Abstract
Background and Purpose
We evaluated deep learning algorithms’ segmentation of acute ischemic lesions on heterogeneous multi-center clinical diffusion-weighted (DWI) datasets and explored the potential role of this tool for phenotyping acute ischemic stroke.
Methods
Ischemic stroke data sets from the MRI-GENetics Interface Exploration (MRI-GENIE) repository consisting of 12 international genetic research centers were retrospectively analyzed using an automated deep learning segmentation algorithm consisting of an ensemble of 3D convolutional neural networks (CNNs). Three ensembles were trained using data from: (1) 267 patients from an independent single-center cohort, (2) 267 patients from MRI-GENIE, and (3) mixture of (1) and (2). The algorithms’ performances were compared against manual outlines from a separate 383 patient subset from MRI-GENIE. Univariable and multivariable logistic regression with respect to demographics, stroke subtypes and vascular risk factors were performed to identify phenotypes associated with large acute DWI volumes and greater stroke severity in 2770 MRI-GENIE patients. Stroke topography was investigated.
Results
The ensemble consisting of a mixture of MRI-GENIE and single-center CNNs performed best. Subset analysis comparing automated and manual lesion volumes in 383 patients found excellent correlation (ρ=0.92, p<0.0001). Median [IQR] DWI lesion volumes from 2770 patients were 3.7 [0.9–16.6] cm3. Patients with small artery occlusion (SAO) stroke subtype had smaller lesion volumes (p<0.0001) and different topography compared to other stroke subtypes.
Conclusions
Automated accurate clinical DWI lesion segmentation using deep learning algorithms trained with multi-center and diverse data is feasible. Both lesion volume and topography can provide insight into stroke etiology with sufficient sample size from “big” heterogeneous multi-center clinical imaging phenotype datasets.
Keywords: computer based model, diffusion-weighted imaging, risk factors, stroke, ischemic, statistical model, Cerebrovascular Disease/Stroke, Ischemic Stroke, Magnetic Resonance Imaging (MRI), Risk Factors
INTRODUCTION
Pooling of multi-center datasets has resulted in recent progress towards understanding the genetic pathways underlying ischemic stroke1 and have identified novel loci associated with ischemic stroke.2, 3 Brain imaging, more precise in characterizing the sequelae of ischemic brain injury than clinical exams, could provide further insight. Analysis of big data repositories required for genome wide association studies and vascular risk factors research will necessitate high-throughput, automated and scalable techniques to measure acute ischemic lesions on diffusion-weighted MRI (DWI) precisely and on a multi-center basis. Importantly, for genetic discovery studies which require the processing of thousands of disparate studies, these algorithms will have to perform accurately on MRI scans that have been obtained for routine clinical purposes, across multiple vendors, not using standardized acquisition protocols.
It has been previously shown that an automated method consisting of an ensemble of 3D convolutional neural networks (CNN) trained on multiparametric DWI data can improve segmentation of acute ischemic lesions compared to single CNNs or CNNs whose segmentations are based on a single diffusion parametric map.4 However, this study’s results were based on training and evaluation data from a single center and one MRI vendor. Performance might be further improved using multi-center training data since a known short-coming of deep learning algorithms trained at one center is that they may be over-trained and fail to perform accurately with multi-center datasets. Another question is to what extent diversity of training data can further improve segmentation results. Here, we investigate the robustness of a single center trained neural network for the task of segmenting DWI lesions from the MRI-GENetics Interface Exploration (MRI-GENIE) study, a large multi-center stroke genetics imaging repository, and compare its performance against an algorithm trained with a data subset from MRI-GENIE. In addition, we investigate whether a more diverse training data set, consisting of data from the single center cohort and MRI-GENIE, can lead to improved performance.
Once we have optimized our segmentation algorithm, we applied it to the entire MRI-GENIE cohort to compare the relationships of the predicted volumes against known cerebrovascular risk factors associated with large DWI lesion volumes to test consistency of our automated results with those previously demonstrated using manual approaches. We will also investigate difference in stroke lesion topography as a function of stroke subtype. We will not explore genetic pathways associated with large DWI volumes since that is beyond the scope of this study.
METHODS
Because of the clinical nature of the data collected for this study, requests to access the dataset from qualified researchers trained in human subject confidentiality protocols may be sent to Natalia Rost (nrost@partners.org). The algorithm used for training the CNNs is OpenSource (available at https://github.com/Kamnitsask/deepmedic).
Subjects
All analyses were retrospectively performed under local institutional review board approval.
Single-Center Cohort
The independent, single-center data consisted of 267 acute ischemic stroke patients who presented at a single academic medical center admitted between 1996–2012 with MRI obtained within 24h since the patient was last known to be well (LKW). All patients had not been treated with revascularization treatment prior to MRI. The training cohort were drawn from repositories for which manual outlines were available which had been drawn for other studies.5–7
MRI-GENIE Cohort
The MRI-GENIE cohort consisted of 3,301 acute ischemic stroke data sets from 12 international centers from the Stroke Genetics Network.1 All patients had signed informed consent and were included if the following were available: clinical MRI, demographics, vascular risk factors, genome-wide genotyping and stroke subtype measured with the causative classification of stroke (CCS) system.8 Revascularization interventions were not reported. Six centers also had NIH Stroke Scale (NIHSS) scores collected from patients within 11 days of admission as part of the Genetics of Ischaemic Stroke Functional Outcome study.9
Manual Outlines were available from 666 subjects that had been drawn by two readers (Readers 1 and 2) for another study involving patients from 8 centers and had been randomly selected10 (Manual Cohort A, N=666). 25 datasets were randomly selected from Manual Cohort A for training Reader 2 (please see Supplemental Methods). Another 25 datasets were randomly selected to compare inter-rater manual agreement and were outlined independently by Readers 1 and 2. A second set of 25 datasets (Manual Cohort B) were randomly selected from a ninth center and outlined by a de novo third reader (Reader 3). All Readers had access to the iDWI, ADC and b0 maps but did not systematically refer to ADC and b0 images and created outlines without using thresholds. All readers drew outlines blinded to the pipeline results.
Manual Cohort B was used to determine which of the 3 ensemble models performed best on unseen data from a “new” site. Using outlines from Manual Cohort A for choosing the best ensemble model would bias results towards the ensemble model trained with MRI-GENIE data since the evaluation cohort would have been drawn by Readers 1 and 2. The ensemble model with the best performance metrics (described below) segmenting Manual Cohort B was then used for remaining analyses.
MRI
Single-Center cohort
All data had been obtained for clinical purposes from 1.5T scanners (General Electric Medical Systems). For the majority of cases, the diffusion-weighting (b-value) was 1000 s/mm2. (Please see Supplemental Methods for details.)
MRI-GENIE cohort
MRI-GENIE data were acquired on 1 T, 1.5 T and 3T MRI equipment from 6 vendors (please see Supplemental Methods for details). All datasets were reviewed for availability of raw diffusion data or at least 2 of the 3 of the following diffusion parametric maps: isotropic trace DWI (iDWI), apparent diffusion coefficient (ADC) and b0 maps. Data were excluded which had insufficient image quality for humans to identify the infarct.
Diffusion Parametric Image calculation
For studies with the raw diffusion data available, the individual high b-value volumes were corrected for eddy current distortions prior to calculation of iDWI (geometric mean of the high b-value acquisitions), and ADC maps (slope of the linear regression fit of the log of the DWI and b0, b-value=0 s/mm2 images).11 If only 2 of the post-processed ADC, iDWI or b0 images were present, the third was calculated from the other two: iDWI=b0 * exp (–b-value*ADC). If the b-value was not encoded in the images, 1000 s/mm2 was assumed.
Automated segmentation with CNN
ADC, iDWI and b0 datasets were up-sampled to 1 mm3 isotropic voxels. A brain mask was created by multiplying the skull-stripped up-sampled b0 image12 with an ADC mask (upsampled ADC map greater than 0). The brain mask was used to normalize ADC, iDWI and b0 values by subtracting the mean intensity values (limited to 1–99% of range to exclude outliers) and dividing by the standard deviation. (Please see Supplemental Figure I).
3D CNNs were trained on a workstation with an NVIDIA Tesla K40 GPU using the DeepMedic (v0.7.0) framework with two pathways.13 Details of the DeepMedic framework can be found in the publication13 and in the Supplemental Methods. By design, the DeepMedic training process involves random sampling of data to avoid over-fitting and retain computational efficiency. Despite training a CNN with the same hyper-parameters and data, this sampling strategy inherently results in variations in the lesions segmentation. To compensate for sampling-induced noise, six independent CNNs were trained and the voxel-wise average of their posterior maps for the lesion class were computed, resampled back to the original image resolution and then thresholded at 50% and masked with the brain mask created at the normalization step (please see Supplemental Figure I).
We trained three ensembles: single-center, MRI-GENIE, and mixed. For the single-center ensemble, we trained 6 CNNs using only data from the single center. We repeated this training of 6 independent CNNs with the same number of subjects (N=267) from MRI-GENIE cohort to create the MRI-GENIE ensemble. We then selected 3 of the CNNs trained on the independent cohort and 3 of the CNNs trained on the MRI-GENIE cohort to create the Mixed ensemble. In this manner, we controlled for performance benefits from an imbalance of number of subjects involved in training across ensembles.
Segmentation Performance Evaluation
Automated segmentation results were compared to manual outlines volumetrically (Spearman’s non-parametric correlation). Voxel-wise performance was assessed by Dice (measure of overlap between automated and manual lesions), sensitivity, and precision (positive predictive value) scores. Dice score, precision and sensitivity were computed as follows: Dice = 2TP/(2*TP+FP+FN); Precision = TP/(TP+FP); Sensitivity = TP/(TP+FN) for which TP=true positives, FP=false positive and FN=false negatives. All metrics range from 0–100%, for which higher values indicate better performances. All voxel-wise metrics were evaluated at 1 mm resolution to reduce confounds from different MRI acquisition resolutions. Performance between the best performing ensemble and its individual CNNs were compared (paired two-sided Wilcoxon signed rank tests). Comparisons were limited to patients for whom DWI lesions were detected by the manual readers and whose data were not used for training.
Statistical analysis
Analyses were conducted using JMP Pro 14.0 (SAS Institute, Cary, NC) unless otherwise noted. Univariable comparisons were done with Wilcoxon two-sample rank sum test for continuous variables, and two-sided Fisher’s Exact Test for categorical variables. Results are described in terms of mean±standard deviation, or median [interquartile range]. For all analyses, p-values < 0.05 were considered statistically significant.
Because of the pooling of multi-center data, heterogeneity of clinical and imaging phenotypes across centers was measured using the I2 metric,14 calculated with the meta package in R (http://meta-analysis-with-r.org). Moderate heterogeneity was defined as I2 between 0.5 and 0.75, and high heterogeneity as I2 > 0.75. Mixed modeling (fitted with restricted maximum likelihood method) was used with center treated as a random effect to investigate the association between segmentation performance and lesion volume, time-to-MRI and MRI acquisition parameters (field-of-view, b-value, field strength and vendor).
One-way analysis of variance followed by post-hoc Wilcoxon rank tests was used to compare lesion volumes as a function of center and CCS scores. Multivariable linear regression for predictors of large lesion volumes was performed by including variables that were significant (p<0.05) on a univariable basis. Genetic research center was included as a random effect in all models. For univariable and multivariable analyses, stroke subtype was dichotomized to SAO or non-SAO.
To investigate the topographical differences between stroke subtypes, b0 data were registered to the ICBM T2 atlas15, 16 (which had been registered to MNI 152 1 mm atlas from FSL17) using non-linear registration techniques.17, 18 The automatically segmented lesions were spatially registered to MNI 152 1 mm atlas using the derived transformation parameters. Lesion incidence maps were calculated for all MRI-GENIE DWI data. Frequency maps as a function of stroke subtype were generated by dividing the incidence map by the number of subjects and compared using cross-correlation (fslcc17).
All figures of MRI data are displayed in radiologic convention. Figures with heat maps were generated using FSLeyes (version 0.27).19 Incidence and frequency map results were displayed using MRIcroGL (http://www.mricro.com).
RESULTS
Subjects
The demographics for the 267 patients in the single-center training cohort are shown in Supplemental Table I. For the independent multi-center dataset, of the 3301 patients available in MRI-GENIE, 531 were excluded for reasons described above (please see Supplemental Figure II). Distribution of the remaining 2770 patients as a proportion of the MRI-GENIE and Stroke Genetics Network cohorts by center are shown in Supplemental Figure III. The mean±SD age was 63.4±14.7 years old (N=2765). Median [IQR] NIHSS score was 3 [2–7] (N=1312), obtained 1 [0–1] days from admission. Time-to-MRI was 1 [0–4] (N=2464) days from admission. Moderate to high heterogeneity across centers was observed across parameters (please see Supplemental Results). Demographics breakdown by center can be found in Supplemental Tables II and III. Demographics for the MRI-GENIE training cohort are provided in Supplemental Table IV.
Segmentation Performance Evaluation
Influence of training data on segmentation performance
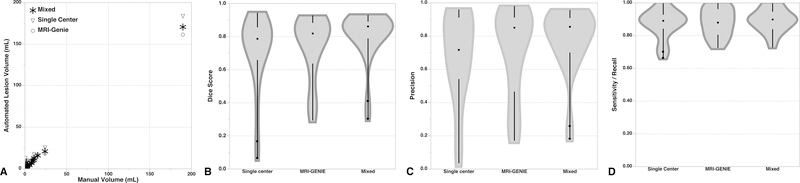
Two cases in Manual Cohort B for which no lesion was detected by the Reader 3 were excluded and analysis was limited to the remaining 23 patients. The distribution of Manual Cohort B volumes was 2.0 [0.9–9.1] cm3. The mixed ensemble model had a Dice score (0.86 [0.79–0.89]) that was significantly superior to that of the single-center cohort ensemble (0.79 [0.66–0.86], p=0.0002) and MRI-GENIE ensemble (0.82 [0.64–0.88], p=0.007) (see Figure 1). The single-center ensemble was comparable to the MRI-GENIE ensemble (p=0.24). The mixed ensemble precision (0.86 [0.7–0.91]) was greater than the single-center ensemble (0.72 [0.54–0.91], p=0.002) but not the MRI-GENIE ensemble (0.85 [0.47–0.91], p=0.09). There was no significant difference in precision between the single-center and MRI-GENIE ensembles (p=0.66). There were no statistically significant differences in terms of sensitivity between the ensembles (mixed: 0.90 [0.84–0.95]; MRI-GENIE: 0.88 [0.81–0.96]; single-center: 0.89 [0.84–0.93]). For the remainder of the analysis, we thus used the mixed ensemble model.
Figure 1:
Evaluation of 3 ensemble segmentations compared to Manual Cohort B outlines. Shown are (A) correlations (Single-Center Ensemble: ρ=0.74, p<0.0001; MRI-GENIE Ensemble: ρ=0.88, p<0.0001; Mixed Ensemble: ρ=0.91, p<0.0001) and violin plots (middle dot are median values, spaces between black lines are IQR) of (B) Dice score (C) precision and (D) sensitivity. See text for details.
Ensemble versus individual CNN models
Manual Cohort A consisted of 666 patients. 16 patients were excluded since no DWI lesion was identified by the readers. 267 patients used for training 3 of the CNNs in the mixed ensemble were excluded for performance metrics analysis to avoid biasing the results. Median [IQR] manual lesion volumes for the remaining 383 subjects were 2.9 [0.8–19.2] cm3. The Ensemble model outperformed all individual CNN models (please see Supplemental Figure IV) in terms of Dice score (0.77 [0.57–0.88]), and precision 0.83 [0.57–0.94]) but not sensitivity (0.82 [0.66–0.91]) for which it underperformed one individual CNN model, performed equivalently to 2 others and outperformed 3. There was one patient for which the ensemble model did not detect any lesion (measured volume was 0.2 cm3), and thus precision could not be calculated for the one patient and excluded in the analysis. For the remainder of the analyses, we therefore focus on the ensemble results.
Effects of MRI acquisition on performance
The median automated lesion volumes for the 383 subjects were 3.9 [0.9–18.3] cm3, with median difference from measured volumes of −0.02 [−0.9–0.8] cm3, and correlated significantly with respect to manual outlines (ρ=0.92, p<0.0001). Mixed modeling with center as a random effect showed that Dice score (p<0.0001), precision (p<0.0001), and sensitivity (p<0.0001) were significantly greater with larger lesion volumes. No differences in performance (p>0.05) were found as a function of time-to-MRI (though very late scans could have poor segmentation results, please see Supplemental Figure V), field-of-view, field strength and vendor. However, there were significant differences for high b-value for Dice (b-value=1500 s/mm2 (N=52): 0.52 [0.30–0.76] vs b-value=1000 s/mm2 (N=331): 0.80 [0.63–0.88], p=0.009) and precision (b-value=1500 s/mm2 (N=52): 0.43 [0.21–0.80] vs b-value=1000 s/mm2 (N=330): 0.86 [0.65–0.94], p=0.02), but not sensitivity (p=0.64).
Inter-rater analysis: Human versus human versus machine
For inter-rater analysis, no statistically significant differences were found between the performance metrics of Reader 2 and Ensemble method for the 25 independent cases in terms of (a) median volumetric differences with Reader 1 (Reader 2: 0.4 [−1.7–4.4]; Ensemble: −1.6 [−3.4–0.9] cm3; p=0.10) (b) Dice score (Reader 2: median 0.86 [IQR 0.81–0.95]; Ensemble: 0.88 [0.78–0.95]; p=0.93), (c) precision (Reader 2: 0.93 [0.83–0.97], Ensemble: 0.94 [0.81–0.96], p=0.95), and (d) sensitivity (Reader 2: 0.86 [0.79–0.93]; Ensemble: 0.90 [0.83–0.94], p=0.11). Figure 2 shows an example of discordant results between Reader 1 and 2 in a subject with acute, subacute and chronic infarcts. The Ensemble model segmented a lesion that encompassed regions intermediate between the two readers.
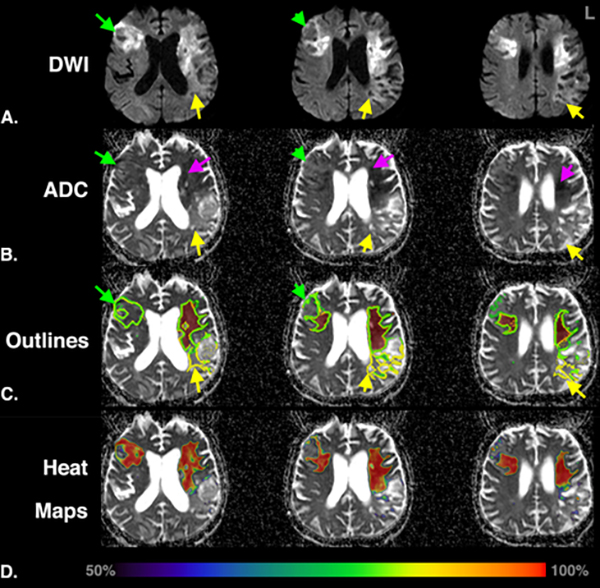
Figure 2:
Example of discordance between Readers 1 and 2 and automated segmentations for a 76-year old man, with Undetermined stroke etiology, imaged 20 days from stroke onset. Shown are (A) iDWI and (B) ADC maps, (C) lesion segmentations (Ensemble: green outline, Reader 1: red region, Reader 2: yellow outline) and (D) ensemble probability (a.k.a. heat) maps for tissue infarction. Regions only outlined by Reader 2 (yellow arrows) have elevated ADC and iDWI. Regions where the Ensemble agreed with Reader 2 (green arrows) have pseudo-normal ADC and elevated DWI. Regions for which both readers and the ensemble model agree have reduced ADC (purple arrows) and highest probability. Lesion segmentation for this subject was complicated by the mixture of acute, subacute and chronic regions of ischemic injury that led to discordant results from similarly trained human readers. This showcases the benefit of automated algorithms that generate consistent reproducible results. Images are in radiological orientation. ADC, Apparent Diffusion Coefficient; iDWI, isotropic trace DWI.
Risk Factors for Large Acute Ischemic Lesion Volumes
The segmentation results using the Ensemble model was 3.7 [0.9–16.6] cm3 across all 2770 patients. Automated lesion volumes between patients with and without non-zero manual outlines were not significantly different (p=0.69). The remainder of the analyses therefore used results from all 2770 patients.
DWI lesion volumes (lnDWIv) differed significantly (p<0.0001) by center and by CCS (Figure 3). Lesion volumes differed significantly in terms of self-identified race (white), AF, and SAO strokes (see Table 1). Lesion volumes were significantly correlated with NIHSS (N=1312, ρ=0.38, p<0.0001), but not age (ρ=−0.03, p=0.08). Results of univariable and multivariable predictors of larger lesion volumes are provided in the Supplemental Results.
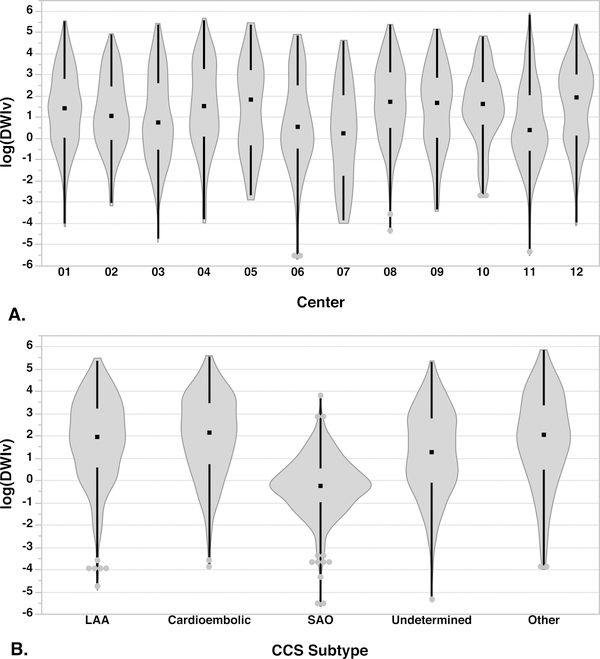
Figure 3:
Distribution of log of the DWI lesion volumes by (A) center and (B) CCS subtype. There was a significant difference between centers in terms of the DWI volumes (p<0.0001). Centers 6, 7 and 11 had the smallest median lesion volumes (< 2 cm3), while Centers 5 and 12 had the largest median volumes (>6 cm3). Small artery occlusion had the smallest median volume compared to all the other subtypes (p<0.0001) followed by Undetermined (p<0.0001).
Table 1:
Differences in automatic DWI lesion volumes (cm3) as a function of demographics and vascular risk factors. The first column (All) shows the number of patients (%) positive for the phenotype out of 2770 patients unless otherwise noted.
| Characteristic | All | Yes | No | P-value |
|---|---|---|---|---|
| Sex, male | 1701 (61.4) | 3.5 [0.9–16.5] | 4.0 [0.9–16.9] | 0.99 |
| Ethnicity, Hispanic (N=2723) | 178 (6.5) | 4.9 [1.0–21.6] | 3.7 [0.9–16.6] | 0.17 |
| Race, white (N=2560) | 2287 (89.3) | 4.1 [1.0–17.9] | 2.5 [0.8–13.4] | 0.02 |
| Medical History | ||||
| Hypertension (N=2749) | 1813 (66.0) | 3.4 [0.9–15.9] | 4.5 [0.9–18.0] | 0.08 |
| Diabetes Mellitus (N=2741) | 643 (23.5) | 3.2 [0.8–13.4] | 4.0 [0.9–17.4] | 0.10 |
| Atrial Fibrillation (N=2736) | 410 (15.0) | 7.3 [1.4–31.4] | 3.3 [0.8–15.1] | <0.0001 |
| Coronary Artery Disease (N=2719) | 491 (18.1) | 4.0 [1.0–16.5] | 3.6 [0.9–16.7] | 0.42 |
| Current or former smoker | 1448 (52.3) | 3.4 [0.9–17.0] | 3.9 [0.9–16.1] | 0.93 |
| Causative Classification of Stroke | ||||
| Large artery atherosclerosis | 599 (21.6) | 7.1 [1.8–25.8] | 2.9 [0.8–14.5] | <0.0001 |
| Cardioembolic | 426 (15.4) | 8.6 [2.1–32.7] | 3.1 [0.8–14.7] | <0.0001 |
| Small Artery Occlusion | 432 (15.6) | 0.8 [0.4–1.7] | 5.4 [1.321.3] | <0.0001 |
| Undetermined | 1112 (40.1) | 3.5 [0.9–15.8] | 3.8 [0.9–17.8] | 0.16 |
| Other | 201 (7.3) | 7.5 [1.6–29.9] | 3.4 [0.9–16.0] | <0.0001 |
Stroke Lesion Topography
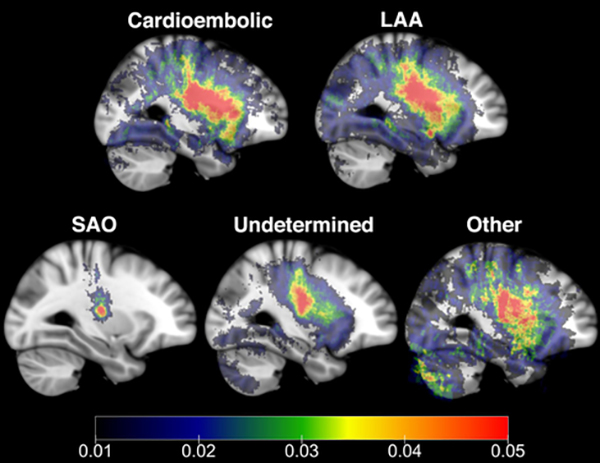
Supplemental Figure VI shows examples of stroke segmentations across locations with similar volumes, demonstrating the importance of knowledge of lesion topography. MRI data from 7 patients could not be non-linearly co-registered due to image artifacts. Stroke lesion incidence maps for remaining 2763 are shown in Figure 4. Figure 5 shows the frequency maps for each stroke subtype. SAO had the greatest difference compared to non-SAO (R=0.39) followed by Other vs non-Other (R=0.89) with the remaining subtypes showing high similarity (LAA vs non-LAA, R=0.95; cardioembolic vs non-cardioembolic, R=0.94; undetermined vs non-undetermined, R=0.94).
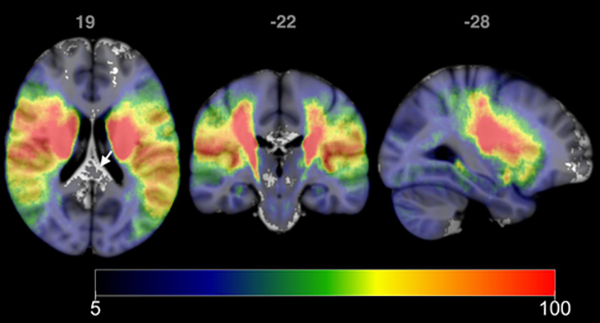
Figure 4:
Incidence maps for 2763 patients. Each voxel represents the number of patients with lesions involving that voxel. Maximum incidence was 295, with incidence shown using the range 5 to 100. Voxels for which less than 5 patients had lesions at that location do not have an overlaid colormap (see arrow). Images are shown in radiological format.
Figure 5:
Frequency maps (incidence map/number of subjects) showing topographical distribution of infarcts across different stroke subtypes – cardioembolic (N=425), LAA (N=597), SAO (N=431), Other (N=200), and Undetermined (N=1110). SAO frequency map had the lowest correlation compared with the other subtypes, followed by Other. Images are shown in radiological format.
DISCUSSION
We have demonstrated accurate fully automated segmentation of ischemic lesions of multi-center clinically acquired DWI data is feasible. These data sets were obtained as part of routine clinical practice involving multiple field strengths, sequences, vendors, and acquisition protocols that spanned over several years, resulting in very heterogeneous populations. We showed that an ensemble algorithm trained with data from a single center can perform comparably well with a model trained with multi-center data. Furthermore, we demonstrated that even better results can be obtained when using an ensemble algorithm combining CNNs trained on diverse data sets. The performance of all models were similar to another study which used CNN approaches to evaluate 361 patients from multiple centers, achieving mean Dice score of 0.67.20 Our study is distinct from the other study20 in that we performed a thorough comparison of the effects of various training datasets on the performance of machine learning algorithms. In addition, the other study developed a CNN that was trained (N=380) and tested (N=361) on data from the same centers, naturally achieving high performance metrics. One of the key questions we sought to address was whether models trained on one center’s DWI data can be transferred to segment DWI data from another center accurately. If the answer was no, for genetic discovery studies that require big datasets, a new algorithm would need to be trained whenever a new center’s dataset was incorporated into a repository. Fortunately, our results suggest that a robust segmentation network developed using a mixture of ensembles trained on diverse datasets can provide accurate segmentation.
We also explored factors inherent to multi-center datasets that can affect the performance of machine learning algorithms. Interestingly, variations in DWI acquisition across vendors did not affect the algorithm performance, with the exception of high b-value. The results of poor performance on data acquired with very high b-values (1500 s/mm2) is not surprising since diffusion data acquired with b-values>1000 s/mm2 are known to exhibit non-mono-exponential behavior leading to non-pathological differences between normal gray and white matter.21 We found that smaller lesion volumes had poorer Dice scores, consistent with previous results using the ensemble approach4 and other methods.20 Even for human readers, the intra-rater and inter-rater volume measurements fluctuate by up to 2.7 cm3.22 The performance of our method may therefore be underestimated since the MRI-GENIE cohort consisted of primarily smaller lesion volumes. Segmentation accuracy did not depend on the timing of the MRI scan with respect to stroke presentation. We mitigated the impact of timing by training with data acquired over a range of times with our mixed ensemble model.
Although our finding that SAO stroke subtypes involve smaller DWI lesion volumes is well known, and stroke subtype classification typically uses imaging, it is reassuring that our automated algorithms generated consistent results. Without having manual lesion volumes available for all 2770 subjects, this finding along with results showing positive correlation of NIHSS with automated DWI segmentations reinforces our confidence in the accuracy of our results. In addition, given our large dataset, we were able to create incidence maps as a function of stroke subtype consisting of at least 200 subjects for each subtype. However, 200 subjects may still be insufficient to test hypotheses, emphasizing the need for even larger imaging repositories and automated pipelines for extracting phenotypic information.
Major strengths of our study are the very large number of patients (N=2770) in our study and their diversity. However, the pooling of multi-center datasets for which data were collected retrospectively can also be considered a limitation due its heterogeneity. For the majority of patients, the b-value of the DWI acquisition was not available and thus a b-value=1000 s/mm2 was assumed, affecting ADC calculation. However, our pipeline normalized input images prior to segmentation and thus mitigated the effects of incorrect b-values. Thus, the heterogeneity of the MRI-GENIE cohort could also be considered a major strength of the study since it allows us to test the generalizability of our approach for disparate datasets.
In conclusion, we have demonstrated the feasibility of automated acute lesion segmentation of multicenter, big stroke data repositories that can be used to facilitate the genetic discovery of acute clinical MRI phenotypes. Although we did not present genetic analysis here, we have laid the foundations for future investigations of single nucleotide polymorphisms associated with large DWI lesions. Whereas the interpretation of genetic association with acute infarct volume may be limited due to variability of imaging endophenotypes linked to a specific ischemic stroke subtype, using advanced genetic analysis approaches such as mendelian randomization may offer disease-specific insights. High-throughput studies investigating the association between imaging phenotypes with genetics, stroke severity and long-term functional outcomes in large multi-center data sets will become realistic using automated tools powered by artificial intelligence.
Supplementary Material
Acknowledgments
We acknowledge the support of NVIDIA Corporation with the donation of the Tesla K40 GPU used for this research.
Sources of Funding:
• NIH: R01NS030678 (AV, DLW), R01NS039987 (BBW), R01NS042733 (BBW, JFM), R01NS059775 (OW), R01NS063925 (OW), R01NS082285 (NSR, OW), R01NS086905 (NSR, OW, SJK), R01NS100178 (BM), R01NS100417 (AV), R01NS103824–01 (AV), R01NS29993 (RLS, TR), P50NS051343 (OW), U01NS069208 (BBW, JR, OW, JWC), U10NS077311 (AV), K23NS064052 (NSR), P41EB015896 (OW), 1S10RR019307 (OW)
• The Swedish state under the agreement between the Swedish government and the county councils, the Avtal om Läkarutbildning och Forskning (ALF) agreement: (AL, CJ, JW)
• Swedish Stroke Association: (AL, CJ, JW)
• Swedish Heart and Lung Foundation: (AL, CJ)
• Lund University: (AL)
• Region Skåne: (AL)
• Skåne University Hospital: (AL)
• Freemasons Lodge of Instruction Eos in Lund: (AL)
• Foundation of Färs & Frosta: (AL)
• Swedish Research Council: (CJ)
• Spanish Ministry of Science and Innovation (PI051737, PI10/02064, PI12/01238, PI15/00451); European Regional Development Fund (EDRF) Red de Investigación Cardiovascular (RD12/0042/0020); Fundació la Marató TV3 (76/C/2011); Recercaixa’13 (JJ086116): (JJC)
• European Research Council: Horizon 2020 MSC Global Fellowship #753896 (MDS)
• Fonds voor Wetenschappelijk Onderzoek: 1841918N (RL)
• Crafoord Foundation: (JW)
• Henry Smith Charity, Qatar National Research Fund, Stroke Association (United Kingdom), Dept of Health (United Kingdom), British Council, United Kingdom-India Education Research Initiative (UKIERI): Bio-Repository of DNA in Stroke (BRAINS)
• American Heart Association: Clinical Research Training Fellowship (MRE); Cardiovascular Genome-Phenome Study (#15GPSPG23770000) (JWC); Discovery Grant supported by the Bayer Group (#17IBDG33700328) (JWC), Uncovering new patterns in cardiovascular disease and stroke (#18UNPG34030160) (OW).
• President’s PhD Scholarship of Imperial College London: (KK)
• Department of Veterans Affairs (United States): Baltimore Research Enhancement Program (JWC)
Footnotes
Disclosures:
Arne Lindgren: Honoraria: Bayer, BMS Pfizer; Advisory Board: Bayer, Astra Zeneca, BMS Pfizer
Jonathan Rosand: Consulting: New Beta Innovations, Pfizer, Boehringer Ingelheim
Ona Wu: Consulting: Penumbra, Inc; Advisory Board Member: Genentech, Inc.
REFERENCES
- 1.Meschia JF, Arnett DK, Ay H, Brown RD Jr., Benavente OR, Cole JW, et al. Stroke genetics network (SiGN) study: Design and rationale for a genome-wide association study of ischemic stroke subtypes. Stroke. 2013;44:2694–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50:524–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.NINDS Stroke Genetics Network, International Stroke Genetics Consortium. Loci associated with ischaemic stroke and its subtypes (SiGN): A genome-wide association study. Lancet Neurol. 2016;15:174–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winzeck S, Mocking SJT, Bezerra R, Bouts MJRJ, Mcintosh EC, Diwan I, et al. Ensemble of convolutional neural networks improves automated segmentation of acute ischemic lesions using multiparametric diffusion-weighted MRI. AJNR Am J Neuroradiol. 2019:40(6):938–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu O, McIntosh E, Bezerra R, Diwan I, Garg P, Mocking S, et al. Prediction of lesion expansion in patients using acute MRI. Stroke. 2012;43:A3319 [Google Scholar]
- 6.Wu O, Schwamm LH, Garg P, Pervez MA, Yoo AJ, Chautinet A, et al. Using MRI as the witness: Multimodal MRI-based determination of acute stroke onset. Stroke. 2010;41:E273–E273 [Google Scholar]
- 7.Wu O, Koroshetz WJ, Østergaard L, Buonanno FS, Copen WA, Gonzalez RG, et al. Predicting tissue outcome in acute human cerebral ischemia using combined diffusion- and perfusion-weighted MR imaging. Stroke. 2001;32:933–942 [DOI] [PubMed] [Google Scholar]
- 8.Giese AK, Schirmer MD, Donahue KL, Cloonan L, Irie R, Winzeck S, et al. Design and rationale for examining neuroimaging genetics in ischemic stroke: The MRI-GENIE study. Neurol Genet. 2017;3:e180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maguire JM, Bevan S, Stanne TM, Lorenzen E, Fernandez-Cadenas I, Hankey GJ, et al. Giscome – genetics of ischaemic stroke functional outcome network: A protocol for an international multicentre genetic association study. European Stroke Journal. 2017;2:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giese AK, Xu H, Schirmer MD, Donahue KL, Dalca AV, Gaynor BJ, et al. Genetics of acute ischemic lesion volume: The MRI-genetics interface exploration (MRI-GENIE) study. Stroke. 2018;49 (Suppl 1):AWMP56 [Google Scholar]
- 11.Sorensen AG, Wu O, Copen WA, Davis TL, Gonzalez RG, Koroshetz WJ, et al. Human acute cerebral ischemia: Detection of changes in water diffusion anisotropy by using MR imaging. Radiology. 1999;212:785–792 [DOI] [PubMed] [Google Scholar]
- 12.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, et al. Efficient multi-scale 3d CNN with fully connected crf for accurate brain lesion segmentation. Med Image Anal. 2017;36:61–78 [DOI] [PubMed] [Google Scholar]
- 14.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.UCLA Brain Mapping Center. ICBM T2 atlas. 2018
- 16.Mazziotta JC, Toga AW, Evans A, Fox P, Lancaster J. A probabilistic atlas of the human brain: Theory and rationale for its development. The international consortium for brain mapping (ICBM). Neuroimage. 1995;2:89–101 [DOI] [PubMed] [Google Scholar]
- 17.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62:782–790 [DOI] [PubMed] [Google Scholar]
- 18.Andersson JLR, Jenkinson M, S. S Non-linear registration, aka spatial normalisation. Technical Report FMRIB Technical Report TR07JA2. 2007 [Google Scholar]
- 19.McCarthy P FSLeyes FSLeyes | Zenodo; Available from: 10.5281/zenodo.1470761. Accessed April 1, 2019. [DOI] [Google Scholar]
- 20.Chen L, Bentley P, Rueckert D. Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks. Neuroimage Clin. 2017;15:633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshiura T, Wu O, Zaheer A, Reese TG, Sorensen AG. Highly diffusion-sensitized MRI of brain: Dissociation of gray and white matter. Magn Reson Med. 2001;45:734–740 [DOI] [PubMed] [Google Scholar]
- 22.Luby M, Bykowski JL, Schellinger PD, Merino JG, Warach S. Intra- and interrater reliability of ischemic lesion volume measurements on diffusion-weighted, mean transit time and fluid-attenuated inversion recovery MRI. Stroke. 2006;37:2951–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.