Abstract
Spatial mapping of proteins in tissues is hindered by limitations in multiplexing, sensitivity, and throughput. Here we report immunostaining with signal amplification by exchange reaction (Immuno-SABER), which achieves highly multiplexed signal amplification via DNA-barcoded antibodies and orthogonal DNA concatemers generated by primer exchange reactions (PER). SABER offers independently programmable signal amplification without in situ enzymatic reactions, and intrinsic scalability to rapidly amplify and visualize a large number of targets when combined with fast exchange cycles of fluorescent imager strands. We demonstrated 5–180-fold signal amplification in diverse samples (cultured cells, and FFPE, cryosectioned or whole mount tissues), and simultaneous signal amplification for 10 different proteins using standard equipment and workflows. We also combined SABER with expansion microscopy to enable rapid, multiplexed super-resolution tissue imaging. Immuno-SABER presents an effective and accessible platform for multiplexed and amplified imaging of proteins with high sensitivity and throughput.
In situ protein imaging using immunofluorescence (IF) maps specific targets within their native environment. Accurate representation of cell type and state requires visualization of multiple markers. However, conventional methods offer limited multiplexing (typically no more than 5) due to spectral overlap of fluorophores. Several recent methods achieve higher multiplexing, but often with decreased sensitivity, throughput, or accessibility. Multiplexed ion beam imaging (MIBI)1,2, imaging mass cytometry (IMC)3, and multiplexed vibrational imaging4 use specialized instruments to point-scan small fields (e.g. ~2–5 min per 50 μm×50 μm); hence are exceedingly slow for large, centimeter scale tissue sections. Fluorescence methods utilizing sequential antibody staining (MxIF5, CycIF6,7, 4i8) offer high accessibility and relatively fast image acquisition, but require multiple slow cycles of primary antibody incubation (typically hours to overnight per cycle), and can take weeks to image tens of targets5–7. Alternatively, DNA-barcoding methods simultaneously apply ten(s) of antibodies, labeled with orthogonal DNA barcodes, followed by fast sequential barcode readout either through rapid binding/unbinding of fluorescent imager strands using DNA-Exchange (DEI)9–11, or by in situ polymerization of fluorescent dNTP analogs (CODEX)12, reducing experimental time to a couple of days. However, to overcome the overlap of antibody host species, high multiplexing requires conjugating DNA strands to primary antibodies, which results in lower signal (due to lack of amplification from secondary antibodies) and decreased sensitivity, especially for low abundance targets in tissues with high autofluorescence and scattering, and low antigen access. Limited signal further elongates acquisition time and reduces throughput. In situ signal amplification is thus critically needed to improve the signal, throughput, and sensitivity. A preferred amplification method should be scalable and compatible with rapid multiplexing, individually tunable for each target to accommodate the high dynamic range of the proteome13, and applicable for spatially overlapping, dense targets. However, existing methods pose various limitations for satisfying these requirements.
Tyramide signal amplification (TSA)14 amplifies the signal through covalent binding of diffusive tyramide molecules in the vicinity of the target. Due to lack of orthogonal chemistries, TSA amplifies one target per round, and necessitates slow rounds of (typically microwave-based) antibody removal for spectral multiplexing15,16. By using orthogonal sequences, DNA-based methods allow simultaneous signal amplification. In rolling circle amplification (RCA)17, a processive polymerase acts on a circular template to synthesize long concatenated repeats. RCA offers high levels of amplification and potential for multiplexing, but in situ enzymatic reaction is hard to control or tune for individual targets18. Both TSA and RCA may also lead to blurring of signals and decreased resolution, respectively due to spreading of the tyramide molecules or the large size of the amplicons (reaching from 250 nm to over ~1 μm radius19,20). Branched DNA assemblies21,22, such as RNAscope23, generate complex tree structures for stable binding of fluorescent DNA strands, whereas Hybridization Chain Reaction (HCR) utilizes the triggered assembly of metastable fluorophore-conjugated hairpins24–26. The structural complexity of existing DNA-assembly based platforms could present potential challenges for designing highly multiplexed orthogonal systems. In practice, simultaneous signal amplification for proteins beyond spectral multiplexing (3–5 targets) remains to be demonstrated27,28.
Here we report Immuno-SABER, a highly multiplexed and individually controllable signal amplification method free of in situ enzymatic reaction (Fig. 1). After staining the sample with multiple DNA-barcoded primary antibodies, SABER entails hybridization of these barcodes to orthogonal single-stranded DNA concatemers, generated in a pre-programmed manner via PER29 (Fig. 1a), which bind multiple fluorophore-bearing imager strands for multiplexed signal amplification30 (Fig. 1b). Rapid, spectrally unlimited multiplexing can be achieved via Exchange-SABER, which combines SABER amplification with rapid exchange cycles of fluorescent imager strands9–11 (Fig. 1c). We validated Immuno-SABER in diverse samples including cultured cells, cryosections, formalin-fixed paraffin-embedded (FFPE) sections and whole mount tissues, and demonstrated independently tunable amplification from 5 to 180-fold, covering the range conventionally achieved by secondary antibodies to TSA. The simple design of SABER amplifiers makes it scalable. We further demonstrate simultaneous signal amplification and imaging of 10 protein targets in tissues. Finally, we combine SABER with expansion microscopy (ExM)31 to achieve rapid, highly multiplexed super-resolution tissue imaging.
Figure 1. Immuno-SABER schematic.
(a) PER mechanism29: (1) A 9-mer primer of sequence a binds to the single stranded a* sequence on the hairpin (* denotes complementarity). (2) The primer is extended by a strand displacing polymerase (e.g. Bst) isothermally and autonomously. The hairpin features a stopper sequence that halts polymerization, which releases the polymerase. (3) The newly synthesized a is displaced from the hairpin through branch migration. (4) The extended primer and the hairpin autonomously dissociate. (5) Repetition of this copy-and-release process produces a long concatemer of a. (b) Immuno-SABER schematic: (1a) Antibodies conjugated with bridge strands are used to simultaneously stain multiple targets. (1b) Primer sequences (green) are independently extended to a controlled length using PER. (2) Concatemers hybridize to the bridge sequence (blue) on the antibody. (3) Fluorophore (purple star)-labeled 20-mer DNA “imager” strands hybridize to the repeated binding sites on the concatemers. Each imager is designed to bind to a dimer of the unit primer sequence. (c) Exchange-SABER schematic: (1) Different biological targets (t.1 to t.n) are labeled with antibodies conjugated to orthogonal bridge strands (b.1 to b.n). (2) Orthogonal pre-extended concatemers are hybridized (via bridge complements b.1* to b.n*) to the bridge strands on the antibodies simultaneously. (3) Target t.1 is visualized by hybridization of imager i.1* to the i.1 sites on the concatemer bound to b.1 on the corresponding antibody, (4) Multiple targets can be imaged sequentially hybridization and dehybridization of orthogonal imagers in multiple rapid exchange cycles. (5) The images are computationally aligned and pseudo-colorized to overlay different targets.
Results
Validation of in situ signal amplification by Immuno-SABER
For in situ signal amplification, Immuno-SABER relies on controlled in vitro synthesis of amplifier concatemers by PER, followed by programmed in situ assembly. PER utilizes a catalytic hairpin for controllable extension of a short primer sequence in an iterative manner (as in Fig. 1a). Primer designs rely on a 3-letter code (made of only A, T, and C nucleotides), where G nucleotides are avoided in the primer sequence and in the reaction mixture29,30. Hence, the C nucleotide after the template on the hairpin acts as a stopper for the polymerase. This simple step-by-step synthesis offers a tight control of the reaction by external parameters (such as hairpin or dNTP concentration, reaction time and temperature) as well as high programmability and yields long DNA concatemers of desired lengths reaching >500 nucleotides (nt)30. For a modular design, we conjugated antibodies with 42-nucleotide DNA sequences (‘bridges’, from the orthogonal library we designed previously30), which enable hybridization of concatemers through their 5’ segments of complementary sequence to the bridges. To preserve antigen recognition, we optimized out protocol to conjugate 1–3 oligos per antibody (see Supplemental Note 1). We also developed an optional DNA-tag and toehold displacement mediated purification strategy (Supplementary Fig. 1).
For application of our in vitro extension and in situ assembly strategy on biological specimens, labeling specificity, resolution and amplification efficiency are the main considerations. First, to evaluate the specificity and preservation of morphology, we performed Immuno-SABER staining in cultured cells for microtubules as a test case for a densely arranged structural protein target. We have observed specific staining, clear tubular morphology and similar staining pattern to conventional immunostaining with fluorophore-conjugated secondary antibodies (Fig. 2a and b). The resolution of the images, as quantified by full-width-half-maximum (FWHM) of the fitted line plots across the thin microtubules, was unaltered compared to the secondary antibody control (Supplementary Fig. 2a–c). These results demonstrated the suitability of Immuno-SABER for in situ labeling of proteins, with high resolution and high specificity even at dense arrangements.
Figure 2. Validation and quantification of signal amplification by Immuno-SABER.
(a) Cultured BS-C-1 cells were immunostained for alpha-tubulin and three conditions were prepared for comparison: Unamplified condition, where (i) unextended primers with single binding site for imager (with Alexa647) was hybridized to the bridge on the antibody, (ii) the extended concatemer was hybridized for signal amplification (linear amplification), (iii) conventional antibody staining was performed with Alexa647-conjugated secondary antibodies. (b) Representative images of each (max projections from confocal z-stack taken with a 63× objective). (c) Representative images for CD8a staining (labeled with Atto488-imager) in human tonsil FFPE sections (single plane large area scans with a 20× objective cropped to show a region of the CD8a+-cell rich interfollicular zone. (d) Cone arrestin staining in mouse retina cryosections (maximum projection of a confocal z-stack taken with a 20× objective). See Online Methods for experimental details. (e) Level of signal amplification by Immuno-SABER was quantified by measuring the background-subtracted mean fluorescence for several regions of interest in the tissues and expressed as fold amplification over unamplified signal level. Conventional secondary antibody amplification was also quantified similarly and shown as reference. For CD8a FFPEs, n = 80 (for unamplified), 100 (for SABER), 94 (for conventional) rectangular ROIs (each covering 0.03–1.20 mm2 tissue regions; consecutive tissue sections are used for the three conditions). For Cone arrestin, n = 6 images from 2 retina samples. Error bar indicates SEM. (f) Immuno-SABER was performed in whole-mount retina sections for Collagen IV and Vimentin. Maximum projections from confocal z-stacks are displayed.
For validation of the labeling strategy in tissue samples and quantification of the signal amplification, we used two types of tissue preparations: 5 μm-thick FFPE human tonsil sections and 40 μm-thick mouse retina cryosections. Both tissue types provide a good validation platform thanks to the morphologically distinct and well-conserved organization of different cell types that can be identified by well-established biomarkers. Retina has a layered organization of different types of cells whereas tonsils feature multiple germinal centers with stereotypic organization of a large number of distinct cell types.
For the FFPE samples, we imaged the T-cell membrane marker CD8a. CD8a+ cells largely are present in the marginal zone (outside of the germinal centers)32. For mouse retina cryosections, we imaged cone arrestin, a specific marker of the cone photoreceptor cells9. Both markers showed signal patterns consistent with the expected distribution of the markers, suggesting high specificity (Fig. 2c–d). For both targets and sample types, Immuno-SABER yielded similar or slightly higher fluorescence signal than conventional fluorophore-conjugated secondary antibody staining using the same fluorophore. We quantified the amplification level as 6.7-fold for CD8a and 19.4-fold for cone arrestin. For comparison, conventional secondary antibody staining yielded 5.3 and 17.5-fold, respectively (Fig. 2e). Overall, our strategy generated ~5–20-fold improvement in the signal level compared to the unamplified control, reaching amplification in the range of secondary antibodies. The degree of in situ signal amplification may depend on multiple factors, including abundance and organization of targets, the antibodies (e.g. clonality, conjugation efficiency), the method of quantification (the unamplified signal level, thresholding, background subtraction), as well as the experimental conditions and properties of the SABER sequences (e.g. concatemer length30).
Despite reaching >500 nt, SABER concatemers can effectively penetrate relatively thick samples. We validated the concatemer penetration capability in whole-mount preparations of mouse retina by successfully staining for the Muller cell marker Vimentin and blood vessel marker Collagen IV, which were both predominantly detected in the 100 μm region from nerve fiber layer to outer plexiform layer of the retina, as expected33 (Fig. 2f and Supplementary 2d). This high penetration may potentially be attributed to SABER concatemers being largely linear DNA structures that are designed in 3-letter code to avoid secondary structures.
Enhancement of signal through branching
To enable further signal enhancement, which would increase the sensitivity for the proteins of lower abundance, or improve the imaging throughput, we developed a sequential amplification strategy where independently extended secondary concatemers can be branched off the primary concatemer to create more binding sites for fluorescent imagers (‘branched SABER’) (Supplementary Fig. 3a). We have performed similar tests to check the effect of an additional amplification round targeting the same proteins in cell culture, human FFPE tonsil tissues and mouse retina cryosections (Supplementary Fig. 3b–f). With one round of branching, we obtained additional 2.8-fold amplification for CD8a in FFPE sections and 8.4-fold amplification for cone arrestin in cryosections over single round (Supplementary Fig. 3d). Microtubule staining in cells indicates that the specificity of labeling and the high-resolution morphology of the target were still preserved with branched SABER (Supplementary Fig. 3e). FWHM estimations demonstrated that the size of the structures was still bound by the diffraction-limit and were not substantially altered (Supplementary Fig. 3f).
To evaluate the preservation of the access to the target and of the variation in overall signal distribution we performed the baseline quantification, and linear and branched amplification on the same sample for a ubiquitous protein (Lamin B) in HeLa cells using DNA-conjugated secondary antibodies and varying lengths of concatemers (Supplementary Fig. 4). We did not detect a significant difference in the baseline signal for concatemers up to 700 nt in length (for linear amplification), suggesting the length range we utilize in our experiments do not cause decreased access (red plots in Supplementary Fig. 4b and g). For reference we also compared the baseline signal to secondary antibodies, which were conjugated on average to 5 fluorophores. We were able to see that in this case signal level was ~5-fold higher than the single-fluorophore baseline, further demonstrating comparable antigen access (Supplementary Fig. 4d). For the increasing concatemer lengths, the theoretically expected amplification trend was obtained across samples with quantitatively consistent intensity values. Blue bars in Supplementary Fig. 4b and g can be compared for more insights; for example for linear amplification, ~2-fold higher signal intensities were obtained with the long primary concatemer (700 nt) versus the short primary concatemer (350 nt).
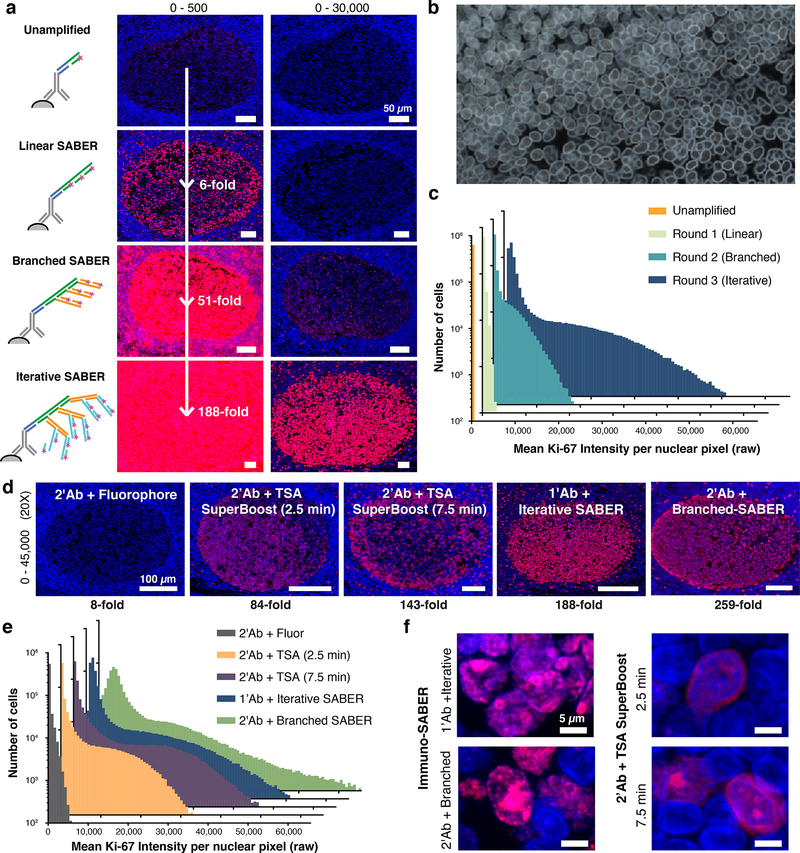
Immuno-SABER allows further improvement of sensitivity by performing multiple branching rounds (‘iterative SABER’). We applied this strategy both in tonsil FFPE sections and mouse retina cryosections, and performed 3 sequential amplification rounds following primary antibody staining. We targeted the proliferating cell nuclear marker Ki-67 that is mainly found in the germinal center (i.e. the dark zone) in tonsil sections, and synaptic marker SV2 that is found mainly in synapses of both outer and inner plexiform layers9. Fig. 3a shows Ki-67-rich germinal centers stained with different amplification levels, displayed at two different contrast scales to allow visual comparison of the signal level.
Figure 3. Immuno-SABER signal can be further amplified through branching.
(a) Nuclear Ki-67 (Alexa647, red) imaging with DAPI (blue) in the Ki-67-rich germinal center of FFPE human tonsil sections shown with up to 3 rounds of amplification (iterative SABER). 16-bit images were scaled to two different maximum pixel values (500 and 30,000) to allow visual comparison. Signals were quantified in each case versus the unamplified sample and the fold changes are provided. (b) Machine-learning based contouring of the nuclei for quantification of signal per cell. See Supplementary Fig. 5a and Online Methods for more information. (c) Mean Ki-67 signal intensity for each nucleus was obtained from automated segmentation and the histogram was plotted for the whole tissue section for each condition. The consecutive sections each contain 636,479–717,176 identified nuclei. (d) Images show germinal centers in FFPE human tonsil sections with Ki-67 labeling (red) with conventional secondary antibody-fluorophore staining, with TSA (HRP-conjugated secondaries) for 2.5 and 7.5 min, with primary antibodies using iterative SABER amplification, or with secondary antibodies using branched SABER amplification. TSA was applied using poly-HRP conjugated secondary antibodies of a commercial SuperBoost Kit with 2.5 or 7.5 min tyramide-Alexa647 incubation. The amplification levels are noted below the images. (e) Histograms to visualize mean nuclear signal level were plotted for the conditions in panel d. The consecutive tissue sections each contain 586,183–717,176 cells. (f) Samples were imaged with a confocal microscope at higher resolution with 63× magnification to evaluate signal blurring. Images with different scaling are displayed in Supplementary Fig. 5f.
To follow the change in the signal level for individual nuclei in the whole section scans, we developed a machine-learning algorithm that automatically annotates DAPI-defined nuclear contours (Fig. 3b and Supplementary Fig. 5a) for nuclear segmentation. This allowed us to assign mean Ki-67 signal intensity to individual nuclei in sections of 550,000–750,000 cells each, and plot histograms for each amplification condition (Fig. 3c). The total nuclear signal for each condition gave a basic quantitative estimate of the amplification level with respect to the unamplified case, yielding 6, 52, and 188-fold respectively for linear, branched and iterative SABER (Fig. 3a). Although the amplification process is exponential in nature, throughout the three rounds the coefficient of variation (standard deviation over mean) for the population remained at a similar level (round 1: 1.76; round 2: 1.65; round 3: 1.55), suggesting that increased amplification level does not create a substantial variation in the distribution.
For these samples, concatemer hybridizations can also be performed for shorter durations (for example with incubation times down to 75 min per round (instead of overnight for the longer primary concatemer, and 3 h for branching as in Fig. 3a) without significant difference in signal level (Supplementary Fig. 5b–c). Iterative-SABER (3-rounds) was also applied for SV2 in thicker mouse retina cryosections generating ~80-fold amplification (Supplementary Fig. 5d).
For such high levels of amplification, catalyzed reporter deposition-based TSA is considered a gold standard with reports of 10–1000-fold amplification14,34,35. This can be further improved 2–10 fold via poly-HRP conjugated secondary antibodies. However, TSA is not suitable for simultaneous multiplexed amplification due to lack of orthogonal chemistries and difficulty in controlling the in situ reaction, and also not ideal for high-resolution imaging due to spreading of the fluorescent tyramide molecules leading to blurring20. To investigate how Immuno-SABER performs in comparison, we first utilized a commercial kit for conventional TSA with mono-HRP conjugated secondary antibodies and Alexa647-tyramide. In retina cryosections, we observed that iterative SABER (with conjugated primary antibodies) could amplify the signal to a higher level than TSA with secondary antibodies (Supplementary Fig. 5e). We also used the commercial SuperBoost™ kit that features poly-HRP conjugated secondary antibodies for Ki-67 staining of FFPE tonsil sections, which yielded up to 143-fold amplification (Fig. 3d–e), Comparatively, both iterative SABER (188-fold, Fig. 3a) with primary antibodies and branched SABER (2-rounds) using secondary antibodies (259-fold) yielded higher signal than TSA (Fig. 3d–e). Where species overlap is not of concern (single or low-plex imaging), linear or branched SABER with secondary antibodies can be performed for increased signal with better-preserved morphology.
At this high amplification level, Immuno-SABER still provided crisp staining and preserved the subcellular morphology, whereas TSA-staining resulted in blurring of the signal and spilling over of the label from the target compartments (Ki-67 is a nuclear protein highly enriched in nucleoli), as signal coming from outside of DAPI labeled nuclei can be observed in the high-magnification confocal images (Fig. 3f and Supplementary Fig. 5f–g). Increasing the tyramide incubation duration to 10 min (2–10 min being the manufacturer recommended optimization range) worsened the blurring (Supplementary Fig. 5f–g).
Simultaneous multiplexed amplification by Immuno-SABER
PER has little inherent restrictions in sequence design outside of preferred single strandedness of the concatemer that is facilitated by the use of 3-letter code, making design of orthogonal sequences straightforward. We previously developed a computational pipeline to design orthogonal PER primer-hairpin pairs with maximum extension efficiency and minimum crosstalk by utilizing in silico simulations using NUPACK36 and designed 50 orthogonal sequences to enable multiplexed imaging29,30. Here, we tested 32 primer sequences and extended each into long DNA concatemers in controlled fashion up to the target length range of 600 to 700-nt (Supplementary Fig. 6a). Since extension efficiency is sequence-dependent, reaction conditions were optimized for each primer by modulating the hairpin concentration and reaction time to obtain concatemers of similar lengths (Supplementary Table 1). All of the tested primers extended into long concatemers in the desired length range, with 31 out of 32 sequences (except #51) yielding a predominant long concatemer band, albeit some heterogeneity in the distribution of shorter reaction products (Supplementary Fig. 6a). Next, we evaluated the orthogonality of detection (crosstalk check) in situ through an imaging-based multi-well plate assay. We targeted α-Tubulin with DNA-conjugated antibodies in cells, and applied each concatemer in separate wells with their cognate imagers or with all other imagers. All 32 sequences yielded specific staining and we observed minimal crosstalk (crosstalk signal/cognate signal = 4%) for only one non-cognate imager-primer pair (Supplementary Fig. 6b–e), suggesting all other sequence combinations are suitable for multiplexed detection.
Next, we sought to validate the applicability of Immuno-SABER for independently programmed multiplexed signal amplification in tissues. We first tested spectral multiplexing on 5 μm human FFPE tonsil sections. FFPE samples are the standard preparations for clinical settings and for archival purposes. However, these preparations suffer from autofluorescence and low antigen access, making amplification of the target signal a necessity. Clinical immunohistochemistry procedures typically utilize amplification methods such as chromatic reactions to improve sensitivity, limiting the multiplexing capability to only one target per section. Multiple thin sections have to be prepared in order to stain different markers, which is not ideal since biopsy-based sample collection is invasive and samples can be scarce. Imaging throughput is another big concern since typically centimeter-scale tissues need to be imaged by whole slide scanning.
After standard preparation protocols for deparaffinization and antigen-retrieval, we simultaneously stained the samples for Ki-67, CD8a, IgA and IgM using antibodies conjugated to orthogonal bridge sequences. Among these targets IgA and IgM are expressed at high levels, whereas Ki-67 and CD8a are more moderately expressed (as a proxy, in the tonsil tissue RNA expression level of IgJ, the joining chain domain for multimeric IgA and IgM was found to be 1,360 tags per million (TPM), while Ki-67 and CD8a expressions are at 15 and 19 TPM, respectively37). We performed whole slide scanning with 5-color spectral multiplexing (DAPI + 4 targets) (Fig. 4a). Immuno-SABER amplification also enables improved imaging throughput by allowing short exposure times. For abundant proteins IgA and IgM, linear amplification provided bright enough signal that permit 1–20 ms exposure times at 20× magnification. For moderately available proteins Ki-67 and CD8a branched SABER enabled 2–10 ms camera exposures and fast, high quality scans of subcellular resolution (Fig. 4a, right).
Figure 4. Immuno-SABER and Exchange-SABER in FFPE human tonsil samples.
(a) Centimeter-scale whole-slide imaging of human tonsil sections with 5-color spectral multiplexing (DAPI + 4 targets). A zoom-in view of the region marked with the grey box showing 4-target imaging with subcellular resolution with a 20× objective and exposure times for each target (auto-exposure setting) is given at the bottom. For IgA and IgM (higher copy number), linear amplification yields high enough signal to achieve auto-exposure times of 1–20 ms under optimized conditions. For Ki-67 and CD8a (lower copy number)47,48 branched amplification (one round of branching) was applied to allow auto-exposure times of 2–10 ms. (b) Higher multiplexing via Exchange-SABER: The schematic for multiplexed imaging workflow where all antibodies are applied simultaneously, followed by simultaneous amplification, and sequential rounds of imaging. IgA and CD3e labeling structures are shown as examples to illustrate the workflow for linear and branched SABER on the same sample. c) Images show a zoom-in view of a germinal center in human FFPE tonsil sections imaged in 7-colors (DAPI + 6-targets) with a single exchange round (round 1: top row; round 2: bottom). 4 of the targets CD8a (Atto488), CD3e (Alexa647), Ki-67 (Alexa750) and PD-1 (Alexa750) were visualized with simultaneous branched SABER amplification, whereas IgA (Atto488) and IgM (Atto565) were visualized with linear amplification.
Next, we utilized Exchange-SABER to increase the multiplexing level and image different cell types in germinal centers with 6 markers (CD8a, CD3e, Ki-67, PD-1, IgA and IgM) (Fig. 4b). SABER amplification was applied simultaneously for all targets via hybridization of concatemer to the orthogonal bridge strands on the antibodies. Simultaneous branching was applied to the 4 less abundant targets (CD3e, CD8a, Ki67 and PD-1) following the primary concatemer hybridization, and 3 markers were imaged per round in 2 rounds as depicted in Fig. 4b. Fig. 4c shows a germinal center with interfollicular zone. With our exchange protocol, efficient imager removal can be performed in 10 min without displacing the concatemers (Supplementary Fig. 7).
Exchange-SABER imaging in mouse retina cryosections
Next, we validated Immuno-SABER in thick (40 μm) cryosections and demonstrated 10-target protein imaging in mouse retina (Fig. 5). We first screened antibodies against a list of targets that have defined staining patterns, including Cone arrestin, SV2, VLP1 (Visinin-like protein 1), Rhodopsin, Calretinin, Protein kinase C alpha (PKCα), Glial fibrillary acidic protein (GFAP), Vimentin, Collagen IV, Calbindin. We then conjugated DNA bridge strands to those antibodies, and validated the specificity and affinity of DNA-conjugated antibodies by comparing the staining patterns from conventional staining using unmodified primary antibodies followed by indirect IF (Supplementary Fig. 8a). As controls for exchange imaging, we compared the pre- and post-washing images for SV2, and found the imager strands were efficiently removed after (Supplementary Fig. 8b), without sample damage or signal loss with correlation coefficient between images >0.95 in 3 wash rounds (Supplementary Fig. 8c).
Figure 5. Exchange-SABER in mouse retina cryosections.
(a) 10 protein targets labeling various retinal cell types were visualized in 40 μm mouse retina cryosections. The markers targeted with Immuno-SABER were Rhodopsin (rod photoreceptors), GFAP (astrocytes), Vimentin (Muller cells9), Collagen IV (blood vessels), three calcium binding proteins49,50 VLP1, Calretinin (found in a subset of amacrine and ganglion cells) and Calbindin (note that although Calbindin was suggested to be also found in a subset of amacrine and ganglion cells, the Calbindin antibody used here mostly labels horizontal cells50), and PKCα (blue cone cells and rod bipolar cells50). The sections were first incubated with all DNA-conjugated antibodies simultaneously. All SABER concatemers were then added simultaneously to the sample, followed by washing and sequential incorporation of the imager strands and multi-round imaging. A z-stack of images was acquired for each target, and DAPI was imaged in every exchange cycle to monitor sample drift. The maximum projected images of each stack were computationally aligned using a subpixel registration algorithm using DAPI as the drift marker9, and pseudo-colored for the overlay presentation. (b) Zoom-in view of the area marked by the white rectangle in a. Three cell subtypes (marked with arrows, I: VLP1+ and Calretinin+, II: VLP1− and Calretinin+, III: VLP1+ and Calretinin−) can be differentiated based on VLP1 and Calretinin expression.
The multiplexed imaging result showed that all targets were successfully captured with expected staining patterns (Fig. 5a). Interestingly, we found that two calcium-binding proteins, VLP1 and Calretinin, together identify three populations of cells, VLP1+/Calretinin−, VLP1−/Calretinin+ and VLP1+/Calretinin+ (Fig. 5b). This observation was confirmed by validating the antibody specificity through comparing the staining patterns of three different antibodies against the same targets and by conventional indirect IF using unconjugated primary antibodies (Supplementary Fig. 8d–f).
Fast multiplexed super-resolution imaging by Expansion-SABER
The spatial resolution of conventional fluorescence microscopy is limited due to diffraction of light. A variety of super-resolution techniques have been developed to overcome this limitation. Expansion Microscopy (ExM), which improves the practical resolution by physically expanding hydrogel embedded samples, enables sub-diffraction imaging of tissues without specialized super-resolution instruments31. However, one challenge for ExM is the dilution of fluorescence signals during physical expansion, which creates a high need for signal amplification. Both indirect IF and HCR have been used to achieve higher signals in ExM with limited multiplexing capability28. A technique termed Magnified Analysis of the Proteome (MAP) was developed to achieve higher multiplexing by combining expansion with repeated antibody staining and retrieval, demonstrating 7 rounds of sequential labeling; however, due to the slow permeation of the antibodies into the thick expanded tissue samples, each round of primary and secondary antibody staining takes 2–9 days, making the approach slow and laborious for high-multiplexing38.
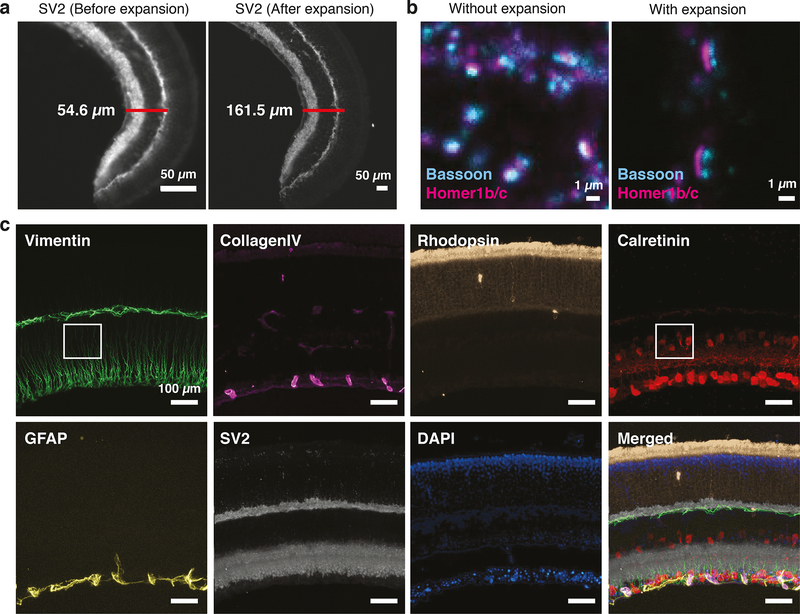
To solve this problem, we combined ExM with SABER (Expansion-SABER). We modified the 5′ end of the SABER concatemers with an acrydite moiety that could be incorporated into polyacrylate hydrogels. Using the original ExM protocol31 with Immuno-SABER for SV2 in mouse retina cryosections, we obtained ~3-fold expansion (Fig. 6a), which is slightly lower than the expansion factor reported before (~4.5-fold) due to the shrinkage of the gel in the ionic gel re-embedding solution or in the imaging buffer with 0.5× PBS (as was observed before for the expansion-FISH protocol31,39. Higher levels of expansion can be attained by the more recent expansion protocols that can achieve 10–20-fold expansion40,41. To validate the sub-diffraction resolution, we imaged a pre-synaptic marker, Bassoon, and a post-synaptic marker, Homer1b/c (which are typically ~160 nm apart42) in fixed primary mouse hippocampal neuron culture (Fig. 6b and Supplementary Fig. 9a). While Bassoon and Homer1b/c were readily separated after expansion, they strongly overlapped without expansion. We next demonstrated Exchange-SABER in expansion samples by visualizing 6 targets (Vimentin, Collagen IV, Rhodopsin, Calretinin, GFAP, SV2) in mouse retina cryosections (Fig. 6c). Individual Muller cells labeled by Vimentin became clearly distinguishable after expansion (Supplementary Fig. 9b). Since the expanded samples reached ~350 μm in thickness we increased both the incubation time and wash time for imagers (to 45 min and 1.5 h, respectively) to achieve optimal imaging quality. We were able to perform 6-target imaging in 2 rounds of imager exchange in 6 h, which is substantially faster than the MAP protocol that would take >3 days38. To further increase the speed, we incorporated an alternative fluorophore removal protocol using imager strands with a disulfide bond between the DNA sequence and the fluorophore, which allow quick removal of the fluorescence signal by reducing agents (e.g. TCEP)43. This approach shortened the signal removal time from 1.5 h to only 10 min (Supplementary Fig. 9c–d).
Figure 6. Multiplexed super-resolution imaging using Expansion-SABER.
(a) 40 μm mouse retina cryosections were stained for SV2 using DNA-conjugated SV2 antibodies, followed by SABER concatemer hybridization. Before and after images were respectively acquired before hydrogel formation, or after hydrogel formation and expansion (~3-fold), using the original expansion protocol31,39. (b) Images of pre- and post-synaptic sites of neuronal synapses in fixed primary mouse hippocampal neuron culture samples with and without expansion (different fields of view are shown). The pre-synaptic sites were labeled with anti-Bassoon antibodies and the post-synaptic sites were labeled with anti-Homer1 antibodies. DNA-conjugated secondary antibodies were used to target Bassoon and Homer1 primary antibodies, followed by SABER concatemers application. (c) ExM imaging of 6 protein targets in the originally 40 μm-thick mouse retina section (expanded ~3-folds) with Exchange-SABER. 2 exchange rounds with Atto488-, Atto565- and Alexa647- conjugated imager strands were performed to visualize all 6 targets in the expanded samples. DAPI was imaged in both rounds to serve as a registration marker. The images are maximum projections of z-stacks, drift-corrected using DAPI channels, and pseudo-colored for presentation. A zoom-in view of the boxed region is available in Supplementary Fig. 9b.
Discussion
Immuno-SABER provides a new highly multiplexed amplification capability to increase the detection sensitivity and tune it independently for multiple targets. To validate its suitability for tissue imaging, we addressed three main considerations. (i) Specific labeling of targets in commonly used sample preparations: We used SABER to label various protein targets (nuclear, cytoplasmic, membrane) in cell and tissue preparations (FFPE, cryosections, expansion samples), and reproduced the expected labeling patterns. (Fig. 2 and Supplementary Fig. 2). (ii) Efficient access to the target and quantifiable amplification without sacrificing resolution: SABER probes can penetrate into whole mount preparations and reach targets at 100-μm depth. Linear amplification with primary antibodies in tissues yielded 5–20-fold signal enhancement (typically at a similar level to indirect IF for the same target) (Fig. 3 and Supplementary Fig. 4). Branched SABER generated ~50-fold amplification (>5-fold higher signal than indirect IF), and iterative SABER produced up to ~180-fold amplification (surpassing TSA) (Fig. 3, and Supplementary Fig. 4–5). At these levels of amplification, SABER did not have a significant impact to the resolution for conventional microscopy and tissue imaging (Fig. 2 and Supplementary Fig. 2–3). This observation still holds for super-resolution imaging with ExM (Fig. 6 and Supplementary Fig. 9). We also validated that the signal level can be further controlled by the concatemer length, and the concatemers (<700-nt for linear, and <450-nt for branching, in cells) do not hinder the access to antigens (Supplementary Fig. 4). (iii) Multiplexing: Through the use of orthogonal concatemers, we showed multiplexed amplification for 10 protein targets in the same sample (Fig. 5), and simultaneous use of linear and branched SABER (Fig. 4). The ability to program and control the amplification fold for each target is valuable for multiplexed imaging of target proteins expressed with a high dynamic range. To lead the way for higher multiplexing, we performed in situ testing of a pool of 32 from our 50 in silico-designed sequences30, where we detected minimal crosstalk for only 1 of the 1024 pairs (Supplementary Fig. 6). Owing to the simplicity of sequence design, it is straightforward to upscale the orthogonal pool size and reach much higher multiplexing levels.
Immuno-SABER is compatible with widely available sample preparation and microscopy platforms, (such as standard wide-field and confocal microscopes or slide scanners), promising easy adoption by research and clinical laboratories. SABER enables higher throughput in two ways. Firstly, it saves sample preparation time by allowing application of multiple primary antibodies in one step, thus alleviate the need for multiple time-consuming antibody staining cycles. This advantage is particularly important for thicker tissue samples such as whole mount and ExM preparations (where we imaged 6 targets in ~3-fold expanded tissues within 3–6 h in contrast to ~4–5 days required in previous protocols such as MAP38). Secondly, SABER saves image acquisition time as high signal level supports reduced camera exposures. A 10–100 fold improvement in image acquisition can deliver 1–2 orders of magnitude enhancement of throughput, which is much needed for discovery-oriented cell/tissue profiling and large-scale mapping projects.
To implement Immuno-SABER for new targets, we recommend aiming to conjugate maximum 1–3 bridge oligos per antibody to preserve the antigen recognition efficiency of the antibody, and to validate specificity of the staining by comparison to unconjugated controls. For relative quantifications, a baseline control can be utilized (on the same sample as in Supplementary Fig. 4, or on different samples as in Fig. 3). As in any signal amplification method, increasing amplification level is expected to yield a wider spread in the population, while moving the mean to higher intensities. For branched amplification, it is theoretically possible that the small differences in the initial binding might lead to larger variations. However, at the population level these factors should have an even effect on the sample (unless there is critical internal heterogeneity, like the target being more buried in some locations, which would bias antibody binding for all IF studies), as suggested by the consistency of the stainings in Supplementary Fig. 4. Advantageously, Immuno-SABER avoids in situ enzymatic reactions, which are hard to reproduce in a quantitative manner since the enzyme activity is harder to control. For SABER, every step relies on simple DNA-DNA binding that can be primarily modulated by salt and formamide concentration, and temperature.
In summary, Immuno-SABER is a simple and effective method for multiplexed and sensitive in situ protein detection with individually programmable signal amplification. Immuno-SABER enables higher-throughput for imaging assays ranging from super-resolution studies to centimeter scale tissue mapping efforts. SABER strategy is also directly applicable for RNA and DNA targets as we recently demonstrated in our SABER-FISH work30. We expect that the unified SABER framework will be useful to a broad range of researchers, complementing single-cell RNAseq analysis with the functional information of protein expression, and will prove valuable for a wide spectrum of potential applications including tissue atlases44,45, tumor/disease profiling, in situ single-cell validation for bulk assays such as flow or mass cytometry, or CITE-Seq46, as well as digital pathology and biomarker screening and discovery.
Online Methods
Technical details for the experiments are given below. We also provide generalized, step-by-step protocols for antibody-DNA conjugation, purification of conjugated antibodies and performing Immuno-SABER in cells. FFPE samples and fixed cryosections in the Supplemental Protocols.
Additionally, we will keep an updated Frequently Asked Questions section available at http://immuno-saber.net and http://saber-imaging.net.
PER sequences and preparation of SABER concatemers
In vitro extension of primers
Concatemer extensions were prepared as described previously30. Typically, 100 μl reactions were prepared in 1× PBS (diluted from the 10× stock, Thermo Fisher AM9625) with final concentrations of: 10 mM MgSO4, 400–1000 units/ml of Bst LF polymerase (NEB M0275L or McLab BPL-300), 600 μM each of dATP/dCTP/dTTP (NEB #M0275L), 100 nM of Clean.G (5’-CCCCGAAAGTGGCCTCGGGCCTTTTGGCCCGAGGCCACTTTCG-3’) hairpin29, 50 nM-1.5 μM hairpin, and water to 90 μl. The Clean.G hairpin has a 5’ stretch of C’s. Pre-incubation with Clean.G helps to get rid of the impurities in the dHTP mixtures (made up of dATP, dTTP, and dCTP), which may have small amounts of dGTP contamination29. After Clean.G incubation for 15 min at 37°C, 10 μl of 10 μM primer was added, and the reaction was incubated for another 1–3 h at 37°C followed by 20 min at 80°C to heat inactivate the polymerase. Reaction products can be stored at −20°C for several months. In our demonstrations, PER products were diluted into concatemer hybridization solutions for binding to the bridge sequences. Alternatively, concatemers can be purified and concentrated using a MinElute (Qiagen #28004) kit with distilled water elution to reduce volume and salt concentration from the reaction condition. Primer sequences and details of the extension conditions utilized for Supplementary Fig. 6 are listed in Supplementary Table 1. These primer sequences were presented as the 3’ tail of the complements of the bridge strands given in Supplementary Table 2, in the format: 5’- bridge*- tt (spacer nucleotides) - 9mer primer sequence - 3’, where * denotes the reverse complement. For the extensions in Supplementary Fig. 6, the 25mer-tester* bridge was used for all the primers. The strands were obtained from IDT. Primers were synthesized and provided with standard desalting. Hairpins were ordered with 3’ inverted dT modification to ensure they cannot be extended. Due to the modification, they were ordered with HPLC purification, which is critical for their performance. Details of the primer and hairpin design criteria are described in our previous work29,30. For primary concatemers, we utilize concatemers reaching the 600–700 base based on empirical experience30.
Gel electrophoresis
After extension, for internal quality control, the lengths of the concatemers were evaluated by diluting 1 μl of in vitro reaction with 19 μl water. For quality control, samples were then run on 1–2% E-Gel EX agarose gels (Thermo Fisher #G402001) for 10 min on the E-gel apparatus (Invitrogen, iBase) alongside a 1 kb Plus DNA Ladder (Invitrogen) and imaged with the SybrGold channel on a Typhoon FLA 9000 scanner.
For the comparison gel in Supplementary Fig. 6, unpurified concatemers were run using 6% TBE-UREA PAGE gels (Thermo Fisher) at 55°C. The gel was pre-run for 1 h before loading the samples. 160 ng Quick-Load Purple Low Molecular Weight DNA Ladder (NEB #N0557S) was loaded as size reference. The reaction products were diluted 1:7 with 2× Urea-Loading Dye, and denatured at 95°C for 5 min. 9 μl from each sample was loaded on the gel. Both samples and the ladder were denatured. Samples were run for 20 min at 75 V, and at 130 V for 1 h. Gels were stained with 1:10,000 SybrGold in 0.5× Tris-Borate-EDTA (TBE) for 30 min and scanned on a Typhoon FLA 9000 scanner.
Imager strands
SABER imager strands are 20mer DNA oligonucleotides with fluorophores on the 5’ end. The imagers used for our experiments also carry a 3’ inverted dT modification, which is optional to include and is not essential for SABER experiments. Imagers were designed to bind the dimers of the primer unit sequence to achieve stable but easily reversible binding that is necessary for DNA-exchange-imaging. Hence, the format of the imager sequence is: 5’ - Fluorophore- tt - primer* - t - primer* - t - Inverted dT - 3’ (t’s are spacer T nucleotides.). They were ordered from IDT with 5’ fluorophore (Atto488, Atto565, Alexa647 or Alexa750), and 3’ inverted dT modification, with HPLC purification. They are named as i.primerID#*. Sequences are listed in Supplementary Table 3.
Branching primers
For stable hybridization of the secondary (branching) concatemers onto the primary concatemers, trimers of the unit repeat sequence were used as bridges, creating a 30-mer hybridization sequence. Hence, branching primers are designed in the format: 5’- p.1* - t -p.1* - t - p.1* - ttt (spacer) - p.2 – 3’, where p.1 is the primer used for the primary concatemer, p.2 is the primer for the secondary concatemer, and t’s are spacer T nucleotides. For secondary concatemers, we utilize extensions <500 bases based on empirical experience.
Similarly, for iterative branching, the tertiary concatemer is designed to use the trimers of primer2 as the bridge, in the format: 5’- p.2* - t - p.2* - t - p.2* - ttt - p.3 – 3’, where p.3 is the primer for the tertiary concatemer and t’s are spacer T nucleotides. For the third round, we utilize extensions <300 bases based on empirical experience.
Antibody-DNA conjugation and purification
Conjugation
The conjugation involves crosslinking of thiol-modified DNA oligonucleotides to lysine residues on antibodies in a non-sequence-specific way. Briefly, 25 μl of 1 mM 5’-thiol-modified DNA oligonucleotides (Integrated DNA Technologies) were activated by 100 mM DTT (Thermo Fisher #20291) for 2 h at RT in dark, and then purified using NAP5 columns (GE Healthcare Life Sciences #17–0853-02) to remove excess DTT. Antibodies formulated in PBS only (or with sodium azide) were concentrated using 0.5 ml 50 kDa Ambicon Ultra Filters (EMD Millipore #UFC510096) to 2 mg/ml and reacted with maleimide-PEG2-succinimidyl ester crosslinkers (Thermo Fisher #22102) for 1.5 h at 4°C (100 μg antibodies: 2.5–3.4 μl of 0.85 mg/ml crosslinker). Antibodies were then purified using 0.5 ml 7 kDA Zeba desalting columns (Thermo Fisher #89883) to remove excess crosslinkers. Activated DNA oligonucleotides were incubated with antibodies (Molar ratio of antibody/ssDNA ∼ 1:11) overnight at 4°C. Final conjugated antibodies were washed using 2 ml 50 kDa Amicon Ultra Filters six times to remove non-reacted DNA oligonucleotides. The list of bridge sequences used for conjugation is provided in Supplementary Table 2. The list of antibodies and the corresponding bridge sequences (IDT) used for each staining, as well as the capture and toehold strands for purification are provided in Supplementary Table 4. Conjugated antibodies were diluted in the 1:50–1:200 for immunostaining. Detailed step-by-step protocol for general use is available in Supplementary Protocols.
Purification
To increase the staining efficiency, conjugated antibodies can be optionally purified using a DNA toehold-mediated affinity pull-down protocol (Supplementary Fig. 1a). For this, 200 μl of high capacity streptavidin agarose (Thermo Fisher #20357) was centrifuged down, washed 3 times using 500 μl PBS, and incubated with 10 μl from1 mM of biotin-labeled binding sequences in 300 μl PBS with 0.1% Triton X-100 for 30 min at RT. The agarose was then washed twice with PBS with 0.1% Triton X-100, followed by blocking with 250 μl blocking buffer (2% BSA + 0.1% Triton in PBS) for 1 h at RT with rotation. The agarose was then centrifuged and resuspended with 200 μl incubation buffer (1% BSA + 0.1% Triton in PBS) containing the DNA-conjugated antibodies, followed by rotation at 4°C for 1 h. The sample was centrifuged at 4°C and washed twice with 200 μl incubation buffer. The bound antibodies were recovered by adding 20 μl of 1 mM toehold strands (listed in Supplementary Table 4) in 200 μl incubation buffer. After centrifugation, the supernatant was collected and the agarose was washed three times with 300 μl washing buffer (PBS + 0.1% Triton), collecting supernatant for each time. The supernatant was pooled together and buffer exchanged using 2 ml 50 kDa Amicon Ultra Filters six times to remove toehold DNA oligonucleotides. Binding sequences and toehold sequences were designed using NUPACK (http://www.nupack.org)36,51,52 and are provided in Supplementary Table 4.
Gel electrophoresis
To examine DNA antibody conjugation, antibodies were denatured in LDS sample buffer (Thermo Fisher #NP0007) without reducing reagents (e.g. DTT or 2-ME) at 90 to 95°C for 3 min, and left to cool down to RT. The samples were run on 3 to 8% Tris-acetate PAGE gels (Thermo Fisher #EA03752BOX) at 80 V for 30 min and 120 V for 3.5 h. The gels were stained with SimplyBlue™ safe stain (Thermo Fisher #LC6060) according to the manufacturer’s manual, and imaged using a Biorad Gel Doc™ EZ imager system. Note that BSA should be avoided in the purification step if the sample needs to be examined using PAGE gels.
Microtubule staining in cell culture and FWHM analysis
Cell culture and staining
BS-C-1 cells (ATCC® CCL-26, African green monkey) were grown in Dulbecco’s modified Eagle medium (Gibco #10564) supplemented with 10% (vol/vol) serum (Gibco #10437), 50 U/ml penicillin, and 50 μg/ml streptomycin (Gibco #15070 and cultured at 37°C in the presence of 5% CO2. They were plated on eight-well ibidi glass-bottom μ-slides (ibidi #80826) and grown until 50–60% confluency. For cell culture experiments in Fig. 2 and Fig. 3, cells were fixed with 4% paraformaldehyde (PFA) for 45 min, and quenched with 100 mM NH4Cl in PBS for 20 min and washed with PBS for 5 min. Cells were then permeabilized and blocked in 2% nuclease-free BSA (AmericanBIO, CAS 9048–46-8) + 0.1%-Triton in PBS for 30 min. Samples were incubated with DNA-conjugated primary antibodies diluted in the incubation buffer made of 0.1% Triton X-100, 2% nuclease-free BSA, 0.2 mg/ml sheared salmon sperm DNA, 0.05% dextran sulfate (Millipore #S4030), 4 mM EDTA (Ambion #AM9260G) in PBS overnight at 4°C, and then washed with PBS with 0.1% Triton X-100 and 2% BSA for 3×10 min. Samples were then washed with PBS 2×5 min and post-fixed using 5 mM BS(PEG)5 (Thermo Fisher #21581) in PBS for 30 min, followed by quenching in 100 mM NH4Cl in PBS for 5 min. The incubation with the primary concatemer was performed at 37°C in 20% formamide (deionized, Ambion #AM9342), 10% dextran sulfate and 0.1% (v/v) Tween-20 in 2× SSC with 0.2 mg/ml sheared salmon sperm DNA for 3 h. The 650-nucleotide long primary concatemers prepared in vitro by PER were diluted in this buffer at 133 nM final primer concentration (primer concentration in the PER mix is considered a proxy for the concatemer concentration after the reaction, since all primers are expected to be extended by the catalytic hairpins that are provided in excess). After concatemer hybridization the samples were washed for 5 min at RT with 45% formamide in PBS and three times for 10 min each with PBS + 0.1% Triton X-100 at 37°C. Branching hybridization was performed at 37°C in 30% formamide, 10% dextran sulfate and 0.1% (v/v) Tween-20 in 2× SSC with 0.2 mg/ml sheared salmon sperm DNA for overnight at 133 nM final concentration of the 450-nt long secondary concatemers. Samples were washed for 5 min at RT with 45% formamide in PBS and 3×10 min each with PBS + 0.1% Triton X-100 at 37°C. Imagers were hybridized at 1–1.5 μM final concentration in PBS + 0.1% Triton X-100 for 1 h at RT (hybridization duration with the imagers can be significantly decreased for faster preparation), followed by a 5 min wash with PBS + 0.1% Triton X-100 and 2×5 min wash with PBS. Samples were stained with 4 μg/ml DAPI (Invitrogen #D1306) in PBS for 10 min and washed twice for 1 min with PBS. Imaging was performed in SlowFade with DAPI (Invitrogen #S36938) embedding medium.
Imaging
For the images exemplified in Fig. 2a, Supplementary Fig. 2a and 3e, a Leica SP5 confocal with a 63×/NA 1.3 glycerol objective was used. A white light laser (470–670 nm) was used at 650 nm for excitation of Alexa647, and a PMT was used for detection.
FWHM analysis
The best focus planes for isolated microtubules were manually selected from z-stacks. 2.5 μm lineplots were drawn across isolated microtubules in this single plane image using Fiji53. Small rectangular background areas were manually drawn to obtain the background values around selected microtubule lines. Average background was subtracted from the lineplot values. Gaussian curves were fit to the background-subtracted values and FWHM was calculated based on the fits and average values were obtained by analyzing 30–45 lineplots from 5 images per condition using a Python script. A similar calculation was performed for 200 nm dark red fluosphere beads (Invitrogen #F8807). Distributions were displayed as box plots. Lineplots with multiple discernible peaks were discarded. Two-sample t-test was performed to check for statistical significance.
Lamin B staining in cell culture for amplification quantification
Staining
HeLa cells (human) were cultured and stained following the general protocol as described above for microtubules, with the following the changes: Before the secondary antibody incubation, blocking was done with in 2% donkey serum + 1.5 % BSA + 0.1% Triton + 0.2 μg/μl sheared salmon sperm DNA for 15 min. Secondaries were also diluted in the same buffer, and stained for 1.5 h at RT. Post-fixation was done for 10 min with 4% PFA. Primary concatemer hybridization: 125 nM for 2 h at 37°C in 20% formamide, 10% dextran sulfate and 0.1% (v/v) Tween-20 in 2× SSC with 0.2 mg/ml sheared salmon sperm DNA. Before and after the concatemer hybridization a block/wash was done for 10 min with 0.1% Triton-X100 in PBS, 0.2 μg/μl salmon sperm DNA+3.3% dextran sulfate for 10 min. Imager hybridization 1 was performed with 1 μm Alexa647-i.27* for 75 min at RT. For linear amplification samples, imager hybridization 2 was performed similarly with Alexa647-i.28*. Branch concatemer hybridization: 100 nM, in the same buffer, overnight at 37°C. For branching samples, imager hybridization 2 and 3 were done with Alexa647-i.30* and Alexa647-i.25*, respectively. Sequences and extension conditions are listed in Supplementary Table 5.
Imaging
21–26 plane z-stacks were imaged using a Zeiss Axio Observer Z1 with a 100×/1.4 oil objective. Alexa647 (Cy5: 590–650,665+) images were acquired with 10% of the LED power with 150 ms exposure, and DAPI (300–400,420–470) with 20% power for 100 ms.
Signal quantification
Maximum projections of the z-stacks were created using FIJI. DAPI images were acquired and used to create nuclear masks in MATLAB and Image Processing Toolbox. Mean Alexa647 signal per pixel were calculated within the mask region for each cell.
SABER application, quantification and analysis on human tonsil FFPE sections
Preparation of formalin-fixed paraffin-embedded (FFPE) tonsil samples and antigen retrieval
Human specimens were retrieved from the archives of the Pathology Department of Beth Israel Deaconess Medical Center under the discarded/excess tissue protocol as approved in Institutional Review Board (IRB) Protocol #2017P000585. We have complied with all relevant ethical regulations. 5 μm sections were cut with a rotary microtome, collected in a water bath at 30˚C, transferred to positively charged glass slides and baked at 60˚C for 2 h. For antigen unmasking, slides were placed on a PT-Link instrument (Agilent), which allows the entire pre-treatment process of deparaffinization, rehydration and epitope retrieval (with citrate buffer) to be combined into a single step. Slides were held at 4˚C in PBS until staining. Antigen-retrieved FFPE sections used in multicolor experiments in Fig. 4 were acquired from Ultivue Inc.
Staining of antigen-retrieved FFPE tonsil samples
After antigen retrieval, sections were optionally stored in PBS at 4˚C for up to 2 weeks. For staining, sections were washed in PBS for 15 min and outlined with a hydrophobic pen (ImmEdge Hydrophobic Barrier PAP Pen, Vector Laboratories #H4000) orenclosed in a removable chamber (ibidi, #80381). At this stage a mild 1 h pre-bleaching with 1% H2O2 in PBS can be optionally applied (for TSA). This step is not necessary for Immuno-SABER. We incorporated this step for the Ki-67 stainings in Fig. 3 and Supplementary Fig. 5, to keep the conditions similar to TSA preparations, which benefit from a pre-bleaching step significantly. Samples were blocked for 1 h with PBS containing 2% BSA and 0.1–0.3% Triton X-100, with 3 buffer exchanges. DNA-conjugated primary antibodies were diluted in the blocking solution supplemented with 0.2 μg/ml sheared salmon sperm DNA, 0.05% dextran sulfate, and optionally 4 mM EDTA, and incubated on the samples overnight at 4˚C in a humidified chamber. The antibodies and respective bridge sequences used for the experiments are listed in Supplementary Table 4. Depending on the antibody this step can be shortened to 1 h when performed at RT or at 37°C. Excess antibodies were washed at RT for 3×15 min with PBS containing 2% BSA and 0.1–0.3% Triton X-100, and 2×5 min with PBS. Bound antibodies were then post-fixed with 5 mM BS(PEG)5 in PBS for 30 min at RT, followed by quenching in 100 mM NH4Cl in PBS for 5 min and washed for 15 min with PBS with 0.1% Triton X-100 at RT. Post-fixation is critical to ensure that the antibodies are not washed away during further labeling and imaging. The incubation with the primary concatemer was performed at 37°C in 20–30% formamide, 10% dextran sulfate and 0.1% (v/v) Tween-20 in 2× SSC with 0.2 mg/ml sheared salmon sperm DNA for 1 h to overnight. Concatemers prepared in vitro by PER were diluted in this buffer at 66–150 nM (1:15 to 1:7.5 dilution) final primer concentration. For multiplexing, all primary concatemers were incubated simultaneously. After concatemer hybridization the samples were washed for 5 min at RT with 45–50% formamide in PBS and three times for 10 min each with PBS + 0.1% Triton X-100 at 37°C. For cases, where further amplification was desired, branching hybridizations were performed similarly (at 37°C in 30% formamide, 10% dextran sulfate and 0.1% Tween-20 in 2× SSC with 0.2 mg/ml sheared salmon sperm DNA for 1 h to overnight). Bridge and primer sequences for each target and experiment are given in Supplementary Table 4. For the unamplified sample, the unextended primer with single imager binding site (equivalent to two repeats of the primer sequence) was incubated at the same concentration instead of the extended concatemer (for CD8a: bc42_2*-tt-p.25-a-p.25-a; for Ki-67, bc42_0*-tt-p.30-a-p.30-a, sequences are given in Supplementary Table 5)
For different tissue types, and combination of targets for multiplexing, experimental conditions may need to be optimized to achieve the best signal level. For primary concatemers we recommend using sequences ≤650 bases, for secondary concatemers ≤450 bases, and for tertiary concatemers ≤250 bases. For iterative amplification the wash temperatures were raised to 42°C and the 45–50% formamide in PBS wash step was performed once as a final wash only at the end of all iterations (rather than after each amplification round).
Fluorophore hybridization and dehybridization
Imagers were hybridized at 1–1.5 μM final concentration in PBS + 0.1% Triton X-100 for 1–2 h at RT, followed by a 5 min wash with PBS + 0.1% Triton X-100 at 37°C and 2×5 min wash with PBS at RT. Elongated hybridization times were used for convenience, for faster protocols, hybridization duration for the imagers can be significantly decreased with similar performance. Samples were stained with 1 μg/ml DAPI in PBS for 10 min and washed 2×1 min with PBS. For multiplexing experiments, samples were imaged in PBS shortly after preparation and coverslips were temporarily secured with Fixogum (Marabu). Coverslipping of the samples for imaging is optional depending on the instrument type. Imagers were removed with 10 min incubation at RT in 50% formamide in PBS, followed by 2×5 min wash with PBS at RT. A new round of imager hybridization was performed as above.
For experiments with a single round of imager hybridization (no multiplexing or only spectral multiplexing) samples were embedded in SlowFade with DAPI, secured with nail polish, and imaged on the same day or embedded with ProLong Diamond (Invitrogen #P36971) and incubated at RT overnight for curing. Also see Supplemental Note 2.
Tyramide signal amplification
For FFPE samples, Anti-Rabbit IgG Alexa647 Tyramide SuperBoost™ Kit (Life Technologies #B40916) was used according to manufacturer’s recommendations. The optional step of 1 h pre-bleaching with 1% H2O2 in PBS was applied. Tyramide-Alexa647 was incubated for 2.5 min, 7.5 min and 10 min. Longer incubation times (≥7.5 min) were observed to cause increased blurring of the signal, making shorter incubation times more favorable (as suggested by the manufacturer).
Fluorescence Imaging
FFPE samples in Fig. 2, 3, and Supplementary Fig. 2, 3, 5b–c were imaged with an Olympus VS-120 system equipped with an Orca R2 monochrome (16 bit) camera using a 20×/0.75 NA air objective, with a pixel size of 0.320 μm and single plane tile scans were acquired. For Fig. 3 staining, exposure times of 35 ms and 30 ms were used for DAPI and Ki-67 respectively. For the CD8a staining in Fig. 1–2, exposure time of 350 ms was used.
For the zoom-in’s in Fig. 3f and Supplementary Fig. 5f a Leica SP5 confocal with 63×/NA 1.3 Glycerol objective were used. A white light laser (470–670 nm) was used at 650 nm for excitation of Alexa647, and a PMT was used for detection.
Multiplex FFPE samples in Fig. 4 and Supplementary Fig. 7b were imaged with a Perkin Elmer Vectra Polaris microscope in whole slide scanning configuration with a 20×/0.80 NA air objective. For Fig. 4c imagers were first hybridized for three of the targets (as depicted in the figure), followed by nuclear DAPI staining, washing, coverslipping and whole section imaging in 5 channels (3 markers plus DAPI and autofluorescence). This was followed by dehybridization as described above. The second set of imagers was similarly hybridized and the sample was reimaged. Images obtained in .qptiff format were converted to .tif with Imaris software and aligned using “Align by line ROI” plugin on Fiji using the DAPI fluorescence in each cycle and overlays for display were assembled in Adobe Photoshop.
Signal quantification
For quantification of signal amplification for CD8a labeling in tonsil samples in the Fig. 2 and Supplementary Fig. 3b,d, rectangular regions of interest (ROIs) covering 0.30–1.2 mm2 tile scans were selected after manual inspection to exclude areas with autofocusing errors or sectioning imperfections, and a customized CellProfiler routine was used first to remove masked autofluroescent structures identified in an independent channel (using global robust thresholding), then to calculate mean fluorescence signal/pixel for cell regions masked via thresholding of the CD8a signal54. For masking of the labeled structures, edges were enhanced by Canny edge finding method with automatic threshold calculation. Then global robust background thresholding was applied. Upper outlier fraction and correction factor for thresholding was adjusted manually to compensate for the difference in overall signal level under different amplification conditions. The background was calculated by averaging the CD8a fluorescence signal of the tissue regions (ensured by presence of DAPI) outside of the masked signal regions (after dilation of the masked pixels). The final intensity value was obtained by subtracting the mean background value for each ROI from the average signal value for masked cells. Amplification fold was calculated by dividing the mean background-subtracted fluorescence by the unamplified sample (for linear SABER, Fig. 2c,e) or by the linear amplification sample (for branched SABER, Supplementary Fig. 3b,d). Figures were prepared for display using OMERO for ROI selection, and scaling55.
Nuclear segmentation and analysis of single-nuclei intensity distribution and quantification of fold-amplification
We annotated the contours of all the nuclei for 80 patches of 128×128 pixels. This dataset was then augmented 8-fold using reflections and rotations, resulting on a dataset of 640 images. For each image, an equal number of non-contour pixels were randomly selected to represent the complementary class in the machine-learning algorithm. We trained a variant of the U-Net model56, adding batch normalization57 and residual learning58, to classify pixels in two classes (class 1: nuclei contours; class 2: background and nuclei interiors). We used our own implementation in TensorFlow (hyperparameters are given in Supplemental Note 3). The approximate center of each nucleus was then identified through the regional maxima of a Gaussian-blurred version of the inverted contour probability class. These regional maxima were used as seed points for applying a marker-controlled watershed transform. Background objects were eliminated based on mean intensity and area. For the fold amplification estimation, mean signal intensity from all the nuclei in each tissue sample were summed up to obtain the total Ki-67 signal level. Then, total signal for each sample was divided by the total signal for the unamplified condition. Total values were not corrected for the variation of the number of cells in each section (tissue samples are consecutive sections from the same tissue). Histograms that show the distribution of Ki-67 signal per individual cells were plotted using Python.
SABER application and quantification on mouse retina cryosections
Sample preparation
All animal procedures complied with all relevant ethical regulations and were in accordance with the National Institute for Laboratory Animal Research Guide for the Care and Use of Laboratory Animals as approved by the Harvard Medical School Committee on Animal Care. Animals were given a lethal dose of sodium pentobarbital (120 mg/kg) (MWI, 710101) and enucleated immediately. Eyes were removed and fixed in PFA for 15 to 30 min. Following dissection, retinas were immersed in 30% sucrose overnight prior to freezing in TFM (EMS, 72592) and cryosectioning at ~30–40 μm. Eight-well ibidi glass-bottom μ-slides were treated with 0.3 mg/ml poly-D-Lysine for at least 30 min, followed by 3 PBS washes. Retina sections were immobilized onto the glass and stored at −20°C. Sections were washed with Tris-buffered saline (TBS) + 0.3% Triton X-100 for 3×10 min washes. Samples were then permeabilized and blocked in 5% normal donkey serum (Jackson ImmunoResearch 017–000-001) + 0.2 mg/ml sheared salmon sperm DNA + 0.1–0.3% Triton X-100 in PBS for 2 h. Samples were incubated with DNA-conjugated primary antibodies diluted in the incubation buffer made of 0.1% Triton X-100, 5% normal donkey serum, 0.2 mg/ml sheared salmon sperm DNA, 0.05% dextran sulfate, 5 mM EDTA in PBS overnight at 4°C, and then washed with PBS with 0.1% Triton X-100, 1% normal donkey serum + 5 mM EDTA for 3×30 min. Samples were then washed with PBS 2×5 min and post-fixed using 5 mM BS(PEG)5 in PBS for 30 min, followed by washing in PBS for 2 min and quenching in 100 mM NH4Cl in PBS or 1× TBS for 10 min.
Immuno-SABER
Extended concatemers were diluted in 1:7.5 to 1:20 (depending on the target density) in incubation buffers for overnight hybridization at RT. Two different concatemer hybridization buffers were used: buffer 1 is 40% formamide + 10% dextran sulfate + 0.1% Triton X-100 + 0.02% sodium azide + 5 mM EDTA in PBS, and buffer 2 is 30% formamide + 10% dextran sulfate + 0.1% Triton X-100 + 0.02% sodium azide + 5 mM EDTA in PBS. Buffer 1 was used for incubation with primary concatemers and buffer 2 was used for branching concatemers. Excess concatemers were washed with 45% formamide + 0.1 % Triton X-100 + 5mM EDTA in PBS for 30 min and twice with 30% formamide + 0.1 % Triton X-100 + 5mM EDTA in PBS for 30 min at RT. For branched conditions, 45% formamide was replaced with 40% formamide for the first wash. For multiplexed imaging experiment, all primary concatemers were incubated simultaneously. Bridge and primer sequences for each target and experiment are given in Supplementary Table 4.
To quantify linear SABER amplification for cone arrestin, the amplification samples were incubated with SABER concatemers extended from 25mer-tester*-tt-p.28. The unamplified samples were hybridized with the unextended primer 25mer-tester*-tt-p.28-a-p.28, which carries one imager binding site (equivalent to two repeats of the primer sequence) instead of the extended concatemer (at the same final concentration). For quantification of branched SABER for cone arrestin, the branching sample was first incubated with the primary concatemers extended from 25mer-tester*-tt-p.28, followed by incubation with secondary concatemers extended from 28*-t-28*-t-28*-ttt-p.25 primer.
For iterative SABER quantification for SV2, the unamplified sample was hybridized with the unextended primer bc42_3*-tt-p.27-a-p.27. The linear amplification sample was hybridized with the primary concatemer extended from bc42_3*-tt-p.27. The branching sample additionally hybridized with the secondary concatemer 27*-t-27*-t-27*-ttt-p.28. The iterative amplification sample was additionally hybridized the tertiary concatemer extended from 28*-t-28*-t-28*-ttt-p.32. Sequences are listed in Supplementary Table 5.
Fluorescence Imaging
Fluorophore-labeled imager strands were diluted in 0.1% Triton X-100 in PBS to ~250 nM-1 μM, and incubated with samples for 30 min, followed by washing using 0.1% Triton X-100 in 0.5× PBS for three times. Samples were left in PBS during image acquisition. For multiplexing, imagers strands were dehybridized by 0.1× PBS + 30% formamide incubation for 3×10 min, followed by 3×5 min washes with 1× PBS to remove the residual formamide before rehybridization of imager strands as above.
For 10-color multiplexing, the entire experiment was done using 7 exchange rounds (round 1: Calbindin and Vimentin; 2: GFAP and PKCα; 3: Collagen IV and Calretinin; 4: Rhodopsin; 5: Vlp1, 6: SV2; 7: Cone arrestin) with an average of 1.5 h per round (that included 30 min for imager hybridization, 15 min for wash of excess imager strands, 15 min for imaging and 30 min for imager strands displacement). It should be noted that the duration for each step is sample dependent with thicker samples requiring longer time to ensure complete penetration and signal removal. To ensure the best signal and simplify the multiplexed experimental design, we allowed excess time for each step, however it is possible to shorten incubation and washing times upon optimization.
All images for mouse retina sections were acquired using a Zeiss Axio Observer with LSM 710 scanning confocal system with a 20×/0.8 NA air objective. The images were 512×512 pixels or 1024×1024 pixels and acquired at acquisition speed 7. Each image was acquired by averaging 2 images. Atto488 was visualized using a 488 nm laser; Atto565 was visualized using a 546 nm laser; Alexa647 was visualized using a 633 nm laser. To remove imager strands, samples were washed three times with 30% formamide + 0.1% Triton X-100 in 0.1× PBS. The samples were left in PBS during imaging. Acquired images were scaled and colorized for display using FIJI53 and Photoshop.
Tyramide signal amplification
For retina cryosections, Alexa647 TSA Kit #6 with HRP-goat anti-mouse IgG (Life Technologies #T20916) was used following manufacturer’s recommendations, without the optional pre-bleaching step and with 7.5 min tyramide incubation.
Quantification
For quantification of signal amplification in retina samples (SV2 and cone arrestin), the mask regions were selected manually using FIJI and mean fluorescence intensity was calculated. The background was calculated by averaging the fluorescence signal of six randomly selected regions outside the retinas. The final fold-amplification values were obtained by subtracting the background value from the average signal value for the condition and normalizing that by the unamplified (Fig. 2d, Supplementary Fig. 5d) or by the linear condition (Supplementary Fig. 3c–d).
SABER application on whole mount retina samples
Sample preparation and staining
Whole mount retina staining was performed on the free-floating samples, which allowed reagents to penetrate from both sides of retinas. The samples preparation was conducted with a similar protocol as above but with longer incubation and wash times. DNA-conjugated primary antibodies were incubated for 40 h at 4°C, and washed for 3 × 1 h. DNA extensions were incubated for 40 h at RT, and washed for 3 × 1 h. Bridge and primer sequences for each target are given in Supplementary Table 4. Fluorescent oligos were incubated for 2 h at RT, followed by 3 × 30 min. For imaging, retinas were flattened by creating 4 incisions and mounted on a glass slide. After flattening, the thickness of whole mount retina samples us typically ~160 to 180 μm.
Imaging
The images were acquired using a Zeiss Axio Observer with LSM 710 scanning confocal system with a 10×/0.45 NA air objective. A z-stack of 98 sections with 1 μm spacing was taken for each target. It should be noted that although the entire whole mount retina is about ~180 μm, the signal from vimentin and collagen IV are typically located in half of the retina section from the nerve fiber layer to the outer plexiform layer.
PER sequence crosstalk analysis
For microtubule staining in Supplementary Fig. 6, BS-C-1 cells were grown in glass-bottomed 96-well plate (Ibidi #89626) with 5,000 cells per well. Cells were fixed with 4% PFA for 15 min, and quenched with 50 mM NH4Cl in PBS for 7 min. Cells were then permeabilized and blocked in 0.1% Triton X-100, 0.1% Tween20, 2% nuclease-free BSA (AmericanBIO, CAS 9048–46-8) and 0.2 mg/ml sheared salmon sperm DNA (Thermo Fisher #AM9680) in PBS for 1 h. Samples were incubated with DNA-conjugated primary antibodies diluted in incubation buffer (0.05% Triton X-100, 0.05% Tween20, 2% nuclease-free BSA, 0.2 mg/ml sheared salmon sperm DNA, 0.05% dextran sulfate (Millipore #S4030), 5 mM EDTA in PBS) overnight at 4°C, and then washed with washing buffer (0.05% Triton X-100, 0.05% Tween-20, 2% nuclease-free BSA, 5 mM EDTA in PBS) for five times (1–2 min for the first two washes and 10 min incubation for the other three washes). Samples were washed with PBS twice and post-fixed using 5 mM BS(PEG)5 in PBS for 1 h, followed by quenching in TBS for 10 min. Concatemer and imager hybridizations were performed as described for the retina cryosections.
For cognate wells, 20 nM corresponding imager strands were incubated, while for crosstalk wells, all other imager strands were incubated with 20 nM for each imager strand. The samples were imaged using Zeiss Axio Observer Z1 with a 20×/0.8 air objective. For signal quantification, the bright field images were acquired and used to create masks in MATLAB. Average fluorescence signals were calculated within the mask region. The background was calculated as the average fluorescence signals outside the cells. The final fluorescence intensity was average fluorescence intensity within cell masks minus background intensity.
Expansion microscopy
The PER primer sequences were modified with acrydite at the 5’-end (IDT), and extended as above. Mouse retina samples were stained with DNA-conjugated antibodies, followed by concatemer hybridization as above for retina cryosections. After washing away excess concatemers, a layer of expandable gel was formed according to the original expansion microscopy protocol31. In brief, samples were incubated monomer solution (1x PBS, 2 M NaCl, 8.625% (w/v) sodium acrylate, 2.5% (w/v) acrylamide, 0.15% (w/v) N,N’-methylenebisacrylamide) with ammonium persulfate (APS) and teramethylethylenediamine (TEMED) on ice with open air for 20 min. A gelation chamber was then constructed by placing a #1 coverglass on each side of the tissue section. The specimens were transferred to a humidified incubator and left at 37°C for 2 h. The samples were then digested using Proteinase K (New England BioLabs, Cat.No: P8107S) at 1:100 dilution in digestion buffer (50 mM Tris (pH 8), 1 mM EDTA, 0.5% Triton X-100, 0.8 M guanidine HCl) at 37 °C overnight. The digested samples were then expanded in excess volumes of de-ionized water. To prevent expanded samples from shrinkage, they were re-embeded in a nonexpandable gel (3% acrylamide, 0.15% N,N′-Methylenebisacrylamide with 0.05% APS, 0.05% TEMED). The gel was placed on a bind-silane treated #1.5 coverglass and immersed in the gel solution on ice for 20 min, followed by gelation at 37°C for 1.5 h. 5 μl Bind-Silane reagent (GE #GE17–1330-01) was diluted with 8 ml of ethanol, 1.8 ml of ddH2O and 200 μl of acetic acid. For coating, coverglasses were washed with ddH20 followed by 100% ethanol, then incubated with bind-silane and air-dried. After re-embedding, imager strands were applied as for normal cryosections, but with longer washing (6×10 min), and were imaged as above in regular retina tissue section imaging. Dehybridization time was also elongated to 6×10 min incubation.
For the primary neuron culture, the culture was grown on a 12 mm diameter round #1 coverslips, and stained with Bassoon and Homer1b/c antibodies, followed by DNA-conjugated anti-mouse and rabbit secondary antibodies. The primary antibodies were stained in 5% Normal Donkey serum with 0.1% Triton X-100, and the secondary antibodies were stained in 5% Normal Donkey serum with 0.1% Triton X-100, 5mM EDTA and 0.2 mg/ml sheared salmon sperm DNA. Concatemer incubations were done as for retina cryosections. The expandable gel (19% (w/v) sodium acrylate, 10% (w/v) acrylamide, 0.05% (w/v) N,N’-methylenebisacrylamide in PBS) was formed by placing the coverslip against a parafilm sheet with 20 μl of expansion gel solution in between. The gel was then digested and expanded as above. The gel was transferred to a coverslip dish (ibidi #81148) and was incubated with the imager strands (Atto488-i.30* and Atto565-i.26*) in 0.5× PBS without re-embedding and left in 0.5× PBS during confocal imaging, which gave a similar expansion factor of ~3 fold. The imager strands were incubated for 30 min and followed by 3×10 min washes using 0.5× PBS. After imaging, the imager strands were washed for 5×10 min, and imaging was performed as described as above for retina cryosections.
For TCEP reduction of fluorophores in expanded samples, the samples were incubated with 50 mM TCEP (646547; Sigma) diluted in 1 × PBS and the fluorescence signal was monitored in a time course of 15 min.
Statistics and reproducibility
Cell experiments in Fig. 2b, Supplementary Fig. 2a–c, 3e–f and 4 were performed a number of times with similar results before the final data were quantified for a single experiment. All retina histology experiments (Fig. 2d–f, 5, 6; Supplementary Fig. 2d, 4d, 9) were conducted at least twice with similar results, except the controls in Supplementary Fig. 8, which were performed as a single dataset with internal controls. The FFPE stainings in Fig. 2, 3, Supplementary 3b–d, and 5 were separately tested in at least two independent optimization experiments, before the quantification on one dataset from the consecutive sections from the same source sample. For FFPE sections, single, double, and triple antibody stainings were performed separately in multiple occasions prior to the same sample 4-color and 6-color multiplexed experiments in Fig. 4 and Supplementary Fig. 7. The in vitro reactions in Supplementary Fig. 6a were individually optimized and each tested multiple times before the combined gel run. Cell experiments in Supplementary Fig. 6 were repeated twice with similar results. The plots in Supplementary Fig. 1c, 2b, 8b, 9d are specific to the image shown, and are representative of the overall tendency of the measurements in the respective dataset. Further internal controls through the optimization phase including negative controls with antibodies or other elements missing, and signals matching expected morphologies further increase our confidence in the consistency and reproducibility of the technique in multiple contexts.
Life Sciences Reporting Summary
Further information on experimental design and antibodies are available in the Life Sciences Reporting Summary.
Data and Software Availability
The data and essential custom scripts for image processing will be made available from the corresponding authors P.Y. (py@hms.harvard.edu), S.K.S. (Sinem.Saka@wyss.harvard.edu), and Y.W. (yuwang01@fas.harvard.edu) upon request. The deep learning algorithm and test dataset for automated identification of nuclear contours in tonsil tissues is available on https://github.com/HMS-IDAC/UNet. The MATLAB code for nuclear segmentation is available on: https://github.com/HMS-IDAC/SABERProbMapSegmentation.
Supplementary Material
Acknowledgements
We thank C. Cepko, P. Sorger, Z.Maliga and J. Lin for discussion. We thank the Neurobiology Department and the Neurobiology Imaging Facility for instrument support. This facility is supported in part by the Neural Imaging Center as part of an NINDS P30 Core Center grant #NS072030. We thank M. Manesse, T. Archivald and D. Bowman for help with the FFPE samples, and I. Goldaracena for comments on the manuscript. We thank S. Wang for providing neuronal cultures. Funding: This work was supported by grants from the National Institutes of Health (under grants NIH Common Fund 1UG3HL145600/HuBMAP, 1R01EB018659, 1U01MH106011, 1DP1GM133052, 1R01GM124401), the Office of Naval Research (under grants N00014–16-1–2410, N00014–16-1–2182, N00014–18-1–2549), the National Science Foundation (under grant CCF-1317291), Harvard Medical School Dean’s Initiative, and Wyss Institute’s Molecular Robotics Initiative to P.Y. G.M.C. was supported by NIH grant (R01NS083898, RM1 HG008525, R01MH113279), and P.S.K. was supported by NIH grant (1R01MH113349). J.Y.K. was supported by a National Science Foundation Graduate Research Fellowship. B.J.B. was supported by a Damon Runyon Cancer Research Foundation Fellowship (HHMI). S.W.L. was supported by HHMI and the National Institutes of Health (grant 5K99EY028215–02). S.K.S. was supported by a long-term postdoctoral fellowship from Human Frontier Science Program (HFSP) (LT000048/2016-L) and an EMBO long-term fellowship (ALTF 1278–2015).
Footnotes
Competing Financial Interests. S.K.S., Y.W., J.Y.K., B.J.B., and P.Y. are inventors for provisional patent applications. P.Y. is a co-founder of Ultivue, Inc and NuProbe Global.
Supplementary Information. Supplementary Information contains nine supplementary figures, five supplementary tables, and three supplementary notes. Detailed step-by-step protocols are available in the Supplementary Protocols.
References
- 1.Angelo M et al. Multiplexed ion beam imaging of human breast tumors. Nat Med 20, 436–442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levenson RM, Borowsky AD & Angelo M Immunohistochemistry and mass spectrometry for highly multiplexed cellular molecular imaging. Lab Invest 95, 397–405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giesen CW, Schapiro HAO, D.; Zivanovic N; Jacobs A; Hattendorf B; Schüffler PJ; Grolimund D; Buhmann JM; Brandt S; Varga Z; Wild PJ; Günther D; Bodenmiller B Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nature Methods 11, 417–425 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Wei L et al. Super-multiplex vibrational imaging. Nature 544, 465–470 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerdes MJ et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A 110, 11982–11987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin JR, Fallahi-Sichani M & Sorger PK Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat Commun 6, 8390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin J-R et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. eLife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gut G, Herrmann MD & Pelkmans L Multiplexed protein maps link subcellular organization to cellular states. Science 361 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Wang Y et al. Rapid Sequential in Situ Multiplexing with DNA Exchange Imaging in Neuronal Cells and Tissues. Nano Lett 17, 6131–6139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schueder F et al. Universal Super-Resolution Multiplexing by DNA Exchange. Angew Chem Int Ed Engl 56, 4052–4055 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jungmann R et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nature methods 11, 313–318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goltsev Y et al. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 174, 968–981 e915 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lundberg E et al. Defining the transcriptome and proteome in three functionally different human cell lines. Mol Syst Biol 6, 450 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bobrow M, Litt GJ, Shaughnessy KJ, Mayer PC & Conlon J The use of catalyzed reporter deposition as a means of signal amplification in a variety of formats. Journal of Immunological Methods 150, 145–149 (1992). [DOI] [PubMed] [Google Scholar]
- 15.Yarilin D et al. Machine-based method for multiplex in situ molecular characterization of tissues by immunofluorescence detection. Sci Rep 5, 9534 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stack EC, Foukas PG & Lee PP Multiplexed tissue biomarker imaging. J Immunother Cancer 4, 9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schweitzer B et al. Immunoassays with rolling circle DNA amplification: a versatile platform for ultrasensitive antigen detection. Proc Natl Acad Sci U S A 97, 10113–10119 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagendran M, Riordan DP, Harbury PB & Desai TJ Automated cell-type classification in intact tissues by single-cell molecular profiling. Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng RZ, K.; Wang L; Sun Y; LI J DNA-Sequence-Encoded Rolling Circle Amplicon for Single-Cell RNA Imaging. CHEM 4, 1373–1386 (2018). [Google Scholar]
- 20.Chen Y et al. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J Cell Biol 217, 4025–4048 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pachl CT, J. A; Kern DG; Sheridan PJ; Fong SJ; Stempien M; Hoo B; Besemer D; Yeghiazarian T; Irvine B; Kolberg J Rapid and precise quantification of HIV-1 RNA in plasma using a branched DNA signal amplification assay. JAIDS Journal of Acquired Immune Deficiency Syndromes 8, 446 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Kern D et al. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol 34, 3196–3202 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang F et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14, 22–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dirks RM & Pierce NA Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci U S A 101, 15275–15278 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi HM et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat Biotechnol 28, 1208–1212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi HMT et al. Third-generationin situhybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Xie W, Kohman RE & Church GM Multiplexed imaging using same species primary antibodies with signal amplification. bioRxiv (2018). [Google Scholar]
- 28.Lin R et al. A hybridization-chain-reaction-based method for amplifying immunosignals. Nat Methods 15, 275–278 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Kishi JY, Schaus TE, Gopalkrishnan N, Xuan F & Yin P Programmable autonomous synthesis of single-stranded DNA. Nat Chem 10, 155–164 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kishi JY et al. SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat Methods 16, 533–544 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen F, Tillberg PW & Boyden ES Expansion microscopy. Science 347, 543–548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nave H, Gebert A & Pabst R Morphology and immunology of the human palatine tonsil. Anat Embryol (Berl) 204, 367–373 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Slijkerman RW et al. The pros and cons of vertebrate animal models for functional and therapeutic research on inherited retinal dystrophies. Prog Retin Eye Res 48, 137–159 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Speel EJ, Hopman AH & Komminoth P Amplification methods to increase the sensitivity of in situ hybridization: play card(s). J Histochem Cytochem 47, 281–288 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Clutter MR, Heffner GC, Krutzik PO, Sachen KL & Nolan GP Tyramide signal amplification for analysis of kinase activity by intracellular flow cytometry. Cytometry A 77, 1020–1031 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zadeh JN et al. NUPACK: Analysis and design of nucleic acid systems. J Comput Chem 32, 170–173 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Uhlen M et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Ku T et al. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat Biotechnol 34, 973–981 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen F et al. Nanoscale imaging of RNA with expansion microscopy. Nat Methods 13, 679–684 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang JB et al. Iterative expansion microscopy. Nat Methods 14, 593–599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Truckenbrodt S et al. X10 expansion microscopy enables 25‐nm resolution on conventional microscopes. EMBO reports 19, e45836 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dani A, Huang B, Bergan J, Dulac C & Zhuang X Superresolution imaging of chemical synapses in the brain. Neuron 68, 843–856 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moffitt JR et al. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc Natl Acad Sci U S A 113, 11046–11051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Regev A et al. The Human Cell Atlas. eLife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snyder MM et al. Mapping the Human Body at Cellular Resolution -- The NIH Common Fund Human BioMolecular Atlas Program. Preprint at https://arxiv.org/abs/1903.07231 (2019). [DOI] [PMC free article] [PubMed]
- 46.Stoeckius M et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takada SE, Evidence EG for an association between CD8 molecules and the T cell receptor complex on cytotoxic T cells. Journal of Immunology 139, 3231–3235 (1987). [PubMed] [Google Scholar]
- 48.Beck M et al. The quantitative proteome of a human cell line. Mol Syst Biol 7, 549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernstein HG et al. Regional and cellular distribution of neural visinin-like protein immunoreactivities (VILIP-1 and VILIP-3) in human brain. J Neurocytol 28, 655–662 (1999). [DOI] [PubMed] [Google Scholar]
- 50.Haverkamp S & Wassle H Immunocytochemical analysis of the mouse retina. J Comp Neurol 424, 1–23 (2000). [PubMed] [Google Scholar]
Methods References
- 51.Pierce MB & Dirks RM A Partition Function Algorithm for Nucleic Acid Secondary Structure Including Pseudoknots. J Comput Chem 24, 1664–1677 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Dirks RM & Pierce NA An algorithm for computing nucleic acid base-pairing probabilities including pseudoknots. J Comput Chem 25, 1295–1304 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Schindelin J et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carpenter AE et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7, R100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allan C et al. OMERO: flexible, model-driven data management for experimental biology. Nat Methods 9, 245–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ronneberger O, Fischer P & Brox T U-Net: Convolutional Networks for Biomedical Image Segmentation. Preprint at https://arxiv.org/abs/1505.04597 (2015).
- 57.Ioffe S & Szegedy C Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. Preprint at https://arxiv.org/abs/1502.03167 (2015).
- 58.He K, Zhang X, Ren S & Sun J Deep Residual Learning for Image Recognition. Preprint at https://arxiv.org/abs/1512.03385 (2015).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and essential custom scripts for image processing will be made available from the corresponding authors P.Y. (py@hms.harvard.edu), S.K.S. (Sinem.Saka@wyss.harvard.edu), and Y.W. (yuwang01@fas.harvard.edu) upon request. The deep learning algorithm and test dataset for automated identification of nuclear contours in tonsil tissues is available on https://github.com/HMS-IDAC/UNet. The MATLAB code for nuclear segmentation is available on: https://github.com/HMS-IDAC/SABERProbMapSegmentation.