Highlights
-
•
Mutants of P450BM3 can metabolise noscapine.
-
•
Noscapine is N-demethylated with high selectivity.
-
•
The metabolites produced are of interest for drug development.
-
•
The profile of metabolites generated resembles that of mammalian CYP3A4.
Abbreviations: P450, cytochrome P450; P450BM3, CYP102A1 from Bacillus megaterium; IPTG, isopropyl β-D-1-thiogalactopyranoside; NADPH, nicotinamide adenine dinucleotide phosphate; MOPS, 3-(N-morpholino)propanesulfonic acid; DMSO, dimethyl sulfoxide; LC-MS/MS, liquid chromatography-tandem mass spectrometry; QTOF, quadrupole time-of-flight; WT, wild-type
Keywords: Cytochrome P450, N-demethylation, Hydroxylation, Site-directed mutagenesis, Biocatalysis
Abstract
Cytochrome P450 enzymes are a promising tool for the late-stage diversification of lead drug candidates and can provide an alternative route to structural modifications that are difficult to achieve with synthetic chemistry. In this study, a library of P450BM3 mutants was produced using site-directed mutagenesis and the enzymes screened for metabolism of the opium poppy alkaloid noscapine, a drug with anticancer activity. Of the 18 enzyme mutants screened, 12 showed an ability to metabolise noscapine that was not present in the wild-type enzyme. Five noscapine metabolites were detected by LC-MS/MS, with the major metabolite for all mutants being N-demethylated noscapine. The highest observed regioselectivity for N-demethylation was 88%. Two hydroxylated metabolites, a catechol and two C-C cleavage products were also detected. P450-mediated production of hydroxylated and N-demethylated noscapine structures may be useful for the development of noscapine analogues with improved biological activity. The variation in substrate turnover, coupling efficiency and product distribution between the active mutants was considered alongside in silico docking experiments to gain insight into structural and functional effects of the introduced mutations. Selected mutants were identified as targets for further mutagenesis to improve activity and when coupled with an optimised process may provide a route for the preparative-scale production of noscapine metabolites.
1. Introduction
Enzymes are emerging as a useful tool for the structural diversification of pharmaceutical drug candidates [1] and at process scale can potentially provide a safer and more environmentally friendly alternative to synthetic chemical approaches. Late-stage diversification is traditionally achieved by modifying a drug lead, such as a natural product, into a range of structural analogues with altered or additional functional groups that are then screened for improved pharmacological properties. This late-stage functionalisation is often limited to a small proportion of structural positions at which synthetic modification is practicable. For example, compounds containing multiple similar functional groups can require extensive protection/deprotection steps to achieve a chemoselective modification, and many C–H bonds are essentially inaccessible to functionalisation. A complementary strategy is to screen a library of natural or engineered enzymes for their ability to biosynthetically modify a drug compound. This approach takes advantage of the wide range of reaction chemistries and regioselectivity exhibited by enzymes and can offer rapid routes to drug analogues that would otherwise be difficult to produce with synthetic approaches, achieved in a single step under mild aqueous conditions.
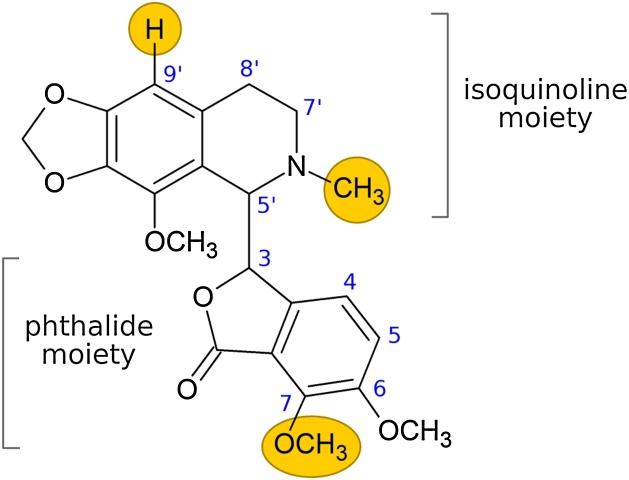
The opium poppy alkaloid noscapine (Fig. 1) has been a target for structural diversification to improve its native anticancer activity [2] and this may benefit from an enzymatic approach. Noscapine interferes with microtubule assembly during tumour cell division [3], although this activity is fairly weak compared to other microtubule-binding chemotherapeutics such as paclitaxel and colchicine. Noscapine does have the advantage, however, of very low toxicity compared to these drugs [4,5], suggesting that structural analogues may have improved anticancer activity and retain a favourable toxicity profile. Most noscapine analogues produced to date carry functional group modifications at the 9′-, 7- and N-positions of the noscapine scaffold (Fig. 1), as these bonds can be selectively targeted using synthetic chemical reactions [2,[6], [7], [8], [9], [10]]. While many of these analogues have been shown to have improved activity compared to the parent drug, none have progressed further than Phase I clinical trials. Investigating structure-activity relationships at alternative positions on the noscapine backbone may therefore provide a route to new classes of analogues with enhanced activity.
Fig. 1.
The structure of noscapine, consisting of phthalide and isoquinoline moieties. The yellow circles highlight positions on the molecule that have been modified by others to produce analogues with enhanced biological activity.
Cytochrome P450 monooxygenase enzymes (P450s) offer a promising alternative to synthetic derivatisation of noscapine and have been used to produce other drug analogues [11]. This approach mimics xenobiotic metabolism in mammals and other higher eukaryotes, where a drug compound is oxidised by microsomal P450s in the liver to produce drug metabolites. These metabolic reactions include hydroxylations, epoxidations or heteroatom dealkylations that may be difficult to achieve with chemical synthetic methods. Mammalian microsomal P450s are known to produce at least nine noscapine metabolites through demethylation, hydroxylation and C-C cleavage mechanisms [[12], [13], [14]] and these compounds may be of interest for the development of noscapine analogues. The scale-up of biosynthetic reactions with microsomal P450s, however, can be difficult due to the inherent instability, insolubility and low activity of these enzymes [15].
An alternative, more practical approach is to use the more stable bacterial fatty acid hydroxylase P450BM3 from Bacillus megaterium, an enzyme that has not yet been investigated for activity on noscapine. This enzyme has several advantages compared to microsomal P450s. P450BM3 is a fusion of a P450 substrate-binding domain and a mammalian-like reductase domain [16] and is therefore ‘self-sufficient’ compared to microsomal P450s, which require at least two discrete enzymes to produce a functional catalyst. P450BM3 is also cytoplasmic, as opposed to the intracellular membrane-bound location of microsomal P450s and can be expressed recombinantly at high levels in bacteria [17]. While the native substrates of P450BM3 are long-chain fatty acids, the wild-type enzyme is capable of metabolising some drug compounds [18]. Its substrate specificity, however, can be significantly broadened through protein mutagenesis. Both directed evolution and rational engineering approaches have been used extensively with P450BM3 to produce mutants capable of synthesising a wide variety of drug metabolites, as well as other industrially important fine chemicals [19,20].
In this study, we have combined P450BM3 mutations known to enhance activity on bulky drug-like substrates to produce a new library of mutant enzymes. The mutant library was then screened for activity to assess the potential utility of P450BM3 mutants in noscapine analogue development and for enzymatic routes of analogue production.
2. Materials and methods
2.1. Chemicals and reagents
Noscapine was provided by Sun Pharmaceutical Industries (Port Fairy, Australia). The N-demethylated noscapine standard (N-nornoscapine) was a kind gift from Peter Scammells at Monash University. Molecular biology enzymes and competent cells were purchased from New England Biolabs (Ipswich, MA, USA). Primers were synthesised by GeneWorks (Thebarton, Australia). Complex growth media components were Oxoid brand (Thermo Fisher Scientific, Scoresby, Australia). Kanamycin, isopropyl β-D-1-thiogalactopyranoside (IPTG), nicotinamide adenine dinucleotide phosphate (NADPH), glucose-6-phosphate and glucose-6-phosphate dehydrogenase were purchased from Sigma-Aldrich (Castle Hill, Australia). Chromatographic separations were performed using deionised water (resistivity 18.2 mΩ; MilliQ) and chromatography-grade methanol (Merck, Bayswater, Australia).
2.2. Cloning and mutagenesis
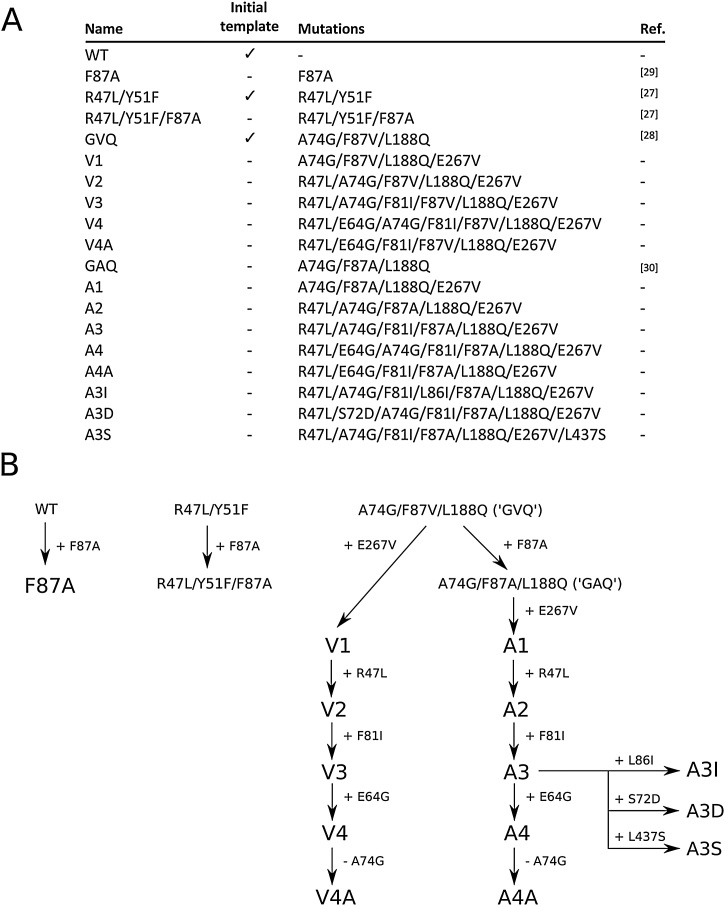
The wild-type (WT) CYP102A1 gene was cloned from Bacillus megaterium ATCC 14581. Cells were cultured in nutrient medium at 30 °C and genomic DNA was extracted with an AxyPrep™ multisource genomic DNA extraction kit (Corning, Corning, NY, USA). The CYP102A1 gene was amplified by PCR using primers that introduced flanking BamHI and XhoI restriction sites (primers BM3_fwd_BamHI and BM3_rev_XhoI; see Supplementary Table S1 for all primer sequences). The amplicon was ligated into the pET28a(+) vector (Merck, Bayswater, Australia) and the plasmid was transformed into chemically competent DH5α E. coli cells for cloning. Correct sequence identity was confirmed by Sanger sequencing (AGRF, Melbourne, Australia) and alignment with the B. megaterium ATCC 14581 sequence in GenBank (accession CP009920.1). Plasmids containing genes for the R47 L/Y51 F and GVQ (A74 G/F87 V/L188Q) variants were kindly provided by Stephen Bell at The University of Adelaide. The mutant genes were amplified by PCR using the primers above and ligated into the pET28a(+) vector. The order of mutant production by sequential mutation and back mutation is shown in Fig. 2B. Mutants F87A, R47 L/Y51 F/F87A and GAQ were created by introducing the F87A mutation into WT, R47 L/Y51 F, and GVQ sequences, respectively, using the Q5 site-directed mutagenesis kit (New England Biolabs) and primer pair F87A_fwd and F87A_rev. The mutant sets V1-V4 and A1-A4 were created by the cumulative addition of E267 V, R47 L, F81I, and E64 G mutations to the GVQ and GAQ templates respectively using the QuikChange multi-site-directed mutagenesis kit (Agilent, Mulgrave, Australia) with single primers for each mutation (Table S1). Variants V4A and A4A were produced by back-mutation of the A74 G mutation in V4 and A4 using the same protocol. The A3I, A3D and A3S mutants were created by the addition of L86I, S72D and L437S mutations independently to the A3 template using the same method. Mutant plasmids were transformed into DH5α F’ Iq cells for cloning. For protein expression, all plasmids were transformed into BL21(DE3) cells to produce glycerol stocks, which were stored at −80 °C.
Fig. 2.
(A) Details of the P450BM3 mutant library used in this study, which consisted of 18 mutants and the wild-type (WT) enzyme. Variants used as starting templates for library construction are indicated in the second column. References are provided for specific combinations of mutations that have been previously described. (B) Mutation workflow used to create the P450BM3 library.
2.3. Protein expression and lysate preparation
In trial expression experiments we noted that expression of the P450BM3 variants was enhanced under microaerobic (low oxygen) conditions, an observation that has been made by others for expression of P450BM3 [17] and mammalian P450 enzymes [21]. A microaerobic protocol was therefore employed for expression as follows. Glycerol stocks were used to inoculate 5 mL of lysogeny broth (LB) supplemented with 30 μg/mL of kanamycin. The cultures were grown overnight at 30 °C and 200 rpm on an orbital shaker and this culture then used to inoculate 100 mL of the same media in a baffled 500 mL flask to give a starting OD600 of ˜0.015. The flasks were shaken at 250 rpm at 37 °C for 2–3 h until the OD600 reached 0.6-0.8. The cells were harvested by centrifugation (3200 x g, 10 °C, 15 min) and the pellets washed twice by resuspension/centrifugation with 10 mL volumes of MOPS minimal medium (as per Neidhardt et al. [22] but with FeSO4 increased to 100 μM) supplemented with 0.2% glucose and 0.5 mM 5-α-aminolevulinic acid. The cells were resuspended in 100 mL of the modified MOPS medium in a 125 mL round-bottomed flask with a filter cap (Corning, NY, USA) to facilitate microaerobic conditions. Prior to induction, the flasks were shaken at 20 °C and 140 rpm for 5–6 h to allow growth to resume. Protein expression was induced with 0.5 mM IPTG and allowed to continue for 12–14 h. The cells were harvested by centrifugation (3200 x g, 4 °C, 15 min), the supernatant was discarded and the pellets were snap frozen in liquid nitrogen, stored at −80 °C and used within two weeks. To prepare cell lysates, the frozen pellets were resuspended in 4 mL of potassium phosphate buffer (50 mM, pH 7.4). The resuspended cells were sonicated on ice (6 mm probe, 30% amplitude, 10 s on, 20 s off for five cycles) using a QSonica Q500 machine (QSonica LLC, Newtown, CT, USA). The lysates were then centrifuged at 18,000 x g at 4 °C for 15 min to remove cell debris.
2.4. P450 quantification
The P450 concentration of the cell lysates was measured using carbon monoxide difference spectroscopy by the method of Omura and Sato [23] with extinction coefficients εΔ450-490 = 91 mM cm−1 and εΔ420-490 = -41 mM cm−1, with simultaneous determination of inactive P420 enzyme using extinction coefficients εΔ450-490 = -11 mM cm−1 and εΔ420-490 = 110 mM cm−1 [24].
2.5. Enzyme activity screening
Activity assays were performed at a 200 μL scale in 96-well polystyrene microtitre plates (Corning). The reaction mixture consisted of potassium phosphate buffer (50 mM, pH 7.4) containing 25 μM noscapine added in DMSO (final DMSO concentration 2%) and sufficient cell lysate to provide a P450 concentration of 500 nM. The reactions were initiated by the addition of 20 μL of an NADPH recycling system providing final concentrations of 0.2 mM NADPH, 1 mM glucose-6-phosphate and 0.5 U/mL glucose-6-phosphate dehydrogenase. For each enzyme variant, a control was included that received an additional 20 μL of buffer instead of the NADPH recycling system. A set of controls using lysate from BL21(DE3) cells containing the empty pET28a(+) vector were also included. Each assay reaction and control were performed in triplicate. The assay plates were pre-incubated at 30 °C for 15 min prior to initiation and the reactions were allowed to proceed at 30 °C for 25 min. To quench the reaction and extract metabolites, a multichannel pipette was used to transfer 150 μL from each well into 300 μL of ice-cold chloroform in a 500 μL deep-well polypropylene 96-well plate (Corning) and the plate was left on ice for 15 min. The plates were centrifuged for 10 min at 4 °C and 3200 x g to separate the phases and 200 μL of the organic phase was transferred to a fresh 96-well plate. Each well was evaporated to dryness under a nitrogen gas stream. The extracted metabolites were reconstituted in 60 μL of 0.1% formic acid and transferred to glass vials for LC–MS/MS analysis.
2.6. LC–MS/MS analysis
Metabolites were analysed by LC–MS/MS at Metabolomics Australia (School of BioSciences, University of Melbourne) using an Agilent 6520 series QTOF mass spectrometer with a dual sprayer ESI source attached to a 1260 Infinity series HPLC system and a G1315C diode array detector. The instrument was operated in positive ion mode using the following conditions: nebuliser pressure 30 psi, gas flow rate 10 L min−1, gas temperature 300 °C, capillary voltage 4000 V, fragmentor 150 V and skimmer 65 V. Metabolites were detected in targeted mode using a collision energy of 20 eV. The targeted list of precursor ion masses and retention times is provided in Table S2. Proposed fragmentations are shown in Fig. S2. Chromatography was performed using a Kinetix® biphenyl column (1.7 μm, 100 Å, 100 x 2.1 mm; Phenomenex, Lane Cove, Australia) maintained at 55 °C and a flow rate of 0.3 mL min−1. The sample injection volume was 20 μL. A binary gradient method was developed where the mobile phase consisted of (A) 0.1% formic acid and 10 mM ammonium formate in water and (B) 0.1% formic acid and 10 mM ammonium formate in methanol. The gradient elution program was as follows: isocratic at 25% solvent B for one minute; 25–50% solvent B gradient over one minute; 50–70% solvent B gradient over nine minutes; stepped to 95% solvent B and held for one minute; then re-equilibration at 25% solvent B for seven minutes. Absorbance was monitored between 190–500 nm and a wavelength of 311 nm was used for relative metabolite quantification, assuming similar extinction coefficients of all detected metabolites at this wavelength. Ultraviolet absorption spectra for all measured metabolites are provided in Fig. S1.
2.7. Measurement of NADPH/noscapine consumption and cofactor coupling efficiency
The coupling of NADPH consumption to product formation was measured for all P450BM3 variants that showed activity in the screening experiments. Assays to determine NADPH consumption were performed in triplicate in 96-well glass-bottom plates (Greiner Bio-One, Frickenhausen, Germany) at a 200 μL scale, containing 200 nM P450 enzyme from cell lysate, 100 μM noscapine added from methanol stock (final MeOH concentration 1%) and approximately 200 μM of NADPH added in a 20 μL volume to initiate the reaction. The exact NADPH concentration was determined by UV spectrophotometry (ε340 = 6.022 mM cm−1) from triplicate control samples containing 180 μL potassium phosphate buffer and 20 μL of NADPH stock. For each enzyme variant, three control samples that received 20 μL buffer instead of NADPH were measured. After initiation of the reactions, the plates were placed into a FLUOstar OPTIMA plate reader (BMG Labtech, Mornington, Australia) and the NADPH concentration was monitored by reading the absorbance at 340 nm. After complete depletion of NADPH in all assay wells, a multichannel pipette was used to transfer 100 μL from each assay or control well into 100 μL of ice-cold acetonitrile in a fresh 96-well plate. The mixtures were left on ice for 15 min and the plates were then centrifuged at 3200 x g for 20 min at 4 °C to remove precipitated protein. A volume of 40 μL of supernatant from each well was then added to 60 μL of 0.5% TFA in glass HPLC vials for analysis.
The product concentrations in these assays were quantified by UHPLC analysis using a method developed on an Agilent 1290 Infinity II system using an Acquity® BEH C18 column (1.7 μm, 130 Å, 100 x 2.1 mm; Waters, Rydalmere, Australia) held at 60 °C. A binary gradient program was used with mobile phase solvents comprised of (A) 0.1% trifluoroacetic acid and (B) methanol at a flow rate of 0.3 mL min−1. A 20 μL injection volume was used with a gradient elution program as follows: isocratic using 20% solvent B for 0.5 min; a gradient of 20–37% solvent B gradient over 1.5 min; isocratic using 37% solvent B for 2.5 min; a gradient of 37–95% solvent B gradient over two minutes; isocratic using 95% solvent B for 0.5 min; then reequilibration using 20% solvent B for seven minutes. The noscapine metabolite peaks were measured at a wavelength of 311 nm and quantified using a noscapine standard curve; the extinction coefficients were assumed to be similar for all measured metabolites at this wavelength (see Fig. S1 for metabolite absorption spectra).
The coupling efficiency was calculated by dividing the total product concentration in the assay samples by the initial NADPH concentration.
2.8. In silico docking
Docking studies were performed using the Schrödinger software package (version 2017.4, Schrödinger, LLC, New York, NY, USA). Default settings were used unless otherwise noted. The crystal structure of the PEG-bound P450BM3 pentamutant R47 L/F81I/F87 V/L188Q/E267 V (PDB ID: 4ZF6 [25]) was used to model the receptor. This variant contains the same mutations as mutant V3 tested in this work with the exception of the A74 G mutation, which is not in the 4ZF6 structure. To examine the effects of the F87A mutation, the Val87 residue in 4ZF6 was changed to Ala87 using the mutagenesis tool in Maestro. The Protein Preparation Wizard was used to prepare the receptor. All water molecules and the bound PEG ligand were removed. The Fe3+ state was selected for the haem group. The noscapine ligand was prepared using LigPrep and docked in a non-protonated state. Docking simulations were performed using the Induced Fit Docking protocol [26]. The grid box centre was selected as the centroid of the ethylene glycol molecule in the active site of the 4ZF6 structure and the box dimensions were set to 22 Å x 22 Å x 22 Å. During docking, the planarity of conjugated pi groups was enhanced. In the initial docking stage, the Leu75 and Val78 residues on the B’-helix were trimmed. The Cys400 residue, which ligates the haem iron, was excluded from side chain refinement. Docking images were generated using PyMOL (version 2.0.7, Schrödinger, LLC).
3. Results & discussion
3.1. Mutant library development
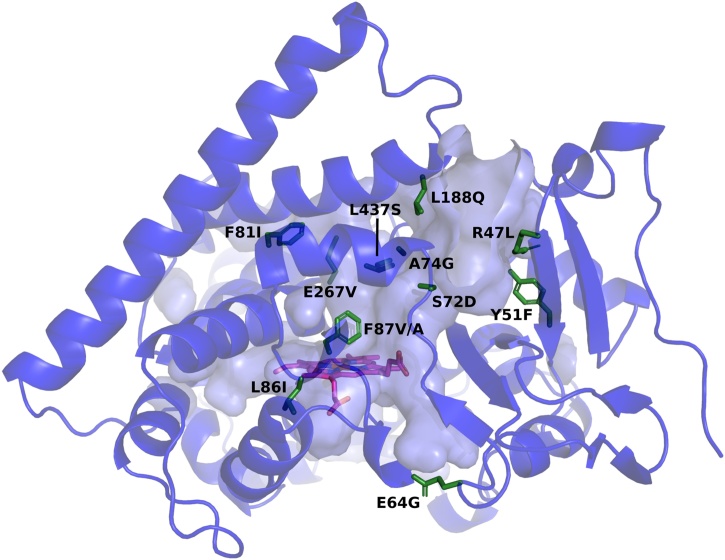
A P450BM3 mutant library, shown in Fig. 2A, was developed with the aim of finding mutants capable of producing useful noscapine metabolites and providing biosynthetic routes to noscapine analogue production. Our approach was to select a range of mutations shown by previous work to enhance P450BM3 oxidation of bulky, drug-like compounds and apply these in various combinations to noscapine, which has not previously been examined as a substrate for P450BM3 mutants. The locations of the twelve mutations at eleven sites are shown in Fig. 3. A library of eighteen mutant variants was produced from combinations of the twelve mutations, together with the wild-type (WT) enzyme. This approach makes use of the significant characterisation of P450BM3 mutations performed over the past two decades and demonstrates how mutations already described in the literature can be applied in new combinations to a selected target of interest. Overall, fourteen of the eighteen mutants used in the study are new to this work and four mutants, including the three starting templates and the F87A single mutant, are specific combinations described previously (Fig. 2A).
Fig. 3.
Structure of wild-type P450BM3 enzyme (PDB: 1BU7 [56]). Wild-type amino acids mutated in this work are coloured green and labelled with the mutations introduced. The haem group is coloured magenta.
The starting templates for the library were the WT enzyme and well-described R47 L/Y51F [27] and GVQ [28] (A74 G/F87 V/L188Q) mutants. The sequence in which mutations were introduced is shown in Fig. 2B. A common Phe87 substitution, F87A [29], was applied to all three templates. The resulting GAQ [30] (A74 G/F87A/L188Q) mutant and original GVQ mutant were then used as templates for sequential introduction of four mutations shown to improve drug-metabolising activity, E267 V, R47 L, F81I and E64G [27,31], leading to two series of four mutants, V1-V4 and A1-A4, which differ only by Phe87 substitution. The Gly74 residue was back-mutated to alanine in two mutants (V4A and A4A) to assess whether the A74 G mutation in the GVQ and GAQ starting templates is beneficial for noscapine metabolism, as this mutation is usually used for hydrocarbon oxidation rather than drug metabolite production. The A3 mutant was also further mutated to give A3I, A3D and A3S to examine the effects of three additional mutations shown by others to enhance activity and/or modify selectivity [31,32]. As most mutations were introduced sequentially, producing a library with a progressively increasing number of mutations, the effect of individual mutations on noscapine metabolism may be specific to the background of mutations into which they were introduced.
3.2. Mutant screening
3.2.1. Mutant activity
The introduced mutations successfully imparted noscapine activity to P450BM3. Twelve enzymes from the panel of eighteen mutants were active on noscapine, producing the metabolites shown in Fig. 4, which were detected by LC–MS/MS. The WT enzyme did not show any turnover of noscapine.
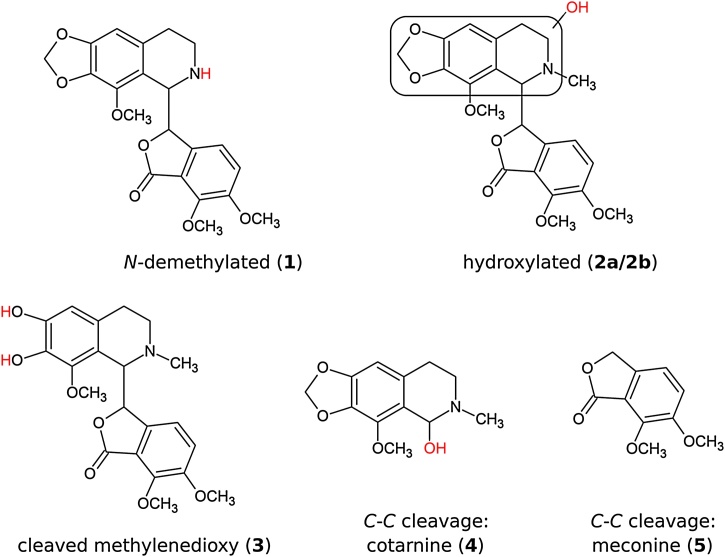
Fig. 4.
Noscapine metabolites detected by LC—MS/MS in the P450BM3 mutant screening experiments. Red text denotes modifications relative to the parent compound. The exact locations of the hydroxyl groups on metabolites 2a and 2b could not be resolved by the LC—MS/MS method used.
The major metabolite produced by all active mutants was N-demethylated noscapine (1), a desirable compound that can only be produced synthetically from noscapine through multiple chemical steps [6,33]. The process of N-demethylation improves the in vitro cytotoxicity against several cancer cell lines compared to unmodified noscapine [7]. This compound is also an essential intermediate in the production of N-substituted noscapine analogues, several of which show further enhanced anticancer activity in vitro [6,7]. The best synthetic route for N-demethylation of noscapine involves the oxidation of noscapine to its N-oxide, which is isolated as a hydrochloric acid salt, followed by treatment with an iron-containing catalyst to yield the final product. The highest reported yield for this process is 74% [6]. A one-pot, biocatalytic N-demethylation, as presented here, may be preferable for larger-scale syntheses, particularly as the highest-yielding synthetic protocols use chlorinated solvents. This enzymatic approach to N-demethylation may also be more broadly applicable to other opiate alkaloids, for which synthetic N-demethylation can be problematic.
Five additional noscapine metabolites, including further compounds of interest, were also detected in the mutant screen. These were: two hydroxylated metabolites (2a/2b), a catechol product resulting from cleavage of the methylenedioxy bridge (3), and two metabolites produced by cleavage of the central C-C bond (cotarnine and meconine, 4 and 5). The MS/MS fragmentation patterns of the hydroxylated metabolites 2a/2b (Fig. S2) indicated that the hydroxyl groups were attached to the isoquinoline moiety of noscapine, although the specific sites could not be resolved by this technique. Small amounts of double-oxidation products resulting from combinations of N-demethylation, hydroxylation and methylenedioxy cleavage were also detected (Table S2; Fig. S2). A UHPLC chromatogram showing noscapine turnover of mutant A3 is provided in Fig. S4.
The hydroxylated metabolites (2a/2b) may be useful precursors for further synthetic modification in the search for noscapine analogues with enhanced anticancer activity, as structure-activity relationships (SARs) have only been studied at the 9′-, 7- and N-positions on the noscapine scaffold. The cleaved methylenedioxy bridge metabolite (3) is potentially less useful as it contains two neighbouring hydroxyl groups, which does not allow for selective functionalisation. Cleavage of the central noscapine C-C bond, producing metabolites 4 and 5, is an undesirable reaction, as it results in loss of anticancer activity [5] and this reaction can be easily achieved by acid hydrolysis [34].
The N-demethylation and methylenedioxy bridge cleavage of drug compounds are common reactions in P450 drug metabolism. While the mechanism of N-demethylation has been debated, evidence now appears to favour the hydrogen atom transfer (HAT) reaction, whereby hydroxylation occurs on the N-methyl group followed by decomposition to the tertiary amine and formaldehyde [35]. Cleavage of the methylenedioxy bridge occurs by hydroxylation of the methylenedioxy carbon, followed by decomposition to the catechol metabolite and formic acid [36]. The C-C cleavage of noscapine, however, is recognised as a rare reaction in mammalian P450 metabolism [37] and is thought to proceed via an amine cation radical [12], as proposed for the peroxidase/H2O2 catalysed cleavage of other isoquinoline alkaloids [38]. It is possible that C-C cleavage of noscapine is mediated by P450BM3 peroxidase-like activity using H2O2 generated by peroxide uncoupling.
The reactivity introduced to P450BM3 by the mutations resembles that observed in mammalian microsomal P450s, with the potential advantage of greater enzyme stability. A recent study of noscapine metabolism by human and mouse microsomal P450s detected nine different metabolites [14], including compounds 1 and 3–5 that were also found here; although only one isoquinoline-hydroxylated metabolite (2a/2b) was reported and the specific hydroxylation site was not determined. Three O-demethylated structures and a phthalide-hydroxylated compound were also described that were not detected here. In this study, all detected sites of metabolism were located on the isoquinoline moiety of noscapine; qualitatively, this metabolite profile most closely resembles that of the human CYP3A4 P450 enzyme, except for the phthalide hydroxylation that was also observed with CYP3A4. Of the mammalian P450 enzymes, P450BM3 shares the greatest sequence homology with CYP3A4 (27%) [18] and similarities in metabolite profiles between CYP3A4 and both WT [18] and mutant P450BM3 [39] have previously been observed, consistent with the findings presented here.
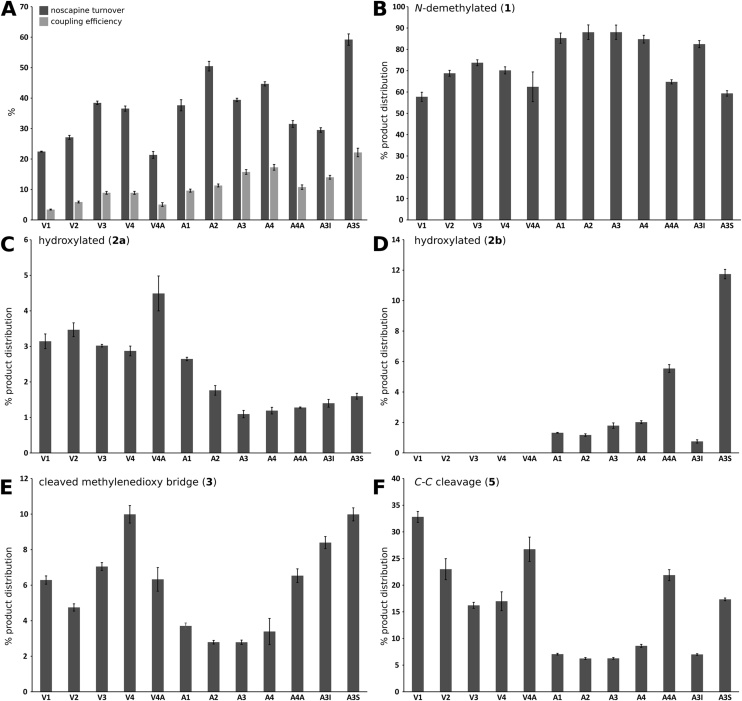
A difference of approximately three-fold was observed in substrate turnover between the most active (A3S) and least active (V4A) variants (59% and 21%, respectively), as shown in Fig. 5, which presents the noscapine turnover (Fig. 5A), coupling efficiency (Fig. 5A) and product distributions for the active mutants in the P450BM3 library (Fig. 5B-F). Numerical results are also tabulated in the Supplementary Material (Table S3).
Fig. 5.
Properties of the screened mutants. (A) Noscapine turnover (dark bars); coupling efficiency (light bars). (B–F) Product distribution percentages for metabolites 1, 2a, 2b, 3, and 5. (F) was generated from data for meconine (5) which shares a similar UV absorption spectrum with the other noscapine metabolites (Fig. S1) and is produced stoichiometrically with 4. The WT enzyme and F87A, R47 L/Y51 F, R47 L/Y51 F/F87A, GVQ, GAQ and A3D did not show detectable activity and are not included. The small amounts of double-oxidation products detected (Table S2; Fig. S2) are not shown. Three replicates were used for each enzyme mutant; error bars are ± 1 SE. Numerical results are tabulated in the Supplementary Material (Table S3).
The dominant reaction catalysed by all active mutants was N-demethylation, with the N-demethylated product 1 forming between 59–88% of the product profile (Fig. 5B), with the highest yield occuring for A2. The C-C cleavage products 4 and 5 collectively varied from 6 to 33% of the product profile (Fig. 5F), while the remaining metabolites 2a, 2b and 3 typically made up less than 10% of the product profile (Fig. 5C, D and E, respectively). The hydroxylated metabolite 2b was produced by only seven mutants. The highest yield of 2a was produced by V3 and the highest yield of 2b by A3S (considering both substrate turnover and selectivity).
The measured coupling efficiencies, defined as the molar ratio of metabolites produced to NADPH turnover, ranged between 3–22% and varied roughly in proportion to noscapine metabolism (Fig. 5A). A high coupling efficency would be desirable for preparative-scale production of noscapine metabolites and should be considered together with the substrate turnover (Fig. 5A) and selectivity for the desired products (Fig. 5B-F). The coupling efficiencies reported here were measured using cell lysates rather than purified enzyme, so some NADPH turnover may be attributable to the non-specific oxidation of lysate components. Nevertheless, the observed coupling efficencies are within the wide range reported in the literature for P450-mediated drug oxidations.
The low aqueous solubility of noscapine measured under the assay conditions applied here (approximately 100 μM in 50 mM potassium phosphate buffer, pH 7.4) precluded the measurement of detailed kinetic and binding data for the enzymes examined. Specifically, a substrate concentration of 100 μM was found to be near or below the km value of the most active mutants, preventing determination of turnover numbers (results not shown).
3.2.2. Effects of specific mutations
Several insights can be gained by examining the effects of the 12 mutations on noscapine metabolism across the 18 enzyme mutants. The L437S mutation produced the variant with the highest noscapine turnover in this study, A3S (Fig. 5A). This mutant showed one of the lowest regioselectivities towards N-demethylation in the screen (59%; Fig. 5B) and produced significantly more of the hydroxylated metabolite 2b than the other mutants (12% of product distribution; Fig. 5D), which may be a property of interest for further development. The A3S mutant also had the highest coupling efficiency of 22%. In a previous study using a similar background of mutations, the L437S mutation enhanced activity on most compounds in a panel of 43 drugs, significantly altering the product distribution in many cases [32]. Leu437 forms part of the β-4 loop that borders the active site cavity (Fig. 5) and the polar side chain introduced by the L437S mutation may stabilise noscapine binding conformations through hydrogen bond interactions.
The E267V mutation was found to be crucial in imparting noscapine-metabolising activity to P450BM3, probably due to conformational changes caused by elimination of the Glu267-Lys440 salt bridge between the I-helix and β4 sheet. The triple-mutants GVQ and GAQ (containing A74G/F87V/L188Q and A74G/F87A/L188Q respectively) did not show any noscapine metabolism under the conditions studied but the addition of E267V to these templates (producing V1 and A1, Fig. 2B) resulted in detectable noscapine metabolism of 21% and 37% respectively (Fig. 4A). Select mutations are known to cause the substrate-free P450BM3 to adopt the structure of the substrate-bound WT enzyme [[40], [41], [42], [43], [44], [45]], a change present in two substrate-free crystal structures containing the E267V mutation (PDB: 4RSN [43] and 5E9Z [44]). It has been suggested that such mutations destabilise the substrate-free WT conformation, lowering the free energy barrier of the transition to the substrate-bound form and causing the transition to occur in the absence of substrate. This produces a “catalytically ready” structure with a significantly expanded substrate range. As two mutations shown to independently cause this rearrangement, D251G [45] and A264E [40], also disrupt interactions of the I-helix, it seems likely that the gain of function conferred by E267V occurs through the same mechanism. This conformational change was also thought to be responsible for the 18-fold increase in diclofenac metabolism when E267V was added to a R47L/Y51F/F87V/I401P template [43].
The mutations F87V and F87A were found to be important for activity, with F87A allowing greater substrate turnover, coupling efficiency, N-demethylation selectivity and the production of an additional hydroxylated metabolite (2b). All active mutants in this study possessed either an F87V or F87A mutation (derived from the GVQ or GAQ template respectively); with five pairs of active mutants that differed only in this respect (F87V/F87A respectively: V1/A1, V2/A2, V3/A3, V4/A4, V4A/A4A). These mutations replace the bulky active-site Phe87 in the WT enzyme (Fig. 3) and have been used by many groups to broaden the substrate range of P450BM3 [20]. Fig. 5A shows that the F87A variants (A1-A4 and A4A) generally had higher noscapine turnover and coupling efficiency than their F87V-containing counterparts (V1-V4 and V4A). There were also notable changes in the product distribution: the F87A mutation was typically associated with improved N-demethylation selectivity, mostly due to a significant decrease in C-C bond cleavage (e.g. 26% decrease between V1 and A1), with the exception of A4A. The F87A mutation also allowed the production of the second hydroxylated metabolite (2b), which was absent in the product profiles of all F87V variants (V1-V4 and V4A; Fig. 5F). The smaller alanine residue in the F87A-containing mutants may allow a closer approach of the noscapine N-methyl group to the haem centre within the enzyme active site, improving the selectivity and coupling efficiency, whilst also allowing hydroxylation of an isoquinoline carbon that is inaccessible when valine is present.
The addition of R47L to V1 and A1, producing V2 and A2 (Fig. 2B), increased both the substrate turnover and coupling efficiency (Fig. 5A). The R47 L mutation is located at the surface of the substrate access channel (Fig. 3). In the wild-type enzyme, the charged Arg47 residue interacts with the carboxyl group of fatty acid substrates [46,47] and its replacement with the neutral leucine residue has been shown to improve activity with hydrophobic substrates, presumably by permitting a higher rate of substrate entry into the access channel [27]. The increase in noscapine turnover seen in Fig. 5A is consistent with this mechanism, however, this change in substrate entry does not explain the change in product distribution seen for V2 (11% increase in N-demethylated noscapine compared to V1; Fig. 5B) or the coupling efficiency improvements seen for both A2 and V2.
The clear benefit of the A74G mutation to noscapine metabolism was shown by the back-mutation of A74G to the wild-type Ala74 in two mutants (producing V4A and A4A; Fig. 2B) and the associated 4–7% drop in coupling efficiency and 10–13% decrease in noscapine turnover (Fig. 5A). The A74 G mutation is at the N-terminus of the short B’-helix, which caps the active site (Fig. 3). The mutation is not included in any available crystal structures and it is unknown if it produces significant structural changes. Removal of the Ala74 side chain may allow increased flexibility of the N-terminal region of the B’-helix through reduced steric interactions [28], improving noscapine accommodation within the active site.
Whilst the F81I mutation has been used to improve P450BM3 activity on several drug substrates [31,32], this had varied effects depending on the F87A/V background applied here. Noscapine turnover increased in combination with F87V (i.e. between mutants V2 and V3) but decreased with F87A (between A2 and A3), yet the coupling efficiency improved in both cases (Fig. 5A). Like the A74G mutation, the F81I mutation reduces hydrophobic interactions of the B’-helix and likely permits greater flexibility of the P450BM3 lid region [25].
The E64G mutation also produced mixed effects, having little significant effect on noscapine turnover and coupling efficiency when combined with the F87V mutation (i.e. between V3 and V4) but increasing turnover and coupling with F87A (between A3 and A4). This mutation is located away from the active site (Fig. 3) and has been suggested to affect dynamic interactions between the haem and reductase domains of P450BM3 [44].
The S72D mutation abolished all noscapine metabolism when introduced into the A3 mutant, producing A3D (Fig. 2B; A3D is excluded from Fig. 5 as it showed no substrate turnover). This is in contrast to a P450BM3 library screen with 43 drugs, where this mutation was shown to generally improve the metabolism of uncharged substrates [32]. Ser72 is located at the junction of the access channel and active site pocket and has been suggested by computational studies to have a hydrogen bond-mediated anchoring effect on some drug substrates during catalysis [48,49]. While noscapine has eight hydrogen bond acceptors and might therefore participate in H-bonding interactions with Ser72, it seems unlikely that eliminating these interactions would alone be responsible for the complete abolition of noscapine metabolism observed here. The introduction of a negatively charged aspartate residue at the entrance to the active site more likely obstructs the bulky and hydrophobic noscapine molecule, preventing noscapine from making a close approach to the haem.
The addition of the L86I mutation to the R47 L/A74 G/F81I/F87A/L188Q/E267 V background of A3, producing A3I, deteriorated both the noscapine turnover and coupling efficiency (Fig. 5A). This is in contrast to the enhanced N-demethylation of 3,4-methylenedioxymethamphetamine (MDMA) [31] observed when L86I was added to a R47 L/F87 V/L188Q/E267 V background. This illustrates the importance of the context in which mutations are introduced.
3.3. In silico docking
Molecular docking studies with selected active P450BM3 mutants and noscapine were performed to rationalise the enzyme activity observed in the screening experiments. An induced-fit docking method was used which allows for movement of side chains in the vicinity of the docked ligand. The PDB structure 4ZF6 [43] (P450BM3 pentamutant R47L/F87V/L188Q/E267V/F81I) was used to model the receptor, as this crystal structure contains similar mutations to the active P450BM3 variants examined in this work, most closely resembling V3 but without the A74G mutation. We repeated the docking after the in silico mutation of Val87 to alanine to test whether the docking poses supported our experimental observations, as the replacement of F87V with F87A caused significant and consistent changes in noscapine turnover, regioselectivity and coupling efficiency (i.e. these measurements were typically higher for A1-A4 compared to V1-V4).
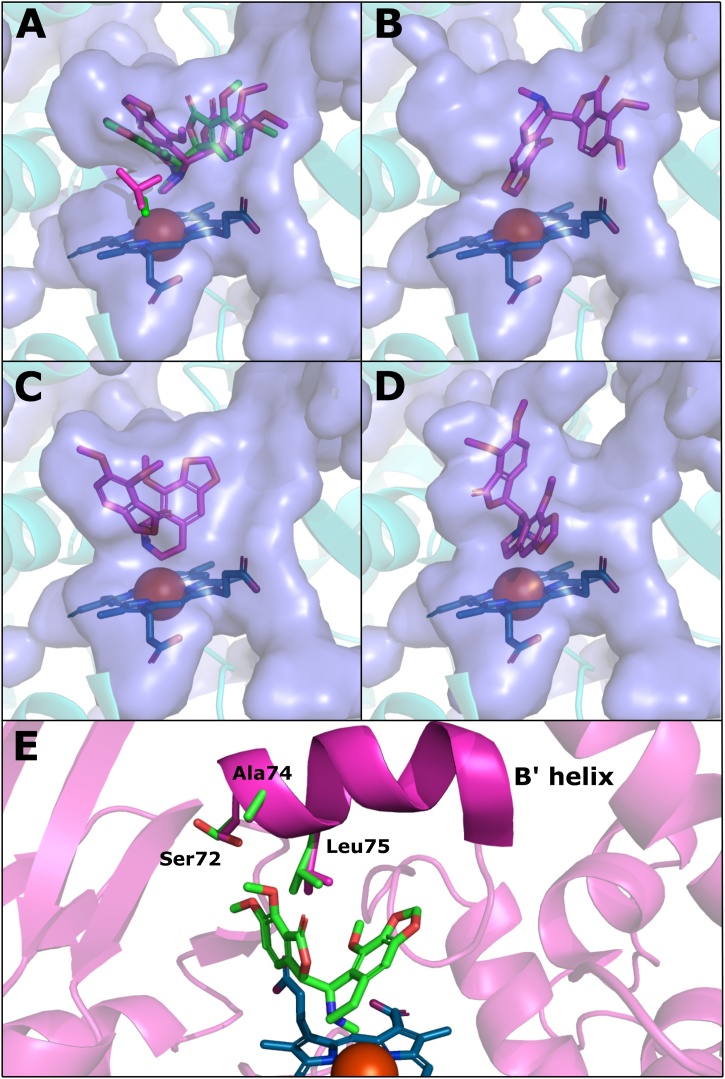
The docking studies produced substrate poses with the noscapine N-methyl group proximal to the haem iron, consistent with N-demethylation (Fig. 6A). Replacement of Val87 with alanine allowed a closer approach of the N-methyl group to the haem (3.1 Å vs 3.4 Å) with the methyl group located more centrally above the iron atom (Fig. 6A), consistent with the improved N-demethylation activity typically seen in the F87A-containing mutants (i.e. A1-A4 versus V1-V4; Fig. 5B). The docking studies also predicted several orientations in which the methylenedioxy carbon was positioned close to the haem iron, an example of which is shown in Fig. 6B. This position is consistent with the production of the cleaved methylenedioxy metabolite (3). The orientation also appeared independent of the amino acid present at position 87. The slightly lower abundance of this metabolite in the product profiles of A1-A4 compared to V1-V4 (Fig. 5E) may be due to improved coupling efficiency of N-demethylation, shifting the product distribution towards N-demethylated noscapine (1).
Fig. 6.
Substrate poses from induced-fit docking studies performed with P450BM3 mutants and noscapine. A P450BM3 pentamutant (R47L/F81I/F87V/L188Q/E267V; PDB: 4ZF6 [25]) was used to model the receptor. Docking runs were performed before and after in silico mutation of F87 V to F87A; the F87A variant is shown unless otherwise noted. The blue surface represents the internal cavity of the active site. The haem group is represented as blue sticks with the iron atom shown as a brown sphere. (A) N-demethylation poses for the F87 V (purple) and F87A (green) variants. The Ala87 and Val87 sidechains are shown in corresponding colours near the haem group. (B) One of the methylenedioxy bridge cleavage poses found. (C) Pose consistent with hydroxylation of the 7′ carbon. (D) Pose consistent with hydroxylation of the 8′ carbon. (E) N-demethylation pose showing greater detail of the Ser72, Ala74 and Leu75 sidechains. Sidechains shown in purple were shifted by the induced-fit protocol from the original positions shown in green. All docking poses had GlideScores between -8.5 and -6.5 kcal/mol.
The two hydroxylated metabolites (2a/2b) observed in the screening experiments were determined to be hydroxylated on the isoquinoline moiety of noscapine although the locations of the hydroxy groups could not be further resolved by LC—MS/MS. In the docking studies, two poses were produced with CH groups of the isoquinoline moiety directed towards the haem: one with the carbon alpha to the nitrogen at position 7′ being closest to the iron (2.9 Å; Fig. 6C) and the other with the β-carbon at the 8′ position closest (3.3 Å, Fig. 6D). These poses are consistent with displacement of the proximal water ligand and initiation of the catalytic cycle, however the actual site of substrate oxidation depends on interactions with the ferryl oxygen of the reactive haem intermediate Compound I, which was not modelled in this work. The docking studies performed here may therefore be of limited use for the prediction of substrate oxidation sites. Other potential hydroxylated products include an N-oxide produced from an orientation similar to N-demethylation, as N-oxygenation has been observed alongside N-demethylation in the metabolism of several tertiary amine drugs by microsomal P450s [50,51], an aromatic hydroxylation at the 9′ position, or a stereoisomer pair at the 7′ or 8′ position.
The docking studies did not find any poses that would be consistent with C-C bond cleavage (i.e. oxidation of the 5′ carbon producing metabolites 2 and 3). This may be because the substrate orientation leading to C-C cleavage fits poorly within the active site or because these metabolites are produced through peroxide uncoupling, as discussed above.
The A74 G mutation, which is not present in the 4ZF6 structure used for docking simulations, was shown by the enzyme screening experiments to significantly improve coupling efficiency and noscapine turnover (mutants V4A vs V4 and A4A vs A4; Fig. 5A). The induced-fit docking algorithm shifted several side chains in the region of Ala74 on the B’-helix, most notably Ser72 and Leu75, to accommodate poses compatible with N-demethylation of noscapine (Fig. 6E), suggesting that a close interaction between noscapine and this region of the active site occurs during substrate binding. Increased flexibility of this ‘cap’ region of the active site likely explains the improvements in turnover and coupling efficiency conferred by the A74 G mutation, as well as the increase in coupling efficiency produced by the F81I mutation (mutants V3 vs V2 and A3 vs A2; Fig. 5A). The close contact between noscapine and this area of the B’-helix can also explain the observation that replacing Ser72 with the larger, charged aspartate residue (S72D) abolished all noscapine activity in the A3 to A3D mutation (A3D is therefore excluded from Fig. 5), as these interactions would likely be unable to occur.
The induced-fit docking studies highlight further mutations that may prove beneficial to P450BM3 activity. The orientation of Leu75 in the N-demethylation pose in Fig. 6E suggests that this residue, positioned between the isoquinoline and phthalide moieties of noscapine, may be a bottleneck during substrate binding. Mutation of Leu75 to a smaller residue, such as alanine, has been shown to improve activity on several drug-like compounds [52,53]; these mutations were not tested here and may improve noscapine activity. Recent molecular dynamics simulations suggest, however, that hydrophobic interactions between Leu75 and some drug substrates are important contributors to binding stability [54], making the interactions between this residue and noscapine difficult to predict.
3.4. Significant metabolites and mutants
In the context of noscapine analogue research, the most interesting metabolites produced in the mutant screen were N-demethylated noscapine (1) and the two hydroxylated metabolites (2a, 2b). The P450BM3 variants with the highest regioselectivity for N-demethylation were A2 and A3 which both produced N-demethylated noscapine with a selectivity of 88% (Fig. 5B). While A3 showed lower noscapine turnover than A2 (39% vs 50%), it was more efficient in terms of cofactor usage (16% vs 11%; Fig. 5A) and is therefore the best candidate for producing N-demethylated noscapine at a preparative scale after further process optimization.
Hydroxylation at either the 7′ or 8′ position of noscapine, as suggested by the docking simulations, adds a new chiral centre to the noscapine scaffold which may be useful in developing more complex analogues that cannot easily be achieved by chemical synthesis. While the largest selectivity for a hydroxylated metabolite was only 12%, observed for metabolite 2b by mutant A3S (Fig. 5D), this mutant had the highest noscapine turnover and one of the highest coupling efficiencies observed (59% and 19%, respectively; Fig. 5A). Whilst the library screened here was relatively small, containing eighteen P450BM3 mutants, it illustrates the efficiency of applying previously described mutations to a new drug target. The introduction of further rational mutations, informed by the results presented here, may be used to further shift the enzyme selectivity towards 2a/2b or other potential metabolites of interest.
The development and scale-up of the biotransformation presented here for the production of 1 or 2a/2b could employ a number of strategies to further optimise yield and reduce operating costs, which would be necessary for the future application of these mutants to preparative-scale N-demethylation. Cofactor recycling or a whole-cell biotransformation approach could be employed to reduce or eliminate the need for addition of NADPH, which is expensive above the small laboratory scale applied here. A substrate supply strategy [55], such as a two-liquid-phase system, could also be used to address the poor aqueous solubility of noscapine and improve product titres.
3.5. Conclusions
The high regioselectivity for N-demethylation exhibited by the P450BM3 mutant A3 developed here provides proof of concept for a potential alternative biocatalytic approach to the chemical synthesis of N-demethylated noscapine (1), a precursor for producing noscapine analogues with enhanced anticancer activity. Two hydroxylated metabolites were observed (2a/2b), with the highest production occurring for mutants V4A (2a) and A3S (2b). These metabolites may have chiral hydroxyl groups at previously uncharacterised positions on the noscapine scaffold and may be useful for the production of new classes of noscapine analogues. The study utilised a small mutant library constructed by combining mutations known to enhance P450BM3 activity on bulky, drug-like substrates, an approach that applies valuable information present in the literature to efficiently produce structural modifications of a desired drug target. The effects of the mutations on P450BM3 activity, together with docking simulations, provide functional insights into P450BM3-mediated drug metabolism. A conformational transition of substrate-free P450BM3 to that of the wild-type, substrate-bound structure, induced by a salt-bridge disruption, appeared critical to metabolism of the non-natural noscapine substrate, as has been observed for other mutant/drug combinations. Noscapine turnover and coupling efficiency also depended on: (1) residue size at position 87, and (2) the flexibility of the active site cap region. The metabolite profile most closely resembled that of the human microsomal P450 CYP3A4 and these mutants could be used as a practical alternative to produce noscapine metabolites for drug metabolism studies. The mutant enzymes presented here also have promise for use in a whole-cell biotransformation process, which once further optimised may allow for the efficient scale-up of metabolite production.
Author contributions
LR performed the experiments. AL assisted with metabolite analysis. DC assisted with modelling work. AL, DC, AJ, TB, GS and SG helped with experimental design and provided technical advice. LR and SG wrote the manuscript with input from all authors.
Funding
This work was supported by Sun Pharmaceutical Industries Ltd and The University of Melbourne. LR was supported by an Australian Government Research Training Program Scholarship.
Declaration of Competing Interest
None.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.btre.2019.e00372.
Contributor Information
Luke Richards, Email: luke.richards@unimelb.edu.au.
Adrian Lutz, Email: adrian.lutz@unimelb.edu.au.
David K. Chalmers, Email: david.chalmers@monash.edu.
Ailsa Jarrold, Email: ailsa.jarrold@sunpharma.com.
Tim Bowser, Email: tim@bowsermail.com.
Geoffrey W. Stevens, Email: gstevens@unimelb.edu.au.
Sally L. Gras, Email: sgras@unimelb.edu.au.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Devine P.N., Howard R.M., Kumar R., Thompson M.P., Truppo M.D., Turner N.J. Extending the application of biocatalysis to meet the challenges of drug development. Nat. Rev. Chem. 2018;2(12):409–421. [Google Scholar]
- 2.DeBono A., Capuano B., Scammells P.J. Progress toward the development of noscapine and derivatives as anticancer agents. J. Med. Chem. 2015;58(15):5699–5727. doi: 10.1021/jm501180v. [DOI] [PubMed] [Google Scholar]
- 3.Ye K.Q., Ke Y., Keshava N., Shanks J., Kapp J.A., Tekmal R.R., Petros J., Joshi H.C. Opium alkaloid noscapine is an antitumor agent that arrests metaphase and induces apoptosis in dividing cells. Proc. Natl. Acad. Sci. U.S.A. 1998;95(4):1601–1606. doi: 10.1073/pnas.95.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winter C.A., Flataker L. Toxicity studies on noscapine. Toxicol. Appl. Pharmacol. 1961;3(1):96–106. doi: 10.1016/0041-008x(61)90013-8. [DOI] [PubMed] [Google Scholar]
- 5.Ke Y., Ye K., Grossniklaus H.E., Archer D.R., Joshi H.C., Kapp J.A. Noscapine inhibits tumor growth with little toxicity to normal tissues or inhibition of immune responses. Cancer Immunol. Immunother. 2000;49(4-5):217–225. doi: 10.1007/s002620000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeBono A.J., Xie J.H., Ventura S., Pouton C.W., Capuano B., Scammells P.J. Synthesis and biological evaluation of N-substituted noscapine analogues. ChemMedChem. 2012;7(12):2122–2133. doi: 10.1002/cmdc.201200365. [DOI] [PubMed] [Google Scholar]
- 7.Manchukonda N.K., Naik P.K., Santoshi S., Lopus M., Joseph S., Sridhar B., Kantevari S. Rational design, synthesis, and biological evaluation of third generation alpha-noscapine analogues as potent tubulin binding anti-cancer agents. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0077970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aneja R., Vangapandu S.N., Lopus M., Chandra R., Panda D., Joshi H.C. Development of a novel nitro-derivative of noscapine for the potential treatment of drug-resistant ovarian cancer and T-cell lymphoma. Mol. Pharmacol. 2006;69(6):1801–1809. doi: 10.1124/mol.105.021899. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J., Gupta K., Aggarwal S., Aneja R., Chandra R., Panda D., Joshi H.C. Brominated derivatives of noscapine are potent microtubule-interfering agents that perturb mitosis and inhibit cell proliferation. Mol. Pharmacol. 2003;63(4):799–807. doi: 10.1124/mol.63.4.799. [DOI] [PubMed] [Google Scholar]
- 10.Anderson J.T., Ting A.E., Boozer S., Brunden K.R., Crumrine C., Danzig J., Dent T., Faga L., Harrington J.J., Hodnick W.F., Murphy S.M., Pawlowski G., Perry R., Raber A., Rundlett S.E., Stricker-Krongrad A., Wang J., Bennani Y.L. Identification of novel and improved antimitotic agents derived from noscapine. J. Med. Chem. 2005;48(23):7096–7098. doi: 10.1021/jm050674q. [DOI] [PubMed] [Google Scholar]
- 11.Fessner N.D. P450 Monooxygenases Enable Rapid Late-Stage Diversification of Natural Products via C−H Bond Activation. ChemCatChem. 2019 doi: 10.1002/cctc.201801829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsunoda N., Yoshimura H. Metabolic fate of noscapine. II. Isolation and identification of novel metabolites produced by C-C bond cleavage. Xenobiotica. 1979;9(3):181–187. doi: 10.3109/00498257909038719. [DOI] [PubMed] [Google Scholar]
- 13.Tsunoda N., Yoshimura H. Metabolic fate of noscapine.; III. Further studies on identification and determination of the metabolites. Xenobiotica. 2008;11(1):23–32. doi: 10.3109/00498258109045268. [DOI] [PubMed] [Google Scholar]
- 14.Fang Z.Z., Krausz K.W., Li F., Cheng J., Tanaka N., Gonzalez F.J. Metabolic map and bioactivation of the anti-tumour drug noscapine. Br. J. Pharmacol. 2012;167(6):1271–1286. doi: 10.1111/j.1476-5381.2012.02067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Reilly E., Kohler V., Flitsch S.L., Turner N.J. Cytochromes P450 as useful biocatalysts: addressing the limitations. Chem. Commun. (Camb.) 2011;47(9):2490–2501. doi: 10.1039/c0cc03165h. [DOI] [PubMed] [Google Scholar]
- 16.Narhi L.O., Fulco A.J. Characterization of a catalytically self-sufficient 119,000-dalton cytochrome P-450 monooxygenase induced by barbiturates in Bacillus megaterium. J. Biol. Chem. 1986;261(16):7160–7169. [PubMed] [Google Scholar]
- 17.Pflug S., Richter S.M., Urlacher V.B. Development of a fed-batch process for the production of the cytochrome P450 monooxygenase CYP102A1 from Bacillus megaterium in E. Coli. J. Biotechnol. 2007;129(3):481–488. doi: 10.1016/j.jbiotec.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 18.Nardo G., Fantuzzi A., Sideri A., Panicco P., Sassone C., Giunta C., Gilardi G. Wild-type CYP102A1 as a biocatalyst: turnover of drugs usually metabolised by human liver enzymes. JBIC J. Biol. Inorg. Chem. 2007;12(3):313–323. doi: 10.1007/s00775-006-0188-4. [DOI] [PubMed] [Google Scholar]
- 19.Di Nardo G., Gilardi G. Optimization of the bacterial cytochrome P450 BM3 system for the production of human drug metabolites. Int. J. Mol. Sci. 2012;13(12):15901–15924. doi: 10.3390/ijms131215901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitehouse C.J., Bell S.G., Wong L.L. P450(BM3) (CYP102A1): connecting the dots. Chem. Soc. Rev. 2012;41(3):1218–1260. doi: 10.1039/c1cs15192d. [DOI] [PubMed] [Google Scholar]
- 21.Vail R.B., Homann M.J., Hanna I., Zaks A. Preparative synthesis of drug metabolites using human cytochrome P450s 3A4, 2C9 and 1A2 with NADPH-P450 reductase expressed in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2005;32(2):67–74. doi: 10.1007/s10295-004-0202-1. [DOI] [PubMed] [Google Scholar]
- 22.Neidhardt F.C., Bloch P.L., Smith D.F. Culture medium for enterobacteria. J. Bacteriol. 1974;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Omura T., Sato R. The Carbon Monoxide-Binding Pigment of Liver Microsomes. I. Evidence for Its Hemoprotein Nature. J. Biol. Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 24.Imai Y., Sato R. Conversion of P-450 to P-420 by neutral salts and some other reagents. Eur. J. Biochem. 1967;1(4) doi: 10.1007/978-3-662-25813-2_57. 419-&. [DOI] [PubMed] [Google Scholar]
- 25.Geronimo I., Denning C.A., Rogers W.E., Othman T., Huxford T., Heidary D.K., Glazer E.C., Payne C.M. Effect of mutation and substrate binding on the stability of cytochrome P450BM3 variants. Biochemistry. 2016;55(25):3594–3606. doi: 10.1021/acs.biochem.6b00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherman W., Day T., Jacobson M.P., Friesner R.A., Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006;49(2):534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 27.Carmichael A.B., Wong L.L. Protein engineering of Bacillus megaterium CYP102. The oxidation of polycyclic aromatic hydrocarbons. Eur. J. Biochem. 2001;268(10):3117–3125. doi: 10.1046/j.1432-1327.2001.02212.x. [DOI] [PubMed] [Google Scholar]
- 28.Li Q.S., Schwaneberg U., Fischer M., Schmitt J., Pleiss J., Lutz-Wahl S., Schmid R.D. Rational evolution of a medium chain-specific cytochrome P-450 BM-3 variant. Biochim. Biophys. Acta. 2001;1545(1-2):114–121. doi: 10.1016/s0167-4838(00)00268-5. [DOI] [PubMed] [Google Scholar]
- 29.Oliver C.F., Modi S., Sutcliffe M.J., Primrose W.U., Lian L.Y., Roberts G.C. A single mutation in cytochrome P450 BM3 changes substrate orientation in a catalytic intermediate and the regiospecificity of hydroxylation. Biochemistry. 1997;36(7):1567–1572. doi: 10.1021/bi962826c. [DOI] [PubMed] [Google Scholar]
- 30.Budde M. 2007. Biocatalysis with Cytochrome P450 Monooxygenases: Towards Selective Oxidation of Terpenes and Fatty Acids. [Google Scholar]
- 31.van Vugt-Lussenburg B.M., Stjernschantz E., Lastdrager J., Oostenbrink C., Vermeulen N.P., Commandeur J.N. Identification of critical residues in novel drug metabolizing mutants of cytochrome P450 BM3 using random mutagenesis. J. Med. Chem. 2007;50(3):455–461. doi: 10.1021/jm0609061. [DOI] [PubMed] [Google Scholar]
- 32.Reinen J., van Leeuwen J.S., Li Y., Sun L., Grootenhuis P.D., Decker C.J., Saunders J., Vermeulen N.P., Commandeur J.N. Efficient screening of cytochrome P450 BM3 mutants for their metabolic activity and diversity toward a wide set of drug-like molecules in chemical space. Drug Metab. Dispos. 2011;39(9):1568–1576. doi: 10.1124/dmd.111.039461. [DOI] [PubMed] [Google Scholar]
- 33.Aggarwal S., Ghosh N.N., Aneja R., Joshi H., Chandra R. A convenient synthesis of aryl-substituted N-Carbamoyl/N-Thiocarbamoyl narcotine and related compounds. Helv. Chim. Acta. 2002;85(8):2458–2462. doi: 10.1002/rcm.661. [DOI] [PubMed] [Google Scholar]
- 34.Ashour A., Hegazy M.A., Moustafa A.A., Kelani K.O., Fattah L.E. Validated stability-indicating TLC method for the determination of noscapine. Drug Test. Anal. 2009;1(7):327–338. doi: 10.1002/dta.46. [DOI] [PubMed] [Google Scholar]
- 35.Li D., Wang Y., Han K. Recent density functional theory model calculations of drug metabolism by cytochrome P450. Coord. Chem. Rev. 2012;256(11-12):1137–1150. [Google Scholar]
- 36.Fukuto J.M., Kumagai Y., Cho A.K. Determination of the mechanism of demethylenation of (methylenedioxy) phenyl compounds by cytochrome P450 using deuterium isotope effects. J. Med. Chem. 1991;34(9):2871–2876. doi: 10.1021/jm00113a028. [DOI] [PubMed] [Google Scholar]
- 37.Bolleddula J., Chowdhury S.K. Carbon-carbon bond cleavage and formation reactions in drug metabolism and the role of metabolic enzymes. Drug Metab. Rev. 2015;47(4):534–557. doi: 10.3109/03602532.2015.1086781. [DOI] [PubMed] [Google Scholar]
- 38.Inubushi Y., Aoyagi Y., Matsuo M. The enzymatic oxidation of isoquinoline alkaloids. Tetrahedron Lett. 1969;10(28):2363–2366. doi: 10.1016/s0040-4039(01)88165-x. [DOI] [PubMed] [Google Scholar]
- 39.van Vugt-Lussenburg B.M., Damsten M.C., Maasdijk D.M., Vermeulen N.P., Commandeur J.N. Heterotropic and homotropic cooperativity by a drug-metabolising mutant of cytochrome P450 BM3. Biochem. Biophys. Res. Commun. 2006;346(3):810–818. doi: 10.1016/j.bbrc.2006.05.179. [DOI] [PubMed] [Google Scholar]
- 40.Joyce M.G., Girvan H.M., Munro A.W., Leys D. A single mutation in cytochrome P450 BM3 induces the conformational rearrangement seen upon substrate binding in the wild-type enzyme. J. Biol. Chem. 2004;279(22):23287–23293. doi: 10.1074/jbc.M401717200. [DOI] [PubMed] [Google Scholar]
- 41.Butler C.F., Peet C., Mason A.E., Voice M.W., Leys D., Munro A.W. Key mutations alter the cytochrome P450 BM3 conformational landscape and remove inherent substrate bias. J. Biol. Chem. 2013;288(35):25387–25399. doi: 10.1074/jbc.M113.479717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitehouse C.J., Yang W., Yorke J.A., Tufton H.G., Ogilvie L.C., Bell S.G., Zhou W., Bartlam M., Rao Z., Wong L.L. Structure, electronic properties and catalytic behaviour of an activity-enhancing CYP102A1 (P450(BM3)) variant. Dalton Trans. 2011;40(40):10383–10396. doi: 10.1039/c1dt10098j. [DOI] [PubMed] [Google Scholar]
- 43.Ren X., Yorke J.A., Taylor E., Zhang T., Zhou W., Wong L.L. Drug oxidation by cytochrome P450BM3 : metabolite synthesis and discovering new P450 reaction types. Chemistry. 2015;21(42):15039–15047. doi: 10.1002/chem.201502020. [DOI] [PubMed] [Google Scholar]
- 44.Capoferri L., Leth R., ter Haar E., Mohanty A.K., Grootenhuis P.D., Vottero E., Commandeur J.N., Vermeulen N.P., Jorgensen F.S., Olsen L., Geerke D.P. Insights into regioselective metabolism of mefenamic acid by cytochrome P450 BM3 mutants through crystallography, docking, molecular dynamics, and free energy calculations. Proteins. 2016;84(3):383–396. doi: 10.1002/prot.24985. [DOI] [PubMed] [Google Scholar]
- 45.Di Nardo G., Dell’Angelo V., Catucci G., Sadeghi S.J., Gilardi G. Subtle structural changes in the Asp251Gly/Gln307His P450 BM3 mutant responsible for new activity toward diclofenac, tolbutamide and ibuprofen. Arch. Biochem. Biophys. 2016;602:106–115. doi: 10.1016/j.abb.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Li H., Poulos T.L. The structure of the cytochrome p450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid. Nat. Struct. Biol. 1997;4(2):140–146. doi: 10.1038/nsb0297-140. [DOI] [PubMed] [Google Scholar]
- 47.Noble M.A., Miles C.S., Chapman S.K., Lysek D.A., MacKay A.C., Reid G.A., Hanzlik R.P., Munro A.W. Roles of key active-site residues in flavocytochrome P450 BM3. Biochem. J. 1999;339(Pt 2):371–379. [PMC free article] [PubMed] [Google Scholar]
- 48.Kim K.H., Kang J.Y., Kim D.H., Park S.H., Park S.H., Kim D., Park K.D., Lee Y.J., Jung H.C., Pan J.G., Ahn T., Yun C.H. Generation of human chiral metabolites of simvastatin and lovastatin by bacterial CYP102A1 mutants. Drug Metab. Dispos. 2011;39(1):140–150. doi: 10.1124/dmd.110.036392. [DOI] [PubMed] [Google Scholar]
- 49.Venkataraman H., Verkade-Vreeker M.C., Capoferri L., Geerke D.P., Vermeulen N.P., Commandeur J.N. Application of engineered cytochrome P450 mutants as biocatalysts for the synthesis of benzylic and aromatic metabolites of fenamic acid NSAIDs. Bioorg. Med. Chem. 2014;22(20):5613–5620. doi: 10.1016/j.bmc.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 50.Eiermann B., Engel G., Johansson I., Zanger U.M., Bertilsson L. The involvement of CYP1A2 and CYP3A4 in the metabolism of clozapine. Br. J. Clin. Pharmacol. 1997;44(5):439–446. doi: 10.1046/j.1365-2125.1997.t01-1-00605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olesen O.V., Linnet K. Studies on the stereoselective metabolism of citalopram by human liver microsomes and cDNA-expressed cytochrome P450 enzymes. Pharmacology. 1999;59(6):298–309. doi: 10.1159/000028333. [DOI] [PubMed] [Google Scholar]
- 52.Le-Huu P., Heidt T., Claasen B., Laschat S., Urlacher V.B. Chemo-, Regio-, and Stereoselective Oxidation of the Monocyclic Diterpenoid β-Cembrenediol by P450 BM3. ACS Catal. 2015;5(3):1772–1780. [Google Scholar]
- 53.Loskot S.A., Romney D.K., Arnold F.H., Stoltz B.M. Enantioselective total synthesis of nigelladine a via late-stage C-H oxidation enabled by an engineered P450 enzyme. J. Am. Chem. Soc. 2017;139(30):10196–10199. doi: 10.1021/jacs.7b05196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geronimo I., Denning C.A., Heidary D.K., Glazer E.C., Payne C.M. Molecular determinants of substrate affinity and enzyme activity of a cytochrome P450BM3 variant. Biophys. J. 2018;115(7):1251–1263. doi: 10.1016/j.bpj.2018.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim P.Y., Pollard D.J., Woodley J.M. Substrate supply for effective biocatalysis. Biotechnol. Prog. 2007;23(1):74–82. doi: 10.1021/bp060314b. [DOI] [PubMed] [Google Scholar]
- 56.Sevrioukova I.F., Li H., Zhang H., Peterson J.A., Poulos T.L. Structure of a cytochrome P450–redox partner electron-transfer complex. Proc. Natl. Acad. Sci. 1999;96(5):1863–1868. doi: 10.1073/pnas.96.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.