Crossovers are essential for the accurate segregation of chromosomes, but it is important that they be properly positioned. Crossovers must not occur too close to the centromere, but reduced crossover density may extend outward for some distance...
Keywords: Drosophila, meiotic recombination, heterochromatin, centromere effect, crossover patterning
Abstract
Crossovers are essential in meiosis of most organisms to ensure the proper segregation of chromosomes, but improper placement of crossovers can result in nondisjunction and aneuploidy in progeny. In particular, crossovers near the centromere can cause nondisjunction. Centromere-proximal crossovers are suppressed by what is termed the centromere effect, but the mechanism is unknown. Here, we investigate contributions to centromere-proximal crossover suppression in Drosophila melanogaster. We mapped a large number of centromere-proximal crossovers, and find that crossovers are essentially absent from the highly repetitive (HR)-heterochromatin surrounding the centromere but occur at a low frequency within the less-repetitive (LR)-heterochromatic region and adjacent euchromatin. Previous research suggested that flies that lack the Bloom syndrome helicase (Blm) lose meiotic crossover patterning, including the centromere effect. Mapping of centromere-proximal crossovers in Blm mutants reveals that the suppression within the HR-heterochromatin is intact, but the distance-dependent centromere effect is lost. We conclude that centromere-proximal crossovers are suppressed by two separable mechanisms: an HR-heterochromatin effect that completely suppresses crossovers in the HR-heterochromatin, and the centromere effect, which suppresses crossovers with a dissipating effect with distance from the centromere.
CROSSOVERS are essential for the proper segregation of homologous chromosomes in meiosis, as evidenced by the fact that chromosomes lacking a crossover frequently segregate improperly in meiosis I (Koehler et al. 1996; Lamb et al. 1996). However, it is not only the presence of crossovers that is important, but also their proper placement along the chromosome, as apparent meiosis II nondisjunction occurs primarily between chromosomes that experienced a centromere-proximal crossover, both for the Drosophila melanogaster X chromosome (Koehler et al. 1996) and for human chromosome 21 (Lamb et al. 1996). Beadle (1932) first reported that meiotic crossovers are reduced near the centromere—a phenomenon now referred to as the centromere effect.
Meiotic recombination is initiated by DNA double-strand breaks (DSBs), each of which can be repaired to give crossover or noncrossover products through a tightly controlled decision (Lake and Hawley 2016). In addition to the centromere effect, interference and assurance also govern crossover patterning. Interference is the phenomenon where one crossover suppresses the occurrence of another crossover nearby [Sturtevant 1913; reviewed in Berchowitz and Copenhaver (2010)]. Assurance is the phenomenon in which each pair of homologous chromosomes almost always receive at least one crossover regardless of size [Mather 1937; reviewed in Jones and Franklin (2006)]. The effect of these crossover patterning phenomena on DSB repair results in the typical crossover distribution where most crossovers occur in the middle to distal end of the chromosome, and are decreased near the centromere. The mechanisms of these phenomena are largely undescribed and remain elusive. In this study, we use Drosophila melanogaster to gain insight into how crossovers are suppressed in centromere-proximal regions.
Approximately one-third of each Drosophila chromosome is composed of highly repetitive, pericentromeric satellite sequence arrays in a heterochromatic state. Heterochromatin may play a role in decreasing crossovers in the pericentric regions by being accessible to proteins that either make DSBs or repair them into crossovers. Support for this comes from cytological studies, where Mehrotra and McKim (2006) observed no DSBs colocalizing with the heterochromatic mark HP1. Additionally, dominant Su(var) (suppressor of position-effect variegation) mutations, that likely cause heterochromatin to assume a more open structure, cause an increase in crossovers within the pericentromeric heterochromatin (Westphal and Reuter 2002). These results support the idea that suppression of crossovers near the centromere is due to exclusion of DSBs from heterochromatin.
Heterochromatin cannot be the only cause of the centromere effect, as early studies on the centromere effect involving chromosome rearrangements in Drosophila show that crossover suppression extends into the euchromatin. Mather (1939) showed that a euchromatic region moved closer to the centromere, but nearer to a smaller amount of heterochromatin, experienced a greater decrease in crossovers than did a region moved slightly farther away from the centromere, but nearer to a larger amount of heterochromatin. He concluded that the decrease in crossovers was due to proximity to the centromere rather than the proximity to heterochromatin. Yamamoto and Miklos (1978) studied X chromosomes in Drosophila that had large deletions of the pericentromeric heterochromatin, and showed that the larger the deletion, the farther the decrease in crossovers spread into the euchromatin. They concluded that centromere-proximal crossover suppression does not depend on the amount of heterochromatin, but on distance from the centromere. Nonetheless, the question still remains whether heterochromatin has the ability to decrease crossovers in adjacent euchromatic regions; we address that question in this work.
Heterochromatin is not homogeneous and may not behave uniformly throughout. In polytene chromosomes, two distinct classes of heterochromatin have been described: alpha-heterochromatin is the small, densely staining region of the chromocenter that is highly underreplicated in this tissue, whereas beta-heterochromatin is more diffusely staining and is moderately replicated (Gall et al. 1971; Ashburner 1980; Lamb and Laird 1987; Miklos and Cotsell 1990). Heterochromatin also is not homogeneous based on sequence composition. Regions of pericentric heterochromatin adjacent to the euchromatin are composed of blocks of transposable elements (TEs) with varying amounts of repeats and interspersed unique sequence. This has made it possible to assemble these regions in the reference genome (Hoskins et al. 2015). Chromatin domains identified in cell lines show that much of this sequence is heterochromatic or transcriptionally silent (Filion et al. 2010; Thurmond et al. 2019). In contrast, sequences closer to the centromere are highly repetitive, consisting largely of blocks of tandemly arrayed satellite sequences. These have not been assembled to the reference genome, but in situ hybridization of satellite sequences on rearranged chromosomes and long-read sequencing have permitted assembly of some satellite arrays (Lohe et al. 1993; Khost et al. 2017). We will refer to the two types of heterochromatin as highly repetitive (HR)-heterochromatin and less-repetitive (LR)-heterochromatin.
In this study, we investigate the role of the two types of heterochromatin and the centromere effect in suppressing pericentromeric crossovers. We show that centromere-proximal crossover suppression is mediated by both a HR-heterochromatin effect that presumably results from exclusion of DSBs from highly repetitive heterochromatin, and a centromere effect that is a meiosis-specific mechanism that decreases with increasing distance from the centromere.
Materials and Methods
Drosophila stocks
Flies were maintained on standard medium at 25°. Mutant alleles that have been previously described include BlmN1 and BlmD2 (McVey et al. 2007). BlmN1/BlmD2 mutants experience maternal-effect lethality, which was overcome using the UAS::GAL4 system with the matα driver as previously described (Kohl et al. 2012).
Phenotypic crossover distribution assay
Crossover distributions were obtained in test crosses of the following females to males carrying all the recessive markers:
Crossovers between px and sp were scored by crossing virgin pxsp / + to homozygous px sp males. Additionally, px bwDsp / + and pxsp / bwD were crossed to pxsp homozygous males for scoring this interval in a bwD background. Crossovers in Blm mutants were scored the same way as chromosome 2L in wild type. Each cross was set up as a single experiment with at least 20 vials that were flipped after 3 days. After a further 3 days, parents were removed from the second set of vials. All progeny were scored for parental and recombinant phenotypes for 5 days from all vials. Genetic distances are given as centiMorgans (cM; equivalent to map units), where cM = (number of crossovers/total number of flies)*100. See Supplemental Material, Table S1 for phenotypic crossover distribution data.
Fine mapping of centromere-proximal crossovers
Crossovers were finely mapped near the centromere using single-nucleotide polymorphisms (SNPs) and indels between isogenized strains. Illumina whole-genome sequencing was performed on each isogenized strain and genomes were assembled to the Dm6 D. melanogaster reference sequence (Hoskins et al. 2015), using BBMap (version 37.93, Bushnell 2014). SNPs and indels were called in comparison to the reference sequence using SAMtools mpileup (Sversion 1.7, Li et al. 2009; Li 2011), and then compared between strains using VCFtools (version 0.1.14, Danecek et al. 2011). Primers were designed to amplify only the wild-type chromosome so that each SNP/indel could be genotyped. See Table S2 for list of primers and locations. Centromere-proximal crossovers were identified by phenotypic markers on each chromosome. For all chromosomes, crosses were set up between a wild-type chromosome and a chromosome with recessive markers; females heterozygous for these were collected and crossed to males homozygous for the recessive markers, and progeny were scored. Crossovers were collected between f and Dp(1;1)y+ on the X chromosome, between b and vg on chromosome 2, and between h and e on chromosome 3. See Tables S3 and S4 for crossover distribution results from fine mapping for wild type and Blm mutant, respectively.
Drosophila whole mount ovary immunofluorescence
About ten 3- to 4-day-old virgins were kept in a vial with yeast paste overnight with a few males. Ovaries were dissected in phosphate-buffered saline (PBS) and incubated for 20 min in fixative buffer (165 µl fresh PBS, 10 µl NP-40, 600 µl heptane, 25 µl 16% paraformaldehyde). Ovaries were washed three times in PBST (1× PBS + 0.1% Tween-20), then incubated in blocking solution (PBST + 1% BSA). Ovaries were then incubated in primary antibody diluted in blocking solution at 4°, then washed three times in PBST and incubated in secondary antibody diluted in blocking solution. After antibody incubation ovaries were washed three times quickly in PBST and mounted with DAPI Fluoromount-G (Thermo Scientific). Antibodies for H3K9me3 (39161; Active Motif) and C(3)G (Anderson et al. 2005) were used.
Generation of fluorescence in situ hybridization probes
BAC clone (BAC PAC RPCI-98 library) DNA was extracted using a MIDI-prep kit (#740410; Clontech). The probe for the bw locus was Clone BACR48M01. BAC DNA was used in a nick-translation reaction to create biotinylated probes. Nick translation reaction: 5 µl 10× DNA Pol I buffer, 2.5 µl dNTP mix (1 mM each of dCTP, dATP, dGTP), 2.5 µl biotin-11-dUTP (1 mM), 5.0 µl 100 mM BME, 10 µl of freshly diluted dDNase I, 1 µl DNA Pol I, 1 µg of template DNA, water up to 50 µl. The reaction was incubated in a thermocycler at 15° for 4 hr. The fluorescence in situ hybridization (FISH) probe was purified using PCR purification kit (Qiagen) and quantified using Qubit (Q32854; Thermofisher), then diluted to 2 ng/µl in hybridization buffer [2× saline-sodium citrate (SSC) buffer, 50% formamide, 10% w/v dextran sulfate, 0.8 mg/ml salmon sperm DNA]. AACAC oligonucleotide probe was obtained from Integrative DNA Technologies (IDT, www.idtdna.com). Sequence: Cy3-(AACAC)7.
Drosophila whole mount ovary IF-FISH
Ovaries were dissected as described above, incubated in fixative buffer for 4 min (100 mM sodium cacodylate (pH 7.2), 100 mM sucrose, 40 mM potassium acetate, 10 mM sodium acetate, 10 mM EGTA, 5% paraformaldehyde), washed four times quickly in 2× SSCT [5 ml 20× saline sodium citrate (SSC), 50 µl Tween-20, up to 50 ml with water], washed 10 min in 2× SSCT + 20% formamide, 10 min 2× SSCT + 40% formamide, then two times 10 min in 2× SSCT + 50% formamide. Ovaries were predenatured by incubating at 37° for 4 hr, 92° for 3 min, 60° for 20 min. Probe(s) was added and ovaries were incubated in a thermocycler at 91° for 3 min then overnight at 37°. Ovaries were then washed with 2× SSCT + 50% formamide at 37° for 1 hr, then in 2× SSCT + 2% formamide for 10 min at room temperature (RT), then in 2× SSCT quickly four times. Ovaries were then incubated in blocking solution (6 mg/ml NGS in 2× SSCT) for 4 hr, then washed quickly three times in 2× SSCT. Ovaries were incubated overnight in primary antibody diluted in 2× SSCT at RT, then washed three times quickly in 2× SSCT, incubated with secondary antibody diluted in 2× SSCT for 2 hr, then washed three times quickly in 2× SSCT. Ovaries were then incubated with streptavidin (1.5 µl of 488-conjugated streptavidin diluted in 98.5 µl detection solution [0.5 ml 1 M Tris, 400 mg BSA, water to 10 ml]) for 1 hr at RT, washed two times quickly in 2× SSCT, 1 hr in 2× SSCT, then 3 hr in 2× SSCT. Ovaries were then mounted in DAPI fluoromount. Primary antibody for C(3)G (Anderson et al. 2005) was used.
Imaging and quantification
Images of whole-mount germaria were taken using a Zeiss LSM710 confocal laser scanning microscope using 40× oil-immersion objective. Images were saved as .czi files and processed using FIJI (ImageJ). Distance between foci for Figure 3 was measured using FIJI. Images were obtained as single 0.5 µm z-slices and distance was measured between foci in both single and combined stacks to account for 3D orientation of foci. Distances were compared using unpaired t-test.
Figure 3.
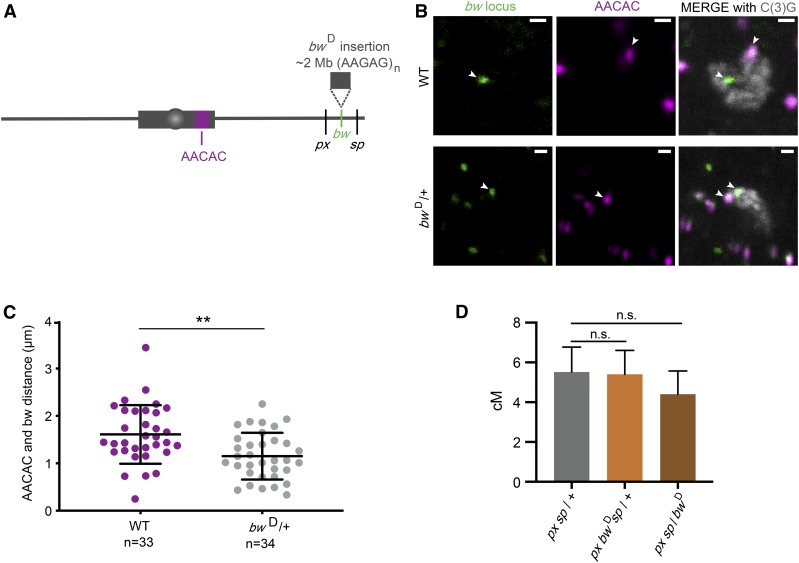
Insertion of a block of heterochromatin does not decrease crossovers. (A) Schematic of the bwD mutation and AACAC locus used for staining. (B) Representative staining for bw locus (left panels), AACAC locus (middle panels), and C(3)G to identify meiotic cells (merged with foci in the right panels). White arrows point to the foci in all images; WT, wild type. (C) Quantification of the distance between foci in wild type (WT) and bw in bwD/+ flies. (** P < 0.001). (D) Recombination between px and sp for pxsp / + (5.5 cM; n = 1287), px bwDsp / + (5.4 cM; n = 1363), and pxsp / bwD (4.4 cM; n = 1197). Error bars represent 95% confidence intervals. (pxsp / + vs. px bwDsp / +, n.s. P = 0.86) (pxsp / + vs. pxsp / bwD, n.s. P = 0.32). For full data set, see Table S6.
Statistical methods and modeling
Fisher’s Exact Test was used to compare total crossovers to total number of flies; two-tailed P values are reported. Proximal crossover distributions in wild type and Blm mutants were compared by both a chi-squared test and a G test for goodness-of-fit, using the wild-type distribution (fraction of crossovers in each interval) as the expected values and Blm as observed. To determine whether crossover distributions are nonrandom, we used deviat (Cirulli et al. 2007; LaFave et al. 2014). We obtained the deviat code from Mohamed Noor and rewrote it for MATLAB (available at https://github.com/sekelsky/deviat). For cM/Mb, 95% confidence intervals were calculated from confidence intervals of the proportion of crossovers in the sample size, using the method of Wilson (1927) implemented at http://www.vassarstats.net/prop1.html.
We conducted an analysis of crossover density using a model averaging approach (Burnham et al. 2011). In this approach, models of varying composition and complexity are weighted according to their ability to fit the data parsimoniously, then averaged to construct predictions and inference. A benefit of this approach is lack of picking one best model when uncertainty exists among a set of candidate models. Similarly, there are no hard P-value cutoffs which can be used to artificially exclude weak, but potentially important variables. All statistical analyses were completed using the R language (version 3.6; R Core Team 2019).
The count of crossovers in each chromosome section was modeled with negative binomial regressions fit using maximum likelihood using the MASS library (version 7.3–51.4; Venables and Ripley 2002). All models use a log link function to relate the linear combination of predictor variables to the mean number of crossovers. All models also include an offset variable (a variable whose slope is assumed to be 1) of the log (# of number of flies × length of chromosome section). This offset accounts for the different sampling involved in each observation and can be thought of changing the model to one fitting the density of crossovers per fly per section. Prior to fitting, all quantitative variables were centered and standardized by dividing by two times the SD of the variable.
The most complex or “global model” included, in addition to the offset, linear additive effects of the density of TEs and gene density and a quadratic response to distance from the centromere (distance from the centromere is calculated as distance from the end of the genome assembly for each chromosome arm):
All subsets of this model that included the quadratic effect of distance, only when there was a linear effect of distance, were fit. Model selection and averaging were conducted using the MuMIn library (version 1.4.36; Barton 2019). We fit all possible submodels of the global. This led to 150 models being fit. We used the corrected Akaike Information criterion (AICc) as our measure of model performance and selected a final model set based on a 95% confidence set and then calculated model averaged estimates of coefficients and their SE. Models that had higher AICc than nested models were excluded based on the recommendation of Richards et al. (2011) to avoid including overly complex models that do not improve model performance.
Data availability
All data necessary for confirming the conclusions in this paper are included in this article and in supplemental figures and tables. Drosophila stocks described in this study are available upon request. We have uploaded Supplemental Material to Figshare. Table S1 includes complete data set for crossovers between phenotypic markers in wild type and Blm. Table S2 includes all primers used for SNP/indel genotyping between isogenized strains of D. melanogaster. Table S3 includes all data for mapping of crossovers using the SNP/indel method for wild type including chromosome, interval size, number of crossovers, # of genes, # TEs, and total number of flies scored for each interval. Table S4 includes all data for mapping of crossovers using the SNP/indel method for Blm mutant. Table S5 has P values for analysis of randomness of crossover distribution. Table S6 includes crossover data between px and sp for wild type and bwD mutants. Table S7 includes model averaged standardized effect sizes for each chromosome. Table S8 includes the 95% confidence set for wild type chromosome analysis. Table S9 includes modeled average parameters for mutant analysis. Table S10 includes 95% confidence set for 2L Blm mutant chromosome analysis. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8865191.
Results
Pericentromeric crossover distribution
To gain a deeper understanding of the centromere effect, we sought to more finely map centromere-proximal crossovers. Crossovers near the centromere have classically been mapped using phenotypic markers in the euchromatin on either side of the centromere. Additionally, whole-genome mapping has been used to more precisely map crossovers within the genome (e.g., Comeron et al. 2012; Miller et al. 2016). However, these methods have caveats that do not allow us to fully understand the distribution of centromere-proximal crossovers. Using phenotypic markers to map crossovers limits resolution to only the most centromere-proximal markers used. Whole-genome mapping provides precise locations of crossovers, but only a handful of centromere-proximal crossovers have been mapped using this method. For example, from whole-genome sequencing of 98 flies, only one crossover was mapped between the markers pr and cn that flank the chromosome 2 centromere (Miller et al. 2016). We therefore developed a method to map a large number of crossovers with more precision than phenotypic mapping allows, allowing us to gain a better understanding of the relationship between crossover distribution in euchromatin and the two types of heterochromatin (LR-heterochromatin and HR-heterochromatin).
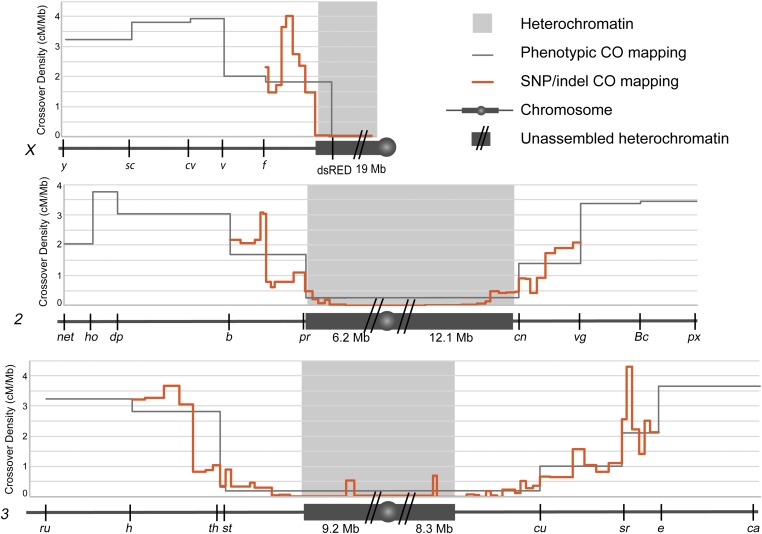
We collected proximal crossovers between isogenized Drosophila chromosomes, then more finely mapped these using SNP and indel markers to intervals that range from 0.23 to 1.9 Mb. We mapped ∼160–300 crossovers per chromosome arm. This mapping shows that crossovers are decreased near the centromere and increase in frequency with distance from the centromere (Figure 1). Interestingly, we see a low frequency of crossovers in the assembled LR-heterochromatin, but crossover frequency goes down to nearly zero in the highly repetitive heterochromatin on every chromosome arm. Of 37,219 total flies scored, only three, all on chromosome 2, inherited a crossover between the most centromere-proximal SNPs/indels used in our mapping. These crossovers may still have occurred within LR-heterochromatin, either proximal to our most proximal markers, or in sequences not included in the genome assembly. Alternatively, they may have been within HR-heterochromatin or unique sequences embedded within HR-heterochromatin. We cannot exclude the possibility that these crossovers are mitotic in origin. The fact that we do see a small number of crossovers in the less-repetitive heterochromatin was surprising because it has been reported that DSBs do not colocalize with heterochromatic markers (Mehrotra and McKim 2006; see Discussion).
Figure 1.
Fine mapping of centromere-proximal crossovers. Chromosomes are represented under each graph (X, 2, 3) with euchromatin (dark gray line), heterochromatin (dark gray box), unmapped heterochromatin (dark gray box with two slashes), and the centromere (dark gray circle). Approximate amount of heterochromatin is displayed underneath chromosome for each chromosome arm (values obtained from Hoskins et al. 2002). Heterochromatin boundaries (light gray blocks) are based on H3K9me2 ChIP array boundaries (Riddle et al. 2011). Phenotypic markers used for mapping crossovers are indicated on each chromosome. Crossover density (centiMorgan/megabase) is plotted for crossovers scored between phenotypic markers (gray line), and for crossovers scored using SNP/indel mapping (orange line). There were no crossovers between the most proximal SNP/indel markers on chromosome X and 3, but there were three crossovers in this region on chromosome 2. Number of crossovers mapped was 160 for chromosome X, 415 for chromosome 2, and 622 for chromosome 3. For full data set, see Tables S1 and S3.
The distribution of crossovers is significantly nonrandom on every arm except the X (Table S5), but generally increases from proximal to distal (see below for modeling). On some arms, most notably 2L and 3R, there are euchromatic intervals that seem to have higher crossover density than flanking intervals (Figure 1). These are unlikely to represent hotspots of the type reported for other model organisms, but rather represent fine-scale variation like that reported by Comeron et al. (2012), who found that warm and cool regions vary between strains, Thus, it would be interesting to repeat our analysis with different chromosome isolates.
Our fine mapping gives a clearer understanding of crossover distribution near the centromere, but to begin understanding the contribution to this distribution, we analyzed a mutant that does not experience centromere-proximal crossover suppression.
Genetic separation of the centromere effect and the HR-heterochromatin effect
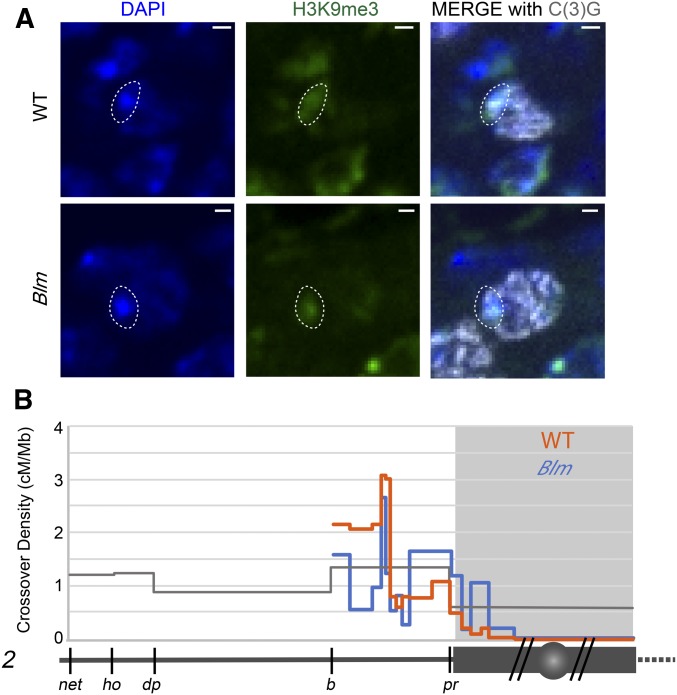
If the centromere effect is genetically controlled, we would anticipate that it should be possible to identify mutants that do not experience suppression of crossovers near the centromere. Hatkevich et al. (2017) identified a mutant that they hypothesized does not experience the centromere effect. Drosophila Blm helicase, like Saccharomyces cerevisiae Sgs1, has been proposed to direct DSBs down the meiotic DSB repair pathway to allow the proper crossover patterning (De Muyt et al. 2012; Zakharyevich et al. 2012; Hatkevich et al. 2017). The conclusion that Blm mutants do not have the centromere effect was based on a flat distribution of crossovers and a measure of the strength of the centromere effect (Hatkevich et al. 2017). That study only mapped crossovers using phenotypic markers in the euchromatin on either side of the centromere, so it was unclear whether the effects of loss of Blm are limited to the assembled parts of the genome (euchromatin and LR-heterochromatin) or extend into the HR-heterochromatin. To answer this question, we mapped proximal crossovers in Blm mutants using the SNP/indel mapping approach. Importantly, Blm mutants appear to have normal heterochromatin in pachytene as evidenced by H3K9me3 staining colocalizing with DAPI dense, heterochromatic regions (Figure 2A).
Figure 2.
Fine mapping of centromere-proximal crossovers in Blm mutants. (A) Heterochromatin staining in wild type and in Blm mutants. Left: DAPI staining of DNA; middle: H3K9me3 staining for heterochromatin; right: merge. The dotted circle outlines the DAPI-bright region that overlaps with heterochromatin, showing that Blm mutants have normal localization of heterochromatin. Images are from a single 0.5 µm z-slice. (B) SNP/indel mapping of crossovers on 2L in wild type (orange) and Blm mutants (blue). Symbols are the same as in Figure 1. For full data sets, see Tables S1 and S4.
SNP/indel mapping of Blm mutants reveals a relatively flat distribution of crossovers throughout the chromosome arm and into the assembled LR-heterochromatic sequence (Figure 2B). This distribution is significantly different than that of wild-type flies (P < 0.0001 by both chi-squared and G tests for goodness of fit). We used deviat (Cirulli et al. 2007; LaFave et al. 2014) to compare the observed distribution of crossovers to that expected if interval size is the sole determinant of number of crossovers (see Materials and Methods; Table S5). In wild-type flies, the crossover distribution is significantly different from expected (P < 0.0001 for every arm except X), but this is not the case in Blm mutants (P = 0.9926). As in wild type, Blm mutants experience no crossovers within the HR-heterochromatin. We conclude that suppression of proximal crossovers can be separated into two phenomena: the HR-heterochromatin effect, defined as the virtual absence of crossovers within highly repetitive heterochromatin, and the centromere effect, which has a dissipating effect with distance from the centromere. We hypothesize that the HR-heterochromatin effect is likely due to the absence of DSBs in this region, whereas the centromere effect is achieved through regulation of DSB repair outcome.
Heterochromatin is not sufficient to produce a centromere effect
We sought to test whether the HR-heterochromatin effect and centromere effect can be separated by measuring recombination around a heterochromatic locus that is distant from the centromere. We did this by using the bwD mutation, which has an insertion of ∼2 Mb of heterochromatin in the bw locus on distal chromosome 2R (Slatis 1955; Dernburg et al. 1996) (Figure 3A). This mutation causes dominant suppression of the bw gene by pairing with its homolog, and causing localization near the pericentromeric heterochromatin of chromosome 2 (Henikoff and Dreesen 1989; Dreesen et al. 1991; Henikoff et al. 1995; Dernburg et al. 1996). We used this tool to address two questions: First, does an insertion of heterochromatin located far from the centromere suppress crossovers in adjacent intervals? Second, does spatial proximity to pericentromeric heterochromatin within the nucleus suppress crossovers?
We first asked whether the heterochromatic insertion of bwD causes nuclear localization of the locus near clustered pericentromeric heterochromatin in meiotic cells in the same fashion as it does in somatic cells. We used a probe for the bw locus and a probe for a repeat in the pericentromeric heterochromatin of chromosome 2 (AACAC), as well as a marker of meiotic cells [C(3)G, a component of the synaptonemal complex (SC), the protein structure that forms between paired homologous chromosomes] (Figure 3B). We then measured the distance between the two foci in meiotic cells and see that the distance between the bw locus and AACAC heterochromatin locus is significantly shorter in bwD compared to wild type (P < 0.001) (Figure 3C). This suggests that the heterochromatic insertion in bwD does localize near the pericentromeric heterochromatin in meiotic cells.
We measured recombination between phenotypic markers on either side of the bw locus, px and sp. The px gene is located at 2R:22.5 Mb; sp is not mapped to the genome but is between or at 2R:24.0 Mb and Kr at 2R:25.2 Mb, so the distance between px and sp is between 1.5 and 2.7 Mb (Thurmond et al. 2018). If the heterochromatic insertion in bwD leads to suppression of crossovers in adjacent regions we would expect to see a decrease in crossovers between px and sp. We assume there are no crossovers within the heterochromatin of the bwD mutation since we measured crossovers in flies heterozygous for this mutation, both in cis and in trans to px and sp. There was no significant difference in number of crossovers in flies with or without bwD (P = 0.86 and P = 0.32; Figure 3D), suggesting that the heterochromatin insertion does not cause a decrease in crossovers in the adjacent regions and that spatial proximity to the pericentromeric heterochromatin compartment of the nucleus does not have a strong effect on crossing over.
Genomic contributions to the centromere effect
The results with bwD suggest that the centromere effect is not due solely to proximity to pericentromeric heterochromatin, so we asked whether other genomic features contribute to the centromere effect. TE density and gene density have been suggested to influence crossover rates genome-wide in other organisms (reviewed in Kent et al. 2017). TEs are middle-repetitive elements found throughout the genome but are most abundant within LR-heterochromatin adjacent to euchromatin (Yamamoto et al. 1990; Carmena and González 1995). Conversely, genes are less abundant in the LR-heterochromatin than in the euchromatin. In Arabidopsis thaliana crossovers are negatively correlated with TE density and positively correlated with gene density (Giraut et al. 2011). Therefore, we searched for correlations between crossover distribution and distance from the centromere, TE density, and gene density.
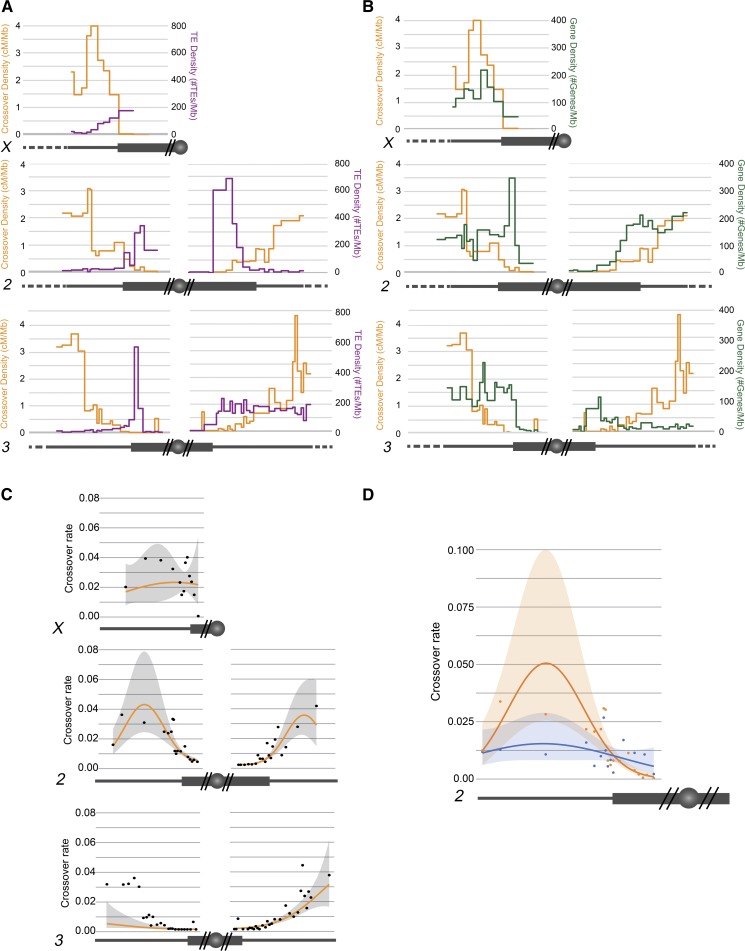
Figure 4, A and B show TE and gene density overlaid with our SNP/indel mapping of proximal crossovers. We modeled how distance from the centromere, TE density, and gene density contribute to the variation seen in crossover distribution (Figure 4C). Two models were selected in the 95% confidence set (Tables S7 and S8). All predictor variables were included in this final set indicating statistically important effects of distance from the centromere, TE density, and gene density that varied across chromosomes. Unless otherwise stated, all effects mentioned have 95% confidence intervals that do not overlap zero. For all chromosomes except X, distance from the centromere had a positive effect and a negative squared distance term. Two chromosome arms, 2R and X, had positive effects of gene density; on 3R, a negative effect was found with 95% confidence intervals just overlapping zero, suggesting a potential negative effect. In general, standardized effect sizes for gene density were lower than for distance from the centromere. For TE density, all chromosomes but X had 95% confidence intervals that did not overlap zero. The effect was dramatically negative in 3R, with a negative standardized effect size of magnitude over three times greater than the next effect size. Other chromosomes had smaller magnitude effect size, being negative for 2L, 3L, and 3R, but positive for 2R. This modeling shows that TE and gene density do decrease the variation seen in the model; however, they contribute less to the model produced than the effect of distance from the centromere. These results support the idea that centromere-proximal crossover distribution is dictated not only by genomic features such as TE or gene density, but that there is some factor suppressing crossover rate that decreases with distance from the centromere.
Figure 4.
Distribution of TE and gene density. Chromosomes are depicted under each graph, as in Figure 1. (A and B) Crossover distribution from SNP/indel mapping (orange) represented on left axis as centimorgan/megabase. (A) Transposable element (TE) density (purple) plotted on right axis as number of TEs/Mb. (B) Gene density (green) plotted on right axis as number of genes/megabase. (C) Crossover rate in relation to distance from the centromere. Observed data are plotted along with modeled marginal relationship with distance from the centromere. For the marginal predictions, gene density, and TE density were set at their mean value across each chromosome. (D) Crossover rate in relation to distance from the centromere for chromosome 2L for Blm mutant and wild type. Observed data are plotted along with modeled marginal relationship with distance from the centromere. For the marginal predictions, gene density, and TE density were set at their mean value across each chromosome. For statistical analyses, see Tables S7–S10.
We applied the same modeling methods to the Blm mutant to understand whether these mutants truly do not have a centromere effect, and to what extent TE and gene density play a role in crossover distribution in Blm mutants (Figure 4D). Two models were selected in the 95% confidence set (Tables S9 and S10). There was no effect of gene density in either wild type or mutant, consistent with analysis of the wild-type chromosomes. In the wild type, all remaining modeled effects (distance, distance2, and TE density) had 95% confidence intervals that did not overlap zero. In the Blm mutant, no effect size had confidence intervals that did not overlap zero, suggesting that none of them were valuable predictors of crossover rate. While we cannot prove zero effect, the best estimated effect of distance in the mutant is less than one-quarter that of the wild type (Table S9). These modeling results support the hypothesis that Blm mutants experience a much weaker centromere effect, if any, and that the crossover distribution in Blm is not demonstrably under the influence of distance from the centromere or chromosome characteristics. Importantly, these results provide more evidence that centromere-proximal crossover suppression is mediated both an HR-heterochromatin effect and another effect whose strength varies with distance to the centromere and is under genetic control.
Discussion
Two contributions to suppression of proximal crossovers
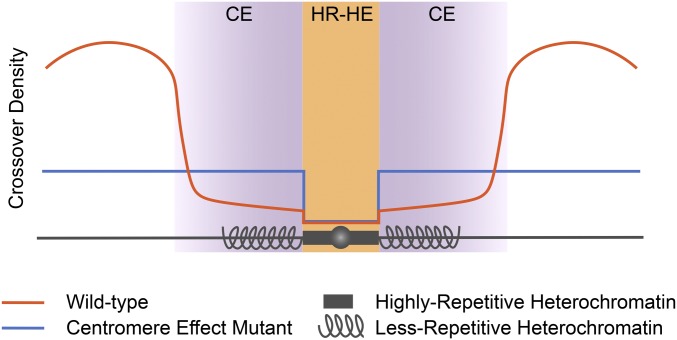
Our mapping of a large number of proximal crossovers in both wild-type flies and Blm mutants leads us to propose a model for centromere-proximal crossover suppression (Figure 5). In this model, crossovers are completely suppressed in HR-heterochromatin due to the absence of DSBs. Adjacent to this region the centromere effect strongly suppresses crossovers, but that suppression dissipates with distance from the centromere until a region in the euchromatin where crossovers rise steeply to peak around the middle of each chromosome arm (orange line). In the Blm mutant (blue line), the HR-heterochromatin effect is still intact, but the centromere effect is lost: crossover density is relatively even throughout the assembled LR-heterochromatin and euchromatin. We conclude that pericentromeric crossover suppression is achieved by both HR-heterochromatin suppression and a centromere effect, and these two processes are separable.
Figure 5.
Model for suppression of centromere-proximal crossovers. The HR-heterochromatin effect (HR-HE) (orange) and centromere effect (CE) (purple) are both responsible for suppressing crossovers in the pericentromeric region. The HR-HE completely suppresses crossovers in the highly repetitive heterochromatin (dark gray box), but does not have an effect outside of this region. The centromere effect starts in the less-repetitive heterochromatin (spiral gray lines), and extends into the euchromatin of each arm (straight gray lines). Representative crossover distribution is shown for wild type (orange) and a centromere effect mutant (blue).
Suppression of crossovers by heterochromatin
Heterochromatin has long been thought to contribute to centromere-proximal suppression of crossovers, but the specifics of where this suppression occurred were unknown. In this study, we used a centromere effect mutant (Blm) that still has apparently normal heterochromatic marks to show that the heterochromatin effect impacts the highly repetitive heterochromatin but not the adjacent less-repetitive heterochromatin. This was a surprising result because a previous cytological study did not detect any colocalization between cytological markers of DSBs and heterochromatin (Mehrotra and McKim 2006). It is possible that, although DSBs do occur within less-repetitive heterochromatin, they are at a lower density than in euchromatin (perhaps by being excluded from TEs) so the sample size in the previous study was insufficient to detect these relatively rare DSBs.
We used bwD to test whether HR-heterochromatin distant from the centromere exerts a centromere effect. The lack of an effect on crossing over between px and sp suggests that HR-heterochromatin is not sufficient to reduce crossovers in flaking regions. We could not assay the effects of homozygosity for bwD because homozygotes were inviable, even for chromosomes that had the nearby markers px and sp recombined onto bwD. Slatis (1955) conducted similar experiments and reported a decrease in crossovers in flies homozygous for bwD. This may suggest that HR-heterochromatin does affect adjacent euchromatin even when distant from the centromere. However, Slatis also reported a decrease in flies heterozygous for bwD, in contrast to our findings; the reasons for this difference are unknown. It might be informative to revisit these studies and map crossovers between px, bwD, and sp more precisely, like we did for proximal crossovers.
Why can crossovers occur within the less-repetitive heterochromatin, but not the highly repetitive heterochromatin? One reason could be differential access of DSB machinery to the DNA. Perhaps the dense packing of HR-heterochromatin does not allow access of the DSB machinery. Additionally, there could be different heterochromatic marks or protein machinery in these regions that differentially regulate DSBs or crossover formation. Carpenter (1975) noted that the SC is morphologically different in pericentromeric heterochromatin vs. euchromatic arms, being less distinct (under the electron microscope) in pericentromeric regions. In Drosophila, the SC is required for most DSBs (Jang et al. 2003); perhaps pericentromeric SC does not support DSB formation. Interestingly, the boundary between heterochromatic and euchromatic SC is gradual rather than distinct (Carpenter 1975), potentially providing in a distance-dependent effect of DSB density. If the centromere effect is related to a gradient in DSB density, then one would expect to see a similar pattern in noncrossover gene conversion events. It is difficult to map noncrossover events in the LR-heterochromatin due to the high density of TEs and correspondingly low density of SNPs and indels. However, the analysis of Miller et al. (2016) concluded that noncrossovers are not subject to the centromere effect. Their analysis suggests that, at least within euchromatin where most of their noncrossovers mapped, the reduction in proximal crossovers is not due to reduced DSB formation, so must be caused by modulating DSB repair outcome (i.e., crossovers vs. noncrossover).
In the Blm mutant, crossovers do not occur within the highly repetitive heterochromatin, but they do occur outside of that boundary at a higher frequency than in wild type. This suggests that DSBs are made in the less-repetitive heterochromatin at a rate similar to that of the euchromatin, but that in wild type they are more frequently being repaired to noncrossovers rather than to crossovers. Westphal and Reuter (2002) reported an increase in centromere-proximal crossovers in Su(var) mutants, which presumably cause heterochromatin to assume a more open structure. This result suggests that the closed structure of heterochromatin can suppress crossovers, in opposition to our result with Blm mutants that have normal heterochromatic marks but allow more crossovers within the LR-heterochromatin. It is possible that the Blm mutation is altering heterochromatin structure in a way that we did not detect cytologically, although this would represent a previously unreported function for Blm helicase. It would be interesting to look at distribution of heterochromatin marks in meiotic cells of wild-type and Blm mutant flies. This is currently not feasible because of the lack of a method to isolate meiotic nuclei from egg chambers in which 16 cells are connected by cytoplasmic bridges. It is possible that the effect in Su(var) mutations is restricted to either LR-heterochromatin or HR-heterochromatin. It would be informative to conduct our SNP/indel mapping on crossovers in these mutants.
It is notable that Blm mutants also experience crossovers on chromosome 4, which normally never has crossovers (Hatkevich et al. 2017). We hypothesized that chromosome 4 does not have crossovers because of a very strong centromere effect, which is lost in Blm mutants (Hartmann and Sekelsky 2017); the results reported here support this hypothesis. It would be interesting to finely map crossovers on chromosome 4 in Blm mutants to determine if there is a flat distribution and see if there is a separable HR-heterochromatin effect on this chromosome as well, though this would require a marker to the left of the centromere.
Recombination and genomic features
The relationship between gene density, TE density, and recombination rate has been a long-standing discussion (reviewed in Kent et al. 2017). It is difficult to parse out these relationships because there are many factors influencing distribution of TEs, genes, and crossovers. It has been argued that the distribution of TEs and genes is, in part, dictated by recombination. For example, higher recombination could be favored in regions of high gene density to promote greater genetic diversity within populations. Conversely, lower recombination rates in regions of high TE density could help to prevent ectopic recombination between similar TE sequences in different genomic locations. The high density of TEs in proximal or heterochromatic regions could actually result from the low recombination rate preventing removal of TEs (Bartolomé and Maside 2004). Recombination might also be directly silenced within TE sequences. Miller et al. (2016) reported that crossovers can occur within TEs, but less frequently than expected. It has been suggested that active silencing of TEs could lead to the silencing or suppression of recombination around those regions (reviewed in Kent et al. 2017). Therefore, it is difficult to determine whether, or how, TE density and gene density affect recombination rates. Our data support results seen previously in that TE density is increased in areas of low recombination and gene density is increased in areas of high recombination. When we factor these variables into models of crossover distribution, we see a strong impact of TE density on crossover rate. One caveat of our studies is that transposable elements have been shown to vary between different strains of Drosophila, and we have based these analyses off the transposable element distribution within the D. melanogaster reference sequence (Ananiev et al. 1984; Rahman et al. 2015). With advances in long-read sequencing technology, it might be possible in the future to do studies similar to ours but in strains in which LR-heterochromatin has been assembled de novo.
In our experiments, one might expect to observe a correlation between gene and crossover density and an inverse correlation between TE and crossover density simply because genes are less dense and TEs more dense in LR-heterochromatin. However, this was not the case in the experiments of Beadle (1932), Mather (1939), and Yamamoto and Miklos (1978), all of whom measured crossovers in euchromatic sequence that was moved closer to the centromere by chromosome rearrangements. In Beadle’s experiments, crossovers were severely reduced in cu to sr and sr to e intervals in flies homozygous for a translocation that moved about two thirds of 3R onto chromosome 4. These reductions could not be due to number of TEs or genes. It is also unlikely the reductions were due to spreading of heterochromatin across the translocation breakpoint, as this would need to have gone all the way through the 6.9 Mb cu to sr interval.
The centromere effect
The classic experiments with chromosome rearrangements discussed above (Beadle 1932; Mather 1939; Yamamoto and Miklos 1978) argue that the centromere effect is a phenomenon that suppresses crossovers in a manner that is inversely related to proximity to the centromere. Our mapping of centromere-proximal crossovers on structurally normal chromosomes (Figure 1) supports this conclusion. Modeling of crossover distributions finds a strong positive contribution of distance from the end of the genome assembly on each arm of the large autosomes (2 and 3), but not on the X (Figure 4). Several considerations required us to use the end of the genome assembly in our modeling, but the X chromosome result suggests that the relevant feature is really distance from the centromere, including HR-heterochromatin. HR-heterochromatin on 2 and 3 ranges from ∼5 to 8 Mb, but the X has ∼11 Mb of HR-heterochromatin, including the rDNA. Thus, the most proximal euchromatin on the X is already far enough from the centromere to experience only a weak centromere effect. The experiments of Yamamoto and Miklos (1978) support this conclusion. Although the nature of the centromere effect remains unknown, our finding that meiotic DSB repair outcome in Blm mutants is blind to proximity to the centromere indicates that this is a regulated meiotic crossover patterning process.
Conclusions
In conclusion, we find that centromere-proximal crossover suppression is a result of two separable mechanisms: an HR-heterochromatin effect that suppresses crossovers in highly repetitive pericentromeric heterochromatin, and the centromere effect that suppresses proximal crossovers in a manner that dissipates with increasing distance from the centromere. The HR-heterochromatin effect is likely due to the absence of DSBs with satellite sequences, but the mechanism of the centromere effect is unknown. This work is the first in-depth examination of the centromere effect since it was first described, and these findings provide the groundwork for future mechanistic studies of the centromere effect.
Acknowledgments
We thank Juan Carvajal-Garcia for comments on the manuscript. This work was supported by a grant from the National Institute of General Medical Sciences to J.S. under award 1R35 GM-118127.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8865191.
Communicating editor: J. Bateman
Literature Cited
- Ananiev E. V., Barsky V. E., Ilyin Y. V., and Ryzic M. V., 1984. The arrangement of transposable elements in the polytene chromosomes of Drosophila melanogaster. Chromosoma 90: 366–377. 10.1007/BF00294163 [DOI] [Google Scholar]
- Anderson L. K., Royer S. M., Page S. L., McKim K. S., Lai A. et al. , 2005. Juxtaposition of C(2)M and the transverse filament protein C(3)G within the central region of Drosophila synaptonemal complex. Proc. Natl. Acad. Sci. USA 102: 4482–4487. 10.1073/pnas.0500172102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., 1980. Some aspects of the structure and function of the polytene chromosomes of the Diptera. Insect Cytogenetics 10: 65–84. [Google Scholar]
- Bartolomé C., and Maside X., 2004. The lack of recombination drives the fixation of transposable elements on the fourth chromosome of Drosophila melanogaster. Genet. Res. 83: 91–100. 10.1017/S0016672304006755 [DOI] [PubMed] [Google Scholar]
- Barton K., 2019. MuMIn: multi-model inference. Available at: https://cran.r-project.org/package=MuMIn. Accessed on April 15, 2019.
- Beadle G. W., 1932. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc. Natl. Acad. Sci. USA 18: 160–165. 10.1073/pnas.18.2.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz L. E., and Copenhaver G. P., 2010. Genetic interference: don’t stand so close to me. Curr. Genomics 11: 91–102. 10.2174/138920210790886835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham K. P., Anderson D. R., and Huyvaert K. P., 2011. AIC model selection and multimodel inference in behavioral ecology: some background, observations, and comparisons. Behav. Ecol. Sociobiol. 65: 23–35. 10.1007/s00265-010-1029-6 [DOI] [Google Scholar]
- Bushnell B., 2014. BBMap. Available at: https://sourceforge.net/projects/bbmap/. Accessed on March 13, 2016.
- Carmena M., and González C., 1995. Transposable elements map in a conserved pattern of distribution extending from beta-heterochromatin to centromeres in Drosophila melanogaster. Chromosoma 103: 676–684. 10.1007/BF00344228 [DOI] [PubMed] [Google Scholar]
- Carpenter A. T., 1975. Electron microscopy of meiosis in Drosophila melanogaster females. I. Structure, arrangement, and temporal change of the synaptonemal complex in wild-type. Chromosoma 51: 157–182. 10.1007/BF00319833 [DOI] [PubMed] [Google Scholar]
- Cirulli E. T., Kliman R. M., and Noor M. A., 2007. Fine-scale crossover rate heterogeneity in Drosophila pseudoobscura. J. Mol. Evol. 64: 129–135. 10.1007/s00239-006-0142-7 [DOI] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., and Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905 10.1371/journal.pgen.1002905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E. et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A., Jessop L., Kolar E., Sourirajan A., Chen J. et al. , 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46: 43–53. 10.1016/j.molcel.2012.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., Broman K. W., Fung J. C., Marshall W. F., Philips J. et al. , 1996. Perturbation of nuclear architecture by long-distance chromosome interactions. Cell 85: 745–759. 10.1016/S0092-8674(00)81240-4 [DOI] [PubMed] [Google Scholar]
- Dreesen T. D., Henikoff S., and Loughney K., 1991. A pairing-sensitive element that mediates trans-inactivation is associated with the Drosophila brown gene. Genes Dev. 5: 331–340. 10.1101/gad.5.3.331 [DOI] [PubMed] [Google Scholar]
- Filion G. J., Van Bemmel J. G., Braunschweig U., Talhout W., Kind J. et al. , 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224. 10.1016/j.cell.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall J. G., Cohen E. H., and Polan M. L., 1971. Repetitive DNA sequences in Drosophila. Chromosoma 33: 319–344. 10.1007/BF00284948 [DOI] [PubMed] [Google Scholar]
- Giraut L., Falque M., Drouaud J., Pereira L., Martin O. C. et al. , 2011. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 7: e1002354 10.1371/journal.pgen.1002354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M. A., and Sekelsky J., 2017. The absence of crossovers on chromosome 4 in Drosophila melanogaster: imperfection or interesting exception? Fly (Austin) 11: 253– 259 10.1080/19336934.2017.1321181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatkevich T., Kohl K. P., McMahan S., Hartmann M. A., Williams A. M. et al. , 2017. Bloom syndrome helicase promotes meiotic crossover patterning and homolog disjunction. Curr. Biol. 27: 96–102. 10.1016/j.cub.2016.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., and Dreesen T. D., 1989. Trans-inactivation of the Drosophila brown gene: evidence for transcriptional repression and somatic pairing dependence. Proc. Natl. Acad. Sci. USA 86: 6704–6708. 10.1073/pnas.86.17.6704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Jackson J. M., and Talbert P. B., 1995. Distance and pairing effects on the brownDominant heterochromatic element in Drosophila. Genetics 140: 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R. A., Smith C. D., Carlson J. W., Carvalho A. B., Halpern A. et al. , 2002. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol 3: RESEARCH0085. 10.1186/gb-2002-3-12-research0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R. A., Carlson J. W., Wan K. H., Park S., Mendez I. et al. , 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25: 445–458. 10.1101/gr.185579.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J. K., Sherizen D. E., Bhagat R., Manheim E. A., and McKim K. S., 2003. Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci. 116: 3069–3077. 10.1242/jcs.00614 [DOI] [PubMed] [Google Scholar]
- Jones G. H., and Franklin F. C., 2006. Meiotic crossing-over: obligation and interference. Cell 126: 246–248. 10.1016/j.cell.2006.07.010 [DOI] [PubMed] [Google Scholar]
- Kent T. V., Uzunović J., and Wright S. I., 2017. Coevolution between transposable elements and recombination. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372: 20160458 10.1098/rstb.2016.0458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khost D. E., Eickbush D. G., and Larracuente A. M., 2017. Single-molecule sequencing resolves the detailed structure of complex satellite DNA loci in Drosophila melanogaster. Genome Res. 27: 709–721. 10.1101/gr.213512.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler K. E., Boulton C. L., Collins H. E., French R. L., Herman K. C. et al. , 1996. Spontaneous X chromosome MI and MII nondisjunction events in Drosophila melanogaster oocytes have different recombinational histories. Nat. Genet. 14: 406–414. 10.1038/ng1296-406 [DOI] [PubMed] [Google Scholar]
- Kohl K. P., Jones C. D., and Sekelsky J., 2012. Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science 338: 1363–1365. 10.1126/science.1228190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFave M. C., Andersen S. L., Stoffregen E. P., Holsclaw J. K., Kohl K. P. et al. , 2014. Sources and structures of mitotic crossovers that arise when BLM helicase is absent in Drosophila. Genetics 196: 107–118. 10.1534/genetics.113.158618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake C. M., and Hawley R. S., 2016. Becoming a crossover-competent DSB. Semin. Cell Dev. Biol. 54: 117–125. 10.1016/j.semcdb.2016.01.008 [DOI] [PubMed] [Google Scholar]
- Lamb M. M., and Laird C. D., 1987. Three euchromatic DNA sequences under-replicated in polytene chromosomes of Drosophila are localized in constrictions and ectopic fibers. Chromosoma 95: 227–235. 10.1007/BF00294779 [DOI] [PubMed] [Google Scholar]
- Lamb N. E., Freeman S. B., Savage-Austin A., Pettay D., Taft L. et al. , 1996. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat. Genet. 14: 400–405. 10.1038/ng1296-400 [DOI] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. 10.1093/bioinformatics/btr509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J. et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe A. R., Hilliker A. J., and Roberts P. A., 1993. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Genetics 134: 1149–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather K., 1937. The determination of position in crossing-over. II. The chromosome length-chiasma frequency relation. Cytologia Fujii Jubilee: 514–526. [Google Scholar]
- Mather K., 1939. Crossing over and heterochromatin in the X chromosome of Drosophila melanogaster. Genetics 24: 413–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Andersen S. L., Broze Y., and Sekelsky J., 2007. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics 176: 1979–1992. 10.1534/genetics.106.070052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S., and McKim K. S., 2006. Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet. 2: e200 10.1371/journal.pgen.0020200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos G. L., and Cotsell J. N., 1990. Chromosome structure at interfaces between major chromatin types: alpha- and beta-heterochromatin. BioEssays 12: 1–6. 10.1002/bies.950120102 [DOI] [PubMed] [Google Scholar]
- Miller D. E., Smith C. B., Yeganeh Kazemi N., Cockrell A. J., Arvanitakis A. V. et al. , 2016. Whole-genome analysis of individual meiotic events in Drosophila melanogaster reveals that noncrossover gene conversions are insensitive to interference and the centromere effect. Genetics 203: 159–171. 10.1534/genetics.115.186486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman R., Chirn G. W., Kanodia A., Sytnikova Y. A., Brembs B. et al. , 2015. Unique transposon landscapes are pervasive across Drosophila melanogaster genomes. Nucleic Acids Res. 43: 10655–10672. 10.1093/nar/gkv1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2019. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. [Google Scholar]
- Richards S. A., Whittingham M. J., and Stephens P. A., 2011. Model selection and model averaging in behavioural ecology: the utility of the IT-AIC framework. Behav. Ecol. Sociobiol. 65: 77–89. 10.1007/s00265-010-1035-8 [DOI] [Google Scholar]
- Riddle N. C., Minoda A., Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B. et al. , 2011. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 21: 147–163. 10.1101/gr.110098.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatis H. M., 1955. A reconsideration of the brown-dominant position effect. Genetics 40: 247–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1913. The linear arrangement of six sex-linked factors in Drosophila, as shown by their mode of association. J. Exp. Biol. 14: 43–59. 10.1002/jez.1400140104 [DOI] [Google Scholar]
- Thurmond J., Goodman J. L., Strelets V. B., Attrill H., Gramates L. S. et al. , 2019. FlyBase 2.0: the next generation. Nucleic Acids Res. 47: D759–D765. 10.1093/nar/gky1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables W. N., and Ripley B. D., 2002. Modern Applied Statistics with S-PLUS, Ed. 4th Springer, New York: 10.1007/978-0-387-21706-2 [DOI] [Google Scholar]
- Westphal T., and Reuter G., 2002. Recombinogenic effects of suppressors of position-effect variegation in Drosophila. Genetics 160: 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E. B., 1927. Probable inference, the law of succession, and statistical inference. J. Am. Stat. Assoc. 22: 209–212. 10.1080/01621459.1927.10502953 [DOI] [Google Scholar]
- Yamamoto M., and Miklos G. L., 1978. Genetic studies on heterochromatin in Drosophila melanogaster and their implications for the functions of satellite DNA. Chromosoma 66: 71–98. 10.1007/BF00285817 [DOI] [PubMed] [Google Scholar]
- Yamamoto M. T., Mitchelson A., Tudor M., O’Hare K., Davies J. A. et al. , 1990. Molecular and cytogenetic analysis of the heterochromatin-euchromatin junction region of the Drosophila melanogaster X chromosome using cloned DNA sequences. Genetics 125: 821–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K., Tang S., Ma Y., and Hunter N., 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347. 10.1016/j.cell.2012.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data necessary for confirming the conclusions in this paper are included in this article and in supplemental figures and tables. Drosophila stocks described in this study are available upon request. We have uploaded Supplemental Material to Figshare. Table S1 includes complete data set for crossovers between phenotypic markers in wild type and Blm. Table S2 includes all primers used for SNP/indel genotyping between isogenized strains of D. melanogaster. Table S3 includes all data for mapping of crossovers using the SNP/indel method for wild type including chromosome, interval size, number of crossovers, # of genes, # TEs, and total number of flies scored for each interval. Table S4 includes all data for mapping of crossovers using the SNP/indel method for Blm mutant. Table S5 has P values for analysis of randomness of crossover distribution. Table S6 includes crossover data between px and sp for wild type and bwD mutants. Table S7 includes model averaged standardized effect sizes for each chromosome. Table S8 includes the 95% confidence set for wild type chromosome analysis. Table S9 includes modeled average parameters for mutant analysis. Table S10 includes 95% confidence set for 2L Blm mutant chromosome analysis. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8865191.