Abstract:
One of the key ambitions of neuroimaging-based pain biomarker research is to augment patient and clinician reporting of clinically relevant phenomena with neural measures for prediction, prognosis, and detection of pain. Despite years of productive research on the neuroimaging of pain, such applications have seen little advancement. However, recent developments in identifying brain-based biomarkers of pain through advances in technology and multivariate pattern analysis provide some optimism. Here, we (1) define and review the different types of potential neuroimaging-based biomarkers, their clinical and research applications, and their limitations and (2) describe frameworks for evaluation of pain biomarkers used in other fields (eg, genetics, cancer, cardiovascular disease, immune system disorders, and rare diseases) to achieve broad clinical and research utility and minimize the risks of misapplication of this emerging technology. To conclude, we discuss future directions for neuroimaging-based biomarker research to achieve the goal of personalized pain medicine.
Keywords: Neuroimaging, MRI, Classification, Prognosis, Prediction, Biomarkers, Chronic pain, Signature, Pattern, Multivariate
1. Introduction
Magnetic resonance imaging (MRI) has opened a window to the brain by allowing noninvasive study of both structure and function. Neuroimaging has elucidated how pain processing is linked within the central nervous system (CNS); how it is disrupted in chronic pain; and how those disruptions occur with the chronification of pain.18,20,62,63,67,75,108 However, to date, functional MRI (fMRI) has provided minimal direct clinical application for pain.
Identifying and validating neuroimaging-based biomarkers and surrogate endpoints for pain would be useful for clinical and research communities in several ways: (1) prognosis (ie, for indicating the likely progression of pain after injury or surgery,5,74 or the progression from chronic pain to high-impact chronic pain57,58,102), (2) identifying likely patient responders to a particular treatment (ie, prediction), (3) identifying a specific pain disorder (ie, diagnosis), (4) identifying targets for therapeutic intervention, and (5) defining surrogate endpoints to augment clinical endpoints and predict clinical benefit (N.B. Food and Drug Administration [FDA] validation of surrogate endpoints requires a different validation process than biomarkers, but are listed here in the context of biomarkers). Valid neuroimaging-based biomarkers of pain would also be useful in providing evidence in the legal system. The benefits and limitations of machine learning and classification techniques that can assess neuroimaging data for meaningful patterns of neural structure and function have been reviewed elsewhere.86 Preliminary brain biomarkers have been identified in individuals experiencing acute and chronic pain.3,11,12,16,50,51,56,61,103,110,111,114 Potential future applications of this technology are exciting; however, the field of brain biomarkers of pain is still in its early phases. As such, we should not prematurely apply neuroimaging biomarkers of pain generally for clinical or legal purposes, until proper validation is performed. Although the task of formal validation is large, we can learn from other fields how to systematically approach the validation of neuroimaging pain biomarkers. Finally, as the field evolves, it is important to consider ethical, social, and legal implications of future validated biomarkers and how or whether they should be used, as previously reviewed.19,21,35
Our goals of this article are to (1) review the different types of potential neuroimaging-based biomarkers, their clinical and research applications, and limitations and (2) describe frameworks used in other fields (eg, genetics, cancer, cardiovascular disease, immune system disorders, and rare diseases) for the future validation of pain biomarkers to achieve broad clinical and research utility and minimize the risks of misapplication of this emerging technology. Although we focus predominantly on MRI-based pain biomarkers, there are a variety of imaging methods to characterize structure (eg, quantitative morphometry, white matter “connectivity,” gray matter ultrastructure, and cytoarchitectonic mapping) and function (eg, physiology [cerebral blood flow], metabolism, receptor distribution, gene and protein expression, electrophysiology, and functional connectivity). Of note, the purpose of this article is not to provide a comprehensive review of brain imaging in clinical and experimental pain or of brain-based biomarkers for pain. For those purposes, there are several recent comprehensive reviews.19,63,64,72,73,98,109,112
2. The BEST way forward in developing neuroimaging biomarkers for pain: definitions, applications, and utility
Pain is a complex physiological and psychological experience,70 making it both subjective and inherently difficult to study and treat. Individual variability in pain perception poses additional challenges to assessing and treating pain.17,29,76 The most common means to determine whether an individual has pain in both research and clinical settings is subjective reporting. For example, rating pain on a scale from 0 to 10 is an extensively validated and useful measure.79 However, in some clinical situations, individuals are unable to report their pain, such as in very young, elderly, infirmed, and unconscious patients. In these cases, objective biomarkers of pain could be helpful.
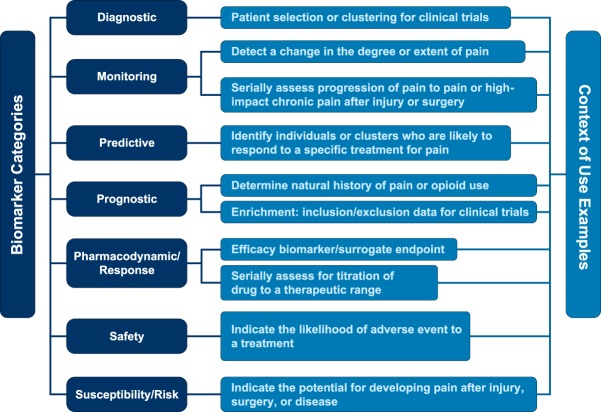
It is of importance to first clearly and precisely define the types of biomarkers to assure effective and unambiguous communication. The 2016 FDA-NIH Biomarker Working Group glossary, BEST (Biomarkers, EndpointS, and other Tools Resource),7 defines a biomarker as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention.” Thus, a biomarker is not an assessment of how an individual feels, functions, or survives—the characteristics of which more appropriately defines a clinical endpoint. The BEST glossary also provides a framework for conceptualizing the multiple types and uses of biomarkers. We draw heavily from the BEST Working Group and provide several of these categories of biomarkers and their potential application to clinical, research, and legal aspects of pain below. Although we focus on neuroimaging biomarkers of pain in this review, the concepts apply to other potential biomarkers of pain (eg, molecular, histologic, radiographic, or physiologic). Figure 1 summarizes the biomarker definitions.
Figure 1.
Biomarker Definitions with Context of Use Examples. Adapted from the 2016 FDA-NIH Biomarker Working Group glossary, BEST (Biomarkers, EndpointS, and other Tools Resource)1. FDA, Food and Drug Administration.
2.1. Diagnostic biomarkers
This type of biomarker detects or confirms the presence of a condition or identifies individuals within a specific subtype of a condition.7 Pertinent to neuroimaging-based pain biomarkers, the research field has primarily focused on developing diagnostic biomarkers. Our group's initial effort was to address a simple question of whether a pattern or signature of brain activity identified in a training set could be used to accurately determine whether individuals (not part of the training set) were experiencing pain to a thermal stimulus.12 For this goal, we trained a linear support vector machine learning algorithm to distinguish painful from nonpainful stimuli with more than 80% classification accuracy. Subsequently, Wager et al.103 demonstrated the ability of using a trained brain signature to distinguish between brain states experiencing painful heat and nonpainful warmth; pain anticipation and pain recall; physical and social pain; physical pain and the empathy of pain49; and the presence or absence of pain self-regulation.110 Other groups have extended these general concepts and methodology to distinguish the presence or absence of various chronic pain states, including but not limited to chronic low back pain,100 fibromyalgia,56 irritable bowel syndrome,53 pelvic pain,3,50 and trigeminal neuralgia114; further details can be found in the following reviews.73,109
Along with a rapid growth in the number of research studies focused on neuroimaging-based pain detection, the field has sparked much controversy. Researchers have debated whether chronic pain is a disease of the brain,95 whether brain-derived diagnostics (ie, identifiers of disease) of chronic pain are plausible or would be beneficial,84 and whether neuroimaging surrogate measures of pain are useful.59,85 Some argue that “brain imaging adds neuroanatomical and neurophysiological information, not validity, to pain reports”95 and question whether “these biomarkers “mark” the pain itself or just the neural causes and correlates of pain?”85 One review95 argues that neuroimaging findings merely reflect one part of a chronic pain condition, which affects the person as a whole (across many body systems and functions, both physiologically and psychologically). Another concern for the use of neuroimaging-based pain biomarkers is the concept of reverse inference in that brain biomarkers may not take into account how selectively an area is activated by the mental process in question (eg, pain).77 Based on these valid concerns, neuroimaging findings should not and cannot be expected to represent the sole source, cause, or experience of pain. Furthermore, we should be cautious in generalizing findings until there are appropriately well-controlled replication studies.
In addition, researchers, clinicians, and ethicists have raised the issue of potentially detrimental outcomes from false negatives leading people truly suffering from pain to be unjustifiably denied treatment or compensation. Similarly, a false positive could subject a person to unnecessary and risky treatments. There is also the concern of false-negative findings compromising doctor–patient, employee–patient, or family–patient trust.21
These concerns are understandable. However, the goal of neuroimaging biomarkers of pain should not be to replace patient self-reporting, but rather to supplement the information from self-reporting. In other words, we do not normally need an MRI to tell us a patient is in pain when we can just ask them. Although the initial studies of diagnostic brain biomarkers of pain were conceptually simple (but methodologically complex), they were necessary to establish the research infrastructure and methods upon which this technology could be expanded to provide true clinical and research utility. We provide some examples of the utility of diagnostic pain biomarkers below.
The practice of pain medicine requires accurate diagnosis of specific pain conditions. Diagnostic biomarkers can help clinicians determine whether a patient has a particular medical condition for which a treatment may be indicated. For example, imagine a 44-year-old woman who falls and injures her wrist. She subsequently develops burning pain in her hand and wrist with swelling and color changes. This presents a clinical challenge to distinguish whether such a presentation is complex regional pain syndrome (CRPS)—a terribly disabling neuropathic pain condition—or simply a delay in healing after injury, as injury itself leads to similar signs and symptoms as CRPS. A brain biomarker to detect the presence or absence of CRPS could be clinically useful to target appropriate treatment in those with CRPS and avoid overtreatment in those without.
As is becoming increasingly appreciated, many chronic pain conditions have subtypes or clusters with markedly different prognoses or responses to a specific treatment. Researchers can use diagnostic biomarkers in clinical trials evaluating chronic pain subtypes to select patients more likely to respond to a specific treatment (ie, to ultimately serve as a predictive biomarker). Finally, the brain regions and networks identified by these biomarkers can serve as therapeutic targets for pharmaceuticals, mind–body interventions, transcranial magnetic stimulation, or deep brain stimulation. For example, Kutch et al. pooled data (n = 1079) across 7 academic sites as part of the NIH Multidisciplinary Approach to the Study of Chronic Pelvic Pain Research Network54 to compare those with urological chronic pelvic pain syndrome (UCPPS) with pain-free controls and individuals with fibromyalgia. A subset of individuals (n = 182) underwent fMRI and structural MRI. Individuals with UCPPS reported pain ranging from localized (pelvic) to widespread (throughout the body). The authors found that individuals with widespread UCPPS had increased brain gray matter volume and functional connectivity involving sensorimotor and insular cortices.50 These results indicate that individuals with localized UCPPS may represent a different phenotype than those with more widespread pain and may respond differentially to treatment. Development of a brain-based biomarker of pain to better classify subtypes of pain may allow for more efficacious targeting of specific treatments.
For diagnostic brain-based biomarkers to have true clinical and research utility, we first need to assess their performance. A perfect diagnostic biomarker test would detect 100% of all patients with a disease or disease subset (ie, 100% sensitivity for individuals with the disease who test positive) and detect 100% of patients who do not have the disease (ie, 100% specificity for people without the disease who test negative). However, no biomarker test has perfect sensitivity and specificity, thereby requiring tradeoffs among these features. Additional measures of diagnostic biomarker performance include positive predictive value (ie, the proportion of those who tested positive who have the disease or condition) and negative predictive value (ie, the proportion of those who tested negative who do not have the disease or condition).7 The diagnostic biomarker must have appropriate analytical validity (ie, the performance of the detection measure itself). For instance, analytical validity must address: (1) the dynamic range of the detection method (will it pick up a positive signal within the range of variability), (2) the precision of the detection method within the full range of the population of interest, and (3) the accuracy of the detection method. This can be a complex problem for imaging biomarkers and especially for signatures which use an algorithm as the actual detection method. Finally, the biomarker must be tested and validated in large samples of the population to which it is intended to be used to assure generalizability. This emphasizes the importance of incorporating base rates or disease prevalence in assessing the generalized performance of a diagnostic biomarker for broad clinical or research purposes.
The concepts of applying epidemiologic base rates, or disease prevalence, to assess the validity of a diagnostic test in a general population are well-founded applications of Bayes theorem. In fact, over 60 years ago, Meehl and Rosen69 described how base rates impact the diagnostic utility of a test, noting that utility is strongly influenced by the base rate of the diagnosis in the population of interest.91 Classic epidemiologic studies have demonstrated that, contrary to intuition, tests with more than 90% sensitivity and specificity can perform poorly overall when the base rate of the condition is low—a phenomenon known as the “base rate fallacy.”82 This issue was raised in an editorial by Robinson et al.,83 in which the authors critique multiple brain imaging studies' results for not including base rates. However, in this stage of research discovery, the use of base rates is premature and should be saved for studies that assess clinical validity in a general population (as described below in the section on ACCE), rather than be used in carefully controlled laboratory environments. The following study illustrates these points. In Ung et al.,100 we applied machine learning techniques to distinguish individuals with chronic low back pain vs carefully matched healthy controls, and then defined the neural correlates responsible for this distinction. The subjects chosen with low back pain were narrowly screened to have little to no significant emotional distress, no current medications, no radicular symptoms, and no other sites of pain. The advantage of this narrow selection was to reduce confounds and allow for scientific discovery. In other words, the application of epidemiologic base rates would have been inappropriate for the purposes of this and other similar studies seeking to develop and refine methods and techniques. It is important to note that these subjects with low back pain bear little resemblance to real-world patients with chronic low back pain seen in a clinical setting, limiting the generalizability of the results. Fortunately, the field is making rapid advances that will soon allow diagnostic brain-based biomarkers to be tested in the general population. Until then, application of these diagnostic biomarkers for clinical, commercial, or legal applications is premature.19
2.2. Prognostic biomarkers
Prognostic biomarkers can indicate an increased (or decreased) likelihood of a future clinical event, disease recurrence, exacerbation of a painful condition, or progression in patients with pain.7 Neuroimaging-based prognostic biomarkers of pain hold great potential in identifying patients likely to develop persistent pain or opioid use after injury. For example, researchers have investigated the use of resting state functional connectivity after an acute back pain injury to predict persistence of pain.5,42 These authors noted that when pain persisted, brain gray matter density decreased; in addition, greater functional connectivity of the nucleus accumbens with prefrontal cortex predicted pain persistence.5,42
Prognostic biomarkers could also be useful in defining the natural history of a painful condition or disease progression. For example, as part of a multicenter clinical trial to characterize chronic pelvic pain, Kutch et al.51 used resting state functional connectivity to significantly predict short-term (3-month) pain reduction in individuals with chronic pelvic pain with 73.1% accuracy (69.2% sensitivity and 75.0% precision). In addition, prognostic biomarkers could aid in targeted clinical trials by selecting patients more likely to have pain conditions at high risk of exacerbation and therefore thought more likely to respond to a particular treatment.
2.3. Susceptibility/risk biomarker
A susceptibility/risk biomarker is one associated with an increased, or decreased, risk of developing a chronic pain condition in an individual who does not yet have that condition. This contrasts with prognostic biomarkers noted above, which are used to indicate an increased or decreased likelihood of a clinical event in an individual who already has the painful condition.
At the time of this writing, we were unable to identify any published research involving neuroimaging to track healthy people without pain and use brain biomarkers to predict who will develop chronic pain. The NIH is planning on funding large scale multicenter trials to identify such susceptibility/risk biomarkers in musculoskeletal pain and after surgery. These trials will include neuroimaging, and the results should yield valuable biomarkers to identify who is vulnerable to the development of chronic pain. Here, we will use examples from other non-neuroimaging fields to illustrate the utility of such biomarkers in predicting risk of developing chronic pain after surgery, injury, disease, or idiopathically.
We and other researchers have identified preoperative risk factors for the development of persistent pain and opioid use after surgery.14,38,40,41 Recently, Hah et al.39 used a k-means clustering approach in 422 patients undergoing surgery to identify risk predictors of who would develop persistence of pain and opioid use and delayed recovery postoperatively. They identified a possible uniform predictor of disparate surgical outcomes long after hospital discharge. With accurate neuroimaging susceptibility/risk biomarkers, clinical trials could test therapeutic interventions in those most likely to develop persistent pain or opioid use, thereby sparing those at less risk from potentially untoward adverse events from unnecessary intervention.
The Orofacial Pain: Prospective Evaluation and Risk Assessment (OPPERA) study was an NIH-funded, multicenter, cross-disciplinary investigation of the development of temporomandibular disorder (TMD).30 The primary goals of the OPPERA study were to identify putative psychological and physiological risk factors, clinical characteristics, and related genetic mechanisms that influence the development of chronic orofacial pain associated with TMD. In this prospective inception cohort study of 3263 individuals with no current or previous experience of TMD, the investigators collected a comprehensive battery of baseline and follow-up measures to predict those individuals who ultimately developed TMD. In addition to multiple psychosocial factors predicting development of TMD, the authors found 6 single-nucleotide polymorphisms as risk factors for chronic TMD.4,92 Through the Helping to End Addiction Long-term (HEAL) project, the NIH is currently initiating a multicenter trial to characterize risk factors and define susceptibility/risk biomarkers (including neuroimaging) to predict which individuals develop persistent pain after surgery. These initiatives and others aimed at identifying susceptibility/risk biomarkers may provide information that would allow for earlier identification of individuals at risk of developing persistent pain conditions and indicate modifiable factors (eg, diet, exercise, and behavioral change) that could be influenced to mitigate or reduce the susceptibility of chronic pain.
2.4. Predictive biomarkers
Predictive biomarkers are used to identify those individuals who are more likely to respond to a specific treatment.7 Biomarkers that can predict treatment responses are critical for the development and success of precision pain medicine approaches.13 Predictive biomarkers are typically used in pharmaceutical and device development to enrich a clinical study population for a subsequent randomized controlled trial (RCT).6,87,99 To date, most research in predictive biomarkers has been in the field of genetics for the prediction of cancer treatment response. For example, in a study of cancer treatments approved by the FDA from 1998 to 2013, researchers demonstrated that a biomarker-based approach to clinical trials of anticancer drugs was associated with improved efficacy and longer progression-free survival relative to conventional trials.87
Two examples from psychiatry illustrate the potential of neuroimaging-based predictive biomarkers to predict treatment responses. Amygdala reactivity and early life stress (ELS) both have been strongly implicated in the mechanisms of depression in animal and human models.23,31,33 Researchers integrated neuroimaging and ELS measures within a controlled trial of antidepressant outcomes.33 They demonstrated that the interaction between ELS and amygdala engagement predicted functional remission on antidepressants with more than 80% cross-validated accuracy. In depressed people exposed to high ELS, a greater likelihood of remission was predicted by amygdala hyperreactivity to socially rewarding stimuli, whereas for those with low-ELS exposure, amygdala hyporeactivity to both rewarding and threat-related stimuli predicted remission.33 Thus, amygdala reactivity and ELS are biobehavioral biomarkers for predicting functional remission in depression.
Like many pain conditions, depression is not a unitary disease; rather, it is a heterogeneous syndrome encompassing subtypes with varied, co-occurring symptoms and divergent treatment responses. Drysdale et al. used neuroimaging in a large multisite sample (n = 1188) to demonstrate that patients with depression can be divided into 4 neurophysiological subtypes defined by distinct patterns of dysfunctional connectivity in limbic and frontostriatal networks.22 The authors used this clustering to develop diagnostic biomarkers with high (82%–93%) sensitivity and specificity for depression subtypes in multisite validation (n = 711) and out-of-sample replication (n = 477) data sets. Interestingly, these subtypes could not be differentiated solely on the basis of clinical features but were associated with differing clinical-symptom profiles. The researchers used these biomarkers to predict responsiveness to transcranial magnetic stimulation therapy (n = 154), thereby identifying the individuals most likely to benefit from targeted neurostimulation therapies.22
Predictive biomarkers could help guide go/no-go decisions in selecting effective analgesics in early human drug development. Borsook et al. outlined the utility of functional imaging to define biomarkers to predict efficacy and safety, determine drug–dose relationships, and provide objective measure of symptom response and disease modification.8–10 A more recent example of this was by Wanigasekera et al.107 who assessed the utility of fMRI with a capsaicin-induced central sensitization (a mechanism relevant to neuropathic pain) to differentiate an effective (gabapentin) from ineffective (ibuprofen) treatment and both from placebo. They found that gabapentin reduced connectivity between the thalamus and secondary somatosensory cortex, whereas ibuprofen did not when compared with placebo. They also determined that the neural activity evoked by hyperalgesia from the right nucleus cuneiformis and the left posterior insula was more sensitive than the behavioral pain scores in detecting a statistically significant difference between gabapentin and placebo. This work built upon the group's previous work to generate and validate a general protocol for neuroimaging-based assessment of drug activity in the CNS that can be used to optimize drug discovery and validation.23
Predictive biomarkers would have obvious utility in the clinical field of pain medicine. Effect sizes for many analgesic efficacy RCTs in chronic pain are modest at best. Nonetheless, within each treatment group, there are often clear responders mixed with nonresponders.13 Neuroimaging-based biomarkers would be valuable in predicting these responders in an RCT.
The neuroimaging biomarker could be used either to select patients for participation or to stratify patients into biomarker-positive and biomarker-negative groups, with the primary endpoint being the effect in the biomarker-positive group. These biomarker enriched clinical trials could be used to identify effective treatments that might otherwise fail if more heterogeneous populations were enrolled.
Predictive pain biomarkers would also inform patient care decisions. Currently, clinicians are faced with a myriad, and increasing number, of treatment choices for pain including pharmacologic, psychological, interventional, physical therapy, complementary and alternative medicine, and self-management approaches. Unfortunately, outside of clinical experience, clinicians often have little information to guide them on treatment decisions. Predictive pain biomarkers could aid clinical decision-making in choosing the best treatment(s) for a specific patient with a specific pain condition under specific environmental circumstances, such as the role of long-term prescribing of opioids in chronic noncancer pain, which is one of the most important pain issues our country is facing.
According to the Centers for Disease Control and Prevention, opioid misuse, abuse, addiction, and associated overdose deaths have reached epidemic levels in the United States.15 More than 90 Americans die daily from opioid overdose.93 This health care crisis exists despite studies showing that long-term use is increasingly associated with significant negative side effects, risk of misuse, abuse, and addiction.90,93,94 However, stable doses of opioids may provide extended pain relief with limited side effects for a subgroup of individuals.47 Consequently, a poor risk/benefit ratio in a large patient population may obscure a positive profile in a subgroup of opioid-responsive chronic pain patients. Perhaps, one of the most valuable applications of predictive pain biomarkers would be in predicting those patients who would respond favorably, and with minimal adverse events, to opioids—and those who are at increased risk of misuse, abuse, diversion, and overdose.
The development of neuroimaging-based predictive biomarkers could transform our care of those in pain. From optimizing clinical trials to aiding clinical care decisions, predictive biomarkers are a critical component of realizing the goal of precision pain medicine.
2.5. Monitoring biomarkers
A monitoring biomarker is used to serially assess the presence, status, or extent of a medical condition, or to provide evidence of a treatment or adverse effect.7 This type of biomarker represents a change in a biomarker value across multiple points in time. As such, this biomarker category is broad and can include other types of biomarkers if they are assessed serially. For example, clinicians could use a neuroimaging monitoring pain biomarker to serially assess the progression of pain to persistent pain or opioid use after surgery or injury or the progression from chronic pain to high-impact chronic pain in an individual.102
2.6. Pharmacodynamic/response biomarkers
A pharmacodynamic/response biomarker is a biomarker whose levels change in response to an exposure to a medical product or an environmental agent. A change in a pharmacodynamic/response biomarker can provide evidence for clinical efficacy or assess an endpoint related to safety concerns. It can also provide clinical decision support for patient management to help determine whether to continue treatment or to adjust dose. In addition, these biomarkers can be useful for pharmaceutical/device development by assessing whether a treatment had a pharmacodynamic/device effect related to a clinical response. Because of the repeated nature of their assessment, pharmacodynamic/response biomarkers are also often considered monitoring biomarkers. For example, tricyclic antidepressants (TCAs) are a class of medications often used to treat chronic neuropathic pain. Clinicians can monitor blood levels of TCAs as a pharmacodynamic/response biomarker and use the results to titrate drug levels to within a therapeutic range.78 Similarly, the corrected QT interval is used as a safety biomarker to assess potential for drugs such as methadone or TCAs to induce torsades de pointes, a potentially fatal arrhythmia.
2.7. Safety biomarkers
Safety biomarkers detect or predict adverse drug or exposure effects.7 Many of the treatments used in pain management, particularly medications, have undesirable and potentially harmful or toxic effects. An example of this is the use of TCAs for treating neuropathic pain, as noted above. Although TCAs can be efficacious for pain, they can also have adverse effects (eg, drowsiness, constipation, cardiac arrhythmias and sudden death, blurred vision, and orthostatic hypotension). A neuroimaging-based safety biomarker would be useful if it could predict which treatments would negatively impact which patients under specific circumstances.
2.8. Need for multimodal biomarkers of pain
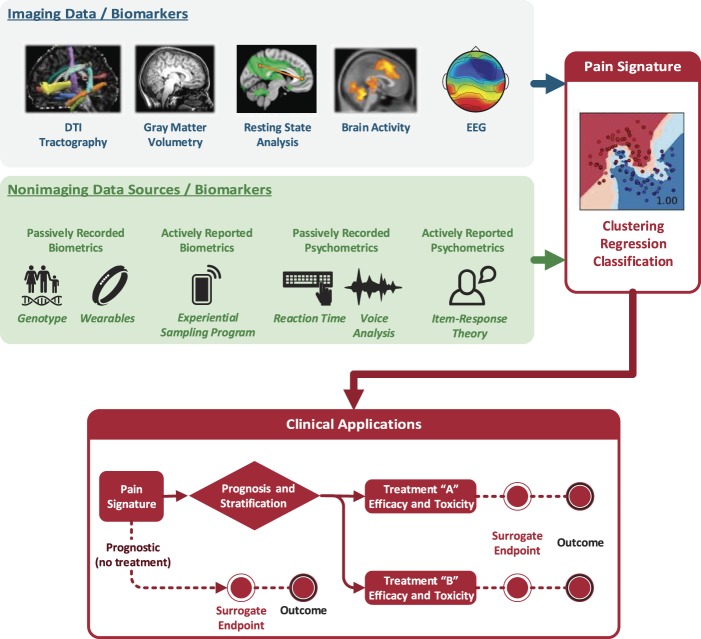
It is clear that neuroimaging alone will not capture all of the variance in models defining diagnostic, predictive, prognostic, and risk biomarkers. More likely, we will need to combine neuroimaging and non-neuroimaging data (eg, behavioral, genotype, phenotype, and longitudinal data) into a multimodal biomarker of pain. Combining measures of genomics and other 'omics, activity monitors, passively recorded psychometrics, and quantitative sensory testing with neuroimaging data could improve the sensitivity and specificity of neuroimaging-based biomarkers. Figure 2 illustrates the integration of multiple potential biomarkers and provides a schematic of how these different types of biomarkers may be applied.
Figure 2.
Multimodal Pain Signatures. Imaging data sources (top, gray box) and nonimaging data sources (middle, green box) can be combined into machine learning algorithms to provide a multimodal signature pattern of pain (right, red box). Various sources of imaging biomarkers include (1) structural changes measured with MRI (eg, diffusion tensor imaging [DTI] of white matter tractography; gray matter volumetry), (2) functional differences measured with fMRI (eg, resting state fMRI networks and functional connectivity between brain regions; brain activity in response to evoked stimulation or during a task), and (3) functional differences measured with non-MRI modalities such as EEG. Nonimaging data sources include, eg, genotype information, biometrics from wearable technology (eg, actigraphy), actively reported biometrics (eg, through handheld devices for recording patients' symptoms throughout the day), psychometrics including reaction time tests and voice analysis (eg, to measure emotional states such as depression or anxiety), and actively reported psychometrics (ie, demographic, psychological, and clinical questionnaires) (middle, green box). The multimodal pain signature can then be used in a variety of biomarker applications (bottom, red box). fMRI, functional MRI.
3. A framework for evaluating neuroimaging-based biomarkers of pain
Neuroimaging-based pain biomarkers must be validated to have true clinical and research utility. Validation is “a process to establish that the performance of a test, tool, or instrument is acceptable for its intended purpose.”7 It is critical to confirm that a neuroimaging pain biomarker measures what it is intended to measure and that it predicts or measures the relevant clinical concept (ie, it has appropriate analytical and clinical validity). A biomarker must also have clinical utility, in that it provides information that assists in the care of patients. An essential first step in evaluating a neuroimaging-based pain biomarker is to precisely define the brain measures it is intended to assay, the pain condition of interest, the purpose of the test, and the population or health care setting in which it is going to be used. In other words, context is paramount in considering the validity and utility of a neuroimaging-based pain biomarker to achieve clinical or research utility.
To formally evaluate a biomarker, we can draw upon other fields that have developed evaluation frameworks. One such framework that researchers have successfully adopted in the field of genetic testing is the ACCE criteria.71,88,115 The ACCE framework was established by the Centers for Disease Control Prevention Office of Public Health Genomics to set a standard path for genetic tests to follow toward clinical and public use.55 After initial research discovery, the framework includes the ACCE acronym stages of (1) Analytic Validity (2) Clinical Validity (3) Clinical Utility, and (4) Ethical, Legal, and Social Impacts37,115 (Fig. 3). The ACCE model includes a standard set of 44 questions to ensure proper application and to safeguard against critical social, ethical, and legal issues.37 Although originally written for the purpose of diagnostic genetic tests, these questions can be adapted to other types of biomarkers listed in Section 2. For example, we have adapted these standard questions to reflect an approach for diagnostic neuroimaging pain biomarkers.
Figure 3.
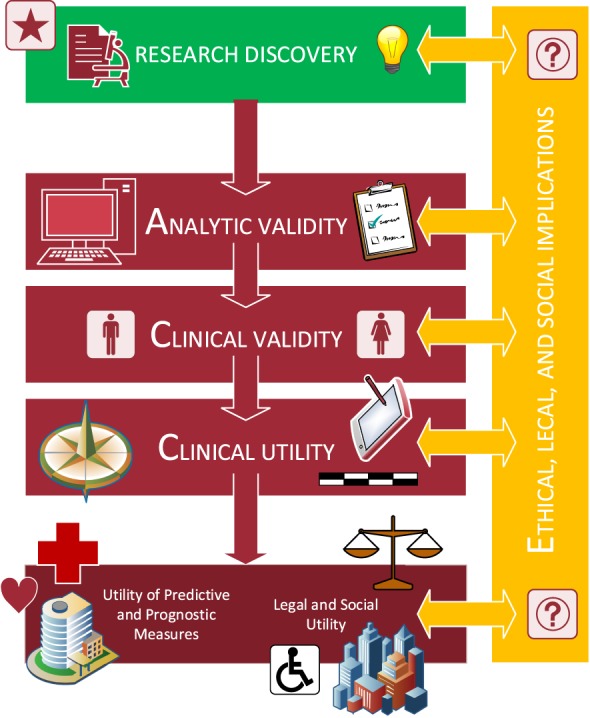
Framework for Evaluating Neuroimaging-based Biomarkers of Pain. A candidate biomarker will need to be vetted through all stages of analytic validity, clinical validity, clinical utility, and ethical, legal, and social implications.
The National Academies of Medicine (NAM) recently completed a report entitled “An Evidence Framework for Genetic Testing.”1 In this report, the authors built upon ACCE and other testing frameworks to refine the notion of analytic validity, clinical validity, and clinical utility and emphasize the importance of integrating societal benefit into the evaluation process.1
In 2010, a landmark report from NAM provided a foundation for evaluating biomarkers and surrogate endpoints.89,104 This report recommended an evaluation process that included: “(1) Analytical validation: analyses of available evidence on the analytical performance of an assay; (2) Qualification (NB: Qualification as used here is synonymous with clinical utility used in ACCE): assessment of available evidence on associations between the biomarker and disease states, including data showing effects of interventions on both the biomarker and clinical outcomes; and (3) Utilization: contextual analysis based on the specific use proposed and the applicability of available evidence to this use. This includes a determination of whether the validation and qualification conducted provide sufficient support for the use proposed.”89,104 These steps are interrelated and not separated in time. Therefore, conclusions in 1 step may require revisions in other steps.
The FDA Center for Drug Evaluation and Research Biomarker Qualification Program's mission is to work with external stakeholders to develop biomarkers as drug development tools.27 Their program outlines a multistep process for a biomarker to be qualified for drug development. This program relies heavily on the BEST resource outlined above as a “living” glossary of terms used in biomarker qualification science and medical product development. The FDA's program relies heavily on defining a specific context of use for the biomarker to be used in drug development. The FDA defines the Context of Use as a complete and precise statement that describes the appropriate use of the biomarker and how the qualified biomarker is applied in drug development and regulatory review. The context of use statement also describes important criteria regarding the circumstances under which the biomarker is qualified. As with the other frameworks above, the FDA Biomarker Qualification Program involves both analytical validation and clinical validation for approval. Here, the FDA defines analytical validation as to “establish that the preanalytical considerations and performance characteristics acceptably support the biomarker's context of use,” while clinical validation is to “establish that the biomarker acceptably identifies, measures, or predicts the concept of interest.” Two important points about the FDA Biomarker Qualification Program is in what it does not do. Specifically, the program does not imply the test/assay has been reviewed by the FDA and cleared or approved for use in patient care. In addition, the qualification does not qualify the biomarker for use in clinical practice.
In summary, there is more commonality with these frameworks than differences. They all require analytical and clinical validity. The differences are more to do with the intended purposes of the framework, with for instance, the FDA Biomarker Qualification Program aimed more at drug development and the others having a broader utility.
3.1. Analytical validity
The analytical validity of a neuroimaging-based biomarker refers to “assessing [an] assay and its measurement performance characteristics and determining the range of conditions under which the assay will give reproducible and accurate data.”105 In this context, an assay is a method to analyze or quantify brain activity or a structure in an individual or animal. Analytical validity is concerned with assessing the performance of a biomarker test in a laboratory setting as opposed to the clinic or general population. Here, we need quality assurance to ensure that the results are reliable and reproducible across scanners, clinical settings, and analysis pipelines.
Pooling of neuroimaging data across centers and scanners will be required to yield the numbers of subjects to demonstrate proper analytical and clinical validity. Multiple neuroimaging research networks have addressed the challenges of pooling multicenter neuroimaging data, as well as developing common informatics, quality control, analysis, and visualization tools and protocols. These networks include the International Consortium for Brain Mapping,68 Alzheimer' Disease Neuroimaging Initiative (ADNI),44 Function Biomedical Informatics Research Network (fBIRN),32 National Institutes of Health Pediatric MRI Database,24 and the Human Connectome Project.60,101 As part of the National Institutes of Health-funded Multi-Disciplinary Approach to the Study of Chronic Pelvic Pain (MAPP), we emulated many of these pioneering research networks in establishing the neuroimaging protocols, databases, analyses, and visualizations used across 5 research centers with different manufacturers and models of MRI scanners. Alger et al. outlined many of the challenges and solutions to neuroimaging across multiple centers and scanners within the MAPP network.2 The MAPP network subsequently published multiple articles demonstrating the feasibility and insights that are possible with large multicenter neuroimaging data sets of pain.3,28,43,48,50–52,54,65,66,96,97,113
The analysis approach and algorithms used in analytical validation are critical. Most approaches are using a multivariate pattern analysis or machine learning approach. An important factor in the context use of such analyses is the embedding of feature selection, classifier optimization, and the estimation of the models' generalizability in a cross-validation scheme. In the cross-validation steps, it is important to avoid information leakage between the training and the test samples to avoid overfitting and inflated estimates of classification accuracy. Moreover, even in the case of correct feature embedding, different cross-validation schemes might lead to different results. In a multivariate pattern analysis meta-analysis for detecting neuroimaging biomarkers of depression, the authors identified that 2-fold cross-validation was associated with higher diagnostic accuracy than 10-fold or leave-one-out cross-validation.45 We will need generalizability of the models across research centers as well as agreement on the identification of the methodological and clinical variables moderating classification success.
The biomarker must also show adequate sensitivity and specificity before it is assessed in subsequent biomarker evaluation steps. This quality assurance will typically include both internal and external control assessments within a structured framework. Analytical validation can also include determining the extent to which data from different tests for the same biomarker may be compared to one another. Highly comparable data strengthen the biomarker and add power to retrospective analyses of data related to the biomarker.89
3.2. Clinical validity
Clinical validity is defined as the “evidentiary process of linking a biomarker with biological processes and clinical endpoints.”89 Clinical validity (1) defines the ability of a neuroimaging pain biomarker to detect or predict the presence or absence of a phenotype or clinical disease or (2) defines a biomarker's ability to predict the effects of interventions on clinical endpoints of interest. If a biomarker-clinical endpoint relationship occurs over several interventions, we can consider the biomarker more generalizable. Candidate biomarkers that are both informative of pathophysiology and highly prognostic (ie, able to identify risk factors for developing disorders and disease predisposition) should pass standards of diagnosticity, interpretability, deployability, and generalizability. Similarly, candidate biomarkers will need to differentiate CNS features associated with different clinical pain conditions—assuming such features exist. Are the CNS features of chronic low back pain the same or different as compared with CRPS, migraine, pelvic pain, or fibromyalgia? Are there biomarker differences in a painful condition that is presumed to be more peripherally vs centrally driven? As noted previously, in a cohort of localized chronic pelvic pain vs pelvic pain plus widespread pain, we identified different neuroimaging biomarkers that distinguish both groups.50 Assuming there are differences in features amongst painful conditions, do they matter regarding prognosis of natural history or prediction of treatment response? In addition, candidate biomarkers will need to account for patient heterogeneity introduced with age, severity and duration of pain, concomitant medications or other treatments, and comorbid conditions such as depression, anxiety, and catastrophizing. In addition, population base rates will need to account for prevalence of chronic pain syndromes,69,83 similar to previous applications for genetic and psychological testing. Formally evaluating the sensitivity, specificity, positive predictive value, and negative predictive value of the biomarker is also pertinent. Researchers must include appropriately selected controls in any formal biomarker evaluation.
3.3. Utilization or clinical utility
Utilization refers to the “contextual analysis based on the specific use proposed and the applicability of available evidence to this use. This includes a determination of whether the validation and qualification conducted provide sufficient support for the use proposed.”89 Clinical utility assesses the likelihood that the biomarker will lead to an improved clinical outcome. Clinical utility helps address the purpose of the biomarker? Do the biomarker findings change clinical management or prognosis? What is the natural history of the disorder? Are there effective interventions based on the biomarker results? Is the biomarker result information useful for family members? Does the biomarker give rise to any ethical, legal, or social consequences? Are there other ways of achieving the same purpose apart from using a neuroimaging biomarker? For example, and as mentioned previously, there is little clinical utility for a neuroimaging pain biomarker to simply determine in a binary manner if a person seen in a clinical setting is in pain or not. We can simply just ask. What is the cost of the neuroimaging biomarker? Is the biomarker cost effective? In these latter 2 questions on cost, we will have to consider that at $500+/hour of scanner time, neuroimaging biomarkers are expensive. For broad use, more cost-effective MRI systems will need to be developed or other imaging modalities used (eg, EEG and functional-near infrared spectroscopy) that are less costly.
3.4. Ethical, legal, and social implications
The NAM and ACCE frameworks formally consider ethical, legal, and social implications of biomarkers. This component of the biomarker evaluation is perhaps the most difficult to address as it is wide ranging.
A neuroimaging biomarker that accurately detects pain could be useful in society for reasons beyond improving patient care. For example, in the legal system, the existence and extent of pain cannot be taken for granted but is at the heart of a dispute. In many personal injury cases, plaintiffs seek damages for ongoing pain that may have little evidence beyond self-report. Even more often—hundreds of thousands of times a year—in disability determinations, workers claim to private disability insurers or to Social Security that pain makes them incapable of working. The courts and administrative agencies cannot always by default accept the claimant's self-report as true, but often, there is little other evidence. While clinicians and researchers usually can “just ask” patients if they are in pain, the incentives involved means the legal system cannot blindly trust their answers, particularly in the American adversary system where there is always “the other side” fighting against any claims. Not only can this lead to people being granted or denied damages or benefits improperly but also the uncertainty involved increases the time and money spent in litigation.46,80 Further legal applications of neuroimaging-based pain biomarkers are beyond the scope of this review, but have been described in the following reviews.19,81
We must also consider several legal concerns regarding pain biomarkers.34 First, what would be necessary before neuroimaging evidence of pain could be accepted into evidence to use in a court or an administrative proceeding? A neuroimaging-based biomarker would have to perform at an acceptable level of accuracy in at least 3 areas: (1) accuracy with respect to demographics and phenotype (eg, men vs women, young vs elderly, mentally ill, and illegal drug users), (2) accuracy with different kinds of pain (eg, acute vs chronic, low back pain, and migraine), and (3) accuracy that cannot be undermined by countermeasures to “fool” the neuroimaging-based biomarker.
In addition to these issues, we must resolve the deeper question of “how accurate would it need to be?” The law does not set standards based on P values or confidence intervals; judges will need to decide whether it is “sufficiently” reliable to help the legal process.36 Second, even if a neuroimaging-based biomarker met the vague standards for accuracy, could a claimant be forced to undergo such a test? Typically, people can be forced to undergo medical examinations at the request of their legal opponents or forfeit their claims. Is it different when the test involves probing an individual's interior mental states vs a vertebral disk? The question whether there is, or should be, some kind of “cognitive liberty” that is free from compulsory intrusion is important, yet undecided.25,26 Although a reliable brain biomarker of pain would clearly be of benefit to the legal system, the additional requirements for fair and just use are complicated, and misguided use could profoundly impact patients' lives beyond their clinical care and outcomes.
Perhaps, the greatest risk may be the premature availability of such tests and their use by patients, doctors, employers, agencies, or judges when they have not been shown to be “accurate enough.” Our system has incentives for individuals to use any evidence they can to try to persuade others, as well as incentives to lead some people to sell goods and services to others without knowing they are effective—and, in the cases of some frauds, while consciously knowing that they are ineffective.
Before and during each of the above evaluation stages, critical questions must be addressed. For example, we must consider the impact of the biomarker on insurance and employment, health care disparities including equity and access, privacy and confidentiality, and stigmatization. As alluded to earlier, there is great potential for misuse and abuse of these neuroimaging-based pain biomarkers with real potential for stigma and discrimination. An early example of this involved broad-based community screening efforts for sickle cell disease in the 1970s. These screening efforts were accompanied by a misunderstanding of the health implications of the carrier state, leading to subsequent insurance and employment discrimination.106 This, and other discrimination events around genetic testing, led to the federal Genetic Information Nondiscrimination Act in 2008, which blocks health insurers and employers from using genetic information (but not, eg, neuroimaging information) in health coverage or employment decisions. The Patient Protection and Affordable Care Act of 2010 added further protections, although their continuation is in some doubt. Nonetheless, in any event, there is still potential for discrimination not covered under these acts including determination of life insurance, mortgage insurance, long-term care insurance, and long-term disability insurance. Importantly, there are few protections in place for the misuse of neuroimaging-based pain biomarkers. As such, these potential issues will require scientists, clinicians, ethicists, attorneys, and patients to be vigilant for such misuse and abuse.
4. Conclusions
We have reviewed the different types of potential neuroimaging-based pain biomarkers, their clinical and research applications, and their limitations. The field of neuroimaging-based biomarkers has advanced rapidly. With this rapid advancement comes a need for structured frameworks and processes to validate them as biomarkers. We have presented such a framework adapted from several successfully applied frameworks in other fields. This model of assuring analytical validity, clinical validity, and clinical utility and accounting for ethical, legal, and social implications can help advance these biomarkers to achieve broad clinical and research utility while minimizing the risks of misapplication of this emerging technology. Neuroimaging pain biomarkers are helping to advance the goal of personalized pain medicine to ultimately aid clinicians and patients to choose the best treatment that both safe and effective.
Disclosures
The authors have no conflict of interest to declare.
This work was supported by NIH grants DA029262 (K24, SCM), DA040154 (K99, KTM), and GM089626 (T32) and the Redlich Pain Research Endowment.
Acknowledgements
The authors thank Dr Ming-Chih J. Kao for designing the figure and Dr Mary Pelleymounter (NIH) for her review and feedback.
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
References
- [1].An evidence framework for genetic testing. Washington: National Academies Press, 2017. [PubMed] [Google Scholar]
- [2].Alger JR, Ellingson BM, Ashe-McNalley C, Woodworth DC, Labus JS, Farmer M, Huang L, Apkarian AV, Johnson KA, Mackey SC, Ness TJ, Deutsch G, Harris RE, Clauw DJ, Glover GH, Parrish TB, Hollander J, Kusek JW, Mullins C, Mayer EA, Investigators MRN. Multisite, multimodal neuroimaging of chronic urological pelvic pain: methodology of the MAPP Research Network. Neuroimage Clin 2016;12:65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bagarinao E, Johnson KA, Martucci KT, Ichesco E, Farmer MA, Labus J, Ness TJ, Harris R, Deutsch G, Apkarian AV. Preliminary structural MRI based brain classification of chronic pelvic pain: a MAPP network study. PAIN 2014;155:2502–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bair E, Ohrbach R, Fillingim RB, Greenspan JD, Dubner R, Diatchenko L, Helgeson E, Knott C, Maixner W, Slade GD. Multivariable modeling of phenotypic risk factors for first-onset TMD: the OPPERA prospective cohort study. J Pain 2013;14:T102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Baliki MN, Petre B, Torbey S, Herrmann KM, Huang L, Schnitzer TJ, Fields HL, Apkarian AV. Corticostriatal functional connectivity predicts transition to chronic back pain. Nat Neurosci 2012;15:1117–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ballman KV. Biomarker: predictive or prognostic? J Clin Oncol 2015;33:3968–71. [DOI] [PubMed] [Google Scholar]
- [7].BEST (biomarkers, EndpointS, and other tools) resource. Silver Spring, 2016. [PubMed] [Google Scholar]
- [8].Borsook D, Becerra L, Hargreaves R. Biomarkers for chronic pain and analgesia. Part 1: the need, reality, challenges, and solutions. Discov Med 2011;11:197–207. [PubMed] [Google Scholar]
- [9].Borsook D, Becerra L, Hargreaves R. Biomarkers for chronic pain and analgesia. Part 2: how, where, and what to look for using functional imaging. Discov Med 2011;11:209–19. [PubMed] [Google Scholar]
- [10].Borsook D, Hargreaves R, Becerra L. Can functional magnetic resonance imaging improve success rates in CNS drug discovery? Expert Opin Drug Discov 2011;6:597–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Brodersen KH, Wiech K, Lomakina EI, Lin CS, Buhmann JM, Bingel U, Ploner M, Stephan KE, Tracey I. Decoding the perception of pain from fMRI using multivariate pattern analysis. Neuroimage 2012;63:1162–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Brown JE, Chatterjee N, Younger J, Mackey S. Towards a physiology-based measure of pain: patterns of human brain activity distinguish painful from non-painful thermal stimulation. PLoS One 2011;6:e24124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bruehl S, Apkarian AV, Ballantyne JC, Berger A, Borsook D, Chen WG, Farrar JT, Haythornthwaite JA, Horn SD, Iadarola MJ. Personalized medicine and opioid analgesic prescribing for chronic pain: opportunities and challenges. J Pain 2013;14:103–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Carroll IR, Hah JM, Barelka PL, Wang CK, Wang BM, Gillespie MJ, McCue R, Younger JW, Trafton J, Humphreys K. Pain duration and resolution following surgery: an inception cohort study. Pain Med 2015;16:2386–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].(CDC) CfDCaP. Understanding the epidemic. Center for Disease Control and Prevention, 2016. [Google Scholar]
- [16].Cheng JC, Rogachov A, Hemington KS, Kucyi A, Bosma RL, Lindquist MA, Inman RD, Davis KD. Multivariate machine learning distinguishes cross-network dynamic functional connectivity patterns in state and trait neuropathic pain. PAIN 2018;159:1764–76. [DOI] [PubMed] [Google Scholar]
- [17].Coghill RC. Individual differences in the subjective experience of pain: new insights into mechanisms and models. Headache 2010;50:1531–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cowen R, Stasiowska MK, Laycock H, Bantel C. Assessing pain objectively: the use of physiological markers. Anaesthesia 2015;70:828–47. [DOI] [PubMed] [Google Scholar]
- [19].Davis KD, Flor H, Greely HT, Iannetti GD, Mackey S, Ploner M, Pustilnik A, Tracey I, Treede RD, Wager TD. Brain imaging tests for chronic pain: medical, legal and ethical issues and recommendations. Nat Rev Neurol 2017;13:624–38. [DOI] [PubMed] [Google Scholar]
- [20].Davis KD, Moayedi M. Central mechanisms of pain revealed through functional and structural MRI. J Neuroimmune Pharmacol 2012;8:518–34. [DOI] [PubMed] [Google Scholar]
- [21].Davis KD, Racine E, Collett B. Neuroethical issues related to the use of brain imaging: can we and should we use brain imaging as a biomarker to diagnose chronic pain? PAIN 2012;153:1555–9. [DOI] [PubMed] [Google Scholar]
- [22].Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, Fetcho RN, Zebley B, Oathes DJ, Etkin A, Schatzberg AF, Sudheimer K, Keller J, Mayberg HS, Gunning FM, Alexopoulos GS, Fox MD, Pascual-Leone A, Voss HU, Casey BJ, Dubin MJ, Liston C. Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med 2017;23:28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Duff EP, Vennart W, Wise RG, Howard MA, Harris RE, Lee M, Wartolowska K, Wanigasekera V, Wilson FJ, Whitlock M, Tracey I, Woolrich MW, Smith SM. Learning to identify CNS drug action and efficacy using multistudy fMRI data. Sci Transl Med 2015;7:274ra216. [DOI] [PubMed] [Google Scholar]
- [24].Evans AC; Brain Development Cooperative G. The NIH MRI study of normal brain development. Neuroimage 2006;30:184–202. [DOI] [PubMed] [Google Scholar]
- [25].Farahany NA. Searching secrets. U Pa L Rev 2011;160:1239–308. [Google Scholar]
- [26].Farahany NA. Incriminating thoughts. Stan L Rev 2012;64:351–408. [Google Scholar]
- [27].FDA Center for Drug Evaluation and Research. Biomarker qualification program. vol. 2019. [Google Scholar]
- [28].Fernandez-Rhodes L, Gong J, Haessler J, Franceschini N, Graff M, Nishimura KK, Wang Y, Highland HM, Yoneyama S, Bush WS, Goodloe R, Ritchie MD, Crawford D, Gross M, Fornage M, Buzkova P, Tao R, Isasi C, Aviles-Santa L, Daviglus M, Mackey RH, Houston D, Gu CC, Ehret G, Nguyen KH, Lewis CE, Leppert M, Irvin MR, Lim U, Haiman CA, Le Marchand L, Schumacher F, Wilkens L, Lu Y, Bottinger EP, Loos RJL, Sheu WH, Guo X, Lee WJ, Hai Y, Hung YJ, Absher D, Wu IC, Taylor KD, Lee IT, Liu Y, Wang TD, Quertermous T, Juang JJ, Rotter JI, Assimes T, Hsiung CA, Chen YI, Prentice R, Kuller LH, Manson JE, Kooperberg C, Smokowski P, Robinson WR, Gordon-Larsen P, Li R, Hindorff L, Buyske S, Matise TC, Peters U, North KE. Trans-ethnic fine-mapping of genetic loci for body mass index in the diverse ancestral populations of the Population Architecture using Genomics and Epidemiology (PAGE) Study reveals evidence for multiple signals at established loci. Hum Genet 2017;136:771–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fillingim RB. Individual differences in pain: understanding the mosaic that makes pain personal. PAIN 2017;158(suppl 1):S11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fillingim RB, Slade GD, Diatchenko L, Dubner R, Greenspan JD, Knott C, Ohrbach R, Maixner W. Summary of findings from the OPPERA baseline case-control study: implications and future directions. J Pain 2011;12:T102–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gibbs RM, Lipnick S, Bateman JW, Chen L, Cousins HC, Hubbard EG, Jowett G, LaPointe DS, McGredy MJ, Odonkor MN, Repetti G, Thomas E, Rubin LL. Toward precision medicine for neurological and neuropsychiatric disorders. Cell Stem Cell 2018;23:21–4. [DOI] [PubMed] [Google Scholar]
- [32].Glover GH, Mueller BA, Turner JA, van Erp TG, Liu TT, Greve DN, Voyvodic JT, Rasmussen J, Brown GG, Keator DB, Calhoun VD, Lee HJ, Ford JM, Mathalon DH, Diaz M, O'Leary DS, Gadde S, Preda A, Lim KO, Wible CG, Stern HS, Belger A, McCarthy G, Ozyurt B, Potkin SG. Function biomedical informatics research network recommendations for prospective multicenter functional MRI studies. J Magn Reson Imaging 2012;36:39–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Goldstein-Piekarski AN, Korgaonkar MS, Green E, Suppes T, Schatzberg AF, Hastie T, Nemeroff CB, Williams LM. Human amygdala engagement moderated by early life stress exposure is a biobehavioral target for predicting recovery on antidepressants. Proc Natl Acad Sci U S A 2016;113:11955–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Greely HT. Reading minds with neuroscience—possibilities for the law. Cortex 2011;47:1254–5. [DOI] [PubMed] [Google Scholar]
- [35].Greely HT. Neuroscience, mindreading, and the courts: the example of pain. J Health Care L Pol'y 2015;18. [Google Scholar]
- [36].Greely HT, Wagner AD. Reference guide on neuroscience. Reference manual on scientific evidence. 3rd ed Federal Judicial Center and National Academies Press, 2011. p. 747–812. [Google Scholar]
- [37].Haddow JE, Palomaki GE. ACCE: a model process for evaluating data on emerging genetic tests. In: Little J, Burke W, Khoury MJ, editors. Human genome epidemiology: a scientific foundation for using genetic information to improve health and prevent disease: Oxford University Press, 2003. p. 217–33. [Google Scholar]
- [38].Hah J, Mackey SC, Schmidt P, McCue R, Humphreys K, Trafton J, Efron B, Clay D, Sharifzadeh Y, Ruchelli G, Goodman S, Huddleston J, Maloney WJ, Dirbas FM, Shrager J, Costouros JG, Curtin C, Carroll I. Effect of perioperative gabapentin on postoperative pain resolution and opioid cessation in a mixed surgical cohort: a randomized clinical trial. JAMA Surg 2018;153:303–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hah JM, Cramer E, Hilmoe H, Schmidt P, McCue R, Trafton J, Clay D, Sharifzadeh Y, Ruchelli G, Goodman S, Huddleston J, Maloney WJ, Dirbas FM, Shrager J, Costouros JG, Curtin C, Mackey SC, Carroll I. Factors associated with acute pain estimation, postoperative pain resolution, opioid cessation, and recovery: secondary analysis of a randomized clinical trial. JAMA Netw Open 2019;2:e190168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hah JM, Mackey S, Barelka PL, Wang CK, Wang BM, Gillespie MJ, McCue R, Younger JW, Trafton J, Humphreys K. Self-loathing aspects of depression reduce postoperative opioid cessation rate. Pain Med 2014;15:954–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hah JM, Sharifzadeh Y, Wang BM, Gillespie MJ, Goodman SB, Mackey SC, Carroll IR. Factors associated with opioid use in a cohort of patients presenting for surgery. Pain Res Treat 2015;2015:829696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hashmi JA, Baliki MN, Huang L, Baria AT, Torbey S, Hermann KM, Schnitzer TJ, Apkarian AV. Shape shifting pain: chronification of back pain shifts brain representation from nociceptive to emotional circuits. Brain 2013;136:2751–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Huang L, Kutch JJ, Ellingson BM, Martucci KT, Harris RE, Clauw DJ, Mackey S, Mayer EA, Schaeffer AJ, Apkarian AV, Farmer MA. Brain white matter changes associated with urological chronic pelvic pain syndrome: multisite neuroimaging from a MAPP case-control study. PAIN 2016;157:2782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jack CR, Jr, Shiung MM, Gunter JL, O'Brien PC, Weigand SD, Knopman DS, Boeve BF, Ivnik RJ, Smith GE, Cha RH, Tangalos EG, Petersen RC. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology 2004;62:591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kambeitz J, Cabral C, Sacchet MD, Gotlib IH, Zahn R, Serpa MH, Walter M, Falkai P, Koutsouleris N. Detecting neuroimaging biomarkers for depression: a meta-analysis of multivariate pattern recognition studies. Biol Psychiatry 2017;82:330–8. [DOI] [PubMed] [Google Scholar]
- [46].Kolber AJ. Pain detection and the privacy of subjective experience. Am J L Med 2007;33:433–56. [DOI] [PubMed] [Google Scholar]
- [47].Krebs EE, Gravely A, Nugent S, Jensen AC, DeRonne B, Goldsmith ES, Kroenke K, Bair MJ, Noorbaloochi S. Effect of opioid vs nonopioid medications on pain-related function in patients with chronic back pain or hip or knee osteoarthritis pain: the SPACE randomized clinical trial. JAMA 2018;319:872–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Krieger JN, Stephens AJ, Landis JR, Clemens JQ, Kreder K, Lai HH, Afari N, Rodríguez L, Schaeffer A, Mackey S, Andriole GL, Williams DA. Relationship between chronic nonurological associated somatic syndromes and symptom severity in urological chronic pelvic pain syndromes: baseline evaluation of the MAPP study. J Urol 2015;193:1254–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Krishnan A, Woo CW, Chang LJ, Ruzic L, Gu X, Lopez-Sola M, Jackson PL, Pujol J, Fan J, Wager TD. Somatic and vicarious pain are represented by dissociable multivariate brain patterns. Elife 2016;5:e15166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kutch JJ, Ichesco E, Hampson JP, Labus JS, Farmer MA, Martucci KT, Ness TJ, Deutsch G, Apkarian AV, Mackey SC, Klumpp DJ, Schaeffer AJ, Rodriguez LV, Kreder KJ, Buchwald D, Andriole GL, Lai HH, Mullins C, Kusek JW, Landis JR, Mayer EA, Clemens JQ, Clauw DJ, Harris RE, Network MR. Brain signature and functional impact of centralized pain: a multidisciplinary approach to the study of chronic pelvic pain (MAPP) network study. PAIN 2017;158:1979–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kutch JJ, Labus JS, Harris RE, Martucci KT, Farmer MA, Fenske S, Fling C, Ichesco E, Peltier S, Petre B, Guo W, Hou X, Stephens AJ, Mullins C, Clauw DJ, Mackey SC, Apkarian AV, Landis JR, Mayer EA; Network MR. Resting-state functional connectivity predicts longitudinal pain symptom change in urologic chronic pelvic pain syndrome: a MAPP network study. PAIN 2017;158:1069–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kutch JJ, Yani MS, Asavasopon S, Kirages DJ, Rana M, Cosand L, Labus JS, Kilpatrick LA, Ashe-McNalley C, Farmer MA, Johnson KA, Ness TJ, Deutsch G, Harris RE, Apkarian AV, Clauw DJ, Mackey SC, Mullins C, Mayer EA. Altered resting state neuromotor connectivity in men with chronic prostatitis/chronic pelvic pain syndrome: a MAPP: research Network Neuroimaging Study. Neuroimage Clin 2015;8:493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Labus JS, Van Horn JD, Gupta A, Alaverdyan M, Torgerson C, Ashe-McNalley C, Irimia A, Hong JY, Naliboff B, Tillisch K, Mayer EA. Multivariate morphological brain signatures predict patients with chronic abdominal pain from healthy control subjects. PAIN 2015;156:1545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Landis JR, Williams DA, Lucia MS, Clauw DJ, Naliboff BD, Robinson NA, van Bokhoven A, Sutcliffe S, Schaeffer AJ, Rodriguez LV, Mackey S, Kusek JW, Mullins C, Clemens JQ. The MAPP research network: design, patient characterization and operations. BMC Urol 2014;14:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Lawson RP, Drevets WC, Roiser JP. Defining the habenula in human neuroimaging studies. Neuroimage 2013;64:722–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lopez-Sola M, Woo CW, Pujol J, Deus J, Harrison BJ, Monfort J, Wager TD. Towards a neurophysiological signature for fibromyalgia. PAIN 2017;158:34–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Mackey S. National pain strategy task force: the strategic plan for the IOM pain report. Pain Med 2014;15:1070–1. [DOI] [PubMed] [Google Scholar]
- [58].Mackey S. Future directions for pain management: lessons from the institute of medicine pain report and the national pain strategy. Hand Clin 2016;32:91–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Mackey SC. Central neuroimaging of pain. J Pain 2013;14:328–31. [DOI] [PubMed] [Google Scholar]
- [60].Marcus DS, Harms MP, Snyder AZ, Jenkinson M, Wilson JA, Glasser MF, Barch DM, Archie KA, Burgess GC, Ramaratnam M, Hodge M, Horton W, Herrick R, Olsen T, McKay M, House M, Hileman M, Reid E, Harwell J, Coalson T, Schindler J, Elam JS, Curtiss SW, Van Essen DC; Consortium WU-MH. Human Connectome Project informatics: quality control, database services, and data visualization. Neuroimage 2013;80:202–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Marquand A, Howard M, Brammer M, Chu C, Coen S, Mourao-Miranda J. Quantitative prediction of subjective pain intensity from whole-brain fMRI data using Gaussian processes. Neuroimage 2010;49:2178–89. [DOI] [PubMed] [Google Scholar]
- [62].Martucci KT, Mackey SC. Imaging pain. Anesthesiol Clin 2016;34:255–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Martucci KT, Mackey SC. Neuroimaging of pain: human evidence and clinical relevance of central nervous system processes and modulation. Anesthesiology 2018;128:1241–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Martucci KT, Ng P, Mackey S. Neuroimaging chronic pain: what have we learned and where are we going? Future Neurol 2014;9:615–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Martucci KT, Shirer WR, Bagarinao E, Johnson KA, Farmer MA, Labus JS, Apkarian AV, Deutsch G, Harris RE, Mayer EA, Clauw DJ, Greicius MD, Mackey SC. The posterior medial cortex in urologic chronic pelvic pain syndrome: detachment from default mode network-a resting-state study from the MAPP Research Network. PAIN 2015;156:1755–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Martucci KT, Shirer WR, Bagarinao E, Johnson KA, Farmer MA, Labus JS, Apkarian AV, Deutsch G, Harris RE, Mayer EA, Clauw DJ, Greicius MD, Mackey S. The posterior medial cortex in urologic chronic pelvic pain syndrome: detachment from default mode network. A resting-state study from the MAPP research network. PAIN 2015;156:1755–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Martucci KT, Weber KA, II, Mackey SC. Altered cervical spinal cord resting state activity in fibromyalgia. Arthritis Rheumatol 2018;71:441–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos Trans R Soc Lond B Biol Sci 2001;356:1293–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Meehl PE, Rosen A. Antecedent probability and the efficiency of psychometric signs, patterns, or cutting scores. Psychol Bull 1955;52:194–216. [DOI] [PubMed] [Google Scholar]
- [70].Merskey H, Bogduk N. Classification of chronic pain: Descriptions of chronic pain syndromes and definitions of pain terms. Seattle: IASP Press, 1994. [Google Scholar]
- [71].Modell SM. Success in public health genomics: beyond the ACCE criteria. Public Health 2013;127:978–80. [DOI] [PubMed] [Google Scholar]
- [72].Morton DL, Sandhu JS, Jones AK. Brain imaging of pain: state of the art. J Pain Res 2016;9:613–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Mouraux A, Iannetti GD. The search for pain biomarkers in the human brain. Brain 2018;141:3290–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Mutso AA, Petre B, Huang L, Baliki MN, Torbey S, Herrmann KM, Schnitzer TJ, Apkarian AV. Reorganization of hippocampal functional connectivity with transition to chronic back pain. J Neurophysiol 2014;111:1065–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Nash P, Wiley K, Brown J, Shinaman R, Ludlow D, Sawyer AM, Glover G, Mackey S. Functional magnetic resonance imaging identifies somatotopic organization of nociception in the human spinal cord. PAIN 2013;154:776–81. [DOI] [PubMed] [Google Scholar]
- [76].Ochsner KN, Ludlow DH, Knierim K, Hanelin J, Ramachandran T, Glover GC, Mackey SC. Neural correlates of individual differences in pain-related fear and anxiety. PAIN 2006;120:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Poldrack RA. Inferring mental states from neuroimaging data: from reverse inference to large-scale decoding. Neuron 2011;72:692–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Preskorn SH. Tricyclic antidepressant plasma level monitoring: an improvement over the dose-response approach. J Clin Psychiatry 1986;47:24–30. [PubMed] [Google Scholar]
- [79].Price DD, McGrath PA, Rafii A, Buckingham B. The validation of visual analogue scales as ratio scale measures for chronic and experimental pain. PAIN 1983;17:45–56. [DOI] [PubMed] [Google Scholar]
- [80].Pustilnik AC. Imaging brains, changing minds: how pain neuroimaging can transform the law. Ala L Rev 2015;66:1099–158. [Google Scholar]
- [81].Pustilnik AC. Legal evidence of subjective states: a brain-based model of chronic pain increases accuracy and fairness in law. Harv Rev Psychiatry 2017;25:279–88. [DOI] [PubMed] [Google Scholar]
- [82].Reinhart A. Statistics done wrong: the woefully complete guide. San Francisco: No Starch Press, 2015. [Google Scholar]
- [83].Robinson M, Boissoneault J, Sevel L, Letzen J, Staud R. The effect of base rate on the predictive value of brain biomarkers. J Pain 2016;17:637–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Robinson ME, Staud R, Price DD. Pain measurement and brain activity: will neuroimages replace pain ratings? J Pain 2013;14:323–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Robinson ME, Staud R, Price DD. Reply to commentary. J Pain 2013;14:334–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Rosa MJ, Seymour B. Decoding the matrix: benefits and limitations of applying machine learning algorithms to pain neuroimaging. PAIN 2014;155:864–7. [DOI] [PubMed] [Google Scholar]
- [87].Rubin R. A precision medicine approach to clinical trials. JAMA 2016;316:1953–5. [DOI] [PubMed] [Google Scholar]
- [88].Sanderson S, Zimmern R, Kroese M, Higgins J, Patch C, Emery J. How can the evaluation of genetic tests be enhanced? Lessons learned from the ACCE framework and evaluating genetic tests in the United Kingdom. Genet Med 2005;7:495–500. [DOI] [PubMed] [Google Scholar]
- [89].Sartorius A, Kiening KL, Kirsch P, von Gall CC, Haberkorn U, Unterberg AW, Henn FA, Meyer-Lindenberg A. Remission of major depression under deep brain stimulation of the lateral habenula in a therapy-refractory patient. Biol Psychiatry 2010;67:e9–e11. [DOI] [PubMed] [Google Scholar]
- [90].Seth P, Scholl L, Rudd RA, Bacon S. Overdose deaths involving opioids, cocaine, and psychostimulants-United States, 2015–2016. Am J Transplant 2018;18:1556–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Simundic AM. Measures of diagnostic accuracy: basic definitions. EJIFCC 2009;19:203–11. [PMC free article] [PubMed] [Google Scholar]
- [92].Smith SB, Mir E, Bair E, Slade GD, Dubner R, Fillingim RB, Greenspan JD, Ohrbach R, Knott C, Weir B, Maixner W, Diatchenko L. Genetic variants associated with development of TMD and its intermediate phenotypes: the genetic architecture of TMD in the OPPERA prospective cohort study. J Pain 2013;14:T91–101 e101–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Substance-Abuse-and-Mental-Health-Services-Administration-(US);-Office-of-the-Surgeon-General-(US). Facing addiction in America: the surgeon general's report on alcohol, drugs, and health. Facing addiction in America: the surgeon general's report on alcohol, drugs, and health. Washington, 2016. [PubMed] [Google Scholar]
- [94].Substance Abuse and Mental Health Services Administration CfBHSaQ. Results from the 2015 National survey on drug use and health: Detailed tables. Rockville, 2016. [PubMed] [Google Scholar]
- [95].Sullivan MD, Cahana A, Derbyshire S, Loeser JD. What does it mean to call chronic pain a brain disease? J Pain 2013;14:317–22. [DOI] [PubMed] [Google Scholar]
- [96].Sutcliffe S, Bradley CS, Clemens JQ, James AS, Konkle KS, Kreder KJ, Lai HHH, Mackey SC, Ashe-McNalley CP, Rodriguez LV. Urological chronic pelvic pain syndrome flares and their impact: qualitative analysis in the MAPP network. Int Urogynecol J 2015;26:1047–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Sutcliffe S, Jemielita T, Lai HH, Andriole GL, Bradley CS, Clemens JQ, Gallop R, Hooton TM, Kreder KJ, Krieger JN, Kusek JW, Labus J, Lucia MS, Mackey S, Naliboff BD, Robinson NA, Rodriguez LV, Stephens-Shields A, van Bokhoven A, Wolin KY, Yan Y, Yang CC, Landis JR, Colditz GA, Network MR. A case-crossover study of urological chronic pelvic pain syndrome flare triggers in the MAPP research network. J Urol 2018;199:1245–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Tracey I, Woolf CJ, Andrews NA. Composite pain biomarker signatures for objective assessment and effective treatment. Neuron 2019;101:783–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].U.S. Department of Health and Human Services FaDAF, (CDER), (CBER), (CDRH). Guidance for industry: enrichment strategies for clinical trials to support approval of human drugs and biological products. In: FDA, editor. Rockville: U.S. Department of Health and Human Services Food and Drug Administration (FDA), 2012. p. 1–42. [Google Scholar]
- [100].Ung H, Brown JE, Johnson KA, Younger J, Hush J, Mackey S. Multivariate classification of structural MRI data detects chronic low back pain. Cereb Cortex 2014;24:1037–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TE, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss SW, Della Penna S, Feinberg D, Glasser MF, Harel N, Heath AC, Larson-Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen SE, Prior F, Schlaggar BL, Smith SM, Snyder AZ, Xu J, Yacoub E; Consortium WU-MH. The Human Connectome Project: a data acquisition perspective. Neuroimage 2012;62:2222–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Von Korff M, Scher AI, Helmick C, Carter-Pokras O, Dodick DW, Goulet J, Hamill-Ruth R, LeResche L, Porter L, Tait R, Terman G, Veasley C, Mackey S. United States national pain strategy for population research: concepts, definitions, and pilot data. J Pain 2016;17:1068–80. [DOI] [PubMed] [Google Scholar]
- [103].Wager TD, Atlas LY, Lindquist MA, Roy M, Woo CW, Kross E. An fMRI-based neurologic signature of physical pain. N Engl J Med 2013;368:1388–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Wagner JA, Ball JR. Implications of the institute of medicine report: evaluation of biomarkers and surrogate endpoints in chronic disease. Clin Pharmacol Ther 2015;98:12–15. [DOI] [PubMed] [Google Scholar]
- [105].Wagner JA, Williams SA, Webster CJ. Biomarkers and surrogate end points for fit-for-purpose development and regulatory evaluation of new drugs. Clin Pharmacol Ther 2007;81:104–7. [DOI] [PubMed] [Google Scholar]
- [106].Wailoo K. Drawing blood: Technology and disease identity in twentieth-century America. Baltimore: Johns Hopkins University Press. [Google Scholar]
- [107].Wanigasekera V, Mezue M, Andersson J, Kong Y, Tracey I. Disambiguating pharmacodynamic efficacy from behavior with neuroimaging: implications for analgesic drug development. Anesthesiology 2016;124:159–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Weber KA, II, Sentis AI, Bernadel-Huey ON, Chen Y, Wang X, Parrish TB, Mackey S. Thermal stimulation alters cervical spinal cord functional connectivity in humans. Neuroscience 2018;369:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Woo CW, Chang LJ, Lindquist MA, Wager TD. Building better biomarkers: brain models in translational neuroimaging. Nat Neurosci 2017;20:365–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Woo CW, Roy M, Buhle JT, Wager TD. Distinct brain systems mediate the effects of nociceptive input and self-regulation on pain. Plos Biol 2015;13:e1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Woo CW, Schmidt L, Krishnan A, Jepma M, Roy M, Lindquist MA, Atlas LY, Wager TD. Quantifying cerebral contributions to pain beyond nociception. Nat Commun 2017;8:14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Woo CW, Wager TD. Neuroimaging-based biomarker discovery and validation. PAIN 2015;156:1379–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Woodworth D, Mayer E, Leu K, Ashe-McNalley C, Naliboff BD, Labus JS, Tillisch K, Kutch JJ, Farmer MA, Apkarian AV, Johnson KA, Mackey S, Ness T, Landis J, Deutsch G, Harris R, Clauw D, Mullins C, Ellingson BM. Unique microstructural changes in the brain associated with urological chronic pelvic pain syndrome (UCPPS) revealed by diffusion tensor MRI, super-resolution track density imaging, and statistical parameter mapping: a MAPP network neuroimaging study. PLoS One 2015;10:e0140250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Zhong J, Chen DQ, Hung PS, Hayes DJ, Liang KE, Davis KD, Hodaie M. Multivariate pattern classification of brain white matter connectivity predicts classic trigeminal neuralgia. PAIN 2018;159:2076–87. [DOI] [PubMed] [Google Scholar]
- [115].Zimmern RL, Kroese M. The evaluation of genetic tests. J Public Health (Oxf) 2007;29:246–50. [DOI] [PubMed] [Google Scholar]