Abstract
We present tetramer-associated T-cell receptor sequencing (TetTCR-Seq), a method to link T cell receptor (TCR) sequences to their cognate antigens in single cells at high throughput. Binding is determined using a library of DNA-barcoded antigen tetramers that is rapidly generated by in vitro transcription and translation. We applied TetTCR-Seq to identify patterns in TCR cross-reactivity with cancer neo-antigens and to rapidly isolate neo-antigen-specific TCRs with no cross-reactivity to the wild-type antigen.
The ability to link T cell antigens, peptides bound by the major histocompatibility complex (pMHC), to T cell receptor (TCR) sequences is essential for monitoring and treating immune-related diseases. Fluorescently labeled T cell antigen oligomers, such as pMHC tetramers, are widely used to identify antigen-binding T cells1. However, spectral overlap limits the number of pMHC tetramer species that can be studies in parallel and the extent of cross-reactivity that can be examined1. CyTOF with isotope-labeled pMHC tetramers can interrogate a larger number of antigen species simultaneously, but its destructive nature prevents the linkage of antigens bound to TCR sequences1.
DNA-barcoded pMHC dextramer technology has been used for the analysis of antigen-binding T cell frequencies to samples of more than 1,000 pMHCs for T cells sorted in bulk2. However, with bulk analysis, information on the binding of single or multiple peptides to individual T cells is lost. In addition, antigen-linked TCR sequences cannot be obtained, which is valuable for tracking antigen-binding T cell lineages in disease settings, TCR-based therapeutics development3, and for uncovering patterns in TCR-antigen recognition4. Another limitation is the high-cost and long-duration associated with synthesizing peptides chemically5, which prevents the quick generation of a pMHC library that can be tailored to specific pathogens or neo-antigens in an individual.
To address these challenges, we developed TetTCR-Seq, for the high-throughput pairing of TCR sequences with potentially multiple pMHC species bound on single T cells. First, we constructed a large library of fluorescently labeled, DNA-barcoded (DNA-BC) pMHC tetramers in a rapid and inexpensive manner using in vitro transcription/translation (IVTT) (Fig. 1a). Next, tetramer-stained cells were single-cell sorted and the DNA-BC and TCRαβ genes were amplified by RT-PCR (Fig. 1b). A molecular identifier (MID) was included in the DNA-BC to provide absolute counting of the copy number for each species of tetramers bound to the cell. Finally, nucleotide-based cell barcodes were used to link multiple peptide specificities with their bound TCRαβ sequences (Fig. 1b). DNA-BC pMHC tetramers are compatible with isolation of rare antigen-binding precursor T cells6, making TetTCR-Seq a versatile platform to analyze both clonally expanded and precursor T cells.
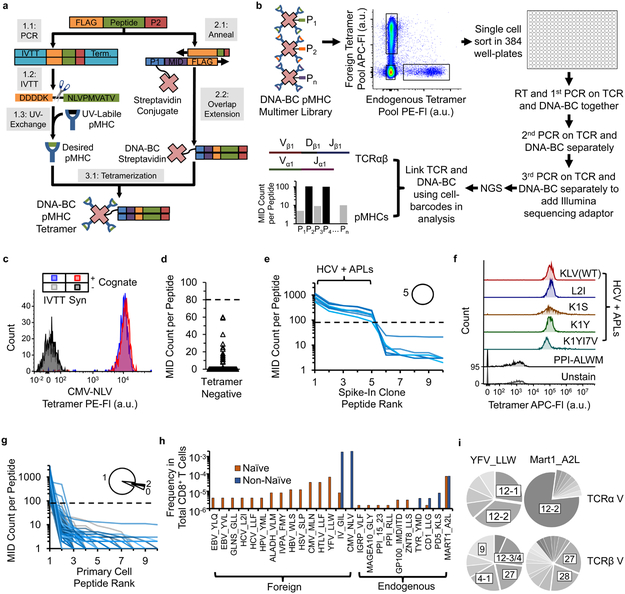
Figure 1: Workflow for generation of DNA-BC pMHC tetramer library and proof-of-concept of using TetTCR-Seq for high-throughput linking of antigen binding to TCR sequences for single T cells.
(a) Workflow for generation of DNA-BC pMHC tetramers. (b) Workflow of TetTCR-Seq. (c) Comparison of staining performance for IVTT and synthetic peptide generated pMHC tetramers on T cell clones. Experiment repeated independently once with similar results. (d) MID counts per peptide detected on single T cells sorted from the Tetramer− fraction in Experiment 1 (768 peptides from 8 cells). Dashed line, MID threshold. (e) Peptide rank curve by MID counts for each of top 10 ranked peptides for single sorted cells from the spike-in clone (8 cells) in Experiment 1. Dashed line is as in (d). Each blue solid line represents the MID counts associated with each of the 96 peptides that can potentially bind on a single cell. Inset, proportion of cells with the indicated number of positively binding peptides. (f) Fluorescent intensity of the HCV-KLV(WT) binding T cell clone, used as spike-in in Experiment 1, stained individually with the indicated pMHC tetramers in a separate experiment. Experiment performed once. (g) Peptide rank curve by MID counts as in (e) for the Tetramer+ primary T cell populations (167 cells) in Experiment 1. Grey solid lines indicate cells with no detected peptides. (h) Calculated frequencies of antigen-binding T cell populations in total CD8+ T cells for peptide with at least 1 detected T cell, separated by phenotype, in Experiment 1. (i) V-gene usage of unique TCR sequences that are specific for YFV_LLW (naïve and non-naïve combined, n = 11 cells for TRAV, n = 15 cells for TRBV) or MART1_A2L (naïve and non-naïve combined, n = 33 cells for TRAV, n = 43 cells for TRBV). P1, P2, and Pn, unique peptide ligands. NGS, next-generation sequencing. Fl, fluorescence intensity. a.u., arbitrary unit. APL, altered peptide ligand.
As constructing large pMHC libraries via UV-mediated peptide exchange using traditionally synthesized peptide is costly with long turnaround times5, we use a set of peptide-encoding oligonucleotides that serve as both the DNA-BCs for identifying antigen specificities and as DNA templates for peptide generation via IVTT (Fig. 1a). The IVTT step only adds a few hours once oligonucleotides are synthesized. This significantly reduces the cost (about 20-fold) and time (2–3 days instead of weeks) compared to synthesizing peptides chemically.
pMHC tetramers generated by our IVTT method have similar performance as the synthetic-peptide counterparts (Fig. 1c and Supplementary Fig. 1 and 2). DNA-BC conjugation did not interfere with staining and has comparable sensitivity to fluorescent readouts (Supplementary Fig. 3 and 4). Six main TetTCR-Seq experiments were performed (Supplementary Fig. 5). We first assessed the ability of TetTCR-Seq to accurately link TCRαβ sequences with pMHC binding from primary CD8+ T cells in human peripheral blood. In Experiment 1, we constructed a 96-peptide library consisting of well documented foreign and endogenous peptides bound to HLA-A2 (Supplementary Table) and isolated dominant pathogen-specific T cells as well as rare precursor antigen-binding T cells from a healthy CMV sero-positive donor (Fig. 1, Supplementary Fig. 6, 7). To test whether TetTCR-Seq can detect cross-reactive peptides, we included a documented HCV wildtype (WT) peptide, HCV-KLV(WT)7, and 4 candidate altered peptide ligands (APL) with 1–2 amino acid (AA) substitutions (Supplementary Table). An established HCV-KLV (WT) T cell clone7 was spiked into the donor’s sample to test its cross-reactive potential.
Bound peptides were classified by their MID counts using two criteria: an MID threshold derived from tetramer negative controls and a ratio of MID counts between the peptides above and below this threshold (Fig. 1d and Supplementary Information). Using this classification scheme, we identified the HCV-KLV(WT) epitope from all spike-in cells sorted (Fig. 1e, Supplementary Fig. 8a). In addition, we discovered that all four APLs were also classified as binders. A separate experiment confirmed the binding of these APLs to the T cell clone (Fig. 1f). These results show that TetTCR-Seq is able to resolve positively-bound peptides in primary T cells and identify up to five cross-reactive peptides per cell.
The majority of primary T cells were classified as binding one peptide (Fig. 1g). This result is expected because the probability of TCR cross-reactivity between similar peptides is higher than disparate ones8, 9. Among the peptides surveyed, we found a high degree of peptide diversity in the foreign-antigen-binding naïve T cells (Fig. 1h). This diversity reduced to two dominant peptides for CMV and influenza in the non-naïve repertoire10 (Fig. 1h). This is expected given the CMV sero-positive status and the high probability of influenza exposure or vaccination for this donor. The majority of cells within the endogenous-antigen-binding population bind MART1-A2L, which corroborates its high documented frequency6, 10 (Fig. 1h). Linked TCR and DNA-BC analysis revealed that TCRα V genes 12–2 and 12–1/12–2 dominate in MART1-A2L and YFV-LLW specific TCRs, respectively (Fig. 1i), which is consistent with other reports11, 12. TetTCR-Seq on a second CMV sero-positive donor (Experiment 2) verified the findings from Experiment 1 (Supplementary Fig. 9). These results highlight the ability of TetTCR-Seq to accurately link pMHC binding with TCR sequences.
Naïve T cells from healthy donors are a useful source of neo-antigen-binding TCRs3. However, most neo-antigens are 1 AA from the WT sequence, meaning that neo-antigen-binding TCRs can potentially cross-react with host cells to cause autoimmunity. Although clinical adverse effects caused by neo-antigen recognizing T cells cross-reacting with endogenous tissue have not been reported, possibly due to the lack of technology development, other forms of cross-reactivity have been reported to cause death in clinical trials13. We next applied TetATCR-Seq to study the extent of cancer antigen cross-reactivity in healthy donor peripheral blood T cells and isolate neo-antigen (Neo)-specific TCRs with no cross-reactivity to wildtype counterpart antigen (WT). In Experiment 3, we surveyed 20 pairs of Neo-WT peptides that bind with high affinity to HLA-A2. pMHC tetramer-based selection of naïve T cells has an inherent risk of selecting T cells reactive to peptides that are not naturally processed. As such, peptides were also chosen based on previous evidence of tumor expression and T cell targeting3, 14-16 (Supplementary Table). Neo and WT pMHC pools were labeled using two separate fluorophores, allowing sorting of three cell populations, Neo+WT−, Neo−WT+, and Neo+WT+ (Fig. 2a and Supplementary Fig. 10).
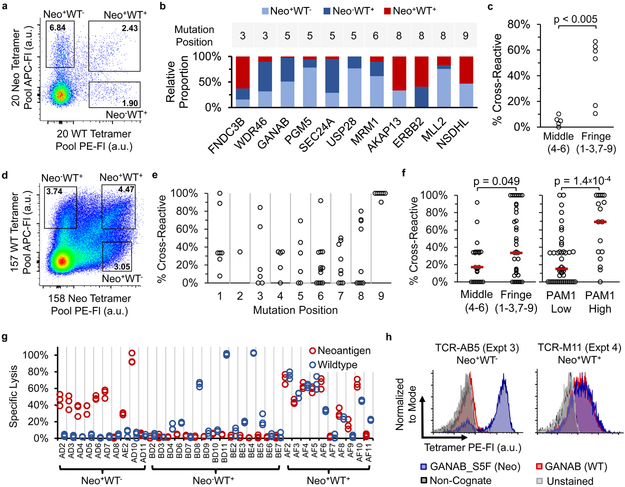
Figure 2: High prevalence of neo-antigen binding T cells that cross-react to WT counterpart peptides and high-throughput isolation of neo-antigen-specific TCRs for multiple specificities in parallel using TetTCR-Seq.
(a-c) Experiment 3, isolation of single Neo and/or WT binding T cells from a healthy donor using a 40 Neo-WT antigen library. (a) DNA-BC pMHC tetramer staining profile of naïve CD8+ T cells from the tetramer pool-enriched fraction. See Supplementary Fig. 10 for gating scheme. (b) Relative proportion of T cells among the three possible antigen binding combinations (Neo+WT−, Neo−WT+, Neo+WT+) for each Neo-WT antigen-pair from Experiment 3. Only antigen-pairs where both peptides were detected in at least one cell and have at least three detected cells in total (149 cells, see Methods) were included. (c) Effect of neoantigen mutation position (indicated in parenthesis) on the proportion of cross reactive T cells from red bars in (b). (5 Neo-WT pairs for middle and 6 for fringe, One-tailed Mann Whitney U-Test). (d-f) Experiment 5 and 6, isolation of Neo and/or WT binding T cells using a 315 Neo-WT antigen library. (d) Staining profile as in (a) for Experiment 5. See Supplementary Fig. 15 for gating scheme. (e) Proportion of cross-reactive T cells for Neo-WT antigen-pairs based on mutation position. Same data filter as (b) is used. (62 Neo-WT pairs from 517 cells). (f) Effect of neoantigen mutation position as in (c) or PAM1 value on the proportion of cross reactive T cells in (e). Red bars denote median. (left-to-right, n = 23, 39, 45, 17 Neo-WT pairs, One-Tailed Mann Whitney U-Test). Alternative analysis using contingency tables are shown in Supplementary Figure 16. (g) LDH cytotoxicity assay on in vitro expanded primary T cell lines sorted using DNA-BC pMHC tetramers as in (a) interacting with T2 cells pulsed with the 20 neo-antigen peptide pool or 20 WT counterpart peptide pool. Each condition was performed in triplicates derived from separate wells of cells. (h) Staining of Jurkat 76 cell line transduced with TCRs from Experiment 3 and 4 with the indicated tetramers. Experiment was performed once. Based on TetTCR-Seq of the original T cells, TCR-AB5 recognized the neo-antigen GANAB_S5F while TCR-M11 recognized both GANAB_S5F and its WT counterpart, GANAB.
T cells with two detected peptide binders accounted for 84% of the Neo+WT+ population, 98% of which belonged to a Neo-WT antigen-pair (Supplementary Fig. 11). Cells in the Neo+WT+ population bound 11 of the 20 Neo-WT antigen-pairs, indicating that Neo-WT cross-reactivity is wide-spread in the precursor T cell repertoire (Fig. 2b). By analyzing the proportion of mono- and cross-reactive T cells from each Neo-WT pair, we observed that neo-antigens with mutations at fringe positions 3, 8, and 9 elicited significantly more cross-reactive T cells than the ones at center positions 4, 5, and 6 (Fig. 2c). This is consistent with observations made by others using alanine substitutions on peptides in a mouse model17. TetTCR-Seq on a separate donor (Experiment 4) showed the same trend (Supplementary Fig. 13), indicating that this property is conserved between donors for the peptides tested.
To test the feasibility of TetTCR-Seq to screen larger libraries, we assembled a 315 Neo-WT antigen-pair library and profiled T cell cross-reactivity in more than 1,000 Tetramer+ CD8+ sorted single T cells from two donors (Experiment 5 and 6, Fig. 2d, and Supplementary Fig. 15,16). ELISA on all 315 pMHC species showed no difference in pMHC UV-exchange efficiency between detected and undetected peptides (Supplementary Fig. 17). Similar to Experiment 3 and 4, neo-antigen mutations in the fringes have elevated percentages of cross-reactive T cells than mutations in the middle (Fig. 2e,f, Supplementary Fig. 16j). Using this larger dataset, we also found that neo-antigen mutations with high PAM1 values, a surrogate for chemical similarity related to evolutionary mutational probability18, have a significantly higher percentage of cross-reactive T cells than those with low PAM1 values (Fig. 2f, ). Thus, in addition to mutation position, WT-binding T cells are more likely to recognize the neo-antigen if the mutated AA is chemically similar to the original. Additional, so far unaccounted variations still exist between peptides, highlighting the necessity for experimental screening against WT cross-reactivity when using neo-antigen based therapy in cancer.
We also assessed the utility of TetTCR-Seq to isolate neo-antigen-specific TCRs with no cross-reactivity to WT. We generated primary cell lines, each derived from 5 sorted cells in the Neo+WT−, Neo−WT+, and Neo+WT+ populations from Experiment 3 and 4. These cells lysed antigen-pulsed target cells in a manner that matched their gating scheme during sorting, independent of the choice of pMHC tetramer fluorophore (Fig. 2g). Further TetTCR-Seq analysis of Neo+WT− and Neo+WT+ T cell lines showed unique TCRs in each cell line targeting a limited range of antigens (Supplementary Fig. 19a and b). Cytotoxicity test confirmed the cross-reactivity of Neo+WT+ cell lines as identified by TetTCR-Seq (Supplementary Fig. 19c). Lastly, the antigen-recognition of Jurkat76 cell lines transduced with TCRs identified from Experiment 3 and 4 matched their original specificities (Fig. 2h, Supplementary Fig. 20). Together, our T cell line and TCR-transduced Jurkat experiments show that TetTCR-Seq is not only capable of identifying cross-reactive TCRs on a large scale but can also identify functionally reactive neo-antigen specific TCRs that are not cross-reactive to WT-peptide in a high-throughput manner, which could be valuable in TCR re-directed adoptive cell transfer therapy3, 19.
In conclusion, we show that TetTCR-Seq can accurately link TCR sequences with multiple antigenic pMHC binders in a high-throughput manner, which can be broadly applied to interrogate antigen-binding T cells in T cell populations, from infection to autoimmune disease and cancer immunotherapy, potentially even for individual patients. With methods emerging for predicting antigenic pMHCs for groups of TCR sequences4, TetTCR-Seq can not only facilitate development in this area but also help to validate informatically predicted antigens. Lastly, TetTCR-Seq can be integrated with single-cell transcriptomics and proteomics to gain further insights into the connections between single T cell phenotype, and TCR sequence and pMHC-binding landscape 20.
Methods
PE/APC-labeled streptavidin conjugation to DNA-linker
Conjugation of the DNA linker to PE and APC-labeled streptavidin was performed following manufacturer’s protocols (Solulink). Excess unconjugated DNA linker was removed by 6 wash steps in a Vivaspin 6 100 kDa protein concentrator (GE Healthcare). Conjugates were concentrated, and then passed through a 0.2 μm centrifugal filter. The molar DNA:protein conjugation ratio was kept between 1:3 to 1:7.
DNA:protein conjugation ratio was determined by absorbance using a 1 mg/ml of PE or APC-labeled streptavidin reference solution. The absorbance of the DNA-streptavidin conjugate was then compared with this standard curve to determine the effective protein concentration of the conjugate. The DNA concentration was determined from the difference in the A260 absorbance between the DNA-streptavidin conjugate and a protein concentration-matched version of the PE/APC streptavidin.
Generation of DNA-barcoded fluorescent streptavidin
Annealing of the peptide-encoding oligonucleotide to the complementary DNA-linker on the DNA-linker PE/APC streptavidin conjugate was done at 55°C for 5 minutes, then cooled to 25°C at −0.1°C/s in the presence of 250 μM dNTP in 1x Cutsmart buffer (NEB). Then, 1 μl of extension mixture consisting of 0.1 μl Cutsmart 10x, and 0.125 μl Klenow Fragment Exo- (5 U/μl, NEB) was added before starting the extension at 37°C for 1 hour. The reaction is stopped by adding EDTA. The final DNA-barcoded fluorescent streptavidin conjugate was stored at 4°C. Corresponds to step 2.1 and 2.2 in Fig. 1a.
In vitro transcription/translation:
Peptide-encoding DNA oligonucleotides were purchased from IDT and Sigma. DNA templates were generated by PCR with 400 μM dNTP, 1.05 μM IVTT_r primer, 1 μM IVTT_f primer, 25 pM DNA oligonucleotide, and 0.0375 U/μl Ex Taq HS DNA Polymerase. The reaction proceeded for 95°C 3 min, then 30 cycles of 95°C 20 s, 52°C 40 s, 72°C 45 s, then 72°C 5 min. The PCR product was diluted with 73.3 μl of water. Corresponds to step 1.1 in Fig. 1a.
20 μl of 1.5x concentrated PURExpress IVTT master mix (NEB) was made, consisting of 10 μl Solution A, 7.5 μl solution B, 0.8 μl of Release Factor 1+2+3 (5 reaction/μl, NEB special order), 0.25 μl Enterokinase (16 U/μl, NEB), 0.25 μl Murine Rnase Inhibitor (40 U/ul, NEB), and 1.2 μl H2O. 1 μl of the diluted PCR product was added to 2 μl of the IVTT master mix on ice and then incubated at 30°C for 4 hours. Corresponds to step 1.2 in Fig. 1a.
pMHC UV exchange and tetramerization:
Biotinylated pMHC containing a UV-labile peptide was directly added to the completed IVTT reaction. pMHC UV exchange and tetramerization follows previously described protocol5, 6 (see Supplementary Information). The UV exchange was performed for 60 minutes on ice, and then incubated at 4°C for at least 12 hours. Confirmation of the quality and concentration of UV-exchanged pMHC monomer was assessed by an ELISA assay as described previously5.
DNA-barcoded fluorescent streptavidin conjugate was then added to its corresponding UV-exchanged pMHC monomer mix at molar ratio of 1:6.7 and incubated at 4°C for 1 hour to produce the final DNA-BC pMHC Tetramer. Corresponds to step 1.3 and 3. 1 in Fig. 1a.
DNA-BC pMHC tetramer pooling
500 μl of staining buffer (PBS, 5 mM EDTA, 2% FBS, 100 ug/ml salmon sperm DNA, 100 μM d-biotin, 0.05% sodium azide) was added to a 100 kDa vivaspin protein concentrator (GE) and incubated for at least 30 minutes. The concentrator was spun at 10,000g and further staining buffer was added until 1 ml of solution ran through the membrane. Immediately prior to cell staining, 0.65 μl of each DNA pMHC tetramer is added to 400 μl of staining buffer, transferred to the concentrator, and then spun at 7,000 g for 10 minutes or longer until the volume reaches ~50 μl. Corresponds to Fig. 1b, left panel.
pMHC tetramer staining and sorting
Human Leukocyte Reduction System (LRS) chambers were obtained from de-identified donors by staff members at We Are Blood with informed consent. The use of LRS chamber from de-identified donors for this study was approved by the Institutional Review Board of the University of Texas at Austin and is compliant with all relevant ethical regulations. Antigen-specific T cell isolation was performed following a previously established protocol6. In brief, CD8+ T cells was isolated from LRS chambers using the RosetteSep™ CD8+ T cell Enrichment Cocktail (STEMCELL) together with Ficoll-paque (GE Healthcare).
Cells were either kept either on ice or at 4°C in refrigerator for the remainder of the experiment. In Experiment 1, 2, 3 and 5, an HCV-KLV(WT) binding clone, pre-stained with BV605-CD8a, was spiked into the main sample. Cells were resuspended into staining buffer containing ~60 nM of each DNA-BC pMHC tetramer and 0.025 mg/ml of BV785-CD8a (RPA-T8) antibody and incubated for 1 hour at 4°C. Cells were washed and then incubated with anti-PE and anti-APC microbeads (Miltenyi). After washing, tetramer-staining cells were enriched using an LS column (Miltenyi). The enriched fraction was eluted off the column and washed into FACS buffer containing 0.05% sodium azide, and stained with AF488-CD3, 7-AAD, BV421-CCR7, BV510-CD45RA, and BV785-CD8a (Biolegend). The tetramer-depleted flowthrough fraction was stained with the same antibody panel. Single cells were sorted using BD FACSAria II into 4 μl lysis buffer following previously published protocol7.
TCR library preparation
Single cell TCR amplification and sequencing was done following published protocol7 with a minor modification. RT was performed on TCR in the same lysis well when DNA-BCs were present. During the 1st PCR amplification, P1 and P2 primer was included in the primer mix at 100 nM final concentration for concurrent amplification of TCR and the DNA-BC (Supplementary Table 10). During subsequent PCRs, TCR and DNA-BC were amplified separately in parallel wells in 384 well-plates. After PCR, multiple cells were pooled, purified by gel electrophoresis and extraction, and then sequenced using Ilumina Mi-seq V2 kit. Sequence reads were analyzed following previous protocol7. Corresponds to Fig. 1b.
DNA-BC library preparation
1 μl of 1st PCR product from the TCR, DNA-BC amplification was combined with 100 nM V1f_rxn2 and V1r_rxn2 primer, and 0.025 U/μl Ex Taq HS (Takara) to 5 μl volume for a 2nd PCR. PCR proceeded at 95ºC 3 minutes, then 10 cycles of 95ºC 20 sec, 55ºC 40sec, and 72ºC 45sec, then 72ºC 5 min. PCR primers include cell barcode sequences to code wells and partial Illumina adaptor as previously described7.
A 3rd PCR was used to add Illumina adaptor using ILLU_f and ILLU_r primers. PCR proceeds at same configuration as the 2nd PCR, but using 5 cycles. Multiple wells are then pooled and purified by gel electrophoresis and gel extraction. Library is sequenced to a depth of at least 6000 reads/cell. Refer to Supplementary Table for primer sequences. Corresponds to Fig. 1b.
DNA-BC sequence processing
Raw reads were filtered based on the constant region of the DNA-BC. Reads were further separated according to cell barcodes. Within each cell barcode, reads with an identical MID sequence were clustered together and a consensus peptide-encoding sequence was built for each cluster. Each cluster represents one MID count.
Clusters were filtered based on the peptide-encoding region to be 25–30 nt in length, and with a Levenshtein distance no greater than 2 from the nearest known DNA-BC sequence. A histogram was then created expressing the percent of total reads belonging to each group of clusters sharing the same read count. Clusters with low read counts, which occur due to sequencing errors, were removed (See Supplementary Fig. 7a-c)21. The clusters were then collected into their corresponding cell and peptide, based on the cell barcode and peptide-encoding DNA sequence, respectively.
Calculation of antigen-binding T cell frequency
The absolute frequency of antigen-binding T cells for peptide ai is calculated as follows:
Total CD8+ T cell count was determined by measuring the fraction of CD8+ T cells in the flow-through using 7-AAD with anti-CD3 and CD8 antibodies, multiplied by the total live cell count of flow-through by a cell counter. In Experiment 2 and 5, counting beads were not added to the sample but the entire enriched fraction was recorded; therefore, we assumed 100% bead recovery in these two experiments. Absolute frequencies could not be calculated for Experiment 6 because counting beads were not added and not all of the enriched fraction was recorded.
Calculation of percent cross-reactive T cells for Experiment 3–6
We calculate the relative proportion of T cells belonging to the Neo+WT+, Neo−WT+, and Neo+WT− antigen-binding cell populations for each Neo-WT antigen-pair using cells with positive antigen detection. We restricted our analysis to cells with the one identified antigen in the Neo−WT+ and Neo+WT− sorted populations and the two identified antigens in the Neo+WT+ sorted population (Supplementary Fig. 11e, 13e, 16i). From this dataset, we performed normalization to account for differences in the frequency and number of cells sorted for the three cell populations. Taking these two normalizations into account, the equation for calculating the relative proportion p of cells binding to peptide a in population b for Experiment 3–4 is:
ai refers to a Neo-WT antigen-pair in the Neo+WT+ population, corresponding WT peptide only in the Neo−WT+ population, and corresponding Neo peptide only in the Neo+WT− population. bj refers to one of the three cell populations Neo+WT−, Neo−WT+, or Neo+WT+. count(ai,bj) refers to the antigen-binding T cell count in cell population bj binding to peptide ai. Relfreq(bj) refers to the percentage of cell population bj taken from the tetramer gating in the tetramer-enriched fraction, which is a measure of the relative cell frequency (Supplementary Fig. 10a). totalsort(bj) is the total number of cells sorted for cell population bj.
The percent cross reactive T cells for any Neo-WT antigen-pair ai is simply p(ai,bNeo+WT+) (same values as red bars in Fig. 2b). While this calculation can be performed for all Neo-WT antigen-pairs, we restricted our analysis to Neo-WT antigen-pairs containing at least 3 cells where both the Neo and WT antigen were detected in at least one cell. PAM1 values for amino acid pairs i and j are calculated by adding the one directional PAM1 values, PAM1ij + PAM1ji, as defined by Wilbur et al18. This was calculated for all Neo-WT antigen pairs (Supplementary Fig. 16k)
An aggregate analysis was performed for experiment 5–6. Since cells are aggregated from these two experiments, we normalized for the cell counts in the three Tetramer+ populations but not the cell frequency because the relative frequency of the three cell populations in both experiments were comparable between one another. The altered equation used for Experiment 5–6 is the following:
T cell lines and functional assay
T cell lines were generated according to previously published protocol6, but using the DNA-BC pMHC tetramer pool. Cells were gated in the same manner as Supplementary Fig. 10 except for the AF488 channel, where CD3-AF488 was replaced by the dump channel CD4,14,16,19,32,56-AF488. 5 cells from the same population (Neo+WT−, Neo−WT+, Neo+WT+) were sorted into each well. Functional status was analyzed 10 – 21 days after re-stimulation.
Functionality was measured and analyzed using the LDH cytotoxicity assay kit (Thermofisher) following manufacturer’s instructions as described previously. For Figure 2g and Supplementary Fig. 18, T2 cells (ATTC) were pulsed with a peptide pool consisting of either the 20 neoantigen peptides (250 μM total, 12.5 μM each peptide) or 20 wildtype peptides (250 μM total, 12.5 μM each peptide). Background cytotoxicity was subtracted by using T2 cells pulsed with HCV-KLV(WT) peptide (250 μM). For supplementary Fig. 19c, T2 cells were pulsed with 12.5 μM of a single peptide or a peptide pool consisting of the 19 indicated neo-antigen or WT peptides at 12.5 μM per peptide. Background cytotoxicity was subtracted by using T2 cells not pulsed with peptide. For each well, 60,000 T cells were incubated with 6,000 peptide-pulsed T2 cells for 4 hours at 37°C. Each condition for each cell line (derived from 5 single sorted cells) was performed in triplicates.
Lentiviral TCR transduction
Lentivirus production and TCR transduction was performed as previously described4 with the following modifications. TCR were synthesized as GenParts (GenScript) and was cloned into pLEX_307 (a gift from David Root via Addgene) under EF-1a promoter. The vector also confers puromycin resistance. All vector sequences were confirmed via Sanger sequencing prior to viral production. 72 hours after transduction, expression of the TCR was analyzed by flow cytometry. Antigen binding of the transduced cells was confirmed by pMHC tetramer and anti-CD3 antibody (Biolegend) staining.
Statistics and Reproducibility
The relevant statistical test, sample size, replicate type, and p values for each figure are found in the figure and/or corresponding figure legend. For functional assays, an LDH value was measured for each of three separate wells of cells, all derived from the same cell lines, that were subjected to the same condition (Fig. 2g and Supplementary Fig. 18, 19c). Representative Experiments 1, 3, and 5 were each repeated once with similar results as described for Experiments 2, 4, and 6, respectively. Refer to the Life Sciences Reporting Summary for additional details on experimental design and reagents.
Supplementary Material
Acknowledgements
We thank Dr. Ben Wendel for helpful discussions and for producing recombinant HLA-A2; Dr. Mark M. Davis and Dr. Huang Huang at Stanford University for helpful discussion of the lentiviral transduction protocol and providing a template TCR construct and HLA-A2 construct; Dr. Wolfgang Uckert at Max Delbruck Center for Molecular Medicine for sharing the Jurkat 76 cell line; Dr. Amy Brock at UT Austin for sharing HEK 293T cell line; Dr. Jizhong Lou at the Institute of Biophysics, Chinese Academy of Sciences, for helping with HCV APL prediction; Patrik Parker, Kushal Patel, and Henry Pan for assistance with initial prototyping; the NIH tetramer center for additional pMHC tetramer reagents. We also thank anonymous blood donors and staff members at We Are Blood for sample collection. This work was supported by NIH Grants R00AG040149 (N.J.), S10OD020072 (N.J.), R33CA225539 (N.J), by NSF CAREER Award 1653866 (N.J.), by the Welch Foundation Grant F1785 (N.J.), by Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation (N.J.), and by National Natural Science Foundation of China major international (regional) joint research project 81220108006 (W.J.) and NSFC-NHMRC joint research grant 81561128016 (W.J). N.J. is a Cancer Prevention and Research Institute of Texas (CPRIT) Scholar and a Damon Runyon-Rachleff Innovator. S.-Q.Z. is a recipient of Thrust 2000—Archie W. Straiton Endowed Graduate Fellowship in Engineering No. 1. A.A.S. is a recipient of the Cockrell School of Engineering fellowship and the Thrust 2000 - Mario E. Ramirez Endowed Graduate Fellowship in Engineering.
Footnotes
Competing Financial Interests
N.J. is a scientific advisor of ImmuDX LLC and Immune Arch Inc. A provisional patent application has been filed by the University of Texas at Austin for the method described here.
Data Availability
All TCR and peptide information is in the Supplementary Tables. Sequencing data can be accessed with the accession number phs001678.v1.p1from dbGaP.
Code Availability
Custom analysis code can be downloaded from GitHub at following link https://github.com/utjianglab/TetTCR.
References
- 1.Newell EW & Davis MM Nat Biotechnol 32, 149–157 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentzen AK et al. Nat Biotech 34, 1037–1045 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Strønen E et al. Science (2016). [Google Scholar]
- 4.Glanville J et al. Nature 547, 94–98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodenko B et al. Nat. Protocols 1, 1120–1132 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Yu W et al. Immunity 42, 929–941 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang SQ et al. Sci Transl Med 8, 341ra377 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birnbaum Michael E. et al. Cell 157, 1073–1087 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bullock TNJ, Mullins DW, Colella TA & Engelhard VH The Journal of Immunology 167, 5824–5831 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Newell EW et al. Nat Biotechnol 31, 623–629 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bovay A et al. Eur J Immunol 48, 258–272 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dietrich P-Y et al. The Journal of Immunology 170, 5103–5109 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Cameron BJ et al. Science Translational Medicine 5, 197ra103–197ra103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen CJ et al. J Clin Invest 125, 3981–3991 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajasagi M et al. Blood 124, 453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carreno BM et al. Science 348, 803–808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson RW et al. Immunity 42, 95–107 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilbur WJ Molecular biology and evolution 2, 434–447 (1985). [DOI] [PubMed] [Google Scholar]
- 19.Dudley ME et al. Science (New York, N.Y.) 298, 850–854 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson VM et al. Nature Biotechnology 35, 936 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Fu GK, Wilhelmy J, Stern D, Fan HC & Fodor SPA Analytical Chemistry 86, 2867–2870 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.