Abstract
Many Gram-negative bacterial species use contact-dependent growth inhibition (CDI) systems to compete with neighboring cells. CDI+ strains express cell-surface CdiA effector proteins, which carry a toxic C-terminal region (CdiA-CT) that is cleaved from the effector upon transfer into the periplasm of target bacteria. The released CdiA-CT consists of two domains. The C-terminal domain is typically a nuclease that inhibits cell growth, and the N-terminal “cytoplasm-entry” domain mediates toxin translocation into the target-cell cytosol. Here, we use nuclear magnetic resonance and circular dichroism spectroscopic approaches to probe the structure, stability and dynamics of the cytoplasm-entry domain from Escherichia coli STEC_MHI813. Chemical shift analysis reveals that the CdiA-CTMHI813 entry domain is composed of a C-terminal helical bundle and a dynamic N-terminal region containing two disulfide linkages. Disruption of the disulfides by mutagenesis or chemical reduction destabilizes secondary structure over the N-terminus, but has no effect on the C-terminal helices. Though critical for N-terminal structure, the disulfides have only modest effects on global thermodynamic stability, and the entry domain exhibits characteristics of a molten globule. We find that the disulfides form in vivo as the entry domain dwells in the periplasm of inhibitor cells prior to target-cell recognition. CdiA-CTMHI813 variants lacking either disulfide still kill target bacteria, but disruption of both bonds abrogates growth inhibition activity. We propose that the entry domain’s dynamic structural features are critical for function. In its molten globule-like state, the domain resists degradation after delivery, yet remains pliable enough to unfold for membrane translocation.
Keywords: bacterial competition, BamA, two-partner secretion, type V secretion system
INTRODUCTION
Bacteria use many strategies to communicate and compete with other microorganisms in the environment. One widespread competitive mechanism is contact-dependent growth inhibition (CDI), in which Gram-negative bacteria use CdiB/CdiA two-partner secretion (TPS) proteins to deliver protein toxins directly into neighboring target cells [1, 2]. CdiB is an outer-membrane β-barrel protein that exports and presents the CdiA effector protein on the surface of CDI+ bacteria. CdiA forms a filament that projects from the inhibitor cell to recognize receptors on the surface of neighboring target bacteria [3–7]. Receptor-interactions initiate a series of conformational changes that ultimately transfer the C-terminal toxin region of CdiA (CdiA-CT) into the target cell [7]. CdiA-CT sequences are highly variable between bacteria, though the region is typically demarcated by a conserved peptide motif, exemplified by the Val-Glu-Asn-Asn (VENN) sequence commonly found in CdiA proteins from the Enterobacteriaceae [1, 8]. CdiA-CT polymorphism corresponds to many distinct toxin activities. For example, CdiA-CTEC93 from Escherichia coli EC93 dissipates the proton gradient across the cytoplasmic membrane of target cells [9], whereas CdiA-CTSTECO31 from E. coli STEC_O31 contains an EndoU RNase domain that cleaves the anticodon loop of tRNAGlu molecules [10]. CDI+ bacteria protect themselves from self-intoxication with specific CdiI immunity proteins, which bind and neutralize cognate toxin domains [1, 2]. Thus, CDI enables bacteria to inhibit the growth of non-isogenic competitors. Because neighboring sibling cells are immune to intoxication, these systems are thought to play a significant role in kin selection and self-nonself discrimination in bacteria [11, 12].
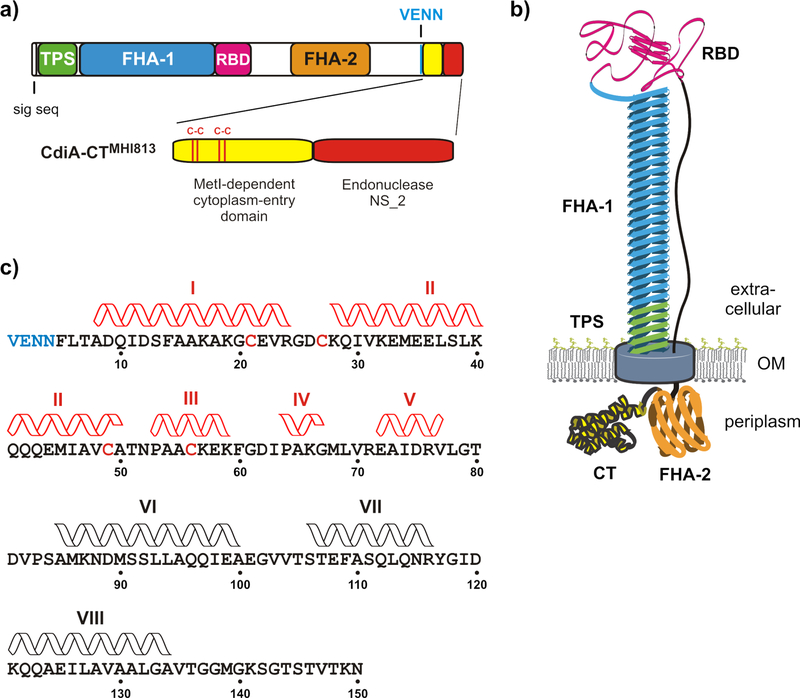
CdiA proteins vary considerably in size and sequence between species, but all share the same general architecture with constituent domains arranged in the order that they act temporally during toxin delivery (Fig. 1a) [1, 2, 7]. The N-terminus of CdiA contains sequences required for export from the inhibitor cell. These include a signal peptide for sec-dependent secretion into the periplasm, and the TPS transport domain, which is required for CdiB-dependent export to the cell surface (Fig. 1a). Remarkably, CdiA export across the outer membrane is halted at a specific site, and the entire C-terminal half of the effector is retained within the periplasm of the inhibitor cell (Fig. 1b). The extracellular portion of CdiA forms a thin filament that projects several hundred Å from the cell surface [7, 13, 14]. This filament corresponds primarily to the FHA-1 domain, which comprises numerous filamentous hemagglutinin (FHA) peptide repeats. These degenerate sequences fold into a right-handed β-helix, with each 19-residue FHA-1 motif extending the filament approximately 4.8 Å [13, 14]. The receptor-binding domain resides at the distal tip of this filament (Fig. 1b), where it is poised to interact with target bacteria. The CdiA chain then reverses course and returns to the cell to sequester its C-terminal half in the periplasm [7]. The periplasmic region includes the toxic CdiA-CT, as well as a second FHA-2 peptide-repeat domain that is distinct from the FHA-1 filament (Figs. 1a & 1b). CdiA remains in this state of arrested secretion until it binds a target bacterium, after which export resumes and the FHA-2 domain is deposited directly onto the target cell. FHA-2 becomes stably associated with target-cell outer membrane and is hypothesized to form a transmembrane conduit to deliver toxin [7]. Once inside the target-cell periplasm, the CdiA-CT is released by cleavage near the VENN sequence. The CdiA-CT region is composed of two domains, each with a distinct function in the CDI pathway. The C-terminal domain is typically a toxic nuclease that inhibits target-cell growth, whereas the N-terminal “cytoplasm-entry” domain guides the nuclease across the target-cell inner membrane into the cytosol [15]. Genetic evidence indicates that cytoplasm-entry domains hijack specific integral membrane proteins to mediate translocation. For example, the entry domain from E. coli STEC_MHI813 requires MetI – a component of the ABC transporter for methionine – to deliver the C-terminal Endonuclease NS_2 domain into target bacteria (Fig. 1a) [15]. Entry domains are often linked to different nuclease domains in CdiA proteins. The CdiA-CT regions from E. coli strains 3006 and NC101 contain unrelated tRNase toxin domains, yet share nearly identical cytoplasm-entry domains that exploit the PtsG glucose transporter [15]. Thus, entry domains are modular and capable of guiding different toxic payloads across the cytoplasmic membrane.
Figure 1. CdiA domain architecture and topology.
(a) CdiA carries an N-terminal signal sequence peptide followed by the two-partner secretion (TPS) transport domain (Pfam: PF05860). The FHA-1 domain is composed of filamentous hemagglutinin (FHA) peptide repeats (PF05594). The centrally located receptor-binding domain (RBD) recognizes E. coli BamA on target bacteria. The FHA-2 domain (PF13332) is composed of a second, distinct class of peptide repeats. The VENN motif demarcating the C-terminal toxin region (CdiA-CT) is indicated. The CdiA-CT region from E. coli MHI813 is composed of an N-terminal entry domain that recognizes MetI, and a C-terminal Endonuclease NS_2 toxin domain (PF13930). (b) Topology of CdiA on the surface of inhibitor cells. The N-terminal TPS domain of CdiA is thought to interact with CdiB and the FHA-1 domain forms a filament capped by the RBD. The CdiA chain returns to the cell to sequester the FHA-2 and CdiA-CT regions in the periplasm. (c) Sequence of the MetI-dependent cytoplasm-entry domain of CdiA-CTMHI813. Domain residues are numbered from Val1 of the VENN motif (blue font). The positions of α-helices determined by ∆13Cα chemical shift analysis are indicated. Disulfide-dependent helices are rendered in red.
Entry domains play a key role in CDI toxin delivery, but the underlying molecular mechanisms are largely unexplored. Though metabolite transporters are commonly hijacked in this process, it seems unlikely that protein toxins could be imported in the same manner as small molecules. This conclusion is consistent with data showing that transport-defective MetI and PtsG proteins still support CDI toxin translocation [15]. Moreover, entry domains exploit several classes of integral membrane proteins including the major facilitator superfamily (MFS), resistance-nodulation-division (RND), phosphotransferase system (PTS) and AAA+ superfamily [15, 16]. These membrane protein families differ from one another in structure and function, further suggesting that they act as passive receptors rather than active translocators during CDI. Though translocation is poorly understood, there is evidence that the proton gradient across the target-cell inner membrane is required for toxin entry [17]. Dissipation of the proton gradient blocks toxins from reaching the cytoplasm, but still allows delivery into the target-cell periplasm. Remarkably, delivered CdiA-CTs can dwell in the periplasm of de-energized target bacteria for up to 20 minutes and still remain competent to enter the cytoplasm once the proton gradient is reestablished [17]. These findings indicate that CDI toxins cross target-cell membranes in discrete steps, and further suggest that the proton gradient powers the final translocation step into the cytosol.
Crystal structures are available for several CdiA-CT nuclease domains, but the cytoplasm-entry domains are usually not resolved in these models [18–21]. Therefore, we have turned to nuclear magnetic resonance (NMR) and circular dichroism (CD) spectroscopy to gain insights into the structure and thermodynamic stability of cytoplasm-entry domains. We find that the MetI-dependent entry domain from Escherichia coli STEC_MHI813 is a dynamic α-helical bundle that exhibits relatively low thermodynamic stability. Two disulfide linkages stabilize α-helices near the N-terminus, and this region becomes largely disordered when the bonds are reduced or disrupted through mutagenesis. These disulfide bonds are important for efficient toxin delivery during CDI, and inspection of several different entry domain families reveals paired Cys residues in the same relative positions as in CdiA-CTMHI813. Taken together, our results suggest that cytoplasm-entry domains are under selective pressure to maintain low global stability. We propose that the dynamic, molten globule-like properties of the CdiA-CTMHI813 entry domain are critical for toxin translocation and are likely a general feature of all cytoplasm-entry domains.
RESULTS
The cytoplasm-entry domain of CdiA-CTMHI813 is α-helical
1H-15N HSQC spectra of the oxidized cytoplasm-entry domain from CdiA-CTMHI813 revealed considerable structural heterogeneity consistent with mixed, non-native disulfide bonds (data not shown). However, the domain exhibited well resolved resonances and favorable spectral dispersion when reduced with dithiothreitol (Fig. S1a). Therefore, we probed the structural features of the CdiA-CTMHI813 entry domain under reducing conditions using triple-resonance HNCACB and HN(CO)CACB experiments [22]. 13Cα resonance assignments were made for 143 of the domain’s 150 residues (Table S3). Differences in 13Cα chemical shift relative to random coil can be used to identify secondary structure elements, with positive and negative ∆13Cα values characteristic of α-helices and β-strands, respectively [23]. The ∆13Cα plot reveals four regions of continuous positive chemical shift (Fig. 2a), suggesting that Ala65-Val77, Ala85-Ala100, Ser106-Arg116 and Lys121-Ala134 form α-helices (Fig. 1c). These helical regions are separated by short stretches of negative ∆13Cα (Fig. 2a), suggesting that Leu78-Val82, Glu101-Thr105 and Tyr117-Asp120 form connecting turns. In contrast, the N-terminal portion (Val1-Phe60) of the reduced domain exhibits small-range ∆13Cα fluctuations (Fig. 2a), which are indicative of a predominately disordered structure. However, we note that moderately positive ∆13Cα values over Ala8-Ala16 and Glu36-Gln42 suggest that these regions may be in rapid exchange between random-coil and α-helix (Fig. 2a).
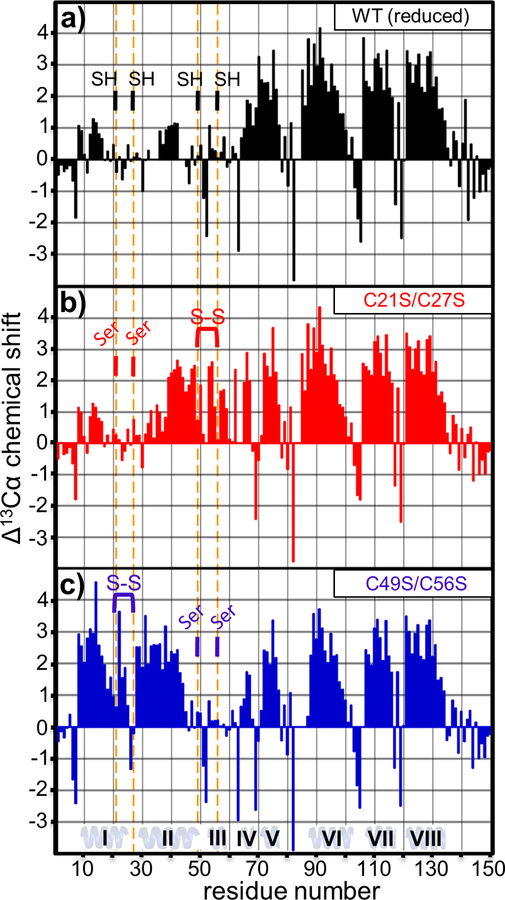
Figure 2. ∆13Cα chemical shift analysis.
The 13Cα differential chemical shift was plotted as a function of residue for: (a) reduced wild-type cytoplasm-entry domain, (b) oxidized C21S/C27S domain and (c) oxidized C49S/C56S domain. Vertical orange lines show the positions of Cys residues or Cys to Ser substitutions. Cys residues are indicated as reduced (SH) or oxidized (S-S).
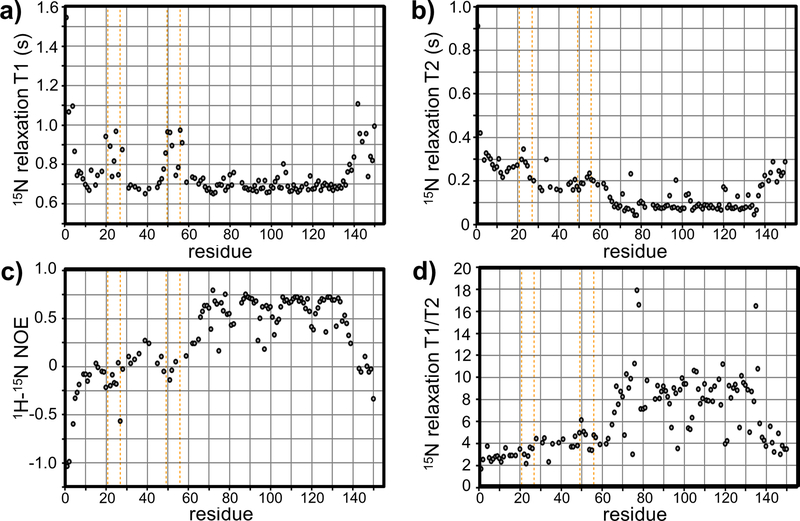
We next measured 15N relaxation parameters (T1, T2, 1H-15N NOE), which reflect the molecular tumbling and internal motions of the entry domain. We obtained 124 T1 and T2 curves for the wild-type domain under reducing conditions (Figs. 3a & 3b). In general, the C-terminal region shows greater T1/T2 ratios (Fig. 3d) and higher 1H-15N NOE values (Fig. 3c) relative to the N-terminal region. These data again suggest that the C-terminal region from Ala65 – Ala134 is structured [24]. Local T2 increases at Asp75, Gln97, Asp120 and Lys121 (Fig. 3b), coupled with decreased 1H-15N NOE and invariant T1 values (Figs. 3c & 3a), suggest that these residues form flexible turns that connect helices [24]. T1 values for the C-terminal helical region (Ala65-Ala134) are relatively constant (Fig. 3a), suggesting that the helices tumble as a unit with an overall correlation time of about 8.7 ns. By contrast, N-terminal residues Val1-Phe60 have significantly lower T1/T2 values (Fig. 3d), which are indicative of increased internal motion. Moreover, decreased 1H-15N NOE values over the N-terminus indicate increased mobility relative to the C-terminal four-helix bundle (Fig. 3c). Within the N-terminal region, residues Gly20-Lys28 and Val48-Lys57 display increased T1 relaxation times (Fig. 3a), suggesting that they constitute highly flexible loops [24]. Residues Gly20-Lys28 and Cys49-Lys57 have relatively uniform 1H-15N NOE values near zero (Fig. 3c), and increased T1 values (Fig. 3a). Taken together with the small ∆13Cα values (Fig. 2a), these data suggest that Gly20-Lys28 and Cys49-Lys57 are in dynamic equilibrium between ordered and unfolded conformations [24]. Given that the T2 values for these regions do not show dramatic rate enhancements (Fig. 3b), the interconversion must be fast enough (>103 s−1) to have little effect on T2.
Figure 3. NMR relaxation parameters.
The wild-type cytoplasm-entry domain was 15N-labeled and reduced with DTT to determine 15N-relaxation parameters. Plots show (a) T1 relaxation, (b) T2 relaxation and (c) 1H15N-NOE values for each backbone amide resonance. T1/T2 ratios are plotted in panel (d). Vertical orange lines show the positions of Cys residues.
Disulfide bonds stabilize N-terminal α-helices
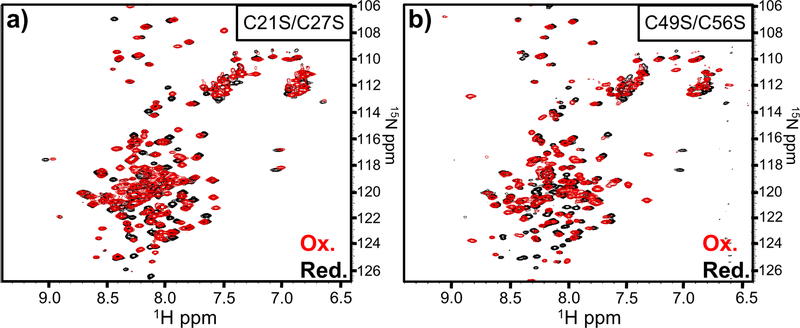
Given that regions surrounding the reduced Cys residues appear to be in dynamic equilibrium between structured and unstructured states, we tested whether disulfide linkages promote N-terminal secondary structure. Because the wild-type domain is not tractable under oxidizing conditions, we generated domain variants that retain either the Cys21-Cys27 or Cys49-Cys56 pair. The resulting C21S/C27S and C49S/C56S domains show well-resolved resonances and good spectral dispersion under both reducing and oxidizing conditions (Figs. 4a, 4b, S1b & S1c). Moreover, 1H-15N HSQC spectra for the reduced C21S/C27S and C49S/C56S domains are nearly identical to that of the wild-type domain under reducing conditions, except for amide resonance shifts corresponding to the Ser substitutions and adjacent residues (Fig. S2). These data suggest that each domain variant adopts the same overall conformation in the absence of disulfide bonds.
Figure 4. NMR analysis of oxidized entry domain variants.
1H-15N HSQC spectra were acquired for entry domains carrying (a) C21S/C27S and (b) C49S/C56S mutations. Red spectra correspond to oxidized forms, and black spectra were acquired under reducing conditions in the presence of 10 mM DTT.
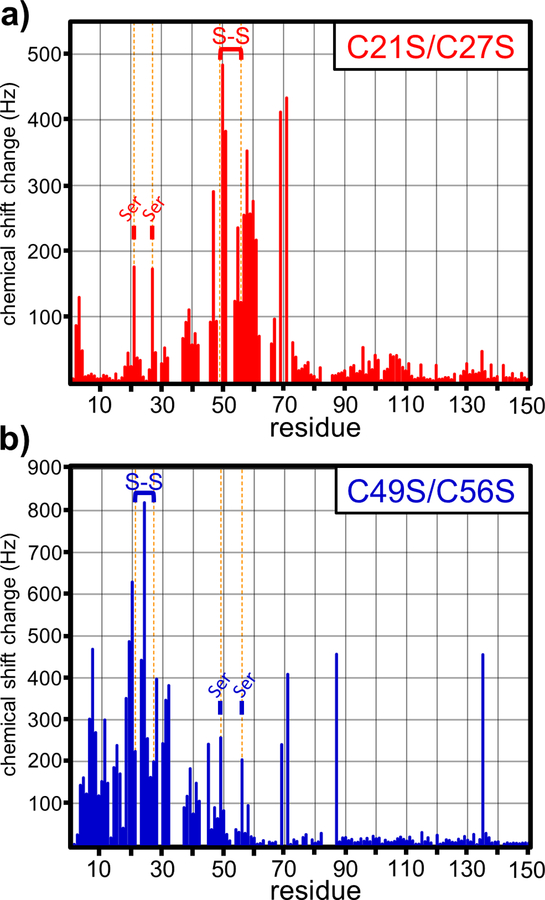
We then examined changes in the 1H-15N HSQC spectra when the C21S/C27S and C49S/C56S domains form disulfides under oxidizing conditions (Figs. 4a & 4b). In general, the most significant chemical shift perturbations occurred within the N-terminal region (Fig. 5). ∆13Cα plots of the oxidized C21S/C27S and C49S/C56S domains indicate that disulfide bonds promote significant α-helix formation over the N-terminus (Figs. 2b & 2c). The Cys49-Cys56 bond in the oxidized C21S/C27S domain stabilizes α-helices over residues Leu39-Ala50 and Pro53-Lys59 (Fig. 2b). Similarly, formation of the Cys21-Cys27 bond in oxidized C49S/C56S induces two continuous stretches of positive ∆13Cα over residues Ala8-Arg24 and Lys28-Glu44 (Fig. 2c). These latter helices appear to be separated by a turn at Asp26 and Cys27, which exhibit negative ∆13Cα values (Fig. 2c). These data suggest that the two disulfides induce the formation of three α-helices at Ala8-Arg24 (helix I), Lys28-Ala50 (helix II) and Pro53-Lys59 (helix III) in the wild-type domain (Fig. 1c). Disulfide bond formation also disrupts the continuous stretch of positive ∆13Cα values observed over residues Ala65-Val77 under reducing conditions. This region appears to be split into two smaller helices of Pro64-Gly67 (helix IV) and Glu72-Val77 (helix V) when either disulfide forms (Figs. 2a, 2b & 2c). Residues Met68-Arg71 appear to undergo a disulfide-dependent transition from α-helix to linker. The amide resonances for Leu69 and Arg71 show particularly large chemical shift changes between reducing and oxidizing conditions (Fig. 5), consistent with a pronounced change in local structure upon disulfide formation. Thus, eight distinct α-helices are observable in the cytoplasm-entry domain. The formation of N-terminal helices I, II and III is disulfide dependent, and these linkages also significantly influence the central IV and V helices. By contrast, helices VI, VII and VIII at the C-terminus of the domain are unaffected by oxidation state.
Figure 5. Disulfides induce local secondary structure formation.
Chemical shift perturbations due to disulfide formation were plotted as a function of residue number for the (a) C21S/C27S and (b) C49S/C56S entry domain variants. Vertical orange lines denote the positions of Cys residues in the wild-type entry domain. Disulfide bonds are indicated as S-S and positions with substitutions indicated with Ser.
The CdiA-CTMHI813 cytoplasm-entry domain has features of a molten globule
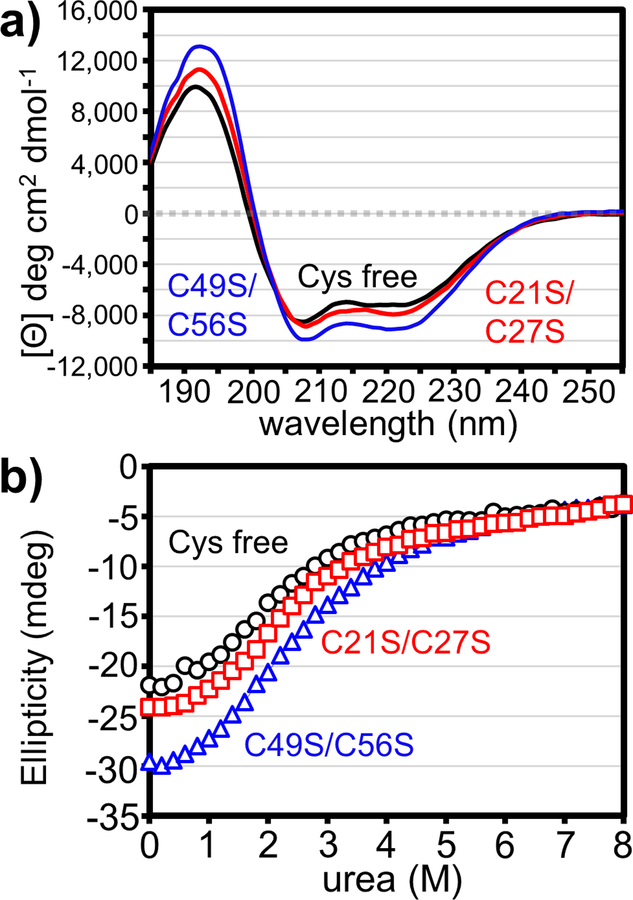
Disulfide-dependent structural features suggest that the entry domain requires these bonds to maintain global stability. To test this hypothesis, we used circular dichroism (CD) spectroscopy to monitor domain stability during chemical denaturation with urea. Because DTT absorbs strongly at the wavelengths used to measure protein secondary structure, we generated a “Cys-free” (C21S/C27S/C49S/C56S) domain to mimic the wild-type domain under reducing conditions. As anticipated from the NMR data, the CD spectra for oxidized C21S/C27S, oxidized C49S/C56S and Cys-free domains are all characteristic of α-helical proteins with minima at 208 and 222 nm (Fig. 6a) [25, 26]. We note that the C49S/C56S domain exhibits more pronounced molar ellipticity than the C21S/C27S and Cys-free domains (Fig. 6a). This suggests that the C49S/C56S domain has greater α-helical content, consistent with the number of helical residues predicted from ∆13Cα analyses (Fig. 2). Though the transitions were not cooperative (Fig. 6b), we fitted a two-state model to estimate the free energies of domain unfolding (∆Gu) (Table 1, Fig. S7). Surprisingly, individual disulfides provide little stability to the cytoplasm-entry domain. For example, the C49S/C56S domain retains the Cys21-Cys27 disulfide that stabilizes helices I and II, yet it has the same ∆Gu as the Cys-free domain lacking this bond (Table 1). Moreover, stable globular proteins typically have ∆Gu values of 5–9 kcal/mol [27], and therefore the entry domains exhibit relatively low thermodynamic stabilities. Low ∆Gu values and non-cooperative unfolding transitions are characteristic of molten globules [28, 29], which contain native-like secondary structure, but have loosely packed hydrophobic cores [30–32]. We note that the entry domain CD spectra show greater ellipticity at 208 nm relative to 222 nm (Fig. 6a), which is often a feature of molten globules (Fig. 6a) [30]. In addition, the murea values for chemical denaturation (0.7–1.0) are fairly low for a protein of this size (expected ~1.7) (Table 1) [33, 34]. This latter parameter is also consistent with a molten globule-like state [30].
Figure 6. Thermodynamic stability of cytoplasm-entry domains.
(a) Circular dichroism spectra of cytoplasm-entry domain variants. Data are presented as mean residue ellipticity for three averaged scans. (b) Chemical denaturation of entry domain variants. Ellipticity at 224 nm was measured as a function of urea concentration.
Table 1.
Stability parameters derived from CD.
| Protein | ΔGu (kcal/mol) | murea (kcal/mol•M) |
|---|---|---|
| Cys free | 1.6 +/− 0.1 | 0.8 +/− 0.05 |
| C21S/C27S | 2.2 +/− 0.1 | 1.0 +/− 0.04 |
| C49S/C56S | 1.6 +/− 0.1 | 0.7 +/− 0.03 |
Cysteines are important for CdiAMHI813 mediated growth inhibition
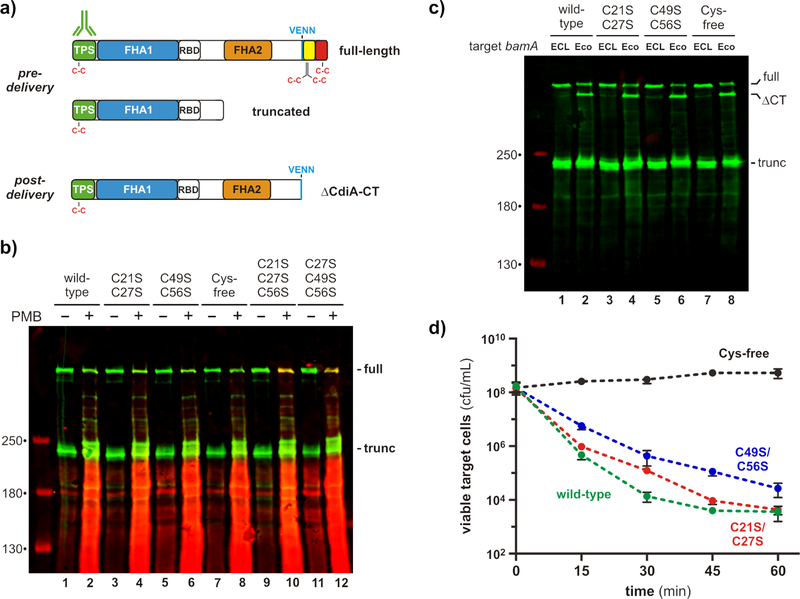
We next asked whether Cys residues in the entry domain form disulfides under physiological conditions. CdiA has an unusual surface topology in which its C-terminal half is held within the periplasm prior to target-cell recognition (Fig. 1b) [7]. Because the periplasm is an oxidizing environment, we reasoned that Cys residues in the entry domain should form disulfides. We tested this prediction by treating cells with maleimide-conjugated fluorescent dye to label reduced Cys residues in vivo. To ensure that CdiA is maintained in the pre-delivery state, we used an E. coli strain that lacks CdiA-receptors and therefore cannot deliver toxin to sibling cells [4]. Because the outer membrane of E. coli is impermeable to maleimide-dye [7], we also treated cells with polymyxin B to allow dye into the periplasm. Total protein was then isolated for immunoblotting with antibodies to the N-terminal TPS transport domain, and the blots were analyzed by two-color fluorimaging to detect dye-labeled CdiA. Immunoblotting showed that CdiA is produced in both full-length and truncated forms as described previously (Figs. 7a & 7b) [7, 35]. Truncated CdiA lacks the FHA-2 and CdiA-CT regions (Fig. 7a), but is still presented on the cell surface where it functions as an adhesin [7, 35]. Neither full-length nor truncated CdiA was labeled with maleimide dye, even when cells were permeabilized with polymyxin B (Fig. 7b, lanes 1 & 2). This result suggests that the eight Cys residues of CdiAMHI813 – two in the N-terminal TPS transport domain, four in the entry domain, and two in the C-terminal Endonuclease NS_2 domain – are all oxidized (Fig. 7a). Similar results were obtained with CdiA proteins that contain C21S/C27S, C49S/C56S and Cys-free versions of the cytoplasm-entry domain (Fig. 7b, lanes 4, 6 & 8). To demonstrate that reduced Cys residues can actually be detected using this approach, we also tested CdiA proteins that contain single Cys residues corresponding to Cys21 and Cys49 of the entry domain. These latter CdiA proteins were only labeled when the cells were permeabilized (Fig. 7b, lanes 10 & 12), consistent with the periplasmic localization of the entry domain. Taken together, these data indicate that Cys21-Cys27 and Cys49-Cys56 form disulfide bonds in vivo.
Figure 7. Entry domain disulfides form in vivo.
(a) Location of Cys residues in the three major forms of CdiA. CdiA is initially produced by inhibitor cells in full-length and truncated forms. Upon binding target bacteria, CdiA is processed at the VENN sequence to release the CdiA-CT region. (b) Maleimide-dye probing for sulfhydryls. Inhibitor cells expressing the indicated CdiA proteins were treated with maleimide-conjugated fluorescent dye in the presence or absence of polymyxin B (PMB). Proteins were analyzed by immunoblotting with antibodies to the N-terminal TPS transport domain of CdiA. Maleimide-dye fluorescence is rendered in red and antibody fluorescence in green. The migration positions of full-length and truncated CdiA are indicated on the right of the blot. (c) CdiA-CT processing. Inhibitor cells expressing the indicated CdiA proteins were incubated with bamAEco target cells or mock-target bamAECL cells. Proteins were analyzed by immunoblotting with antibodies to the N-terminal TPS transport domain of CdiA. The migration position of full-length, processed (∆CT) and truncated CdiA are indicated on the right of the blot. (d) Target-cell inhibition. Inhibitor cells expressing the indicated CdiA proteins were incubated with bamAEco target cells in shaking broth, and viable target cells were enumerated as colony forming units (cfu) per mL as a function of time. Data are presented as the average ± standard deviation for three independent experiments.
The CdiA-CT region is cleaved from full-length CdiA after transfer into the target-cell periplasm (Fig. 7a) [7]. To determine whether entry domain disulfides influence this processing, we used immunoblot analysis to monitor CdiA-CT cleavage in response to target-cell recognition. Wild-type CdiA is processed efficiently when inhibitors are mixed with target bacteria that express the bamA gene from E. coli (bamAEco), which encodes the receptor for CdiA (Fig. 7c, lane 2). By contrast, the CdiA-CT region is not cleaved during co-culture with mock target cells that express bamA from Enterobacter cloacae (bamAECL) (Fig. 7c, lane 1), because the BamAECL protein is not recognized as a receptor by CdiA [4]. Mutation of Cys residues within the cytoplasm-entry domain has no discernable effect on processing, and even the Cys-free domain was efficiently cleaved from CdiA (Fig. 7c, lane 8), demonstrating that the N-terminal α-helices are not required for CdiA-CT cleavage.
Finally, we tested whether the disulfides are required for toxin delivery into the cytoplasm of target bacteria. We incubated inhibitor cells with target bacteria at a 1:1 ratio and tracked viable target cells over 60 min of co-culture. Target bacteria were killed rapidly by the wild-type CdiA, with viable target cells decreasing 100-fold after 15 min and ~104-fold after 45 min (Fig. 7d). CdiA proteins carrying the C21S/C27S and C49S/C56S mutations killed target bacteria with somewhat slower kinetics, but the number of viable targets was about the same as wild-type after 60 min of co-culture (Fig. 7d). By contrast, CdiA carrying the Cys-free cytoplasm-entry domain showed no growth inhibition activity (Fig. 7d). Thus, the CdiA-CTMHI813 entry domain requires at least one disulfide linkage to maintain function, though the identity of the stabilizing bond does not appear to be critical.
DISCUSSION
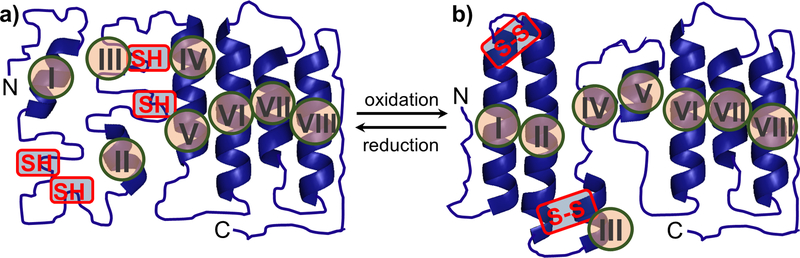
CdiA proteins collectively deliver a variety of nuclease toxins into Gram-negative bacteria [8, 10, 18, 20, 36–38]. Delivery requires two discrete translocation steps that pass the toxin domain across the outer and inner membranes of target cells. Recent work suggests that toxin transfer into the target-cell periplasm is mediated by the FHA-2 domain of CdiA, which is proposed to form a conduit through the outer membrane of target bacteria [7]. Once inside the periplasm, the CdiA-CT region is cleaved for subsequent transport into the target-cell cytoplasm. This latter translocation step requires the cytoplasm-entry domain, which exploits integral membrane proteins to guide the toxin into the cytosol [15]. Here, we show that the cytoplasm-entry domain of CdiA-CTMHI813 is predominately α-helical and contains a dynamic N-terminal region that is organized by two disulfide bonds. The bond linking Cys21 and Cys27 stabilizes N-terminal helices I and II, and the Cys49-Cys56 linkage extends helix II and promotes the formation of helix III (Fig. 8). When these disulfides are reduced, the N-terminal region undergoes rapid interconversion between disordered and α-helical conformations. The C-terminal portion of the entry domain forms a relatively cohesive helical bundle that folds independently from the dynamic N-terminal region. Though we were unable to directly examine the wild-type entry domain under oxidizing conditions, our data strongly suggest that it contains both disulfides and is composed of eight α-helices (Fig. 8). Moreover, the disulfide-stabilized form is physiologically relevant, because the entry domain resides in the oxidizing environment of the inhibitor-cell periplasm prior to delivery. Therefore, the disulfides must not impede export through CdiB, nor do they interfere with translocation into target bacteria. In fact, these bonds are clearly important for function, because CdiA-CTMHI813 containing the Cys-free entry domain is unable to inhibit target bacteria in competition co-culture. Taken together, these results indicate that one or more of the disulfide-stabilized helical elements is critical for efficient toxin delivery.
Figure 8. A model of cytoplasm-entry domain secondary structure.
(a) Reduced. (b) Oxidized.
Crystal structures are available for several CdiA-CT•CdiI complexes, but none of the models contains an intact cytoplasm-entry domain [10, 18, 20, 21, 36–38]. The results presented here provide one possible explanation for these observations, because the dynamic, molten globule-like features of the CdiA-CTMHI813 entry domain suggest that it should be difficult to crystallize. In addition, the domain’s low thermodynamic stability could render it sensitive to limited proteolysis, which is often used to generate compact complexes for crystallography. Indeed, mass spectrometry and SDS-PAGE analyses have shown that entry domains are often cleaved from the CdiA-CT in the crystallization liquor and consequently are not present in CdiA-CT•CdiI crystals [10, 18]. The CdiA-CT•CdiIo11EC869 complex from E. coli strain EC869 is a notable exception to this trend [20, 39]. The C-terminal half of the YciB-dependent entry domain is resolved in the CdiA-CT•CdiIo11EC869 structure, where it forms a helical bundle that packs against the toxic DNase domain [20]. These van der Waals contacts presumably prevented cleavage from the nuclease domain during limited proteolysis, but the functional significance of inter-domain interactions is not clear. Related YciB-dependent entry domains are associated with predicted ParB and BECR toxins in other CdiA proteins, and these domains share no sequence homology with the Zn2+-dependent DNase found in CdiA-CTo11EC869 (Fig. S3). Moreover, the YciB-dependent entry domain is able to guide a heterologous tRNase toxin from Burkholderia pseudomallei 1026b into E. coli target bacteria [15, 36]. The latter chimeric CdiA-CT is not found in any naturally occurring CdiA protein, further arguing that the entry domain need not evolve specific interactions with its toxic cargo. Similarly, the MetI-dependent entry domain analyzed here is linked to at least seven toxin families in various CdiA proteins (Fig. S4). Most of these toxins lack functional annotations, though CdiA from Pectobacterium carotovorum PCC21 carries an obvious RNase T1 domain; and the toxin from Pectobacterium atrosepticum SCRI1043 is homologous to a characterized RNase domain that cleaves 16S rRNA (Fig. S4) [18]. Collectively, these observations illustrate how cytoplasm-entry and toxic nuclease domains are rearranged naturally to generate novel delivery modules.
Entry domains have diversified to exploit several import pathways, but they probably share common structural features required for translocation function. We note that the C-terminal regions of MetI- and YciB-dependent entry domains both fold into α-helical bundles (Fig. S5a) [20]. Moreover, all characterized entry domains contain paired Cys residues in the N-terminal region (Fig. S5b). This conservation is particularly striking given that enterobacterial CdiA proteins – which typically range from ~2,700 to over 6,000 residues – are often otherwise devoid of Cys residues [1, 7]. Taken together with data showing that at least one disulfide is required for CdiA-CTMHI813-mediated cell killing, these observations suggest that paired Cys residues are generally important for entry domain function. Our results show that disulfides are not required for transfer into the target-cell periplasm, nor do they contribute to CdiA-CT processing after delivery. Perhaps these bonds protect the released CdiA-CT fragment from peptidases in the target-cell periplasm. (Fig. S6) CdiA-CT processing is thought to occur within, or adjacent to, the VENN motif [7], and therefore the disulfides are well-positioned to stabilize the new free N-terminus (Fig. S4). According to this model, paired Cys residues are conserved across many different entry domains because disulfide linkages are an effective and parsimonious way to protect the released CdiA-CT from degradation.
Proteins are typically secreted in an unfolded state, and therefore must be prevented from attaining their final tertiary structures prior to export [40]. Many proteins destined for export are bound by the cytosolic SecB chaperone, which delays folding to ensure the proteins remain competent for translocation through SecYEG [41–44]. Given that the CdiA-CT region must cross four membranes as it passes between inhibitor and target cells, it is perhaps not surprising that the cytoplasm-entry domain unfolds so readily. Further, the molten globule-like properties of the entry domain could very well be critical for function. We note that the entry domain can dwell in the periplasm of inhibitor cell for several hours, yet it remains poised for rapid export once a target cell is encountered [7, 17]. Therefore, the entry domain must unfold on-demand, because the β-barrel lumen of CdiB is only wide enough to accommodate an individual α-helix [45]. Low global stability presumably also contributes to transport across target-cell membranes, though these processes remain poorly understood. Toxin translocation into the target-cell cytoplasm is particularly enigmatic, but several studies on colicin import provide a possible framework for this phenomenon. Colicins are diffusible antibacterial proteins that bind to specific receptors on E. coli and transfer their C-terminal toxin domains into the cell [46]. Like CdiA, colicins carry a variety of toxins that either degrade nucleic acids or form ion-conducting pores in the cell membrane. Upon encountering the cytoplasmic membrane, colicin pore-forming domains appear to adopt a molten globule-like state that promotes the insertion of hydrophobic core helices α8 and α9 into the lipid bilayer [47–49]. Kleanthous and coworkers have found that the nuclease domains of colicins E3 and E9 also adopt a molten globule state when mixed with anionic lipids and have proposed that these conformational changes enable autonomous translocation into the cytosol [50–52]. The loosely packed core of the entry domain could promote the same process, perhaps allowing an individual helix to enter the membrane. However, entry domains are not particularly hydrophobic, though predicted helices VI and VIII from CdiA-CTMHI813 are somewhat amphipathic. This overall lack of hydrophobicity may explain why entry domains hijack integral membrane proteins during translocation [15]. According to this model, interactions between the entry domain and its receptor facilitate insertion into the membrane. Although our study focused on the cytoplasm-entry domain, nuclease domain cargoes are undoubtedly subject to the same thermodynamic constraints. Indeed, our unpublished data show that toxin domains have relatively low unfolding energies. Moreover, many different CDI toxin domains retain nuclease activity after chemical denaturation and refolding [8, 10, 18, 20, 36–38, 53–55]. Thus, both domains of the CdiA-CT region undergo facile unfolding transitions, consistent with the ability to be translocated across multiple membranes.
The evolutionary pressure for cytoplasm-entry domains of low thermodynamic stability is easy to rationalize, but it is less obvious why these domains almost always contain paired Cys residues that presumably form disulfide linkages. The data presented here show that each disulfide has a modest effect on global domain stability, perhaps indicating that the bonds act locally to protect the free N-terminus of CdiA-CTMHI813 from peptidases. However, N-terminal helix stability does not precisely correlate with translocation efficiency. Helices I and II are destabilized in the C21S/C27S domain, yet this variant supports about the same level of target-cell killing as wild-type (see Fig. 7d). Such robustness is difficult to reconcile with the conservation of disulfides across entry domains (Fig. S5b). One possible explanation is that the disulfide bonds function as redox switches that control the folding state of the N-terminal region. Such disulfide switches have been identified and characterized in a number of other secreted proteins [56]. In this model, disulfides are important for the structural integrity of the CdiA-CT immediately after release into the target-cell periplasm, but the bonds are eventually reduced to initiate membrane translocation. Though the periplasm is an oxidizing environment, Gram-negative bacteria express periplasmic protein disulfide isomerases capable of reducing these bonds [57, 58]. Moreover, respiring bacteria contain reduced ubiquinol and menaquinol dissolved in the cytoplasmic membrane [59]. In principle, electrons from these quinols could be used to reduce the disulfides [60, 61]. This latter possibility is particularly intriguing because it would ensure that reduction occurs at the periplasmic surface of the cell membrane. Thus, it is tempting to speculate that receptor interactions bring the entry domain into close proximity with the cytoplasmic membrane, where the disulfides are subsequently reduced to initiate membrane insertion.
MATERIALS AND METHODS
Plasmid constructions
All plasmids and oligonucleotide primers are listed in Supplementary Tables S1 and S2, respectively. A DNA fragment encoding the cytoplasm-entry domain from CdiAMHI813 (NCBI Reference Sequence: WP_001383049.1) was amplified from plasmid pCH11446 using primers CH4211/CH3734 and ligated to plasmid pET21d using NcoI/XhoI restriction sites to generate plasmid pCH12234. By convention, E. coli CdiA-CT residues are numbered from Val1 of the VENN peptide motif (Fig. 1c) [18, 53]. Cysteine to serine substitutions were generated by overlap-extension PCR using primers CH4051, CH4052, CH4053 and CH4054. The resulting PCR products containing C21S/C27S, C49S/C56S and C21S/C27S/C49S/C56S mutations were ligated to plasmid pET21d at the NcoI/XhoI restriction sites to produce plasmids pCH13794, pCH15123 and pCH15122, respectively. The constructions of plasmids pZS21::bamAEco and pZS21::bamAECL have been described previously [4].
Chimeric CdiA expression plasmids were constructed by allelic exchange of the counter-selectable pheS* marker from plasmid pCH10163 as described [20]. The various cdiA-CT/cdiIMHI813 sequences were PCR amplified using primers CH3174/CH3175, then combined with upstream and downstream homology regions amplified from the cdiAEC93 gene. The cdiAEC93 upstream homology fragment was amplified with primers DL1527/DL2470, and the downstream fragment amplified with primers DL1663/DL2368. The three PCR products (cdiA-CT/cdiIMHI813, upstream cdiAEC93 and downstream cdiAEC93) were then fused to each other using overlapping-extension PCR [62] using primers DL1527/DL2368. The final DNA products (100 ng) were electroporated together with plasmid pCH10163 (300 ng) into E. coli DY378 cells [63]. Clones with recombinant plasmids were selected on yeast extract glucose-agar supplemented with 33 µg/mL chloramphenicol and 10 mM D/L-p-chlorophenylalanine.
Protein expression, purification, and oxidation
Entry domains were over-produced in E. coli Rosetta cells (Novagen) grown at 37 °C with shaking in either LB media or M9 minimal media supplemented with 15NH4Cl [99% 15N] and/or D-glucose [U-13C6-99%]. Cells were grown to OD600 ~0.3 and expression induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Cell were harvested by centrifugation after 6 h of culture. Cell pellets were re-suspended in 50 mM sodium phosphate (pH 8.0), 300 mM sodium chloride, 10 mM imidazole, 10 mM β-mercaptoethanol (β-ME) and broken by passage through a French pressure cell. Cell debris was removed by centrifugation at 14,600 x g in a Beckman JA-20 rotor for 1 h. Supernatants were loaded onto a Sepharose Fast Flow Ni2+ column (GE Healthcare) and the column washed with seven column volumes of lysis buffer supplemented with 35 mM imidazole. Domains were eluted in lysis buffer supplemented with 250 mM imidazole. Purified proteins were dialyzed against 50 mM sodium phosphate (pH 6.5), 0.02% NaN3 and concentrated with 10 kDa cut-off Centriprep centrifugal filter (Millipore). Oxidation was assessed by SDS-PAGE under reducing and non-reducing conditions.
Circular dichroism (CD) spectroscopy
Circular dichroism measurements were acquired on a Jasco J-1500 spectrophotometer coupled to an EXOS Koolance thermoregulator. Spectra were acquired with 10 μM cytoplasm-entry domain in 20 mM sodium phosphate (pH 6.5) from 180–280 nm in a 1 mm path-length quartz cuvette. Chemical denaturation measurements were made with 2.5 μM cytoplasm-entry domain in 20 mM sodium phosphate (pH 6.5) in the presence of increasing concentrations of urea. The measurements were performed in a 1 cm path-length quartz cuvette. We monitored the CD signal at 224 nm. Thermodynamic parameters were derived by linear extrapolation [64], and model fitting was performed in the Mathematica software suite (Wolfram Research).
NMR spectroscopy
NMR experiments were performed with a Varian Inova 600 MHz spectrometer at 30 °C. NMR data was processed with the nmrPipe software package [65], and assignments were made with ANSIG3.3 [66]. Sequential backbone and side chain assignments were obtained from measurements of 15N, 13C labeled protein samples from the following two- and three-dimensional experiments: 1H-15N HSQC, 1H-13C HSQC, HNCACB, and HBCBCACONH. Disulfide bonds were reduced with 10 mM dithiothreitol (DTT) for 2 h prior to NMR data acquisition. Resonance assignments were made for most residues of the reduced WT protein, oxidized C21S/C27S protein, and oxidized C49S/C56S protein (Table S3).
Relaxation measurements
Measurements of T1 and T2 values for 15N nuclei used the pulse sequences of Farrow et al. [67]. T1 measurements were performed with relaxation delays of 11, 55, 110, 220, 440, 660, 880, 1320, and 1650 ms. T2 measurements were acquired with relaxation delays of 16.61, 33.22, 49.82, 66.43, 83.04, 99.65, 116.3, 132.9, 149.5, 166.1, 182.7, and 199.3 ms. Recycle delays were 1.5 s for both T1 and T2 measurements. T1 and T2 values were obtained by least-squares fitting of peak heights to a single exponential decay function. Steady-state 1H-15N NOE values were determined from spectra recorded in the presence and absence of 3 s of proton saturation. Cross peak heights were used to determine the heteronuclear NOE in the 1H-15N HSQC spectra. The measurements were performed in triplicate using a refocused 1H-15N HSQC pulse sequence with gradient coherence selection.
Dye-labeling and immunoblotting
E. coli CH9591 (EPI100 ∆bamA::cat pZS21::bamAECL) cells carrying CdiAMHI813 expression plasmids were suspended at OD600 ~ 0.05 in LB media supplemented with 66 µg/mL chloramphenicol and grown to mid-log phase at 37 °C. Cultures were then treated with spectinomycin (100 µg/mL) for 20 min to block protein synthesis. Cells were harvested by centrifugation and re-suspended at OD600 ~ 0.3 in 1.0 mL of 1x phosphate buffered saline supplemented with 1 mM MgSO4 (PBS-Mg) to probe intact cells, or 100 µg/mL polymyxin B in PBS-Mg to probe permeabilized cells. IRDye680LT-maleimide (LI-COR) was added to a final concentration of 40 µM for intact cells and 120 µM for permeabilized cells. Labeling reactions were incubated in the dark at room temperature for 15 min, then quenched with 6 mM β-ME. Cells were collected by centrifugation and washed with PBS-Mg supplemented with 6 mM β-ME. Cell pellets were re-suspended in urea-lysis buffer [50% urea, 150 mM NaCl, 20 mM Tris-HCl (pH 8.0)] and subjected to a freeze-thaw cycle to extract proteins for SDS-PAGE and immunoblotting. Urea-soluble proteins (5 µL) were resolved by SDS-PAGE on Tris-tricine 6% polyacrylamide gels, then blotted to low-fluorescence PVDF membranes for immunoblotting with polyclonal antisera raised against the N-terminal TPS transport domain of CdiA [35]. Fluorescence signals from the 800CW-conjugated goat anti-rabbit IgG secondary antibody and 680LT-maleimide dye were detected and visualized using a LI-COR Odyssey infrared imager.
Toxin delivery and competition co-cultures assays
To monitor CdiA-CTMHI813 processing, E. coli CH9591 inhibitor strains carrying CdiAMHI813 expression plasmids were mixed at a 1:2 ratio with E. coli CH9604 (EPI100 ∆bamA::cat pZS21::bamAEco) or E. coli CH9591 (bamAECL mock targets) for 20 min in LB media supplemented with 100 µg/mL spectinomycin to block new protein synthesis. Cells were collected by centrifugation and proteins extracted with urea-lysis buffer. Urea-soluble proteins (5 µL) were analyzed by immunoblotting with antisera to the CdiA TPS transport domain. Cell killing assays were performed using rifampicin-resistant E. coli MC4100 target cells. Target cells were mixed at a 1:1 ratio in LB media with inhibitor strains that express CdiAMHI813 variants. Competition co-cultures were incubated at 37 °C with shaking, and samples were withdrawn after 0, 15, 30, 45 and 60 min to enumerate viable target cells as colony forming units (cfu) on LB-agar supplemented with rifampicin. Mean viable target-cell counts from three independent experiments are reported together with the standard deviation.
Biological Magnetic Resonance Data Bank Accession Numbers
CdiA-CTMHI813 entry domain WT (reduced): BMRB 27753
CdiA-CTMHI813 entry domain C21S/C27S: BMRB 27754
CdiA-CTMHI813 entry domain C49S/C56S: BMRB 27755
Supplementary Material
HIGHLIGHTS.
Antibacterial contact dependent inhibition toxins have conserved pairs of cysteines in their cytoplasm entry domains.
Cytoplasm entry domain cysteine pairs form disulfides that are important for translocation into the target cell cytoplasm.
Disulfide formation promotes helical stability in an otherwise disordered protein region.
Disulfides are formed in the periplasm of the toxin-delivering cell and do not affect CdiA expression or processing.
ACKNOWLEDGMENTS
We thank Calvin Lin, Victor Passanisi, Arslan Harmon, Nicholas Wilson and Zainab Noorsher for assistance with protein purification and characterization. This work was supported by grants GM117930 (C.S.H.) and AI121789 (C.S.H. and F.W.D.) from the National Institutes of Health.
REFERENCES
- [1].Willett JL, Ruhe ZC, Goulding CW, Low DA, Hayes CS. Contact-Dependent Growth Inhibition (CDI) and CdiB/CdiA Two-Partner Secretion Proteins. J Mol Biol 2015;427:3754–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Jones AM, Low DA, Hayes CS. Can’t you hear me knocking: contact-dependent competition and cooperation in bacteria. Emerg Top Life Sci 2017;1:75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Aoki SK, Malinverni JC, Jacoby K, Thomas B, Pamma R, Trinh BN, et al. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol Microbiol 2008;70:323–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ruhe ZC, Wallace AB, Low DA, Hayes CS. Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species. MBio 2013;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Beck CM, Willett JL, Cunningham DA, Kim JJ, Low DA, Hayes CS. CdiA Effectors from Uropathogenic Escherichia coli Use Heterotrimeric Osmoporins as Receptors to Recognize Target Bacteria. PLoS Pathog 2016;12:e1005925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ruhe ZC, Nguyen JY, Xiong J, Koskiniemi S, Beck CM, Perkins BR, et al. CdiA Effectors Use Modular Receptor-Binding Domains To Recognize Target Bacteria. MBio 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ruhe ZC, Subramanian P, Song K, Nguyen JY, Stevens TA, Low DA, et al. Programmed Secretion Arrest and Receptor-Triggered Toxin Export during Antibacterial Contact-Dependent Growth Inhibition. Cell 2018;175:921–33 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, et al. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 2010;468:439–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Aoki SK, Webb JS, Braaten BA, Low DA. Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli. J Bacteriol 2009;191:1777–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Michalska K, Quan Nhan D, Willett JLE, Stols LM, Eschenfeldt WH, Jones AM, et al. Functional plasticity of antibacterial EndoU toxins. Mol Microbiol 2018;109:509–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Anderson MS, Garcia EC, Cotter PA. Kind discrimination and competitive exclusion mediated by contact-dependent growth inhibition systems shape biofilm community structure. PLoS Pathog 2014;10:e1004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Garcia EC, Anderson MS, Hagar JA, Cotter PA. Burkholderia BcpA mediates biofilm formation independently of interbacterial contact-dependent growth inhibition. Mol Microbiol 2013;89:1213–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Makhov AM, Hannah JH, Brennan MJ, Trus BL, Kocsis E, Conway JF, et al. Filamentous hemagglutinin of Bordetella pertussis. A bacterial adhesin formed as a 50-nm monomeric rigid rod based on a 19-residue repeat motif rich in beta strands and turns. J Mol Biol 1994;241:110–24. [DOI] [PubMed] [Google Scholar]
- [14].Kajava AV, Cheng N, Cleaver R, Kessel M, Simon MN, Willery E, et al. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol Microbiol 2001;42:279–92. [DOI] [PubMed] [Google Scholar]
- [15].Willett JL, Gucinski GC, Fatherree JP, Low DA, Hayes CS. Contact-dependent growth inhibition toxins exploit multiple independent cell-entry pathways. Proc Natl Acad Sci U S A 2015;112:11341–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Koskiniemi S, Garza-Sanchez F, Edman N, Chaudhuri S, Poole SJ, Manoil C, et al. Genetic analysis of the CDI pathway from Burkholderia pseudomallei 1026b. PLoS One 2015;10:e0120265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ruhe ZC, Nguyen JY, Beck CM, Low DA, Hayes CS. The proton-motive force is required for translocation of CDI toxins across the inner membrane of target bacteria. Mol Microbiol 2014;94:466–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Beck CM, Morse RP, Cunningham DA, Iniguez A, Low DA, Goulding CW, et al. CdiA from Enterobacter cloacae delivers a toxic ribosomal RNase into target bacteria. Structure 2014;22:707–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Johnson PM, Beck CM, Morse RP, Garza-Sanchez F, Low DA, Hayes CS, et al. Unraveling the essential role of CysK in CDI toxin activation. Proc Natl Acad Sci U S A 2016;113:9792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Morse RP, Nikolakakis KC, Willett JL, Gerrick E, Low DA, Hayes CS, et al. Structural basis of toxicity and immunity in contact-dependent growth inhibition (CDI) systems. Proc Natl Acad Sci U S A 2012;109:21480–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Morse RP, Willett JL, Johnson PM, Zheng J, Credali A, Iniguez A, et al. Diversification of beta-Augmentation Interactions between CDI Toxin/Immunity Proteins. J Mol Biol 2015;427:3766–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yamazaki T, Lee W, Arrowsmith CH, Muhandiram DR, Kay LE. A Suite of Triple-Resonance Nmr Experiments for the Backbone Assignment of N-15, C-13, H-2 Labeled Proteins with High-Sensitivity. J Am Chem Soc 1994;116:11655–66. [Google Scholar]
- [23].Wishart DS, Sykes BD. The C-13 Chemical-Shift Index - a Simple Method for the Identification of Protein Secondary Structure Using C-13 Chemical-Shift Data. Journal of Biomolecular Nmr 1994;4:171–80. [DOI] [PubMed] [Google Scholar]
- [24].Farrow NA, Zhang OW, Formankay JD, Kay LE. Comparison of the Backbone Dynamics of a Folded and an Unfolded Sh3 Domain Existing in Equilibrium in Aqueous Buffer. Biochemistry-Us 1995;34:868–78. [DOI] [PubMed] [Google Scholar]
- [25].Kelly SM, Price NC. The application of circular dichroism to studies of protein folding and unfolding. Biochim Biophys Acta 1997;1338:161–85. [DOI] [PubMed] [Google Scholar]
- [26].Greenfield N, Davidson B, Fasman GD. The use of computed optical rotatory dispersion curves for the evaluation of protein conformation. Biochemistry-Us 1967;6:1630–7. [DOI] [PubMed] [Google Scholar]
- [27].Pace CN. Conformational stability of globular proteins. Trends Biochem Sci 1990;15:14–7. [DOI] [PubMed] [Google Scholar]
- [28].Demarest SJ, Deechongkit S, Dyson HJ, Evans RM, Wright PE. Packing, specificity, and mutability at the binding interface between the p160 coactivator and CREB-binding protein. Protein Sci 2004;13:203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kjaergaard M, Teilum K, Poulsen FM. Conformational selection in the molten globule state of the nuclear coactivator binding domain of CBP. P Natl Acad Sci USA 2010;107:12535–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Vassilenko KS, Uversky VN. Native-like secondary structure of molten globules. Bba-Protein Struct M 2002;1594:168–77. [DOI] [PubMed] [Google Scholar]
- [31].Dolgikh DA, Gilmanshin RI, Brazhnikov EV, Bychkova VE, Semisotnov GV, Venyaminov SY, et al. Alpha-Lactalbumin - Compact State with Fluctuating Tertiary Structure. Febs Letters 1981;136:311–5. [DOI] [PubMed] [Google Scholar]
- [32].Mok KH, Nagashima T, Day IJ, Hore PJ, Dobson CM. Multiple subsets of side-chain packing in partially folded states of alpha-lactalbumins. P Natl Acad Sci USA 2005;102:8899–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Myers JK, Pace CN, Scholtz JM. Denaturant M-Values and Heat-Capacity Changes - Relation to Changes in Accessible Surface-Areas of Protein Unfolding. Protein Sci 1995;4:2138–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol 1986;131:266–80. [DOI] [PubMed] [Google Scholar]
- [35].Ruhe ZC, Townsley L, Wallace AB, King A, Van der Woude MW, Low DA, et al. CdiA promotes receptor-independent intercellular adhesion. Mol Microbiol 2015;98:175–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Johnson PM, Gucinski GC, Garza-Sanchez F, Wong T, Hung LW, Hayes CS, et al. Functional Diversity of Cytotoxic tRNase/Immunity Protein Complexes from Burkholderia pseudomallei. J Biol Chem 2016;291:19387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Batot G, Michalska K, Ekberg G, Irimpan EM, Joachimiak G, Jedrzejczak R, et al. The CDI toxin of Yersinia kristensenii is a novel bacterial member of the RNase A superfamily. Nucleic Acids Res 2017;45:5013–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Michalska K, Gucinski GC, Garza-Sanchez F, Johnson PM, Stols LM, Eschenfeldt WH, et al. Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs. Nucleic Acids Res 2017;45:10306–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Poole SJ, Diner EJ, Aoki SK, Braaten BA, t’Kint de Roodenbeke C, Low DA, et al. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet 2011;7:e1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dalbey RE, Kuhn A. Protein traffic in Gram-negative bacteria--how exported and secreted proteins find their way. FEMS Microbiol Rev 2012;36:1023–45. [DOI] [PubMed] [Google Scholar]
- [41].Collier DN, Bankaitis VA, Weiss JB, Bassford PJ Jr. The antifolding activity of SecB promotes the export of the E. coli maltose-binding protein. Cell 1988;53:273–83. [DOI] [PubMed] [Google Scholar]
- [42].Liu G, Topping TB, Cover WH, Randall LL. Retardation of Folding as a Possible Means of Suppression of a Mutation in the Leader Sequence of an Exported Protein. Journal of Biological Chemistry 1988;263:14790–3. [PubMed] [Google Scholar]
- [43].Hardy SJ, Randall LL. A kinetic partitioning model of selective binding of nonnative proteins by the bacterial chaperone SecB. Science 1991;251:439–43. [DOI] [PubMed] [Google Scholar]
- [44].Topping TB, Randall LL. Chaperone SecB from Escherichia coli mediates kinetic partitioning via a dynamic equilibrium with its ligands. Journal of Biological Chemistry 1997;272:19314–8. [DOI] [PubMed] [Google Scholar]
- [45].Clantin B, Delattre AS, Rucktooa P, Saint N, Meli AC, Locht C, et al. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science 2007;317:957–61. [DOI] [PubMed] [Google Scholar]
- [46].Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, et al. Colicin biology. Microbiol Mol Biol Rev 2007;71:158–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schendel SL, Cramer WA. On the nature of the unfolded intermediate in the in vitro transition of the colicin E1 channel domain from the aqueous to the membrane phase. Protein Sci 1994;3:2272–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zakharov SD, Lindeberg M, Griko Y, Salamon Z, Tollin G, Prendergast FG, et al. Membrane-bound state of the colicin E1 channel domain as an extended two-dimensional helical array. Proc Natl Acad Sci U S A 1998;95:4282–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].van der Goot FG, Gonzalez-Manas JM, Lakey JH, Pattus F. A ‘molten-globule’ membrane-insertion intermediate of the pore-forming domain of colicin A. Nature 1991;354:408–10. [DOI] [PubMed] [Google Scholar]
- [50].Mosbahi K, Lemaitre C, Keeble AH, Mobasheri H, Morel B, James R, et al. The cytotoxic domain of colicin E9 is a channel-forming endonuclease. Nat Struct Biol 2002;9:476–84. [DOI] [PubMed] [Google Scholar]
- [51].Mosbahi K, Walker D, James R, Moore GR, Kleanthous C. Global structural rearrangement of the cell penetrating ribonuclease colicin E3 on interaction with phospholipid membranes. Protein Sci 2006;15:620–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mosbahi K, Walker D, Lea E, Moore GR, James R, Kleanthous C. Destabilization of the colicin E9 Endonuclease domain by interaction with negatively charged phospholipids: implications for colicin translocation into bacteria. J Biol Chem 2004;279:22145–51. [DOI] [PubMed] [Google Scholar]
- [53].Diner EJ, Beck CM, Webb JS, Low DA, Hayes CS. Identification of a target cell permissive factor required for contact-dependent growth inhibition (CDI). Genes Dev 2012;26:515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Jones AM, Garza-Sanchez F, So J, Hayes CS, Low DA. Activation of contact-dependent antibacterial tRNase toxins by translation elongation factors. Proc Natl Acad Sci U S A 2017;114:E1951–E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Nikolakakis K, Amber S, Wilbur JS, Diner EJ, Aoki SK, Poole SJ, et al. The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems. Mol Microbiol 2012;84:516–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hogg PJ. Disulfide bonds as switches for protein function. Trends Biochem Sci 2003;28:210–4. [DOI] [PubMed] [Google Scholar]
- [57].Kadokura H, Katzen F, Beckwith J. Protein disulfide bond formation in prokaryotes. Annu Rev Biochem 2003;72:111–35. [DOI] [PubMed] [Google Scholar]
- [58].Messens J, Collet JF. Pathways of disulfide bond formation in Escherichia coli. Int J Biochem Cell Biol 2006;38:1050–62. [DOI] [PubMed] [Google Scholar]
- [59].Bishop DHL, Pandya KP, King HK. Ubiquinone and Vitamin K in Bacteria. Biochem J 1962;83:606–&. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bader M, Muse W, Ballou DP, Gassner C, Bardwell JC. Oxidative protein folding is driven by the electron transport system. Cell 1999;98:217–27. [DOI] [PubMed] [Google Scholar]
- [61].Bader MW, Xie T, Yu CA, Bardwell JC. Disulfide bonds are generated by quinone reduction. J Biol Chem 2000;275:26082–8. [DOI] [PubMed] [Google Scholar]
- [62].Aiyar A, Xiang Y, Leis J. Site-directed mutagenesis using overlap extension PCR. Methods Mol Biol 1996;57:177–91. [DOI] [PubMed] [Google Scholar]
- [63].Thomason L, Court DL, Bubunenko M, Costantino N, Wilson H, Datta S, et al. Recombineering: genetic engineering in bacteria using homologous recombination. Current protocols in molecular biology / edited by Frederick M Ausubel [et al. 2007;Chapter 1:Unit 1 16. [DOI] [PubMed] [Google Scholar]
- [64].Pace CN, Shaw KL. Linear extrapolation method of analyzing solvent denaturation curves. Proteins 2000;Suppl 4:1–7. [DOI] [PubMed] [Google Scholar]
- [65].Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 1995;6:277–93. [DOI] [PubMed] [Google Scholar]
- [66].Kraulis PJ. Ansig - a Program for the Assignment of Protein H-1 2d-Nmr Spectra by Interactive Computer-Graphics. J Magn Reson 1989;84:627–33. [Google Scholar]
- [67].Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR 1994;4:727–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.