Xue et al. describe a phenomenon in maize and its nearest wild relative, teosinte, by which backcross progeny of a specific teosinte and maize exhibit a sickly whole-plant phenotype involving changes in morphology, vigor, and viability...
Keywords: Zea mays, maize, teosinte, epigenetic, transposable element (TE), CNVs, sRNAs
Abstract
In the course of generating populations of maize with teosinte chromosomal introgressions, an unusual sickly plant phenotype was noted in individuals from crosses with two teosinte accessions collected near Valle de Bravo, Mexico. The plants of these Bravo teosinte accessions appear phenotypically normal themselves and the F1 plants appear similar to typical maize × teosinte F1s. However, upon backcrossing to maize, the BC1 and subsequent generations display a number of detrimental characteristics including shorter stature, reduced seed set, and abnormal floral structures. This phenomenon is observed in all BC individuals and there is no chromosomal segment linked to the sickly plant phenotype in advanced backcross generations. Once the sickly phenotype appears in a lineage, normal plants are never again recovered by continued backcrossing to the normal maize parent. Whole-genome shotgun sequencing reveals a small number of genomic sequences, some with homology to transposable elements, that have increased in copy number in the backcross populations. Transcriptome analysis of seedlings, which do not have striking phenotypic abnormalities, identified segments of 18 maize genes that exhibit increased expression in sickly plants. A de novo assembly of transcripts present in plants exhibiting the sickly phenotype identified a set of 59 upregulated novel transcripts. These transcripts include some examples with sequence similarity to transposable elements and other sequences present in the recurrent maize parent (W22) genome as well as novel sequences not present in the W22 genome. Genome-wide profiles of gene expression, DNA methylation, and small RNAs are similar between sickly plants and normal controls, although a few upregulated transcripts and transposable elements are associated with altered small RNA or methylation profiles. This study documents hybrid incompatibility and genome instability triggered by the backcrossing of Bravo teosinte with maize. We name this phenomenon “hybrid decay” and present ideas on the mechanism that may underlie it.
PLANT breeders seek to develop improved varieties through crosses among different individuals of a species followed by selection. New combinations of alleles can lead to improved performance, allowing development of elite varieties. In many plant species, the direct combination of genetic information in the two parents can lead to hybrid vigor (heterosis). In other cases, there can be deleterious consequences of crossing individuals that is often referred to as hybrid incompatibility. This phenomenon has been particularly well-studied for crosses between members of related species (Bomblies and Weigel 2007; Rieseberg and Blackman 2010; Fishman and Sweigart 2018). Even within a species there are examples of combinations that can lead to reduced vigor or fertility (Bomblies 2010). In some classic examples, hybrid incompatibility is caused by chromosomal rearrangements such as inversions or translocations that can result in partial sterility (Fishman and Sweigart 2018).
Maize geneticists have a long history of crossing maize (Zea mays ssp. mays) and its wild relatives, the annual Mexican teosintes (Z. mays ssp. parviglumis or Z. mays ssp. mexicana) for a variety of reasons. Beadle studied chromosome pairing in maize-teosinte hybrids and the inheritance of domestication traits in a maize-teosinte F2 population (Beadle 1932, 1972). Kermicle studied pollen-pistil incompatibility in maize-teosinte hybrids and their derivatives (Kermicle 2006). Multiple QTL studies have mapped the genes controlling domestication traits in maize-teosinte hybrid populations (e.g., Doebley and Stec 1991; Briggs et al. 2007). Other studies have utilized populations derived from maize × teosinte crosses to map and identify key domestication genes including teosinte branched 1 (tb1), teosinte glume architecture (tga1), and prolificacy 1.1 (prol1.1) (Doebley et al. 1997; Wang et al. 2005; Wills et al. 2013). Finally, two projects assayed the effects of teosinte chromosome segments introgressed into maize on a variety of domestication and agronomic traits (Studer and Doebley 2012; Liu et al. 2016). Although there are a few known chromosomal inversions that are polymorphic in teosinte and maize (Fang et al. 2012) and a few polymorphic factors for pollen-pistil compatibility that can prevent hybrid formation (Lu et al. 2014), there are no reports of a severe loss of vigor or viability among maize-teosinte hybrids or their descendant lines.
During the construction mapping populations to study the effects of teosinte alleles, we noted an unusual sickly syndrome in the backcross progeny resulting from crosses of maize with teosinte from near the Valle de Bravo in the state of Mexico, hereafter “Bravo” teosinte. The initial F1 hybrids are normal, but a sickly phenotype is observed in the Backcross 1 (BC1) and is more pronounced in subsequent backcross generations. Once the sickly phenotype appears in a lineage, healthy plants are never recovered by additional backcrosses to the normal maize parent. The inheritance pattern for the hybrid decay syndrome does not depend upon inheritance of specific chromosomal segments from the teosinte parent and does not segregate in backcross populations. We documented genomic instability in these backcross populations with increased copy number for specific sequences some with homology to transposable elements (TEs). De novo assembly of transcripts identifies a collection of upregulated sequences including some that have little or no similarity to sequences in the genomes of the maize recurrent parents (W22 or B73). Although the global patterns of DNA methylation and small RNA (sRNA) production are similar between sickly and normal plants, some TEs and sequences that are upregulated in sickly plants show altered methylation and sRNA profiles. Our observations suggest that crosses between Bravo teosinte and maize can trigger genomic instability that is inherited in all progeny. We name this phenomenon “hybrid decay,” a transgenerational decline in vigor and viability triggered in backcross populations of Bravo teosinte with maize.
Materials and Methods
Plant materials, RNA preparation, and DNA preparation
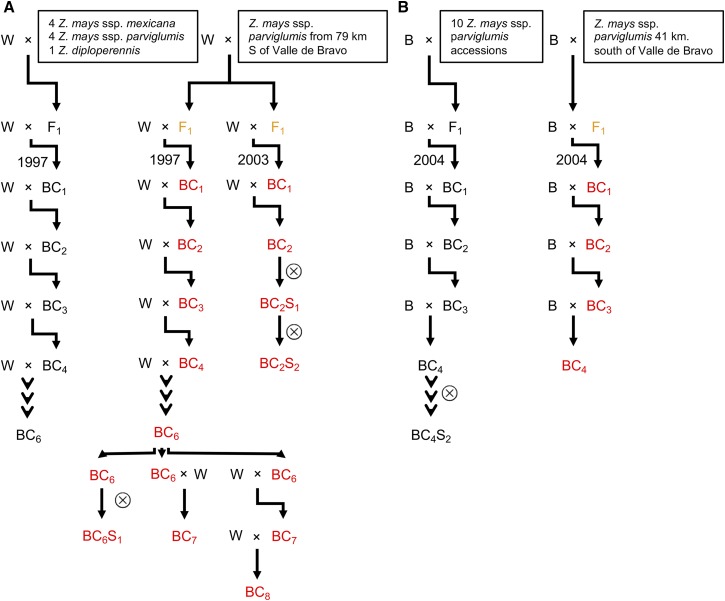
For backcrosses to W22, a Bravo teosinte accession (Beadle and Kato Site 6 from 79 km south of Valle de Bravo) was used, and a Blanco teosinte accession (Beadle and Kato Site 4 from 1 mile south of the town of Palo Blanco, Guerrero) was used at the normal control. For backcrosses to B73, another Bravo teosinte accession (PI384063; Beadle and Kato Site 7, 41 km south of Valle de Bravo) was used, and a Teloloapan teosinte accession (PI384065) was used as the normal control. The W22 backcrosses for both Bravo teosinte and the Blanco teosinte control were generated as part of a previously published project (Studer and Doebley 2012). The B73 backcrosses for both Bravo teosinte and the Teloloapan control were generated as part of another previously published project (Liu et al. 2016). The BC lines derived from both Bravo teosinte accessions were not included in these published papers because of the unexpected hybrid decay phenomenon, however they were constructed as part of those projects as were the BC lines for the teosinte controls. For the current project, the W22 BC6 lines were crossed with their recurrent parent to produce BC7 (and subsequent BC8) seeds.
To sample plant tissues for genomic analyses, seeds were treated with fungicide and placed on multiple layers of damp germination paper and covered by one layer of wet germination paper. The tray was placed under fluorescent lights at the room temperature. When plants reached ∼5 cm height, they were transferred to small pots containing soil in the growth chamber with an 11-hr light and 13-hr dark cycle until the V1 to V2 seedling stage. Then whole above-ground seedlings were harvested and stored at −80° before DNA and RNA extraction for genotype-by-sequencing (GBS), whole-genome bisulfite sequencing (WGBS), whole-genome sequencing (WGS), RNA-sequencing (RNA-seq), and/or small-RNA sequencing (sRNA-seq). High-quality DNA was extracted using CTAB protocol (Doebley and Stec 1991) with a modification to remove RNA with RNase A and purify DNA again with phenol and chloroform. Total RNA was extracted from seedlings using a standard TRIzol protocol (Invitrogen, Carlsbad, CA).
Phenotypic data collection and analysis
W22 and backcross lines were grown at the University of Wisconsin West Madison Agricultural Research Station (Madison, WI) in summer of 2014, 2016, and 2017. For each BC line and W22 used for phenotype investigation, a population of 72 seeds was planted in six blocks with one plot for each line. Each plot was 3.6 m long and 0.9 m wide, and was sown with 12 seeds. All of the BC1 plants were evaluated for plant height (length of the primary stalk from the ground to the node of top leaf) and the height of the top ear node (height from ground to base of top ear). Advanced backcross lines (mainly BC6 and BC7, and partial for BC8) were evaluated for 12 traits. Three plant architecture traits were investigated: plant height, the height of top ear node, and ear barrenness. Five primary ear morphology traits were investigated: cupules per rank (number of cupules in a single rank from base to the tip of the ear), kernel row number around the ear, ear diameter, ear length, and seed set rate. Three primary tassel traits were investigated: tassel branch number, pollen-shedding or not (BC7 and BC8), and days to anthesis (BC7 and BC8). One seedling trait was investigated: primary root length (BC8). Student’s t-test was used for plant height, the height of first ear node, ear diameter, ear length, seed setting rate, tassel branch number, and root length, which have normality distributions. Mann–Whitney–Wilcoxon test was used for cupule per rank, kernel row number, days to anthesis, and pollen-shedding, which were not normally distributed. The seeds used for measuring primary root length were treated with fungicide, rolled up in the germination paper, and placed in an incubator in the dark at 37° for 2 days. All statistical analyses were performed using the R package for Statistical Computing.
SNP genotyping methods
The 96-plex libraries were constructed according to GBS protocol (Elshire et al. 2011). Each DNA sample was digested with ApeKI restriction endonuclease, and ligated to barcode adaptors. All samples were then pooled together for PCR to increase the fragment pool. Single-end sequencing (100 bp reads) of 96-plex library per flowcell channel was performed on the Illumina HiSeq2000/2500. On average, ∼2 million reads were collected from each sample, resulting in roughly 0.1× coverage of the maize genome (0.2× to 0.3× after accounting for two or three replicates). The resulting Illumina FASTQ files were processed with the TASSEL-GBS pipeline for SNP calling (Glaubitz et al. 2014). The reads were trimmed and aligned to the B73 reference genome (AGPv2). Polymorphisms were called under ZeaGBSv2.7 Build, which contained 955,690 SNPs derived from >60,000 samples (http://www.panzea.org/).
Fluorescence in situ hybridization
For germination and root tip harvest, captan-treated seeds were germinated at 28° in moist vermiculite. After 3 or 4 days, the distal 1–1.5 cm segment was harvested from primary roots 3.5–5.5 cm in length. Excised root tips were transferred immediately to a humidified, 0.6-ml microcentrifuge tube with a hole in the lid and treated with nitrous oxide gas (160 psi, 2.5 hr) to stop development at metaphase (Kato 1999). Roots were subsequently fixed for 10–12 min in 90% ice-cold acetic acid, rinsed twice with cold 70% ethanol, transferred to a new tube of 70% ethanol, and stored at −20°. Because the primary roots from Bravo and Blanco teosinte lines were thin and produced few metaphase spreads, in a subsequent planting, the germinated seeds were transferred to six-packs containing Pro-Mix BX and allowed to grow for ∼13 days before processing the roots as described above. A balanced fertilizer and a water-soluble iron chelate were applied during the growth period.
The chromosome morphology of two or three roots from each seed stock was examined using fluorescence in situ hybridization (FISH; Kato et al. 2004). The Cent4 probe used was re-designed to remove homology to 180-bp knob heterochromatin (Lamb et al. 2007). Probe concentrations were as described in Birchler et al. (2007), with the exception that the blue and green probes were labeled with coumarin-5-dUTP (custom synthesis; Perkin Elmer Life and Analytical Sciences) and fluorescein-12-dUTP (Perkin Elmer, Norwalk, CT), respectively. Detailed protocols are available in Kato et al. (2011). Images were acquired using an Olympus BX61 fluorescence microscope fitted with a Cool-1300QS CCD camera (VDS Vosskühler) and FISHView EXPO 4.5 software (Applied Spectral Imaging). Image background was increased or decreased using the Curves function of Adobe Photoshop CS3 while maintaining original signal strength as much as possible.
WGS data generation, data processing, and alignments
DNA concentration was verified using the Qubit dsDNA HS Assay Kit (Life Technologies, Grand Island, NY). Samples were prepared according the TruSeq Nano DNA LT Library Prep Kit (Illumina, San Diego, CA) with minor modifications. Samples were sheared using a Covaris M220 Ultrasonicator (Covaris, Woburn, MA), and were size selected for an average insert size of 350 bp using solid-phase reversible immobilization (SPRI) bead-based size exclusion. Quality and quantity of the finished libraries were assessed using an Agilent DNA1000 chip and Qubit dsDNA HS Assay Kit, respectively. Seven libraries were normalized to 2 nM and then pooled into one lane. Cluster generation was performed using the Illumina Rapid PE Cluster Kits v2 and the Illumina cBot. Single-read 100 bp sequencing was performed, using Rapid v2 SBS chemistry on an Illumina HiSeq2500 sequencer. All reads were trimmed with Cutadapt/1.8.1 (Martin 2011) to remove the Illumina TruSeq Universal adapted as well as requiring a minimum read length of 30 and a phred score of 10. Reads were aligned to the W22 maize reference genome and the de novo transcript assemblies using Bowtie2/2.2.4 (Langmead and Salzberg 2012). Uniquely mapping reads were further processed into 100 bp windows across the maize genome for each sample and counts per million were calculated. CNVseq (Xie and Tammi 2009) was used to identify regions of copy number variation between the Bravo BC samples and the W22 sample. The BravoBC1 and BravoBC6 bam files containing uniquely aligning reads were merged to generate a single BravoBC bam file. This BravoBC file was used as the test sample in CNVseq with W22 as the reference sample. CNVseq input criteria used included a log2 cutoff of 2, P-value cutoff of 0.001, and a window size of 1 kb. CNVseq output was further filtered to include only those regions with a log2 fold change (log2FC) > 3, resulting in 436 unique regions. HTseq/0.5.3 (Anders et al. 2015) was used to count the number of reads mapping to these specific regions across all samples. Additional filtering included the removal of regions containing insufficient coverage across all non-Bravo BC samples as well as a calculated log2FC value for Bravo BC vs. Blanco BC1, BlancoBC6, and Blanco Teosinte > 3, resulting in 19 unique regions.
RNA-seq data generation, data processing alignment, and analysis
RNA was quantified and quality tested by NanoDrop and Agilent RNA NanoChip. Samples were prepared according to the TruSeq Stranded mRNA Library Prep (Illumina). One microgram of RNA was transferred to a final volume of 50 μl with nuclease-free water, polyA selected, and fragmented for 6 min. First-strand (with random hexamers) and second-strand complementary DNA (cDNA) and adenylate 3′ ends were synthesized, universal and multiple indexing adapters (contain unique 6-bp indices barcode sequences) were ligated to the ends of the double-stranded cDNA, and the DNA fragments were enriched by 11 cycles of PCR based on the adapter sequences primers. Quality and quantity of the finished libraries were assessed using an Agilent DNA1000 chip and Qubit dsDNA HS Assay Kit, respectively. Eight indexed DNA libraries are normalized to 10 nM and then pooled into one lane of an Illumina HiSeq2000/2500 sequencer (100 bp, single end). Approximately 20 million reads were generated for each sample (Supplemental Material, Table S1). Raw reads were trimmed using cutadapt version 8.1.1 using the -m 30 -q 10–quality base = 33 options. All reads were aligned to W22 genome sequence (Springer et al. 2018) or the de novo transcript assemblies using Tophat2 (Kim et al. 2015). Four mismatches, a minimum intron size of 5 bp, and a maximum intron size of 60,000 bp were used for alignment. More than 80% of reads were mapped on to the reference (Table S1). Transcript quantification was performed with HTSeq-count (Anders et al. 2015), using the maize W22 gene annotation. Principal component analysis (PCA) analysis was performed with the plotPCA function (Wickham 2016) using the variance stabilizing transformation matrix for RNA counts which were calculated by the R/Bioconductor package DESeq2 (Love et al. 2014). Differentially expressed genes were identified using DESeq2 and filtered using a fold-change cutoff of 2 and a false discovery rate (FDR)-adjusted P-value cutoff of <0.05. TE family expression was quantified as in Anderson et al. (2018). Briefly, the W22 TE annotation file was modified to resolve nested TEs using RTrackLayer (Lawrence et al. 2009) and used as input to HTSeq-count. The SAM output was then parsed using a custom script where mapped reads are assigned to TE families when they mapped uniquely to a single TE or when multimapped but hit only a single TE family. Differential expression (DE) analysis was performed using DESeq2, using the same contrasts and cutoffs as used to call differentially expressed genes.
RNA-seq de novo transcriptome assembly
A de novo assembly of the Bravo backcross transcriptome was generated using 52 M reads concatenated from three Bravo backcross RNA-seq libraries (Table S1). Illumina TruSeq adapters were removed with cutadapt v1.7.1 (Martin 2011) and only reads ≥70 bp retained. Reads were then filtered with FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html) v0.0.14 fastx_artifacts_filter and fastq_quality_trimmer to remove artifacts and retain only reads Q33 or greater; final library quality was assessed with fastqc v0.11.7. De novo assembly was performed with Trinity (Grabherr et al. 2011) version r20140717 with a–min_contig_length of 200 bp. Relative expression of each “gene” isoform was estimated with RSEM v1.3.0 (using bowtie2 v2.3.0 for alignment) using the Trinity utility script align_and_estimate_abundance.pl from Trinity version r20140717. For differential expression testing a SuperTranscript reference (Davidson et al. 2017) was generated using the Trinity utility script (Haas et al. 2013) Trinity_gene_splice_modeler.py from Trinity version v2.6.6. Differentially expressed transcripts were identified by mapping RNA-seq reads to the transcriptome assembly using hisat2 (Kim et al. 2015) v2.1.0, retaining unique-mapping reads, and running DEseq2. The differentially expressed de novo transcripts were then analyzed for putative coding regions using TransDecoder-v5.3.0 by retaining ORFs ≥100 amino acids or regions with homology to protein domains. Protein homology was identified using blast+ (Camacho et al. 2009) v2.7.1 blastp against ether Swissprot (ftp://ftp.ncbi.nlm.nih.gov/blast/db/swissprot.tar.gz; downloaded May 31, 2018) or UniRef90 (ftp://ftp.uniprot.org/pub/databases/uniprot/uniref/uniref90/uniref90.fasta.gz; downloaded May 31, 2018) databases and using HMMER v 3.1b2 (Johnson et al. 2010) hmmscan to search the Pfam-A database (ftp://ftp.ebi.ac.uk/pub/databases/Pfam/current_release/Pfam-A.hmm.gz; downloaded May 31, 2018).
sRNA-seq data generation, data processing alignment, and analysis
Samples were prepared according to the TruSeq Small RNA Library Prep Kit (Illumina). One microgram of RNA was transferred to a final volume 6 μl with nuclease-free water. The 3′ (containing the unique six-base indexes barcode sequences) and 5′ adapters were ligated, and the reverse transcription was performed followed by amplification (11 cycles). PCR cDNA products were further purified using 6% TBE gel and gel bands corresponding to molecular sizes of 145–160 bp were excised. A total of 24 sRNA libraries (Table S1) were pooled into one lane of Illumina HiSeq2500 sequencer for sequencing (single-end reads of 50-bp) at University of Wisconsin (Madison, WI). Raw reads were first demultiplexed and passed to FastQC for initial quality control. Reads from sRNA-seq contain the 3′ sequencing adapter because the read is longer than the molecule that is sequenced. Thus, the high-quality clean reads were processed using cutadapt v1.15 (Martin 2011) to remove the 3′ adapter sequence (TGGAATTCTCGGGTGCCAAGG). Any reads that did not contain an adapter were discarded, and only 18∼42 nt long reads were retained for subsequent analyses. Adaptor-free reads from ribosomal RNA, transfer RNA, small nucleolar RNA, and small nuclear RNA fragments were removed by aligning them against the corresponding genomic sequences of Z. mays genome using Bowtie v1.1.2 (Langmead et al. 2009) allowing for two mismatches. The remaining reads were mapped to the W22 (https://www.maizegdb.org/genome/genome_assembly/Zm-W22-REFEREN CE-NRGENE-2.0) and B73 genomes (ftp://ftp.gramene.org/pub/gramene/release-58/fasta/zea_mays/dna/) for sRNA samples from Doebley and Flint-Garcia Laboratories, respectively. At this step, ShortStack v3.4 was used with parameters “-bowtie_m 1000 -ranmax 50 -mmap u -mismatches 0,” which used local genomic context to guide decisions on proper placements of multimapped sRNA-seq reads (Johnson et al. 2016). To calculate and compare sRNA abundance in different samples, the genome was tiled into 500-bp windows and reads whose 5′ end nucleotides fall within a window were counted. sRNA with a size between 19 and 26 nt were selected and sRNA abundance for each window were calculated as reads per million of total reads. Differential comparisons of expression abundance were conducted by the R package “DESeq2” (Love et al. 2014).
WGBS data generation, data processing alignment, and analysis
Genomic DNA was sheared to 200–300 bp fragments, which were then subject to end repair, A-tailing, adapter ligation, and dual-SPRI size selection using KAPA library preparation kit (KK8232), following manufacturer’s instructions. The size (250–450 bp) selected library was then treated with bisulfite to convert unmethylated cytosine to uracil using Zymo EZ DNA methylation lightning kit (D5031). The converted DNA was then PCR-amplified using KAPA HiFi HotStart Uracil + (KK2801) with the following program: 95° for 2 min, eight times of 98° for 30 sec, 60° for 30 sec, 72° for 4 min, and a final extension at 72° for 10 min. To increase PCR efficiency, the bisulfite converted DNA was split into two equal parts and two parallel PCR reactions were performed, the final PCR products were combined and cleaned up together using SPRI beads. Library quality was checked using Agilent Bioanalyzer to ensure that the libraries size is in the right range (200–700 bp with a peak ∼300 bp). Library quantity was checked using Picogreen to make sure that the final concentration is >2 nM. Libraries that passed quality control were then equally pooled and sequenced across multiple lanes on an Illumina HiSeq2500 machine. For each library, 150–252 M paired-end 125 bp reads were generated (Table S1). Adapter sequences were trimmed and read quality was assessed using Trim Galore! with the default parameters and paired-end reads mode. Reads that passed quality control were mapped to maize W22 genome using BSMAP (Xi and Li 2009), allowing at most five mismatches. Nucleotides with a quality score <20 were trimmed from 3′ end of reads. Reads that are uniquely mapped and that are properly paired were used to extract methylation status at individual cytosine using methratio.py, which is included in the BSMAP package. At this stage, duplicate reads due to PCR bias were removed. Also, nucleotides in the overlapped part of paired hits are only counted once instead of twice. The final output of methratio.py contains the number of methylated and unmethylated reads for each cytosine. This file was used to create 100 bp nonoverlapping sliding windows across the maize chromosomes for each of the three cytosine contexts, CG, CHG, and CHH (H = A, C, or T). Within each 100 bp window and for each sequence context, the total number of methylated/unmethylated reads for each cytosine was summed and used to derive methylation levels of the 100 bp window, [#C/(#C+#T)]. Differential methylation region (DMR) calling was performed between the Bravo BC lines (BravoBC1 and BravoBC6) and W22 to identify context-specific DMRs. We required CG/CHG DMRs to have a minimum of three symmetrical CG/CHG sites (six Cs total for the two strands) and a minimum methylation difference of 60% between the two samples. DMRs were called based on 100 bp bins with >2× coverage in both samples. Coverage was defined as the ratio of the total number of times a cytosine was covered by the total number of cytosines of that specific context.
For PCA, the average DNA methylation level was determined for 100 bp nonoverlapping windows for all three sequence contexts (CG, CHG, and CHH). The matrix of CG DNA methylation levels for each 100 bp window for the seven genotypes was used to perform a principle components analysis (R package prcomp).
Data availability
The WGBS data are available under the National Center for Biotechnology Information BioProject accession number PRJNA526266, the RNA-seq data are available under accession number PRJNA528342, the WGS data are available under accession number PRJNA528290, and the sRNA-seq data are available under accession number PRJNA528352. The Sequence Read Archive accession numbers are available in Table S1. The GBS data reported in this paper have been deposited in the FigShare database (https://figshare.com/s/a882aff235818fc1762c). Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7882973.
Results
Discovery of a hybrid decay phenomenon in maize × Bravo populations
Two projects assayed the effects of teosinte chromosomal segments that had been transferred into maize by backcross breeding (Studer and Doebley 2012; Liu et al. 2016). These two projects used teosinte accessions collected from various geographic locations (Figure S1). Two of the accessions of Z. mays ssp. parviglumis came from near Valle de Bravo in the state of Mexico. These two accessions have typical teosinte characteristics (Figure S2); however, observations from these projects revealed that the crosses of maize lines with teosinte from the Valle de Bravo region result in a loss of vigor and fertility in backcross generations. We use the term “sickly syndrome” for the collection of abnormal traits that appear in the backcross plants and the term “hybrid decay” for this phenomenon and its unknown causal mechanism. While F1 hybrids of maize and Bravo teosinte are phenotypically normal, some sickly plants appear in the BC1, and all plants in the BC2 and later backcross generations are sickly. We did not observe a notable increase in severity beyond the BC2 generation.
The initial observation of hybrid decay was made during a project assessing the phenotypic effects of allelic diversity within teosinte for the domestication gene tb1 (Studer and Doebley 2012). Ten accessions of teosinte were crossed and then backcrossed into W22 (recurrent and female parent) for multiple generations, including five Z. mays ssp. parviglumis, four Z. mays ssp. mexicana, and one Z. diploperennis (Figure 1A). The F1 plants of all crosses exhibited phenotypes typical of maize × teosinte F1 hybrids. The backcross populations from all teosinte accessions other than the Bravo teosinte accession exhibited the expected variation in morphological traits that distinguish maize and teosinte, but the plants of these populations were otherwise healthy and produced viable offspring. However, in crosses of the Bravo teosinte accession (Beadle and Kato Site 6 from 79 km south of Valle de Bravo), multiple phenotypic abnormalities, including reduced stature, abnormal floral morphology, and reduced fertility, were observed in the BC2 and subsequent backcross generations (Figure 1A and Figure 2). Moreover, all backcross plants from multiple independent, single seed descent backcross lineages derived from the Bravo accession exhibit this sickly syndrome, suggesting the phenotype is not due to a single chromosomal region from Bravo segregating in a Mendelian fashion.
Figure 1.
Origins of the hybrid decay syndrome within the pedigrees of materials produced in this study. The crossing scheme is shown to illustrate the pedigree of the materials and the appearance of the hybrid decay syndrome. The crossing schemes utilized in the Doebley laboratory (A) and Flint-Garcia laboratory (B) are shown. The female parent of each cross is listed first (W indicates W22 and B indicates B73 in the pedigrees). The left portions of (A and B) illustrate the crossing scheme used for multiple other accessions as a single pedigree and in all cases these individuals did not display the phenotypic abnormalities of hybrid decay. Any generations that exhibit the appearance of the hybrid decay syndrome are shown using red text. The generations that have progeny in subsequent generations that exhibit the hybrid decay syndrome are shown using orange text but were not carefully assessed for phenotypes.
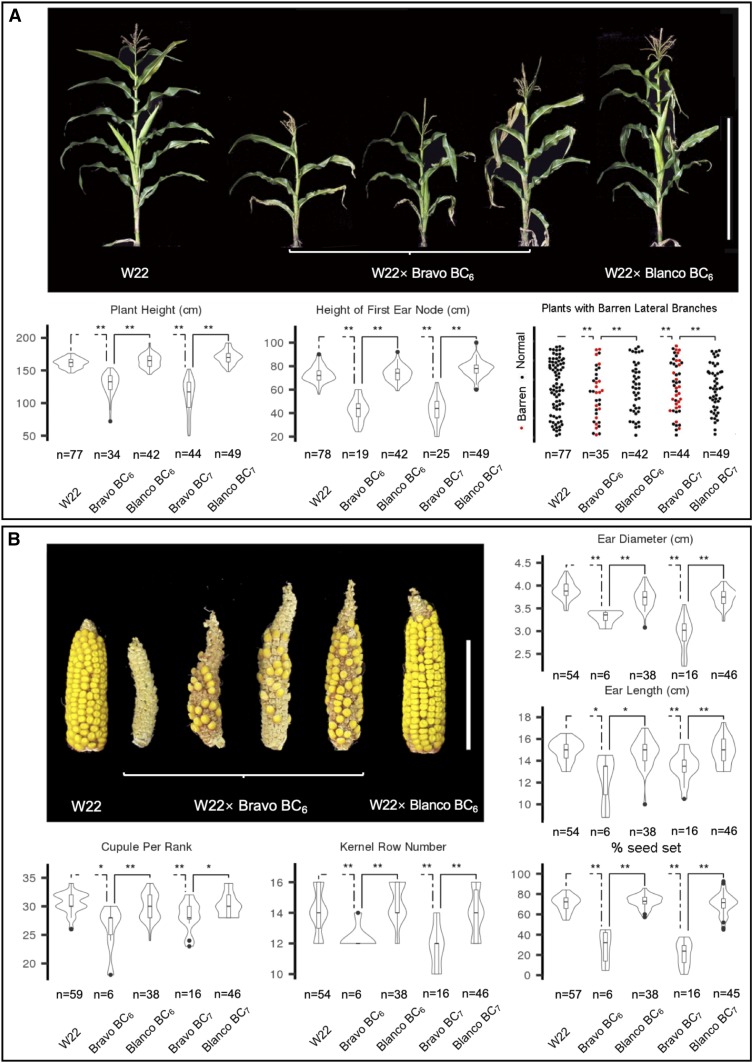
Figure 2.
Phenotypes associated with the hybrid decay. Whole plants (A) and ears (B) from W22 × Bravo BC4 or a control accession W22 × Blanco BC4 are shown. In (C), examples of BC6S1 plants derived from Blanco or Bravo accessions are shown. Bars, 1 m (A and C) and 10 cm (B).
Similar observations were made in an independent project attempting to generate BC2S3 recombinant inbred lines from W22 and Bravo teosinte from the same F1 individual as used by Studer and Doebley (2012) (Figure 1A). In this project, roughly 2000 plants from the BC1 generation were grown (Briggs et al. 2007) and many of these BC1 plant showed aberrant phenotypes. The sickly syndrome became more apparent in the BC2 generation with all plants being clearly affected. Self-pollination of the BC2 plants yielded only sickly plants with no healthy segregants observed.
An independent set of crosses was performed to generate BC4 near-isogenic lines (NILs) between Z. mays ssp. parviglumis and maize inbred B73 (Liu et al. 2016). One of 11 accessions used for this study exhibited sickly phenotypes similar to that described above (Figure 1B). The sickly NIL population was derived from a teosinte accession (PI 384063; Beadle and Kato Site 7) collected 41 km south of Valle de Bravo (Figure S1), just 38 km from the location of the accession discussed above. In the lineages that exhibit the sickly syndrome, all plants were affected and there was no evidence for the syndrome to segregate. Difficulty in generating a sufficient number of progenies prevented advancing the sickly population beyond the BC3 generation, while the other 10 NIL populations were created without problems.
In summary, these three experiments indicated that there is a previously unobserved phenomenon, hybrid decay, that appears in Bravo teosinte backcrosses to maize that causes a sickly syndrome that is transmitted from affected plants to all offspring across all subsequent generations of backcrossing and selfing.
Confirmation and quantification of the hybrid decay syndrome
The independent discovery by two of our groups of hybrid decay with teosinte from the Valle de Bravo offered only anecdotal evidence. Therefore, we decided to confirm the occurrence of hybrid decay in crosses of maize and individuals of Bravo teosinte and compare these crosses to control crosses between maize and individuals from another teosinte population. For the control teosinte, we used an accession collected from 1 mile south of the town of Palo Blanco, Guerrero by Beadle and Kato (Site 4), “Blanco” teosinte. Blanco teosinte had previously exhibited normal phenotypic outcomes in crosses with W22 maize (Studer and Doebley 2012). We collected quantitative data on several phenotypes as a measure of the sickly syndrome.
First, we assessed whether the sickly syndrome could be documented in the BC1 of crosses of one of the Bravo accessions (Beadle and Kato Site 6) as well as Blanco teosinte (Beadle and Kato Site 4). The BC1 generation derived from the Bravo teosinte are shorter and have lower heights for the top ear relative to BC1 plants derived from the Blanco teosinte (Figure S3). Next, to provide a detailed description of the sickly syndrome while minimizing the effects of the segregation of genetic loci from maize and teosinte, we assessed a number of quantitative traits in the advanced W22 × Bravo backcross lines (BC6–8) compared to advanced BC lines derived from the cross of W22 and Blanco teosinte. These data showed that Bravo BC6 has a number of phenotypic abnormalities as compared to Blanco BC6 plants (Figure 3). Plant height and the height of the first ear node are significantly reduced in the Bravo BC6 and BC7 plants (Figure 3A). Nearly 50% of the plants have barren lateral branches in the Bravo BC6 and BC7 generations, while barrenness was never observed in W22 or the Blanco BC lines (Figure 3A). The ears of the Bravo BC6 and BC7 plants have reduced diameter, length, kernel row number, and seed set relative to the Blanco and W22 controls (Figure 3B). The seed set is nearly threefold lower for the Bravo BC materials. The Bravo plants have reduced tassel branch number, some male sterility, and flower later than the controls (Figure S4A). The Bravo BC8 seeds also have reduced seedling root length at 2 days after germination as compared to Blanco BC8 seeds and W22 (Figure S4B).
Figure 3.
Phenotypic characterization of hybrid decay. (A) Plant morphology: examples of W22, W22 × Bravo BC6, and W22 × Blanco BC plants with violin plots for plant height, height of first ear node and proportion of plants with barren lateral branches. (B) Ear traits: ear diameter, ear length, cupules per rank, kernel row number, and seed set with representative ears. Student’s t-test was used for plant height, the height of top ear node, ear diameter, ear length, and seed setting rate. Mann–Whitney–Wilcoxon test was used for cupule per rank, and kernel row number. The number of plants measured for each trait (n) is listed for each plot. The two-dashed lines indicate comparisons between W22 × Bravo BC lines and W22, while solid lines indicate comparisons between W22 × Bravo BC lines and W22 × Blanco BC lines. * P < 0.05, ** P < 0.01.
Meiotic drive cannot explain hybrid decay
Meiotic drive (Lindholm et al. 2016), the preferential transmission of one allele from a heterozygote to all offspring, is a possible mechanism for hybrid decay. Under this mechanism, a sickly syndrome–inducing allele would be transmitted to all offspring. A meiotic drive–based mechanism for hybrid decay would predict that a specific sequence from Bravo was retained through all BCs and was causal for the sickly syndrome. To test this hypothesis, we performed GBS for several individuals that exhibit the sickly syndrome as well as control plants (Figure S5A). Seeds from several different Bravo BC6 ears (3763, 3771 and 3772) that represent different backcrosses from a common Bravo BC4 line (Figure S5B) were screened. The BC6 plants tend to have one or two teosinte introgression regions per line, but we did not observe any specific teosinte introgression segments common to all Bravo BC6 plants as predicted by a meiotic drive mechanism (Figure S5A). This result suggests the sickly syndrome is not due to preferential inheritance of a specific locus through a meiotic drive like mechanism.
Hybrid decay is not associated with karyotypic alterations
To test if there are any cytological manifestation of hybrid decay, we produced a FISH karyotype of the Blanco and Bravo teosintes, their W22 BC6 descendants, and the recurrent W22 parent (Figure S6). The two teosinte karyotypes are distinct from each other (Albert et al. 2010) and are heterozygous for some sites. The distinctiveness of the teosinte karyotypes is expected. Both BC6 Bravo-derived plants have chromosomes typical of the W22 parent (Figure S6). The Bravo and Blanco BC6 progenies are comparable and show no evidence of chromosomal aberrations or abnormalities that distinguish the Bravo BC6 from the Blanco BC6 or W22.
Hybrid decay is not dependent on transmission through male and female parents
The crossing schemes that uncovered hybrid decay were based on backcrossing using the recurrent maize line as the female (pistil) parent and the sickly plants as the male (pollen) parent. The mechanism of hybrid decay could potentially depend upon paternal transmission of genetic factors from sickly plants via their pollen. To test whether the sickly syndrome could be transmitted through both the male and female parents, the Bravo and Blanco BC6 lines were reciprocally crossed with W22. Plant height and height of the top ear node were equally reduced in the lines when the sickly Bravo BC6 line was used as either the male or female parent (Figure S7). This result suggests that once hybrid decay has been established, the sickly phenotype is not dependent on transmission through either the male or female parent. However, the direction of the original cross to create the F1 with maize as the female parent could be required to initiate hybrid decay.
Genome changes in Bravo BC plants
We hypothesized that crossing Bravo teosinte and maize triggered genome instability such as the activation of TEs. In order to assess changes in copy number for genomic sequences in the Bravo BC plants, we assessed the WGS read depth for 1 kb windows using low-coverage WGS data aligned to the W22 genome (Table S1). The average read depth of the Bravo BC1 and Bravo BC6 plants relative to W22 (Figure 4A) reveals a handful of genomic regions with substantial (>10-fold) increases in copy number in the Bravo BC plants. The higher read depth for these regions likely indicates copy number gains, but does not necessarily mean that there are additional copies at this location. These additional copies could be located elsewhere in the genome.
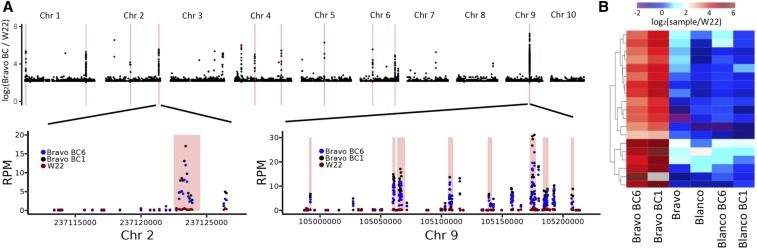
Figure 4.
Genome content changes associated with the hybrid decay. (A) The read depth ratio (log2) for the Bravo BC (average of BC1 and BC6) relative to W22 is plotted for 1 kbp bins across the genome. Only bins with differences over fourfold difference are shown. The red lines indicate regions that are found to exhibit consistently higher read depth in the Bravo BC materials relative to W22, Blanco, and Blanco BC lines based on CNVseq (P < 0.01). For a region on chromosome 2 and a cluster of significant regions on chromosome 9, we show the reads per million (RPM) values for all windows with data in these regions. The red shaded regions indicate the regions identified as significant based on CNVseq and the color of the points indicates the genotypes. (B) For each of these 19 regions, we determined the relative read depth of each sample relative to W22 and performed hierarchical clustering. The log2 (sample/W22) is indicated by the color. Dark blue indicates no change in read depth relative to W22, while red indicates higher read depth relative to W22.
To test which regions with aberrant read depth ratios in the Bravo BC plants relative to W22 were statistically significant, we used CNVseq (Xie and Tammi 2009). There are 19 regions that passed this test and all exhibit increased read depth in the Bravo BC lines relative to the other genotypes (Figure 4 and Table S2). A similar analysis was performed to determine whether a similar number of regions of variable copy number would be identified in the Blanco BC plants but we did not identify any regions with significant increase in copy number in the Blanco BC materials. The regions that exhibit gains in read depth in the Bravo BC plants have 8- to 60-fold changes (Figure 4B) and are 1000–6500 bp in length. The most prominent example is on chromosome 9 where there are eight significant CNVseq regions within a 50 kb region (Table S2). The Bravo BC6 sample used for this analysis contains an introgression from Bravo in the middle of chromosome 7, but is otherwise homozygous for chromosomal segments derived from W22, therefore these are not likely to represent introgressed Bravo segments.
These 19 regions that exhibit consistently higher read depth in the Bravo BC samples also exhibit several notable features. First, they do not have high read depth in the sampled Bravo teosinte individual or W22 (Figure 4B and Table S2). This result suggests that these sequences were amplified in copy number during the backcrossing. Second, several of these sequences overlap annotated genes or transposons within the W22 genome. However, the boundaries of the regions showing copy number changes rarely align precisely with the boundaries of annotated features such as transposons or genes, limiting the ability to infer potential mechanisms for increased copy number. Third, visual inspection of the alignments in several of these regions suggests the higher copy is a sequence with homology but not identity to these regions (Figure S8). This inference is based on the observation that the elevation in read depth is variable within a single region and the observation that there are multiple SNPs relative to the W22 reference genome that are present in the majority of reads that are aligned to a region (Figure S8). In conclusion, our analysis of read mapping to the W22 genome identified sequences with similarity to some regions of the W22 genome that were increased in copy number during the backcrossing.
Gene expression changes in Bravo BC plants
To identify changes in transcript abundance in sickly plants, we monitored steady-state transcript abundance using RNA-seq in seedling tissue of individuals from Bravo-derived sickly plants and controls for both the W22 and B73 backcross series (Table S1). For the W22 × Bravo series, the W22 × Bravo BCs, the W22 × Blanco BCs (controls), and the Bravo, Blanco, and W22 parents were assayed. For the B73 × Bravo series, we used a teosinte control from near the town of Teloloapan, Guerrero, Mexico (accession number PI384065) for which the BCs with B73 did not show the sickly syndrome (Liu et al. 2016). For this series, we assayed the B73 × Bravo BCs, the B73 × Teloloapan BCs (controls), and the Bravo, Teloloapan, and B73 parents.
To understand the impact of hybrid decay on the transcriptome, we performed two analyses. First, a PCA of the RNA-seq data for all genotypes revealed that the plants fall into three clusters: W22 and its BCs, B73 and its BCs, and all pure teosintes (Figure S9A). Thus, the lines with the sickly syndrome cluster with their recurrent parent related lines, suggesting that this syndrome does not induce major differences in the W22 or B73 transcriptomes at the seedling stage. Second, to search for genes that are differentially expressed in sickly plants, DESeq2 (Love et al. 2014) was used to contrast the six sickly samples (W22 Bravo BC1 and BC6 and B73 Bravo BC1 and BC2) with the 14 nonsickly samples, including the W22, B73, Bravo, Blanco, Teloloapan, and control BC plants. This analysis identified a total of 18 differentially expressed genes (log2FC > 1 and FDR < 0.05), all of which are upregulated in the Bravo BC sickly plants (Figure S10 and Table S3). Thus, while the transcriptomes of plants with hybrid decay are not radically altered, there is a select set of 18 genes that are upregulated in these plants. A similar analysis performed to identify differentially expressed genes in the Blanco BC plants relative to other materials did not identify any genes with significant differences.
The 18 genes identified have some notable features. First, they do not appear to be randomly located around the genome but rather there is a cluster of five genes within a single 1 Mbp region on chromosome 9 (Table S3). Second, gene ontology analysis does not show a specific functional category to which the 18 belong; indeed, 10 of the 18 are hypothetical proteins with no known function. Third, only 8 of these 18 W22 genes have a collinear homolog annotated in the B73 genome and only 5 of the 18 genes have a collinear homolog based on comparison with sorghum (Table S3). Genes that lack a collinear homolog in other maize lines (and sorghum or other grasses) are typically nonessential. There are relatively few examples of mutants in such genes that have a detectable phenotypic effect (Schnable 2015). Thus, although these 18 genes cannot be excluded as candidates for the causal agents of hybrid decay, their sequence homology provides no insight into how they might cause it.
The RNA-seq alignment data for each locus revealed a few key details (see examples in Figure S11). First, only 4 of the 18 genes exhibit read mapping patterns suggestive of expected transcription throughout the gene and proper splicing. Second, for 14 of the 18 genes, the increase in transcript abundance does not match the predicted structure of the W22 transcript, rather they exhibit expression only for a portion of the gene, often overlapping an intron (Figure S11). Third, the majority of reads that map to these 18 genes have sequence polymorphisms relative to the W22 genome. This observation suggests that the increased expression is likely from sequences from the Bravo parent with homology to several regions of W22, but is not likely due to transcription of sequences that are present in W22.
Identification of novel transcripts present within Bravo BC plants
The observation that some of the 18 W22 genes with increased expression were similar to, but distinct from, W22 genome sequences led us to investigate the possibility of novel transcripts in these plants. We performed a de novo assembly of transcripts from the RNA-seq data for sickly plants. The reads from the W22 Bravo BC1 and BC6 samples were pooled and used for transcript assembly by Trinity (Haas et al. 2013). We obtained 73,145 assembled transcript contigs (with 93,592 isoforms) from this assembly with an N50 of 1358 bp. Then, a PCA using the read counts per transcript was performed and revealed that PC2 separates the sickly plants from other related individuals (Figure S9B). The ability to distinguish sickly from normal plants with a PCA using these novel transcripts implies that the sickly phenotype involves transcripts that are not part of the W22 gene set. Next, to identify specific sequences that are differentially expressed in the sickly plants relative to the other genotypes, all RNA-seq reads from all samples were aligned to assembled transcripts. Differential expression analysis identified 59 transcripts that have significant differences in expression between the six sickly samples relative to the 14 nonsickly samples (Figure 5, Dataset S1, and Table S4).
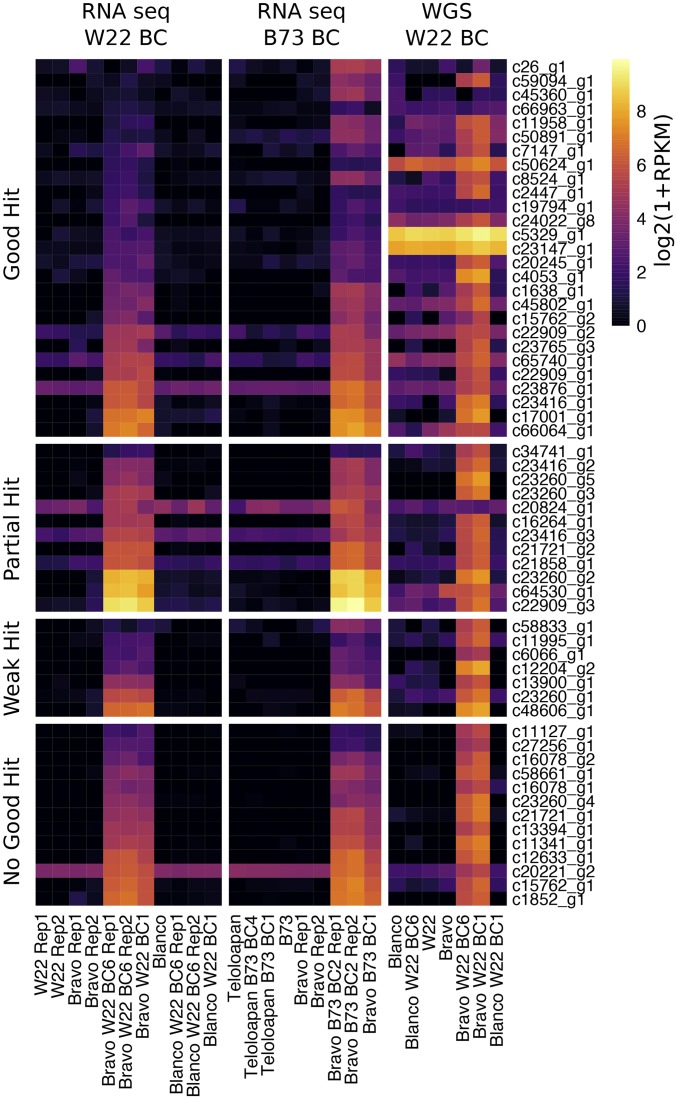
Figure 5.
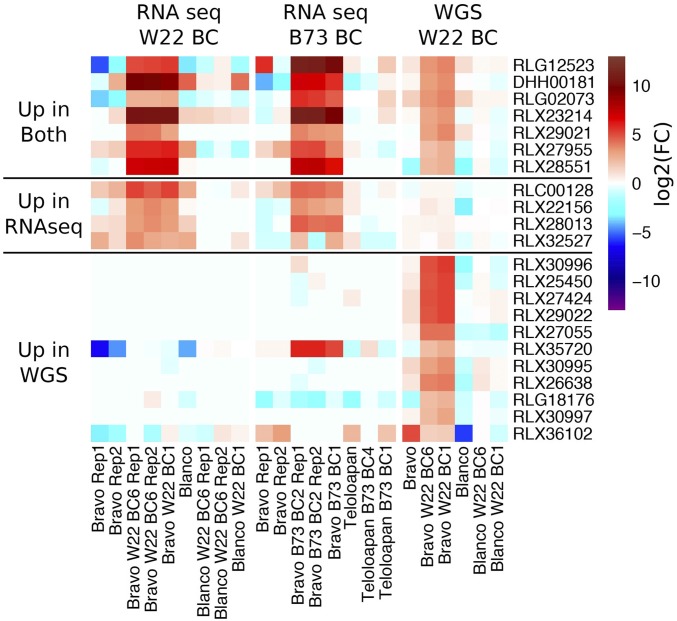
Expression and read depth for de novo transcripts with altered expression in sickly plants. The de novo assembled transcripts (from W22 Bravo BC1 and BC6 RNA-seq samples) were used to perform differential expression analysis. A set of 59 transcripts with altered expression was identified. These transcripts were classified based on their alignments to the W22 genome. Each group of transcripts is ordered by FPKM expression in the Bravo BC6 Rep1 sample and a heat map is used to visualize the expression level. In addition, the RPKM values based on unique alignments of the WGS data to all de novo transcripts are also visualized as a heat map using the legend on the right.
We proceeded to characterize these 59 transcripts that are highly expressed in seedling tissue of the sickly genotypes (Table S4). To determine whether these transcripts represent genes that are present in the W22 genome, genes that are highly similar to W22, or novel sequences, the highest expressed isoform of each transcript was aligned to the W22 genome using BLAST. There are 27 transcripts with high similarity to the W22 genome (>95% identity and >50% of the length of the transcript aligning to the genome). There are 12 transcripts with partial alignments to the W22 genome (>95% identity but <50% of the transcript aligns to the genome). The remaining 20 transcripts have either weak BLAST hits (seven transcripts with <95% identity) or no significant alignments to the W22 genome (13 transcripts). The annotations for the genomic regions that the 39 transcripts with good or partial hits to W22 were assessed. There are 10 transcripts that at least partially overlap annotated genes (including 8 of the 18 W22 genes we identified). The remaining 29 transcripts included 16 that overlap annotated retrotransposons (RL), 3 that overlap helitron (DH) elements, and 10 that align to unannotated regions between genes and TEs. In many instances, these transcripts only overlap a portion of these features (Table S4) and likely do not represent transcription of the full annotated gene or TE although there are some examples of the transcript containing the full features.
There are questions about the potential functions that might be encoded in these upregulated transcripts. The coding potential and domains present within these transcripts were assessed. Only about half of the highly expressed transcripts have coding potential and 13 of these produce putative proteins that contain domains with significant similarity to PFAM domains (Table S4). There was little evidence for a common function that was present in these transcripts. There are examples of specific enzymatic activities (amino transferase, glycosyl transferase, peptidase) or domains of unknown function. One of the more interesting observations was the presence of several long transcripts (c23147_g1_i2 and c23260_g1_i7) that contain regions annotated as having retrotransposon functions (integrase/reverse transcriptase) and another transcript (c13394_g1_i3) with a transposase related domain. The observations that several of the transcripts contain domains related to TEs and that some transcripts align to regions annotated as TEs suggest there is upregulation and activation of TEs in hybrid decay.
Correspondence between upregulated genes/transcripts and WGS copy number variants
Since the RNA-seq expression data for both the 18 W22 genes and 59 novel transcripts suggested that something more complicated than simple gene upregulation was occurring, and since the WGS data indicated that some regions of the genome had elevated copy number in sickly plants, we compared the RNA-seq fold-change (log2) values to that for the WGS data. The read depth of each of these genes/transcripts was assessed by aligning the WGS data to the 18 W22 genes and the 59 de novo transcripts and determining FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values (Tables S3 and S4). First, Figure S10 shows that the 18 upregulated genes in sickly plants exhibit higher read depth in the whole-genome shotgun data, suggesting increased copy number at the DNA level. Second, the majority of 59 novel transcripts have a higher WGS read depth in sickly plants relative to nonsickly plants (Figure 5 and Table S4). There are 41 transcripts with at least 10-fold increase in read depth in the Bravo BC plants and another 11 transcripts with 3- to 10-fold increases in read depth. It is notable that the majority of these regions exhibit relatively low copy in the Bravo teosinte individual that was sequenced (Figure 5). In conclusion, there is a set of genes/transcripts that are highly expressed and increased in copy number in the sickly plants and many of these sequences are not present in the W22 genome, suggesting a proliferation of sequences donated by the Bravo teosinte parent.
Correspondence of upregulated sequences to transposons
To investigate the potential upregulation of TEs from the W22 genome in the Bravo BC plants, we mapped the RNA-seq reads to transposons using a mapping strategy that counts the number of RNA-seq reads per transposon family rather than per locus (Anderson et al. 2018). To enable comparisons between the RNA-seq and WGS datasets, we tested for a difference in counts per TE family between sickly Bravo BCs (BC1 and BC6) and normal Blanco BCs (BC1 and BC6) (Tables S5 and S6). This analysis identified 11 TE families with significant increases in expression (Table S5) and 18 TE families with increase in read depth for WGS data (Table S6). Seven of the TE families were identified as having significant increases in both expression and genomic read depth. A comparison of the expression levels and genomic read depth for each of these families in all samples reveals that the increases in expression and/or read depth are limited to the Bravo BC families (Figure 6). Unclassified LTR retrotransposons (RLX) were the most common families that exhibited increase. However, there was also a Helitron family (DHH) and two gypsy-like retrotransposons families (RLG) with increases in both expression and copy number (Tables S5 and S6). There are no TE families with an elevation of expression or genomic copy number in the normal Blanco BCs. Thus, at least 22 TE families show evidence of activation in sickly plants.
Figure 6.
Changes in expression and copy number for some transposable element (TE) families. TE families that exhibit increases in expression level or read depth were identified by comparing the W22 Bravo BC1/BC6 lines with W22 Blanco BC1/BC6 controls. A clustered heat map is used to visualize expression and copy number relative to W22 for these TE families in all samples. The TE families were divided into three groups, the seven TE families that exhibit significant increase in both RNA-seq and WGS data, four families with significant increase in RNA-seq but not in WGS data, and the 11 families with significant increase in WGS data but not in RNA-seq.
DNA methylation changes in plants with the hybrid decay syndrome
To test for changes in DNA methylation in sickly plants, we generated WGBS data for the W22 BC materials to search for unique DNA methylation patterns in sickly plants. The analysis of DNA methylation patterns was restricted to alignments of reads to the W22 genome. This approach provides a survey for the majority of genome present in the W22 BC samples. A comparison of the global DNA methylation patterns by PCA reveals that the Bravo BC plants are closely related to W22 with no evidence for genome-wide repatterning of DNA methylation (Figure S12). To identify loci with altered DNA methylation, the methylome of the Bravo BC1 and BC6 plants was compared to the W22, Bravo teosinte and Blanco BC6 plants to identify CG and CHG differentially methylated regions (DMRs) with >60% difference in DNA methylation levels for 100 bp tiles. This analysis identified 167 CG DMRs (Table S7) and 260 CHG DMRs (Table S8). Reduced methylation accounted for 75.4% of CG DMRs and 62.3% of CHG DMRs. There are CG DMRs near 11 of the 19 regions with elevated copy number. There are also CG DMRs located within 2 kb for 19 of the 46 upregulated transcripts that had alignments to the W22 genome (Table S4).
sRNA abundance in plants with the hybrid decay syndrome
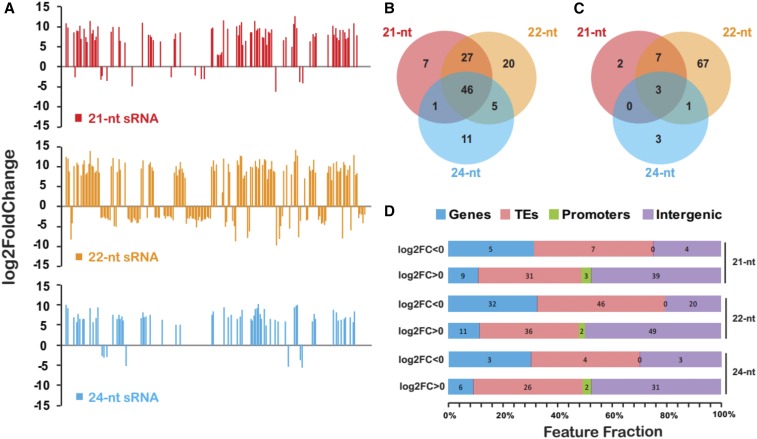
We analyzed the genome-wide changes of sRNAs using sRNA-seq at the seedling stage of the sickly Bravo BC plants and normal control plants for both W22 and B73 background (Table S1). We did not observe global changes of sRNAs between normal and sickly plants in either genetic background (Figures S13 and S14), suggesting that the sickly phenotype is not associated with major disruption of sRNAs. To identify genomic regions with differential levels of sRNA abundance between normal and sickly plants, we quantified sRNA expression by normalizing against total reads for each size class of sRNAs per 500-bp window along the W22 and B73 genomes. A total of 200 and 112 regions (FDR ≤ 0.01 and fold change ≥ 2) with differential sRNA abundance were identified in the W22 and B73 genomes, respectively (Table S9 and S10). Most of the differentially expressed sRNA regions showed strongly directional bias to significantly increased sRNA levels in sickly Bravo BC plants in both W22 and B73 background, especially for 21 and 24-nt sRNAs (Figure 7A, Figure S15A, Tables S9 and S10). More than half of these regions exhibit upregulation for multiple sizes of sRNAs simultaneously in sickly plants as compared to normal control plants (Figure 7, A and B and Figure S15, A and B). Although we also observed that some of regions in sickly maize with W22 genetic background specifically produced decreased 22-nt sRNA compared with the normal plants, this is not the case for the B73 genetic background (Figure 7, A and C and Figure S15A). Thus, we concluded that a group of sRNAs produced in the seedlings that will later exhibit a sickly phenotype might contribute to hybrid decay in Bravo BC plants.
Figure 7.
Feature analyses of differentially expressed sRNA regions in sickly Bravo BC plants compared with controls. (A) Graph of differentially expressed sRNA regions one-by-one, showing the bias that most of differentially expressed sRNA regions produced increased 21-nt, 22-nt, and 24-nt sRNA in sickly plants as compared to control plants. The y-axis indicates the log2 transformation of fold changes, with positive value representing the generation of increased sRNA in sickly Bravo BC plants compared with the normal controls. (B) Almost half of differentially expressed sRNA regions with increased sRNA in sickly plants produced three classes (21-nt, 22-nt, and 24-nt) of sRNAs concurrently. (C) Most of differentially expressed sRNA regions with decreased sRNA in sickly plants only produced 22-nt sRNAs. (D) Overlap profiling of differentially expressed sRNA regions with W22 genomic features, including genes, transposable elements (TEs), 2-kb promoter regions of annotated genes, and intergenic regions. The log2FC > 0 means that increased sRNAs were generated in sickly Bravo BC plants compared with the normal controls. The log2FC < 0 means that reduced sRNAs were produced in sickly Bravo BC plants compared with the normal controls.
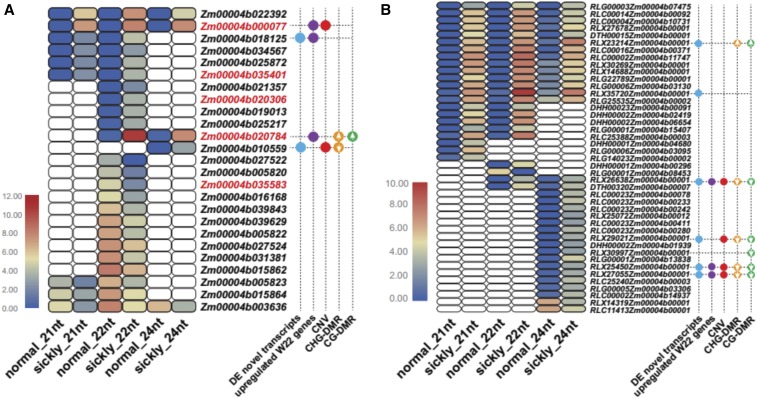
An analysis of the differential sRNA generating loci showed overlap with genomic features, such as TEs, gene body, and promoter regions of annotated W22 or B73 genes (Figure 7D and Figure S15C). Thus, we reanalyzed the sRNA changes with TEs or genes as units of measurement (Tables S11–S14). A total of 25 annotated W22 genes showed significantly differential accumulation of sRNAs between normal and sickly plants (Figure 8A and Table S11). Similarly, we detected 13 annotated B73 genes with a different amount of sRNAs generated between normal and sickly plants (Figure S16 and Table S13). Most of these genes exhibit significant variation for 22-nt sRNA levels (Figure 8A). Only five genes with colinear homology show altered sRNA profiles in both W22 and B73; four that produced increased and one that produced decreased sRNAs in sickly plants (Figure 8A, Figure S15A, Tables S11 and S13). We also detected some TEs with altered sRNA generation in sickly plants compared with normal plants (Figure 8B, Tables S12 and S14). Interestingly, almost all of these TEs consistently produced more sRNA in sickly plants than in normal plants for both W22 and B73 background (Figure 8B and Figure S15B). Strikingly, four of these TEs also showed either increased copy number variant (CNV), upregulated expression of novel transcripts, or reduced CG and/or CHG DNA methylation around them (Figure 8B). We speculate that hybrid decay affects DNA methylation around these TEs so that they and neighboring regions express novel transcripts or produce extra sRNAs. The sRNAs were also aligned to the 59 novel transcripts identified as upregulated based on RNA-seq data (Table S15). A subset of these sequences exhibit abundant sRNAs in the Bravo BC plants.
Figure 8.
Annotated W22 genes (A) and transposable elements (TEs) (B) that potentially generated significantly different amount of sRNAs between sickly and control plants and their comparisons with other features of genes or novel transcripts expression, CG or CHG DNA methylations, and copy number. Genes with red fonts indicate that the corresponding B73 homolog genes also produced more sRNA in sickly plants with B73 genetic background (Figure S16). The points with different colors stand for genes or TEs that are located near the novel transcripts (blue), upregulated W22 genes (purple), increased copy number regions (red), and increased or decreased CG (orange) or CHG (green) methylation regions in sickly plants with W22 background.
Discussion
Hybrid decay is a transgenerational epigenetic phenomenon observed in the backcross progeny of certain teosinte individuals from near the Valle de Bravo with W22 or B73. Abnormal, sickly phenotypes appear in the BC1 generation and a pronounced sickly phenotype is manifest in more advanced BC generations. The phenotypic effects of hybrid decay are pleiotropic, affecting plant stature, the ear, the tassel, and root growth. A remarkable feature of hybrid decay is that the Bravo and maize parent lines are themselves normal phenotypically, indicating that hybrid decay is due to an interaction between these two genomes and not a feature of either one. We also observed that this syndrome once established can be transmitted to offspring through either the male or female gametes. The inheritance of the hybrid decay is non-Mendelian and epigenetic in that it is transmitted from parent to all progeny and does not segregate among the progeny. Once the syndrome is initiated in a lineage, normal plants are never recovered in any progeny of subsequent generations.
Hybrid decay alters the genome of affected plants in multiple ways. First, there are some genomic sequences with homology to 19 regions of the W22 genome that have strongly elevated copy numbers in sickly plants. Eight of these sequences are clustered in a 50 kb segment of chromosome 9. Second, these amplified sequences do not have exact sequence identity to the W22 genome but show sequence polymorphisms that distinguish them from W22, suggesting that their origin may be from the Bravo genome. Third, there are 18 W22 genes that are upregulated in plants with the sickly syndrome, and many of these genes share sequence homology with the amplified genomic sequences and some also map to the same region on chromosome 9. Fourth, sickly plants express 59 novel transcripts, all of which are upregulated in sickly plants as compared to controls. Some of these transcripts share homology with the 18 differentially expressed W22 genes or the genomic sequences that have elevated copy number in sickly plants. Fifth, there are 22 TE families that have either elevated expression, elevated WGS read depth or both. Most of these are retrotransposons. Sixth, there are 231 DMRs between sickly plants and heathy controls. Many of these regions, which show reduced methylation in sickly plants, are also associated elevated copy number in sickly plants. Seventh, there are several hundred genomic regions with differential sRNA production, most of these have elevated levels of sRNAs in sickly plants. Finally, these multiple genomic features of hybrid decay are correlated such that some W22 genes and TEs involved show multiple anomalous features.
A key feature of hybrid decay is that after six generations of backcrossing to maize, it is expected that the genome of Bravo-derived plants will be largely identical to that of the recurrent parent with only small segments of residual Bravo DNA as regions of heterozygosity. Both FISH and GBS marker data confirm that this is true. However, we also detected novel transcripts in the sickly BC6 plants that are not present in the recurrent parent. CNVs in the sickly plants demonstrate that some genomic sequences are being amplified. While these CNVs are similar to sequences present in the W22 genome, they are clearly not derived from the W22 genome based on the presence of polymorphisms. Moreover, many of the novel transcripts have low homology to the W22 genome. Together, these observations suggest that non-W22 sequences have proliferated in the genome of the Bravo BC plants, that these sequences can be highly expressed and associated with alterations in sRNA production and DNA methylation. Some of these sequences have homology to TEs and it is possible that transposon activation is involved in this genomic instability.
Some unanswered questions
A central question for future research will be to identify the sequence(s) in Bravo teosinte and/or maize that act as the driver(s) that triggers hybrid decay. If there are specific sequences or DNA elements that trigger hybrid decay, why do they not initiate this phenomenon in the pure Bravo or the maize parent? Is there a protective factor (or epigenetic state) that silences the drivers in Bravo teosinte that is not transmitted from the F1 to the BC generations? Are the drivers activated in the F1 such that its gametes are set to trigger hybrid decay or is this phenomenon initiated in the zygote of the BC1? Would a Bravo teosinte × W22 F2 population exhibit hybrid decay or even segregate for it? What are the steps involved from the initiation of hybrid decay to the likely downstream effects that we describe including elevated expression of novel transcripts, CNVs, amplification of TEs and changes in the methylation and sRNA profiles? We observed a significant concentration of genomic alteration associated with a region on chromosome 9. Is there something special about this region regarding hybrid decay? Regarding the CNVs, are they the result of tandem duplication or elements dispersed throughout the genome? The lack of knowledge about the genomic distribution of these sequences that have increased in copy number limits our ability to visualize some of the changes in expression, sRNAs or DNA methylation in a physical genomic context. Some of the changes in expression or methylation could be the result of behavior of a subset of the copies that are found at uncharacterized locations within the genomes of plants that exhibit the sickly phenotype.
There remain other questions to be answered. Namely, is Bravo teosinte polymorphic in this regard such that some Bravo teosinte individuals would not trigger hybrid decay? Are there other teosinte populations that can trigger hybrid decay? And finally, our crosses were all made using maize as the female parent, would hybrid decay also have been triggered if maize had been used as the pollen parent and teosinte as the female parent?
Thoughts on the underlying mechanism
We suspect that the activation and amplification of TEs from the Bravo genome in the Bravo BC plants underlie hybrid decay and that these TEs might have escaped genome surveillance during hybridization. Many TEs in plant genomes are kept silent by DNA methylation at CG, CHG, and CHH contexts (Law and Jacobsen 2010; Matzke et al. 2015; Cuerda-Gil and Slotkin 2016). De novo methylation occurs through a mechanism known as RNA-directed DNA methylation (RdDM), in which small interfering RNAs (siRNAs) derived from TEs guide the deposition of DNA methylation at homologous sequences (Law and Jacobsen 2010; Matzke et al. 2015; Cuerda-Gil and Slotkin 2016). Once established, methylation at CG contexts can be maintained independently of RdDM, but methylation at CHG and particularly CHH contexts requires RdDM to be maintained (Law and Jacobsen 2010; Matzke et al. 2015; Cuerda-Gil and Slotkin 2016). De novo CHH methylation has been shown to be essential in preventing new bursts of transposition in Arabidopsis (Marí-Ordóñez et al. 2013; Cavrak et al. 2014). In this context, it may be significant that a CHH methyltransferase (Zm00004b000077 in W22; Zm00001d027329 in B73) is among the genes with elevated messenger RNA and sRNA levels in sickly plants (Figure 8A and Figure S16A).
During plant development, DNA methylation is reprogramed and reinforced in the germ cells and during embryogenesis. The central cell but not the egg cell undergoes active demethylation, resulting in the activation of TEs and the production of siRNAs in the endosperm. The siRNAs are thought to move from the endosperm into the embryo to cause de novo DNA methylation at homologous TEs (Hsieh et al. 2009; Bouyer et al. 2017). Studies in Arabidopsis show that CHH methylation in embryos increases during embryogenesis, reaching full methylation levels in mature embryos (Bouyer et al. 2017; Kawakatsu et al. 2017). CHH methylation levels decline subsequently in plant development because the RdDM machinery is highly expressed only in meristematic tissues (Hsieh et al. 2009; Bouyer et al. 2017). Thus, embryogenesis is a key period when DNA methylation is reinforced. We speculate that, in the F1 embryos of the maize × Bravo cross, certain TEs in the Bravo genome cannot be properly targeted by maternal siRNAs derived from maize TEs in the endosperm due to sequence diversification. These TEs are thus amplified and inserted into the maize genome, as revealed by CNVseq in this study. In post-embryonic F1 plants, although the TEs give rise to siRNAs, which are likely derived from RNA polymerase II–generated transcripts from the TEs, the siRNAs are unable to cause DNA methylation. Potential reasons include low levels of expression of RNA polymerase V (Pol V), which is required for siRNA-mediated DNA methylation, and inability of Pol V to access active chromatin at the TEs (note that DNA methylation promotes the recruitment of Pol V to chromatin in Arabidopsis). In subsequent BCs, the TEs continue to escape surveillance due to the lack of maternal, maize-derived siRNAs that can target them, resulting in their further amplification and remobilization.
How can TE activation and remobilization cause hybrid decay? Two possible mechanisms are proposed here. First, in our study, we observed sRNAs of 21–24 nt that are specifically produced from the strongly amplified TEs or TE homologs in sickly plants (Figure 8). We speculate that these “foreign” or “ectopic” sRNAs may repress the expression of certain maize genes to cause the sickly phenotype. For example, the 24-nt siRNAs might target some maize sequences and cause DNA methylation, which in turn could repress the expression of genes near these sites. The 21-nt siRNAs may target maize transcripts to cause post-transcriptional gene silencing through RNA cleavage. The 22-nt siRNAs could even lead to the biogenesis of secondary siRNAs from target transcripts, and the secondary siRNAs may have additional targets in the genome, thus amplifying the effects. In summary, the downregulation of target genes by the ectopic siRNAs leads to the sickly phenotype. Although downregulation of gene expression was not observed with our RNA-seq data from seedling tissues, one cannot rule out that this occurs in specific tissues or cells that are crucial for development, such as meristems. Alternatively, the ectopic siRNAs could lead to translational repression of maize messenger RNAs, an effect that is not reflected at the transcriptome level. Second, the activated TEs could lead to the upregulation of nearby genes to cause the sickly phenotype. It has been documented that the epigenetic states of TEs can affect the expression of nearby genes, especially when the TEs are in promoters or introns of genes (Cui and Cao 2014; Ito and Kakutani 2014). In this study, we observed upregulation of 18 W22 genes as well as 59 novel transcripts that are probably from the Bravo genome. These protein products may have adverse effects on plant development. Additionally, there may be specific upregulated sequences that contribute to hybrid decay. We note that one LTR retrotransposon family (RLX23214) with increased copy number in sickly plants carries a sequence homologous to a CHH methyltransferase gene (Zm00004b000077 in W22; Zm00001d027329 in B73), and sRNAs map to regions of both the TE and the gene. Although knockouts of this gene do not have an obvious phenotypic effect in maize inbreds (Li et al. 2014), the gene is an active component of silencing in reproductive tissues (Garcia-Aguilar et al. 2010) and provides a compelling target for continued study of the mechanistic basis of hybrid decay.
Finally, we note that our observations of hybrid decay with teosinte are similar to a form of hybrid dysgenesis observed in maize. When Zapalote Chico, a maize landrace grown by the Zapotec people of Oaxaca, is crossed to other maize germplasm, offspring show low vigor or sterility (Gutiérrez-Nava et al. 1998). This hybrid dysgenesis is due to the activation of a Mutator transposon in crosses with non-Zapalote Chico maize germplasm (Gutiérrez-Nava et al. 1998). As in hybrid decay, the initial cause is unknown, but the outcome—differential TE activity and methylation—is similar. Understanding the epigenetic and genetic similarities of systems of hybrid decay and hybrid dysgenesis may help to understand the prevalence of these systems, and if they play a role in reproductive isolation and divergence between populations and species.
Acknowledgments
We thank Xuehua Zhong and Chin Jian Yang for helpful discussions regarding this work, and Eric Rentmeester, Adam Mittermaier, Elizabeth Buschert, and Jonathan Giesler for technical assistance. We thank the University of Wisconsin Biotechnology Center DNA Sequencing Facility for providing next-generation sequencing consultation and services. This research was supported by the National Science Foundation grants IOS-1238014 (J.F.D. and S.F.-G.), IOS-1237931 (to N.M.S.), and IOS-1444514 (J.A.B.), and by USDA-NIFA (United States Department of Agriculture National Institute for Food and Agriculture) grant 2016-67013-24747 (to N.M.S.) and USDA-ARS base funds (to S.F.-G.).
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7882973.
Communicating editor: A. Paterson
Literature Cited
- Albert P. S., Gao Z., Danilova T. V., and Birchler J. A., 2010. Diversity of chromosomal karyotypes in maize and its relatives. Cytogenet. Genome Res. 129: 6–16. 10.1159/000314342 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., and Huber W., 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S. N., Zynda G. J., Song J., Han Z., Vaughn M. W. et al. , 2018. Subtle perturbations of the maize methylome reveal genes and transposons silenced by chromomethylase or RNA-directed DNA methylation pathways. G3 (Bethesda) 8: 1921–1932. 10.1534/g3.118.200284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G. W., 1932. Studies of Euchlaena and its hybrids with Zea. Z. Indukt. Abstamm. Vererbungsl. 62: 291–304. [Google Scholar]
- Beadle G. W., 1972. The mystery of maize. Field Mus. Nat. Hist. Bull. (Chicago) 43: 2–11. [Google Scholar]
- Birchler J. A., Albert P. S., and Gao Z., 2007. Stability of repeated sequence clusters in hybrids of maize as revealed by FISH. Trop. Plant Biol. 1: 34–39. 10.1007/s12042-007-9001-y [DOI] [Google Scholar]
- Bomblies K., 2010. Doomed lovers: mechanisms of isolation and incompatibility in plants. Annu. Rev. Plant Biol. 61: 109–124. 10.1146/annurev-arplant-042809-112146 [DOI] [PubMed] [Google Scholar]
- Bomblies K., and Weigel D., 2007. Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat. Rev. Genet. 8: 382–393. 10.1038/nrg2082 [DOI] [PubMed] [Google Scholar]
- Bouyer D., Kramdi A., Kassam M., Heese M., Schnittger A. et al. , 2017. DNA methylation dynamics during early plant life. Genome Biol. 18: 179 10.1186/s13059-017-1313-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs W. H., McMullen M. D., Gaut B. S., and Doebley J., 2007. Linkage mapping of domestication loci in a large maize teosinte backcross resource. Genetics 177: 1915–1928. 10.1534/genetics.107.076497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J. et al. , 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavrak V. V., Lettne N., Jamge S., Kosarewicz A., Bayer L. M. et al. , 2014. How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet. 10: e1004115 10.1371/journal.pgen.1004115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuerda-Gil D., and Slotkin R. K., 2016. Non-canonical RNA-directed DNA methylation. Nat. Plants 2: 16163 (erratum: Nat. Plants 3: 16211) 10.1038/nplants.2016.163 [DOI] [PubMed] [Google Scholar]
- Cui X., and Cao X., 2014. Epigenetic regulation and functional exaptation of transposable elements in higher plants. Curr. Opin. Plant Biol. 21: 83–88. 10.1016/j.pbi.2014.07.001 [DOI] [PubMed] [Google Scholar]
- Davidson N. M., Hawkins A. D. K., and Oshlack A., 2017. SuperTranscripts: a data driven reference for analysis and visualisation of transcriptomes. Genome Biol. 18: 148 (erratum: Genome Biol. 18: 160) 10.1186/s13059-017-1284-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., and Stec A., 1991. Genetic analysis of the morphological differences between maize and teosinte. Genetics 129: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J., Stec A., and Hubbard L., 1997. The evolution of apical dominance in maize. Nature 386: 485–488. 10.1038/386485a0 [DOI] [PubMed] [Google Scholar]
- Elshire R. J., Glaubitz J. C., Sun Q., Poland J. A., Kawamoto K. et al. , 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6: e19379 10.1371/journal.pone.0019379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Pyhäjärvi T., Weber A. L., Dawe R. K., Glaubitz J. C. et al. , 2012. Megabase-scale inversion polymorphism in the wild ancestor of maize. Genetics 191: 883–894. 10.1534/genetics.112.138578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L., and Sweigart A. L., 2018. When two rights make a wrong: the evolutionary genetics of plant hybrid incompatibilities. Annu. Rev. Plant Biol. 69: 707–731. 10.1146/annurev-arplant-042817-040113 [DOI] [PubMed] [Google Scholar]
- Garcia-Aguilar M., Michaud C., Leblanc O., and Grimanelli D., 2010. Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22: 3249–3267. 10.1105/tpc.109.072181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaubitz J. C., Casstevens T. M., Lu F., Harriman J., Elshire R. J. et al. , 2014. TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One 9: e90346 10.1371/journal.pone.0090346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A. et al. , 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Nava M., Warren C. A., León P., and Walbot V., 1998. Transcriptionally active MuDR, the regulatory element of the mutator transposable element family of Zea mays, is present in some accessions of the Mexican land race zapalote Chico. Genetics 149: 329–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D. et al. , 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8: 1494–1512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T. F., Ibarra C. A., Silva P., Zemach A., Eshed-Williams L. et al. , 2009. Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454. 10.1126/science.1172417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., and Kakutani T., 2014. Control of transposable elements in Arabidopsis thaliana. Chromosome Res. 22: 217–223. 10.1007/s10577-014-9417-9 [DOI] [PubMed] [Google Scholar]
- Johnson L. S., Eddy S. R., and Portugaly E., 2010. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11: 431 10.1186/1471-2105-11-431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N. R., Yeoh J. M., Coruh C., and Axtell M. J., 2016. Improved placement of multi-mapping small RNAs. G3 (Bethesda) 6: 2103–2111. 10.1534/g3.116.030452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A., 1999. Air drying method using nitrous oxide for chromosome counting in maize. Biotech. Histochem. 74: 160–166. 10.3109/10520299909047968 [DOI] [PubMed] [Google Scholar]
- Kato A., Lamb J. C., and Birchler J. A., 2004. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101: 13554–13559. 10.1073/pnas.0403659101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A., Lamb J. C., Albert P. S., Danilova T., Han F. et al. , 2011. Chromosome painting for plant biotechnology. Methods Mol. Biol. 701: 67–96. 10.1007/978-1-61737-957-4_4 [DOI] [PubMed] [Google Scholar]
- Kawakatsu T., Nery J. R., Castanon R., and Ecker J. R., 2017. Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 18: 171 10.1186/s13059-017-1251-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle J. L., 2006. A selfish gene governing pollen-pistil compatibility confers reproductive isolation between maize relatives. Genetics 172: 499–506. 10.1534/genetics.105.048645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., and Salzberg S. L., 2015. HISAT: a fast-spliced aligner with low memory requirements. Nat. Methods 12: 357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J. C., Danilova T., Bauer M. J., Meyer J. M., Holland J. J. et al. , 2007. Single-gene detection and karyotyping using small-target fluorescence in situ hybridization on maize somatic chromosomes. Genetics 175: 1047–1058. 10.1534/genetics.106.065573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J. A., and Jacobsen S. E., 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. 10.1038/nrg2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Gentleman R., and Carey V., 2009. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics 25: 1841–1842. 10.1093/bioinformatics/btp328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Eichten S. R., Hermanson P. J., Zaunbrecher V. M., Song J. et al. , 2014. Genetic perturbation of the maize methylome. Plant Cell 26: 4602–4616. 10.1105/tpc.114.133140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm A. K., Dyer K. A., Firman R. C., Fishman L., Forstmeier W. et al. , 2016. The ecology and evolutionary dynamics of meiotic drive. Trends Ecol. Evol. 31: 315–326. 10.1016/j.tree.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Liu Z., Cook J., Melia-Hancock S., Guill K., Bottoms C. et al. , 2016. Expanding maize genetic resources with predomestication alleles: maize–teosinte introgression populations. Plant Genome 9 10.3835/plantgenome2015.07.0053 [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., and Anders S., 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Kermicle J. L., and Evans M. M., 2014. Genetic and cellular analysis of cross-incompatibility in Zea mays. Plant Reprod. 27: 19–29. 10.1007/s00497-013-0236-5 [DOI] [PubMed] [Google Scholar]
- Marí-Ordóñez A., Marchais A., Etcheverry M., Martin A., Colot V. et al. , 2013. Reconstructing de novo silencing of an active plant retrotransposon. Nat. Genet. 45: 1029–1039. 10.1038/ng.2703 [DOI] [PubMed] [Google Scholar]
- Martin M., 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Matzke M. A., Kanno T., and Matzke A. J. M., 2015. RNA-Directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 66: 243–267. 10.1146/annurev-arplant-043014-114633 [DOI] [PubMed] [Google Scholar]
- Rieseberg L. H., and Blackman B. K., 2010. Speciation genes in plants. Ann. Bot. 106: 439–455. 10.1093/aob/mcq126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J. C., 2015. Genome evolution in maize: from genomes back to genes. Annu. Rev. Plant Biol. 66: 329–343. 10.1146/annurev-arplant-043014-115604 [DOI] [PubMed] [Google Scholar]
- Springer N. M., Anderson S. N., Andorf C. M., Ahern K. R., Bai F. et al. , 2018. The maize W22 genome provides a foundation for functional genomics and transposon biology. Nat. Genet. 50: 1282–1288. 10.1038/s41588-018-0158-0 [DOI] [PubMed] [Google Scholar]
- Studer A. J., and Doebley J. F., 2012. Evidence for a natural allelic series at the maize domestication locus teosinte branched1. Genetics 191: 951–958. 10.1534/genetics.112.138479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Nussbaum-Wagler T., Li B., Zhao Q., Vigouroux Y. et al. , 2005. The origin of the naked grains of maize. Nature 436: 714–719. 10.1038/nature03863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H., 2016. ggplot2: Elegant Graphics for Data Analysis. Springer, Heidelberg, Germany. [Google Scholar]
- Wills D. M., Whipple C. J., Takuno S., Kursel L. E., Shannon L. M. et al. , 2013. From many, one: genetic control of prolificacy during maize domestication. PLoS Genet. 9: e1003604 10.1371/journal.pgen.1003604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y., and Li W., 2009. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10: 232 10.1186/1471-2105-10-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., and Tammi M. T., 2009. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics 10: 80 10.1186/1471-2105-10-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The WGBS data are available under the National Center for Biotechnology Information BioProject accession number PRJNA526266, the RNA-seq data are available under accession number PRJNA528342, the WGS data are available under accession number PRJNA528290, and the sRNA-seq data are available under accession number PRJNA528352. The Sequence Read Archive accession numbers are available in Table S1. The GBS data reported in this paper have been deposited in the FigShare database (https://figshare.com/s/a882aff235818fc1762c). Supplemental material available at FigShare: https://doi.org/10.25386/genetics.7882973.