Abstract
Chronic pain is an endemic problem involving both peripheral and brain pathophysiology. Although biomarkers have revolutionized many areas of medicine, biomarkers for pain have remained controversial and relatively underdeveloped. With the realization that biomarkers can reveal pain-causing mechanisms of disease in brain circuits and in the periphery, this situation is poised to change. In particular, brain pathophysiology may be diagnosable with human brain imaging, particularly when imaging is combined with machine learning techniques designed to identify predictive measures embedded in complex data sets. In this review, we explicate the need for brain-based biomarkers for pain, some of their potential uses, and some of the most popular machine learning approaches that have been brought to bear. Then, we evaluate the current state of pain biomarkers developed with several commonly used methods, including structural magnetic resonance imaging, functional magnetic resonance imaging and electroencephalography. The field is in the early stages of biomarker development, but these complementary methodologies have already produced some encouraging predictive models that must be tested more extensively across laboratories and clinical populations.
Keywords: Biomarkers, Pain, Neuroimaging, MRI, EEG, MVPA, Machine learning
1. Introduction
Pain is the primary reason why people seek health care and is the top source of disability in the United States.95 Chronic pain is a disease in its own right36,134 and, if untreated, can lead to depression,87 insomnia, depressed immune function, substance abuse,87 impaired cognitive function,2 and costs to families and caregivers.41
One might expect chronic pain to be diminishing over time, as medical diagnoses become more sophisticated and research brings new treatments to bear. Unfortunately, this does not seem to be the case. In fact, the prevalence of chronic pain is increasing.32,35,63,64 Multiple factors may be driving this increase, including obesity, changes in work demands, increased rates of depression and anxiety, aging populations, and increased symptom awareness.38,42 Regardless, little progress has been made in uncovering the physiological basis of pain in individual patients, although this could potentially drive more effective, individualized treatment.
In many fields, biomarkers have been developed that point to specific structural, biochemical, or other pathophysiological mechanisms, from oncology to cardiology to internal medicine. Echocardiograms and cardiac biochemical markers are routinely used to diagnose heart disease.50 Diabetes can be diagnosed with plasma glucose tests.50 Imaging is routinely used to help diagnose stroke, neoplasms, embolisms, and other causes of disease. In some fields, such as cancer, traditional assessments are increasingly complemented by biomolecular assays that can indicate the effectiveness of specific molecular treatment.50
Pain, however, has few biomarkers that are widely used in clinical practice.153 Some biomarkers are intended to track pain intensity and complement self-reports as a way of assessing the incidence or intensity of pain. Others are intended to reveal underlying pathobiological conditions that cause pain. As we argue below, this latter type is what is most badly needed. However, adequate biomarkers for pain-causing pathology are unavailable for most forms of pain. For example, structural magnetic resonance imaging (sMRI) of the spine is frequently used to diagnose conditions leading to low back pain. This may be useful in specific cases—eg, for identifying subpopulations with particular treatable pathologies—but they are now widely recognized to have poor diagnostic validity for pain.17,18,25
Part of the problem is that pain is complex, involving physical, psychological, emotional, and social aspects.16,150 Perhaps consequently, a large proportion of pain is idiopathic, with no known physical or structural cause. A collection of animal studies has shown that postinjury pain may be maintained by sensitization of an array of nervous system pathways, from spinal sensitization69 to sensitization in nuclei deep within the brain—the amygdala,19,27,99 nucleus accumbens,76,112,123 and medial prefrontal cortex.76,131 In some cases, brain changes potentiate descending pain facilitation, amplifying spinal cord responses to noxious events.81,131 Thus, in addition to peripheral pathology, chronic pain involves hidden pathology in the central nervous system, which has not been accessible to study in humans until the recent advent of noninvasive imaging.1,43,136 Accordingly, there is substantial interest in developing neuroimaging-based biomarkers that can tell us more about (1) how pain is constructed in the brain, (2) what biological varieties of pain there may be, and—crucially for patients—(3) what form of treatable pathophysiology an individual patient with chronic pain may have.
The development of such biomarkers has, however, been controversial. A thoughtful contingency of scholars and ethicists have rightly pointed out that relying on biological surrogate measures for pain rather than patients' self-reports would set a dangerous precedent and be disastrous for those whose pain is denied because it does not register in a biomarker-based test.28 However, the space of uses of potential biomarkers is large, and these ethical concerns apply only to one of many use cases for biomarkers defined by the U.S. Food and Drug Administration (FDA)—use as a “surrogate endpoint” to replace symptom reports.50 The FDA defines a range of other uses for biomarkers, from determining risk of future pain progression (prognostic biomarkers) to tracking whether a drug is exerting its intended pharmacodynamic effects (response biomarkers), or even predicting whether a treatment will be helpful for an individual patient (predictive biomarkers). For many of these categories, biomarkers need not track symptoms directly to be useful, as long as they reveal biological processes related to the generation or maintenance of pain.
In certain cases, a surrogate endpoint is necessary. Infants, like those with severe cognitive impairments and dementia, cannot tell us how they feel, which makes adequate treatment difficult.77,100,119 Early-life pain also increases later pain sensitivity and chronic pain risk.29,104,148 Even in these cases, we need not use biomarkers as surrogate endpoints, but rather as additional confirmatory measures, part of a broader pattern of behaviors (eg, infant cries and facial expressions) that can help people and their care providers determine the best course of action.
However, the most compelling use of brain biomarkers is in detecting pathophysiology and defining new biologically-based diagnoses of pain disorders. Imagine a patient whose back injury has healed, but whose pain persists due to sensitization in parabrachial–amygdala pathways. Back surgery would be a poor choice, as it is unlikely to help and may even make the situation worse: 20% to 40% of patients experience increased long-term pain and disability after surgery.3,101,126 Thus, a neuroimaging-based biomarker for parabrachial–amygdala sensitization could be a useful predictive biomarker for back surgery.
Accordingly, a number of recent funding initiatives are directed at development of biomarkers for pain. Some, like the U.S. National Institutes of Health's “Helping to End Addiction Long-Term” (HEAL) initiative, take a multipronged approach. Some HEAL funding programs focus on preclinical pain markers. Others, like the Acute to Chronic Pain Signatures program, focus on human prognostic biomarkers, with imaging- and tissue “omic”–based biomarkers both playing essential roles.
Here, we review studies that have advanced the field of brain biomarker development. Hundreds of studies have contributed to our understanding of the brain bases of pain,1,34 but we restrict our review to studies that develop brain models suitable for diagnosing the presence of pain, predict its intensity in individual people, or predict treatment outcomes. In addition, the studies we review attempt to validate their predictions on new, out-of-sample individuals from the same or different populations. These models generally use multiple brain features to form a prediction of pain incidence or intensity, based on the idea that pain encoding is distributed across multiple brain systems.
In addition, we restrict the scope of the review to several commonly used methods: sMRI and functional MRI (fMRI) and electroencephalography (EEG). Functional MRI is further separated into task-related (eg, painful stimulation–evoked) and resting-state fMRI (rs-fMRI). These methods are complementary, and each has its unique strengths and use cases. Structural MRI relies on relatively standardized acquisition methods available at virtually every major hospital and can identify stable changes that confer risk of chronic pain7,89 or result from pain-inducing injuries.125 Functional MRI can track within-person fluctuations in pain over time, yielding insights into the brain systems most closely associated with the experience of pain itself or associated behaviors. Electroencephalography is the most cost-effective measure of the 3 and can yield millisecond-level information about the timing of pain-related signals and about pain-associated brain oscillations.105 Both rs-fMRI and EEG can yield measures of stable person-level characteristics, through studies of individual differences in stimulus-evoked responses, fMRI connectivity, or patterns of EEG coherence.
1.1. Types of biomarkers
A biomarker is “a defined characteristic that is measured as an indicator of normal biological processes, pathogenic processes, or responses to an exposure or intervention, including therapeutic interventions.”50 The FDA recently developed a glossary in which different types of biomarkers have been defined. As we describe below, pain researchers have developed biomarkers that could be purposed for several of the use cases defined by the FDA, including diagnostic, prognostic, predictive, susceptibility/risk, and surrogate endpoint biomarkers.
A diagnostic biomarker is a measure indicative of a certain condition or disease.50 Applications include confirming the presence of pain or a chronic pain condition, or a specific pain condition. A predictive biomarker is used to predict the response to a treatment (ie, drug, device, or therapy) or an environmental agent,50 including both beneficial and adverse effects.
Diagnostic and predictive biomarkers can be used together to stratify patients, ie, to redefine pain subtypes based on biological categories or “biotypes.” The value of a diagnosis is largely in its ability to guide treatment. “Pain” and “no pain” may not be useful clinical categories, in the sense that “pain sufferers” are not a homogenous population, and there is no one treatment for “pain.” Perhaps surprisingly, more specific types of pain such as “knee osteoarthritis” may also have less diagnostic value than we commonly assume because “arthritis” is a description of symptoms rather than a disease mechanism. It may be caused by issues with localized tissue (eg, knee cartilage), a systemic inflammatory condition such as rheumatoid arthritis, or other systemic processes causing chronic widespread pain. These potential causes have different underlying mechanisms and should be treated differently.
Biomarkers need not identify current pain or disability to be useful—some of the most important uses involve predicting who is likely to develop chronic pain in the future and intervene before it is too late. In some cases, this may be as simple as avoiding surgery if the risks of postsurgical chronic pain are high. Prognostic biomarkers are designed to track future reoccurrence or progression of a disease.50 Prognostic biomarkers apply to people who already have an illness; they could, for example, be used to predict those likely to transition from acute to chronic pain. In healthy populations, susceptibility markers identify individuals at risk of a certain condition or disease.50
The final use case for biomarkers is surrogate endpoints, which are variables intended to reflect an outcome of interest that is a potential substitute or adjunct (supporting) measure of a disease state. Some biological or behavioral measures have been so strongly and consistently linked to disease that they can serve as the basis for validating a new treatment. Examples include forced expiratory volume in 1 second (FEV1) for asthma, serum creatinine in kidney disease, bacterial counts for antiseptics, and blood pressure for cardiovascular disorders.51 Surrogate endpoints generally require a long progression of validation on increasingly large and diverse samples.
We have argued that pain biomarkers should not be used as surrogate endpoints to falsify patients' reports.28,147 This is partly because pain may arise from diverse brain mechanisms, some of which we can measure and others which we cannot. A patient with real pain may nonetheless show brain patterns atypical of pain due to, for example, reorganization after damage. A much stronger case can be made for supplementing existing pain measures, for example, as part of a multimodel pain assessment program. However, we do not rule out the possibility that in the future, brain measures may be precise enough and sufficiently well validated that they could serve as surrogate measures for treatments. If, for example, a reliable biomarker could be developed for human parabrachial–amygdala hypersensitization to normally innocuous stimuli, treatments that reduce such hypersensitization might one day be considered valuable in their own right, even if that hypersensitivity is only a small part of any given patient's total pain and dysfunction.
1.2. Criteria for evaluating biomarkers
There has been considerable debate about whether pain biomarkers should be used for clinical and other (eg, legal) purposes.28 One productive way forward is to treat the use of biomarkers as an empirical matter: define criteria that should be met for a biomarker to be considered valid and useful, and evaluate biomarkers against them. This will allow us to examine biomarkers of various types—behavior, blood-based, cerebrospinal fluid-based, and brain-based—on a level playing field. There are many such criteria, and some, such as cost-effectiveness and the potential for misapplication in current health care environments, extend beyond scientific considerations. Here, we limit the discussion to a partial list of scientific criteria. For further discussion, see Refs. 28, 151, and 153.
1.2.1. Transparency and usability
A biomarker should have clear, standardized procedures for applying it to new cases.151 If the model is a spatiotemporal pattern to be applied across MRI voxels or EEG leads, for example, it is crucial to define precisely which voxels or leads are involved, and to what degree. In many cases, a written description of the measure will be inadequate, and electronic files defining the spatiotemporal patterns to be applied, along with data preprocessing and scaling steps, will be required. We recently reviewed nearly 600 MRI-based models that used machine learning to develop biomarkers for various brain disorders.153 Only a fraction of those models have a shared or shareable procedure for applying them to new cases. Without such procedures, it is difficult to imagine how they will be independently validated and applied.
1.2.2. Sensitivity and effect size
Sensitivity and specificity, and the related characteristics positive predictive value (PPV) and negative predictive value (NPV), are the basic metrics that characterize diagnostic performance. Sensitivity is the likelihood that a biomarker will yield a positive test result if a latent condition (eg, pain) is present, also called the “hit rate” or “recall” for the test. Formally, this can be expressed as P(marker+ |pain), the probability of observing a marker conditional on pain. The marker might be the expression of a continuous brain response after applying some cutoff threshold.147 Although sensitivity is defined for binary events, it is directly and positively related to the effect size of the relationship between the brain measure and pain; thus, for continuous measures, the correlation between the intensity of the marker signal and outcome is an analogue of sensitivity.151
1.2.3. Specificity
Specificity, also called precision, is the probability that a biomarker will respond in the absence of a condition. For pain, this can be expressed as P(marker− |no pain) or as 1 minus the false alarm rate. This is often defined based on disease-free people in the medical literature, but it is also applied to differential diagnosis. When it comes to brain states, there are many distinct states and experiences that are potentially confusable with pain. Specificity can be defined and quantified relative to a specific set of alternatives, and testing various plausible alternatives is a long-term proposition that requires multiple studies. For example, we and others have tested a biomarker for evoked pain, called the Neurologic Pain Signature (NPS), against a number of other, potentially confusable conditions (reviewed in Ref. 147; discussed further below). Although it is specific relative to (ie, does not respond to) many salient, arousing affective stimuli, there will likely be some classes of nonpainful stimuli or mental states that do activate the marker to some degree. These can both inform us as to which conditions share common neural substrates with pain and provide boundary conditions on its usefulness.
The diagnostic utility of a biomarker is more directly related to its PPV and NPV. The PPV is the likelihood that the underlying latent condition is present given a positive test result, P(pain|marker+). It can be calculated from the sensitivity, specificity, and prevalence (or “base rate”) of a disorder. The PPV is highly sensitive to prevalence and specificity. For example, in a disease that affects 1% of the population, even if sensitivity and specificity are both 98%, the PPV is only 33%. That is, a positive biomarker test only implies a 33% chance of having the underlying condition. If the sensitivity drops to 90%, there is little impact (PPV = 31%), but if the specificity drops to 90%, the PPV drops to 9%. Thus, testing and optimizing for specificity is crucially important in biomarker development.28,65
1.2.4. Generalizability
Inevitably, the conditions under which a biomarker is applied will differ from those under which it was developed in some ways. Generalizability refers to whether a prediction will hold when applied to a test data set or condition that differs from the original training set. Generalization can be assessed across individuals, variations in testing procedures and analysis pipelines, equipment (eg, different scanners), and populations (for a more extensive discussion, see Ref. 65). As the test conditions vary from the training conditions, diagnostic accuracy invariably decreases, although some biomarkers are more generalizable than others. For example, we have tested the NPS in 34 unique cohorts of participants from collaborators worldwide (counting only published results to date153,162) and validated its generalizability across multiple types of somatic and visceral pain (see below).
Many machine learning based studies use cross-validation to assess generalization to out-of-sample participants. The idea is to randomly split the participants into training (eg, 80% of participants) and testing (eg, 20% of participants), often stratifying on outcome and/or other variables. A biomarker is developed on the training data, which may involve selecting or combining across multiple variables to achieve maximum accuracy, and then, the final marker is tested on the held-out test sample. Cross-validation is a well-established safeguard against bias and overoptimistic accuracy estimates, but it also has limitations and can fail.143 In our survey of machine learning based neuroimaging biomarkers for clinical conditions, cross-validation was used in nearly all articles, but only a small subset of articles (about 9%) tested their marker in an independent cohort.153 Assessing generalizability across multiple sources of variation will be crucial as translational efforts move forward,28 and some recent efforts have been aimed explicitly at optimizing generalizability.65
Another important aspect of generalizability is ecological validity. To be translationally useful, biomarkers developed in research laboratories should be applicable to clinical or other appropriate settings.
1.2.5. Interpretability and explainability
A biomarker should be interpretable in several senses (see Ref. 153 for more discussion). First, it should have convergent validity with other methods, eg, human electrophysiology, lesion studies, and invasive techniques in animal studies (eg, optogenetics, chemogenetics, and imaging).28 This type of external validation is important for confidence that a biomarker is biologically meaningful and is underpinned by plausible mechanisms. It is also a crucial aspect of falsifiability. Second, for biomarkers to be credible and trusted by users, it is advantageous if the principles underlying their predictions can be explained (eg, in terms of crucial brain regions, systems, or neurochemicals).
2. Multivariate pattern analysis and machine learning analysis
Multivariate pattern analysis (MVPA) and machine learning have been often used to construct biomarkers. Multivariate pattern analysis is a set of methods that model task or mental states (eg, pain) using distributed patterns of neural activity.54 In univariate approaches, tasks or states are predictors, and brain signals are the outcomes to be explained—usually one voxel at a time. In MVPA approaches, mental states are assumed to reflect combinations of brain signals working together. Machine learning is a complementary concept. Machine learning comprises a set of algorithms, data selection methods and processing procedures developed to identify predictive models from complex, multivariate data. In the MVPA space, encoding refers to how single voxels encode task features or mental states, and decoding refers to the process of making predictions about such features or states from brain data.98 Biomarkers are essentially decoding models.
The features of a data set are variables used to train a model. Many kinds of brain features can be used. In fMRI, signals might be task-evoked activity in a set of voxels, activity in components extracted with independent component analysis (ICA), fluctuation energy at certain frequencies, functional or effective connectivity across a set of regions, graph theoretic properties such as global network efficiency, and more. In sMRI, features could include local gray matter density estimates with voxel-based morphometry, cortical thickness, bending energy, gray matter volume in various structures, and other measures. For EEG and magnetoencephalography (MEG), features can include stimulus-evoked potentials, energy at various oscillation frequencies, the amplitude and phase of coherence measures across sensors, and activity in latent sources.105 Often, features are selected or combined together into higher-level units during machine learning analysis.
Compared with univariate analyses, optimized MVPA patterns often have dramatically larger effect sizes, and thus increased sensitivity, in relation to tasks and mental states.57,65 This is because most mental states are accomplished by distributed networks—signal in multiple brain areas is relevant.153 When this is true, models that capture those distributed signals will outperform those based on local signals. In addition, MVPA patterns have shown much greater specificity as well.57 Although single voxels are not very selective for individual tasks or mental states, different tasks can produce distinct patterns of activity across voxels, even when those voxels are all activated by multiple tasks.154 Suppose that a set of voxels each include neural populations that respond to 2 tasks. Thus, they are both activated by both tasks. However, the density of neurons dedicated to task 1 and task 2 will vary across voxels. This will influence the relative level of activity across voxels and allow the tasks to be discriminated based on the observed multivoxel patterns.
2.1. Analysis choices in biomarker construction
There are a wide variety of potential choices to be made when performing MVPA, including which outcomes (tasks or mental states) to predict and which features to include. Outcomes are generally either categorical (eg, a stimulus class, subject response, or disease status) or continuous (eg, measures of pain or function, or age), and can vary within-person, between-person, or both.
Feature selection is also a critical part of the process. One fundamental choice concerns the spatial scope of the analysis. Many early applications of predictive modeling were applied within individual brain regions, particularly in the visual system, to “decode” object features based on local topography.55,61 For translational purposes, it has become popular to build models that include features distributed across the whole brain. This integrates all the measures available across the brain, and sometimes even across multiple types of images, into a single predictive model.
The traditional wisdom has been that constructing such maps is not feasible because the number of features (eg, voxels) exceeds the number of observations (eg, subjects or trials), causing problems with model overfitting and interpretability. However, statistical techniques, including kernel form regression or classification, dimension reduction, and penalization, can help stabilize maps even when large numbers of voxels are included in the model. Also, techniques such as cross-validation and multistudy prospective testing permit valid and essentially unbiased tests of model performance. If effect sizes are large and brain activity or related measures are robustly related to the outcome, then predictive maps with high accuracy can be estimated even using small samples.21
Another important consideration is how to deal with confounds. Drug use, comorbid pathology, age, sex, and head movement are specific concerns for clinical decoding studies. Some of these might be part of the disorder and not easily separated. For example, a prognostic biomarker for chronic postsurgical pain may involve co-occurring factors such as fear of pain and depression. However, it could still serve as a useful biomarker. In addition, some biomarkers might reflect consequences rather than causes of pain but still be useful. For example, motor cortical connectivity changes might result from reduced mobility but still correlate with pain. What is key in these situations is to understand, to the degree possible, which biomarkers are causally related to pain pathogenesis, and which may be more closely related to other co-occurring variables.
Several studies have investigated ways to control for confounds and/or test whether they are likely driving relationships between biomarkers and outcomes.26,111,128,133 Some helpful procedures include: (1) regressing out the confound within the cross-validation loop; this is important because doing this outside the loop might create dependence and lead to pessimistic performances128; (2) testing whether a biomarker relates more strongly to the outcome of interest (eg, pain) than any potential co-occurring variables (eg, sleep loss or drug use); (3) testing the mediation between variables, eg, if a biomarker mediates the relationship between sleep loss and pain, it is related to pain even when controlling for sleep loss; (4) during training, identify biomarkers unrelated to co-occurring variables by stratifying samples and matching these on confounds; and (5) disaggregate some variables, such as sex, and test whether predictions are better within subgroups than across the whole population.
One confound that deserves special attention in decoding analyses is head motion.86 Head motion might have large influences on decoding performance (eg, predicting pain condition vs control where patients with pain might have more difficulties to lie still in the scanner). There are several ways to mitigate this, including behavioral training before scanning,86 real-time feedback during scanning, and postprocessing methods such as scrubbing,109 aCompCor,96 ICA-AROMA,129 RETROICOR,46 and more (for comparisons, see Refs. 102 and 118).
2.2. Types of algorithms
2.2.1. Classification algorithms
Classification is a supervised learning technique used to establish rules for identifying the category/class to which a new data point will fall under. Common techniques include support vector machines (SVMs), k-nearest neighbors, and Gaussian naive Bayes.
Many classification algorithms seek to find a hyperplane that separates observations in the feature space by category/class. In this setting, the distance between the hyperplane and the closest data points on either side is referred to as the margin. Support vector machines find the hyperplane that has the largest margin. A quadratic programming algorithm is used to estimate the coefficients that maximize the margin. Support vector machines are effective in high-dimensional spaces—eg, with whole-brain patterns.
The k-nearest neighbor algorithm is both simple and effective. Classification of a new data point is performed by searching through the entire training set for the K most similar instances (neighbors) and performing a simple majority vote of their category/class. The algorithm is simple to implement, robust to noisy training data, and effective with large training data sets. Finding the neighbors can be difficult in very high-dimensional data (eg, many voxels), which can negatively affect the algorithm's performance.
Gaussian naive Bayes is another simple yet powerful algorithm for classification. It involves using Bayes' theorem to compute the conditional probability for each class given each of a set of input features (eg, voxels) is treated independently. It is called “naive” because it assumes that each feature is independent. This is a strong and often unrealistic assumption, but in many cases, trying to model the complex dependencies across features can be counterproductive for prediction. Thus, the approach is effective for a large range of complex problems. It requires a small amount of training data to estimate the necessary parameters and is fast compared to more complex methods.
2.2.2. Regression algorithms
Regression algorithms are used to predict the value of a continuous outcome variable, given the values of a feature vector. Common techniques include multiple linear regression and regression trees. Because of the number of features exceeds the number of observations, penalized regression techniques are often used in practice.
This involves building prior knowledge and constraints into the regression equation (ie, the cost function) to encourage desirable characteristics. For example, L1 penalization, used in LASSO regression, constrains the absolute value of regression coefficients and promotes sparsity (nonzero weights on only a few features). L2 penalization, used in ridge regression, constrains the geometric mean of the coefficients. A key difference between these 2 approaches is that while ridge regression shrinks all of the model coefficients towards zero, LASSO shrinks the coefficients corresponding to the less important features to zero, thereby removing them from the model. Thus, LASSO can be used for feature selection when there are a large number of features. Elastic net regression combines both types of penalties into a single model. The consequence of using this combination is to effectively shrink coefficients (like in ridge regression), while setting some coefficients to zero (as in LASSO). This approach tends to perform better than LASSO when features are correlated with one another, as in brain imaging. In this setting, LASSO tends to choose only one of the correlated features, while setting the rest to zero.
In addition to operating on voxels, dimension-reduction steps before regression can extract components, which then become predictors. Principal component analysis, ICA, and latent factor analysis are all examples (see, eg, the LASSO-PCR algorithm described in Refs. 146 and 147). This is advantageous when working with brain data because the data decomposition can capture covariation across voxels, reducing the problem of arbitrary selection of voxels within a correlated set found with LASSO. Operating on components can also increase model interpretability.
2.2.3. Decision trees
Decision trees are another important class of predictive modeling algorithms and can be used for both regression and classification. They are used to segment feature space (eg, values on a set of brain voxels) into a number of smaller regions associated with particular outcome values. Tree methods are both nonparametric and nonlinear. They are easy to learn and fast for making predictions, and are accurate for a broad range of problems. Random forests are a type of additive model that makes predictions by combining decisions from a sequence of decision trees. Each tree is constructed independently using a different random subsample of the data.
2.2.4. Decoding models and multivariate extensions
The most straightforward applications of all the algorithms described above use the algorithms as decoding models. Brain states can be represented as a vector of features using individual voxels,21,147 a collection of regions of interest (ROIs), temporal or spatial frequencies, or patterns of connectivity.31,117 Correlations across these features are usually accounted for in some way (eg, this covariance is modeled in the regression process). In all the cases above, a multivariate model of the brain is used to predict a univariate outcome, usually a task or behavior thought to index a mental state. This goal matches the goal of biomarker development.
In some cases, one might wish to identify patterns that predict combinations of task or behavioral variables without specifying in advance what those combinations are. For example, one might wish to model differences across 4 different types of painful stimuli without prior knowledge of which distinctions the brain “cares about.” Or, one might want to predict a latent behavioral variable that is a combination of correlated variables—eg, pain intensity, affect, and interference measures—without prespecifying how those variables should be combined. Techniques including partial least squares, canonical correlation, or semiblind ICA are all extensions that can “decode” multiple outcomes simultaneously.93
2.2.5. Encoding–decoding models
Encoding–decoding models are another extension of the modeling framework described above. In the encoding part of the model, a set of features describing a stimulus are used to predict the activity in each individual voxel.98 For example, a visual stimulus might be decomposed into a set of features using Gabor filters,62 words into semantic features,93 and speech into acoustic features.103 The voxel's activity is regressed on these features, providing a tuning curve for the voxel in the feature space. This is repeated for all voxels, much as in standard univariate mapping. The encoding model can be validated by predicting the brain maps evoked by new, out-of-sample test stimuli.62 To make predictions about the task/behavior a person is experiencing, the decoding part of the model takes a brain image and generates the most likely task/behavioral features given activity in each voxel, aggregated across voxels into a single overall prediction.66 Thus, overall, encoding–decoding models add considerable flexibility in modeling stimulus–brain relationships.
2.2.6. Deep learning
Deep learning is part of a family of machine learning methods based on multilayer neural networks. It exploits hierarchical feature representations learned directly from the raw data, instead of using features designed using domain-specific knowledge. Neural networks are related to data compression approaches and other techniques described above, and can be formally equivalent or nearly so to these techniques depending on the way networks are constructed. For example, a 2-layer network consisting of an input layer (eg, with one node per brain voxel) and an output layer with one node per psychological category can implement a linear classifier such as logistic regression. Deep neural networks contain one or more intermediate (or “hidden”) layers, providing a series of hierarchical, usually nonlinear transformations of the input data. These networks differ from other machine learning techniques in that the hidden layers encode complex features learned from the data, thereby achieving increasingly higher levels of abstraction and complexity.
There are 2 major classes of deep learning models that differ in how information is propagated through the network. Feedforward networks propagate information in a single direction, going from the input to the output layer. Recurrent networks contain feedback connections that allow the information layer or higher-level layers to affect lower-level representations. In addition, recent efforts add memory features, allowing activity from past inputs to persist and affect the current activity and output. An example is long short-term memory networks. Another widely used development is the addition of convolutional layers, which have connections that are constrained so that they map a space of representations from one layer to a single unit in the next layer, allowing the model to generalize across a space of lower-level representations.
3. Pain biomarkers: state of the field
To provide a picture of current work on neuroimaging-based biomarkers for pain, we searched for articles on PubMed (through December 31, 2018) using 3 different search terms (“biomarker,” “MVPA,” and “machine learning”) combined with “pain” and “neuroimaging” (eg, pain AND neuroimaging AND machine learning). Other measures considering biomarkers, including behavioral measures and facial expressions, are reviewed in detail by Lötsch and Ultsch.85 Articles were grouped by the imaging method, and the 4 most widely used techniques were selected for this review: fMRI, rs-fMRI, sMRI, and EEG (n = 50 studies). Studies that did not include a proper cross-validation method were excluded (n = 3), as this is one of the basic steps and requirements to validate a predictive model. In total, 47 studies were included. Figure 1 presents an overview of the studies and clearly indicates an increase in the use of machine learning techniques for pain prediction, as in many fields. Compared to the number of machine learning based models in other fields, including Alzheimer, Parkinson, autism, attention deficit hyperactivity disorder, and others,153 the number remains relatively small.
Figure 1.
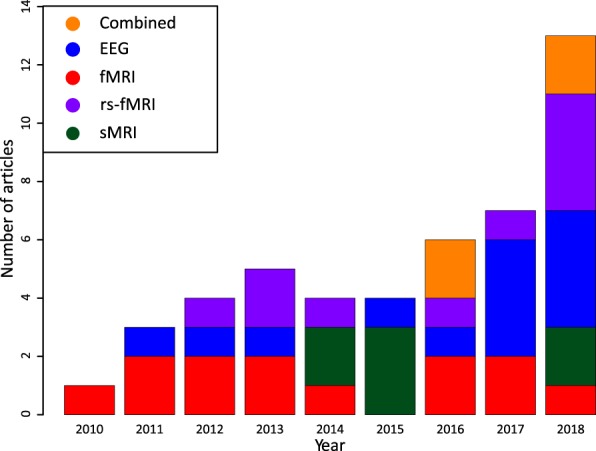
Timeline of machine learning articles for pain: a timeline showing the number of published articles per neuroimaging technique or combinations of techniques for pain studies investigating biomarkers (47 in total). Studies include the use of EEG, task fMRI (denoted fMRI), rs-fMRI, sMRI, or a combination of techniques (denoted combined) and use a cross-validation method for their predictive model. EEG, electroencephalography; fMRI, functional magnetic resonance imaging; rs-fMRI, resting-state functional magnetic resonance imaging; sMRI, structural magnetic resonance imaging.
3.1. Functional magnetic resonance imaging
3.1.1. Evoked pain
Although decoding in fMRI was already used in the early 2000s, mostly in vision research, it was not until 2010 that the first article was published predicting pain.56,90 Marquand et al.90 demonstrated the feasibility of predicting subjective heat pain intensity from whole-brain fMRI volumes using Gaussian process regression. This provided a relatively rare example of the use of machine learning to predict a continuous outcome. A second study predicted pain intensities using a regularization algorithm, induced by an injection with an ascorbic acid.110 Further developments were shown by Brown et al.,13 who used a second test set as a form of prospective validation. They first measured fMRI activity during painful and nonpainful thermal stimulation and trained an SVM that was used to classify pain in a cross-validation sample, with accuracy of 86.6%, and a hold-out test sample, with accuracy of 74.6%. The whole-brain SVM model included positive weights in regions known to receive nociceptive input—chiefly the mid-insula, anterior cingulate cortex, and somatosensory cortex—which is an important neuroanatomical validation. The whole-brain model also outperformed models based on individual ROIs, suggesting that distributed models are helpful.
The conclusion that distributed predictive models are helpful was supported by Brodersen et al.,12 who used whole-brain activity before and during near-threshold laser stimulation to predict whether a stimulus would be experienced as painful or not (accuracy was 57.6% and 61.4%, respectively).12 They found that several individual regions were predictive of pain (eg, right and left primary somatosensory cortex and right insula), but considering multiple areas together significantly improved the prediction accuracy.
Like Marquand et al.,90 Cecchi et al.20 used a regression model to predict pain, this time combining machine learning with a dynamic nonlinear psychophysical model.20 The psychophysical model captured the transformation of noxious input into pain, accounting for nonlinear and time-delayed effects of the rate of change and stimulus history (including “offset analgesia”-type effects). This continuous signal was then predicted using fMRI time series data. This study illustrates the advantages of combining machine learning and dynamic psychophysical models.
With the exception of Brown et al.,13 studies to this point had focused on within-person prediction12,20,90,110—which means that the brain model differed across individuals—without attempting to develop a biomarker tracking pain intensity that could be applied to new individuals. In addition, these studies did not test specificity relative to other types of nonpainful sensory and emotional events. In 2013, Wager et al.147 developed a regression model that predicted pain intensity across individuals and across 4 separate studies.147 The model was named the NPS as a way of providing a label that could indicate when the same model (eg, the same, pre-trained regression weights) was being used in subsequent studies. The NPS was trained and initially tested on a cross-validation sample and tested prospectively on 3 subsequent studies. It showed high sensitivity and specificity (94% or more) for discriminating pain from nonpainful warmth, pain anticipation, and pain recall when applied to new individuals. It also discriminated pain from the “social pain” induced by viewing stimuli related to romantic rejection, which had previously been found to activate many “pain-processing” areas,37 including the insula, anterior cingulate cortex, and secondary somatosensory cortex.68 Finally, the NPS response was suppressed by the opiate remifentanil but unaffected by a placebo manipulation (open vs hidden remifentanil, which affected pain reports), showing differential responses to pharmacological and psychological interventions.
Importantly, it was not claimed that the NPS was a model for all pain under all circumstances, but rather a model of a brain system that contributes to pain experience and report, likely alongside other psychological and brain processes. The existence of other components has been borne out by a number of other studies since.8,11,82,155,156,162 Some psychological manipulations, however, do appear to influence NPS responses.60,82
A later modeling effort identified a “signature” intended to capture additional variability related to psychological influences and decision-making processes.156 Models were trained to predict pain after controlling for stimulus intensity and the previously developed NPS, and a group model (named the Stimulus Intensity-Independent Pain Signature-1 [SIIPS-1]) was constructed. This model was positively associated with pain in 98% of the participants and mediated influences of expectancy cues and perceived control in 2 independent, prospective hold-out studies.
Further studies have continued to identify the specificity of the NPS across different conditions, showing no responses to aversive pictures,21 observations of others in pain,67 and pain anticipation.67,82 Studies have also shown generalization to multiple types of evoked pain, including thermal, mechanical, laser, electrical, and visceral (rectal distension67,162). The NPS also shows moderately high test–retest reliability, comparable with, but somewhat lower than the reliability for self-reported pain.152
In parallel, other studies have used fMRI decoding for specific purposes and to develop new methods.138 In an interesting study, Liang et al.79 showed that visual, tactile, and auditory stimuli evoke distinct patterns of activity in primary sensory cortices corresponding to all 3 modalities.79 Thus, primary visual activity could provide above-chance decoding of whether a stimulus is somatosensory or auditory. This fits with a body of recent work, showing that brain information is distributed much more broadly than many of us initially assumed.
3.1.2. Chronic pain
A recurring theme in chronic pain research is the idea that patients exhibit long-term brain reorganization that makes them react differently to evoked pain. Several early studies used evoked responses to predict whether individuals experienced chronic pain. For example, Baliki et al.6 found that patients with chronic low back pain (cLBP) showed reduced responses to painful stimulus offset in the nucleus accumbens. Although the model was not cross-validated, they did show that the effect held up in a subsequent scanning run from those participants. Callan et al.15 used fMRI during evoked electrical stimulation on the back to classify patients with cLBP vs healthy controls with 92.3% accuracy in a cross-validation sample. Likewise, Harper et al.52 applied pressure pain to patients with temporomandibular disorder (TMD) and healthy controls. They were not able to classify patients from controls above-chance based on fMRI activity, however, they were able to discriminate pressure pain from rest and discriminate between facial pain (involved in TMD) and thumb pain (a control area) in patients with TMD but not in healthy controls. Thus, the study is a nice illustration of how positive controls (basic positive findings for pressure vs rest) can help make null findings (patient vs control) more useful.
Another pain disorder that has received attention is fibromyalgia. Using fMRI data during a visual stimulation task, researchers were able to distinguish between patients with fibromyalgia and healthy controls with 82% accuracy.53 Increased visual sensitivity in patients was also correlated with their pain intensity. This suggests that fibromyalgia may involve sensory abnormalities beyond pain—an idea borne out in subsequent studies.83,84 In one study, López-Solà et al.84 found that patients with fibromyalgia both showed increased NPS responses to pressure pain and altered fMRI responses to basic visual and auditory stimuli, captured by an SVM classifier.84 These features were combined into a model that classified patients from matched controls with 93% cross-validated accuracy.
3.2. Evaluation
The use of fMRI for acute pain has allowed for a diverse range of methods, classifiers, and pain stimuli (Table 1). Both pain intensity scores90 and low and high pain is investigated using thermal or laser stimuli.12,13,147 More recently, chronic pain has been investigated, with promising results.15,84 Interestingly, fMRI has been used primarily for diagnostic purposes, and a priority for the future is the development of prognostic and predictive biomarkers. In addition, models have focused on pain but neglected other outcomes, including functionality, resistance to distraction under pain, and other pain-relevant outcomes.
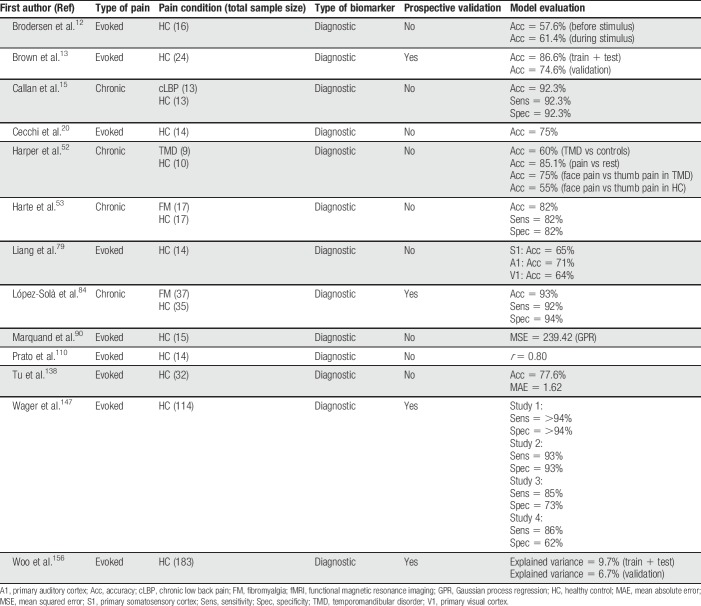
Table 1.
Summary of all fMRI articles discussed in this review.
Most models show good classification performance, and some have been validated in related samples13,84 or tested for generalizability to new samples.147,156 Some models predicting evoked pain, particularly the NPS, have been extensively validated across samples, but evoked pain models predicting clinical pain have not been validated in independent samples. This is a priority for future work. In terms of interpretability, activation of the insula, anterior cingulate, secondary somatosensory cortex, and thalamus are recurring themes, demonstrating some convergence. However, whether the models produce consistent or divergent brain patterns is difficult to ascertain, and more direct model comparisons are needed. In summary, a range of evoked pain models exist, and the most promising models should be tested further, particularly for utility across clinical pain conditions.
3.3. Resting-state functional magnetic resonance imaging
3.3.1. Chronic pain
Functional connectivity measures provide an appealing way to characterize individual differences without relying on experimental tasks. It has been used to differentiate patients with pain from controls in subacute back pain, functional dyspepsia, fibromyalgia, migraine, neuropathic pain, and chronic pelvic pain (CPP). Most studies have applied machine learning procedures to identify patterns of pairwise connectivity that differentiate patients vs controls. Some studies have also used graph theoretic measures or other higher-order summary properties. A few studies have also begun to develop prognostic biomarkers.
In functional dyspepsia, several studies have combined functional connectivity with machine learning procedures applied to pairwise connectivity patterns. In one study, connectivity changes that were correlated with symptom scores were used as features for classification, resulting in 88% accuracy in an independent test set, relying mostly on features in the limbic/paralimbic system and prefrontal cortex.97 Liu et al.80 classified patients vs controls by subjecting regional homogeneity values to SVM analysis, with 87% cross-validated accuracy.80 Regional homogeneity measures the similarity of synchronization between the time series of a voxel and its closest voxels.158 This is an example of using a higher-order property that can be extracted from fMRI; however, it may also be very sensitive to head movement. Ruling out confounds, including movement, is an ongoing issue and will become more and more important as translational efforts progress.
In an attempt at differential diagnosis across disorders, another study attempted to find differences in functional connectivity in areas involved in the salience network and default mode network between patients with fibromyalgia, rheumatoid arthritis, and healthy controls.130 A predefined model was not able to successfully classify the different groups. Exploratory analyses identified a model with diagnostic accuracy up to 78.8%, but this may be overoptimistic because of model selection bias; further studies are necessary to investigate the best-performing model in new test subjects.
Functional connectivity in rs-fMRI has also been shown to provide reasonable classification accuracy between healthy controls and patients with migraine.24 A diagonal quadratic discriminative analysis resulted in an overall accuracy of 81% based on 6 pain-related areas.
Although most studies have used static connectivity (averaging across time), some have begun to use dynamic connectivity measures to predict pain. Dynamic connectivity estimates associations among brain voxels at each time point in a time series, providing an expanded set of features for machine learning (at a cost in signal-to-noise). Cheng et al.23 used elastic net regression to predict state and trait neuropathic pain from both static and dynamic functional connectivity measures. They found stronger associations with trait pain (ρ = 0.72) and found that the most predictive features of these models were dynamic. Relatedly, another recent study used rs-fMRI to investigate low-frequency oscillations (LFOs) in patients with chronic pain.114 Aberrations in LFOs were found to be predictive of trait pain intensity, but not state pain intensity.
The studies above used data collected from a single site and thus did not test generalizability across cohorts and scanners. One recent study of chronic back pain used 3 independent data sets, along with graph theoretic measures, to classify patients with chronic back pain vs controls. Models based on both SVM and deep learning resulted in above-chance, although modest, accuracy (68% and 64% accuracy, respectively).88 These accuracy levels are likely a realistic reflection of the state of the art for rs-fMRI–based identification of patients with pain. In addition, analysis of the network changes captured by the models identified a reorganization of connectivity modules centered on sensorimotor cortical regions. This provides some input into a current ambiguity in the field about the relative importance of somatosensory vs limbic (eg, frontostriatal) systems in pain chronification. However, more systematic model comparisons will be needed to identify the systems crucial for the performance of complex connectivity-based models.
3.3.2. Prognostic and predictive biomarkers
Prognostic biomarkers are rare, although they are an intensive focus of current funding efforts. Other approaches that are currently used (ie, besides machine learning) may further develop and become useful predictive biomarkers.10 In the first example, to the best of our knowledge, of an imaging-based prognostic biomarker for pain, Baliki et al.7 found that functional connectivity in frontostriatal circuits predicted the transition from subacute to chronic back pain 1 year later, with an area under the receiver operator curve score (comparable with accuracy for present purposes) of 0.81.7 Subsequently, Kutch et al.71 predicted 3-month symptom change in patients with urologic CPP syndrome in the multisite MAPP study.71 Functional connectivity data identified 73.1% of patients correctly as improvers or nonimprovers. However, this did not predict longer-term (ie, 6 and 12 months) chronicity. Finally, another innovative study used graph theoretic measures to predict the magnitude of placebo responses to treatment for knee osteoarthritis.132 The right dorsolateral prefrontal cortex was more strongly connected to other regions in strong placebo responders. This effect was tested in an independent cohort and had an area under the curve of 0.95—a high value that indicates that the study should be tested for generalizability and reproducibility in other laboratories.
3.4. Evaluation
These studies, summarized in Table 2, reveal generally modest accuracy in case–control classification and prognosis, although some promising models warrant further testing. Analyses of specificity are markedly absent from the literature, and the development of models designed to generalize across sites has only just begun.23,88 Most studies are case–control diagnostic studies, but it is encouraging that prognostic and predictive biomarkers are entering the space.7,71,132 In terms of interpretability, it is difficult to assess convergence in findings across studies because (1) the connectivity measures and models are complex, involving large numbers of contributing brain voxels; (2) nearly every study used a different analytic method; and (3) there is reasonably good coverage of multiple disorders, but too few studies in any disorder category to extract meaningful patterns from the whole. This is an important shortcoming that should be systematically addressed. Nonetheless, brain systems that appear to be important include alterations in connectivity in (1) frontostriatal circuits associated with the “default mode,” often in the form of hyperconnectivity, and (2) the somatosensory cortex, often increased connectivity with other brain regions and “default mode” regions in particular.
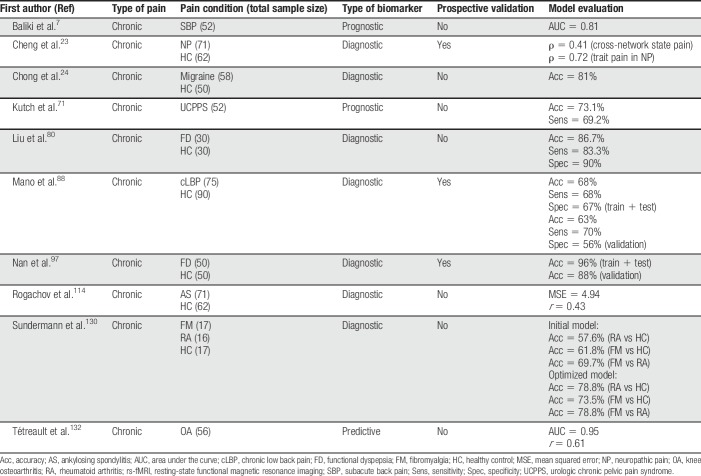
Table 2.
Summary of all rs-fMRI articles discussed in this review.
3.5. Structural magnetic resonance imaging
3.5.1. Chronic pain
Structural MRI has been used to characterize and predict the incidence of chronic visceral pain, musculoskeletal pain, and migraine (for recent reviews on these topics, see Refs. 9 and 127). As with fMRI, most studies perform case–control diagnostic classification.
A number of previous studies investigated case–control differences but did not assess person-level classification. A first study to do so used an SVM classifier to distinguish patients with cLBP from healthy controls.139 Gray matter density estimates from T1 MRIs distinguished the 2 groups with 76% accuracy. Areas important for classification included the secondary somatosensory cortex and motor areas. Using similar approaches, Bagarinao et al.4 were able to distinguish between individuals with CPP and healthy controls with 73% accuracy. Robinson et al.113 showed that it was possible to classify patients with fibromyalgia and healthy controls based on brain volumes, with a decision tree as best classifier performing at 75.5% accuracy. Mood and pain intensity self-report measures outperformed neuroimaging classification (96% accuracy), which is expected as fibromyalgia is defined largely based on pain self-reports; however, this discrepancy illustrates how far brain-based models have to go to fully capture the neurological variations underlying established behavioral measures.
A subsequent study investigated a morphological signature for irritable bowel syndrome.72 Using estimates of gray matter volume, surface area, mean cortical thickness, and mean curvature, they tried to predict whether subjects belonged to patients with irritable bowel syndrome or healthy controls. A sparse partial least squares discriminant analysis was applied onto the data and discriminated patients from healthy controls with 70% accuracy.
Mean cortical thickness, surface area, and volume estimates measured with sMRI were used to distinguish patients with migraine from healthy controls using a diagonal quadratic discriminative analysis.124 This resulted in an overall accuracy of 68% when taking chronic and episodic migraine together. Comparing chronic migraine vs healthy controls and episodic migraine vs healthy controls resulted in classification accuracies of 86.3% and 67.2%, respectively. Patients with chronic vs episodic migraine were distinguished with 84.2% accuracy.
Recently, a study used whole-brain gray matter to discriminate between subjects with primary dysmenorrhea and healthy controls with an accuracy of 75.4% and 70.2% in a separate validation set.22
There are several mechanisms that modulate pain such as pain catastrophizing and cognitive control. Fear of pain has been found to be an important contributor to the development of chronic pain.149 One study used gray matter volume in healthy subjects to predict fear of pain scores with a correlation of r = 0.41149—however, this analysis was “exploratory,” as it was influenced by selection of voxels outside the cross-validation loop. Such studies could be a valuable addition to current progression models of chronic pain conditions where psychological processes play a large role.14
3.6. Evaluation
As with rs-fMRI, classification accuracies for patients with chronic pain vs controls are modest, in the 70% to 80% range (Table 3). This actually reflects much larger effects than are typical in standard brain mapping studies. For example, a “large” effect size of d = 0.8 is required to achieve a modest 2-group classification of 66% (if variables are normally distributed), and 80% classification requires a very large effect size of d = 1.6. Effect sizes of d = 0.5 are typical of standard brain mapping studies.106 However, it is unclear whether accuracy values in this range will be useful in translational settings. With 80% sensitivity and specificity (the balanced accuracy in 2-choice binary classification is both its sensitivity and specificity), the PPV of a relatively common disorder affecting 5% of the population is only 17%.
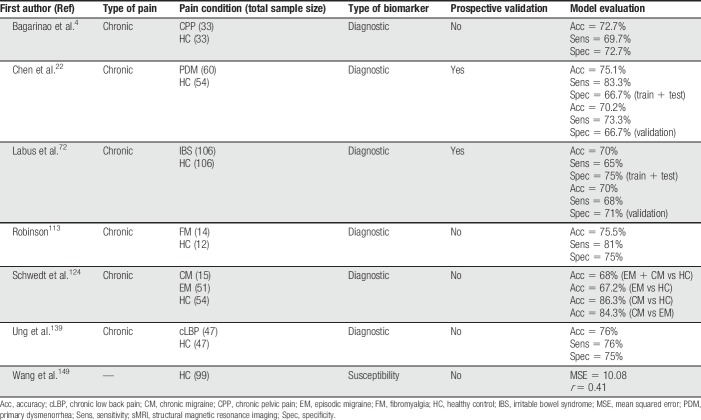
Table 3.
Summary of all sMRI articles discussed in this review.
In addition, sensitivity, specificity, and generalizability must be considered. Future efforts need to focus on comparing brain features across models. As with rs-fMRI, the most consistent changes appear to be located in the medial prefrontal cortex (associated with the “default mode” network) and the somatosensory cortex.
Finally, in most cases, case–control classification biomarkers are not likely to directly reflect or correlate with symptoms such as pain.72 Further efforts should characterize what biobehavioral features of people with chronic pain are being captured by the model. Variables such as age, head movement (which can affect sMRI), socioeconomic status, drug and medication use, and more are very difficult to control for adequately using linear regression, and large-sample studies are likely to be necessary to understand what these pain-related models are capturing.
3.7. Electroencephalography
3.7.1. Evoked pain
Pain-decoding evidence from EEG first appeared in 2012, with a study decoding an individual's sensitivity to pain.122 Subjects received equally strong laser stimuli but reported large differences in pain ratings, showing individual variability in pain perception. An SVM was trained on time–frequency decompositions of the EEG signal, classifying if a subject was pain sensitive or insensitive with 83% accuracy. In the study by Huang et al.,58 participants received short laser heat pulses, and laser-evoked potentials were used to distinguish between low and high pain intensity and predict continuous pain ratings. Accuracy was higher within subjects (86.3%) than between subjects (80.3%). This is an expected effect of interindividual nuisance variability unrelated to pain, and some groups have attempted to normalize the scale of EEG data5 or extract interstimulus EEG features to reduce interindividual noise.78
In an example of a predictive biomarker, Gram et al.48 found that machine learning on EEG data collected during a cold-pressor test was able to predict responders vs nonresponders to opioid treatment with 72% accuracy. Conventional group–based analysis did not show any group differences, whereas the SVM was able to predict individuals' opioid analgesia.
Another promising direction is multimodal classification based on combined brain and autonomic signals, as autonomic responses alone can track pain intensity well in unbiased tests.45 Lancaster et al.74 decoded pain from combined EEG and physiological data (pulse and skin conductance). Using sparse logistic regression, they were able to classify thermal pain stimuli and multimodal sensory stimuli with an average accuracy of 70%. Within-subject accuracy reached as high as 79%.
Some studies have reported high classification accuracy for high vs low pain. Misra et al.92 selected pain-related features from a time–frequency analysis—theta and gamma power in the prefrontal cortex and lower beta power in the contralateral sensorimotor cortex—and used them to classify high vs low pain heat stimuli with 89.6% accuracy. Likewise, Vijayakumar et al.144 used tonic thermal stimuli to mimic chronic pain aspects. A random forest model was able to classify pain in 10 different levels with an accuracy of 89.5%. Most information could be decoded from the gamma band, although all frequency bands contributed to pain classification. For studies claiming high accuracy in particular, prospective tests on new samples, and replication by independent laboratories, are needed.
Converging evidence that low peak alpha frequency and/or power are important was provided by Furman et al.,44 who found that peak alpha frequency was correlated with later sensitivity to capsaicin-potentiated heat pain in a subsequent test (r = 0.55).
3.7.2. Chronic pain
Early research showed that patients with chronic pancreatitis (a form of visceral pain) showed differences in spectral EEG after administration of pregabalin and placebo.49 Based on these features, an SVM was able to classify patients into a pregabalin- or placebo-receiving group with 85.7% accuracy. This study showed the possibility of using EEG as a response (eg, pharmacodynamic) biomarker.
As with evoked pain, chronic pain has been associated with elevated EEG theta frequency energy and reduced alpha energy. In a large study, Vanneste et al.142 used resting-state EEG to assess thalamocortical dysrhythmia, a characteristic comprised in part of slowing of alpha frequencies into the theta range (cf. Ref. 44). This may occur across disorders, including neuropathic pain, tinnitus, and depression, with spatial patterns varying across disorders.142 A predictive model differentiated patients with neuropathic pain from healthy controls with 92.5% accuracy. Moreover, different disorders were associated with different spatial patterns, and the model correctly classified most subjects in multiway classification, providing some evidence for diagnostic specificity. As with other cases, independent replication without altering the predictive weights would help validate this high-accuracy finding.
There are also examples of predictive and prognostic biomarkers in the EEG literature. In an example of a predictive biomarker, preoperative EEG during a cold-pressor test was used to predict postoperative pain treatment after hip replacement, with 65% accuracy.47 No differences were found in conventional between-group analyses of responders and nonresponders. Vuckovic et al.145 developed a prognostic biomarker based on resting-state EEG. Many patients with a spinal cord injury later develop central neuropathic pain. A linear discriminant analysis classifier and artificial neural network classified patients who would develop central neuropathic pain within 6 months with 86% and 83% accuracy, respectively, providing a potential presymptomatic substrate for early intervention.
3.8. Evaluation
Electroencephalography has been used for diagnostic, predictive, and prognostic biomarkers in evoked and chronic pain; this diversity of applications reflects benefits related to its low cost and portability (Table 4). However, current models are all separate studies and it is unclear how the results converge and whether models rely on similar features. Furthermore, it is unclear how these compare between different chronic pain conditions. In addition, prospective validation and tests of generalizability are very rare. The most promising models should be validated, and further research is necessary.
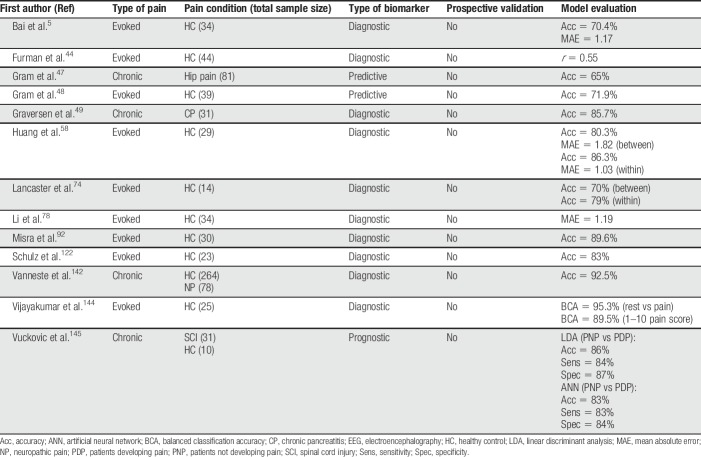
Table 4.
Summary of all EEG articles discussed in this review.
3.9. Multimodal neuroimaging and multiple data sources
Few studies have attempted to combine the discussed techniques into multimodal classifiers. One group investigated evoked pain with both EEG and fMRI,137 using both stimulus-evoked and prestimulus activity. Time–frequency EEG patterns and BOLD-fMRI patterns before and after a laser-evoked pain stimulation showed reliable classification of low and high pain intensities and better performance than solely stimulus-evoked activity (83.5% vs 78.2%).
A step towards integrating different modalities was investigated by Zhang et al.159 They used rs-fMRI and sMRI to predict migraine. The model used rs-fMRI features related to amplitude of LFOs and regional homogeneity and sMRI regional gray matter volume. An SVM with a multikernel strategy yielded an accuracy of 83.7%.
This line was continued in a recent study combining functional connectivity from rs-fMRI, regional cerebral blood flow from arterial spin labeling, and high-frequency heart rate variability.75 Back pain was exacerbated with maneuvers to induce low and high pain states in patients with cLBP. Combined features resulted in an accuracy of 92.5% of within-patient classification of high and low states. The model also predicted individual differences in maneuver-induced pain (r = 0.63).
A multimodal predictive biomarker was developed by Vachon-Presseau et al.,141 who predicted placebo response using questionnaires, sMRI, and rs-fMRI in patients with cLBP. Data from questionnaires yielded an accuracy of 72% in predicting placebo pill responders and nonresponders, whereas sMRI and rs-fMRI failed to achieve significance. Questionnaire data predicted placebo response magnitude (r2 = 0.31), as did rs-fMRI to a lesser degree (r2 = 0.13), although the latter did not generalize to data collected at other visits. sMRI did not predict placebo pill response. Table 5 shows a summary of the above described studies.
Table 5.
Summary of all combined techniques discussed in this review.
An important other development will be the use of multiple data sources and imaging modalities. Previous research has already shown that physiological responses such as heart rate, skin conductance, pupil dilation, and body temperature can be used to predict pain with a performance comparable with neuroimaging methods (for a review, see Ref. 85). Different aspects of pain may be reflected in brain and physiological responses, thus representing nonoverlapping information and adding in the performance.74 For chronic pain conditions, neuroimaging may be combined with physiologic measures such as physical functioning157 or urine metabolomics for neuropathic pain.40 It would be advantageous to pursue this direction, as few studies have investigated a combination of data sources and this could lead to converging evidence. Classifiers such as SVM also perform well with multiple data types, which makes it easy to use several data sources.13
3.10. Other methods
Besides the discussed neuroimaging methods above, other methods such as arterial spin labeling, functional near-infrared spectroscopy, and diffusion tensor imaging may be promising as well, but less research has been performed using these methods. Functional near-infrared spectroscopy, for example, has found to be effective in classifying high and low evoked pain.108,115 Some systems are portable and relatively inexpensive, which makes functional near-infrared spectroscopy an interesting candidate for biomarkers.39 Arterial spin labeling is promising as a way of measuring stable blood flow during rest or tonic pain states.135 Arterial spin labeling scans have been used to differentiate between presurgical and postsurgical states.100 Diffusion tensor imaging studies have been used to classify healthy controls and patients with trigeminal neuralgia160,161 and predict treatment responders.59 Finally, a study using decoding in magnetoencephalography was able to predict high and low pain scores in subjects with primary dysmenorrhea.70 Future studies will reveal more of the possibilities of these neuroimaging methods.
4. Discussion
In this review, we described a variety of pain-predictive models using fMRI, rs-fMRI, sMRI, and EEG, complementing other more restricted reviews.9,116 Although many of these models show great promise, further steps need to be taken to improve biomarkers. High-accuracy models must be tested across research groups with prospective hold-out samples. Cross-validation is only a partial solution because it is still possible to inadvertently overfit models and capitalize on chance.143 Overfitting is a substantial problem in decoding models. There are many possible steps and manipulations in the analysis pipeline, which could result in p-hacking and overfitting. Some of the discussed results might also be guilty of this. There are very few tests of specificity or attempts to train models with high specificity and generalizability. Developing prognostic and predictive biomarkers in particular will also require larger samples.
Increasing sample size and testing sensitivity and specificity across disorders will be greatly facilitated by data-sharing initiatives, including the Pain and Interoception Imaging Network (PAIN) repository,73 OpenPain (principal investigator: A. Vania Apkarian), UK Biobank,91 and OpenfMRI.107 Open data platforms will also aid in the problem of overfitting, making reproducibility, validation, and generalization easier to investigate. In addition, it is important to share models, so that their performance can be evaluated across contexts and samples.
Recently, researchers have identified 4 depression biotypes based on patterns of fMRI connectivity. Two of these responded more favorably to a brain stimulation treatment.33 However, whether this will hold up on validation, and whether other studies can use the same training methods,30 remains to be seen. Clearly, there is a need for continued, multistudy validation of established biomarkers across laboratories.
Ultimately, cooperation and competition initiatives may be necessary for replication and validation of biomarkers in new data sets.120 This could be performed by aggregating data across sites (as in the MAPP consortium71 and Placebo Imaging Consortium162) or through competitions that hold test data “in escrow” to prevent groups from overfitting the training data (eg, Kaggle competitions).121
Furthermore, it is important to actively assess the convergent validity of biomarkers. Models are often not directly comparable, and it is unclear how results and models from different studies fit together, and how they form a coherent, cumulative understanding. The gap between animal and human studies is large (and growing), and models should increasingly use results and concepts from animal neuroscience to constrain and corroborate human predictive models.140
The field will likely develop many more biomarkers the coming years. It would be helpful to evaluate these new articles and recommend state-of-the-art studies. Less optimistic views of the current developments should also be considered.94 Important points to evaluate in new studies could include (1) sample size; (2) use of validated or standardized methodology; (3) adequate analysis and correction for potential movement and clinical confounds; (4) transparent and shareable models; (5) neuroscientific explanation and external validation; (6) independent cohort(s) for validation and/or generalization; and (7) data/tool open availability at the time of publication, among others. Attention to these criteria will help the field to curate and promote state-of-the-art approaches and move the field towards biomarkers useful both in understanding the neural bases of pain and in translational applications.
Disclosures
T.D. Wager is on the Scientific Advisory Board of Curable Health, Inc, has grants from the National Institute of Mental Health (NIMH) and National Institute on Drug Abuse (NIDA), has consulted for GSK, performed contract work for PainQX, and is collaborating with WaviMed and Cliexa. M.A. Lindquist has grants from the National Institute of Health and has consulted for CHDI. M.M. van der Miesen has no conflicts of interest.
This research was supported by grants R01 MH076136, R01 DA035484, and R01 DA046064 (T.D.W.), and R01 EB026549 (M.A.L. and T.D.W.).
Acknowledgements
The authors thank Glenn van der Lande and Guido van Wingen for valuable discussions and comments on earlier drafts.
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
References
- [1].Apkarian AV, Bushnell MC, Treede RD, Zubieta JK. Human brain mechanisms of pain perception and regulation in health and disease. Eur J Pain 2005;9:463–84. [DOI] [PubMed] [Google Scholar]
- [2].Apkarian AV, Sosa Y, Krauss BR, Thomas PS, Fredrickson BE, Levy RE, Harden RN, Chialvo DR. Chronic pain patients are impaired on an emotional decision-making task. PAIN 2004;108:129–36. [DOI] [PubMed] [Google Scholar]
- [3].Baber Z, Erdek MA. Failed back surgery syndrome: current perspectives. J Pain Res 2016:979–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bagarinao E, Johnson KA, Martucci KT, Ichesco E, Farmer MA, Labus J, Ness TJ, Harris R, Deutsch G, Apkarian AV, Mayer EA, Clauw DJ, Mackey S. Preliminary structural MRI based brain classification of chronic pelvic pain: a MAPP network study. PAIN 2014;155:2502–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bai Y, Huang G, Tu Y, Tan A, Hung YS, Zhang Z. Normalization of pain-evoked neural responses using spontaneous EEG improves the performance of EEG-based cross-individual pain prediction. Front Comput Neurosci 2016;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Baliki MN, Geha PY, Fields HL, Apkarian AV. Predicting value of pain and analgesia: nucleus accumbens response to noxious stimuli changes in the presence of chronic pain. Neuron 2010;66:149–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Baliki MN, Petre B, Torbey S, Herrmann KM, Huang L, Schnitzer TJ, Fields HL, Apkarian AV. Corticostriatal functional connectivity predicts transition to chronic back pain. Nat Neurosci 2012;15:1117–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Becker S, Gandhi W, Pomares F, Wager TD, Schweinhardt P. Orbitofrontal cortex mediates pain inhibition by monetary reward. Soc Cogn Affect Neurosci 2017;12:651–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Boissoneault J, Sevel L, Letzen J, Robinson M, Staud R. Biomarkers for musculoskeletal pain conditions: use of brain imaging and machine learning. Curr Rheumatol Rep 2017;19:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bosma RL, Cheng JC, Rogachov A, Kim JA, Hemington KS, Osborne NR, Raghavan LV, Bhatia A, Davis KD. Brain dynamics and temporal summation of pain predicts neuropathic pain relief from ketamine infusion. Anesthesiology 2018;129:1015–24. [DOI] [PubMed] [Google Scholar]
- [11].Bräscher AK, Becker S, Hoeppli ME, Schweinhardt P. Different brain circuitries mediating controllable and uncontrollable pain. J Neurosci 2016;36:5013–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Brodersen KH, Wiech K, Lomakina EI, Lin CS, Buhmann JM, Bingel U, Ploner M, Stephan KE, Tracey I. Decoding the perception of pain from fMRI using multivariate pattern analysis. Neuroimage 2012;63:1162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Brown JE, Chatterjee N, Younger J, Mackey S. Towards a physiology-based measure of pain: patterns of human brain activity distinguish painful from non-painful thermal stimulation. PLoS One 2011;6:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bushnell MC, Čeko M, Low LA. Cognitive and emotional control of pain and its disruption in chronic pain. Nat Rev Neurosci 2013;14:502–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Callan D, Mills L, Nott C, England R, England S. A tool for classifying individuals with chronic back pain: using multivariate pattern analysis with functional magnetic resonance imaging data. PLoS One 2014;9:e98007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Carlino E, Frisaldi E, Benedetti F. Pain and the context. Nat Rev Rheumatol 2014;10:348–55. [DOI] [PubMed] [Google Scholar]
- [17].Carragee EJ, Alamin TF, Carragee JM. Low-pressure positive discography in subjects asymptomatic of significant low back pain illness. Spine (Phila Pa 1976) 2006;31:505–9. [DOI] [PubMed] [Google Scholar]
- [18].Carragee EJ, Alamin TF, Miller JL, Carragee JM. Discographic, MRI and psychosocial determinants of low back pain disability and remission: a prospective study in subjects with benign persistent back pain. Spine J 2005;5:24–35. [DOI] [PubMed] [Google Scholar]
- [19].Carrasquillo Y, Gereau RW. Activation of the extracellular signal-regulated kinase in the amygdala modulates pain perception. J Neurosci 2007;27:1543–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cecchi GA, Huang L, Hashmi JA, Baliki M, Centeno MV, Rish I, Apkarian AV. Predictive dynamics of human pain perception. PLoS Comput Biol 2012;8:e1002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chang LJ, Gianaros PJ, Manuck SB, Krishnan A. A sensitive and specific neural signature for picture-induced negative affect. PLoS Biol 2015;13:e1002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen T, Mu J, Xue Q, Yang L, Dun W, Zhang M, Liu J. Whole-brain structural magnetic resonance imaging—based classification of primary dysmenorrhea in pain-free phase: a machine learning study. PAIN 2019;160:734–41. [DOI] [PubMed] [Google Scholar]
- [23].Cheng JC, Rogachov A, Hemington KS, Kucyi A, Bosma RL, Lindquist MA, Inman RD, Davis KD. Multivariate machine learning distinguishes cross-network dynamic functional connectivity patterns in state and trait neuropathic pain. PAIN 2018;158:1764–76. [DOI] [PubMed] [Google Scholar]
- [24].Chong CD, Gaw N, Fu Y, Li J, Wu T, Schwedt TJ. Migraine classification using magnetic resonance imaging resting-state functional connectivity data. Cephalalgia 2017;37:828–44. [DOI] [PubMed] [Google Scholar]
- [25].Chou R, Atlas SJ, Stanos SP, Rosenquist RW. Nonsurgical interventional therapies for low back pain: a review of the evidence for an American pain society clinical practice guideline. Spine (Phila Pa 1976) 2009;34:1094–1109. [DOI] [PubMed] [Google Scholar]
- [26].Chyzhyk D, Varoquaux G, Thirion B, Milham M. Controlling a confound in predictive models with a test set minimizing its effect. International Workshop on Pattern Recognition in Neuroimaging, PRNI 2018;2018:1–4.
- [27].Corder G, Ahanonu B, Grewe BF, Wang D, Schnitzer MJ, Scherrer G. An amygdalar neural ensemble that encodes the unpleasantness of pain. Science 2019;363:276–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Davis KD, Flor H, Greely HT, Iannetti GD, MacKey S, Ploner M, Pustilnik A, Tracey I, Treede RD, Wager TD. Brain imaging tests for chronic pain: medical, legal and ethical issues and recommendations. Nat Rev Neurol 2017;13:624–38. [DOI] [PubMed] [Google Scholar]
- [29].Denk F, McMahon SB, Tracey I. Pain vulnerability: a neurobiological perspective. Nat Neurosci 2014;17:192–200. [DOI] [PubMed] [Google Scholar]
- [30].Dinga R, Schmaal L, Penninx B, van Tol MJ, Veltman DJ, van Velzen L, van Der Wee N, Marquand A. Evaluating the evidence for biotypes of depression: attempted replication of Drysdale et al. NeuroImage Clin 2019;22:101796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Dosenbach NUF, Nardos B, Cohen AL, Fair DA, Power JD, Church JA, Nelson SM, Wig GS, Vogel AC, Lessov-Schlaggar CN, Barnes KA, Dubis JW, Feczko E, Coalson RS, Pruett JR, Jr, Barch DM, Petersen SE, Schlaggar BL. Prediction of individual brain maturity using fMRI. Science 2010;329:1358–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Downie AS, Hancock MJ, Rzewuska M, Williams CM, Lin CWC, Maher CG. Trajectories of acute low back pain: a latent class growth analysis. PAIN 2015;157:225–34. [DOI] [PubMed] [Google Scholar]
- [33].Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, Fetcho RN, Zebley B, Oathes DJ, Etkin A, Schatzberg AF, Sudheimer K, Keller J, Mayberg HS, Gunning FM, Alexopoulos GS, Fox MD, Pascual-Leone A, Voss HU, Casey BJ, Dubin MJ, Liston C. Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med 2017;23:28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Duerden EG, Albanese MC. Localization of pain-related brain activation: a meta-analysis of neuroimaging data. Hum Brain Mapp 2013;34:109–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Dunn KM, Campbell P, Jordan KP. Long-term trajectories of back pain: cohort study with 7-year follow-up. BMJ Open 2013;3:e003838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].EFIC. Declaration on Pain. Available at: https://www.europeanpainfederation.eu/about-efic/efic-declaration-on-pain/ n.d.
- [37].Eisenberger NI. Social pain and the brain: controversies, questions, and where to go from here. Annu Rev Psychol 2015;66:601–29. [DOI] [PubMed] [Google Scholar]
- [38].Fayaz A, Croft P, Langford RM, Donaldson LJ, Jones GT. Prevalence of chronic pain in the UK: a systematic review and meta-analysis of population studies. BMJ Open 2016;6:e010364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ferrari M, Quaresima V. A brief review on the history of human functional near-infrared spectroscopy (fNIRS) development and fields of application. Neuroimage 2012;63:921–35. [DOI] [PubMed] [Google Scholar]
- [40].Finco G, Locci E, Mura P, Massa R, Noto A, Musu M, Landoni G, D'Aloja E, De-Giorgio F, Scano P, Evangelista M. Can urine metabolomics be helpful in differentiating neuropathic and nociceptive pain? A proof-of-concept study. PLoS One 2016;11:e0150476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Flor H, Turk DC, Scholz OB. Impact of chronic pain on the spouse: marital, emotional and physical consequences. J Psychosom Res 1987;31:63–71. [DOI] [PubMed] [Google Scholar]
- [42].Freburger JK, Holmes GM, Agans RP, Jackman AM, Darter JD, Wallace AS, Castel LD, Kalsbeek WD, Carey TS. The rising prevalence of chronic low back pain. Arch Intern Med 2009;169:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Friebel U, Eickhoff SB, Lotze M. Coordinate-based meta-analysis of experimentally induced and chronic persistent neuropathic pain. Neuroimage 2011;58:1070–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Furman AJ, Meeker TJ, Rietschel JC, Yoo S, Muthulingam J, Prokhorenko M, Keaser ML, Goodman RN, Mazaheri A, Seminowicz DA. Cerebral peak alpha frequency predicts individual differences in pain sensitivity. Neuroimage 2018;167:203–10. [DOI] [PubMed] [Google Scholar]
- [45].Geuter S, Gamer M, Onat S, Büchel C. Parametric trial-by-trial prediction of pain by easily available physiological measures. PAIN 2014;155:994–1001. [DOI] [PubMed] [Google Scholar]
- [46].Glover GH, Li T, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR Magn Reson Med 2000;44:162–7. [DOI] [PubMed] [Google Scholar]
- [47].Gram M, Erlenwein J, Petzke F, Falla D, Przemeck M, Emons MI, Reuster M, Olesen SS, Drewes AM. Prediction of postoperative opioid analgesia using clinical-experimental parameters and electroencephalography. Eur J Pain 2017;21:264–77. [DOI] [PubMed] [Google Scholar]
- [48].Gram M, Graversen C, Olesen AE, Drewes AM. Machine learning on encephalographic activity may predict opioid analgesia. Eur J Pain 2015;19:1552–61. [DOI] [PubMed] [Google Scholar]
- [49].Graversen C, Olesen SS, Olesen AE, Steimle K, Farina D, Wilder-smith OHG, Bouwense SAW, Van Goor H, Drewes AM. The analgesic effect of pregabalin in patients with chronic pain is reflected by changes in pharmaco-EEG spectral indices. Br J Clin Pharmacol 2011;73:363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Group F-NBW. BEST (biomarkers, endpoints, and other tools) resource. Bethesda: Silver Spring Food Drug Adm (US), Co-published by Natl Institutes Heal (US), 2016. [PubMed] [Google Scholar]
- [51].Group F-NBW. Table of surrogate endpoints that were the basis of drug approval or licensure. 2018. Available at: https://www.fda.gov/drugs/development-resources/table-surrogate-endpoints-were-basis-drug-approval-or-licensure. [Google Scholar]
- [52].Harper DE, Shah Y, Ichesco E, Gerstner GE, Peltier SJ. Multivariate classification of pain-evoked brain activity in temporomandibular disorder. Pain Rep 2016;1:e572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Harte SE, Ichesco E, Hampson JP, Peltier SJ, Schmidt-Wilcke T, Clauw DJ, Harris RE. Pharmacologic attenuation of cross-modal sensory augmentation within the chronic pain insula. PAIN 2016;157:1933–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Haxby JV. Multivariate pattern analysis of fMRI: the early beginnings. Neuroimage 2012;62:852–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Haxby JV, Gobbini MI, Furey ML, Ishai A, Schouten JL, Pietrini P. Distributed and overlapping representations of face and objects in ventral temporal cortex. Science 2001;293:2425–31. [DOI] [PubMed] [Google Scholar]
- [56].Haynes JD, Rees G. Decoding mental states from brain activity in humans. Nat Rev Neurosci 2006;7:523–34. [DOI] [PubMed] [Google Scholar]
- [57].Hebart MN, Baker CI. Deconstructing multivariate decoding for the study of brain function. Neuroimage 2018;180:4–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Huang G, Xiao P, Hung YS, Zhang ZG, Hu L. A novel approach to predict subjective pain perception from single-trial laser-evoked potentials. Neuroimage 2013;81:283–93. [DOI] [PubMed] [Google Scholar]
- [59].Hung PSP, Chen DQ, Davis KD, Zhong J, Hodaie M. Predicting pain relief: use of pre-surgical trigeminal nerve diffusion metrics in trigeminal neuralgia. Neuroimage Clin 2017;15:710–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jepma M, Koban L, van Doorn J, Jones M, Wager TD. Behavioural and neural evidence for self-reinforcing expectancy effects on pain. Nat Hum Behav 2018;2:838–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kamitani Y, Tong F. Decoding the visual and subjective contents of the human brain. Nat Neurosci 2005;8:679–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kay KN, Naselaris T, Prenger RJ, Gallant JL. Identifying natural images from human brain activity. Nature 2008;452:352–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Kongsted A, Kent P, Axen I, Downie AS, Dunn KM. What have we learned from ten years of trajectory research in low back pain? BMC Musculoskelet Disord 2016;17:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kongsted A, Kent P, Hestbaek L, Vach W. Patients with low back pain had distinct clinical course patterns that were typically neither complete recovery nor constant pain. A latent class analysis of longitudinal data. Spine J 2015;15:885–94. [DOI] [PubMed] [Google Scholar]
- [65].Kragel PA, Koban L, Barrett LF, Wager TD. Review representation, pattern information, and brain Signatures : from neurons to neuroimaging. Neuron 2018;99:257–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kriegeskorte N. Pattern-information analysis: from stimulus decoding to computational-model testing. Neuroimage 2011;56:411–21. [DOI] [PubMed] [Google Scholar]
- [67].Krishnan A, Woo CW, Chang LJ, Ruzic L, Gu X, López-Solà M, Jackson PL, Pujo J, Fan J, Wager TD. Somatic and vicarious pain are represented by dissociable multivariate brain patterns. Elife 2016;5:e15166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kross E, Berman MG, Mischel W, Smith EE, Wager TD. Social rejection shares somatosensory representations with physical pain. Proc Natl Acad Sci U S A 2011;108:6270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kuner R, Flor H. Structural plasticity and reorganisation in chronic pain. Nat Rev Neurosci 2016;18:20–30. [DOI] [PubMed] [Google Scholar]
- [70].Kuo PC, Chen YT, Chen YS, Chen LF. Decoding the perception of endogenous pain from resting-state MEG. Neuroimage 2017;144:1–11. [DOI] [PubMed] [Google Scholar]
- [71].Kutch JJ, Labus JS, Harris RE, Martucci KT, Farmer MA, Fenske S, Fling C, Ichesco E, Peltier S, Petre B, Guo W, Hou X, Stephens AJ, Mullins C, Clauw DJ, Mackey SC, Apkarian AV, Landis JR, Mayer EA. Resting-state functional connectivity predicts longitudinal pain symptom change in urologic chronic pelvic pain syndrome: a MAPP network study. PAIN 2017;158:1069–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Labus JS, Van Horn JD, Gupta A, Alaverdyan M, Torgerson C, Ashe-McNalley C, Irimia A, Hong JY, Naliboff B, Tillisch K, Mayer EA. Multivariate morphological brain signatures predict patients with chronic abdominal pain from healthy control subjects. PAIN 2015;156:1545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Labus JS, Naliboff B, Kilpatrick L, Liu C, Ashe-McNalley C, Dos Santos IR, Alaverdyan M, Woodworth D, Gupta A, Ellingson BM, Tillisch K, Mayer EA. Pain and Interoception Imaging Network (PAIN): a multimodal, multisite, brain-imaging repository for chronic somatic and visceral pain disorders. Neuroimage 2017;124:1232–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lancaster J, Mano H, Callan D, Kawato M, Seymour B. Decoding acute pain with combined EEG and physiological data. International IEEE/EMBS Conference Neural Engineering (NER) 2017:521–4.
- [75].Lee J, Mawla I, Kim J, Loggia ML, Ortiz A, Jung C, Chan ST, Gerber J, Schmithorst VJ, Edwards RR, Wasan AD, Berna C, Kong J, Kaptchuk TJ, Gollub RL, Rosen BR, Napadow V. Machine learning-based prediction of clinical pain using multimodal neuroimaging and autonomic metrics. PAIN 2019;160:550–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Lee M, Manders TR, Eberle SE, Su C, D'amour J, Yang R, Lin HY, Deisseroth K, Froemke RC, Wang J. Activation of corticostriatal circuitry relieves chronic neuropathic pain. J Neurosci 2015;35:5247–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Li D, Puntillo K, Miaskowski C. A review of objective pain measures for use with critical care adult patients unable to self-report. J Pain 2008;9:2–10. [DOI] [PubMed] [Google Scholar]
- [78].Li L, Huang G, Lin Q, Liu J, Zhang S, Zhang Z. Magnitude and temporal variability of inter-stimulus EEG modulate the linear relationship between laser-evoked potentials and fast-pain perception. Front Neurosci 2018;12:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Liang M, Mouraux A, Hu L, Iannetti GD. Primary sensory cortices contain distinguishable spatial patterns of activity for each sense. Nat Commun 2013;4:1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Liu P, Qin W, Wang J, Zeng F, Zhou G, Wen H, von Deneen KM, Liang F, Gong Q, Tian J. Identifying neural patterns of functional dyspepsia using multivariate pattern analysis: a resting-state fMRI study. PLoS One 2013;8:e68205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Liu Y, Latremoliere A, Li X, Zhang Z, Chen M, Wang X, Fang C, Zhu J, Alexandre C, Gao Z, Chen B, Ding X, Zhou J, Zhang Y, Chen C, Wang KH, Woolf CJ, He Z. Touch and tactile neuropathic pain sensitivity are set by corticospinal projections. Nature 2018;561:547–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].López-Solà M, Koban L, Wager TD. Transforming pain with prosocial meaning. Psychosom Med 2018;80:814–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].López-Solà M, Pujol J, Wager TD, Garcia-Fontanals A, Blanco-Hinojo L, Garcia-Blanco S, Poca-Dias V, Harrison BJ, Contreras-Rodríguez O, Monfort J, Garcia-Fructuoso F, Deus J. Altered functional magnetic resonance imaging responses to nonpainful sensory stimulation in fibromyalgia patients. Arthritis Rheumatol 2014;66:3200–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].López-Solà M, Woo CW, Pujol J, Deus J, Harrison BJ, Monfort J, Wager TD. Towards a neurophysiological signature for fibromyalgia. PAIN 2017;158:34–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Lötsch J, Ultsch A. Machine learning in pain research. PAIN 2017;159:623–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Makowski C, Lepage M, Evans AC. Head motion: the dirty little secret of neuroimaging in psychiatry. J Psychiatry Neurosci 2018;43:180022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Manchiakanti L, Giordano J, Boswell MV, Fellows B, Manchukonda R, Pampati V. Psychological factors as predictors of opioid abuse and illicit drug use in chronic pain patients. J Opioid Manag 2007;3:89–100. [DOI] [PubMed] [Google Scholar]
- [88].Mano H, Kotecha G, Leibnitz K, Matsubara T, Nakae A, Shenker N, Shibata M, Voon V, Yoshida W, Lee M, Yanagida T, Kawato M, Rosa MJ, Seymour B. Classification and characterisation of brain network changes in chronic back pain: a multicenter study. Wellcome Open Res 2018;3:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Mansour AR, Baliki MN, Huang L, Torbey S, Herrmann KM, Schnitzer TJ, Apkarian AV. Brain white matter structural properties predict transition to chronic pain. PAIN 2013;154:2160–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Marquand A, Howard M, Brammer M, Chu C, Coen S, Mourão-Miranda J. Quantitative prediction of subjective pain intensity from whole-brain fMRI data using Gaussian processes. Neuroimage 2010;49:2178–89. [DOI] [PubMed] [Google Scholar]
- [91].Miller KL, Alfaro-Almagro F, Bangerter NK, Thomas DL, Yacoub E, Xu J, Bartsch AJ, Jbabdi S, Sotiropoulos SN, Andersson JLR, Griffanti L, Douaud G, Okell TW, Weale P, Dragonu I, Garratt S, Hudson S, Collins R, Jenkinson M, Matthews PM, Smith SM. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat Neurosci 2016;19:1523–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Misra G, Wang W, Archer DB, Roy A, Coombes SA. Automated classification of pain perception using high-density electroencephalography data. J Neurophysiol 2017;117:786–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Mitchell TM, Shinkareva SV, Carlson A, Chang K, Malave VL, Mason RA, Just MA. Associated with the meanings of nouns. Science 2008;320:1191–5. [DOI] [PubMed] [Google Scholar]
- [94].Mouraux A, Iannetti GD. The search for pain biomarkers in the human brain. Brain 2018;141:3290–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Murray CJL, Lopez AD. Measuring the global burden of disease. N Engl J Med 2013;369:448–57. [DOI] [PubMed] [Google Scholar]
- [96].Muschelli J, Nebel MB, Caffo BS, Barber AD, Pekar JJ, Mostofsky SH. Reduction of motion-related artifacts in resting state fMRI using aCompCor. Neuroimage 2014;96:22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Nan J, Liu J, Li G, Xiong S, Yan X, Yin Q, Zeng F, von Deneen KM, Liang F, Gong Q, Qin W, Tian J. Whole-brain functional connectivity identification of functional dyspepsia. PLoS One 2013;8:e65870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Naselaris T, Kay KN, Nishimoto S, Gallant JL. Encoding and decoding in fMRI. Neuroimage 2011;56:400–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Neugebauer V, Li W, Bird GC, Han JS. The amygdala and persistent pain. Neuroscientist 2004;10:221–34. [DOI] [PubMed] [Google Scholar]
- [100].O'Muircheartaigh J, Marquand A, Hodkinson DJ, Krause K, Khawaja N, Renton TF, Huggins JP, Vennart W, Williams SCR, Howard MA. Multivariate decoding of cerebral blood flow measures in a clinical model of on-going postsurgical pain. Hum Brain Mapp 2015;36:633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Parker SL, Mendenhall SK, Godil SS, Sivasubramanian P, Cahill K, Ziewacz J, McGirt MJ. Incidence of low back pain after lumbar discectomy for herniated disc and its effect on patient-reported outcomes. Clin Orthop Relat Res 2015;473:1988–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Parkes L, Fulcher B, Yücel M, Fornito A. An evaluation of the efficacy, reliability, and sensitivity of motion correction strategies for resting-state functional MRI. Neuroimage 2018;171:415–36. [DOI] [PubMed] [Google Scholar]
- [103].Pasley BN, David SV, Mesgarani N, Flinker A, Shamma SA, Crone NE, Knight RT, Chang EF. Reconstructing speech from human auditory cortex. PLoS Biol 2012;10:e1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Pineda R, Neil J, Dierker D, Smyser C, Wallendorf M, Kidokoro H, Reynolds L, Rogers C, Mathur A, Van Essen D, Inder T. Alterations in brain structure and neurodevelopmental outcome in preterm infants hospitalized in different neonatal intensive care unit environments. J Pediatr 2015;164:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Ploner M, May ES. EEG and MEG in pain research—current state and future perspectives. PAIN 2018;159:206–211. [DOI] [PubMed] [Google Scholar]
- [106].Poldrack RA, Baker CI, Durnez J, Gorgolewski KJ, Matthews PM, Munafò MR, Nichols TE, Poline JB, Vul E, Yarkoni T. Scanning the horizon: towards transparent and reproducible neuroimaging research. Nat Rev Neurosci 2017;18:115–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Poldrack RA, Barch DM, Mitchell JP, Wager TD, Wagner AD, Devlin JT, Cumba C, Koyejo O, Milham MP. Toward open sharing of task-based fMRI data: the OpenfMRI project. Front Neuroinform 2013;7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Pourshoghi A, Zakeri I, Pourrezaei K. Application of functional data analysis in classification and clustering of functional near-infrared spectroscopy signal in response to noxious stimuli. J Biomed Opt 2016;21:101411. [DOI] [PubMed] [Google Scholar]
- [109].Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage 2014;84:320–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Prato M, Favilla S, Zanni L, Porro CA, Baraldi P. A regularization algorithm for decoding perceptual temporal profiles from fMRI data. Neuroimage 2011;56:258–67. [DOI] [PubMed] [Google Scholar]
- [111].Rao A, Monteiro JM, Mourao-Miranda J. Predictive modelling using neuroimaging data in the presence of confounds. Neuroimage 2017;150:23–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Ren W, Centeno MV, Berger S, Wu Y, Na X, Liu X, Kondapalli J, Apkarian AV, Martina M, Surmeier DJ. The indirect pathway of the nucleus accumbens shell amplifies neuropathic pain. Nat Neurosci 2016;19:220–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Robinson ME, O'Shea AM, Craggs J, Price DD, Letzen JE, Staud R. Comparison of machine classification algorithms for fibromyalgia: neuroimages versus self-report. J Pain 2015;16:472–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Rogachov A, Cheng JC, Hemington KS, Bosma RL, Kim J, Osborne NR, Inman RD, Davis KD. Abnormal low-frequency oscillations reflect trait-like pain ratings in chronic pain patients revealed through a machine learning approach. J Neurosci 2018;38:7293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Rojas RF, Huang X, Ou K. Toward a functional near-infrared spectroscopy-based monitoring of pain assessment for nonverbal patients. J Biomed Opt 2017;22:106013. [DOI] [PubMed] [Google Scholar]
- [116].Rosa MJ, Seymour B. Decoding the matrix: benefits and limitations of applying machine learning algorithms to pain neuroimaging. PAIN 2014;155:864–7. [DOI] [PubMed] [Google Scholar]
- [117].Rosenberg MD, Finn ES, Scheinost D, Papademetris X, Shen X, Constable RT, Chun MM. A neuromarker of sustained attention from whole-brain functional connectivity. Nat Neurosci 2015;19:165–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Satterthwaite TD, Elliott MA, Gerraty RT, Ruparel K, Loughead J, Calkins ME, Eickhoff SB, Hakonarson H, Gur RE, Wolf DH, Gur RC. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage 2013;64:240–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Schnakers C, Zasler ND. Pain assessment and management in disorders of consciousness. Curr Opin Neurol 2007;20:620–6. [DOI] [PubMed] [Google Scholar]
- [120].Schuller B, Batliner A, Steidl S, Seppi D. Recognising realistic emotions and affect in speech: state of the art and lessons learnt from the first challenge. Speech Commun 2011;53:1062–87. [Google Scholar]
- [121].Schuller B, Steidl S, Batliner A. The INTERSPEECH 2009 emotion challenge. Proceedings Annual Conference of the International Speech Communication Association, INTERSPEECH 2009:312–15.
- [122].Schulz E, Zherdin A, Tiemann L, Plant C, Ploner M. Decoding an individual's sensitivity to pain from the multivariate analysis of EEG data. Cereb Cortex 2012;22:1118–23. [DOI] [PubMed] [Google Scholar]
- [123].Schwartz N, Temkin P, Jurado S, Lim BK, Heifets BD, Polepalli JS, Malenka RC. Decreased motivation during chronic pain requires long-term depression in the nucleus accumbens. Science 2014;345:535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Schwedt TJ, Chong CD, Wu T, Gaw N, Fu Y, Li J. Accurate classification of chronic migraine via brain magnetic resonance imaging. Headache 2015;55:762–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Seminowicz DA, Laferriere AL, Millecamps M, Yu JSC, Coderre TJ, Bushnell MC. MRI structural brain changes associated with sensory and emotional function in a rat model of long-term neuropathic pain. Neuroimage 2009;47:1007–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Skolasky RL, Wegener ST, Maggard AM, Riley LH. The impact of reduction of pain after lumbar spine surgery: the relationship between changes in pain and physical function and disability. Spine (Phila Pa 1976) 2014;39:1426–32. [DOI] [PubMed] [Google Scholar]
- [127].Smith A, López-Solà M, McMahon K, Pedler A, Sterling M. Multivariate pattern analysis utilizing structural or functional MRI—in individuals with musculoskeletal pain and healthy controls: a systematic review. Semin Arthritis Rheum 2017;47:418–31. [DOI] [PubMed] [Google Scholar]
- [128].Snoek L, Miletić S, Scholte HS. How to control for confounds in decoding analyses of neuroimaging data. Neuroimage 2019;184:741–60. [DOI] [PubMed] [Google Scholar]
- [129].Sokil MB, Lyashuk OL, Dovbush AP. ICA-AROMA: a robust ICA-based strategy for removing motion artifacts from fMRI data. INMATEH Agric Eng 2016;48:119–24. [Google Scholar]
- [130].Sundermann B, Burgmer M, Pogatzki-Zahn E, Gaubitz M, Stüber C, Wessolleck E, Heuft G, Pfleiderer B. Diagnostic classification based on functional connectivity in chronic pain: model optimization in fibromyalgia and rheumatoid arthritis. Acad Radiol 2014;21:369–77. [DOI] [PubMed] [Google Scholar]
- [131].Tan LL, Pelzer P, Heinl C, Tang W, Gangadharan V, Flor H, Sprengel R, Kuner T, Kuner R. A pathway from midcingulate cortex to posterior insula gates nociceptive hypersensitivity. Nat Neurosci 2017;20:1591–601. [DOI] [PubMed] [Google Scholar]
- [132].Tétreault P, Mansour A, Vachon-Presseau E, Schnitzer TJ, Apkarian AV, Baliki MN. Brain connectivity predicts placebo response across chronic pain clinical trials. PLoS Biol 2016;14:e1002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Todd MT, Nystrom LE, Cohen JD. Confounds in multivariate pattern analysis: theory and rule representation case study. Neuroimage 2013;77:157–65. [DOI] [PubMed] [Google Scholar]
- [134].Tracey I, Bushnell MC. How neuroimaging studies have challenged us to rethink: is chronic pain a disease? J Pain 2009;10:1113–20. [DOI] [PubMed] [Google Scholar]
- [135].Tracey I, Johns E. The pain matrix: reloaded or reborn as we image tonic pain using arterial spin labelling. PAIN 2010;148:359–60. [DOI] [PubMed] [Google Scholar]
- [136].Tracey I, Mantyh PW. The cerebral signature for pain perception and its modulation. Neuron 2007;55:377–91. [DOI] [PubMed] [Google Scholar]
- [137].Tu Y, Tan A, Bai Y, Hung YS, Zhang Z. Decoding subjective intensity of nociceptive pain from pre-stimulus and post-stimulus brain activities. Front Comput Neurosci 2016;10:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Tu YH, Fu ZN, Tan A, Huang G, Hu L, Hung YS, Zhang ZG. A novel and effective fMRI decoding approach based on sliced inverse regression and its application to pain prediction. Neurocomputing 2018;273:373–84. [Google Scholar]
- [139].Ung H, Brown JE, Johnson KA, Younger J, Hush J, Mackey S. Multivariate classification of structural MRI data detects chronic low back pain. Cereb Cortex 2014;24:1037–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Upadhyay J, Geber C, Hargreaves R, Birklein F, Borsook D. A critical evaluation of validity and utility of translational imaging in pain and analgesia: utilizing functional imaging to enhance the process. Neurosci Biobehav Rev 2018;84:407–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Vachon-Presseau E, Berger SE, Abdullah TB, Huang L, Cecchi GA, Griffith JW, Schnitzer TJ, Apkarian AV. Brain and psychological determinants of placebo pill response in chronic pain patients. Nat Commun 2018;9:3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Vanneste S, Song JJ, De Ridder D. Thalamocortical dysrhythmia detected by machine learning. Nat Commun 2018;9:1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Varoquaux G. Cross-validation failure: small sample sizes lead to large error bars. Neuroimage 2018;180:68–77. [DOI] [PubMed] [Google Scholar]
- [144].Vijayakumar V, Case M, Shirinpour S, He B. Quantifying and characterizing tonic thermal pain across subjects from EEG data using random forest models. IEEE Trans Biomed Eng 2017;64:2988–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Vuckovic A, Jose V, Gallardo F, Jarjees M, Fraser M, Purcell M. Prediction of central neuropathic pain in spinal cord injury based on EEG classifier. Clin Neurophysiol 2018;129:1605–17. [DOI] [PubMed] [Google Scholar]
- [146].Wager TD, Atlas LY, Leotti LA, Rilling JK. Predicting individual differences in placebo analgesia: contributions of brain activity during anticipation and pain experience. J Neurosci 2011;31:439–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Wager TD, Atlas LY, Lindquist MA, Roy M, Woo CW, Kross E. An fMRI-based neurologic signature of physical pain. N Engl J Med 2013;368:1388–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Walker SM, Beggs S, Baccei ML. Persistent changes in peripheral and spinal nociceptive processing after early tissue injury. Exp Neurol 2016;275:253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Wang X, Baeken C, Fang M, Qiu J, Chen H, Wu GR. Predicting trait-like individual differences in fear of pain in the healthy state using gray matter volume. Brain Imaging Behav 2018:1–6. [DOI] [PubMed] [Google Scholar]
- [150].Wiech K, Ploner M, Tracey I. Neurocognitive aspects of pain perception. Trends Cogn Sci 2008;12:306–13. [DOI] [PubMed] [Google Scholar]
- [151].Woo CW, Wager TD. Neuroimaging-based biomarker discovery and validation. PAIN 2015;156:1379–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Woo CW, Wager TD. What reliability can and cannot tell us about pain report and pain neuroimaging. PAIN 2016;157:511–13. [DOI] [PubMed] [Google Scholar]
- [153].Woo CW, Chang LJ, Lindquist MA, Wager TD. Building better biomarkers: brain models in translational neuroimaging. Nat Neurosci 2017;20:365–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Woo CW, Koban L, Kross E, Lindquist MA, Banich MT, Ruzic L, Andrews-Hanna JR, Wager TD. Separate neural representations for physical pain and social rejection. Nat Commun 2014;5:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Woo CW, Roy M, Buhle JT, Wager TD. Distinct brain systems mediate the effects of nociceptive input and self-regulation on pain. PLoS Biol 2015;13:e1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Woo CW, Schmidt L, Krishnan A, Jepma M, Roy M, Lindquist MA, Atlas LY, Wager TD. Quantifying cerebral contributions to pain beyond nociception. Nat Commun 2017;8:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Younger J, McCue R, Mackey S. Pain outcomes: a brief review of instruments and techniques. Curr Pain Headache Rep 2009;13:39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Zang Y, Jiang T, Lu Y, He Y, Tian L. Regional homogeneity approach to fMRI data analysis. Neuroimage 2004;22:394–400. [DOI] [PubMed] [Google Scholar]
- [159].Zhang Q, Wu Q, Zhang J, He L, Huang J, Zhang J, Huang H, Gong Q. Discriminative analysis of migraine without aura: using functional and structural MRI with a multi-feature classification approach. PLoS One 2016;11:e0163875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Zhang Y, Mao Z, Cui Z, Ling Z, Pan L, Liu X, Zhang J, Yu X. Diffusion tensor imaging of axonal and myelin changes in classical trigeminal neuralgia. World Neurosurg 2018;112:e597–607. [DOI] [PubMed] [Google Scholar]
- [161].Zhong J, Chen DQ, Hung PS, Hayes DJ, Liang KE, Davis KD, Hodaie M. Multivariate pattern classification of brain white matter connectivity predicts classic trigmenial neuralgia. PAIN 2018;159:2076–87. [DOI] [PubMed] [Google Scholar]
- [162].Zunhammer M, Bingel U, Wager TD. Placebo effects on the neurologic pain signature: a meta-analysis of individual participant functional magnetic resonance imaging data. JAMA Neurol 2018;75:1321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]