Abstract
Limited antifungal diversity and availability are growing problems for the treatment of fungal infections in the face of increasing drug resistance. The echinocandins, one of the newest classes of antifungal drugs, inhibit production of a crucial cell wall component. However, these compounds do not effectively inhibit the growth of the opportunistic fungal pathogen Cryptococcus neoformans, despite potent inhibition of the target enzyme in vitro. Therefore, we performed a forward genetic screen to identify cellular processes that mediate the relative tolerance of this organism to the echinocandin drug caspofungin. Through these studies, we identified 14 genetic mutants that enhance caspofungin antifungal activity. Rather than directly affecting caspofungin antifungal activity, these mutations seem to prevent the activation of various stress-induced compensatory cellular processes. For example, the pfa4Δ mutant has defects in the palmitoylation and localization of many of its target proteins, including the Ras1 GTPase and the Chs3 chitin synthase, which are both required for caspofungin tolerance. Similarly, we have confirmed the link between caspofungin treatment and calcineurin signaling in this organism, but we suggest a deeper mechanism in which caspofungin tolerance is mediated by multiple pathways downstream of calcineurin function. In summary, we describe here several pathways in C. neoformans that contribute to the complex caspofungin tolerance phenotype in this organism.
Keywords: antifungal, chitin, calcineurin, signalosome, Ras
INVASIVE fungal diseases primarily affect people with immune system defects, resultingin significant morbidity and mortality in these vulnerable patient populations (Park et al. 2009; Pyrgos et al. 2013; Rajasingham et al. 2017). Limited therapeutic options and availability are major challenges for effective treatment of systemic fungal infections, particularly in regions where fungal infection rates are highest (Loyse et al. 2013; Perfect and Bicanic 2014). Historically, it has been difficult to identify novel antifungal agents that are not also toxic to humans, since many cellular processes are highly conserved between humans and fungi. In the search for novel antifungal drugs, identification of fungi-specific cellular processes has been a major focus. The fungal cell wall represents a key structure for fungal viability, growth, and host evasion (Latgé 2007; Doering 2009; Gow and Hube 2012; O’Meara et al. 2013; Esher et al. 2018). Thus, compounds that target the production and maintenance of the fungal cell wall, with little to no effect on the human host, would make exciting and specific antifungal agents.
Echinocandins are cell wall-targeting antifungal compounds that have been identified and synthesized from natural products (Denning 2003; Letscher-Bru and Herbrecht 2003). These compounds inhibit the synthesis of β-1,3-glucan, a crucial cell wall component for many fungi (Taft et al. 1988; Kurtz et al. 1994; Kurtz and Douglas 1997; Maligie and Selitrennikoff 2005). Echinocandin antifungals, such as caspofungin, micafungin, and anidulafungin, are used extensively in clinical settings for the treatment of infections caused by diverse fungi. However, echinocandins do not have potent antifungal activity against the fungal pathogen Cryptococcus neoformans, whose growth is only inhibited at high levels of caspofungin that are not clinically achievable in patients (Abruzzo et al. 1997; Bartizal et al. 1997; Espinel-Ingroff 1998). The fact that this drug is so ineffective against this fungus is surprising for a number of reasons. First, the gene that encodes the β-1,3-glucan synthase catalytic subunit in C. neoformans, FKS1, is essential in this organism (Thompson et al. 1999). Additionally, the C. neoformans enzyme is highly sensitive to caspofungin in vitro, even potentially at lower concentrations than species that are clinically susceptible to these drugs, such as Aspergillus species (Maligie and Selitrennikoff 2005). Based on these data, caspofungin could be expected to be an effective inhibitor of C. neoformans growth.
Given these observations, several investigators have tried to explain the discrepancy between the high sensitivity of the target enzyme’s activity and the high tolerance of the organism to caspofungin. Recent work using a fluorescently tagged form of caspofungin has suggested that intracellular concentrations of caspofungin are low in wild-type C. neoformans (Huang et al. 2016). Therefore, the cell wall or the polysaccharide capsule could prevent the accessibility of caspofungin, a high-molecular weight drug, to its target enzyme. Alternatively, caspofungin could be entering the cell but be rapidly eliminated by degradation or through the action of a multidrug resistance pump. We hypothesized that interrupting specific C. neoformans cell processes would render this fungus more susceptible to caspofungin, even at these relatively low effective concentrations. To this end, we screened through two targeted gene deletion collections to identify C. neoformans mutants that were hypersensitive to caspofungin relative to the wild-type strain. We also demonstrate possible mechanisms for how the cellular processes controlled by these genes might affect echinocandin resistance in C. neoformans.
Materials and Methods
Strains, media, and growth conditions
The collections used for the caspofungin sensitivity screen consist of both the 2008 Madhani and 2015 Madhani plate collections, which were purchased from the Fungal Genetics Stock Center (Liu et al. 2008; Chun and Madhani 2010). The wild-type (WT) strain used in this study is the clinical strain H99 (Perfect et al. 1980). Newly generated strains, as well as strains from alternate sources, are listed in Supplemental Material, Table S1. A similar screen using a subset of these mutant strains was also recently performed (Huang et al. 2016).
Strains were maintained on yeast extract, peptone, and dextrose (YPD) agar (10% yeast extract, 20% peptone, 2% dextrose, and 20% Bacto agar), and overnight cultures were incubated in YPD liquid medium. Drug susceptibility testing was performed in yeast nitrogen base medium (1× YNB + 2% glucose) (Pfaller et al. 1990; Jessup et al. 1998). Cultures for microscopy were prepared in synthetic complete (SC) medium (1× YNB + 1× complete amino acids + 2% glucose).
Strain creation
To generate the new and independent mutant strains used in this study, targeted gene deletion constructs were designed to replace the entire open reading frame with the neomycin (NEO) or nourseothricin (NAT) dominant selectable markers. Each knockout construct was generated using PCR overlap-extension and a split selectable marker as described previously (Davidson et al. 2002; Kim et al. 2012). All constructs were transformed into the C. neoformans H99 strain by biolistic transformation, as previously described (Toffaletti et al. 1993). All deletion primers used in this study can be found in Table S2A.
Upon transformation, strains were selected on YPD medium containing either NAT or NEO. Deletion mutants were checked by a combination of positive and negative confirmation PCRs demonstrating replacement of the WT locus with the mutant allele, followed by Southern blotting to confirm single integration of the deletion constructs (data not shown).
Plasmids used in this study can be found in Table S3. Cloning primers can be found in Table S2B. The overexpressed GFP-RHO1 construct was generated by cloning the RHO1 gene plus its native terminator into the BamHI site of the pCN19 vector, which contains the histone H3 promoter and GFP with the NAT selection marker. The endogenous FKS1-GFP construct was engineered by cloning the following into the pUC19 vector: the FKS1 gene (without promoter), the GFP gene, the FKS1 terminator, and the NEO marker flanked by genomic sequences to target this construct to the FKS1 locus. These plasmids were transformed as described, and selected on NEO or NAT, and transformants were confirmed by a positive PCR to document the presence of the introduced allele. In the case of the eFKS1-GFP construct, PCRs to confirm integration of the construct into the endogenous FKS1 locus were performed.
Caspofungin sensitivity primary screen
A small pilot screen revealed that the calcineurin B subunit mutant was hypersensitive to caspofungin compared to the WT, agreeing with published data (Del Poeta et al. 2000). This strain became the standard against which to measure other potentially caspofungin-susceptible strains. To determine the optimal conditions under which to perform the caspofungin sensitivity screen, we assessed WT vs. cnb1Δ growth in YNB medium at caspofungin concentrations between 5 and 50 µg/ml with shaking at 150 rpm at 30° (Cancidas; Merck). We identified 15 μg/ml caspofungin to be a concentration at which the cnb1Δ mutant strain was markedly impaired for growth and the WT grew robustly. We then screened the strain collections of 3880 isolates, first preincubating in YPD liquid medium for 16 hr with shaking (150 rpm) at 30°. Cultures were diluted 1:10 in 96-well plates containing either YNB or YNB + caspofungin (15 μg/ml). Strains were incubated with shaking (150 rpm) at 30° for 24 hr, and growth was assessed by measuring OD600 on a FLUOStar Optima plate reader (BMG Labtech). Plates were also pin-replicated to YPD plates to assess strain viability after incubation with caspofungin.
After screening, strains were divided into four groups based on caspofungin susceptibility: (1) strains that were inviable postcaspofungin treatment; (2) strains that were viable after caspofungin treatment, but that had significantly decreased growth (OD600 that was >2 SD less than the average WT OD600); (3) strains that did not have significantly different growth from WT in caspofungin medium; and (4) strains that did not grow in YNB.
Disc-diffusion secondary screen
To confirm the caspofungin sensitivity of the mutants identified in the above screen, each mutant was incubated overnight in 150 µl of YPD in a 96-well plate with shaking, along with the WT and cna1Δ mutant strains as controls. Strains were diluted 1:200 in PBS, then 75 µl per strain was spread onto YNB agar in 6-well plates. Sterile filter discs were placed in the center of each well and 5 μl of 7.5 mg/ml caspofungin was added to each disc. Plates were incubated at 30° for 3 days. After incubation, plates were imaged and zones of inhibition were measured. Mutant phenotypes were classified as WT-like, cna1Δ-like, or intermediate.
Minimal inhibitory concentration assay
Minimal inhibitory concentration (MIC) assays were performed according to modified Clinical Laboratory Standards Institute standard methods for broth microdilution testing of antifungal susceptibility (Pfaller et al. 1990; Clinical and Laboratory Standards Institute 2008). In brief, cells were diluted in phosphate-buffered saline (PBS) to an OD600 of 0.25, then diluted 1:100 in YNB medium. Caspofungin was diluted in PBS. Next, 2× working stocks of caspofungin were prepared in YNB medium, then caspofungin was serially diluted twofold in 100 μl YNB medium, following which 100 µl of 1:100 dilution of cells was added to diluted drug in 96-well plates. The final concentration range of caspofungin was 200–0.39 µg/ml. Plates were incubated for 48 hr without shaking at 30° or 35°. After 48 hr, OD600 was measured on a FLUOStar Optima plate reader. MIC50 values were calculated by calculating relative growth using (drug-treated OD600/untreated OD600), with MIC50 corresponding to a ≥ 50% decrease in relative growth.
Checkerboard assay
Checkerboard assays to assess antifungal drug synergy using the fractional inhibitory concentration index (FIC) for combinations of compounds were performed as described (National Committee for Clinical Laboratory Standards 1992; Franzot and Casadevall 1997). In brief, the WT strain H99 was inoculated from plated colonies into PBS at an OD600 of 0.25, and subsequently diluted 1:100 into RPMI medium. Nikkomycin Z stock was diluted in PBS (nikkomycin Z; Sigma [Sigma Chemical], St. Louis, MO). Manumycin A, clorgyline, and 2-bromopalmitate (2BP) were diluted in DMSO (manumycin A, Bioviotica; clorgyline, Sigma; and 2BP, Sigma). Caspofungin was diluted for a final concentration range between 100 and 1.5625 μg/ml, and the test drugs were diluted to the following final concentration ranges: nikkomycin Z 400 to 0.78125 μg/ml, manumycin A 40 to 0.078 μM, 2BP 400 to 0.78125 µM, and clorgyline 100 to 1.5625 µM. Assays were incubated at 30 and 37°. FIC index values for a combination of compounds A and B were calculated as:
where an FIC index value of < 1.0 is considered synergistic (with < 0.5 considered strongly synergistic), additive if the value was 1.0, autonomous if the value was between 1.0 and 2.0, and antagonistic if the FIC index was > 2.0 (Franzot and Casadevall 1997).
Whole-genome sequencing, alignment, and variant calling
Whole-genome sequencing was performed on both the WT background strain and the msh1Δ mutant strain, by the Duke Center for Genome and Computational Biology Genome Sequencing Shared Resource using an Illumina MiSeq instrument. Paired-end libraries were sequenced with read lengths of 251 bases. Reads were aligned to the version 3 H99 genome (Janbon et al. 2014) using BWA-MEM with default settings (Li and Durbin 2009). The Genome Analysis Toolkit (GATK) best practices pipeline (McKenna et al. 2010) was used in combination with SAMtools (Li et al. 2009) and Picard to realign reads before SNP calling using the UnifiedGenotyper component of GATK with the haploid ploidy setting. The resulting variant call formats were filtered using VCFtools (Danecek et al. 2011) and annotated for variant effect using SnpEff (Cingolani et al. 2012). Heterozygous calls were removed as presumed mismapped repetitive regions. Raw reads are available on the National Center for Biotechnology Information (NCBI) Sequence Read Archive under accession number PRJNA501913.
Chitin and chitosan assay
The chitin and chitosan contents of C. neoformans cell walls were assessed as described (Banks et al. 2005). Briefly, cells were incubated overnight in YPD. Cultured cells were then diluted to an OD of 0.8 in SC or SC + 15 μg/ml caspofungin, and incubated with shaking at 30° for 6 hr. Cells were divided and lyophilized, then either mock treated or treated with acetic anhydride to acetylate chitosan to form chitin. Cell walls were then digested with 5 mg/ml chitinase for 72 hr. Monomer levels of N-acetylglucosamine were assessed by a p-dimethylaminobenzaldehyde colorimetric assay and read on a FLUOStar Optima plate reader. Acetic anhydride samples represented levels of both chitin and chitosan in the cell wall, while untreated samples represented chitin alone. Chitosan levels were calculated as the difference between the acetic anhydride-treated and the untreated samples. Data were analyzed using a two-way ANOVA, followed by Student’s t-tests to determine statistical significance.
Cell wall staining and microscopy
Cells were prepared for cell wall staining as described (Ost et al. 2017). WT cells were cultured overnight in YPD medium. Overnight cultures were diluted to an OD600 of 1 in 15 ml SC or SC plus caspofungin (5, 10, or 20 µg/ml caspofungin), and incubated with shaking at 30°. At the indicated timepoints, 1 ml aliquots of each culture were collected and stained with calcofluor white (CFW). Cells were pelleted at 5000 rpm for 2 min, then resuspended in 100 µl PBS + 25 μg/ml CFW and incubated in the dark at room temperature for 10 min. Cells were then washed two times with PBS and resuspended in 50 μl PBS for imaging. Strains were imaged on a Zeiss ([Carl Zeiss], Thornwood, NY) Axio Imager A1 fluorescence microscope equipped with an Axio-Cam MRM digital camera to capture both DIC and fluorescent images. Cell wall-staining fluorescence intensity was analyzed using Fiji software, and the mean gray values were analyzed (Schindelin et al. 2012). Data presented represent the average fluorescence values. Data were analyzed using a two-way ANOVA, followed by Student’s t-tests to determine statistical significance.
For fluorescent fusion protein microscopy, strains were incubated in SC medium for 18 hr at 30° with shaking. These cultures were pelleted at 3000 rpm for 5 min, and resuspended in SC or SC + 15 μg/ml caspofungin. Strains were incubated with shaking at 30° for 90 min, with aliquots collected for imaging at 15-min intervals. Aliquots were incubated with NucBlue Live Ready Probes reagent for 5 min, pelleted at 5000 rpm for 2 min, then resuspended in 50 μl SC (Thermo Fisher Scientific). Strains were imaged on a Zeiss Axio Imager A1 fluorescence microscope equipped with an Axio-Cam MRM digital camera to capture both DIC and fluorescent images. For the Fks1-GFP and GFP-Rho1 localization experiments, cultures were incubated for 18 hr at 30° with shaking in SC medium. These cultures were normalized to an OD600 of 2.0 in SC plus 0, 5, 10, or 15 μg/ml caspofungin, and imaged at 20-min intervals for 1.5 hr.
RNA preparation and quantitative real-time PCR
The WT strain was grown in YPD for 18 hr at 30° with shaking. Cells were then inoculated at an OD600 of 1.5 into 5 ml SC, SC + 10 µg/ml caspofungin, or SC + 15 μg/ml caspofungin. Cultures were incubated at 30° with shaking for 90 min, then cells were harvested by centrifugation at 3000 rpm for 5 min and lyophilized. RNA was isolated using a RNeasy Plant Mini Kit (QIAGEN, Valencia, CA), with the addition of bead beating for 1 min prior to lysis and on-column DNase treatment (QIAGEN). cDNA was prepared using the AffinityScript QPCR cDNA synthesis kit using oligo-dT primers to bias for mRNA transcripts (Agilent Genomics). Quantitative real-time PCR was performed using PowerUp SYBR Green Master mix (Applied Biosystems, Foster City, CA) on a QuantStudio 6 Flex system. Real-time PCR primers are listed in Table S2C (Esher et al. 2018). Data were analyzed using a two-way ANOVA, followed by Student’s t-tests to determine statistical significance.
Data availability
Strains and plasmids are available upon request. File S1 contains a list and descriptions of all supplemental files. Sequence data are available at the NCBI Sequence Read Archive under the accession number PRJNA501913. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8085290
Results
Initial screen for processes contributing to caspofungin tolerance in C. neoformans
We performed a forward genetic screen of targeted deletion mutants to identify cellular processes that contribute to C. neoformans tolerance to caspofungin treatment. Using two screening methods in sequence, we screened 3880 mutants for altered growth during caspofungin treatment at 30° (Liu et al. 2008; Chun and Madhani 2010). We specifically chose this permissive incubation temperature for our initial screens to potentially capture pathways that may be required for thermotolerance, and for which mutant strains would therefore not be viable at higher temperatures. Our screening methods yielded 54 mutants that appeared to have reduced caspofungin tolerance relative to the WT strain. We then performed broth microdilution assays to compare caspofungin susceptibility to the WT strain, as well as to a cna1Δ strain with a mutation in the calcineurin A subunit gene. Strains with altered calcineurin function are known to be more susceptible to caspofungin (Del Poeta et al. 2000; Kraus et al. 2003). Of the mutants identified in the primary screen, 14 were confirmed to have caspofungin susceptibility similar to or greater than the cna1Δ mutant strain (Table 1). This strict screening criterion enabled us to focus our studies exclusively on more highly drug-susceptible mutants (Table 1). The remaining strains displayed only minimal increases in sensitivity to caspofungin and they were not tested further. Of note, we identified the cnb1Δ calcineurin regulatory subunit mutant, which phenocopies the cna1Δ mutant, as a caspofungin-hypersensitive mutant in our screen, validating our screening methods (Odom et al. 1997; Del Poeta et al. 2000; Fox et al. 2001; Kraus et al. 2003).
Table 1. Caspofungin MICs for screen hits.
| Gene locus tag | Gene product mutated | MIC (µg/ml) |
|---|---|---|
| CNAG_00375 | SAGA complex histone acetyltransferase (Gcn5) | 12.5 |
| CNAG_02682 | Hypothetical protein (Msh1 homolog) | 0.78 |
| CNAG_05070 | Sulfite reductase (NADPH) hemoprotein, β-component | 0.78 |
| CNAG_06902 | Hypothetical protein | 12.5 |
| CNAG_02236 | Type 2A-like serine/threonine protein phosphatase (Ppg1) | 0.78 |
| CNAG_03981 | Palmitoyltransferase Pfa4 | 6.25 |
| CNAG_03841 | Hypothetical protein | 12.5 |
| CNAG_02292 | Copper chaperone Lys7 | 12.5 |
| CNAG_04992 | Hypothetical protein | 12.5 |
| CNAG_03080 | Fatty acid elongase | 12.5 |
| CNAG_07636 | Chitin synthase regulator (Csr2) | 3.125 |
| CNAG_02891 | Endoplasmic reticulum rhodanese-like protein (Rdl2) | 12.5 |
| CNAG_01717 | Cell differentiation protein Rcd1 | 12.5 |
| CNAG_00888 | Calcineurin B subunit | 3.125 |
MIC, minimal inhibitory concentration. SAGA: Spt3-Ada2-Gcn5 complex.
Within the list of sensitive mutants, we identified multiple biological processes that seem to be important for caspofungin tolerance based on gene ontology (GO) analysis (see Table S4 for full GO analysis) (Stajich et al. 2012). Of note, the msh1Δ mutant strain displayed a very low caspofungin MIC. This strain has a defect in a predicted homolog of a MutS mismatch repair protein most likely functioning on mitochondrial DNA (Sia and Kirkpatrick 2005). Interestingly, independently created C. neoformans msh1Δ mutants did not display the caspofungin-susceptible phenotype of the original mutant, suggesting that secondary mutations in this strain were the cause of increased drug susceptibility. Indeed, whole-genome sequencing of the original msh1Δ mutant strain revealed several additional mutations in both the nuclear and mitochondrial genomes, as predicted for a strain with potential defects in DNA repair mechanisms (Table S5).
Calcineurin signaling plays a role in caspofungin tolerance in C. neoformans
In previous in vitro studies, the calcineurin inhibitor FK506 has demonstrated synergistic interactions with caspofungin against C. neoformans (Del Poeta et al. 2000). In our caspofungin sensitivity screen, we identified a mutant of the calcineurin B regulatory subunit, cnb1Δ, to be highly sensitive to caspofungin, as seen in previous work (Kraus et al. 2003). Indeed, mutants of both the calcineurin A catalytic and B regulatory subunits—cna1Δ and cnb1Δ, respectively—exhibit an eightfold increase in caspofungin sensitivity when compared to the WT strain (Table 1 and Table 2). Therefore, we strove to identify the mechanism by which this phosphatase is facilitating caspofungin tolerance in C. neoformans by examining the activation of a known target of calcineurin signaling in C. neoformans, the Crz1 transcription factor.
Table 2. Additional caspofungin MICs at 30°.
| Gene locus tag | Gene product mutated | MIC (µg/ml) |
|---|---|---|
| WT | NA | 25 |
| CNAG_04796 | Calcineurin A subunit | 3.125 |
| CNAG_01744 | Crz1 transcription factor | 12.5 |
| CNAG_05581 | Chitin synthase 3 | 3.125 |
| CNAG_00293 | Ras1 GTPase | 25 |
| CNAG_05348 | Cdc42 GTPase | 25 |
| CNAG_05968 | Cdc420 GTPase | 25 |
| CNAG_04243 | Cdc24 guanine nucleotide exchange factor | 25 |
| CNAG_06165 | Ste20 PAK kinase | 25 |
| CNAG_04761 | Ras2 GTPase | 25 |
| CNAG_05998 | Rac2 GTPase | 25 |
| CNAG_02883 | Rac1 GTPase | 25 |
| CNAG_05925 | Septin Cdc3 | 25 |
| CNAG_01740 | Septin Cdc12 | 25 |
| CNAG_05970 | PAK kinase Pak1 | 25 |
MIC, minimal inhibitory concentration; WT, wild-type. PAK: p21-activated protein kinase.
The Crz1 transcription factor is activated through dephosphorylation by calcineurin, and Crz1 mediates many of the known transcriptional effects of this calcineurin signaling (Lev et al. 2012). We assessed the effect of caspofungin on Crz1 nuclear localization by examining mCherry-tagged Crz1 fusion protein (Crz1-mCherry) colocalization with the GFP-Nop1 nucleolar marker, as well as with DAPI staining (Chow et al. 2017). In untreated cells, Crz1-mCherry remains localized in the cytosol. We determined that Crz1-mCherry localizes to the nucleus after 45 min of incubation in 15 µg/ml of caspofungin (Figure 1, A and B). The nuclear localization of Crz1-mCherry suggests that Crz1 is likely being activated under these conditions. However, the crz1Δ mutant displayed a caspofungin MIC more similar to WT than to the cna1Δ or cnb1Δ mutant strains (Table 2). Together, these results suggest that, although Crz1 is being activated during caspofungin treatment, caspofungin tolerance is mediated in part through calcineurin-dependent but Crz1-independent targets. Therefore, we assessed the caspofungin susceptibility of several strains with loss-of-function mutations in downstream calcineurin targets (Table S6) (Park et al. 2016). Unlike the cna1Δ or cnb1Δ calcineurin subunit mutants, none of the tested strains deficient in individual downstream calcineurin effector genes displayed altered caspofungin susceptibility. These data are consistent with emerging reports that C. neoformans calcineurin acts through both Crz1-dependent and -independent processes (Lev et al. 2012; Chow et al. 2017). Alternatively, there may be functional redundancy among the calcineurin effectors with respect to caspofungin susceptibility.
Figure 1.
Crz1 is activated and localizes to the nucleus in response to caspofungin treatment. (A) A strain expressing Crz1-mCherry and GFP-Nop1 (nucleolar marker) was incubated in SC medium with either 0 or 15 µg/ml caspofungin for 45 min, then stained with NucBlue Live Cell nuclear stain for 5 min and imaged on a Zeiss AxioVision epifluorescence microscope. Bar, 5 μm. (B) Percentage of cells displaying nuclear Crz1-mCherry localization as measured by colocalization with GFP-Nop1. Results represent percentage nuclear localization from three independent experiments. Error bars represent SEM. * P-value < 0.05.
The Pfa4 palmitoyltransferase plays a role in caspofungin tolerance through regulation of its target proteins
The Pfa4 palmitoyltransferase is required for the addition of palmitoyl groups to various C. neoformans proteins (Nichols et al. 2015; Santiago-Tirado et al. 2015). This post-translational modification is necessary for the proper localization and function of these target proteins. In our MIC assays, we documented a fourfold increase in caspofungin susceptibility of the pfa4∆ strain at 30°. Given the temperature sensitivity of this mutant strain (Nichols et al. 2015; Santiago-Tirado et al. 2015), we would not have isolated it in screens at higher incubation temperatures. The caspofungin sensitivity of the pfa4∆ mutant was more pronounced at 35°, a temperature at which the pfa4Δ mutant grows but with a modest growth delay compared to WT (Table 1 and Table 3) (Nichols et al. 2015; Santiago-Tirado et al. 2015).
Table 3. Additional caspofungin MICs at 35°.
| Gene locus tag | Gene product mutated | MIC (µg/ml) |
|---|---|---|
| H99 | N/A | 25 |
| CNAG_00293 | Ras1 GTPase | 6.25 |
| CNAG_05348 | Cdc42 GTPase | 12.5 |
| CNAG_05968 | Cdc420 GTPase | 25 |
| CNAG_04243 | Cdc24 guanine nucleotide exchange factor | 12.5 |
| CNAG_06165 | Ste20 PAK kinase | 25 |
| CNAG_04761 | Ras2 GTPase | 25 |
| CNAG_05998 | Rac2 GTPase | 25 |
| CNAG_02883 | Rac1 GTPase | 25 |
| CNAG_05925 | Septin Cdc3 | 12.5 |
| CNAG_01740 | Septin Cdc12 | 6.25 |
| CNAG_05970 | PAK kinase Pak1 | 25 |
| CNAG_03981 | Palmitoyltransferase Pfa4 | 0.39 |
MIC, minimal inhibitory concentration.
Additionally, since Pfa4-mediated palmitoylation also seems to play a role in caspofungin tolerance in C. neoformans, we assessed whether there might be synergy between caspofungin and inhibitors of palmitoyltransferases. 2BP is a competitive inhibitor of palmitoyltransferases with antifungal activity against Aspergillus fumigatus (Jennings et al. 2008; Fortwendel et al. 2012). In contrast, we found that 2BP displayed limited antifungal activity alone against C. neoformans, with an MIC of 25–50 µM. However, 2BP displayed synergy with caspofungin at both 30 and 37° (Table 4). This observed pharmacologic synergy between caspofungin and 2BP is consistent with the synthetic effect on growth inhibition between caspofungin and the pfa4Δ mutation.
Table 4. FIC indices in combination with caspofungin.
| Drug tested in combination | Combination drug MIC | FIC index (30°) | Drug relationship | Combination drug MIC | FIC index, (37°) | Drug relationship |
|---|---|---|---|---|---|---|
| Manumycin A | 5 µM | 0.508–0.75 | Synergistic | 1.25 µM | 0.562–0.75 | Synergistic |
| 2BP | 25–50 µM | 0.266–0.625 | Synergistic to strongly synergistic | 12.5 µM | 0.188–0.531 | Synergistic to strongly synergistic |
| Nikkomycin Z | 100 µg/ml | 1 | Additive | ND | ND | ND |
| Clorgyline | 25–50 µM | 0.25–0.281 | Strongly synergistic | 25–50 µM | 0.18–0.625 | Synergistic to strongly synergistic |
FIC, fractional inhibitory concentration; MIC, minimal inhibitory concentration; ND, not determined.
Since Pfa4 is responsible for the regulation of various functions within the cell, it is likely that this caspofungin sensitivity is due to dysregulation of one or more Pfa4 palmitoylation targets. Moreover, the additional growth and drug-sensitivity phenotypes at 35° are in concordance with known targets of Pfa4 being required for full thermotolerance in C. neoformans (Nichols et al. 2015). Therefore, the caspofungin susceptibility caused by mutations in several Pfa4-regulated gene products was assessed.
The thermotolerance arm of the Ras signaling pathway is required for caspofungin tolerance
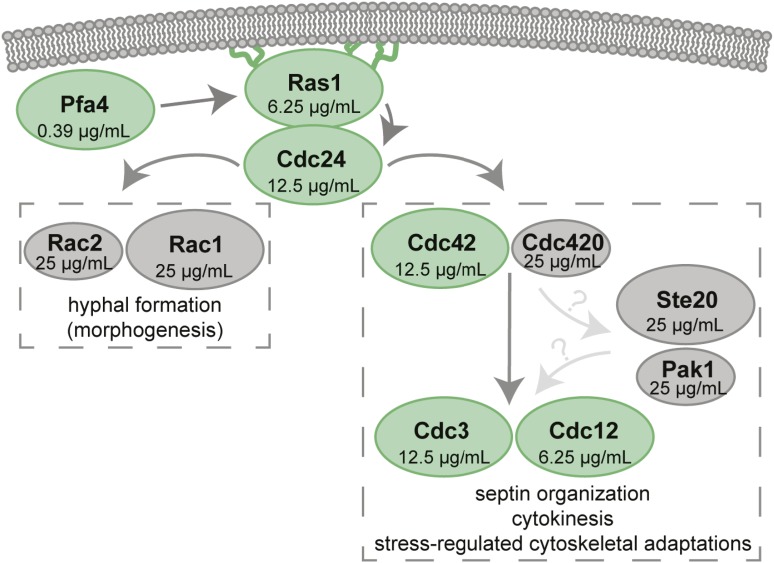
Pfa4 palmitoylates the C. neoformans Ras1 GTPase, and this post-translational modification is required for the proper subcellular localization of Ras1, as well as its function (Nichols et al. 2015). To determine whether the caspofungin susceptibility of the pfa4Δ mutant is reflected in this downstream target pathway, we assessed caspofungin susceptibility of the ras1Δ mutant, as well as for mutants in the Ras1 morphogenesis (mediated by Rac proteins) and Ras1 thermotolerance subpathways (mediated by Cdc24/Cdc42 and the septin proteins) (Figure 2) (Waugh et al. 2002; Nichols et al. 2007; Ballou et al. 2009, 2013a,b). The ras1∆ mutant is fourfold more sensitive to caspofungin than WT. Similar increases in caspofungin susceptibility were also noted for the cdc42Δ and cdc24Δ mutants, as well as the cdc3Δ and cdc12Δ septin mutants, which are further downstream effectors of the C. neoformans Ras thermotolerance pathway (Nichols et al. 2007; Ballou et al. 2009, 2013a). These proteins mediate dynamic actin cytoskeletal changes associated with budding and cell division, and a robust septin complex is required for C. neoformans to grow in the presence of cell stresses such as elevated temperature. Therefore, each of these signaling proteins is required for cryptococcal viability at elevated temperatures and other states of cell stress. Accordingly, the changes in caspofungin susceptibility in the corresponding mutant strains were only observed at 35°, not 30°, indicating that the elements of this pathway are necessary for caspofungin tolerance under conditions at which the pathway is activated (Table 2 and Table 3). In contrast, the Rac1 and Rac2 proteins are not required for caspofungin tolerance. These Ras1-mediated GTPases are involved in a distinct signaling pathway controlling morphological transitions, such as hyphal formation during mating (Ballou et al. 2013b).
Figure 2.
Ras1 signaling partner mutants are differentially affected by caspofungin treatment. Model of Ras signaling in C. neoformans. Paired paralogs are represented as the major (larger oval) and minor (smaller oval) paralog (Ballou et al. 2013a). Caspofungin minimal inhibitory concentrations at 35° for each component are presented within each oval. The green ovals represent mutants that displayed increased caspofungin susceptibility, while the gray ovals display wild-type caspofungin susceptibility.
Although specific inhibition of fungal Ras activity is limited by the highly conserved nature of this protein, recent investigations have explored inhibitors of Ras-modifying enzymes as antifungal agents (Hast et al. 2011; Selvig et al. 2013; Esher et al. 2016; Pianalto and Alspaugh 2016). In addition to palmitoylation, Ras-like GTPases also require the post-translational addition of lipophilic prenyl groups to C-terminal cysteine residues for their proper localization and function. These lipid additions are catalyzed by protein-farnesyltransferases and/or protein geranylgeranyltransferases (Palsuledesai and Distefano 2014). Additionally, protein-farnesyltransferase inhibitors (FTIs) could be affecting the function of not only Ras proteins, but other farnesylated proteins. Therefore, we assessed the pharmacological synergy between caspofungin and the FTI manumycin A (Hast et al. 2011). Manumycin A treatment has been demonstrated previously to disrupt the plasma membrane localization of GFP-Ras1 in C. neoformans, consistent with its biochemical activity as an FTI (Hast et al. 2011). In checkerboard MIC assays, manumycin A displayed synergy with caspofungin, with FIC indices ranging from 0.508 to 0.75 at 30°, and from 0.562 to 0.75 at 37° (Table 4) (Franzot and Casadevall 1997). Given the limited intrinsic antifungal activity of this first-generation FTI, these results suggest that Ras inhibitors with greater anticryptococcal activity might be promising coadministered agents to augment the effect of caspofungin against C. neoformans.
Chitin synthesis and the Chs3 chitin synthase are involved in the response to caspofungin treatment
One of the most prominent targets of Pfa4 palmitoyltransferase activity is the Chs3 chitin synthase, which is responsible for the biosynthesis of chitin that is destined to become chitosan in the cell wall (Banks et al. 2005; Baker et al. 2007). In addition to the pfa4Δ mutant strain, our screen also identified the chitin synthase regulator mutant strain csr2Δ as a highly caspofungin-sensitive strain (Table 1). Csr2 and Chs3 coregulate chitin synthesis in C. neoformans (Banks et al. 2005). Given the altered caspofungin tolerance of two mutants predicted to affect the function of the Chs3 chitin synthase, we assessed the caspofungin susceptibility of a chs3∆ strain to determine if misregulation of this protein, and a lack of proper chitin and chitosan deposition, might contribute to the caspofungin susceptibility of the pfa4Δ mutant strain. The chs3Δ mutant strain was eightfold more sensitive to caspofungin than the WT strain (Table 2). These results are consistent with observations in A. fumigatus and Candida species in which compensatory increases in cell wall chitin are induced upon exposure of these fungi to caspofungin (Walker et al. 2008, 2012; Fortwendel et al. 2010; Verwer et al. 2012). To further explore the interaction of caspofungin treatment and a response involving chitin biosynthesis in the cell wall, we used the chito-oligomer-binding dye CFW to assess changes in C. neoformans cell wall chitin content in response to caspofungin treatment. We observed a dose-dependent increase in CFW staining for cells treated with caspofungin compared to untreated cells, with the most significant difference being at the highest concentration of caspofungin (Figure 3, A and B). Accordingly, when we assessed cell wall chitin and chitosan composition using an in vitro colorimetric assay (Banks et al. 2005), we found that C. neoformans cells treated with 15 µg/ml caspofungin have an ∼2.5-fold increase in both chitin and chitosan compared to untreated cells (Figure 3C).
Figure 3.
Cell wall chitin and chitosan levels increase during CSF treatment. (A) Quantification of CFW staining of C. neoformans wild-type cells treated with CSF. Cells were incubated in SC medium at 30° with 0, 5, or 20 µg/ml CSF over 24 hr. At each timepoint, cells were harvested and stained with CFW to assess total cell wall chito-oligomer content, then imaged on a Zeiss AxioVision epifluorescence microscope. Images were masked for fluorescence and average fluorescent intensity for each cell was quantified using ImageJ. Each timepoint represents quantification of > 80 cells over at least three images. Error bars represent SEM. Statistics performed were two-way ANOVA, followed by pair-wise Student’s t-tests. * P < 0.05 and *** P < 0.001. (B) Representative images of cells treated as above with 0, 5, or 20 µg/ml CSF, and then stained for chitin content with CFW at 4 hr post-CSF treatment. (C) Quantification of cell wall chitin and chitosan using a DMAB colorimetric assay. Cells were incubated in SC ± 15 µg/ml CSF at 30° for 6 hr, then harvested and the assay performed as described. Error bars represent SEM. Statistics performed were two-way ANOVA, followed by pair-wise Student’s t-tests. * P < 0.05 and *** P < 0.001. CFW, calcofluor white; CSF, caspofungin; DMAB, p-dimethylaminobenzaldehyde; GlcNAc, N-acetylglucosamine; ns, not significant.
Since the chs3Δ chitin synthase mutant strain is so susceptible to the effects of caspofungin, we hypothesized that the coadministration of caspofungin with the chitin synthase inhibitor nikkomycin Z might result in a similar synergistic antifungal effect. We tested antifungal drug interactions using a checkerboard MIC assay, using varying concentrations of each drug in combination. In this assay, we did not observe synergy between caspofungin and nikkomycin Z, one of the few known inhibitors of chitin synthesis in some fungi (Gaughran et al. 1994; Li and Rinaldi 1999). The FIC index, a marker of drug interaction, was 1.0, suggesting an additive effect between the two compounds rather than drug synergy (Table 4). However, historically, nikkomycin Z has not been an effective inhibitor of C. neoformans growth, potentially due to the redundancy in chitin synthase genes in this organism (Li and Rinaldi 1999; Kraus et al. 2003; Banks et al. 2005). Eight chitin synthases have been identified in C. neoformans, with both distinct and overlapping functions (Banks et al. 2005). While the specific biochemical target of nikkomycin Z has yet to be identified in C. neoformans, treatment with nikkomycin Z activates the Pkc1 cell wall integrity pathway, suggesting that nikkomycin Z inhibits cell wall biosynthesis (Kraus et al. 2003). Given the very limited antifungal activity of nikkomycin Z against C. neoformans and our new genetic studies, we propose that there is potential for synergy between caspofungin and future, more effective chitin synthase inhibitors, or other cell wall biosynthesis inhibitors.
Cell wall gene expression is altered during caspofungin treatment
Given the caspofungin-induced changes in cell wall chitin content, we explored whether the expression of cell wall biosynthesis genes was altered in response to caspofungin treatment. We assessed the transcript abundance for genes involved in the synthesis of chitin (CHS1, CHS2, CHS3, CHS4, CHS5, CHS6, CHS7, and CHS8), chitosan (CDA1, CDA2, and CDA3), α-1,3-glucan (AGS1), β-1,3-glucan (FKS1), and β-1,6-glucan (KRE6 and SKN7) (Thompson et al. 1999; Banks et al. 2005; Reese et al. 2007; Gilbert et al. 2010; Esher et al. 2018). Many of these cell wall biosynthesis genes demonstrate altered regulation in response to caspofungin treatment (Figure 4). CHS1, CHS2, CHS4, CHS7, SKN1, and CDA1 all exhibited significant increases in expression, especially at the highest concentration of caspofungin tested in this experiment. These results may reflect some of the compensatory processes seen in other fungi, such as Candida species and A. fumigatus, in which increased cell wall chitin has been proposed to be a mechanism for these fungi to overcome the effects of caspofungin treatment (Walker et al. 2008; Fortwendel et al. 2010). However, we also noted the potential involvement of more diverse cell wall components. Additionally, the upregulation of SKN1 suggests a potential role for β-1,6-glucan synthesis in the caspofungin tolerance mechanism for C. neoformans. In light of previous data showing that caspofungin exposure results in a decrease in β-1,6-glucan staining via immunoelectron microscopy, there is the potential that the upregulation of β-1,6-glucan biosynthetic genes could represent transcriptional compensation for secondary cell wall effects after drug treatment (Feldmesser et al. 2000). Interestingly, CHS5, CHS6, CDA2, and CDA3 all displayed decreased expression during caspofungin treatment. Therefore, though not all cell wall-associated genes are upregulated in response to caspofungin, there seems to be a decisive alteration in the expression of cell wall biosynthesis and modification genes, reflecting a coordinated compensatory response to the cell wall inhibitor caspofungin.
Figure 4.
Cell wall biosynthesis gene expression is altered in response to CSF. Wild-type C. neoformans cells were incubated in SC medium containing 0, 10, or 15 µg/ml CSF for 90 min, followed by RNA purification and quantitative real-time PCR for the indicated target genes, using the GPD1 gene as an internal control. The results represent average values for biological triplicate samples. Statistical significance was assessed using two-way ANOVA, followed by Student’s t-test for pair-wise comparisons. * P < 0.05, ** P < 0.01, *** P < 0.001, and **** P < 0.0001. CSF, caspofungin.
Fks1-GFP localization is not altered during caspofungin treatment
Since we did not observe statistically significant differences in FKS1 transcript levels after exposure to caspofungin, we assessed whether treatment with this drug has a direct effect on the cellular localization of the β-1,3-glucan synthase. We examined the localization of the two components of the complex: the Fks1 catalytic subunit and the Rho1 regulatory subunit, each as a fusion protein tagged with GFP. We treated C. neoformans strains expressing either endogenous Fks1-GFP or overexpressed GFP-Rho1 in a gradient of caspofungin concentrations, and assessed localization over 1 hr of treatment (Figure S1). Both Fks1-GFP and GFP-Rho1 localized to endomembranes, including structures that resemble perinuclear endoplasmic reticulum staining (Pianalto et al. 2018). Similarly, both proteins also localized to the outer edge of the cell. Fks1-GFP, in particular, appeared to localize to the plasma membrane, but in distinct patches rather than along the entire cell surface. Therefore, there was no significant difference in Fks1-GFP or GFP-Rho1 localization in response to caspofungin treatment.
Inhibition of efflux pumps increases potency of caspofungin
Recent studies suggest that caspofungin is not efficiently maintained at high intracellular concentrations in C. neoformans (Huang et al. 2016). Therefore, we hypothesized that this organism might be preventing the intracellular accumulation of caspofungin through the activity of one or more efflux pumps. To address this hypothesis, we performed a drug interaction assay to determine whether caspofungin potency might be enhanced in the presence of clorgyline, a monoamine oxidase inhibitor that also acts as an inhibitor of ATP-binding cassette (ABC) transporters in other fungal species (Holmes et al. 2012). This pharmacological approach was pursued since there are > 50 predicted ABC transporters and > 150 predicted major facilitator superfamily proteins encoded in the C. neoformans genome (Loftus et al. 2005; Holmes et al. 2016). We found that the efflux inhibitor clorgyline and caspofungin demonstrated a strong synergistic interaction, with an FIC index ranging from 0.25 to 0.281 at 30° and 0.18–0.625 at 37° (Table 4). These data suggest that drug efflux might be a contributor to the innate tolerance of C. neoformans to caspofungin treatment.
Discussion
Despite the medical importance of fungal infections, few novel antifungal drugs have been introduced in recent years. Echinocandins, one of the most recently approved classes of antifungal drugs, were especially promising given their limited toxicity profile and novel mechanism of action. However, these agents display limited efficacy against several fungal classes, including the thermally dimorphic fungi and Cryptococcus species (Abruzzo et al. 1997; Bartizal et al. 1997; Espinel-Ingroff 1998).
Our screen, using a collection of C. neoformans mutant strains, revealed that cryptococcal tolerance to the echinocandin drug caspofungin is likely a multifaceted phenomenon. We identified several pathways and processes important for full tolerance of caspofungin, including those associated with cell surface integrity, such as chitin biosynthesis and the phosphatase Ppg1, or general stress tolerance, such as calcineurin. Indeed, one of the most caspofungin-sensitive mutants that we identified was a ppg1Δ mutant. Ppg1 is a phosphatase that has been shown to be necessary for full cell wall integrity, even under nonstress conditions, but that is also required for survival in cell wall stress conditions (Gerik et al. 2005). However, Ppg1 is not a part of the traditional cell wall integrity MAP kinase pathway in C. neoformans. Interestingly, we did not see hypersensitivity among mutants in the cell wall integrity MAPK pathway, which suggests that noncanonical cell wall integrity signaling could be playing a role in the response to caspofungin-induced cell wall stress (Gerik et al. 2005). We also found multiple proteins that have potential antioxidant roles, such as an Rdl2 Rhodanese homolog and a putative Lys7 homolog, which typically partners with superoxide dismutase 1 (Culotta et al. 1997; Orozco et al. 2012). We chose to focus our initial evaluation on strains with mutations in cellular pathways previously implicated in C. neoformans pathogenesis.
Another recent study assaying C. neoformans mutants for caspofungin sensitivity identified several other processes that were not identified in this study. The differences in the caspofungin-susceptible C. neoformans mutants identified in the two different studies are likely explained by variations in experimental conditions. First, the two screens used overlapping but different mutant libraries for the screens, as well as different cutoffs for defining altered caspofungin susceptibility. Also, we specifically chose to perform our screens at 30° to identify mutants that might have thermotolerance defects; for example, mutants such as the calcineurin subunit cnb1Δ mutant would not have been discovered at a higher temperature given its intrinsic inability to grow at higher temperatures.
In their study, Huang et al. determined that mutation of the Cdc50 lipid flippase β-subunit increased the sensitivity of this organism to caspofungin and fluconazole (Huang et al. 2016). By using a fluorescently labeled caspofungin molecule, they also observed that the cdc50Δ mutant strain displayed increased drug uptake compared to WT strains, suggesting that the altered membrane integrity and lipid content in the mutant strain might allow better penetration of the drug into the cell, and, therefore, higher efficacy. This observation implies that overcoming limitations of entry into or accumulation of the drug in the cryptococcal cell might enhance fungal cell killing. To further explore the importance of drug efflux, we assessed the effect of the efflux pump inhibitor clorgyline on caspofungin activity, and we found strong synergy between these two compounds. We specifically employed clorgyline as a pharmacological inhibitor of ABC transporters to address the potential functional redundancy of the large number of C. neoformans genes likely to encode efflux pumps (Loftus et al. 2005; Holmes et al. 2016).
In validation of our screening methods, we identified the calcineurin B subunit mutant as a hypersensitive strain. Calcineurin signaling is required for full virulence of C. neoformans and is involved in the response to elevated temperatures, as well as the response to caspofungin and other cellular stresses (Odom et al. 1997; Cruz et al. 2000; Del Poeta et al. 2000; Fox et al. 2001). Similarly, calcineurin signaling has been shown to be required for paradoxical growth in the presence of high concentrations of caspofungin in Candida species and A. fumigatus (Walker et al. 2008, 2012; Fortwendel et al. 2010; Lee et al. 2011; Juvvadi et al. 2015). In C. neoformans, calcineurin is known to regulate one transcription factor, Crz1, and many calcineurin-dependent cellular processes are likely mediated by Crz1 (Lev et al. 2012). Importantly, recent work identified several genes involved in cell wall biosynthesis and remodeling that are transcriptionally upregulated by Crz1 and Cna1 in response to thermal stress (Chow et al. 2017). However, this work also revealed that a significant number of genes are regulated differentially between the cna1Δ and the crz1Δ transcriptomes, suggesting that other targets of Cna1 activity could have effects on the transcriptional response to stress. Additional work demonstrated that calcineurin likely has multiple downstream targets in addition to Crz1, including proteins that localize to stress granules and P-bodies, and are involved in post-transcriptional responses to thermal stress (Park et al. 2016). Our work suggests that the calcineurin-regulated response to caspofungin likely involves both the activity of the Crz1 transcription factor as well as Crz1-independent calcineurin targets. Indeed, it has also been demonstrated in C. albicans that Crz1 is only partially responsible for the calcineurin-mediated echinocandin tolerance seen in that organism (Singh et al. 2009). Future work could probe Crz1-independent calcineurin targets for roles in caspofungin tolerance in this organism.
We also tested multiple processes regulated by the Pfa4 palmitoyltransferase to explore whether the drug-susceptible phenotype of the palmitoyltransferase mutant could be attributed to dysfunction of one or more of its downstream targets. Interestingly, we determined that the C. neoformans Ras1-mediated thermotolerance pathway is required for tolerance to caspofungin treatment in this organism. This signaling pathway responds to extracellular cues to control the activation of the Cdc42 GTPase, a protein that directs actin cytoskeleton polarization as well as directional growth and budding (Ballou et al. 2009). The septin proteins Cdc3 and Cdc12 are required for cytokinesis through their role in septum formation. Additionally, Cdc3 and Cdc12 are necessary for normal cell morphogenesis at elevated temperatures, and the localization of these proteins is mediated by Cdc42. In the absence of any of these pathway proteins, the cryptococcal cell is growth-impaired under many stressful conditions, such as mammalian physiological temperatures (Nichols et al. 2007; Ballou et al. 2009, 2013a). Although mutants of genes encoding two members of this pathway, RAS1 and CDC42, were present in the collections tested in this study, their growth phenotypes are only apparent at elevated temperatures. As a result, these mutants would not be expected to display caspofungin susceptibility at the lower temperatures tested, emphasizing the importance of screening for drug sensitivity in multiple incubation conditions.
Since Ras and Ras-related proteins are required for pathogenesis in many fungal systems, there have been concerted efforts to identify small molecules that inhibit Ras protein function. One of the most promising directions includes targeting protein prenyltransferases, enzymes that add lipid moieties to the C-terminal regions of Ras-like GTPases, directing the localization of these proteins to cellular membranes, which are their sites of function (Vallim et al. 2004; Nichols et al. 2009; Fortwendel et al. 2012; Esher et al. 2016). Prenylation inhibitors are predicted to alter the localization and function of many cellular proteins in addition to Ras, potentially having broad antiproliferative effects in divergent cell types. Structural studies have identified fungal-specific features of farnesyltransferase enzymes, suggesting that antifungal specificity could be engineered into FTIs (Hast et al. 2011; Mabanglo et al. 2014). Antifungal susceptibility testing demonstrated in vitro synergy between the activities of caspofungin and the FTI manumycin A, the basis of which was identified in our screen. Importantly, the currently available FTIs tested have limited efficacy as antifungal compounds when used alone. However, as newer and more potent antifungal FTIs are identified, these agents might be used as adjunctive therapies to enhance the effect of caspofungin.
We also determined that chitin biosynthesis is upregulated during caspofungin treatment and that the Chs3 chitin synthase is required for caspofungin tolerance. As caspofungin inhibits the biosynthesis of the cell wall component β-1,3-glucan, upregulation of other cell wall biosynthesis or cell wall-modifying enzymes may compensate for altered glucan content. Indeed, in A. fumigatus, cell wall chitin deposition, as well as the expression of chitin synthase genes, increases during caspofungin treatment (Fortwendel et al. 2009, 2010). Additionally, Candida clinical isolates with naturally increased levels of chitin, as well as strains grown in chitin-inducing conditions, demonstrate increased survival during caspofungin treatment (Walker et al. 2008, 2012; Lee et al. 2011). A similar phenomenon appears to be occurring in C. neoformans: during caspofungin treatment, we measured increased levels of chitin both by CFW staining and by an in vitro biochemical quantification of chitin monomers. There is a similar increase in chitosan levels. As Cryptococcus species typically display higher levels of chitosan in the cell wall than other pathogenic fungal species (Banks et al. 2005), these inducible cell wall changes in chitin/chitosan content likely represent a conserved mechanism by which C. neoformans adapts to caspofungin treatment. Additionally, the increased baseline chitosan levels in the C. neoformans cell wall may also contribute to its innate tolerance to this drug.
In conclusion, several varied cellular processes were represented among the strains with enhanced caspofungin susceptibility. It is possible that the loss of some of these processes enhances the intracellular accumulation of the drug, preventing the low concentrations of caspofungin in C. neoformans cells suggested in prior studies. Alternatively, compromising these cellular pathways may render the C. neoformans cell susceptible to lower effective concentrations of this drug. As new fungal-specific inhibitors are developed for processes such as Ras protein localization and chito-oligomer synthesis, these agents may provide promising new directions for combination antifungal therapy.
Acknowledgments
The authors would like to thank the Madhani laboratory and National Institutes of Health (NIH) funding R01 AI-100272 for the deletion mutant collection in C. neoformans, and Joseph Heitman for support of and insight into this project. We dedicate this paper to the memory of Patricia J. Pukkila, a committed scientist, wonderful mentor, and long-time editor of Genetics. This work was supported by NIH grants R01 AI-074677 (J.A.A.) and P01 AI-104533, as well as a National Defense Science and Engineering grant to K.M.P. awarded by the Department of Defense, Office of Scientific Research, 32 CFR 168a. K.M.P. was also supported by a Duke University Bass Instructional Fellowship.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8085290.
Communicating editor: A. Gladfelter
Literature Cited
- Abruzzo G. K., Flattery A. M., Gill C. J., Kong L., Smith J. G., et al. , 1997. Evaluation of the echinocandin antifungal MK-0991 (L-743,872): efficacies in mouse models of disseminated aspergillosis, candidiasis, and cryptococcosis. Antimicrob. Agents Chemother. 41: 2333–2338. 10.1128/AAC.41.11.2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker L. G., Specht C. A., Donlin M. J., Lodge J. K., 2007. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot. Cell 6: 855–867. 10.1128/EC.00399-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou E. R., Nichols C. B., Miglia K. J., Kozubowski L., Alspaugh J. A., 2009. Two CDC42 paralogues modulate Cryptococcus neoformans thermotolerance and morphogenesis under host physiological conditions. Mol. Microbiol. 75: 763–780. 10.1111/j.1365-2958.2009.07019.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou E. R., Kozubowski L., Nichols C. B., Alspaugh J. A., 2013a. Ras1 Acts through duplicated Cdc42 and Rac proteins to regulate morphogenesis and pathogenesis in the human fungal pathogen Cryptococcus neoformans. PLoS Genet. 9: e1003687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou E. R., Selvig K., Narloch J. L., Nichols C. B., Alspaugh J. A., 2013b. Two Rac paralogs regulate polarized growth in the human fungal pathogen Cryptococcus neoformans. Fungal Genet. Biol. 57: 58–75. 10.1016/j.fgb.2013.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks I. R., Specht C. A., Donlin M. J., Gerik K. J., Levitz S. M., et al. , 2005. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryot. Cell 4: 1902–1912. 10.1128/EC.4.11.1902-1912.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartizal K., Gill C. J., Abruzzo G. K., Flattery A. M., Kong L., et al. , 1997. In vitro preclinical evaluation studies with the echinocandin antifungal MK-0991 (L-743,872). Antimicrob. Agents Chemother. 41: 2326–2332. 10.1128/AAC.41.11.2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow E. W. L., Clancey S. A., Billmyre R. B., Averette A. F., Granek J. A., et al. , 2017. Elucidation of the calcineurin-Crz1 stress response transcriptional network in the human fungal pathogen Cryptococcus neoformans. PLoS Genet. 13: e1006667 10.1371/journal.pgen.1006667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun C. D., Madhani H. D., 2010. Applying genetics and molecular biology to the study of the human pathogen Cryptococcus neoformans. Methods Enzymol. 470: 797–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute, 2008 Reference method for broth dilution antifungal susceptibility testing of yeasts; Approved Standard–Third Edition. CLSI document M27–A3. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- Cruz M. C., Sia R. A. L., Olson M., Cox G. M., Heitman J., 2000. Comparison of the roles of calcineurin in physiology and virulence in serotype D and serotype A strains of Cryptococcus neoformans. Infect. Immun. 68: 982–985. 10.1128/IAI.68.2.982-985.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culotta V. C., Klomp L. W. J., Strain J., Casareno R. L. B., Krems B., et al. , 1997. The copper chaperone for superoxide dismutase. J. Biol. Chem. 272: 23469–23472. 10.1074/jbc.272.38.23469 [DOI] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R. C., Blankenship J. R., Kraus P. R., de Jesus Berrios M., Hull C. M., et al. , 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148: 2607–2615. 10.1099/00221287-148-8-2607 [DOI] [PubMed] [Google Scholar]
- Del Poeta M., Cruz M. C., Cardenas M. E., Perfect J. R., Heitman J., 2000. Synergistic antifungal activities of bafilomycin A1, fluconazole, and the pneumocandin MK-0991/caspofungin acetate (L-743,873) with calcineurin inhibitors FK506 and L-685,818 against Cryptococcus neoformans. Antimicrob. Agents Chemother. 44: 739–746. 10.1128/AAC.44.3.739-746.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D. W., 2003. Echinocandin antifungal drugs. Lancet 362: 1142–1151. 10.1016/S0140-6736(03)14472-8 [DOI] [PubMed] [Google Scholar]
- Doering T. L., 2009. How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans. Annu. Rev. Microbiol. 63: 223–247. 10.1146/annurev.micro.62.081307.162753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esher S. K., Ost K. S., Kozubowski L., Yang D.-H., Kim M. S., et al. , 2016. Relative contributions of prenylation and postprenylation processing in Cryptococcus neoformans pathogenesis. mSphere 1: e00084-15 10.1128/mSphere.00084-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esher S. K., Ost K. S., Kohlbrenner M. A., Pianalto K. M., Telzrow C. L., et al. , 2018. Defects in intracellular trafficking of fungal cell wall synthases lead to aberrant host immune recognition. PLoS Pathog. 14: e1007126 10.1371/journal.ppat.1007126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinel-Ingroff A., 1998. Comparison of in vitro activities of the new triazole SCH56592 and the echinocandins MK-0991 (L-743,872) and LY303366 against opportunistic filamentous and dimorphic fungi and yeasts. J. Clin. Microbiol. 36: 2950–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmesser M., Kress Y., Mednick A., Casadevall A., 2000. The effect of the echinocandin analogue caspofungin on cell wall glucan synthesis by Cryptococcus neoformans. J. Infect. Dis. 182: 1791–1795. 10.1086/317614 [DOI] [PubMed] [Google Scholar]
- Fortwendel J. R., Juvvadi P. R., Pinchai N., Perfect B. Z., Alspaugh J. A., et al. , 2009. Differential effects of inhibiting chitin and 1,3-β-D-glucan synthesis in Ras and calcineurin mutants of Aspergillus fumigatus. Antimicrob. Agents Chemother. 53: 476–482. 10.1128/AAC.01154-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortwendel J. R., Juvvadi P. R., Perfect B. Z., Rogg L. E., Perfect J. R., et al. , 2010. Transcriptional regulation of chitin synthases by calcineurin controls paradoxical growth of Aspergillus fumigatus in response to caspofungin. Antimicrob. Agents Chemother. 54: 1555–1563. 10.1128/AAC.00854-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortwendel J. R., Juvvadi P. R., Rogg L. E., Asfaw Y. G., Burns K. A., et al. , 2012. Plasma membrane localization is required for RasA-mediated polarized morphogenesis and virulence of Aspergillus fumigatus. Eukaryot. Cell 11: 966–977. 10.1128/EC.00091-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox D. S., Cruz M. C., Sia R. A. L., Ke H., Cox G. M., et al. , 2001. Calcineurin regulatory subunit is essential for virulence and mediates interactions with FKBP12–FK506 in Cryptococcus neoformans. Mol. Microbiol. 39: 835–849. 10.1046/j.1365-2958.2001.02295.x [DOI] [PubMed] [Google Scholar]
- Franzot S. P., Casadevall A., 1997. Pneumocandin L-743,872 enhances the activities of amphotericin B and fluconazole against Cryptococcus neoformans in vitro. Antimicrob. Agents Chemother. 41: 331–336. 10.1128/AAC.41.2.331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughran J. P., Lai M. H., Kirsch D. R., Silverman S. J., 1994. Nikkomycin Z is a specific inhibitor of Saccharomyces cerevisiae chitin synthase isozyme Chs3 in vitro and in vivo. J. Bacteriol. 176: 5857–5860. 10.1128/jb.176.18.5857-5860.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerik K. J., Donlin M. J., Soto C. E., Banks A. M., Banks I. R., et al. , 2005. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol. Microbiol. 58: 393–408. 10.1111/j.1365-2958.2005.04843.x [DOI] [PubMed] [Google Scholar]
- Gilbert N. M., Donlin M. J., Gerik K. J., Specht C. A., Djordjevic J. T., et al. , 2010. KRE genes are required for β-1,6-glucan synthesis, maintenance of capsule architecture and cell wall protein anchoring in Cryptococcus neoformans. Mol. Microbiol. 76: 517–534. 10.1111/j.1365-2958.2010.07119.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow N. A. R., Hube B., 2012. Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 15: 406–412. 10.1016/j.mib.2012.04.005 [DOI] [PubMed] [Google Scholar]
- Hast M. A., Nichols C. B., Armstrong S. M., Kelly S. M., Hellinga H. W., et al. , 2011. Structures of Cryptococcus neoformans protein farnesyltransferase reveal strategies for developing inhibitors that target fungal pathogens. J. Biol. Chem. 286: 35149–35162. 10.1074/jbc.M111.250506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A. R., Keniya M. V., Ivnitski-Steele I., Monk B. C., Lamping E., et al. , 2012. The monoamine oxidase A inhibitor clorgyline is a broad-spectrum inhibitor of fungal ABC and MFS transporter efflux pump activities which reverses the azole resistance of Candida albicans and Candida glabrata clinical isolates. Antimicrob. Agents Chemother. 56: 1508–1515. 10.1128/AAC.05706-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A. R., Cardno T. S., Strouse J. J., Ivnitski-Steele I., Keniya M. V., et al. , 2016. Targeting efflux pumps to overcome antifungal drug resistance. Future Med. Chem. 8: 1485–1501. 10.4155/fmc-2016-0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Liao G., Baker G. M., Wang Y., Lau R., et al. , 2016. Lipid flippase subunit Cdc50 mediates drug resistance and virulence in Cryptococcus neoformans. MBio 7: e00478-16 10.1128/mBio.00478-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon G., Ormerod K. L., Paulet D., Byrnes E. J., III, Yadav V., et al. , 2014. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 10: e1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings B. C., Nadolski M. J., Ling Y., Baker M. B., Harrison M. L., et al. , 2008. 2-Bromopalmitate and 2-(2-hydroxy-5-nitro-benzylidene)-benzo[b]thiophen-3-one inhibit DHHC-mediated palmitoylation in vitro. J. Lipid Res. 50: 233–242. 10.1194/jlr.M800270-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessup C. J., Pfaller M. A., Messer S. A., Zhang J., Tumberland M., et al. , 1998. Fluconazole susceptibility testing of Cryptococcus neoformans: comparison of two broth microdilution methods and clinical correlates among isolates from Ugandan AIDS patients. J. Clin. Microbiol. 36: 2874–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juvvadi P. R., Muñoz A., Lamoth F., Soderblom E. J., Moseley M. A., et al. , 2015. Calcium-mediated induction of paradoxical growth following caspofungin treatment is associated with calcineurin activation and phosphorylation in Aspergillus fumigatus. Antimicrob. Agents Chemother. 59: 4946–4955. 10.1128/AAC.00263-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M. S., S. Y. Kim, K. W. Jung, and Y. S. Bahn, 2012 Targeted gene disruption in Cryptococcus neoformans using double-joint PCR with split dominant selectable markers. Methods Mol. Biol. 845: 67–84. [DOI] [PubMed] [Google Scholar]
- Kraus P. R., Fox D. S., Cox G. M., Heitman J., 2003. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol. Microbiol. 48: 1377–1387. 10.1046/j.1365-2958.2003.03508.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz M. B., Douglas C. M., 1997. Lipopeptide inhibitors of fungal glucan synthase. J. Med. Vet. Mycol. 35: 79–86. 10.1080/02681219780000961 [DOI] [PubMed] [Google Scholar]
- Kurtz M. B., Heath I. B., Marrinan J., Dreikorn S., Onishi J., et al. , 1994. Morphological effects of lipopeptides against Aspergillus fumigatus correlate with activities against (1,3)-β-D-glucan synthase. Antimicrob. Agents Chemother. 38: 1480–1489. 10.1128/AAC.38.7.1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latgé J.-P., 2007. The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 66: 279–290. 10.1111/j.1365-2958.2007.05872.x [DOI] [PubMed] [Google Scholar]
- Lee K. K., MacCallum D. M., Jacobsen M. D., Walker L. A., Odds F. C., et al. , 2011. Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob. Agents Chemother. 56: 208–217. 10.1128/AAC.00683-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letscher-Bru V., Herbrecht R., 2003. Caspofungin: the first representative of a new antifungal class. J. Antimicrob. Chemother. 51: 513–521. 10.1093/jac/dkg117 [DOI] [PubMed] [Google Scholar]
- Lev S., Desmarini D., Chayakulkeeree M., Sorrell T. C., Djordjevic J. T., 2012. The Crz1/Sp1 transcription factor of Cryptococcus neoformans is activated by calcineurin and regulates cell wall integrity. PLoS One 7: e51403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. K., Rinaldi M. G., 1999. In vitro antifungal activity of nikkomycin Z in combination with fluconazole or itraconazole. Antimicrob. Agents Chemother. 43: 1401–1405. 10.1128/AAC.43.6.1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu O. W., Chun C. D., Chow E. D., Chen C., Madhani H. D., et al. , 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135: 174–188. 10.1016/j.cell.2008.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus B. J., Fung E., Roncaglia P., Rowley D., Amedeo P., et al. , 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307: 1321–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyse A., Thangaraj H., Easterbrook P., Ford N., Roy M., et al. , 2013. Cryptococcal meningitis: improving access to essential antifungal medicines in resource-poor countries. Lancet Infect. Dis. 13: 629–637. 10.1016/S1473-3099(13)70078-1 [DOI] [PubMed] [Google Scholar]
- Mabanglo M. F., Hast M. A., Lubock N. B., Hellinga H. W., Beese L. S., 2014. Crystal structures of the fungal pathogen Aspergillus fumigatus protein farnesyltransferase complexed with substrates and inhibitors reveal features for antifungal drug design. Protein Sci. 23: 289–301. 10.1002/pro.2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maligie M. A., Selitrennikoff C. P., 2005. Cryptococcus neoformans resistance to echinocandins: (1,3) β-glucan synthase activity is sensitive to echinocandins. Antimicrob. Agents Chemother. 49: 2851–2856. 10.1128/AAC.49.7.2851-2856.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Committee for Clinical Laboratory Standards, 1992 Reference Method for Broth Dilution Antifungal Susceptibility Testing for Yeasts: Proposed Standard M27-P. National Committee for Clinical Laboratory Standards, Villanova, PA. [Google Scholar]
- Nichols C. B., Perfect Z. H., Alspaugh J. A., 2007. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 63: 1118–1130. 10.1111/j.1365-2958.2006.05566.x [DOI] [PubMed] [Google Scholar]
- Nichols C. B., Ferreyra J., Ballou E. R., Alspaugh J. A., 2009. Subcellular localization directs signaling specificity of the Cryptococcus neoformans Ras1 protein. Eukaryot. Cell 8: 181–189. 10.1128/EC.00351-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols C. B., Ost K. S., Grogan D. P., Pianalto K., Hasan S., et al. , 2015. Impact of protein palmitoylation on the virulence potential of Cryptococcus neoformans. Eukaryot. Cell 14: 626–635. 10.1128/EC.00010-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom A., Muir S., Lim E., Toffaletti D. L., Perfect J., et al. , 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 16: 2576–2589. 10.1093/emboj/16.10.2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara T. R., Holmer S. M., Selvig K., Dietrich F., Alspaugh J. A., 2013. Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. MBio 4: e00522-12 10.1128/mBio.00522-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco H., Matallana E., Aranda A., 2012. Oxidative stress tolerance, adenylate cyclase, and autophagy are key players in the chronological life span of Saccharomyces cerevisiae during winemaking. Appl. Environ. Microbiol. 78: 2748–2757. 10.1128/AEM.07261-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ost K. S., Esher S. K., Leopold Wager C. M., Walker L., Wagener J., et al. , 2017. Rim pathway-mediated alterations in the fungal cell wall influence immune recognition and inflammation. MBio 8: e02290-16 10.1128/mBio.02290-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsuledesai C. C., Distefano M. D., 2014. Protein prenylation: enzymes, therapeutics, and biotechnology applications. ACS Chem. Biol. 10: 51–62. 10.1021/cb500791f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B. J., Wannemuehler K. A., Marston B. J., Govender N., Pappas P. G., et al. , 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23: 525–530. 10.1097/QAD.0b013e328322ffac [DOI] [PubMed] [Google Scholar]
- Park H.-S., Chow E. W. L., Fu C., Soderblom E. J., Moseley M. A., et al. , 2016. Calcineurin targets involved in stress survival and fungal virulence. PLoS Pathog. 12: e1005873 10.1371/journal.ppat.1005873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect J. R., Bicanic T., 2014. Cryptococcosis diagnosis and treatment: what do we know now. Fungal Genet. Biol. 78: 49–54. 10.1016/j.fgb.2014.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect J. R., Lang S. D. R., Durack D. T., 1980. Chronic cryptococcal meningitis: a new experimental model in rabbits. Am. J. Pathol. 101: 177–194. [PMC free article] [PubMed] [Google Scholar]
- Pfaller M. A., Rinaldi M. G., Galgiani J. N., Bartlett M. S., Body B. A., et al. , 1990. Collaborative investigation of variables in susceptibility testing of yeasts. Antimicrob. Agents Chemother. 34: 1648–1654. 10.1128/AAC.34.9.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianalto K. M., Alspaugh J. A., 2016. New horizons in antifungal therapy. J. Fungi (Basel) 2: 26 10.3390/jof2040026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pianalto K. M., Ost K. S., Brown H. E., Alspaugh J. A., 2018. Characterization of additional components of the environmental pH-sensing complex in the pathogenic fungus Cryptococcus neoformans. J. Biol. Chem. 293: 9995–10008. 10.1074/jbc.RA118.002741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrgos V., Seitz A. E., Steiner C. A., Prevots D. R., Williamson P. R., 2013. Epidemiology of Cryptococcal Meningitis in the US: 1997–2009. PLoS One 8: e56269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasingham R., Smith R. M., Park B. J., Jarvis J. N., Govender N. P., et al. , 2017. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect. Dis. 17: 873–881. 10.1016/S1473-3099(17)30243-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese A. J., Yoneda A., Breger J. A., Beauvais A., Liu H., et al. , 2007. Loss of cell wall alpha(1–3) glucan affects Cryptococcus neoformans from ultrastructure to virulence. Mol. Microbiol. 63: 1385–1398. 10.1111/j.1365-2958.2006.05551.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Tirado F. H., Peng T., Yang M., Hang H. C., Doering T. L., 2015. A single protein S-acyl transferase acts through diverse substrates to determine Cryptococcal morphology, stress tolerance, and pathogenic outcome. PLoS Pathog. 11: e1004908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvig K., Ballou E. R., Nichols C. B., Alspaugh J. A., 2013. Restricted substrate specificity for the geranylgeranyltransferase-I enzyme in Cryptococcus neoformans: implications for virulence. Eukaryot. Cell 12: 1462–1471. 10.1128/EC.00193-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia E. A., Kirkpatrick D. T., 2005. The yeast MSH1 gene is not involved in DNA repair or recombination during meiosis. DNA Repair (Amst.) 4: 253–261. 10.1016/j.dnarep.2004.10.002 [DOI] [PubMed] [Google Scholar]
- Singh S. D., Robbins N., Zaas A. K., Schell W. A., Perfect J. R., et al. , 2009. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 5: e1000532 10.1371/journal.ppat.1000532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich J. E., Harris T., Brunk B. P., Brestelli J., Fischer S., et al. , 2012. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res. 40: D675–D681. 10.1093/nar/gkr918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft C. S., Stark T., Selitrennikoff C. P., 1988. Cilofungin (LY121019) inhibits Candida albicans (1–3)-β-D-glucan synthase activity. Antimicrob. Agents Chemother. 32: 1901–1903. 10.1128/AAC.32.12.1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. R., Douglas C. M., Li W., Jue C. K., Pramanik B., et al. , 1999. A glucan synthase FKS1 homolog in Cryptococcus neoformans is single copy and encodes an essential function. J. Bacteriol. 181: 444–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffaletti D. L., Rude T. H., Johnston S. A., Durack D. T., Perfect J. R., 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175: 1405–1411. 10.1128/jb.175.5.1405-1411.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallim M. A., Fernandes L., Alspaugh J. A., 2004. The RAM1 gene encoding a protein-farnesyltransferase β-subunit homologue is essential in Cryptococcus neoformans. Microbiology 150: 1925–1935. 10.1099/mic.0.27030-0 [DOI] [PubMed] [Google Scholar]
- Verwer P. E. B., van Duijn M. L., Tavakol M., Bakker-Woudenberg I. A. J. M., van de Sande W. W. J., 2012. Reshuffling of Aspergillus fumigatus cell wall components chitin and β-Glucan under the influence of caspofungin or nikkomycin Z alone or in combination. Antimicrob. Agents Chemother. 56: 1595–1598. 10.1128/AAC.05323-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L. A., Munro C. A., de Bruijn I., Lenardon M. D., McKinnon A., et al. , 2008. Stimulation of chitin synthesis rescues Candida albicans from Echinocandins. PLoS Pathog. 4: e1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L. A., Gow N. A. R., Munro C. A., 2012. Elevated chitin content reduces the susceptibility of Candida species to caspofungin. Antimicrob. Agents Chemother. 57: 146–154. 10.1128/AAC.01486-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh M. S., Nichols C. B., DeCesare C. M., Cox G. M., Heitman J., et al. , 2002. Ras1 and Ras2 contribute shared and unique roles in physiology and virulence of Cryptococcus neoformans. Microbiology 148: 191–201. 10.1099/00221287-148-1-191 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. File S1 contains a list and descriptions of all supplemental files. Sequence data are available at the NCBI Sequence Read Archive under the accession number PRJNA501913. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.8085290