Abstract
Enterovirus D68 (EV-D68) undergoes structural transformation between mature, cell-entry intermediate (A-particle) and empty forms throughout its life cycle. Structural information for the various forms and antibody-bound capsids will facilitate the development of effective vaccines and therapeutics against EV-D68 infection, which causes childhood respiratory and paralytic diseases worldwide. Here, we report the structures of three EV-D68 capsid states representing the virus at major phases. We further describe two original monoclonal antibodies (15C5 and 11G1) with distinct structurally defined mechanisms for virus neutralization. 15C5 and 11G1 engage the capsid loci at icosahedral three-fold and five-fold axes, respectively. To block viral attachment, 15C5 binds three forms of capsids, and triggers mature virions to transform into A-particles, mimicking engagement by the functional receptor ICAM-5, whereas 11G1 exclusively recognizes the A-particle. Our data provide a structural and molecular explanation for the transition of picornavirus capsid conformations and demonstrate distinct mechanisms for antibody-mediated neutralization.
Enterovirus D68 (EV-D68), a member of human enterovirus species D, was first isolated from children with respiratory infections in 19621. It was considered an obscure pathogen until more frequent clusters of infections were recognized during the last decade in Europe, North America and Japan2–4. In particular, an outbreak in the United States in 2014 accounted for 1,153 cases and 14 deaths5,6. EV-D68 replicates primarily in the human respiratory tract causing bronchiolitis and pneumonia, and has been associated with fever, cough and wheezing. Importantly, it has also been associated with severe neurological complications such as paralysis, acute flaccid myelitis and cranial nerve dysfunction7,8. Although EV-D68 poses a substantial public health threat, the dearth of virology and immunology information on this emerging picornavirus hinders the design and development of effective vaccines and therapeutics. The crystal structures of the mature EV-D68 virion and its complexes with a potential drug or sialic acid receptor have recently been reported9,10 and the major features of EV-D68 are similar to those of other known picornaviruses, such as poliovirus, human rhinovirus (HRV) and enterovirus A71 (EV71)11–14. The capsid of EV-D68 contains 60 copies of each of four proteins VP1 to VP4, arranged with pseudo T = 3 symmetry9. The capsid accommodates deep surface depressions (‘canyons’) around each icosahedral five-fold axis, which can be bound by the sialic acid receptor. Such binding can trigger a series of conformational changes of the capsid, such as collapse of the pocket and conformational changes at receptor-binding sites, leading to an earlier intermediate state that is different from the classical uncoating intermediates (so-called ‘A-particles’) of other picornaviruses10,15–17. In addition to sialic acid, intercellular adhesion molecule 5 (ICAM-5) has been identified as a functional receptor for EV-D68, but the binding site and the in vivo role of ICAM-5 remain unclear10,18.
It is well known that picornaviruses generally undergo structural transitions of their capsids and four major particle forms can be observed during the life cycle: procapsid, mature virion, A-particle and empty particle11–13,15,17. Such particle variation is necessary for viral infection and is a structural form of immune flummox that represents an obstacle for vaccine development against EV-D6819,20. As powerful tools for investigating neutralizing mechanisms, neutralizing antibodies against picornavirus EV71 and HRV have been shown to promote premature uncoating of virions and genome release21,22. However, more detailed neutralization mechanisms related to whether neutralizing antibodies can cross-react with viruses at different conformational states of the life cycle, or interfere with dynamic transitions between states have not been extensively studied.
Here we show two EV-D68-specific neutralizing antibodies, 15C5 and 11G1, that exhibit different binding properties and neutralizing mechanisms. Near-atomic-resolution cryogenic electron microscopy (cryoEM) structures of procapsid, mature virion, A-particle and three neutralizing antibody-bound viral complexes reveal that the neutralizing antibody 11G1 specifically binds the A-particle, but the neutralizing antibody 15C5 binds and triggers the transformation of mature virions into classical A-particles, thus mimicking host receptor interactions. These results provide atomic-level details and insights into the process of virus entry and potential mechanisms of antibody neutralization, and inform a structure-based rationale for the design of EV-D68 vaccines and therapeutics.
Results
ICAM-5-induced EV-D68 A-particle shares similar structure to procapsid.
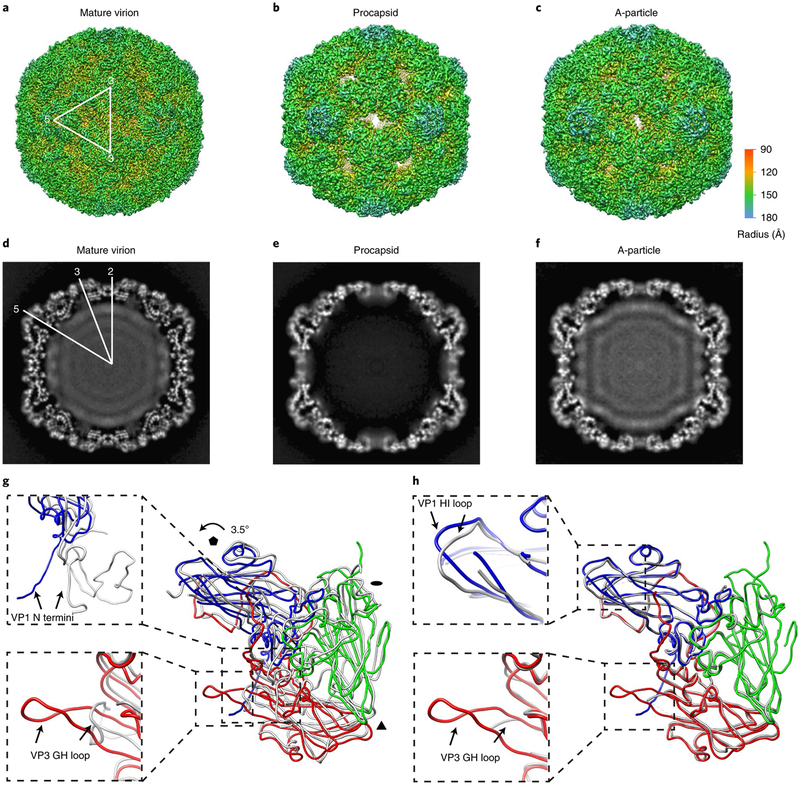
A clinical EV-D68 strain (GenBank accession no. KM881710) isolated from the 2014 outbreak in the United States was used for this analysis. Like other picornaviruses, the viral culture of EV-D68 in vitro yields two major types of particle: procapsid and mature virion (Supplementary Fig. 1a,b). On addition of the functional receptor ICAM-5, viral genomic RNA in mature virions was exposed and vulnerable to degradation at a significantly lower temperature (about 30 °C) than in untreated mature virions (about 55 °C) based on a particle stability thermal release assay (Supplementary Fig. 1e). These data suggest that ICAM-5 triggered transformation of EV-D68 mature virions to A-particles (A-particle_i5) in vitro (Supplementary Fig. 1c–e). We then determined the cryoEM structures of these three capsid forms at near-atomic reslolutions (3.8, 3.4 and 4.0 Å for the procapsid, mature virion and A-particle_i5, respectively; Fig. 1a–c and Supplementary Figs. 2 and 3). Of note, approximately 4% of particles were excluded during image processing selection on mature virions (Supplementary Fig. 2). Three-dimensional (3D) reconstruction of the excluded particles yielded a 4.4-Å-resolution map of A-particles essentially identical to ICAM-5-induced A-particles (correlation coefficient value of 0.99; Supplementary Fig. 4). Therefore, this minor class of particles derived from the supernatant of virus-infected cells is now referred to as A-particles triggered by an unknown stimulus (A-particle_us). The density map derived from the major class of particles demonstrated that EV-D68 mature virions feature a compactly packed capsid with a maximum diameter of ~320 Å along its five-fold axis (Fig. 1a). Compared to the mature virion, the procapsid and A-particle resemble the capsid structure (correlation coefficient 0.94), but share expanded diameters of ~330 Å along five-fold axes and open two-fold channels (Fig. 1a–c). The density attributable to encapsidated genome is present in both mature virions and A-particles, but absent in procapsids (Fig. 1d–f). However, the genome in the A-particle seems more organized and interacts more directly with the inner surface of the capsid (Fig. 1f), which implies that it is poised for release into the host cell. To our surprise, no interpretable density of cellular receptor ICAM-5 was observed in 2D/3D classifications or final 3D reconstruction of ICAM-5-triggered A-particles, indicating that the binding of ICAM-5 with the capsid might be transient with low affinity, or the receptor-binding site may be lost after the transformation.
Fig. 1 |. The cryoEM structures of EV-D68 particles.
a–c, Iso-contoured views (along the icosahedral two-fold axis) of cryoEM maps (radially coloured) of the mature virion (a), procapsid (b) and A-particle (c). One icosahedral asymmetric unit is marked by a white triangle in a. The procapsid and A-particle show capsids with open two-fold channels, which are closed in the mature virion. d–f, Central sections of the corresponding maps displayed in the upper row. The densities of encapsidated genomic RNA are present in both the mature virion (d) and A-particle (f), but absent in the procapsid (e). g,h, Superpositions of protomers of A-particle protomer (VP1: blue; VP2: green; VP3: red) with that of mature virion (grey) (g) or procapsid (grey) (h). The orientation of the protomers is marked by black polygons indicating icosahedral five-, three- and two-fold axes, respectively. The colour scheme for the capsid proteins will be kept the same in all figures unless noted otherwise. The regions of the most significant differences between models are highlighted with magnified views in the dashed boxes: VP1 N termini (upper box) and VP3 GH loops (lower box) between the A-particle and the mature virion (g); VP1 HI loops (upper box) and VP3 GH loops (lower box) between the A-particle and the procapsid (h).
The density maps of all three types of particle show clear delineation of protein backbones and resolve side chains of most amino-acid residues, which allowed us to build their atomic models (Supplementary Table 1). Overall, our atomic model of the mature virion (clinical strain) is similar to that of the crystal structure of the EV-D68 mature virion (Fermon strain)9, with a root mean squared deviation (r.m.s.d.) of 1.07 Å for all Cα atoms in the overlay of each protomer (Supplementary Figs. 5 and 6 and Supplementary Table 2). By contrast, structural comparison between protomers of the mature virion and the A-particle revealed conformational changes in the mature virion triggered by ICAM-5. Like some other picornaviruses15,16,22,23, the particle diameter expands ~3% and each protomer rotates anticlockwise by 3.5° with the pivot point located at the corner of VP3 close to the icosahedral three-fold axis (Fig. 1g, Supplementary Fig. 7a,b and Supplementary Video 1). The ICAM-5-triggered A-particle also shares some common features with the classical A-particles of other picornaviruses, such as collapsed pocket-factor-binding regions, externalized VP1 amino termini and lost VP4s, and open two-fold-axis channels (Fig. 1g and Supplementary Fig. 7c,d). Apart from these, the VP3 GH loop also undergoes a major conformational shift and forms a β-hairpin structure (Fig. 1g). Despite the different protein components, the capsid shells of the procapsid (comprised of VP0, VP1 and VP3) and the A-particle (constituted by VP1, VP2 and VP3) share a similar structure based on an r.m.s.d. of 0.73 Å for all Cα atoms between corresponding protomers. Superposition of the two models illustrates recognizable differences including a ~4 Å shift of VP1 HI loops and disordered versus identifiable density of VP3 GH loops in the procapsid and the A-particle, respectively (Fig. 1h). Since VP1 loops in some picornaviruses function as immunogenic targets for neutralizing antibodies13,17,24, we postulate that the distinct conformations of VP1 HI loops in EV-D68 A-particles and procapsids have rendered antigenicity differences.
Anti-EV-D68 neutralizing antibodies 15C5 and 11G1 confer protective efficacies in vivo.
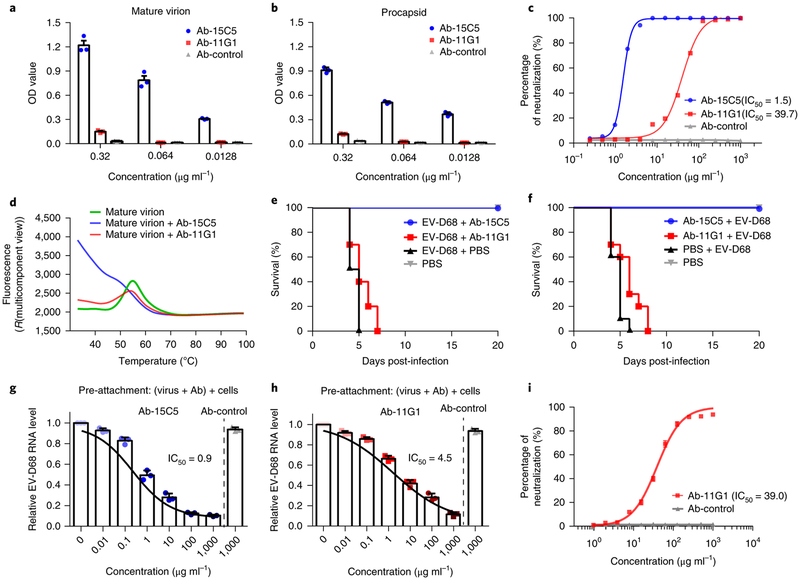
To better explore the antibody-mediated neutralization of EV-D68 and antigenic profiles at different stages in its life cycle, we generated monoclonal antibodies in mice immunized with supernatant from virus cultures. Two representative neutralizing antibodies, named 15C5 and 11G1, were selected for further functional and structural analysis. In binding assays, 15C5 shows high binding efficiency for both mature virions and procapsids in a concentration-dependent manner, whereas the binding of 11G1 to either type of particle is relatively weak (Fig. 2a,b). In virion-based neutralization assays, 15C5 showed 30-fold higher neutralizing potency than 11G1, manifested by half-maximum inhibitory concentration (IC50) values of 1.5 versus 39.7 μg ml−1 (Fig. 2c). These findings suggested that the two neutralizing antibodies neutralize via distinct mechanisms. Furthermore, in the RNA-binding fluorescent assay, the accessibility of the dyes to mature virions with or without the presence of 11G1 remains at about the same level, suggesting minimum genome exposure except at non-physiological temperatures from 50 °C to 60 °C. In contrast, the binding of 15C5 to the EV-D68 mature virion significantly increases the accessibility of the dyes to the genomic RNA and prominent fluorescence was detected at 37 °C (Fig. 2d). Taken together, these data suggest that 15C5 binding to mature virions induces conformational changes in the capsid, and 11G1 does not neutralize virus by binding to mature virions.
Fig. 2 |. Characterization of the neutralizing antibodies 15C5 and 11G1.
a,b, Binding efficiencies of the neutralizing antibodies 15C5 and 11G1 to EV-D68 mature virion (a) or procapsid (b) evaluated with binding ELISA. The values are expressed as mean ± s.d. The experiments were independently repeated in triplicate, c, Neutralization efficiencies of the neutralizing antibodies 15C5 and 11G1 against EV-D68 virus plotted as a function of their concentrations (IC50 titres are noted), d, Particle stability thermal release assay of EV-D68 mature virions and their complexes with either 15C5 or 11G1. The fluorescence value indicates the level of genome exposure relative to temperature. The fluorescence traces are shown for EV-D68 mature virions (green line) as well as their complexes with 15C5 (blue line) or 11G1 (red line). The experiments were independently repeated in triplicate. e,f, Antibody-mediated protection against EV-D68 infections. Groups of 1-day-old BALB/c mice (10 mice per group) were treated with either 15C5 or 11G1 24 h after (e) or 12 h before (f) infection with EV-D68 (106 TCID50 per mouse, i.p.). The mortality of each group was monitored and recorded daily after infection. g,h, The amount of cell-bound EV-D68 viruses detected by quantitative real-time RT-PCR in the presence of the neutralizing antibody 15C5 (g) or 11G1 (h) added before the viruses. The values are expressed as mean ± s.d. The experiments were independently repeated in triplicate, i, Post-attachment neutralization assay of theneutralizing antibody 11G1 shows comparable neutralization capacity to that of regular conditions. The values are expressed as mean ± s.d. The experiments were repeated in triplicate. The IC50 was calculated by nonlinear regression fitting curves using GraphPad Prism version 7.0.
The protective efficacy against mortality for 15C5 or 11G1 used to treat infection was measured in vivo in 1-day-old mice that were challenged with the lethal dose of a clinical EV-D68 strain. To evaluate the therapeutic efficacy, we administered either antibody to mice at 24 h post-infection. The survival rate of the mice in the 15C5-treated (60 μg g−1) group was 100%, and 11G1 treatment resulted in reduced illness severity and delayed mortality compared to the control group (Fig. 2e). For prophylaxis, mice were administered the same dosage of either 15C5 or 11G1 at 12h before virus challenge. Mice pre-treated with 15C5 showed a 100% survival rate, and those with 11G1 had delayed onset of mortality when compared to the control group (Fig. 2f).
To further explore the neutralization mechanisms of 15C5 and 11G1, a cell-based PCR with reverse transcription (RT-PCR) assay17 was used to quantify the ability of these antibodies to block virus particles from attaching to host cells. When pre-mixed with EV-D68 virus, either 15C5 or 11G1 can reduce the number of particles bound to host cells. 15C5 demonstrated higher potency than 11G1 for inhibiting viral attachment and the effect was concentration-dependent (Fig. 2g,h). Since 11G1 cannot bind to mature virions effectively, we performed a post-attachment neutralization assay to investigate whether 11G1 can neutralize virus after attachment of EV-D68 to the cell. As shown in Fig. 2i, 11G1 could neutralize viruses in a post-attachment manner with an IC50 value of 39.0 μg ml−1 that is comparable to the neutralization potency (IC50 = 39.7 μg ml−1) under regular conditions (Fig. 2c).
CryoEM structures of 15C5 or 11G1 complexes with EV-D68 and 15C5-triggered structural transformation.
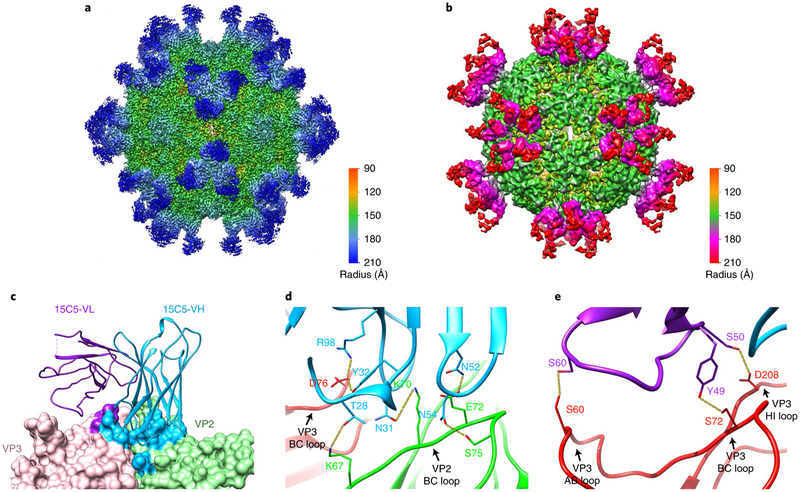
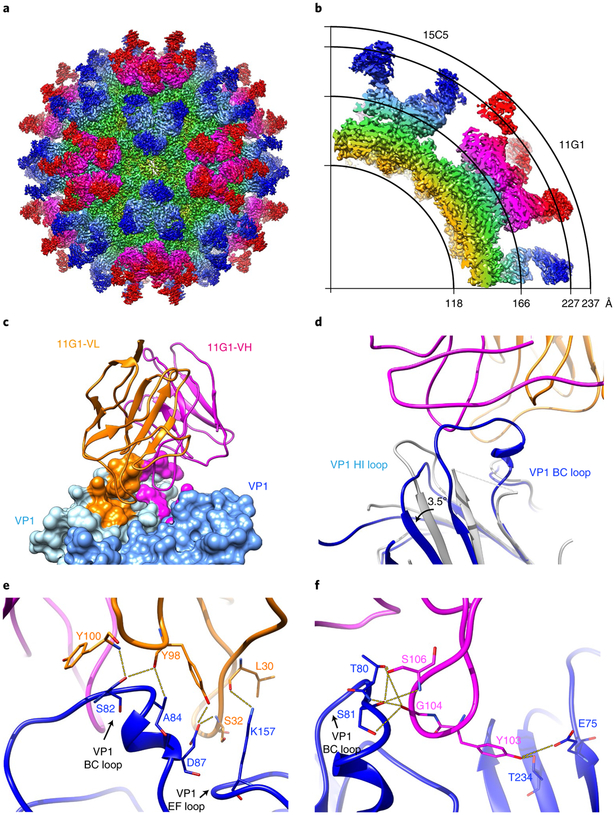
The binding profiles of 15C5 and 11G1 to three forms of EV-D68 capsid were further evaluated with cryoEM and 2D classification using Relion software25. The data indicated that 15C5 efficiently binds to all three types of particle, and that 11G1 binds only to A-particles, exhibiting little or no binding to mature virions or procapsids (Supplementary Fig. 8). In addition, the density of 11G1 binding to A-particles is less than that of 15C5, suggesting that 11G1 binding is not saturating. 3D reconstructions of EV-D68 mature virion in complex with 15C5 (EV-D68-M:15C5) and A-particles_us in complex with 11G1 (EV-D68-A:11G1) were determined at resolutions of 3.6 Å and 7.2 Å, respectively (Fig. 3a,b and Supplementary Fig. 9a–f). The density maps show that 15C5 and 11G1 target the virus capsid around its three-fold and five-fold vertices, respectively, and thus up to 60 copies of 15C5 or 11G1 Fabs can bind to each virus particle (Fig. 3a,b). Mimicking a property of ICAM-5, the binding of 15C5 triggers the opening of two-fold channels in mature virions and reorganization of the genome (Supplementary Fig. 10).
Fig. 3 |. CryoEM structures of immune complexes EV-D68-M:15C5 and EV-D68-A:11G1.
a,b, Iso-contoured views of cryoEM maps (radially coloured) of the immune complexes EV-D68-M:15C5 (a) and EV-D68-A:11G1 (b). Groups of three 15C5 Fabs and five 11G1 Fabs bind at each three-fold and five-fold axis, respectively, c, Surface representation shows the interaction interface between Fab 15C5 and the capsid. Each Fab 15C5 binds across VP2 and VP3 from two adjacent protomers. Footprints of the VH and VL of the Fab on the capsid are coloured with light blue and purple, respectively. d,e, Close-up views of the interaction interface between the capsid and either the heavy (d) or light chain (e) of Fab 15C5. Note that the heavy chain binds across the VP2 BC loop and the VP3 BC loop from two adjacent protomers as shown in d. Potential hydrogen bonds and salt bridges are marked by yellow dashed lines.
We next built an atomic model of the EV-D68-M:15C5 complex, which includes three major capsid proteins (VP1, VP2 and VP3) and the variable domain of 15C5 Fab, but excludes the constant domain that has a disordered density in the cryoEM map (Supplementary Fig. 11a). Each Fab has a footprint that extends across two adjacent protomers, and its binding buries a total surface area of ~915 Å2 on the capsid (Fig. 3c). The heavy chain of Fab 15C5 binds across the VP2 BC loop and the VP3 BC loop from two adjacent protomers, accounting for ~76% of the buried area (Fig. 3d), and the light chain interacts with the AB, BC and HI loops of the same VP3 that binds the heavy chain (Fig. 3e). The Fab–capsid interaction establishes an elaborate hydrogen-bonding network including eight bonds formed between complementarity-determining regions H1, H3, L2 and VP3 AB, BC, HI loops and VP2 BC, HI loops (Fig. 3d,e and Supplementary Table 3), and several van der Waals interactions are observed between the 15C5 Fab and the capsid (Supplementary Table 3). In addition, escape mutants were generated by propagating the viruses in the presence of 15C5. All 15C5-resistant mutants had an amino-acid change at position 74 (L74S), which is located at the VP3 BC loop and is a critical contact residue in the virus–antibody interaction (Supplementary Table 3).
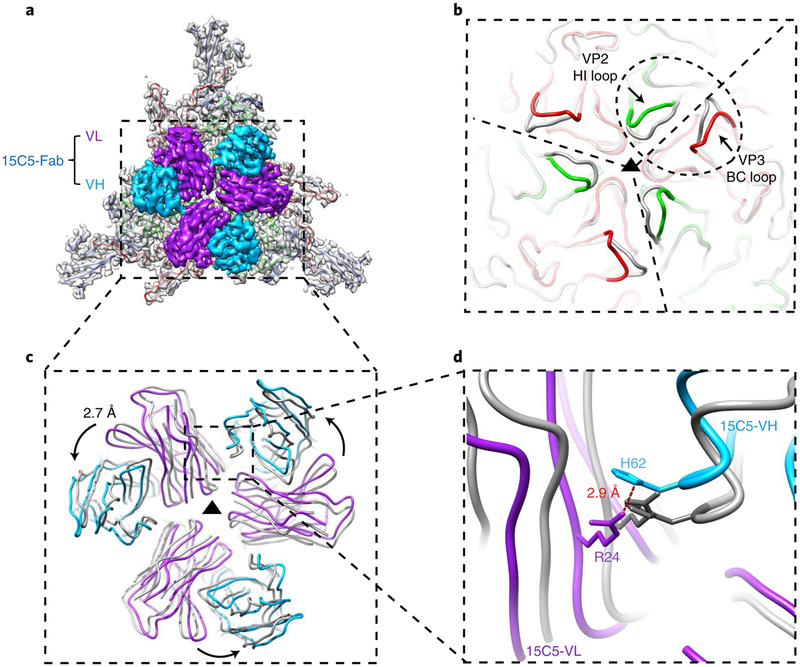
The capsid in the immune complex EV-D68-M:15C5 exhibits a notably different structure from that of the EV-D68 mature virion, but similar to ICAM-5-induced A-particles (the r.m.s.d. between protomers from two respective models is 0.48 Å; Supplementary Fig. 11b). Moreover, binding of three 15C5 Fabs at each three-fold axis occurs with variable regions packed tightly together (Fig. 4a). Thus, we used the 3 three-fold-related asymmetric units of EV-D68-M:15C5 for individually superimposing onto the model of the mature virion (Fig. 4b,c). The overlay showed subtle conformation changes in the VP2 HI loop (with the Cot atom shifted by 3.2 Å) and the VP3 BC loop (with the Cα atom shifted by 3.8 Å), and we believe the changes are necessary to accommodate the binding of adjacent 15C5 Fabs since both loops were directly involved in the interaction (Fig. 4b). The comparison also revealed that the three Fabs became closer to each other in the superimposed model than in the original model of the 15C5 immune complex (Fig. 4c). Arg 24L and His 62H are the two nearest residues (distance of 2.9 Å) between neighbouring heavy and light chains in adjacent Fabs, and are distant from the virus–antibody binding interface in the original immune-complex model. In the superimposed models, these two residues exhibit a steric clash (Fig. 4d). To accommodate the physical proximity of the adjacent Fabs, we hypothesize that when 15C5 binds mature viri-ons, the VP2 HI loop and the VP3 BC loop are pushed apart to trigger the conformational change from mature virion to A-particle via a cascade of molecular rearrangements and allosteric effects. This results in an anticlockwise movement of ~2.7 Å of each Fab along the three-fold axis (Fig. 4c,d).
Fig. 4 |. Superposition of mature virion with or without the binding of 15C5 shows details of 15C5-induced conformational changes.
a, Outside view of the density attributable to the 3 three-fold-related asymmetric units segmented out of the cryoEM map of EV-D68-M:15C5. The capsid density (grey) is fitted with atomic models of three major capsid proteins and, for clarity, only the variable domains of three Fabs are shown, b, The 3 three-fold-related asymmetric units of EV-D68-M:15C5 (coloured wire diagram) are individually superimposed onto the models of the mature virion (grey). The scope of the epitope region is marked by the dashed ellipse. The significant conformational changes occurring at the VP2 HI loop and the VP3 BC loop after the binding of Fabs are highlighted. c,d, Atomic models of the three Fabs in a (coloured wire diagram) together with three superimposed Fabs (grey). The superimposed Fabs, which show a 2.7 Å movement and became closer when compared to the original model of the 15C5 immune complex, represent the theoretical binding of Fabs to the mature virion at the same epitope prior to conformational changes of the capsid. The close-up view (d) shows that the two nearest residues (distance of 2.9 Å) between neighbouring heavy and light chains are His 62H and Arg 24L derived from two adjacent Fabs. These two amino acids exhibit a steric clash in the superimposed models (grey).
CryoEM structure of EV-D68 bound with both neutralizing antibodies 15C5 and 11G1.
We next sought to build the double-antibody complex EV-D68-M:15C5:11G1 by adding 11G1 to the EV-D68-M:15C5 complex. A triple immune complex was successfully generated and its cryoEM density map was reconstructed at a final resolution of 3.5 Å (Supplementary Fig. 9g–i). A total of 60 copies of 15C5 and 60 copies of 11G1 concurrently bound to the transformed A-particle at icosahedral three-fold and five-fold vertices, respectively (Fig. 5a,b). The densities attributable to 11G1 were dramatically improved in this complex when compared with those in the EV-D68-A:11G1 complex (Supplementary Fig. 8), which indicates that 11G1 may favour A-particles converted from mature virions triggered by 15C5 rather than ICAM-5.
Fig. 5 |. The cryoEM structure of EV-D68 in complex with 15C5 and 11G1.
a, Iso-contoured views of the cryoEM map (radially coloured) of the immune complex EV-D68-M:15C5:11G1. The EV-D68 capsid is bound with 60 copies of Fab 15C5 at three-fold axes and 60 copies of Fab 11G1 at five-fold axes, respectively, b, A quarter of the central section of the cryoEM map in a; the radii are marked by the arcs, c, Surface representation shows the interaction interface of Fab 11G1. Each Fab 11G1 (heavy chain: magenta; light chain: orange) binds across two neighbouring VP1s (light blue and blue) of the capsid. Footprints of the VH and VL of the Fab on the capsid are coloured with magenta and orange, d, A close-up view (wire diagram) of the interaction interface between 11G1 and the capsid shows the conformational changes (indicated with black arrows) of the VP1 BC loop and HI loop before (grey) and after (coloured) binding with 11G1. e,f, Close-up views of the interaction interfaces between the capsid and the light (e) or heavy chain (f) of Fab 11G1. Potential hydrogen bonds are marked by yellow dashed lines.
In the triple immune complex, the interaction between 15C5 and the capsid is essentially identical to that in the EV-D68-M:15C5 complex (Supplementary Figs. 11c,d and 12). Nevertheless, the atomic model allowed us to further refine the epitope for 11G1.11G1 interacts with BC, DE, EF and HI loops and two β-strands of VP1, which buries a total surface area of ~970 Å2 on the capsid (Fig. 5c and Supplementary Fig. 12). Notably, the VP1 BC loop is anchored and well resolved by the binding of 11G1 (Fig. 5d), whereas the loop is disordered in the other structures of all three forms of individual particles as well as the EV-D68-M:15C5 immune complex. In addition, the binding of 11G1 induces minor conformational changes of the VP1 BC and HI loops that both rotate about 3.5° away from the Fab relative to the VP1 framework region when compared with those in the EV-D68-M:15C5 complex (Fig. 5d). The interaction interface between 11G1 and the capsid involves 3 (L1, L3 and H3) of the 6 complementarity-determining regions, which forms 14 hydrogen bonds and several van der Waals contacts with the VP1s (Fig. 5e,f and Supplementary Table 4). Among these hydrogen bonds, two (Tyr 103H–Glu 75 and Tyr 103H–Thr 234) are formed by each 11G1 Fab binding VP1 across adjacent protomers. To further confirm the epitope recognized by 11G1, virus escape mutants were generated under the immune pressure of 11G1; the resultant 11G1-resistant viruses harboured a mutation at position 229 (Q229K) or position 234 (T234A) of VP1, both of which are located in the virus–antibody binding interface (Supplementary Table 4).
Neutralizing antibodies 15C5 and 11G1 inhibit viral infection via distinct mechanisms.
Picornaviruses enter cells via receptor-mediated endocytosis where A-particles may form within the endosome of the infected cell26,27. However, on the basis of previous work17,28–31 and the structures of three capsid forms in this study, the mature virion interacts with cellular receptors anchored on the surface of host cells, which may lead to transformation into the A-particle conformation before endocytosis, and would be considered as an alternative route for virus entry (Supplementary Fig. 13). The binding of 15C5 can also induce the conformational change similar to that induced by ICAM-5 binding, indicating that the 15C5-binding sites (VP2 HI loop and VP3 BC loop) may serve as sensors to trigger capsid transformation. Unlike ICAM-5, which binds to virus particles in a transient manner, 15C5 engages mature virions with high efficiency (Fig. 3a) and efficiently blocks the subsequent A-particle from attaching to the cell surface (Fig. 2g) thereby neutralizing viral infection before attachment (Fig. 2c,f). In contrast, 11G1 exclusively recognizes the A-particle rather than the mature virion, which suggests that 11G1 neutralizes with assistance from other factors, such as ICAM-5 or 15C5-like antibodies. The post-attachment inhibition effect of 11G1 (Fig. 2i) suggests that 11G1 neutralizes viruses after attachment, but before endocytosis. The neutralizing antibody 11G1 binds two adjacent protomers from neighbouring VP1s (Fig. 5c) and may potentially achieve neutralization by interfering with genome release from the A-particles into host cells (Supplementary Fig. 13).
Taken together, 15C5 binds the three-fold axis of the viral capsid, triggers the transformation of mature virions to A-particles, stays bound, and blocks binding to host cells. The neutralizing antibody 11G1 binds the five-fold axis, but only on the A-particle. Therefore, the two antibodies examined in this study neutralize EV-D68 in spatially, temporally and functionally distinct manners, and potentially block virus infection at different stages of the EV-D68 life cycle.
Discussion
Picornaviruses initiate infection by mature virions binding the cellular receptor, triggering endocytosis and conformational transformation to A-particles, leading to the release of viral genome into host cell cytosol23,32,33. High-resolution structural analysis of mature virions, A-particles and their complexes with cellular receptors is critical for understanding the molecular and structural basis of this dynamic uncoating process15,34. Structural investigation of EV71 particles has suggested a sensor-adaptor (VP1 GH loop) triggering mechanism for virus uncoating13. In our study, near-atomic-resolution structures of the EV-D68 procapsid, mature virion and A-particles (unknown stimulus-induced, ICAM-5-induced or 15C5-induced) and their immune complexes offer a framework for understanding the mechanics of EV-D68 particle transformation and uncoating, and mechanisms of virus neutralization (Supplementary Fig. 13). We demonstrated that the EV-D68 mature virion can initiate cell entry by binding to a cellular receptor ICAM-5 that triggers the formation of a necessary intermediate (A-particle). This transformation results in a reorganization of the RNA genome, suggesting that this step may be critical for release of genome into the host cell and initiation of infection. We also demonstrated two distinct antibody-mediated mechanisms of viral neutralization. Binding of 15C5 at the three-fold axes induces a conformational change of EV-D68 mature virions to A-particles that is a prerequisite for subsequent genome release16,35,36. In addition to triggering the conformational change to A-particles, 15C5 also inhibits attachment of A-particles to cell surface receptors and thereby neutralizes viral infection. Structural analysis on immune complexes showed that 15C5 recognizes a quaternary epitope that bridges across VP3 and the VP2 from two adjacent protomers, which may lock the capsid at an intermediate stage and prevent further conformational change and the uncoating process. Notably, this neutralization mechanism is different from some other reported antibodies against EV71 or HRV, which have been shown to induce premature viral uncoating and genome release21,22. 11G1 appears to neutralize enterovirus by a unique post-attachment mechanism. After cell engagement by mature virions triggers transformation to the A-particle, 11G1 is able to bind at a step before endocytosis and neutralize virus infection. The specificity and structural complementarity of these two antibodies and distinct timing and neutralizing activity suggest that the two antigenic sites recognized by 15C5 and 11G1 may serve as excellent targets for the design of vaccines and antiviral therapeutics against EV-D68.
From an immunological viewpoint, an epitope (such as of 15C5) located on the mature virion can escape neutralization in vivo by genetic variation under the pressure of the neutralizing antibody response. However, an epitope (such as of 11G1) revealed transiently during the capsid transition to an A-particle conformation may be less susceptible to escape mutation. We propose that A-particles, bearing both the 15C5-like epitope and the unique 11G1-like epitope, may be the preferred antigenic form for the design of vaccine candidates against the canonical native virion. Our study revealed a potential approach for generating stable EV-D68 A-particles by exposing isolated virions to ICAM-5 or 15C5. The atomic-level definition of immune complexes provides a basis for confirming antigenic integrity of candidate vaccines. Therefore, the antibodies 15C5 and 11G1 provide important tools for structure-based vaccine design and could potentially be developed as therapeutic interventions for EV-D68 infections.
Methods
Cells and viruses.
Human RD (Rhabdomyosarcoma) cells were obtained from the American Type Culture Collection (ATCC, CCL-136) and maintained in Minimal Essential Medium (Gibco) supplemented with 10% fetal bovine serum (Gibco). The cells were routinely tested and confirmed negative for contamination with mycoplasma. The EV-D68 strain STL-2014–12 (GenBank accession no. KM881710) infectious clone was constructed in our laboratory and propagated in RD cells. Virus were stored in aliquots at −80 °C.
Virus production and purification.
EV-D68 was propagated in RD cells at a multiplicity of infection of 0.2 at 33 °C. Virus was collected 3 days after infection, and then centrifuged at 7,000g for 30 min to remove cell debris and precipitated by using 7% (w/v) polyethylene glycol 8000 (PEG 8000) and 0.2 M NaCl in 0.1 M phosphate buffer (PB, pH 7.4) at 4°C for 12 h. After centrifugation, virus was resuspended and then sedimented through a linear 15–45% (w/v) sucrose density gradient at 140,000g for 3 h at 4°C using a Beckman SW41 Ti rotor. Two fractions were collected and independently dialysed against PB (pH 7.4) and concentrated by an Ultra-4 centrifugal concentrator. The quantity and quality of EV-D68 virions were examined by negative-staining electron microscopy.
Neutralization assay.
RD cells were prepared in Minimal Essential Medium supplemented with 2% fetal bovine serum and seeded at 1 × 104 cells per well into 96-well plates. For neutralization assay, monocolonal antibodies were serially diluted by twofold dilutions (the initial concentration of 1 mg ml−1) and incubated with an equal volume of EV-D68 (100 TCID50) at 33 °C for 1 h; then the virus/monocolonal antibody mixture was added to 96-well plates pre-seeded with RD cells. For post-attachment neutralization assay, EV-D68 (100 TCID50) was added to 96-well plates pre-seeded with RD cells and incubated at 4 °C for 1 h. Then, serially twofold-diluted 11G1 (the initial concentration of 1 mg ml−1) was added to the virus/cell plate. The 96-well plates were incubated at 33 °C for 3–5 days. The cytopathic effects (CPEs) were observed under a microscope, and the neutralization titres were read as the highest dilution of cells protected from 50% CPE, taken as the average of the triplicates. The relative cell viability (%) was measured by a MTT assay. The percentage of neutralization of the antibody was calculated with the following formula: percentage of neutralization = (ODtest − ODvirus control)/(ODcell control − ODvirus control) × 100%. The EV-D68 titre (TCID50) was calculated using the Reed–Muench method.
Binding enzyme-linked immunosorbent assay.
The purified EV-D68 mature virions and procapsids were adsorbed at 50 ng per well in PB (pH 7.4) to enzyme-linked immunosorbent assay (ELISA) plates (96-well) at 4 °C overnight. Each well was blocked with BSA-PBS (1% bovine serum albumin in PBS) at 37 °C for 2 h, after washing with PBST (0.05% Tween-20 in PBS). Antibodies, ICAM-5 (ICAM-5-Fc; R&D, 1950-M5–050) or IgG1-Fc (R&D, 110-HG-100) were added at various concentrations and incubated at 37 °C for 1 h. Each well was then incubated with GAM–HRP or GAH–HRP secondary antibody at 1:5,000 dilutions at 37 °C for 30 min. After colour development using TMB chromogen substrate and the reaction stopped with 2 M H2SO4, absorbance was measured at A450/620 nm.
Particle stability thermal release assay.
Thermofluor experiments37 were performed with an MX2005p RT–PCR instrument (Agilent). The environment-sensitive fluorescent dye SYTO9 (Invitrogen, S34854) was used to detect the presence of RNA. Each 50 μl reaction was set up in a thin-walled PCR plate, containing 1.0 μg of EV-D68 mature virion and 5 μM SYTO9 in PBS. The temperature was ramped from 25 to 99 °C with the fluorescence level recorded in triplicate at 0.5 °C intervals. In addition, a similar assay was also performed on virus–monoclonal antibody or virus–ICAM-5 complexes with 1.0 μg of EV-D68 mature virion pre-incubated with either 2.5 μg of antibody or 3 μg of ICAM-5 protein. The RNA release (Tr) and melting temperature (Tm) were taken as the minimums of the negative first derivative of the RNA exposure and protein denaturation curves, respectively.
In vivo protection test.
BALB/c mice were obtained from the Slac Laboratory Animal Co. Animal experiments were approved by Xiamen University Laboratory Animal Center (XMULAC). All procedures were conducted in accordance with animal ethics guidelines and approved protocols. All mice used in the study were randomly assigned to the different groups. Experiments were not performed in a blinded fashion. Groups of neonatal BALB/c mice (n = 10, gender-random) were inoculated intraperitoneally (i.p.) with 50 μl of neutralizing antibodies (60 μg g−1 body weight) or PBS buffer at 24 h after or 12 h before infection i.p. with 50 μl of EV-D68 (106 TCID50 per mouse). The mice in the control groups were treated with 50 μl of PBS. Daily monitoring of body weight and clinical disease continued in mice 20 days post infection.
Pre-attachment inhibition assay.
The virus pre-attachment assays were conducted as previously described17. In brief, EV-D68 virus was mixed with serially diluted monoclonal antibody 15C5 or 11G1 at 4 °C for 1 h before the virus attached to pre-seeded cells in 96-well plates. After three washes with cold PBS to remove unbound viruses, total cellular RNA was extracted using the QIAamp Mini viral RNA Extraction Kit (Qiagen). The amount of cell-bound virus RNA was determined by quantitative real-time RT–PCR. The analysis of relative levels of EV-D68 RNA in different samples was performed by the comparative 2-ΔΔCT method38.
Preparation of antibodies and antibody fragments.
To obtain the anti-EV-D68 monocolonal antibodies 15C5 and 11G1, BALB/c mice were immunized with the supernatant of virus culture emulsified in complete Freund’s adjuvant and boosted twice at two-week intervals with supernatant in incomplete Freund’s adjuvant. Then spleen cells from immunized mice were fused with myeloma cells (Sp2/0Ag-14) and hybridoma supernatant was screened, using ELISA and neutralization assay against EV-D68. Antibodies were purified from mouse ascitic fluid by protein A affinity chromatography. The variable region sequences of antibodies were determined by cloning and sequencing of their complementary DNA. To obtain antibody Fab fragments, monoclonal antibody 15C5 or 11G1 was digested with 1 ‰ (w/w) papain in 0.02 M PB (pH 7.0), containing 0.03 M l-Cys and 0.05M EDTA at 37 °C for 10–12 h. After adding 30 mM iodoacetamide, the resulting antibody fragment (Fab) was purified via DEAE column (TOSOH) chromatography.
Generation and sequencing of the escape mutant viruses.
15C5 or 11G1 (200 μg ml−1) was incubated with 1 × 107 TCID50 of EV-D68 for 1 h at room temperature, followed by 2 h at 37 °C. The mixtures were added to RD cells and incubated at 37 °C until CPE developed. After three freeze–thaw cycles, the cultures were clarified by centrifugation. The second and third round of infection in the presence of 15C5 or 11G1 was performed with the antibody concentration of 800 μg ml−1. The obtained 15C5- or 11G1-resistant viruses were purified from plaques and then cloned into RD cells. The P1 region of the mutated viral genome was amplified by RT–PCR and subsequently verified by DNA sequencing.
CryoEM sample preparation and data acquisition.
EV-D68 A-particle was prepared by the mixture of mature virion with ICAM-5 at a molar ratio of five ICAM-5 to every viral particle. EV-D68 immune complexes were prepared by the mixture of mature virion with Fab 15C5 or 11G1 or both at a molar ratio of 72 Fab to every viral particle. All of the mixtures were incubated for 30 min at 37 °C. Aliquots (3 μl) of purified procapsid, mature virion, ICAM-5-induced A-particle or immune complexes were deposited onto glow-discharged holey carbon Quantifoil Cu grids (R2/2, 200 mesh, Quantifoil Micro Tools) inside an FEI Mark IV Vitrobot at a humidity level of 100%. After 6 s blotting, the grids were plunge-frozen into liquid ethane cooled by liquid nitrogen. Specimens were examined under low-dose conditions at 300 kV with a FEI Tecnai F30 transmission electron microscope. All images were recorded on a Falcon II direct electron detector in the 17-frame video mode at a nominal magnification of 93,000, corresponding to a pixel size of 1.128 Å, and with the defocus ranging from 1.5 to 3.0 μm. The total electron dose was set to 25 e− Å−2 and the exposure time was 1 s. FEI software EPU was used for all data collection; micrographs with drift and astigmatism were discarded. In total, 774 (mature virion), 677 (procapsid), 1,792 (A-particle), 2,414 (EV-D68-M:15C5), 189 (EV-D68-A:11G1) and 1,785 (EV-D68-M:15C5:11G1) micrographs were selected with a defocus range of 0.7–2.8, 0.9–3.6, 0.8–3.8, 0.7–4.1, 1.7–4.2 and 1.0–4.5 μm respectively, for further image processing.
Image processing.
Video frame alignment and contrast transfer function estimation of each aligned micrograph were carried out with the programs Motioncor239 and Gctf40. A total of 45,822 (mature virion), 14,265 (procapsid), 25,045 (A-particle), 105,346 (EV-D68-M:15C5), 414 (EV-D68-A:11G1) and 143,304 (EV-D68-M:15C5:11G1) particles were auto-picked by the Autopick session of software Relion 2.025. Initial 3D models of each of the six types of particle were generated with a random model method using AUTO3DEM41. Two rounds of reference-free 2D classifications, two rounds of unsupervised 3D classifications and final 3D reconstruction were all performed with Relion 2.0. A total of 11,938 (mature virion), 13,849 (procapsid), 9,625 (A-particle), 58,540 (EV-D68-M:15C5), 375 (EV-D68-A:11G1) and 102,512 (EV-D68-M:15C5:11G1) particles were included in final 3D reconstructions, respectively. The resolutions of the final maps were estimated on the basis of the gold-standard FSC curve with a cutoff at 0.14342. Local resolution variations were estimated using ResMap43.
Model building and refinement.
Ab initio model building of all three particles and the immune complexes EV-D68-M:15C5 and EV-D68-M:15C5:11G1 was carried out in Coot44. The X-ray model of EV-D68 (PDB code 4wm8) was used for modelling the EV-D68 mature virion. The atomic models of CVA6 (PDB code 5×s4) and its immune complex (PDB code 5×s7) served as templates to build models of the procapsid, A-particle and immune complexes of EV-D68. Briefly, these templates were manually fitted into the segmented volume (including an asymmetric unit) of the final cryoEM maps of viral particles and immune complexes, respectively. Sequence assignment was guided by aromatic and other residues with bulky side chains. Atomic positions of residues were manually built and modified using Coot and further refined using the module phenix. real_space_refine in PHENIX44,45. After five cycles of refinement, problematic regions, Ramachandran outliers and poor rotamers were corrected in Coot. Then the resulting models were fitted into the density of the other six subunits in an asymmetric unit, which were executed as a whole for real-space model optimization, ensuring that clashes from neighbouring asymmetric units were avoided and optimized. Model statistics including bond lengths, bond angles, all atom clashes, rotamer statistics, Ramachandran plot statistics and so on were closely inspected with Coot during the whole process. Finally, additional validation of the models was performed with Molprobity46. The sequence alignments were carried out using Clustal W and Clustal X version 2 on the EBI server and rendered using ESPript software47. Density-map-based visualization, segmentation and video generation were then performed with Chimera48. Fab densities in the difference maps were projected on a stereographic sphere using RIVEM49. The Fab–capsid interactions were analysed using the software CCP450 and the PISA server (www.ebi.ac.uk/pdbe/pisa) with the donor to acceptor distances ≤4 Å for hydrogen-bond and salt-bridge interactions and ≤ 3.5 Å for other contacts.
Accession codes.
The sequence of the VP gene of EV-D68 (this study), EV-D68 (Fermon strain), EV-71, CV-A16, CV-A6 and CV-B3 can be found as GenBank accession no. KM881710, AY426531.1, FJ600325, FJ198212, KR706309 and M88483, respectively.
Data availability
The atomic coordinates of the mature virion, procapsid, A-particle_i5, EV-D68-M:15C5 and EV-D68-M:15C5:11G1 have been deposited in the Protein Data Bank (accession numbers 6AJ0, 6AJ3, 6AJ2, 6AJ7 and 6AJ9, respectively). The cryoEM maps of the mature virion, procapsid, A-particle_us, A-particle_i5, EV-D68-M:15C5, EV-D68-A:11G1 and EV-D68-M:15C5:11G1 have been deposited in the Electron Microscopy Data Bank (accession numbers EMDB-9629, EMDB-9632, EMDB-9635, EMDB-9631, EMDB-9633, EMDB-9636 and EMDB-9634, respectively).
Supplementary Material
Acknowledgements
This work was supported by a grant from the National Science and Technology Major Projects for Major New Drugs Innovation and Development (no. 2018ZX09711003-005-003), the National Science and Technology Major Project of Infectious Diseases (no. 2017ZX10304402-002-003), the National Natural Science Foundation of China (no. 81401669 and 81801646) and the Natural Science Foundation of Fujian Province (no. 2015105073). This work was supported in part by funding by the National Institutes of Health (grants R37-GM33050, GM071940, DE025567 and AI094386). We acknowledge the use of instruments at the Electron Imaging Center for Nanomachines supported by UCLA and by instrumentation grants from the NIH (1S10RR23057 and 1U24GM116792) and NSF (DBI-1338135 and DMR-1548924). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript. We also thank L. Wang and K. M. Morabito for proofreading the manuscript.
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41564-018-0275-7.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Schieble JH, Fox VL & Lennette EH A probable new human picornavirus associated with respiratory diseases. Am. J. Epidemiol 85, 297–310 (1967). [DOI] [PubMed] [Google Scholar]
- 2.Ikeda T et al. Acute respiratory infections due to enterovirus 68 in Yamagata, Japan between 2005 and 2010. Microbiol. Immunol 56, 139–143 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Kaida A et al. Distinct genetic clades of enterovirus D68 detected in 2010, 2013, and 2015 in Osaka City, Japan. PLoS ONE 12, e0184335 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knoester M et al. Upsurge of enterovirus D68, the Netherlands, 2016. Emerg. Infect. Dis 23, 140–143 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Midgley CM et al. Severe respiratory illness associated with enterovirus D68 - Missouri and Illinois, 2014. MMWR. Morb. Mortal. Wkly Rep. 63, 798–799 (2014). [PMC free article] [PubMed] [Google Scholar]
- 6.Holm-Hansen CC, Midgley SE & Fischer TK Global emergence of enterovirus D68: a systematic review. Lancet Infect. Dis 16, e64–e75 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Messacar K et al. A cluster of acute flaccid paralysis and cranial nerve dysfunction temporally associated with an outbreak of enterovirus D68 in children in Colorado, USA. Lancet 385, 1662–1671 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Imamura T & Oshitani H Global reemergence of enterovirus D68 as an important pathogen for acute respiratory infections. Rev. Med. Virol 25, 102–114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y et al. Structure and inhibition of EV-D68, a virus that causes respiratory illness in children. Science 347, 71–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y et al. Sialic acid-dependent cell entry of human enterovirus D68. Nat. Commun 6, 8865 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogle JM, Chow M & Filman DJ Three-dimensional structure of poliovirus at 2.9 A resolution. Science 229, 1358–1365 (1985). [DOI] [PubMed] [Google Scholar]
- 12.Rossmann MG et al. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317, 145–153 (1985). [DOI] [PubMed] [Google Scholar]
- 13.Wang X et al. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat. Struct. Mol. Biol 19, 424–429 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plevka P, Perera R, Cardosa J, Kuhn RJ & Rossmann MG Crystal structure of human enterovirus 71. Science 336, 1274 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren J et al. Picornavirus uncoating intermediate captured in atomic detail. Nat. Commun 4, 1929 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shingler KL et al. The enterovirus 71 A-particle forms a gateway to allow genome release: a cryoEM study of picornavirus uncoating. PLoS Pathog. 9, e1003240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L et al. Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody. Nat. Commun 8, 505 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei W et al. ICAM-5/Telencephalin is a functional entry receptor for enterovirus D68. Cell Host Microbe 20, 631–641 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Zhang YX et al. A highly conserved amino acid in VP1 regulates maturation of enterovirus 71. PLoS Pathog. 13, e1006625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chong P et al. Immunological and biochemical characterization of coxsackie virus A16 viral particles. PLoS ONE 7, e49973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plevka P et al. Neutralizing antibodies can initiate genome release from human enterovirus 71. Proc. Natl Acad. Sci. USA 111, 2134–2139 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong Y et al. Antibody-induced uncoating of human rhinovirus B14. Proc. Natl Acad. Sci. USA 114, 8017–8022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee H et al. The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs. Sci. Adv 2, e1501929 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye X et al. Structural basis for recognition of human enterovirus 71 by a bivalent broadly neutralizing monoclonal antibody. PLoS Pathog. 12, e1005454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergelson JM & Coyne CB Picornavirus entry. Adv. Exp. Med. Biol 790, 24–41 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Johnson JE & Vogt PK Cell entry by non-enveloped viruses. Curr. Top. Microbiol. Immunol 343, v–vii (2010). [PubMed] [Google Scholar]
- 28.Curry S, Chow M & Hogle JM The poliovirus 135S particle is infectious. J. Virol 70, 7125–7131 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaplan G, Freistadt MS & Racaniello V R. Neutralization of poliovirus by cell receptors expressed in insect cells. J. Virol 64, 4697–4702 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsang SK, McDermott BM, Racaniello VR & Hogle JM Kinetic analysis of the effect of poliovirus receptor on viral uncoating: the receptor as a catalyst. J. Virol 75, 4984–4989 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baggen J, Thibaut HJ, Strating J & van Kuppeveld FJM The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Microbiol 16, 368–381 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Bubeck D, Filman DJ & Hogle JM Cryo-electron microscopy reconstruction of a poliovirus-receptor-membrane complex. Nat. Struct. Mol. Biol 12, 615–618 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergelson JM et al. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275, 1320–1323 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Belnap DM et al. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl Acad. Sci. USA 97, 73–78 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levy HC, Bostina M, Filman DJ & Hogle JM Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J. Virol 84, 4426–4441 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Organtini LJ, Makhov AM, Conway JF, Hafenstein S & Carson SD Kinetic and structural analysis of coxsackievirus B3 receptor interactions and formation of the A-particle. J. Virol 88, 5755–5765 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walter TS et al. A plate-based high-throughput assay for virus stability and vaccine formulation. J. Virol. Methods 185, 166–170 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ & Schmittgen TD Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang K Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan X, Sinkovits RS & Baker TS AUTO3DEM–an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol 157, 73–82 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheres SH & Chen S Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swint-Kruse L & Brown CS Resmap: automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics 21, 3327–3328 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gouet P, Robert X & Courcelle E ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31, 3320–3323 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pettersen EF et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 49.Xiao C & Rossmann MG Interpretation of electron density with stereographic roadmap projections. J. Struct. Biol 158, 182–187 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collaborative Computational Project, Number 4. TheCCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates of the mature virion, procapsid, A-particle_i5, EV-D68-M:15C5 and EV-D68-M:15C5:11G1 have been deposited in the Protein Data Bank (accession numbers 6AJ0, 6AJ3, 6AJ2, 6AJ7 and 6AJ9, respectively). The cryoEM maps of the mature virion, procapsid, A-particle_us, A-particle_i5, EV-D68-M:15C5, EV-D68-A:11G1 and EV-D68-M:15C5:11G1 have been deposited in the Electron Microscopy Data Bank (accession numbers EMDB-9629, EMDB-9632, EMDB-9635, EMDB-9631, EMDB-9633, EMDB-9636 and EMDB-9634, respectively).