Abstract
Contractile arrays of actin and myosin II filaments drive many essential processes in nonmuscle cells, including migration and adhesion. Sequential organization of actin and myosin along one dimension is followed by expansion into a two-dimensional network of parallel actomyosin fibers, in which myosin filaments are aligned to form stacks. The process of stack formation has been studied in detail. However, factors that oppose myosin stack formation have not yet been described. Here, we show that tropomyosins act as negative regulators of myosin stack formation. Knockdown of any or all tropomyosin isoforms in rat embryonic fibroblasts resulted in longer and more numerous myosin stacks and a highly ordered actomyosin organization. The molecular basis for this, we found, is the competition between tropomyosin and alpha-actinin for binding actin. Surprisingly, excessive order in the actomyosin network resulted in smaller focal adhesions, lower tension within the network, and smaller traction forces. Conversely, disordered actomyosin bundles induced by alpha-actinin knockdown led to higher than normal tension and traction forces. Thus, tropomyosin acts as a check on alpha-actinin to achieve intermediate levels of myosin stacks matching the force requirements of the cell.
INTRODUCTION
The actomyosin cytoskeleton is responsible for cell shape and for generating the forces that propel numerous essential processes, such as cell division, cell migration, and embryonic morphogenesis (Zaidel-Bar et al., 2015). Specialized actomyosin structures are assembled in a regulated manner at the appropriate time and place to perform the necessary mechanical work (Agarwal and Zaidel-Bar, 2019). A key factor affecting the function of actomyosin structures is their architecture. The length, orientation, and connectivity of actin filaments (F-actin) determines the type of interactions they have with myosin II bipolar filaments and the forces generated as a result of myosin II motor activity (Koenderink and Paluch, 2018). The architecture, in turn, is determined by the sites of actin polymerization as well as numerous other actin-binding proteins, such as actin cross-linkers and severing proteins (Ennomani et al., 2016). Prominent actomyosin structures found in fibroblasts cultured on stiff substrates, as well as in myofibroblasts in dermal wounds, myoepithelial cells in secreting glands, and endothelial cells under high shear stress are stress fibers (Kreis and Birchmeier, 1980; Franke et al., 1984; Tomasek et al., 2002; Gudjonsson et al., 2005). As their name implies, stress fibers generate tension, which can be used for cell body translocation and rear retraction during migrating, to contract a tissue or remodel the extracellular matrix (Pellegrin and Mellor, 2007; Kassianidou and Kumar, 2015). The structure of stress fibers in nonmuscle cells is somewhat reminiscent of sarcomeres in muscle myofibrils (Weber and Groeschel-Stewart, 1974; Lazarides, 1975; Gordon, 1978). Repetitive units of nonmuscle myosin II bipolar filaments, associated with the minus ends of F-actin and tropomodulin, are interleaved by alpha-actinin-rich regions of F-actin, where actin polymerization takes place (Hu et al., 2017). In contrast with muscle myofibrils, stress fibers are dynamic structures, with both myosin II and actin subunits rapidly turning over and the filaments flowing centripetally (Verkhovsky et al., 1995; Hotulainen and Lappalainen, 2006; Burnette et al., 2011). Many years ago it was observed that adjacent parallel stress fibers can interact with each other, matching up their myosin II filaments to create a two-dimensional lattice (Verkhovsky et al., 1995; Svitkina et al., 1997). The lattice is made of stacks of myosin oriented orthogonally to the F-actin fibers. More recently, the process of myosin concatenation to form myosin stacks has been studied in more detail using live structured illumination microscopy (Fenix et al., 2016; Beach et al., 2017; Hu et al., 2017). Two mechanisms have been described: sideways expansion of an existing myosin filament by splitting, and long-range movement of misaligned myosin filaments to bring them into close proximity and registration. Both mechanisms were shown to depend on myosin activity and actin dynamics (Beach et al., 2017; Hu et al., 2017). Depletion of the formin Fmnl3, the actin-severing protein cofilin1, or the actin cross-linker alpha-actinin-4 all result in disruption of myosin stack formation (Hu et al., 2017; Kemp and Brieher, 2018). Myosin-18B, which colocalizes with myosin II in stress fibers, was found to be enriched at the ends of myosin II stacks (Jiu et al., 2019). Deletion of myosin-18B resulted in severe defects in myosin II stack formation, suggesting that it is essential for their assembly and/or stability. Moreover, myosin-18B knockout cells displayed abnormal morphogenesis, migration, and ability to exert forces to the environment (Jiu et al., 2019). Although several proteins required for myosin stack formation have now been identified, it is not yet known whether there are negative regulators that act to counterbalance the process of stack formation. A more general open question is whether there could be “excessive order” in the actomyosin network and if so what would be its functional consequences.
RESULTS
Tropomyosin negatively regulates myosin stack formation in fibroblasts
In search of regulators of myosin II stack formation, we performed a small interfering RNA (siRNA) screen of contractome components in REF52 cells. After candidate gene knockdown (KD), cells were immunolabeled for myosin and F-actin and their cytoskeletal organization was examined by structured illumination microscopy (SIM). The majority of gene KDs either had no effect or resulted in myosin stack disruption (Supplemental Table S1). Only one family of genes, when knocked down, resulted in an increase in myosin stack formation compared with control siRNA cells. These were the tropomyosin genes: Tpm1, Tpm2, Tpm3, and Tpm4. Knocking down any of the tropomyosin genes on their own, as well as depleting all tropomyosins at once, resulted in a dramatic increase in the length of myosin stacks and the emergence of an extremely ordered actomyosin organization not normally observed in fibroblasts (Figure 1A). Quantification of the KD efficiency by quantitative reverse transcription PCR (qRT-PCR) showed that Tpm1 was depleted by 55% and all other tropomyosins were depleted by more than 80% (Supplemental Figure S1A). Reexpression of siRNA-insensitive Tpm3.1 in Tmp3 KD cells completely reverted the phenotype to wild type, suggesting the observed phenotype was caused specifically by tropomyosin KD (Supplemental Figure S1, B and C). Overexpression of Tpm3.1 in wild-type cells, on the other hand, led to disruption of myosin stack formation (Figure 1B). To quantify the effects of tropomyosin depletion or overexpression on myosin organization we first measured myosin intensity along the length of actin fibers by generating intensity line profiles, such as those shown in Figure 2A. Analysis of more than 70 line profiles from more than 15 cells showed that both the frequency and amplitude of myosin peaks along the stress fibers significantly increased in tropomyosin-KD cells compared with control. The mean frequency of peaks in total tropomyosin KD was 44 ± 2% higher than control and the mean amplitude was 53 ± 5% higher, suggesting that tropomyosin interferes with the alignment of myosin filaments within a single stress fiber (Figure 2B). Next, we measured the length of myosin stacks orthogonal to the actin filaments. We did this automatically, by segmenting a thresholded image and then using a custom MATLAB code that identified myosin structures perpendicular to actin stress fibers. This analysis, performed on more than 10 images from each condition, revealed a significant (P < 0.001) increase in the number of myosin stacks longer than 0.5 μm when tropomyosin levels were reduced by tpm3 or total tropomyosin KD; and a significant (P < 0.01) decrease in myosin stack length when tropomyosin levels were increased by overexpression (Figure 2C). Taken together, these results demonstrate that all tropomyosin isoforms have an inhibitory effect on the ordered organization of myosin into discrete domains along stress fibers and into stacks between adjacent fibers.
FIGURE 1:
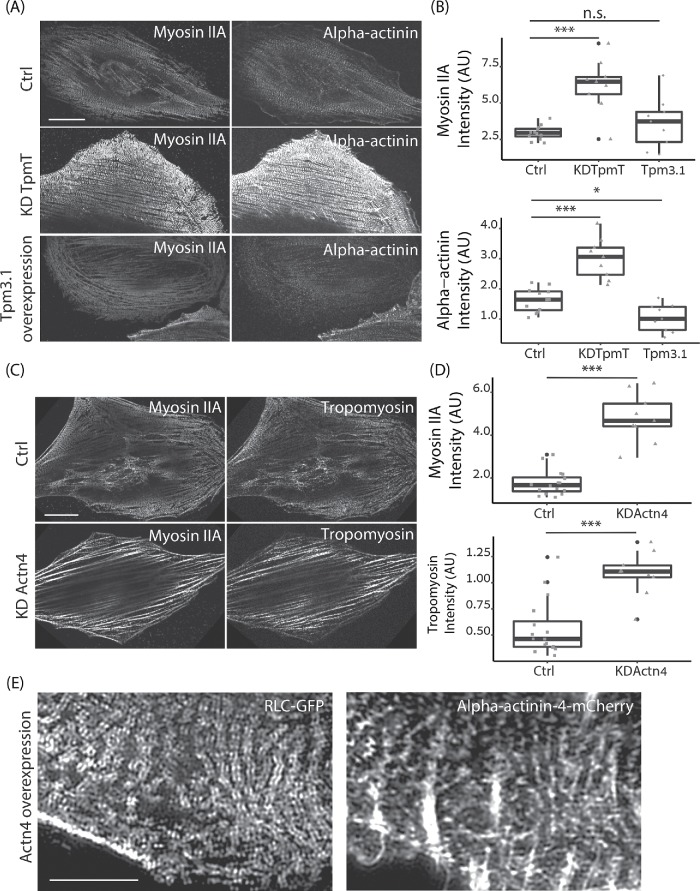
Organization of myosin II filaments in REF52 cells depleted for tropomyosin. (A) Representative images of REF52 cells transfected with nontargeting siRNA (Ctrl), siRNA against tropomyosin 1 (Tpm1), tropomyosin 2 (Tpm2), tropomyosin 3 (Tpm3), tropomyosin 4 (Tpm4), and a combination of tropmyosin 1, 2, 3, and 4 (TpmT), F-actin labeled with phalloidin and immunolabeled for myosin IIA. (B) Representative image of myosin IIA immunolabeled REF52 cells overexpressing tropomyosin 3.1 (Tpm3.1 OE). Images were taken with a SIM microscope. Scale bar is 10 µm.
FIGURE 2:
Analysis of myosin organization along and orthogonal to stress fibers. (A) Line scan across myosin stacks is shown in a representative image immunolabeled for myosin IIA (left). Representative profiles of line scanning for Ctrl, TpmT KD, and Tpm3.1 overexpression are presented (right). (B) Graphs of mean amplitude and peak frequency for different KD groups and Tpm3.1 overexpression. The number of line scans is n = 90 (Ctrl), n = 124 (KD Tpm3), n = 93 (KD TpmT), and n = 71 (Tpm3.1 OE). The images for analysis were taken with a W1 spinning-disk microscope. (C) Representative myosin IIA image (immunostaining) and its thresholded image to identify the length of myosin stack (left). The number of myosin stacks longer than 500 nm identified for different groups (middle). Average lengths of myosin stack per image are shown for different groups (right). The number of images is n = 18 (Ctrl), n = 11 (KD Tpm3), n = 24 (KD TpmT), and n = 10 (Tpm3.1 OE). The images for analysis were taken with a W1 spinning-disk microscope.
Tropomyosin inhibits myosin stack formation through its competition with alpha-actinin
Given the importance of actin cross-linking by alpha-actinin for myosin stack formation (Hu et al., 2017) and the well-established competition between tropomyosin and alpha-actinin over binding F-actin (Drabikowski and Nowak, 1968; Zeece et al., 1979; Gateva et al., 2017), we hypothesized that the effects of tropomyosin on myosin stack formation could be explained by its antagonistic relationship with alpha-actinin. To test this idea, we immunolabeled control, total tropomyosin KD, and Tpm3.1 overexpressing cells for myosin II and alpha-actinin and imaged them using a spinning-disk microscope (Figure 3A). We found that the increase in myosin stack formation in the tropomyosin-KD condition was accompanied by a strong increase in alpha-actinin localization along stress fibers, while overexpression of Tpm3.1, which led to disruption of myosin stacks but did not reduce average myosin levels, went hand in hand with a strong decrease in alpha-actinin intensity, as quantified in Figure 3B. We also performed the inverse experiment in which we depleted alpha-actinin from cells by siRNA and measured the level of tropomyosin 3 associated with the actomyosin cytoskeleton. As shown in Figure 3C and quantified in Figure 3D, the loss of myosin stacks and formation of thick and disorganized actomyosin bundles in Actn4 KD cells was accompanied by a twofold increase in tropomyosin associated with these structures. Moreover, overexpression of alpha-actinin led to an increase in myosin stacks similar to the phenotype of tropomyosin KD, imaged using SIM (Figure 3E). These observations support the idea that tropomyosin’s negative influence on myosin stack formation is mediated through its competition with alpha-actinin. Consistent with this, the double KD of tropomyosin and alpha-actinin did not result in increased myosin stack formation, but rather resulted in a phenotype similar to that of alpha-actinin KD (Supplemental Figure S1, D and E).
FIGURE 3:
Competition between alpha-actinin and tropomyosin for actin filaments and its effect on the organization of myosin II filaments. (A) Representative images of immunolabeled myosin IIA and alpha-actinin in Ctrl, TpmT KD, and Tpm3.1 OE cells. Scale bar is 20 µm. (B) Quantification of fluorescence intensity of myosin IIA and alpha-actinin in the stress fibers of Ctrl, TpmT KD, and Tpm3.1 OE cells. The statistical differences are shown in the graphs. The number of cells n = 12 (Ctrl), n = 9 (KD TpmT), and n = 9 (Tpm3.1 OE). (C) Representative images of immunolabeled myosin IIA and tropomyosin 3 in Ctrl and KD Actn4 cells. Scale bar is 20 µm. (D) Quantification of fluorescence intensity of tropomyosin and myosin IIA in the stress fibers of Ctrl and Actn4 KD cells. The statistical differences are shown in the graphs. The number of cells n = 16 (Ctrl), n = 9 (KD Actn4). (E) Representative image of myosin II A (RLC-GFP) and alpha-actinin-4 (alpha-actinin-4 mCherry) in cells overexpressing alpha-actinin-4. The scale bar is 5 µm. For A–D, the representative images and images for intensity analysis were acquired on a W1 spinning-disk microscope. For E, the representative images were obtained on an N-SIM microscope.
Intriguingly, quantification of relative mRNA levels by qRT-PCR, after siRNA treatment, revealed that antagonism between tropomyosin and alpha-actinin also exists at the transcriptional level. KD of Tpm1 or Tpm4 led to an increase in transcription of alpha-actinin 1 and 4, while KD of alpha-actinin 1 or 4 led to a dramatic increase in the expression of Tpm1 and a small increase in expression of Tpm4 (Supplemental Figure S2).
Tropomyosin KD does not change myosin stack dynamics
To examine the consequence of increased myosin stack formation on the dynamics of the actomyosin network, we performed live superresolution imaging of control and total tropomyosin-KD cells (Supplemental Movie 1). Consistent with previous reports, myosin filaments were observed to incorporate into the actin network of the lamella during its recurrent retractions, thereby forming new stress fibers (Burnette et al., 2011). Once incorporated into stress fibers, myosin was observed to move simultaneously in two directions: along the stress fiber and inward toward the cell center. The first movement involved shrinking of the distance between myosin stacks along the fiber and the latter was the consequence of translocation of the entire stress fiber moving inward, consistent with our previous results (Hu et al., 2017). These movements were visualized by temporally color-coded images and quantified using particle image velocimetry (PIV; Supplemental Figure S3A). Stack formation dynamics were more prominent in tropomyosin-KD cells, because they produce significantly longer myosin stacks. However, once formed, the myosin stacks in tropomyosin-KD cells moved in the same pattern as in the control cells. The average speed of myosin calculated by PIV was also similar for both (Supplemental Figure S3B). Thus, the increased order in the actomyosin network does not appear to change myosin stack dynamics after their formation.
Movie S1.
Dynamics of myosin II in control and total tropomyosin KD REF52 cells. Control cells (A) were transfected with non-targeting siRNA and RLC-GFP to mark myosin II. Tropomyosin knockdown cell (B) were transfected with siRNAs Tpm1, Tpm2, Tpm3, Tpm4, and with RLC-GFP to mark Myosin II. The cells were filmed for 29.5 minutes with 30 second intervals between frames using live-SR super-resolution mode in W1 spinning-disk microscope. The scale bar is 10 μm. The display rate is 15 fps.
Highly ordered actomyosin is associated with smaller focal adhesions and lower tension and traction forces
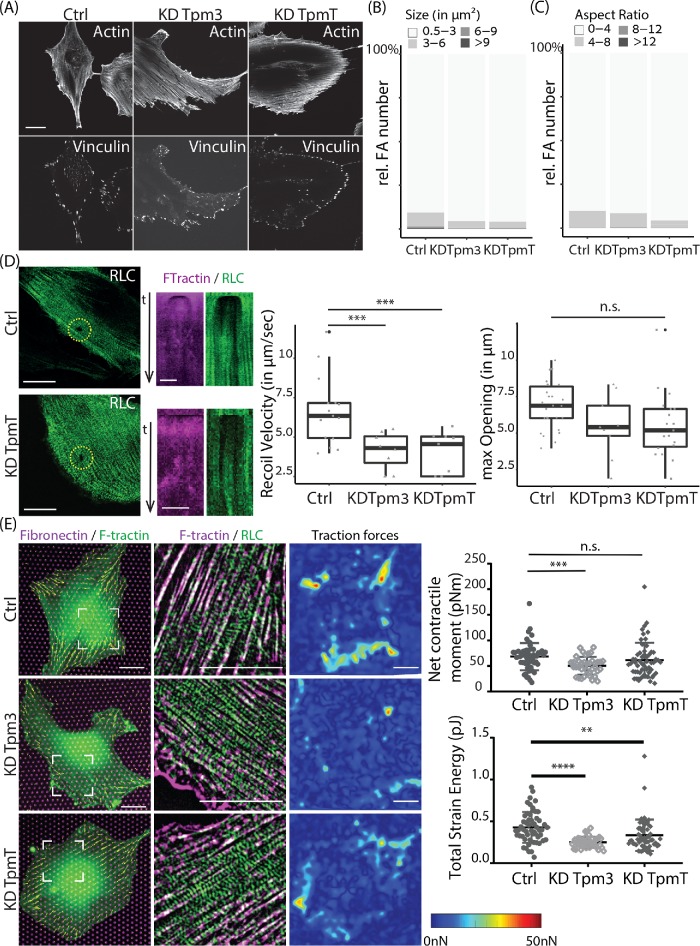
We next turned to investigate how increased order within the actomyosin cytoskeleton, caused by tropomyosin depletion, affects cell architecture and force generation. Analysis of cell area showed an increase in cell area upon total tropomyosin depletion. However, KD of tropomyosin 3 alone did not change cell area, indicating that cell area was not affected by the increase in myosin alignment throughout the cell (Supplemental Figure S3, C and D). Because contractile actomyosin fibers are normally anchored at focal adhesions (FAs), we examined the number, size, shape, and distribution of FAs in control and tropomyosin-KD cells by vinculin immunolabeling and quantitative image analysis (Figure 4A). Interestingly, the highly ordered actomyosin network of Tpm3 or total tropomyosin-KD cells was associated with FAs that were smaller and rounder compared with control cells (Figure 4, B and C). The number of FAs per cell area was not significantly different between control and tropomyosin-KD cells (unpublished data). To probe tension within the actomyosin network we performed laser ablations of stress fibers and monitored the recoiling of the actin edges by high-speed video microscopy. We followed the dynamics of the opening and used a custom MATLAB code to calculate the recoil velocity and maximum opening (as detailed in Materials and Methods). As shown in Figure 4D, both initial recoil velocity and the maximum opening were significantly smaller in tropomyosin-KD cells compared with control fibroblasts, suggesting that the actomyosin networks in tropomyosin-KD cells were under less tension. To corroborate these results we plated control and tropomyosin depleted cells on flexible elastomeric micropillar arrays and tracked pillar deflections to calculate traction forces (Figure 4E; Trichet et al., 2012). On the basis of the analysis of 39 cells, we found the average net contractile moment of Tpm3 KD cells (50 ± 17 pN•m) to be significantly lower (P < 0.001) than that of control siRNA cells (68 ± 27 pN•m). The total strain energy of both Tpm3 KD (0.25 ± 0.07 pJ) and tropomyosin total (TpmT) KD cells (0.33 ± 0.19 pJ) was significantly smaller (P < 0.0001) than that of control cells (0.43 ± 0.19 pJ). Altogether, these results demonstrate that in the absence of tropomyosin there is an increase in myosin stack formation and a decrease in FA size, actomyosin tension, and traction forces.
FIGURE 4:
Focal adhesion characteristics and force measurements in REF52 cells depleted for tropomyosin. (A) F-actin stained with phalloidin and focal adhesions labeled with vinculin antibody in cells transfected with nontargeting siRNA (Ctrl), tropomyosin-3 siRNA (Tpm3), and siRNA against all tropmyosins (TpmT). Scale bar is 20 µm. (B) Distribution of focal adhesions size. (C) Distribution of focal adhesion aspect ratio. Images analyzed: n = 14 (Ctrl), n = 26 (KD Tpm3), and n = 20 (KD TpmT). The images were acquired on a W1 spinning-disk microscope. (D) Representative images of myosin regulator light chain (RLC-GFP) at time point of laser ablation are shown in green for Ctrl and TpmT KD cells. Scale bar is 20 µm. Associated kymographs of F-Tractin-tdTomato (magenta) and RLC-GFP (green) along a line drawn crossing the gap are displayed in the next row. Scale bar is 5 µm. Recoil velocity and maximum opening are characterized. The differences between groups are shown in the graph. The number of images quantified for opening: n = 26 (Ctrl), n = 10 (KD Tpm3), and n = 20 (KD TpmT). The number of movies quantified for recoil velocity: n = 15 (Ctrl), n = 8 (KD Tpm3), and n = 9 (KD TpmT). The images were obtained on a Nikon A1R confocal microscope. (E) Traction forces of cells. Left, merge representative images of fibronectin-Atto-647N (magenta), F-tractin-tdTomato (green), and traction force vectors for each pillar (yellow). Middle, merge and zoomed image (position indicated on left image) of RLC-GFP (green) and F-tractin-tdTomato (magenta) with a top-hat filter applied. Right, interpolated traction force map. Scale bar is 20 μm. (Graphs) Net contractile moment exerted by the cells on the substrate and total strain energy are plotted. The images were taken with an Olympus IX-81 inverted widefield fluorescence microscope.
Increased disorder in actomyosin networks is associated with higher tension and larger traction forces
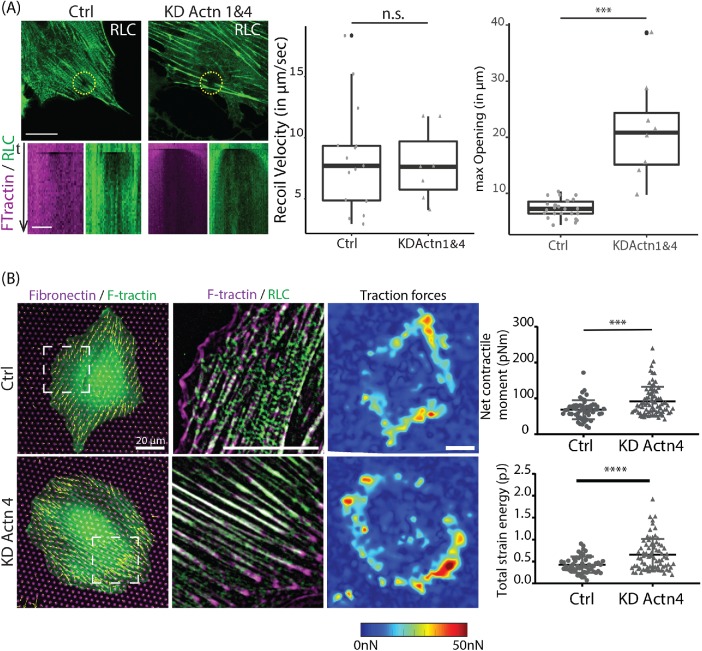
The results presented so far suggested an inverse relationship between myosin stack formation and tension within the actomyosin network, because increasing myosin stacks by tropomyosin KD decreased contractile forces compared with control cells. To test this inverse relationship also in the other extreme, we examined myosin stack formation and force generation in alpha-actinin KD conditions. Consistent with our previous results and other publications (Hu et al., 2017; Kemp and Brieher, 2018), depletion of either alpha-actinin-4 or alpha-actinin-1&4 resulted in the formation of thick stress fibers (Figure 5A). Analysis of mean myosin stack length and the number of stacks longer than 0.5 μm showed a dramatic decrease in both parameters in Actn4 and Actn1&4 KD cells compared with the control (Figure 5B). Line profile analysis along the stress fibers revealed that the frequency of myosin peaks in Actn1&4 KD cells was reduced by 39% ± 3% compared with control, and the mean amplitude of myosin peaks was reduced by 20% ± 1.6% (Figure 5C). Concomitant with the decrease in myosin order the Actn1&4 KD cells exhibited a more than threefold increase in the number of FAs normalized by cell area and their FAs were of larger size and longer than FAs in control cells (Figure 5, D and E). Moreover, laser ablation experiments showed a significantly higher recoil velocity in Actn1&4 KD cells compared with control cells (Figure 6A). In line with this, micropillar experiments measured significantly higher traction forces produced by alpha-actinin-4 KD cells (Figure 6B). On the basis of the analysis of 77 cells, we found that the average net contractile moment of Actn4 KD cells (92 ± 40 pN•m) was significantly higher (P < 0.001) than that of control siRNA cells (68 ± 27 pN•m). The total strain energy for Actn4 KD cells (0.65 ± 0.36 pJ) was also significantly larger than that of control cells (0.43 ± 0.19 pJ).
FIGURE 5:
Depletion of alpha-actinin increases disorder of myosin II filaments along with increases in focal adhesion size, actomyosin tension, and traction forces. (A) Representative images of REF52 cells transfected with nontargeting siRNA (Ctrl), α-actinin-4 siRNA (KD Actn4), and the double KD of α-actinin-1 and -4 (KD Actn1&4), and immunolabeled for myosin IIA. Scale bar is 10 µm. (B) Quantification of myosin stack length and number of stacks longer than 500 nm identified. Images analyzed: n = 18 (Ctrl), n = 28 (KD Actn4), and n = 9 (KD Actn1&4). (C) Line scanning across myosin is shown in representative images immunolabeled for myosin IIA. Representative intensity profiles for Ctrl, KD Actn4, and KD Actn1&4 are presented. Number of line scans analyzed: n = 90 (Ctrl), n = 85 (KD Actn4), and n = 89 (KD Actn1&4). (D) Representative images of actin stained with phalloidin and immunolabeled vinculin shown for Ctrl, KD Actn4, and KD Actn1&4 cells. Scale bar is 20 µm. (E) Quantification of focal adhesions in Ctrl, KD Actn4, and KD Actn1&4 cells. Images analyzed: n = 14 (Ctrl), n = 22 (KD Actn4), and n = 8 (KD Actn1&4). The images were taken with a W1 spinning-disk microscope.
FIGURE 6:
Depletion of alpha-actinin leads to higher tension and larger traction forces. (A) Representative images of myosin regulator light chain (RLC-GFP) at time point of laser ablation are shown in green for Ctrl and Actn1&4 KD cells. Scale bar is 20 µm. Associated kymographs of F-tractin-tdTomato (magenta) and RLC-GFP (green) along a line drawn crossing the gap are displayed in the next row. Scale bar is 5 µm. Maximum opening and recoil velocity are characterized. The differences between groups are shown in the graph. The number of images quantified for opening: n = 26 (Ctrl), n = 8 (KD Actn1&4). The number of movies quantified for recoil velocity: n = 15 (Ctrl), n = 7 (KD Actn1&4). The images were taken with a Nikon A1R confocal microscope. (B) Traction force measurements for Ctrl (n = 55) and KD Actn4 cells (n = 77). Left, representative merged images of fibronectin-Atto-647N (magenta), F-tractin-tdTomato (green), and traction force vectors for each pillar (yellow). Middle, merge and zoomed image (position indicated on left image) of RLC-GFP (green) and F-tractin-tdTomato (magenta) with a top-hat filter applied. Right, interpolated traction force map. Scale bar is 20 μm. Net contractile moment exerted by the cells on the substrate and total strain energy are plotted. The differences between groups are shown in the statistical graphs. Note that the data for control cells are the same data as in Figure 4E. The images were taken with an Olympus IX-81 inverted widefield fluorescence microscope.
DISCUSSION
This study explores a novel characteristic of the contractile actomyosin system of nonmuscle cells: the ability to regulate the degree of order in the distribution of myosin II filaments. While striated muscles have highly ordered nonchanging organization with constant length of sarcomeres and either registered or nonregistered organization of sarcomeres in neighboring myofibrils, the system of stress fibers (ventral or circumferential) in nonmuscle cells varies in the degree of periodicity in organization of myosin filament domains along the stress fibers, as well as in the degree of registry in the mutual positioning of neighboring stress fibers. Thus, nonmuscle cells can regulate not only the myosin filament assembly and activity, but also the order of their long-range organization.
Here, we found a generic principle of regulation of myosin filament ordering based on coordinated function of a positive regulator, alpha-actinin4, and proteins of the tropomyosin family, working as negative regulators. The interplay between these two types of actin-binding regulatory molecules and myosin filaments is depicted in the schematic Figure 7. As was observed previously and extensively confirmed in our experiments, alpha-actinin-4 and tropomyosin(s) compete with each other for the binding to actin filaments, so that depletion of one resulted in augmentation in association of another with the actin bundles. Each of them, in addition, may compete with myosin filaments for association with the actin filaments, as can be inferred from the fact that KD of either tropomyosin(s) or alpha-actinin-4 increased the amount of myosin associated with the actin bundles. Finally, alpha-actinin is indispensable for the ordering of myosin filaments associated with actin bundles and for stack formation. Thus, KD of tropomyosin(s) promotes myosin filament ordering by creating a high concentration of the myosin filament and alpha-actinin along the actin filaments, whereas the overexpression of tropomyosin(s) interferes with ordering via displacement of alpha-actinin. Similarly, the depletion of alpha-actinin makes ordering impossible, but promotes the formation of actin filament bundles with high levels of myosin II and tropomyosin, which are highly contractile.
FIGURE 7:
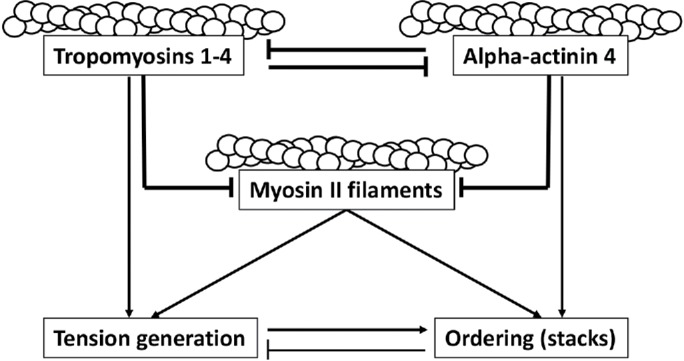
Schematic model of the reciprocal regulation of actomyosin structure and contractility by alpha-actinin-4 and tropomyosins. Tropomyosin 1-4, alpha-actinin 4, and myosin II filaments all compete for binding along F-actin. The balance between them dictates the degree of order (myosin stacks) and amount of tension generated by the actomyosin network, with alpha-actinin 4 promoting more order and less tension, whereas tropomyosin promotes less order and higher tension. The drawn lines reflect our phenotypic observations and do not imply direct interactions.
The increase in force produced by Actn4 KD cells is consistent with a previous study (Shao et al., 2010) that showed an increase in myosin light chain 2 expression after Actn4 KD. Whether the increase in force we measured is due to an increase in myosin light chain expression or due to the increased disorder in actomyosin organization, or both, remains to be tested. A low level of contractility of highly ordered myosin II filament arrays (and consequent decrease of focal adhesion size) in tropomyosin-KD or alpha-actinin overexpressing cells can hardly be explained by just depletion or displacement of tropomyosin(s). It is more plausible that actin filaments highly cross-linked with alpha-actinin resist the contractile forces generation by myosin filaments. Some tropomyosin isoforms can activate myosin IIA (Barua et al., 2014; Hundt et al., 2016; Gateva et al., 2017). Therefore, the reduction in traction forces upon tropomyosin depletion could also be due to a reduction in myosin activity because of the absence of tropomyosin. One might argue that the increase in myosin stacks is the result of decreased tension and not the other way around, but this is not likely because direct inhibition of myosin activity results in loss of myosin stacks (Hu et al., 2017).
Optimal contractility probably requires an optimal level of cross-linking and therefore does not grow monotonously with the increase of myosin filament order.
Although it is well established that different tropomyosins have specific functions (Gunning et al., 2015; Gateva et al., 2017; Jansen and Goode, 2019), in our experiments the KDs of each of four tropomyosin genes produced a similar effect on myosin filament order, strongly enhancing it. Does this mean that functions of these different tropomyosins are nonredundant in our assays? Or, if they are redundant, is the degree of myosin filament order so sensitive to the total amount of tropomyosins that removal of the products of only one gene of four is sufficient to change it? We do not know the answer to these questions yet.
The simple model outlined above, based on an interplay driven by “steric competition” between two regulatory proteins, is probably only a first approximation of the true complexity in the cell. A plethora of regulatory factors can modulate the assembly and subsequent ordering of the myosin filaments. Among them are myosin light and heavy chain kinases and phosphatases, myosin II tail-binding protein S100 A4, aforementioned myosin-18, etc. It is possible that tropomyosin(s) work as scaffold proteins recruiting these regulatory factors (including negative regulators of ordering and stacking) into the proximity of myosin filaments. Similarly, alpha-actinin could recruit accessory factors promoting myosin filament ordering; for example, actin polymerization-depolymerization regulators. The molecular interactions that can mediate such “second level” of regulation should be elucidated in future studies. Moreover, transcription regulation interplay between tropomyosins, alpha-actinins, and myosin II is also possible, as our data show. The mechanism of such interplay also deserves further studies.
In summary, while the competition between alpha-actinin and tropomyosin for binding F-actin is well established, our study reveals how this competition plays out at the larger scale of actomyosin network architecture. Given the importance of actomyosin organization for cellular processes such as cell adhesion, polarity, and migration, the reciprocal regulation we described here has important ramifications for basic cell behavior.
MATERIALS AND METHODS
Cell culture
The rat embryonic fibroblasts (REF52 cells) cell line was established by the group of W. Topp at Cold Spring Harbor Laboratory (McClure et al., 1982). REF52 cells were cultured in DMEM (11965092; Invitrogen) containing 10% fetal bovine serum (10082147; Invitrogen) at 37°C in a 5% CO2 humidified incubator. Cells were free of mycoplasma as tested by a PCR-based method (Goding, 1996).
siRNA KD
REF52 cells were transfected with ON-TARGETplus Rat siRNA SMARTpool library (Dharmacon) or a nontargeting control pool siRNA (D-001810-10-05; Dharmacon) at a concentration of 20 µM using electroporation (Neon; Invitrogen) following the manufacturer’s protocol. After 48 h, the cells were replated on 10 μg/ml fibronectin (11080938001; Roche) coated glass coverslips (No. 1.5H, 017650; Marienfeld) and incubated overnight before fixation and immunofluorescence staining. siRNA sequences are available in Supplemental Table 1.
qRT-PCR analysis
REF52 cells were transfected with the indicated siRNA or plasmid. After 48–72 h RNA was extracted using the Monarch Total RNA Miniprep kit (NEB; Cat. T2010) and transcribed into cDNA using the Lunascript kit (NEB; Cat. E3010). RT-PCR was performed using Fast SYBR Green Master Mix (Thermo; Cat. 4385612) and the primer indicated in Supplemental Table S1.
Results were analyzed in R using the pcr package (M. Ahmed, Version 1.1.2, 2018).
Plasmid transfection
Cells were transfected using electroporation (Neon; Invitrogen) following the standard protocol. The following plasmids were used for overexpression experiments: F-Tractin-tdTomato (gift from M. J. Schell, Uniformed Services University, Bethesda, MD); tropomyosin 3.1-NeonGreen (gift from Peter Gunning, University of New South Wales, Australia); α-actinin-4-mCherry (gift from M. Murata-Hori, The Institute for Stem Cell Biology and Regenerative Medicine, India); and myosin regulatory light chain (RLC)-GFP (gift from Wendy A. Wolf and Rex L. Chisholm, Northwestern University, Chicago, IL). After transfection, overexpression cells were plated on 10 μg/ml fibronectin-coated glass coverslips and incubated overnight before immunofluorescence staining and imaging.
Immunofluorescence antibody staining
Anti-vinculin (dilution 1:200; Sigma-Aldrich; V9131), anti-myosin-IIA tail domain (dilution 1:1000; Sigma-Aldrich; M8064), anti-α-actinin-1&4 (dilution 1:200; Sigma-Aldrich; A5044), anti-tropomyosin 3 (CG3, mouse monoclonal antibody; gift from Peter Gunning, University of New South Wales, Australia), Alexa-Fluor 488 conjugated phalloidin (dilution 1:1000; Molecular Probes; A12379), and Alexa-Fluor conjugated secondary antibodies (dilution 1:400; Molecular Probes) were used for immunofluorescence staining.
Cells were fixed in warm 4% paraformaldehyde for 15 min at 37°C, washed with 1X phosphate-buffered saline (PBS), permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) for 5 min at room temperature, blocked with 1% bovine albumin serum (A7906; Sigma-Aldrich) for 1 h, immunofluorescence stained with primary antibodies overnight at 4°C, then incubated with secondary antibodies for 1 h at room temperature followed by three times wash with 1X PBS.
Cell imaging using SIM
To apply SIM, high-precision glass coverslips with thickness 170 µm (No. 1.5H; 017650; Marienfeld) were deep cleaned and used for the cells plating as described in a previous study (Hu et al., 2017). Fixed cells were imaged by dual color 3D-SIM (Nikon). The SIM images were taken with dual color (laser 488 and laser 561) SIM mode using a 100× oil (NA 1.49) objective.
Live superresolution used to generate data in Supplemental Figure S3A was performed on a W1 spinning disk with a live-SR module (Gataca Systems). This module is an optically demodulated structured illumination technique with online processing that uses the pinhole patterns of Nipkow disks to enhance high spatial frequencies. It brings the resolution down to 120 nm while allowing fast imaging.
Cell imaging using W1 spinning-disk microscope
A W1 spinning-disk microscope (Roper Scientific) or live-SR (spinning-disk-based structured illumination superresolution; York et al., 2013) was used to take images for intensity-based quantification or for live cell dynamics analysis. Laser line wavelengths 488, 561, and 642 nm were used. The objective Plan Apo 100× oil NA 1.45 and camera Prime95B (Photometrics) are used for structured illumination live-SR.
Laser ablation with Nikon A1R confocal microscope
Laser ablation was performed using a UV laser (355 nm; 1 kHz repetition rate; PowerChip, Teem Photonics) with a 60× objective (NA 1.4; Nikon) in a Nikon A1R confocal microscope (Hara et al., 2016). The UV laser was introduced into the microscope through a custom-made UV dichroic mirror with a mechanical shutter controlled by a homemade ImageJ plug-in. An UV laser power of 50 nW measured at the back aperture of the objective with an exposure of 2 s was used for locally ablating actin stress fibers. F-Tractin-tdTomato and RLC-GFP images were taken using a Galvano confocal scanner with frame average, resulting in 3–4 s per frame. The images were recorded four frames before ablation, 2 s ablation time, and then continue imaging for ∼5 min.
Traction force microscopy
Traction force microscopy was performed using fabricated micropillar arrays in polydimethylsiloxane (PDMS; Gupta et al., 2015) with 2.1 μm diameter and 4.1 μm height arranged in a triangular lattice with a 4 μm center-to-center spacing. PDMS was cured at 80°C for 2 h to obtain Young’s modulus of 2 MPa, thus the pillar stiffness k = 59 nN/μm (Schoen et al., 2010). A solution containing 50 μg/ml fibronectin (Roche) and 1 μg/ml Atto-647N conjugated fibronectin (Sigma-Aldrich; #76508) in Dulbecco’s phosphate buffered saline (DPBS) was added to flat PDMS stamps and incubated for at least 1 h at room temperature, washed once in ultrapure water, and dried with N2. The micropillars were treated in an ultraviolet ozone cleaner (BioForce Nanosciences; UV Ozone ProCleaner Plus) for 15 min, and then the fibronectin-coated PDMS was stamped onto the micropillars. A solution containing 0.2% Pluronic F-127 in ultrapure water was added to the micropillars for 1 h, washed three times in DPBS, then seeded with cells in complete growth medium and allowed to adhere for at least 5 h. The media was exchanged with Leibovitz’s L-15 complete media and the cells were imaged at 37°C using an inverted widefield fluorescence microscope (Olympus IX-81, UPLSAPO 60XW/1.2NA objective, X-Cite 120Q mercury lamp, Photometrics CoolSNAP EZ CCD camera). Pillar deflections d were measured using a custom-built MATLAB program, the force on a pillar is F = kd, the strain energy of a pillar is 1/2*kd2, and the total strain energy is a summation of all pillars located within the cell. The net contractile moment (Butler et al., 2002) μ was calculated as the summation over all pillars p of the scalar product of traction force vector F with the vector of the pillar position pointing toward the cell center of mass r:
 |
Imaging analysis
In Figures 2A and 5C a 100 px (11 µm) line was drawn over stress fibers associated myosin in images taken with a W1 spinning-disk microscope and the intensity plotted using Fiji (Schindelin et al., 2012). The local maxima of each line plot were calculated using a custom-made MATLAB code. Mean amplitudes were plotted using R (R Core Team, 2016; Højsgaard and Halekoh, 2018). In Figures 2C and 5B and Supplemental Figure S1, B and D, images containing both actin and myosin channels were loaded into a custom-made MATLAB-based program. After thresholding, actin filaments were segmented followed by the identification of myosin stacks perpendicular (angle >60°) to the segmented actin filaments. All measurements were normalized to the mean of the control group. The identified myosin stack measurements were plotted using the ggplot package in R (Wickham, 2016).
In Figure 3, mean intensity on stress fibers within whole cell are measured for W1 spinning-disk images. In ImageJ, myosin, alpha-actin, or tropomyosin 3 images are auto thresholded, then the mean intensity is measured on the threshold image.
In Supplemental Figure S2, to analyze the movement of myosin filament stacks in control and tropomyosin total KD REF52 cells obtained by a live-SR W1 spinning-disk microscope, we first use Temporal-color code (LUT Rainbow RGB) in ImageJ to present the temporal motion of myosin filaments. Then PIV analysis was used to quantify the quiver and speed of movement. We applied MatPIV 1.6 (Sveen, 2004), a free MATLAB toolbox, to calculate the quiver speed map and absolute speed value map through whole cell on each time point. The averages of the absolute speed value during a 30-min-long period are presented in a spectrum color map. The mean values of the average speed map are calculated and plotted in a statistical graph. The drift of images during acquisition was corrected with the plug-in “Image Stabilizer” in Fiji before the above-mentioned quantifications.
FAs were analyzed as previously described (Chen et al., 2018). In short, vinculin fluorescence was background subtracted and contrast adjusted to identify FAs. The actin fluorescence was used to generate the distance map accordingly. To calculate the distance of the individual FA both images were combined. The process was automated using a home-written ImageJ macro. Plots were generated using the ggplot package in R. R was also used for statistical testing.
Laser ablations were analyzed as previously described (Priti et al., 2018), only in movies where complete ablation was confirmed in the actin channel. Briefly, we manually tracked the distance between the two stress fiber edges at each time point using the Fiji plug-in MTrackJ (Meijering et al., 2012) and the data were fitted by an exponential curve using a custom-made MATLAB-based program. The recoil velocity was calculated as the derivative of the exponential function at the effective time of laser cut. The maximum opening was measured on kymographs generated using the KymographBuilder plug-in in Fiji.
Quantifications for Figures 2, 3, 4, D and E, 5, and 6 are displayed as boxplots (25% Quartile, Median, 75% Quartile and outliers) in black with an overlay of the individual data points in gray. Quantification for Figures 4, B and C, and 5E are displayed as stacked bar graphs normalized to 100%.
Statistical analysis
For Figures 2, B and C, 3, B and D, 4, D and E, and 5, B and C, and Supplemental Figure 3D, significances were tested using analysis of variance and a subsequent Tukey HSD test. For Figure 6, A and B, and Supplemental Figures 1, B–E, and 3B, significances were tested using a Student’s t test. For Figures 4, B and C, and 5E, significances were tested using a Chi2 test. For all graphs significances are indicated as follows: P < 0.0001, ****; P < 0.001, ***; P < 0.01, **; P < 0.05, *.
Supplementary Material
Acknowledgments
We thank Gianluca Grenci, Mukund Gupta, and Felix Margadant for micropillar setup and force detection software, Yusuke Toyama for help with laser ablation, Hui-Ting Ong for MATLAB coding, Zhongwen Chen for help with focal adhesion analysis, and Priti Agarwal with recoil velocity measurement. B.L. is supported by LABEX “Who am I?,” Agence Nationale de la Recherche “POLCAM” (Grant no. ANR-17-CE13-0013) and the ERC under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreements no. 617233. H.G. is funded by Israel Science Foundation Grant no. 1293/17. A.D.B. is supported by the National Research Foundation, Prime Minister’s Office, Singapore, and the Ministry of Education under the Research Centres of Excellence program through the Mechanobiology Institute (ref. no. R-714-006-006-271), Grant no. 7120790101 EU-H2020-MSCA-ITN, and a Maimonides Israeli-France grant from Israeli Ministry of Science, Technology and Space at the Weizmann Institute. This article is dedicated to Wen Yuting and her son Darwin in loving memory of Guo (Gary) Zhenhuan. Gary, a passionate biologist and true friend, left us prematurely.
Abbreviations used:
- Actn1&4
both alpha-actinin-1 and alpha-actinin-4
- Actn4
alpha-actinin-4
- Ctrl
control
- FA
focal adhesion
- KD
knockdown
- PDMS
polydimethylsiloxane
- qRT-PCR
quantitative reverse transcription PCR
- Tpm
tropomyosin
- TpmT
tropomyosin total.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-02-0082) on June 19, 2019.
REFERENCES
- Agarwal P, Zaidel-Bar R. (2019). Principles of actomyosin regulation in vivo. Trends Cell Biol , 150–163. [DOI] [PubMed] [Google Scholar]
- Barua B, Nagy A, Sellers JR, Hitchcock-DeGregori SE. (2014). Regulation of nonmuscle myosin II bby tropomyosin. Biochemistry , 4015–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach JR, Bruun KS, Shao L, Li D, Swider Z, Remmert K, Zhang Y, Conti MA, Adelstein RS, Rusan NM, et al (2017). Actin dynamics and competition for myosin monomer govern the sequential amplification of myosin filaments. Nat Cell Biol , 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette DT, Manley S, Sengupta P, Sougrat R, Davidson MW, Kachar B, Lippincott-Schwartz J. (2011). A role for actin arcs in the leading-edge advance of migrating cells. Nat Cell Biol , 371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. (2002). Traction fields, moments, and strain energy that cells exert on their surroundings. Am J Physiol Cell Physiol , C595–C605. [DOI] [PubMed] [Google Scholar]
- Chen Z, Oh D, Biswas KH, Yu CH, Zaidel-Bar R, Groves JT. (2018). Spatially modulated ephrinA1:EphA2 signaling increases local contractility and global focal adhesion dynamics to promote cell motility. Proc Natl Acad Sci USA , E5696–E5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabikowski W, Nowak E. (1968). The interaction of alpha-actinin with F-actin and its abolition by tropomyosin. Eur J Biochem , 209–214. [DOI] [PubMed] [Google Scholar]
- Ennomani H, Letort G, Guerin C, Martiel JL, Cao W, Nedelec F, De La Cruz EM, Thery M, Blanchoin L. (2016). Architecture and connectivity govern actin network contractility. Curr Biol , 616–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenix AM, Taneja N, Buttler CA, Lewis J, Van Engelenburg SB, Ohi R, Burnette DT. (2016). Expansion and concatenation of non-muscle myosin IIA filaments drive cellular contractile system formation during interphase and mitosis. Mol Biol Cell , 1465–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke RP, Grafe M, Schnittler H, Seiffge D, Mittermayer C, Drenckhahn D. (1984). Induction of human vascular endothelial stress fibres by fluid shear stress. Nature , 648–649. [DOI] [PubMed] [Google Scholar]
- Gateva G, Kremneva E, Reindl T, Kotila T, Kogan K, Gressin L, Gunning PW, Manstein DJ, Michelot A, Lappalainen P. (2017). Tropomyosin isoforms specify functionally distinct actin filament populations in vitro. Curr Biol , 705–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goding JW. (1996). Monoclonal Antibodies: Principles and Practice, 3rd ed., New York: Academic Press. [Google Scholar]
- Gordon WE., 3rd (1978). Immunofluorescent and ultrastructural studies of “sarcomeric” units in stress fibers of cultured non-muscle cells. Exp Cell Res , 253–260. [DOI] [PubMed] [Google Scholar]
- Gudjonsson T, Adriance MC, Sternlicht MD, Petersen OW, Bissell MJ. (2005). Myoepithelial cells: their origin and function in breast morphogenesis and neoplasia. J Mammary Gland Biol Neoplasia , 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning PW, Hardeman EC, Lappalainen P, Mulvihill DP. (2015). Tropomyosin—master regulator of actin filament function in the cytoskeleton. J Cell Sci , 2965–2974. [DOI] [PubMed] [Google Scholar]
- Gupta M, Sarangi BR, Deschamps J, Nematbakhsh Y, Callan-Jones A, Margadant F, Mege RM, Lim CT, Voituriez R, Ladoux B. (2015). Adaptive rheology and ordering of cell cytoskeleton govern matrix rigidity sensing. Nat Commun , 7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y, Shagirov M, Toyama Y. (2016). Cell boundary elongation by non-autonomous contractility in cell oscillation. Curr Biol , 2388–2396. [DOI] [PubMed] [Google Scholar]
- Højsgaard S, Halekoh U. (2018). doBy: Groupwise Statistics, LSmeans, Linear Contrasts, Utilities. doBy version 4.6–2 https://rdrr.io/cran/doBy/. [Google Scholar]
- Hotulainen P, Lappalainen P. (2006). Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol , 383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Dasbiswas K, Guo Z, Tee YH, Thiagarajan V, Hersen P, Chew TL, Safran SA, Zaidel-Bar R, Bershadsky AD. (2017). Long-range self-organization of cytoskeletal myosin II filament stacks. Nat Cell Biol , 133–141. [DOI] [PubMed] [Google Scholar]
- Hundt N, Steffen W, Pathan-Chhatbbar S, Taft MH, Manstein DJ. (2016). Load-dependent modulation of non-muscle myosin 2A function by tropomyosin 4.2. Sci Rep , 20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen S, Goode BL. (2019). Tropomyosin isoforms differentially tune actin filament length and disassembly. Mol Biol Cell , 671–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiu Y, Kumari R, Fenix AM, Schaible N, Liu X, Varjosalo M, Krishnan R, Burnette DT, Lappalainen P. (2019). Myosin-18B promotes the assembly of myosin II stacks for maturation of contractile actomyosin bundles. Curr Biol , 81–92 e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassianidou E, Kumar S. (2015). A biomechanical perspective on stress fiber structure and function. Biochim Biophys Acta , 3065–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp JP, Jr, Brieher WM. (2018). The actin filament bundling protein alpha-actinin-4 actually suppresses actin stress fibers by permitting actin turnover. J Biol Chem , 14520–14533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenderink GH, Paluch EK. (2018). Architecture shapes contractility in actomyosin networks. Curr Opin Cell Biol , 79–85. [DOI] [PubMed] [Google Scholar]
- Kreis TE, Birchmeier W. (1980). Stress fiber sarcomeres of fibroblasts are contractile. Cell , 555–561. [DOI] [PubMed] [Google Scholar]
- Lazarides E. (1975). Tropomyosin antibody: the specific localization of tropomyosin in nonmuscle cells. J Cell Biol , 549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure DB, Hightower MJ, Topp WC. (1982). Effect of SV40 transformation on the growth factor requirements of the rat embryo cell line REF52 in serum-free medium. Cold Spring Harb Conf Cell Proliferation , 345–364. [Google Scholar]
- Meijering E, Dzyubachyk O, Smal I. (2012). Methods for cell and particle tracking. Methods Enzymol , 183–200. [DOI] [PubMed] [Google Scholar]
- Pellegrin S, Mellor H. (2007). Actin stress fibres. J Cell Sci , 3491–3499. [DOI] [PubMed] [Google Scholar]
- Priti A, Ong HT, Toyama Y, Padmanabhan A, Dasgupta S, Krajnc M, Zaidel-Bar R. (2018). Syncytial germline architecture is actively maintained by contraction of an internal actomyosin corset. Nat Commun , 4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2016). R: A Language and Environment for Statistical Computing, Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen I, Hu W, Klotzsch E, Vogel V. (2010). Probing cellular traction forces by micropillar arrays: contribution of substrate warping to pillar deflection. Nano Lett , 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Wang JH, Pollak MR, Wells A. (2010). alpha-actinin-4 is essential for maintaining the spreading, motility and contractility of fibroblasts. PLoS One , e13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveen JK. (2004). An Introduction to MatPIV v. 1.6.1, Oslo, Norway: Department of Mathematics, University of Oslo, Eprint No. 2.
- Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. (1997). Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J Cell Biol , 397–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. (2002). Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol , 349–363. [DOI] [PubMed] [Google Scholar]
- Trichet L, Le Digabel J, Hawkins RJ, Vedula SR, Gupta M, Ribrault C, Hersen P, Voituriez R, Ladoux B. (2012). Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness. Proc Natl Acad Sci USA , 6933–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhovsky AB, Svitkina TM, Borisy GG. (1995). Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J Cell Biol , 989–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K, Groeschel-Stewart U. (1974). Antibody to myosin: the specific visualization of myosin-containing filaments in nonmuscle cells. Proc Natl Acad Sci USA , 4561–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2016). ggplot2: Elegant Graphics for Data Analysis, New York: Springer-Verlag. [Google Scholar]
- York AG, Chandris P, Nogare DD, Head J, Wawrzusin P, Fischer RS, Chitnis A, Shroff H. (2013). Instant super-resolution imaging in live cells and embryos via analog image processing. Nat Methods , 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R, Zhenhuan G, Luxenburg C. (2015). The contractome—a systems view of actomyosin contractility in non-muscle cells. J Cell Sci , 2209–2217. [DOI] [PubMed] [Google Scholar]
- Zeece MG, Robson RM, Bechtel PJ. (1979). Interaction of alpha-actinin, filamin and tropomyosin with F-actin. Biochim Biophys Acta , 365–370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.