the membrane proximal domain of long KASH domains including Nesprin1–3 and ANC1, can interact with the lipid membrane (Figures 1 and 4). The interaction of KASH with the lipid membrane could provide additional anchorage points, which would potentially provide even higher LINC stability under force. In agreement with these findings, we found that the membrane proximal domain of ANC-1 is required for nuclear anchorage in C. elegans and a shorter KASH domain is only partially functional to anchor nuclei. Although our results suggest that this membrane proximal EEDY domain is important for the function of longer KASH proteins, it is important to note that in mammals, Nesprin4 lacks this membrane proximal EEDY motif (Figure 1). Nesprin4 is known to be important for microtubule-dependent nuclear positioning (Roux et al., 2009; Horn et al., 2013a). However, compared with Nesprin1 and Nesprin2, the dynamics of Nesprin4 are poorly understood and the implications of missing the EEDY motif between the cysteine and the transmembrane helix are unclear and require further investigations.
Why would different LINC-dependent processes require KASH domains of various lengths? There are several differences between the various processes mediated by different SUN–KASH pairs. One major difference is the time scale of the SUN–KASH interaction, which is only a few minutes for KASH5 during meiosis prophase I, and UNC-83 during nuclear migration in development. On the other hand, Nesprin2 and ANC-1 are required for their respective functions for up to several days. Therefore, one explanation is that the additional interactions between the membrane proximal domains of longer KASH domains, with SUN or the lipid membrane, facilitates the more long-term functions of the LINC complexes that employ KASH proteins with longer KASH domains. Unexpectedly, we also found that the longer KASH domain of ANC-1 can significantly inhibit the migratory function of UNC-83 (Figure 5, A and D), which suggests that the addition of a membrane proximal domain to short KASH proteins can also inhibit their function. These results further confirm that the specific length of the KASH domain is important for specific LINC complex functions. An undesirable formation of disulfide bonds between SUN and the membrane proximal domain of longer KASH, or potential interactions of the EEDY motif of the membrane proximal domain of KASH with the membrane may be the mechanism by which longer KASH domains inhibit functions that require shorter KASH domains, but this would require further testing. In this work, we discuss the differences in the luminal KASH domains in the regulation of LINC complexes. However, the differences between SUN proteins are also important to note. Although the KASH binding sites of SUN1 and SUN2 are highly conserved (Lei et al., 2009; Hennen et al., 2017, 2018; Jahed et al., 2018), there are several structural differences in these proteins that could lead to distinct functions of SUN1 and SUN2 (Jahed et al., 2018; Xu et al., 2018). The pairing of KASH domains with distinct SUN domains could further diversify the functions and regulations of LINC complexes and would be intriguing to investigate further.
It is important to note that there are also several other important differences between the functions of ANC-1 and UNC-83 in nuclear anchorage and migration. For example, ANC-1 anchors the LINC complex to the actin cytoskeleton during nuclear anchorage, whereas UNC-83 is linked to microtubules through motor proteins. Therefore, the magnitude and direction of forces generated during these two distinct processes may also vary and may explain the higher force tolerance of longer KASH proteins for their respective functions.
Finally, our MD results imply that forces are more efficiently transmitted from KASH to the CC regions of SUN in longer KASH domains (Figure 2). Our simulations show that forces on the CC region of SUN can induce conformational changes in the CC domains (Figure 7). Interestingly, studies have shown that the CC domain of SUN2 is not a stable trimeric coiled coil in solution, due to structural defects such as hydrophilic residues in its interhelical packing core (Nie et al., 2016). Our simulations suggest that tensile forces on the LINC complex may induce conformational changes in the interhelical packing core of the CC domains, and hence, the conformation of these regions is likely different under force than that observed in solution.
In summary, we show that the presence or absence of a membrane proximal domain in KASH proteins plays an important role in the specific functions of LINC under force.
MATERIALS AND METHODS
Model of SUN2-KASH2 complex
We downloaded the solved structure of the human SUN2 trimer in complex with three KASH peptides of human Nesprin2 from the Protein Data Bank (PDB ID: 4DXS; Sosa et al., 2012) and visualized using visual molecular dynamics (VMD) software (Humphrey et al., 1996). The KASH peptides in this solved structure were 23 residues long and this modeled structure was accordingly labeled as S2K2_0to-23 in the text. A structural model for a shorter KASH (S2K2_0to-17) in complex with SUN2 was developed by residue deletion in VMD software (Humphrey et al., 1996). The model for KASH in complex with cysteine mutant SUN2 (S2K2_0to-23_C-23A) was also developed as before using the mutator tool in VMD (Jahed et al., 2015; Cain et al., 2018). To apply tensile forces on the SUN–KASH complex we attached a dummy atom to the position of the center of mass of the N-terminal residue of the respective KASH peptide at position –23 for S2K2_0to-23 and S2K2_0to-23_C-23A, or position –17 for S2K2_0to-17, via a virtual spring. The position of the C-terminal residue of SUN2 was also fixed in all models. We then measured the forces between the dummy atom and C-23 using NAno-scale Molecular Dynamics (NAMD; Phillips et al., 2005) as the dummy atom was moved at a constant velocity of 0.05 m/s. A moving window average was applied to the force data using the filter function in R with convolution. All plots were prepared using R software.
Model of KASH in the membrane
Peptides of various lengths were modeled using the Phyre2 Protein Fold Recognition Server in Intensive mode (Kelley et al., 2015). The sequence of the transmembrane domain of Nesprin2 was used for all models, which preceded the KASH domains of KASH2, ANC-1, KASH5, and UNC-83 (sequences are shown in Figure 3B). The transmembrane domain was predicted as an alpha helix for all models and all KASH domains were predicted to be disordered as expected. The modeled KASH peptides were anchored to a diacylglycerol and phospholipid membrane (POPC), which was modeled using the VMD membrane plug-in. To remove any overlap between the modeled disordered KASH domains and the lipids, we first fixed all residues of the alpha helical transmembrane domains of all models and applied a constant velocity to the end residue of each KASH peptide at a constant velocity of 0.05 m/s. The alpha helical transmembrane domains of all four KASH peptides were then inserted perpendicular to the lipid membrane and overlapping lipids were removed. The system was then solvated and ionized and all waters in the membrane region were deleted. Because we assembled the system manually, several minimization and equilibration steps were taken before the final simulations presented in the Results section. First, we performed a simulation in which we fixed all atoms in the system except lipid tails to obtain a fluid-like lipid bilayer (Humphrey et al., 1996; Phillips et al., 2005). Second, we performed a minimization and equilibration step in which harmonics constraints were applied to the four KASH proteins permitting lipids, water, and ions to adapt to the shape of the proteins. Third, the constraints on the proteins were removed and the full system was equilibrated for 60 ns. The average RMSD values of each KASH protein were monitored during equilibration and plateaued in the 60-ns simulation times. In the final simulations, the end residues of the transmembrane domains were fixed in place to resemble the immobile large cytoplasmic domains of KASH proteins, and simulations were conducted for 180 ns.
MD simulations
MD simulations were performed using NAMD scalable MD with the CHARMM force field (Phillips et al., 2005). Periodic boundary conditions were applied in all three directions. To calculate long-range electrostatic interactions during MD simulations with periodic boundary conditions, particle mesh Ewald (PME) was used with a 1-Å maximum space between grid points. Simulations were performed at a constant temperature of 310 K and a constant pressure of 1 atm using the Langevin piston method and Hoover’s method during minimization and equilibration.
Trajectory analyses
Dynamical residue cross-correlation heatmaps and atomic RMSF over MD trajectories were evaluated using the rmsf() function in the R Bio3D package (Grant et al., 2006). Total nonbonded interaction energies (electrostatic and van der Waals) were calculated using VMD and NAMD energy with the cutoff for nonbonded interactions set to 12 Å, and using a switching function with a switching distance of 10 Å (Phillips et al., 2005). All plots were generated using the plot() or heatmap.2() functions of the R gplot package.
C. elegans strains and CRISPR/Cas9 editing
C. elegans were cultured on nematode growth medium plates spotted with OP50 bacteria (Stiernagle, 2006). Strains used are listed in Table 1. Some strains were provided by the Caenorhabditis Genetics Center, funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). Knock-in strains were constructed by CRISPR/Cas9 genome editing using the dpy-10 co-CRSPR technique (Arribere et al., 2014; Paix et al., 2016). The anc-1(yc54[unc-83kash]), unc-83(yc55[unc-83::gfp::anc-1kash]), and anc-1(yc60[ΔD-N]) alleles were generated by injecting CRISPR guide RNAs (crRNAs; Table 2; synthesized by IDT or Dharmacon) precomplexed with purified Cas9 protein (UC Berkeley QB3) and universal tracrRNA (IDT or Dharmacon) along with repair templates as single-strand DNA oligonucleotide (ssODN) or double-strand DNA (dsDNA; Table 2; IDT or Dharmacon) into C. elegans gonads (Paix et al., 2015, 2016). crRNA and repair templates against dpy-10 were coinjected to identify animals where the Cas9 was active (Arribere et al., 2014; Paix et al., 2016). Edited animals were identified by PCR and restriction digests. The edited strains were backcrossed to UD522 (Cain et al., 2018) to lose dpy-10 mutations and introduce ycEx249[pcol-19nls::gfp::lacZ, pmyo-2mCherry] to mark hypodermal nuclei with GFP.
TABLE 1:
C. elegans strains in this study.
| Strain | Genotype | Reference |
|---|---|---|
| N2 | Wild type | Brenner, 1974 |
| UD399 | unc-84(n369) X; ycIs10[pcol-10nls::gfp::lacZ] | Bone et al., 2014 |
| UD473 | unc-83(yc26[unc-83::gfp::kash+LoxP])V | Bone et al., 2016 |
| UD522 | ycEx249[pcol-19nls::gfp::lacZ, pmyo-2mCherry] | Cain et al., 2018 |
| UD532 | unc-84(n369) X; ycEx249[pcol-19nls::gfp::lacZ, pmyo-2mCherry] | Cain et al., 2018 |
| UD538 | anc-1(e1873) I; ycEx249[pcol-19nls::gfp::lacZ, pmyo-2mCherry] | Cain et al., 2018 |
| UD582 | unc-83(yc55[unc-83::gfp::anc-1kash+LoxP])V; ycEx249[pcol-19nls::gfp::lacZ, pmyo-2mCherry] | This study |
| UD588 | anc-1(yc54[unc-83kash])I; ycEx249[pcol-19nls::gfp::lacZ, pmyo-2mCherry] | This study |
| UD589 | anc-1(yc60[ΔD-N])I; ycEx249[pcol-19nls::gfp::lacZ, pmyo-2mCherry] | This study |
| UD594 | anc-1(yc54[unc-83kash])I; unc-83(yc55[unc-83::gfp::anc-1kash+LoxP])V; ycEx249[pcol-19nls::gfp::lacZ, pmyo-2mCherry] | This study |
| UD597UD605 | unc-83(yc55[unc-83::gfp::anc-1kash+LoxP])V; ycIs10[pcol-10nls::gfp::lacZ]unc-84(yc35[unc-84::(C953A)::gfp])X; ycIs10[pcol-10nls::gfp::lacZ] | This studyThis study |
| UD604 | unc-83(yc55[unc-83::gfp::anc-1kash+LoxP])V unc-84(yc35[unc-84::(C953A)::gfp])X; ycIs10[pcol-10nls::gfp::lacZ] | This study |
| UD603 | unc-83(yc26[unc-83::gfp::kash];ycI9[Pcol-10nls::gfp::lacZ] | This study |
TABLE 2:
crRNA and repair templates used in this study.
| Gene target | crRNA sequence | DNA repair template sequence | Starting strain | New strain | Reference |
|---|---|---|---|---|---|
| anc-1 | acuuaugggagccgcuuguu | caggtttgtttatattttttaatattaataactaatgtctctcattttcaggcactgcttgttctcctcatgggagctgcctgccttgttttcggaaaaccatttggtccgcatgtaacctatgtgaatggaccaccaccggtttaatctttaattttttattttcattactattcactattgtttcattcatcatgaacctgcccccatacatcccagttg | N2 | UD580 | This study |
| unc-83 | ccgcauguaaccuaugugaa | ctggcagcgctcgcagcgattttctattatcacgtgccacattgcgacgacgagtactgttgccaacttctcaataatttcgctaaaagttttgacccttcgctagaattcgtaaacgggccaccaccattttaactgaatcatcagtattctgattgaaatccc | UD473 | UD581 | This study |
| anc-1 | caguacucgucgucgcaaug | caggcactgcttgttctacttatgggagccgcttgtttggttccacactgttttgctaagagttttgacccttcgctagaattcgtaaacgggccaccaccattttaatc | N2 | UD592 | This study |
| dpy-10 | gcuaccauaggcaccacgag | cacttgaacttcaatacggcaagatgagaatgactggaaaccgtaccgcatgcggtgcctatggtagcggagcttcacatggcttcagaccaacagcctat | Co-CRISPR | Arribere et al., 2014 | |
Nuclear migration and anchorage assays in C. elegans
Nuclear migration in embryonic hyp7 precursors was quantified by counting nuclei abnormally localized in the dorsal cord of L1 larvae as previously described (Starr et al., 2001; Bone et al., 2014). Hyp7 nuclear anchorage was assayed in adult animals expressing nuclear GFP from ycEx249[pcol-19nls::gfp::lacZ, pmyo-2mCherry]; nuclei were scored as clustered if one nucleus contacted another along the longitudinal axis of the worm (Cain et al. 2018). Contacts between nuclei on the perpendicular axis were not counted, as the marker could not distinguish seam cell nuclei in proximity to hyp7 nuclei from clusters of hyp7 nuclei. Only nuclei situated between the pharynx and the anus were counted. Only one lateral side of each animal was scored.
Acknowledgments
This work was supported by the National Science Foundation through grant CMMI-1538707 and the National Institutes of Health through grant R01 GM073874. In addition, this research used resources of the National Energy Research Scientific Computing Center (NERSC), a Department of Energy Office of Science user facility supported by the Office of Science of the United States Department of Energy under contract no. DE-AC02-05CH11231.
Abbreviations used:
- CC
coiled-coil
- ELEC
electrostatic
- INM
inner nuclear membrane
- KASH
Klarsicht, ANC-1, SYNE homology
- LINC
linker of the nucleoskeleton and cytoskeleton
- MD
molecular dynamics
- NE
nuclear envelope
- ONM
outer nuclear membrane
- RMSF
root-mean-square fluctuations
- SUN
Sad-1 and UNC-84.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-02-0079) on April 17, 2019.
REFERENCES
- Arribere JA, Bell RT, Fu BXH, Artiles KL, Hartman PS, Fire AZ. (2014). Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics , 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenovic PT, Conway DE. (2018). The LINC complex. Methods Mol Biol , 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenovic PT, Ramachandran I, Bathula K, Zhu R, Narang JD, Noll NA, Lemmon CA, Gundersen GG, Conway DE. (2016). Nesprin-2G, a component of the nuclear LINC complex, is subject to myosin-dependent tension. Biophys J , 34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone CR, Chang Y-T, Cain NE, Murphy SP, Starr DA. (2016). Nuclei migrate through constricted spaces using microtubule motors and actin networks in C. elegans hypodermal cells. Development , 4193–4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone CR, Tapley EC, Gorjanacz M, Starr DA. (2014). The Caenorhabditis elegans SUN protein UNC-84 interacts with lamin to transfer forces from the cytoplasm to the nucleoskeleton during nuclear migration. Mol Biol Cell , 2853–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974). Caenorhabditis elegans. Methods , 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain NE, Jahed ZSchoenhofen AValdez VAElkin BHao HHarris NJHerrera LAWoolums BMMofrad MRK, et al. (2018). Conserved SUN-KASH interfaces mediate LINC complex-dependent nuclear movement and positioning. Curr Biol , 3086–3097.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain NE, Tapley EC, McDonald KL, Cain BM, Starr DA. (2014). The SUN protein UNC-84 is required only in force-bearing cells to maintain nuclear envelope architecture. J Cell Biol , 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. (2006). Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol , 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson HN, Herrera LA, Brandt JN, Cain NE, Hermann GJ, Starr DA. (2018). The LINC complex. Methods Mol Biol , 163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson HN, Ly N, Meyerzon M, Starr DA. (2010). UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev Biol , 237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady RM, Starr DA, Ackerman GL, Sanes JR, Han M. (2005). Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc Natl Acad Sci USA , 4359–4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA, Caves LSD. (2006). Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics , 2695–2696. [DOI] [PubMed] [Google Scholar]
- Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. (2006). SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol , 3738–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennen J, Hur KH, Saunders CA, Luxton GWG, Mueller JD. (2017). Quantitative brightness analysis of protein oligomerization in the nuclear envelope. Biophys J , 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennen J, Saunders CA, Mueller JD, Luxton GWG. (2018). Fluorescence fluctuation spectroscopy reveals differential SUN protein oligomerization in living cells. Mol Biol Cell , 1003–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn HF, Brownstein ZLenz DRShivatzki SDror AADagan-Rosenfeld OFriedman LMRoux KJKozlov SJeang KT, et al. (2013a). The LINC complex is essential for hearing. J Clin Invest , 740–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn HF, Kim DI, Wright GD, Wong ESM, Stewart CL, Burke B, Roux KJ. (2013b). A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J Cell Biol , 1023–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. (1996). VMD—visual molecular dynamics. J Mol Graph , 33–38. [DOI] [PubMed] [Google Scholar]
- Jahed Z, Fadavi D, Vu UT, Asgari E, Luxton GWG, Mofrad MRK. (2018). Molecular insights into the mechanisms of SUN1 oligomerization in the nuclear envelope. Biophys J , 1190–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahed Z, Mofrad MR. (2019). The nucleus feels the force, LINCed in or not! Curr Opin Cell Biol , 114–119. [DOI] [PubMed] [Google Scholar]
- Jahed Z, Mofrad MRK. (2018). Mechanical LINCs of the nuclear envelope: where SUN meets KASH. Extrem Mech Lett , 99–103. [Google Scholar]
- Jahed Z, Shams H, Mofrad MRK. (2015). A disulfide bond is required for the transmission of forces through SUN-KASH complexes. Biophys J , 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahed Z, Soheilypour M, Peyro M, Mofrad MRK. (2016). The LINC and NPC relationship—it’s complicated! J Cell Sci , 3219–3229. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. (2015). The Phyre2 web portal for protein modelling, prediction and analysis. Nat Protoc , 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Birendra K, Roux KJ. (2015). Making the LINC: SUN and KASH protein interactions. Biol Chem , 295–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K, Zhang X, Ding X, Guo X, Chen M, Zhu B, Xu T, Zhuang Y, Xu R, Han M. (2009). SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc Natl Acad Sci USA , 10207–10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. (2011). The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem , 26743–26753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee MD, Rillo RS, Anderson A, Starr DA. (2006). UNC-83 is a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell , 1790–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie S, Ke H, Gao F, Ren J, Wang M, Huo L, Gong W, Feng W. (2016). Coiled-coil domains of SUN proteins as intrinsic dynamic regulators. Structure , 80–91. [DOI] [PubMed] [Google Scholar]
- Nishioka Y, Imaizumi H, Imada J, Katahira J, Matsuura N, Hieda M. (2016). SUN1 splice variants, SUN1_888, SUN1_785, and predominant SUN1_916, variably function in directional cell migration. Nucleus , 572–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I. (2005). The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci , 3419–3430. [DOI] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Rasoloson D, Seydoux G. (2015). High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics , 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Schmidt H, Seydoux G. (2016). Cas9-assisted recombineering in C. elegans: genome editing using in vivo assembly of linear DNAs. Nucleic Acids Res , e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. (2005). Scalable molecular dynamics with NAMD. J Comput Chem , 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, Burke B. (2009). Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci USA , 2194–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa BA, Kutay U, Schwartz TU. (2013). Structural insights into LINC complexes. Curr Opin Struct Biol , 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa BA, Rothballer A, Kutay U, Schwartz TU. (2012). LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell , 1035–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Han M. (2002). Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science , 406–409. [DOI] [PubMed] [Google Scholar]
- Starr DA, Hermann GJ, Malone CJ, Fixsen W, Priess JR, Horvitz HR, Han M. (2001). Unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development , 5039–5050. [DOI] [PubMed] [Google Scholar]
- Stiernagle T. (2006). Maintenance of C. elegans. WormBook, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Li W, Ke H, Feng W. (2018). Structural conservation of the autoinhibitory domain in SUN proteins. Biochem Biophys Res Commun , 1337–1343. [DOI] [PubMed] [Google Scholar]
- Yu J, Lei K, Zhou M, Craft CM, Xu G, Xu T, Zhuang Y, Xu R, Han M. (2011). KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum Mol Genet , 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]