Abstract
MicroRNAs (miRNAs) associate with argonaute (AGO) proteins to post-transcriptionally modulate the expression of genes involved in various cellular processes. Herein, we show that loss of the Caenorhabditis elegans AGO gene alg-2 results in rapid and significantly increased germ cell apoptosis in response to DNA damage inflicted by ionizing radiation (IR). We demonstrate that the abnormal apoptosis phenotype in alg-2 mutant animals can be explained by reduced expression of mir-35 miRNA family members. We show that the increased apoptosis levels in IR-treated alg-2 or mir-35 family mutants depend on a transient hyperactivation of the C. elegans ERK1/2 MAPK ortholog MPK-1 in dying germ cells. Unexpectedly, MPK-1 phosphorylation occurs downstream of caspase activation and depends at least in part on a functional cell corpse-engulfment machinery. Therefore, we propose a refined mechanism, in which an initial proapoptotic stimulus by the core apoptotic machinery initiates the engulfment process, which in turn activates MAPK signaling to facilitate the demise of genomically compromised germ cells.
Keywords: microRNAs, apoptosis, DNA damage, MAPK signaling
APOPTOSIS describes the process of programmed cell death required to eliminate aberrant and damaged cells during development, or in response to stress. Upon DNA damage, proper regulation of apoptosis induction is essential to protect the organism from tissue deterioration and tumor formation (Thompson 1995). The Caenorhabditis elegans germline has proven to be a valuable model system to study the processes that regulate apoptosis in response to genotoxicity inflicted by ionizing radiation (IR) (Gumienny et al. 1999; Gartner et al. 2000). According to the canonical model, DNA-damage checkpoint signaling triggers phospho-activation of the C. elegans p53-like protein CEP-1, which in turn selectively promotes apoptosis in late-pachytene germ cells by mediating the transcriptional upregulation of the two BH3-only genes egl-1 and ced-13 (Gartner et al. 2000; Derry et al. 2001; Schumacher et al. 2001, 2005b; Hofmann et al. 2002). Both CEP-1 targets antagonize the antiapoptotic Bcl-2 protein CED-9, which resides on the mitochondrial surface and normally restrains the proapoptotic Apaf-1 adaptor protein CED-4 from apoptosome formation and subsequent activation of the caspase homolog CED-3 (Yuan and Horvitz 1990; Wu et al. 1997; Xue and Horvitz 1997; Conradt and Horvitz 1998; del Peso et al. 1998; Yang et al. 1998; Gumienny et al. 1999; Hofmann et al. 2002; Conradt and Xue 2005; Schumacher et al. 2005b). However, this canonical model has been challenged by cytological studies that have demonstrated that instead of being sequestered by CED-9 at mitochondria of living cells, the CED-4 protein localizes predominantly to the perinuclear space (Pourkarimi et al. 2012). Therefore, it remains mechanistically unclear how cells initiate apoptosis downstream of CEP-1 and its transcriptional targets. Numerous factors and pathways have been identified to impinge on the regulation of DNA damage-induced apoptosis, in parallel or downstream, of CEP-1 and EGL-1/CED-13, indicating that upregulation of BH3-only genes alone might be insufficient to fully commit a cell to apoptosis upon genotoxic stress (Reddien et al. 2007; Schertel and Conradt 2007; Greiss et al. 2008).
MAPK signaling is a major driver of gene expression in eukaryotic cells and mediates fundamental cellular processes including cell growth, differentiation, and death (L’Allemain 1994; Tibbles and Woodgett 1999). In C. elegans, the extracellular signal-regulated kinase (ERK1/2) MAPK ortholog MPK-1 represents the terminal regulator of a highly conserved signaling cascade that involves the Ras GTPase LET-60, the Raf MAPK kinase kinase LIN-45, and the MAPK kinase Mek ortholog MEK-2 that ultimately phosphorylates MPK-1 (Lackner et al. 1994; Wu and Han 1994; Sundaram 2013). In the C. elegans germline, MPK-1 drives meiotic germ cell progression, physiological germ cell death, and oocyte maturation (Church et al. 1995; Gumienny et al. 1999; Lee et al. 2007). MPK-1 furthermore becomes phosphorylated in late-pachytene and diplotene germ cells in response to DNA damage where it is required to ensure proper apoptosis induction, presumably by mediating CEP-1/p53 expression and activity (Rutkowski et al. 2011). However, the molecular mechanisms that trigger MPK-1 phosphorylation in response to DNA damage are not well understood (Rutkowski et al. 2011; Ermolaeva et al. 2013). In addition, the essential function of MPK-1 in driving germ cells through meiotic prophase I complicates the study of its exact role and relevance during the killing process of late-pachytene germ cells (Lee et al. 2007).
MicroRNAs (miRNAs) are single-stranded, endogenously transcribed RNA molecules, 19–25 nt in length, that associate with specialized argonaute (AGO) proteins as well as other associated factors to post-transcriptionally silence gene expression by complementarily binding to the 3′-UTR of target mRNAs (Bartel 2004; Friedman et al. 2009; Ha and Kim 2014). In mammalian systems, miRNAs have been identified to modulate the expression of various DNA damage-response and apoptosis factors, and thus have been linked to cell transformation and tumor progression. (Cimmino et al. 2005; Raver-Shapira et al. 2007; Welch et al. 2007; Zhang et al. 2007; Kato et al. 2009; Zhou et al. 2010; Kumar et al. 2011; Lujambio and Lowe 2012; Zang et al. 2012; Kwon et al. 2013).
In C. elegans, three AGO proteins—ALG-1, ALG-2, and ALG-5 (AGO-like gene 1/2/5)—act during miRNA biogenesis and function (Grishok et al. 2001; Tops et al. 2006; Yigit et al. 2006; Brown et al. 2017). In contrast to alg-1-deficient worms, alg-2 loss-of-function mutant animals have no obvious developmental defects and were recently reported to even display a prolonged life span (Grishok et al. 2001; Aalto et al. 2018). Furthermore, adult alg-2 mutant animals display only a mild reduction of overall germline integrity (Bukhari et al. 2012). We chose to utilize the germline of alg-2-deficient animals as a model system to investigate how miRNAs modulate DNA damage-induced apoptosis in vivo. We observed that an alg-2(ok304) mutant strain displayed a rapid and substantial, yet transient, increase of DNA double-strand break (DSB)-induced germ cell apoptosis that depended on the presence of CEP-1 and the EGL-1 protein. By means of next-generation sequencing (NGS), we show that alg-2 mutant animals display significantly reduced expression levels of most mir-35 family members, a miRNA family that has been recently demonstrated to control apoptosis in somatic cells during development in C. elegans by targeting egl-1 mRNA (Sherrard et al. 2017). mir-35 family deletion mutants phenocopied the alg-2 mutant apoptosis defect in response to DNA damage. The elevated apoptosis levels in alg-2 and mir-35 family mutant worms were associated with a simultaneous hyperphosphorylation of the ERK1/2 MAPK homolog MPK-1 within the entire germline “loop”, including dying late-pachytene cells. MPK-1 was essential for IR-induced apoptosis in the alg-2 mutant, but was only phosphorylated downstream of the induction of cell death via CED-3 and initiation of the corpse engulfment machinery. We suggest that MPK-1 signaling functions downstream of the initial activation of the core apoptotic machinery to regulate the life or death decision during DNA damage-induced apoptosis.
Materials and Methods
General methods and strains
All C. elegans strains were maintained on NGM agar plates seeded with Escherichia coli OP50 and were grown at 20°, as previously described (Brenner 1974). C. elegans N2 Bristol was used as wild-type (WT).
The following mutant strains and transgenic lines were used: WM53: alg-2(ok304), MT14119: mir-35-41(nDf50), MT13231: mir-36-41(nDf48), DW102: brc-1(tm1145), TJ1: cep-1(gk138), BJS705: alg-2(ok304);cep-1(gk138), MT8735: egl-1(n1084n3082), PP2096: egl-1(n1084n3082);ced-13(tm536), BJS781: alg-2(ok304);egl-1(n1084n3082), BJS782: alg-2(ok304);egl-1(n1084n3082);ced-13(tm536), VC199: sir-2.1(ok434), BJS622: alg-2(ok304);sir-2.1(ok434), SD939: mpk-1(ga111) unc-79(e1068), MT8666: mek-2(n1989), BJS608: alg-2(ok304);mpk-1(ga111) unc-79(e1068), BJS711: alg-2(ok304);mek-2(n1989), MT4770: ced-9(n1950), BJS726: alg-2(ok304);ced-9(n1950), MT1522: ced-3(n717), WS733: ced-3(op149), MT7408: ced-3(n2438), BJS780: alg-2(ok304);ced-3(n717), BJS834: alg-2(ok304);ced-3(op149), BJS835: alg-2(ok304);ced-3(n2438), BJS836: alg-2(ok304);ced-1(e1735);ced-5(n1812), NL4517: alg-2(ok304);pkIs2256 [alg-2::HA + rol-6(su1006)], MD701: bcIs39[P(lim-7)ced-1::GFP + lin-15(+)], BJS838: alg-2(ok304); bcIs39[P(lim-7)ced-1::GFP + lin-15(+)], BJS839: alg-2(ok304);mpk-1(ga111); bcIs39[P(lim-7)ced-1::GFP + lin-15(+)], PHX1272: egl-1(syb1272[egl-1::V5]), BJS872: alg-2(ok304);egl-1(syb1272[egl-1::V5]), and BJS873: mir-35-41(nDf50);egl-1(syb1272[egl-1::V5]). The strain PHX1272 was obtained from SunyBiotech, China.
Apoptosis scoring using DIC microscopy
After synchronizing worms at the late-L4 larval stage by individual picking, animals were kept at 20° until they reached day 1 of adulthood. Worms were irradiated with the indicated IR-doses using a Biobeam 8000 with 137Cs radionucleotide source (Gamma-Service Medical GmbH, Leipzig, Germany) or were kept untreated to serve as negative controls. After irradiation, worms were allowed to recover at 20° for 4 hr (if not indicated otherwise) before being anesthetized with 5 mM levamisol on 2% agarose pads. Apoptotic germ cell death was assessed using the Nomarski differential interference contrast (DIC) settings on Zeiss Axio Imager M1/2 microscopes (Zeiss [Carl Zeiss], Thornwood, NY), as previously described in Gumienny et al. (1999) and Gartner et al. (2004).
Acridine orange staining of apoptotic corpses
Worms were synchronized at the late-L4 larval stage on 35-mm agar dishes seeded with OP50 and were kept at 20° until they reached day 1 of adulthood. Adult animals were exposed to γ-radiation or remained untreated to serve as negative controls. Worms were subsequently allowed to recover at 20° for 4 hr. For acridine orange (AO) staining, 10 µl of 2% AO stock solution (Sigma [Sigma Chemical], St. Louis, MO) were mixed with 1 ml M9 buffer and 200 µl of the diluted AO solution were added onto each plate. Worms were kept on the AO-treated plates for 1 hr at room temperature (RT) in the dark before being washed three times with M9. To remove superfluous AO stain, worms were subsequently transferred onto fresh 35-mm plates seeded with OP50 and were kept at RT in the dark for another 45 min. Finally, animals were anesthetized with 5 mM levamisol on 2% agarose pads and AO staining was assessed on a Zeiss Axio Imager M1 fluorescent microscope.
Immunofluorescence in dissected C. elegans germlines
Worms were synchronized at the late-L4 larval stage by picking and were kept at 20° until they reached day 1 of adulthood. Adult animals were exposed to γ-radiation or remained untreated to serve as negative controls. Worms were allowed to recover at 20° for the indicated time spans before being dissected in buffer solution containing 20 mM NaN3 with syringe needles to expose germlines. Tissues were fixed for 5 min with 4% paraformaldehyde solution on adhesive HistoBond + slides (Paul Marienfeld GmbH & Co. KG, Lauda-Königshofen, Germany) before freeze cracking on dry ice. Slides were subsequently fixed for 1 min in ice-cold methanol at −20° and washed three times in PBS-T (PBS-Tween), followed by a 30-min blocking step with 10% donkey serum in PBS-T. For staining against SIR-2.1 and V5, slides were first blocked for 20 min with Image-iT FX signal enhancer (Thermo Fisher/Invitrogen, Carlsbad, CA) before blocking for another 15–30 min with 10% donkey serum in PBS-T. The following primary antibodies were used in this study: mouse monoclonal anti-HA antibody [catalog number (Cat. No.): 12CA5, dilution 1:100 in PBS-T containing 10% donkey serum; Merck/Sigma], rabbit polyclonal anti-HA antibody (Cat. No.: H6908, dilution 1:100 in PBS-T containing 10% donkey serum; Merck/Sigma), rabbit polyclonal anti-RAD-51 (Cat. No.: 29480002, dilution 1:350 in PBS-T containing 10% donkey serum; Novus Biologicals, Centennial, CO), mouse V5-Tag antibody, clone SV5-Pk1 (Cat. No.: MCA1360, dilution 1:100 in PBS-T containing 10% donkey serum; Bio-Rad, Hercules, CA), mouse monoclonal anti-MAPK, activated (diphosphorylated ERK1 and 2) (Cat. No.: M9692, dilution 1:100 in PBS-T containing 10% donkey serum; Merck/Sigma), rabbit polyclonal anti-phospho-p42/44 (Erk1/2) (Thr202/Tyr204) (Cat. No.: 9101, dilution 1:100 in PBS-T containing 10% donkey serum; Cell Signaling, Danvers, MA), rabbit polyclonal anti-Sir-2.1 (Cat. No.: PA1-16933, dilution 1:100 in PBS-T containing 10% donkey serum; Thermo Fisher/Invitrogen), mouse anti-GFP Living Colors A.v. Monoclonal Antibody (JL-8) (Cat. No.: 632380, dilution 1:100 in PBS-T containing 10% donkey serum; Takara Bio Clontech, Mountain View, CA). Primary antibody incubation was always performed at 4° overnight. Incubation was followed by three washes with PBS-T. Secondary antibody incubation was done in the dark for 2 hr at room temperature. The following secondary antibodies were used: AlexaFluor 488 donkey anti-mouse IgG (Cat. No.: A21202, dilution 1:500 in PBS-T containing 10% donkey serum; Thermo Fisher/Invitrogen) and AlexaFluor 594 donkey anti-rabbit IgG (Cat. No.: A21207, dilution 1:350 in PBS-T containing 10% donkey serum; Thermo Fisher/Invitrogen). After three final washes in PBS-T, slides were mounted with DAPI Fluoromount-G (SouthernBiotech, Birmingham, AL) and sealed with nail polish. Representative images were taken using a Zeiss Meta 710 confocal microscope and Zen 2009 software. Images were subsequently processed and analyzed using the ImageJ software package. The allocation of phospho-MPK-1 signal into qualitative categories was achieved by assessing the stained germlines under a Zeiss Axio Imager M1 fluorescence microscope at 10× and 40× magnification.
RNA interference against sir-2.1
RNA interference (RNAi) clones against sir-2.1 and the empty vector (EV) control were obtained from the C. elegans RNAi collection (Ahringer), distributed by Source BioScience. Bacterial cultures were grown overnight at 37° in LB medium supplemented with 100 µg/ml ampicillin. The next day, these starter cultures were used to inoculate fresh cultures that were subsequently grown to an OD600 of ∼0.6. For induction of double-stranded RNA expression, cultures were incubated with 1 mM IPTG at 37° for an additional 3 hr. Induced cultures were subsequently seeded on NGM agar plates containing 100 µg/ml ampicillin and 3 mM IPTG. For RNAi-mediated knockdown experiments, worms were transferred to plates seeded either with sir-2.1 RNAi or EV control bacteria, and kept at 20° for two generations. The F1 generation was subsequently synchronized at the late-L4 stage. After 24 hr, day-1 adult germlines were dissected and stained with a SIR-2.1 antibody as described above.
Homologous recombination-directed repair assay
The experiment was performed as previously described in Johnson et al. (2013). Worms were synchronized at the late-L4 stage by picking and were kept for 24 hr at 20° until reaching day 1 of adulthood. For each genotype and condition, five adult animals were transferred to 35-mm NGM plates seeded with a drop of OP50 as food source and were allowed to lay eggs for a time period of 2 hr at 20°. Animals were subsequently removed from plates and eggs were immediately irradiated with 0, 20, or 40 Gy of γ-radiation before being shifted to 20°. After 24 hr, the numbers of hatched vs. nonhatched eggs were scored and percentages of embryonic survival were calculated. The brc-1(tm1145) mutant strain was included to serve as a positive control with impaired homologous recombination (HR)-directed repair capacity. Each experiment was performed in biological triplicates.
Scoring DNA damage-induced cell cycle arrest of mitotic germ cells
The experiment was performed as previously described in Gartner et al. (2004). Worms were synchronized at the late-L4 larval stage by picking and subsequently exposed to 0, 30, 60, and 90 Gy of γ-radiation. At 16 hr post-IR, 20 worms per condition were anesthetized with 5 mM levamisole solution on 2% agarose pads. The number of mitotic cells was scored in a defined region with an area of 3.125 × 6.25 µm at the distal end of the C. elegans germline using Nomarski DIC microscopy on a Zeiss Axio Imager M1. To facilitate precise and consistent scoring of mitotic germ nuclei among different germlines at all focal planes, a netmicrometer was applied to the microscope ocular (d = 26 mm, 12.5 × 12.5/5;10; Zeiss).
Monitoring of gene expression using real-time quantitative PCR
For whole-worm real-time quantitative PCR (RT-qPCR), 200 worms per genotype and condition were synchronized at the L4 larval stage by picking, and kept for 24 hr at 20° until they reached day 1 of adulthood. Animals were subsequently exposed to γ-radiation or remained untreated to serve as controls. After 2 hr at 20°, worms were washed three times with M9 buffer and lysed in 1 ml TRIzol reagent (Thermo Fisher) containing 0.7 mm zirconia/silica beads (Biospec Products, Bartlesville, OK). Afterward, animals were disrupted with a Precellys24 (Bertin, Montigny-le-Bretonneux, France). The RNeasy Mini kit (QIAGEN, Valencia, CA) was used for RNA isolation according to the manufacturers’ specifications, except for the use of 1-bromo-3-chloropropane (Sigma) instead of chloroform for phase separation. Reverse transcription into cDNA was performed using Superscript III (Thermo Fisher/Invitrogen). For RT-qPCR, cDNA was mixed with SYBR Green I (Merck/Sigma) and Platinum Taq polymerase (Thermo Fisher/Invitrogen) and Bio-Rad CFX96 real-time PCR machines were used. All qPCR reactions were done in biological triplicates and in technical replicates. For data analysis, the second-derivative maximum method was used and the induction of target cDNA was calculated as described by Pfaffl (2001):
miRNA NGS
First, 1000 synchronized WT and alg-2(ok304) L1 larvae were allowed to grow for 72 hr at 20° until they reached day 1 of adulthood. Worms were treated with 90 Gy of γ-radiation or remained untreated to serve as controls. After 2.5 hr at 20°, worms were washed three times in M9 buffer before being lysed in 1 ml TRIzol reagent (Thermo Fisher) containing 0.7 mm zirconia/silica beads (Biospec Products). Afterward, animals were disrupted with a Precellys24 (Bertin). Whole-RNA isolation was done using 1-bromo-3-chloropropane (Sigma) for phase separation and the mirVana miRNA isolation kit (Thermo Fisher), according to the manufacturers’ specifications. Sequencing of miRNAs was performed at the Cologne Center for Genomics using an Illumina HiSeq 4000. RNA sequencing data were processed through the QuickNGS analysis pipeline, version 1.2.7, based on Ensembl release 87 as described in Wagle et al. (2015). The data files are deposited under the Gene Expression Omnibus (GEO) accession code GSE124606.
Statistical analysis
All statistical tests, as well as error bar descriptions and sample sizes for each experiment, are detailed in the respective figure legends or are indicated in the graphs themselves. Randomization was not performed because the group allocation was guided by the genotype of the respective mutant worms. Worms of a given genotype were randomly selected from large strain populations for each experiment without any preconditioning.
Data availability
The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article and its figures. The file “Supplemental information” contains figure legends and descriptions of all supplemental files. New strains used in this study will be made available at the Caenorhabditis Genetics Center (University of Minnesota). miRNA expression data are available at the GEO with the accession code GSE124606. Supplemental Material available at FigShare: https://doi.org/10.25386/genetics.8852180.
Results
alg-2 mutant animals exhibit a burst of elevated germ cell apoptosis in response to IR
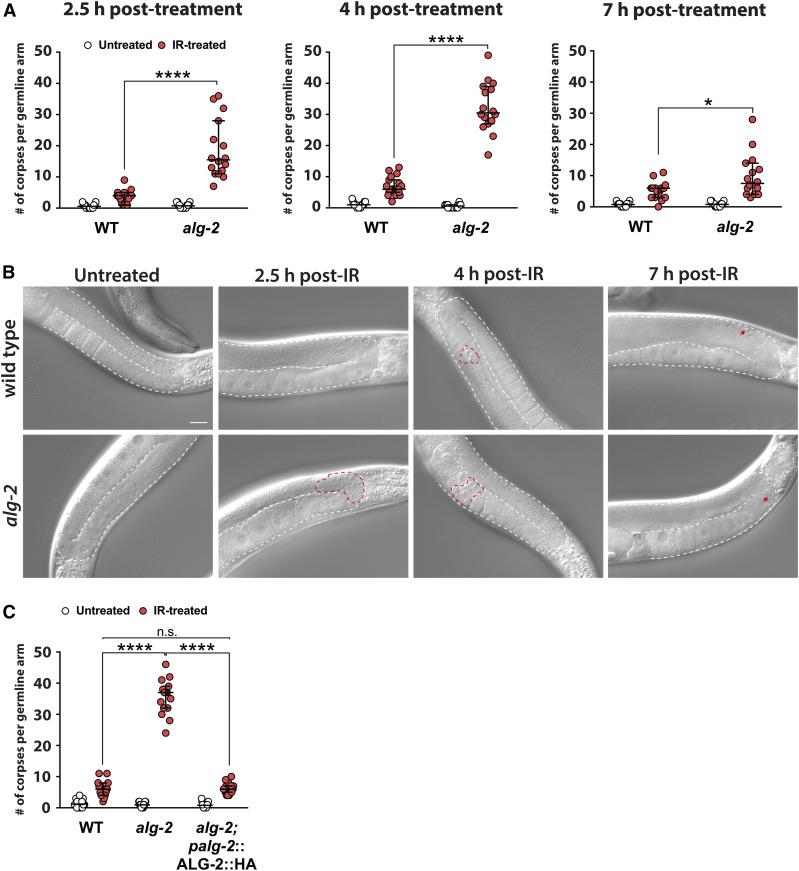
To understand to what extent the microRNA pathway contributes to DNA damage-induced apoptosis in C. elegans, we used Nomarski DIC microscopy to assess apoptosis of late-pachytene germ cells over a time span of 7 hr in day-1 adult WT worms and in alg-2(ok304) deletion mutants treated with 90 Gy of γ-radiation (Figure 1, A and B). At 2.5 hr after IR treatment, the germlines of WT worms displayed a mild increase of germ cell apoptosis within the distal gonad loop. In contrast, we observed a considerable number of early apoptotic cell corpses in the loop of the alg-2 mutant germlines (Figure 1A, left and Figure 1B, highlighted by red dashed line). At 4 hr post-IR treatment, the scored cell death levels of the alg-2 mutant animals were up to five times higher compared to those of the WT worms (Figure 1A, middle). Notably, the majority of the apoptotic cells in alg-2 mutant gonads had at this time point migrated to the proximal end of the germline, an observation commonly made in mutant strains exhibiting abnormally high apoptosis numbers (Figure 1B). These results suggest that in the alg-2 mutant animals, IR-induced cell death occurs mainly within 2–3 hr post-treatment and results in high levels of persistent corpses that evade the engulfment machinery. In addition, the proximal half of IR-treated alg-2 mutant germlines displayed severe disorganization 4 hr post-IR, including malformed oocytes surrounded by cell corpses (Figure 1B). Thus, we conclude that the high number of apoptotic cells in the alg-2 mutant worms has detrimental effects on proper oocyte maturation and general germline integrity. However, these effects were transient as at 7 hr post-IR, alg-2 germlines had regained a normal morphology and their overall cell corpse levels were similar to those of the WT animals (Figure 1, A and B, right). The latter finding demonstrated that, given sufficient time, cell corpse clearance was functional in the alg-2 mutant worms. In contrast to a previous report (Vasquez-Rifo et al. 2012), we did not observe significantly increased physiological germ cell apoptosis in the absence of DNA damage-inducing treatment in the alg-2 mutant animals (Figure 1A), showing that the enhanced cell death in this mutant strain was caused exclusively by the IR treatment. We confirmed the elevated number and persistent presence of apoptotic corpses in the proximal germline half of IR-exposed alg-2 animals via acridine AO staining 4 hr post-treatment (Supplemental Material, Figure S1A). Of note, AO staining failed to properly differentiate between individual corpses under conditions of excessive apoptosis (Figure S1A). Therefore, we proceeded to rely on conventional DIC microscopy to faithfully quantify apoptosis levels in the alg-2 mutant strain. Finally, at 4 hr post-IR treatment, the proximal germline of alg-2 mutant but not WT animals contained numerous germ cell nuclei displaying a strong DAPI staining signal that is characteristic of apoptotic pachytene cells (Figure S1B).
Figure 1.
alg-2(ok304) mutants display transiently elevated apoptosis of late-pachytene germ cells upon IR. (A) Quantification of apoptotic corpse numbers revealed elevated germ cell apoptosis in alg-2(ok304) mutant animals following IR treatment (90 Gy) at different time points. (B) Representative DIC images of IR-treated and untreated WT and alg-2(ok304) mutant germlines taken at the indicated time points. Bar, 20 µm. At 2.5 hr post-IR, considerable numbers of early apoptotic corpses were visible in the gonad loops (highlighted by red dashed line) of alg-2(ok304) mutant animals. At 4 hr post-IR, the majority of corpses had evaded engulfment and migrated to the proximal germline (highlighted by red dashed line). After a further 3 hr, the alg-2(ok304) mutant germlines regained a healthy morphology and exhibited only a few apoptotic cells (highlighted by red arrows). (C) An ALG-2::HA transgene fully rescued the alg-2(ok304) mutant apoptosis phenotype. (A and C) Representative graphs of one out of three independent experiments; medians with 95% C.I.s are shown, a minimum of 15 germlines per genotype and condition were scored. **** P < 0.0001 and n.s. = P > 0.05; Mann–Whitney nonparametric test. #, number; IR, ionizing radiation; n.s., not significant; WT, wild-type.
We next wondered whether an ALG-2::HA rescue transgene under the control of its own promoter could restore normal apoptosis levels in response to IR when expressed in an alg-2 mutant background. Indeed, alg-2(ok304);alg-2p:ALG-2::HA animals showed similar apoptosis levels as WT worms (Figure 1C), corroborating that the observed apoptosis phenotype occurred specifically due to disruption of the alg-2 gene. We confirmed the cytoplasmic expression of the ALG-2::HA fusion protein in alg-2(ok304);palg-2:ALG-2::HA germ cells by staining dissected germlines with two distinct anti-HA antibodies (Figure S2). Importantly, no cytoplasmic HA signal was detectable in WT or alg-2 mutant germlines. Unexpectedly, both antibodies also stained germ cell nuclei throughout the germlines of alg-2(ok304);palg-2:ALG-2::HA animals. However, since nuclear staining was also observable in WT and alg-2 mutant germlines, we conclude that the nuclear signal resulted from unspecific binding of the HA antibodies while the cytoplasmic staining was highly specific to the expression of the HA epitope (Figure S2).
alg-2 mutant worms display a mild decrease in HR-directed repair efficiency
We wondered whether the elevated apoptosis phenotype of IR-treated alg-2 mutant animals could be explained by inefficient DNA DSB repair due to impaired miRNA activity. The C. elegans germline relies on faithful DSB repair via HR (Clejan et al. 2006). Several studies have demonstrated that HR-directed repair efficiency can be assessed in C. elegans by irradiating early-stage embryos within the first 3 hr after being laid and subsequent scoring of their survival rates (Clejan et al. 2006; Johnson et al. 2013). As previously reported, mutations in genes with known functions in HR-dependent DSB repair, such as brc-1(tm1145), lead to high rates of embryonic lethality in this assay (Figure S3A) (Johnson et al. 2013). Interestingly, we observed a very subtle, yet highly reproducible, decrease in alg-2 mutant embryo survival compared to that of WT animals after IR, which suggests a possible defect in HR-directed repair (Figure S3A). To elaborate on this finding, we dissected germlines from irradiated and untreated control adult WT as well as alg-2 mutant animals 16 hr after treatment with 30 Gy of IR, and stained for persistent RAD-51 foci. RAD-51 binds to sites of DSBs, where it forms nucleoprotein filaments that facilitate the invasion into the homologous DNA strand, which is required for DSB repair (Pellegrini et al. 2002; Alpi et al. 2003). Indeed, a small, yet statistically significant increase in the number of IR-induced RAD-51 foci was detectable in alg-2 mutant germlines compared to the number detected in WT animals, indicative of a mild defect in HR-directed repair (Figure S3B). Taken together, our data suggest a putative regulatory role of ALG-2 and its associated miRNAs in the modulation of DSB repair via HR. However, we suspected that the DNA repair defects observed in the alg-2 mutant animals were too subtle to explain the abnormal level and timing of IR-induced cell death displayed by this mutant.
In addition to apoptosis of late-pachytene germ cells in the germline loop, DNA-damage checkpoint signaling can also trigger cell cycle arrest of mitotically proliferating germ cells located in the distal germline tip (Gartner et al. 2000, 2004). Cell cycle arrest occurs at G2 stage and can last for up to 20 hr (Bailly and Gartner 2013). Arrested germ cells cease to proliferate but still grow in size. To test whether loss of alg-2 not only promotes DNA damage-induced apoptosis but also affects cell cycle arrest, we scored the number of germ cells in a predefined area at the distal germline end. In line with previous studies, IR treatment of WT late-L4 larvae led to a dose-dependent decrease in germ cell numbers within the mitotic zone after 16 hr (Figure S3C) (Gartner et al. 2000). The alg-2 mutant strain showed a similar response to IR but exhibited slightly higher numbers of germ cells in the mitotic zone upon treatment with 30 Gy of IR compared to WT worms (Figure S3C). This finding indicates that alg-2 mutant animals display a mild delay in cell cycle arrest at low IR doses, which is in its extent, however, not comparable to their abnormal apoptosis phenotype. Thus, we concluded that the excessive levels of DNA damage-induced apoptosis in alg-2 mutant animals cannot be explained by a general defect in DNA-damage checkpoint activation. Therefore, we hypothesized that ALG-2 directly impinges on the regulation of apoptosis in response to genotoxic stress.
DNA damage-induced cell death in alg-2 mutant worms depends on CEP-1/p53 and the BH3-only protein EGL-1
Loss of functional CEP-1/p53 activity completely abolishes DNA damage-induced germ cell apoptosis but does not affect physiological cell death numbers (Derry et al. 2001; Schumacher et al. 2001). To determine whether the elevated IR-induced apoptosis phenotype observed in alg-2 mutant animals requires CEP-1, we crossed alg-2(ok304) animals with worms homozygous for the cep-1(gk138) deletion. As expected, IR-treated alg-2;cep-1 double mutants no longer exhibited increased cell death levels in the germline, demonstrating that the elevated apoptosis phenotype of the alg-2 mutant worms depends entirely on cep-1 (Figure 2A). In response to DNA damage, CEP-1 induces the expression of the two BH3-only genes egl-1 and ced-13 (Hofmann et al. 2002; Schumacher et al. 2005b). Introduction of the egl-1(n1084n3082) loss-of-function allele into the alg-2 mutant background was sufficient to eliminate the induction of DNA damage-dependent apoptosis (Figure 2B). Additional deletion of the second BH3-only protein-coding gene ced-13 in an alg-2(ok304);egl-1(n1084n3082);ced-13(tm536) triple mutant did not further suppress apoptosis levels in response to IR (Figure 2B). Consequentially, we wondered whether the increased IR-induced apoptosis phenotype of alg-2 animals correlated with enhanced CEP-1 transcriptional activity. Thus, we performed RT-qPCR with RNA isolated from whole worms to assess egl-1 and ced-13 transcription levels, in adult WT and alg-2 mutant animals 2 hr post-IR. As expected, egl-1 and ced-13 mRNA levels were increased in IR-treated WT animals compared to those of untreated worms (Figure 2C). This upregulation depended entirely on the cep-1 gene, thus confirming the specificity of our RT-qPCRs (Figure 2C). By comparing the IR-treated alg-2 mutant animals with untreated alg-2 worms, we observed that the apparent DNA damage-dependent induction of egl-1 and ced-13 transcription was weaker in the alg-2-deficient background compared to that in WT worms (Figure 2C, left). To directly compare the IR-induced egl-1 and ced-13 mRNA levels between WT and alg-2 animals, we also calculated the fold changes of egl-1 and ced-13 transcripts in IR-treated WT, alg-2, and cep-1 mutant worms relative to the respective transcript levels in untreated WT worms. This additional analysis further confirmed that the egl-1 and ced-13 mRNA levels did not significantly differ between the IR-treated WT and alg-2 mutant animals (Figure 2C, right). Instead, our RT-qPCRs suggest that alg-2 mutant animals exhibit increased baseline egl-1 and ced-13 transcription in the absence of genotoxic stress. In conclusion, the abnormal DNA damage-induced apoptosis phenotype in the alg-2 mutant animals does not correlate with elevated egl-1 and ced-13 mRNA levels, suggesting that ALG-2 impinges on the apoptotic response in parallel or downstream of CEP-1 activity.
Figure 2.
mir-35 family is downregulated in alg-2(ok304) mutants and regulates DNA damage-induced apoptosis. (A) cep-1(gk138) suppressed elevated DNA damage-induced apoptosis in alg-2(ok304) mutant animals. Apoptosis was scored 4 hr post-IR treatment (90 Gy). Representative graph from one out of three independent experiments; medians with 95% C.I.s are shown, a minimum of 15 germlines per genotype and condition were scored. **** P < 0.0001; Mann–Whitney nonparametric test. (B) egl-1(n1084n3082) suppressed elevated DNA damage-induced apoptosis in the alg-2(ok304) mutant animals. Representative graph from one out of three independent experiments, medians with 95% C.I.s are shown, a minimum of 15 germlines per genotype and condition were scored. n.s. = P > 0.05 and **** P < 0.0001; Mann–Whitney nonparametric test. (C) RT-qPCR results showing the mRNA levels of the CEP-1 targets egl-1 and ced-13, in WT and alg-2(ok304) mutant animals 2 hr post-IR (90 Gy). mRNA levels in IR-treated animals were either compared to their respective untreated controls (left) or relative to the transcript levels in untreated WT animals (right). mRNA levels were normalized to act-1 and tbg-1 expression, error bars represent SD between three biological replicates. n.s. = P > 0.05, ** P < 0.01, and **** = P < 0.0001; Student’s t-test. (D) RNA sequencing revealed that mir-35 family members are strongly downregulated in alg-2(ok304) mutant worms compared to WT. (E) The two different mir-35 family mutants mir-36-41(nDf48) and mir-35-41(nDf50) displayed strongly increased apoptosis levels 4 hr post-IR (90 Gy). Representative graph from one out of at least three independent experiments, medians with 95% C.I.s are shown, a minimum of 15 germlines per genotype and condition were scored, **** P < 0.0001; Mann–Whitney nonparametric test. (F) RT-qPCR results showing the mRNA levels of the CEP-1 targets egl-1 and ced-13, in WT and mir-35-41(nDf50) mutant animals 2 hr post-IR (90 Gy). mRNA levels in IR-treated animals were either compared to their untreated controls (left) or relative to the transcript levels of untreated WT animals (right). mRNA levels were normalized to act-1, tbg-1, and lmn-1 expression, error bars represent SD between three biological replicates. n.s. = P > 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001; Student’s t-test). #, number; IR, ionizing radiation; n.s., not significant; RT-qPCR, real-time quantitative PCR; WT, wild-type.
ALG-2 regulates IR-induced apoptosis via the mir-35 family of microRNAs
We next wished to determine which microRNAs might be dysregulated in alg-2 mutant animals and could be responsible for the elevated DNA damage-induced apoptosis. We performed miRNA sequencing with adult WT as well as alg-2 loss-of function mutants 2.5 hr after exposure to 90 Gy of IR or mock treatment. Remarkably, expression of only a single miRNA was significantly altered by at least twofold in WT worms upon IR treatment (Table S1). In general, the miRNA transcriptome remained surprisingly stable upon IR in both WT as well as alg-2 mutant animals (Tables S1 and S3); however, under both untreated and IR-treated conditions, the levels of seven out of the eight mir-35 family members were profoundly reduced in alg-2(ok304) mutant animals when compared to WT (Figure 2D and Tables S2 and S4).
Loss of the entire mir-35 family results in embryonic lethality (Alvarez-Saavedra and Horvitz 2010; McJunkin and Ambros 2014). To assess the role of mir-35-42 in DNA damage-induced apoptosis, we therefore worked with the deletion strains mir-35-41(nDf50) and mir-36-41(nDf48), which fail to generate seven and six members of this miRNA family, respectively. Both mutant strains exhibited significantly elevated numbers of apoptotic germ cell corpses within the proximal germline 4 hr post-IR and thus strongly resembled the IR-sensitivity phenotype of the alg-2 mutant animals (Figure 2E). Consistent with our alg-2 data, the comparison of the egl-1 and ced-13 mRNA levels between IR-treated and untreated mir-35-41 mutant worms suggested a weaker transcriptional response to genotoxic stress than that observed in WT animals (Figure 2F, left); however, when compared to untreated WT worms, the egl-1 transcript levels were higher in IR-treated mir-35-41 mutant worms than in IR-exposed WT animals (Figure 2F, right). In contrast, ced-13 mRNA abundance was not significantly different between mir-35-41 mutant animals and WT worms upon IR (Figure 2F, right). Therefore, our results suggest that the elevated apoptosis levels in IR-treated alg-2 mutant germlines are a consequence of reduced mir-35 family levels.
EGL-1 protein is expressed in late-pachytene germ cells upon IR exposure
Many miRNA target-prediction websites suggest egl-1 translation to be repressed by the mir-35-42 cluster of miRNAs. Consistently, several studies recently reported that the mir-35 family regulates apoptosis during C. elegans embryonic development and upon genotoxic stress by specifically targeting the 3′-UTR of egl-1 mRNA (Wu et al. 2010; Sherrard et al. 2017; Tran et al. 2019).
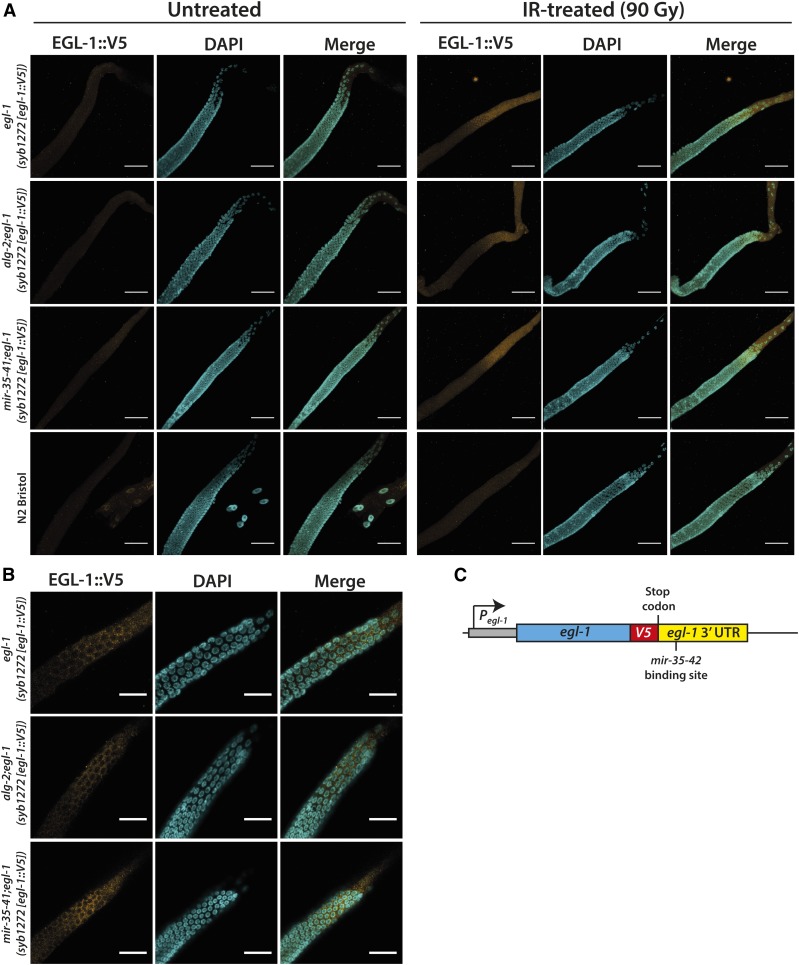
Therefore, we wished to confirm whether ALG-2 and the mir-35 family indeed regulate DNA damage-induced apoptosis by modulating EGL-1 protein abundance in dying germ cells. Unfortunately, attempts to generate an EGL-1-specific antibody have thus far failed, possibly due to the small size of the EGL-1 protein. In contrast to the egl-1 mRNA, no studies have yet directly investigated the EGL-1 protein in vivo. Therefore, we employed a clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-generated knock-in strain that has a V5 protein tag inserted into the endogenous egl-1 locus immediately upstream of the stop codon and the egl-1 3′-UTR (Figure 3C). egl-1(syb1272 [egl-1::V5]) knock-in animals exhibited a mild, nevertheless clear, apoptotic response toward IR-induced genotoxic stress, which was enhanced upon loss of alg-2 (Figure S4), indicating that the V5-tagged EGL-1 protein was functional in vivo. We dissected germlines of day-1 adult egl-1(syb1272 [egl-1::V5]) knock-in worms 2.5 hr post-IR or mock treatment, and stained for the EGL-1::V5 protein with a V5-Tag antibody. We did not detect a noticeable EGL-1::V5 signal in untreated germlines. In contrast, IR treatment triggered a marked induction of EGL-1 protein in late-pachytene germ cells and the rest of the germline loop that could be clearly observed using confocal microscopy (Figure 3A). In contrast, the midpachytene region of the germline remained free of EGL-1. As expected, the EGL-1 protein was found exclusively within the cytoplasm of germ cells and did not enter their nuclei (Figure 3B). Importantly, we did not detect any EGL-1::V5 signal in IR-treated N2 Bristol WT animals that did not carry the V5 insert, thus confirming the V5 antibody specificity (Figure 3A). We next crossed the egl-1(syb1272 [egl-1::V5]) knock-in strain into alg-2(ok304) and mir-35-41(nDf50) mutant animals to stain for endogenous EGL-1::V5 protein expression in these genetic backgrounds. No EGL-1::V5 protein was detectable in untreated alg-2(ok304);egl-1(syb1272 [egl-1::V5]) or mir-35-41(nDf50);egl-1(syb1272 [egl-1::V5]) germlines (Figure 3A). As expected, IR treatment resulted in a clear increase of EGL-1::V5 protein in late-pachytene germ cells and the gonad loop of the two mutant strains. However, unexpectedly, loss of alg-2 did not result in a noticeably elevated EGL-1::V5 protein induction when compared to IR-treated egl-1(syb1272 [egl-1::V5]) animals. mir-35-41 mutant worms also exhibited only a surprisingly mild increase of late-pachytene EGL-1::V5 signal compared to that of WT or alg-2 animals (Figure 3A). Therefore, our results suggest that the anticipated dysregulation of EGL-1 in alg-2 or mir-35-41 mutant worms was more subtle than initially expected, and thus likely not to be a major cause for the increased DNA damage-induced apoptosis in these two mutant strains.
Figure 3.
EGL-1 protein accumulates in late-pachytene germ cells upon IR treatment. (A) EGL-1::V5 expression was induced to comparable extents in germline loops of WT and alg-2(ok304) mutant animals, and was mildly elevated in mir-35-41(nDf50) worms 2.5 hr after IR exposure (90 Gy). Representative images were obtained from Z-stacks taken with a confocal microscope. Bars, 40 µm. (B) Close-up of late-pachytene germline zones revealed cytoplasmic EGL-1::V5 expression in response to IR-treatment (90 Gy). Representative images from a single focal plane. Bars, 40 µm. (C) Schematic representation of the clustered regularly interspaced short palindromic repeats/Cas9-generated egl-1(syb1272 [egl-1::V5]) knock-in. IR, ionizing radiation; WT, wild-type.
The protein deacetylase SIR-2.1 partially contributes to DNA damage-induced apoptosis in alg-2 mutant animals
Our immunofluorescence (IF) experiments with the EGL-1::V5 knock-in strains revealed that even in a WT genetic background, a considerable amount of late-pachytene germ cells exhibited cytoplasmic EGL-1::V5 induction upon IR exposure (Figure 3, A and B). Intriguingly, the number of EGL-1::V5-expressing pachytene germ cells therefore only partially correlated with the rather low levels of apoptosis scored in IR-treated WT germlines (Figure 1A). Therefore, we hypothesized that EGL-1 induction upon genotoxic stress alone may be insufficient to fully commit a cell to apoptotic death. Instead, EGL-1 expression might initially prime germ cells for apoptosis before additional signaling events are required to finalize cell death execution. Therefore, we wondered whether ALG-2/mir-35-42 might fine-tune apoptosis via alternative, EGL-1-independent pathways.
Upon DNA damage, the protein deacetylase SIR-2.1 translocates from the nuclei of dying pachytene cells into the cytoplasm, thereby briefly colocalizing with CED-4. This colocalization could be of functional importance, as SIR-2.1 might contribute to CED-4 activation, and thus trigger apoptosome formation and initiation of DNA damage-induced apoptosis independently of CEP-1/EGL-1 (Greiss et al. 2008). Accordingly, a sir-2.1 deletion fully eliminates IR-induced apoptosis in WT animals without affecting egl-1 transcription (Greiss et al. 2008). To test whether SIR-2.1 is indeed essential for the abnormal apoptotic response of alg-2-deficient worms to genotoxic stress, we first irradiated WT and alg-2 mutant worms with 90 Gy of IR, and dissected their gonads 2.5 hr afterward to stain germ cells with a SIR-2.1 antibody. As previously reported (Greiss et al. 2008), we found SIR-2.1 to be normally located in germ cell nuclei in both the alg-2 mutant as well as WT animals, and a notable number of DAPI stained WT late-pachytene germ cell nuclei lost their SIR-2.1 signal in response to IR (Figure S5A). RNAi-mediated knockdown of sir-2.1 nearly abolished the SIR-2.1 signal in WT germlines, thus confirming the specificity of our antibody staining (Figure S5C). IR treatment of alg-2 mutant animals resulted in the disappearance of the nuclear SIR-2.1 signal from almost the entire late-pachytene zone, leaving nothing more than an indistinct background signal in that particular region of the germline (Figure S5A). We wondered whether this dramatic loss of nuclear SIR-2.1 in IR-treated alg-2 germlines was actively contributing to cell death execution or was a passive consequence of nuclear breakdown downstream of caspase activation. Therefore, we crossed the alg-2(ok304) mutation into a ced-3(n717) loss-of function mutant background. The ced-3(n717) allele completely abrogates the activity of the C. elegans caspase CED-3 (Ellis and Horvitz 1986) and fully eliminated apoptosis in the alg-2 mutant animals (Figure 6C). Unexpectedly, loss of functional ced-3 prevented the cytoplasmic translocation of SIR-2.1 from late-pachytene germ cell nuclei in the alg-2 mutant animals (Figure S5A), suggesting that SIR-2.1 translocation in response to DNA damage occurs downstream of CED-3 initiation in this mutant. In addition, the sir-2.1(ok434) deletion, which suppressed DNA damage-induced apoptosis in WT animals (Greiss et al. 2008), only partially alleviated the elevated apoptosis phenotype of the alg-2 mutant worms (Figure S5B). The interpretation of this partial rescue was further complicated by a noticeably reduced germline size in alg-2;sir-2.1 double-mutant animals (data not shown). Collectively, our data suggest that SIR-2.1 might only partially contribute to DNA damage-induced apoptosis in alg-2 mutant worms.
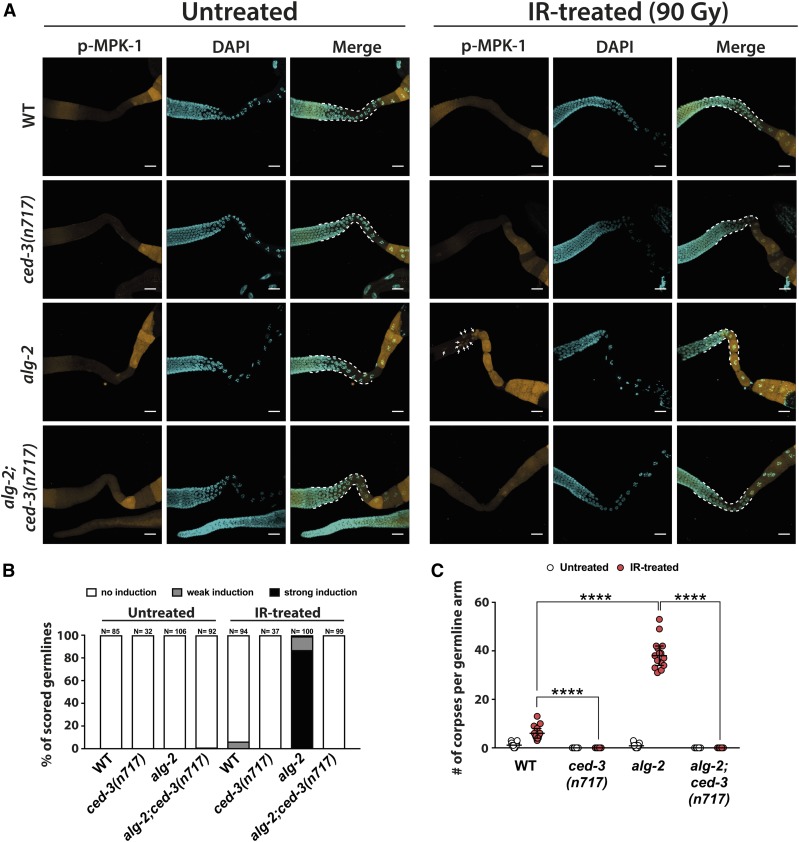
Figure 6.
MPK-1 phosphorylation in alg-2(ok304) germline loops occurs downstream of CED-3 activity. (A) IR-induced MPK-1 phosphorylation was completely abolished in gonad loops of alg-2(ok304);ced-3(n717) double mutants. Germlines were dissected 2.5 hr post-IR treatment (90 Gy). Representative images were obtained from Z-stacks taken with a confocal microscope, loop areas are highlighted by white dashed lines. Bars, 20 µm. (B) Quantification of WT, ced-3(n717), alg-2(ok304), and alg-2(ok304);ced-3(n717) mutant germlines displaying induced MPK-1 phosphorylation in the late-pachytene zone or the entire gonad loop at 2.5 hr post-IR. Phospho-MPK-1 signals were assessed at 10× and 40× magnification with a fluorescent microscope. Graph summarizes data from two independent experiments. Sample sizes are indicated. (C) ced-3(n717) completely abolished apoptosis in both WT and alg-2(ok304) mutant germlines. Apoptosis was scored 4 hr post-IR treatment (90 Gy). Representative graph from one out of three independent experiments, medians with 95% C.I.s are shown, a minimum of 15 germlines per genotype and condition was scored. **** P < 0.0001; Mann–Whitney nonparametric test. #, number; IR, ionizing radiation; WT, wild-type.
Apoptosis phenotype in alg-2 and mir-35-41 mutants is associated with elevated IR-inducible phosphorylation of the MPK-1/ERK1/2 MAPK in the gonad loop
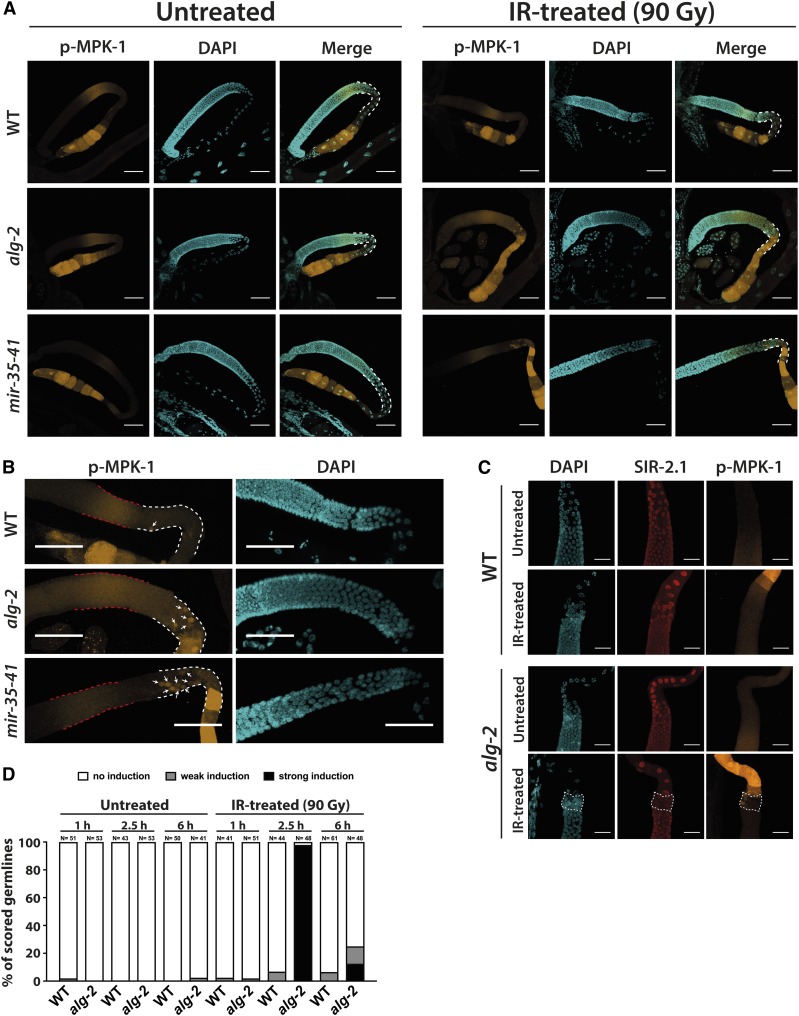
It was previously suggested that transient activation of ERK MAPK signaling in late-pachytene and early-diplotene germ cells located within the gonad loop is required for DNA damage-induced apoptosis in C. elegans (Rutkowski et al. 2011). We wondered whether the elevated germ cell apoptosis levels in IR-treated alg-2 or mir-35-41 deletion strains are accompanied by noticeably stronger activation of the C. elegans ERK1/2 homolog MPK-1 within the gonad loop. Hence, we performed IF staining to detect active, phosphorylated MPK-1 in dissected germlines after γ-radiation using a specific antibody that recognizes the conserved phosphorylation sites of the two mammalian ERK isoforms as well as of C. elegans MPK-1. As previously described, phosphorylated MPK-1 was predominantly detected in germ cells that are in the midpachytene stage of meiosis prophase I as well as in mature oocytes (Church et al. 1995; Lee et al. 2007). We confirmed that under untreated control conditions, the late-pachytene zone and the rest of the loop area of WT, alg-2, and mir-35-41 mutant germlines was nearly free of phosphorylated MPK-1 (Figure 4, A and B and Figure S6) (Church et al. 1995; Lee et al. 2007). Surprisingly, we did not detect a profound phospho-MPK-1 induction in WT germline loops 2.5 hr post-IR treatment. Only occasionally, confocal microscopy revealed that single WT late-pachytene germ cells exhibited MPK-1 activation upon DSB infliction. Importantly, these phospho-MPK-1-positive, late-pachytene cells were usually spatially separated from the phospho-MPK-1 signal in the midpachytene germline zone and the exhibited signal strength did not correlate with the intensity of the midpachytene signal (Figure 4, A and B and Figure S6). In both alg-2 and mir-35-41 mutant germlines, IR treatment resulted in unusually strong MPK-1 hyperphosphorylation throughout the entire germline loop, ranging from the late-pachytene zone until early diakinesis. In both mutant strains, MPK-1 phosphorylation occurred in individual late-pachytene cells and was clearly separate from the active MPK-1 within the midpachytene area. Accordingly, we also observed MPK-1 hyperphosphorylation in IR-exposed alg-2 or mir-35-41 mutant gonad loops, even when no phospho-MPK-1 signal was detectable in the midpachytene region (Figure 4, A and B and Figure S6). Costaining of alg-2 mutant germlines with DAPI as well as antibodies against SIR-2.1 and phospho-MPK-1 further confirmed that MPK-1 was not equally phosphorylated throughout the late-pachytene zone where dying germ cells lost their nuclear SIR-2.1 signal but was rather activated sporadically, in selected, individual cells (Figure 4C).
Figure 4.
ERK/MPK-1 is hyperactivated in alg-2 and mir-35-41 mutant germline loops upon IR treatment. (A) IR-treated alg-2(ok304) and mir-35-41(nDf50) mutant germlines exhibited strong MPK-1 phosphorylation 2.5 hr after IR (90 Gy) within the entire gonad loop area (highlighted by white dashed lines). (B) Close-up images of IR-treated WT, alg-2(ok304), and mir-35-41(nDf50) mutant pachytene zones suggested that the activation of MPK-1 in late-pachytene germ cells (white arrows) occurred independently of the phospho-MPK-1 signal in the midpachytene area (red dashed lines). The germline loop is highlighted by white dashed lines. (A and B) Germlines were dissected 2.5 hr post-IR (90 Gy). Representative images were obtained from Z-stacks taken with a confocal microscope. Bars, 40 µm. (C) IR-induced MPK-1 phosphorylation occurred in individual dying late-pachytene germ cells in the alg-2(ok304) mutant gonad. Germlines were dissected 2.5 hr post-IR (90 Gy) or after mock treatment, and were subsequently stained for DAPI, SIR-2.1, and phospho-MPK-1. Representative images were obtained from Z-stacks taken with a confocal microscope. Bars, 20 µm. (D) Quantification of WT and alg-2(ok304) mutant germlines displaying MPK-1 phosphorylation in the late-pachytene zone, or the entire gonad loop, at 1, 2.5, and 6 hr post-IR. Phospho-MPK-1 signals were assessed at 10× and 40× magnification with a fluorescent microscope. Sample sizes are indicated. IR, ionizing radiation; WT, wild-type.
We continued to characterize the role of MPK-1 phosphorylation in the context of apoptosis by focusing on the alg-2 mutant germline. First, we wanted to determine the timing and degree of IR-inducible MPK-1 activation. Therefore, we dissected WT and alg-2 mutant gonads at different time points after IR treatment, and scored the number of germlines according to the signal strength of the anti-phospho-MPK-1 antibody signal specifically inside the loop area. To do this, we established three distinct, qualitative categories of phospho-MPK-1 signal. The first group, referred to as “no induction,” included all germlines with varying degrees of midpachytene phospho-MPK-1 signal but no distinct and robust MPK-1 activation inside the late-pachytene zone, or the rest of the gonad loop. The second category, named “weak induction,” encompassed all germlines where fluorescence microscopy revealed at least a faint phospho-MPK-1 induction in the gonad loop or in single late-pachytene germ cells. Finally, germlines displaying MPK-1 hyperphosphorylation throughout the entire loop were summarized in the third group called “strong induction.” At 1 hr post-IR treatment, no change in MPK-1 activation was detectable in WT or alg-2 germlines (Figure 4D). After ∼2.5 hr, MPK-1 was robustly phosphorylated within the entire gonad loop of nearly all tested alg-2 mutant worms. Therefore, this IR-induced MPK-1 hyperphosphorylation coincided precisely with the onset of late-pachytene germ cell apoptosis in the IR-treated alg-2 mutant animals (Figure 4D). While a small fraction of the scored WT germlines exhibited clear MPK-1 phosphorylation in late-pachytene germ cells 2.5 hr post-IR, none of the IR-treated WT germlines exhibited MPK-1 hyperphosphorylation throughout the entire loop area similar to that of the alg-2 mutant. After 6 hr, MPK-1 was only rarely activated in both WT and alg-2 germline loops (Figure 4D).
Thus, we conclude that the occurrence of apoptosis in IR-treated alg-2(ok304) mutant germlines after 2.5 hr coincides with strong phosphorylation of the ERK MAPK MPK-1 in a fraction of dying late-pachytene cells as well as the rest of the gonad bend. Accordingly, the alg-2(ok304) germline can be exploited to provide further mechanistic insights into the role of Ras-ERK signaling during DNA damage-induced apoptosis.
MPK-1/ERK1/2 phosphorylation in the gonad loop is required for DNA damage-induced apoptosis in alg-2 mutant worms
Intrigued by the strong MPK-1 phosphorylation within the gonad bend of IR-treated alg-2(ok304) and mir-35-41(nDf50) mutant animals, we decided to exploit the alg-2(ok304) allele to better understand the mechanistic function of MPK-1 signaling during germ cell apoptosis. The study of ERK MAPK signaling in the germline is complicated due to the requirement of proper ERK signaling for the transition of germ cells through meiosis prophase I and oocyte maturation (Church et al. 1995; Lee et al. 2007). Accordingly, complete disruption of the ERK MAPK cascade will severely impair overall germline integrity and conceal possible effects of MPK-1 on germ cell apoptosis. To avoid this issue, we used weak loss-of-function alleles that only partially affect the ERK MAPK cascade. First, we crossed the hypomorph mek-2(n1989) allele into the alg-2(ok304) mutant. mek-2 encodes the homolog of the human MAPKK MEK that directly phosphorylates and thus activates MPK-1/ERK (Kornfeld et al. 1995; Sundaram 2013). Second, we utilized the mpk-1(ga111) temperature-sensitive allele, which results in a valine-to-glycine substitution within the MEK-2 MAPKK-binding site that prevents normal MPK-1 activation and leads to hermaphrodite sterility when kept at 25° (Lackner and Kim 1998). However, at the semipermissive temperature of 20°, mpk-1(ga111) mutant animals produce viable progeny and show no obvious mutant phenotypes. When irradiated with an IR intensity of 90 Gy, germ cell apoptosis levels of alg-2(ok304);mpk-1(ga111) and alg-2(ok304);mek-2(n1989) double mutants were significantly lower compared to those of alg-2 single mutants (Figure S7). In line with previous findings, the mild reduction of MPK-1 or MEK-2 activity at 20° had little effect on WT apoptosis levels (Rutkowski et al. 2011). However, we realized that at this IR dose, apoptosis numbers of both double mutants exhibited strong variability between experiments (Figure S7). Therefore, we decided to expose these weak ERK signaling mutants to a reduced IR-dose of only 30 Gy. Despite the use of such a comparatively low IR dose, we could still detect a prominent phospho-MPK-1 signal within the loop of alg-2 mutant germlines 2.5 hr after treatment (Figure 5, A and B). Both mpk-1(ga111) at 20° as well as mek-2(n1989) suppressed MPK-1 activation in the alg-2 mutant germline bend, while barely affecting overall germline integrity (Figure 5, A and B). In addition, 4 hr after exposure to 30 Gy of IR, both alleles significantly suppressed the severe apoptosis phenotype of the alg-2 mutant without variation between experiments (Figure 5C). Again, at 20° the weak effects of mpk-1(ga111) and mek-2(n1989) had no discernable consequences on IR-induced apoptosis levels in the WT background.
Figure 5.
MPK-1/ERK is hyperphosphorylated and required for elevated DNA damage-induced apoptosis in alg-2(ok304) mutant animals. (A) The mild ERK signaling loss-of-function alleles mpk-1(ga111) and mek-2(n1989) prevented MPK-1 phosphorylation in IR-treated (30 Gy) alg-2(ok304) mutant gonad loops. Germlines were dissected 2.5 hr post-treatment. Representative images were obtained from Z-stacks taken with a confocal microscope, germline loop regions are highlighted by dashed lines. Bars, 20 µm. (B) Quantification of WT, alg-2(ok304), alg-2(ok304);mpk-1(ga111), and alg-2(ok304);mek-2(n1989) mutant germlines displaying induced MPK-1 phosphorylation in the late-pachytene zone or the entire gonad loop at 2.5 hr post-IR. Phospho-MPK-1 signals were assessed at 10× and 40× magnification with a fluorescent microscope. Sample sizes are indicated. (C) The mpk-1(ga111) and mek-2(n1989) alleles alleviated DNA damage-induced germ cell apoptosis in alg-2(ok304) mutant animals. Apoptosis was scored 4 hr post-IR treatment (30 Gy). Representative graph from one out of three independent experiments, medians with 95% C.I.s are shown, a minimum of 15 germlines per genotype and condition were scored. **** P < 0.0001; Mann–Whitney nonparametric test. #, number; IR, ionizing radiation; WT, wild-type.
Taken together, these findings support earlier reports that active MPK-1 in the gonad bend is required to trigger DNA damage-induced apoptosis.
MPK-1 phosphorylation in response to IR occurs downstream of the core apoptotic machinery
It was previously suggested that MPK-1 activation in response to IR regulates the induction of apoptosis by impacting CEP-1/p53 expression and activity (Rutkowski et al. 2011). We wondered whether, conversely, MPK-1 activity could also be subject to regulation via CEP-1. To our surprise, deletion of the cep-1 gene in alg-2(ok304);cep-1(gk138) double mutants not only rescued the apoptosis phenotype of the alg-2(ok304) single mutant (Figure 2A), but also completely abolished MPK-1 phosphorylation in the gonad loop. In fact, loss of cep-1 appeared to diminish MPK-1 phosphorylation throughout the germline (Figure S8, A and B). This finding indicated a proapoptotic function of MPK-1 downstream of CEP-1/p53. More intriguingly, the ced-9(n1950) gain-of-function allele of the antiapoptotic Bcl-2 homolog CED-9, which completely abolished IR-induced germ cell apoptosis in both WT and alg-2 mutant animals, also blocked MPK-1 activation in the gonad bend (Figure S9), suggesting a role of MPK-1 even further downstream in the apoptosis cascade. Finally, we assessed MPK-1 activation in alg-2 mutant animals that lacked the functional C. elegans caspase ced-3. alg-2(ok304);ced-3(n717) double mutants completely failed to induce apoptosis in the germline (Figure 6C) and did not display phosphorylated MPK-1 in the gonad bend in response to γ-radiation (Figure 6, A and B). This finding indicates that MPK-1 promotes apoptosis downstream of the core apoptotic machinery. To understand whether phosphorylation of MPK-1 is directly linked to CED-3 enzymatic activity, we crossed alg-2(ok304) with two previously described weak ced-3 loss-of-function alleles to partially block or delay the onset of apoptosis execution, while possibly still allowing for MPK-1 activation. ced-3(op149) has been shown to suppress and/or delay the onset of embryonic cell death by on average 75% (Hoeppner et al. 2001). The ced-3(n2438) allele appears to affect CED-3 multidimerization by altering a glycine residue located on the CED-3 heterodimer–heterodimer interface, also leading to delayed and reduced embryonic apoptosis (Shaham et al. 1999; Hoeppner et al. 2001). When scoring IR-induced germ cell apoptosis in alg-2(ok304);ced-3(op149) and alg-2(ok304);ced-3(n2438) double mutants, we observed that the germline of the former did not display any cell death 4 hr after IR treatment with 90 Gy, whereas the latter exhibited apoptotic levels similar to a cep-1 loss-of-function mutant (Figure S10C). These results suggest that cell death signaling in the germline is regulated more stringently than in embryonic somatic cells, since mutant ced-3 alleles that are considered weak during embryonic cell death were sufficient to nearly abolish apoptosis in the germline. As expected, both alleles completely prevented MPK-1 phosphorylation in alg-2 mutant germline loops upon IR (Figure S10, A and B). Taken together, these data demonstrate that MPK-1 is phosphorylated downstream of CED-3 to promote DNA damage-induced cell death in the alg-2 mutant germline.
IR-induced MPK-1 phosphorylation is associated with apoptotic cell corpse engulfment
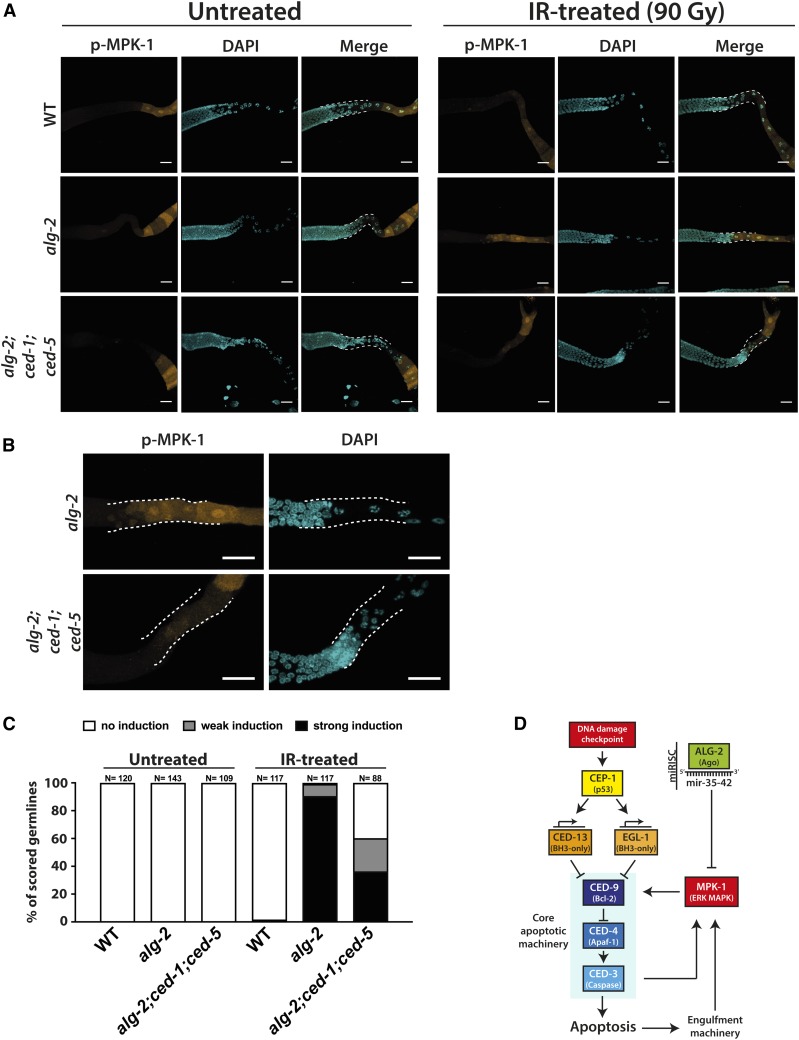
In the C. elegans germline, apoptotic corpses are engulfed and removed via two distinct, yet partially overlapping, pathways. The first pathway involves the conserved mEGF10 gene ced-1, the GULP homolog ced-6, and the ATP-binding cassette transporter gene ced-7. The second pathway depends on the CrkII adaptor homolog CED-2, Dock180-like CED-5, the Rac GTPase ortholog CED-10, and the engulfment and cell motility protein CED-12 (Hedgecock et al. 1983; Ellis et al. 1991; Liu and Hengartner 1998; Wu and Horvitz 1998a,b; Albert et al. 2000; Reddien and Horvitz 2000, 2004; Gumienny et al. 2001; Zhou et al. 2001; Conradt and Xue 2005). Individually, deletion of all of these genes leads to increased levels of persistent cell corpses in the germline. By combining loss-of-function mutations from both pathways, phagocytosis can be even further compromised (Reddien and Horvitz 2004). Previous reports have provided evidence that apoptotic cell engulfment does not only serve to remove dead cells but may also have a function during the actual killing process itself (Hoeppner et al. 2001; Reddien et al. 2001; Johnsen and Horvitz 2016). To address the possibility that the engulfment machinery could contribute to the execution of apoptosis by initiating the Ras-ERK signaling cascade downstream of CED-3 in dying cells, we introduced the ced-1(e1735) and ced-5(n1812) mutant alleles into the alg-2(ok304) mutant background to simultaneously impair both engulfment pathways. We then dissected germlines of untreated and IR-treated alg-2(ok304);ced-1(e1735);ced-5(n1812) triple mutants, and performed IF staining to test for MPK-1 phosphorylation. Indeed, despite a considerable number of accumulating late-pachytene germ cells in the gonad loop, > 60% of alg-2(ok304);ced-1(e1735);ced-5(n1812) triple-mutant germlines displayed no or only weak phospho-MPK-1 induction throughout the gonad bend (Figure 7, A–C).
Figure 7.
MPK-1 phosphorylation in alg-2(ok304) germline loops occurs downstream of the apoptotic corpse engulfment machinery. (A) IR-induced MPK-1 phosphorylation was partially suppressed in gonad loops of alg-2(ok304);ced-1(e1735);ced-5(n1812) triple-mutant animals. Germlines were dissected 2.5 hr post-IR treatment (90 Gy). Representative images were obtained from Z-stacks taken with a confocal microscope, loop areas are highlighted by white dashed lines. Bars, 20 µm. (B) Close-up images of the IR-treated alg-2 and alg-2;ced-1;ced-5 mutant germline loops (highlighted by white dashed lines) shown in (A). Loss of functional engulfment machinery strongly impaired the activation of MPK-1 in alg-2 mutant germline loops upon IR exposure. The given alg-2;ced-1;ced-5 example demonstrates what was considered a weak phospho-MPK-1 induction. Bars, 20 µm. (C) Quantification of WT, alg-2(ok304), and alg-2(ok304);ced-1(e1735);ced-5(n1812) mutant germlines displaying induced MPK-1 phosphorylation in the late-pachytene zone or the entire gonad loop at 2.5 hr post-IR. Phospho-MPK-1 signals were assessed at 10× and 40× magnification with a fluorescent microscope. Graph summarizes data from two independent experiments. Sample sizes are indicated. (D) Proposed model of ALG-2/mir-35-42-regultated apoptosis upon DNA damage detection. IR, ionizing radiation; WT, wild-type.
Next, we crossed the alg-2(ok204) allele into a CED-1::GFP reporter strain. ced-1 encodes a transmembrane protein expressed on the surface of phagocytizing sheath cells that surround and engulf the dying cell (Hedgecock et al. 1983; Zhou et al. 2001). Thus, the CED-1::GFP transgene indirectly labels dying cells by predominantly marking the engulfing sheath cell during earlier stages of the apoptosis process. We exploited this fact as a means to elaborate on a possible relationship between ERK/MPK-1 phosphorylation and the engulfment machinery in the alg-2(ok304) mutant background upon IR. At 2.5 hr post-IR treatment, we detected numerous late-pachytene germ cells in the alg-2 mutant that were either partially or fully enclosed by a CED-1::GFP-labeled membrane (Figure S11). Unexpectedly, only a small fraction of CED-1::GFP-labeled pachytene cells exhibited strong MPK-1 phosphorylation. In contrast, the majority of the cells that were fully circularized by CED-1::GFP showed either weak MPK-1 induction or were completely free of phosphorylated MPK-1 signal, indicating that once successfully engulfed, the dying cell no longer requires active MPK-1. In line with this interpretation, 4 hr post-IR, when late apoptotic corpses are accumulating at the proximal end of the alg-2 mutant germline and have mostly lost the CED-1::GFP signal, phosphorylated MPK-1 was evidently no longer detectable (Figure S12).
Taken together, these results indicate that ALG-2/mir-35-42 regulate MPK-1 activity. MPK-1 phosphorylation is triggered upon initial DNA damage-induced apoptosome and engulfment machinery activation, and in turn is required to fully commit damaged germ cells to undergo apoptotic elimination.
Discussion
The AGO protein ALG-2 regulates DNA damage-induced apoptosis in the C. elegans germline via the mir-35 family
In this study, we report that disruption of the miRNA AGO gene alg-2 leads to highly elevated levels of DNA damage-induced germ cell apoptosis. We could demonstrate that induction of apoptosis in the alg-2(ok304) mutant strain occurred very rapidly after IR and affected the majority of late-pachytene germ cells. The increased apoptosis in alg-2(ok304) mutant animals was unexpectedly not associated with elevated transcription or protein levels of the direct CEP-1/p53 target EGL-1. Nevertheless, numbers of apoptotic cells were restored to baseline once cep-1 or egl-1 gene functions were disrupted. We subsequently showed by miRNA sequencing that in alg-2 mutant animals, levels of most mir-35 family members were sharply reduced, which is consistent with previous findings (Brown et al. 2017). Accordingly, the alg-2 mutant apoptosis phenotype could be reproduced in mir-35-42 family mutants. The increased apoptotic corpse numbers in IR-treated mir-35-41 mutant animals partly correlated with elevated egl-1 mRNA levels, consistent with previously published studies that reported the mir-35-42 miRNA family as able to fine-tune the regulation of apoptosis by targeting egl-1 transcripts (Sherrard et al. 2017; Tran et al. 2019). miRNA-mediated apoptosis regulation at the level of BH3-only proteins homologous to EGL-1 has been documented in mammalian cells [Su et al. 2009; Kole et al. 2011; reviewed in Sionov et al. (2015)]. Thus, our findings provide further evidence for an evolutionarily conserved mechanism where miRNAs fine-tune the apoptosis process in the presence of genotoxic stress.
ALG-2/mir-35-42 promote apoptosis execution by regulating CEP-1/EGL-1-independent pathways
The use of a CRISPR/Cas9-generated EGL-1::V5 knock-in strain allowed us to visualize, for the first time, the IR-induced expression of endogenous EGL-1 protein in late-pachytene germ cells; however, loss of neither alg-2 nor mir-35-41 robustly altered the detected EGL-1::V5 protein signal to a degree that would have been expected based on the published observations using an egl-1 3′-UTR reporter, and egl-1 mRNA in situ hybridizations (Sherrard et al. 2017; Tran et al. 2019). In addition, we were surprised to detect a considerable amount of EGL-1::V5-positive germ cells in IR-treated WT animals. These findings provide new evidence for an important, yet poorly understood concept: CEP-1-mediated induction of EGL-1 alone might be insufficient to fully activate the C. elegans core apoptotic machinery upon genotoxic stress and may merely prime pachytene germ cells for apoptosis. Consequentially, additional signaling pathways are required to impinge on the regulation of apoptosis to fully commit a cell to programmed death. This idea was first proposed in a study that provided evidence for a potential, CEP-1-independent role of SIR-2.1 during CED-4 activation and was further reinforced by evidence that the canonical model of the apoptosome in C. elegans is in fact inconsistent with cytological localization data (Greiss et al. 2008; Pourkarimi et al. 2012). Indeed, several publications have reported factors that appear to promote DNA damage-induced apoptosis downstream of EGL-1 (Reddien et al. 2007; Schertel and Conradt 2007; Greiss et al. 2008; Eberhard et al. 2013).
Are ALG-2 and the mir-35 family involved in the fine-tuning of some of these regulatory pathways? Our own data suggest that sir-2.1 is not essential for the increased germ cell apoptosis in IR-treated alg-2 mutant worms. Instead, loss of both alg-2 and mir-35 severely dysregulated ERK/MPK-1 MAPK signaling in the germline loops of IR-treated animals, and genetic impairment of MPK-1 activity nearly rescued the abnormal IR-induced apoptosis phenotype of alg-2 mutant worms. Mammalian MAPK signaling has long been implicated in the regulation of apoptosis in response to genotoxic stress by modulating the expression and activity of various members of the core apoptotic machinery (Verheij et al. 1998; Gjerset et al. 1999; Lei et al. 2002; Tang et al. 2002; Lei and Davis 2003; Viktorsson et al. 2003; Choi et al. 2006; Kim et al. 2008; Liu et al. 2008). In C. elegans, MPK-1 might promote DNA damage-induced apoptosis through various mechanisms. It was previously suggested that MPK-1 may impinge on the activity of the C. elegans p53 homolog CEP-1 (Rutkowski et al. 2011). MPK-1 might also influence CEP-1 expression in late-pachytene germ cells via a mechanism that remains unclear but might, to some extent, involve the cep-1 translational repressor GLD-1 (Schumacher et al. 2005a; Rutkowski et al. 2011). Alternatively, MPK-1 has been suggested to regulate IR-induced apoptosis downstream of CEP-1, most likely at the level of CED-9 (Eberhard et al. 2013).
Our own IF experiments revealed that MPK-1 becomes selectively phosphorylated in individual late-pachytene germ cells of IR-treated animals (Figure 4, A and B). This punctual MPK-1 activation upon genotoxic stress was mostly independent of the position or intensity of the midpachytene phospho-MPK-1 signal. Therefore, we define DNA damage-dependent MPK-1 activation as a qualitative trait that is primarily characterized by the specific stage of a germ cell.
How could the mir-35 family affect MPK-1 activity upon DNA damage? A recent study proposed the nucleoside-diphosphate kinase NDK-1 as a possible mir-35 target required for DNA damage-induced apoptosis (Tran et al. 2019). Indeed, NDK-1 was previously found to modulate MPK-1 activation downstream of the ERK MAPK kinase kinase LIN-45/Raf (Masoudi et al. 2013). Taking these and prior studies (Rutkowski et al. 2011; Eberhard et al. 2013) into consideration, we therefore suggest ERK/MPK-1 MAPK signaling to be an essential proapoptotic pathway that might promote DNA damage-induced apoptosis in parallel to or, more likely, downstream of EGL-1.
Ras-ERK MAPK signaling promotes DNA damage-induced apoptosis downstream of the core apoptotic machinery
Previous work demonstrated that MPK-1 hyperactivation due to elevated LET-60/Ras activity or loss of the MPK-1 phosphatase LIP-1 sensitizes C. elegans for DNA damage-induced apoptosis. At a semipermissive temperature, the mpk-1(ga111) allele is sufficient to restore normal apoptosis levels in these genetic backgrounds, while not significantly affecting apoptosis signaling in the WT worms (Rutkowski et al. 2011); however, mutations in the genes encoding LET-60 or LIP-1 directly affect the MPK-1 MAPK signaling pathway in C. elegans, thus are not suitable for the study of the mechanistic processes that lead to MPK-1 activation in consequence to DNA damage. Therefore, the alg-2(ok304) mutant strain provides us with a valuable tool to elaborate on this issue. Our genetic data clearly showed that activation of MPK-1 in response to IR occurred downstream of the caspase CED-3. Several studies published in the 1990s reported that executioner caspases, such as caspase-3 or caspase-7, could cleave and activate the p38/JNK MAPKKK MEKK-1 in vitro, also upon infliction of genotoxic stress. In turn, the obtained cleavage product reinforced caspase activity within the dying cell, possibly via a positive feedback loop. However, this feedback loop seemingly did not involve the activation of terminal p38 or JNK MAPKs (Cardone et al. 1997; Widmann et al. 1998a,b). Two other studies provided biochemical data showing that apoptosis of endothelial cells is associated with phosphorylation of p38 downstream of caspase-3 (Nakagami et al. 2001; Lee and Lo 2003). Similarly, the ERK MAPKKK Raf-1 was reported to be a caspase substrate during apoptosis execution in hematopoietic cells. caspase-9-mediated Raf-1 cleavage at position D279 resulted in the generation of a C-terminal fragment with increased kinase activity that could stimulate ERK hyperphosphorylation in vitro and promoted a negative feedback loop to prevent cell death (Cornelis et al. 2005). It would be interesting to test whether the C. elegans caspase CED-3 also directly cleaves and activates members of the ERK/MPK-1 cascade to initiate MAPK signaling.
Our IF experiments further suggest an intricate relationship between the phosphorylation of MPK-1 and the engulfment machinery downstream of the initiation of apoptosis: IR-induced MPK-1 phosphorylation in alg-2 mutant germ cells was noticeably impaired in an engulfment-defective genetic background, while complete envelopment by an engulfing membrane tended to quench MPK-1 phosphorylation in most apoptotic cells. The apoptosis-promoting activity of the engulfment machinery is well documented during C. elegans embryonic development; particularly under conditions of reduced and/or delayed CED-3 caspase activity, abrogation of the engulfment machinery further promotes the survival of embryonic cells normally destined to undergo apoptosis (Hoeppner et al. 2001; Reddien et al. 2001). In several instances, loss of functional engulfment machinery has even allowed the recovery of early-stage apoptotic corpses into morphologically healthy, living cells during development (Hoeppner et al. 2001). A more recent study coined the term of “assisted suicide” to describe the requirement for neighboring engulfing cells to facilitate the apoptotic death of B.al/rapaav embryonic cells that resist caspase-induced apoptotic signaling (Johnsen and Horvitz 2016). It has been suggested that in embryos, the engulfment machinery promotes the polar localization of apoptotic key players in mother cells, which results in asymmetric cell division and the generation of daughter cells that undergo apoptosis (Chakraborty et al. 2015; Mishra et al. 2018). In the C. elegans germline, engulfment of apoptotic corpses is mediated by somatic gonad sheath cells (Gumienny et al. 1999; Gartner et al. 2008). Sheath cells can promote physiological germline apoptosis noncell autonomously by stimulating the Ephrin receptor VAB-1 on the surface of the cell destined to die (Li et al. 2012). Therefore, there is also a possibility that the somatic gonad might promote DNA damage-induced germ cell apoptosis upon activation of the engulfment machinery via receptor-mediated ERK/MPK-1 activation in the dying cell. At least during some cases of C. elegans embryonic cell death, the EGF-like ligand LIN-3 binds to and stimulates the EGF receptor LET-23, which in turn triggers the LET-60/Ras-MPK-1/ERK pathway to promote apoptosis (Jiang and Wu 2014). Additional genetic studies might thus further elucidate the precise mode of MPK-1 activation downstream of CED-3 in IR-treated worms.
Once activated upon IR, how does MPK-1 promote apoptosis? Considering the aforementioned in vitro data, we favor the possibility of a positive feedback loop that reinforces an initial apoptotic stimulus. Dying cells might initiate ERK/MPK-1 MAPK signaling at some point early on during the killing process, either as a direct result of CED-3-mediated cleavage or upon activation of the engulfment machinery by phagocytizing sheath cells. MPK-1 might in turn be required to evoke full apoptosome activity and ultimately apoptotic cell death, possibly via direct stimulation of the core apoptotic machinery (Figure 7D). In mammals, ERK1/2 MAPK signaling is indeed capable of directly phosphorylating core apoptotic factors and thereby of modulating apoptosis rates. For instance, the mammalian CED-9 ortholog Bcl-2 can be subject to regulation by phosphorylation (Chang et al. 1997) and binds Raf-1 MAPK kinase kinase at the surface of mitochondria (Wang et al. 1996). Bcl-2 phosphorylation by ERK1/2 at Ser87 abrogates its antiapoptotic function (Tamura et al. 2004). The closest human ortholog of CED-3 represents caspase-9, the activity of which can be blocked by ERK-mediated phosphorylation at Thr125 (Allan et al. 2003; Martin et al. 2008). Future biochemical studies might help to verify similar ERK target sites in C. elegans core apoptotic factors that could support our hypothesis of an MPK-1-mediated apoptosis-enhancing feedback loop downstream of caspase signaling and the engulfment machinery. Alternatively, MPK-1 might also stimulate many processes downstream of the core apoptotic machinery that contribute to the apoptotic demise of a genomically compromised cell. For instance, a recent study reported that the engulfment machinery promotes a second phase of cellular shrinkage during physiological germ cell apoptosis that involves both nucleoplasmic and cytoplasmic compaction (Raiders et al. 2018). It would be interesting to investigate whether MPK-1 activation might be required for the execution of this process.
Future work with the alg-2 mutant might help to further delineate the regulatory mechanisms of DNA damage-induced apoptosis in C. elegans. Given the involvement of MAPK signaling in the DNA damage response and apoptosis in higher organisms, it is conceivable that those mechanisms might play important roles in the apoptotic response during DNA damage-inflicting chemo- and radiation therapy.
Acknowledgments
We thank the Cologne Excellence Cluster for Cellular Stress Responses in Ageing-Associated Diseases (CECAD) imaging and bioinformatics facilities, and the Cologne Center for Genomics. Worm strains were provided by the National Bioresource Project (supported by The Ministry of Education, Culture, Sports, Science and Technology, Japan), the Caenorhabditis Genetics Center (funded by the National Institutes of Health National Center for Research Resources, US), and the C. elegans Gene Knockout Project at the Oklahoma Medical Research Foundation (part of the International C. elegans Gene Knockout Consortium). M.A.D. received a fellowship from the Cologne Graduate School of Ageing Research. N.S. received a fellowship from the Cologne Cardiovascular Research Center. B.S. acknowledges funding from the Deutsche Forschungsgemeinschaft (SCHU 2494/3-1, SCHU 2494/7-1, CECAD, SFB 829, SFB 670, KFO 286, KFO 329, and GRK2407), the Deutsche Krebshilfe (70112899), and the European Cooperation in Science and Technology (COST) action (BM1408). The authors declare no competing interests.
Author contributions: M.A.D. performed all experiments and analyzed the data. N.S. assisted with experiments and data analysis. B.S. coordinated the project and, together with M.A.D., designed the study and wrote the paper.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8852180.
Communicating editor: V. Reinke
Literature Cited
- Aalto A. P., Nicastro I. A., Broughton J. P., Chipman L. B., Schreiner W. P., et al. , 2018. Opposing roles of microRNA Argonautes during Caenorhabditis elegans aging. PLoS Genet. 14: e1007379 10.1371/journal.pgen.1007379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M. L., Kim J. I., Birge R. B., 2000. Alphavbeta5 integrin recruits the CrkII-Dock180-rac1 complex for phagocytosis of apoptotic cells. Nat. Cell Biol. 2: 899–905. 10.1038/35046549 [DOI] [PubMed] [Google Scholar]
- Allan L. A., Morrice N., Brady S., Magee G., Pathak S., et al. , 2003. Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat. Cell Biol. 5: 647–654. 10.1038/ncb1005 [DOI] [PubMed] [Google Scholar]
- Alpi A., Pasierbek P., Gartner A., Loidl J., 2003. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112: 6–16. 10.1007/s00412-003-0237-5 [DOI] [PubMed] [Google Scholar]
- Alvarez-Saavedra E., Horvitz H. R., 2010. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 20: 367–373. 10.1016/j.cub.2009.12.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly A., Gartner A., 2013. Germ cell apoptosis and DNA damage responses. Adv. Exp. Med. Biol. 757: 249–276. 10.1007/978-1-4614-4015-4_9 [DOI] [PubMed] [Google Scholar]
- Bartel D. P., 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K. C., Svendsen J. M., Tucci R. M., Montgomery B. E., Montgomery T. A., 2017. ALG-5 is a miRNA-associated Argonaute required for proper developmental timing in the Caenorhabditis elegans germline. Nucleic Acids Res. 45: 9093–9107. 10.1093/nar/gkx536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari S. I., Vasquez-Rifo A., Gagne D., Paquet E. R., Zetka M., et al. , 2012. The microRNA pathway controls germ cell proliferation and differentiation in C. elegans. Cell Res. 22: 1034–1045. 10.1038/cr.2012.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone M. H., Salvesen G. S., Widmann C., Johnson G., Frisch S. M., 1997. The regulation of anoikis: MEKK-1 activation requires cleavage by caspases. Cell 90: 315–323. 10.1016/S0092-8674(00)80339-6 [DOI] [PubMed] [Google Scholar]
- Chakraborty S., Lambie E. J., Bindu S., Mikeladze-Dvali T., Conradt B., 2015. Engulfment pathways promote programmed cell death by enhancing the unequal segregation of apoptotic potential. Nat. Commun. 6: 10126 10.1038/ncomms10126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B. S., Minn A. J., Muchmore S. W., Fesik S. W., Thompson C. B., 1997. Identification of a novel regulatory domain in Bcl-X(L) and Bcl-2. EMBO J. 16: 968–977. 10.1093/emboj/16.5.968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. Y., Kim M. J., Kang C. M., Bae S., Cho C. K., et al. , 2006. Activation of Bak and Bax through c-abl-protein kinase Cdelta-p38 MAPK signaling in response to ionizing radiation in human non-small cell lung cancer cells. J. Biol. Chem. 281: 7049–7059. 10.1074/jbc.M512000200 [DOI] [PubMed] [Google Scholar]
- Church D. L., Guan K. L., Lambie E. J., 1995. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development 121: 2525–2535. [DOI] [PubMed] [Google Scholar]
- Cimmino A., Calin G. A., Fabbri M., Iorio M. V., Ferracin M., et al. , 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 102: 13944–13949 [corrigenda: Proc. Natl. Acad. Sci. USA 103: 2464 (2006)]. 10.1073/pnas.0506654102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clejan I., Boerckel J., Ahmed S., 2006. Developmental modulation of nonhomologous end joining in Caenorhabditis elegans. Genetics 173: 1301–1317. 10.1534/genetics.106.058628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt B., Horvitz H. R., 1998. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 93: 519–529. 10.1016/S0092-8674(00)81182-4 [DOI] [PubMed] [Google Scholar]
- Conradt B., Xue D., 2005. Programmed cell death (October 06, 2005), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.32.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis S., Bruynooghe Y., Van Loo G., Saelens X., Vandenabeele P., et al. , 2005. Apoptosis of hematopoietic cells induced by growth factor withdrawal is associated with caspase-9 mediated cleavage of Raf-1. Oncogene 24: 1552–1562. 10.1038/sj.onc.1208401 [DOI] [PubMed] [Google Scholar]
- del Peso L., Gonzalez V. M., Nunez G., 1998. Caenorhabditis elegans EGL-1 disrupts the interaction of CED-9 with CED-4 and promotes CED-3 activation. J. Biol. Chem. 273: 33495–33500. 10.1074/jbc.273.50.33495 [DOI] [PubMed] [Google Scholar]
- Derry W. B., Putzke A. P., Rothman J. H., 2001. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science 294: 591–595. 10.1126/science.1065486 [DOI] [PubMed] [Google Scholar]
- Eberhard R., Stergiou L., Hofmann E. R., Hofmann J., Haenni S., et al. , 2013. Ribosome synthesis and MAPK activity modulate ionizing radiation-induced germ cell apoptosis in Caenorhabditis elegans. PLoS Genet. 9: e1003943 10.1371/journal.pgen.1003943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis H. M., Horvitz H. R., 1986. Genetic control of programmed cell death in the nematode C. elegans. Cell 44: 817–829. 10.1016/0092-8674(86)90004-8 [DOI] [PubMed] [Google Scholar]
- Ellis R. E., Jacobson D. M., Horvitz H. R., 1991. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics 129: 79–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolaeva M. A., Segref A., Dakhovnik A., Ou H. L., Schneider J. I., et al. , 2013. DNA damage in germ cells induces an innate immune response that triggers systemic stress resistance. Nature 501: 416–420. 10.1038/nature12452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R. C., Farh K. K., Burge C. B., Bartel D. P., 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19: 92–105. 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A., Milstein S., Ahmed S., Hodgkin J., Hengartner M. O., 2000. A conserved checkpoint pathway mediates DNA damage–induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5: 435–443. 10.1016/S1097-2765(00)80438-4 [DOI] [PubMed] [Google Scholar]
- Gartner A., MacQueen A. J., Villeneuve A. M., 2004. Methods for analyzing checkpoint responses in Caenorhabditis elegans. Methods Mol. Biol. 280: 257–274. [DOI] [PubMed] [Google Scholar]
- Gartner A., Boag P. R., Blackwell T. K., 2008. Germline survival and apoptosis (September 4, 2008), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.145.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjerset R. A., Lebedeva S., Haghighi A., Turla S. T., Mercola D., 1999. Inhibition of the Jun kinase pathway blocks DNA repair, enhances p53-mediated apoptosis and promotes gene amplification. Cell Growth Differ. 10: 545–554. [PubMed] [Google Scholar]
- Greiss S., Hall J., Ahmed S., Gartner A., 2008. C. elegans SIR-2.1 translocation is linked to a proapoptotic pathway parallel to cep-1/p53 during DNA damage-induced apoptosis. Genes Dev. 22: 2831–2842. 10.1101/gad.482608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A., Pasquinelli A. E., Conte D., Li N., Parrish S., et al. , 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. 10.1016/S0092-8674(01)00431-7 [DOI] [PubMed] [Google Scholar]
- Gumienny T. L., Lambie E., Hartwieg E., Horvitz H. R., Hengartner M. O., 1999. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126: 1011–1022. [DOI] [PubMed] [Google Scholar]
- Gumienny T. L., Brugnera E., Tosello-Trampont A. C., Kinchen J. M., Haney L. B., et al. , 2001. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107: 27–41. 10.1016/S0092-8674(01)00520-7 [DOI] [PubMed] [Google Scholar]
- Ha M., Kim V. N., 2014. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15: 509–524. 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- Hedgecock E. M., Sulston J. E., Thomson J. N., 1983. Mutations affecting programmed cell deaths in the nematode Caenorhabditis elegans. Science 220: 1277–1279. 10.1126/science.6857247 [DOI] [PubMed] [Google Scholar]
- Hoeppner D. J., Hengartner M. O., Schnabel R., 2001. Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature 412: 202–206. 10.1038/35084103 [DOI] [PubMed] [Google Scholar]
- Hofmann E. R., Milstein S., Boulton S. J., Ye M., Hofmann J. J., et al. , 2002. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr. Biol. 12: 1908–1918. 10.1016/S0960-9822(02)01262-9 [DOI] [PubMed] [Google Scholar]
- Jiang H. S., Wu Y. C., 2014. LIN-3/EGF promotes the programmed cell death of specific cells in Caenorhabditis elegans by transcriptional activation of the pro-apoptotic gene egl-1. PLoS Genet. 10: e1004513 10.1371/journal.pgen.1004513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen H. L., Horvitz H. R., 2016. Both the apoptotic suicide pathway and phagocytosis are required for a programmed cell death in Caenorhabditis elegans. BMC Biol. 14: 39 10.1186/s12915-016-0262-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N. M., Lemmens B. B., Tijsterman M., 2013. A role for the malignant brain tumour (MBT) domain protein LIN-61 in DNA double-strand break repair by homologous recombination. PLoS Genet. 9: e1003339 10.1371/journal.pgen.1003339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Paranjape T., Müller R. U., Nallur S., Gillespie E., et al. , 2009. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene 28: 2419–2424 (erratum: Oncogene 28: 3008). 10.1038/onc.2009.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. J., Lee K. H., Lee S. J., 2008. Ionizing radiation utilizes c-Jun N-terminal kinase for amplification of mitochondrial apoptotic cell death in human cervical cancer cells. FEBS J. 275: 2096–2108. 10.1111/j.1742-4658.2008.06363.x [DOI] [PubMed] [Google Scholar]
- Kole A. J., Swahari V., Hammond S. M., Deshmukh M., 2011. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 25: 125–130. 10.1101/gad.1975411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld K., Guan K. L., Horvitz H. R., 1995. The Caenorhabditis elegans gene mek-2 is required for vulval induction and encodes a protein similar to the protein kinase MEK. Genes Dev. 9: 756–768. 10.1101/gad.9.6.756 [DOI] [PubMed] [Google Scholar]
- Kumar M., Lu Z., Takwi A. A., Chen W., Callander N. S., et al. , 2011. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene 30: 843–853. 10.1038/onc.2010.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J. E., Kim B. Y., Kwak S. Y., Bae I. H., Han Y. H., 2013. Ionizing radiation-inducible microRNA miR-193a-3p induces apoptosis by directly targeting Mcl-1. Apoptosis 18: 896–909. 10.1007/s10495-013-0841-7 [DOI] [PubMed] [Google Scholar]
- Lackner M. R., Kim S. K., 1998. Genetic analysis of the Caenorhabditis elegans MAP kinase gene mpk-1. Genetics 150: 103–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner M. R., Kornfeld K., Miller L. M., Horvitz H. R., Kim S. K., 1994. A MAP kinase homolog, mpk-1, is involved in ras-mediated induction of vulval cell fates in Caenorhabditis elegans. Genes Dev. 8: 160–173. 10.1101/gad.8.2.160 [DOI] [PubMed] [Google Scholar]
- L’Allemain G., 1994. Deciphering the MAP kinase pathway. Prog. Growth Factor Res. 5: 291–334. 10.1016/0955-2235(94)90011-6 [DOI] [PubMed] [Google Scholar]
- Lee M. H., Ohmachi M., Arur S., Nayak S., Francis R., et al. , 2007. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics 177: 2039–2062. 10.1534/genetics.107.081356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. R., Lo E. H., 2003. Interactions between p38 mitogen-activated protein kinase and caspase-3 in cerebral endothelial cell death after hypoxia-reoxygenation. Stroke 34: 2704–2709. 10.1161/01.STR.0000096540.40826.BA [DOI] [PubMed] [Google Scholar]
- Lei K., Davis R. J., 2003. JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc. Natl. Acad. Sci. USA 100: 2432–2437. 10.1073/pnas.0438011100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K., Nimnual A., Zong W. X., Kennedy N. J., Flavell R. A., et al. , 2002. The Bax subfamily of Bcl2-related proteins is essential for apoptotic signal transduction by c-Jun NH(2)-terminal kinase. Mol. Cell. Biol. 22: 4929–4942. 10.1128/MCB.22.13.4929-4942.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Johnson R. W., Park D., Chin-Sang I., Chamberlin H. M., 2012. Somatic gonad sheath cells and Eph receptor signaling promote germ-cell death in C. elegans. Cell Death Differ. 19: 1080–1089. 10.1038/cdd.2011.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Mao W., Ding B., Liang C. S., 2008. ERKs/p53 signal transduction pathway is involved in doxorubicin-induced apoptosis in H9c2 cells and cardiomyocytes. Am. J. Physiol. Heart Circ. Physiol. 295: H1956–H1965. 10.1152/ajpheart.00407.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. A., Hengartner M. O., 1998. Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans. Cell 93: 961–972. 10.1016/S0092-8674(00)81202-7 [DOI] [PubMed] [Google Scholar]
- Lujambio A., Lowe S. W., 2012. The microcosmos of cancer. Nature 482: 347–355. 10.1038/nature10888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. C., Allan L. A., Mancini E. J., Clarke P. R., 2008. The docking interaction of caspase-9 with ERK2 provides a mechanism for the selective inhibitory phosphorylation of caspase-9 at threonine 125. J. Biol. Chem. 283: 3854–3865. 10.1074/jbc.M705647200 [DOI] [PubMed] [Google Scholar]
- Masoudi N., Fancsalszky L., Pourkarimi E., Vellai T., Alexa A., et al. , 2013. The NM23–H1/H2 homolog NDK-1 is required for full activation of Ras signaling in C. elegans. Development 140: 3486–3495. 10.1242/dev.094011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McJunkin K., Ambros V., 2014. The embryonic mir-35 family of microRNAs promotes multiple aspects of fecundity in Caenorhabditis elegans. G3 (Bethesda) 4: 1747–1754. 10.1534/g3.114.011973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N., Wei H., Conradt B., 2018. Caenorhabditis elegans ced-3 caspase is required for asymmetric divisions that generate cells programmed to die. Genetics 210: 983–998. 10.1534/genetics.118.301500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagami H., Morishita R., Yamamoto K., Yoshimura S. I., Taniyama Y., et al. , 2001. Phosphorylation of p38 mitogen-activated protein kinase downstream of bax-caspase-3 pathway leads to cell death induced by high D-glucose in human endothelial cells. Diabetes 50: 1472–1481. 10.2337/diabetes.50.6.1472 [DOI] [PubMed] [Google Scholar]
- Pellegrini L., Yu D. S., Lo T., Anand S., Lee M., et al. , 2002. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 420: 287–293. 10.1038/nature01230 [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W., 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourkarimi E., Greiss S., Gartner A., 2012. Evidence that CED-9/Bcl2 and CED-4/Apaf-1 localization is not consistent with the current model for C. elegans apoptosis induction. Cell Death Differ. 19: 406–415. 10.1038/cdd.2011.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiders S. A., Eastwood M. D., Bacher M., Priess J. R., 2018. Binucleate germ cells in Caenorhabditis elegans are removed by physiological apoptosis. PLoS Genet. 14: e1007417 10.1371/journal.pgen.1007417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., et al. , 2007. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell 26: 731–743. 10.1016/j.molcel.2007.05.017 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Horvitz H. R., 2000. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat. Cell Biol. 2: 131–136. 10.1038/35004000 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Horvitz H. R., 2004. The engulfment process of programmed cell death in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 20: 193–221. 10.1146/annurev.cellbio.20.022003.114619 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Cameron S., Horvitz H. R., 2001. Phagocytosis promotes programmed cell death in C. elegans. Nature 412: 198–202. 10.1038/35084096 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Andersen E. C., Huang M. C., Horvitz H. R., 2007. DPL-1 DP, LIN-35 Rb and EFL-1 E2F act with the MCD-1 zinc-finger protein to promote programmed cell death in Caenorhabditis elegans. Genetics 175: 1719–1733. 10.1534/genetics.106.068148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski R., Dickinson R., Stewart G., Craig A., Schimpl M., et al. , 2011. Regulation of Caenorhabditis elegans p53/CEP-1-dependent germ cell apoptosis by Ras/MAPK signaling. PLoS Genet. 7: e1002238 10.1371/journal.pgen.1002238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertel C., Conradt B., 2007. C. elegans orthologs of components of the RB tumor suppressor complex have distinct pro-apoptotic functions. Development 134: 3691–3701. 10.1242/dev.004606 [DOI] [PubMed] [Google Scholar]
- Schumacher B., Hofmann K., Boulton S., Gartner A., 2001. The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr. Biol. 11: 1722–1727. 10.1016/S0960-9822(01)00534-6 [DOI] [PubMed] [Google Scholar]
- Schumacher B., Hanazawa M., Lee M. H., Nayak S., Volkmann K., et al. , 2005a Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell 120: 357–368. 10.1016/j.cell.2004.12.009 [DOI] [PubMed] [Google Scholar]
- Schumacher B., Schertel C., Wittenburg N., Tuck S., Mitani S., et al. , 2005b C. elegans ced-13 can promote apoptosis and is induced in response to DNA damage. Cell Death Differ. 12: 153–161 (erratum: Cell Death Differ. 12: 532). 10.1038/sj.cdd.4401539 [DOI] [PubMed] [Google Scholar]
- Shaham S., Reddien P. W., Davies B., Horvitz H. R., 1999. Mutational analysis of the Caenorhabditis elegans cell-death gene ced-3. Genetics 153: 1655–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrard R., Luehr S., Holzkamp H., McJunkin K., Memar N., et al. , 2017. miRNAs cooperate in apoptosis regulation during C. elegans development. Genes Dev. 31: 209–222. 10.1101/gad.288555.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sionov R. V., Vlahopoulos S. A., Granot Z., 2015. Regulation of bim in health and disease. Oncotarget 6: 23058–23134. 10.18632/oncotarget.5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H., Trombly M. I., Chen J., Wang X., 2009. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 23: 304–317. 10.1101/gad.1749809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M. V., 2013. Canonical RTK-Ras-ERK signaling and related alternative pathways (July 1, 2013), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.80.2, http://www.wormbook.org. 10.1895/wormbook.1.80.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y., Simizu S., Osada H., 2004. The phosphorylation status and anti-apoptotic activity of Bcl-2 are regulated by ERK and protein phosphatase 2A on the mitochondria. FEBS Lett. 569: 249–255. 10.1016/j.febslet.2004.06.003 [DOI] [PubMed] [Google Scholar]
- Tang D., Wu D., Hirao A., Lahti J. M., Liu L., et al. , 2002. ERK activation mediates cell cycle arrest and apoptosis after DNA damage independently of p53. J. Biol. Chem. 277: 12710–12717. 10.1074/jbc.M111598200 [DOI] [PubMed] [Google Scholar]
- Thompson C. B., 1995. Apoptosis in the pathogenesis and treatment of disease. Science 267: 1456–1462. 10.1126/science.7878464 [DOI] [PubMed] [Google Scholar]
- Tibbles L. A., Woodgett J. R., 1999. The stress-activated protein kinase pathways. Cell. Mol. Life Sci. 55: 1230–1254. 10.1007/s000180050369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tops B. B., Plasterk R. H., Ketting R. F., 2006. The Caenorhabditis elegans Argonautes ALG-1 and ALG-2: almost identical yet different. Cold Spring Harb. Symp. Quant. Biol. 71: 189–194. 10.1101/sqb.2006.71.035 [DOI] [PubMed] [Google Scholar]
- Tran, A. T., E. M. Chapman, M. N. Flamand, B. Yu, S. J. Krempel et al., 2019 MiR-35 buffers apoptosis thresholds in the C. elegans germline by antagonizing both MAPK and core apoptosis pathways. Cell Death Differ. DOI: 10.1038/s41418-019-0325-6. DOI: 10.1038/s41418-019-0325-6 10.1038/s41418-019-0325-6 [DOI] [PMC free article] [PubMed]
- Vasquez-Rifo A., Jannot G., Armisen J., Labouesse M., Bukhari S. I., et al. , 2012. Developmental characterization of the microRNA-specific C. elegans Argonautes alg-1 and alg-2. PLoS One 7: e33750 10.1371/journal.pone.0033750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheij M., Ruiter G. A., Zerp S. F., van Blitterswijk W. J., Fuks Z., et al. , 1998. The role of the stress-activated protein kinase (SAPK/JNK) signaling pathway in radiation-induced apoptosis. Radiother. Oncol. 47: 225–232. 10.1016/S0167-8140(98)00007-3 [DOI] [PubMed] [Google Scholar]
- Viktorsson K., Ekedahl J., Lindebro M. C., Lewensohn R., Zhivotovsky B., et al. , 2003. Defective stress kinase and Bak activation in response to ionizing radiation but not cisplatin in a non-small cell lung carcinoma cell line. Exp. Cell Res. 289: 256–264. 10.1016/S0014-4827(03)00264-7 [DOI] [PubMed] [Google Scholar]
- Wagle P., Nikolic M., Frommolt P., 2015. QuickNGS elevates next-generation sequencing data analysis to a new level of automation. BMC Genomics 16: 487 10.1186/s12864-015-1695-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. G., Rapp U. R., Reed J. C., 1996. Bcl-2 targets the protein kinase Raf-1 to mitochondria. Cell 87: 629–638. 10.1016/S0092-8674(00)81383-5 [DOI] [PubMed] [Google Scholar]
- Welch C., Chen Y., Stallings R. L., 2007. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene 26: 5017–5022. 10.1038/sj.onc.1210293 [DOI] [PubMed] [Google Scholar]
- Widmann C., Gerwins P., Johnson N. L., Jarpe M. B., Johnson G. L., 1998a MEK kinase 1, a substrate for DEVD-directed caspases, is involved in genotoxin-induced apoptosis. Mol. Cell. Biol. 18: 2416–2429. 10.1128/MCB.18.4.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmann C., Gibson S., Johnson G. L., 1998b Caspase-dependent cleavage of signaling proteins during apoptosis. A turn-off mechanism for anti-apoptotic signals. J. Biol. Chem. 273: 7141–7147. 10.1074/jbc.273.12.7141 [DOI] [PubMed] [Google Scholar]
- Wu D., Wallen H. D., Inohara N., Nunez G., 1997. Interaction and regulation of the Caenorhabditis elegans death protease CED-3 by CED-4 and CED-9. J. Biol. Chem. 272: 21449–21454. 10.1074/jbc.272.34.21449 [DOI] [PubMed] [Google Scholar]
- Wu E., Thivierge C., Flamand M., Mathonnet G., Vashisht A. A., et al. , 2010. Pervasive and cooperative deadenylation of 3′UTRs by embryonic microRNA families. Mol. Cell 40: 558–570. 10.1016/j.molcel.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Han M., 1994. Suppression of activated Let-60 ras protein defines a role of Caenorhabditis elegans Sur-1 MAP kinase in vulval differentiation. Genes Dev. 8: 147–159. 10.1101/gad.8.2.147 [DOI] [PubMed] [Google Scholar]
- Wu Y. C., Horvitz H. R., 1998a The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell 93: 951–960. 10.1016/S0092-8674(00)81201-5 [DOI] [PubMed] [Google Scholar]
- Wu Y. C., Horvitz H. R., 1998b C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature 392: 501–504. 10.1038/33163 [DOI] [PubMed] [Google Scholar]
- Xue D., Horvitz H. R., 1997. Caenorhabditis elegans CED-9 protein is a bifunctional cell-death inhibitor. Nature 390: 305–308. 10.1038/36889 [DOI] [PubMed] [Google Scholar]
- Yang X., Chang H. Y., Baltimore D., 1998. Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science 281: 1355–1357. 10.1126/science.281.5381.1355 [DOI] [PubMed] [Google Scholar]
- Yigit E., Batista P. J., Bei Y., Pang K. M., Chen C. C., et al. , 2006. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell 127: 747–757. 10.1016/j.cell.2006.09.033 [DOI] [PubMed] [Google Scholar]
- Yuan J. Y., Horvitz H. R., 1990. The Caenorhabditis elegans genes ced-3 and ced-4 act cell autonomously to cause programmed cell death. Dev. Biol. 138: 33–41. 10.1016/0012-1606(90)90174-H [DOI] [PubMed] [Google Scholar]
- Zang Y. S., Zhong Y. F., Fang Z., Li B., An J., 2012. MiR-155 inhibits the sensitivity of lung cancer cells to cisplatin via negative regulation of Apaf-1 expression. Cancer Gene Ther. 19: 773–778. 10.1038/cgt.2012.60 [DOI] [PubMed] [Google Scholar]
- Zhang B., Pan X., Cobb G. P., Anderson T. A., 2007. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 302: 1–12. 10.1016/j.ydbio.2006.08.028 [DOI] [PubMed] [Google Scholar]
- Zhou M., Liu Z., Zhao Y., Ding Y., Liu H., et al. , 2010. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J. Biol. Chem. 285: 21496–21507. 10.1074/jbc.M109.083337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Hartwieg E., Horvitz H. R., 2001. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104: 43–56. 10.1016/S0092-8674(01)00190-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article and its figures. The file “Supplemental information” contains figure legends and descriptions of all supplemental files. New strains used in this study will be made available at the Caenorhabditis Genetics Center (University of Minnesota). miRNA expression data are available at the GEO with the accession code GSE124606. Supplemental Material available at FigShare: https://doi.org/10.25386/genetics.8852180.