Abstract
Streptococcus pneumoniae (pneumococcus, Spn) colonizes the human nasopharynx asymptomatically but can cause infections such as otitis media, and invasive pneumococcal disease such as community-acquired pneumonia, meningitis, and sepsis. Although the success of Spn as a pathogen can be attributed to its ability to synthesize and regulate capsular polysaccharide (CPS) for survival in the host, the mechanisms of CPS regulation are not well-described. Recent studies from our lab demonstrate that deletion of a putative polyamine biosynthesis gene (ΔcadA) in Spn TIGR4 results in the loss of the capsule. In this study, we characterized the transcriptome and metabolome of ΔcadA and identified specific mechanisms that could explain the regulatory role of polyamines in pneumococcal CPS biosynthesis. Our data indicate that impaired polyamine synthesis impacts galactose to glucose interconversion via the Leloir pathway which limits the availability of UDP-galactose, a precursor of serotype 4 CPS, and UDP-N-acetylglucosamine (UDP-GlcNAc), a nucleotide sugar precursor that is at the intersection of CPS and peptidoglycan repeat unit biosynthesis. Reduced carbon flux through glycolysis, coupled with altered fate of glycolytic intermediates further supports impaired synthesis of UDP-GlcNAc. A significant increase in the expression of transketolases indicates a potential shift in carbon flow toward the pentose phosphate pathway (PPP). Higher PPP activity could constitute oxidative stress responses in ΔcadA which warrants further investigation. The results from this study clearly demonstrate the potential of polyamine synthesis, targeted for cancer therapy in human medicine, for the development of novel prophylactic and therapeutic strategies for treating bacterial infections.
Keywords: Streptococcus pneumoniae, polyamines, capsule, Leloir pathway, glycolysis, peptidoglycan, pentose phosphate pathway
Introduction
Streptococcus pneumoniae (pneumococcus) is a Gram-positive bacterial pathogen that can cause infections, such as sinusitis, otitis media, meningitis, septicemia, and most commonly, community-acquired pneumonia (Musher, 1992; Greenwood, 1999). Phase variation enables Spn to modulate CPS expression during nasopharyngeal colonization or invasion of sterile sites (Manso et al., 2014; Li J. et al., 2016). The pneumococcal capsule interferes with opsonization and phagocytosis, and is also the basis for classification of nearly 100 identified serotypes (Geno et al., 2015). Capsule biosynthesis in pneumococcus generally follows the canonical Wzy-dependent pathway, which is common to the majority of the serotypes including serotype 4 (TIGR4) which is the focus of this study. In Wzy-dependent mechanism, CPS repeat units are built on the inner side of the cytoplasmic membrane, transported to the outer side by a flippase, further polymerized by a Wzy polymerase, and covalently linked to the cell wall PG (Eberhardt et al., 2012). Although pneumococcal capsules are well characterized, the description of molecular mechanisms that regulate CPS biosynthesis is limited.
The capsule biosynthesis locus is located between the dexB and aliA genes in the pneumococcal genome and constitutes a single operon with genes cpsA-O, depending on serotype. Transcriptional regulation of CPS expression is reported to involve the first four genes cpsA–cpsD of the cps locus (Morona et al., 2004; Hanson et al., 2011; Ghosh et al., 2018), the promoter sequence upstream of cpsA (Wen et al., 2015), and competence protein (ComE) (Zheng et al., 2017). In addition to transcriptional regulation, recent studies identified effects on central metabolism that impact CPS. Uracil deprivation and impaired pyruvate oxidase, an important enzyme in the production of acetyl-CoA, have both been shown to result in reduced CPS (Echlin et al., 2016; Carvalho et al., 2018). An arginine/ornithine antiporter, arcD, has also been reported to regulate CPS biosynthesis by unknown mechanisms (Gupta et al., 2013). A comprehensive description of the regulatory framework for the expression of this critical virulence factor in pneumococci is still a work in progress.
Polyamines such as spermidine, putrescine, and cadaverine are ubiquitous, polycationic, aliphatic hydrocarbons with amino groups that regulate a number of cellular processes (Bae et al., 2018). We have shown that isogenic deletion of cadA, a gene that encodes a putative lysine decarboxylase that catalyzes the conversion of lysine to cadaverine resulted in an attenuated phenotype in murine models of pneumococcal colonization, pneumonia, and sepsis (Shah et al., 2011). Characterization of S. pneumoniae TIGR4 ΔcadA showed a loss of the capsule (Nakamya et al., 2018), which in part, could be due to transcriptional downregulation of cps4A, the first gene in the cps operon. Expression proteomics analysis of ΔcadA indicated a shift in central metabolism that could limit the availability of precursors for CPS synthesis. However, the limited proteome coverage failed to identify specific molecular mechanisms of CPS regulation.
To identify pneumococcal pathways responsive to polyamines that regulate CPS synthesis, we compared the transcriptomic and metabolomic profiles of S. pneumoniae TIGR4 and ΔcadA strains. Our results show that the ability to convert UDP-Glu to UDP-Gal through Leloir pathway was reduced in ΔcadA. Observed changes in the expression of genes in the amino and nucleotide sugar metabolism are expected to result in reduced intracellular levels of UDP-GlcNAc, a precursor for three N-acetylated sugars in the serotype 4 capsule repeat unit. Impaired Leloir pathway and UDP-GlcNAc synthesis will limit the availability of CPS precursors. Changes in the metabolism in ΔcadA, such as reduced glycolytic activity, altered UDP-GlcNAc metabolism and lysine synthesis could impact the assembly of PG and ultimately the cell wall which provides attachment for the capsule. In summary, using RNA-Seq and metabolomics, we identified specific mechanisms in polyamine synthesis deficient pneumococci, which could limit the availability of precursors for CPS and PG synthesis. This study is the first report on the impact of altered polyamine metabolism, often reported as a target for cancer therapy in human medicine, on bacterial pathogenesis. The results from this study clearly demonstrate the potential of targeting polyamine synthesis in bacteria for the development novel prophylactic and therapeutic strategies.
Materials and Methods
Bacterial Strains and Growth Conditions
Streptococcus pneumoniae serotype 4 strain TIGR4 and ΔcadA were used in this study. All strains were grown in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY) or on 5% sheep blood agar plates (BAP) in 5% CO2. Generation of ΔcadA and initial characterization of CPS are described elsewhere (Nakamya et al., 2018). All strains were grown to mid-log phase (O.D600 nm 0.4) and cells were pelleted, for extraction of total RNA and metabolites. Colony forming units (CFUs) were enumerated to ensure comparable number of working cells.
RNA Sequencing
Total RNA was isolated and purified from mid-log phase cultures of TIGR4 and ΔcadA grown in THY (n = 3) using the RNeasy Midi Kit and QIAcube (Qiagen, Valencia, CA, United States). The quality was analyzed by an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). Libraries for RNA-Seq were prepared with the KAPA RNA Hyper Kit with RiboErase (KAPA Biosystem, Wilmington, MA, United States) with 5 μg RNA as input. The workflow consists of rRNA removal, cDNA generation, end repair to generate blunt ends, A-tailing, adaptor ligation, and PCR amplification. Different adaptors were used for multiplexing samples in one sequencing run. Library concentrations and quality were measured using Qubit ds DNA HS Assay Kit (Life Technologies, Carlsbad, CA, United States) and Agilent Tapestation (Agilent Technologies, Santa Clara, CA, United States). Sequencing was performed on an Illumina Hiseq 3000 for a single read 50 run. Data quality check was done on Illumina SAV. De-multiplexing was performed with Illumina Bcl2fastq2 v 2.17. Analysis to remove failed reads, mapping of the short sequence reads to Spn TIGR4 reference genome, and identification of differentially expressed genes were performed with RNA-Seq tool of CLC Genomic Workbench 11.0.1 (Qiagen, Valencia, CA, United States). Briefly, single end reads of both wild type and mutant were mapped to the S. pneumoniae TIGR4 genome using CLC proprietary read mapper, counted with EM estimation algorithm, and differentially expressed genes were identified based on the fold change generated by the edgeR algorithm. Fold change with false discovery rate (FDR) ≤ 0.05 was accepted to be significant. RNA-Seq raw data and metadata reported in this study are available at NCBI GEO with the accession number GSE130511. The differentially expressed genes were analyzed by integrating multiple bioinformatics resources such as KEGG (Kanehisa et al., 2012), UniProt (The UniProt Consortium, 2018), STRING (Szklarczyk et al., 2019), DAVID (Huang da et al., 2009), and EcoCyc (Keseler et al., 2017) to infer biological functions altered in Spn serotype 4 harboring a gene deletion in a putative lysine decarboxylase.
UPLC–HRMS Untargeted Metabolomics Analysis
Streptococcus pneumoniae TIGR4 and ΔcadA strains were cultured in THY (mid-log phase, ∼109 CFU, n = 5) and transferred onto a 0.2 μm Whatman polycarbonate membrane by vacuum filtration. The membranes were flash-frozen in liquid nitrogen and stored at −80°C until further use. Metabolites from bacteria collected on membrane were extracted by placing the membranes into petri dishes with 1.3 ml of extraction solvent pre-chilled to 4°C (40:40:20 HPLC grade methanol, acetonitrile, and water with 0.1% formic acid). The extraction was allowed to proceed for 15 min at −20°C. The membranes were flipped over and rinsed with the extraction solvent. The extraction solvent was transferred to 2.0 ml centrifuge tubes and an additional 300 μl of extraction solvent was added to each membrane. The membranes were compressed and rinsed to extract the remaining cells and the extraction solvent was transferred to a centrifuge tube. The centrifuge tubes with extraction solution were centrifuged for 5 min (16,100 × g) at 4°C to remove cellular debris and the supernatant was transferred to new 2.0 ml tubes. The residual cells were resuspended with 100 μl of extraction solvent. Extraction was allowed to proceed for another 15 min at −20°C, centrifuged for 5 min (16,100 × g) at 4°C to remove any remaining cells and the supernatant was collected. Tubes containing ∼1.7 ml of the total supernatant were dried under a stream of N2 and solid residue was resuspended in 300 μl of sterile water and transferred to 300 μl autosampler vials. Samples were immediately placed in autosampler trays for mass spectrometric analysis.
Samples placed in an autosampler tray were kept at 4°C. A 10 μl aliquot was injected through a Synergi 2.5-micron reverse-phase Hydro-RP 100, 100 × 2.00 mm LC column (Phenomenex, Torrance, CA, United States) kept at 25°C. The eluent was introduced into the MS via an electrospray ionization source conjoined to an ExactiveTM Plus Orbitrap Mass Spectrometer (Thermo Scientific, Waltham, MA, United States) through a 0.1 mm internal diameter fused silica capillary tube. The mass spectrometer was run in full scan mode with negative ionization mode with a window from 85 to 1000 m/z with a method adapted from Lu et al. (2010). The samples were run with a spray voltage of 3 kV. The nitrogen sheath gas was set to a flow rate of 10 psi with a capillary temperature of 320°C. Automatic gain control (AGC) target was set to 3e6. The samples were analyzed with a resolution of 140,000 and a scan window of 85–800 m/z from 0 to 9 min and 110–1000 m/z from 9 to 25 min. Solvent A consisted of 97:3 water:methanol, 10 mM tributylamine, and 15 mM acetic acid. Solvent B was methanol. The gradient from 0 to 5 min is 0% B, from 5 to 13 min is 20% B, from 13 to 15.5 min is 55% B, from 15.5 to 19 min is 95% B, and from 19 to 25 min is 0% B with a flow rate of 200 μl/min.
Files generated by Xcalibur (RAW) were converted to the open-source mzML format (Martens et al., 2011) via the open-source msconvert software as part of the ProteoWizard package (Chambers et al., 2012). Maven (mzroll) software, Princeton University (Melamud et al., 2010; Clasquin et al., 2012) was used to automatically correct the total ion chromatograms based on the retention times for each sample (Clasquin et al., 2002; Melamud et al., 2010). Metabolites were manually identified and integrated using known masses (±5 ppm mass tolerance) and retention times (Δ ≤ 1.5 min). Unknown peaks were automatically selected via Maven’s automated peak detection algorithms. We used a database of 275 metabolites verified using exact m/z and known retention times, expanded from the original database of Lu et al. (2010) as additional standards become available. The statistical analysis on metabolite peak intensity post CFU normalization was done by MetaboAnalyst 4.0 (Chong et al., 2018). We used quantile normalization that has been reported to be highly efficient in normalizing metabolite variations from mass spectrometry to normalize our data (Lee et al., 2012). Significant differences in metabolite peak intensity between ΔcadA and TIGR4 were identified by a T-test at an adjusted FDR of 0.05 (Li et al., 2017). Sparse partial least squares-discriminant analysis (sPLS-DA) was used for the statistical data presentation. We used KEGG (Kanehisa et al., 2012) and EcoCyc (Keseler et al., 2017) to infer metabolic pathways in S. pneumoniae TIGR4 represented by our untargeted metabolomics data.
Measurement of Pyruvic Acid
To measure the glycolytic pathway activity, we estimated the concentration of secreted pyruvate, the end product of glycolysis, using the Pyruvic Acid Assay Kit (Megazyme, Bray, Ireland) following the manufacturer’s protocols. Briefly, mid-log phase cultures of TIGR4 and ΔcadA were pelleted at 6000 × g for 5 min. Ten microliters of supernatant was added to a mixture of 240 μl distilled water, 20 μl of assay buffer, and 10 μl of assay solution 2 containing NADH. Ten microliters of provided assay standard solution and 10 μl of distilled water were used in place of supernatant for the standard mix and blank mix, respectively. The mixtures were incubated for 2 min at room temperature (25°C). Addition of 2 μl of solution 3 containing D-LDH was used in initiating the pyruvate quantification reaction. After a reaction time of 3 min, the absorbance was read at 340 nm. Excreted pyruvate was estimated using the formula below:
where ΔAsample is the absorbance of the sample, b is the y-intercept, and m is the slope (from the linear equation generated from the assay).
where ΔA is the absorbance measurement.
The amount of pyruvate produced by S. pneumoniae TIGR4 and ΔcadA strains was normalized using estimated pneumococcal total protein according to the BCA method (Smith et al., 1985), and the result presented as pyruvate (ng)/protein (μg).
Estimation of Surface Exposed Phosphocholine
Fluorescence-activated cell sorting (FACS) comparison of surface exposed phosphocholine (PC) levels between the wild type and ΔcadA strains was performed (Shainheit et al., 2014) to validate our reported loss of CPS and higher expression of choline binding proteins (CBPs) in ΔcadA (Nakamya et al., 2018). Briefly, 300 μl of mid-exponential-growth-phase bacteria was pelleted and washed in 1× PBS. Pellets were resuspended in 100 μl of unconjugated IgA, Kappa from murine myeloma anti- PC (Sigma–Aldrich, St. Louis, MO, United States) at 1:100 in 1× PBS and incubated on ice for 30 min. The binding reaction was stopped with 500 μl of 1× PBS and centrifuged at 4,000 × g for 5 min. Pellets were resuspended in 100 μl of phycoerythrin (PE)-conjugated rat anti-mouse IgA secondary antibody (ThermoFisher Scientific, Waltham, MA, United States) at 1:100 in PBS and incubated at 4°C, in the dark, for 30 min. Staining reactions were stopped with 500 μl PBS, and products were pelleted and resuspended in 300 μl of 2% paraformaldehyde (PFA). Samples were collected (10,000 events), analyzed, and plotted using an Attune Acoustic Focusing Cytometer (Life Technology, Foster City, CA, United States).
Results
Effect of cadA Deletion on Pneumococcal Gene Expression
RNA-Seq-based comparative transcriptome analysis of S. pneumoniae TIGR4 and ΔcadA identified significant changes in the expression of 432 genes, of which 179 were downregulated and 253 were upregulated in ΔcadA. Biological functions and pathways represented by genes responsive to the impaired putative lysine decarboxylase gene are discussed in the following sections and shown in Tables 1–4.
TABLE 1.
Differentially expressed genes in ΔcadA from capsule biosynthesis pathways.
| Locus | Fold | ||
| Gene | tag | Description | change |
| Aga | SP_1898 | Alpha-galactosidase | –2.7 |
| galK | SP_1853 | Galactokinase | –5.0 |
| galT2 | SP_1852 | Galactose-1-phosphate uridylyltransferase 2 | –5.9 |
| nanB | SP_1687 | Sialidase B | –3.7 |
| SP_2167 | SP_2167 | Putative L-fuculose kinase fucK | 4.1 |
| fucA | SP_2166 | L-Fuculose phosphate aldolase | 3.7 |
| SP_2168 | SP_2168 | Putative fucose operon repressor | 1.9 |
| SP_0321 | SP_0321 | PTS system, IIA component | –2.4 |
| SP_0323 | SP_0323 | PTS system, IIB component | –3.7 |
| SP_0324 | SP_0324 | PTS system, IIC component | –3.7 |
| SP_0325 | SP_0325 | PTS system, IID component | –3.5 |
| glgB | SP_1121 | 1,4-Alpha-glucan branching enzyme | 2.1 |
| glgC | SP_1122 | Glucose-1-phosphate adenylyltransferase | 1.7 |
| glgD | SP_1123 | Glycogen biosynthesis protein | 1.6 |
| glgA | SP_1124 | Glycogen synthase | 1.7 |
| malP | SP_2106 | Maltodextrin phosphorylase | –2.1 |
| malQ | SP_2107 | 4-Alpha-glucanotransferase | –2.8 |
| exp5 | SP_0758 | PTS system glucose-specific EIICBA component | –1.6 |
| SP_1795 | SP_1795 | Putative sucrose-6-phosphate hydrolase | –3.0 |
| glmS | SP_0266 | Glutamine-fructose-6-phosphate aminotransferase | –1.6 |
| nagA | SP_2056 | N-acetylglucosamine-6-phosphate deacetylase | 2.9 |
| nagB | SP_1415 | Glucosamine-6-phosphate deaminase | 3.9 |
| pyrR | SP_1278 | Bifunctional protein PyrR | –1.9 |
| SP_1163 | SP_1163 | Putative acetoin dehydrogenase, beta subunit | –1.5 |
| SP_1164 | SP_1164 | Putative acetoin dehydrogenase, alpha subunit | –1.4 |
TABLE 4.
Differentially expressed two component system genes in ΔcadA.
| Fold | ||||
| Gene | Locus | change | TCS | Role |
| comDE | SP_2235/6 | −1.7/−2.0 | TCS12 | Competence and virulence |
| ciaRH | SP_0798/9 | −1.7/−1.8 | TCS05 | Competence, virulence, and antibiotics |
| SP_2000/1 | SP_2000/1 | −1.5/−1.7 | TCS11 | No known impact on virulence |
| SP_0661 | SP_0661 | 1.3 | TCS09 | Transition from lung to blood |
| SP_2193 | SP_2193 | 1.4 | TCS06 | Transcriptional regulator of CbpA |
| SP_1632/3 | SP_1632/3 | 1.6/1.4 | TCS01 | No known impact on virulence |
Impaired Polyamine Synthesis Reduces the Expression of Genes That Regulate Intracellular Concentrations of Precursors for Capsular Polysaccharide Synthesis
Streptococcus pneumoniae TIGR4 genes responsive to impaired polyamine synthesis have functions that are directly involved in the metabolism and transport of CPS precursors (Table 1). The acetylated sugars in the repeat unit of serotype 4 polysaccharide structure are synthesized from the following nucleotide sugar precursors: UDP-Gal, UDP-ManNAc, UDP-FucNAc, and UDP-GalNAc (Jones et al., 1991; Geno et al., 2015). Our results show that the expression of genes involved in the synthesis and availability of sugar precursors in the repeat unit is significantly altered in ΔcadA. Expression of alpha-galactosidase (aga), which cleaves galactose from raffinose and melibiose (Rosenow et al., 1999), is downregulated (Table 1). Galactokinase (galK) and galactose-1-phosphate uridylyltransferase 2 (galT2), the genes encoding key enzymes of Leloir pathway, a predominant route for galactose catabolism and generation of UDP-Gal, are significantly downregulated. We observed downregulation of sialidase B (nanB), an enzyme that catalyzes the cleavage of terminal sialic acid from host glycoconjugates to ultimately generate UDP-ManNAc. Genes that encode fuculose kinase and fuculose phosphate aldolase are upregulated in ΔcadA. These enzymes are responsible for the degradation of fucose, a sugar component of CPS repeat unit structure. Glycerone 3-phosphate produced from fucose degradation can be converted by a triosephosphate isomerase directly to G3P, an intermediate of glycolysis and PPP (Higgins et al., 2014). Our data also show the upregulation of a putative fucose operon repressor (SP_2168) that could inhibit fucose utilization. All genes encoding the phosphotransferase system (PTS) IIA-D components specific for the import of N-acetylgalactosamine, a sugar that is part of the CPS repeat unit, are downregulated.
Genes that encode enzymes that ultimately control the intracellular concentrations of UDP-GlcNAc are differentially expressed in ΔcadA (Table 1 and Figure 1). Expression of a glycogen synthesis operon (glgBCDA) which converts glucose 1-phosphate to glycogen was upregulated, while the expression of malP, which reverses this anabolic process was downregulated. This in turn will limit the availability of glucose for the biosynthesis of UDP-GlcNAc, an important substrate for epimerases that catalyze the biosynthesis of the sugars in the repeat unit of CPS. UDP-GlcNAc is converted to UDP-ManNAc and UDP-GalNAc by UDP-N-acetylglucosamine-2-epimerase and UDP-Glu 4-epimerase, respectively (Kay et al., 2016; Chen et al., 2018). UDP-GlcNAc can also be converted to UDP-FucNAc in a multi-step reaction involving SP_0358-60 protein (Kay et al., 2016). Genes that are involved in the degradation of maltose to glucose (malQ), import of glucose into the cell (exp5), and the generation of glucose 6-phosphate and fructose 6-phosphate from sucrose (SP_1795), are downregulated in ΔcadA. Glucose, glucose 6-phosphate, and fructose 6-phosphate, the first three intermediates of glycolysis, are precursors for UDP-GlcNAc. We also observed downregulation of glmS which encodes a glutamine – fructose-6-phosphate aminotransferase that converts fructose 6-phosphate to GlcN6P in UDP-GlcNAc biosynthesis, and upregulation of nagA and nagB, also at the protein level (Nakamya et al., 2018), that convert UDP-GlcNAc intermediates to fructose 6-phosphate. The net effect of gene expression changes described could be reduced concentrations of the saccharides that constitute the repeat unit that ultimately results in reduced CPS in ΔcadA (Figure 1).
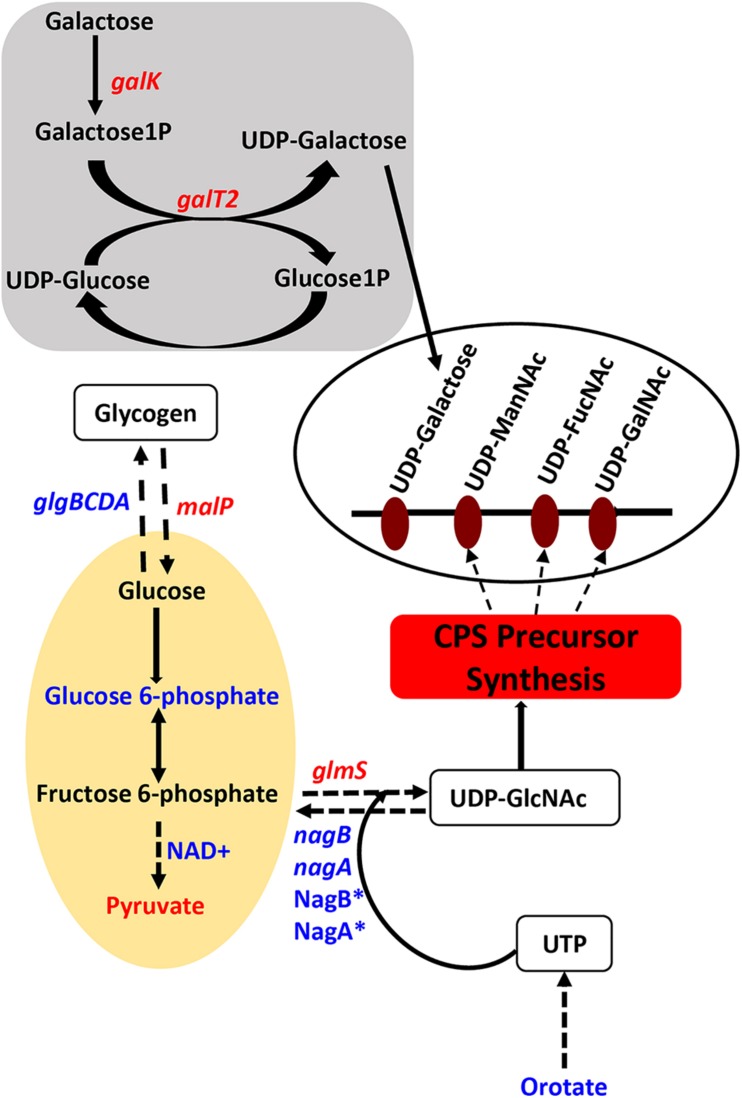
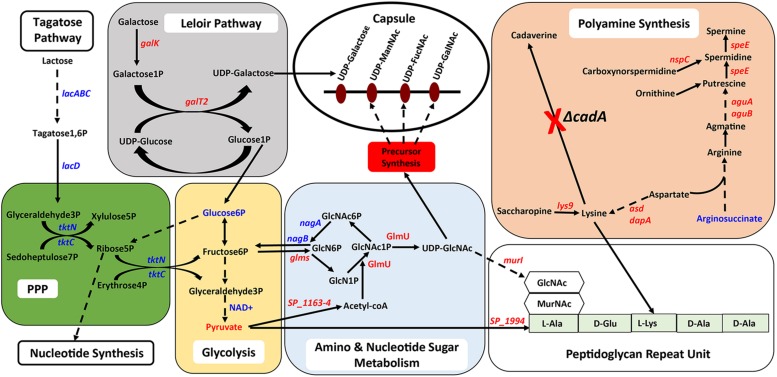
FIGURE 1.
Impaired polyamine synthesis in S. pneumoniae impacts the availability of precursors for capsule (CPS) synthesis. Characterization of a putative lysine decarboxylase deletion strain of S. pneumoniae TIGR4 by RNA-Seq and metabolomics identified changes (blue represents increase and red represents decrease) in the genes (italicized) and metabolites in the deletion strain relative to the wild type that could explain the previously reported loss of the capsule. The deletion strain is impaired in the interconversion of UDP-Glu and UDP-Gal via the Leloir pathway (gray square) due to the reduced expression of galk and galT2 genes, that can reduce the availability of UDP-Gal precursor of the repeat unit of the serotype 4 capsule (open white oval). Carbon flux through glycolysis (yellow oval) is reduced in ΔcadA confirmed by the accumulation of glucose 6-phosphate and NAD+, intermediates of the pathway and reduced levels of the end product pyruvate. Enzymatic reactions are shown as black arrows and multi-step reactions are represented by a broken arrow. Upregulation of N-acetylglucosamine-6- phosphate deacetylase (NagA∗) and GlcN6P deaminase (NagB∗) that could degrade intermediates of UDP-GlcNAc, at the protein level, was earlier reported and was observed at the RNA level in this study. Reduced synthesis of UDP-GlcNAc due to reduced expression of glmS and increased degradation due to higher expression of nagA and nagB will limit the availability of this nucleotide sugar substrate for epimerases that synthesize all three acetylated UDP-sugars of the repeat unit. Accumulation of orotate indicates impaired ability to synthesize UTP, a precursor for UDP-GlcNAc synthesis. The net effect of these changes in metabolism in ΔcadA results in the reduced availability of precursors for CPS synthesis (red rectangle).
We further identified downregulation of pyrR, a gene that encodes a bifunctional enzyme that regulates the interconversion of uracil and uridine monophosphate (UMP). Uracil is known to be a metabolic regulator of CPS biosynthesis in pneumococcus (Carvalho et al., 2018). In addition, UMP is the precursor for UTP that is essential for the activation of saccharides in the repeat unit as it provides UDP required to form UDP-sugars. There was also lower expression of the pyruvate dehydrogenase complex (SP_1163 and SP_1164) responsible for the production of acetyl-CoA that is needed for the acetylation of sugars in the repeat unit of TIGR4.
Impaired Polyamine Synthesis Regulates the Expression of Genes Involved in the Synthesis of Peptidoglycan and Choline-Binding Proteins
In previous work, we reported that deletion of cadA, which encodes a putative lysine decarboxylase, resulted in significantly reduced expression of penicillin-binding protein 2X (Pbp2X) and choline kinase (Pck), proteins involved in PG and teichoic acid biosynthesis (Nakamya et al., 2018). In this study, we identified reduced expression of genes encoding a glutamine transporter complex: glnH, glnP, and glnQ in ΔcadA (Table 2). Glutamine is known to be directly involved in the crosslinking of PG. It serves as an important cofactor in the amidation of glutamate to iso-glutamine in the PG disaccharide–pentapeptide repeat unit, a substrate of penicillin binding protein (Zapun et al., 2013). Genes that encode proteins involved in biosynthesis of amino acids lysine (asd, dapA, and lys9), alanine (SP_1994), and D-Glu from L-glutamate (murI), that are components of the PG repeat unit, are all downregulated in polyamine synthesis-deficient pneumococci. Reduced expression of genes that control intracellular concentrations of UDP-GlcNAc described in the previous section could also negatively impact the PG and, ultimately, cell wall biosynthesis. UDP-GlcNAc is an essential intermediate molecule in PG biosynthesis. It is one of the disaccharide components of the PG repeat unit, and also a precursor for the biosynthesis of MurNAc, the second sugar of the repeat unit (Figure 2). These results indicate that polyamine biosynthesis regulates pneumococcal cell wall biosynthesis, a structure to which the pneumococcal capsule is attached (Larson and Yother, 2017; Figure 2).
TABLE 2.
Differentially expressed genes in ΔcadA involved in PG and choline binding protein synthesis, and carbon utilization pathways.
| Locus | Fold | ||
| Gene | tag | Description | change |
| Peptidoglycan biosynthesis | |||
| glnH | SP_0609 | Amino acid ABC transporter, amino acid-binding protein | –1.6 |
| glnP | SP_0607 | Amino acid ABC transporter, permease protein | –1.5 |
| glnQ | SP_0610 | Amino acid ABC transporter, ATP-binding protein | –1.7 |
| Asd | SP_1013 | Aspartate-semialdehyde dehydrogenase | –5.4 |
| dapA | SP_1014 | 4-hydroxy-tetrahydrodipicolinate synthase | –4.8 |
| lys9 | SP_0919 | Saccharopine dehydrogenase | –26.4 |
| SP_1994 | SP_1994 | Aminotransferase, class I | –1.4 |
| murI | SP_1881 | Glutamate racemase | –1.7 |
| Choline binding protein synthesis | |||
| pspA | SP_0117 | Pneumococcal surface protein A | 2.3 |
| cbpA | SP_2190 | Choline binding protein A | 1.8 |
| cbpI | SP_0069 | Choline binding protein I | 1.5 |
| pcpA | SP_2136 | Choline binding protein PcpA | 2.3 |
| Carbon utilization pathways | |||
| SP_0877 | SP_0877 | PTS system, fructose-specific IIABC components | 2.6 |
| SP_0645 | SP_0645 | Putative PTS system, IIA component | 3.1 |
| SP_0646 | SP_0646 | Putative PTS system, IIB component | 2.6 |
| SP_0647 | SP_0647 | Putative PTS system, IIC component | 2.6 |
| lacD | SP_1190 | Tagatose 1,6-diphosphate aldolase | 1.9 |
| lacC | SP_1191 | Tagatose-6-phosphate kinase | 1.9 |
| lacB | SP_1192 | Galactose-6-phosphate isomerase subunit | 1.7 |
| lacA | SP_1193 | Galactose-6-phosphate isomerase subunit | 1.8 |
| tktC | SP_2127 | Transketolase, C-terminal subunit | 97.9 |
| tktN | SP_2128 | Transketolase, N-terminal subunit | 94.1 |
| SP_2129 | SP_2129 | Putative PTS system, IIC component | 81.5 |
| SP_2130 | SP_2130 | Putative PTS system, IIB component | 68.7 |
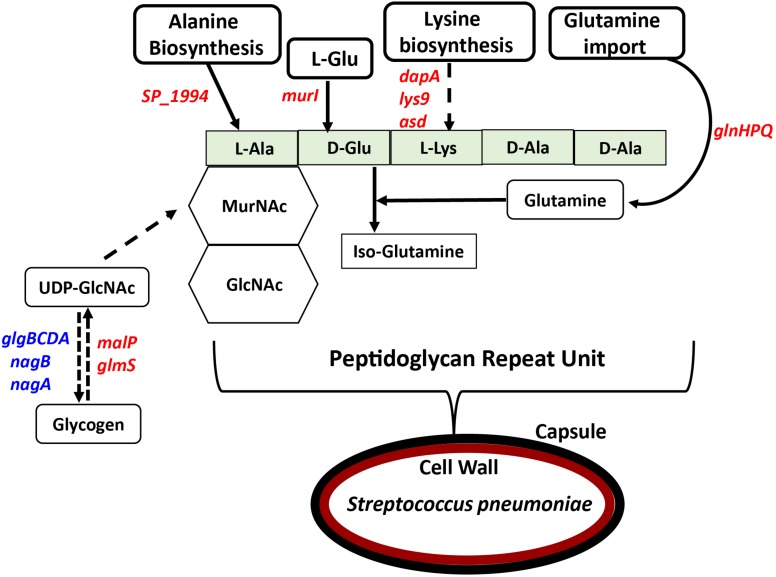
FIGURE 2.
Impact of impaired polyamine synthesis on PG biosynthesis. The PG repeat unit structure in pneumococci constitutes a disaccharide (hexagon) composed of GlcNAc and MurNAc that is linked to an L-Ala, D-Glu, L-Lys, D-Ala, and D-Ala pentapeptide (light green rectangle). RNA-Seq-based comparative transcriptome analysis of ΔcadA impaired in polyamine synthesis and the wild type S. pneumoniae TIGR4 identified gene (italicized) expression changes (increase in blue and decrease in red) that could impact the PG repeat unit synthesis and cross-linking. Enzymatic reactions are shown as black arrows and multi-step reactions are represented by a broken arrow. Reduced synthesis of UDP-GlcNAc due to reduced expression of malP and glmS, and its increased breakdown due to higher expression of nagA, nagB, and the glgBCDA operon, will impact PG synthesis. Reduced expression of SP_1994 that converts pyruvate to alanine, murI that converts L-Glu to D-Glu, and lysine biosynthesis genes (dapA, lys9, and asd) will limit the availability of these three amino acids in PG pentapeptide synthesis. Crosslinking of the PG repeat unit requires conversion of D-Glu to iso-glutamine using glutamine as a co-factor. Impaired glutamine import, due to reduced expression of glnHPQ was observed in ΔcadA. The overall impact of polyamine synthesis impairment is altered cell wall biosynthesis (red oval), due to specific effects on PG synthesis. The cell wall provides the point of structural attachment for the capsule (black oval) in S. pneumoniae.
Impaired polyamine synthesis impacts expression of genes encoding pneumococcal surface protein A, CBPs A and I, and PcpA which were upregulated in ΔcadA. These data suggest that deletion of cadA does not only mediate loss of the capsule and expose CBPs on the cell surface, but also enhance the expression of CBPs. Our earlier report indicated that there is no significant difference in the ability of ΔcadA to colonize the murine nasopharynx with respect to the parent strain (Shah et al., 2011). In this study, the observed upregulation of expression of CBPs could represent a response to genetic cue by pneumococcal defense machinery to enhance adherence and colonization, to overcome the expected clearance by opsonophagocytosis due to the reported loss of the capsule. Comparison of surface exposed PC using an IgA anti-PC antibody and FACS assay indicates higher signal for PC with ΔcadA compared to the wild-type strain (Supplementary Figure 1), validating the reported increase in the expression of CBPs at the RNA and protein levels (Nakamya et al., 2018).
Impaired Polyamine Synthesis Results in Increased Carbon Flux Through PPP
Proteomics analysis of the polyamine synthesis deficient strain ΔcadA indicated a putative shift in central metabolism favoring PPP (Nakamya et al., 2018). Expression of genes responsive to cadA deletion include those that belong to carbon utilization pathways (Table 2). We observed upregulation of fructose-specific PTS system IIABC components, which catalyze the conversion of fructose to fructose 1-phosphate that ultimately generates G3P, an intermediate of glycolysis/PPP. This upregulation could limit the availability of fructose for the synthesis of UDP-GlcNAc. Deletion of cadA also resulted in the upregulation of putative PTS system IIA-C component genes and lacDCBA, genes involved in the import and conversion of galactose via tagatose to G3P. The net effect of these gene expression changes is expected to increase G3P levels in ΔcadA (Figure 3), an intermediate of glycolysis and PPP. Activation of PPP and utilization of G3P in this pathway are supported by upregulation of transketolase genes, both C- and N-terminal subunits tktC and tktN, which encode these enzymes of PPP, consistent with our earlier report of significant upregulation of transketolases at the protein level (Nakamya et al., 2018). Putative PTS system, IIBC components, which play a role in the conversion of L-ascorbate to xylulose-5-phosphate, an intermediate of PPP, are also upregulated, supporting higher PPP activity in ΔcadA compared to the wild-type strain, and corroborate our earlier findings of a shift in central carbon flux at the protein level (Nakamya et al., 2018).
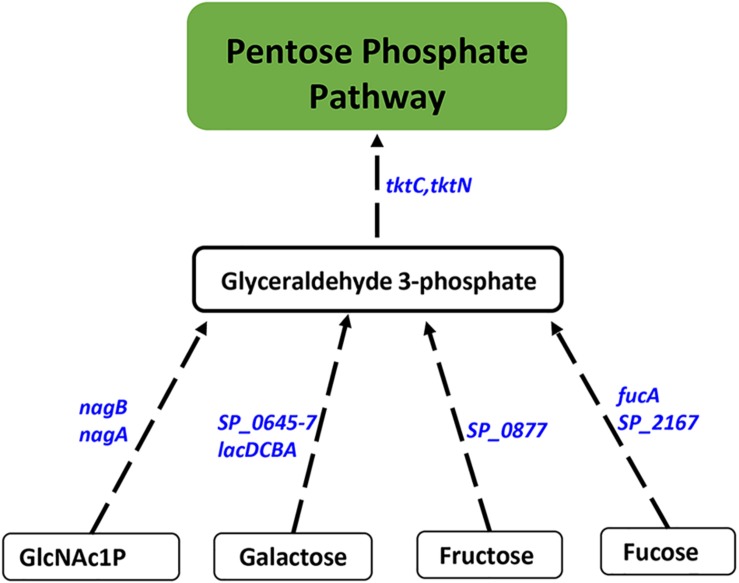
FIGURE 3.
Impaired polyamine synthesis results in increased carbon flow through the PPP. RNA-Seq-based transcriptome analysis of S. pneumoniae TIGR4 that harbors a gene deletion in a putative lysine decarboxylase identified significant upregulation of genes (italicized, blue) in carbohydrate metabolism that indicate increased G3P, an intermediate of glycolysis, and its utilization in PPP. Enzymatic reactions are shown as black arrows and multi-step reactions are represented by a broken arrow. Increased expression of genes in metabolic pathways catalyzing the conversion of galactose (SP_0645-7, lacDCBA), fructose (SP_0877), fucose (fucA, SP_2167), and GlcNAc1P (nagA, nagB) to G3P with concomitant higher expression of transketolase C and N subunits (tktC, tktN) supports the increased metabolic flux through PPP.
Impaired Polyamine Synthesis Alters the Expression of Pneumococcal Virulence Factors and Stress Response
Expression of genes encoding branched chain amino acid transporters livH, livM, livG, and livF is downregulated in ΔcadA (Table 3). These transporters have been shown to contribute to pneumococcal virulence in a murine model of pneumonia and septicemia (Basavanna et al., 2009). We also observed downregulation of metQ that encodes methionine ABC transporter lipoprotein in ΔcadA. Although the exact mechanism is not known, loss of methionine transporter MetQ has been shown to lead to an attenuated phenotype in a murine model of pneumococcal pneumonia and it is reported to delay invasive disease (Saleh et al., 2014). We observed decreased expression of trehalose PTS system, IIABC components, and putative dextran glucosidase in polyamine synthesis deficient pneumococci (Table 3) which could negatively impact oxidative stress responses and the availability of glucose. Trehalose is a disaccharide containing two molecules of glucose. It is known to act as a free radical scavenger and protect yeast against oxidative stress generated by H2O2 and iron (Benaroudj et al., 2001; Jiang et al., 2018). We also observed increase in the expression of the manganese ABC transporter operon (psaBCA), a pneumococcal virulence factor. Evidence of role of manganese in complexes with antioxidants and other biomolecules in the detoxification of reactive oxygen species has been well documented (Culotta and Daly, 2013). Increased expression of the manganese transporter could constitute changes in oxidative stress response in ΔcadA.
TABLE 3.
Differentially expressed genes of known virulence factors, stress response, and polyamine biosynthesis in ΔcadA.
| Locus | Fold | ||
| Gene | tag | Description | change |
| Virulence | |||
| livH | SP_0750 | Branched-chain amino acid ABC transporter, permease protein | –1.5 |
| livM | SP_0751 | Branched-chain amino acid ABC transporter, permease protein | –1.6 |
| livG | SP_0752 | Branched-chain amino acid ABC transporter, ATP-binding protein | –1.7 |
| livF | SP_0753 | Branched-chain amino acid ABC transporter, ATP-binding protein | –1.6 |
| metQ | SP_0149 | Lipoprotein | –1.7 |
| Stress response | |||
| SP_1883 | SP_1883 | Putative dextran glucosidase | –28.1 |
| SP_1884 | SP_1884 | Trehalose PTS system, IIABC components | –26.2 |
| psaB | SP_1648 | Manganese ABC transporter, ATP-binding protein | 2.7 |
| psaC | SP_1649 | Putative manganese ABC transporter, permease protein | 2.7 |
| psaA | SP_1650 | Manganese ABC transporter substrate-binding lipoprotein | 2.7 |
| Polyamine biosynthesis | |||
| aguA | SP_0921 | Putative agmatine deiminase | –25.7 |
| aguB | SP_0922 | Carbon–nitrogen hydrolase family protein | –19.7 |
| nspC | SP_0920 | Carboxynorspermidine decarboxylase | –27.5 |
| speE | SP_0918 | Polyamine aminopropyltransferase | –41.5 |
Impact of Impaired Polyamine Synthesis on the Expression of Two Component Regulatory Systems
Two component regulatory systems (TCSs) are composed of a histidine kinase sensor protein and a regulatory response protein that are involved in signal transduction and adaptation of living organisms to a changing environment. A total of 13 TCSs, commonly denoted as TCS01–13, and 1 orphan regulator not coupled to a histidine kinase have been identified in pneumococcal genomes (Lange et al., 1999; Throup et al., 2000). Pneumococcal TCSs are implicated in adaptive responses to the changes in the host environment ranging from different anatomical sites, competency, environmental stress, antimicrobials, and nutrition availability that ultimately regulate pneumococcal pathogenesis (Gomez-Mejia et al., 2018). Expression of TCS01, TCS05, TCS06, TCS09, TCS11, and TCS12 is significantly altered in ΔcadA with respect to TIGR4 suggesting a possible role for polyamines whether upstream or downstream of TCSs (Table 4).
Metabolic Profile of Putative Lysine Decarboxylase Deficient S. pneumoniae
Impact of cadA Deletion on Metabolites in Glycolysis and Capsule Biosynthesis Pathways
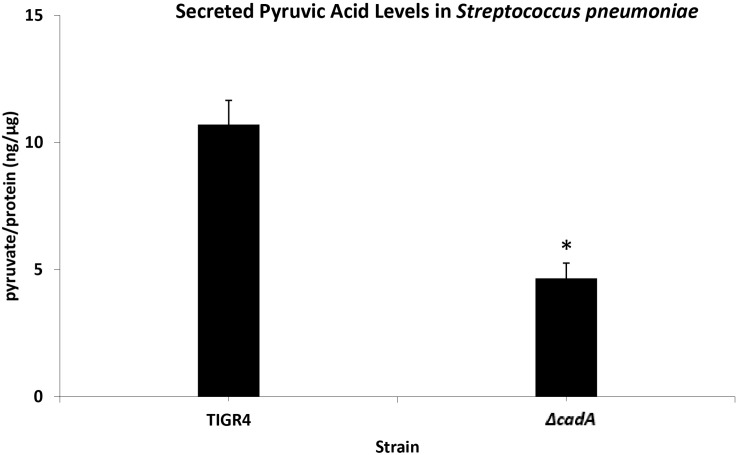
Untargeted metabolomics of TIGR4 and ΔcadA strains identified significant differences in the concentration of 10 metabolites (Table 5 and Figure 4). We observed accumulation of glucose 6-phosphate, an intermediate of glycolysis, indicating that flux through glycolysis is interrupted in ΔcadA (Figure 5). Accumulation of glucose 6-phosphate reduces the available fructose 6-phosphate that can be converted to UDP-GlcNAc, which is a precursor of repeat units in type 4 CPS (Figure 5). The levels of NAD+ are higher in ΔcadA suggesting that glucose in multi-step reactions through G3P is not being converted to pyruvate. NAD+ is required for the interconversion of G3P and 1,3-bisphosphoglycerate in the glycolytic pathway for generating pyruvate (Figure 5). Inhibition of glycolysis, a pathway that provides precursors for the biosynthesis of three out of four saccharides of the capsule repeat unit in ΔcadA could explain reduced CPS in this strain. Pyruvate is the terminal product of glycolysis and comparison of secreted pyruvate (Figure 6) showed that its concentration is significantly low in ΔcadA (4.66 ± 1.91 ng pyruvate/μg protein) compared to TIGR4 (10.71 ± 2.68 ng pyruvate/μg protein). Reduced levels of pyruvate in ΔcadA confirm reduced glycolytic activity in polyamine synthesis impaired pneumococci and support transcriptome analysis that indicates reduced availability of precursors for CPS synthesis. An additional mechanism that also has implications for CPS synthesis is the accumulation of orotate in ΔcadA, which suggests negative regulation of UTP synthesis. Orotate is an intermediate in the de novo biosynthesis of uracil, and uracil can yield UTP in a series of reversible reactions via the salvage pathway. UTP is essential to produce UDP-sugars which are substrates for the enzymes of CPS synthesis. Uracil is proposed to be a signaling molecule for CPS biosynthesis (Carvalho et al., 2018). Changes in the intracellular metabolite concentrations support the role of polyamine synthesis in modulating capsule biosynthesis.
TABLE 5.
Significant fold changes in the intracellular levels of metabolites of ΔcadA.
| Metabolites | Fold change |
| Xanthosine | 4.2 |
| Trehalose 6-phosphate | –2.5 |
| IMP | 2.5 |
| Glucose-6-phosphate | 2.2 |
| Orotate | 2.1 |
| L-Argininosuccinate | 2.0 |
| Ophthalmate | 1.8 |
| N-Acetylglutamine | 1.5 |
| Salicylate | 1.5 |
| NAD+ | 2.0 |
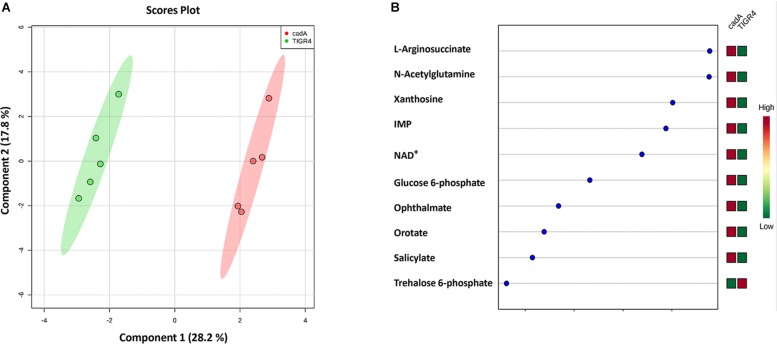
FIGURE 4.
Comparison of the metabolite profiles of S. pneumoniae TIGR4 and ΔcadA. Mass spectrometry-based untargeted metabolomics analysis (n = 5) was performed with the wild type and polyamine synthesis deficient strain ΔcadA. Peak intensity values of identified metabolites were analyzed by Metaboanalyst 4.0 to identify significant changes in expression. (A) Clustering of samples within the sPLS-DA principal component of the wild type strain TIGR4 (green) and deletion mutant strain ΔcadA (pink) indicate significant metabolic differences between the two strains. (B) sPLS-DA loading plot of TIGR4 and ΔcadA with top ranked metabolites with significant differences in intracellular concentrations between the strains is shown.
FIGURE 5.
Intersection between polyamine biosynthesis, carbohydrate metabolism, and the capsule in pneumococci. Interconnected metabolic pathways in a putative lysine decarboxylase synthesis deficient strain of S. pneumoniae TIGR4 (ΔcadA) analyzed by RNA-Seq and metabolomics identified specific molecular mechanisms that could result in reduced intracellular levels of the constituents of the serotype four capsule and PG repeat structures. Enzymatic reactions are shown as black arrows and multi-step reactions are represented by a broken arrow. Changes in gene (italicized) and metabolite levels in ΔcadA relative to the wild-type strain are shown (red represents decrease and blue represents increase). Deletion of cadA results in reduced expression of genes encoding biosynthesis pathways of polyamines such as putrescine, spermidine, and spermine and precursor amino acid lysine. Accumulation of arginosuccinate, an intermediate for the biosynthesis of arginine supports the notion of overall reduction in polyamine synthesis in ΔcadA. Impaired polyamine synthesis renders pneumococci incapable of UDP-Glu to UDP-Gal interconversion, i.e., Leloir pathway (gray square), that could reduce the carbon flux through glycolysis (yellow rectangle). Despite increased carbon flux toward G3P, a glycolytic intermediate, through tagatose metabolism, changes in the levels of metabolites of glycolysis indicate reduced carbon flux. RNA-Seq data indicated that G3P is routed through the PPP (green square), and enhanced PPP activity will result in increased nucleotide synthesis. Changes in amino and nucleotide sugar metabolism (blue square) will result in reduced intracellular levels of UDP-GlcNAc, a precursor for three N-acetylated sugars in the serotype 4 capsule repeat unit (precursor nucleotide sugars shown in the open oval). Impaired Leloir pathway and UDP-GlcNAc synthesis will limit the availability of CPS precursors. Changes in metabolism in ΔcadA, such as reduced glycolytic activity, altered UDP-GlcNAc metabolism and lysine synthesis impacts the assembly of the PG repeat unit (open rectangle) disaccharide and pentapeptide (light green rectangle). Polymerization of PG repeat units generates the cell wall which provides attachment for the capsule.
FIGURE 6.
Measurement of secreted pyruvic acid from Spn TIGR4 and ΔcadA. Secreted pyruvate in a putative lysine decarboxylase deficient ΔcadA strain and Spn TIGR4 was measured using Pyruvic Acid Assay Kit (Megazyme, Bray, Ireland) according to the manufacturer’s protocol. Results show that secreted pyruvate levels in ΔcadA were ∼60% lower compared to the wild type strain. This data suggest that the glycolytic pathway is impaired in the polyamine synthesis deficient pneumococci. ∗indicate significance at P ≤ 0.05.
Impact of cadA on Metabolites Involved in Stress Response and Polyamine Biosynthesis
Metabolomics analysis also showed that the concentration of trehalose 6-phosphate (T6P) is reduced in ΔcadA (Table 4). Trehalose has been shown to protect enzymes against the damaging effect of H2O2 and heat (Asad et al., 2011). It has also been reported to have a protective role against oxidative stress in pathogenic mycobacteria (Kalscheuer and Koliwer-Brandl, 2014). Consequently, the putative lysine decarboxylase gene cadA could be crucial for pneumococcal stress responses indirectly through the regulation of trehalose metabolism. The level of L-arginosuccinate, an intermediate of synthesis of arginine, a precursor for the biosynthesis of putrescine and spermidine, was higher in the mutant strain. This observation supports the predicted reduced synthesis of putrescine and spermidine in ΔcadA by reduced expression of polyamine biosynthesis genes such as aguA, aguB, speE, and nspC identified by RNA-Seq (Table 3 and Figure 5). According to the database of prokaryote operons (DOOR) (Mao et al., 2014), aguA, aguB, speE, and nspC constitute a single operon and could be under the regulation of cadA that is immediately upstream of this operon in the same direction of transcription.
Discussion
Polyamines are ubiquitous polycationic molecules found in all three domains of life (Michael, 2016). They are involved in diverse biological processes ranging from protein synthesis, nucleic acid stability, cellular growth, stress responses, and pathogenicity (Gevrekci, 2017). Intracellular polyamine concentrations are tightly regulated by transport (uptake/efflux), biosynthesis, and catabolism. Polyamine-mediated modulation of host–pathogen interactions has been leveraged and successfully targeted in the design of therapeutics, primarily against protozoan pathogens (Phillips, 2018), specifically targeting the polyamine biosynthesis pathway for the treatment of human African trypanosomiasis (Yun et al., 2010). Recent advances in cancer research have implicated modulation of polyamine synthesis and transport in chemotherapy/prevention due to the requirement of polyamines in cell proliferation (Flynn and Hogarty, 2018).
Limited serotype coverage of the existing pneumococcal vaccines coupled with genomic plasticity and enhanced antimicrobial resistance mandate the development of novel treatment and prevention strategies (Berical et al., 2016; Balsells et al., 2017; Cherazard et al., 2017). Although a large number of potential protein vaccine candidates highly conserved in most pneumococcal serotypes such as PspA, PsaA, PhpA, PhtB, PcsB, and StkP are reported in the scientific literature, clinical trials are pending (Pichichero et al., 2016). Regulation of polyamine transport and synthesis has enormous potential for the discovery of novel interventions for pneumococcal colonization and invasive diseases. We and others reported that polyamine transport protein, PotD, is a potential vaccine candidate as immunization with this protein provides protection against systemic pneumococcal infection (Shah and Swiatlo, 2006) and colonization (Shah et al., 2009; Converso et al., 2017) in mice. We reported that impaired polyamine transport (ΔpotABCD) and synthesis (ΔspeE, ΔcadA) results in attenuation of virulence of S. pneumoniae in murine models of colonization, pneumonia, and sepsis (Shah et al., 2011). Delving further into the role of polyamine biosynthesis in pneumococcal pathogenesis, for the first time, we demonstrated that impaired polyamine synthesis results in a reduced capsule in S. pneumoniae and likely explains the observed attenuation in vivo (Nakamya et al., 2018).
In this study, we characterized the transcriptome and metabolome of pneumococcal serotype 4 strain impaired in polyamine synthesis and identified specific mechanisms which polyamines could employ, individually or collectively, in regulating capsule biosynthesis. We identified the Leloir pathway of galactose catabolism, as a crucial mechanism impaired in ΔcadA (Table 1 and Figure 1). Leloir pathway is the only pathway for the interconversion of galactose and glucose (Frey, 1996). Impairment of Leloir pathway limits the availability of UDP-Gal, a precursor for one of the sugars that constitute the serotype 4 capsule repeat unit. Furthermore, galactose is reported to be the most abundant sugar in the host nasopharynx (Blanchette et al., 2016), and the ability to metabolize this important monosaccharide directly correlates to pneumococcal colonization and virulence (Paixao et al., 2015). Therefore, regulation of galactose catabolism in a polyamine-dependent manner could impact pneumococcal virulence. Deletion of catabolite control protein A (ΔccpA) in the serotype 2 background has been reported to alter carbon utilization pathways and capsule attachment to the cell wall when cultured in a chemically defined medium with galactose as a carbon source (Carvalho et al., 2011). Therefore, CcpA-regulated galactose metabolism is important for the attachment of pneumococcal CPS to the cell wall. In THY growth medium that provides diverse carbon sources, although RNA-Seq did not identify significant changes in the expression of ccpA, our earlier report identified a significant downregulation of CcpA at the protein level in ΔcadA (Nakamya et al., 2018), indicating that CcpA could be part of the polyamine gene regulatory network.
Additional mechanisms identified in this study that could abrogate CPS biosynthesis include impaired glycolysis that could alter amino sugar biosynthesis, and potential upregulation of PPP that could collectively result in reduced levels of UDP-GlcNAc and ultimately limit the availability of acetylated UDP-sugars for CPS synthesis. The terminal product of glycolysis, pyruvate, and pyruvate dehydrogenase complex (SP_1163 and SP_1164) genes that encode enzymes for the conversion of pyruvate to acetyl-CoA are reduced and downregulated in ΔcadA, respectively (Table 1 and Figure 5). Reduced pyruvate will affect CPS production since pyruvate itself is a component of the serotype 4 capsule via its attachment to galactose (Kay et al., 2016) and its conversion to acetyl-CoA is essential for the acetylation of monosaccharides in the capsule repeat unit. Downregulation of pyruvate oxidase (spxB) and the pyruvate dehydrogenase complex resulted in reduced levels of acetyl-CoA and CPS in serotype 4 pneumococci (Echlin et al., 2016). Reduced levels of pyruvate in ΔcadA reported in this study cannot be attributed to transcriptional downregulation of pyruvate oxidase, as we did not identify any significant changes in the expression of spxB by qRT-PCR.
Gene expression changes in ΔcadA indicate that polyamines can negatively regulate PG synthesis in pneumococci (Table 2 and Figure 2). Since the capsule is tethered to the cell wall of pneumococcus via direct glycosidic linkage between glucose and UDP-GlcNAc in PG (Larson and Yother, 2017), changes in PG could result in the reported loss of the capsule in ΔcadA. Changes in PG have implications for the susceptibility of pneumococcus to antibiotics such as penicillin that target cell wall synthesis. Expression of two component system TCS05 (ciaR) that controls and enhances pneumococcal resistance to cell wall targeting antibiotics such as β-lactams (Guenzi et al., 1994) and its sensor ciaH was downregulated in ΔcadA. Reduced expression of ciaRH combined with the downregulation of PBP2x protein reported earlier (Nakamya et al., 2018) indicates that polyamines could be modulating susceptibility to antibiotics in pneumococcus. Expression of TCS01 was upregulated in ΔcadA, while TCS11 expression was downregulated. TCS01 and TCS11 are predicted to be involved in resistance to antibiotics (Gomez-Mejia et al., 2018). Significant decrease in the expression of TCS12 (comDE) (Table 4) that controls pneumococcal competence and virulence, and increased expression of TCS06, a transcriptional regulator of CbpA (Ma and Zhang, 2007), could contribute to the loss of in vivo fitness (Shah et al., 2011) and upregulation of CBP expression at the RNA (Table 2) and protein levels (Nakamya et al., 2018). The six TCSs identified in this study (Table 4) as differentially expressed in ΔcadA represent ∼46% of the 13 TCSs in pneumococcal genomes. Therefore, polyamines could be master regulators in pneumococci via their interaction with TCSs. Alternatively, polyamines could be part of the downstream regulatory network of the TCSs. Deciphering this complex polyamine/TCS regulatory network is necessary for rational vaccine/small molecule intervention strategies that target polyamine metabolism.
The E. coli polyamine modulon that constitutes genes whose expression is upregulated by polyamines at the level of translation is well characterized (Igarashi and Kashiwagi, 2015). Twenty genes identified as part of the polyamine modulon in E. coli are reported to enhance cell proliferation and viability, biofilm formation, and ability to detoxify reactive oxygen species (Igarashi and Kashiwagi, 2018). However, altered polyamine metabolism-mediated regulation of gene expression is a recently identified area of pneumococcal physiology, with implications for pneumococcal pathogenesis. With the reported link of CcpA to regulation of TCS07 and TCS12 (Carvalho et al., 2011), establishment of CcpA as pneumococcal global regulator of carbohydrate metabolism (Iyer et al., 2005), polyamine-mediated changes in expression of CcpA (Nakamya et al., 2018), and altered galactose metabolism reported in this study, ccpA could constitute a polyamine modulon in pneumococcus that warrants future research and validation.
This study clearly demonstrates that polyamine synthesis is required for CPS production in pneumococci. Polyamine biosynthesis genes are well conserved in all pneumococcal genomes (Shah et al., 2011) and are necessary for survival in vivo. Therefore, targeting this pathway is an attractive avenue for discovering novel therapeutics and constitutes an anti-virulence strategy that could offer serotype-independent coverage without impacting nasopharyngeal colonization, i.e., disarm, but not eradicate, pneumococci (Lujan and Gallego, 2016). This approach is similar to other anti-virulence strategies such as the use of epigallocatechin gallate, the most abundant constituent of green tea, to regulate pneumococcal virulence factors pneumolysin and sortase (Song et al., 2017) and Chalcone, a natural phenol to inhibit the activity of sortase in Listeria monocytogenes (Li H. et al., 2016). Another polyphenol, fisetin, has been reported to interfere with the binding of listeriolysin O to the cholesterol receptor and lead to eventual elimination of cytolytic activity of L. monocytogenes (Wang et al., 2015). Fursultiamine hydrochloride, a derivative of thiamine, has been successfully employed to transcriptionally regulate toxin and hemolysin virulence factors in Vibrio vulnificus (Imdad et al., 2018). Future studies focusing on uncovering the regulatory network of pneumococcal polyamine homeostasis are warranted for leveraging the therapeutic potential of this important metabolic pathway for limiting the spread of pneumococci.
Data Availability
The datasets generated for this study can be found in the NCBI GEO, accession number GSE130511.
Author Contributions
BN conceived, supervised, and designed the experiments. MA performed the experiments and drafted the manuscript. LS and MN performed the experiments. JT supervised the FACS analysis. BN and ES finalized the draft. All authors approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Metabolomic extraction and mass spectrometric analyses were performed at the Biological and Small Molecule Mass Spectrometry Core, University of Tennessee, Knoxville, Knoxville, TN, United States, with the assistance of Dr. Shawn R. Campagna, Dr. Hector F. Castro, and Joshua B. Powers.
Abbreviations
- CPS
capsular polysaccharide
- D-Ala
D-alanine
- D-Glu
D-glutamate
- G3P
glyceraldehyde 3-phosphate
- GlcN1P
glucosamine 1-phosphate
- GlcN6P
glucosamine 6-phosphate
- GlcNAc
N-acetylglucosamine
- GlcNAc1P
N-acetylglucosamine 1-phosphate
- GlcNAc6P
N-acetylglucosamine 6-phosphate
- L-Ala
L-alanine
- L-Lys
L-lysine
- MurNAc
N-acetylmuramic acid
- PG
peptidoglycan
- PPP
pentose phosphate pathway
- Spn
Streptococcus pneumoniae
- UDP
uridine diphosphate
- UDP-FucNAc
UDP-N-acetylfucosamine
- UDP-Gal
UDP-galactose
- UDP-GalNAc
UDP-N-acetylgalactosamine
- UDP-Glc
UDP-glucose
- UDP-GlcNAc
UDP-N-acetylglucosamine
- UDP-ManNAc
UDP-N-acetylmannosamine.
Footnotes
Funding. This work was supported by NIH COBRE-P20GM103646 (Phase I and II) grant.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01996/full#supplementary-material
References
- Asad S., Torabi S. F., Fathi-Roudsari M., Ghaemi N., Khajeh K. (2011). Phosphate buffer effects on thermal stability and H2O2-resistance of horseradish peroxidase. Int. J. Biol. Macromol. 48 566–570. 10.1016/j.ijbiomac.2011.01.021 [DOI] [PubMed] [Google Scholar]
- Bae D. H., Lane D. J. R., Jansson P. J., Richardson D. R. (2018). The old and new biochemistry of polyamines. Biochim. Biophys. Acta Gen Subj. 1862 2053–2068. 10.1016/j.bbagen.2018.06.004 [DOI] [PubMed] [Google Scholar]
- Balsells E., Guillot L., Nair H., Kyaw M. H. (2017). Serotype distribution of Streptococcus pneumoniae causing invasive disease in children in the post-PCV era: a systematic review and meta-analysis. PLoS One 12:e0177113. 10.1371/journal.pone.0177113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavanna S., Khandavilli S., Yuste J., Cohen J. M., Hosie A. H., Webb A. J., et al. (2009). Screening of Streptococcus pneumoniae ABC transporter mutants demonstrates that LivJHMGF, a branched-chain amino acid ABC transporter, is necessary for disease pathogenesis. Infect. Immun. 77 3412–3423. 10.1128/IAI.01543-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaroudj N., Lee D. H., Goldberg A. L. (2001). Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J. Biol. Chem. 276 24261–24267. 10.1074/jbc.m101487200 [DOI] [PubMed] [Google Scholar]
- Berical A. C., Harris D., Dela Cruz C. S., Possick J. D. (2016). Pneumococcal vaccination strategies. An update and perspective. Ann. Am. Thor. Soc. 13 933–944. 10.1513/AnnalsATS.201511-778FR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette K. A., Shenoy A. T., Milner J., II, Gilley R. P., McClure E., Hinojosa C. A., et al. (2016). Neuraminidase A-Exposed galactose promotes Streptococcus pneumoniae biofilm formation during colonization. Infect. Immun. 84 2922–2932. 10.1128/IAI.00277-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S. M., Kloosterman T. G., Kuipers O. P., Neves A. R. (2011). CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae. PLoS One 6:e26707. 10.1371/journal.pone.0026707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S. M., Kloosterman T. G., Manzoor I., Caldas J., Vinga S., Martinussen J., et al. (2018). Interplay between capsule expression and uracil metabolism in Streptococcus pneumoniae D39. Front. Microbiol. 9:321. 10.3389/fmicb.2018.00321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., et al. (2012). A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. L., Han D. L., Zhai Y. F., Wang J. H., Wang Y. F., Chen M. (2018). Characterization and mutational analysis of two UDP-Galactose 4-Epimerases in Streptococcus pneumoniae TIGR4. Biochemistry 83 37–44. 10.1134/S0006297918010054 [DOI] [PubMed] [Google Scholar]
- Cherazard R., Epstein M., Doan T. L., Salim T., Bharti S., Smith M. A. (2017). Antimicrobial resistant Streptococcus pneumoniae: prevalence, mechanisms, and clinical implications. Am. J. Ther. 24 e361–e369. 10.1097/MJT.0000000000000551 [DOI] [PubMed] [Google Scholar]
- Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., et al. (2018). MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46 W486–W494. 10.1093/nar/gky310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasquin M. F., Melamud E., Rabinowitz J. D. (2002). LC-MS Data Processing with MAVEN: A Metabolomic Analysis and Visualization Engine. Hoboken, NJ: John Wiley & Sons, Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasquin M. F., Melamud E., Rabinowitz J. D. (2012). LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr. Protoc. Bioinformatics Chater14:Unit14.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converso T. R., Goulart C., Rodriguez D., Darrieux M., Leite L. C. (2017). Systemic immunization with rPotD reduces Streptococcus pneumoniae nasopharyngeal colonization in mice. Vaccine 35 149–155. 10.1016/j.vaccine.2016.11.027 [DOI] [PubMed] [Google Scholar]
- Culotta V. C., Daly M. J. (2013). Manganese complexes: diverse metabolic routes to oxidative stress resistance in prokaryotes and yeast. Antioxid. Redox. Signal. 19 933–944. 10.1089/ars.2012.5093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt A., Hoyland C. N., Vollmer D., Bisle S., Cleverley R. M., Johnsborg O., et al. (2012). Attachment of capsular polysaccharide to the cell wall in Streptococcus pneumoniae. Microb. Drug Resist. 18 240–255. 10.1089/mdr.2011.0232 [DOI] [PubMed] [Google Scholar]
- Echlin H., Frank M. W., Iverson A., Chang T. C., Johnson M. D., Rock C. O., et al. (2016). Pyruvate oxidase as a critical link between metabolism and capsule biosynthesis in Streptococcus pneumoniae. PLoS Pathog 12:e1005951. 10.1371/journal.ppat.1005951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn A. T., Hogarty M. D. (2018). Myc, oncogenic protein translation, and the role of polyamines. Med Sci 6:E41. 10.3390/medsci6020041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey P. A. (1996). The leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 10 461–470. 10.1096/fasebj.10.4.8647345 [DOI] [PubMed] [Google Scholar]
- Geno K. A., Gilbert G. L., Song J. Y., Skovsted I. C., Klugman K. P., Jones C., et al. (2015). Pneumococcal capsules and their types: past, present, and future. Clin. Microbiol. Rev. 28 871–899. 10.1128/CMR.00024-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevrekci A. Ö. (2017). The roles of polyamines in microorganisms. World J. Microbiol. Biotechnol. 33:204. [DOI] [PubMed] [Google Scholar]
- Ghosh P., Luong T. T., Shah M., Thach T. T., Choi S., Lee S., et al. (2018). Adenylate kinase potentiates the capsular polysaccharide by modulating Cps2D in Streptococcus pneumoniae D39. Exp. Mol. Med. 50:116. 10.1038/s12276-018-0141-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Mejia A., Gamez G., Hammerschmidt S. (2018). Streptococcus pneumoniae two-component regulatory systems: the interplay of the pneumococcus with its environment. Int. J. Med. Microbiol. 308 722–737. 10.1016/j.ijmm.2017.11.012 [DOI] [PubMed] [Google Scholar]
- Greenwood B. (1999). The epidemiology of pneumococcal infection in children in the developing world. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354 777–785. 10.1098/rstb.1999.0430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenzi E., Gasc A. M., Sicard M. A., Hakenbeck R. (1994). A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol. Microbiol. 12 505–515. 10.1111/j.1365-2958.1994.tb01038.x [DOI] [PubMed] [Google Scholar]
- Gupta R., Yang J., Dong Y., Swiatlo E., Zhang J. R., Metzger D. W., et al. (2013). Deletion of arcD in Streptococcus pneumoniae D39 impairs its capsule and attenuates virulence. Infect. Immun. 81 3903–3911. 10.1128/IAI.00778-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson B. R., Lowe B. A., Neely M. N. (2011). Membrane topology and DNA-binding ability of the Streptococcal CpsA protein. J. Bacteriol. 193 411–420. 10.1128/JB.01098-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins M. A., Suits M. D., Marsters C., Boraston A. B. (2014). Structural and functional analysis of fucose-processing enzymes from Streptococcus pneumoniae. J. Mol. Biol. 426 1469–1482. 10.1016/j.jmb.2013.12.006 [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Igarashi K., Kashiwagi K. (2015). Modulation of protein synthesis by polyamines. IUBMB Life 67 160–169. 10.1002/iub.1363 [DOI] [PubMed] [Google Scholar]
- Igarashi K., Kashiwagi K. (2018). Effects of polyamines on protein synthesis and growth of Escherichia coli. J. Biol. Chem. 293 18702–18709. 10.1074/jbc.TM118.003465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imdad S., Chaurasia A. K., Kim K. K. (2018). Identification and validation of an antivirulence agent targeting HlyU-Regulated virulence in vibrio vulnificus. Front. Cell. Infect. Microbiol. 8:152. 10.3389/fcimb.2018.00152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R., Baliga N. S., Camilli A. (2005). Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187 8340–8349. 10.1128/jb.187.24.8340-8349.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Liu G. L., Chi Z., Hu Z., Chi Z. M. (2018). Genetics of trehalose biosynthesis in desert-derived Aureobasidium melanogenum and role of trehalose in the adaptation of the yeast to extreme environments. Curr. Genet. 64 479–491. 10.1007/s00294-017-0762-z [DOI] [PubMed] [Google Scholar]
- Jones C., Currie F., Forster M. J. (1991). N.m.r. and conformational analysis of the capsular polysaccharide from Streptococcus pneumoniae type 4. Carbohydr. Res. 221 95–121. 10.1016/0008-6215(91)80051-n [DOI] [PubMed] [Google Scholar]
- Kalscheuer R., Koliwer-Brandl H. (2014). Genetics of Mycobacterial Trehalose Metabolism. Microbiol. Spectr. 2:MGM2-0002-2013. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. (2012). KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40 D109–D114. 10.1093/nar/gkr988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay E. J., Yates L. E., Terra V. S., Cuccui J., Wren B. W. (2016). Recombinant expression of Streptococcus pneumoniae capsular polysaccharides in Escherichia coli. Open Biol. 6:150243. 10.1098/rsob.150243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler I. M., Mackie A., Santos-Zavaleta A., Billington R., Bonavides-Martinez C., Caspi R. (2017). The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 45 D543–D550. 10.1093/nar/gkw1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange R., Wagner C., de Saizieu A., Flint N., Molnos J., Stieger M., et al. (1999). Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237 223–234. 10.1016/s0378-1119(99)00266-8 [DOI] [PubMed] [Google Scholar]
- Larson T. R., Yother J. (2017). Streptococcus pneumoniae capsular polysaccharide is linked to peptidoglycan via a direct glycosidic bond to beta-D-N-acetylglucosamine. Proc. Natl. Acad. Sci. U.S.A. 114 5695–5700. 10.1073/pnas.1620431114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Park J., Lim M. S., Seong S. J., Seo J. J., Park S. M., et al. (2012). Quantile normalization approach for liquid chromatography-mass spectrometry-based metabolomic data from healthy human volunteers. Anal. Sci. 28 801–805. 10.2116/analsci.28.801 [DOI] [PubMed] [Google Scholar]
- Li B., Tang J., Yang Q., Li S., Cui X., Li Y., et al. (2017). NOREVA: normalization and evaluation of MS-based metabolomics data. Nucleic Acids Res. 45 W162–W170. 10.1093/nar/gkx449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Chen Y., Zhang B., Niu X., Song M., Luo Z., et al. (2016). Inhibition of sortase A by chalcone prevents Listeria monocytogenes infection. Biochem. Pharmacol. 106 19–29. 10.1016/j.bcp.2016.01.018 [DOI] [PubMed] [Google Scholar]
- Li J., Li J. W., Feng Z., Wang J., An H., Liu Y., et al. (2016). Epigenetic switch driven by DNA inversions dictates phase variation in Streptococcus pneumoniae. PLoS Pathog 12:e1005762. 10.1371/journal.ppat.1005762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W. Y., Clasquin M. F., Melamud E., Amador-Noguez D., Caudy A. A., Rabinowitz J. D. (2010). Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal. Chem. 82 3212–3221. 10.1021/ac902837x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan M., Gallego M. (2016). Pneumococcal vaccination: should we kill the enemy or just disarm it? Clin. Infect. Dis. 62 148–149. 10.1093/cid/civ805 [DOI] [PubMed] [Google Scholar]
- Ma Z., Zhang J. R. (2007). RR06 activates transcription of spr1996 and cbpA in Streptococcus pneumoniae. J. Bacteriol. 189 2497–2509. 10.1128/jb.01429-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manso A. S., Chai M. H., Atack J. M., Furi L., De Ste Croix M., Haigh R., et al. (2014). A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat. Commun. 5:5055. 10.1038/ncomms6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Ma Q., Zhou C., Chen X., Zhang H., Yang J., et al. (2014). DOOR 2.0: presenting operons and their functions through dynamic and integrated views. Nucleic Acids Res. 42 D654–D659. 10.1093/nar/gkt1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens L., Chambers M., Sturm M., Kessner D., Levander F., Shofstahl J., et al. (2011). mzML-a community standard for mass spectrometry data. Mol. Cell. Proteomics 10:R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamud E., Vastag L., Rabinowitz J. D. (2010). Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 82 9818–9826. 10.1021/ac1021166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael A. J. (2016). Polyamines in eukaryotes, bacteria, and archaea. J. Biol. Chem. 291 14896–14903. 10.1074/jbc.R116.734780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morona J. K., Miller D. C., Morona R., Paton J. C. (2004). The effect that mutations in the conserved capsular polysaccharide biosynthesis genes cpsA, cpsB, and cpsD have on virulence of Streptococcus pneumoniae. J. Infect. Dis. 189 1905–1913. 10.1086/383352 [DOI] [PubMed] [Google Scholar]
- Musher D. M. (1992). Infections caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunity, and treatment. Clin. Infect. Dis 14 801–807. [DOI] [PubMed] [Google Scholar]
- Nakamya M. F., Ayoola M. B., Park S., Shack L. A., Swiatlo E., Nanduri B. (2018). The role of cadaverine synthesis on pneumococcal capsule and protein expression. Med. Sci. 6:E8. 10.3390/medsci6010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paixao L., Oliveira J., Verissimo A., Vinga S., Lourenco E. C., Ventura M. R., et al. (2015). Host glycan sugar-specific pathways in Streptococcus pneumoniae: galactose as a key sugar in colonisation and infection [corrected]. PLoS One 10:e0121042. 10.1371/journal.pone.0121042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M. A. (2018). Polyamines in protozoan pathogens. J. Biol. Chem. 293 18746–18756. 10.1074/jbc.TM118.003342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichichero M. E., Khan M. N., Xu Q. (2016). Next generation protein based Streptococcus pneumoniae vaccines. Hum. Vaccin. Immunother. 12 194–205. 10.1080/21645515.2015.1052198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenow C., Maniar M., Trias J. (1999). Regulation of the α-Galactosidase activity in Streptococcus pneumoniae: characterization of the raffinose utilization system. Genome Res. 9 1189–1197. 10.1101/gr.9.12.1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh M., Abdullah M. R., Schulz C., Kohler T., Pribyl T., Jensch I., et al. (2014). Following in real time the impact of pneumococcal virulence factors in an acute mouse pneumonia model using bioluminescent bacteria. J. Vis. Exp. 84:51174. 10.3791/51174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P., Briles D. E., King J., Hale Y., Swiatlo E. (2009). Mucosal immunization with polyamine transport protein D (PotD) protects mice against nasopharyngeal colonization with Streptococcus pneumoniae. Exp. Biol. Med. 234 403–409. 10.3181/0809-RM-269 [DOI] [PubMed] [Google Scholar]
- Shah P., Nanduri B., Swiatlo E., Ma Y., Pendarvis K. (2011). Polyamine biosynthesis and transport mechanisms are crucial for fitness and pathogenesis of Streptococcus pneumoniae. Microbiology 157(Pt 2), 504–515. 10.1099/mic.0.042564-0 [DOI] [PubMed] [Google Scholar]
- Shah P., Swiatlo E. (2006). Immunization with polyamine transport protein PotD protects mice against systemic infection with Streptococcus pneumoniae. Infect. Immun. 74 5888–5892. 10.1128/iai.00553-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shainheit M. G., Mule M., Camilli A. (2014). The core promoter of the capsule operon of Streptococcus pneumoniae is necessary for colonization and invasive disease. Infect. Immun. 82 694–705. 10.1128/IAI.01289-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., et al. (1985). Measurement of protein using bicinchoninic acid. Anal. Biochem. 150 76–85. 10.1016/0003-2697(85)90442-7 [DOI] [PubMed] [Google Scholar]
- Song M., Teng Z., Li M., Niu X., Wang J., Deng X. (2017). Epigallocatechin gallate inhibits Streptococcus pneumoniae virulence by simultaneously targeting pneumolysin and sortase A. J. Cell Mol. Med. 21 2586–2598. 10.1111/jcmm.13179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A. L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47 D607–D613. 10.1093/nar/gky1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium. (2018). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46:2699. 10.1093/nar/gky092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throup J. P., Koretke K. K., Bryant A. P., Ingraham K. A., Chalker A. F., Ge Y., et al. (2000). A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35 566–576. 10.1046/j.1365-2958.2000.01725.x [DOI] [PubMed] [Google Scholar]
- Wang J., Qiu J., Tan W., Zhang Y., Wang H., Zhou X., et al. (2015). Fisetin inhibits Listeria monocytogenes virulence by interfering with the oligomerization of listeriolysin O. J. Infect. Dis. 211 1376–1387. 10.1093/infdis/jiu520 [DOI] [PubMed] [Google Scholar]
- Wen Z., Sertil O., Cheng Y., Zhang S., Liu X., Wang W. C., et al. (2015). Sequence elements upstream of the core promoter are necessary for full transcription of the capsule gene operon in Streptococcus pneumoniae strain D39. Infect. Immun. 83 1957–1972. 10.1128/IAI.02944-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun O., Priotto G., Tong J., Flevaud L., Chappuis F. (2010). NECT is next: implementing the new drug combination therapy for Trypanosoma brucei gambiense sleeping sickness. PLoS Negl. Trop. Dis. 4:e720. 10.1371/journal.pntd.0000720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun A., Philippe J., Abrahams K. A., Signor L., Roper D. I., Breukink E., et al. (2013). In vitro reconstitution of peptidoglycan assembly from the Gram-positive pathogen Streptococcus pneumoniae. ACS Chem. Biol. 8 2688–2696. 10.1021/cb400575t [DOI] [PubMed] [Google Scholar]
- Zheng Y., Zhang X., Wang X., Wang L., Zhang J., Yin Y. (2017). ComE, an essential response regulator, negatively regulates the expression of the capsular polysaccharide locus and attenuates the bacterial virulence in Streptococcus pneumoniae. Front. Microbiol. 8:277. 10.3389/fmicb.2017.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the NCBI GEO, accession number GSE130511.