Summary
The mucosa is colonized with commensal Neisseria. Some of these niches are sites of infection for the STD pathogen Neisseria gonorrhoeae (Ngo). Given the antagonistic behavior of commensal bacteria towards their pathogenic relatives, we hypothesized that commensal Neisseria may negatively affect Ngo colonization. Here, we report that commensal species of Neisseria kill Ngo through a mechanism based on genetic competence and DNA methylation state. Specifically, commensal-triggered killing occurs when the pathogen takes up commensal DNA that contains a methylation pattern it does not recognize. Indeed, any DNA will kill Ngo if it can enter the cell, is differentially methylated, and has homology to the pathogen genome. Consistent with these findings, commensal Neisseria elongata accelerates Ngo clearance from the mouse in a DNA uptake-dependent manner. Collectively, we propose that commensal Neisseria antagonizes Ngo infection through a DNA-mediated mechanism, and that DNA is a potential microbicide against this highly drug resistant pathogen.
Graphical Abstract
eTOC Blurb
Kim et al report that the STD pathogen Neisseria gonorrhoeae is killed when it takes up DNA released by related commensal bacteria. Neisseria gonorrhoeae employs the DNA uptake system to obtain plasmids conferring antibiotic resistance and immune evasion tactics, thereby suggesting that this system may be the pathogen’s Achilles Heel.
Introduction
Our normal flora (commensals, mutualists) is essential for bodily function. It provides us with essential nutrients and maintains homeostasis of many metabolic processes (An et al., 2014; Backhed et al., 2004; Brestoff and Artis, 2013; Flint and Eskin, 2012; Hooper et al., 2002; Ley et al., 2006; Musso et al., 2011; Nicholson et al., 2012; Tremaroli and Backhed, 2012). It is essential for the development of the gut and immune system (Abt et al., 2012; Belkaid and Hand, 2014; Brestoff and Artis, 2013; Carabotti et al., 2015; Diaz Heijtz et al., 2011; Ivanov et al., 2009; Sommer and Backhed, 2013).
The microbial flora, hereafter termed commensals, also protects the body against colonization by pathogenic bacteria. The probiotic Escherichia coli strain Nissle 1917 outcompetes pathogenic E. coli O157:H7 for carbohydrates and Salmonella Typhimurium for iron (Deriu et al., 2013; Maltby et al., 2013). Staphylococcus epidermidis secretes a protease that inhibits biofilm formation and nasal colonization by Staphylococcus aureus, and Staphylococcus lugdenensis produces an antibiotic peptide that inhibits Staphylococcus aureus colonization in a mouse model of skin infection (Iwase et al., 2010; Zipperer et al., 2016). The presence of Neisseria lactamica in the nasal cavity is correlated with a reduced risk of infection by Neisseria meningitidis, possibly through inducing cross-reactive antibodies (Cartwright et al., 1987; Deasy et al., 2015; Evans et al., 2011; Trotter et al., 2007).
Over 12 species of commensal Neisseria colonize many regions of the human mucosa, including the nasopharynx, endocervix and rectum (Diallo et al., 2016; Knapp and Clark, 1984; Liu et al., 2015). As these sites are occasionally infected by the sexually transmitted pathogen Neisseria gonorrhoeae (Ngo), and in light of the fact that some commensals inhibit colonization of related pathogenic species, we tested the hypothesis that commensal Neisseria antagonize Ngo in vitro and in vivo, using commensal Neisseria elongata (Nel) and a mouse model of Ngo cervico-vaginal (lower genital tract) infection (Jerse, 1999; Jerse et al., 2011).
We report that Nel significantly reduces the viability of Ngo in vitro. The toxic agent is Nel DNA in the supernate. Ngo, a genetically competent organism, is killed when it takes up Nel DNA; Ngo mutants defective in DNA uptake are resistant to killing. Lines of evidence demonstrate that any DNA will kill Ngo, provided that it can enter the cell and its methylation pattern differs from the one in pathogen DNA. Ngo lower genital tract infection can be modeled in the BALB/c mouse (Jerse, 1999; Jerse et al., 2011). Consistent with these in vitro findings, Nel accelerates the clearance of Ngo from mice and a DNA uptake mutant of Ngo is resistant to accelerated clearance. Finally, we show that commensal Neisseria DNA also kills the pathogen N. meningitidis. Our findings reveal a weapon in bacterial interspecies warfare that is based on genetic competence and DNA methylation state. They highlight the potential of DNA as an antimicrobial agent for treating Ngo, a pathogen that has rapidly become resistant to nearly all the antibiotics used clinically for its treatment (Tomberg et al., 2010; Unemo et al., 2012; Unemo and Nicholas, 2012; Unemo and Shafer, 2014).
Results
Nel interacts physically with Ngo and kills the pathogen in vitro
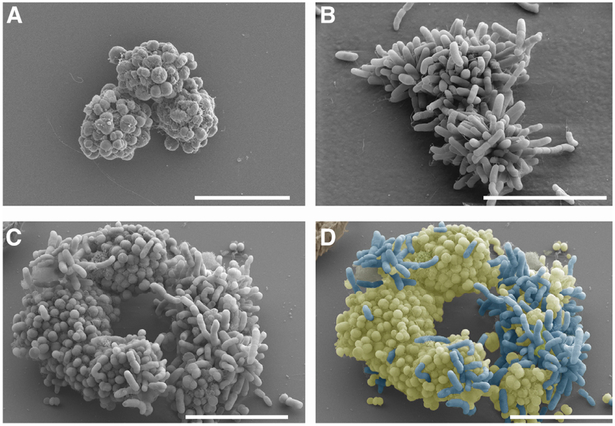
Cultured alone, piliated Ngo and Nel cells actively aggregate into biofilm precursors known as microcolonies (Higashi et al., 2011; Merz et al., 1999). To determine whether commensal and pathogen physically interact, planktonic Nel 29315 and Ngo MS11 (Table S1) cells were cultured alone or together on coverslips for 5 h, and viewed by Scanning Electron Microscopy (SEM). Representative images are shown in Fig 1. The coccoid Ngo cells formed microcolonies with members of its own species (Fig 1 A). The majority of Nel cells, which are short rods, behaved similarly (Fig 1 B). Nel and Ngo microcolonies often abutted each other, and solitary Nel and Ngo cells were occasionally seen attached to or partially inserted into microcolonies of the other species (Fig 1 C, D). How commensal and pathogen differentiate each other is unknown.
Fig 1. Nel and Ngo microcolonies associate with each other.
Scanning electron micrograph of (a) Ngo and (b) Nel cultured alone, and (c, d) Ngo and Nel cultured together for 5 h. The image in (c) was pseudocolored to discriminate Ngo (cocci; yellow) from Nel (rods; blue). Starting CFUs of each species: 5×107. Scale bars: 10 μM.
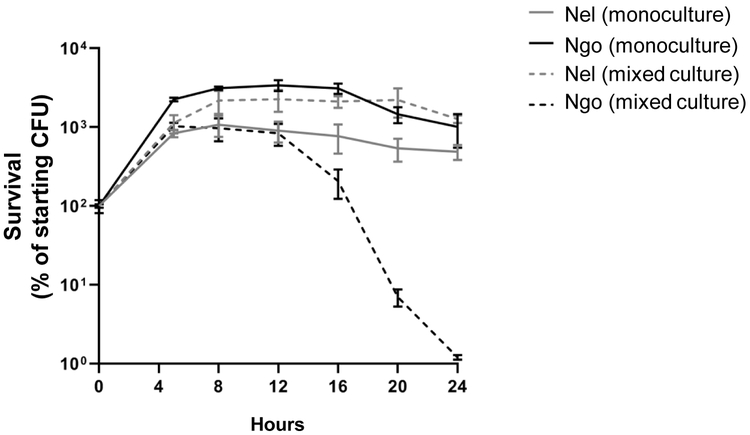
Neither Nel 29315 nor Ngo MS11 lost viability when the two species are co-cultured for short periods (8 h) (Higashi et al., 2011). To determine whether Nel and Ngo affected each other long term, commensal and pathogen were cultured alone or together, for 24 h and their CFUs were determined by plating on selective agar. As reported, Nel CFUs in the monoculture reached stationary phase at 6 to 8 h (Rendon et al., 2013). Its growth was unaffected by the presence of Ngo (Fig 2). Ngo CFUs in the monoculture peaked at a similar time. In the presence of Nel, however, Ngo viable counts began to decline at 16 h; by 24 h they were 3 logs lower than those in the monoculture.
Fig 2. Ngo loses viability during prolonged co-culture with Nel.
Ngo and Nel were cultured alone (~5×107 starting CFU) or together (~5×107 CFU each organism) in 6-well microtiter plates. At the indicated times, CFUs of each organism was determined by plating on selective agar. n=3. Error bars: SEM. Limit of detection (LOD): 10 CFUs. Under these conditions, Nel divides approximately every hour, with growth peaking at 6-8 h (Tonjum et al., 1995).
The experiment was repeated with Ngo strains D006 and D020, low passage strains isolated relatively recently from patients attending a county health department STD clinic (Fig S1). In the presence of Nel, D006, D020 and MS11 CFUs were significantly reduced compared to CFUs recovered from monocultures. Like MS11, D006 and D020 did not affect Nel viability. These results show Nel kills lab and fresh isolates of Ngo.
Nel accelerates the clearance of Ngo in a mouse model of lower genital tract infection
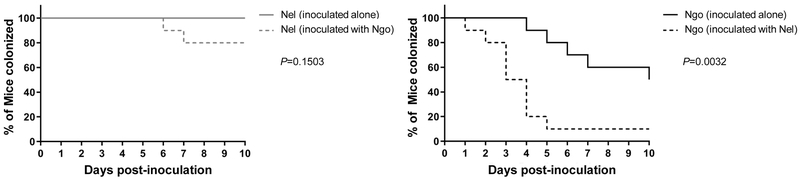
To determine whether Nel affects Ngo colonization in vivo, an experiment was performed using the female BALB/c mouse model of lower genital tract infection (Jerse, 1999; Jerse et al., 2011). Nel and Ngo were inoculated into the vagina of BALB/c mice either alone or mixed in equal numbers (Fig 3). Commensal and pathogen CFUs were determined daily by plating vaginal swabs on selective agar. Nel colonized 80-100% of mice for 10 consecutive days following inoculation, with no statistical difference in the percent of mice colonized or number of Nel recovered from mice that were or were not co-inoculated with Ngo (Fig 3, left panel). When inoculated alone, Ngo colonized 80% of the mice for 5 days and then declined to 50% by day 10. However, the presence of Nel significantly accelerated the clearance of Ngo from the animals (Fig 3, right panel). In mice inoculated with Nel and Ngo, 50% of the animals were culture-positive for Ngo on day 4, compared to 90% of animals inoculated only with Ngo. By day 10, only 10% of mice in the Nel+Ngo group were colonized with Ngo compared to 50% of mice inoculated with Ngo alone (P=0.0032; log-rank test). The average daily burdens of Nel and Ngo in the vagina, shown in Fig S2A-B, also illustrate the reduction in Ngo burden in co-inoculated mice (P=0.0013, repeated measures ANOVA). Nel and Ngo CFUs recovered from individual mice from each experimental group are shown in Fig S2C-F.
Fig 3. Nel accelerates the clearance of WT Ngo from mice.
Female BALB/c mice were inoculated vaginally with 106 CFUs of Nel, Ngo, or a mixture containing 106 CFU of each species. Vaginal swabs were plated on selective agar for 7 consecutive days to quantitate Ngo and Nel CFUs. Y-axis shows the percentage of mice culture-positive for Nel (left panel) and Ngo (right panel) at each time point, plotted as Kaplan Meier curves and analyzed by the Log Rank test. n= 10 mice/group.
Nel kills Ngo through DNA it released into the medium
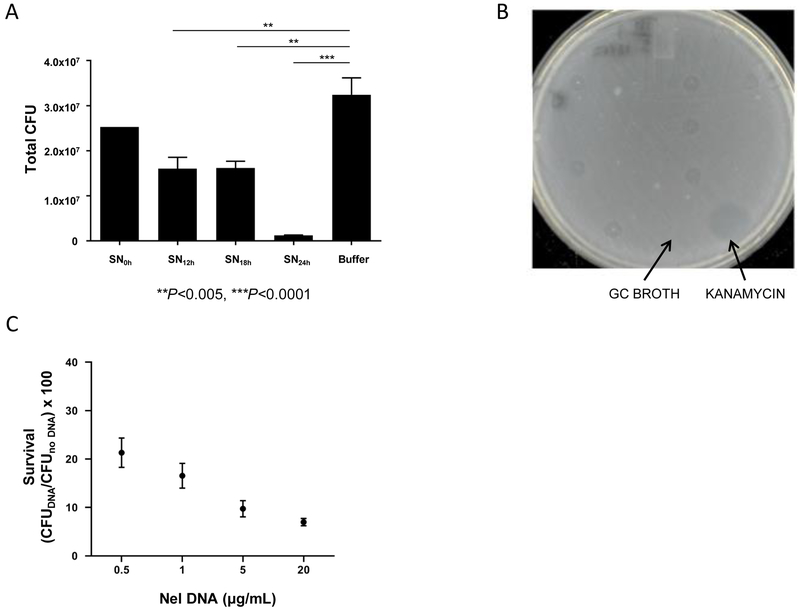
To determine whether Nel killing of Ngo requires cell contact, Ngo CFUs were determined after a 5-h incubation with Nel cell-free supernatants (SN) (Fig 4A). SN from 12 h and 18 h cultures significantly reduced Ngo viability compared to the GCB medium control (P<0.005, One-way ANOVA with Tukey’s Multiple Comparison Test). The 24-h supernatant had the most dramatic effect, reducing Ngo CFUs by 27-fold (P<0.0001, One-way ANOVA with Tukey's Multiple Comparison Test). This suggests killing occurred through a component(s) that Nel released into the medium. The adverse effect of Nel supernatants on Ngo was unlikely to be caused by nutrient depletion, as the supernatants were diluted with an equal volume of fresh medium before the assay.
Fig 4. Nel DNA kills Ngo.
(A) Ngo CFUs recovered after a 5 h incubation with cell-free Nel supernates (SN) harvested at the indicated times from liquid cultures. n=3. Error bars: SEM. (**P<0.005, ***P<0.0001; One-way ANOVA with Tukey’s Multiple Comparison Test). Nel DNA concentration in 16, 20, and 24 h SN were 0.31, 0.81, and 2.72 μg/mL, respectively (n=1, see Materials and Methods).
(B) A representative agar plate from a spot assay used to identify the toxic component in cell-free Nel supernates. Undiluted Nel supernates (5 μL each) were spotted onto a lawn of Ngo cells. Clearance, or kill, zones on the lawn after overnight incubation served as the readout for toxicity of the sample for Ngo. Negative control: GC broth. Positive control: Kanamycin (20 μg/mL).
(C) CFUs of Ngo recovered after a 4 h incubation with purified Nel DNA at the indicated final concentrations. Survival is calculated as CFUs of Ngo incubated with DNA normalized to CFUs of Ngo incubated with sterile buffer. Starting Ngo CFU: 5×105. n=4. Error bars represent SEM. LOD: 10 CFUs.
A spot assay (Dillard, 2011) was developed to provide a more rapid means of identifying the toxic compound(s) in Nel supernatants. Ngo cells were spread over an agar plate; supernatants were spotted on the lawn and the plate was incubated overnight. A zone of clearance on the lawn served as the readout for toxicity (Fig 4B). Kanamycin and GC broth served as the positive and negative controls, respectively.
Supernatants from a 24-h culture of Nel were digested with various enzymes and tested for toxicity to Ngo using the spot assay. DNAse I abolished the toxicity of the supernatant for Ngo, while heat-inactivated DNAse I, and native and heat-inactivated Proteinase K and RNAse were still toxic (Table S2). This suggests that DNA is the killing agent. To verify that Nel DNA is the toxic component, Ngo was incubated with purified Nel chromosomal DNA, and its CFUs determined. Results from this liquid assay show Nel DNA killed Ngo in a dose-dependent manner (Fig 4C).
Ngo DNA uptake/transformation mutants resist killing by Nel DNA
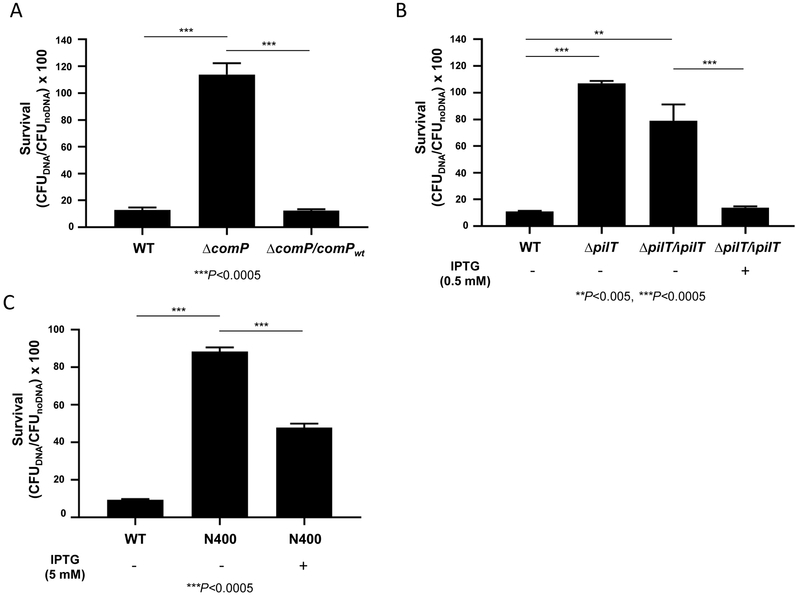
We tested the hypothesis that Nel DNA kills Ngo when it is taken up by the pathogen. All Neisseria species examined to date are naturally competent and readily take up DNA in a sequence-specific manner. DNA uptake involves the binding of ComP, a Type IV pilus-associated protein, to a 10-base pair DNA Uptake Sequence (DUS10; 5' GCCGTCTGAA 3') that is abundant in Neisseria genomes; in Ngo there is ~1 DUS/kb of DNA (Berry et al., 2013; Elkins et al., 1991a; Marri et al., 2010). DNA uptake also requires the Type IV pilus retraction motor PilT (Craig and Li, 2008; Merz et al., 2000; Wolfgang et al., 1998a). Ngo ΔcomP and ΔpilT transform at 3 to 5 log lower frequency than the WT parent (Wolfgang et al., 1998b; Wolfgang et al., 1999). In the spot assay, Ngo ΔcomP was resistant to Nel DNA killing, while the complemented ΔcomP strain was as sensitive to killing as the WT parent (Table S3). In the liquid assay, all Ngo ΔcomP, ΔpilT, and 79% of uninduced ΔpilT/ipilT cells survived exposure to Nel DNA. By contrast, only 11-13% of WT Ngo cells, 12% of complemented comP cells and 14% of induced ΔpilT/ipilT cells survived Nel DNA killing (P<0.0005 WT vs ΔcomP, ΔcomP vs ΔcomP/comPwt, WT vs ΔpilT and uninduced ΔpilT/ipilT vs IPTG induced ΔpilT/ipilT, One-way ANOVA with Tukey's Multiple Comparison Test) (Fig 5A, B).
Fig 5. Ngo competence mutants resist killing by Nel DNA.
CFUs of Ngo DNA uptake/transformation mutants incubated for 4 h with purified Nel DNA (20 μg/mL). WT: Ngo MS11. (A) ΔcomP: isogenic DNA uptake mutant deleted of the comP ORF; ΔcomP/comPWT: complemented ΔcomP strain. (B) ΔpilT: isogenic DNA uptake mutant deleted of the pilT ORF; ΔpilT/ipilT: ΔpilT complemented with WT pilT under control of the IPTG-inducible lac promoter, mock-induced with medium or induced with IPTG during overnight growth on agar and during the liquid killing assay. (C) N400: Ngo MS11 containing recA under control of the IPTG-inducible lac promoter (Tonjum et al., 1995) mock-induced with medium or induced with IPTG during overnight growth on agar and during the liquid killing assay. (A-C) Starting CFU: 5×105. n=3-4 (P<0.0005 WT vs ΔcomP; ΔcomP vs ΔcomP/comPWT; WT vs ΔpilT; uninduced ΔpilT/ipilT vs IPTG-induced ΔpilT/ipilT; WT vs mock-induced N400; mock-induced N400 vs IPTG-induced N400; One-way ANOVA with Tukey’s Multiple Comparison Test). LOD: 10 CFUs.
DNA taken up by Neisseria recombines with homologous sequences in the genome in a RecA-dependent manner, and Ngo ΔrecA is deficient in DNA transformation (Koomey and Falkow, 1987; Koomey et al., 1987). We determined whether recA affected the sensitivity of Ngo to DNA killing, using strain N400, in which recA is controlled by the IPTG-inducible lac promoter (Tonjum et al., 1995). N400 survived DNA killing in the absence of IPTG, and became sensitive in the presence of IPTG (P<0.0005 WT vs uninduced N400, and uninduced N400 vs IPTG-induced N400; One-way ANOVA with Tukey’s Multiple Comparison Test) (Fig 5C). The resistance of N400, ΔcomP, and ΔpilT to DNA killing strongly suggests that the killing mechanism involves the DNA uptake and transformation machinery. They argue against a scenario in which DNA mediated the clumping of Ngo cells.
Ngo ΔcomP resists Nel-accelerated clearance from the mouse
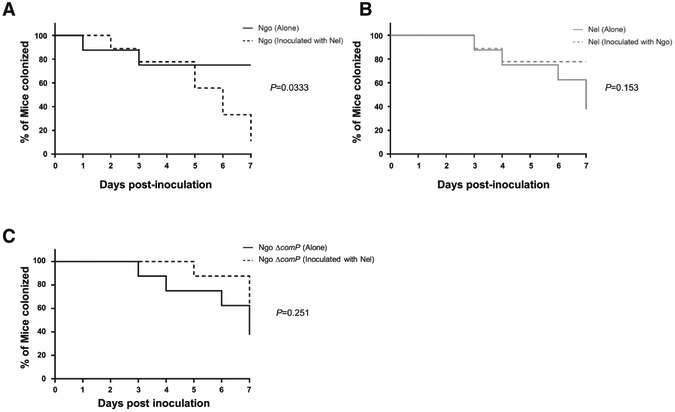
Ngo ΔcomP was tested for its ability to resist Nel clearance from the lower genital tract of the mouse. ΔcomP was inoculated into the vagina alone or with Nel in equal numbers. CFUs of each strain were quantitated daily. As observed above, Nel accelerated the clearance of WT Ngo (P=0.0333, log-rank test) (Fig 6A). Furthermore, the duration of recovery of Nel from the mouse vagina was similar regardless of whether it was inoculated alone or together with Ngo, indicating Ngo did not influence Nel colonization (P=0.153; log-rank test) (Fig 6B). In contrast, Nel did not accelerate the clearance of Ngo ΔcomP (P=0.251, log-rank test) (Fig 6C), strongly suggesting that Nel accelerates Ngo clearance only if the pathogen is competent for DNA uptake. The average daily burdens of Nel and Ngo in the vagina (Fig S3 A-D) also shows a lower Ngo burden in Ngo+Nel co-inoculated mice but not in NgoΔcomP+Nel co-inoculated mice, although the difference in the average number of Ngo recovered from mice inoculated with only Ngo or Ngo+Nel approached but did not achieve significance (P=0.07, repeated measures ANOVA). This result may be due to animal variability within the Ngo+Nel group (Fig S3 E-J), with one mouse highly colonized throughout the experiment (Fig S3, panel I).
Fig 6. Nel does not accelerate the clearance of competence mutant ΔcomP from mice.
Female BALB/c mice were inoculated vaginally with 106 CFUs of Nel, Ngo ΔcomP, or a mixture containing 106 CFU of each species. Vaginal swabs were plated on selective agar for 7 consecutive days to quantitate Ngo and Nel CFUs. Y-axis shows the percentage of mice culture-positive for each strain over 7 days. (A) WT Ngo CFUs. (B) Nel CFUs (C) Ngo ΔcomP. n= 7-9 mice/group. (Data were analyzed by the Log-rank test).
Nel DNA does not kill Ngo via a "toxic" locus
To determine whether Nel DNA toxicity is conferred by a specific gene/genetic element, a library of Bacterial Artificial Chromosomes (BACs) containing 20-50 kb inserts of Nel DNA was constructed in E. coli using the vector pBeloBAC11. The inserts were sequenced and verified by restriction analysis, and DNAs from several BACs with unique inserts were tested for their ability to kill Ngo in the liquid assay. All tested BAC DNAs, but not empty vector, were toxic to Ngo (Table S4). Overlapping fragments of the insert in the BAC clone pBeloBAC11(6.1), subcloned into pUC19, also killed Ngo. It is unlikely that there is a toxic locus in all tested BACs and pBeloBAC11(6.1) subclones, because these clones did not have sequences in common except for the DUS.
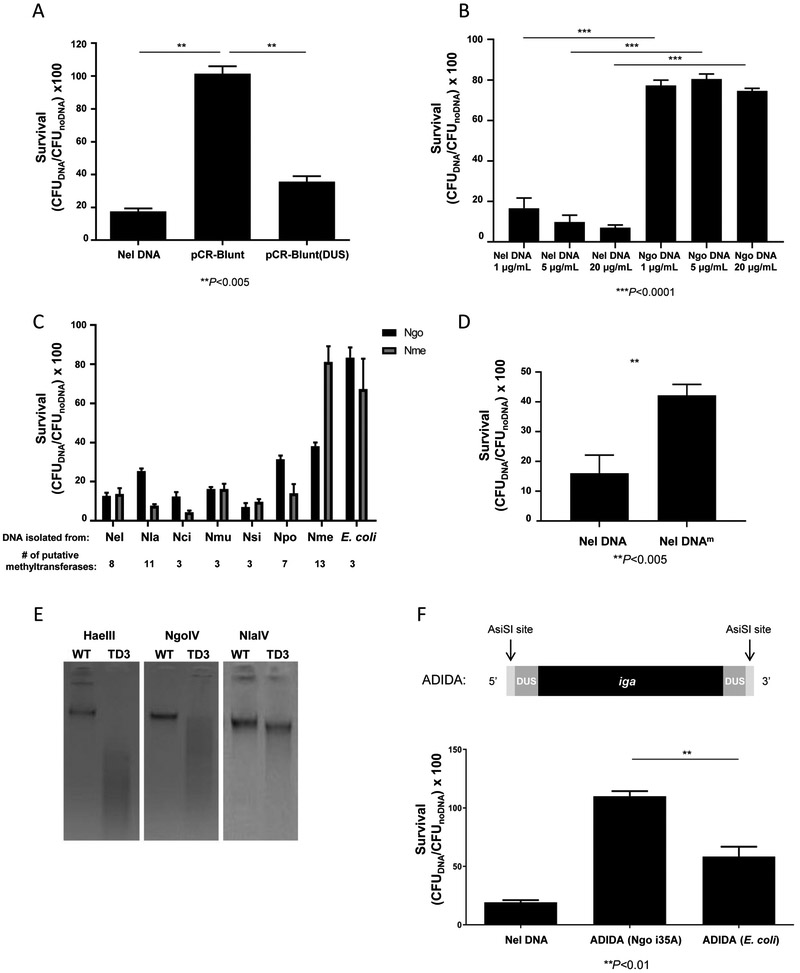
We determined whether the DUS is a toxic sequence. Analysis of Ngo and N. meningitidis (Nme) genomes showed that they have an extended DUS (DUS12; 5' ATGCCGTCTGAA 3'). DUS10 and DUS12 provide a similar function in DNA uptake but DUS12 confers a slight increase in the transformation efficiency of Ngo and Nme (Ambur et al., 2007; Duffin and Seifert, 2010). Thus, DUS12 was used in our assays. E. coli plasmid pCR-Blunt DNA did not kill Ngo unless it contained a DUS12 (Fig 7A). DUS10 and DUS12 are abundant in Neisseria genomes (Ambur et al., 2007; Marri et al., 2010); in the Ngo chromosome, there is ~1 DUS10 and ~0.7 DUS12 per kb of DNA. Yet, Ngo is not killed when incubated with its own DNA (Fig 7B). Taken in context with the BAC cloning data, these results strongly suggest that the DUS is not a toxic sequence.
Fig 7. Ngo is killed when it takes up DNA with a different methylation pattern.
(A) CFUs of Ngo recovered after a 4 h incubation with Nel chromosomal DNA and DNA from plasmids pCR-Blunt and pCR-Blunt (DUS) replicated in E. coli DH5α (5 μg/mL). Survival is calculated as CFU of Ngo incubated with DNA normalized to CFU of Ngo incubated with buffer. Starting Ngo CFU: 5×105. n=3. Error bars: SEM. (**P<0.005; Student’s t-test). LOD: 10 CFUs.
(B) CFUs of Ngo recovered after a 4 h incubation with Nel or Ngo DNA at the final concentrations indicated. Starting Ngo CFU: 5×105 n=3. Error bars: SEM. (***P<0.0001; Student’s t-test). LOD: 10 CFUs.
(C) CFUs of Ngo recovered after a 4 h incubation with DNA (5 μg/mL) from E. coli and the Neisseria species indicated. Nla: N. lactamica, Nci: N. cinerea, Nmu: N. mucosa, Nsi: N. sicca, Npo: N. polysaccharea, and Nme: N. meningitidis. n=3. LOD: 10 CFUs.
(D) CFUs of Ngo recovered after a 4 h incubation with 5 μg/mL of unmodified Nel DNA or Nel DNA whose cytosines were methylated at GpC and CpG sequences using M.CviPI and M.SssI methyltransferases (Nel DNAm). n=3. Error bars: SEM. (A-D): Starting Ngo CFU: 5×105 (**P<0.005; Student’s t-test). LOD: 10 CFUs.
(E) Ngo WT and TD3 DNA digested with HaeIII, NgoIV, and NlaIV restriction enzymes that cleave unmethylated GGCC, GCCGGC, and GGNNCC sequences, respectively.
(F) CFUs of Ngo recovered after a 4 h incubation with (left to right) Nel chromosomal DNA (20 μg/mL), ADIDA replicated in Ngo i35A, and ADIDA replicated in E. coli (1 μg/mL each). Percent survival was calculated as noted. Starting CFU: 5×104. Error bar: SEM. (**P<0.01; Student’s t-test). LOD: 10 CFUs.
The methylation state of the DNA determines its toxicity for Ngo
The lack of evidence for a toxic locus in Nel DNA led us to determine whether an epigenetic feature of the DNA is responsible for killing Ngo. Bacteria modify their DNA by means of methyltransferases that covalently link methyl groups to bases in specific sequences (Noyer-Weidner M., 1992). Among bacteria, the Ngo genome is one of the most heavily methylated; Ngo strains encode/express 14-19 DNA methyltransferases (Blow et al., 2016; Roberts et al., 2015; Stein et al., 1995) (http://tools.neb.com/genomes/index.php?page=N). PacBio Single Molecule, Real-Time (SMRT) sequencing of Ngo FA1090 DNA revealed the methylation patterns consistent with the activity of 10 methyltransferases (Blow et al., 2016; Roberts et al., 2015). In Ngo MS11, the activity of at least 7 of these methyltransferases has been demonstrated experimentally (Gunn et al., 1992) (Table S5). Commensal species of Neisseria, by contrast, encode fewer DNA methyltransferases. In particular, Nel isolates encode 7-10 predicted methyltransferases. Single Molecule, Real-Time (SMRT) sequencing of Nel 29315 DNA indicates 3 methyltransferases are active in this strain (Table S5). These methyltransferases modify sequence motifs that are not modified in Ngo (Table S5). These findings indicate Ngo and Nel DNA have distinct methylation patterns.
That commensal Neisseria encode fewer DNA methyltransferases than Ngo (Roberts et al., 2015)(http://tools.neb.com/genomes/index.php?page=N) led us to test their chromosomal DNA for toxicity to Ngo. DNA from N. lactamica (Nla), N. cinerea (Nci), N. mucosa (Nmu), N. sicca (Nsi), N. polysaccharea (Npo), and Nme all significantly reduced Ngo viability (Fig 7C). The putative number of DNA methyltransferases specific to the Neisseria strains used in this study is listed under the strain names. E. coli DH5α expresses only 3 methyltransferases (Table S5). In contrast to commensal Neisseria DNA however, E. coli DNA, with 1000-fold fewer copies of the DUS, did not affect Ngo viability. These results are consistent with the hypothesis that Ngo is killed when it takes up DNA whose methylation state is different from its own.
We determined whether modifying Nel DNA to partially mimic the Ngo methylation pattern would abolish its toxicity for Ngo. Several Ngo DNA methyltransferases modify cytosines in CpG and GpC motifs (Table S5). The cytosines in CpG and GpC sequences in Nel DNA were methylated in vitro using M.CviPI and M.Sssl, respectively; methylation was verified by confirming the resistance of the modified DNA to digestion by the cognate restriction enzymes (Fig S4). Modified DNA was significantly less toxic to Ngo than unmodified DNA (P<0.005; Student's t-test, Fig 7D). This suggests the toxicity of DNA is determined, at least in part, by differences in the methylation state of the incoming DNA and the recipient Ngo cell.
Ngo is not killed when incubated with its own DNA (Fig. 7B). If the methylation state of the incoming DNA is at the root of the toxicity, then undermethylated Ngo DNA would kill more efficiently than native Ngo DNA. To test this hypothesis, we constructed Ngo TD3, a mutant deleted of the ngoII, ngoIV, and ngoV restriction/modification (R/M) loci. The mutations were verified by sequencing, and loss of methyltransferase activity was verified by confirming the susceptibility of TD3 DNA to digestion with the cognate restriction enzymes (Fig 7E). As expected, HaeIII (an isoschizomer of NgoII, with 4783 sites in Ngo genome) digested TD3 DNA into smaller fragments than NgoIV (1600 sites) or NlaIV (an isoschizomer of NgoV, 1937 sites) (Fig 7E). In addition, the partial digestion of TD3 by NlaIV is likely explained by the activity of methyltransferase NgoIII, which modifies sequences within the NlaIV site (Table S5, Fig 7E). TD3 DNA was slightly more toxic to Ngo than WT DNA (Ngo survival in the presence of WT Ngo DNA: 89.7% +/− 2.10 SEM; of TD3 DNA: 79.8% +/− 1.34 SEM; P<0.0182, Student's t-test). In TD3, only 3 R/M loci were deleted, which likely explains the mild difference in toxicity of its DNA.
As a final test of the importance of methylation in determining the toxicity of DNA for Ngo, a DNA fragment of Ngo origin with either an E. coli or Ngo methylation signature was tested for toxicity to Ngo. The 4kb iga gene in Ngo was modified by insertion of a recognition site for restriction enzyme AsiSI and a DUS12 into its 5' and 3' ends (Fig 7F, top panel). In this mutant, Ngo i35A, the order of this locus is 5' AsiSI-DUS-iga-DUS-AsiSI 3' (ADIDA for short). There are few AsiSI sites in the WT Ngo chromosome; the smallest AsiSI fragment is >15kb. In i35A, the smallest AsiSI fragment is the ~4kb ADIDA. i35A DNA was digested with AsiSI and separated in an agarose gel; DNA migrating at 4 kb, which contains only ADIDA, was purified.
Concurrently, ADIDA was replicated in E. coli DH5α, which expresses 3 DNA methyltransferases (Table S5), and the insert was gel purified. ADIDA (E. coli) and ADIDA (Ngo) would have distinct methylation patterns since their hosts express DNA methylases with different specificities. This was verified by restriction analysis. iga contains NgoI and NgoV recognition sites. As expected, the ADIDA (E. coli) was susceptible to digestion by HaeII and NlaIV (isoschizomers of NgoI and NgoV), whereas ADIDA (Ngo) was resistant to restriction (Fig S5).
ADIDA with E. coli and i35A methylation signatures were evaluated for their ability to kill Ngo. ADIDA (E. coli) killed Ngo significantly more efficiently than ADIDA (Ngo i35A) (P<0.01; Student's t-test; Fig 7F, bottom panel). ADIDA (E. coli) did not kill Ngo as efficiently as Nel chromosomal DNA, the positive control, most likely because it was used at a lower concentration in the assay and has a much lower sequence complexity than Nel chromosomal DNA (see model in Discussion). These experiments demonstrate that Ngo is killed when it takes up DNA with a methylation pattern it does not recognize.
Commensal DNA also kills N. meningitidis (Nme)
Nme, the only other Neisseria species that is pathogenic to humans, is also genetically competent and takes up DNA in a DUS-dependent manner (Rotman and Seifert, 2014). We determined whether commensal Neisseria DNAs are able to kill Nme 8013 in the liquid assay. Nel, Nla, Nci, Nmu, Nsi and Npo DNAs, which have fewer putative DNA methyltransferases than Nme DNA, significantly reduced the viability of this pathogen (Fig 7C, gray bars). Thus, the DNA methylation-based killing mechanism described for Ngo appears to operate in Nme as well.
Discussion
Commensals have been shown to inhibit the colonization of pathogens through protein-based killing mechanisms or outcompeting them for nutrients. Here, we presented evidence that commensal species of Neisseria kill pathogenic Neisseria through a mechanism that is based on genetic competence and DNA methylation state.
Commensal Neisseria elongata (Nel) kills pathogen Neisseria gonorrhoeae (Ngo) through DNA it releases into its surroundings. Ngo is a naturally competent organism that preferentially takes up DNA containing Neisseria-specific DNA Uptake Sequences DUS10 (5' GCCGTCTGAA 3') and DUS12 (5' ATGCCGTCTGAA 3') (Ambur et al., 2007; Duffin and Seifert, 2010; Elkins et al., 1991b; Goodman and Scocca, 1988; Graves et al., 1982). Ngo is killed when it takes up chromosomal DNA purified from Nel and other commensal Neisseria, and Ngo mutants defective in DNA uptake/transformation resist DNA killing. These findings indicate that DNA is toxic to Ngo, but only if it is taken up by the pathogen. Consistent with these in vitro findings, Nel accelerates the clearance of Ngo from the vagina of mice in a DNA uptake-dependent manner.
The Ngo genome is one of the most heavily methylated of bacterial genomes, encoding 14-19 predicted DNA methylases (Blow et al., 2016; Roberts et al., 2015; Stein et al., 1995) (http://tools.neb.com/genomes/index.php?page=N). Nel DNA modified to partially mimic the Ngo methylation pattern kills the pathogen less efficiently than unmodified DNA. Conversely, DNA from Ngo TD3, which is deleted of 3 of its 7 restriction/modification loci, is more toxic to Ngo than WT Ngo DNA. Importantly, an Ngo sequence (ADIDA) with an E. coli methylation pattern kills the pathogen significantly more efficiently than the same sequence with the Ngo methylation signature. These findings indicate that the toxicity of DNA for Ngo is determined, at least in part, by differences in the methylation state of the incoming DNA and the host genome. They demonstrate that any DNA will kill Ngo, provided it is taken up by the pathogen, and its methylation state differs from that of the recipient cell.
The resistance of the Ngo recA mutant to the bactericidal effect of DNA provides a clue to the killing mechanism. In prokaryotes, RecA plays a central role in homologous recombination, binding to single stranded (ss) DNA to form pre-synaptic filaments, searching for sequences homologous to the ssDNA, and initiating synapse formation at regions of homology (Lee et al., 2017; Renkawitz et al., 2014). Neisseria genomes have large regions of sequence homology; DNA taken up by Ngo recombines with their homologues in the chromosome in a RecA-dependent manner (Bennett et al., 2012; Koomey and Falkow, 1987; Koomey et al., 1987; Marri et al., 2010; Mehr and Seifert, 1998).
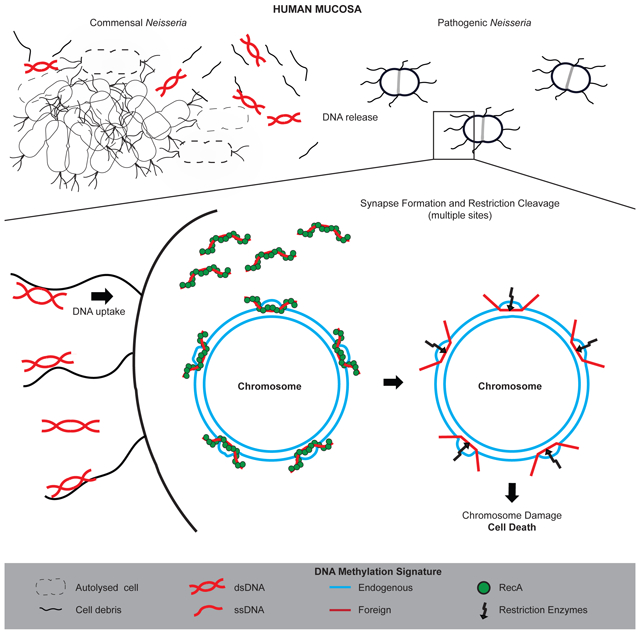
Based on these and other observations, we propose a model for how Ngo is killed when it takes up commensal Neisseria DNA. Commensal DNA is converted to ss form as it enters the Ngo cytoplasm (Chaussee and Hill, 1998). RecA binds to the ssDNA and begins to form synaptic joints at homologous sites in the chromosome (Lee et al., 2017; Renkawitz et al., 2014). In these structures, the commensal DNA would have a foreign methylation signature. Restriction enzyme(s) cleave at the duplexes with mismatched methylation, destroying chromosome integrity. Consistent with this model, all sources of toxic DNA identified in this study have multiple recognition sites for Ngo MS11 restriction/modification enzymes (Table S6).
Some restriction enzymes are able to cleave hemi-methylated DNA (Gruenbaum et al., 1981); whether Ngo restriction enzymes have the ability to cleave duplexes with mismatched methylation patterns remains to be tested. Restriction enzymes and their cognate modification systems must be tightly coordinated to preserve cell viability; in the few systems studied, control occurs at the transcriptional and posttranslational levels (Loenen et al., 2014a; Loenen et al., 2014b). Little is known about how restriction/modification systems are coordinated in Neisseria. We postulate that Ngo chromosome integrity is determined by a race between methylases and restriction enzymes at synapses with mismatched methylation patterns, and by the amount and sequence complexity of DNA taken into the cell, as synapses formed at numerous sites by these DNA strands would overwhelm repair enzymes.
How much DNA Ngo takes up in a short period of time is not known. The highly competent Ngo expresses multiple Tfp, so it is reasonable to assume that many if not all Tfps are capable of taking up DNA concurrently. Multiple segments of commensal Neisseria DNA homologous to different regions of the Ngo chromosome entering the cell would initiate recombination at multiple homologous sites in the chromosome, making these sites sensitive to restriction cleavage.
In the context of this model, the toxicity of E. coli plasmid pCR-Blunt(DUS) to Ngo was initially puzzling, as the DUS does not have Ngo restriction/modification sites, and the vector does not have extensive homology with the Ngo genome. A closer examination of pCR-Blunt revealed short sequences (9-20 bp) with 100% nucleotide homology to ~14000 sites throughout the pathogen genome (Table S7; see Method Details). The same was observed for vectors pBeloBAC11 and pUC19, used respectively for cloning Nel DNA and subcloning of BAC inserts. Many of the short homology tracts in these vectors contain sequence motifs recognized by Ngo MS11 restriction/modification systems (Table S7). Sequence homology as short as 8 bp is sufficient to initiate RecA-mediated synapse formation (Qi et al., 2015). We predict that once pCR-BLUNT(DUS) enters Ngo (enabled by the DUS), these short homology tracts serve as sites of synapse formation. In the case of the pBeloBAC11 and pUC19 recombinants, these short tracks of homology, together with Nel sequences in the inserts, would initiate formation of synaptic structures.
How does Nel DNA find its way into the environment? Ngo extrudes its DNA using Type IV Secretion System (T4SS) proteins AtlA and ParB, encoded in the Gonococcal Genetic Island (GGI) (Dillard and Seifert, 2001; Ramsey et al., 2011). GGI and T4SS are in the commensal Neisseria bacilliformis (Pachulec et al., 2014), but neither atlA nor parB are in 15 other commensal Neisseria tested (Dillard and Seifert, 2001).
Commensal DNA is more likely released into the environment through autolysis. During growth, approximately 4% of Ngo cells autolyse each generation, releasing DNA into the medium (Kohler et al., 2007). Autolysis requires AmiC, a N-acetylmuramyl-L-alanine-amidase, and LtgA, a lytic transglycosylase. Commensal Neisseria, including Nel, encode orthologs of these enzymes (Table S8) (Garcia and Dillard, 2006; Hebeler and Young, 1975, 1976; Kohler et al., 2007). While our liquid assays indicate that relatively high concentrations of Nel DNA are required to kill Ngo, our SEM images show Nel and Ngo microcolonies physically contact each other. At these Nel-Ngo interfaces the concentration of extracellular DNA is likely to be very high. Importantly, the mouse studies strongly suggest that extracellular Nel DNA can reach a high enough local concentration to antagonize Ngo in vivo.
How does Ngo establish a foothold in the body and induce infection if it is killed by resident commensal Neisseria? The available evidence suggests that Ngo is a weak pathogen. Only a subset of individuals exposed to Ngo becomes infected. The transmission rate of Ngo ranges from 20% (female to male) to 70% (male to a female) (Holmes et al., 1970; Lin et al., 1998). In human challenge studies, 30-90% of males inoculated with Ngo become infected (Cohen et al., 1994; Schmidt et al., 2001; Schneider et al., 1995). Host factors including genetics and sex affect susceptibility to Ngo infection (Densen, 1989; Holmes et al., 1970; Lin et al., 1998). Our study strongly suggests that microbiota composition and commensal Neisseria abundance also influence the risk for Ngo infection.
The DNA-based mechanism of tribal warfare presented here also appears to be relevant to pathogen Neisseria meningitidis (Nme). Carriage of commensal Neisseria is inversely correlated with Nme carriage (Deasy et al., 2015; Diallo et al., 2016). Specifically, commensal Neisseria lactamica (Nla) protects against Nme colonization of the nasopharynx (Cartwright et al., 1987; Deasy et al., 2015; Evans et al., 2011). This protective effect was postulated to be due to the presence of cross-reactive antibodies to the pathogen (Oliver et al., 2002); however, natural bactericidal antibodies against Nme have been shown to predate Nla carriage (Trotter et al., 2007). Our observation that commensal Neisseria DNA also kills Nme raises the possibility that commensal Neisseria residing in the nasopharynx may inhibit Nme colonization through the DNA-based killing mechanism described for Ngo.
Our model and co-culture results suggest that Ngo and Nme DNA would not kill commensal Neisseria. Ngo and Nme DNA are more heavily methylated than commensal Neisseria DNA, and the presence of Ngo does not affect the viability of Nel, a genetically competent organism (Higashi et al., 2011). Consistent with this, Ngo DNA does not kill Nel (Fig S6). Several other pathogens are naturally competent for DNA transformation, among them Bacteriodes, Streptococcus pneumonia, Acinetobacter baylyi, Haemophilus influenzae, Helicobacter pylori, and Vibrio cholerae (Blokesch, 2016). Future work will determine whether members of these genera antagonize each other using a similar DNA-based killing mechanism.
Finally, our findings highlight the potential of developing a non-traditional DNA-based method for treating gonorrhea. Ngo causes over 100 million new infections each year worldwide (Organization, 2012). Moreover, gonorrhea enhances HIV transmission (Malott et al., 2013). Ngo has rapidly developed resistance to virtually all antibiotics used for its treatment (Unemo et al., 2012; Unemo and Shafer, 2014), prompting the Centers for Disease Control to declare this pathogen an urgent public health threat (https://www.cdc.gov/std/gonorrhea/arg/default.htm). The huge capacity of Ngo to take up exogenous DNA has allowed the pathogen to rapidly acquire antibiotic resistance determinants. Ironically, DNA uptake may also be its Achilles Heel.
STAR★METHODS
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Magdalene So (somaggie@email.arizona.edu).
Experimental model and subject details
Bacterial strains and maintenance
Table S1 lists the bacterial strains used in this study. Ngo MS11 and Nel subsp. glycolytica ATCC 29315 were used throughout the study unless otherwise indicated. Piliated, Opa non-expressing MS11 was used throughout the study; at the beginning of each experiment these phenotypes were checked by monitoring colony morphology. In some experiments, MS11 ΔpilT (Dietrich et al., 2009) was used. Construction of the complemented derivative of ΔpilT (ΔpilT/ipilT) is described below. N400 is a derivative of MS11 in which recA is under control of the IPTG-inducible lac promoter (Tonjum et al., 1995). For extraction of chromosomal DNA, the following strains were used: E. coli DH5α, N. lactamica ATCC 23970, N. cinerea ATCC 14685, N. mucosa ATCC 25996, N. sicca ATCC 29256, N. polysaccharea ATCC32768, and Neisseria meningitidis 8013. Ngo clinical isolates D006 and D0020 were isolated from the urethra of symptomatic male patients at Durham County STD clinic (Durham, NC, USA). Unless stated otherwise, Neisseria strains were grown on GCB agar containing Kellogg’s supplements I and II (Kellogg et al., 1963) and antibiotics when appropriate (see below). E. coli was grown on LB medium or agar. All bacterial strains were maintained at 37° with 5% CO2. Bacterial density was calculated using the following formulas. Ngo MS11 and Nme 8013: OD600 1.4 = 1×109 CFUs; Nel 29315: OD600 1.0 = 1.2×109 CFUs.
Mouse model
A modification of the female mouse model of experimental Ngo genital tract infection (Jerse et al., 2011) was used to determine whether Nel inhibited Ngo in vivo. Female BALB/c mice (6 to 8 weeks old; National Cancer Institute or Charles River) in the anestrus or diestrus stage of the estrous cycle were identified by vaginal smear and treated with Premarin (Pfizer) on days −2, 0, and +2 and antibiotics to reduce the overgrowth of commensals that occurs under the influence of estrogen (streptomycin, 2.4 mg, BID; vancomycin, 0.4 mg, BID; and trimethoprim, 0.4 g/liter of drinking water) (Jerse et al., 2011). Mice were housed in Thorn units in an ABSL-2 room in the absence of male mice other than sentinel mice. This was to prevent changes to mice hormonal state by male pheromones. Food and water, cages and bedding were autoclaved to reduce the commensal flora load of mice. Animals were checked twice daily by facility personnel and twice daily by the lab staff during the experiments. All experiments were conducted at the Uniformed Services University according to guidelines established by the Association for the Assessment and Accreditation of Laboratory Animal Care using a protocol approved by the University’s Institutional Animal Care and Use Committee.
Method Details
Scanning electron microscopy
Ngo and Nel (5×107 CFUs of each organism) were cultured alone or in a 1:1 ratio on glass coverslips in a 6-well microplate for 5 h. Bacteria were imaged by scanning electron microscopy (Higashi et al., 2011). Samples were washed gently in PBS and fixed successively in PBS containing glutaraldehyde (2.5% wt/vol), osmium tetroxide (1% wt/vol) and uranyl acetate (2% wt/vol). Samples were washed in PBS and dehydrated by successive immersions in ethanol at the following concentrations: 15%, 30%, 50%, 70%, 80%, 90%, 95% (vol/vol, in water), and 100%. The samples were critical point dried and sputter coated with platinum (Higashi et al., 2011). Images were obtained using a Hitachi S-4800 Field-Emission Scanning Electron Microscope.
Co-culture experiments
Nel and Ngo cells harvested from 15 h agar plates (in log phase) were resuspended in GC broth with Kellogg's Supplements I and II (Kellogg et al., 1963). Cells were adjusted to approximately the same density and either grown separately (~5×107 CFUs) or together (~5×107 CFUs each strain) in 1 ml total volume in 6-well microplate dishes at 37°C, 5% CO 2. At the indicated times, the cultures were harvested and serial dilutions made with GC broth were plated on LB agar for Nel CFUs, and GCB agar containing vancomycin (3 μg/mL), colistin (7.5 μg/mL), and nystatin (12.5 μg/mL) (Jacobs and Kraus, 1975) for Ngo CFUs. Plates were incubated overnight and colony forming units were quantitated. The same plates incubated for 48 h did not increase CFU counts. CFU at each time point was calculated from the average of triplicate wells from 3 independent. For co-culture experiments with Ngo clinical isolates D006 and D0020, 0.5 ml of fresh supplemented GC broth was added to each well every 24 h to replenish nutrients. CFU at each time point was calculated from the average of triplicate wells from one experiment.
Construction of mutants and complemented strains
To construct ΔcomP and complemented ΔcomP (ΔcomP/comP), the comP open reading frame (ORF) was replaced with the kanamycin resistance gene (kan). Primers comP_MS11_F and comP_MS11_R (Table S1) containing Ngo comP flanking sequences were used to amplify kan from plasmid pNBNeiKan (Weyand et al., 2016). WT was transformed with the amplicon by spot transformation (Dillard, 2011). Transformants were selected on GCB supplemented agar containing kanamycin (30 μg/ml). To construct the complemented comP strain, a copy of comP and its native promoter was inserted between the iga and trpB sites in WT Ngo MS11 and this region was cloned into the SacI and NdeI sites of pMR68 to generate plasmid pcomPc. The plasmid was then introduced into Ngo by spot transformation (Dillard, 2011). Transformants were selected on GCB supplemented agar containing erythromycin (10 μg/mL). The comP insertion was verified by PCR and sequencing using primers listed in Table S1. In a verified comP clone, the native comP was removed as described above. The resulting Ngo ΔcomP/comP strain was confirmed by PCR and sequencing using primers listed in Table S1.
To construct the complemented strain of ΔpilT (ΔpilT/ipilT), pilT was amplified from WT MS11 with primers MR616 and 617 using Phusion polymerase and the amplicon was cloned into the ClaI and SalI sites of pKH37 (Kohler et al., 2007). The insert was verified by sequencing and the plasmid was transformed into E. coli DH5α. The plasmid was then introduced into Ngo ΔpilT by electroporation as described (Dillard, 2011). The electroporated cells were allowed to recover on GCB agar for 6 h, collected in 1 ml of GC broth, vortexed for 1 min, and plated on GBC agar containing chloramphenicol (Cm; 10 mg/mL). Cm resistant colonies were selected to determine if ipilT had inserted between the lctP and aspC locus, first by determining the size of the PCR products produced by primers MR618 and 619, then by sequencing. The optimal IPTG concentration needed to induce PilT was determined by Western blotting using anti-PilT antibodies of WT and ΔpilT/ipilT cells grown for 14 h on GCB agar containing 0, 0.05, 0.1, and 0.5 mM IPTG. The IPTG concentration that best approximated WT PilT level was 0.5 mM.
Competitive mouse infection experiments
Mice were inoculated vaginally on day 0 with 20 μl of a phosphate-buffered saline (PBS) suspension containing similar numbers of Nel mixed with piliated Ngo, Ngo ΔcomP, or Ngo ΔcomP/comPWT (total CFU ~ 2 ×106 per 20 μl inoculum) (n= 8-10 mice per group). Control groups received 106 CFU of Nel alone or each Ngo strain alone. Vaginal swabs collected daily for 7-10 days post-inoculation were quantitated by culturing on GCB agar with VCNT supplement (Sigma) and 100 μg/ml streptomycin (Sm) for Ngo CFUs, and culturing on LB agar with 100 μg/ml Sm for Nel CFUs. A total of 3 experiments were performed: one with Ngo alone, one comparing Ngo and Ngo ΔcomP, and one comparing Ngo, Ngo ΔcomP, and Ngo ΔcomP/comPWT. Thus, all strains were tested at least twice except for Ngo ΔcomP/comPWT, which was tested once. For randomization, the mice were randomly divided into five animals per group in all experiments. All stages of the study were blinded except collection of vaginal swabs. The technicians who quantitatively cultured and counted the plates were only aware of which samples potentially contain Ngo, Nel or both. This was necessary because the types of plates used to culture Ngo and Nel are different. These individuals, however, did not know which samples contained Ngo, Ngo ΔcomP, or Ngo ΔcomP/comPWT. A 10-day infection period was used in initial experiments; a 7-day study endpoint was used in experiments in which Ngo ΔcomP mutant was tested. This change in the protocol was necessary due to the logistical challenge of adding two additional experimental groups of animals to the experiment.
Sample-sizes for all experiments were determined as follows. Based on a preliminary result, we estimated 80% and 30% colonization frequency in control (Ngo alone) and experimental groups, respectively. Under this estimation, we chose 8-10 animals per group as this sample size has 80% power to predict a difference at a level of P < 0.05.
For statistical analysis, Kaplan Meyer curves were created to compare duration of recovery between control and experimental groups. Differences in the percentage of mice colonized with each species over the 7-day period were analyzed by the Log Rank test. The number of Ngo and Nel CFUs recovered over time was analyzed by a repeated measures ANOVA. The limit of detection (20 CFU/ml of vaginal swab suspension) was used for cultures from which no Ngo or Nel were isolated. No animal had died or needed euthanization (exclusion criteria for data/subjects) during the experiment.
Assaying toxicity of Nel supernatants for Ngo
Nel at a starting density of 2 × 106 CFUs/mL were grown in GC broth with Kellogg’s supplements I and II for 0, 12, 18, and 24 h at 37°C, 5% CO2. Cells were pelleted by centrifugation and the supernatants collected. Supernatants were filtered through a 0.22 μM PVDF filter unit pre-blocked with supplemented GC broth containing 50 mg/mL BSA, then being washed with supplemented GC broth. An equal volume of fresh supplemented GC broth was added to the filtered supernatant and used as the assay broth. Ngo was added to the assay broth at 2 × 107 CFUs/mL and 0.5 mL was added to each well of a 12 well culture plate. As a negative control, a parallel set of cultures was initiated in which Ngo was incubated with filtered supplemented GC broth with an equal volume of fresh supplemented GC broth. Cultures were incubated for 5 h at 37°C, 5% CO2. Ngo CFUs were quantitated by plating serial dilutions on GCB + VCN agar plates. Plates were incubated overnight and CFUs were quantitated. CFUs were calculated from averages of triplicate wells from 3 independent experiments. The same plates incubated for 48 h did not yield increased CFUs.
Quantitating DNA in supernates of Nel liquid cultures
Nel (5×106 CFU/mL) was inoculated into GCB broth containing Supplements I, II and 5 mM NaHCO3, and grown in a shaking incubator at 37°C, 220 RPM. Cells were harvested at 16 h and 24 h, and pelleted by centrifugation at 3000 rcf for 30 min, and the supernates (SNs) were passed through a 0.22 μM PES Steriflip filter. DNA was extracted from 500 μL of each SN using phenol, phenol-chloroform-isoamylalcohol 25:24:1, and chloroform. The DNA was precipitated with isopropanol (1:1 volume) and NaOAc was added to a final concentration of 1M. The DNA pellets were washed 2x with ice cold 70% EtOH and solubilized in 20 μL TE. Separately, known amounts of purified Nel DNA (0, 400, 1000, 2000, 4000, or 10000 ng) were added to 500 uL of supplemented GCB + NaHCO3. The DNA from these standards was extracted using the same protocol. 5 μL of each DNA sample was separated by electrophoresis in a 0.7% agarose gel. Densitometric measurements from the DNA standards (arbitrary units) were used to generate the standard curve (Y=1.489*X+108.22 (R2=0.9889)), where Y is the densitometric measurement and X is the starting amount of DNA in ng. Densitometric measurements from DNA extracted from Nel SNs were substituted as Y values in the standard curve to determine the starting amount of DNA (X). DNA concentration was determined by normalizing X to the starting volume of 500 μL.
Spot assay
Ngo grown on GCB agar plates for 16 h was collected with a sterile Dacron swab and suspended in liquid GC broth to OD600 of 0.2. 100 μL of the bacterial suspension was spread evenly on a GCB agar plate. 5 uL of DNA (20 ng/μL) or Nel supernatant was spotted onto defined sections of the plate, and the liquid was allowed to dry at 37°C for 5-10 min. The plate was incubated at 37°, 5% CO 2 for 12-16 h and the zones of clearance were recorded. 5 μl each of kanamycin (20 μg/ml) and GC broth were spotted on the agar to serve as positive and negative controls, respectively.
DNA liquid culture killing assay
Ngo, Nel and Nme (5×105 CFUs each) were suspended in GC broth containing Kellogg’s Supplements I and II and MgSO4 (5 mM), and seeded into 24-well microplates. DNA was added to the wells at the indicated concentrations, and the plates were incubated for 4 h at 37°, 5% CO 2. The bacteria were harvested with a P1000 pipette, and serially diluted in liquid GC broth. The serial dilutions were plated onto GCB agar containing Kellogg’s Supplements I and II and the plates incubated overnight before colony forming units were counted. CFU values used for survival (%) calculations were taken from the average of triplicate wells from 3-4 independent experiments. In one outlier experiment, Ngo survival (%) was more than two standard deviations below the mean when incubated with 0.5 μg/mL of DNA. Data from this experiment were excluded.
Extraction of chromosomal DNA
Neisseria spp. were grown on GCB agar with Kellogg’s supplements I and II, and E. coli on LB agar plates, for 16 to 18 h at 37°C, 5% CO 2. Cells from one plate were collected with a sterile Dacron swab into 500 μL of GC lysis buffer (0.5 M NaCl, 10 mM EDTA, 50 mM Tris, pH 8.0) with 1% SDS and 1 mg/μL RNAse A (Qiagen). Cells were allowed to lyse at RT for 5 min. DNA from the lysates were extracted sequentially with an equal volume of phenol, phenol-chloroform-isoamyl alcohol (25:24:1), and chloroform. For each extraction step, lysate-organic solvent mixtures were vortexed (1 min) and transferred to phase lock gels. The mixtures were centrifuged at 16,000 x g for 5 min and the aqueous (upper) phases were collected into 15 mL conical tubes. DNA was precipitated with 5 volumes of molecular biology grade 100% ethanol and 2.5 M ammonium acetate at 4°C for 30 min. Precipitated DNA was washed twice with 70% ethanol and dissolved in TE buffer (10 mM Tris, 0.1 mM EDTA, pH 8.0). To assess DNA purity, the NanoDrop spectrophotometer was used to determine the ratio of absorbance at 260 nm to 280 nm. When needed, DNA was subjected to a second RNAse treatment (Thermo), followed by phenol-chloroform purification. The OD260/OD280 ratio of DNA preparations ranged from 1.8 to 2.0.
Construction of Bacterial Artificial Chromosomes
For preparation of inserts, 15 μg of Nel chromosomal DNA was partially digested with 10 units of SphI restriction enzyme for 15 min at 37°C. Digested DNA was separated in a 1% agarose gel, in 0.5x TBE buffer, using clamped homogenous electric fields (CHEF) electrophoresis system at 1-50 sec linear ramp, 6 V/cm for 14 h. Regions of the gel containing 20-60 kb fragments were excised and the DNA was eluted using Model 422 Electro-Eluter per vendor specifications. Eluted DNA was further size selected using 0.6% megabase agarose at 50V for 4 h (1x TAE buffer), to exclude fragments smaller than 20 kb. For preparation of vector DNA, 8 ng of pBeloBAC11 was digested with 10 units of SphI at 37°C for 1 h. Dig ested pBeloBAC11 was incubated with 180 ng of inserts, prepared above, in the presence of 5 μL of T4 ligase, per vendor instruction, at 16°C overnight. The ligation mixtures were desalted on 0.1 M glucose/1 % agarose cones for 90 min on ice (Atrazhev and Elliott, 1996). Desalted DNA was electroporated (325 DC V, 4 kΩ, 330 μF) into E.coli Turbo Electrocompetent 5alpha. After recovery in SOC medium at 37°C for 1 h with continuous shaking, electroporate d E. coli cells were plated on LB agar containing chloramphenicol (12.5 μg/mL), X-gal (80 mg/mL) and IPTG (100 μg/mL). White colonies were picked and grown in liquid LB containing 12.5 μg/mL chloramphenicol at 37°C for 16 h with continuous shaking. Bacterial Artificial Chromosomes (BACs) were extracted using NucleoBond Midi columns.
Restriction enzyme analysis of pBeloBAC11 inserts
5 μg of purified BACs were digested with 20 units of NotI or SphI restriction enzymes. Digested DNA was separated by 0.6% megabase agarose at 50V for 4 h. To determine the expected fragment sizes, BAC inserts were sequenced with M13 primers and the sequences matched to the Nel reference genome (Accession # NZ_CP007726.1).
Subcloning of pBeloBAC11(6.1)
5 μg of purified BAC6.1 DNA was partially digested with 2 units of PstI restriction enzyme at 37°C for 15 min. Digested DNA was separated in a 0.7% agarose gel (1x TAE buffer) at 90V for 1 h. The region of the gel containing 5-10 kb fragments was excised and the DNA was extracted using gel extraction kit (Qiagen). To prepare vector DNA, 10 ng of pUC19 was digested with 10 units of PstI at 37°C for 1 h. Digested pUC19 was incubated with 30 ng of inserts prepared above, in the presence of 2 μL of T4 ligase, per vendor specifications. Competent E. coli DH5α was transformed with 4 μL of ligation mixture. Transformants, after recovery in SOC medium at 37°C for 1 h with continuous shaking, were selected on LB agar containing ampicillin (100 μg/mL). Plasmids were extracted from selected colonies and sequenced using M13 primers listed in Table S1.
Construction of Ngo TD3 (deletion of the ngoII, ngoIV, and ngoV restriction/modification loci)
For each restriction/modification (RM) locus, a plasmid containing sequences immediately flanking the ORFs was constructed as follows. ORFs and flanking sequences were PCR amplified with primers listed in Table S1, and the amplicons were digested with following restriction enzymes (NEB): ClaI and NaeI (ngoII), PmeI (ngoIV), and AseI (ngoV). Digested products were blunted with 1 unit of DNA polymerase fragment Klenow at RT for 15 min, per vendor specifications. Pieces of DNA containing the flanking regions of each ORF (identified on a 0.7% agarose gel based on fragment sizes) were extracted and ligated with T4 ligase per vendor specifications. Ligation products were cloned into pCR-Blunt with T4 ligase, generating plasmids pCR-Blunt (ngoII-fs), pCR-Blunt (ngoIV-fs), and pCR-Blunt (ngoV-fs). To generate single RM mutants, WT MS11 was spot transformed with 500 ng of each plasmid using an established protocol (Dillard, 2011). Deletion of each RM system was verified by PCR and sequencing using primers listed in Table S1. To generate the triple RM mutant, TD3, MS11 ΔngoIV was sequentially spot transformed with 500 ng of pCR-Blunt (ngoV-fs) and pCR-Blunt (ngoII-fs). At each step, deletion of RM systems was verified by PCR and sequencing using primers listed in Table S1. Loss of methyltransferase activities in TD3 was verified by confirming the susceptibility of TD3 DNA to digestion with cognate restriction enzymes HaeIII, NgoIV, and BamHI.
Analysis of Ngo chromosome for short sequence homology to plasmids pCR-Blunt, pBeloBAC11 and pUC19
NCBI Nucleotide Blast program with blastn algorithm was used to align pBeloBAC11 and pUC19 query sequences to the Ngo MS11 (taxid: 528354) chromosomal sequence. Algorithm parameters were set for alignment of short <20 bp sequences: max target sequence of 20000 adjusted for short input sequences with expected threshold of 1000 and word size of 7. Match and mismatch scores were set to 1 and −4, respectively. Scoring costs for existence and extension of gap were set to 5 and 2, respectively. These parameters were selected to minimize the number of alignments containing mismatches or gaps. All queries resulted in approximately 14000 hits. This number was similar even for alignments of concatenated query sequences (pCR-Blunt concatenated to pUC19, etc.) against the Ngo sequence, indicating that ~14000 is the maximum number of aligned sequences that is returned by blast. The actual number homologous regions between these plasmids and the Ngo chromosome therefore likely exceed the number reported by blast. Alignments containing mismatches and gaps were removed from further analysis. Sequence alignments were imported into the Unix program Vim and the number of recognition sequences from MS11 restriction modification systems was counted via regular expression searches.
Testing a DNA fragment with an E. coli or Ngo methylation signature for its ability to kill Ngo
For this experiment, the iga gene in Ngo was modified as follows. A DNA fragment containing a site for the AsiSI restriction enzyme, a DUS12 and the first 980 bp of the 5' coding sequence of iga was synthesized (Integrated DNA Technologies, San Jose, CA, USA) (Table S1). A second fragment containing the terminal 687 bp of the iga coding sequence, 293 bp of the iga downstream flanking sequence, a DUS and an AsiSI site was also synthesized. The sequence order of the two fragments is AsiSI-DUS-5'iga and 3'iga-DUS-AsiSI. Each fragment was cloned into pCR-Blunt, generating pCR-Blunt (iga5) and pCR-Blunt (iga3), respectively. The iga locus in Ngo was converted to AsiSI-DUS-iga-DUS-AsiSI (ADIDA) by sequential transformation of WT bacteria with pCR-Blunt (iga5) and pCR-Blunt (iga3) DNA (500 ng each), generating mutant Ngo i35A. All construction steps were verified by sequencing (Table S1) and confirmed by restriction digestion. The WT Ngo genome contains 9 AsiSI sites; the shortest AsiSI fragment is >15 kb. i35A contains two new AsiSI sites, flanking iga, and AsiSI digestion releases the 4kb ADIDA fragment.
The ADIDA fragment was isolated from Ngo i35A for killing assays. Chromosomal DNA from i35A (1 mg) was digested with AsiSI (500 units, 37°C for 4 h), and the fragments were separated in a preparatory 0.7% agarose gel. The gel region containing 4 kb DNA was excised and the DNA purified using a gel extraction kit (Thermo Fisher).
The ADIDA locus in Ngo i35A was cloned into plasmid pCR-Blunt by PCR amplification using primers listed in Table S1 and the amplified DNA was transformed into E. coli DH5α, generating recombinant pCR-Blunt (ADIDA). The insert from pCR-Blunt (ADIDA) was purified from the vector by AsiSI digestion, agarose gel separation and extraction as described above.
ADIDA fragments derived from E. coli and i35A (1 μg/mL each) were compared for their ability to kill Ngo using the liquid killing assay described above. Nel DNA (20 μg/ml) served as the positive control. CFU was calculated from the averages of triplicate wells from 3 independent experiments.
Construction of pCR-Blunt(DUS)
The entirety of pCR-Blunt was PCR amplified with primers DUS-pCR-Blunt_F and pCR-Blunt_R (Table S1). DUS was added via DUS-pCR_Blunt_F, which contains a DUS12 (ATGCCGTCTGAA). PCR amplicons were circularized with T4 ligase, per vendor specifications, and transformed into chemically competent E. coli DH5α. Transformants were allowed to recover in SOC medium at 37° for 1 h, and selected on LB agar containing 50 μg/mL of kanamycin. DUS in the resulting plasmid pCR-Blunt(DUS) was confirmed by sequencing using M13 primers listed in Table S1.
In vitro methylation of DNA
DNA was incubated with CpG and GpC methyltransferases (M.CviPI and M.SssI, respectively) per vendor specifications. Briefly, 100 μg of DNA was incubated with 20 μL of methyltransferase in nuclease-free water in a total volume of 500 μL (1x NEB methyltransferase buffer). S-adenosylmethionine (SAM) was added to a final concentration of 640 uM. Methylation was verified by digesting 1 μg DNA with 20 units of HaeIII or BstUI at 37°C for 1 h.
Determination of modifications in Neisseria and E. coli DNA
Total DNA was extracted from Nel using a Wizard Genomic DNA Purification Kit. Whole-genome sequencing was conducted using the PacBio RSII platform (Pacific Biosciences, Menlo Park, CA) with P6-C4 chemistry. Each isolate was sequenced using one SMRT cell. Sequencing reads were then assembled using the hierarchical genome assembly process (HGAP3, SMRTAnalysis 2.3.0) workflow, which included consensus polishing using Quiver (Chin et al., 2013). The resulting assembled genomes contained 1–7 contigs each. Methylation analyses were performed using the Modification and Motif Analysis pipeline in SMRT Portal, and motifs and associated modified bases were identified and characterized.
Quantification and Statistical Analysis
Statistical differences between two study groups were evaluated using unpaired, two-tailed Student’s t-test, log-rank test, or ANOVA with Tukey’s Post Test correction using GraphPad Prism 5.0 software. Statistical tests applied and the P values are indicated in the figure legends and the Results section. For in vitro assays, n indicates the number of independent experiments. For in vivo experiments, n indicates the number of mice used. N values are listed in the figure legends. In all figures, error bars represent standard error of the mean (SEM). Limit of Detection (LOD) is indicated by dotted lines, when appropriate. Statistical comparisons with P values <0.05 are defined as statistically significant.
Supplemental Figure Legends
Fig S1. Nel kills Ngo clinical isolates D006 and D020 in mixed culture, Related to Fig 2.
CFUs of Ngo clinical isolates D006 and D0020 and lab strain MS11 cultured alone (~5×107 CFUs total CFUs) or in the presence of Nel (~5×107 total CFUs each strain). Time of first plating: 6 h post-inoculation. Level of detection: 10 CFUs.
Fig S2. CFU/mL of vaginal swab suspension in mice colonized by Ngo and Nel, Related to Fig 3.
Average number of (A) Ngo or (B) Nel CFUs recovered from 1 ml vaginal swab suspensions from mice inoculated vaginally with 106 CFU of Nel or Ngo alone or a mixture containing 106 CFU of each species. (n = 8-9 mice/group). P=0.0013 for the difference in recovery of Ngo from mice inoculated with Ngo alone or Ngo+Nel (repeated measures ANOVA). The number of (C, E) Nel or (D, F) Ngo CFU isolated from vaginal swab suspensions of individual mice inoculated with (C) Nel alone, (D) Ngo alone or (E, F) Ngo + Nel. Dotted line indicates level of detection.
Fig S3. CFU/ml from mice inoculated with WT or ComP-deficient Ngo, or either Ngo strain plus Nel, Related to Fig 6.
(A-D) Average number of Nel or Ngo CFUs recovered from 1 ml vaginal swab suspensions from mice inoculated vaginally with 106 CFU of Nel, WT Ngo or the NgoΔcomP mutant alone or a mixture containing 106 CFU of Nel and either Ngo strain. (n = 8-9 mice/group). P=0.007 for the difference in recovery of WT Ngo from mice inoculated with WT Ngo alone or WT Ngo+Nel (repeated measures ANOVA). The number of (E,H) Nel, (F,I) WT Ngo, or (G,J) NgoΔcomP CFU isolated from individual mice inoculated with (E) Nel alone, (F) WT Ngo alone, (G) Ngo ΔcomP alone, (H,I) WT Ngo + Nel or (J) NgoΔcomP + Nel. Dotted line indicates level of detection.
Fig S4. Restriction digests of Nel DNA confirm methylation of GpC and CpG sequences, Related to Fig 7.
Purified Nel DNA was incubated with buffer or M.CviPI and/or M.SssI methyltransferases, and subsequently digested with HaeIII or BstUI, restriction enzymes which cleave only unmethylated GGCC and CGCG sequence, respectively.
Fig S5. iga DNA from E. coli but not Ngo i35A is fragmented by Ngo restriction enzymes, Related to Fig 7.
ADIDA DNA fragment derived from Ngo i35A (left) and E. coli K-12 (right) digested with HaeII or NlaIV (isoschizomers of NgoI and NgoV) and separated in a 0.7% agarose gel. (−) indicates incubation with buffer alone.
Fig S6. Nel is not killed by Nel or Ngo DNA, Related to Fig 7.
Nel and Ngo CFUs recovered after a 4 h incubation with chromosomal DNA purified from Nel and Ngo (5 μg/mL). Starting CFUs: 5×105. n=3. Error bars represent SEM. LOD: 10 CFUs. (**P<0.01; Student’s t-test).
Table S1. Bacterial strains and primers used in this study, Related to STAR Methods.
Table S2. Identification of DNA as the toxic compound in N. elongata supernatant, Related to Fig 4.
Table S3. Resistance of N. gonorrhoeae DNA uptake mutants to killing by N. elongata DNA, Related to Fig 5.
Table S4. Toxicity of DNA from BACs and BAC subclones for N. gonorrhoeae, Related to Fig 7.
Table S5. Modifications in Nel 29315 (this study), Ngo FA1090 (Blow et al., 2016, Srikhanta et al. 2009), Ngo MS11 (Stein et al., 1992) and E. coli K-12 (Marinus and Løbner-Olesen, 2014) DNA, Related to Fig 7.
Table S6. Toxic DNAs in this study contain sequences recognized by Ngo MS11 R/M systems, Related to Discussion, Fig 4 and Fig 7.
>Table S7. Short regions of homology (9-20 bp) between E. coli plasmids and Ngo MS11 chromosome contain recognition sequences from Ngo MS11 restriction modification (R/M) systems, Related to Discussion and Fig 7.
Table S8. Orthologs of N-acetylmuramyl-L-alanine-amidase AmiC and lytic transglycosylase LtgA in commensal Neisseria, Related to Discussion, Fig 2 and Fig 3.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Enzymes | ||
| HaeIII | New England Biolabs (NEB) (Ipswich, MA, USA) | Cat# R0108S |
| NgoIV | NEB | Cat# R0564S |
| NlaIV | NEB | Cat# R0126S |
| AsiSI | NEB | Cat# R0630S |
| BamHI | NEB | Cat# R0136S |
| NotI | NEB | Cat# R1089S |
| SphI | NEB | Cat# R1082S |
| PstI | NEB | Cat# R0140S |
| BstUI | NEB | Cat# R0518S |
| HaeII | NEB | Cat# R0107S |
| SacI | NEB | Cat# R0156S |
| NdeI | NEB | Cat# R0111S |
| ClaI | NEB | Cat# R0197S |
| SalI | NEB | Cat# R0138S |
| PmeI | NEB | Cat# R0560S |
| AseI | NEB | Cat# R0526S |
| M.CviPI | NEB | Cat# M0227L |
| M.SssI | NEB | Cat# M0226L |
| DNAse I | Invitrogen (Carlsbad, CA, USA) | Cat# 18047019 |
| Proteinase K | Sigma (St. Louis, MO,USA) | Cat# P2308 |
| RNAse A | Qiagen (Hilden, Germany) | Cat# 19101 |
| RNAse A, DNAse and protease-free | Thermo Fisher (Waltham, MA, USA) | Cat# EN0531 |
| T4 ligase | Promega (Madison, WI, USA) | Cat# M0226L |
| Phusion Polymerase | Thermo Fisher | Cat# F531L |
| DNA Polymerase I Klenow Fragment | Promega | Cat# M2201 |
| Bacterial strains (Table S1) | ||
| E. coli (DH5α) | Lab collection | N/A |
| E. coli Turbo Electrocompetent 5alpha | NEB | Cat# 2984H |
| N. gonorrhoeae (MS11) | (Segal et al., 1985) | N/A |
| ΔpilT | (Dietrich et al., 2009) | N/A |
| ΔpilT/ipilT | This study | N/A |
| ΔcomP | This study | N/A |
| ΔcomP/comPWT | This study | N/A |
| TD3 | This study | N/A |
| N400 | (Tonjum et al., 1995) | N/A |
| i35A | This study | N/A |
| N. elongata | ATCC | N/A |
| N. lactamica | ATCC | N/A |
| N. cinerea | ATCC | N/A |
| N. mucosa | ATCC | N/A |
| N. sicca | ATCC | N/A |
| N. polysaccharea | ATCC | N/A |
| N. meningitidis | Lab collection | N/A |
| Oligonucleotides | ||
| Primers used for cloning and sequencing (Table S1) | This study | N/A |
| DNA fragments (Table S1) | This study | N/A |
| Plasmids | ||
| pCR-Blunt | Thermo Fisher | Cat# K270020 |
| pUC19 | AddGene (Watertown, MA, USA) | Cat# 50005 |
| pBeloBAC11 | NEB | Cat# E4154 |
| pKH37 | (Kohler et al., 2007) | N/A |
| pNBNeiKan | (Weyand et al., 2016) | N/A |
| pMR68 | (Ramsey et al., 2012) | N/A |
| Growth Media | ||
| GC medium base | Difco Laboratories Inc (Detroit, MI, USA) | Cat# 228920 |
| GCB containing Kellogg’s Supplements I and II | (Kellogg et al., 1963) | N/A |
| GCB containing vancomycin, colistin, nystatin | (Jacobs and Kraus, 1975) | N/A |
| Glutaraldehyde | Electron Microscopy Sciences (EMS) (Hatfield, PA, USA) | Cat# 16000 |
| Osmium tetroxide | EMS | Cat# 19100 |
| Uranyl acetate | EMS | Cat# 22400 |
| Chemicals | ||
| IPTG | GoldBio (Olivette, MO, USA) | Cat# I2481C50 |
| X-gal | Research Products International (RPI) (Mount Prospect, IL, USA) | Cat# B71800 |
| S-adenosylmethionine (SAM) | NEB | Cat# B9003S |
| BSA | Sigma | Cat# A2153 |
| Ampicilin | GoldBio | Cat# 69523 |
| Kanamycin | GoldBio | Cat# K120SL25 |
| Erythromycin | Sigma | Cat# E5389 |
| Chloramphenicol | Sigma | Cat# C0378 |
| Vancomycin | RPI | Cat# V065500 |
| Colistin | RPI | Cat# C70700 |
| Nystatin | RPI | Cat# N82020 |
| Ethanol | Sigma | Cat# 459836 |
| Isopropanol | Sigma | Cat# I9516 |
| Phenol | Sigma | Cat# 108952 |
| Isoamyl alcohol | Sigma | Cat# 123513 |
| Tissue Culture Plates | ||
| Microplate dish (6-well) | Corning (Corning, NY, USA) | Cat# 0720083 |
| Microplate dish (12-well) | Corning | Cat# 087721B |
| Microplate dish (24-well) | Corning | Cat# 353847 |
| Animal Model: Mice | ||
| Female BALB/c (6-8 weeks old) | National Cancer Institute and Charles River (Jerse et al., 2011) | Cat# Strain_028 |
| Other lab reagents | ||
| 0.22 μM PES Steriflip filter | Millipore (Burlington, MA, USA) | Cat# SE1M179M6 |
| 0.22 μM filter | Millipore | Cat# SLGV033RS |
| Nucleobond Midi Columns | Macherey-Nagel (Düren, Germany) | Cat# 740410 |
| Agarose | Sigma | Cat# A2576 |
| Megabase Agarose | Bio-Rad (Hercules, CA, USA) | Cat# 1613108 |
| Phase Lock Gels | VWR (Radnor, PA, USA) | Cat# 2302820 |
| Gel Extraction Kit | Qiagen | Cat# 28706 |
| Gel Extraction Kit | Thermo Fisher | Cat# K0691 |
| Wizard Genomic DNA Purification Kit | Promega | Cat# A1120 |
| Dacron Swabs | Thermo Fisher | Cat# 164KS01 |
| CHEF Electrophoresis System | Bio-Rad | Cat# 1703670 |
| Model 422 Electro-Eluter | Bio-Rad | Cat# 1652976 |
| NanoDrop spectrophotometer | Thermo Fisher | RR_ID:SCR_015804 |
| Software and algorithms | ||
| Blastn | NCBI | RR_ID:SCR_007190 https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| GraphPad Prism 5.0 | GraphPad (San Diego,CA, USA) | RR_ID: SCR_002798 https://www.graphpad.com |
| SMRT-Analysis | PacBio (Menlo Park, CA, USA) | RR_ID: SCR_002942 https://www.pacb.com/products-and-services/analytical-software/smrt-analysis/ |
| Quiver | (Chin et al., 2013) | N/A https://github.com/PacificBiosciences/GenomicConsensus |
| Vim Text Editor | Vim | N/A https://www.vim.org/ |
Highlights.
Commensal Neisseria kill STD pathogen N. gonorrhoeae by releasing DNA in the environment
Killing requires DNA entry and recombination and a foreign DNA methylation pattern
Commensal N. elongata accelerates clearance of N. gonorrhoeae from the mouse vagina
A N. gonorrhoeae DNA uptake mutant resists this clearance
Acknowledgements
We thank members of the M. So lab and B. Fane and M. K. Johnson for their thoughtful discussions and suggestions, and Jason Pilligua for technical assistance with the mouse competitive infection studies. This work was supported in part by NIH grant AI111944 awarded to M. So.
Footnotes
Declaration of Interests
We declare that authors Magdalene So, Won Jong Kim, Dusting Higashi, Maira Goytia and Ann E. Jerse have submitted applications for following patents related to the work presented here: UA15-042 titled “Compositions and methods for treating gonorrhea” and addendum UA18-253 titled "A novel DNA-based antimicrobial compound for treating infections caused by Neisseria gonorrhoeae and Neisseria meningitidis."
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, et al. (2012). Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambur OH, Frye SA, and Tonjum T (2007). New functional identity for the DNA uptake sequence in transformation and its presence in transcriptional terminators. J Bacteriol 189, 2077–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, and Kasper DL (2014). Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 156, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atrazhev AM, and Elliott JF (1996). Simplified desalting of ligation reactions immediately prior to electroporation into E. coli. Biotechniques 21, 1024. [DOI] [PubMed] [Google Scholar]
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, and Gordon JI (2004). The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A 101, 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, and Hand TW (2014). Role of the microbiota in immunity and inflammation. Cell 157, 121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JS, Jolley KA, Earle SG, Corton C, Bentley SD, Parkhill J, and Maiden MC (2012). A genomic approach to bacterial taxonomy: an examination and proposed reclassification of species within the genus Neisseria. Microbiology 158, 1570–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry JL, Cehovin A, McDowell MA, Lea SM, and Pelicic V (2013). Functional analysis of the interdependence between DNA uptake sequence and its cognate ComP receptor during natural transformation in Neisseria species. PLoS genetics 9, e1004014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokesch M (2016). Natural competence for transformation. Curr Biol 26, 3255. [DOI] [PubMed] [Google Scholar]
- Blow MJ, Clark TA, Daum CG, Deutschbauer AM, Fomenkov A, Fries R, Froula J, Kang DD, Malmstrom RR, Morgan RD, et al. (2016). The Epigenomic Landscape of Prokaryotes. PLoS genetics 12, e1005854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brestoff JR, and Artis D (2013). Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol 14, 676–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabotti M, Scirocco A, Maselli MA, and Severi C (2015). The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Annals of gastroenterology 28, 203–209. [PMC free article] [PubMed] [Google Scholar]
- Cartwright KA, Stuart JM, Jones DM, and Noah ND (1987). The Stonehouse survey: nasopharyngeal carriage of meningococci and Neisseria lactamica. Epidemiology and infection 99, 591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussee MS, and Hill SA (1998). Formation of single-stranded DNA during DNA transformation of Neisseria gonorrhoeae. Journal of bacteriology 180, 5117–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10, 563–569. [DOI] [PubMed] [Google Scholar]
- Cohen MS, Cannon JG, Jerse AE, Charniga LM, Isbey SF, and Whicker LG (1994). Human experimentation with Neisseria gonorrhoeae: rationale, methods, and implications for the biology of infection and vaccine development. J Infect Dis 169, 532–537. [DOI] [PubMed] [Google Scholar]
- Craig L, and Li J (2008). Type IV pili: paradoxes in form and function. Curr Opin Struct Biol 18, 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deasy AM, Guccione E, Dale AP, Andrews N, Evans CM, Bennett JS, Bratcher HB, Maiden MC, Gorringe AR, and Read RC (2015). Nasal Inoculation of the Commensal Neisseria lactamica Inhibits Carriage of Neisseria meningitidis by Young Adults: A Controlled Human Infection Study. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 60, 1512–1520. [DOI] [PubMed] [Google Scholar]
- Densen P (1989). Interaction of complement with Neisseria meningitidis and Neisseria gonorrhoeae. Clinical microbiology reviews 2 Suppl, S11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, and Raffatellu M (2013). Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell host & microbe 14, 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diallo K, Trotter C, Timbine Y, Tamboura B, Sow SO, Issaka B, Dano ID, Collard JM, Dieng M, Diallo A, et al. (2016). Pharyngeal carriage of Neisseria species in the African meningitis belt. The Journal of infection 72, 667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, and Pettersson S (2011). Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A 108, 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich M, Mollenkopf H, So M, and Friedrich A (2009). Pilin regulation in the pilT mutant of Neisseria gonorrhoeae strain MS11. FEMS Microbiol Lett 296, 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillard JP (2011). Genetic Manipulation of Neisseria gonorrhoeae. Current protocols in microbiology Chapter 4, Unit4A 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillard JP, and Seifert HS (2001). A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Molecular microbiology 41, 263–277. [DOI] [PubMed] [Google Scholar]
- Duffin PM, and Seifert HS (2010). DNA uptake sequence-mediated enhancement of transformation in Neisseria gonorrhoeae is strain dependent. Journal of bacteriology 192, 4436–4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins C, Thomas CE, Seifert HS, and Sparling PF (1991a). Species-specific uptake of DNA by gonococci is mediated by a 10-base- pair sequence. J Bacteriol 173, 3911–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins C, Thomas CE, Seifert HS, and Sparling PF (1991b). Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. Journal of bacteriology 173, 3911–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CM, Pratt CB, Matheson M, Vaughan TE, Findlow J, Borrow R, Gorringe AR, and Read RC (2011). Nasopharyngeal colonization by Neisseria lactamica and induction of protective immunity against Neisseria meningitidis. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America 52, 70–77. [DOI] [PubMed] [Google Scholar]
- Flint J, and Eskin E (2012). Genome-wide association studies in mice. Nat Rev Genet 13, 807–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia DL, and Dillard JP (2006). AmiC functions as an N-acetylmuramyl-l-alanine amidase necessary for cell separation and can promote autolysis in Neisseria gonorrhoeae. Journal of bacteriology 188, 7211–7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman SD, and Scocca JJ (1988). Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci U S A 85, 6982–6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JF, Biswas GD, and Sparling PF (1982). Sequence-specific DNA uptake in transformation of Neisseria gonorrhoeae. Journal of bacteriology 152, 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Cedar H, and Razin A (1981). Restriction enzyme digestion of hemimethylated DNA. Nucleic Acids Res 9, 2509–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn JS, Piekarowicz A, Chien R, and Stein DC (1992). Cloning and linkage analysis of Neisseria gonorrhoeae DNA methyltransferases. J Bacteriol 174, 5654–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebeler BH, and Young FE (1975). Autolysis of Neisseria gonorrhoeae. Journal of bacteriology 122, 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebeler BH, and Young FE (1976). Mechanism of autolysis of Neisseria gonorrhoeae. Journal of bacteriology 126, 1186–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi DL, Biais N, Weyand NJ, Agellon A, Sisko JL, Brown LM, and So M (2011). N. elongata produces type IV pili that mediate interspecies gene transfer with N. gonorrhoeae. PLoS One 6, e21373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KK, Johnson DW, and Trostle HJ (1970). An estimate of the risk of men acquiring gonorrhea by sexual contact with infected females. Am J Epidemiol 91, 170–174. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Midtvedt T, and Gordon JI (2002). How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr 22, 283–307. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase T, Shinji H, Tajima A, Sato F, Tamura T, Iwamoto T, Yoneda M, and Mizunoe Y (2010). Isolation and identification of ATP-secreting bacteria from mice and humans. J Clin Microbiol 48, 1949–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs NF Jr., and Kraus SJ (1975). Comparison of hemoglobin-free culture media and thayer-martin medium for the primary isolation of Neisseria gonorrhoeae. J Clin Microbiol 1, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerse AE (1999). Experimental gonococcal genital tract infection and opacity protein expression in estradiol-treated mice. Infect Immun 67, 5699–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerse AE, Wu H, Packiam M, Vonck RA, Begum AA, and Garvin LE (2011). Estradiol-Treated Female Mice as Surrogate Hosts for Neisseria gonorrhoeae Genital Tract Infections. Front Microbiol 2, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DS Jr., Peacock WL Jr., Deacon WE, Brown L, and Pirkle DI (1963). Neisseria Gonorrhoeae. I. Virulence Genetically Linked to Clonal Variation. Journal of bacteriology 85, 1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp J, and Clark V (1984). Anaerobic Growth of Neisseria gonorrhoeae Coupled to Nitrite Reduction. Infect Immun 46, 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler PL, Hamilton HL, Cloud-Hansen K, and Dillard JP (2007). AtlA functions as a peptidoglycan lytic transglycosylase in the Neisseria gonorrhoeae type IV secretion system. Journal of bacteriology 189, 5421–5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koomey JM, and Falkow S (1987). Cloning of the recA gene of Neisseria gonorrhoeae and construction of gonococcal recA mutants. J Bacteriol 169, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koomey M, Gotschlich EC, Robbins K, Bergstrom S, and Swanson J (1987). Effects of recA mutations on pilus antigenic variation and phase transitions in Neisseria gonorrhoeae. Genetics 117, 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AJ, Sharma R, Hobbs JK, and Walti C (2017). Cooperative RecA clustering: the key to efficient homology searching. Nucleic Acids Res 45, 11743–11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, and Gordon JI (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023. [DOI] [PubMed] [Google Scholar]
- Lin JS, Donegan SP, Heeren TC, Greenberg M, Flaherty EE, Haivanis R, Su XH, Dean D, Newhall WJ, Knapp JS, et al. (1998). Transmission of Chlamydia trachomatis and Neisseria gonorrhoeae among men with urethritis and their female sex partners. J Infect Dis 178, 1707–1712. [DOI] [PubMed] [Google Scholar]
- Liu G, Tang CM, and Exley RM (2015). Non-pathogenic Neisseria: members of an abundant, multi-habitat, diverse genus. Microbiology 161, 1297–1312. [DOI] [PubMed] [Google Scholar]
- Loenen WA, Dryden DT, Raleigh EA, and Wilson GG (2014a). Type I restriction enzymes and their relatives. Nucleic Acids Res 42, 20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenen WA, Dryden DT, Raleigh EA, Wilson GG, and Murray NE (2014b).Highlights of the DNA cutters: a short history of the restriction enzymes. Nucleic Acids Res 42, 3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malott RJ, Keller BO, Gaudet RG, McCaw SE, Lai CC, Dobson-Belaire WN, Hobbs JL, St Michael F, Cox AD, Moraes TF, et al. (2013). Neisseria gonorrhoeae-derived heptose elicits an innate immune response and drives HIV-1 expression. Proc Natl Acad Sci U S A 110, 10234–10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS, and Conway T (2013). Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PloS one 8, e53957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marri PR, Paniscus M, Weyand NJ, Rendon MA, Calton CM, Hernandez DR, Higashi DL, Sodergren E, Weinstock GM, Rounsley SD, et al. (2010). Genome sequencing reveals widespread virulence gene exchange among human Neisseria species. PloS one 5, e11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehr IJ, and Seifert HS (1998). Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Molecular microbiology 30, 697–710. [DOI] [PubMed] [Google Scholar]
- Merz AJ, Enns CA, and So M (1999). Type IV pili of pathogenic neisseriae elicit cortical plaque formation in epithelial cells. Mol Microbiol 32, 1316–1332. [DOI] [PubMed] [Google Scholar]
- Merz AJ, So M, and Sheetz MP (2000). Pilus retraction powers bacterial twitching motility. Nature 407, 98–102. [DOI] [PubMed] [Google Scholar]
- Musso G, Gambino R, and Cassader M (2011). Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu Rev Med 62, 361–380. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, and Pettersson S (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. [DOI] [PubMed] [Google Scholar]
- Noyer-Weidner M, T.T.A. (1992). Methylation of DNA in prokaryotes in DNA methylation: molecular biology & biological significance. eds Jost JP, Saluz HP (Birkhauser, Basel, Switzerland: ). 39–108. [Google Scholar]
- Oliver KJ, Reddin KM, Bracegirdle P, Hudson MJ, Borrow R, Feavers IM, Robinson A, Cartwright K, and Gorringe AR (2002). Neisseria lactamica protects against experimental meningococcal infection. Infect Immun 70, 3621–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization, W.H. (2012). Emergence of multi-drug resistant Neissreia gonorrhoeae - Threat of global rise in untreatable sexually transmitted infections.
- Pachulec E, Siewering K, Bender T, Heller EM, Salgado-Pabon W, Schmoller SK, Woodhams KL, Dillard JP, and van der Does C (2014). Functional analysis of the Gonococcal Genetic Island of Neisseria gonorrhoeae. PLoS One 9, e109613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Redding S, Lee JY, Gibb B, Kwon Y, Niu H, Gaines WA, Sung P, and Greene EC (2015). DNA sequence alignment by microhomology sampling during homologous recombination. Cell 160, 856–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey ME, Hackett KT, Kotha C, and Dillard JP (2012). New complementation constructs for inducible and constitutive gene expression in Neisseria gonorrhoeae and Neisseria meningitidis. Appl Environ Microbiol 78, 3068–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey ME, Woodhams KL, and Dillard JP (2011). The Gonococcal Genetic Island and Type IV Secretion in the Pathogenic Neisseria. Front Microbiol 2, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendon MA, Hockenberry AM, McManus SA, and So M (2013). Sigma factor RpoN (sigma54) regulates pilE transcription in commensal Neisseria elongata. Mol Microbiol 90, 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkawitz J, Lademann CA, and Jentsch S (2014). Mechanisms and principles of homology search during recombination. Nat Rev Mol Cell Biol 15, 369–383. [DOI] [PubMed] [Google Scholar]
- Roberts RJ, Vincze T, Posfai J, and Macelis D (2015). REBASE--a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res 43, D298–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman E, and Seifert HS (2014). The genetics of Neisseria species. Annu Rev Genet 48, 405–431. [DOI] [PubMed] [Google Scholar]
- Schmidt KA, Schneider H, Lindstrom JA, Boslego JW, Warren RA, Van de Verg L, Deal CD, McClain JB, and Griffiss JM (2001). Experimental gonococcal urethritis and reinfection with homologous gonococci in male volunteers. Sex Transm Dis 28, 555–564. [DOI] [PubMed] [Google Scholar]
- Schneider H, Cross AS, Kuschner RA, Taylor DN, Sadoff JC, Boslego JW, and Deal CD (1995). Experimental human gonococcal urethritis: 250 Neisseria gonorrhoeae MS11mkC are infective. J Infect Dis 172, 180–185. [DOI] [PubMed] [Google Scholar]
- Segal E, Billyard E, So M, Storzbach S, and Meyer TF (1985). Role of chromosomal rearrangement in N. gonorrhoeae pilus phase variation. Cell 40, 293–300. [DOI] [PubMed] [Google Scholar]
- Sommer F, and Backhed F (2013). The gut microbiota--masters of host development and physiology. Nat Rev Microbiol 11, 227–238. [DOI] [PubMed] [Google Scholar]
- Stein DC, Gunn JS, Radlinska M, and Piekarowicz A (1995). Restriction and modification systems of Neisseria gonorrhoeae. Gene 157, 19–22. [DOI] [PubMed] [Google Scholar]
- Tomberg J, Unemo M, Davies C, and Nicholas RA (2010). Molecular and structural analysis of mosaic variants of penicillin-binding protein 2 conferring decreased susceptibility to expanded-spectrum cephalosporins in Neisseria gonorrhoeae: role of epistatic mutations. Biochemistry 49, 8062–8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonjum T, Freitag NE, Namork E, and Koomey M (1995). Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Molecular microbiology 16, 451–464. [DOI] [PubMed] [Google Scholar]
- Tremaroli V, and Backhed F (2012). Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249. [DOI] [PubMed] [Google Scholar]
- Trotter C, Findlow J, Balmer P, Holland A, Barchha R, Hamer N, Andrews N, Miller E, and Borrow R (2007). Seroprevalence of bactericidal and anti-outer membrane vesicle antibodies to Neisseria meningitidis group B in England. Clinical and vaccine immunology : CVI 14, 863–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M, Golparian D, Nicholas R, Ohnishi M, Gallay A, and Sednaoui P (2012). High-level cefixime- and ceftriaxone-resistant Neisseria gonorrhoeae in France: novel penA mosaic allele in a successful international clone causes treatment failure. Antimicrobial agents and chemotherapy 56, 1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M, and Nicholas RA (2012). Emergence of multidrug-resistant, extensively drug-resistant and untreatable gonorrhea. Future microbiology 7, 1401–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M, and Shafer WM (2014). Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clinical microbiology reviews 27, 587–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyand NJ, Ma M, Phifer-Rixey M, Taku NA, Rendon MA, Hockenberry AM, Kim WJ, Agellon AB, Biais N, Suzuki TA, et al. (2016). Isolation and characterization of Neisseria musculi sp. nov., from the wild house mouse. Int J Syst Evol Microbiol 66, 3585–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M, Lauer P, Park HS, Brossay L, Hebert J, and Koomey M (1998a). PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Molecular microbiology 29, 321–330. [DOI] [PubMed] [Google Scholar]
- Wolfgang M, Park HS, Hayes SF, van Putten JP, and Koomey M (1998b). Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc Natl Acad Sci U S A 95, 14973–14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M, van Putten JP, Hayes SF, and Koomey M (1999). The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Molecular microbiology 31, 1345–1357. [DOI] [PubMed] [Google Scholar]
- Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, Burian M, Schilling NA, Slavetinsky C, Marschal M, et al. (2016). Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535, 511–516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. Nel kills Ngo clinical isolates D006 and D020 in mixed culture, Related to Fig 2.
CFUs of Ngo clinical isolates D006 and D0020 and lab strain MS11 cultured alone (~5×107 CFUs total CFUs) or in the presence of Nel (~5×107 total CFUs each strain). Time of first plating: 6 h post-inoculation. Level of detection: 10 CFUs.
Fig S2. CFU/mL of vaginal swab suspension in mice colonized by Ngo and Nel, Related to Fig 3.
Average number of (A) Ngo or (B) Nel CFUs recovered from 1 ml vaginal swab suspensions from mice inoculated vaginally with 106 CFU of Nel or Ngo alone or a mixture containing 106 CFU of each species. (n = 8-9 mice/group). P=0.0013 for the difference in recovery of Ngo from mice inoculated with Ngo alone or Ngo+Nel (repeated measures ANOVA). The number of (C, E) Nel or (D, F) Ngo CFU isolated from vaginal swab suspensions of individual mice inoculated with (C) Nel alone, (D) Ngo alone or (E, F) Ngo + Nel. Dotted line indicates level of detection.
Fig S3. CFU/ml from mice inoculated with WT or ComP-deficient Ngo, or either Ngo strain plus Nel, Related to Fig 6.
(A-D) Average number of Nel or Ngo CFUs recovered from 1 ml vaginal swab suspensions from mice inoculated vaginally with 106 CFU of Nel, WT Ngo or the NgoΔcomP mutant alone or a mixture containing 106 CFU of Nel and either Ngo strain. (n = 8-9 mice/group). P=0.007 for the difference in recovery of WT Ngo from mice inoculated with WT Ngo alone or WT Ngo+Nel (repeated measures ANOVA). The number of (E,H) Nel, (F,I) WT Ngo, or (G,J) NgoΔcomP CFU isolated from individual mice inoculated with (E) Nel alone, (F) WT Ngo alone, (G) Ngo ΔcomP alone, (H,I) WT Ngo + Nel or (J) NgoΔcomP + Nel. Dotted line indicates level of detection.
Fig S4. Restriction digests of Nel DNA confirm methylation of GpC and CpG sequences, Related to Fig 7.
Purified Nel DNA was incubated with buffer or M.CviPI and/or M.SssI methyltransferases, and subsequently digested with HaeIII or BstUI, restriction enzymes which cleave only unmethylated GGCC and CGCG sequence, respectively.
Fig S5. iga DNA from E. coli but not Ngo i35A is fragmented by Ngo restriction enzymes, Related to Fig 7.
ADIDA DNA fragment derived from Ngo i35A (left) and E. coli K-12 (right) digested with HaeII or NlaIV (isoschizomers of NgoI and NgoV) and separated in a 0.7% agarose gel. (−) indicates incubation with buffer alone.
Fig S6. Nel is not killed by Nel or Ngo DNA, Related to Fig 7.
Nel and Ngo CFUs recovered after a 4 h incubation with chromosomal DNA purified from Nel and Ngo (5 μg/mL). Starting CFUs: 5×105. n=3. Error bars represent SEM. LOD: 10 CFUs. (**P<0.01; Student’s t-test).
Table S1. Bacterial strains and primers used in this study, Related to STAR Methods.
Table S2. Identification of DNA as the toxic compound in N. elongata supernatant, Related to Fig 4.
Table S3. Resistance of N. gonorrhoeae DNA uptake mutants to killing by N. elongata DNA, Related to Fig 5.
Table S4. Toxicity of DNA from BACs and BAC subclones for N. gonorrhoeae, Related to Fig 7.
Table S5. Modifications in Nel 29315 (this study), Ngo FA1090 (Blow et al., 2016, Srikhanta et al. 2009), Ngo MS11 (Stein et al., 1992) and E. coli K-12 (Marinus and Løbner-Olesen, 2014) DNA, Related to Fig 7.
Table S6. Toxic DNAs in this study contain sequences recognized by Ngo MS11 R/M systems, Related to Discussion, Fig 4 and Fig 7.
>Table S7. Short regions of homology (9-20 bp) between E. coli plasmids and Ngo MS11 chromosome contain recognition sequences from Ngo MS11 restriction modification (R/M) systems, Related to Discussion and Fig 7.
Table S8. Orthologs of N-acetylmuramyl-L-alanine-amidase AmiC and lytic transglycosylase LtgA in commensal Neisseria, Related to Discussion, Fig 2 and Fig 3.