Abstract
Stabilization of the MYC oncoprotein by KRAS signaling is a requirement for growth of pancreatic ductal adenocarcinomas (PDAC). However, current understanding of how MYC protein stability is regulated in MYC-dependent cancers such as KRAS-mutant PDAC is incomplete. We developed a flow cytometry-based assay, screened a library of >800 protein kinase inhibitors, and identified compounds that promoted either stability or degradation of MYC. We validated compounds that stabilize or destabilize MYC, and then focused on one compound, UNC10112785, that induced significant loss of MYC protein. We determined that this compound is a potent CDK9 inhibitor with a novel scaffold, and that it causes MYC loss through both transcriptional and post-translational mechanisms. We discovered that CDK9 regulates MYC protein stability through a novel KRAS-independent mechanism involving direct phosphorylation of MYC Ser62. Our studies present the application of an innovative screening strategy that can be adapted to identify novel regulators of protein stability.
One Sentence Summary:
We applied a MYC oncoprotein degradation screen and identified CDK9 as a novel regulator of MYC protein degradation.
INTRODUCTION
In 2017, pancreatic cancer surpassed breast cancer to become the third leading cause of cancer deaths in the U.S (1). By around 2020, pancreatic cancer is projected to surpass colorectal cancer and become the second leading cause of U.S. cancer deaths (2). Currently, the 5-year overall survival rate is at an abysmal 8% (1). Despite a well-defined genetic profile of pancreatic ductal adenocarcinoma (PDAC) (3), clinically effective targeted therapies remain to be developed, with current treatments limited to conventional cytotoxic drugs (4).
The main genetic driver of PDAC initiation, progression and maintenance is mutational activation of the KRAS oncogene, which is found in ~95% of PDAC (3). Although KRAS was the first cancer gene identified in human cancers over 35 years ago (5), the effort to effectively target RAS-driven cancers is still ongoing (6, 7).
The interplay and interdependency of the RAS and the MYC oncogenes in driving cancer development and maintenance is well-established. This association was first demonstrated when it was shown that MYC overexpression was necessary to support RAS transformation of rodent fibroblasts (8). MYC expression is frequently increased in many cancers, most commonly by gene amplification or increased gene transcription (9). Subsequent studies in genetically engineered mouse models of cancer demonstrated the essential role of MYC in KRAS-driven oncogenesis (10, 11) and genetic suppression of MYC impairs KRAS-driven PDAC (12–15). Moreover, overexpression of MYC alone was sufficient to phenocopy mutant KRAS and drive development of metastatic PDAC (16). Thus, targeting MYC could be a novel therapeutic strategy for MYC-dependent cancers such as KRAS-mutant PDAC. However, similar to RAS, MYC has also been considered undruggable due to a surface topology that lacks deep pockets for design of potent small molecule binders (17).
The MYC transcription factor drives a multitude of proliferative and pro-growth phenotypes (9). Strategies to inhibit MYC function have included inhibition of MYC transcription (for example, bromodomain inhibitors, JQ1) (18, 19), inhibition of MYC/MAX dimerization (20, 21), targeting expression of MYC-regulated genes (22) or MYC-associated metabolic vulnerabilities (23). Of these strategies, only bromodomain inhibitors have entered clinical trials, but their relative lack of selectivity for MYC transcription remains a concern (24).
Mutationally activated KRAS promotes increased MYC expression by stimulating MYC gene transcription and by promoting MYC protein stability (14, 25). KRAS effector signaling promotes MYC protein stability through ERK mitogen-activated protein kinase phosphorylation on MYC residue Ser62 (26). Phosphorylated Ser62 then facilitates GSK3β phosphorylation of MYC at Thr58, and subsequent dephosphorylation of Ser62 by the tumor suppressor PP2A promotes E3 ligase FBXW7-dependent MYC degradation. KRAS signaling through the PI3K effector pathway, leading to activation of AKT and concomitant inactivation of GSK3β, represents a second effector signaling mechanism by which KRAS can regulate MYC protein stability. Pharmacologic inhibition of SET, a negative regulator of PP2A, increased MYC degradation and impaired PDAC tumorigenic growth, supporting the therapeutic value of targeting MYC protein degradation (27).
Our previous studies found that KRAS regulation of MYC protein stability in KRAS-mutant PDAC involved both ERK-dependent and -independent mechanisms but not PI3K-AKT signaling (14, 25). To further elucidate the mechanisms by which KRAS regulates MYC protein stability, we developed and applied a MYC protein degradation screen in KRAS-mutant PDAC cells (14). To identify novel protein kinase-dependent mechanisms that regulate MYC protein stability, we then screened the Published Kinase Inhibitor Set (PKIS) of ATP-competitive protein kinase inhibitors (28, 29). This approach, together with two other screening strategies, identified a MEK5-ERK5 compensatory mechanism induced by inhibition of KRAS-ERK1/2 function (14). In this study, we now focus on the methodology for application of the screen and the experimental strategies to validate kinase inhibitors that either stabilize MYC protein or promote its degradation. Our evaluation of one compound that stimulated loss of MYC protein identified cyclin-dependent kinase 9 (CDK9) as a novel regulator of MYC protein stability.
RESULTS
Establishment of a MYC protein degradation screen
We have described our generation and validation of a MYC degradation reporter for use in a cell-based screen to identify protein kinases that regulate MYC protein stability (14). We utilized the pGPS-LP lentiviral reporter plasmid in which a CMV promoter regulates expression of a single bicistronic mRNA transcript that encodes both DsRed and EGFP-tagged proteins, separated by an internal ribosome entry site (IRES) (30). To construct a reporter capable of monitoring MYC protein expression, we introduced the cDNA sequence encoding human MYC into pGPS-LP to encode an EGFP-MYC fusion protein (designated GPS-MYC; Fig. 1A, (14)). We then stably infected the KRAS-mutant PDAC cell line MIA PaCa-2 with the GPS-MYC vector and established populations of cells stably expressing DsRed-IRES-EGFP-MYC (hereafter referred to as GPS-MYC cells). Importantly, EGFP has a long half-life, but partially adopts the degradation characteristics of MYC when expressed as a fusion protein (see below). Further, since DsRed is expressed from the same transcript as EGFP-MYC, we can normalize for fluctuations in transcription and proteasome activity on a per cell basis. Thus, we can infer the relative stability of MYC on a per cell basis by examining the EGFP/DsRed ratio (31).
Fig. 1. Validation of a MYC degradation reporter.
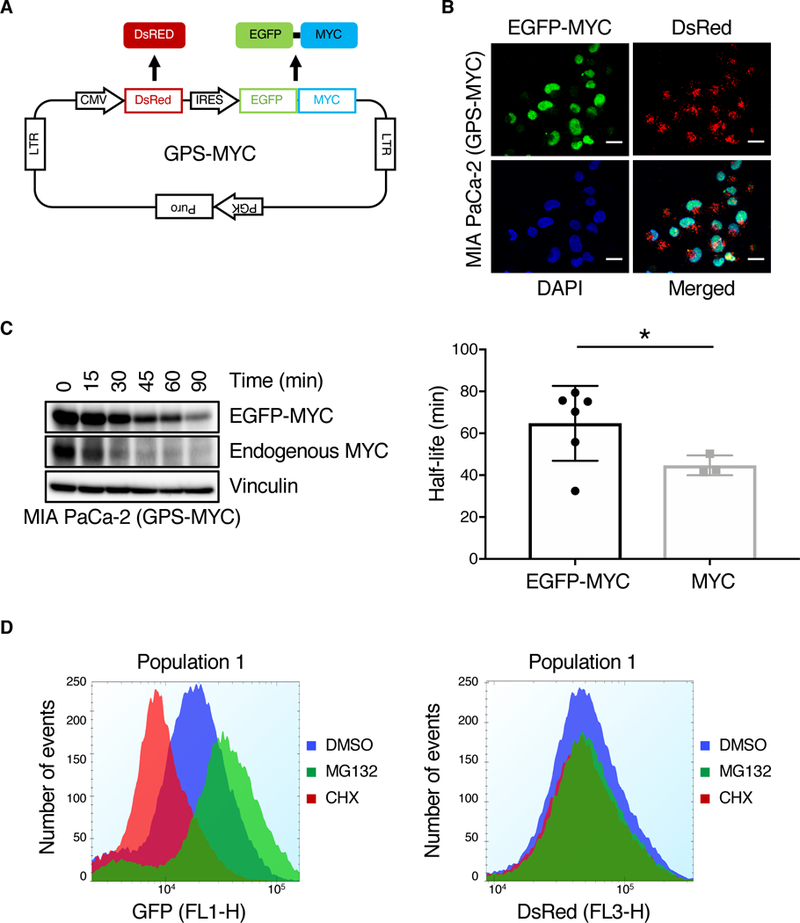
(A) Overview of the GPS-MYC vector. (B) Confocal images of GPS-MYC cells to determine EGFP-MYC subcellular localization to the nucleus, which was visualized by DAPI staining. Scale bar = 20 μm. (C) GPS-MYC cells were treated with CHX for the indicated times and EGFP-MYC and MYC levels were measured by immunoblotting (left panel). The half-lives of EGFP-MYC and endogenous MYC were calculated by fitting the data to a one-phase decay curve. (D) GPS-MYC cells were treated with MG132 and cycloheximide for 6 hours and EGFP and DsRed (F) levels were measured by flow cytometry. All data are representative of at least three independent experiments.
In this study, we first determined whether the exogenously expressed EGFP-MYC displayed properties of endogenous MYC. Like endogenous MYC, EGFP-MYC localization was restricted to the nucleus (Fig. 1B). We showed previously that acute suppression of KRAS caused similar loss of endogenous MYC and EGFP-MYC (14). Although the half-life of EGFP-MYC is slightly longer than that of endogenous MYC (Fig. 1C), demonstrating that EGFP-MYC protein stability is not regulated exactly like endogenous MYC, we reasoned that compounds potent enough to cause loss of the more stable EGFP-MYC would also be able to cause loss of the less stable endogenous MYC. In the GPS-MYC cells, DsRed serves as an internal control for differential expression levels of the cassette amongst GPS-MYC cells. Importantly, treatment of GPS-MYC cells with either cycloheximide (CHX) to inhibit protein synthesis or MG132 to inhibit proteasome-dependent protein degradation altered the EGFP but not the DsRed signal (Fig. 1D). Thus, the ratio of EGFP/DsRed signals provides an accurate determination of MYC protein stability in GPS-MYC cells.
After validating GPS-MYC cells as a cell model for monitoring MYC protein stability, we optimized the assay in a 384-well format using liquid handing automation (Fig. 2A). We first evaluated several sources of commercially available 384-well plates for retention of GPS-MYC cells during the automated washing steps in the screen. Although all the plates were cell culture-treated, not all of them retained cells well during the washing process. Next, because MIA PaCa-2 cells have a propensity to aggregate, we optimized the type and volume of cell dissociation reagents as well as shake speed and frequency during the screen to ensure that cells remained in a single cell suspension throughout the assay. Since MYC has a short half-life in MIA PaCa-2 cells (t1/2 ~50 min) (32), a potential confounding assay variable could be differences in the EGFP-MYC signal over the course of assaying an entire plate. To minimize this potential variable, we optimized the sip time (analysis time per well). At the conclusion of our optimization efforts, the assay duration for each 384-well plate was ~45 min, during which the EGFP and DsRed signals remained constant (Fig. 2B). Since the expression level of EGFP-MYC was only ~5-fold above that of endogenous MYC, the resulting EGFP signal was relatively weak, and, coupled with a high background signal, the dynamic range between the vehicle (DMSO) and CHX controls was also relatively low (~2-fold). Fixation of GPS-MYC cells reduced the EGFP signal even further, rendering the dynamic range too small, so we used live cells in the assay. Despite the relatively narrow dynamic range of the assay between the CHX and DMSO controls, the Z-factors were consistently greater than 0.7, indicating that the assay was robust. All of our screening was then done on the optimized conditions as described in the Materials and Methods section.
Fig. 2. Optimization of a MYC degradation screen.
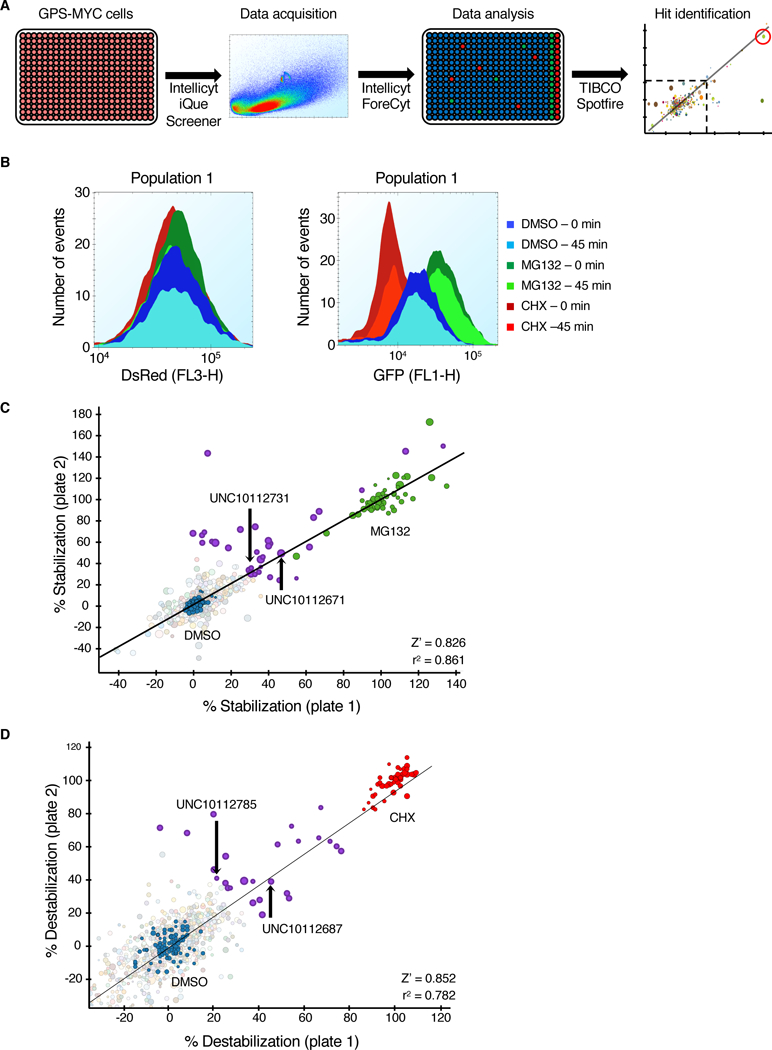
(A) Schematic of the GPS-MYC screen. (B) GPS-MYC cells were treated with vehicle alone (DMSO), or with MG132 or CHX for 6 hours and analyzed on an Intellicyt iQue Screener. The data presented are from the first two and last two wells of each control, representing the beginning (0 min) and the end (45 min) of the assay. (C) The GPS-MYC screen was run in duplicate. Data were normalized to control DMSO (blue circles, 0% stabilization) and MG132 (green circles, 100% stabilization), and hits were determined by a cutoff of a 30% average stabilization of the two replicates. The circles representing the hits are shown in purple. The circle size is proportional to the number of events per well. Stabilizing compounds that were evaluated in the paper are labeled in the graph. (D) Same as (C), except the data were normalized to control DMSO (blue circles, 0% destabilization) and CHX (red circles, 100% destabilization). Destabilizing compounds evaluated in the paper are indicated.
Given the important nature of protein kinases in regulating MYC protein stability, we screened the GPS-MYC cells with the Published Kinase Inhibitor Set (PKIS) of ATP-competitive protein kinase inhibitors (28, 29, 33) to identify novel kinases that regulate MYC protein stability in PDAC. PKIS was generated to maximize structural diversity and the spectrum of kinases inhibited, and the activity profiles of the compounds is published (28, 29, 33), enabling target hypothesis generation as soon as hits are discovered. In total, we screen GPS-MYC cells with this ~800 compound set, treating cells for 6 hours at a concentration of 20 μM, using the Intellicyt iQue Screener. The screen was run in duplicate to ensure reproducibility.
To process the primary screening data, we first separated the data into two parts, percent stabilization and percent destabilization. We calculated percent stabilization by normalizing the results to the DMSO (0%) and MG132 (100%) treatment controls, and we similarly calculated percent destabilization by normalizing the results to the DMSO (0%) and CHX (100%) treatment controls. This enabled determination of the percent effect of the test compounds compared to maximum stabilization or destabilization of EGFP-MYC with MG132 and CHX, respectively. With the cut-off for a hit set at a 30% stabilization or destabilization, we identified 30 compounds that stabilized EGFP-MYC (Fig. 2C, Data File S1) and 26 compounds that destabilized EGFP-MYC (Fig. 2D, Data File S1). Supporting the validation of the GPS-MYC screen to identify regulators of MYC protein stability, we found that UNC10112687 (SB-590885-AAD), which inhibits BRAF and CRAF (29), was identified as a compound that destabilized MYC, whereas a GSK3α/β inhibitor, UNC10112671 (SB-739452) stabilized MYC (fig. S1, A and B). These results are expected because RAF activation of ERK phosphorylates MYC at Ser62 and blocks degradation, whereas GSKβ phosphorylates MYC at Thr58 and promotes degradation (26).
UNC10112731 increases levels of endogenous MYC protein
We prioritized the top stabilizing compounds based on published specificity profiles (28, 29, 33), novelty of the kinases inhibited, and ease of synthesis, and selected and resynthesized UNC10112731 (GW694590A) (29) for further validation (fig. S1C). Consistent with its ability to enhance MYC protein stability, in the initial screen UNC10112731 increased EGFP-MYC without affecting DsRed (Fig. 3A). In a secondary validation screen, treatment of GPS-MYC cells with UNC10112731 showed a dose-dependent increase in EGFP-MYC and endogenous MYC levels when measured by flow cytometry (Fig. 3B) and immunoblot analyses (Fig. 3C). Next, we sought to determine if UNC10112731 treatment also stabilized endogenous MYC protein in other PDAC lines, and found that endogenous MYC protein was increased to varying degrees (2- to 20-fold) in three of eight cell lines analyzed (Fig. 3D). As expected from its identification using the GPS-MYC reporter, this effect was not due to increased transcription (Fig. 3E). Interestingly, the published kinase targets of UNC10112731 include the KIT and PDGFRα receptor tyrosine kinases (28) (Fig. 3F and fig. S1C). However, treatment of PDAC cells with the multi-tyrosine kinase inhibitors imatinib and amuvatinib, both of which have activities against KIT and PDGFRα (34), did not increase endogenous MYC in a dose-dependent manner (Fig. 3G). Therefore, we speculated that the mechanism whereby UNC10112731 stabilizes MYC protein may involve inhibition of additional kinases not identified in the kinase profiling reported for the PKIS compounds.
Fig. 3. MYC degradation screen identifies a compound that stabilizes MYC protein.
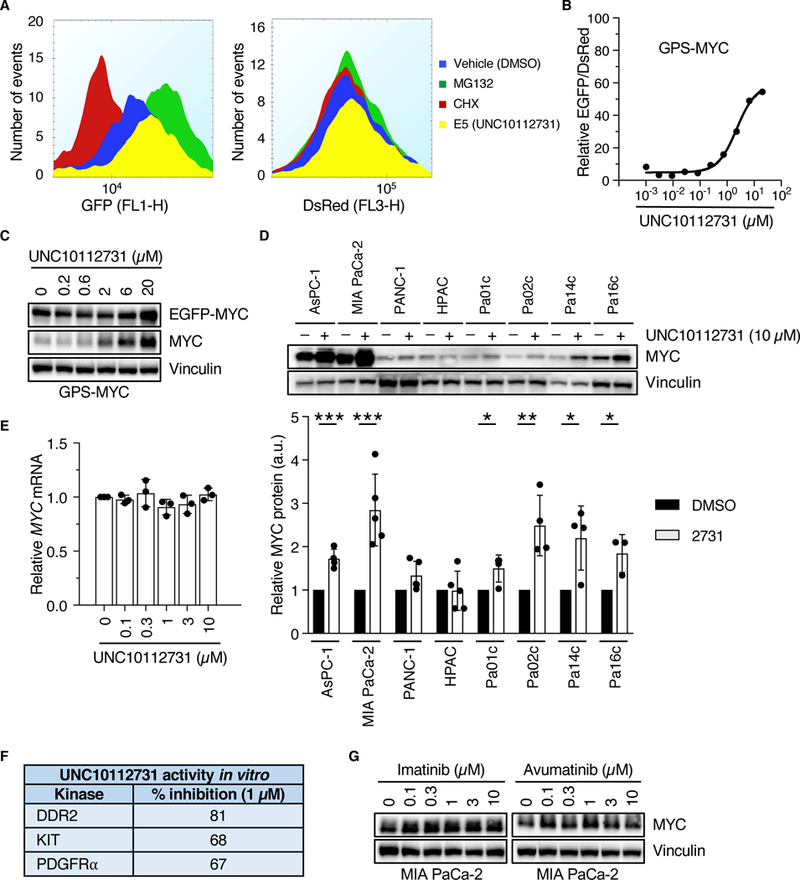
(A) GPS-MYC cells were treated with 20 μM UNC10112731 for 6 hr, and EGFP and DsRed intensities were measured by flow cytometry. Data are from the GPS-MYC screen. (B, C) GPS-MYC cells were treated with increasing concentrations of UNC10112731 for 6 hours and EGFP and DsRed intensities were measured by flow cytometry (B) or immunoblotting (C). (D) KRAS-mutant PDAC cell lines were treated for 6 hours with UNC10112731 and MYC protein levels were measured by immunoblotting (upper panel), with quantitation by densitometry relative to vehicle control (lower panel). Statistical significance was measured by t-test. *p<0.05, **p<0.01, ***p<0.001. (E) MIA PaCa-2 cells were treated for 6 hours with UNC10112731 and MYC mRNA levels were measured by qPCR. MYC mRNA levels were normalized to GAPDH mRNA levels. (F) Kinase selectivity of UNC10112731 as described previously (29). G) MIA PaCa-2 cells were treated for 6 hours with the indicated compounds and MYC protein levels were measured by immunoblot. All data are representative of at least three independent experiments. Data in (D) are presented as means ± SD.
UNC10112785 causes loss of endogenous MYC protein
Six of the destabilizer hits from the primary screen shared the same 7-azaindole pharmacophore (fig. S1, D to H), so we synthesized one of those compounds, UNC10112785 (Fig. 4A), for further analysis. We found that treatment of GPS-MYC cells with UNC10112785 did not affect DsRed levels (Fig. 4B) but did decrease the levels of EGFP-MYC and endogenous MYC (Fig. 4C). Analyses in additional KRAS-mutant PDAC cell lines showed loss of endogenous MYC in all eight cell lines evaluated (Fig. 4D). Examination of the published specificities of five of the 7-azaindole compounds revealed that the shared kinase targets included KIT and DYRK1A/1B (Fig. 4E, and fig. S1, D to G). However, we found that commercially available inhibitors of either KIT (imatinib and amuvatinib) or DYRK1A/1B (AZ191 and TC-S 7004) failed to cause a dose-dependent loss of endogenous MYC protein (Fig. 3G, 4F), suggesting that, like UNC10112731, UNC10112785 may also have additional kinase targets that regulate MYC protein expression.
Fig. 4. UNC10112785 drives MYC protein loss.
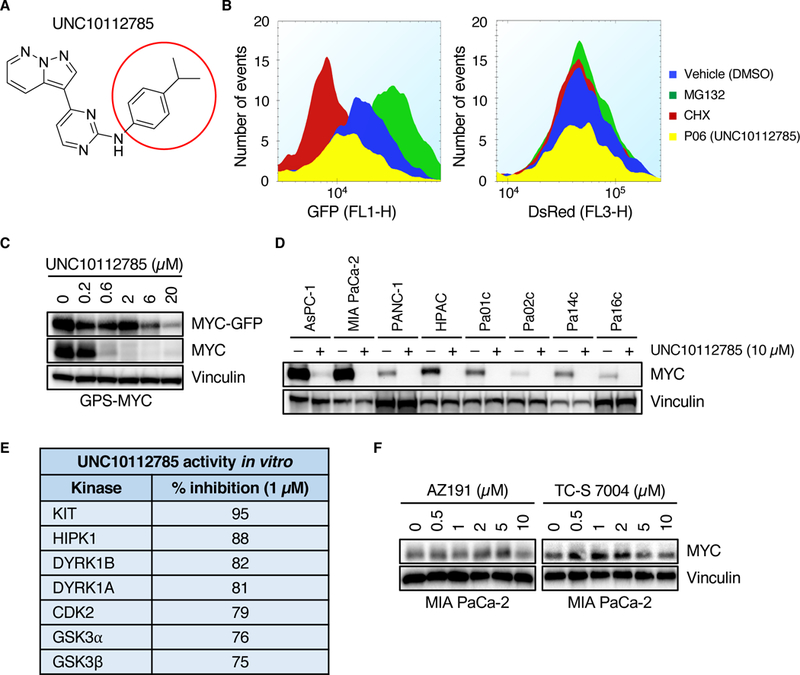
(A) Chemical structure of UNC10112785. The red circle indicates the position at which different analogs were synthesized. (B) GPS-MYC cells were treated with 20 μM UNC10112785 for 6 hours and EGFP and DsRed intensity was measured by flow cytometry. Data from the GPS-MYC screen. (C) GPS-MYC cells were treated with UNC10112785 for 6 hours and EGFP-MYC and MYC levels were measured by immunoblotting. (D) PDAC cells were treated with UNC10112785 and MYC protein levels were measured by immunoblot. (E) Kinase selectivity of UNC10112785 as described previously (29). (F) MIA PaCa-2 cells were treated for 6 hours with the indicated compounds and MYC protein levels were measured by immunoblot. All data are representative of at least three independent experiments.
In 2017, a quantitative, mass spectrometry-based chemical proteomic assay was used to profile the specificity and potency of 243 clinical kinase inhibitors (35). The sensitive technology of this assay, which evaluates full length kinases expressed in a cellular environment, enables detection of more kinases (~300) than the kinome coverage of typical in vitro kinase profiling panels. Moreover, it has been shown to be more relevant to inhibitor potency and selectivity assessment than traditional kinase profiling using recombinant proteins. The profiling of PKIS compounds was performed in vitro at only a few concentrations against an incomplete panel of recombinant protein kinases: 196 for PKIS1 and 232 for PKIS2 (28, 29). We reasoned that the kinase targets of UNC10112785 involved in regulation of MYC stability may not have been included in the original profiling of PKIS compounds. Therefore, to further evaluate the kinase selectivity profile of UNC10112785, we applied the Multiplex Kinase Inhibitor Bead (MIB)-Mass Spectrometry (MS) chemical proteomic assay (36) in MIA PaCa-2 cells (fig. S2A). This assay is based on kinase inhibitor competition assays to assess kinome-wide inhibitor specificity, similar to that described previously (35, 37).
MIB-MS in MIA-PaCa2 cells revealed that UNC10112785 appeared to inhibit CDK8 and the closely related paralog CDK19 (80% identity), and to a lesser degree CDK9 (Fig. 5A). These kinases were not included in the panel evaluated previously with PKIS compounds (28, 29). To validate these activities, we chose CDK8 and CDK19, as well as additional kinases identified by PKIS (Fig. 4E) or MIB-MS (Fig. 5A) for biochemical analyses using recombinant kinases (Table 1). We found that the kinases most potently inhibited in vitro by UNC10112785 were CDK8, CDK19 and CDK9, with IC50s of 1.05, 2.67 and 19.9 nM, respectively, and ~10-fold selectivity over other kinases analyzed.
Fig. 5. Inhibition of CDK9 by UNC10112785 drives MYC loss.
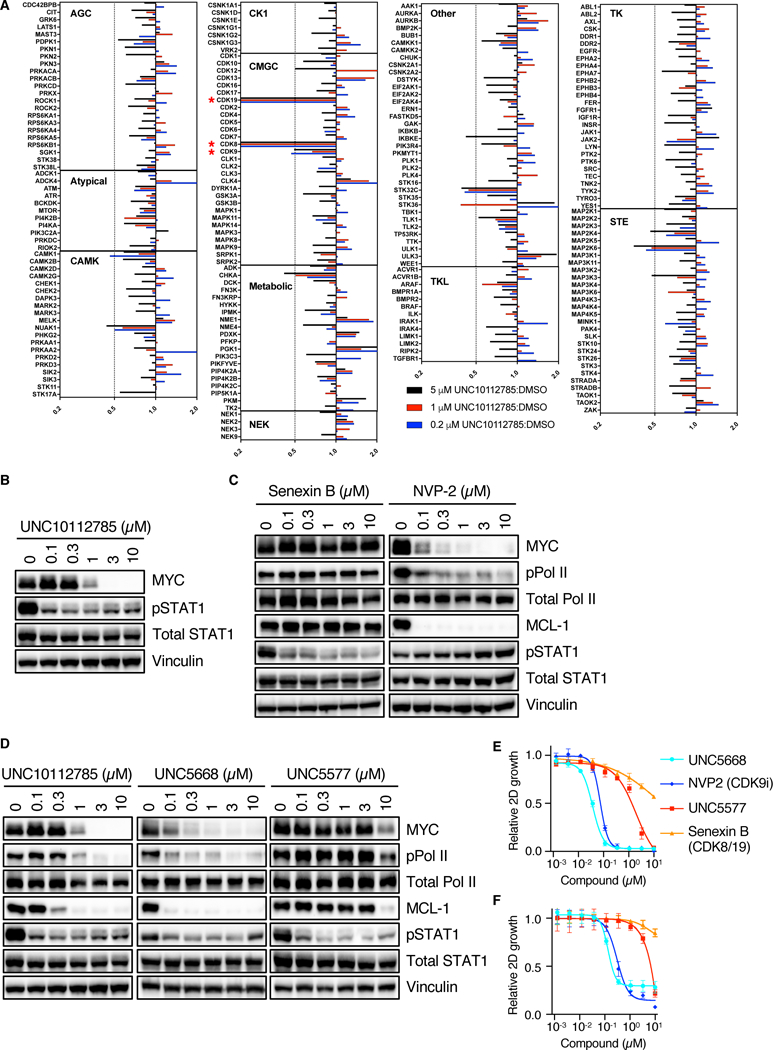
(A) MIA PaCa-2 cells were treated for 1 hour with different concentrations UNC10112785, after which they were lysed and applied to the MIBs column. Kinases were eluted from the column and identified and quantified by LC/MS as described in Methods. Bars to the left of center line indicate kinases reduced after compound addition. Data are from one experiment. (B) MIA PaCa-2 cells were treated with UNC10112785, and CDK8/19 inhibition was measured by pSTAT1 levels. (C, D) MIA PaCa-2 cells were treated with the indicated compounds for 6 hours and CDK8/19 and CDK9 inhibition was measured by pSTAT1 and pPol II, respectively. (E) MIA PaCa-2 cells were treated for 72 hours with the indicated compoundsCell proliferation was measured by cell count and normalized to cells treated with vehicle control (DMSO). (F) MIA PaCa-2 cells suspended in 3% agarose were treated for 72 hours with the indicated compounds. Cell proliferation was measured by alamarBlue and normalized to cells treated with vehicle (DMSO). All data are representative of at least three independent experiments unless otherwise indicated. Data in (E) and (F) are presented as means ± SD.
Table 1.
Biochemical profiling of UNC10112785
| Kinase | Assay Type | IC50 (nM) | %Max |
|---|---|---|---|
| CDK19/cyclin C | LanthaScreen binding | 1.05 | 100 |
| CDK8/cyclin C | LanthaScreen binding | 2.67 | 104 |
| CDK9 (inactive) | LanthaScreen binding | 7.65 | 105 |
| CDK9/cyclin K | LanthaScreen binding | 19.9 | 103 |
| CDK19 (inactive) | LanthaScreen binding | 82.9 | 100 |
| DYRK1B | Z-Lyte | 174 | 96 |
| DYRK1A | Z-Lyte | 284 | 92 |
| KIA | Z-Lyte | 500 | 88 |
| HIPK1 (MYAK) | Z-Lyte | 539 | 95 |
| MAP2K6 (MKK6) | LanthaScreen binding | 2250 | 65 |
| CAMKK1 (CAMKKA) | LanthaScreen binding | 4190 | 59 |
| MAP3K3 (MEKK3) | LanthaScreen binding | 7580 | 52 |
| STK32C (YANK3) | LanthaScreen binding | 9620 | 46 |
Consistent with our MIB-MS and biochemical analyses, we found that treatment of MIA PaCa-2 cells with UNC10112785 led to a reduction in phosphorylation of STAT1 at S727 (pSTAT1), a marker of CDK8/19 activity (Fig. 5B) (38). However, pSTAT1 was maximally reduced by 100 nM, whereas MYC loss was not observed until 1 μM, suggesting that CDK8/19 inhibition may not be the basis for MYC loss. To address this possibility, we treated MIA PaCa-2 cells with Senexin B, a structurally distinct potent and selective CDK8/19 inhibitor (39), which resulted in a reduction in pSTAT1 but not MYC protein levels (Fig. 5C). Senexin B is not a CDK9 inhibitor, as indicated by the absence of a reduction in MCL-1, a well-validated transcriptional target of CDK9 (40). Thus, we conclude that, although UNC10112785 is a potent inhibitor of CDK8/19 in vitro and in cells, MYC loss was not caused by inhibition of CDK8/19.
Inhibition of CDK9 drives MYC loss caused by UNC10112785
Since UNC10112785 inhibited CDK9 in the MIB-MS analyses, and this was verified by biochemical analyses, we next addressed the possibility that inhibition of CDK9 was responsible for UNC10112785-driven loss of MYC. First, we evaluated the activity of a structurally distinct CDK9-selective inhibitor, NVP2 (41). NVP2 treatment reduced phosphorylation of RNA polymerase II (pPol II) at S2, another marker of CDK9 activity (40), as well as protein levels of MCL-1 and MYC. However, NVP2 did not reduce pSTAT1, indicating that it inhibited CDK9 but not CDK8/19 (Fig. 5C).
We then synthesized 10 structurally related analogues of UNC10112785 to determine the relative contributions of their CDK8/19 and CDK9 inhibitory activities to driving MYC loss (Table S1). Some analogues lost the ability to decrease MYC, whereas others were more potent at this than the parent compound (Fig. 5D and fig. S2B). Retention of the ability to suppress pPol II and MCL-1 (CDK9 inhibition) correlated with the loss of MYC, whereas some analogues retained the ability to inhibit pSTAT1 (CDK8/19 inhibition) yet lost the ability to cause loss of MYC. These data support the possibility that UNC10112785 inhibition of CDK9 rather than CDK8/19 is the basis for its reduction of MYC protein.
To further explore the relative contribution of CDK8/19 versus CDK9 inhibition to loss of MYC, we chose two analogues for additional characterization. While both analogues retained low nM activities against CDK8/19 (fig. S2C), the potency of UNC5668 to inhibit CDK9 in vitro was increased by 7.5-fold compared with parent compound UNC10112785, whereas that of UNC5577 was decreased by 5.3-fold. MIB-MS inhibitor competition analyses verified their altered CDK9 activities in MIA PaCa-2 cells (fig. S2D). Consistent with their relative CDK9 potencies in vitro, we found that UNC5668 exhibited 10-fold increased potency over UNC10112785 in cells. Treatment of MIA PaCa-2 cells with 100 nM of UNC5668 suppressed both CDK9 signaling (pPol II and MCL-1) and MYC protein levels (Fig. 5D). In contrast, treatment with 100 nM UNC5577 suppressed CDK8/19 signaling (pSTAT1) but not pPol II, MCL-1 or MYC (Fig. 5D). Finally, since we showed previously that MYC is essential for the growth of PDAC cell lines (14), we next evaluated the ability of these analogues to suppress cell proliferation. Consistent with their ability to reduce MYC levels, all compounds with CDK9 but not CDK8/19 inhibitory activity reduced proliferation of MIA PaCa-2 cells on plastic (Fig. 5E) and colony formation in soft agar (Fig. 5F), at concentrations where potent inhibition of CDK9 signaling was observed. Taken together, these data indicate that inhibition of CDK9 rather than CDK8/19 is responsible for UNC10112785-driven MYC loss.
CDK9 promotes phosphorylation of MYC on Ser62
CDK9 can regulate MYC transcription by direct phosphorylation of Pol II (40). Consistent with this, we found that treatment of PDAC cells with UNC5668 decreased MYC mRNA (Fig. 6A) as well as MYC protein levels (Fig. 6B). Consistent with our identification of a CDK9 inhibitor as a MYC destabilizer in the GPS-MYC assay, concurrent inhibition of the proteasome using MG132 was able to partially rescue the loss of MYC protein (Fig. 6B, upper panel). Thus, CDK9 regulates both MYC transcription and MYC protein stability, Given these two effects of CDK9 on MYC levels, we then speculated that inhibiting the proteasome would be able to rescue MYC protein levels only in the short term, because suppression of transcription would eventually result in suppression of MYC protein regardless of proteasomal activity. Consistent with this possibility, we found that MG132 clearly rescued MYC protein levels at early time points (1 and 2 hours) but not at later time points (4 and 6 hours, Fig. 6B). Thus, inhibition of CDK9 both suppresses MYC transcription and promotes MYC protein degradation.
Fig. 6. CDK9 regulates MYC protein stability through phosphorylation of Ser62.
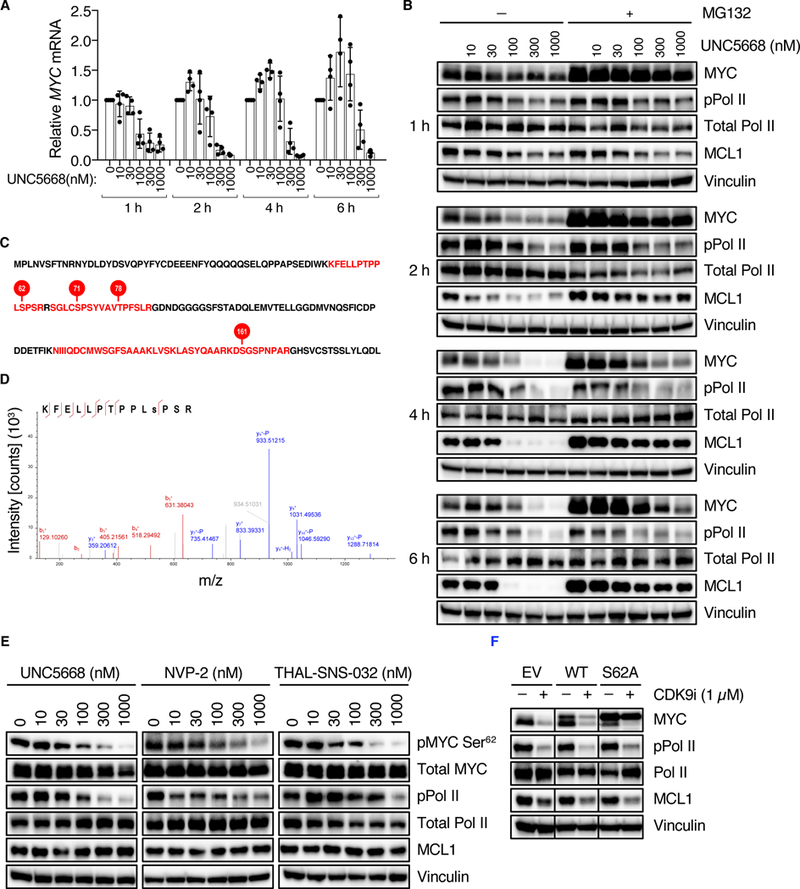
(A) MIA PaCa-2 cells were treated with UNC5668. MYC mRNA levels were measured by qPCR and normalized to BACT mRNA levels. (B) MIA PaCa-2 cells were treated with UNC5668 in the presence or absence of MG132 for the indicated concentrations and times, and MYC protein levels were measured by immunoblot. (C) Recombinant CDK9 and MYC were incubated in the presence of ATP. After separation by SDS-PAGE, the band corresponding to MYC was excised and analyzed by LC/MS. MYC protein sequence: shown in red are phosphorylated peptides, and phosphorylated residues are indicated by red circles. The first 182 amino acids of MYC are shown. Data are from one experiment. (D) Tandem MS/MS spectrum for MYC peptide containing Ser62. (E) MIA PaCa-2 cells were treated for 30 min with the indicated compounds and pMYC Ser62 and total MYC levels were measured by immunoblot. (F) MIA PaCa-2 cells stably expressing MYC proteins were treated with the indicated compounds for 2 hours and MYC levels were measured by immunoblot. Blots are representative of five independent experiments. Other data are representative of at least three independent experiments unless otherwise indicated. Data in (A) are presented as means +/− SD.
Scansite analysis (42) of the MYC protein sequence revealed several putative serine-proline (SP) CDK9 consensus motifs. Therefore, we reasoned that CDK9 might control MYC protein stability through direct phosphorylation. To address this possibility, we performed an in vitro kinase assay coupled with mass spectrometry and found that CDK9 phosphorylates MYC on several residues in vitro, including Ser62 (Fig. 6, C and D). To determine if CDK9 can also promote MYC phosphorylation at Ser62 in cells, we treated MIA PaCa-2 cells for 30 min with three structurally distinct CDK9 inhibitors. Consistent with the in vitro kinase assay results, we found that inhibition of CDK9 activity in PDAC cells, including by THAL-SNS-032 targeted CDK9 degradation (43), decreased MYC phosphorylation at Ser62 (Fig. 6E and fig. S3A) while not affecting total MYC levels. Since Ser62 phosphorylation attenuates degradation of MYC (26), we conclude that CDK9 can enhance MYC protein stability at least in part by promoting Ser62 phosphorylation. In support of this possibility, we found that exogenous expression of the MYC S62A phospho-deficient mutant partially rescued MYC degradation caused by CDK9i (Fig. 6F and fig. S3B). That MYC S62A did not fully prevent MYC loss caused by CDK9i may indicate that there are additional phosphorylation sites, possibly including those we identified by mass spectrometry (Fig. 6C), that also contribute to CDK9 regulation of MYC protein stability.
Since we previously identified ERK5 as a regulator of MYC degradation through phosphorylation at MYC Ser62 phosphorylation (14), we wanted to eliminate the possibility that CDK9 regulated MYC stability through MEK5-ERK5 signaling. Unlike CDK9i, the ERK5 inhibitor XMD8–92 (ERK5i) did not reduce phosphorylation of the CDK9 substrate Pol II, and conversely CDK9i did not reduce phosphorylation of the MEK5 substrate ERK5 (Fig. 7A). Furthermore, MIB-MS analysis of UNC5668 did not demonstrate a decrease in MEK5/MAP2K5 activity (Fig. 5A). Thus, we conclude that CDK9 and MEK5-ERK5 comprise distinct signaling networks that each regulate MYC protein phosphorylation and stability.
Fig. 7. CDK9 and MEK5-ERK5 comprise distinct signaling mechanisms that regulate MYC Ser62 phosphorylation and stability.
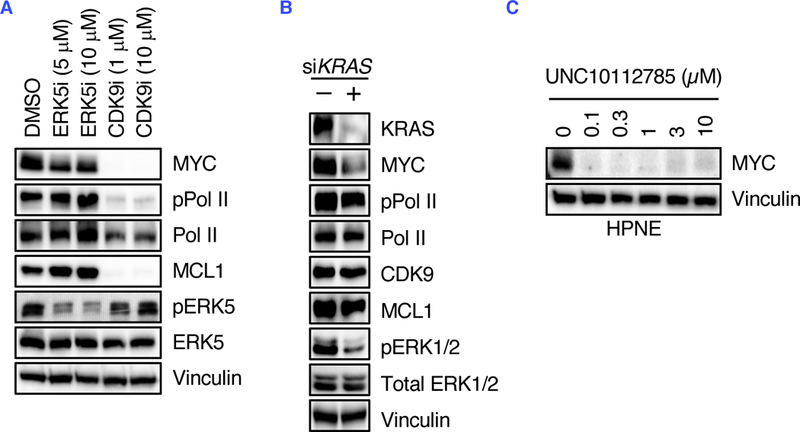
(A) MIA PaCa-2 cells were treated for 6 hours with XMD8–92 (ERK5i) and UNC5668 (CDK9i), and ERK5 and CDK9 signaling were measured by immunoblotting for the indicated proteins. (B) MIA PaCa-2 cells were treated with KRAS siRNA for 24 hours and CDK9 signaling was measured by immunoblotting for the indicated proteins. (C) HPNE cells were treated for 6 hours with the indicated concentrations of UNC10112785 and MYC levels were measured by immunoblotting for the indicated proteins. Data are representative of at least three independent experiments.
Finally, we previously determined that MEK5 activity comprised a compensatory activity caused by KRAS suppression (14). We therefore determined if CDK9 activity is also linked with mutant KRAS activity. Whereas acute KRAS suppression in KRAS-mutant MIA PaCa-2 cells caused an increase in MEK5 activity (14), no increase in CDK9 activity was observed (Fig. 7B). Additionally, CDK9i treatment of HPNE immortalized human pancreatic ductal cells, that are KRAS wild-type, also caused a reduction in MYC (Fig. 7C). We conclude that, unlike MEK5, CDK9 activity is not regulated by mutant KRAS activity.
DISCUSSION
Although RAS and MYC are well-validated cancer drivers implicated in the growth of a diverse spectrum of cancers, both have largely been considered ‘undruggable’ cancer targets (17). Renewed efforts have identified promising directions for inhibiting RAS (6), but the development of selective anti-MYC therapies remains a significant challenge (44, 45). One promising but relatively underexplored strategy to target MYC involves disruption of the signaling mechanisms that promote MYC protein stability (26). For example, we previously found that mutant KRAS prevents MYC degradation in PDAC through both ERK-dependent and -independent signaling mechanisms (14). These findings prompted us to identify additional protein kinase-dependent mechanisms that regulate MYC protein stability in KRAS-mutant PDAC. Targeting of these kinases to promote MYC degradation may then serve as a novel, indirect therapeutic strategy in KRAS-driven cancers. In this study, we developed a MYC degradation screen in the KRAS-mutant PDAC cell line MIA PaCa-2 and identified kinase inhibitors that either increased or decreased MYC protein levels. We then used these compounds as tools to identify new protein kinase-dependent mechanisms that regulate MYC protein stability. We identified CDK9 as a novel regulator of MYC protein stability through enhanced phosphorylation at Ser62, a modification that prevents MYC degradation (26). We propose that our screening approach can be adapted to investigate the mechanisms that regulate the degradation of other clinically important proteins.
Our GPS-MYC screen identified multiple kinase inhibitors that either stabilize or destabilize MYC protein. However, the dynamic range of the assay as currently constituted is better suited for identifying stabilizing compounds than destabilizing compounds. Thus, this assay could more easily be adapted to identify novel compounds that stabilize important proteins such as tumor suppressors. Although stabilizing compounds would not be useful for therapeutically targeting oncogenic proteins such as MYC, they are nonetheless valuable tools for understanding signaling mechanisms that regulate MYC activity in PDAC and other cancers.
While our GPS-MYC screen illustrates the potential power of this strategy to identify novel regulators of MYC protein stability, it also reveals some challenges associated with using the PKIS library of protein kinase inhibitors. For example, a number of the strongest stabilizers that we identified targeted multiple kinases, making delineation of the relevant kinase(s) a daunting task. Similarly, in our characterization of UNC10112731 as a compound that stabilized MYC protein, we were not able to ascribe this activity to the published targets of this compound. Instead, we suspect that additional kinase(s) may be responsible for stabilizing MYC. We faced a similar challenge in our identification of the protein kinase(s) that account for the ability of UNC10112785 to cause loss of MYC protein. Initially, we focused on a set of MYC-destabilizing compounds that shared the same core chemotype, with the rationale that they targeted the same kinase. Analysis of the published PKIS data available for their specificities led us to investigate the role of the DYRK1A/1B family of kinases. After failing to validate DYRK1A/1B upon using additional commercially available DYRK1A/1B inhibitors, we then speculated that UNC10112785 had activities not identified previously. To address this possibility, we applied MIB-MS analyses that identified potent activity against CDK8 and the related paralog CDK19, and against CDK9, all of which are kinases that were not included in the original PKIS biochemical profiling dataset. We then performed in vitro assays with recombinant kinases and validated that UNC10112785 exhibited potent inhibition of CDK8/19 and CDK9 in vitro. Further evaluation of this compound in PDAC lines demonstrated its ability to inhibit CDK8/19 and CDK9. As demonstrated in another study (35), we suggest that cell-based proteomic profiling of protein kinase inhibitor specificity is an effective approach to provide a more comprehensive determination of the cellular targets of protein kinase inhibitors. Based on our studies, we recommend that researchers using the PKIS set for screening should not rely solely on the published specificity dataset and instead should also perform further independent profiling of inhibitor specificity in their studies. Ongoing profiling of the PKIS libraries using wide-spectrum kinase profiling in live cells (46) will also help to address this concern for future researchers.
We were initially surprised that we identified CDK9 as a regulator of MYC protein stability, since to date there has been no evidence of such regulation. Instead, CDK9 is known to be part of the P-TEFb complex that is recruited to the MYC promoter by BRD4, where CDK9 then phosphorylates Pol II to stimulate transcriptional elongation and MYC expression (47). Since MYC expression from our GPS-MYC reporter is controlled from a heterologous CMV promoter that is not subject to the same mechanisms that control endogenous MYC transcription, our reporter is able to reveal nontranscriptional mechanisms that regulate MYC protein levels. Collectively, our data are in agreement with CDK9 regulation of MYC transcription, and additionally suggest that CDK9 may directly phosphorylate MYC on Ser62 to block MYC protein degradation. Ser62 can also be phosphorylated upon KRAS activation of ERK1/2, and we previously identified ERK5 as another kinase that can phosphorylate MYC Ser62 (14). Thus, the roster of kinases that can promote MYC protein stability through the Thr58/Ser62 phosphodegron continues to expand.
There has been a renewed interest in developing selective CDK9 inhibitors for cancer treatment (48–51), due in large part to their inhibition of MCL-1 and MYC transcription. We now propose that destabilization of MYC protein is a third anti-tumor activity of CDK9 inhibitors. This supports their clinical evaluation in MYC-dependent cancers, particularly where MYC expression is deregulated, such as in KRAS-mutant PDAC. In conclusion, our study presents the development and application of a MYC protein degradation screen which we used to identify novel regulators of MYC protein stability. We propose that this screen can be readily adapted to study the stability of many other important proteins.
MATERIALS AND METHODS
Cell culture
Human pancreatic cancer cell lines were obtained from ATCC (MIA PaCa-2, PANC-1, HPAC, AsPC-1) or were a gift from Dr. Jason Fleming at MD Anderson Cancer Center (Pa01c, Pa02c, Pa14c, Pa16c). Cells were maintained in DMEM (GPS-MYC, MIA PaCa-2, PANC-1, HPAC, Pa01c, Pa02c, Pa14c, Pa16c) or RPMI-1640 (AsPC-1) supplemented with 10% fetal bovine serum (FBS) and antibiotics (Pen Strep; Sigma 15070063). STR (short tandem repeat) analyses were performed to verify cell line identity, and all cell lines tested negative for mycoplasma contamination.
Antibodies and reagents
The following antibodies were used, from Cell Signaling Technology: anti-MYC (5605), anti-Pol II (14958), anti-pSTAT1Ser727 (8826), anti-STAT1 (14995), anti-pERK (4370), anti-ERK (9201), anti-CDK9 (2316); from AbCam: anti-pMYC Ser62 (ab106952), anti-pPol II Ser2 (ab5095); from Sigma-Aldrich: anti-KRAS (WH0003845M1), anti-vinculin (V9131) from Santa Cruz Biotechnology: anti-MCL1 (sc-12756). The protein kinase inhibitors NVP-2 (HY-12214A), amuvatinib (HY-10206) and imatinib (HY-15463) were obtained from MedChemExpress, TC-S 7004 (5088) from Tocris Bioscience, and AZ 191 (5232) from Sigma-Aldrich. MG132 (M7449) and cycloheximide (C7698) were also obtained from Sigma-Aldrich. THAL-SNS-032 was a gift from Dr. Nathanael Gray (Harvard/DFCI). Synthetic schemes for UNC10112731 and UNC10112785 and derivatives are shown in Supplemental Methods.
Expression vectors
To generate the GPS-MYC plasmid, the human MYC DNA was amplified by PCR using the following Gateway cloning primers:
attB1:5’-GGGGACAAGTTTGTACAAAAAAGCAGGCTTAGAAGGAGATAGAACCATGCCCCTCAACGTTAGCTTCAC-3’
attB2:5’-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTACGCACAAGAGTTCCGTAGC-3’ The resultant PCR product was cloned into pDONR223 (Addgene) with the BP reaction and subsequently cloned into the pGPS-LP vector with the LR reaction.
MIA PaCa-2 GPS-MYC Screen
Lentivirus particles were generated by transfecting HEK293T cells with GPS-MYC and the packaging plasmids pMD2.G and pSPAX2 (Addgene) using Fugene 6 transfection reagent (Promega). MIA PaCa-2 cells were infected with lentivirus and selected with 2 μg/ml puromycin to establish mass populations of cells stably expressing EGFP-MYC (designated GPS-MYC cells). To minimize variability during the screening and validation processes, large amounts of GPS-MYC cultures were expanded and then frozen at a concentration of 106 cells/ml in Recovery Cell Culture Freezing Medium (Gibco). Cells were thawed and allowed to grow for two days prior to the screen, and then were plated at 20,000 cells/well into a clear-bottom, white 384-well plate (Corning 3707) using a Multidrop Combi Reagent Dispenser (Thermo Fisher). The following day, growth medium was removed from the assay plate and replaced with inhibitor-containing growth medium using a Multimek Automated Liquid Handler (NanoScreen).
To prepare the assay plates, 1 μl DMSO was added to columns 1 and 2, 1 μl of 375 μg/ml cycloheximide (final concentration 7.5 μg/ml) was added to column 23, and 1 μl of 187.5 μM MG132 (final concentration 3.75 μM) was added to column 24. Columns 2–22 contained PKIS (Published Kinase Inhibitor Sets) compounds. Fifty μl DMEM supplemented with 2% FBS was added per well, and 20 μl of the resultant PKIS compound-containing media were added to each plate using a Multimek Automated Liquid Handler (NanoScreen).
After 6 hours the plates were washed with 75 μl PBS per well, 10 μl Accumax Cell Dissociation Solution (Innovative Cell Technologies) was added per well, and the plates were centrifuged briefly (5 s at 500 rpm). After 20 min incubation at 37ºC, 10 μl DMEM supplemented with 2% FBS was added to each well to quench the dissociation reaction. All liquids were added with a Multidrop Combi Reagent Dispenser (Thermo Fisher) at slow speed. The plates were then analyzed on an iQue Screener PLUS (Intellicyt) using medium sip speed, 4 s sip time, shaking initially at 2000 rpm for 30 s and then at 2000 rpm for 10 s after every 12 wells. The data were imported into the CICBDD database (ScreenAble Solutions) and each compound was normalized to control wells. The normalized data were then imported into TIBCO Spotfire for visualization and analysis. Some analyses were also performed in ForeCyt (Intellicyt).
Immunoblotting
Cells were lysed in 1% Triton buffer (25 mM TRIS, pH 7.4, 100 mM NaCl, 10% glycerol, 1 mM EDTA, 1% Triton-X 100), supplemented with protease (Roche) and phosphatase inhibitors (Sigma) for 10 min, with brief vortexing. Lysates were clarified by centrifugation at 18,000 x g for 15 min at 4ºC, and protein concentrations were measured using the Bradford reagent. Standard immunoblotting procedures were performed as we described previously (14).
Confocal microscopy
Cells were plated on MatTek dishes (MatTek Corporation) and fixed for 5 min in ice-cold paraformaledehye (4%), washed 3X with cold PBS, stained with DAPI (0.1 μg/ml) for 10 min, and washed 3X with cold PBS. Images were collected on an Olympus FV1000 microscope using a 40X objective.
Gene silencing
All siRNA experiments were performed using 10 nM siRNA and Lipofectamine RNAiMAX (Life Technologies) according to the manufacturer’s protocol.
Quantitative real-time PCR
mRNA was isolated using an RNeasy RNA isolation kit (Qiagen) and reverse transcription was performed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Samples were run on a QuantStudio 6 Flex (Applied Biosystems) and analyzed using the ddCT method.
Growth assays
To measure anchorage-dependent growth, cells were plated at low density in 96 well plates and allowed to attach overnight. Inhibitors were then added with a D300e Digital Dispenser (Tecan). After 72 hours, cells were labeled with calcein AM (Thermo Fisher) according to the manufacturer’s protocol, and counted on a SpectraMax MiniMax 300 Imaging Cytometer (Molecular Devices).
To measure anchorage-independent growth, soft agar colony formation assays were performed. In a 96-well plate, wells were coated with 50 μl 2% SeaPrep Agarose (Lonza) mixed with cell culture medium. Next, 100 μl of 5000 cells mixed with 1% SeaPrep Agarose were added to each well and allowed to solidify. Fifty μl of medium were added to each well, and drugs were dispensed with a D300e Digital Dispenser (Tecan). After 72 hours, the alamarBlue cell viability reagent (Thermo Fisher) was added and incubated for 3 hours. Fluorescence was quantified on a SpectraMax MiniMax 300 Imaging Cytometer (Molecular Devices).
Multiplexed Inhibitor Beads – Mass Spectrometry (MIB-MS)
Lysates were prepared and MIB-MS was performed as described previously (36). Briefly, cells were harvested on ice in lysis buffer [50 mM HEPES (pH 7.5), 0.5% Triton X-100, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10 mM sodium fluoride, 2.5 mM sodium orthovanadate, 1X protease inhibitor cocktail (Roche), 1% phosphatase inhibitor cocktail 2 (Sigma-Aldrich), and 1% of phosphatase inhibitor cocktail 3 (Sigma-Aldrich)]. Lysates were sonicated for 3×10 sec on ice and centrifuged at 10,000 x g for 10 min at 4°C. Supernatant was collected and syringe-filtered through a 0.2 μm SFCA membrane (Corning). Approximately 5 mg of lysate was brought to 1 M NaCl and passed through a column of multiplexed inhibitor-conjugated beads (MIBs) consisting of Sepharose-conjugated UNC2147A, CTx-0294885, UNC8088A, purvalanol B, PP58 and VI16832. The kinase-bound MIBs were washed with 5 ml of high-salt buffer and 5 ml of low-salt buffer [50 mM HEPES (pH 7.5), 0.5% Triton X-100, 1 mM EDTA, 1 mM EGTA, and 10 mM sodium fluoride, and 1 M NaCl or 150 mM NaCl, respectively]. The columns were washed a final time with 1 ml 0.1% SDS wash buffer [50 mM HEPES (pH 7.5), 0.5% Triton X-100, 1 mM EDTA, 1 mM EGTA, 10 mM sodium fluoride, 150 mM NaCl, 0.1% SDS] then eluted with 1 ml of elution buffer [0.5% SDS, 1% 2 mercaptoethanol, and 0.1 M Tris (pH 6.8)] (100°C, 10 min). Eluted kinases were reduced (dithiothreitol) and alkylated (iodoacetamide) then concentrated with Amicon 10k Ultra centrifugal filters (Millipore). Detergent was removed from the eluate by chloroform/methanol precipitation. Protein pellets were resuspended in 50 mM HEPES (pH 8.0) and digested overnight at 37ºC with sequencing grade modified trypsin (Promega). Digested peptides were desalted using PepClean C18 spin columns (Thermo Scientific) and filtered with centrifugal filter columns (Nest). Peptides were extracted and dried by vacuum centrifugation. All peptide samples were stored at −80˚C until further analysis.
LC-MS/MS analysis was performed as described below. Raw data files were processed using MaxQuant version 1.5.3.17 and searched against the reviewed Uniport human database (containing 20,203 entries), using Andromeda within MaxQuant. Enzyme specificity was set to trypsin, up to two missed cleavage sites were allowed, carbamidomethylation of Cys was set as a fixed modification and oxidation of Met was set as a variable modification. A 1% FDR was used to filter all data. Match between runs was enabled (2 min match time window, 20 min alignment window), and a minimum of two peptides was required for label-free quantitation using the LFQ intensities. Further analyses were performed in Perseus version 1.6.0.2 and R.
In vitro MYC phosphorylation
Recombinant MYC (Abcam; ab169901) at 1000 nM was incubated with 50 nM recombinant CDK9 together with cyclin T1 required for kinase activity (Thermo Fisher; PV4131) and 100 μM ATP for 30 min at 30°C. The reaction was stopped by adding SDS sample buffer and boiling for 5 min at 95°C. After separation by SDS-PAGE, the gel was stained for 30 min with SimplyBlue Coomassie (Thermo Fisher) and destained overnight in water. Protein bands corresponding to MYC were excised and the proteins were reduced, alkylated, and in-gel digested with trypsin overnight at 37˚C. Peptides were extracted and dried by vacuum centrifugation. All peptide samples were stored at −80˚C until further analysis.
LC-MS/MS analysis was performed as described below. Raw data files were processed using Proteome Discoverer version 2.1 (Thermo Scientific). Peak lists were searched against a reviewed Uniprot human database, appended with a common contaminants database, using Sequest. The following parameters were used to identify tryptic peptides for protein identification: 10 ppm precursor ion mass tolerance; 0.02 Da product ion mass tolerance; up to two missed trypsin cleavage sites. Carbamidomethylation of Cys was set as a fixed modification and oxidation of Met and phosphorylation of Ser, Thr and Tyr were set as variable modifications. The phosphoRS node was used to localize the sites of phosphorylation. Peptide false discovery rates (FDR) were calculated by the Percolator node using a decoy database search and data were filtered using a 1% FDR cutoff.
LC-MS/MS analysis
Samples were analyzed by LC/MS/MS using an Easy nLC 1000 coupled to a QExactive HF mass spectrometer (Thermo Scientific). Samples were injected onto an Easy Spray PepMap C18 column (75 μm id × 25 cm, 2 μm particle size; Thermo Scientific) and separated over a 45 min or 2 hour method. The gradient for separation consisted of 5–32% mobile phase B at a 250 nl/min flow rate, where mobile phase A was 0.1% formic acid in water and mobile phase B consisted of 0.1% formic acid in ACN. The QExactive HF was operated in data-dependent mode where the 15 most intense precursors were selected for subsequent fragmentation. Resolution for the precursor scan (m/z 400–1600) was set to 120,000 with a target value of 3 × 106 ions. MS/MS scans resolution was set to 15,000 with a target value of 2 × 104 ions. The normalized collision energy was set to 27% for HCD. Peptide match was set to preferred, and precursors with unknown charge or a charge state of 1 and ≥ 8 were excluded.
Supplementary Material
Acknowledgments:
We thank Dr. Nathanael Gray for THAL-SNS-032. We also thank the UNC Flow Cytometry Core Facility and Dr. Nancy Cheng of the UNC CICBDD for help in optimizing the GPS-MYC assay. We thank Dr. Naim Rashid for ensuring proper statistical analyses were performed. Research reported in this publication was supported in part by the North Carolina Biotech Cancer Institutional Support Grant 2015-IDG-1001.
Funding: The UNC Flow Cytometry Core Facility and the Michael Hooker Proteomics Center are supported in part by P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center. K.H.P. was supported by the UNC University Cancer Research Fund. D.R.B. was supported by NCI fellowships (T32CA071341 and F31CA216965). A.V.V. was supported by fellowships from the NCI (T32CA009156) and American Cancer Society (128014-PF-15–059-01-TBE). R.G.H. was supported by a grant from Debbie’s Dream Foundation. This work was supported by grants (C.J.D. and A.D.C.) from the NIH/NCI (CA42978, CA179193, CA175747, CA199235) and the Pancreatic Cancer Action Network-AACR. C.J.D. was also supported by a grant from the DoD (W81XWH-15–1-0611) and from the Lustgarten Pancreatic Cancer Foundation (388222). Work carried out in the CICBDD was also supported by the University Cancer Research Fund.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: Plasmids generated for this study will be deposited at Addgene. The PKIS compound set is available from the Structural Genomics Consortium (SGC) at UNC (SGC-UNC@unc.edu).
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A, Cancer statistics, 2018. CA Cancer J Clin 68, 7–30 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Rahib L et al. , Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 74, 2913–2921 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Waters AM, Der CJ, KRAS: the critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb Perspect Med 8, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryan DP, Hong TS, Bardeesy N, Pancreatic adenocarcinoma. N Engl J Med 371, 2140–2141 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Cox AD, Der CJ, Ras history: The saga continues. Small GTPases 1, 2–27 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papke B, Der CJ, Drugging RAS: Know the enemy. Science 355, 1158–1163 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Ledford H, Cancer: The Ras renaissance. Nature 520, 278–280 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Land H, Parada LF, Weinberg RA, Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature 304, 596–602 (1983). [DOI] [PubMed] [Google Scholar]
- 9.Dang CV, MYC on the path to cancer. Cell 149, 22–35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soucek L et al. , Modelling Myc inhibition as a cancer therapy. Nature 455, 679–683 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soucek L et al. , Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev 27, 504–513 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazur PK et al. , Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A 107, 13438–13443 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saborowski M et al. , A modular and flexible ESC-based mouse model of pancreatic cancer. Genes Dev 28, 85–97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaseva AV et al. , KRAS Suppression-Induced Degradation of MYC Is Antagonized by a MEK5-ERK5 Compensatory Mechanism. Cancer Cell 34, 807–822 e807 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walz S et al. , Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511, 483–487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin WC et al. , Dormant cancer cells contribute to residual disease in a model of reversible pancreatic cancer. Cancer Res 73, 1821–1830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dang CV, Reddy EP, Shokat KM, Soucek L, Drugging the ‘undruggable’ cancer targets. Nat Rev Cancer 17, 502–508 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Filippakopoulos P et al. , Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mazur PK et al. , Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat Med 21, 1163–1171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berg T et al. , Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc Natl Acad Sci U S A 99, 3830–3835 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stellas D et al. , Therapeutic effects of an anti-Myc drug on mouse pancreatic cancer. J Natl Cancer Inst 106, (2014). [DOI] [PubMed] [Google Scholar]
- 22.Kota J et al. , Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137, 1005–1017 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gouw AM, Toal GG, Felsher DW, Metabolic vulnerabilities of MYC-induced cancer. Oncotarget 7, 29879–29880 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrieu G, Belkina AC, Denis GV, Clinical trials for BET inhibitors run ahead of the science. Drug Discov Today Technol 19, 45–50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayes TK et al. , Long-Term ERK Inhibition in KRAS-mutant pancreatic cancer is associated with MYC degradation and senescence-like growth suppression. Cancer Cell 29, 75–89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farrell AS, Sears RC, MYC degradation. Cold Spring Harb Perspect Med 4, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farrell AS et al. , Targeting inhibitors of the tumor suppressor PP2A for the treatment of pancreatic cancer. Mol Cancer Res 12, 924–939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drewry DH et al. , Progress towards a public chemogenomic set for protein kinases and a call for contributions. PLoS One 12, e0181585 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elkins JM et al. , Comprehensive characterization of the Published Kinase Inhibitor Set. Nat Biotechnol 34, 95–103 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Emanuele MJ et al. , Global identification of modular cullin-RING ligase substrates. Cell 147, 459–474 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yen HC, Elledge SJ, Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 322, 923–929 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Vaseva AV et al. , KRAS suppression-induced degradation of MYC is antagonized by a MEK5-ERK5 compensatory mechanism. Cancer Cell in press, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drewry DH, Willson TM, Zuercher WJ, Seeding collaborations to advance kinase science with the GSK Published Kinase Inhibitor Set (PKIS). Curr Top Med Chem 14, 340–342 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tibes R et al. , A phase I, first-in-human dose-escalation study of amuvatinib, a multi-targeted tyrosine kinase inhibitor, in patients with advanced solid tumors. Cancer Chemother Pharmacol 71, 463–471 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Klaeger S et al. , The target landscape of clinical kinase drugs. Science 358, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duncan JS et al. , Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149, 307–321 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krulikas LJ et al. , Application of integrated drug screening/kinome analysis to identify inhibitors of gemcitabine-resistant pancreatic cancer cell growth. SLAS Discov 23, 850–861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rzymski T, Mikula M, Wiklik K, Brzozka K, CDK8 kinase--An emerging target in targeted cancer therapy. Biochim Biophys Acta 1854, 1617–1629 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Porter DC et al. , Cyclin-dependent kinase 8 mediates chemotherapy-induced tumor-promoting paracrine activities. Proc Natl Acad Sci U S A 109, 13799–13804 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonawane YA et al. , Cyclin dependent kinase 9 inhibitors for cancer therapy. J Med Chem 59, 8667–8684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, Gray NS, SnapShot: kinase inhibitors i. Mol Cell 58, 708 e701 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Obenauer JC, Cantley LC, Yaffe MB, Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res 31, 3635–3641 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olson CM et al. , Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol 14, 163–170 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitfield JR, Beaulieu ME, Soucek L, Strategies to inhibit Myc and their clinical applicability. Front Cell Dev Biol 5, 10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen H, Liu H, Qing G, Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther 3, 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vasta JD et al. , Quantitative, wide-spectrum kinase profiling in live cells for assessing the effect of cellular ATP on target engagement. Cell Chem Biol 25, 206–214 e211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franco LC, Morales F, Boffo S, Giordano A, CDK9: A key player in cancer and other diseases. J Cell Biochem 119, 1273–1284 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Dey J et al. , Voruciclib, a clinical stage oral CDK9 inhibitor, represses MCL-1 and sensitizes high-risk Diffuse Large B-cell Lymphoma to BCL2 inhibition. Sci Rep 7, 18007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bragelmann J et al. , Systematic kinase inhibitor profiling identifies cdk9 as a synthetic lethal target in NUT midline carcinoma. Cell Rep 20, 2833–2845 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Narita T et al. , Cyclin-dependent kinase 9 is a novel specific molecular target in adult T-cell leukemia/lymphoma. Blood 130, 1114–1124 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Yin T et al. , A novel CDK9 inhibitor shows potent antitumor efficacy in preclinical hematologic tumor models. Mol Cancer Ther 13, 1442–1456 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


